Abstract
MicroRNAs (miRNAs) are post-transcriptional regulators of gene expression that play critical roles in animal development and physiology, though functions for most miRNAs remain unknown. Worms with reduced miRNA biogenesis due to loss of Drosha or Pasha/DGCR8 activity are sterile and fail to ovulate, indicating that miRNAs are required for the process of oocyte maturation and ovulation. Starting with this penetrant sterile phenotype and using new strains created to perform tissue specific RNAi, we characterize the roles of the C. elegans Pasha, pash-1, and two miRNA-specific Argonautes, alg-1 and alg-2, in somatic gonad cells and in germ cells in the regulation of ovulation. Conditional loss of pash-1 activity resulted in a reduced rate of ovulation and in basal and ovulatory sheath contractions. Similarly, knockdown of miRNA-specific Argonautes in the cells of the somatic gonad by tissue-specific RNAi results in a reduction of the ovulation rate and in basal and ovulatory sheath contractions. Reduced miRNA pathway gene activity resulted in a range of defects, including oocytes that were pinched upon entry of the oocyte into the distal end of the spermatheca in about 42% of the ovulation events observed following alg-1 RNAi. This phenotype was not observed on worms exposed to control RNAi. In contrast, knockdown of alg-1 and alg-2 in germ cells results in few defects in oocyte maturation and ovulation. These data identify specific steps in the process of ovulation that require miRNA pathway gene activity in the somatic gonad cells.
Keywords: microRNA, ovulation, somatic gonad, germ cells, C. elegans
Introduction
MicroRNAs (miRNAs) are ~22 nt small non-coding RNAs that function to repress the translation of target mRNAs, typically through binding to sites in their 3′ untranslated region (UTR) (Bartel, 2009). Through their association with Argonaute proteins, they serve as guide molecules for activity of the miRNA-induced silencing complex (miRISC) (Ha and Kim, 2014). Worms have two Argonautes that are required in the miRISC, but not other small interfering RNA (siRNA) pathways, and are encoded by alg-1 and alg-2 (Grishok et al., 2001; Hutvágner et al., 2001). Most mature miRNAs are generated through the canonical miRNA biogenesis pathway, consisting of a nuclear processing step, to generate a stem-loop pre-miRNA structure, and a cytoplasmic processing step, to generate the mature, active miRNA. Nuclear processing requires the RNase III enzyme Drosha, along with its cofactor, DGCR8/Pasha, whereas cytoplasmic processing requires the Dicer RNase III enzyme (Ha and Kim, 2014). Dicer processes both miRNAs and siRNAs while Drosha/DGCR8/Pasha is only known to process miRNAs (Grishok et al., 2001; Hutvágner et al., 2001; Ketting et al., 2001).
Although miRNAs are essential for worm, fly, fish and mouse development (Bernstein et al., 2003; Giraldez et al., 2005; Grishok et al., 2001; Ketting et al., 2001; Lee et al. 2004; Wienholds et al., 2003), the identification of specific biological functions and direct downstream targets for miRNAs remains a critical gap in our knowledge. Individual miRNAs for which functions have been described in C. elegans include the pioneering lin-4 and let-7, which act to regulate larval developmental timing. The identification of lin-4 and let-7 was achieved through the strong, penetrant phenotypes induced by their mutation (Lee et al., 1993; Reinhart et al., 2000). However, most loss of function mutations in individual miRNA genes do not result in readily observable mutant phenotypes (Miska et al., 2007). One approach to identify and characterize miRNA-regulated processes is to examine the effects of inhibiting the activity of the miRNA biogenesis pathway. Loss of the miRNA specific Argonaute genes alg-1 and alg-2 during early development causes embryonic lethality, demonstrating an essential role for miRNAs during embryogenesis (Grishok et al., 2001).
Interestingly, worms that have maternal, but not zygotic, activity of miRNA biogenesis genes, including drsh-1 (Drosha), pash-1 (Pasha/DGCR8) and dcr-1 (Dicer), are sterile with endomitotic oocytes, indicating strong, penetrant ovulation defects (Denli et al., 2004; Grishok et al., 2001; Knight and Bass, 2001). In addition, mice with conditional loss of Dicer activity in the somatic cells of the gonads (Nagaraja et al., 2008), and of Pasha ortholog DGCR8 activity in the female reproductive tract are sterile (Kim et al., 2016). These results indicate that miRNAs are required in the mouse somatic gonad for normal fertility. In worms, Dicer/dcr-1 activity is required in the somatic gonad for fertility (Drake et al., 2014) and Argonaute/alg-1 acts in the somatic distal tip cell to control germline proliferation (Bukhari et al., 2012). However, the specific events of oocyte maturation and ovulation that require miRNA activity in the somatic gonad cells in worms remain unknown.
A role for miRNAs in the control of ovulation in germ cells is less clear. While translational regulation is essential for meiotic maturation in animals (Mendez and Richter, 2001), the activity of miRNAs may not be required for germ cell development in all organisms (Ma et al., 2010). In worms, Dicer is phosphorylated and localized to the nucleus during most of oogenesis in worms, thereby preventing its normal cytoplasmic processing role (Drake et al., 2014). However, miRNA biogenesis likely functions at an early stage of germ cell development because mature, processed miRNAs are present in oocytes (Gu et al., 2009; McEwen et al., 2016). It is clear that some maternal miRNAs that are present in oocytes, including the miR-35 family, are essential for early development (Alvarez-Saavedra and Horvitz, 2010). Mosaic analysis indicates that Dicer activity is not essential in the germ line for the processes of oocyte maturation and ovulation to occur (Drake et al., 2014). However, it remains possible that maternal miRNAs act in the oocyte to more finely regulate the processes of oocyte maturation and ovulation.
Notably, the differences in miRNA abundance between worms that have zygotic deletion of Dicer and wild-type animals were found to be modest (Drake et al., 2014; Grishok et al., 2001, Knight and Bass 2001). This is surprising because Dicer is required for the cytoplasmic processing of miRNAs. A possible explanation for the continued presence of miRNAs in the dcr-1 zygotic mutants is presence of maternal Dicer activity. Because of the modest reduction of miRNA levels in a zygotic dcr-1 mutant, we analyzed the function of the miRNA specific Argonautes, alg-1 and alg-2, to determine if miRNAs can act to regulate oocyte maturation and ovulation, since these proteins act downstream of Dicer and are necessary for miRISC activity.
In worms, ovulation is a complex, rhythmic behavior that is regulated by multiple signaling pathways between the soma and the germ cells. The gonad arms contain germ cells that divide mitotically at the distal end and mature into oocytes as they reach the most proximal position of the gonad arm (Greenstein, 2005). The somatic distal tip cell (DTC) controls the mitotic zone and ten somatic sheath cells surround the rest of the germ cells with the six most proximal sheath cells capable of contraction. The spermatheca is also contractile with a constricted distal end preventing the oocyte from entering the spermatheca until ovulation (McCarter et al., 1999). Major sperm protein (MSP) is released from sperm and interacts with receptors on the somatic sheath cells to initiate contractions of the proximal sheath cells and activate meiotic maturation in the oocyte. Upon meiotic resumption, the oocyte signals to the sheath cells and spermatheca resulting in an increase in sheath contraction rate and intensity, termed ovulatory contractions, and dilation of the distal end of the spermatheca. The mature oocyte is thus propelled into the spermatheca where it is fertilized (Iwasaki et al., 1996; McCarter et al., 1999; Yin et al., 2004). Both sheath cell contraction and spermatheca dilation are dependent upon IP3-mediated calcium release (Clandinin et al., 1998). Meiotic maturation is the rate-limiting step in the production of embryos (McCarter et al., 1999). Thus, the rate of ovulation typically reflects the rate of meiotic maturation.
In order to investigate the role of miRNAs in specific events of oocyte maturation and ovulation more directly in both germ cells and the somatic gonad, we assessed the effects of conditional knockdown of miRNA pathway genes, including pash-1 and the two miRNA specific Argonautes, alg-1 and alg-2, on ovulation events, including ovulation rate, sheath cell contractility, and movement of oocyte through the spermatheca. We found that pash-1 as well as alg-1 and alg-2 activities are not essential in germ cells for ovulation. However, alg-1 and alg-2 are important in the cells of the somatic gonad for control of sheath cell contraction and dilation of the distal end of the spermatheca. Also, our data indicates that alg-1 and alg-2 activity in the somatic gonad acts to maintain the rate of meiotic maturation in the oocyte.
