Abstract
The genome of D. radiodurans harbors genes for structural and regulatory proteins of Kdp ATPase, in an operon pattern, on Mega plasmid 1. Organization of its two-component regulatory genes is unique. Here we demonstrate that both, the structural as well as regulatory components of the kdp operon of D. radiodurans are expressed quickly as the cells experience potassium limitation but are not expressed upon increase in osmolarity. The cognate DNA binding response regulator (RR) effects the expression of kdp operon during potassium deficiency through specific interaction with the kdp promoter. Deletion of the gene encoding RR protein renders the mutant D. radiodurans (ΔRR) unable to express kdp operon under potassium limitation. The ΔRR D. radiodurans displays no growth defect when grown on rich media or when exposed to oxidative or heat stress but shows reduced growth following gamma irradiation. The study elucidates the functional and regulatory aspects of the novel kdp operon of this extremophile, for the first time.
Introduction
Potassium (K+) is essential for physiological functions such as regulation of intracellular pH, transmembrane electrical potential and turgor pressure in all living organisms [1]. K+ homeostasis is critical for adaptation to several biotic or abiotic stresses in bacteria [1–6]. Bacteria accumulate 0.1–0.6 M K+ intracellularly from trace amounts (0.1–10 mM) of this cation present in the environment [7] through a variety of low and high affinity K+ transporters. In E. coli, low affinity K+ transporters Trk and Kup are constitutively expressed and maintain physiological concentration of potassium in the cell [8] while high affinity ATP-dependent K+ uptake system (Km 2 μM), is effected by an inducible KdpATPase expressed under K+ limiting conditions (active below 5 mM K+) [5]. Ktr, a fourth constitutively expressed K+ uptake transporter, is absent in E. coli but encoded by several other microbes [9].
The homologs of kdp operon and its two-component signaling system are present in Gram-positive as well as Gram-negative bacteria and archaeal species [10] wherein kdp operon expression is stimulated by low K+ concentration and high osmolarity. In addition, its expression has also been coupled with regulation of virulence genes and pathogenesis in Staphylococcus aureus, Salmonella typhimurium, Yersinia pestis, mycobacteria etc. [11, 12] Expression of kdp operon is also associated with high NaCl tolerance in S. aureus [12], phosphate limitation in E. coli [13], or drought stress in Anabaena 7120 [14]. Thus, kdp operon forms a part of basic metabolism and also contributes to survival under a variety of stressful conditions.
Gram-positive D. radiodurans exhibits extreme resistance to gamma radiation as well as to desiccation [15], owing to its customized DNA damage repair system, enzymatic/non-enzymatic antioxidants and metabolites. A number of elegant studies reported differential expression/levels of transcriptome, small RNAs, proteome, antioxidants, metabolites etc. during the phase of DNA repair [16–21]. K+ is necessary for the stability of replication process and maintaining genome integrity in cells [22, 23], both the functions actively carried out by D. radiodurans while recovering from DNA damage. Thus, role of K+ homeostasis in the stress resistances of D. radiodurans needs detailed exploration.
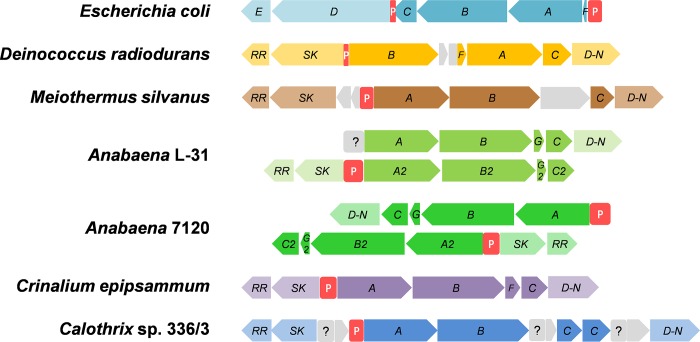
The genome of D. radiodurans shows presence of two K+ uptake systems, namely, Ktr and K+-transporting ATPase (Kdp). The organization of kdp genes in D. radiodurans is distinct from well studied kdp operon in E. coli, especially in the two component regulatory genes (Fig 1). In D. radiodurans, structural genes kdpBAC (DR_B0083, DR_B0086, DR_B0087, respectively) were present in one operon along with a naturally truncated regulatory component kdpD-N (representing N-terminal half of the E. coli kdpD, DR_B0088) (Fig 1). The remaining regulatory components, sensor kinase (SK, representing C-terminal half of the E. coli kdpD, DR_B0082) and the response regulator (RR, DR_B0081) were present in a separate operon, in an opposite orientation, immediately upstream of the kdpBACD-N operon (Fig 1) [24]. Homologs of such truncated kdpD genes are found in the genomes of Deinococcus-Thermus group and cyanobacteria (Fig 1), that share close evolutionary relationship [25]. In cyanobacteria, that harbor 2 separate kdp operons in yet another distinct organization of regulatory components (Fig 1), only one kdp operon responds to potassium limitation, high osmolarity or desiccation [14, 26–28]. However, the role and regulation of kdp operon in the normal physiology and stress responses of D. radiodurans remains completely unexplored.
Fig 1. Comparison of organization of kdp operon in bacteria.
Organization of kdp operon in E. coli (model organism), representative examples from Deinococcus-Thermus group and cyanobacteria (as depicted in KEGG database [29]). The promoter regions are shown with red boxes denoted by the letter ‘P’. Undefined promoter region is shown with a gray box and question mark. A, B, C, D, E and F denote kdpA, kdpB, kdpC, kdpD, kdpE and kdpF genes, respectively. RR, SK and D-N denote response regulator, sensor kinase and N-terminal half of kdpD gene, respectively.
In this study, we report that the kdp operon of D. radiodurans is induced following K+ limitation but does not respond to high osmolarity, unlike E. coli Kdp ATPase. We also demonstrate that regulation of kdp operon is effected through the cognate response regulator (RR) protein. The knockout mutant of RR (ΔRR) displayed kdp expression null phenotype even under potassium limiting conditions. In rich media, the growth of ΔRR mutant was similar to wild type D. radiodurans. Survival response of ΔRR mutant to other relevant stresses was also evaluated.
