Abstract
Cpf1 is a new class II family of CRISPR-Cas RNA-programmable endonucleases with unique features that make it a very attractive alternative or complement to Cas9 for genome engineering. Using constitutively expressed Cpf1 from Francisella novicida, the present study demonstrates that FnCpf1 can mediate RNA-guided DNA cleavage at targeted genomic loci in the popular model and industrial yeast Saccharomyces cerevisiae. FnCpf1 very efficiently and precisely promoted repair DNA recombination with efficiencies up to 100%. Furthermore, FnCpf1 was shown to introduce point mutations with high fidelity. While editing multiple loci with Cas9 is hampered by the need for multiple or complex expression constructs, processing itself a customized CRISPR array FnCpf1 was able to edit four genes simultaneously in yeast with a 100% efficiency. A remarkable observation was the unexpected, strong preference of FnCpf1 to cleave DNA at target sites harbouring 5′-TTTV-3′ PAM sequences, a motif reported to be favoured by Cpf1 homologs of Acidaminococcus and Lachnospiraceae. The present study supplies several experimentally tested guidelines for crRNA design, as well as plasmids for FnCpf1 expression and easy construction of crRNA expression cassettes in S. cerevisiae. FnCpf1 proves to be a powerful addition to S. cerevisiae CRISPR toolbox.
INTRODUCTION
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) systems are adaptive immune systems widely distributed across bacteria and archaea, designed to destroy DNA of invading mobile genetic elements (1). These immune systems, in which endonucleases are guided by single stranded RNA to find their target DNA, have been turned into powerful genome editing tools over the past few years (2,3). The rapid implementation of CRISPR-based DNA editing systems has tremendously improved molecular toolboxes for a broad spectrum of organisms, ranging from simple prokaryotes to metazoan animals (4). By increasing the speed of genetic engineering, CRISPR-based systems have already impacted the field of microbial biotechnology (5–7). The push towards sustainable alternatives to oil-derived chemicals requires the construction of powerful microbial cell factories that can produce new chemicals, using unnatural substrates at high yields and rates, under harsh industrial conditions. Constructing such advanced cell factories requires extensive and fast genetic engineering strategies, that enable to test various designs in search of the optimal genetic configuration. Even the tractable and genetically accessible model and industrial yeast Saccharomyces cerevisiae has rapidly adopted CRISPR-aided DNA editing, making it a standard practice for strain construction (8–10).
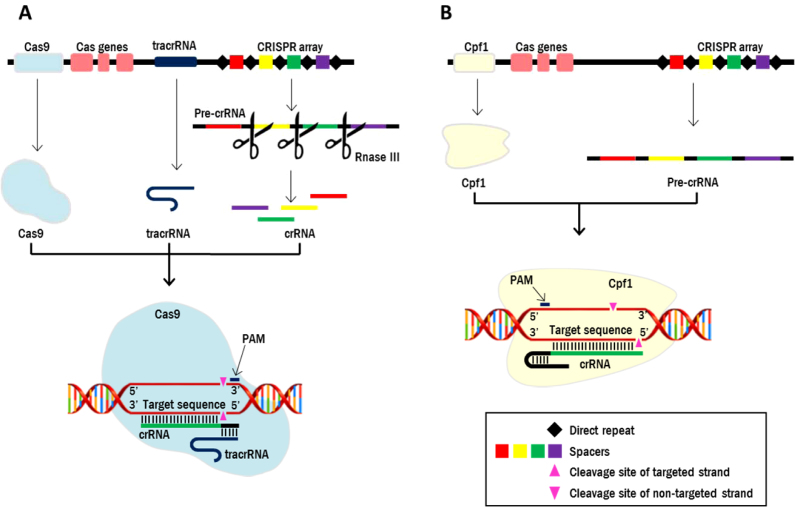
Two classes of CRISPR systems have been identified based on the architecture of the CRISPR locus (11–13). Class II, to which the very popular Streptococcus pyogenes Cas9 (SpCas9) belongs, has been favoured for heterologous genome editing mainly due to the structural simplicity of its endonuclease formed of a single subunit (2). Like all CRISPR-based systems, SpCas9 requires an RNA molecule, called CRISPR-RNA (crRNA) to guide the nuclease towards the editing site (Figure 1). In addition, to target and edit DNA, SpCas9 requires another RNA fragment, the trans-activating RNA (tracrRNA), that binds to the crRNA and to SpCas9 (Figure 1). In native systems, the DNA sequences encoding crRNAs (also called spacers) are co-localised in a CRISPR array, in which they are separated by repeated DNA motifs called the Direct Repeats (DRs, Figure 1). In their native system, the repeats of the precursor crRNA transcript base pair with the ‘anti-repeat’ part of tracrRNA, and these dsRNA helices are recognized and cleaved by RNaseIII (14). For efficient heterologous editing, the CRISPR system is generally simplified by expressing the crRNA already connected to the tracrRNA in a chimeric single guide RNA (sgRNA), and each chimeric sgRNA is expressed from its own promoter, thereby avoiding the requirement of an RNase for processing the precursor CRISPR-RNA transcript (2) (Figure 1). These sgRNAs have been shown to be functional in a wide variety of organisms, including S. cerevisiae, and extensive studies have delivered a number of basic principles to guide the design of crRNAs for efficient SpCas9-mediated DNA editing (15). However, SpCas9-based editing has some shortcomings. For instance, as all known CRISPR endonucleases, SpCas9 can only cut DNA located near a PAM (Protospacer Adjacent Motif) sequence meant to distinguish self from non-self DNA in native immune systems (16–18). The SpCas9 PAM sequence 5′-NGG-3′ is G-rich and located at the 3′ end of the protospacer (18). While this PAM is rather frequently distributed across the yeast genome (ca. 53 unique genomic targets per 1000 bp, which is the average size of S. cerevisiae genes, (8)), it is not always available in the area where editing is desired, more particularly in AT-rich regions. Furthermore, for reasons not yet fully understood, the efficiency of DNA editing varies greatly as a function of the targeted sequence, which further reduces the number of available ‘active’ PAM sequences. Also, chimeric guide RNAs with individual expression systems are not compatible with multiplex, high-throughput genome editing. The highest number of multisite editing reported so far in yeast is six, but it requires complex plasmid construction for individual expression of each sgRNA and simultaneous transformation of three plasmids, which could be greatly simplified and accelerated using the native CRISPR array systems (9). Non-chimeric gRNAs, based on native CRISPR systems, have been shown to enable SpCas9-mediated DNA cleavage in S. cerevisiae, however their performance for multiplexing is so far an order magnitude lower than that of chimeric systems (19). In S. cerevisiae, alternative systems involving ribozymes have been shown to enable dCas9-mediated transcriptional regulation (20), however their efficiency for multisite genome editing has not been explored yet.
Figure 1.
Schematic representation of DNA interference by the Cas9 and Cpf1 endonucleases. As compared to Cas9, Cpf1 does not require a tracrRNA, has a T-rich PAM sequence located at the 5′ end of the protospacer, is capable to mature its own crRNA array, cleaves DNA distal from the PAM and generates staggered ends.
Cpf1, a new family of class II CRISPR bacterial endonucleases was recently identified (21) and shown to mediate heterologous DNA editing in bacteria, as well as in plant and mammalian cells (22–27). This enzyme family, recently renamed Cas12a and tentatively classified as Type V-A (12), presents some characteristics reminiscent of Cas9, but also some very distinct and attractive features. Cpf1 variants from three bacteria, Francisella novicida (FnCpf1), Acidaminococcus sp. BV3L6 (AsCpf1) and Lachnospiraceae bacterium (LbCpf1) have been studied most intensively. Belonging to class II as Cas9, Cpf1 operates as single protein. Resolution of the crystal structure of LbCpf1 and AsCpf1 has shown that Cpf1 and Cas9 share a bi-lobed structure with a central channel in which the RNA-DNA heteroduplex is bound (28–30). However proteins of the Cpf1 family lack HNH domains, and a single RuvC nuclease domain seems to be responsible for cleavage of both DNA strands. In addition they contain a Nuc domain, but current models predict that it is most likely not directly involved in DNA cleavage (30,31). Cpf1 and Cas9 display more striking differences both in structure and function. The Cpf1 PAM is T-rich, and described as 5′-TTTN-3′ (or 5′-TTTV-3′ (32)) for AsCpf1 and LbCpf1, and 5′-TTN-3′ for FnCpf1, and is located at the 5′ end of the protospacer (27). Contrary to Cas9, Cpf1 cleaves DNA distal from the PAM and generates staggered ends (27) (Figure 1). More remarkably, Cpf1 does not require a tracrRNA and is the first known CRISPR endonuclease that harbours a distinct endoribonuclease domain (30,33) (Figure 1). Cpf1 matures the CRISPR-RNA array itself and therefore does not require the activity of an additional RNase (Figure 1). These features propel Cpf1 as an attractive system for multiplex genome editing.