Materials and Methods
Strains and maintenance
C. elegans strains were grown on NGM plates seeded with E. coli strain AMA1004 at 20°C unless otherwise indicated. Strains used are listed in Table S1.
RNAi by feeding
All bacteria for RNAi experiments were isolated from the Ahringer RNAi library (Kamath et al., 2003). To knock down alg-1 activity, the X-6D15 clone (Source Bioscience) was used. alg-1 and alg-2 are 80% identical at the nucleotide level, this level of similarity is within a range where partial cross-interference in RNAi assays is expected, therefore it is likely that alg-1 knockdown results in knockdown of alg-2 (Grishok et al., 2001; Schubert et al., 2000). RNAi bacteria were cultured in Luria-Bertani (LB) broth supplemented with 100 μg/ml ampicillin, and 50 μg/ml tetracycline. Overnight cultures of RNAi bacteria were used to seed NGM plates supplemented with 1mM IPTG and 100 μg/ml ampicillin. Plates were kept at room temperature for 24 hours to allow for induction of dsRNA expression. Worms were transferred to RNAi plates at the L4 stage and F1 progeny were analyzed as young adults. Alternatively, for post-embryonic RNAi, worms were transferred at approximately the L2 stage and subsequently analyzed as young adults, approximately 42 hours later. Bacteria that have the pPD129.36/L4440 plasmid, which is an empty RNAi plasmid, was used as a negative control for RNAi.
Microscopy
Nomarski DIC and epifluorescence microscopy was performed using a Nikon 80i compound microscope equipped with a CoolSNAP HQ2 monochrome camera (Roper Scientific, CA). Images were captured with a 60× Plan Apo objective lens using Elements software (Nikon). For time-lapse observations of sheath contraction and ovulation, day 1 young adult worms were anesthetized for 20–25 min in M9 solution with 0.1% tricaine and 0.01% tetramisole (Sigma-Aldrich, St. Louis, MO) before viewing (McCarter et al., 1997). Anesthetized worms were mounted on a 2% agarose pad. For germline RNAi experiments using worms expressing the pie-1prom∷rde-1(+) transgene, day 1 young adult worms were immobilized by placing worms on 2 μl of Polybead Microspheres 0.10μm (Polysciences) on a 5% agarose pad. A small amount of petroleum jelly was placed around the coverslip to prevent desiccation. Only worms that displayed movement indicating viability on the pad were analyzed for ovulation events.
Ovulation rate assay
The method used to calculate ovulation rate was modified from McCarter et al. (1999). The ovulation rate is calculated as the number of ovulation events per gonad arm per hour, which reflects the rate of meiotic maturation. Day 1 young adults were placed onto individual plates and viewed using a Nikon SMZ-1500 stereomicroscope to determine the initial number of embryos present in the uterus. Worms were then transferred to a 20°C incubator for an average of 3 hours; at which point the final number of embryos inside the uterus was counted. The number of ovulations for each individual worm was determined by counting the number of embryos or live progeny produced. These values were placed into the formula [(Final number of embryos in uterus − Initial number of embryos in uterus) + number of progeny or embryos produced]/[(2 gonad arms) (3 hours)] = number of ovulation events per hour per gonad arm.
In vivo analysis of sheath cell contractions
Time-lapse DIC images of the proximal gonad and oocytes were captured (10 frames/sec) during individual ovulation events prior to rounding of the proximal oocyte through entry of the fertilized embryo into the uterus. Worms were observed until ovulation occurred or for 60 minutes. If the proximal oocyte did not resume meiosis and no ovulation occurred within 60 minutes, they were categorized as failing to resume meiosis. Worms in which the oocyte resumed meiosis but failed to fully enter or exit the spermatheca were observed for a minimum of 10 minutes post nuclear envelope breakdown or entry into the spermatheca, respectively. To measure the rate of gonadal sheath contractions, the number of lateral displacements were counted, focusing on one side of the −1 oocyte that included the junction to −2 oocyte. The rate of basal contractions was calculated during a 3–5 minute interval prior to the initiation of ovulatory contractions. The rate of ovulatory contractions was determined by the maximum number of contractions that occurred in the −3 to 0 minute period, with time 0 indicating when the oocyte was fully in the spermathecal (McCarter et al., 1999).
Construction of transgenic strains
Gibson assembly (NEB) was used to generate the pCR4 plasmid containing the inx-8prom∷rde-1(+)∷unc-543′utr transgene in the pCFJ909 plasmid backbone for use in MiniMOS insertion (Frøkjaer-Jensen et al., 2014). PCR amplification of a 1067 bp genomic fragment upstream of the inx-8 start site was performed using primers AA1225 and AA1229 and N2 genomic DNA as a template. Primer sequences are provided in Table S2. PCR amplification of rde-1(+)∷unc-543′utr was performed using AA1052 and AA1222 primers and the pXXY2004.1 plasmid (Espelt et al., 2005) as a template. pXXY2004.1 was kindly provided by Dr. Keith Nehrke (University of Rochester, NY). PCR products (0.05 pmoles) were mixed with a 5.1 kb PstI-SpeI pCFJ909 fragment (0.05 pmoles) in a Gibson assembly reaction (NEB). pCR4 construction was confirmed by sequencing. To generate strains that contain the inx-8prom∷rde-1(+)∷unc-543′utr transgene inserted in the genome, an injection mix containing pCR4 (10ng/ul), along with coinjection plasmids pGH8 (10ng/ul), pCFJ90 (2.5ng/ul), pCFJ104 (10ng/ul), pMA122 (5 ng/ul) and pCFJ601 (50 ng/ul) was injected into unc-119(ed3) worms. Worms that contained a transgene insertion were obtained as described in Frøkjaer-Jensen et al. (2014). Progeny were screened for rescue of the Unc phenotype and for the absence of coinjection marker expression (detected by RFP expression), which indicated the presence of an extrachromosomal array. For the xwTi1 inx-8prom∷rde-1(+)∷unc-543′utr insertion, inverse PCR was performed to determine the genomic location (Table S1). Worms with inx-8prom∷rde-1(+)∷unc-543′utr transgenes were crossed with rde-1(ne219)V mutant worms using standard genetic approaches, using PCR to follow the xwTi1 insertion (AA1309 with Ocf1588, and AA1310 with AA1309 primers). The presence of the ne219 allele was confirmed by sequencing.
MultiSite Gateway cloning (Life Technologies) was used to generate the pCR6 plasmid containing the pie-1prom∷rde-1(+)∷tbb-23′utr transgene. First, PCR was used to amplify the rde-1(+) coding region using primers AA1282 and AA1283 with pXXY2004.1 as a template. The necessary attB1 and attB2 sequences were added using PCR primers. The PCR product was recombined with the Gateway donor vector pDONR221 to make the pCR5 rde-1(+) entry clone. pCR5 construction was confirmed by sequencing. To generate the pie-1prom∷rde-1(+)∷tbb-23′utr transgene, pCR5 along with entry clones pCM1.127 and pCM1.36, which supplied the pie-1 promoter and the tbb-2 3′ UTR sequences (Addgene plasmid #21384, #17249), respectively, were recombined with destination vector pCFJ907 to generate pCR6. pCR6 junctions were confirmed by sequencing. rde-1(ne219) mutant worms were injected with pCR6 (10ng/μl) along with coinjection plasmids as described above. Worms that contained a transgene insertion were obtained as described in Frøkjaer-Jensen et al. (2014). Progeny were screened for resistance to G418 (Invitrogen), which was added to NGM plates to a final concentration of 0.4 mg/ml, along with the absence of coinjection marker expression, which indicated the presence of an extrachromosomal array. Inverse PCR was performed to determine the genomic location of the xwTi5 insertion (Table S1).
