Abstract
The lung is the most common extra-abdominal site of metastasis in colorectal cancer (CRC), in which circular RNA (circRNA) may have a crucial role. Therefore, the present study detected circRNA expression to identify novel targets to further study lung metastasis in CRC. In the present study, total RNA was extracted from CRC tissues of patients with and without lung metastasis to perform high-throughput microarray assay in order to detect differentially expressed circRNA. Following this, gene ontology (GO) and pathway analyses of the genes producing differentially expressed circRNA were performed to predict the function of circRNA using standard enrichment computational methods. Additionally, the circRNA/microRNA (miRNA) interactions were constructed with bioinformatics methods to predict the binding of miRNA with circRNA. In the CRC tissues from patients with lung metastasis, 431 circRNA were detected to be differentially expressed, including 192 upregulated and 239 downregulated over 2-fold compared with the CRC tissues without metastasis. Furthermore, GO analysis revealed that the genes producing upregulated circRNA were involved in DNA repair, while the genes producing downregulated circRNA were enriched in signal transduction. By pathway analysis, it was identified that the genes producing downregulated circRNA were involved in the nuclear factor-κB and Wnt signaling pathway in the CRC tissues from patients with lung metastasis compared with the CRC tissues without metastasis. In addition, it was demonstrated that hsa_circRNA_105055, hsa_circRNA_086376 and hsa_circRNA_102761 could commonly bind with miR-7 regulating target genes PRKCB, EPHA3, BRCA1 and ABCC1. The findings of the present study may provide a novel perspective on circRNA and lay a foundation for future research of potential roles of circRNA in CRC with lung metastasis.
Keywords: colorectal cancer, lung metastasis, circular RNA, gene ontology analysis, pathways analysis
Introduction
Colorectal cancer (CRC), as the third most common cancer affecting the gastrointestinal tract, is a common type of cancer worldwide and is associated with a high mortality rate (1–3). The majority of cases of mortality from CRC occur in patients with metastases from the primary cancer (4). In patients with CRC, ~10% have lung metastasis (5,6); the lungs are the second most common site of metastasis in CRC, following the liver (7). Currently, the diagnosis of CRC in patients with pulmonary metastases depends on radiology and biopsy (8). However, radiology-guided biopsy has its limitation of sensitivity (9). Therefore, to improve the survival of patients with CRC, it is necessary to explore novel biomarkers and the molecular mechanism of metastatic progression in CRC that may be useful for earlier diagnosis and treatment of patients with lung metastases.
It has been reported that non-coding RNA, including transfer RNA (tRNA), microRNA (miRNA), long non-coding RNA and circular RNA (circRNA), are involved in biological and pathological processes (10–12). In contrast to tRNA, miRNA and long non-coding RNA, circRNA are less well characterized. circRNA were first identified in RNA viruses as early as the 1970s (13); however, the majority of these were misconstrued as 'splicing rubbish' (14). Since a study that firstly ascribed an actual function to one of these circRNA molecules (15), the potential of circRNA has elevated the scientific community's awareness of these molecules (16). With the development of RNA deep sequencing technology and bioinformatics, study has revealed that large numbers of circRNA are endogenous, abundant, conserved and stable in mammalian cells (15,17–22), and regulate gene expression at the transcriptional or post-transcriptional level by interacting with miRNA or other molecules (23). Given that circRNA interact with miRNA to regulate their target genes, circRNA may be involved in diseases correlated with miRNA (24). Recently, circRNA have been reported to mediate cancer progression (12), and the global abundance of circRNA was demonstrated to be lower in CRC than in normal tissue (25). However, to date, little is known about the genome-wide expression and function analysis of circRNA in CRC tissues of patients with lung metastasis.
Therefore, the present study examined differentially expressed circRNA in tissues from patients with CRC with and without lung metastasis so as to identify novel diagnostic and prognostic markers in patients with CRC with lung metastasis.
Materials and methods
Study approval
All samples were collected from patients between January, 2013 and December, 2015 in the Department of Gastrointestinal and Hernia Surgery, the First Affiliated Hospital of Kunming Medical University (Kunming, China). The project was reviewed and approved by the Ethics Committee of the First Affiliated Hospital of Kunming Medical University. All patients included in the present study provided written informed consent prior to surgery.
Samples
Primary CRC samples were obtained from 3 male patients (age, 66.33±7.37 years) with CRC and lung metastasis (Exp A) and 3 male patients (age, 64.67±11.93 years) with CRC without metastasis (Ctrl A), respectively. All patients were histologically confirmed to have CRC based on colonoscopy (26) and did not receive any other forms of therapy at the time of enrollment. At the time of surgery, all tissue samples were immediately frozen in liquid nitrogen and stored at −80°C for further use.
RNA extraction and quality control
According to the manufacturer's protocol, total RNA was extracted from each sample using a homogenizer and TRIzol regent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Subsequently, the quantification and quality of purified RNA were assessed using a NanoDrop ND-1000 (NanoDrop Technologies; Thermo Fisher Scientific, Inc.). The integrity of RNA was assessed by electrophoresis on a denaturing 1.5% agarose gel.
Microarray analysis
Sample labeling and human circRNA array hybridization were performed according to the manufacturer's protocol (Arraystar, Inc., Rockville, MD, USA). Briefly, total RNA were digested with RNase R (Epicentre; Illumina, Inc., San Diego, CA, USA) to remove linear RNA and enrich circRNA. Subsequently, the enriched circRNA were amplified and transcribed into fluorescent cRNA utilizing a random priming method (Arraystar Super RNA Labeling kit; Arraystar, Inc.). The labeled cRNA were purified using an RNeasy mini kit (Qiagen GmbH, Hilden, Germany). The concentration and specific activity of the labeled cRNA (pmol Cy3/µg cRNA) were measured using a NanoDrop ND-1000. A total of 1 µg of each labeled cRNA was fragmented by adding 5 µl 10X blocking agent and 1 µl 25X fragmentation buffer (Arraystar Super RNA Labeling kit; Arraystar, Inc.). Subsequently, the mixture was heated at 60°C for 30 min, and finally 25 µl 2X hybridization buffer was added to dilute the labeled cRNA. Following this, 50 µl hybridization solution was dispensed into the gasket slide and assembled to the circRNA expression microarray slide. The slides were incubated for 17 h at 65°C in an Agilent Hybridization Oven (Agilent Technologies, Inc., Santa Clara, CA, USA). The hybridized arrays were washed using Agilent wash buffer 1 and wash buffer 2, fixed and scanned using the Agilent Scanner G2505C (both from Agilent Technologies, Inc.).
