Abstract
Recent advances in genome-wide association studies (GWAS) across autoimmune and immune-mediated diseases have augmented our understanding of pathogenic mechanisms underlying these diseases. This has further highlighted their heterogeneous nature, both within and between diseases. Furthermore, varying responses to therapy have also served to underline the importance of this heterogeneity in the manner in which these diseases are diagnosed and treated. Here we discuss our current understanding of the shared pathways of autoimmunity, including the tumor necrosis factor (TNF), major histocompatibility complex (MHC), interleukin 23 receptor (IL23R) and protein tyrosine phosphatase non-receptor type 22 (PTPN22) pathways. In addition, we summarize effective specific therapies tested across major autoimmune diseases, highlighting the insight they have provided into disease mechanisms and their implications for potential future improvements.
Although Paul Ehrlich proposed in 1901 that damaging self-reactivity or ‘horror autoxicus’ was impossible, this notion was soon contradicted by data from many sources. First, paroxysomal nocturnal hemoglobinuria was shown to be due to anti–red cell antibodies, and subsequently anti-thyroid antibodies were identified in individuals with thyroiditis. Sir Frank McFarlane Burnet’s ‘Clonal Selection Theory’ in 1957 suggested clear notions of how autoimmune diseases might arise from somatic mutations in antigen receptors, leading to ‘forbidden clones’ that were mistakenly not deleted during lymphocyte development.
GWASs for associations of genetic variants with various autoimmune diseases have informed our comparative understanding of disease mechanisms and revealed pathways that might yield potential drug targets. Furthermore, there has been a recent acceleration in the development of effective new therapies in autoimmune diseases by using knowledge that predates the recent advances in genetics. By helping advance knowledge of the molecular mechanisms of disease pathogenesis, genetics and genomics might help define novel therapeutic targets. Genetics and genomics might also be useful in some instances for clinical stratification.
Here we explore shared pathways of autoimmunity and recent advances in our molecular understanding of GWAS signals. We also summarize the effects of therapeutic agents tested across major autoimmune diseases and the pathophysiologic insights obtained from those tests, and we consider potential future therapeutic efforts.
Autoimmune disease phenotypes
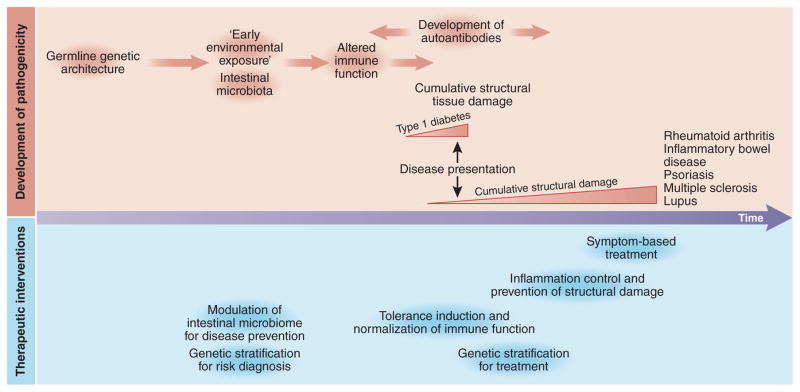
It is now apparent that many chronic inflammatory and destructive diseases are autoimmune, including rheumatoid arthritis, Graves’ disease, Hashimoto’s thyroiditis, and Sjogren’s syndrome, which each affect about 1% of the world’s population. In addition, autoimmune diseases also comprise less-common diseases such as type 1 diabetes, multiple sclerosis, Crohn’s disease, vitiligo, pernicious anemia, primary biliary cirrhosis, systemic lupus erythematosus, and ankylosing spondylitis. Over 80 autoimmune diseases have now been identified. These diseases are distinguished by their primary target organ (joints, skin—psoriasis; central nervous system—multiple sclerosis; intestine—inflammatory bowel disease (IBD); pancreas—type 1 diabetes mellitus), time course of disease presentation relative to tissue damage and major genetic associations (Table 1). For type 1 diabetes mellitus and autoimmune thyroid disease, extensive tissue destruction antedates disease presentation; therefore, for both of these diseases, the primary therapy remains hormone replacement, as opposed to anti-inflammatory agents (Fig. 1).
Table 1.
Major genetic association signals across autoimmune diseases
| MHC class | IL23R | PTPN22 | CTLA4a | |
|---|---|---|---|---|
| Type 1 diabetes | Class II | Arg620Trp | Non-coding | |
| Juvenile idiopathic arthritis | Class II | Arg620Trp | ||
| Autoimmune thyroid disease | Class II | Arg620Trp | Non-coding | |
| Rheumatoid arthritis | Class II | Arg620Trp | Non-coding | |
| Multiple sclerosis | Class II | |||
| Celiac disease | Class II | Non-coding | ||
| Systemic lupus erythematosis | Class II | Arg620Trp | ||
| Psoriatic arthritis | Class I | Distinct alleles | ||
| Psoriasis | Class I | Arg381Gln | ||
| Ankylosing spondylitis | Class I | Arg381Gln | ||
| Inflammatory bowel disease | Class II | Arg381Gln | Arg620Trp |
Underlined and bolded codons represent the allele associated with disease. Red color distinguishes the PTPN22 association with arginine rather than tryptophan.
Non-coding variants associated with the CTLA4 region may be distinct between diseases.
Figure 1.
Timeline of pathogenicity and therapeutic interventions in autoimmune diseases. Altered immune function antedates disease presentation across autoimmune diseases. In some cases, most notably for type 1 diabetes, substantial structural damage has already occurred by the time of disease presentation. In such cases, therapeutic options are restricted to symptomatic treatment, such as hormone replacement (insulin, thyroid hormones). For many other autoimmune diseases, therapeutic paradigms involve earlier treatment, control of inflammation and prevention of structural damage. In the future, induction of immune tolerance, and normalization of immune function may be accomplished, guided by genetic stratification for both treatment and prevention of disease.
In autoimmune diseases, germline genetic variation accounts for only a fraction of disease trait variability, typically less than 20%. Most GWAS signals are driven by non-coding mutations that modulate gene expression (Box 1). Because autoimmunity most commonly presents in late childhood to early adulthood, additional phenotypic variability probably arises from factors that arise during development, such as autoantibody formation and epigenetic programming, as well as from environmental factors such as the intestinal microbiome and tobacco use. However, the integration of shared and distinguishing features of autoimmune diseases provides a conceptual framework within which to accelerate the development and application of novel therapeutic agents.
Box 1. Molecular understanding of non-coding associations.
GWASs have defined the overall landscape of genetic polymorphisms in autoimmune diseases. However, in only a minority of disease-associated loci can a single, causal allele be identified that statistically accounts for the association signal in the region110. It is estimated that 90% of causal alleles are non-coding in nature, with a substantial fraction of these presumed to exert their pathogenic effects through modulation of gene expression. Early studies indicate that GWAS loci are indeed enriched for expression quantitative trait loci (eQTL)111. The potential value of mapping disease-associations with eQTLs is that it could provide more specific insight into disease mechanism. However eQTL are extraordinarily context specific, varying by cell and tissue type, time course and activation conditions. In a recent study in human macrophages stimulated by either lipopolysaccharide or IFN-γ, it was estimated that as many as 80% of transcripts are associated with eQTL112. Thus it is important to map disease-associated alleles with an eQTL observed in a relevant condition in order to provide pathophysiologic insights.