While intensively studied in higher eukaryotes, Cpf1-aided genome editing has been comparatively underexplored in the microbial kingdom. Thus far, Cpf1-mediated DNA cleavage has only been demonstrated in two bacteria, Escherichia coli and Corynebacterium glutamicum (22,25), and has not been established in lower eukaryotes. The goal of the present study was firstly to evaluate FnCpf1 functionality for targeted genome editing in S. cerevisiae. Secondly, we explored ways to improve the efficiency of genome editing by FnCpf1 and thereby propose design principles and offer plasmids for efficient DNA cleavage in baker's yeast. Finally, the present work demonstrates that FnCpf1 can edit multiple genomic loci simultaneously with high efficiency.
MATERIALS AND METHODS
Strains and cultivation techniques
All S. cerevisiae strains used in this study belong to the CEN.PK genetic background and are listed in Table 1 (34,35). Yeast cultures were grown in 500 ml shake flasks containing 100 ml of medium at 30°C with 200 rpm agitation. Complex and nonselective media contained 10 g l−1 yeast extract, 20 g l−1 peptone and 20 g l−1 glucose (YPD). When selection was required, YPD was supplemented with 200 mg l−1 G418. Synthetic medium containing 3 g l−1 KH2PO4, 0.5 g l−1 MgSO4·7H2O, 5 g l−1 (NH4)2SO4, 1 ml l−1 of a trace element solution, and 1 ml l−1 of a vitamin solution as previously described (36) and supplemented with 20 g l−1 glucose was used for culture propagation (SMG). When selection on SMG with G418 was required (NH4)2SO4 was replaced with 3 g l−1 K2SO4 and 2.3 g l−1 filter-sterilized urea to maintain a stable pH (37). For plasmid propagation, E. coli XL1-Blue cells (Agilent Technologies, Santa Clara, CA, USA) were cultivated in Lysogeny broth (LB) medium supplied with ampicillin (100 mg l−1) or kanamycin (50 mg l−1) at 37 °C with 180 rpm agitation. Solid media were obtained by addition of 20 g l−1 agar.
Table 1. List of yeast strains used in this study.
| Strain name | Genotype | Origin |
|---|---|---|
| CEN.PK113–7D | MATa MAL2–8c SUC2 | (34) |
| CEN.PK113–5D | MATa ura3–52 | (34) |
| IMX1139 | MATa ura3–52 sga1Δ::TEF1p::Fncpf1::CYC1t::KlURA3 | This study |
| IME384 | MATa ura3–52 pUDE731 | This study |
| IME385 | MATa ura3–52 pUD706 | This study |
| IMX1511 | MATa ura3–52 sga1Δ::TEF1p::Fncpf1::CYC1t::KlURA3 Δhis4 + pUDE713 | This study |
| IMX1512 | MATa ura3–52 sga1Δ::TEF1p::Fncpf1::CYC1t::KlURA3 Δade2 Δhis4 + pUDE709 | This study |
| IMX1522 | MATa ura3–52 sga1Δ::TEF1p::Fncpf1::CYC1t::KlURA3 Δcan1 + pUDE721 | This study |
| IMX1523 | MATa ura3–52 sga1Δ::TEF1p::Fncpf1::CYC1t::KlURA3 Δcan1 + pUDE722 | This study |
| IMX1524 | MATa ura3–52 sga1Δ::TEF1p::Fncpf1::CYC1t::KlURA3 Δpdr12 + pUDE723 | This study |
| IMX1525 | MATa ura3–52 sga1Δ::TEF1p::Fncpf1::CYC1t::KlURA3 Δpdr12 + pUDE724 | This study |
| IMX1526 | MATa ura3–52 sga1Δ::TEF1p::Fncpf1::CYC1t::KlURA3 Δpdr12 + pUDE725 | This study |
| IMX1535 | MATa ura3–52 sga1Δ::TEF1p::Fncpf1::CYC1t::KlURA3 Δade2 Δcan1 Δhis4 Δpdr12 + pUDE735 | This study |
Frozen stocks of S. cerevisiae and E. coli were prepared by addition of the sterile glycerol (30% v/v) to exponentially grown cultures and were stored as frozen aliquots at –80 °C.
Molecular biology techniques
PCR reactions for diagnostic purposes were performed using DreamTaq DNA polymerase (Thermo Fisher Scientific, Walthman, MA, USA) according to manufacturer's instructions. When high fidelity amplification was needed, Phusion® High-Fidelity DNA polymerase (Thermo Fisher Scientific) was used according to supplier's instructions. Oligonucleotides were ordered from Sigma Aldrich (St Louis, MO, USA) with PAGE or desalted purity depending on the purpose. DNA fragments were separated on agarose gels and were excised when purification of the fragment was required (Zymoclean, Zymo Research, Irvine, CA, USA). Bacterial plasmids were isolated using Sigma GenElute Plasmid kit (Sigma-Aldrich). When plasmid purification from yeast was required, Zymoprep Yeast Plasmid Miniprep II Kit was used (Zymo Research). Restriction digestion with DpnI for removal of circular templates (Thermo Fisher Scientific) was performed as recommended in the instruction manual. E. coli chemical transformations were performed following manufacturer's recommendations (Agilent Technologies).
Gene deletions were confirmed by diagnostic PCR and Sanger sequencing (Baseclear, Leiden, Netherlands).
Construction of a S. cerevisiae strain with genomic integration of Fncpf1
The integration construct consisted of two linear DNA fragments, one containing the Fncpf1 expression cassette and the other harbouring the KlURA3 marker, which were assembled in vivo in yeast and integrated into the SGA1 locus (Supplementary Figure S1). To construct the Fncpf1 expression cassette, the human codon-optimized F. novicida cpf1 tagged with C-terminal nuclear localization signal (NLS) and 3xHA tag was PCR-amplified from pY004 (Addgene plasmid #69976, https://www.addgene.org/69976/, (27)) using primers 10141 and 10144 (Table 2, Supplementary Table S1). The plasmid p414-TEF1p-cas9-CYC1t (Addgene plasmid #43802) backbone was amplified with primers 10145 and 10146. The amplified Fncpf1 and p414-TEF1p-cas9-CYC1t fragments were assembled using NEBuilder® HiFi DNA Assembly Master Mix (New England BioLabs, Ipswich, MA, USA) resulting in plasmid pUDC175 (Table 2). The newly constructed TEF1p::Fncpf1::CYC1t expression unit was amplified from pUDC175 with Phusion® High-Fidelity DNA Polymerase (ThermoFischer Scientific) and primers 10147 and 10189 (Supplementary Table S1) which introduced a short homology to the SGA1 chromosomal locus and an homology the co-transformed fragment respectively. The KlURA3 integration fragment was PCR-amplified with Phusion® High-Fidelity DNA Polymerase (ThermoFischer Scientific) using primers 10190 and 10192 which introduced an homology to the Fncpf1 fragment and an homology to the chromosomal SGA1 locus respectively, and using pMEL10 as template (Table 2, Supplementary Table S1). Two micrograms of each integration fragment were transformed to S. cerevisiae CEN.PK113–5D (MATa ura3–52, Table 1) using the lithium acetate transformation protocol (38). Transformants were selected on SMG plates. Correct assembly and integration of the cassette in the SGA1 locus were verified via PCR with primers listed in Supplementary Table S1. After a second restreaking, a single colony isolate was selected, named IMX1139 (Table 1), and its genome was sequenced.
Table 2. List of plasmids used in this study.