Data analysis
Scanned images were imported into Agilent Feature Extraction software (version 11.0.1.1; Agilent Technologies, Inc.) for raw data extraction. When comparing two groups of profile differences (such as CRC with lung metastasis vs. without lung metastasis), the fold change (i.e. the ratio of the group averages) between the groups for each circRNA is computed. The statistical significance of the difference may be conveniently estimated by a t-test. circRNA with fold changes ≥2 and P-values of <0.05 were selected as being significantly differentially expressed.
Gene ontology (GO) analysis
GO analysis (geneontology.org) may be used to construct meaningful annotation of gene products in a wide variety of organisms. GO contains three domains, including biological process (BP), cellular components (CC) and molecular function (MF). The -log10 (P-value) indicates the enrichment score representing the significance of GO term enrichment among genes producing differentially expressed circRNA. Fold enrichment represents the proportion of the changes in genes as proportions in the GO database in this function (such as, the greater the proportion, the more reliability, which is seen as the more significant changes in BP, CC or MF).
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
KEGG (genome.jp/kegg/) was utilized to harvest pathway clusters covering our knowledge of the molecular interaction and reaction networks in genes producing differentially expressed circRNA.
Annotation for circRNA/miRNA interactions
In the present study, circRNA/miRNA interactions were predicted with Arraystar's home-made miRNA target prediction software (Arraystar, Inc.) based on TargetScan (targetscan.org/vert_71/) and miRanda (microrna.org/microrna/home.do). The differentially expressed circRNA within all the comparisons were annotated in detail with the circRNA/miRNA interaction information.
Results
Overview of differentially expressed circRNA in the tissues of patients with CRC and lung metastasis
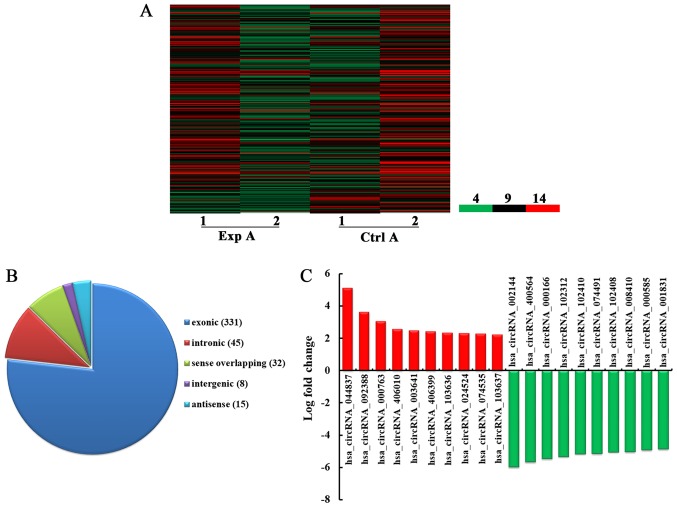
High-throughput microarray is an efficient approach for studying the biological function of RNA. As demonstrated in Fig. 1A, a total of 431 circRNA were detected to be differentially expressed with fold change ≥2.0, P<0.05 and false discovery rate <0.05 in the Exp A group compared with the Ctrl A group. Among them, 192 and 239 circRNA were upregulated and downregulated, respectively. Among the dysregulated circRNA, in the light of their relation with their coding genes, the circRNA were classified into five categories: 331 were exonic, 45 were intronic, 32 were sense overlapping, eight were intergenic and 15 were antisense (Fig. 1B). According to the log fold change, the top 10 upregulated and downregulated circRNA were selected (Fig. 1C).
Figure 1.
Overview of altered circRNA in the Exp A and Ctrl A groups. (A) Heat map of differentially expressed circRNA of CRC samples. Column 1 and 2 in Exp A represent the expression of circRNA in Exp A, while column 1 and 2 in Ctrl A represent the expression of corresponding circRNA in Ctrl A. 4, the lowest normalized intensity of each sample (log2 transformed); 9, middle normalized intensity of each sample (log2 transformed); 14, the highest normalized intensity of each sample (log2 transformed). (B) The circRNA were classified into five types according to the relationship of the genomic loci with their associated coding genes. (C) The top 10 upregulated and downregulated circRNA based on the log fold change. Exp A, CRC tissues from patients with lung metastasis; Ctrl A, CRC tissues from patients without metastasis; circRNA, circular RNA; CRC, colorectal cancer.
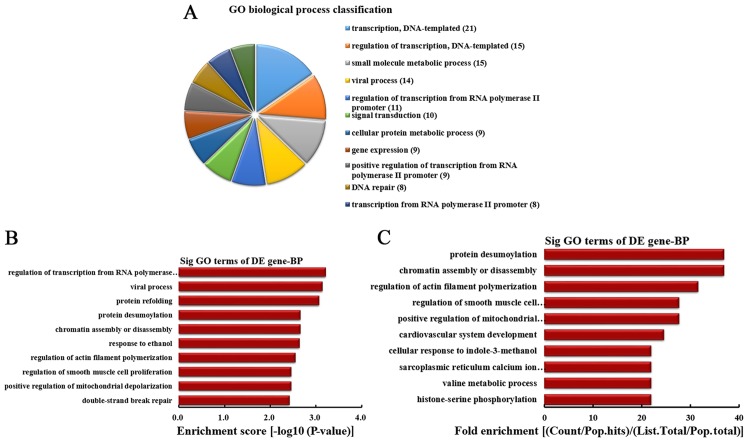
BP analysis of the genes producing upregulated circRNA
According to the number of linear counterparts of differentially overexpressed circRNA involved in BPs, the top 10 BPs were classified (Fig. 2A). Among them, 21 linear counterparts of differentially overexpressed circRNA were involved in transcription, DNA-templated, 15 were involved in regulation of transcription, DNA-templated, 15 were involved in small molecule metabolic process, 14 were involved in viral process, 11 were involved in the regulation of transcription from RNA polymerase II promoter, 10 were involved in signal transduction, 9 were involved in cellular protein metabolic process, 9 were involved in gene expression, 9 were involved in positive regulation of transcription from RNA polymerase II promoter and 8 were involved in DNA repair. According to the enrichment score, the top 10 BPs were selected (Fig. 2B). These included regulation of transcription from RNA polymerase II promoter, viral process, protein refolding, protein desumoylation, chromatin assembly or disassembly, response to ethanol, regulation of actin filament polymerization, regulation of smooth muscle cell proliferation, positive regulation of mitochondrial depolarization and double-strand break repair. Additionally, according to the fold enrichment, the top 10 BPs were selected (Fig. 2C). These included protein desumoylation, chromatin assembly or disassembly, regulation of actin filament polymerization, regulation of smooth muscle cell proliferation, positive regulation of mitochondrial depolarization, cardiovascular system development, cellular response to indole-3-methanol, sarcoplasmic reticulum calcium ion transport, valine metabolic process and histone-serine phosphorylation.
