Abstract
Background
Echinococcus granulosus is a harmful cestode parasite that causes cystic echinococcosis in humans as well as various livestock species and wild animals. Calmodulin (CaM), a Ca2+ sensor protein, is widely expressed in eukaryotes and mediates a variety of cellular signaling activities.
Methods
In the present study, the cDNA encoding CaM in Echinococcus granulosus (rEgCaM) was successfully cloned and the molecular and biochemical characterizations carried out. The antigenicity and immunoreactivity of rEgCaM was detected and the preliminary enzyme-linked immunosorbent assay (ELISA)-based serodiagnostic potential of EgCaM was assessed. The locations of this protein in the adult worm and larval stage, and the mRNA expression in different states of E. granulosus protoscoleces (PSCs) were defined clearly. Moreover, the Ca2+-binding properties of EgCaM were measured.
Results
rEgCaM is a highly conserved calcium-binding protein, consisting of 149 amino acids. Immunoblotting analysis revealed that rEgCaM could be identified using E. granulosus infected sheep serum. The use of rEgCaM as an antigen was evaluated by indirect ELISA which exhibited a high sensitivity (90.3%), but low specificity (47.1%). rEgCaM was ubiquitously expressed in protoscoleces and adults of E. granulosus, as well as in the germinal layer of the cyst wall. The mRNA expression level of rEgCaM was increased from the start of H2O2 exposure and then gradually decreased because of the increased apoptosis of PSCs. In electrophoretic mobility tests and 1-anilinonaphthalene-8-sulfonic acid assays, rEgCaM showed a typical characteristic of a calcium-binding protein.
Conclusions
To our knowledge, this is the first report on CaM from E. granulosus and rEgCaM is likely to be involved in some important biological function of E. granulosus as a calcium-binding protein.
Keywords: Echinococcus granulosus, Calmodulin, Ca2+-binding protein, Immunohistochemical localization, Quantitative real-time PCR
Background
Cystic echinococcosis (CE), also called hydatid disease, is a serious zoonotic parasitic disease caused by the larval form of Echinococcus granulosus and is an important public health issue in both developed and developing countries [1, 2]. The larval stage of E. granulosus infects livestock and humans, while the adult worm parasitizes the small intestine of canids. The metacestode larva is a unilocular, fluid-filled cyst which includes a germinal layer, a laminated layer, and an external layer derived from dead host cells and fibrosis. CE results in an estimated 1–3 million disability-adjusted life years (DALYs) globally per annum and 20–90% of CE prevalence is observed in domestic animals, leading to an annual economic loss of approximately US$3 billion [3]. The World Health Organization included echinococcosis as one of the 17 neglected tropical diseases in its strategic plan from 2008 to 2015 [4].
Calmodulin (CaM), a small calcium sensor protein, is one of the most evolutionarily ancient proteins in eukaryotes [5]. The functions of CaM include Ca2+ binding and conversion of Ca2+ signals though downstream proteins to regulate various physiological processes, such as muscle contraction, metabolism and cell motility [6, 7]. The structure of CaM is characterized by two globular heads joined by an extended α-helical linker; each of the globular heads includes two EF-hands domains that can bind to a calcium ion (Ca2+) [8, 9]. After binding to Ca2+, CaM changes its conformation, exposing more hydrophobic residues in order to interact with diverse target proteins [10, 11]. In Caenorhabditis elegans, 56 Ca2+-bound-calmodulin binding proteins were identified using mRNA-display, including heat shock proteins, myosin family members, CaM-dependent kinases, protein phosphatases and phosphodiesterases [12].
Although CaM has been widely studied and well-characterized in many organisms, it has not been cloned or characterized in Echinococcus spp.. Only in Echinococcus multilocularis (a species that has a close genetic relationship with E. granulosus), was CaM predicted as a potential drug target with a high score and available chemical leads (known drugs or approved compounds) [13]. In the present study, E. granulosus calmodulin (EgCaM) was identified and characterized. The antigenicity and immunoreactivity of rEgCaM were detected and the preliminary enzyme-linked immunosorbent assay (ELISA)-based serodiagnostic potential of EgCaM was assessed. The locations of this protein in the adult worm and larval stage, and its mRNA expression in different states of protoscoleces (PSCs) were clearly defined. Moreover, the Ca2+-binding properties of EgCaM were assessed.
Methods
Animals and parasites
Echinococcus granulosus protoscoleces (PSCs) and cyst walls were isolated aseptically using a previously reported method from liver hydatid cysts of naturally infected cattle presented for routine slaughter in an abattoir in Qinghai Province, China [14]. Two thousand PSCs were cultured in 1 ml of Roswell Park Memorial Institute (RPMI) 1640 medium with 10% bovine serum albumin (BSA; Hyclone, Logan, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin (Sigma-Aldrich, St. Louis, USA). Adult worms were obtained from a 5-month-old dog after 35 days after artificial infection with PSCs. Four New Zealand white rabbits were prepared to produce polyclonal antibodies.
Sera
Sera from 31 sheep naturally infected with E. granulosus were isolated, and 24 negative sera were isolated from healthy sheep with no cysts at autopsy. All samples were collected from the slaughterhouse in Xinjiang Province. Sera (seven samples) from sheep naturally infected with Taenia multiceps and sera (10 samples) from goats naturally infected with Cysticercus tenuicollis were collected from the slaughterhouse in Sichuan Province.