Materials and methods
Bacterial strains and growth conditions
All the strains used in this study are listed in Table 1. Wild type D. radiodurans strain R1 ATCC BAA-816 or mutant constructed therein, were grown under standard growth conditions [32°C with 150 rpm agitation]. The cells were cultured either in TGY (1% Bacto Tryptone, 0.5% Bacto Yeast Extract, 0.1% glucose) broth, or in minimal medium adapted from previous studies [30–32] with modifications. The composition of the minimal medium was: 20mM Na/K phosphate buffer, 0.5% glucose, 0.05% casamino acids, macronutrients: 2.5mM (NH4)2SO4, 0.1mM CaCl2, 5μM FeSO4, 0.5mM MgCl2, micronutrients: 2nM CuSO4, 50nM MnCl2, 10nM ZnSO4, 50nM CoCl2, nicotinic acid (1μg/ml), biotin (1μg/ml). Liquid or solid (1.5% Bacto agar) media were supplemented with 10 μg/ml kanamycin when required. The plasmid constructs were maintained or propagated in E. coli JM109 while E. coli BL21(DE3)pLysS strain was used for the over-expression of recombinant proteins (Table 1). All E. coli strains were grown at 37°C either in LB (Luria-Bertani) broth with agitation (150 rpm), or on LB agar plates supplemented with 50μg/ml kanamycin or 100μg/ml carbenicillin, when necessary. Alternatively, E. coli BL21(DE3)pLysS cells harboring pET29b vector with desired gene were grown in auto-induction medium [33] at 20°C with 150 rpm agitation.
Table 1. Bacterial strains, plasmids and primers used in this study.
| Bacterial strains | ||
| Strain | Description | Reference/ Source |
| Deinococcus radiodurans | ||
| R1 | Wild type strain ATCC BAA-816 | [15] |
| ΔRR | ΔDR_B0081 D. radiodurans R1 strain, Kanr | This study |
| E. coli | ||
| JM109 | F′traD36 proAB+ lacIq lacZΔM15/Δ(lac-proAB) glnV44 e14- gyrA96 recA1 relA1 endA1 thi-1 hsdR17 mcrB+ | New England Biolabs |
| JM110 | F′traD36 lacIq lacZΔM15 proAB+ rpsL thr leu thi lacY galK galT ara tonA tsx dam dcm glnV44 Δ(lac-proAB) | New England Biolabs |
|
E. coli BL21 (DE3) pLysS |
F– ompT gal dcm lon hsdSB(rB- mB-) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]), Kanr. | Novagen |
| Plasmids | ||
| Plasmids | Description | Reference |
| pUC4K | 3.91 kb, Kanr | Amersham |
| pBlueScript | 2.96 kb phagemid cloning vector; Ampr | Stratagene |
| pET29b | 5.4 kb, pBR322 origin, Kanr | Novagen |
| pET-kdpB | 7.4 kb, NdeI/XhoI fragment of DR_B0083 ORF (2028 bp) cloned in NdeI/XhoI restricted pET29b, Kanr | |
| pET-skdpB | 6.3 kb, NdeI/XhoI fragment of soluble portion of DR_B0083 ORF (914 bp) cloned in NdeI/XhoI restricted pET29b, Kanr | This study |
| pET-RR | 6.0 kb, NdeI/XhoI fragment of DR_B0081 ORF (630 bp) cloned in NdeI/XhoI restricted pET29b, Kanr | This study |
| pΔRR1 | 4.08 kb, pBS carrying 0.54 kb of sequence flanking the 5′ and 3′ ends of DR_B0081 (DR_B0081-up and DR_B0081-dn); for the purpose of deleting the chromosomal copy of DR_B0081, Ampr. | This study |
| pΔRR2 | 5.33 kb, pΔRR1 with Kanr cassette inserted between DR_B0081-up and DR_B0081-dn, Ampr, Kanr. | This study |
| Primers | ||
| Primer | Sequence | Reference |
| Primers for cloning DR_B0083 (kdpB) and DR_B0081 (RR) genes | ||
| kdpB-F | 5’- TTTCCGGGTACCCATATGACCACGGCCCCTCAG—3’ (KpnI, NdeI) | This study |
| kdpB-R | 5'- AACGCCCTCGAGTGACATCAATCCACC -3' (XhoI) | This study |
| skdpB-F | 5'-TATAACATATGGATAGGGCTTTGCAG-3' (NdeI) | This study |
| skdpB-R | 5'-TCGTTCTCGAGGGAAAAAGTGGTCAGGGC-3'(XhoI) | This study |
| RR-F | 5’-TAAACGGTACCCATATGCCTGACCCGGTGGGC-3’ (KpnI, NdeI) | This study |
| RR-R | 5’-ATATACTCGAGCAGCAGCCCCGCCCG-3’ (XhoI) | This study |
| Primers for DR_B0081 (RR) knockout mutagenesis | ||
| RR-up-F | 5’-ACACAGGTACCAGCTGAACTACGTAGCGA-3’ (KpnI) | This study |
| RR-up-R | 5’-ACACAGATATCTGGCCTTTTGTTGTGGAT-3’ (EcoRV) | This study |
| RR-dn-F | 5’-AGATAGATATCAGCAGGATGCCCACCGGG-3’ (EcoRV) | This study |
| RR-dn-R | 5’-AGATAGGATCCGCCAACGAAACGCTGCTC-3’ (BamHI) | This study |
| Primers for promoter cloning | ||
| P200bp-F | 5'-AATATAAGCTTCGGCGGCGGGAACAGCTC-3' (HindIII) | This study |
| P200bp-R | 5'-AATATGATATCCCTGGTCGCGCGGCGAGA-3' (EcoRV) | This study |
| P38-F | 5'-AACGGCGCTATGAGTCTTTCTTCTTTCCGGTGGCGGCC-3’ | This study |
| P38-R | 5'-GGCCGCCACCGGAAAGAAGAAAGACTCATAGCGCCGTT-3’ | This study |
| Primers for sequencing | ||
| kdpB-548-F_Seq | 5'- CATCGTCATTCAAATCACCTC -3' | This study |
| kdpB-1098-F_Seq | 5'- GAGTTCATCGAATTCACCGCC -3' | This study |
| kdpB-1620-F_Seq | 5'- AACATGGTGGATTTGGACAGC -3' | This study |
| Primers for non-specific DNA | ||
| 0906RTF | 5’-TTTATCCACGCCAACACCTA-3’ | This study |
| 0906RTR | 5’-GGCCTTGATGAGGTTCTTGT-3’ | This study |
Stress conditions
D. radiodurans cells or its mutant was grown overnight in TGY medium. The cells were washed twice in either TGY, 20 mM K+ phosphate supplemented minimal medium (hereafter referred to as K20 medium) or 20 mM sodium phosphate supplemented minimal medium (hereafter referred to as K0 medium) and the washed cells were inoculated in TGY, K20 medium or K0 medium, respectively, at an initial cell density of OD600 = 0.5/ml. The cultures were incubated at 32°C with 150 rpm agitation. The cells resuspended in K0 medium experienced K+ limitation while the cells resuspended in K20 medium or TGY served as controls. For ionic or osmotic stress, K1 medium (1:20 dilution of K20 medium in K0 medium) was supplemented with either 0.1M NaCl or 0.2M sucrose. For gamma irradiation experiments, overnight grown cells of wild type or mutant D. radiodurans cells were resuspended in fresh TGY medium (OD600 = 3.0), 2 μl aliquots of the cell suspensions and their serial dilutions were spotted onto the TGY plate. The plate was exposed to 5 kGy gamma irradiation (Gamma Cell 5000, Bhabha Atomic Research Centre, dose rate: 1.85 kGy/hr). Following stress, the plate was incubated at 32° C for 24 h for recovery before the result was scored. For heat and oxidative stresses, the overnight grown cells of wild type or mutant D. radiodurans cells were resuspended in fresh TGY medium (OD600 = 0.5) and exposed to either 42° C or 100 mM H2O2 for 1h with 150 RPM agitation. Further, the cells were concentrated (OD600 = 3.0) and 2 μl aliquots of the cell suspensions and their serial dilutions were spotted onto the TGY plate. The plate was incubated at 32° C for 24 h for recovery before the result was scored.