Related to gene expression changes, enrichment of GWAS signals has been observed for regions with specific regulatory chromatin states, cross-species conserved elements, DNA-accessible regions and histone marks. Generally, GWAS signal enrichment has been greater for cell-specific active enhancers (for example, H3K27ac peaks) than for promoter-associated peaks (for example, H3K4me3, H3K9ac). Disease and trait-associated polymorphisms are enriched in cell- and tissue-specific epigenetic marks, consistent with plausible mechanisms of disease pathogenesis. For example, autoimmune diseases have mapped to immune cell–specific marks, and metabolic diseases (for example, lipid regulation) have mapped with liver-specific histone marks, respectively. Of note, IBD GWAS signals enrich for both immune cell– and gastrointestinal-specific marks, indicating multiple contributing cell types in driving disease113.
More precise cell subset definitions of the active enhancer landscape have provided additional insight into autoimmune disease pathogensis95. The greatest enrichment of GWAS signals is observed within active transcriptional regions for a variety of CD4+ effector T cells (for example, TH17, TH1, TH2) for numerous autoimmune diseases including multiple sclerosis, celiac disease, primary biliary cirrhosis, type 1 diabetes, rheumatoid arthritis and inflammatory bowel disease. Generally, enrichment scores of GWAS signals within epigenetic marks for Treg cells were lower, with the greatest enrichment observed for multiple sclerosis. B cell enrichment scores were highest in multiple sclerosis, primary biliary cirrhosis, systemic lupus erythematosis, followed by rheumatoid arthritis, with lower B cell enrichment observed in classically seronegative diseases such as inflammatory bowel disease, psoriasis and ankylosing spondylitis. Interestingly, relatively low enrichment scores were observed in peripheral blood monocytes95. We speculate that this reflects the highly contextual and plastic nature of tissue macrophages and their enhancer landscape114. Whether more significant GWAS signal enrichment is observed with epigenetic and expression data from tissue-relevant macrophages should be the subject of future studies.
Within-disease heterogeneity
Many autoimmune diseases include the expression of autoantibodies, although autoantibodies are not a requirement. However, not all self-proteins are autoantigens, and not all potential epitopes are autoantigenic. What makes a self-protein autoantigenic is not clear. Sometimes autoantigens have additional properties; for example, they may interact with toll-like receptors (TLRs) or chemokine receptors1,2, but usually these properties are not known. Equally unclear is why the presence of autoantibodies and the development of autoimmune disease are not more closely linked. Some autoantibodies, such as antibodies to the thyroid stimulating hormone (TSH) receptor, are agonistic, resulting in the excessive stimulation of the thyroid gland present in Graves’ disease3. Others, such as anti-DNA antibodies, are mostly non-pathogenic. Clearly there are some important steps that need to be understood in the development of autoimmune disease aside from the generation of autoantibodies. One of these steps is immunoglobulin (Ig) class switching, because initially autoantibodies are IgM, but they mature into higher-affinity IgG autoantibodies. For example, IgM anti-dsDNA antibodies are less pathogenic than their IgG counterparts in systemic lupus erythematosus4.
Within a single disease, there is considerable variation in clinical manifestations and severity. For example, in rheumatoid arthritis, there is marked heterogeneity in the age of onset. Furthermore, as a person with this disease ages, there is heterogeneity in the number of affected joints and their distribution; in some more severe cases, people develop extra-articular complications, such as nodules and lung fibrosis. There is considerable heterogeneity in the speed of progression and in the extent of joint damage, with the degree of cartilage and bone damage varying. Some, but not all, phenotypic heterogeneity is related to known factors, such as ‘seropositivity’, with rheumatoid factor (RF) and anti-citrullinated antibody (ACPA) often being associated with more severe disease than seronegative arthritis; seropositive disease also has different human leukocyte antigen (HLA) associations5.
Related to disease heterogeneity, disease concordance rates in identical twins are surprisingly low, ranging from 15% in rheumatoid arthritis to 24% in systemic lupus erythematosus, and a bit higher in type 1 diabetes, with estimates averaging approximately 30% (refs. 6,7). Why this is is not understood, but it suggests an important role for subtle differences in the environment in the development of autoimmune disease. It is known that between twins, differences in epigenetic marks increase with age8. This discordance in disease development in identical twins also makes clear the difficulties of predicting disease; the popular concept that knowing a patient’s DNA sequence will reveal their disease risk is thus clearly naive.
Heterogeneity of autoimmunity is also very apparent in inbred animal models; inbred mice within the same cage are analogous to identical twins reared together. In some animal models, such as in mouse models of collagen-induced arthritis, a minority of the mice might get the disease. Hence the issue of autoimmune disease being multi-factorial and not being just due to genetics is clear, and future studies should be designed to incorporate non-genetic factors in a longitudinal manner.
Shared pathways of autoimmunity
Genetic and non-genetic approaches have provided insights into the shared pathways of autoimmune and immune-mediated diseases and hence the identification of pathways that are likely to represent valid therapeutic targets. An important new insight from recent genetic studies is that loss-of-function alleles that decrease disease risk highlight potentially ideal therapeutic targets, as they represent a naturally occurring manner in which to validate a target. That these loss-of-function, disease-protective alleles exist in healthy individuals suggests that blocking a particular pathway might be both a safe and effective treatment at the earliest possible stages of disease pathogenesis. One example of the efficacy of this approach has involved the identification of loss-of-function alleles in proprotein convertase subtilisin/kexin type 9 (PCSK9), which is associated with both decreased LDL cholesterol levels and death from coronary artery disease6,. Soon after these genetic associations were identified, multiple agents to block PCSK9 expression were rapidly developed and demonstrated marked efficacy in phase 2 (ref. 9) and 3 (refs. 10–12) studies; however, rare neurocognitive events (less than 1% of cases) were observed more frequently in the treatment arm10. Other forms of cholesterol-lowering therapy such as statins may also predispose people to neuropsychiatric and cognitive problems13.
MHC
The genetic associations within the MHC with autoimmune disease provided the first experimental evidence of genetic predisposition to autoimmune diseases in the 1970s (ref. 14). The relevance and mechanism of MHC as a major risk factor across all forms of autoimmunity in humans and mice became clear when it was elucidated that MHC molecules on antigen-presenting cells (APCs) bind peptides and present them to appropriate T cells.
GWAS that genotype a dense map of markers in case-control cohorts have highlighted a dominant role for the MHC region in autoimmune disease15,16. The MHC region includes genes with diverse immune function in addition to genes encoding the classical antigen-presenting molecules. It is the most genetically diverse region in the genome owing to the effects of natural selection. For most autoimmune diseases, genetic associations within the MHC region are the most significant, and there are multiple independent MHC alleles that can predispose individuals to autoimmune disease. The predominant MHC associations with autoimmune disease can be generally classified as MHC class I or class II predominant (Table 1). The role of the cell surface MHC molecules is to present peptide antigens to T lymphocytes. Class I and class II MHC molecules are distinguished by their expression and antigen sources. Class I molecules are expressed on all cells, whereas class II expression is restricted to professional APCs such as dendritic cells and B cells. Regarding antigen source, class I antigens are derived from cytosolic sources, whereas class II antigens are derived from extracellular proteins. Associations of the MHC with genetic disease are of primary importance in contrasting class I (psoriatic arthritis, ankylosing spondylitis) with class II (rheumatoid arthritis, juvenile idiopathic arthritis) joint disease (Table 1).
Although there are many loci in the MHC that can predispose an individual to multiple autoimmune diseases and disease subtypes, for the majority of loci, precise causal alleles have not thus far been defined. Within a given locus, causal alleles may be different between diseases, reflecting either genetic differences in antigen recognition (MHC region) or differences in cell-specific enhancers, which may provide precise insights into the pathogenic and protective cell subsets for particular diseases.