| Plasmid | Genotypea | Assemby method | Reference |
|---|---|---|---|
| p414-TEF1p-Cas9-CYC1t | CEN6/ARS4 ampRTRP1 TEF1p::Spcas9-CYC1t | (8) | |
| pMEL10 | 2 μm ampRKlURA3 SNR52p::gRNA-CAN1.Y::SUP4t | (9) | |
| pROS13 | 2 μm ampR KanMX SNR52p ::gRNA-CAN1.Y gRNA-ADE2.Y ::SUP4t | (9) | |
| pRS416 | CEN6/ARS4 ampRURA3 | (62) | |
| PY004 | ampRFncpf1 | (27), Addgene #69976 | |
| pUDC175 (Addgene #103019) | CEN6/ARS4 ampRTRP1 TEF1p::Fncpf1::CYC1t | In vivo | This study |
| pUD520 | KanR SNR52p::crADE2–1.L::SUP4t | GenArt | This study |
| pUD521 | KanR SNR52p::crADE2–2.L::SUP4t | GenArt | This study |
| pUD438 | KanR SNR52p::crADE2–3.L::SUP4t | GenArt | This study |
| pUD522 | KanR SNR52p::crADE2–4.L::SUP4t | GenArt | This study |
| pUD523 | KanR SNR52p::crADE2–5.L::SUP4t | GenArt | This study |
| pUD524 | KanR SNR52p::crADE2–6.L::SUP4t | GenArt | This study |
| pUD550 | KanR SNR52p::crCAN1–1.L::SUP4t | GenArt | This study |
| pUD439 | KanR SNR52p::crCAN1–1.crADE2–3.L::SUP4t | GenArt | This study |
| pUD440 | KanR SNR52p::crCAN1–1.crHIS4–1.crPDR12–1.crADE2–3.L::SUP4t | GenArt | This study |
| pUD552 | KanR SNR52p::crADE2–3.S::SUP4t | GenArt | This study |
| pUD605 | 2 μm KanMX ampRSNR52p::crADE2–1.L::SUP4t | In vivo | This study |
| pUD606 | 2 μm KanMX ampRSNR52p::crADE2–2.L::SUP4t | In vivo | This study |
| pUD627 | 2 μm KanMX ampRSNR52p::crADE2–3.L::SUP4t | In vivo | This study |
| pUD607 | 2 μm KanMX ampRSNR52p::crADE2–4.L::SUP4t | In vivo | This study |
| pUD608 | 2 μm KanMX ampRSNR52p::crADE2–5.L::SUP4t | In vivo | This study |
| pUD609 | 2 μm KanMX ampRSNR52p::crADE2–6.L::SUP4t | In vivo | This study |
| pUD628 (Addgene #103018) | 2 μm KanMX ampRSNR52p::crADE2–3.S::SUP4t | In vivo | This study |
| pUD629 | 2 μm KanMX ampRSNR52p::crCAN1–1.S::SUP4t | In vivo | This study |
| pUD630 | 2 μm KanMX ampRSNR52p::crCAN1–1.crADE2–3.S::SUP4t | In vivo | This study |
| pUDE712 | 2 μm KanMX ampRSNR52p::crHIS4–2.S::SUP4t | In vitro | This study |
| pUDE713 | 2 μm KanMX ampRSNR52p::crHIS4–3.S::SUP4t | In vitro | This study |
| pUDE714 (Addgene #103021) | 2 μm KanMX ampRSNR52p::crHIS4–4.S::SUP4t | In vitro | This study |
| pUDE708 | 2 μm KanMX ampRSNR52p::crADE2–3.crHIS4–2.S::SUP4t | In vitro | This study |
| pUDE709 | 2 μm KanMX ampRSNR52p::crADE2–3.crHIS4–3.S::SUP4t | In vitro | This study |
| pUDE710 (Addgene #103020) | 2 μm KanMX ampRSNR52p::crADE2–3.crHIS4–4.S::SUP4t | In vitro | This study |
| pUDE720 | 2 μm KanMX ampRSNR52p::crCAN1–2.S::SUP4t | In vitro | This study |
| pUDE721 | 2 μm KanMX ampRSNR52p::crCAN1–3.S::SUP4t | In vitro | This study |
| pUDE722 (Addgene #103022) | 2 μm KanMX ampRSNR52p::crCAN1–4.S::SUP4t | In vitro | This study |
| pUDE723 | 2 μm KanMX ampRSNR52p::crPDR12–2.S::SUP4t | In vitro | This study |
| pUDE724 (Addgene #103023) | 2 μm KanMX ampRSNR52p::crPDR12–3.S::SUP4t | In vitro | This study |
| pUDE725 | 2 μm KanMX ampRSNR52p::crPDR12–4.S::SUP4t | In vitro | This study |
| pUDE735 (Addgene #103024) | 2 μm KanMX ampRSNR52p::crCAN1–4.crHIS4–4.crPDR12–3.crADE2–3.S::SUP4t | In vitro | This study |
| pUD706 | 2 μm ampRKlURA3 | In vivo | This study |
| pUDE731 (Addgene #103008) | 2 μm ampRKlURA3 TEF1p::Fncpf1::CYC1t | In vitro | This study |
aThe presence of an S or a L following the crRNA name indicates that the direct repeats in the CRISPR array are either Short (19 nt) or Long (36 nt), respectively.
The reference number of plasmids deposited to Addgene is indicated next to the plasmid name between brackets when relevant.
Construction of a S. cerevisiae strain expressing FnCpf1 from a multicopy plasmid
A multicopy plasmid encoding Fncpf1 was constructed by Gibson assembly of the pMEL10 backbone, obtained by amplification of pMEL10 using primers 2055 and 4173 (Supplementary Table S1), and the Fncpf1 expression cassette (amplified with primers 5976 and 2629 using pUDC175 as a template (Supplementary Table S1)). Plasmid assembly was confirmed by PCR analysis using primers 2376 and 10408 (Supplementary Table S1) and restriction digestion analysis using FastDigest PdmI (Thermo Fisher Scientific). The resulting plasmid was named pUDE731 (Table 2). 500 ng of pUDE731 were transformed to CEN.PK113–5D (MATa ura3–52, Table 1) using the lithium acetate transformation protocol (38). To obtain an empty plasmid used as control for pUDE731, PCR-amplified pMEL10 backbone (primers 2055/4173) and repair oligo made with primers 12269/12270 were cotransformed into CEN.PK113–5D for in vivo assembly. Transformants containing pUDE731 and the in vivo assembled empty plasmid were selected on SMG plates and checked using diagnostic PCR with primers 2376 and 10408 on genomic DNA prepared as previously described (39). A clone carrying pUDE731 and showing the expected bands was additionally confirmed by Sanger sequencing of a DNA fragment containing the Fncpf1 expression cassette, amplified using primers 2750/2376 and 4661 (Supplementary Table S1). This strain was named IME384 (Table 1). A transformant shown to carry the empty plasmid by PCR was further characterized by restriction analysis. The strain was named IME385 (Table 1) and the verified empty plasmid pUD706 (Table 2).
Selection of target sites, design of crRNA arrays
In first instance, to knock-out the targeted genes (ADE2, HIS4, PDR12 and CAN1) spacers were designed following several criteria: (i) both strands of the coding region of the target genes were screened for the presence of a PAM of 5′-TTN-3′. For every PAM found, 25nt downstream were selected as potential target sequence; (ii) sequences containing poly-T stretches longer than six were discarded due to the possibility of premature RNA polymerase III transcripts formation (8,40); (iii) spacers exhibiting similarity with other chromosomal loci determined by the BLASTn webtool (41) were considered as possible off-targets and were excluded; iv) target sequences fulfilling the three first criteria were screened for their AT content and secondary structure of the mature crRNA. The crRNA structure was analysed using the RNA fold web server (42), only open RNA secondary structures were favoured, as they might allow efficient interaction with FnCpf1.
As several spacers designed with these criteria did not promote efficient FnCpf1-mediated DNA editing, new design principles were defined and tested as described in the Results section.
Construction of crRNA expression plasmids
The crRNA expression cassettes systematically comprised the RNA polymerase III dependent SNR52 promoter, the target sequence(s) flanked by direct repeats and the SUP4 terminator. crRNA arrays were either ordered as linear synthetic fragments (IDT-BVBA, Leuven, Belgium) and directly assembled into a plasmid backbone, or synthetized by GenArt on plasmids (Regensburg, Germany) with further assembly. Two types of direct repeats were tested, a long repeat of 36 nt (GTCTAAGAACTTTAAATAATTTCTACTGTTGTAGAT) and a short repeat of 19 nt (AATTTCTACTGTTGTAGAT).
crRNA expression constructs were obtained using two different methods. crRNA expression plasmids were initially constructed by in vivo assembly of four fragments (Table 2) (43). For this purpose, a mixture containing a DNA fragment with the ampR marker, a 2 micron fragment for yeast propagation, a KanMX marker cassette and the synthetized linear crRNA array was transformed in IMX1139. Each fragment was PCR-amplified using template plasmids pRS416 for ampR, pROS13 for 2μm and kanMX, with primers pairs 2054/2055, 10224/10225, 10313/10314, respectively (Supplementary Table S1). These primers incorporated orthogonal sequences (Synthetic Homologous Recombination sequences, SHR, (43)) to each fragment, thereby enabling their assembly by homologous recombination in yeast. Primer pair 10477/10478 was used to amplify the crRNA arrays from a corresponding plasmid synthetized by GeneArt (Table 2), while incorporating SHR’s (Supplementary Table S1). Fragments were digested by DpnI (Thermo Fisher Scientific) and gel-purified prior to transformation. For transformation 100 fmol of 2μm and KanMX fragments and 200 fmol of crRNA fragment and ampR were supplied (44). The plasmids constructed using this method were named pUD605 to pUD609 and pUD627 to pUD630 (Table 2).
To evaluate the effect on DNA delivery on FnCpf1 efficiency, crRNA plasmids pUD627 and pUD628 (Table 2) constructed by in vivo assembly were extracted from S. cerevisiae transformants. The extracted plasmids were checked by restriction analysis using FD PstI and FD PvuI. Additionally, the spacer region was Sanger sequenced with the primer pair 10477/10478. For transformation, 500 ng of the corresponding plasmid was transformed in IMX1139.