Bioinformatic analysis
The open reading frame (ORF) was identified using an ORF finder tool and analyzed using BLASTp at NCBI. Proteomics tools on the ExPaSy website (http://www.expasy.org/) were used to analyze the physicochemical parameters, signal peptide and transmembrane regions of the amino acid sequence. A phylogenetic tree (neighbor-joining tree) for calmodulin was constructed using MEGA v. 5.
Cloning, expression and purification of recombinant EgCaM (rEgCaM)
Total parasite RNA from PSCs was extracted using a commercial kit (Cowin Biotech, Beijing, China) following the manufacturer’s instructions. cDNA was synthesized using 1 μg of total RNA as the template for the Reverse Transcription System (Thermo Fisher, Waltham, USA). The sequence encoding calmodulin was amplified by polymerase chain reaction (PCR) using the following specific primers: sense primer 5′-CGG GAT CCA TGG CTG ACC AAC TTA CA-3′ and antisense primer 5′-CCC TCG AGC TAC TTC GAC TGC ATC ATC-3′, which introduce BamHI and XhoI restriction enzyme sites (underlined), respectively. The PCR protocol included 30 cycles of 95 °C for 30 s, 62 °C for 30 s, and 72 °C for 1 min. After purification, the PCR product was ligated into the T&A cloning vector and transformed into Escherichia coli DH5α. Plasmid DNA digested using BamHI and XhoI restriction enzymes was cloned into expression vector pET-28a (Novagen, Madison, USA) and transformed into E. coli BL21 (DE3) (Cowin Biotech, Beijing, China). An E. coli clone with the correct DNA sequence was cultured in Luria Bertani broth, and expression of rEgCaM was induced using 1 mM isopropyl-1-thio-β-D -galactopyranoside (IPTG) at 37 °C for 6 h. The rEgCaM with a poly-histidine tag was affinity purified from bacterial lysate using Ni-NTA His-tag resin (Qiagen, Hilden, Germany). The purified rEgCaM was analyzed by 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The final concentration of purified rEgCaM was determined using a BCA protein assay kit (Beyotime, Jiangsu, China).
Preparation of polyclonal antibodies against rEgCaM
Four rabbits were used to produce the polyclonal antibodies. Rabbit sera were collected before immunization to provide a reagent for negative controls. For the first immunization, 200 μg of rEgCaM emulsified with an equal volume of Freund’s complete adjuvant (Sigma-Aldrich) was injected subcutaneously. The second and third injections to boost immunization were given by mixing 100 μg of protein with an equal volume of Freund’s incomplete adjuvant at 2-week intervals. Two weeks after the final injection, rabbit antisera were collected. The antibody titer was determined by enzyme-linked immuno-sorbent assay (ELISA). Immunoglobulin G (IgG) was further isolated from the antisera using a Protein G-Sepharose column (Bio-Rad, Richmond, USA).
Western blotting
Purified rEgCaM was separated by SDS-PAGE and subsequently transferred onto a polyvinylidene difluoride membrane (Millipore, Schwalbach, Germany). The membranes were blocked with 5% (w/v) skimmed milk at room temperature for 2 h, and then incubated with E. granulosus positive sheep sera or anti-rEgCaM rabbit sera (1:150 v/v dilutions with 0.01 M PBS) overnight at 4 °C. After washing with TBST (40 mM Tris-HCl, 0.5 M NaCl, 0.5% Tween-20, pH 7.4) the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rat antibody for 1 h. Immunoreactive bands were detected using diaminobenzidine (DAB) reagent (Tiangen, Beijing, China) according to the manufacturer’s instructions. Total PSCs extract was used to detect the specificity and sensitivity of rabbit anti-rEgCaM IgG, used the method described above. Non-infected sheep and pre-immunized rabbit sera were used as negative controls.
Development of an indirect ELISA
Ninety-six-well microtiter plates were coated with 100 μl of two-fold diluted rEgCaM antigen (ranging from 1:20–1:2560) diluted in 0.1 M carbonate buffer (pH 9.6) at 4 °C overnight. After washing three times with phosphate-buffered saline-Tween 20 (PBST) to remove non-adsorbed antigen, the plates were blocked with 5% skimmed milk (w/v) in PBS at 37 °C for 1 h. Positive and negative sera isolated from sheep were two-fold diluted in PBS ranging from 1:20 to 1:640 (3.2 μg/well to 0.1 μg/well) and incubated at 37 °C for 1 h. After washing, 100 μl of rabbit anti-sheep HRP-conjugated antibody (diluted to 1:3000 with PBS) was added to each well and incubation continued at 37 °C for 1 h. After a final wash, the enzyme reaction was visualized by the addition of 3,3′,5,5′-tetramethylbenzidine (TMB) at room temperature for 15 min and stopped with Stop Solution (0.5 M phosphoric acid). The OD value (optical density) was measured at 450 nm using a microplate reader.
Evaluation and statistical analysis
The best dilutions of rEgCaM antigen and sera were determined, and then 62 sheep sera (31 for E. granulosus-positive, 7 for T. multiceps-positive and 24 for negative control) and 10 goat sera (for C. tenuicollis-positive) were serodiagnosed using the indirect ELISA described as above. Echinococcus granulosus-positive and negative sera were used in all plates, acting as the intra-plate controls. The sensitivity of this method was assessed by the percentage value of ELISA positive and true positives, while the specificity was evaluated by the cross-reaction with T. multiceps and C. tenuicollis-positive sera.
The negative cut-off was defined as the mean value +3× standard deviations (SD) from the OD values obtained from 24 negative sera. The significance of comparisons between test sera groups was estimated by ANOVA (SPSS Inc., Chicago, IL, USA).