TNF
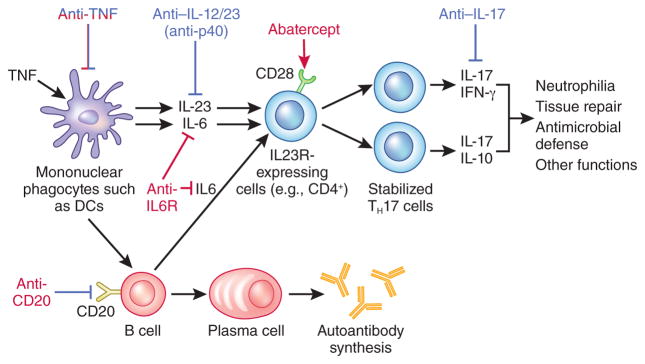
The identification that cytokines (especially proinflammatory cytokines that initiate inflammatory and immune responses) have a central role in autoimmunity and could be individually targeted for therapeutic benefit was a key conceptual advance in the understanding of autoimmune diseases. Studies in tissues associated with autoimmune diseases such as the rheumatoid synovium, which was studied owing to its accessibility, helped to identify marked upregulation of many pro-inflammatory cytokines, including TNF, interleukin (IL)-1β, granulocyte macrophage colony-stimulating factor (GM-CSF), interferon-γ (IFN-γ), IL-2 and IL-6 (ref. 17). Crucially, it was subsequently shown in synovial cultures taken from patients with rheumatoid arthritis that monoclonal antibodies against TNF downregulated IL-1β secretion, and subsequently the release of many other pro-inflammatory cytokines (Fig. 2) (refs. 18,19). Rather than suggesting a system of overlapping, highly redundant inflammatory pathways that cause inflammation, these studies supported a model of a delicately balanced, hierarchical cytokine network or cascade dependent on TNF that resulted in inflammation. It is of interest in this context that TNF appears to ‘raise the alarm’, as it is released rapidly after almost any stressful stimulus, and it acts to recurit the ‘firefighters’, recruiting leukocytes to sites of stress.
Figure 2.
Levels of therapeutic targeting across autoimmunity. Targetable pathways of rheumatoid arthritis and psoriasis are shown. Therapies that are effective in rheumatoid arthritis are shown in red, and therapies effective in psoriasis are shown in blue. The broad efficacy of anti-TNF therapy across many diseases reflects its central role in the cytokine network. In contrast, a more limited range of diseases have demonstrated positive results to blockade with anti–IL-12/23. IL-23 perpetuates and stabilizes IL23R-expressing pathogenic cells, including CD4+ TH17 cells. IL-17, as produced by TH17 cells, promotes neutrophilia, as well as enhances antimicrobial defense and tissue repair. Antagonism of IL6R and CTLA4 fusion proteins preventing CD28 co-stimulation are effective in the treatment of rheumatoid arthritis, as are B cell ablation approaches, such as by CD20-specific antibody administration, which are approved for the treatment of rheumatoid arthritis.
After the success of clinical trials using anti-TNF therapy in rheumatoid arthritis20, related diseases such as Crohn’s disease21,22 and psoriasis were also tested and shown to respond to this treatment. It was then determined that in this group of diseases, including rheumatoid arthritis, juvenile rheumatoid arthritis, Crohn’s disease, ulcerative colitis23, psoriasis24, psoriatic arthritis25 and ankylosing spondylitis26,27, that TNF expression is an important rate-limiting step in disease pathogenesis, such that anti-TNF therapy is effective in the majority of such patients. Consistent with the beneficial effect of TNF blockade in rheumatoid arthritis and Crohn’s disease, mice overexpressing TNF were demonstrated to have gut- and joint-associated immunopathologies28.
With regard to the importance of genetics and genomics in planning target discovery, it is important to note that defining TNF as a target occurred totally independently of these technologies. Furthermore, in disease tissue microarrays or in GWASs, the TNF pathway does not show any genetic association, even with diseases in which anti-TNF therapy is very effective. This is important because it indicates that the high-throughput technologies based on DNA or RNA sequencing will miss important therapeutic targets. No single approach can be used to address such an important problem as the definition of therapeutic targets.
The IL12/23 pathway
Autoimmunity was long thought to be driven by IFN-γ–expressing CD4+ T helper 1 (TH1) cells29. However, the loss of IFN-γ increased inflammation in a mouse model of multiple sclerosis, as it did in a model of arthritis questioning this role for TH1 cells in autoimmunity30, and IFN-γ–specific antibody treatment increases arthritis activity in animal and human studies31,32.
Subsequent studies implicated CD4+ TH17 cells in a variety of mouse models of autoimmunity33,34; the clinical relevance of these findings was validated by human genetic studies implicating the IL-23 pathway in autoimmune disease pathogenesis35; this pathway is essential for amplifying and stabilizing the pathogenic function of TH17 cells36. Among the IL-23 pathway genes, there are multiple associations of SNPs within IL23R with multiple immune diseases, such as IBD37, psoriasis38,39, ankylosing spondylitis40 and psoriatic arthritis41 have been reported. (Of note, the associated IL23R polymorphisms in psoriatic arthritis are distinct from those reported in the other diseases.) The most notable genetic association of IL23R with most of these autoimmune diseases is the coding polymorphism Arg381Gln: the minor glutamine allele (observed in approximately one out of seven individuals with European ancestry) confers a two- to threefold increase in protection, compared to the more-common arginine allele, against developing IBD37. This protective glutamine allele is a loss-of-function mutation resulting in decreased numbers of IL-23–dependent CD4+ TH17 and CD8+ Tc17 cells in human cells42–44.
Monoclonal antibodies against p40, the cytokine subunit common to IL-12 and IL-23 are approved for the treatment of psoriasis45 and psoriatic arthritis46, and phase 2 studies in IBD have shown promising results47. Given the protective, loss-of-function allele in IL23R, it is possible that selective targeting of IL-23 may be equally or more efficacious than combined IL12/23 targeting. Monoclonal antibodies blocking the IL-23–specific p19 subunit have demonstrated promising results in Crohn’s disease48 and psoriasis49. With respect to the interpretation of therapeutic responses to blocking IL-17 across autoimmune diseases, the role of TH17 cells in autoimmune disease pathology is complex and is discussed later.
PTPN22 association with seropositive autoimmune diseases
In addition to the MHC, a major genetic association with multiple autoimmune diseases identified before the GWAS era involved the Arg620Trp polymorphism in PTPN22. Specifically, the less-common tryptophan allele is associated with a large number of seropositive autoimmune diseases such as rheumatoid arthritis, type 1 diabetes, systemic lupus erythematosis and autoimmune thyroid disease50. No association is observed with multiple sclerosis, and the opposite Arg620 allele is associated with Crohn’s disease51. The tryptophan allele disrupts the binding of PTPN22 to CSK (c-Src tyrosine kinase), resulting in alterations in thresholds for T and B cell receptor signaling52. CSK normally downregulates T cell receptor signaling, and mice lacking PTPN22 have increased T cell activation53. However, the effect of the tryptophan allele on lymphocyte responsiveness in humans remains controversial, with both increased and decreased T cell and B cell receptor signaling reported54–57.