Results
Sterility in miRNA biogenesis mutants has been previously observed. We start by showing that the basis of sterility in drsh-1(ok369) mutant worms is the failure of ovulation, accompanied by and likely due to the absence of sheath contractions. Then we demonstrate the consequences that the knockdown of the pash-1 gene has for various steps of ovulation. Finally, we use tissue specific knockdown of miRISC genes alg-1 and alg-2 in somatic gonad and germ cells, as a way to determine the function of miRNA pathway genes that act further down the miRNA pathway in the regulation of oocyte maturation and ovulation.
miRNA-specific biogenesis genes are required for ovulation
While sterility has been described for miRNA biogenesis mutants, including dcr-1 and drsh-1 mutants (Denli et al., 2004; Grishok et al., 2001; Knight and Bass, 2001), specific defects in the process of ovulation have not been described. drsh-1(ok369) homozygous mutants were derived from balanced, heterozygous worms and therefore had maternal activity of drsh-1(+), allowing for worms to complete embryonic and larval development. Zygotic drsh-1 mutant worms have a sterile phenotype with endomitotic oocytes (Denli et al., 2004). To examine this defect further, ovulation rate analysis and video microscopy was performed. Zygotic drsh-1 mutants produced no progeny (Fig. S1A) and had few, if any, ovulation events (Fig. 1A). In agreement, video microscopy revealed essentially no gonadal sheath contractions in ~1 hour of observation (data not shown). These results provide quantitative analysis of the observed sterile phenotype and further support a requirement for miRNA biogenesis genes in the process of ovulation.
Figure 1. miRNA pathway genes are required for the regulation of the ovulation rate, somatic sheath contractions and distal spermathecal dilation during ovulation.
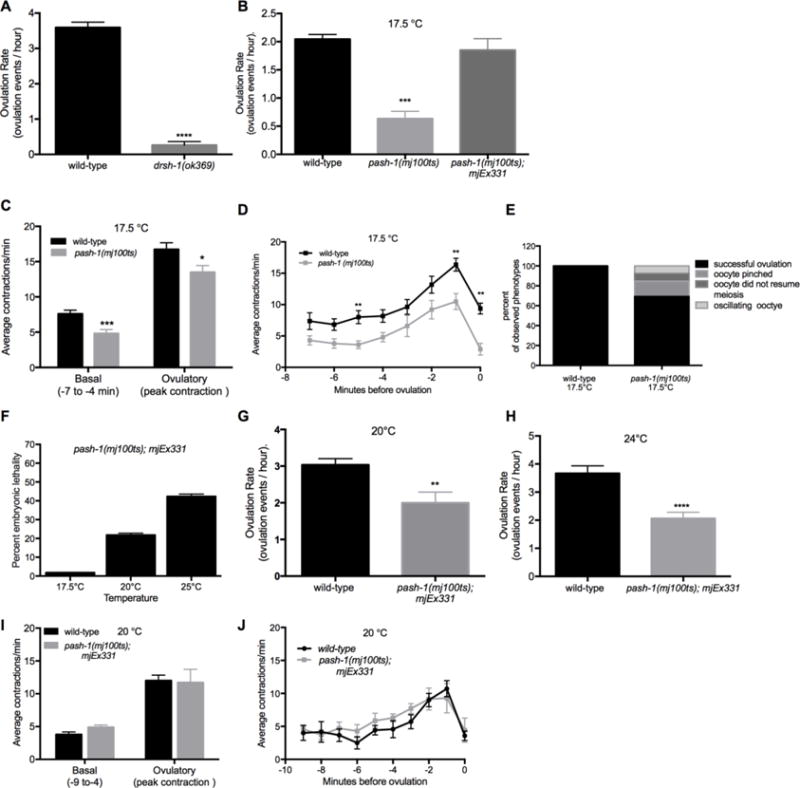
(A–B) Ovulation rates were measured by total progeny and embryo production in populations of wild-type, miRNA biogenesis mutant worms and miRNA biogenesis mutant worms expressing a rescue array at different temperatures. (A) Ovulation rates (ovulation events/hour) for wild-type and drsh-1(ok369) worms at normal growing conditions of 20° C. drsh-1(ok369) were zygotic mutants, collected from heterozygous hermaphrodites and therefore had maternal drsh-1 activity (n = 14–30). (B) Wild-type, pash-1(mj100ts) and pash-1(mj100ts) carrying an extrachromosomal rescue array, mjEx331, grown at the permissive temperature of 17.5°C ovulation rates (ovulation events/hour) (n = 10–25). (C–E) Individual ovulation events were analyzed using time-lapse microscopy. (C) Average basal and ovulatory sheath contractions were determined for wild-type and pash-1(mj100ts) mutants grown at 17.5°C. Average basal contractions were determined using the −7 to −4 minute interval with time 0 corresponding to when the oocyte is inside the spermatheca. Average peak ovulatory contractions were determined using the single highest contraction rate at 17.5°C (n = 10). (D) Wild-type and pash-1(mj100ts) sheath contraction rates were analyzed as single minute intervals from −7 minutes until 0 minutes at 17.5°C (n = 10). (E) A summary of ovulation phenotypes observed for individual ovulation events in wild-type (n=10) and pash-1(mj100ts) (n =13 events) worms. (F–J) pash-1(mj100ts) mutant worms carrying an extrachromosomal rescue array, mjEx331, were grown at different temperatures (17.5–25°C). Worms grown at restrictive temperatures lost activity from the pash-1(mj100ts) allele. The rescue array is only expressed in the soma due to transgene silencing. (F) Embryonic lethality was measured at 17.5°C, 20°C and 25°C (n > 200). (G–H) The ovulation rate (ovulation events/hour) was measured by counting total progeny and embryos produced for wild-type and pash-1(mj100ts), mjEx331 worms grown at the restrictive temperatures of 20°C and 24°C respectively. (I–J) Sheath contractility was analyzed for individual ovulatory events using time-lapse microscopy in wild-type and pash-1(mj100ts), mjEx331 worms. The average rate of basal contractions was determined from the −9 to −4 minute interval. Time 0 corresponds to when the oocyte is inside the spermatheca. The average rate of ovulatory sheath contraction was determined by using the single highest contraction rate observed in individual worms for wild-type and pash-1(mj100ts), mjEx331 worms at 20°C (n=7). pash-1(mj100ts); mjEx331 worms grown at 20°C. There were no ovulation defects observed for wild-type worms grown at 20°C (n = 10, data not shown), or for pash-1(mj100ts); mjEx331 worms grown at 20°C (n = 7, data not shown). Error bars indicate SEM. Statistical analysis was performed using unpaired, non-parametric, t-test * p < 0.05, *** p < 0.001, **** p < 0.0001.
To identify the specific steps in ovulation for which miRNA biogenesis is required, we used the mj100 conditional allele of the pash-1 gene, which results in reduced, but not eliminated, miRNA biogenesis activity. We used pash-1(mj100ts) worms, with and without the mjEx331 (eft-3prom∷pash-1(+)) extrachromosomal rescue array (Lehrbach et al., 2012). pash-1(mj100ts) mutant worms are viable and develop essentially normally at 15°C but are not viable at 25°C, with severe defects observed in miRNA biogenesis (Lehrbach et al., 2012). It is important to note that pash-1(mj100ts) worms display an early aging phenotype with shortened lifespan and altered metabolism at the restrictive temperature of 25°C (Lehrbach et al., 2012). It is possible that worms may display a weaker early aging phenotype at the intermediate temperatures. To minimize any potential indirect effects from early aging, all analysis was performed in young adult worms, within 24 hours of the L4 molt.
Compared to wild-type worms, pash-1(mj100ts) mutants had significantly reduced brood size (Fig. S1A) and ovulation rate at 15° C (data not shown), and these defects were even more pronounced at the intermediate temperature of 17.5° C (Fig. 1B). The temperature of 17.5°C was chosen for further analysis because worms showed a reduced ovulation rate but remained viable. When worms are grown at 17.5° C, pash-1 activity is expected to be moderately reduced (Lehrbach et al., 2012) so that phenotypes observed demonstrate the effects of a reduction, but not an absence, of miRNAs. When pash-1(mj100ts) mutant worms are grown at 20°C or higher, the worms are not viable, therefore analysis of ovulation events was not possible at elevated temperatures.
Next, we analyzed whether decreased pash-1 activity resulted in other observable ovulation phenotypes. Video microscopy of pash-1(mj100ts) worms grown at 17.5°C was performed to observe individual ovulation events (n = 13). Gonadal sheath cells were monitored and found to have a reduced rate for basal and ovulatory contractions (Fig. 1C–D) compared to wild-type. There were no ovulation defects observed in wild-type worms grown at 17.5°C (Fig. 1E). In contrast, in over 30% of the recorded ovulation events in pash-1(mj100ts) worms ovulation defects were observed (Fig. 1E). In 15% of the recorded ovulation events, the proximal oocyte was pinched by the distal end of the spermatheca while entering the spermatheca (Fig. 1E and Fig. 2D–F). In addition, in 8% of the ovulation events the oocyte failed to resume meiosis, and in another 8% of the events the oocyte oscillated in and out of the spermatheca (Fig. 1E and Fig. 2G–I). These data indicate that miRNAs are required either in the somatic cells or in the germ cells for regulation of sheath cell contractions and spermatheca dilation in the process of ovulation.