Immunohistochemical localization of EgCaM
Fresh adult worms, PSCs and cyst walls of E. granulosus were fixed with 4% paraformaldehyde, embedded in paraffin wax, and then sliced into 5 μm thick sections. The sections were dewaxed, rehydrated, treated to inactivate endogenous peroxidase activity and incubated in 5% BSA in phosphate buffered saline (PBS) for 1 h at room temperature. Then, the sections were then incubated with anti-rEgCaM rabbit IgG or native rabbit IgG (1:500 v/v dilutions in PBS) overnight at 4 °C. After washing three times with PBS, the sections were then incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (1:200 v/v dilution in 0.1% Evan’s Blue) for 1 h at 37 °C in the dark. After washing three times with PBS, glycerine was added to the sections and images were observed under a fluorescence microscope.
rEgCaM mRNA expression of PSCs in different states
PSCs were cultured in 12-well microplates and incubated with 5 mM H2O2 for 6 h at 37 °C to induce cell apoptosis and death of PSCs. Every hour, PSCs were collected and stored at −80 °C for further study.
Total RNA was extracted and the corresponding cDNA was obtained as described above. To quantify the transcript level of rEgCaM in different states of PSCs, quantitative real-time reverse transcription PCR were conducted [15]. The primers for rEgCaM were 5′-GAA GGA TAC CGA TAG TGA GGA AGA-3′ and 5′-ATC ATT TCG TCA ACC TCC TCG TC-3′. The primers of the housekeeping gene ACTB (encoding β-actin) were 5′-ATG GTT GGT ATG GGA CAA AAG G-3′ and 5′-TTC GTC ACA ATA CCG TGC TC-3′. The data were calculated using the 2-∆∆CT method.
Ca2+-binding properties of rEgCaM
To investigate the Ca2+-binding properties of rEgCaM, denaturing and native gel electrophoresis were used. rEgCaM (0.5 mM) was incubated with CaCl2 (50 mM) and EDTA (50 mM), respectively, on ice for 1 h. The control was untreated rEgCaM protein. Subsequently, equal volumes of SDS gel loading buffer were added and the samples were heated in boiling water for 5 min, followed by separation on 15% denaturing polyacrylamide gels. Native gel electrophoresis followed a similar procedure but without SDS, and the mixture was directly loaded onto the gels (not heated). Proteins were visualized by staining with Coomassie Blue.
ANS fluorescence
1-anilinonaphthalene-8-sulfonic acid (ANS) is a widely used fluorescent probe. rEgCaM (0.5 mM) and ANS (3 mM; Sigma-Aldrich, USA) in PBS were incubated with 50 mM CaCl2 and 50 mM EDTA, respectively, on ice for 1 h. rEgCaM alone (0.5 mM) was the control. The results were measured in a Spectra Max M5 microplate fluorometer at 25 °C with 380 nm as the excitation wavelength for ANS. The emission wavelength scans were collected between 400 and 700 nm. The experiments were repeated three times.
Results
Sequence analysis of calmodulin
The cDNA sequence of EgCaM had an open reading frame of 450 bp (GenBank: KR153481) encoding a protein of 149 amino acids with a theoretical molecular weight of 16.8 kDa (isoelectric point, pI = 4.09). The instability index was calculated as 33.43. No signal peptide or transmembrane regions were found in the deduced amino acid sequence. BLASTp showed that the amino acid sequence of EgCaM shared 84.9–100% identity with CaMs from E. multilocularis, Hymenolepis microstoma, Fasciola hepatica, Schistosoma japonicum, Caenorhabditis elegans, Toxocara canis, Plasmodium falciparum, Trypanosoma cruzi, Homo sapiens and Mus musculus (Fig. 1). Four Ca2+-binding domains were located at residues 21–32, 57–68, 94–106 and 130–142 in EgCaM. A phylogenetic tree showed the relationship of EgCaM with calmodulin from other parasites and hosts; EgCaM clustered with the calmodulins from E. multilocularis and H. microstoma, but not with the other calmodulins (Fig. 2).
Fig. 1.
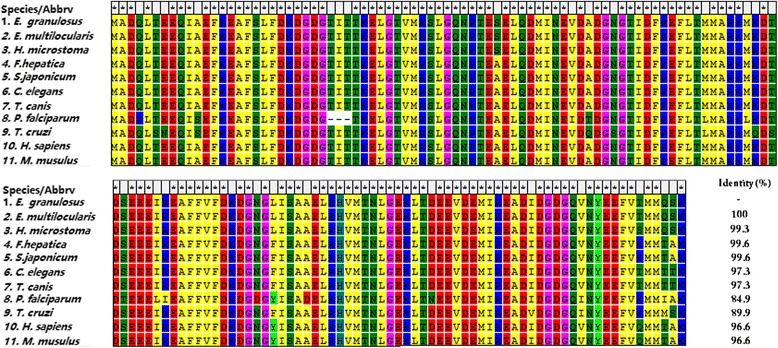
Multiple alignment of the deduced amino acid sequence of Echinococcus granulosus calmodulin with sequences of related proteins from other parasites and hosts. Echinococcus multilocularis (CDS37648.1), Hymenolepis microstoma (CDS28106.1), Fasciola hepatica (CAL91032.1), Schistosoma japonicum (AAW27335.1), Caenorhabditis elegans (NP_503386.1), Toxocara canis (KHN73369.1), Plasmodium falciparum (AAA29509.1), Trypanosoma cruzi (XP_808093.1), Homo sapiens (NP_001734.1), and Mus musculus (NP_033920.1)
Fig. 2.