Cloning of DR_B0083 (complete or partial) and DR_B0081 ORFs, and over-expression and purification of corresponding proteins
Genomic DNA of D. radiodurans was prepared as per the protocol reported previously [34]. The DR_B0083 (kdpB), DR_B0083/826-1740bp (skdpB), DR_B0081 (RR) ORFs were PCR amplified from the genomic DNA of D. radiodurans using gene-specific primers listed in Table 1. The PCR products were restriction digested using NdeI/XhoI and cloned into pET-29b plasmid vector at the identical sites. Correct clone was ascertained by DNA sequencing. The resultant constructs pET-kdpB, pET-skdpB or pET-RR were transformed in E. coli BL21(DE3)pLysS and expression of corresponding proteins was induced in auto-induction medium at 20°C. The cells were harvested after 15h of growth and the over-expressed proteins (sKdpB and RR) were purified by affinity chromatography using Ni-nitrilotriacetic acid (Ni+2-NTA)-agarose resin (Qiagen, Germany), as per the manufacturer’s protocol. The purified RR protein was used for promoter interaction studies. The sKdpB and RR proteins were purified by gel elution method and used to generate polyclonal antibodies in rabbit, at a commercial facility of Bangalore Genei (India).
Deletion of DR_B0081 (RR) gene and confirmation of the knockout mutant
The ΔDR_B0081 mutant was constructed as per the procedure described previously [35]. In brief, mutagenesis strategy involved complete replacement of DR_B0081 ORF with an aph cassette that conferred kanamycin resistance. For this purpose, the upstream (RR-up) and downstream (RR-dn) sequences (540 bp each) of DR_B0081 were PCR amplified using primer pairs RR-up-F/R and RR-dn-F/R, respectively (Table 1). The PCR amplified RR-up and RR-dn DNA fragments were restriction digested with KpnI/EcoRV and EcoRV/BamHI, respectively and were ligated to the KpnI/BamHI sites of pBlueSkript vector to obtain plasmid pΔRR1. To obtain aph cassette, plasmid pUC4K was restriction digested with HincII to release the 1252 bp DNA fragment harboring aph cassette. The aph cassette was cloned into the EcoRV site of pΔRR1 plasmid, by blunt end ligation. The resultant construct pΔRR2, with aph cassette inserted between the RR-up and RR-dn sequences, was used to transform WT D. radiodurans. Kanamycin resistant transformants were selected on TGY plates containing 10μg/ml kanamycin. The ΔRR knockout mutant was confirmed by PCR and western blot analysis of RR protein.
Immuno-detection of KdpB and RR proteins
The cellular extracts were prepared by sonication (Branson Digital Sonifier, Model 250). Cell debris was removed by centrifugation (10000 rpm for 10 min). The supernatant was centrifuged at 100000g for 1h. The supernatant was collected as cytosolic fraction. The pellet was washed 2 times and then collected as membrane fraction. The cellular proteins extracts, cytosolic or membrane fractions from stressed or unstressed cells were resolved by 12% SDS-PAGE and electro-blotted onto nitrocellulose membrane (Amersham Biosciences, India). The presence of KdpB or RR proteins was probed with primary polyclonal anti-D. radiodurans-sKdpB or anti-D. radiodurans-RR antibodies (1:10,000 dilution), respectively, followed by goat anti-rabbit IgG tagged to alkaline phosphatase (Sigma, 1:5000 dilution). The western blots were developed using NBT-BCIP solution (Roche, Germany). The immuno-detected protein bands which appeared on the membranes were quantified using CLIQS 1D PRO (Total Labs, UK).
Electrophoretic mobility shift assay (EMSA)
The promoter region (200 bp, DNA sequence immediately upstream of DR_B0083 i.e. kdpB gene) of kdp operon was PCR amplified using specific primers (Table 1). Alternatively, the intergenic region (38 bp) between DR_B0082 (sensor kinase) and DR_B0083 (kdpB) was generated in vitro by two primer (P38-F/ P38-R) annealing (Table 1). The DNA fragments were labeled with DIG-ddUTP (Digoxigenin) as per manufacturer’s protocol (Roche, India). DIG labeled promoter fragment (45 ng of DIG-labeled 200 bp DNA) was incubated with indicated concentrations of RR protein in binding buffer [20 mM HEPES pH 7.6,10 mM (NH4)2SO4, 30 mM KCl, 1 mM EDTA, 1 mM DTT and 0.2% Tween20 (v/v)] at 37°C for 1 h. RR protein was phosphorylated using acetyl phosphate as per the procedure detailed earlier [28]. The DNA–protein complexes were resolved on 10% native polyacrylamide gel at 50 V in 0.5X TBE gel running buffer. The resolved DNA–protein complexes were electro-blotted onto nylon membrane for 30 mins. The nylon membrane was cross-linked with UV, probed with anti-DIG antibody and subsequently developed using colorimetric substrate, NBT-BCIP. Each experiment was repeated three times. The bands which appeared on membranes were quantified using gel quant software (Biochem lab solutions) and the KD value for RR protein was obtained by fitting the data to Hill’s equation [36]. Specificity of RR interaction was confirmed by titration of DNA-protein complexes with unlabeled target-specific or non-specific DNA. Further, to determine specific interaction of RR protein with the kdp promoter, EMSA was carried out either by replacing kdp promoter fragment with non-specific DNA or by replacing RR with BSA protein. Non-specific DNA was a DNA sequence (153) bp from DR_0906 gene amplified using primers listed in Table 1.
Bioinformatic analyses
BLASTP [37] and ClustalW [38] tools were used to analyze sequence similarities and for phylogenetic analyses of the proteins encoded by kdp operon of D. radiodurans. Presence of E. coli like gene promoter elements in the upstream sequence of kdp operon was analyzed using BPROM software [39]. Phobius software was used to predict presence of a transmembrane domain in KdpB protein [40].
Results and discussion
The kdp operon of D. radiodurans responds to K+ limitation
To ascertain if the kdp operon of D. radiodurans responds to K+ limitation, we cloned the genes for soluble domain of KdpB (sKdpB, amino acids 277–573 of KdpB protein) and response regulator protein, and over-expressed and purified the corresponding proteins and raised polyclonal antibodies (S1 Fig). The genes for full length kdpB and sensor kinase were also cloned but expression of corresponding proteins could not be achieved in E. coli despite several attempts.
