Abstract
DNA tetrahedron as the simplest 3D DNA nanostructure has been applied widely in biomedicine and biosensing. Herein, we design and fabricate a series of circular assemblies of DNA tetrahedron with high purity and decent yields. These circular nanostructures are confirmed by endonuclease digestion, gel electrophoresis and atomic force microscopy. Inspired by rotary protein motor, we demonstrate these circular architectures can serve as a stator for a rotary DNA motor to achieve the circular rotation. The DNA motor can rotate on the stators for several cycles, and the locomotion of the motor is monitored by the real-time fluorescent measurements.
INTRODUCTION
In biological system, polypeptide subdomains can self-assemble into complex protein motors to achieve lots of functions such as cargo transport, molecular recognition and cell locomotion (1). Inspired by the biological machinery, many artificial molecular motors have been designed and fabricated to mimic the motions of protein motors (2). ATP synthase (3) and the bacterial flagellar motor (4) are two rotary protein machines which achieve rotating torque via chemical fuels. It is still challenging to make synthetic nanomachines to mimic these rotary motors (5).
Recently, DNA nanotechnology where DNA is used as material to construct nanostructures, has shown its power in various nanodevice fabrication (6–12). Several DNA nanomachines has been built to achieve the circular rotating motion (13–22). For example, 180o rotation was achieved either driven by strand displacement reaction (13) or cation triggering (19); a clockwise or anticlockwise walking route was accomplished on a bipedal stepper (14); most recently, a mechanically interlocked nanomotor was constructed to realize random rotation (20). Even though, there is still large space left to further improve the DNA based rotary motor, because the previous designs are relatively complicate, and the controllability of rotating angle/step is low. A minor change in the rotating step need to redesign the whole DNA sequences. In fact, nature is quite smart and already realize the diversity in rotation by assembling different number of subunits with similar functions. Hence, the artificial rotary motor can be fabricated with the similar manner, that is, assembling the building block/subunit with certain size and similar function together, to get an integral nanomachine. Hence, it is crucial to choose a suitable building block/subunit.
DNA tetrahedron, which was first fabricated by Goodman and Turberfield in 2004 (23,24), is one of the simplest 3D nanostructures that is assembled entirely by short oligonucleotides (< 100 nt). DNA tetrahedron has been applied in many fields since it was created due to its unique advantages. For example: (i) The structure is stable enough even inside the cell. Hence, it has been taken as a promising capsule for drug/gene delivery and bioimaging both in vitro and in vivo (25–31). (ii) 3D tetrahedron is structurally rigid. Therefore, it can be used as scaffold to assemble metal nanoparticles or proteins (32–34). Furthermore, DNA tetrahedron has also been used as spacers in biosensors to improve the detection sensitivity (35–39). (iii) DNA tetrahedron is easy to assemble with a high yield and low cost. Four oligonucleotide strands that are achievable on automated DNA synthesis and a simple one-pot annealing step is facile enough to form the structure quantitatively (23,24,40). (iv) The tetrahedron can be easily modified both on the vertex and the edge for different functionalities (41,42). The pioneer works show that DNA tetrahedron could be an ideal building block for constructing higher-order structures (43,44). Wang et al. achieved tetrahedron dimer and trimer through the hybridization of single strands extended from the vertexes (43). Focke's group also assembled a dimer structure by loop hybridization (DNA-DNA kissing strategy) (44). However, advanced assemblies with DNA tetrahedron as subunit have not been achieved yet.
In this work, we explored the possibility of using DNA tetrahedron to construct complex nanostructures. A series of circular assemblies were constructed via a stepwise self-assembly strategy. These circular nanostructures were validated by endonuclease digestion, gel electrophoresis and atomic force microscopy (AFM). Moreover, a rotary DNA motor was achieved by assembling four tetrahedrons with different track strands into a circular stator. The locomotion of the motor was monitored by the real-time fluorescent measurement.
MATERIALS AND METHODS
Materials
DNA strands were purchased from Sangon (Shanghai, China), and the sequences are listed in the supporting information. Tris(hydroxymethyl)aminomethane hydrochloride (Tris) was purchased from Bio-Rad (Singapore); MgCl2 were from Quality Reagent Chemical (Singapore); EDTA were from USB biochemical (USA); agarose was purchased from Bio-Rad (Singapore); 100bp DNA ladder was from Thermo Fisher (Singapore); Endonuclease MspI and BspHI were purchased from New England Biolabs (Singapore). Ultrapure water with 18.2 MΩ·cm was used in all experiments.