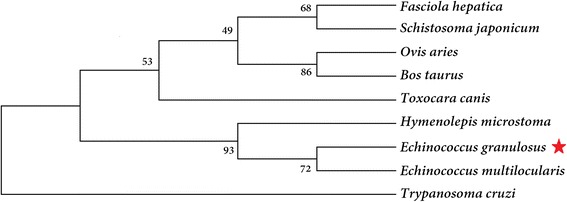
Phylogenetic tree of calmodulin. The tree was built using the neighbor-joining method in MEGA v. 5. The bootstrap values are shown at the branching points (1000 replications)
Expression, purification and western blotting analysis of EgCaM
The pET28a(+) plasmid containing the EgCaM cDNA was confirmed by double digestion and DNA sequencing. Soluble EgCaM with a His-tag was expressed in Escherichia coli BL21 after 4 h of induction with 1 mM IPTG at 37 °C. The purified rEgCaM produced a single band of approximately 20 kDa (including the His-tag), which agreed with the predicted molecular weight (Fig. 3). In western blotting, rEgCaM could react with E. granulosus positive sheep sera and anti-rEgCaM rabbit sera. The specific band was visible, which was not observed following incubation of the membrane with sera of non-infected sheep or with pre-immunized rabbit sera (Fig. 3). In addition, total PSCs extract was blotted with rabbit anti-rEgCaM IgG and showed a protein of approximately 16.0 kDa. The size of this protein was similar to the theoretical molecular weight of EgCaM. It was concluded that EgCaM had a good antigenicity and immunoreactivity.
Fig. 3.
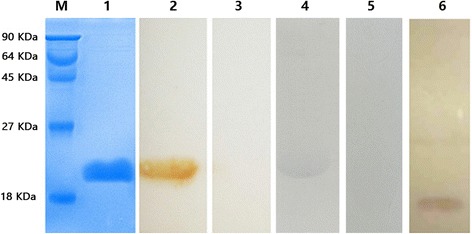
Purification and western blotting analysis of recombinant E. granulosus calmodulin (rEgCaM). Lane M: protein molecular weight markers; Lane 1: purified rEgCaM; Lane 2: rEgCaM was probed with E. granulosus-positive sheep sera; Lane 3: rEgCaM probed with sera of non-infected sheep; Lane 4: rEgCaM probed with anti-rEgCaM rabbit sera; Lane 5: rEgCaM probed with pre-immunized rabbit sera; Lane 6: total E. granulosus protoscoleces (PSCs) extract probed with rabbit anti-rEgCaM IgG
Indirect ELISA
Building on the good antigenicity and immunoreactivity of EgCaM, the preliminary serodiagnostic potential of EgCaM based on indirect ELISA was assessed. The optimal concentration of the EgCaM antigen was 1.6 μg/well and the best dilution of the sera was 1:160. The cut-off value of the EgCaM-ELISA was 0.472 (mean = 0.354, SD = 0.040) which was inferred from the E. granulosus-negative sera of sheep. Based on the cut-off value, a total of 38 sheep sera (31 for E. granulosus-positive and seven for T. multiceps-positive) and 10 goat sera (C. tenuicollis-positive) were tested. Twenty-eight sera samples from sheep infected with E. granulosus were detected as positive, indicating a sensitivity of 90.3% (28/31). In the cross-reaction assay, the OD values of five T. multiceps-positive sera isolated from sheep and three C. tenuicollis-positive sera isolated from goat were lower than the cut-off value. This indicated that the specificity of this assay was 47.1% (8/17).
Immunohistochemical localization of EgCaM in parasite sections
Anti-rEgCaM rabbit IgG was used to detect the native protein in protoscoleces, cyst walls and adult worms using immunofluorescence analysis (Fig. 4). Specific immunofluorescence was detected in almost all tissues, including the tegument and parenchymal region of the protoscoleces, the tegument and inner body of adult worms, as well as the germinal layer of cyst wall, suggesting that rEgCaM was ubiquitously expressed in all of the tissues of E. granulosus except for the laminated layer of the cyst wall. No specific fluorescence was observed in any sections when native rabbit IgG was used.
Fig. 4.
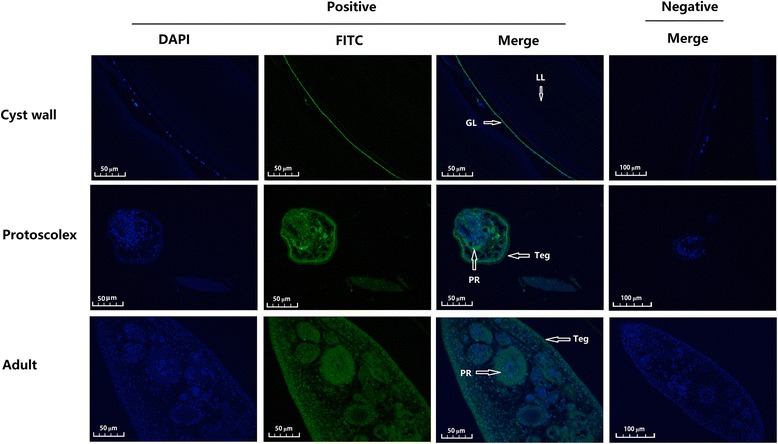
Immunohistochemical localization of E. granulosus calmodulin (EgCaM) in sections of E. granulosus. Anti-recombinant EgCaM (rEgCaM) rabbit IgG was used as the primary antibody and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG was used as the secondary antibody to detect the native protein in protoscoleces, the germinal layer, and adult worms. The nuclear DNA was stained with 2-(4-amidinophenyl)-1H–indole-6-carboxamidine (DAPI). Pre-immune rabbit sera was used as negative control. Images of protoscoleces are magnified at ×400; germinal layer and adult worm are magnified at ×200. Abbreviations: LL, laminated layer; GL, germinal layer; Teg, tegument; PR, parenchymal region
mRNA expression of rEgCaM in PSCs treated with H2O2
The mRNA expression of rEgCaM in PSCs treated with 5 mM H2O2 was assessed by quantitative real-time PCR (qPCR) and normalized using the level of the ACTB mRNA.