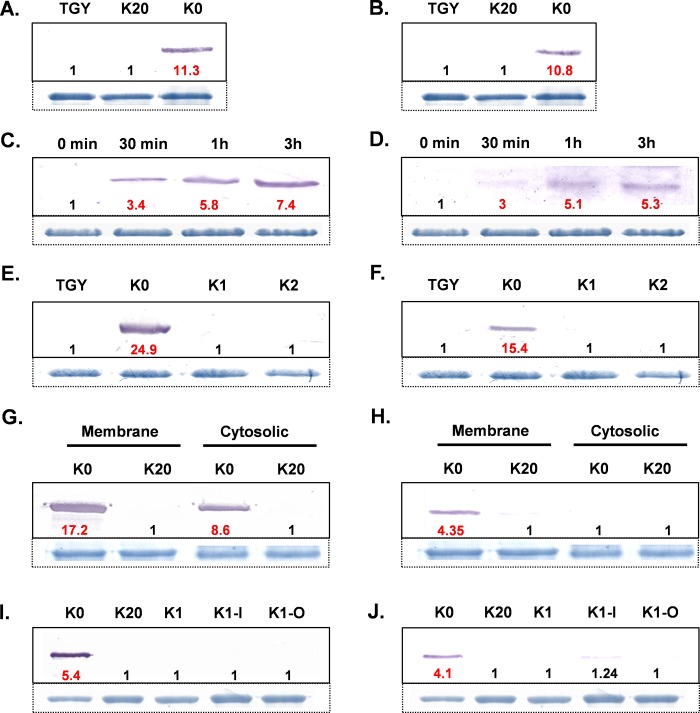
We probed the induction of KdpB as well as RR proteins in response to K+ limitation. The KdpB as well as RR proteins were undetectable in D. radiodurans cells grown in TGY medium or K20 medium, but the cells incubated in K0 medium showed prominent induction of both KdpB and RR proteins (Fig 2A and 2B). The result shows that the kdp operon of D. radiodurans responds to K+ limitation. The induction of RR protein post K+ limitation is intriguing since in Anabaena L-31, the basal level of RR remains unaffected by K+ limitation [28], while in E. coli as well as in Clostridium acetobutylicum the expression of KdpE increases as a result of synthesis of a polycistronic mRNA encompassing kdpABCDE genes and a read-through effect [41, 42]. Further, we checked the kinetics of gene expression and observed that both the KdpB and RR proteins were synthesized within 30 minutes of transfer of D. radiodurans cells from TGY medium to K0 medium (Fig 2C and 2D). The response to K+ limitation was further assessed at different concentrations of K+. Both KdpB as well as RR proteins were induced in K0 medium but not in K1 medium (1 mM K+ phosphate added to K0 medium) (Fig 2E and 2F), indicating that unlike E. coli, the kdp operon of D. radiodurans could be induced only below 1 mM K+ concentration. Similar results were observed for Anabaena L-31 earlier [28]. While most of the KdpB protein localized in membrane (Fig 2G), the RR protein was entirely localized in the membranes (Fig 2H). RR is expected to be a cytosolic protein as it does not possess membrane anchoring or spanning domains. RR protein, when over-expressed in heterologous E. coli, localized entirely in the cytoplasm. In D. radiodurans, DNA is present in the nucleoid [43] and about 70% of the DNA is associated with the membranes [44]. The localization of RR protein in the membrane may be due to its possible interaction with kdp operon signaling proteins (KdpD-N and SK) localized in the membrane and its functionality can be facilitated by association of DNA with the membranes.
Fig 2. KdpB and RR expression under different growth conditions.
Expression of KdpB (A) or RR (B) proteins in D. radiodurans cells incubated in TGY, K20 or K0 media. Time course of induction of KdpB (C) or RR (D) proteins in D. radiodurans cells following shift from TGY to K0 medium. Expression of KdpB (E) or RR (F) proteins in D. radiodurans cells incubated in TGY, K0, K1 or K2 media. Localization of KdpB (G) or RR (H) proteins in D. radiodurans cells incubated in K20 or K0 media. Expression of KdpB (I) or RR (J) proteins in D. radiodurans cells grown either in K20 or K0 media, or exposed to ionic (-I, 0.1M NaCl) or osmotic (-O, 0.2M sucrose) stresses in K1 medium. The cellular proteins (100 μg) were resolved by 12% SDS-PAGE, electroblotted onto nitrocellulose membrane and immuno-stained using anti-KdpB or anti-RR antibodies as detailed in materials and methods. The top most protein band (~ 125 kDa) in the corresponding Coomassie stained gel is shown below Fig 2A, 2B, 2C, 2D, 2E, 2F, 2I and 2J, as loading control. For membrane or cytosolic protein extracts, 63 kDa protein band or 44 kDa protein band, respectively, are shown as loading controls (See S2 Fig for details on loading controls). Red bold numbers below the KdpB or RR immuno-stained bands indicate fold increase in their levels over the lanes in which these bands were not observed (denoted by 1).
The kdp operon of D. radiodurans does not respond to ionic or osmotic stresses
In addition to response to K+ limitation, the kdp operon is also induced by high osmolarity in bacteria [45–47]. In E. coli K12, the expression of kdp operon was about 10-fold higher in response to K+ limitation as compared to its expression in response to increased osmolarity by 0.4M NaCl, which in turn is about 10-fold higher compared to its expression in response to 0.6M sucrose stress [48]. Expression of kdp operon was originally proposed to be in response to changes in turgor pressure, but was but later debated [49–51]. Here we show that the kdp operon of D. radiodurans does respond to K+ limitation but does not respond at all to increased osmolarity. We changed the osmolarity of the K1 medium either by adding 0.1M NaCl or 0.2M sucrose and checked induction of KdpB and RR proteins. Neither KdpB nor RR protein were induced upon addition of NaCl or sucrose (Fig 2I and 2J). The kdp operon of Anabaena L-31, signaling components of which are similar to those of kdp operon of D. radiodurans, does respond to NaCl stress but does not respond to sucrose [27], although, kdp operon of another cyanobacterium Anabaena torulosa does respond to sucrose [26]. In contrast, the kdp operon of D. radiodurans appears to be highly specific for K+ limitation alone, and free from the influences of osmolarity.