Understanding heterogeneity in autoimmune disease through therapy response
Heterogeneity in anti-TNF responses. The finding that anti-TNF therapy, while the most profitable drug class and the standard of care in eight diseases is not effective in multiple sclerosis and, in fact, may worsen disease is a clue to the pathogenesis of this disease, the mechanisms of which remain largely unresolved. GWAS results are largely unrevealing in this regard; although the TNF signature in anti-TNF–responsive diseases can be gleaned from some associations (for example, associations with TNFAIP3, which downregulates TNF effects15), mutations associated with a TNF signature are not as common in autoimmune disease as those associated with other pathways. (Table 1). GWAS-identified TNF association signatures would certainly be an insufficient basis for selecting between alternative therapies for individual patients. GWASs of anti-TNF responses have demonstrated genetic associations with CD84 (ref. 58), but this finding has not yet been confirmed in independent studies. Furthermore, it is worth pointing out that the dominant contribution to anti-TNF nonresponsiveness as determined by this study is not genetic; the CD84-associated SNPs accounted for only 2.6% of the variance in anti-TNF responses58. Importantly, there is no clear cut boundary between responders and nonresponders to anti-TNF treatment; it is a continuum, and the degree of response depends on the timing of therapy, with the greatest response if therapy commences within 6 months of diagnosis59,60. The most telling evidence for the nongenetic basis of low responsiveness is that patients that do not respond at a given point in time, so-called nonresponders, have a 50% chance of responding within 6 months to a different or same anti-TNF therapy. The specific factors that determine TNF responsiveness are not well understood, but the actual presence of TNF in joints in RA at the time of treatment is important for responsiveness59,60.
Targeting the IL-12/IL-23 pathway in psoriasis
Psoriasis is a major success story in precision therapeutic targeting. This is due to clear GWAS results combined with the fact that that disease status and the response to therapy can be precisely followed (including with serial tissue sampling). Currently approved biologic therapies for plaque psoriasis include, in chronological order of approval, anti-TNF61, anti-p40 (anti–IL-12/23) (ref. 62) and, most recently, anti–IL-17 therapies63. Serial sampling studies of psoriatic skin lesions from patients that responded favorably to anti-TNF treatment have shown that the downregulation of pro-inflammatory cytokines begins with IL-1β and IL-8, with decreases in IL-23 observed later64.
Because the result of GWASs have associated multiple components of the IL-23 pathway with psoriasis, including, IL23R and IL23A (p19) (ref. 65), one might speculate that an important way in which anti-TNF therapy acts, particularly in psoriasis, is by downregulation of IL-23. With the approval of anti–IL-12/23, a key question is whether therapies that target upstream targets such as TNF or more-focused therapies targeting downstream components would be more effective. In a large randomized, controlled trial of individuals with moderate to severe psoriasis, anti-IL-12/23 (p40) demonstrated superior efficacy to the less-potent anti-TNF therapy in psoriasis (etanercept)45. It remains to be determined whether even more focused approaches, such as IL-23–specific therapies (for example, anti-IL-23A or p19), which would not affect IL-12 signaling, will be equally or more efficacious than approaches that block both cytokines. They might be predicted to be safer.
Another issue is whether the targeting of inflammatory mediators produced by IL-23–responsive cells would be effective. In fact, blocking either IL-17A alone63 or both IL-17A and IL-17F by anti-IL17R (ref. 66) agents is effective in psoriasis in humans. The clinical and molecular response to IL-17 inhibition was rapid compared to anti-TNF or anti-IL12/23 blockade, with near-resolution of symptoms of within six weeks63,66. IL-17 acts by enhancing neutrophilia, tissue remodeling and repair and production of antimicrobial proteins67. These results suggest an essential and relatively downstream effector role for IL-17 in disease pathogenesis68. In all autoimmune diseases, treatment of acute flares and maintenance of disease remission are often conceptually separated. Whether blockade of IL-17 versus IL-23 has differential efficacy in the treatment of flares and maintenance of long-term remission will need to be studied.
Tissue-specific considerations for therapy
Given its marked efficacy in psoriasis, and the efficacy in IBD of anti–IL-12/23 (ustekinumab) in phase 2 studies47, it was particularly disappointing that anti–IL-17 treatment not only was ineffective in IBD, but also it demonstrated a trend toward worsened disease compared to placebo69. To an extent, this result was predicted by studies in mice lacking IL-17A, which have an accelerated wasting disease in the CD45Rb high-transfer model of colitis70. The intestine has a baseline tolerance to resident microbiota (mediated by IL-10 (ref. 71) and TGF-β1 (refs. 72,73), among many mediators), but it also maintains a robust defense capacity against pathogens and against breaching of the usual anatomic and functional containment of intestinal microbiota74–76. There is a crucial role for IL-10 secreted by TH17 cells in the maintenance of intestinal tolerance71,77, and thus it may be speculated that the worsening of intestinal inflammation observed with anti–IL-17 treatment in IBD69 might result from impaired function of protective TH17 cells.
This disparity in therapeutic effectiveness might be explained by the fact that tissue-specific TH17 cell subsets have evolved to combat specific pathogens. For example, Candida albicans–specific human TH17 cell clones produce IL-17 and IFN-γ, but not IL-10, whereas Staphylococcus aureus (a typical skin pathogen)-specific TH17 cells produce IL-17 and are able to produce IL-10 upon re-stimulation78. Thus, tissue-specific autoimmune disease differences may reflect corresponding tissue- or organ-specific differences in typically encountered infectious pathogens (Box 2). Anti–IL-12/23 therapies (ustekinumab) have shown positive results for ankylosing spondylitis in a small, open-label study79, and have also been approved for the treatment of psoriatic arthritis80; both of these are diseases have been linked with mutations in the MHC class I region and the IL-23 signaling pathway. Furthermore, for both psoriatic arthritis66,81 and ankylosing spondylitis82, early clinical trials blocking the IL-17 pathway have demonstrated promising results (Table 2) (ref. 83). This therapeutic effectiveness in blocking IL-17 in psoriatic arthritis is despite differences in skin IL-17 and joint IFN-γ expression signatures84.
Box 2. Evolving host-microbiome relationships.
Any increases in the incidence of autoimmune diseases measurable in decades must reflect environmental changes that have evolved relatively recently in human history115. The human genetic architecture evolved in response to natural selection. Genetic variation which provides protection against historically significant pathogens may, in a different time and environment confer increased risk to a distinct disease115,116.
Changes in factors such as diet, sanitation and use of antibiotics can affect the content of the intestinal microbiome117. There is increasing evidence that the changing intestinal microbiome can drive increasing incidence of autoimmune diseases118. For example, expansion of intestinal Prevotella copri was correlated with newly diagnosed rheumatoid arthritis119. Correlational observations in humans are further supported by studies in model organisms which demonstrate how genetic factors (for example, innate immune deficiency in Myd88-deficient mice) can alter the intestinal microbiome, which in turn, modulates risk for the development of autoimmunity (for example, type 1 diabetes)120. A long-term goal would be to modulate host-microbiome interactions to decrease risk for autoimmune diseases in high-risk individuals.
There is also overlap between genes contained within loci that have been linked to increased risk of developing IBD, and between genes that are implicated in the predisposition to mycobacterial infection51. Ashkenazi Jewish individuals have a higher prevalence of IBD, and they have historically been relatively protected against dying from tuberculosis121. Thus one might speculate that the higher prevalence IBD in the Ashkenazim is due to positive selection for genetic variants that protect against tuberculosis, but in a different time and environment (for example, distinct intestinal microbiome composition), confer increased risk for IBD.
One might speculate that the plethora of loci identified by GWAS of autoimmune diseases reflects a matching abundance of functional polymorphisms positively selected for in response to historically significant infectious pathogens122. While generally well-tolerated with a favorable side-effect profile, blockade of TNF is associated with modestly increased risk for infections generally123,124. Of particular note, anti-TNF therapy has been associated with reactivation of latent tuberculosis125, such that testing for Mycobacterium tuberculosis
Table 2.
Efficacy of therapeutic agents across autoimmune diseases
| TNF | IL12/23 | IL17 | IL6 | CTLA4 | Anti-CD20 | |
|---|---|---|---|---|---|---|
| Rheumatoid arthritis | ||||||
| Juvenile idiopathic arthritis* | ||||||
| Multiple sclerosis | ||||||
| Psoriasis | ||||||
| Psoriatic arthritis | ||||||
| Ankylosing spondylitis | ||||||
| Inflammatory bowel disease | ||||||
| Specific agents | Infliximab, adalimumab, etanercept, certolizumab | Ustekinumab | Secukinumab, brodalumab | Tocilizumab | Abatacept | Rituximab |
Dark green cells, approved indications; light green cells, preliminary positive findings; red cells, shown to worsen disease; gray cells, no evidence for efficacy.