As shown in Fig. 5, the mRNA expression level of rEgCaM was increased at the start of H2O2 exposure and the level was upregulated by 4-fold after 2 h. The expression gradually decreased because of the increased apoptosis of the PSCs (Fig. 5).
Fig. 5.
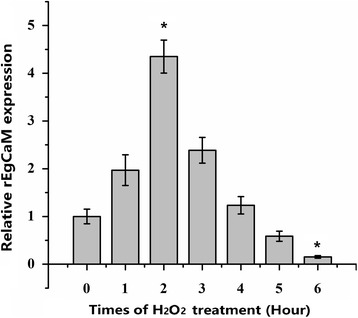
Relative expression levels of recombinant E. granulosus calmodulin mRNA (rEgCaM) in PSCs treated with H2O2 for different times. The mRNA expression of rEgCaM in PSCs treated with 5 mM H2O2 for 0, 1, 2, 3, 4, 5 and 6 h were assessed by qPCR analysis and normalized to the levels of the β-actin gene. Data are presented as the mean ± SD of triplicate experiments. Statistically significant differences between the 0 h group (as the control) and the other groups were determined using Student’s t-test (2 h: t (7) = 3.832, P = 0.005; 6 h: t (7) = -2.792, P = 0.000) (*P < 0.05*)
Biochemical characterization of rEgCaM
rEgCaM in the presence of Ca2+ demonstrated increased electrophoretic mobility on SDS-PAGE, compared with protein the treated with EDTA (Fig. 6a). On native polyacrylamide gels, the electrophoretic mobility of rEgCaM with Ca2+ was slower than that of protein exposed to EDTA (Fig. 6b). In SDS-PAGE and native gels, the control protein (untreated rEgCaM) showed the same mobility as rEgCaM in the presence of Ca2+, suggesting that the purified protein bound Ca2+ during the processes of expression or purification.
Fig. 6.
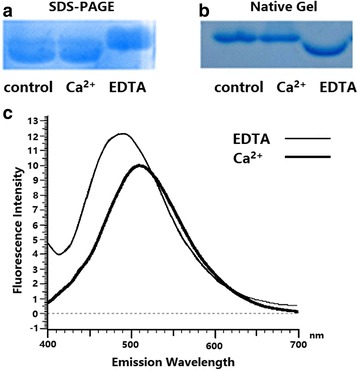
Analysis of Ca2+-binding properties of recombinant E. granulosus calmodulin (rEgCaM). The mobility of recombinant protein in the presence and absence of Ca2+ was compared on both SDS-PAGE (a) and native gel (b) electrophoresis. c 1-anilinonaphthalene-8-sulfonic acid (ANS) fluorescence emission spectra of rEgCaM
The maximum ANS fluorescence emission wavelength of rEgCaM in the presence of EDTA was 509 nm. The fluorescence intensity was enhanced when rEgCaM bound Ca2+ and the emission maximum moved to 490 nm (Fig. 6c), indicating that rEgCaM underwent conformational changes after binding Ca2+.
Discussion
Calmodulin, as a dynamic Ca2+ sensor, is present in all eukaryotic cells and mediates a variety of cellular signaling activities, such as regulation of gene expression, enzymatic activities and mitochondrial events, modulation of ion channel activities, and specific mechanisms of synaptic transmission [16–18]. Although calmodulin has been widely studied and well characterized in many organisms, there are little data on the molecular and biochemical characterization of CaM in E. granulosus [19–23]. A number of calcium-binding “CaM-like” proteins have been identified in E. granulosus, but none of these has a “classical” CaM sequence or structure. CaM is typically a protein of 16 kDa, comprising two globular domains connected by a flexible alpha helix hinge [19]. The cDNA sequence encoding E. granulosus calmodulin was identified as encoding a “classical” CaM and uploaded to GenBank (accession code: KR153481). In the present study, we successfully expressed the “classical” CaM of E. granulosus in E. coli, characterized its locations in E. granulosus sections, assessed its mRNA expression of PSCs in different states, and assessed its serodiagnostic potential. Furthermore, we also determined the bioinformatic features and the ability of the protein to bind Ca2+ of rEgCaM.
Four Ca2+-binding domains in EgCaM at amino acid positions 21–32, 57–68, 94–106 and 130–142 demonstrated that this protein belongs to the EF-hand calcium-binding protein (CaBP) family, which contain two or more EF-hands domains [24]. Additionally, the amino acid sequence of EgCaM showed high identity with calmodulins from cestodes, trematodes, nematodes, protozoons and mammals (Fig. 2). Calmodulin was previously characterized as a highly conserved protein from the parasite Schistosoma mansoni; the alignment of both SmCaM1 and SmCaM2 shared 97–98% identity with other CaMs from mammals, flatworms and insects [19]. These data indicated that calmodulin has remained highly conserved across species during evolution. Moreover, mutations and the composition of the calmodulin intergenic spacer in Leishmania species have been studied as a molecular target that might have taxonomic value [25]. Moreover, Karabinos [26] found that CaM-like proteins of C. elegans arose from a CaM ancestor through repeated gene duplications, fusions and sequence divergence. In E. granulosus, CaM-like proteins showed a low identity with EgCaM; therefore, it would be meaningful to explore the relationships between CaM and CaM-like genes when more homologs of cestodes are included in the future.