Figure 2. Ovulation defects observed following knockdown of miRNA pathway gene activity.
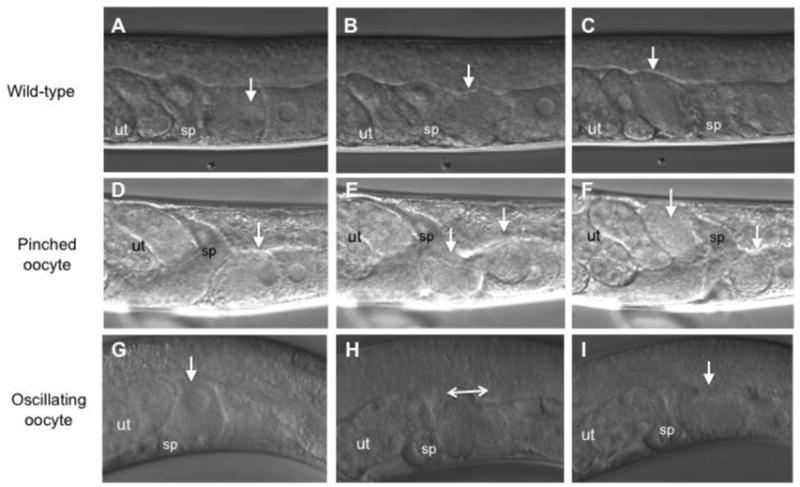
Individual ovulation events were analyzed using time-lapse Nomarski DIC microscopy and representative micrographs show selected observed phenotypes. Animals are oriented with the dorsal side up. The oocytes housed in the gonad arm are toward the right and the uterus is toward the left. The ovulating oocyte is indicated by a white arrowhead. (A–C) Wild-type ovulation event. (A) The proximal oocyte resumed meiosis as shown by the initiation of breakdown of the nuclear envelope. B) The proximal oocyte subsequently entered the spermatheca. C) After transit through the spermatheca, the fertilized oocyte was observed inside uterus. (D–E) Ovulation event with pinching of the ovulating oocyte by the distal spermatheca is shown. rrf-1(ok589) exposed to alg-1 post embryonic RNAi (D) The proximal oocyte resumed meiosis. (E) The proximal oocyte was pinched by distal spermatheca as the oocyte entered the spermatheca. Two arrowheads show the two sides of the pinched oocyte. (F) Pinching resulted in an oocyte fragment in the uterus and a fragment in the proximal somatic gonad indicated by arrowheads. (G–I) Ovulation event with oscillation of the oocyte into and out of the spermatheca in a pash-1(mj100) worm grown at 17.5 °C G) The proximal oocyte resumed meiosis. (H) The proximal oocyte was observed to enter the spermatheca and then oscillate back and forth between the spermatheca and proximal gonad arm. (I) The ovulating oocyte never transited through the spermatheca but rather remained in the proximal gonad arm. sp, spermatheca. ut, uterus
In order to determine if germ cells require pash-1 activity for ovulation, pash-1(mj100ts) worms with the mjEx331 rescuing extra-chromosomal array were analyzed at elevated temperatures. Because extra-chromosomal arrays are typically silenced in the germ line, the mjEx331 array provides somatic, but not germ cell, pash-1(+) rescuing activity. First, compared to wild-type worms, pash-1(mj100ts) with mjEx331 at 17.5° C had no significant difference in the rate of ovulation (Fig. 1B). This result indicates that expression of wild-type pash-1(+) activity in somatic cells is sufficient to restore ovulation rate at the intermediate temperature of 17.5° C (Fig. 1B). To validate knockdown of maternal miRNAs in the germline, we analyzed the rate of embryonic lethality in pash-1(mj100ts);mjEx331 grown at elevated temperatures. It is known that loss of maternal miRNAs, including the mir-35 family, results in a fully penetrant embryonic lethality phenotype (Alvarez-Saavedra and Horvitz, 2010). Embryonic lethality increased with temperature in pash-1(mj100ts);mjEx331 worms (Fig. 1F), suggesting reduced levels of maternal miRNAs in the oocyte. pash-1(mj100ts) worms without the rescuing array that are shifted to the restrictive 25°C show 100% embryonic lethality and mature miR-35 is not detectable (Lehrbach et al., 2012). Because the observed embryonic lethality phenotype was not fully penetrant in pash-1(mj100ts);mjEx331 at 24°C (Fig 1F), it is likely that there some activity of the pash-1(ts) allele is still present in worms grown at 24°C for many generations.
Next we tested ovulation rates for the pash-1(mj100ts);pash-1∷gfp with the mjEx331 rescue array at elevated temperatures (Fig. 1G–H). It was not possible to analyze pash-1(mj100ts) worms without the rescue array at elevated temperatures because viability is reduced when worms are grown at 20°C or higher. The ovulation rate was reduced in pash-1(mj100ts);mjEx331 grown at 20°C and even further at 24°C compared to wild-type worms (Fig. 1G–H). The ovulation rate of worms grown at 24°C was reduced from 3.7 ovulation events/hour in wild-type to 2.1 (p < 0.0001) in the pash-1(mj100ts);mjEx331 (Fig. 1H). The decrease in the ovulation rate of the pash-1(mj100ts) with the mjEx331 rescue array at elevated temperatures suggests a possible role for miRNAs in germ cells in the process of ovulation, or alternatively is a result of incomplete pash-1 (+) somatic rescue of the array.
Video microscopy was performed to analyze ovulation events (n = 13) in pash-1(mj100ts);mjEx331 worms with somatic pash-1(+) rescue. Worms were analyzed at 20° C. In spite of the decreased ovulation rate (Fig. 1G), there were no observable differences in the rate of basal or ovulatory contractions compared to wild-type (Fig. 1I–J). In addition, there were no defects in transit through the spermatheca observed during ovulation events in pash-1(mj100ts); mjEx331 (n=13 successful ovulation events). Together, our results suggest a role for pash-1 activity in somatic cells and germ cells for the ovulation rate, and in somatic cells for sheath contractions and transit of the oocyte through the spermatheca.
miRNA-specific Argonaute activity is required in somatic gonad cells, but not germ cells, for ovulation
Having identified ovulation rate, sheath contractions, and distal spermatheca defects in the pash-1(mj100ts) worms, we next investigated the role for miRNA specific Argonautes in the cells of the somatic gonad during ovulation by knocking down alg-1 and alg-2. The miRISC functions downstream of Pasha/pash-1 in the miRNA pathway and is required for miRNA activity. C. elegans have 27 Argonaute genes (Youngman and Claycomb, 2014). Out of these, only alg-1 and alg-2, have been shown to be required for the miRNA pathway, (Grishok et al., 2001; Hutvágner et al., 2004), and not RNAi (Grishok et al., 2001). To validate the RNAi knockdown of alg-1, we exposed L4-stage alg-2(ok304) mutant worms to alg-1 RNAi and verified 100% embryonic lethality (data not shown), in parallel with every RNAi experiment. First, tissue specific knockdown of alg-1 was performed in the cells of the somatic gonad. Expression of alg-1 and alg-2 has been analyzed extensively (Tops et al., 2006; Vasquez-Rifo et al., 2012). alg-1 is expressed from early embryogenesis to adulthood in most, if not all, cells (Tops et al., 2006) and both alg-1 and alg-2 are expressed in the cells of the somatic gonad (Vasquez-Rifo et al., 2012).