Treatment responses are distinct between systemic and polyarticular forms of juvenile idiopathic arthritis
The success of targeting the IL-23 or IL-17 pathways in these diseases contrasts with the effectiveness of these therapies in rheumatoid arthritis, in which similar positive findings have thus far not been reported. This may reflect the fundamentally distinct genetic architecture of rheumatoid arthritis85,86, characterized by associations with MHC class II, the absence of associations with IL23R and the presence of associations with PTPN22 (Table 1). The dichotomy between MHC class I– and class II–predominant diseases reflects fundamentally distinct pathogeneses, and thus distinct optimal therapeutic approaches.
Blocking T cell activation: abatacept and autoimmune diseases
Given the dominant role of MHC class II associations across autoimmune diseases, it would seem logical that modulating TCR activation might be useful therapeutically. The outcome of engagement of TCRs with peptide-bearing MHC molecules on APCs is modulated by additional signaling partner interactions. These ‘second signal’ partnerships can activate T cells (for example, CD28 expressed on T cells paired with B7 molecules on APCs) or inhibit T cell activation (for example, CTLA4-B7) in nature. CTLA4 (encoding cytotoxic T-lymphocyte associated protein 4) is expressed on T cells, and polymorphisms in the CTLA4 gene region are associated with a variety of autoimmune diseases, notably type 1 diabetes87, autoimmune thyroid disease88, rheumatoid arthritis85, and celiac disease89 (Table 1). Rare, heterozygous CTLA4 mutations result in a functional haploinsufficiency, with dysregulation of FoxP3+ regulatory T cells (Treg cells) and systemic autoimmunity, although this results in incomplete genetic penetrance and a wide range of disease manifestations90,91.
Rheumatoid arthritis is the only autoimmune disease demonstrating an association with CTLA4 in which biologics are routinely administered (Table 1). Interestingly, rheumatoid arthritis is also the major approved indication for treatment with abatacept, a high-affinity CTLA4 fusion protein (comprising the CTLA4 extracellular domain plus an IgG1 Fc fragment) that blocks second signals (e.g., CD28 co-stimulation) required for T cell activation92 (Table 2). In contrast, well-powered studies of IBD demonstrated a lack of efficacy for abatacept in Crohn’s disease or ulcerative colitis93. It may be speculated that this lack of efficacy occurs because effector memory T cells, predominant in the gut, are less dependent on CD28 for co-stimulation94.
B cell depletion approaches
Whereas autoantibodies are a characteristic feature of many autoimmune diseases, it is important to distinguish between examples of primary pathogenicity versus autoantibody expression that reflect the secondary effects of increased immune activity (for example, through CD4+ T cell help). GWAS signals are enriched in both B cell gene expression95 and enhancer96 landscapes. Among B cell enhancer regions, GWAS signals are most enriched in lupus, followed by rheumatoid arthritis. Germline genetic variation identified by GWASs reflects primary, driving events, and so it may be inferred that B cells are pathogenic in these GWAS-signal-enriched diseases.
Rheumatoid arthritis is the major approved indication for B cell depletion approaches, such as through anti-CD20 (rituximab)97 (Fig. 2). In relapsing-remitting multiple sclerosis, another GWAS-signal B cell enhancer–enriched disease, rituximab treatment resulted in fewer inflammatory brain lesions and clinical relapses98 (Table 2), but it has not been approved by the US Food and Drug Administration (FDA) for this use. In addition, ritiuximab has demonstrated modest efficacy in ankylosing spondylitis in anti-TNF–naive patients, but has not been approved by the FDA for this thus far99,100. In systemic lupus erythematosus, rituximab, while very effective in some patients, was also only of modest overall efficacy and so was not approved by the FDA. In contrast, no efficacy was observed with rituximab in ulcerative colitis in a phase 2 study101. These findings are consistent with the absence of enrichment for ulcerative colitis GWAS signals in B cell enhancer regions95. Taken together, this would indicate that the autoantibodies commonly observed in ulcerative colitis (for example, pANCA) are secondary effects and not primary drivers of disease. There is ongoing research on other B cell targets, but none has reached clinical approval apart from Belimumab, an antibody specific to B-lymphocyte stimulator, which is a fully human monoclonal antibody. Belimumab is approved for systemic lupus erythematosus by the FDA, but has shown modest efficacy102,103.
Conclusions and future directions
Heterogeneity in disease pathophysiology and therapeutic responses is driven by genetic, developmental and environmental factors. Genetic discovery through GWASs over the past several years has identified a striking overlap of genetic loci between autoimmune, chronic inflammatory diseases. The immediate work that needs completion includes a comprehensive dissection of similarities and differences in the precise allelic architecture at these loci, integrated with tissue- and context-specific data on epigenetics and gene expression, all within the larger context of trying to define the effect of the environment on the predisposition to disease, which is poorly understood apart from, for example, smoking. These nongenetic aspects can be explored in a variety of ways, one of the most powerful being by epidemiology. Prospective epidemiological studies involving very large patient cohorts followed over the long term will be needed to try to determine which environmental factors might be important. These types of studies are now being designed through efforts such as the Precision Medicine Initiative104. Also critical in this regard is the serial sampling of relevant human tissues in the context of therapeutic interventions. It is incumbent that complete and timely reporting of results from therapeutic interventions in humans, both positive and negative, be mandated upon those responsible for clinical trials. Biomedical discovery is classically based on defining unifying principles of broad applicability. Similarly, the practice of medicine has traditionally been based on repeated observations and definitions of similarities between patients that guide treatment decisions on the basis of prior experience. It is increasingly clear, however, that the full realization of the promise of genetic and genomic discovery will require a paradigm shift embracing complexity, heterogeneity, and information-rich decision matrices personalized to individual patients105, their diets, microbiomes, past medical histories, and other environmental factors.
With regard to drug development, an additional practical consideration is the immunogenicity of biologic agents, exacerbated with intermittent administration, which reduces efficacy and increases adverse side effects106. With the changing economic landscape in health care, the costs and long-term efficacy of specific biologic targeting107 will increasingly be compared with small molecules, which are generally associated with lower rates of immunogenicity, but also variable target specificity; regrettably, they are not necessarily cheaper (e.g., tofacitanib).
Central to this new era of precision medicine will be a long-term, longitudinal view of research partners including, importantly, the patients (Fig. 1). The invaluable medical prescription, primum non nocere (first do no harm), has in the past resulted in a symptom-based treatment approach in autoimmune diseases. However, as new more effective therapies are developed, the goals of attaining control of inflammation and preventing structural tissue damage are increasingly being realized. For diseases such as type 1 diabetes, because a substantial amount of structural damage antedates disease presentation, symptom-based treatment (here, application of insulin) remains the cornerstone of therapy. For all autoimmune diseases, future goals involve earlier intervention, induction of immune tolerance, and possibly normalization of immune function. Critical to this progress will be defining how early microbial exposures modulate the immune system, for both early diagnosis and disease prevention. However, the major current hurdle is to consider which therapeutic combinations might be both safe and effective in autoimmune diseases. This is not a trivial task; premature attempts to develop combinations, for example using etanercept and anakinra108,109 have yielded challenging results, namely no increase in efficacy but a marked increase in infections. But defining safe and effective combinations is possible. In rheumatoid arthritis, more than 50%, and in many reports, almost 70% of patients on TNF blockade are also treated with low-dose methotrexate. Targeting pathways that maintain disease chronicity but that are not involved in host defense against infection might help get us closer to a cure. The multiple clues from GWASs might help progress on this path.