Recently, increasing numbers of studies have used recombinant protein antigens in serological diagnosis methods to detect animals suspected of being infected [27–29]. It was important to evaluate the serodiagnostic potential of this protein to promote further research. In our study, western blotting analysis of EgCaM showed that this protein had a good antigenicity and immunoreactivity. However, even though sensitivity was high, the use of rEgCaM in the serodiagnostic ELISA lacked utility because of its low specificity. This might reflect the high conservation of CaM. In conclusion, the results revealed that rEgCaM was not a suitable as a diagnostic antigen.
Immunohistochemical localizations showed that rEgCaM was ubiquitously expressed in the larva, germinal layer and adult worm sections of E. granulosus. For example, EgCaM was expressed in the germinal layer, which is one of the most physiologically active regions of the cysts. It is possible that an unknown Ca2+-dependent mechanism occurs in this region. The distribution of EgCaM in the tegument of the adult stage indicated that the Ca2+ signaling pathway could function between E. granulosus and its host. Interestingly, a similar result was also reported in another flatworm Clonorchis sinensis [22]. Calcineurin, a Ca2+-calmodulin activated serine-threonine protein phosphatase, has been studied in protoscoleces and it was suggested that calcineurin is specifically involved in exocytic activity [30]. EgCaM was also expressed in the tegument tissues and parenchymal region of protoscoleces stage, implying that calmodulin is associated with exocytic activity in protoscoleces. These data indicated that EgCaM might play a crucial role in the growth and development of the parasite.
In other species, it has been proven that calcium ions and CaM are involved in abiotic stress responses including drought, salt stress, cold and heat stimuli [31–33]. However, there are few studies on the function of CaM in parasites dealing with abiotic stress. During development and proliferation in the host, E. granulosus must cope with oxidants and reactive oxygen species (ROS) derived from their own metabolism and from the host’s activated immune cells [34]. To further study the function of CaM in E. granulosus under the oxidative stress, PSCs were treated with 5 mM H2O2 for the indicated times up to 6 h at 37 °C. The mRNA expression level of rEgCaM was increased from the start of H2O2 exposure. The result suggested a role for EgCaM in the defense against oxidative stress. With increasing exposure time, the activity and metabolic rate of PSCs declined and the apoptosis or death of PSCs was enhanced. The mRNA expression of rEgCaM also decreased. Thus, it was demonstrated that CaM is related to the growth and metabolism of PSCs. The results suggested that calmodulin plays an indispensable role in many physiological functions in E. granulosus.
EgCaM is an EF-hand calcium binding protein with four Ca2+-binding domains, displaying changes in its electrophoretic mobility in the absence and presence of Ca2+. rEgCaM presented a typical Ca2+-induced electrophoretic mobility shift in this study (Fig. 6). Grab et al. [35] studied the effect of different divalent cations on migration through SDS-PAGE of purified canine and porcine brain calmodulins. Under denaturing condition, in the presence of Ca2+, the protein migrated much faster, which was consistent with our previous study [35]. He et al. [36] also came to the same conclusion when they invested CaM from Sarcoptes scabiei. In our study, rEgCaM in the presence of Ca2+ demonstrated increased electrophoretic mobility on SDS-PAGE, compared with the protein treated with EDTA (Fig. 6a). On native polyacrylamide gels, the electrophoretic mobility of rEgCaM with Ca2+ was slower than that of protein exposed to EDTA (Fig. 6b) which presented a typical Ca2+-induced electrophoretic mobility shift. These results were consistent with previous studies of CaBPs in C. sinensis and Fasciola hepatica [20, 22, 32]. Moreover, some calmodulins with mutations in their serine and tyrosine residues retain these characteristics [37]. One of the biochemical features of calmodulin is conformational change, exposing more hydrophobic residues on the protein surface after Ca2+ binding [38]. rEgCaM displayed this characteristic, which was verified using an ANS fluorescence assay. When Ca2+ was bound to rEgCaM, ANS fluorescence was enhanced, and the ANS fluorescence emission wavelength shifted from 509 nm to 490 nm.
Conclusions
In this study, E. granulosus calmodulin was cloned, expressed and characterized. The antigenicity and immunoreactivity of rEgCaM were detected using western blotting and indirect ELISA. The results showed that this protein had a good antigenicity and immunoreactivity, but was not a suitable potential diagnostic antigen. rEgCaM was ubiquitously expressed in protoscoleces and adults of E. granulosus, as well as the germinal layer of cyst wall. The mRNA expression level of rEgCaM increased from the start of H2O2 exposure and then gradually decreased due to the increased apoptosis of PSCs. The Ca2+-binding properties of EgCaM were measured and rEgCaM presented a typical Ca2+-induced electrophoretic mobility shift. In conclusion, our study demonstrated that CaM might play an indispensable role in many physiological functions in E. granulosus.
Acknowledgements
The authors thank Junyang Zhu, Min Yan and Xuefeng Yan for their helpful assistance in some experimental steps. We are also extremely grateful to teachers and classmates at the Laboratory of Animal Infectious Disease and Microarray, Laboratory of Animal Diseases and Environmental Hazards of Sichuan Province in Sichuan Agricultural University for kindly allowing us to conduct the protein purification experiments and use their fluorescence microscope in their laboratories, respectively. We also thank the native English speaking scientists of Elixigen Company (Huntington Beach, California) for editing our manuscript.