RR protein induces kdp operon under K+ limitation
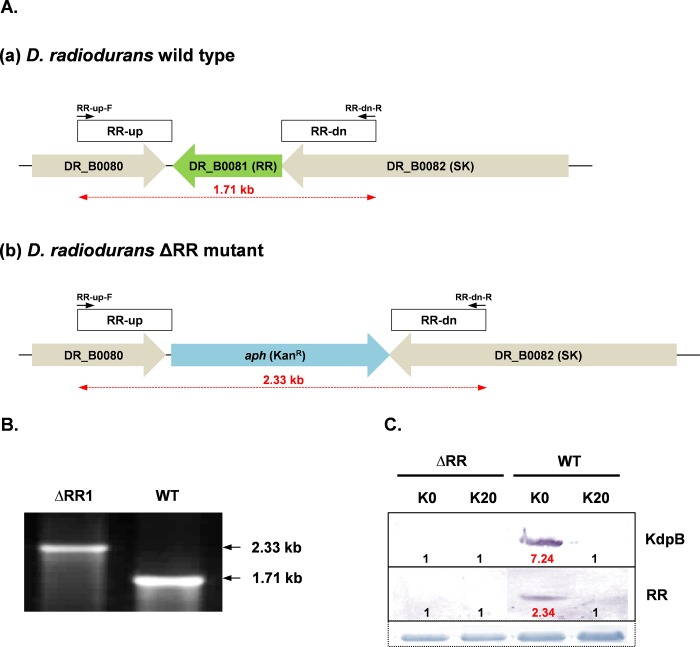
Comparison of the RR protein encoded by DR_B0081 gene of D. radiodurans revealed 23.9% identity with KdpE of E. coli. BLAST analysis also revealed homology of RR protein with similar two-component response regulators from Deinococcus-Thermus group and cyanobacteria. Therefore, it was pertinent to ascertain if the RR protein was indeed the response regulator of the kdp operon of D. radiodurans. We constructed a knockout mutant of RR by replacing the DR_B0081 gene with aph gene conferring kanamycin resistance, by site directed mutagenesis, as per the strategy outlined in Fig 3A. The knockout mutant was confirmed by PCR analysis. When RR-up-F/RR-dn-R primer pair was used, 2.33 kb and 1.71 kb bands were amplified from the genomic DNA of the prospective RR knockout mutant and wild type D. radiodurans, respectively, thereby confirming that the 0.63 kb DR_B0081 gene had been replaced by 1.25 kb aph cassette (Fig 3A and 3B). The mutant thus obtained failed to induce KdpB upon K+ limitation. KdpB as well as RR proteins were induced in wild type D. radiodurans under similar culture conditions (Fig 3C). The data confirmed that the RR protein encoded by DR_B0081 gene was indeed responsible for expression of Deinococcal kdp operon under potassium limitation.
Fig 3. Construction and confirmation of ΔRR mutant.
(A) Schematic representation of the RR gene (DR_B0081) in wild type D. radiodurans (a) and its replacement with kanamycin resistance cassette (aph) in ΔRR D. radiodurans (b). The primers used for the PCR confirmation of the mutant are shown. (B) Confirmation of complete deletion of RR gene in ΔRR D. radiodurans as compared to wild type D. radiodurans, using primer pair shown in Fig 3A. (C) Expression of KdpB or RR proteins in wild type or ΔRR D. radiodurans cells incubated in K0 or K20 media. Details of immuno-staining, loading controls and fold change levels were same as described in legend to Fig 2.
RR protein binds to an intergenic DNA sequence between structural and signaling genes of kdp operon
[Tn]-rich region present in the kdp promoter region in E. coli has been shown to bind response regulator KdpE [52, 53]. A similar [Tn]-rich region is present in the intergenic region (38 bp) between the kdpB and SK genes of D. radiodurans and in the upstream sequences of kdp operons in other bacteria as well [53, 54] (Fig 4A). Thus, the 38 bp intergenic region harbouring [Tn]-rich sequence is a potential site for regulation of kdp operon. This short sequence did not possess any sequence elements essential for gene expression as analysed by BPROM software. However, several of the Deinococcal promoters are devoid of the typical E. coli type sequence elements but are still expressed constitutively [55]. Here, we chose a short (38 bp) as well as longer sequence (200 bp) for our study. We determined the binding of RR protein to the promoter sequence of kdpBACD operon (PkdpB-38/ PkdpB-200). Retardation in the mobility of PkdpB-200 incubated with increasing concentrations of RR protein confirmed binding of RR to PkdpB-200 (Fig 4B). We obtained a similar profile of retardation in the mobility of PkdpB-200 incubated with phosphorylated RR protein (S3 Fig). The apparent equilibrium dissociation constant (KD) was calculated to be 3.62 ± 0.56 μM, which signifies the amount of protein required for 50% binding to PkdpB-200. The Hill coefficient value was determined to be 1.36 ± 0.17. Thus, RR showed very low or no co-operativity in binding (Fig 4C). Similarly, the mobility of PkdpB-38 was also retarded in the presence of increasing concentrations of RR protein (S4 Fig), confirming that this small intergenic region indeed provided the site for RR binding. To probe specific interaction of RR protein with PkdpB-200, competitive EMSA was carried out by titrating the DIG-labelled PkdpB-RR complexes with unlabeled PkdpB-200. With increasing concentration of unlabelled PkdpB-200, the labelled PkdpB-200 DNA was released from the PkdpB-200-RR complexes, where the labelled free DNA was clearly visible following competition with increasing amounts of unlabeled specific DNA (Fig 4D). Non-specific interactions of RR with nsDNA or of PkdpB-200 with any protein were ruled out by either incubating RR protein with PkdpB-200 or with nsDNA, or incubating BSA with PkdpB-200 or with nsDNA. DNA-protein complexes were observed only when PkdpB-200 was incubated with RR (Fig 4E), thereby confirming the specific interaction of RR with PkdpB-200.
Fig 4. [Tn]-rich region present in the kdp promoter region in various bacteria.
(A) [Tn]-rich region is shown in pale blue box. The number after the [Tn]-rich region indicate the number of bases between the [Tn]-rich region and the start codon. [Tn]-rich and [An]-rich KdpE binding site of E. coli and M. smegmatis, respectively, are shown in blue boxes. [Tn]-rich sequences in the upstream regions of kdpB gene in other bacteria are shown in red bold letters. (B) Binding of RR protein to the PkdpB-200 of D. radiodurans. The indicted concentrations of RR protein were incubated with PkdpB-200 promoter (45 ng of DIG-labeled 200 bp dsDNA) at 37°C for 1 h and the DNA–protein complexes were resolved by 10% native PAGE. The amount of DNA-protein complexes were estimated using GelQuant software. Substrate DNA and DNA-RR complex are shown by “—” and “←”, respectively, while wells of the gels are marked by asterisk. (C) The representative graph for DNA protein complexes. The data-points were fitted into Hill’s equation (dotted line) to determine KD value. (D) Titration of RR-promoter complexes with unlabeled promoter DNA. (E) Interaction of RR or non-specific protein BSA with PkdpB-200 (specific target) or non-specific DNA sequence. For (D) and (E), the DNA-protein complexes were resolved as described in legend to Fig 4B.