We generated two strains for tissue specific RNAi; the first targeted the entire somatic gonad, and the second the sheath cells of the somatic gonad. The first strain included a transgene with a wild-type copy of the rde-1 gene, regulated by the mir-786 promoter, which is expressed in the somatic gonadal sheath cells, spermatheca, uterus, as well as in the posterior cells of the intestine (Kemp et al., 2012). This transgene was expressed from an extrachromosomal array in an alg-2(ok304); rde-1(ne219) genetic background. The alg-2(ok304) mutation was included to further reduce miRNA specific Argonaute activity. The second strain contained a transgene with the inx-8 promoter driving the expression of wild-type copy of the rde-1 gene in the somatic gonadal sheath cells. The somatic innexin inx-8 gene is expressed in the proximal sheath cells as a component of the gap junctions that connect sheath cells to developing oocytes (Starich et al., 2014). Therefore, this promoter was chosen to test for the function of miRNA activity specifically in somatic sheath cells. The ability of the inx-8 promoter to drive the expression of rde-1(+) in the sheath cells was validated by analyzing inx-8prom∷rde-1(+) transgenic worms that had ubiquitous expression of gfp. These worms were exposed to gfp RNAi, and it was found to successfully knockdown gfp in the sheath cells (Table S3 and Fig. S2).
The rde-1(ne219);inx-8prom∷rde-1(+) transgenic worms and the rde-1(ne219) control strain were exposed to alg-1 RNAi and a control RNAi, while the alg-2(ok304);rde-1(ne219); mir-786prom∷rde-1(+) and the alg-2(ok304);rde-1(ne219) control strain were exposed to post-embryonic alg-1 RNAi and a control RNAi (see methods). Compared to alg-2(ok304);rde-1(ne219) controls, alg-2(ok304);rde-1(ne219); mir-786prom∷rde-1(+) worms showed a significantly reduced rate of ovulation from a rate of 3.2 ovulation events/hour following control RNAi to 0.9 ovulation events/hour following alg-1 RNAi (Fig. 3A). There was also a significant decrease in overall brood size (p < 0.0001, Fig. S1).
Figure 3.
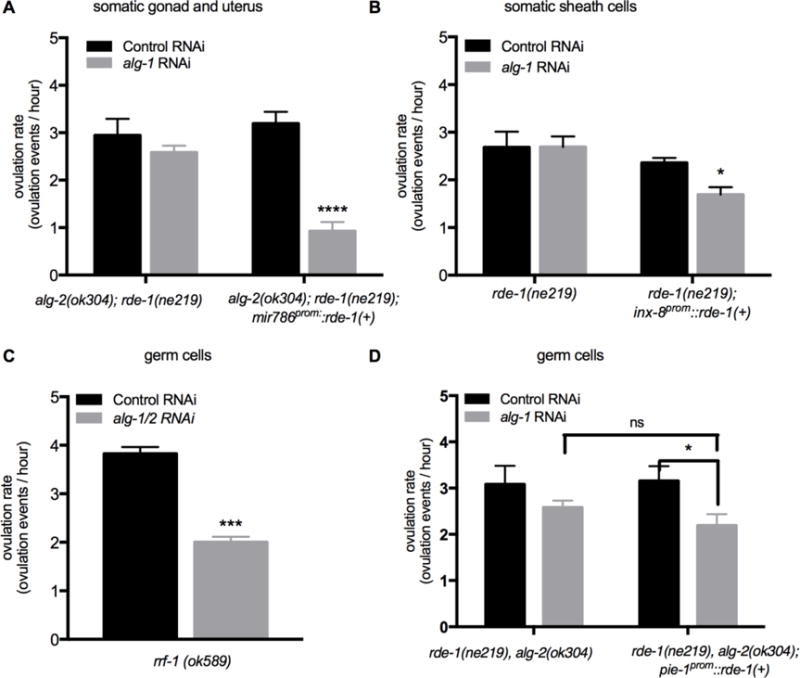
miRNA-specific Argonautes alg-1 and alg-2 are required in the somatic gonad, but likely not in the germ cells, for the regulation of ovulation rate. Ovulation rate was analyzed following RNAi to knockdown alg-1 or a control RNAi (empty vector). For each group, 10–20 young adult worms were analyzed. To verify alg-1 knockdown, we exposed alg-2(ok304) L4 stage worms to alg-1 RNAi and confirmed that they exhibited 100% embryonic lethality. (A) Ovulation rates (ovulation events/hour) measured by total progeny and embryo production in alg-2(ok304); rde-1(ne219) and alg-2(ok304);rde-1(ne219); mir-786prom∷rde-1(+) following post-embryonic RNAi. The mir-786 promoter drives the expression of rescuing rde-1(+) activity throughout the somatic gonad. (B) Ovulation rates (ovulation events/hour) measured by total progeny and embryo production in rde-1(ne219) control strain and rde-1(ne219); inx-8prom∷rde-1(+) following RNAi. The inx-8 promoter drives expression of rescuing rde-1(+) activity in the somatic sheath cells. (C–D) Tissue specific gene knockdown in germ cells was performed using rrf-1(ok589) mutants and rde-1(ne219); alg-2(ok304); pie-1prom∷rde-1(+) transgenic worms following RNAi to knockdown alg-1 or with control (empty vector) RNAi. (C) Ovulation rates (ovulation events/hour) for rrf-1(ok589) worms following alg-1 or control (empty vector) RNAi (n = 10–20). (D) Ovulation rates (ovulation events/hour) for rde-1(ne219); alg-2(ok304) control strain and rde-1(ne219); alg-2(ok304); pie-1prom∷rde-1(+) transgenic worms following post-embryonic alg-1 or control RNAi. The pie-1 promoter drives rescuing rde-1(+) expression in germ cells. Error bars indicate SEM. Statistical analysis was performed using unpaired, non-parametric, t-test for C and two-way ANOVA, Tukey’s multiple comparison for A–B and D. * p < 0.05, *** p < 0.001, *** p < 0.001, **** p < 0.0001
Ovulation also requires the normal development and physiology of germ cells to proceed normally. Signals from sperm, including MSP, act to trigger meiotic maturation in the proximal oocyte, while signals from the oocyte act to trigger increased sheath contractility and dilation of the distal end of the spermatheca (Iwasaki et al., 1996; McCarter et al., 1999). The decrease in the ovulation rate observed in pash-1(mj100ts) worms with the rescue array, lead us to further assess the role of miRNAs in the germ cells. Our approach was to knockdown miRNA-specific Argonautes. First, knockdown of alg-1 was performed in rrf-1 mutant worms. rrf-1 encodes an RNA dependent RNA polymerase that is required for RNAi in somatic tissue (McCarter et al., 1999; Sijen et al., 2001; Yin et al., 2004). rrf-1 mutants are sensitive to RNAi in the germline, but are resistant in somatic cells (Sijen et al., 2001). However, rrf-1 mutants have been found to display RNAi in some somatic tissues including the intestine and the hypodermis, but, importantly, no RNAi was observed in cells of the somatic gonad (Kumsta and Hansen, 2012). RNAi was performed in rrf-1 worms at the L4 stage and their F1 progeny were analyzed as young adults. In rrf-1(ok589) mutants, knockdown of alg-1 by RNAi resulted in a reduced ovulation rate (Fig. 3C), supporting the results observed in the pash-1(ts) worms with the rescue array.
Because the phenotypes observed upon alg-1 knockdown in the rrf-1(ok589) mutants may reflect indirect involvement of somatic tissues, a tissue specific RNAi strain was constructed, using the pie-1 promoter to drive rde-1(+) activity, followed by the tbb-2 3′UTR. The pie-1 gene encodes a zinc finger protein that is essential for germline cell fate (Reese et al., 2000). The pie-1 promoter allows expression in all germ cells (D’Agostino et al., 2006). This transgene was expressed in an alg-2(ok304); rde-1(ne219) genetic background. For these experiments, post-embryonic RNAi was performed starting in L2 stage worms in an effort to avoid potential knockdown in any somatic lineages (Reece-Hoyes et al., 2007). The ability of the pie-1 promotor to drive expression of rde-1(+) in germ cells, and therefore restore RNAi sensitivity, was validated using a control RNAi experiments (Table S3). Exposure to alg-1 RNAi beginning at the L2 stage in alg-2(ok304); rde-1(ne219); pie-1prom∷rde-1(+) hermaphrodite worms, resulted in 100% embryonic lethality indicating a strong, penetrant reduction in miRNA biogenesis in the germline. When worms of this genotype were exposed to post-embryonic alg-1 RNAi, a decreased rate of ovulation was observed compared to control RNAi (p = 0.0474) (Fig. 3D). However, the rate of ovulation was not significantly different from rde-1(ne219);alg-2(ok304) without the pie-1prom∷rde-1(+) transgene (Fig. 3D) indicating that the modest effect could be attributed to a variation in the rde-1(ne219);alg-2(ok304) background and was not due to the specific knockdown of miRNA biogenesis in the germline. In addition, there was no significant decrease in the total number of embryos produced (Fig. S1D). These results indicate that germ cells do not require alg-1 and alg-2 to maintain the normal ovulation rate.