A second set of plasmids was assembled in vitro using NEBuilder® HiFi DNA Assembly Master Mix (New England Biolabs) targeting either a single locus HIS4 (crHIS4–2, crHIS4–3 and crHIS4–4), ADE2 (crADE2–3), CAN1 (crCAN1–2, crCAN1–3, crCAN1–4), PDR12 (crPDR12–2, crPDR12–3, crPDR12–4) or targeting multiple loci (crADE2–3.crHIS4–2, crADE2–3.crHIS4–3, crADE2–3.crHIS4–4, and crCAN1–4.crHIS4–4 .crPDR12–3.crADE2–3). To this end, a linear fragment with crRNA array was assembled with a PCR-amplified fragment of pUD628 (primers 5793 and 11940). crRNA array and plasmid backbone harboured 60 nt homology flanks to promote assembly of the two fragments. Correct plasmid assembly was confirmed by diagnostic PCR and Sanger sequencing. The plasmids were named pUDE708 to pUDE714, pUDE720 to pUDE725 and pUDE735 (Table 2). For transformation to IMX1139, 500 ng of plasmid DNA were used, with the exception of the transformations presented in Figure 7 for which 2 μg were used. pUD628 (Addgene #103018), pUDE714 (Addgene #103021), pUDE722 (Addgene #103022) and pUDE724 (Addgene #103023) carrying crADE2–3, crHIS4–4, crCAN1–4 and crPDR12–3, respectively for single deletion, pUDE710 (Addgene #103020) carrying crADE2–3 and crHIS4–4 for double deletion, and pUDE735 (Addgene #103024) carrying the quadruple arrays combining crCAN1–4, crHIS4–4, crPDR12–3 and crADE2–3 are available from Addgene (Table 2). These plasmids carry crRNAs framed by short DRs of 19 nt. Also, pUDC175 (Addgene #103019) and pUDE731 (Addgene #103008), centromeric and episomal plasmids respectively, harbouring Fncpf1 for expression in S. cerevisiae, are available from Addgene (Table 2).
Figure 7.
Multiplex genome editing by FnCpf1 in S. cerevisiae. (A) composition of CRISPR arrays for single deletion of CAN1 and PDR12, and quadruple deletion of ADE2, CAN1, HIS4, and PDR12. Three different crRNAs were tested for CAN1 and PDR12. 19-nt direct repeats were used and CRISPR plasmids were assembled in vitro using Gibson assembly. (B) Fraction of transformants with single deletion using single arrays (plasmids used: pUDE720 to pUDE725). (C) Fraction of clones with triple (3D) and quadruple deletion (4D) after transformation with the quadruple array (pUDE735). No transformants without deletion, or with single or double deletion were found. Two strains were tested for multiplex genome editing, IMX1139 with genomic integration of Fncpf1 and IME384 in which Fncpf1 is carried by a multicopy plasmid. B, C: deletion was quantified by diagnostic PCR (Supplementary Figure S6). The number of transformants checked by PCR is indicated between brackets. Plating was performed just after transformation, without additional incubation.
Strain construction through FnCpf1-mediated genome editing
The crRNA array expression plasmids or plasmid fragments were transformed to IMX1139 or IME384 expressing FnCpf1. 1 μg of 120 bp dsDNA repair DNA was co-transformed to enable repair of the edited genomic locus by homologous recombination. As exception, 2 μg of repair DNA were co-transformed in the experiments shown in Figure 7. To assess crRNA efficiency an identical transformation omitting the repair DNA fragment was systematically performed. The repair DNA fragment was generated by annealing in a 1:1 ratio two complementary 120 nt oligonucleotides that were initially heated at 95°C and then cooled down to room temperature (9). Transformed cells were plated on solid YPD plates supplemented with G418. In the case of IME384, transformants were selected on SMG with G418 and urea as a nitrogen source, supplemented with 20 mg l−1 adenine and 125 mg l−1 of histidine. When extended incubation was tested, 100 μl of the transformed cells were first recovered on YPD for 24–48 h before plating. Duplicate transformations were performed for each experiment and dilutions of 10−1, 10−2 and 10−3 were plated.
Whole genome sequencing
The genome of IMX1139 was sequenced using MiSeq (Illumina, San Diego, CA, USA) with MiSeq® Reagent Kit v3 with 2 × 300 bp read length. Genomic DNA was extracted using the Genomic DNA kit (Qiagen, Hilden, Germany). Extracted DNA was quantified by BR ds DNA kit using Qubit spectrophotometer (Invitrogen, Carlsbad, CA, USA) and mechanically sheared with the M220 ultrasonicator (Covaris, Woburn, MA, USA) using settings aiming at 550 bp average size. DNA libraries were prepared using the TruSeq DNA PCR-Free Library Preparation Kit according to the manufacturer's instructions (Illumina). qPCR quantification of libraries was done with the KAPA Library Quantification Kit for Illumina platforms (Kapa Biosystems, Wilmington, MA, USA) on a Rotor-Gene Q PCR cycler (Qiagen). Sequence reads of genomic DNA were mapped onto the CEN.PK113–7D reference strain sequence (35) and on the unique integrated Fncpf1-KlURA3 contig using the Burrows–Wheeler Alignment tool (BWA) and further processed using SAMtools (45,46). The sequencing raw data are available at NCBI (https://www.ncbi.nlm.nih.gov/bioproject/) under the Bioproject number PRJNA394199.
Growth rate measurements
To evaluate the potential toxicity of FnCpf1 expression in S. cerevisiae, IMX1139 (expressing Fncpf1 from a chromosomal locus), IME384 (expressing Fncpf1 from a multicopy plasmid pUDE731), IME385 (containing the empty multicopy plasmid pUD706) and CEN.PK113–7D (Table 1) were grown in SMG medium in shake-flask culture. Growth was monitored by measuring optical density (660 nm) at regular time intervals using Libra S11 spectrophotometer (Biochrom, Cambridge, UK). The maximum specific growth rates were calculated from duplicate shake-flask cultures.
RESULTS
FnCpf1 expression from genomic DNA is not toxic for S. cerevisiae
FnCpf1-mediated genome editing in S. cerevisiae requires three parts, (i) the endonuclease Cpf1, (ii) the crRNA that will guide Cpf1 to the targeted DNA site, and (iii) a small, double stranded DNA fragment that will elicit repair of the double strand DNA cleavage caused by FnCpf1 via homologous recombination and thereby restore chromosome integrity (repair DNA). A yeast strain carrying a single copy of the Fncpf1 gene from Francisella novicida U112 integrated in its genome was therefore constructed (Supplementary Figure S1). A Fncpf1 allele that was codon-optimized for expression in human and fused at its C-terminus with the nuclear localization signal (27) was cloned between the strong and constitutive TEF1 promoter and the CYC1 terminator. Together with the URA3 gene from Kluyveromyces lactis, the Fncpf1 expression cassette was integrated in the SGA1 locus on chromosome IX of S. cerevisiae strain CEN.PK113–5D. PCR analysis and whole genome sequencing of a selected transformant, renamed IMX1139, confirmed the correct integration, copy number and sequence for Fncpf1 (Supplementary Figure S1). Moreover, whole genome sequencing also revealed the absence of unwanted mutations or chromosomal rearrangements in IMX1139.
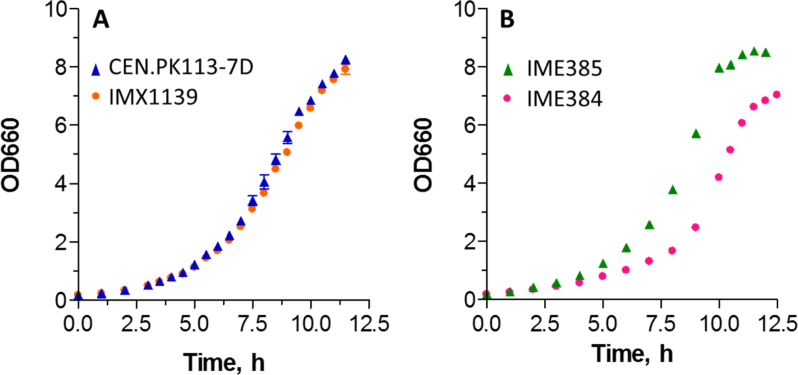
The impact of Fncpf1 and its translation product on growth of S. cerevisiae was assessed. The prototrophic IMX1139 grew as fast as the isogenic control strain CEN.PK113–7D in chemically defined medium with glucose as sole carbon source at 30°C (specific growth rate of 0.41 ± 0.01 h−1 and 0.42 ± 0.01 h−1 for IMX1139 and CEN.PK113–7D, respectively), revealing that FnCpf1 expression had no detectable impact on S. cerevisiae physiology (Figure 2A).
Figure 2.
Specific growth rate of strains expressing FnCpf1 and their control strains. A: IMX1139, expressing FnCpf1 constitutively from its genomic DNA, and its congenic control strain CEN.PK113–7D. B: IME384, expressing FnCpf1 from a multicopy plasmid (pUDE731) and its congenic control strain IME385 containing the same multicopy plasmid but without FnCpf1 (pUD706). The strains were cultivated in shake-flask on chemically defined medium with glucose as sole carbon source. The data points represent the average and mean deviation of two independent culture replicates.
To further explore the potential toxicity of FnCpf1, a strain expressing FnCpf1 from a multicopy plasmid, using the same strong, constitutive promoter as the one used for IMX1139, was constructed. When grown in shake-flask, this strain, IME384, displayed a substantial decrease in specific growth rate (24% decrease). IME384 grew at 0.29 ± 0.00 h−1 while its isogenic control strain IME385 (carrying the corresponding empty plasmid) grew at a specific growth rate of 0.38 ± 0.00 h−1 (Figure 2B), demonstrating the toxicity of FnCpf1 at extreme expression levels.