Figure 2.
BP analysis of the genes producing upregulated circRNA. (A) BP classification based on the number of linear counterparts of upregulated circRNA. (B) The top 10 BPs according to the enrichment score. (C) The top 10 BPs according to the fold enrichment. BP, biological process; circRNA, circular RNA; GO, gene ontology; DE, differentially expressed; Sig, significant.
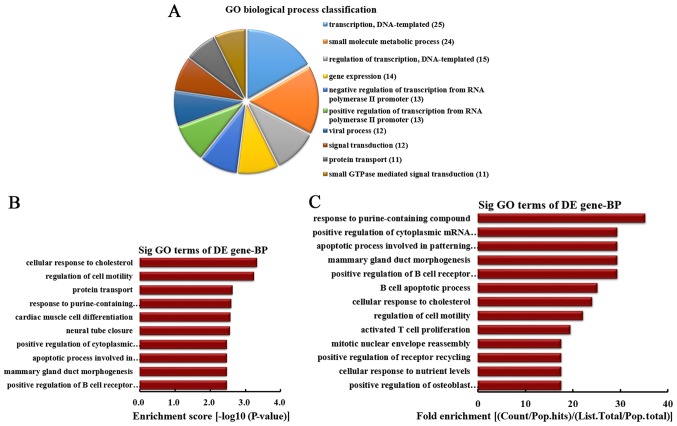
BP analysis of the genes producing downregulated circRNA
According to the number of linear counterparts of down-regulated circRNA involved in BPs, the top 10 BPs were classified (Fig. 3A). Among them, 25 linear counterparts of differentially downregulated circRNA were involved in transcription, DNA-templated, 24 were involved in small molecule metabolic process, 15 were involved in regulation of transcription, DNA-templated, 14 were involved in gene expression, 13 were involved in negative regulation of transcription from RNA polymerase II promoter, 13 were involved in positive regulation of transcription from RNA polymerase II promoter, 12 were involved in viral process, 12 were involved in signal transduction, 11 were involved in protein transport and 11 were involved in small GTPase-mediated signal transduction. According to the enrichment score, the top 10 BPs were selected (Fig. 3B). These included cellular response to cholesterol, regulation of cell motility, protein transport, response to purine-containing compound, cardiac muscle cell differentiation, neural tube closure, positive regulation of cytoplasmic mRNA processing body assembly, apoptotic process involved in patterning of blood vessels, mammary gland duct morphogenesis and positive regulation of B cell receptor signaling pathway. Furthermore, according to the fold enrichment, the top 13 BPs were selected (Fig. 3C). These included response to purine-containing compound, positive regulation of cytoplasmic mRNA processing body assembly, apoptotic process involved in patterning of blood vessels, mammary gland duct morphogenesis, positive regulation of B cell receptor signaling pathway, B cell apoptotic process, cellular response to cholesterol, regulation of cell motility, activated T cell proliferation, mitotic nuclear envelope reassembly, positive regulation of receptor recycling, cellular response to nutrient levels and positive regulation of osteoblast proliferation.
Figure 3.
BP analysis of the genes producing downregulated circRNA. (A) BP classification based on the number of linear counterparts of downregulated circRNA. (B) The top 10 BPs according to the enrichment score. (C) The top 13 BPs according to the fold enrichment. BP, biological process; circRNA, circular RNA; GO, gene ontology; DE, differentially expressed; Sig, significant.
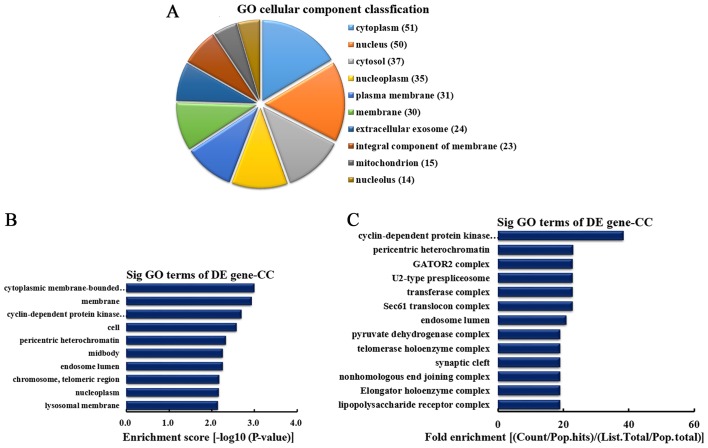
CC analysis of the genes producing upregulated circRNA
According to the number of linear counterparts of differentially overexpressed circRNA identified in CC, the top 10 CC categories were classified (Fig. 4A). Among them, 51 linear counterparts of differentially overexpressed circRNA were involved in the cytoplasm, 50 were involved in the nucleus, 37 were involved in the cytosol, 35 were involved in the nucleoplasm, 31 were involved in plasma membrane, 30 were involved in the membrane, 24 were involved in the extracellular exosome, 23 were involved in the integral component of the membrane, 15 were involved in the mitochondrion and 14 were involved in the nucleolus. According to enrichment score, the top 10 CCs were selected (Fig. 4B). These included cytoplasmic membrane-bounded vesicle, membrane, cyclin-dependent protein kinase activating kinase holoenzyme complex, cell, pericentric heterochromatin, midbody, endosome lumen, chromosome (telomeric region), nucleoplasm and lysosomal membrane. Furthermore, according to the fold enrichment, the top 13 CCs were selected (Fig. 4C). These included cyclin-dependent protein kinase activating kinase holoenzyme complex, pericentric heterochromatin, GATOR2 complex, U2-type prespliceosome, transferase complex, Sec61 translocon complex, endosome lumen, pyruvate dehydrogenase complex, telomerase holoenzyme complex, synaptic cleft, nonhomologous end joining complex, elongator holoenzyme complex and lipopolysaccharide receptor complex.