alg-1 and alg-2 are required in the somatic gonad for sheath contraction and spermatheca dilation
We used our tissue specific RNAi strains to investigate whether reduction in alg-1 and alg-2 activity in cells of the somatic gonad and germ cells would result in reduced sheath contractions and distal spermathecal dilation, as observed in pash-1(mj100ts) mutants. Video microscopy of alg-2(ok304);rde-1(ne219); mir-786prom∷rde-1(+) worms demonstrated that the average basal contractions were reduced from 4.2 to 2.7 contractions/minute (p < 0.05) while the average ovulatory contractions had a larger reduction from 16.7 to 9.1 contractions/min. (p < 0.0001) following alg-1 RNAi compared to control RNAi (Fig. 4A–B). Similar, but more severe defects to pash-1(ts) mutants were observed during video microscopy of individual ovulation events (n = 12): 25% showed successful ovulations (Fig. 2A–C, Fig. 4C), 42% showed pinching by the distal spermatheca (Fig. 2D–F, Fig. 4C), 25% failed to resume meiosis and did not ovulate within 1 hour of observation (Fig. 4C), and 8% entered but failed to exit the spermatheca (Fig. 4C). alg-2(ok304);rde-1(ne219); mir-786prom∷rde-1(+) worms on control RNAi showed few ovulation defects: 92% showed successful ovulations (Fig. 2A–C, Fig. 4C), and 8% entered but failed to exit the spermatheca (Fig. 4C, n = 12). These data support a role for miRNAs acting in the somatic gonad to regulate the rate of meiotic maturation, rate of sheath cell contractions, and the dilation of the distal spermatheca.
Figure 4. miRNA-specific Argonautes alg-1 and alg-2 are required in the somatic gonad for sheath contractility and distal spermathecal dilation.
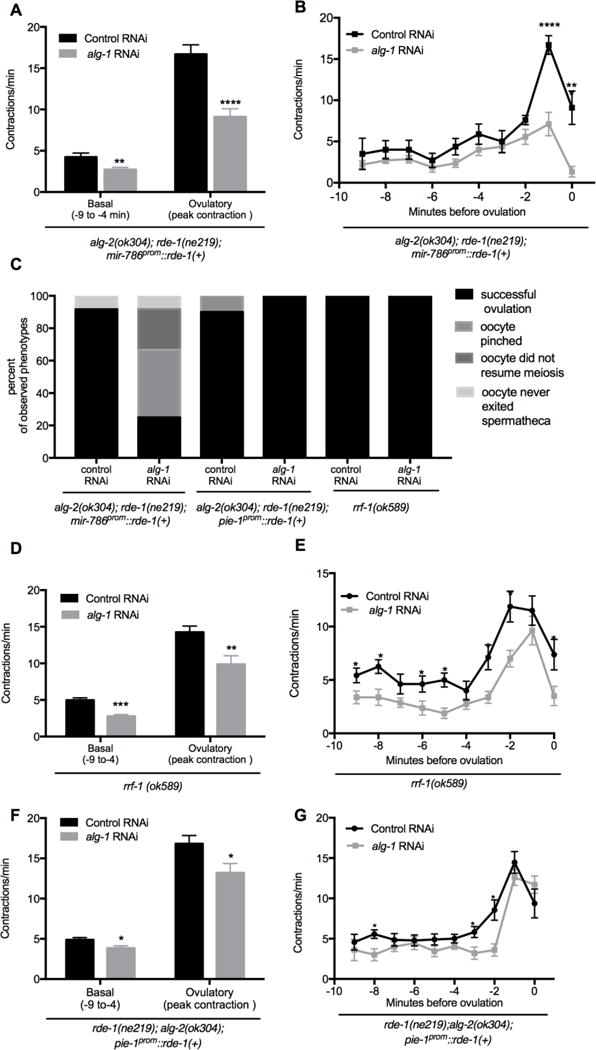
(A–B) Sheath contractility was analyzed during individual ovulatory events using time-lapse microscopy in alg-2(ok304); rde-1(ne219); mir-786prom∷rde-1(+) following post-embryonic alg-1 and control RNAi (n = 12). (B) The average rate of basal contractions was determined from the −9 to −4 minute interval with time 0 corresponding to when the oocyte is inside the spermatheca. Average peak ovulatory sheath contraction rate was determined by using the single highest contraction rate observed in individual worms. The average peak ovulatory contractions were determined by using the single highest contraction rate observed in individual worms. (B) Contraction are shown as single minute intervals prior to ovulation. (C) A summary of ovulation phenotypes observed for individual ovulation events in rde-1(ne219); alg-2(ok304); mir786prom∷rde-1(+) (n=12), rde-1(ne219); alg-2(ok304); pie-1prom∷rde-1(+) (n=10), and rrf-1(ok589) (n=10) mutant worms following control and alg-1 RNAi. For descriptions of ovulation defects, see Figure 2. (D–G) alg-1 and control (empty vector) RNAi was performed on rrf-1 and post-embryonically on rde-1(ne219); alg-2(ok304); pie-1prom∷rde-1(+) mutant worms. (D) The average rate of basal contractions was determined using the −9 to −4 minute interval with time 0 corresponding to when the oocyte is inside the spermatheca. The average ovulatory sheath contraction rate was determined by using the single highest contraction rate observed in individual worms (n = 10). (E) Contraction rates are shown in single minute intervals prior to ovulation. (F) The average rate of basal contractions was determined using the −9 to −4 minute interval. The average ovulatory sheath contraction rate was determined by using the single highest contraction rate observed in individual worms (n = 10). (G) Contraction rates are shown in single minute intervals prior to ovulation for rde-1(ne219); alg-2(ok304); pie-1prom∷rde-1(+) worms following RNAi. Error bars indicate SEM. Statistical analysis was performed using unpaired, non-parametric, t-test for (A–B, D–G). * p < 0.05, *** p < 0.001, *** p < 0.001, **** p < 0.0001
To assess the role of alg-1 and alg-2 in germ cells in individual ovulation events, we used our tissue specific RNAi strains that allow for knockdown in germ cells but not in somatic gonad cells. In rrf-1(ok589) mutants, knockdown of alg-1 by RNAi resulted in reduced basal and ovulatory sheath contractions (Fig. 4D–E), however the pattern of sheath contractions closely mirrored that of worms placed on control RNAi. Importantly, rrf-1(ok589) worms exposed to alg-1 RNAi and control RNAi showed wild-type ovulation events (Fig. 2A–C, Fig. 4C) with no defects in oocyte transit through the spermatheca observed (n>10, Fig. 4C).
We next performed video microscopy on the alg-2(ok304); rde-1(ne219); pie-1prom∷rde-1(+)) worms to on both alg-1 and control RNAi. A modest decrease in the rates for basal and ovulatory contractions was observed following alg-1 RNAi, but the pattern of sheath contractions mirrored even more closely those of worms exposed to control RNAi (Fig. 4F–G). Importantly, knocking down alg-1 in the alg-2(ok304); rde-1(ne219); pie-1prom∷rde-1(+)) mutants did not result in any defects in oocyte transit through the spermatheca (n > 10, Fig. 4C). We conclude that miRNA pathway genes are not essential in germ cells for the process of ovulation in C. elegans.
Discussion
Worms with mutation in genes involved in the miRNA-specific biogenesis pathway including drsh-1 and pash-1, are sterile (Denli et al., 2004; Grishok et al., 2001) and we observed essentially no successful ovulations. However, the functional roles of miRNAs in the complex process of ovulation remain unknown. Using a conditional pash-1(mj100ts) mutant and new strains created to perform tissue specific RNAi, we characterized the roles of miRNA-specific biogenesis gene pasha in the whole worm and Argonautes alg-1 and alg-2 in somatic gonad cells and in germ cells. The knockdown of the miRNA-specific Argonautes interferes with the miRNA biogenesis pathway downstream of previously characterized miRNA biogenesis mutants (Denli et al., 2004; Drake et al., 2014; Grishok et al., 2001; Knight and Bass, 2001) and impacts the activity of the miRNAs. We conclude that alg-1 and alg-2 and likely miRNA activity, is not essential in germ cells for ovulation, but is critical in the cells of the somatic gonad for proper sheath cell contraction and distal spermathecal dilation. In addition, results indicate that miRNAs may function in cells of the somatic gonad for the control of meiotic maturation in oocytes as evident by the decrease in ovulation rate, which requires oocyte maturation to occur.