FnCpf1 is capable of RNA-mediated targeted genomic DNA editing in S. cerevisiae
To supply the crRNA to IMX1139 and promote FnCpf1-mediated DNA cleavage, crRNA expression cassettes carrying the constitutive SNR52 promoter, a single 25-nt spacer surrounded by two direct repeats of 36 nt from Francisella novicida (27) and the SUP4 terminator were synthetized (Figure 3A). To easily monitor FnCpf1 activity, the spacer was designed to target ADE2, a gene essential for adenine biosynthesis, deletion of which results in adenine auxotrophy and in red colouring of colonies (47). The 5′-TTN-3′ PAM previously defined for FnCpf1 (27) was used to select the targeted DNA sequence. The plasmid carrying this crRNA expression cassette was assembled in yeast using in vivo assembly (43), (Figure 3A) by transforming the following four fragments to yeast: (i) the crRNA expression cassette, (ii) a yeast selection marker, (iii) a yeast autonomous origin of replication and (iv) a selection marker together with origin of replication for expression in E. coli. These four fragments of the CRISPR plasmid were transformed to yeast together with the DNA fragment (i.e. repair DNA) meant to promote repair of the chromosomal cleavage caused by FnCpf1.
Figure 3.
Efficiency of ADE2 editing by FnCpf1. (A) Design of the CRISPR plasmid harbouring the CRISPR array for in vivo assembly in yeast. SHR, homologous sequence for recombination (43). (B) AT content and position in the coding region of ADE2 of the crRNAs. (C) Comparison of the genome editing efficiency of six crRNA with various AT content and target sequence (grey bars). The genome editing efficiency was also measured when cells were incubated after transformation in liquid medium for 48 h (black bars). The efficiency is calculated as the number of red colonies divided by the total number of colonies on the transformation plates in the presence of repair DNA fragments. Values represent the average and standard deviation of two biological and two technical replicates. (Plasmids used: pUD605 to pUD609, Table 2).
While various software algorithms are available to guide crRNA design for SpCas9 in S. cerevisiae (9,48–50) for maximal cleavage efficiency and specificity, design principles for the newly discovered FnCpf1 are still being explored. Both the AT content of the gRNA and the site of cleavage are important for efficient genome editing by SpCas9 (51). Therefore, six crRNA with AT contents ranging from 36% to 84% and targeting sequences spread across the whole coding sequence of ADE2 were chosen (crADE2–1 to crADE2–6, Figure 3B, Table 3 and Supplementary Table S2). PCR analysis of the cleavage site confirmed that DNA was cleaved as expected and correctly repaired via homologous recombination by the supplied repair DNA fragment (Figure 3C and Supplementary Figure S2). The six crRNAs led to very different editing efficiencies ranging from below 1% to 37%. However, similar efficiencies were obtained for AT contents ranging from 36 to 72% (28 ± 2% and 29 ± 4% respectively), revealing that FnCpf1 was not sensitive to large variations in AT content within this range. While the 84% AT content could explain why the efficiency of this crADE2–6 was very low, also crADE2–2 and crADE2–4 with 44% and 60% AT content hardly led to genome editing. These results suggested that other factors than AT content did affect the FnCpf1 endonuclease activity.
Table 3. Attributes of the spacers used in this study.
| Targeted gene | crRNA name | 5′ to 3′ sequence (PAM) | AT content (%) | Position from ATG |
|---|---|---|---|---|
| ADE2 (1716 nt) | crADE2–1 | T(TTA)CGGGCACACCGATGACAGGAAGTGG | 36 | 1438 |
| crADE2–2 | T(TTT)CGGCGTACAAAGGACGATCCTTCAG | 44 | 723 | |
| crADE2–3 | T(TTC)CCGGTTGTGGTATATTTGGTGTGGA | 52 | 743 | |
| crADE2–4 | T(TTA)CATTCAATTGTGCAAATGCCTAGAG | 60 | 1498 | |
| crADE2–5 | T(TTA)ATTTGGGATGTTTTACTTGAAGATT | 72 | 247 | |
| crADE2–6 | T(TTG)ATTAAATGCTCTTTTTGAATATATT | 84 | 317 | |
| CAN1 (1773 nt) | crCAN1–1 | T(TTA)TTTGGTCTATCAAAGAACAAGTTGG | 64 | 1204 |
| crCAN1–2 | CTT(TTC)ATTGGTTTATCCACACCTCTGACCA | 64 | 322 | |
| crCAN1–3 | CAT(TTC)AAGGTACTGAACTAGTTGGTATCAC | 60 | 893 | |
| crCAN1–4 | GTT(TTG)CCACATATCTTCAACGCTGTTATCT | 60 | 1123 | |
| HIS4 (2400 nt) | crHIS4–1 | G(TTG)CCCAATGTAAGGAGATTGTGTTTGC | 56 | 1514 |
| crHIS4–2 | T(TTC)TCCAATCAATTCATGGTAAAACAAA | 72 | 328 | |
| crHIS4–3 | T(TTA)CTAAAGATTCTAGCCCCACCAAACC | 52 | 730 | |
| crHIS4–4 | T(TTA)GCATCTTGGCTAGCAATGAACAGAG | 52 | 227 | |
| PDR12 (4536 nt) | crPDR12–1 | A(TTC)CATTTATGAAATATGAAGCTGGTGC | 64 | 1847 |
| crPDR12–2 | CAT(TTC)GTCGAGATCGAACCATGACGATGAT | 52 | 39 | |
| crPDR12–3 | GTT(TTA)GCACAAAGAATCAATATGGGTGTCA | 60 | 2674 | |
| crPDR12–4 | CAT(TTC)GCATATAAGCATGCTTGGAGAAATT | 62 | 2269 |
NB: in the text and in Table 2, a letter is added at the end of the crRNA name listed in this table to indicate whether the crRNA is framed by short (S, 19 nt) or long (L, 36 nt) direct repeats.
To increase the editing efficiency that was overall relatively low, cells were incubated after transformation in liquid medium for 48 hours. This incubation did successfully increase the editing efficiency up to 78 ± 4% for the three crRNAs that gave the highest efficiencies right after transformation (crADE2–1, crADE2–3 and crADE2–5), but did not improve the efficiency for the other three crRNAs (Figure 3C). As the double-stranded repair DNA supplied to cells is rapidly degraded by nucleases in the hours following transformation, new DNA cuts resulting from FnCpf1 activity during prolonged incubation of cells in liquid medium can only result in repair via non-homologous end joining (NHEJ). However, PCR analysis and sequencing of ten colonies with the red phenotype after 48 hours incubation revealed that the DNA cleavage caused by FnCpf1 was exclusively repaired by integration of the supplied repair DNA via homologous recombination (Supplementary Figure S2). During prolonged incubation, in the absence of repair DNA and due to the low occurrence of DNA repair by NHEJ in S. cerevisiae, failure to repair the double strand DNA cleavage caused by FnCpf1 results in cell death. The surviving cells, i.e. cells that have already performed ADE2 editing and repair by homologous recombination shortly after transformation, appeared to be enriched in the culture.
Direct repeat length has a strong impact on FnCpf1-mediated genome editing in S. cerevisiae
It has been shown in several hosts that shorter DR can improve efficiency of genome editing by FnCpf1 (52). New CRISPR cassettes were synthetized with DR of 19 nt instead of the 36 nt previously used, framing the crADE2–3 spacer targeting ADE2 (crADE2–3.S, in which the letter S after the crRNA name denotes short DRs in contrast with L that denotes a long DRs (36 nt)). Shortening the DR length had a marked impact on editing efficiency as transformation plates were covered with red colonies, and white colonies were virtually absent, leading to knockout efficiencies of 100% (Table 4). In addition, transformation with CRISPR cassettes with 36-nt DR typically led to the formation of a substantial number of white colonies in the absence of repair DNA (typically 30–40 colonies per 100 colonies counted in the presence of repair DNA in experiments presented in Figure 3). In these colonies the selection marker was present, but FnCpf1 was not able to cleave DNA. When using CRISPR cassettes with 19-nt DR, hardly any colonies were observed when repair DNA was omitted from the transformation mix.
Table 4. ADE2 editing efficiency of FnCpf1 for interruption and point mutation using long (36 nt) and short (19 nt) direct repeats.
| Protospacer | DR length | Mutation type | Plasmid assemblya | Genome editing efficiency |
|---|---|---|---|---|
| crADE2–3 (52% AT) | 36 nt | Deletion | in vivo | 37 ± 2%b |
| 36 nt | Deletion | Pre-assembled | 19 ± 6%b | |
| 19 nt | Deletion | Pre-assembled | 100%2 | |
| 19 nt | Point mutation | Pre-assembled | 100%c |
aPre-assembled plasmids were purified from yeast cells after in vivo assembly and re-used for transformation to yeast.
b Efficiency calculated as the number of red colonies divided by the total number of colonies on the transformation plates in the presence of repair DNA fragments. Values represent the average and standard deviation of two biological and two technical replicates.
c Efficiency calculated by dividing the number of colonies with the correct point mutation over the total number of colonies tested.