Relevance of kdp operon to stress resistance of D. radiodurans
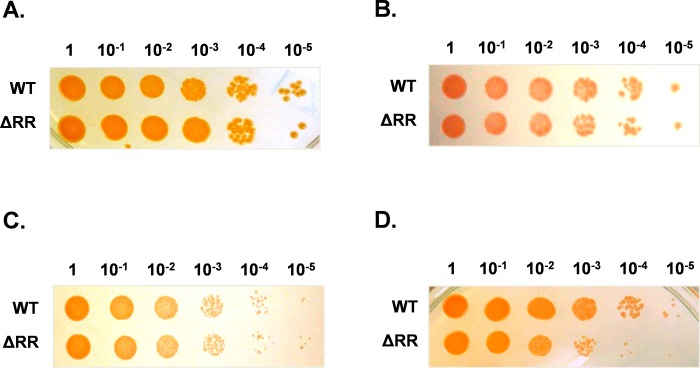
Since kdp operon is reported to give survival advantage under stressful conditions in both Gram-positive as well as Gram-negative bacteria [4–12], we checked if presence of inducible kdp operon offered any advantage to D. radiodurans during stress. We used a kdp expression null ΔRR mutant to evaluate survival fitness following different stresses. The ΔRR mutant did not show any growth defect, as compared to wild type D. radiodurans, when grown in TGY medium under standard growth conditions (Fig 5A), or when exposed to 100 mM H2O2 (Fig 5B) or 42° C heat stress (Fig 5C). However, survival of ΔRR mutant was reduced, as compared to wild type D. radiodurans, following 5 kGy gamma irradiation (Fig 5D).
Fig 5.
Survival of wild type and ΔRR mutant of D. radiodurans (A) under standard growth conditions, (B) exposed to 100 mM H2O2, (C) exposed to 42° C or (D) exposed to 5 kGy gamma irradiation.
The study, for the first time, reports functional characterization of the kdp operon of the radioresistant extremophile D. radiodurans, having a distinct operon organization. Unique features of the kdp operon of D. radiodurans include (a) induction of both, the structural and regulatory components, of the kdp operon under extreme potassium limitation, (b) lack of stimulation by increased osmolarity either by NaCl or sucrose, (c) expression of kdp operon through specific interaction of unphosphorylated RR protein with kdp promoter, and (d) confers a minor survival advantage to the organism while recovering from gamma radiation stress.
Supporting information
The gel image shows sKdpB and RR proteins were overexpressed in E. coli BL21(DE3)pLysS (30 μg protein/lane) and purified sKdpB and RR proteins (5 μg protein/lane) in lanes 1–2 and 3–4, respectively. Molecular weight marker (SDS-7, Sigma) are shown in lane M. The purified RR protein was used for promoter interaction studies. The sKdpB and RR protein bands (shown by arrows on right hand side) were purified by gel elution method and were further used for generation of polyclonal antibodies in rabbit.
(TIF)
Whole cell protein extract (Lane 1), cytosolic fraction (Lane 2) and membrane fraction (Lane 3) were resolved by 12% SDS-PAGE. Molecular weight markers (P7712L, NEB) are shown in lane M. The 125 kDa (S-layer protein, DR_2577, *), 63 kDa (ABC transporter-binding protein, DR_1571, **) and 44 kDa (Elongation factor Tu, DR_0309, ***) were used as loading controls for whole cell protein extract, membrane fraction and cytosolic fraction, respectively. For details on the protein identities, please see reference No. 16.
(TIF)
The indicted concentrations of phosphorylated RR protein were incubated with PkdpB-200 promoter (45 ng of DIG-labeled 200 bp dsDNA) at 37°C for 1 h and the DNA–protein complexes were resolved by 10% native PAGE. Substrate DNA and DNA-RR complexes are shown by “—” and “←”, respectively, while wells of the gels are marked by asterisk.
(TIF)
The indicted concentrations of RR protein were incubated with PkdpB-38 promoter (45 ng of DIG-labeled 38 bp dsDNA) at 37°C for 1 h and the DNA–protein complexes were resolved by 12% native PAGE. Substrate DNA and DNA-RR complexes are shown by “—” and “←”, respectively, while wells of the gels are marked by asterisk.
(TIF)
Data Availability
All relevant data are within the paper.
Funding Statement
The work was funded by Department of Atomic Energy, India.
References
- 1.Gries CM, Bose JL, Nuxoll AS, Fey PD, Bayles KW. The Ktr potassium transport system in Staphylococcus aureus and its role in cell physiology, antimicrobial resistance and pathogenesis. Molecular Microbiology. 2013;89(4):760–73. 10.1111/mmi.12312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Epstein W. The Roles and Regulation of Potassium in Bacteria. Progress in Nucleic Acid Research and Molecular Biology. 2003;75:293–320. 10.1016/S0079-6603(03)75008-9 [DOI] [PubMed] [Google Scholar]
- 3.Su J, Gong H, Lai J, Main A, Lu S. The Potassium Transporter Trk and External Potassium Modulate Salmonella enterica Protein Secretion and Virulence. Infection and Immunity. 2009;77(2):667–75. 10.1128/IAI.01027-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Checchetto V, Segalla A, Sato Y, Bergantino E, Szabo I, Uozumi N. Involvement of Potassium Transport Systems in the Response of Synechocystis PCC 6803 Cyanobacteria to External pH Change, High-Intensity Light Stress and Heavy Metal Stress. Plant and Cell Physiology. 2016;57(4):862–77. 10.1093/pcp/pcw032 [DOI] [PubMed] [Google Scholar]
- 5.Ballal A, Basu B, Apte SK. The Kdp-ATPase system and its regulation. Journal of Biosciences. 2007;32(3):559–68 [DOI] [PubMed] [Google Scholar]
- 6.Ochrombel I, Ott L, Krämer R, Burkovski A, Marin K. Impact of improved potassium accumulation on pH homeostasis, membrane potential adjustment and survival of Corynebacterium glutamicum. Biochimica et Biophysica Acta (BBA)—Bioenergetics. 2011;1807(4):444–50. 10.1016/j.bbabio.2011.01.008 [DOI] [PubMed] [Google Scholar]
- 7.McLaggan D, Naprstek J, Buurman ET, Epstein W. Interdependence of K+ and glutamate accumulation during osmotic adaptation of Escherichia coli. Journal of Biological Chemistry. 1994;269(3):1911–7. [PubMed] [Google Scholar]
- 8.Epstein W, Kim BS. Potassium Transport Loci in Escherichia coli K-12. Journal of Bacteriology. 1971;108(2):639–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gründling A. Potassium Uptake Systems in Staphylococcus aureus: New Stories about Ancient Systems. mBio. 2013;4(5). 10.1128/mBio.00784-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heermann R, Jung K. K+ Supply, Osmotic Stress, and the KdpD/KdpE Two-component System Two-component systems in bacteria Caister Academic Press, Norwich, UK: 2012:181–98. [Google Scholar]