Figure 4.
CC analysis of the genes producing upregulated circRNA. (A) CC classification based on the number of linear counterparts of upregulated circRNA. (B) The top 10 CCs according to the enrichment score. (C) The top 13 CCs according to the fold enrichment. CC, cellular component; circRNA, circular RNA; GO, gene ontology; DE, differentially expressed; Sig, significant.
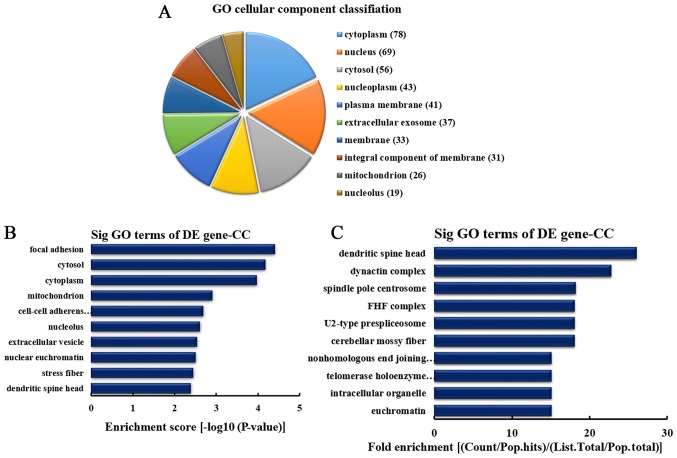
CC analysis of the genes producing downregulated circRNA
According to the number of linear counterparts of differentially downregulated circRNA identified in CCs, the top 10 CCs were classified (Fig. 5A). Among them, 78 linear counterparts of differentially downregulated circRNA were involved in the cytoplasm, 69 were involved in the nucleus, 56 were involved in the cytosol, 43 were involved in the nucleoplasm, 41 were involved in the plasma membrane, 37 were involved in the extracellular exosome, 33 were involved in the membrane, 31 were involved in the integral component of the membrane, 26 were involved in mitochondrion and 19 were involved in the nucleolus. According to the enrichment score, the top 10 CC categories were selected (Fig. 5B). These included focal adhesion, cytosol, cytoplasm, mitochondrion, cell-cell adherens junction, nucleolus, extracellular vesicle, nuclear euchromatin, stress fiber and dendritic spine head. Additionally, according to the fold enrichment, the top 10 CC categories were selected (Fig. 5C). These included dendritic spine head, dynactin complex, spindle pole centrosome, Fuse Toes complex, U2-type prespliceosome, cerebellar mossy fiber, nonhomologous end joining complex, telomerase holoenzyme complex, intracellular organelle and euchromatin.
Figure 5.
CC analysis of the genes producing downregulated circRNA. (A) CC classification based on the number of linear counterparts of downregulated cir-cRNA. (B) The top 10 CCs according to the enrichment score. (C) The top 10 CCs according to the fold enrichment. CC, cellular component; circRNA, circular RNA; GO, gene ontology; DE, differentially expressed; Sig, significant; FHF complex, Fuse Toes complex.
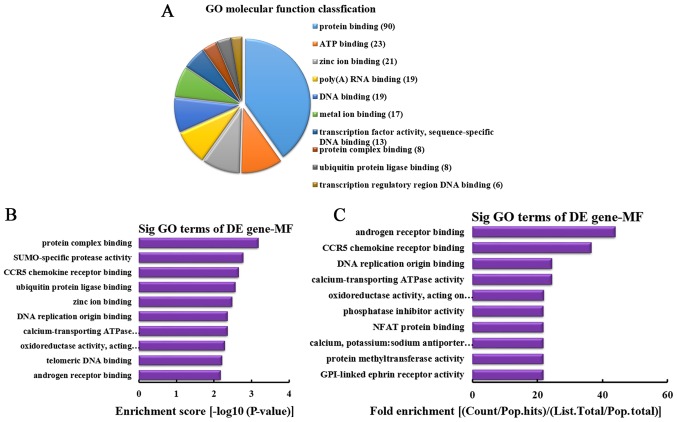
MF analysis of the genes producing upregulated circRNA
According to the number of linear counterparts of differentially overexpressed circRNA identified in MF, the top 10 MFs were classified (Fig. 6A). Among them, 90 linear counterparts of differentially overexpressed circRNA were involved in protein binding, 23 were involved in ATP binding, 21 were involved in zinc ion binding, 19 were involved in poly(A) RNA binding, 19 were involved in DNA binding, 17 were involved in metal ion binding, 13 were involved in transcription factor activity, sequence-specific DNA binding, 8 were involved in protein complex binding, 8 were involved in ubiquitin protein ligase binding and 6 were involved in transcription regulatory region DNA binding. According to the enrichment score, the following top 10 MFs were selected (Fig. 6B): Protein complex binding, SUMO-specific protease activity, C-C chemokine receptor type 5 (CCR5) chemokine receptor binding, ubiquitin protein ligase binding, zinc ion binding, DNA replication origin binding, calcium-transporting ATPase activity, oxidoreductase activity [acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen], telomeric DNA binding and androgen receptor binding. Additionally, according to the fold enrichment, the following top 10 MFs were identified (Fig. 6C): Androgen receptor binding, CCR5 chemokine receptor binding, DNA replication origin binding, calcium-transporting ATPase activity, oxidoreductase activity [acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen], phosphatase inhibitor activity, nuclear factor (NF) of activated T cells protein binding, calcium, potassium: sodium antiporter activity, protein methyltransferase activity and glycosylphosphatidylinositol-linked ephrin receptor activity.
Figure 6.