Apparatus
UV absorbance was measured on a Shimadzu UV-1800 spectrophotometer. Size exclusion chromatography (SEC) was run on Shimadzu UFLC system. Agarose gel was run on the Bio-Rad horizontal electrophoresis system (Wide Mini-Sub Cell GT Cell). AFM images were obtained on a Bruker Multimode 8 SPM equipped with a liquid cell. DLS were undertaken on Malvern Zetasizer Nano ZSP instrument. Real time fluorescent spectrum was obtained on Shimadzu RF-5301 Spectrofluorophotometer with the software Labsolutions RF.
Construction of single tetrahedron
The DNA strand was dissolved in ultrapure H2O to make a stock solution, and the concentration was determined with UV absorbance at 260 nm. Four strands with equal molar quantities were mixed in TEM buffer (10 mM Tris–HCl, pH 8.0, 1mM EDTA, 20 mM MgCl2) to a concentration of 100 nM for each strand. The aforementioned assembly solutions were kept at 95 °C for 10 mins, and then cooled down to room temperature in atmosphere for 2 h. The tetrahedrons were characterized on 6% native polyacrylamide gel electrophoresis (PAGE).
Construction of circular and lattice structures
The tetrahedrons constructed above were used directly without further purification. The concentration for each tetrahedron was taken as 100 nM.
Circular dimer
The two monomer T11 and T1’1’ were mixed with equal molar quantity, e.g. 100 nM × 200 μl, and incubated overnight at room temperature. The crude product was ready for further purification and characterization.
Circular trimer
200 μl T12 were mixed with 200 μl T12’ and incubated overnight at room temperature to form a dimer first. Then the dimer was further hybridized with 200 μl T1’1’ for another 12 h to get the final product. The crude product was ready for further purification and characterization.
Circular tetramer
200 μl T12 were hybridized with 200 μl T12’ overnight to make a Dimer 1. At the same time, 200 μl T1’2 were hybridized with 200 μl T1’2’ to make a Dimer 2. The final circular tetramer was obtained by mixing and incubating Dimers 1 and 2 for another 12 h. The crude product was ready for further purification and characterization.
Circular pentamer
200 μl T12’ were hybridized with 200 μl T1’2’ overnight to make a dimer. At the same time, 400 μl T1’2 were hybridized with 200 μl T11 to make a trimer. The dimer and trimer were purified by SEC first, and then mixed with equal molar quantity and incubated for another 12 h to get the final circular pentamer.
Circular hexamer:
400 μl T1’2 were hybridized with 200 μl T11 overnight to make a Trimer 1. At the same time, 400 μl T1’2’ were hybridized with 200 μl T11 to make a Trimer 2. The Trimers 1 and 2 were purified by SEC first, and then mixed with equal molar quantity and incubated for another 12 h to get the final circular hexamer.
Lattice hexamer
The structure was fabricated by a circular tetramer and a linear dimer. The tetramer was constructed as the step descripted above with four tetrahedron T12, T12’, T1’2-O, T1’2’-O. Here, the name T1’2-O and T1’2’-O indicates that one more overhang is extended from the S2 strand of T1’2 and T1’2’ for further hybridization with the dimer. The dimer was formed by mixing T2-O’ and T2’-O’. The circular tetramer and dimer were purified by SEC, and then mixed with equal molar quantity and incubated for another 12 h to get the final lattice hexamer.
Lattice octamer
The structure was made of two circular tetramers, 1 and 2. The tetramer was constructed as the step descripted above. Tetramer 1 was assembled with four tetrahedron T12, T12’, T1’2-O, T1’2’-O, and tetramer 2 was fabricated with T12, T12’, T1’2-O’, T1’2’-O’. After SEC purification, the two tetramers were mixed with equal molar quantity and incubated for another 12 h to get the final lattice octamer.
SEC purification and characterization
The SEC column (Phenomenex, BioSep-SEC-S 3000 300 × 7.8 mm) was used and the chromatograph was monitored at 260 nm. 50–500 μl of sample as-prepared or concentrated by Amicon Ultra-0.5 ml Centrifugal Filters-100K was injected into the UFLC system, and washed under isocratic elution in mobile phase (10 mM Tris–HCl, pH 7.3, 450 mM NaCl) with a flow rate of 0.5 ml/min. The target product peak was collected and concentrated by the Amicon Ultra-0.5 ml Centrifugal Filters-100K at 12 000 g to a desired volume. The yield of the product was calculated by integrating the peak area.