For experiments with shorter direct repeats, CRISPR plasmids were first pre-assembled by in vivo assembly, then purified from the yeast strains before being transformed to cells in which the genome editing efficiency was monitored. Conversely, genome editing efficiency in experiments shown in Figure 3 was tested directly in cell populations in which the CRISPR plasmids were directly assembled in vivo. To check whether the aforementioned improved efficiency resulted from utilization of pre-assembled plasmid and not from shorter DR, we repeated ADE2 editing with crADE2–3.L using 36 nt direct repeats, but this time with a pre-assembled CRISPR plasmid. Efficiency was not improved, and even slightly decreased using pre-assembled plasmids, confirming that shorter direct repeats were responsible for the strongly enhanced FnCpf1-mediated genome editing (Table 4).
FnCpf1 is an efficient tool to insert point mutations
To take genome editing one step further, FnCpf1 was assessed for in vivo site directed mutagenesis in S. cerevisiae. A 120 nt repair fragment was designed, carrying a two nucleotide change to mutate the PAM and incorporate a premature TAA stop codon in the middle of ADE2 coding sequence, thereby leading to a shortened ADE2 transcript and hence an inactive phosphoribosylaminoimidazole carboxylase. Mutation of the PAM aimed at preventing further cleavage of ADE2 by FnCpf1. The red colour of the obtained colonies indicated that the transformants were effectively targeted and sequencing of the ADE2 locus confirmed the integration of the premature stop codon in the PAM in all tested transformants (Table 4 and Figure 4). FnCpf1-mediated genome editing therefore very efficiently generated point mutations at a user-specified location in the genome of S. cerevisiae.
Figure 4.
Confirmation of FnCpf1-mediated introduction of a point mutation in ADE2. Sanger sequencing of the genomic DNA locus targeted for FnCpf1-mediated point mutation in seven randomly selected transformants. The control is the genomic DNA of the congenic strain CEN.PK113–7D. (Plasmid used: pUD628 carrying crADE2–3.S, Table 2).
Efficient simultaneous editing of two genomic targets by FnCpf1
To test double and quadruple deletion, CRISPR arrays targeting ADE2 and CAN1 or ADE2, CAN1, HIS4 and PDR12 were synthetized (Supplementary Figure S3). All spacers had similar AT content ranging from 52% to 64% (Table 3). The crRNA targeting ADE2 was systematically located at the last position of the array before the terminator and long repeats (36 nt) were used. Unexpectedly, no FnCpf1-mediated deletion was observed for CAN1 either using single, double or quadruple CRISPR array (Supplementary Figure S3). Similarly, diagnostic PCR revealed that neither HIS4 nor PDR12 were deleted when using the quadruple CRISPR array (Supplementary Figure S3). PCR analysis would fail to identify FnCpf1 editing if the cleavage was not repaired via homologous recombination but rather by non-homologous end joining, as the latter would lead to short indels that can only be identified by sequence analysis. However, none of the sequenced transformants (20 transformants from the plates with repair DNA and 10 from the plates without repair for each targeted gene) carried indels at the targeted locus, revealing that crCAN1.L, crHIS4–1.L and crPDR12.L failed to induce FnCpf1-mediated genome editing (Supplementary Figure S4). This lack of DNA editing by FnCpf1 was confirmed at a larger scale by phenotypic analysis of transformants. Remarkably, however, ADE2 was successfully deleted whether the crRNA was carried by the single, double or quadruple crRNA array (Supplementary Figure S3). Moreover, the efficiency of ADE2 deletion was not substantially reduced when four loci (28 ± 4% efficiency) were targeted as compared to single locus targeting (36 ± 2% efficiency; Supplementary Figure S3).
As several crRNAs failed to promote FnCpf1-mediated gene deletion, we designed a series of three crRNAs targeting the HIS4 gene (Table 3). These three new crRNAs named crHIS4–2, crHIS4–3 and crHIS4–4 were tested for single deletion, as well as for double deletion, in combination with crADE2–3.S (Figure 5A). For this experiment, short direct repeats of 19 nucleotides were used, and the plasmids carrying the crRNAs were assembled in vitro, prior to transformation. Deletion was checked by diagnostic PCR (Supplementary Figure S5). As shown in the previous experiments, crADE2–3.S led to very efficient FnCpf1-mediated editing of ADE2 when using a single target, but remarkably, when targeting both ADE2 and HIS4, crADE2–3.S also promoted ADE2 deletion with 100% efficiency with any of the crRNAs targeting HIS4 (Figure 5B). crHIS4–2.S, crHIS4–3.S and crHIS4–4.S displayed different editing efficiencies for single locus targeting, with HIS4–2.S being unable to guide FnCpf1 for editing, while the latter two crRNAs resulted in HIS4 deletion with 86% and 100% efficiency, respectively. When combined with crADE2–3.S for double targeting, crHIS4–2.S failed to promote gene deletion, while 25% of the tested clones displayed double deletion when using crHIS4–3.S (Figure 5B). 100% of the tested transformants displayed a double ADE2 HIS4 deletion when using crADE2–3.S and crHIS4–4.S, without requirement of extended incubation, thereby demonstrating that FnCpf1 does have the potential to very efficiently promote multisite homologous recombination-mediated DNA editing.
Figure 5.
FnCpf1-mediated editing of single and double genomic targets. (A) Composition of CRISPR arrays for single and double deletion of ADE2 and HIS4. 19-nt direct repeats were used and CRISPR plasmids were assembled in vitro using Gibson assembly. (B) Fraction of transformants with single or double deletion as measured by diagnostic PCR (Supplementary Figure S5), following the design described in A. The number of transformants checked by PCR is indicated between brackets. (Plasmids used: pUD628, pUDE712 to pUDE714, pUDE708 to pUDE710). Plating was performed just after transformation, without additional incubation.
Refining the guidelines for crRNA design for predictable and efficient multiplex genome editing up to four targets in S. cerevisiae
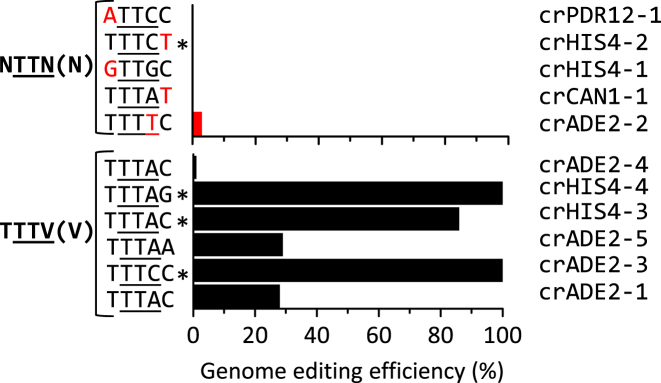
Remarkably, seven out of the twelve tested crRNA guides resulted in no or extremely low (below 3%) genome editing efficiencies. For these crRNAs, sequence analysis of the targeted sites revealed the complete absence of DNA editing by FnCpf1. Comparing the PAM of these crRNAs strikingly revealed that the PAM of efficient crRNAs shared characteristics that have been shown to strongly enhance DNA editing efficiency with Cpf1 from Acidaminococcus (AsCpf1) and Lachnospiraceae bacterium (LbCpf1) (26,27,31). These two Cpf1 variants favour a 5′-TTTV-3′ PAM (V = A/G/C), which differs from the reported FnCpf1 PAM (5′-NTTN-3′) by a strong preference for a thymidine at the 5′ position of the PAM, and by a marked decrease in efficiency in the presence of thymidine at the 3′ end (31). The same study revealed that thymidine is strongly disfavoured in the first position after the PAM. Remarkably, out of the six crRNAs with AT content within acceptable range (44–72%) that failed to promote genome editing in S. cerevisiae, five do not meet the criteria defined for AsCpf1 and LbCpf1 (Figure 6). Two have a thymidine in the first position after the PAM (TTTA-T, crCAN1; TTTC-T, crHIS4–2), two do not harbor thymidine in the 1st position of the PAM (GTTG-C, crHIS4–1; ATTC-C, crPDR12) and one has a thymidine in the last position of the PAM (TTTT-C, crADE2–2). These results suggested that FnCpf1 preferred 3′-TTTV-5′ as PAM, and the absence of thymidine as first base after the PAM, when expressed in S. cerevisiae. We used these new criteria to design crRNAs targeting CAN1 and PDR12. Out of the six new crRNAs, five were able to promote FnCpf1-mediated genome editing (Figure 7), demonstrating that the new design criteria increased the predictability of genome editing by FnCpf1.
Figure 6.
Overview of PAM sequences of the crRNAs used in this study and their efficiency for genome editing. Only crADE2–6, which had an extreme AT content (84%) is not represented. Efficiency calculated as indicated in Figures 3 and 5. * indicates arrays containing 19-nt repeats instead of 36.
As crRNAs able to efficiently target four different loci in yeast genomic DNA were available, we explored their ability to target these four loci simultaneously. An array carrying crCAN1–4, crHIS4–4, crPDR12–3 and crADE2–3 was synthetized (with short DR, pUDE735) and transformed to IMX1139 (carrying a chromosomal copy of Fncpf1), as well as to IME384, a strain expressing FnCpf1 from a multicopy vector (Figure 7). Both strains showed an extremely high level of quadruple deletion, as 88% and 100% of the tested cloned displayed four simultaneous deletions in IMX1139 and IME384 respectively (Figure 7, Supplementary Figure S6). Remarkably, the efficiency of DNA editing was not affected by positioning of the crRNA on the array as the deletion efficiency of ADE2 was 100% in all tested arrays, and all four targets were equally well edited. It is noteworthy that the number of colonies obtained for quadruple multiplexing was low. While transformation for single deletion resulted in ca. 200 transformants per plate, in identical conditions transformations with quadruple arrays yielded a 20-fold lower number of transformants. The low number of colonies obtained and tested did not allow to draw conclusions on a potential impact of FnCpf1 expression level on genome editing efficiency.