MF analysis of the genes producing upregulated circRNA. (A) MF classification based on the number of linear counterparts of upregulated circRNA. (B) The top 10 MFs according to the enrichment score. (C) The top 10 MFs according to the fold enrichment. MF, molecular function; circRNA, circular RNA; GO, gene ontology; DE, differentially expressed; Sig, significant; NFAT, nuclear factor of activated T cells; CCR5, C-C chemokine receptor type 5; GPI, glycosylphosphatidylinositol.
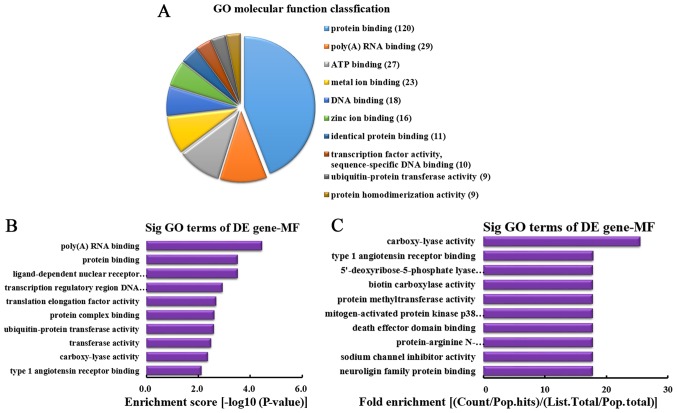
MF analysis of the genes producing downregulated circRNA
According to the number of linear counterparts of differentially downregulated circRNA identified in MF, the top 10 MFs were classified (Fig. 7A). Among them, 120 linear counterparts of differentially downregulated circRNA were involved in protein binding, 29 were involved in poly(A) RNA binding, 27 were involved in ATP binding, 23 were involved in metal ion binding, 18 were involved in DNA binding, 16 were involved in zinc ion binding, 11 were involved in identical protein binding, 10 were involved in transcription factor activity, sequence-specific DNA binding, 9 were involved in ubiquitin-protein transferase activity and 9 were involved in protein homodimerization activity. According to the enrichment score, the following top 10 MF categories were selected (Fig. 7B): Poly(A) RNA binding, protein binding, ligand-dependent nuclear receptor transcription coactivator activity, transcription regulatory region DNA binding, translation elongation factor activity, protein complex binding, ubiquitin-protein transferase activity, transferase activity, carboxy-lyase activity and type 1 angiotensin receptor binding. According to the fold enrichment, the following top 10 MFs were selected (Fig. 7C): Carboxy-lyase activity, type 1 angiotensin receptor binding, 5′-deoxyribose-5-phosphate lyase activity, biotin carboxylase activity, protein methyltransferase activity, mitogen-activated protein kinase p38 binding, death effector domain binding, protein-arginine N-methyltransferase activity, sodium channel inhibitor activity and neuroligin family protein binding.
Figure 7.
MF analysis of the genes producing downregulated circRNA. (A) MF classification based on the number of linear counterparts of downregulated circRNA. (B) The top 10 MFs according to the enrichment score. (C) The top 10 MFs according to the fold enrichment. MF, molecular function; circRNA, circular RNA; GO, gene ontology; DE, differentially expressed; Sig, significant.
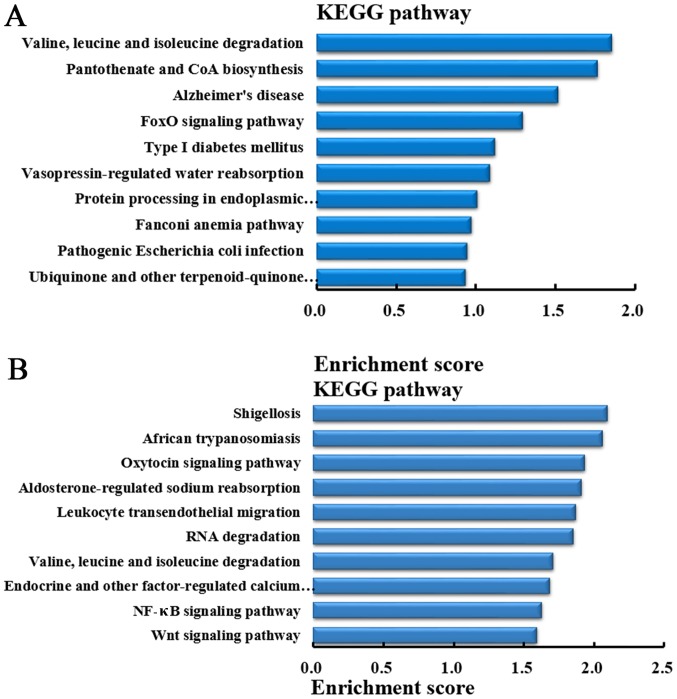
Pathway analysis of the genes producing differentially expressed circRNA
In order to understand the pathways that differentially expressed circRNA involved in lung metastasis in CRC, KEGG pathway analysis was performed to analyze the genes producing differentially expressed circRNA. According to the enrichment score, the top 10 pathways in which the genes producing upregulated circRNA were involved in are demonstrated in Fig. 8A. While, based on the enrichment score, the top 10 pathways in which the genes producing downregulated circRNA were involved are demonstrated in Fig. 8B.
Figure 8.
Pathway analysis of the genes producing differentially expressed circRNA. (A) The top 10 upregulated pathways according to the enrichment score. (B) The top 10 downregulated pathways according to the enrichment score. circRNA, circular RNA; KEGG, Kyoto Encyclopedia of Genes and Genomes; NF-κB, nuclear factor-κB.
Candidate circRNA as potential miRNA sponges
To investigate the circRNA/microRNA interactions, the binding sites of circRNA were predicted. As demonstrated in Table I, it was identified that hsa_circRNA_105055 (upregulated), hsa_circRNA_086376 (downregulated) and hsa_circRNA_102761 (downregulated) could bind with miR-7 regulating target genes PRKCB, EPHA3, BRCA1 and ABCC1. Additionally, hsa_circRNA_102348 and hsa_circRNA_102562 could bind with miR-410 regulating target genes PDHX, ATM and ABCC1.
Table I.