In contrast to drsh-1 zygotic mutants, we did not observe a penetrant sterile phenotype in any of our experimental conditions. This likely reflects the conditional or partial knockdown of miRNA biogenesis. This could be due, in part, to incomplete knockdown of Argonaute activity. Also, worms have 27 Argonaute genes, including one that is closely related to ALG-1/2, T23D8.7/hpo-24, therefore it remains possible that additional Argonaute proteins could function to mediate miRNA regulation in the germline (Youngman and Claycomb, 2014), though such activity has never been described. In addition, worms with the mir-786prom∷rde-1(+) transgene expressed rde-1(+) activity from an extrachromosomal array, which can have some mosaicism in transgene expression. Lastly, worms with the pash-1(mj100ts) allele likely have some residual protein activity at the semi-permissive temperature of 17.5°C.
miRNAs in germ cells may not have an essential role in ovulation
miRNAs are found in germ cells across the animal kingdom including worms, flies, mice, and cows (Gilchrist et al., 2016; Gu et al., 2009; Lee et al., 2014; Ma et al., 2010; McEwen et al., 2016). In worms, the mir-35 family is one of several miRNA families found to be highly enriched in germ cells, specifically in oocytes (Gu et al., 2009; McEwen et al., 2016). Like other miRNAs that are expressed in oocytes, the mir-35 family is required for embryonic development but has no known function in oocyte formation or maturation (McJunkin and Ambros, 2014). In previous work with zygotic dcr-1(0), levels of miRNAs in the germline were surprisingly high, leaving the possibility that they are functioning even in the dcr-1(0) genetic background. In this work, we take an alternative approach by reducing the activity of miRNA-specific genes alg-1 and alg-2 in the germ cells. ALG-1 and ALG-2 act downstream of Dicer in the biogenesis pathway and function in the activity of the miRNA silencing as a core component of the miRISC. This approach allowed us to examine the function of miRNAs in germ cells in the process of oocyte maturation and ovulation.
Post-transcriptional control of gene expression is essential in the C. elegans germline for the development of mature oocytes. Analysis of reporter transgene expression demonstrated that the 3′UTRs of mRNAs in the germline and developing oocytes play a larger role than the promoter regions (Merritt et al., 2008). RNA binding proteins and miRNAs can both function through sites in the 3′ UTR to control translation. Many RNA binding proteins are essential for normal germline development, including PUF proteins, GLD-1, and NOS-3 (Lee and Schedl, 2006). To test whether miRNAs found in germ cells act as 3′ UTR regulators in the process of ovulation, we knocked down miRNA biogenesis in these cells. There was strong embryonic lethality in both our pie-1prom∷rde-1(+) and our pash-1(ts);mjEx331 rescue mutants indicating strong, penetrant knockdown of miRNA activity. Surprisingly, despite this strong knockdown, we did not observe similarly strong or penetrant ovulation defects. Our study therefore indicates that germ cell miRNAs do not play an essential role for the control of oocyte maturation or ovulation, but are required soon after ovulation for early embryonic development.
The observation that miRNA activity is largely dispensable in the germline prior fertilization is consistent with research in mice and worms, but inconsistent with research in flies. In mice, gene regulation by miRNAs is inactive in both oocytes and early embryos before the maternal-to-zygote transition (Svoboda, 2010). Also in mice, oocytes with a deletion in the miRNA-specific Dgcr8 exhibit normal maturation, and deletion of both maternal and zygotic Dgcr8 alleles does not result in any pre-implantation development phenotypes. In addition, limited miRNA-directed mRNA degradation occurs in mouse oocytes (Ma et al., 2010). In worms, sterility of dcr-1(0) mutants is due to lack of DCR-1 in the soma, not the germline (Drake et al., 2014). Dicer function is inhibited until the end of oogenesis and resumes just before fertilization (Drake et al., 2014).
Ovulation events that require ALG-1 And ALG-2 in the somatic gonad involve IP3-receptor channels
The basal and the more intense ovulatory sheath contractions that propel the mature oocyte into the spermatheca require calcium release via IP3 receptor channels (McCarter et al., 1999; Yin et al., 2004). Upon meiotic resumption, the oocyte produces LIN-3/EGF that interacts with the LET-23/EGFR on the distal spermatheca cells, causing dilation, likely by initiating IP3-dependent calcium release (Bui and Sternberg, 2002; Clandinin et al., 1998; Yin et al., 2004). After fertilization, directional constriction of the spermatheca propels the embryo into the uterus, and this constriction also requires calcium release through IP3 receptors (Kovacevic et al., 2013).
Reduced activity of the IP3 signaling pathway produces ovulation defects similar to those observed when we reduced miRNA biogenesis in the somatic gonad. Worms with reduced IP3-dependent calcium release exhibit a decrease in both basal and ovulatory sheath contractions (Yin et al., 2004). Worms exposed to plc-3 RNAi and itr-1(sa73) mutants exhibit pinching of the proximal oocyte by the distal spermatheca during ovulation. We observed a similar pinching of the proximal oocyte in worms with reduced ALG-1 and ALG-2 in the somatic gonad. (The phenotypes were not identical. The itr-1(sa73) mutant’s distal spermatheca was found to dilate and constrict several times during ovulation (Denli et al., 2004; Grishok et al., 2001; Knight and Bass, 2001; Yin et al., 2004), which was not the case in our study. These observations suggest a possible role for miRNAs in the regulation of the IP3 signaling pathway in somatic gonad cells.
Previously, we identified a role of mir-786 in the control of the IP3 mediated rhythmic behavior of defecation (Kemp et al., 2012). mir-786 shows high expression in the somatic gonad (Kemp et al., 2012) and its deletion causes synthetic sterility in a sensitized genetic background (Brenner et al., 2010; Nagaraja et al., 2008), suggesting that miR-786 is one of the miRNAs that control ovulation from the somatic gonad. However, loss of mir-786 alone does not result in strong ovulation defects (unpublished data) indicating that additional miRNAs act in sheath cells and the spermatheca to control ovulation.
ALG-1 and ALG-2 Function in the Somatic Gonad to Regulate the Rate of Meiotic Maturation
We are using the rate of ovulation and a readout for the rate of meiotic maturation. The control of meiotic maturation involves signaling between MSP and Gαs receptors on the somatic sheath cells, which activate the adenylate cyclase (acy-4) –PKA pathway. The activation is required for resumption of meiosis in the proximal oocyte (Govindan et al., 2009). ALG-1 and ALG-2 together with a set of miRNAs expressed in the distal tip cell of the somatic gonad maintain germ cell proliferation, oocyte abundance and brood size (Bukhari et al., 2012). Therefore the decrease in ovulation rate and brood size that we observed following knock down of ALG-1 and ALG-2 in the somatic gonad may result from reduced miRNA activity in the distal tip cell. It is possible that miRNAs function in the sheath cells to directly or indirectly control the activation of acy-4 and the induction of meiotic maturation of the proximal oocyte.
Supplementary Material
Highlights.
miRNAs in germ cells may not have an essential role in ovulation.
miRNAs in the somatic gonad regulate ovulation events that are mediated by IP3-dependant calcium release.
miRNAs may Function in the somatic gonad to regulate the rate of meiotic maturation.