Funding
This work was supported by a grant from the Key Technology R&D Program of Sichuan Province, China (No.2015NZ0041; http://www.scst.gov.cn/). The funder had no role in study design, data collection and analysis, decision to publish or reparation of the manuscript.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Abbreviations
- ANS
1-anilinonaphthalene-8-sulfonic acid
- CE
Cystic echinococcosis
- ELISA
Enzyme-linked immuno-sorbent assay
- PBS
Phosphate-buffered saline
- PSCs
E. granulosus protoscoleces
- rEgCaM
Recombinant E. granulosus calmodulin
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
Authors’ contributions
NW and GYY conceived the study. NW and XQZ designed the study and wrote the first version of the manuscript. XQZ and XJS participated in its design and coordination and performed the statistical analysis. XBG and WML conceived the study and collected the experimental material. YX and XRP collected and analyzed the raw data. GYY is responsible for this study, participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animals involved in this paper were purchased from the Laboratory Animal Center of Sichuan Agricultural University. All procedures were carried out strictly according to the Guide for the Care and Use of Laboratory Animals and with approval from the Animal Ethics Committee of Sichuan Agricultural University (Ya’an, China) (Approval No. 2013–028).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ning Wang, Email: wangningzhuhui@hotmail.com.
Xiuqin Zhong, Email: xiuqinzhong@hotmail.com.
Xingju Song, Email: 18728153735@163.com.
Xiaobin Gu, Email: 448070309@qq.com.
Weiming Lai, Email: 462591336@qq.com.
Yue Xie, Email: 496605886@qq.com.
Xuerong Peng, Email: 916769407@qq.com.
Guangyou Yang, Email: guangyou1963@aliyun.com.
References
- 1.Eckert J, Deplazes P. Biological, epidemiological, and clinical aspects of echinococcosis, a zoonosis of increasing concern. Clin Microbiol Rev. 2004;17:107–135. doi: 10.1128/CMR.17.1.107-135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson JAM, Gray DJ, Clements ACA, Barnes TS, McManus DP, Yang YR. Environmental changes impacting Echinococcus transmission: research to support predictive surveillance and control. Glob Chang Biol. 2013;19:677–688. doi: 10.1111/gcb.12088. [DOI] [PubMed] [Google Scholar]
- 3.Torgerson PR, MacPherson CNL. The socioeconomic burden of parasitic zoonoses: global trends. Vet Parasitol. 2011;182:79–95. doi: 10.1016/j.vetpar.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Craig PS, McManus DP, Lightowlers MW, Chabalgoity JA, Garcia HH, Gavidia CM, et al. Prevention and control of cystic echinococcosis. Lancet Infect Dis. 2007;7:385–394. doi: 10.1016/S1473-3099(07)70134-2. [DOI] [PubMed] [Google Scholar]
- 5.Friedberg F, Rhoads AR. Evolutionary aspects of calmodulin. IUBMB Life. 2001;51:215–221. doi: 10.1080/152165401753311753. [DOI] [PubMed] [Google Scholar]
- 6.Haeseleer F, Imanishi Y, Sokal I, Filipek S, Palczewski K. Calcium-binding proteins. Intracellular sensors from the calmodulin superfamily. Biochem Bioph Res Co. 2002;290:615–623. doi: 10.1006/bbrc.2001.6228. [DOI] [PubMed] [Google Scholar]
- 7.Berridge MJ, Lipp P, Bootman MD. Berridge M, Lipp P, Bootman M. The versatility and universality of calcium signaling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 8.Babu YS, Sack JS, Greenhough TJ, Bugg CE, Means AR, Cook WJ. Three-dimensional structure of calmodulin. Nature. 1985;315:37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- 9.Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax A. Solution structure of calcium-free calmodulin. Nat Struct Biol. 1995;2:768–776. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 10.Zhang M, Yuan T. Molecular mechanisms of calmodulin's functional versatility. Biochem Cell Biol. 1998;76:313–323. doi: 10.1139/o98-027. [DOI] [PubMed] [Google Scholar]
- 11.Gopalakrishna R, Anderson WB. The effects of chemical modification of calmodulin on Ca2+-induced exposure of a hydrophobic region. Separation of active and inactive forms of calmodulin. BBA-Mo Cell Res. 1985;844:265–9. doi: 10.1016/0167-4889(85)90099-0. [DOI] [PubMed] [Google Scholar]
- 12.Shen X, Valencia C, Gao W, Cotten S, Dong B, Huang B, Liu R. Ca(2+)/Calmodulin-binding proteins from the C. elegans proteome. Cell Calcium. 2008;43:444–456. doi: 10.1016/j.ceca.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Tsai IJ, Zarowiecki M, Holroyd N, Garciarrubio A, Sanchezflores A, Brooks KL, et al. The genomes of four tapeworm species reveal adaptations to parasitism. Nature. 2013;496:57–63. doi: 10.1038/nature12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song X, Hu D, Yan M, Wang Y, Wang N, Gu X, Yang G. Molecular characteristics and serodiagnostic potential of dihydrofolate reductase from Echinococcus granulosus. Sci Rep. 2017;7:514. doi: 10.1038/s41598-017-00643-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu D, Song X, Xie Y, Zhong X, Wang N, Zheng Y, et al. Molecular insights into a tetraspanin in the hydatid tapeworm Echinococcus granulosus. Parasit Vectors. 2015;8:311. doi: 10.1186/s13071-015-0926-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dick IE, Tadross MR, Liang H, Lai HT, Yang W, Yue DTA. Modular switch for spatial Ca2+ selectivity in the calmodulin regulation of CaV channels. Nature. 