Potential and common miRNA sponging by circRNA.
| circRNA | Common sponging miRNA | miRNA target genes |
|---|---|---|
| hsa_circRNA_105055 | miR-7 | PRKCB |
| hsa_circRNA_086376 | EPHA3 | |
| hsa_circRNA_102761 | BRCA1 ABCC1 |
|
| hsa_circRNA_102348 | miR-410 | PDHX |
| hsa_circRNA_102562 | ATM ABCC1 |
miRNA, microRNA; circRNA, circular RNA.
Discussion
CRC is one of the most frequent malignant tumors in developed countries (27,28). Of the patients with CRC, ~50% eventually develop distant metastases, such as pulmonary metastasis, resulting in poor outcomes even when these patients have resectable primary tumors (29). Therefore, it is important to investigate the molecular mechanism involved in CRC with pulmonary metastasis. In the present study, it was identified that abnormal circRNA were expressed in CRC tissues from patients with pulmonary metastasis compared with CRC tissues without pulmonary metastasis. GO, KEGG pathway and circRNA/miRNA interaction analyses were conducted to determine the potential function and regulatory mechanisms of circRNA in CRC tissues from patients with pulmonary metastasis.
First, through high-throughput microarray, it was identified that 431 circRNA were differentially expressed in CRC tissues from patients with pulmonary metastasis compared with CRC tissues without pulmonary metastasis. Among them, 192 and 239 circRNA were upregulated and down-regulated, respectively. Among the abnormally expressed circRNA, in the light of their relation with their coding genes, the circRNA were classified into five categories: 331 were exonic, 45 were intronic, 32 were sense overlapping, 8 were intergenic and 15 were antisense. Previous study indicated that 39 circRNA were significantly differentially expressed between normal colon mucosa and CRC samples, 11 of them were upregulated in cancer and 28 were downregulated (25). Circ_001569 has been demonstrated to be upregulated, while Cir-ITCH has been indicated to be typically downregulated in CRC in comparison with peritumoral tissue (30,31). The expression of circRNA were altered in primary and metastatic sites of epithelial ovarian cancer (32). Furthermore, study has indicated that 469 circRNA were markedly differentially expressed between bladder carcinoma and normal tissues with a fold change ≥2.0 and P<0.05, among which 285 circRNA were upregulated and 184 were downregulated (33). To the best of our knowledge, the present study is the first to report the differentially expressed circRNA in CRC tissues from patients with pulmonary metastasis.
Then, GO analysis of the genes producing differentially expressed circRNA was performed to investigate the potential enriched GO of differentially expressed circRNA. It was demonstrated that the genes producing upregulated circRNA were involved in DNA repair. It has been reported that DNA repair commonly appeared in CRC (34–36), and the carriers of allele A in DNA repair genes XRCC1 were connected with a higher risk of disseminated CRC (37). The present study also revealed that the genes producing downregulated circRNA were enriched in signal transduction. Previous study has indicated that various signal transduction pathways were important in genetic programming and growth control of metastatic CRC and CRC (38–41). Therefore, these studies combined with the present study demonstrated that metastatic CRC may result from the interaction effects of multiple circRNA involved in regulation of multi-systems.
Additionally, KEGG pathway analysis was performed to analyze the genes producing differentially expressed circRNA. It was demonstrated that the genes producing downregulated circRNA were involved in the NF-κB and Wnt signaling pathway in the CRC tissues from patients with lung metastasis compared with the CRC tissues without metastasis. Previous study indicated that Akt/protein kinase B (PKB) was a major antiapoptotic pathway that was frequently hyperactivated in the majority of cancer types (42), and the antiapoptotic NF-κB signaling pathway was one of the downstream targets of activated Akt/PBK and controlled the expression of genes and cellular processes involved in cell proliferation, oncogenesis, angiogenesis, and apoptosis (43,44). Furthermore, a previous study also indicated a critical role for aberrant Wnt signaling in CRC that was involved in diverse cellular processes, such as cell migration (45).
In addition, bioinformatics methods were performed in the present study to predict circRNA/microRNA interactions. The results demonstrated that hsa_circRNA_105055 (upregulated), hsa_circRNA_086376 (downregulated) and hsa_circRNA_102761 (downregulated) may bind with miR-7 regulating target genes PRKCB, EPHA3, BRCA1 and ABCC1. Additionally, hsa_circRNA_102348 and hsa_circRNA_102562 may bind with miR-410 regulating target genes PDHX, ATM and ABCC1. Previous study indicated that miR-7 could inhibit CRC cell proliferation and induce apoptosis by targeting XRCC2 (46). PRKCB and EPHA3 were differentially expressed in CRC tissues (47–49). It has also been reported that BRCA1 and ERCC1 mRNA were associated with lymph node metastasis in Chinese patients with CRC (50). BRCA1 gene overexpression was correlated with radiation response in CRC (51), and BRCA1 mutation may be associated with early onset of CRC (52). Study has also indicated that the expression of ABCC1 was altered in association with colon carcinogenesis (53). For miR-410, previous study revealed that miR-410 could regulate apoptosis by targeting Bak1 in human CRC cells (54). PDHX may be targeted by miR-26a to regulate glucose metabolism of CRC, which inhibited the conversion of pyruvate to acetyl coenzyme A in the citric acid cycle (55). ATM may be negatively regulated by miR-203 to induce oxaliplatin resistance in CRC (56). To the best our knowledge, the present study is the first to report that hsa_circRNA_105055, hsa_circRNA_086376 and hsa_circRNA_102761 may bind with miR-7 regulating target genes PRKCB, EPHA3, BRCA1 and ABCC1, and hsa_circRNA_102348 and hsa_ circRNA_102562 may bind with miR-410 regulating target genes PDHX, ATM and ABCC1. However, these binding interactions need to be verified through functional analysis in future study.
In conclusion, the present study demonstrated that 431 circRNA were differentially expressed in CRC tissues from patients with pulmonary metastasis compared with tissues without metastasis. By utilizing GO, KEGG pathway and circRNA/miRNA interaction analyses, the present study revealed that hsa_circRNA_105055 (upregulated), hsa_circRNA_086376 (downregulated) or hsa_circRNA_102761 (downregulated) may regulate the pulmonary metastasis of CRC through binding with miR-7 to regulate PRKCB (the target gene of miR-7) that is involved in the NF-κB or Wnt signaling pathway. However, this requires further study. The present findings may provide a novel perspective on circRNA and lay a foundation for future research of potential roles of circRNA in CRC with pulmonary metastasis.