Acknowledgments
Some strains used in this study were obtained from the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We thank the lab of Eric Miska for sharing their pash-1(mj100ts); mjEx331 strain with us. We thank Katherine Maniates for her discussion and comments. This work was supported by NIH grant R15 GM084451.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nature Genetics. 2003;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Brenner JL, Jasiewicz KL, Fahley AF, Kemp BJ, Abbott AL. Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr Biol. 2010;20:1321–1325. doi: 10.1016/j.cub.2010.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui YK, Sternberg PW. Caenorhabditis elegans inositol 5-phosphatase homolog negatively regulates inositol 1,4,5-triphosphate signaling in ovulation. Mol Biol Cell. 2002;13:1641–1651. doi: 10.1091/mbc.02-01-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari SIA, Vasquez-Rifo A, Gagné D, Paquet ER, Zetka M, Robert C, Masson J-Y, Simard MJ. The microRNA pathway controls germ cell proliferation and differentiation in C. elegans. Cell Res. 2012;22:1034–1045. doi: 10.1038/cr.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clandinin TR, DeModena JA, Sternberg PW. Inositol trisphosphate mediates a RAS-independent response to LET-23 receptor tyrosine kinase activation in C. elegans. Cell. 1998;92:523–533. doi: 10.1016/s0092-8674(00)80945-9. [DOI] [PubMed] [Google Scholar]
- D’Agostino I, Merritt C, Chen P-L, Seydoux G, Subramaniam K. Translational repression restricts expression of the C. elegans Nanos homolog NOS-2 to the embryonic germline. Dev Biol. 2006;292:244–252. doi: 10.1016/j.ydbio.2005.11.046. [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BBJ, Plasterk RHA, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Drake M, Furuta T, Suen KM, Gonzalez G, Liu B, Kalia A, Ladbury JE, Fire AZ, Skeath JB, Arur S. A requirement for ERK-dependent Dicer phosphorylation in coordinating oocyte-to-embryo transition in C. elegans. Dev Cell. 2014;31:614–628. doi: 10.1016/j.devcel.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelt MV, Estevez AY, Yin X, Strange K. Oscillatory Ca2+ signaling in the isolated Caenorhabditis elegans intestine: role of the inositol-1,4,5-trisphosphate receptor and phospholipases C beta and gamma. J Gen Physiol. 2005;126:379–392. doi: 10.1085/jgp.200509355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C, Davis MW, Sarov M, Taylor J, Flibotte S, LaBella M, Pozniakovsky A, Moerman DG, Jorgensen EM. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat Methods. 2014;11:529–534. doi: 10.1038/nmeth.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist GC, Tscherner A, Nalpathamkalam T, Merico D, LaMarre J. MicroRNA Expression during Bovine Oocyte Maturation and Fertilization. Int J Mol Sci. 2016;17:396. doi: 10.3390/ijms17030396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308(5723):833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Govindan JA, Nadarajan S, Kim S, Starich TA, Greenstein D. Somatic cAMP signaling regulates MSP-dependent oocyte growth and meiotic maturation in C. elegans. Development. 2009;136:2211–2221. doi: 10.1242/dev.034595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein D. Control of oocyte meiotic maturation and fertilization. WormBook. 2005:1–12. doi: 10.1895/wormbook.1.53.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D, Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, Chen CCG, Chaves DA, Duan S, Kasschau KD, Fahlgren N, Yates JR, III, Mitani S, Carrington JC, Mello CC. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki K, McCarter J, Francis R, Schedl T. emo-1, a Caenorhabditis elegans Sec61p gamma homologue, is required for oocyte development and ovulation. J Cell Biol. 1996;134:699–714. doi: 10.1083/jcb.134.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kemp BJ, Allman E, Immerman L, Mohnen M, Peters MA, Nehrke K, Abbott AL. miR-786 Regulation of a Fatty-Acid Elongase Contributes to Rhythmic Calcium-Wave Initiation in C. elegans. Curr Biol. 2012;22:2213–2220. doi: 10.1016/j.cub.2012.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Kim HR, Kim H, Yang SC, Park M, Yoon JA, Lim HJ, Hong SH, DeMayo FJ, Lydon JP, Choi Y, Lee DR, Song H. Deficiency in DGCR8-dependent canonical microRNAs causes infertility due to multiple abnormalities during uterine development in mice. Sci Rep. 2016;6:20242. doi: 10.1038/srep20242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic I, Orozco JM, Cram EJ. Filamin and Phospholipase C-ε Are Required for Calcium Signaling in the Caenorhabditis elegans Spermatheca. PLoS Genet. 2013;9:e1003510. doi: 10.1371/journal.pgen.1003510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumsta C, Hansen M. C. elegans rrf-1 mutations maintain RNAi efficiency in the soma in addition to the germline. PLoS ONE. 2012;7:e35428. doi: 10.1371/journal.pone.0035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Choi Y, Kim K, Jin H, Lim J, Nguyen TA, Yang J, Jeong M, Giraldez AJ, Yang H, Patel DJ, Kim VN. Adenylation of Maternally Inherited MicroRNAs by Wispy. Mol Cell. 2014;56:696–707. doi: 10.1016/j.molcel.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Schedl T. RNA-binding proteins. WormBook. 2006:1–13. doi: 10.1895/wormbook.1.79.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lehrbach NJ, Castro C, Murfitt KJ, Abreu-Goodger C, Griffin JL, Miska EA. Post-developmental microRNA expression is required for normal physiology, and regulates aging in parallel to insulin/IGF-1 signaling in C. elegans. RNA. 2012;18:2220–2235. doi: 10.1261/rna.035402.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Flemr M, Stein P, Berninger P, Malik R, Zavolan M, Svoboda P, Schultz RM. MicroRNA activity is suppressed in mouse oocytes. Curr Biol. 2010;20:265–270. doi: 10.1016/j.cub.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol. 1999;205:111–128. doi: 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev Biol. 1997;181:121–143. doi: 10.1006/dbio.1996.8429. [DOI] [PubMed] [Google Scholar]
- McEwen TJ, Yao Q, Yun S, Lee CY, Bennett KL. Small RNA in situ hybridization in Caenorhabditis elegans, combined with RNA-seq, identifies germline-enriched microRNAs. Dev Biol. 2016:1–39. doi: 10.1016/j.ydbio.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McJunkin K, Ambros V. The embryonic mir-35 family of microRNAs promotes multiple aspects of fecundity in Caenorhabditis elegans. G3 (Bethesda) 2014;4:1747–1754. doi: 10.1534/g3.114.011973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja AK, Andreu-Vieyra C, Franco HL, Ma L, Chen R, Han DY, Zhu H, Agno JE, Gunaratne PH, DeMayo FJ, Matzuk MM. Deletion of Dicer in somatic cells of the female reproductive tract causes sterility. Mol Endocrinol. 2008;22:2336–2352. doi: 10.1210/me.2008-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Shingles J, Dupuy D, Grove CA, Walhout AJM, Vidal M, Hope IA. Insight into transcription factor gene duplication from Caenorhabditis elegans Promoterome-driven expression patterns. BMC Genomics. 2007;8:27. doi: 10.1186/1471-2164-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese KJ, Dunn MA, Waddle JA, Seydoux G. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol Cell. 2000;6:445–455. doi: 10.1016/s1097-2765(00)00043-5. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Schubert C, Lin R, Devries C, Plasterk R, Priess J. MEX-5 and MEX-6 Function to Establish Soma/Germline Asymmetry in Early C. elegans Embryos. Mol Cell. 2000;5:671–682. doi: 10.1016/S1097-2765(00)80246-4. [DOI] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Starich TA, Hall DH, Greenstein D. Two Classes of Gap Junction Channels Mediate Soma-Germline Interactions Essential for Germline Proliferation and Gametogenesis in Caenorhabditis elegans. Genetics. 2014;198:1127–1153. doi: 10.1534/genetics.114.168815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P. Why mouse oocytes and early embryos ignore miRNAs? RNA Biol. 2010;7:559–563. doi: 10.4161/rna.7.5.12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tops BBJ, Plasterk RHA, Ketting RF. The Caenorhabditis elegans Argonautes ALG-1 and ALG-2: almost identical yet different. Cold Spring Harb Symp Quant Biol. 2006;71:189–194. doi: 10.1101/sqb.2006.71.035. [DOI] [PubMed] [Google Scholar]
- Vasquez-Rifo A, Jannot G, Armisen J, Labouesse M, Bukhari SIA, Rondeau EL, Miska EA, Simard MJ. Developmental characterization of the microRNA-specific C. elegans Argonautes alg-1 and alg-2. PLoS ONE. 2012;7:e33750. doi: 10.1371/journal.pone.0033750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Koudijs MJ, van Eeden FJM, Cuppen E, Plasterk RHA. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nature Genetics. 2003;35(3):217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- Yin X, Gower NJD, Baylis HA, Strange K. Inositol 1,4,5-trisphosphate signaling regulates rhythmic contractile activity of myoepithelial sheath cells in Caenorhabditis elegans. Mol Biol Cell. 2004;15:3938–3949. doi: 10.1091/mbc.E04-03-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman EM, Claycomb JM. From early lessons to new frontiers: the worm as a treasure trove of small RNA biology. Front Genet. 2014;5:416. doi: 10.3389/fgene.2014.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