Acknowledgments
J.H.C. is supported by US National Institutes of Health grants U01 DK62429, U01 DK062422, R01 DK092235, SUCCESS, by he Helmsley Charitable Trust and by the Sanford J. Grossman Charitable Trust.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Hradetzky S, et al. The human skin-associated autoantigen alpha-NAC activates monocytes and dendritic cells via TLR-2 and primes an IL-12-dependent Th1 response. J Invest Dermatol. 2013;133:2289–2292. doi: 10.1038/jid.2013.161. [DOI] [PubMed] [Google Scholar]
- 2.Harlow L, Fernandez I, Soejima M, Ridgway WM, Ascherman DP. Characterization of TLR4-mediated auto-antibody production in a mouse model of histidyl-tRNA synthetase-induced myositis. Innate Immun. 2012;18:876–885. doi: 10.1177/1753425912446714. [DOI] [PubMed] [Google Scholar]
- 3.Kopp P. The TSH receptor and its role in thyroid disease. Cell Mol Life Sci. 2001;58:1301–1322. doi: 10.1007/PL00000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross GL, Barland P, Grayzel AI. The immunoglobulin class of anti-DNA antibodies: detection by a fluorometric immunoassay: clinical and pathological correlations in SLE. J Rheumatol. 1978;5:373–383. [PubMed] [Google Scholar]
- 5.Klareskog L, Catrina AI, Paget S. Rheumatoid arthritis. Lancet. 2009;373:659–672. doi: 10.1016/S0140-6736(09)60008-8. [DOI] [PubMed] [Google Scholar]
- 6.Salvetti M, Ristori G, Bomprezzi R, Pozzilli P, Leslie RD. Twins: mirrors of the immune system. Immunol Today. 2000;21:342–347. doi: 10.1016/s0167-5699(00)01658-3. [DOI] [PubMed] [Google Scholar]
- 7.Bogdanos DP, et al. Twin studies in autoimmune disease: genetics, gender and environment. J Autoimmun. 2012;38:J156–J169. doi: 10.1016/j.jaut.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Fraga MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein EA, et al. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. N Engl J Med. 2012;366:1108–1118. doi: 10.1056/NEJMoa1105803. [DOI] [PubMed] [Google Scholar]
- 10.Sabatine MS, et al. Efficacy and safety of evolocumab in reducing lipids and cardiovascular events. N Engl J Med. 2015;372:1500–1509. doi: 10.1056/NEJMoa1500858. [DOI] [PubMed] [Google Scholar]
- 11.Robinson JG, et al. Efficacy and safety of alirocumab in reducing lipids and cardiovascular events. N Engl J Med. 2015;372:1489–1499. doi: 10.1056/NEJMoa1501031. [DOI] [PubMed] [Google Scholar]
- 12.Blom DJ, et al. A 52-week placebo-controlled trial of evolocumab in hyperlipidemia. N Engl J Med. 2014;370:1809–1819. doi: 10.1056/NEJMoa1316222. [DOI] [PubMed] [Google Scholar]
- 13.McDonagh J. Statin-related cognitive impairment in the real world: you’ll live longer, but you might not like it. JAMA Intern Med. 2014;174:1889. doi: 10.1001/jamainternmed.2014.5376. [DOI] [PubMed] [Google Scholar]
- 14.Dick HM. HLA and disease. Introductory review. Br Med Bull. 1978;34:271–274. doi: 10.1093/oxfordjournals.bmb.a071510. [DOI] [PubMed] [Google Scholar]
- 15.Cho JH, Gregersen PK. Genomics and the multifactorial nature of human autoimmune disease. N Engl J Med. 2011;365:1612–1623. doi: 10.1056/NEJMra1100030. [DOI] [PubMed] [Google Scholar]
- 16.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–673. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 17.Feldmann M, Brennan FM, Maini RN. Role of cytokines in rheumatoid arthritis. Annu Rev Immunol. 1996;14:397–440. doi: 10.1146/annurev.immunol.14.1.397. [DOI] [PubMed] [Google Scholar]
- 18.Brennan FM, Chantry D, Jackson A, Maini R, Feldmann M. Inhibitory effect of TNF-α antibodies on synovial cell interleukin-1 production in rheumatoid arthritis. Lancet. 1989;2:244–247. doi: 10.1016/s0140-6736(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 19.Gregory AP, et al. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature. 2012;488:508–511. doi: 10.1038/nature11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maini R, et al. Infliximab (chimeric anti-tumour necrosis factor α monoclonal antibody) versus placebo in rheumatoid arthritis patients receiving concomitant methotrexate: a randomised phase III trial. ATTRACT Study Group. Lancet. 1999;354:1932–1939. doi: 10.1016/s0140-6736(99)05246-0. [DOI] [PubMed] [Google Scholar]
- 21.Present DH, et al. Infliximab for the treatment of fistulas in patients with Crohn’s disease. N Engl J Med. 1999;340:1398–1405. doi: 10.1056/NEJM199905063401804. [DOI] [PubMed] [Google Scholar]
- 22.Targan SR, et al. A short-term study of chimeric monoclonal antibody cA2 to tumor necrosis factor α for Crohn’s disease. Crohn’s Disease cA2 Study Group. N Engl J Med. 1997;337:1029–1035. doi: 10.1056/NEJM199710093371502. [DOI] [PubMed] [Google Scholar]
- 23.Rutgeerts P, et al. Infliximab for induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2005;353:2462–2476. doi: 10.1056/NEJMoa050516. [DOI] [PubMed] [Google Scholar]
- 24.Chaudhari U, et al. Efficacy and safety of infliximab monotherapy for plaque-type psoriasis: a randomised trial. Lancet. 2001;357:1842–1847. doi: 10.1016/s0140-6736(00)04954-0. [DOI] [PubMed] [Google Scholar]
- 25.Mease PJ. Tumour necrosis factor (TNF) in psoriatic arthritis: pathophysiology and treatment with TNF inhibitors. Ann Rheum Dis. 2002;61:298–304. doi: 10.1136/ard.61.4.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorman JD, Sack KE, Davis JC., Jr Treatment of ankylosing spondylitis by inhibition of tumor necrosis factor α. N Engl J Med. 2002;346:1349–1356. doi: 10.1056/NEJMoa012664. [DOI] [PubMed] [Google Scholar]
- 27.Sfikakis PP. Behcet’s disease: a new target for anti-tumour necrosis factor treatment. Ann Rheum Dis. 2002;61(Suppl 2):ii51–ii53. doi: 10.1136/ard.61.suppl_2.ii51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 29.Kuchroo VK, et al. T cell response in experimental autoimmune encephalomyelitis (EAE): role of self and cross-reactive antigens in shaping, tuning, and regulating the autopathogenic T cell repertoire. Annu Rev Immunol. 2002;20:101–123. doi: 10.1146/annurev.immunol.20.081701.141316. [DOI] [PubMed] [Google Scholar]
- 30.Ferber IA, et al. Mice with a disrupted IFN-γ gene are susceptible to the induction of experimental autoimmune encephalomyelitis (EAE) J Immunol. 1996;156:5–7. [PubMed] [Google Scholar]
- 31.Lemmel EM, Obert HJ, Hofschneider PH. Low-dose γ-interferon in treatment of rheumatoid arthritis. Lancet. 1988;1:598. doi: 10.1016/s0140-6736(88)91403-1. [DOI] [PubMed] [Google Scholar]