Acknowledgments
The present study was supported by the Project of Department of Health in Yunnan Province (grant no. 2014NS124), the China Guanghua Foundation, the General Joint Project of Yunnan Provincial Science and Technology Department and Kunming Medical University (grant no. 2015FB024) and the Major Project of Yunnan Provincial Bureau of Education (grant no. ZD2015010).
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 4.Shah SA, Haddad R, Al-Sukhni W, Kim RD, Greig PD, Grant DR, Taylor BR, Langer B, Gallinger S, Wei AC. Surgical resection of hepatic and pulmonary metastases from colorectal carcinoma. J Am Coll Surg. 2006;202:468–475. doi: 10.1016/j.jamcollsurg.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 5.Rotolo N, De Monte L, Imperatori A, Dominioni L. Pulmonary resections of single metastases from colorectal cancer. Surg Oncol. 2007;16(Suppl 1):S141–S144. doi: 10.1016/j.suronc.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Goya T, Miyazawa N, Kondo H, Tsuchiya R, Naruke T, Suemasu K. Surgical resection of pulmonary metastases from colorectal cancer. 10-year follow-up. Cancer. 1989;64:1418–1421. doi: 10.1002/1097-0142(19891001)64:7<1418::AID-CNCR2820640709>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 7.Galandiuk S, Wieand HS, Moertel CG, Cha SS, Fitzgibbons RJ, Jr, Pemberton JH, Wolff BG. Patterns of recurrence after curative resection of carcinoma of the colon and rectum. Surg Gynecol Obstet. 1992;174:27–32. [PubMed] [Google Scholar]
- 8.McAuliffe JC, Qadan M, D'Angelica MI. Hepatic resection, hepatic arterial infusion pump therapy, and genetic biomarkers in the management of hepatic metastases from colorectal cancer. J Gastrointest Oncol. 2015;6:699–708. doi: 10.3978/j.issn.2078-6891.2015.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen D, Sun Q, Cheng X, Zhang L, Song W, Zhou D, Lin J, Wang W. Genome-wide analysis of long noncoding RNA (lncRNA) expression in colorectal cancer tissues from patients with liver metastasis. Cancer Med. 2016;5:1629–1639. doi: 10.1002/cam4.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattick JS, Taft RJ, Faulkner GJ. A global view of genomic information - moving beyond the gene and the master regulator. Trends Genet. 2010;26:21–28. doi: 10.1016/j.tig.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Malouf GG, Zhang J, Yuan Y, Compérat E, Rouprêt M, Cussenot O, Chen Y, Thompson EJ, Tannir NM, Weinstein JN, et al. Characterization of long non-coding RNA transcriptome in clear-cell renal cell carcinoma by next-generation deep sequencing. Mol Oncol. 2015;9:32–43. doi: 10.1016/j.molonc.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S, Xu D, Lin HK, Gong Z. Circular RNAs in cancer: Novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5:472–480. [PMC free article] [PubMed] [Google Scholar]
- 13.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 15.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 16.Perkel JM. Assume nothing: The tale of circular RNA. Biotechniques. 2013;55:55–57. doi: 10.2144/000114061. [DOI] [PubMed] [Google Scholar]
- 17.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 21.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 23.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 24.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation - exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorson AG, Christensen MA, Davis SJ. The role of colonoscopy in the assessment of patients with colorectal cancer. Dis Colon Rectum. 1986;29:306–311. doi: 10.1007/BF02554117. [DOI] [PubMed] [Google Scholar]
- 27.Jemal A, Thun MJ, Ries LA, Howe HL, Weir HK, Center MM, Ward E, Wu XC, Eheman C, Anderson R, et al. Annual report to the nation on the status of cancer, 1975–2005, featuring trends in lung cancer, tobacco use, and tobacco control. J Natl Cancer Inst. 2008;100:1672–1694. doi: 10.1093/jnci/djn389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yiu HY, Whittemore AS, Shibata A. Increasing colorectal cancer incidence rates in Japan. Int J Cancer. 2004;109:777–781. doi: 10.1002/ijc.20030. [DOI] [PubMed] [Google Scholar]
- 29.Lin PC, Lin JK, Lin CC, Wang HS, Yang SH, Jiang JK, Lan YT, Lin TC, Li AF, Chen WS, et al. Carbohydrate antigen 19-9 is a valuable prognostic factor in colorectal cancer patients with normal levels of carcinoembryonic antigen and may help predict lung metastasis. Int J Colorectal Dis. 2012;27:1333–1338. doi: 10.1007/s00384-012-1447-1. [DOI] [PubMed] [Google Scholar]
- 30.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PLoS One. 2015;10:e0131225. doi: 10.1371/journal.pone.0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmed I, Karedath T, Andrews SS, Al-Azwani IK, Mohamoud YA, Querleu D, Rafii A, Malek JA. Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget. 2016;7:36366–36381. doi: 10.18632/oncotarget.8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang M, Zhong Z, Lv M, Shu J, Tian Q, Chen J. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget. 2016;7:47186–47200. doi: 10.18632/oncotarget.9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karam RA, Al Jiffry BO, Al Saeed M, Abd El Rahman TM, Hatem M, Amer MG. DNA repair genes polymorphisms and risk of colorectal cancer in Saudi patients. Arab J Gastroenterol. 2016;17:117–120. doi: 10.1016/j.ajg.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 35.de Freitas IN, de Campos FG, Alves VA, Cavalcante JM, Carraro D, Coudry Rde A, Diniz MA, Nahas SC, Ribeiro U., Jr Proficiency of DNA repair genes and microsatellite instability in operated colorectal cancer patients with clinical suspicion of lynch syndrome. J Gastrointest Oncol. 2015;6:628–637. doi: 10.3978/j.issn.2078-6891.2015.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scarbrough PM, Weber RP, Iversen ES, Brhane Y, Amos CI, Kraft P, Hung RJ, Sellers TA, Witte JS, Pharoah P, et al. A cross-cancer genetic association analysis of the DNA repair and DNA damage signaling pathways for lung, ovary, prostate, breast, and colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2016;25:193–200. doi: 10.1158/1055-9965.EPI-15-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dimberg J, Skarstedt M, Slind Olsen R, Andersson RE, Matussek A. Gene polymorphism in DNA repair genes XRCC1 and XRCC6 and association with colorectal cancer in Swedish patients. APMIS. 2016;124:736–740. doi: 10.1111/apm.12563. [DOI] [PubMed] [Google Scholar]