Endonuclease digestion experiment
The circular dimer, trimer and tetramer incorporated with the digestion site were purified by SEC and dispersed to NEBuffer 4 (50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, 1 mM DTT, pH 7.9) by buffer exchanging with Amicon Ultra-0.5 ml Centrifugal Filters-100K. 50 μl of DNA sample (10 ng/μl) was incubated with 0.4 μl of BSA (Bovine Serum Albumin, 20 mg/ml), 2 μl of MspI (20, 000 units/ml), or BspHI (10 000 units/ml) or both of two enzymes at 37°C for 1 h. 3 μl SDS (sodium dodecyl sulfate, 5 wt%) was added to the sample which was treated with BspHI to disrupt binding. The enzyme digested samples were loaded onto 2.5% agarose gel directly for further analysis.
AFM characterization of different structures
Freshly cleaved mica was treated with 0.1% (v/v) APTES ((3-Aminopropyl) triethoxysilane) aqueous solution for 10 min to make the surface positively charged, and then washed with 2 ml ultrapure water and dried by compress nitrogen. 40 μl of SEC purified DNA structure with a concentration of 2 nM in TEM buffer was dropped onto the mica surface and incubated for 5 min. Then the sample was ready for measurement under the ScanAsyst in fluid mode with the ScanAsyst fluid+ probe (Bruker, Singapore).
Real time fluorescent monitoring of the rotary DNA motor
Before the experiment, the four components (stator, rotor, fuel strand and anti-fuel strand) were treated as following: The stator (circular tetramer) was purified by SEC and concentrated to 100 nM in TEM buffer. Two strands of rotor were mixed in TEM buffer to make a partial duplex structure with a concentration of 20 μM. The fuel strands (F1–F4) and anti-fuel strands (A1–A4) were dissolved into TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8.0) to make a 40 μM stock solution.
The experiment was carried out at room temperature. The excitation and emission slit were set to be 3 and 5 nm respectively, and the excitation/emission wavelength for Texas red, FAM, Cy5 and HEX were set to be 590/611, 490/520, 640/662 and 530/555 nm respectively. 100 μl of purified stator was transferred to a cuvette, and then 0.5 μl of rotor was added. Then the real-time fluorescent monitoring at four different channels started. Two minutes later, 0.275 μl of fuel strands and anti-fuel strands were added successively and mixed by rapid pipetting. Two rotation cycles were achieved during 8 h of monitoring.
RESULTS
Design principle of the circular assemblies of DNA tetrahedrons
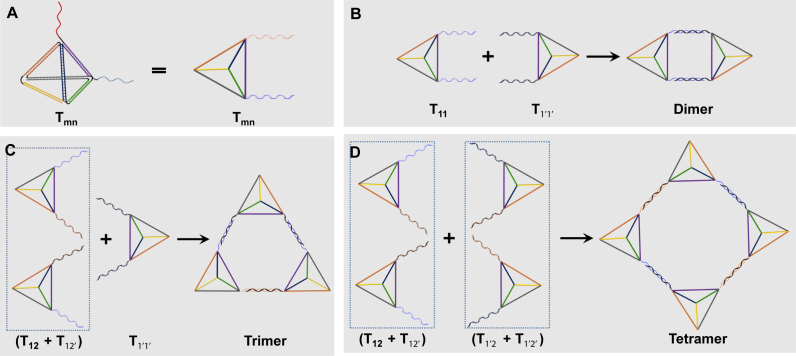
The tetrahedron monomers were designed according to the literature (23,24): each tetrahedron was made of 4 single DNA strands (S1-S4), with 20 base pairs in each edge. In order to make higher-order circular structures, desired overhangs located at the vertex point with 17-base long are extended from the same end of each strands. Two bases of the overhang sequence adjacent to the tetrahedron vertex are used as spacers, and the rest 15 bases are to hybridize with the overhangs on other tetrahedrons. The numbers and locations of overhangs can be further adjusted based upon the design of nanostructures. In principle, eight homo or hetero overhangs can be introduced to one tetrahedron, making the design of more diverse structures possible. In this work, at least overhangs on two different vertexes of each tetrahedron are required for constructing the circular assemblies. Here, the tetrahedrons are named as Tmn, with m and n representing the two overhangs of different sequences (Figure 1A, Supplementary Tables S1 and S2).