- 32.Williams RO, Williams DG, Feldmann M, Maini RN. Increased limb involvement in murine collagen-induced arthritis following treatment with anti-interferon-γ. Clin Exp Immunol. 1993;92:323–327. doi: 10.1111/j.1365-2249.1993.tb03399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cua DJ, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 34.Ahern PP, et al. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity. 2010;33:279–288. doi: 10.1016/j.immuni.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 36.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Duerr RH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nair RP, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-κB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cargill M, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80:273–290. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reveille JD, et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat Genet. 2010;42:123–127. doi: 10.1038/ng.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowes J, et al. Dense genotyping of immune-related susceptibility loci reveals new insights into the genetics of psoriatic arthritis. Nat Commun. 2015;6:6046. doi: 10.1038/ncomms7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarin R, Wu X, Abraham C. Inflammatory disease protective R381Q IL23 receptor polymorphism results in decreased primary CD4+ and CD8+ human T-cell functional responses. Proc Natl Acad Sci USA. 2011;108:9560–9565. doi: 10.1073/pnas.1017854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Meglio P, et al. The IL23R R381Q gene variant protects against immune-mediated diseases by impairing IL-23-induced Th17 effector response in humans. PLoS ONE. 2011;6:e17160. doi: 10.1371/journal.pone.0017160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pidasheva S, et al. Functional studies on the IBD susceptibility gene IL23R implicate reduced receptor function in the protective genetic variant R381Q. PLoS ONE. 2011;6:e25038. doi: 10.1371/journal.pone.0025038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Griffiths CE, et al. Comparison of ustekinumab and etanercept for moderate-to-severe psoriasis. N Engl J Med. 2010;362:118–128. doi: 10.1056/NEJMoa0810652. [DOI] [PubMed] [Google Scholar]
- 46.Gottlieb A, Narang K. Ustekinumab in the treatment of psoriatic arthritis: latest findings and clinical potential. Ther Adv Musculoskelet Dis. 2013;5:277–285. doi: 10.1177/1759720X13501021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandborn WJ, et al. Ustekinumab induction and maintenance therapy in refractory Crohn’s disease. N Engl J Med. 2012;367:1519–1528. doi: 10.1056/NEJMoa1203572. [DOI] [PubMed] [Google Scholar]
- 48.OP025. A randomized, double-blind placebo-controlled phase 2a induction study of MEDI2070 (anti-p19 antibody) in patients with active Crohn’s disease who have failed anti-TNF antibody therapy. J Crohns Colitis. 2015;9(Suppl 1):S15–S16. [Google Scholar]
- 49.Krueger JG, et al. Anti-IL-23A mAb BI 655066 for treatment of moderate-to-severe psoriasis: Safety, efficacy, pharmacokinetics, and biomarker results of a single-rising-dose, randomized, double-blind, placebo-controlled trial. J Allergy Clin Immunol. 2015 doi: 10.1016/j.jaci.2015.01.018. [DOI] [PubMed]
- 50.Criswell LA, et al. Analysis of families in the Multiple Autoimmune Disease Genetics Consortium (MADGC) Collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet. 2005;76:561–571. doi: 10.1086/429096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanford SM, Bottini N. PTPN22: the archetypal non-HLA autoimmunity gene. Nat Rev Rheumatol. 2014;10:602–611. doi: 10.1038/nrrheum.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hasegawa K, et al. PEST domain-enriched tyrosine phosphatase (PEP) regulation of effector/memory T cells. Science. 2004;303:685–689. doi: 10.1126/science.1092138. [DOI] [PubMed] [Google Scholar]
- 54.Arechiga AF, et al. Cutting edge: the PTPN22 allelic variant associated with autoimmunity impairs B cell signaling. J Immunol. 2009;182:3343–3347. doi: 10.4049/jimmunol.0713370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vang T, et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet. 2005;37:1317–1319. doi: 10.1038/ng1673. [DOI] [PubMed] [Google Scholar]
- 56.Zhang J, et al. The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness. Nat Genet. 2011;43:902–907. doi: 10.1038/ng.904. [DOI] [PubMed] [Google Scholar]
- 57.Zikherman J, et al. PTPN22 deficiency cooperates with the CD45 E613R allele to break tolerance on a non-autoimmune background. J Immunol. 2009;182:4093–4106. doi: 10.4049/jimmunol.0803317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui J, et al. Genome-wide association study and gene expression analysis identifies CD84 as a predictor of response to etanercept therapy in rheumatoid arthritis. PLoS Genet. 2013;9:e1003394. doi: 10.1371/journal.pgen.1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Breedveld F. The value of early intervention in RA–a window of opportunity. Clin Rheumatol. 2011;30(Suppl 1):S33–S39. doi: 10.1007/s10067-010-1638-5. [DOI] [PubMed] [Google Scholar]
- 60.Goekoop-Ruiterman YP, et al. Clinical and radiographic outcomes of four different treatment strategies in patients with early rheumatoid arthritis (the BeSt study): a randomized, controlled trial. Arthritis Rheum. 2005;52:3381–3390. doi: 10.1002/art.21405. [DOI] [PubMed] [Google Scholar]
- 61.Leonardi CL, et al. Etanercept as monotherapy in patients with psoriasis. N Engl J Med. 2003;349:2014–2022. doi: 10.1056/NEJMoa030409. [DOI] [PubMed] [Google Scholar]
- 62.Krueger GG, et al. A human interleukin-12/23 monoclonal antibody for the treatment of psoriasis. N Engl J Med. 2007;356:580–592. doi: 10.1056/NEJMoa062382. [DOI] [PubMed] [Google Scholar]
- 63.Langley RG, et al. Secukinumab in plaque psoriasis–results of two phase 3 trials. N Engl J Med. 2014;371:326–338. doi: 10.1056/NEJMoa1314258. [DOI] [PubMed] [Google Scholar]
- 64.Gottlieb AB, et al. TNF inhibition rapidly down-regulates multiple proinflammatory pathways in psoriasis plaques. J Immunol. 2005;175:2721–2729. doi: 10.4049/jimmunol.175.4.2721. [DOI] [PubMed] [Google Scholar]
- 65.Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361:496–509. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- 66.Mease PJ, et al. Brodalumab, an anti-IL17RA monoclonal antibody, in psoriatic arthritis. N Engl J Med. 2014;370:2295–2306. doi: 10.1056/NEJMoa1315231. [DOI] [PubMed] [Google Scholar]
- 67.Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 2010;140:845–858. doi: 10.1016/j.cell.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 68.Krueger JG. Hiding under the skin: a welcome surprise in psoriasis. Nat Med. 2012;18:1750–1751. doi: 10.1038/nm.3025. [DOI] [PubMed] [Google Scholar]
- 69.Hueber W, et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut. 2012;61:1693–1700. doi: 10.1136/gutjnl-2011-301668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O’Connor W, Jr, et al. A protective function for interleukin 17A in T cell–mediated intestinal inflammation. Nat Immunol. 2009;10:603–609. doi: 10.1038/ni.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kühn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 72.Li MO, Flavell RA. TGF-β: a master of all T cell trades. Cell. 2008;134:392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007;26:579–591. doi: 10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 74.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fung TC, Artis D, Sonnenberg GF. Anatomical localization of commensal bacteria in immune cell homeostasis and disease. Immunol Rev. 2014;260:35–49. doi: 10.1111/imr.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sonnenberg GF, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Glocker EO, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zielinski CE, et al. Pathogen-induced human TH17 cells produce IFN-γ or IL-10 and are regulated by IL-1β. Nature. 2012;484:514–518. doi: 10.1038/nature10957. [DOI] [PubMed] [Google Scholar]
- 79.Poddubnyy D, Hermann KG, Callhoff J, Listing J, Sieper J. Ustekinumab for the treatment of patients with active ankylosing spondylitis: results of a 28-week, prospective, open-label, proof-of-concept study (TOPAS) Ann Rheum Dis. 2014;73:817–823. doi: 10.1136/annrheumdis-2013-204248. [DOI] [PubMed] [Google Scholar]