- 38.Győrffy B, Stelniec-Klotz I, Sigler C, Kasack K, Redmer T, Qian Y, Schäfer R. Effects of RAL signal transduction in KRAS- and BRAF-mutated cells and prognostic potential of the RAL signature in colorectal cancer. Oncotarget. 2015;6:13334–13346. doi: 10.18632/oncotarget.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu X, Li C, Wang YK, Jiang K, Gai XD. Sorbitol induces apoptosis of human colorectal cancer cells via p38 MAPK signal transduction. Oncol Lett. 2014;7:1992–1996. doi: 10.3892/ol.2014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji H, Greening DW, Barnes TW, Lim JW, Tauro BJ, Rai A, Xu R, Adda C, Mathivanan S, Zhao W, et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics. 2013;13:1672–1686. doi: 10.1002/pmic.201200562. [DOI] [PubMed] [Google Scholar]
- 41.Nowakowska-Zajdel E, Mazurek U, Stachowicz M, Niedworok E, Fatyga E, Muc-Wierzgoń M. Cellular signal transduction pathways by leptin in colorectal cancer tissue: Preliminary results. ISRN Endocrinol. 2011;2011:575397. doi: 10.5402/2011/575397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marte BM, Downward J. PKB/Akt: Connecting phosphoinositide 3-kinase to cell survival and beyond. Trends Biochem Sci. 1997;22:355–358. doi: 10.1016/S0968-0004(97)01097-9. [DOI] [PubMed] [Google Scholar]
- 43.Mayo MW, Baldwin AS. The transcription factor NF-kappaB: Control of oncogenesis and cancer therapy resistance. Biochim Biophys Acta. 2000;1470:M55–M62. doi: 10.1016/s0304-419x(00)00002-0. [DOI] [PubMed] [Google Scholar]
- 44.Hidalgo M, Rowinsky EK. The rapamycin-sensitive signal transduction pathway as a target for cancer therapy. Oncogene. 2000;19:6680–6686. doi: 10.1038/sj.onc.1204091. [DOI] [PubMed] [Google Scholar]
- 45.Qi J, Yu Y, Akilli Öztürk Ö, Holland JD, Besser D, Fritzmann J, Wulf-Goldenberg A, Eckert K, Fichtner I, Birchmeier W. New Wnt/β-catenin target genes promote experimental metastasis and migration of colorectal cancer cells through different signals. Gut. 2016;65:1690–1701. doi: 10.1136/gutjnl-2014-307900. [DOI] [PubMed] [Google Scholar]
- 46.Xu K, Chen Z, Qin C, Song X. miR-7 inhibits colorectal cancer cell proliferation and induces apoptosis by targeting XRCC2. Onco Targets Ther. 2014;7:325–332. doi: 10.2147/OTT.S59364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Zheng T. Screening of hub genes and pathways in colorectal cancer with microarray technology. Pathol Oncol Res. 2014;20:611–618. doi: 10.1007/s12253-013-9739-5. [DOI] [PubMed] [Google Scholar]
- 48.Slaby O, Sachlova M, Brezkova V, Hezova R, Kovarikova A, Bischofová S, Sevcikova S, Bienertova-Vasku J, Vasku A, Svoboda M, et al. Identification of microRNAs regulated by isothiocyanates and association of polymorphisms inside their target sites with risk of sporadic colorectal cancer. Nutr Cancer. 2013;65:247–254. doi: 10.1080/01635581.2013.756530. [DOI] [PubMed] [Google Scholar]
- 49.Huhn S, Bevier M, Pardini B, Naccarati A, Vodickova L, Novotny J, Vodicka P, Hemminki K, Försti A. Colorectal cancer risk and patients' survival: Influence of polymorphisms in genes somatically mutated in colorectal tumors. Cancer Causes Control. 2014;25:759–769. doi: 10.1007/s10552-014-0379-1. [DOI] [PubMed] [Google Scholar]
- 50.Yuanming L, Lineng Z, Baorong S, Junjie P, Sanjun C. BRCA1 and ERCC1 mRNA levels are associated with lymph node metastasis in Chinese patients with colorectal cancer. BMC Cancer. 2013;13:103. doi: 10.1186/1471-2407-13-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang MY, Wang JY, Chang HJ, Kuo CW, Tok TS, Lin SR. CDC25A, VAV1, TP73, BRCA1 and ZAP70 gene overexpression correlates with radiation response in colorectal cancer. Oncol Rep. 2011;25:1297–1306. doi: 10.3892/or.2011.1193. [DOI] [PubMed] [Google Scholar]
- 52.Sopik V, Phelan C, Cybulski C, Narod SA. BRCA1 and BRCA2 mutations and the risk for colorectal cancer. Clin Genet. 2015;87:411–418. doi: 10.1111/cge.12497. [DOI] [PubMed] [Google Scholar]
- 53.Kobayashi M, Funayama R, Ohnuma S, Unno M, Nakayama K. Wnt-β-catenin signaling regulates ABCC3 (MRP3) transporter expression in colorectal cancer. Cancer Sci. 2016;107:1776–1784. doi: 10.1111/cas.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu C, Zhang A, Cheng L, Gao Y. miR 410 regulates apoptosis by targeting Bak1 in human colorectal cancer cells. Mol Med Rep. 2016;14:467–473. doi: 10.3892/mmr.2016.5271. [DOI] [PubMed] [Google Scholar]
- 55.Chen B, Liu Y, Jin X, Lu W, Liu J, Xia Z, Yuan Q, Zhao X, Xu N, Liang S. MicroRNA-26a regulates glucose metabolism by direct targeting PDHX in colorectal cancer cells. BMC Cancer. 2014;14:443. doi: 10.1186/1471-2407-14-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Y, Wan G, Spizzo R, Ivan C, Mathur R, Hu X, Ye X, Lu J, Fan F, Xia L, et al. miR-203 induces oxaliplatin resistance in colorectal cancer cells by negatively regulating ATM kinase. Mol Oncol. 2014;8:83–92. doi: 10.1016/j.molonc.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]