2008;451:830–834. doi: 10.1038/nature06529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wayman GA, Lee YS, Tokumitsu H, Silva A, Soderling TR. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008;59:914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aich A, Shaha C. Novel role of calmodulin in regulating protein transport to mitochondria in a unicellular eukaryote. Mol Cell Biol. 2013;33:4579–4593. doi: 10.1128/MCB.00829-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taft AS, Yoshino TP. Cloning and functional characterization of two calmodulin genes during larval development in the parasitic flatworm Schistosoma mansoni. J Parasitol. 2011;97:72–81. doi: 10.1645/GE-2586.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell SL, McFerran NV, Moore CM, Tsang Y, Glass P, Hoey EM, et al. A novel calmodulin-like protein from the liver fluke, Fasciola hepatica. Biochimie. 2012;94:2398–2406. doi: 10.1016/j.biochi.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 21.Alvarado ME, Wasserman M. Calmodulin expression during Giardia intestinalis differentiation and identification of calmodulin-binding proteins during the trophozoite stage. Parasitol Res. 2012;110:1371–1380. doi: 10.1007/s00436-011-2637-4. [DOI] [PubMed] [Google Scholar]
- 22.Zhou JJ, Sun JF, Huang Y, Zhou CH, Liang P, Zheng MH, et al. Molecular identification, immunolocalization, and characterization of Clonorchis sinensis calmodulin. Parasitol Res. 2013;112:1709–1717. doi: 10.1007/s00436-013-3329-z. [DOI] [PubMed] [Google Scholar]
- 23.Ginger ML, Collingridge PW, Brown RW, Sproat R, Shaw MK, Gull K. Calmodulin is required for paraflagellar rod assembly and flagellum-cell body attachment in trypanosomes. Protist. 2013;164:528–540. doi: 10.1016/j.protis.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Grabarek Z. Structural basis for diversity of the EF-hand calcium-binding proteins. J Mol Biol. 2006;359:509–525. doi: 10.1016/j.jmb.2006.03.066. [DOI] [PubMed] [Google Scholar]
- 25.Miranda A, Samudio F, Castillo J, Calzada JE. The calmodulin intergenic spacer as molecular target for characterization of Leishmania species. Parasit Vectors. 2014;7:35. doi: 10.1186/1756-3305-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karabinos A. Molecular evolution of the multiple calmodulin-like cal genes in C. elegans and in nematodes. Dev Genes Evol. 2016;226:1–13. doi: 10.1007/s00427-016-0558-z. [DOI] [PubMed] [Google Scholar]
- 27.Zhang WB, Li J, Li Q, Yang D, Zhu B, You H, et al. Identification of a diagnostic antibody-binding region on the immunogenic protein EpC1 from Echinococcus granulosus and its application in population screening for cystic echinococcosis. Clin Exp Immunol. 2007;149:80–86. doi: 10.1111/j.1365-2249.2007.03386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maity A, Bhattacharya D, Batabyal S, Chattopadhyay S, Bera AK. Echinococcus granulosus antigen B for serodiagnosois of cystic echinococcosis in buffalo (Bubalus bubalis) Buffalo Bull. 2014;33:94–100. [Google Scholar]
- 29.Pagnozzi D, Biosa G, Addis MF, Mastrandrea S, Masala G, Uzzau S. An easy and efficient method for native and immunoreactive Echinococcus granulosus antigen 5 enrichment from hydatid cyst fluid. PLoS One. 2014;9:e104962. doi: 10.1371/journal.pone.0104962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolao MC, Cumino AC. Biochemical and molecular characterization of the calcineurin in Echinococcus granulosus larval stages. Acta Trop. 2015;146:141–151. doi: 10.1016/j.actatropica.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 31.Reddy ASN, Ali GS, Celesnik H, Day IS. Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell. 2011;23:2010–2032. doi: 10.1105/tpc.111.084988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang T, Syang C. A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J Biol Chem. 2010;285:7119–7126. doi: 10.1074/jbc.M109.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galon Y, Nave R, Boyce JM, Nachmias D, Knight MR, Fromm H. Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 2008;582:943–948. doi: 10.1016/j.febslet.2008.02.037. [DOI] [PubMed] [Google Scholar]
- 34.Cheng Z, Zhu S, Wang L, Liu F, Tian H, Pengsakul T, Wang Y. Identification and characterisation of Emp53, the homologue of human tumor suppressor p53, from Echinococcus multilocularis: its role in apoptosis and the oxidative stress response. Int J Parasitol. 2015;45:517–526. doi: 10.1016/j.ijpara.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 35.Grab DJ, Berzins K, Cohen RS, Siekevitz P. Presence of calmodulin in postsynaptic densities isolated from canine cerebral cortex. J Biol Chem. 1979;254:8690–8696. [PubMed] [Google Scholar]
- 36.He R, Shen N, Lin H, Gu X, Lai W, Peng X, Yang G. Molecular characterization of calmodulin from Sarcoptes scabiei. Parasitol Int. 2017;66:1–6. doi: 10.1016/j.parint.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Stateva SR, Salas V, Benaim G, Menéndez M, Solís D, Villalobo A. Characterization of phospho-(tyrosine)-mimetic calmodulin mutants. PLoS One. 2015;10:e0120798. doi: 10.1371/journal.pone.0120798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikura M. Calcium binding and conformational response in EF-hand proteins. Trends Biochem Sci. 1996;21:14–17. doi: 10.1016/S0968-0004(06)80021-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.