DISCUSSION
In the present study, FnCpf1 expressed as single copy from the SGA1 locus using the strong and constitutive TEF1p promoter did not affect yeast physiology. Using the same cloning and expression strategy, Cas9 expression was similarly found to be neutral towards yeast growth (9). Conversely, higher expression levels of FnCpf1, mediated by expression from a multicopy plasmid, substantially impaired growth of S. cerevisiae, as previously reported for Cas9 (53,54). Generally, a stable integrated copy of Cpf1 is preferred, since this allows growth of strains on complex media and, when multiple rounds of transformation are required, efficient recycling of crRNA carrying plasmids.
Guided by earlier work performed in vitro and in vivo, the initial design of the crRNAs used in this study was based on a 5′-TTN-3′ PAM, a spacer of 25 nucleotides and direct repeats of 36 nucleotides (25,27,55). This design led to genome editing in S. cerevisiae with maximum efficiencies around 40%. Most influential for genome editing was the size reduction of the direct repeats from 36 to 19 nucleotides, as previously shown in mammalian cells (52), which consistently resulted in efficiencies of 100% for several targeted sites. The present work also demonstrated that FnCpf1 can be used for single nucleotide mutagenesis. While several studies reported that FnCpf1 is less efficient or even inactive for genome editing as compared to its orthologues AsCpf1 and LbCpf1 (for instance in rice (56), or in human cells (27,57)), the present study demonstrated that Cpf1 from Francisella novicida could efficiently and precisely cleave S. cerevisiae genome, thereby promoting homology directed repair.
A surprising outcome of this work was the clear and strong preference of FnCpf1 for crRNAs with 5′-TTTV-3′ as PAM, and without thymidine as first base after the PAM, when expressed in S. cerevisiae. These preferences are shared with its close relative AsCpf1 and LbCpf1. Structural studies of Cpf1 variants showed that the PAM duplex is bound to a groove formed by the WED, REC1, and PI domains (28,30). In this groove, the PAM duplex is recognized by Cpf1 by a combination of interactions with specific amino acids and by shape readout mechanisms (28,30). FnCpf1, LbCpf1 and AsCpf1 are remarkably well conserved in this region, and all amino acids suggested to be important for the 5′-TTTV-3′ PAM recognition by AsCpf1 are conserved in FnCpf1 (28,30,31). Also, a recent study on engineering AsCpf1 PAM specificity identified key amino acid residues that are also conserved in FnCpf1 (58). The high homology between FnCpf1 and its orthologs suggested that it might also favour a 5′-TTTV-3′ PAM. Because editing of human cells by FnCpf1 initially was reported to be relatively inefficient (27), only a few studies reported its application for genome editing. In many of these studies the PAM sequence was fortuitously preceded by a thymidine. For instance, in the study by Fonfara et al., in which the FnCpf1 PAM was relaxed from 5′-TTN-3′ to 5′-YTN-3′, the plasmid used to evaluate the PAM preference in vivo carried a thymidine located 5′ to the PAM (25). High throughput studies also suggest a slight preference for a thymidine preceding the 5′-YTN-3′ PAM for FnCpf1 (27,55). Altogether these observations seem to support the 5′-TTTV-3′ PAM preference found for FnCpf1 in the present study. This hypothesis should be further explored by a more systematic study of the PAM requirement for FnCpf1 in S. cerevisiae.
Based on the present results we recommend to apply the following criteria for crRNA design for Cpf1-based editing: (i) 5′-TTTV-3′ PAM, (ii) no thymidine in the first position of the crRNA spacer sequence, (iii) AT content between 30% and 70%, (iv) direct repeats of 19 nucleotides. Still, two crRNAs with optimal PAM and first position of the crRNA sequence (TTTA(C) for crADE2–4 and TTTC(A) for CAN1–2) did not lead to genome editing. As already observed for Cas9, other factors can also influence the efficiency of CRISPR endonuclease such as the presence of proteins or genomic DNA secondary structures that prevent access of the endonuclease to the targeted genomic locus.
While using Cpf1 for single locus targeting already offers substantial advantages, such as the possibility to target AT-rich regions or to combine Cpf1 with other CRISPR-Cas enzymes such as Cas9, its major strength resides in its potential to edit the crRNA array itself, combined with the simplicity and short size of the crRNA array. In S. cerevisiae, applications of Cas9 for multisite editing remains rather limited, either because of the need of complicated DNA constructs in the case of a chimeric guide RNA, or because of low efficiency when CRISPR arrays are used (9,19,53,59,60). While ribozymes can compensate for the absence of crRNA cleavage by Cas9 in various organisms (61), their efficiency for multiplex genome editing has not been explored in S. cerevisiae yet. Furthermore, crRNA arrays equipped with ribozymes require complex DNA assembly or expensive custom DNA synthesis, as each expression unit, composed of two different ribozymes (typically Hammer Head and HDV) and of a single guide RNA, is 211 nt long (61). While 844 bp crRNA arrays are required to target four genes with artificial ribozyme and single guide RNA constructs using Cas9, simple, native 176 nt arrays suffice to promote quadruple genomic locus editing with FnCpf1 with 100% efficiency. FnCpf1 genome editing efficiency was not affected by the position of the crRNA on the array or by the number of protospacers when using up to four targets. The number of colonies obtained with quadruple crRNA arrays was strongly decreased as compared to single or double arrays. Overexpressing FnCpf1 using a multicopy plasmid did not increase the number of colonies obtained after transformation, suggesting that FnCpf1 was not a limiting factor for genome editing. This decrease in number of transformants can be explained by several factors, such as the decreased probability of the occurrence of multisite DNA cuts and repairs with increasing number of targets. In view of the absence of detectable benefit of expressing FnCpf1 from a multicopy plasmid for single or multisite editing up to four targets and of the toxicity of overexpression of FnCpf1, we advise to use single copy genomic integration of Fncpf1 for genome editing in S. cerevisiae. Despite the low number of transformants obtained with multiplexing, which can be experimentally addressed, genome editing with FnCpf1 was remarkably efficient.
In conclusion, FnCpf1 is a powerful addition to the CRISPR toolbox in S. cerevisiae. The plasmid carrying Fncpf1 framed by the TEF1 promoter and CYC1 terminator, as well as the plasmids expressing crRNAs for single and quadruple targeting of ADE2, CAN1, HIS4 and PDR12, as well as double ADE2 and HIS4 targeting are available, and can be obtained through Addgene. Furthermore the tools supplied in this study provide an experimental foundation to easily express any crRNA. Cloning in pUD628 of 176 nt dsDNA fragment obtained by annealing of two long oligonucleotides allows the facile construction of crRNA arrays of up to four spacer sequences and expands the application of FnCpf1 for editing the entire yeast genome.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Pilar de la Torre for sequencing IMX1139, Marcel van den Broek for bioinformatics support for whole genome sequence analysis and Mark Bisschops for critically reading the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
AdLibYeast ERC consolidator [648141 to P.D.L.]; European Union's Horizon 2020 Framework Programme for Research and Innovation; project: Model-Based Construction and Optimisation of Versatile Chassis Yeast Strains for Production of Valuable Lipid and Aromatic Compounds [720824 to J.M.D.]. Funding for open access charge: ERC consolidator [648141].
Conflict of interest statement. None declared.