- 80.McInnes IB, et al. Efficacy and safety of ustekinumab in patients with active psoriatic arthritis: 1-year results of the phase 3, multicentre, double-blind, placebo-controlled PSUMMIT 1 trial. Lancet. 2013;382:780–789. doi: 10.1016/S0140-6736(13)60594-2. [DOI] [PubMed] [Google Scholar]
- 81.McInnes IB, et al. Efficacy and safety of secukinumab, a fully human anti-interleukin-17A monoclonal antibody, in patients with moderate-to-severe psoriatic arthritis: a 24-week, randomised, double-blind, placebo-controlled, phase II proof-of-concept trial. Ann Rheum Dis. 2014;73:349–356. doi: 10.1136/annrheumdis-2012-202646. [DOI] [PubMed] [Google Scholar]
- 82.Baeten D, et al. Anti-interleukin-17A monoclonal antibody secukinumab in treatment of ankylosing spondylitis: a randomised, double-blind, placebo-controlled trial. Lancet. 2013;382:1705–1713. doi: 10.1016/S0140-6736(13)61134-4. [DOI] [PubMed] [Google Scholar]
- 83.Schett G, Elewaut D, McInnes IB, Dayer JM, Neurath MF. How cytokine networks fuel inflammation: Toward a cytokine-based disease taxonomy. Nat Med. 2013;19:822–824. doi: 10.1038/nm.3260. [DOI] [PubMed] [Google Scholar]
- 84.Belasco J, et al. Comparative genomic profiling of psoriatic arthritis synovium versus skin lesions. Arthritis Rheumatol. 2015;67:934–944. doi: 10.1002/art.38995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Eyre S, et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet. 2012;44:1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Onengut-Gumuscu S, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cooper JD, et al. Seven newly identified loci for autoimmune thyroid disease. Hum Mol Genet. 2012;21:5202–5208. doi: 10.1093/hmg/dds357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Trynka G, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kuehn HS, et al. Immune dysregulation in human subjects with heterozygous germline mutations in CTLA4. Science. 2014;345:1623–1627. doi: 10.1126/science.1255904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zeissig S, et al. Early-onset Crohn’s disease and autoimmunity associated with a variant in CTLA-4. Gut. 2014 doi: 10.1136/gutjnl-2014-308541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Genovese MC, et al. Abatacept for rheumatoid arthritis refractory to tumor necrosis factor alpha inhibition. N Engl J Med. 2005;353:1114–1123. doi: 10.1056/NEJMoa050524. [DOI] [PubMed] [Google Scholar]
- 93.Sandborn WJ, et al. Abatacept for Crohn’s disease and ulcerative colitis. Gastroenterology. 2012;143:62–69. doi: 10.1053/j.gastro.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 94.Kaser A. Not all monoclonals are created equal—lessons from failed drug trials in Crohn’s disease. Best Pract Res Clin Gastroenterol. 2014;28:437–449. doi: 10.1016/j.bpg.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 95.Hu X, et al. Integrating autoimmune risk loci with gene-expression data identifies specific pathogenic immune cell subsets. Am J Hum Genet. 2011;89:496–506. doi: 10.1016/j.ajhg.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Farh KK, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Edwards JC, et al. Efficacy of B-cell-targeted therapy with rituximab in patients with rheumatoid arthritis. N Engl J Med. 2004;350:2572–2581. doi: 10.1056/NEJMoa032534. [DOI] [PubMed] [Google Scholar]
- 98.Hauser SL, et al. B-cell depletion with rituximab in relapsing-remitting multiple sclerosis. N Engl J Med. 2008;358:676–688. doi: 10.1056/NEJMoa0706383. [DOI] [PubMed] [Google Scholar]
- 99.Wendling D, et al. Rituximab treatment for spondyloarthritis. A nationwide series: data from the AIR registry of the French Society of Rheumatology. J Rheumatol. 2012;39:2327–2331. doi: 10.3899/jrheum.120201. [DOI] [PubMed] [Google Scholar]
- 100.Song IH, et al. Different response to rituximab in tumor necrosis factor blocker-naive patients with active ankylosing spondylitis and in patients in whom tumor necrosis factor blockers have failed: a twenty-four-week clinical trial. Arthritis Rheum. 2010;62:1290–1297. doi: 10.1002/art.27383. [DOI] [PubMed] [Google Scholar]
- 101.Leiper K, et al. Randomised placebo-controlled trial of rituximab (anti-CD20) in active ulcerative colitis. Gut. 2011;60:1520–1526. doi: 10.1136/gut.2010.225482. [DOI] [PubMed] [Google Scholar]
- 102.Kandala NB, et al. Belimumab: a technological advance for systemic lupus erythematosus patients? Report of a systematic review and meta-analysis. BMJ Open. 2013 doi: 10.1136/bmjopen-2013-002852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fattah Z, Isenberg DA. Recent developments in the treatment of patients with systemic lupus erythematosus: focusing on biologic therapies. Expert Opin Biol Ther. 2014;14:311–326. doi: 10.1517/14712598.2014.871256. [DOI] [PubMed] [Google Scholar]
- 104.Collins FS. Reengineering translational science: the time is right. Sci Transl Med. 2011;3:90cm17. doi: 10.1126/scitranslmed.3002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Baert F, et al. Influence of immunogenicity on the long-term efficacy of infliximab in Crohn’s disease. N Engl J Med. 2003;348:601–608. doi: 10.1056/NEJMoa020888. [DOI] [PubMed] [Google Scholar]
- 107.Shankar G, Shores E, Wagner C, Mire-Sluis A. Scientific and regulatory considerations on the immunogenicity of biologics. Trends Biotechnol. 2006;24:274–280. doi: 10.1016/j.tibtech.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 108.Weinblatt M, et al. Selective co-stimulation modulation using abatacept in patients with active rheumatoid arthritis while receiving etanercept: a randomised clinical trial. Ann Rheum Dis. 2007;66:228–234. doi: 10.1136/ard.2006.055111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Genovese MC, et al. Combination therapy with etanercept and anakinra in the treatment of patients with rheumatoid arthritis who have been treated unsuccessfully with methotrexate. Arthritis Rheum. 2004;50:1412–1419. doi: 10.1002/art.20221. [DOI] [PubMed] [Google Scholar]
- 110.MacArthur DG, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fairfax BP, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343:1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lavin Y, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 116.Pritchard JK, Di Rienzo A. Adaptation—not by sweeps alone. Nat Rev Genet. 2010;11:665–667. doi: 10.1038/nrg2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Scher JU, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wen L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jacobs J. The Jewish Encyclopedia: A Guide to its Contents, an Aid to its Use. Funk & Wagnalls; 1906. [Google Scholar]
- 122.Quintana-Murci L, Clark AG. Population genetic tools for dissecting innate immunity in humans. Nat Rev Immunol. 2013;13:280–293. doi: 10.1038/nri3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Aaltonen KJ, et al. Systematic review and meta-analysis of the efficacy and safety of existing TNF blocking agents in treatment of rheumatoid arthritis. PLoS ONE. 2012;7:e30275. doi: 10.1371/journal.pone.0030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lichtenstein GR, et al. Serious infection and mortality in patients with Crohn’s disease: more than 5 years of follow-up in the TREAT registry. Am J Gastroenterol. 2012;107:1409–1422. doi: 10.1038/ajg.2012.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Keane J, et al. Tuberculosis associated with infliximab, a tumor necrosis factor α-neutralizing agent. N Engl J Med. 2001;345:1098–1104. doi: 10.1056/NEJMoa011110. [DOI] [PubMed] [Google Scholar]