- 11.Freeman ZN, Dorus S, Waterfield NR. The KdpD/KdpE Two-Component System: Integrating K+ Homeostasis and Virulence. PLOS Pathogens. 2013;9(3):e1003201 10.1371/journal.ppat.1003201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price-Whelan A, Poon CK, Benson MA, Eidem TT, Roux CM, Boyd JM, et al. Transcriptional Profiling of Staphylococcus aureus During Growth in 2 M NaCl Leads to Clarification of Physiological Roles for Kdp and Ktr K+ Uptake Systems. mBio. 2013;4(4). 10.1128/mBio.00407-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schramke H, Laermann V, Tegetmeyer HE, Brachmann A, Jung K, Altendorf K. Revisiting regulation of potassium homeostasis in Escherichia coli: the connection to phosphate limitation. MicrobiologyOpen. 2017;6(3):e00438 10.1002/mbo3.438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katoh H, Asthana RK, Ohmori M. Gene Expression in the Cyanobacterium Anabaena sp. PCC7120 under Desiccation. Microbial Ecology. 2004;47(2):164–74. 10.1007/s00248-003-1043-6 [DOI] [PubMed] [Google Scholar]
- 15.Battista JR. AGAINST ALL ODDS:The Survival Strategies of Deinococcus radiodurans. Annual Review of Microbiology. 1997;51(1):203–24. 10.1146/annurev.micro.51.1.203 . [DOI] [PubMed] [Google Scholar]
- 16.Basu B, Apte SK. Gamma Radiation-induced Proteome of Deinococcus radiodurans Primarily Targets DNA Repair and Oxidative Stress Alleviation. Molecular & Cellular Proteomics. 2012;11(1). 10.1074/mcp.M111.011734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daly MJ, Gaidamakova EK, Matrosova VY, Kiang JG, Fukumoto R, Lee D-Y, et al. Small-Molecule Antioxidant Proteome-Shields in Deinococcus radiodurans. PLOS ONE. 2010;5(9):e12570 10.1371/journal.pone.0012570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Misra HS, Khairnar NP, Barik A, Indira Priyadarsini K, Mohan H, Apte SK. Pyrroloquinoline-quinone: a reactive oxygen species scavenger in bacteria. FEBS Letters. 2004;578(1–2):26–30. 10.1016/j.febslet.2004.10.061 [DOI] [PubMed] [Google Scholar]
- 19.Ujaoney AK, Padwal MK, Basu B. Proteome dynamics during post-desiccation recovery reveal convergence of desiccation and gamma radiation stress response pathways in Deinococcus radiodurans. Biochimica et Biophysica Acta (BBA)—Proteins and Proteomics. 2017;1865(9):1215–26. 10.1016/j.bbapap.2017.06.014 [DOI] [PubMed] [Google Scholar]
- 20.Luan H, Meng N, Fu J, Chen X, Xu X, Feng Q, et al. Genome-Wide Transcriptome and Antioxidant Analyses on Gamma-Irradiated Phases of Deinococcus radiodurans R1. PLOS ONE. 2014;9(1):e85649 10.1371/journal.pone.0085649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai C-H, Liao R, Chou B, Contreras LM. Transcriptional Analysis of Deinococcus radiodurans Reveals Novel Small RNAs That Are Differentially Expressed under Ionizing Radiation. Applied and Environmental Microbiology. 2015;81(5):1754–64. 10.1128/AEM.03709-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durand A, Sinha AK, Dard-Dascot C, Michel B. Mutations Affecting Potassium Import Restore the Viability of the Escherichia coli DNA Polymerase III holD Mutant. PLOS Genetics. 2016;12(6):e1006114 10.1371/journal.pgen.1006114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes FM Jr, Cidlowski JA. Potassium is a critical regulator of apoptotic enzymes in vitro and in vivo. Advances in Enzyme Regulation. 1999;39(1):157–71. 10.1016/S0065-2571(98)00010-7 [DOI] [PubMed] [Google Scholar]
- 24.White O, Eisen JA, Heidelberg JF, Hickey EK, Peterson JD, Dodson RJ, et al. Genome Sequence of the Radioresistant Bacterium Deinococcus radiodurans R1. Science. 1999;286(5444):1571–7. 10.1126/science.286.5444.1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta RS, Johari V. Signature Sequences in Diverse Proteins Provide Evidence of a Close Evolutionary Relationship Between the Deinococcus-Thermus Group and Cyanobacteria. Journal of Molecular Evolution. 1998;46(6):716–20. 10.1007/pl00006352 [DOI] [PubMed] [Google Scholar]
- 26.Alahari A, Ballal A, Apte SK. Regulation of Potassium-Dependent Kdp-ATPase Expression in the Nitrogen-Fixing Cyanobacterium Anabaena torulosa. Journal of Bacteriology. 2001;183(19):5778–81. 10.1128/JB.183.19.5778-5781.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ballal A, Apte SK. Differential Expression of the Two kdp Operons in the Nitrogen-Fixing Cyanobacterium Anabaena sp. Strain L-31. Applied and Environmental Microbiology. 2005;71(9):5297–303. 10.1128/AEM.71.9.5297-5303.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ballal A, Apte SK. Characterization of a response regulator protein that binds to Anabaena sp. strain L-31 kdp-promoter region. Archives of Biochemistry and Biophysics. 2008;474(1):65–71. 10.1016/j.abb.2008.02.017 [DOI] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 2000;28(1):27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venkateswaran A, McFarlan SC, Ghosal D, Minton KW, Vasilenko A, Makarova K, et al. Physiologic Determinants of Radiation Resistance in Deinococcus radiodurans. Applied and Environmental Microbiology. 2000;66(6):2620–6. 10.1128/aem.66.6.2620-2626.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holland AD, Rothfuss HM, Lidstrom ME. Development of a defined medium supporting rapid growth for Deinococcus radiodurans and analysis of metabolic capacities. Applied Microbiology and Biotechnology. 2006;72(5):1074–82. 10.1007/s00253-006-0399-1 [DOI] [PubMed] [Google Scholar]
- 32.Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Venkateswaran A, et al. Accumulation of Mn(II) in Deinococcus radiodurans Facilitates Gamma-Radiation Resistance. Science. 2004;306(5698):1025–8. 10.1126/science.1103185 [DOI] [PubMed] [Google Scholar]
- 33.Studier FW. Stable Expression Clones and Auto-Induction for Protein Production in E. coli In: Chen YW, editor. Structural Genomics: General Applications. Totowa, NJ: Humana Press; 2014. p. 17–32. [DOI] [PubMed] [Google Scholar]
- 34.Battista JR, Park M-J, McLemore AE. Inactivation of Two Homologues of Proteins Presumed to Be Involved in the Desiccation Tolerance of Plants Sensitizes Deinococcus radiodurans R1 to Desiccation. Cryobiology. 2001;43(2):133–9. 10.1006/cryo.2001.2357 [DOI] [PubMed] [Google Scholar]
- 35.Basu B, Apte SK. A novel serralysin metalloprotease from Deinococcus radiodurans. Biochimica et Biophysica Acta (BBA)—Proteins and Proteomics. 2008;1784(9):1256–64. 10.1016/j.bbapap.2008.05.009 [DOI] [PubMed] [Google Scholar]