REFERENCES
- 1. Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P.. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007; 315:1709–1712. [DOI] [PubMed] [Google Scholar]
- 2. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van der Oost J. Molecular biology. New tool for genome surgery. Science. 2013; 339:768–770. [DOI] [PubMed] [Google Scholar]
- 4. Hsu P.D., Lander E.S., Zhang F.. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014; 157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Estrela R., Cate J.H.. Energy biotechnology in the CRISPR-Cas9 era. Curr. Opin. Biotechnol. 2016; 38:79–84. [DOI] [PubMed] [Google Scholar]
- 6. Vervoort Y., Linares A.G., Roncoroni M., Liu C., Steensels J., Verstrepen K.J.. High-throughput system-wide engineering and screening for microbial biotechnology. Curr. Opin. Biotechnol. 2017; 46:120–125. [DOI] [PubMed] [Google Scholar]
- 7. Lee J.H., Wendisch V.F.. Production of amino acids—genetic and metabolic engineering approaches. Bioresour. Technol. 2017; 245:1587–1575. [DOI] [PubMed] [Google Scholar]
- 8. DiCarlo J.E., Norville J.E., Mali P., Rios X., Aach J., Church G.M.. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013; 41:4336–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mans R., van Rossum H.M., Wijsman M., Backx A., Kuijpers N.G., van den Broek M., Daran-Lapujade P., Pronk J.T., van Maris A.J., Daran J.M.. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 2015; 15:fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee M.E., DeLoache W.C., Cervantes B., Dueber J.E.. A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth. Biol. 2015; 4:975–986. [DOI] [PubMed] [Google Scholar]
- 11. Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H. et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015; 13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shmakov S., Smargon A., Scott D., Cox D., Pyzocha N., Yan W., Abudayyeh O.O., Gootenberg J.S., Makarova K.S., Wolf Y.I. et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017; 15:169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mohanraju P., Makarova K.S., Zetsche B., Zhang F., Koonin E.V., van der Oost J.. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016; 353:aad5147. [DOI] [PubMed] [Google Scholar]
- 14. Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E.. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011; 471:602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van der Oost J. Beat their swords into ploughshares. Microb. Biotechnol. 2015; 8:34–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deveau H., Barrangou R., Garneau J.E., Labonte J., Fremaux C., Boyaval P., Romero D.A., Horvath P., Moineau S.. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008; 190:1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bolotin A., Quinquis B., Sorokin A., Ehrlich S.D.. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005; 151:2551–2561. [DOI] [PubMed] [Google Scholar]
- 18. Mojica F.J., Diez-Villasenor C., Garcia-Martinez J., Almendros C.. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009; 155:733–740. [DOI] [PubMed] [Google Scholar]
- 19. Bao Z., Xiao H., Liang J., Zhang L., Xiong X., Sun N., Si T., Zhao H.. Homology-integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae. ACS Synth. Biol. 2015; 4:585–594. [DOI] [PubMed] [Google Scholar]
- 20. Deaner M., Mejia J., Alper H.S.. Enabling graded and large-scale multiplex of desired genes using a dual-mode dCas9 activator in Saccharomyces cerevisiae. ACS Synth Biol. 2017; 6:1931–1943. [DOI] [PubMed] [Google Scholar]
- 21. Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015; 163:759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang Y., Qian F., Yang J., Liu Y., Dong F., Xu C., Sun B., Chen B., Xu X., Li Y. et al. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat. Commun. 2017; 8:15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim Y., Cheong S.A., Lee J.G., Lee S.W., Lee M.S., Baek I.J., Sung Y.H.. Generation of knockout mice by Cpf1-mediated gene targeting. Nat. Biotechnol. 2016; 34:808–810. [DOI] [PubMed] [Google Scholar]
- 24. Hur J.K., Kim K., Been K.W., Baek G., Ye S., Hur J.W., Ryu S.M., Lee Y.S., Kim J.S.. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat. Biotechnol. 2016; 34:807–808. [DOI] [PubMed] [Google Scholar]
- 25. Fonfara I., Richter H., Bratovic M., Le Rhun A., Charpentier E.. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016; 532:517–521. [DOI] [PubMed] [Google Scholar]
- 26. Endo A., Masafumi M., Kaya H., Toki S.. Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci. Rep. 2016; 6:38169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015; 163:759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamano T., Nishimasu H., Zetsche B., Hirano H., Slaymaker I.M., Li Y., Fedorova I., Nakane T., Makarova K.S., Koonin E.V. et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell. 2016; 165:949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dong D., Ren K., Qiu X., Zheng J., Guo M., Guan X., Liu H., Li N., Zhang B., Yang D. et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature. 2016; 532:522–526. [DOI] [PubMed] [Google Scholar]
- 30. Stella S., Alcon P., Montoya G.. Structure of the Cpf1 endonuclease R-loop complex after target DNA cleavage. Nature. 2017; 546:559–563. [DOI] [PubMed] [Google Scholar]
- 31. Swarts D.C., van der Oost J., Jinek M.. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol. Cell. 2017; 66:221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim H.K., Song M., Lee J., Menon A.V., Jung S., Kang Y.M., Choi J.W., Woo E., Koh H.C., Nam J.W. et al. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat. Methods. 2017; 14:153–159. [DOI] [PubMed] [Google Scholar]
- 33. Fontana L., Partridge L., Longo V.D.. Extending healthy life span–from yeast to humans. Science. 2010; 328:321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Entian K.D., Kötter P.. Stansfield I, Stark MJR. Yeast Gene Analysis. 2007; 36:2nd edn, Amsterdam: Academic Press, Elsevier; 629–666. [Google Scholar]
- 35. Nijkamp J.F., van den Broek M., Datema E., de K.S., Bosman L., Luttik M.A., Daran-Lapujade P., Vongsangnak W., Nielsen J., Heijne W.H. et al. De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb. Cell Factor. 2012; doi:10.1186/1475-2859-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Verduyn C., Postma E., Scheffers W.A., van Dijken J.P.. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992; 8:501–517. [DOI] [PubMed] [Google Scholar]
- 37. Pronk J.T. Auxotrophic yeast strains in fundamental and applied research. Appl. Environ. Microbiol. 2002; 68:2095–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gietz R.D., Schiestl R.H.. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007; 2:35–37. [DOI] [PubMed] [Google Scholar]
- 39. Looke M., Kristjuhan K., Kristjuhan A.. Extraction of genomic DNA from yeasts for PCR-based applications. Biotechniques. 2011; 50:325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Braglia P., Percudani R., Dieci G.. Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J. Biol. Chem. 2005; 280:19551–19562. [DOI] [PubMed] [Google Scholar]
- 41. Coordinators N.R. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2013; 41:D8–D20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kuijpers N.G., Solis-Escalante D., Bosman L., van den Broek M., Pronk J.T., Daran J.M., Daran-Lapujade P.. A versatile, efficient strategy for assembly of multi-fragment expression vectors in Saccharomyces cerevisiae using 60 bp synthetic recombination sequences. Microb. Cell Fact. 2013; 12:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kuijpers N.G., Chroumpi S., Vos T., Solis-Escalante D., Bosman L., Pronk J.T., Daran J.M., Daran-Lapujade P.. One-step assembly and targeted integration of multigene constructs assisted by the I-SceI meganuclease in Saccharomyces cerevisiae. FEMS Yeast Res. 2013; 13:769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li H., Durbin R.. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010; 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S. . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dorfman B.Z. The isolation of adenylosuccinate synthetase mutants in yeast by selection for constitutive behavior in pigmented strains. Genetics. 1969; 61:377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jakociunas T., Bonde I., Herrgard M., Harrison S.J., Kristensen M., Pedersen L.E., Jensen M.K., Keasling J.D.. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab. Eng. 2015; 28:213–222. [DOI] [PubMed] [Google Scholar]
- 49. Ronda C., Pedersen L.E., Hansen H.G., Kallehauge T.B., Betenbaugh M.J., Nielsen A.T., Kildegaard H.F.. Accelerating genome editing in CHO cells using CRISPR Cas9 and CRISPy, a web-based target finding tool. Biotechnol. Bioeng. 2014; 111:1604–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Labun K., Montague T.G., Gagnon J.A., Thyme S.B., Valen E.. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016; 44:W272–W276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang T., Wei J.J., Sabatini D.M., Lander E.S.. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014; 343:80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zetsche B., Heidenreich M., Mohanraju P., Fedorova I., Kneppers J., DeGennaro E.M., Winblad N., Choudhury S.R., Abudayyeh O.O., Gootenberg J.S. et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 2017; 35:31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ryan O.W., Skerker J.M., Maurer M.J., Li X., Tsai J.C., Poddar S., Lee M.E., DeLoache W., Dueber J.E., Arkin A.P. et al. Selection of chromosomal DNA libraries using a multiplex CRISPR system. Elife. 2014; 3, doi:10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Generoso W.C., Gottardi M., Oreb M., Boles E.. Simplified CRISPR-Cas genome editing for Saccharomyces cerevisiae. J. Microb. Methods. 2016; 127:203–205. [DOI] [PubMed] [Google Scholar]
- 55. Leenay R.T., Maksimchuk K.R., Slotkowski R.A., Agrawal R.N., Gomaa A.A., Briner A.E., Barrangou R., Beisel C.L.. Identifying and visualizing functional PAM diversity across CRISPR-Cas systems. Mol. Cell. 2016; 62:137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang M., Mao Y., Lu Y., Tao X., Zhu J.K.. Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol. Plant. 2017; 10:1011–1013. [DOI] [PubMed] [Google Scholar]
- 57. Kim D., Kim J., Hur J.K., Been K.W., Yoon S.H., Kim J.S.. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016; 34:863–868. [DOI] [PubMed] [Google Scholar]
- 58. Gao L., Cox D.B.T., Yan W.X., Manteiga J.C., Schneider M.W., Yamano T., Nishimasu H., Nureki O., Crosetto N., Zhang F.. Engineered Cpf1 variants with altered PAM specificities. Nat.Biotechnol. 2017; 35:789–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Horwitz A.A., Walter J.M., Schubert M.G., Kung S.H., Hawkins K., Platt D.M., Hernday A.D., Mahatdejkul-Meadows T., Szeto W., Chandran S.S. et al. Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas. Cell Syst. 2015; 1:88–96. [DOI] [PubMed] [Google Scholar]
- 60. Wong A.S., Choi G.C., Cui C.H., Pregernig G., Milani P., Adam M., Perli S.D., Kazer S.W., Gaillard A., Hermann M. et al. Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:2544–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gao Y., Zhao Y.. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J. Integr. Plant Biol. 2014; 56:343–349. [DOI] [PubMed] [Google Scholar]
- 62. Sikorski R.S., Hieter P.. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989; 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.