- 36.Hill AV. The possible effects of the aggregation of the molecules of hæmoglobin on its dissociation curves. The Journal of Physiology. 1910;40:iv—vii. [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–10. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 38.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 39.Solovyev V, Salamov A. Automatic Annotation of Microbial Genomes and Metagenomic Sequences. In Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies (Ed. R.W. Li). Nova Science Publishers. 2011:61–78. [Google Scholar]
- 40.Käll L, Krogh A, Sonnhammer ELL. A Combined Transmembrane Topology and Signal Peptide Prediction Method. Journal of Molecular Biology. 2004;338(5):1027–36. 10.1016/j.jmb.2004.03.016 [DOI] [PubMed] [Google Scholar]
- 41.Polarek JW, Williams G, Epstein W. The products of the kdpDE operon are required for expression of the Kdp ATPase of Escherichia coli. Journal of Bacteriology. 1992;174(7):2145–51. 10.1128/jb.174.7.2145-2151.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Treuner-Lange A, Kuhn A, Dürre P. The kdp system of Clostridium acetobutylicum: cloning, sequencing, and transcriptional regulation in response to potassium concentration. Journal of Bacteriology. 1997;179(14):4501–12. 10.1128/jb.179.14.4501-4512.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eltsov M, Dubochet J. Fine Structure of the Deinococcus radiodurans Nucleoid Revealed by Cryoelectron Microscopy of Vitreous Sections. Journal of Bacteriology. 2005;187(23):8047–54. 10.1128/JB.187.23.8047-8054.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burrell AD, Feldschreiber P, Dean CJ. DNA-membrane association and the repair of double breaks in X-irradiated Micrococcus radiodurans. Biochimica et Biophysica Acta (BBA)—Nucleic Acids and Protein Synthesis. 1971;247(1):38–53. 10.1016/0005-2787(71)90805-7 [DOI] [PubMed] [Google Scholar]
- 45.Frymier JS, Reed TD, Fletcher SA, Csonka LN. Characterization of transcriptional regulation of the kdp operon of Salmonella typhimurium. Journal of Bacteriology. 1997;179(9):3061–3. 10.1128/jb.179.9.3061-3063.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heermann R, Jung K. The complexity of the ‘simple’ two-component system KdpD/KdpE in Escherichia coli. FEMS Microbiology Letters. 2010;304(2):97–106. 10.1111/j.1574-6968.2010.01906.x [DOI] [PubMed] [Google Scholar]
- 47.Epstein W. The KdpD Sensor Kinase of Escherichia coli Responds to Several Distinct Signals To Turn on Expression of the Kdp Transport System. Journal of Bacteriology. 2016;198(2):212–20. 10.1128/jb.00602-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heermann R, Zigann K, Gayer S, Rodriguez-Fernandez M, Banga JR, Kremling A, et al. Dynamics of an Interactive Network Composed of a Bacterial Two-Component System, a Transporter and K+ as Mediator. PLOS ONE. 2014;9(2):e89671 10.1371/journal.pone.0089671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asha H, Gowrishankar J. Regulation of kdp operon expression in Escherichia coli: evidence against turgor as signal for transcriptional control. Journal of Bacteriology. 1993;175(14):4528–37. 10.1128/jb.175.14.4528-4537.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamann K, Zimmann P, Altendorf K. Reduction of Turgor Is Not the Stimulus for the Sensor Kinase KdpD of Escherichia coli. Journal of Bacteriology. 2008;190(7):2360–7. 10.1128/JB.01635-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laimins LA, Rhoads DB, Epstein W. Osmotic control of kdp operon expression in Escherichia coli. Proceedings of the National Academy of Sciences. 1981;78(1):464–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugiura A, Nakashima K, Tanaka K, Mizuno T. Clarification of the structural and functional features of the osmoregulated kdp operon of Escherichia coli. Molecular Microbiology. 1992;6(13):1769–76. 10.1111/j.1365-2958.1992.tb01349.x [DOI] [PubMed] [Google Scholar]
- 53.Sugiura A, Nakashima K, Mizuno T. Sequence-directed DNA Curvature in Activator-binding Sequence in the Escherichia coli kdpABC Promoter. Bioscience, Biotechnology, and Biochemistry. 1993;57(2):356–7. 10.1271/bbb.57.356 [DOI] [PubMed] [Google Scholar]
- 54.Ali MK, Li X, Tang Q, Liu X, Chen F, Xiao J, et al. Regulation of Inducible Potassium Transporter KdpFABC by the KdpD/KdpE Two-Component System in Mycobacterium smegmatis. Frontiers in Microbiology. 2017;8(570). 10.3389/fmicb.2017.00570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anaganti N, Basu B, Apte SK. In situ real-time evaluation of radiation-responsive promoters in the extremely radioresistant microbe Deinococcus radiodurans. Journal of Biosciences. 2016;41(2):193–203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The gel image shows sKdpB and RR proteins were overexpressed in E. coli BL21(DE3)pLysS (30 μg protein/lane) and purified sKdpB and RR proteins (5 μg protein/lane) in lanes 1–2 and 3–4, respectively. Molecular weight marker (SDS-7, Sigma) are shown in lane M. The purified RR protein was used for promoter interaction studies. The sKdpB and RR protein bands (shown by arrows on right hand side) were purified by gel elution method and were further used for generation of polyclonal antibodies in rabbit.
(TIF)
Whole cell protein extract (Lane 1), cytosolic fraction (Lane 2) and membrane fraction (Lane 3) were resolved by 12% SDS-PAGE. Molecular weight markers (P7712L, NEB) are shown in lane M. The 125 kDa (S-layer protein, DR_2577, *), 63 kDa (ABC transporter-binding protein, DR_1571, **) and 44 kDa (Elongation factor Tu, DR_0309, ***) were used as loading controls for whole cell protein extract, membrane fraction and cytosolic fraction, respectively. For details on the protein identities, please see reference No. 16.
(TIF)
The indicted concentrations of phosphorylated RR protein were incubated with PkdpB-200 promoter (45 ng of DIG-labeled 200 bp dsDNA) at 37°C for 1 h and the DNA–protein complexes were resolved by 10% native PAGE. Substrate DNA and DNA-RR complexes are shown by “—” and “←”, respectively, while wells of the gels are marked by asterisk.
(TIF)
The indicted concentrations of RR protein were incubated with PkdpB-38 promoter (45 ng of DIG-labeled 38 bp dsDNA) at 37°C for 1 h and the DNA–protein complexes were resolved by 12% native PAGE. Substrate DNA and DNA-RR complexes are shown by “—” and “←”, respectively, while wells of the gels are marked by asterisk.
(TIF)
Data Availability Statement
All relevant data are within the paper.