Figure 1.
The schematic illustration of the circular structure design with the DNA tetrahedron as the building blocks. (A) The tetrahedron monomer (Tmn) with two overhang sequences (m, n) extended from two vertices, respectively. (B–D) The self-assembly procedures for circular dimer, trimer and tetramer by using tetrahedrons with different overhangs.
For all the nanostructures in this work, a universal tetrahedron core was used for all Tmn, and only the 17-base overhangs need to be redesigned according to the higher-order nanostructures. Hence, the sequence design is significantly simplified and special computer software is usually not required. In order to involve as few overhang sequences as possible, a stepwise assembly method was used. Only two counterparts participated in each step to achieve high assembly yield and simplify the purification. For example, there will be three steps to form a tetramer. Step 1: Different building blocks with two overhangs each, Tmn, are annealed with high yield. Step 2: Two linear dimers were formed by two Tmn with one pair of complementary overhangs. Step 3: Tetramers were assembled by the two linear dimers via second pair of overhangs (Figure 1D). In this case, only two pairs of overhangs (1, 1’, 2 and 2’) are needed for the construction of a series of circular assemblies. Overhangs n and n’ are complementary to each other. The stepwise assembly processes of various circular Tmn architectures were illustrated in Figure 1 and Supplementary Figure S1, and the detailed assembly procedures are shown in the MATERIAL AND METHODS Section.
Construction and characterization of the circular dimer, trimer and tetramer
In order to assemble the dimer, trimer and tetramer, six tetrahedrons named T11, T1’1’, T12, T12’, T1’2 and T1’2’ were assembled with the strand design in Supplementary Tables S1 and S2. The tetrahedrons were characterized by polyacrylamide gel electrophoresis (PAGE) (Supplementary Figure S2) and size exclusion chromatography (SEC) (Figure 2A, red curve). Both characterizations showed the high purity of the desired Tmn (>95%), indicating that they can be used directly for the following circular structure construction.
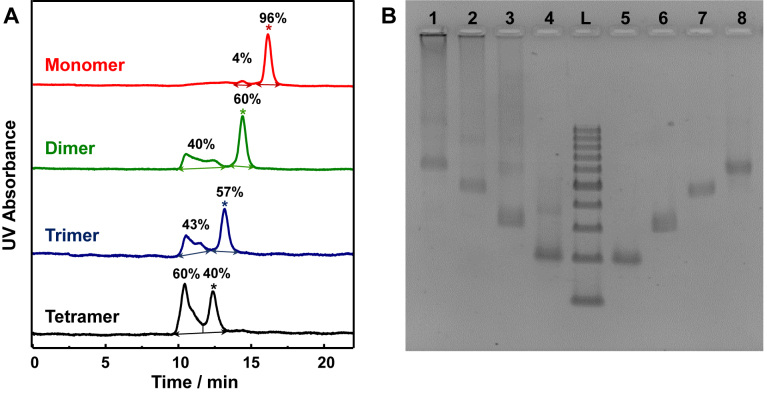
Figure 2.
SEC and 2.5% agarose gel electrophoresis of the tetrahedron monomer, dimer, trimer and tetramer. (A) The SEC profile of tetrahedron monomer (red line), dimer (green line), trimer (blue line) and tetramer (black line). The peaks of target products are labeled with asterisk, and the yields are indicated for each peak. (B) The 2.5% agarose gel electrophoresis of the tetrahedron monomer, dimer, trimer, and tetramer before and after SEC purification. Lane L: DNA ladder with 100–1000 base pairs; Lane 1–4: tetramer, trimer, dimer and monomer before purification; Lanes 5–8: monomer, dimer, trimer and tetramer respectively after SEC purification.
Circular dimer, trimer and tetramer were assembled according to the aforementioned method (Figure 1). By using the agarose gel electrophoresis (Supplementary Figure S3), target bands can be observed in the crude products. The mobility of these products perfectly matched with the theoretical size of each entity (141, 282, 423 and 564 base pairs for the tetrahedron monomer, dimer, trimer and tetramer, respectively) according to the 100–1000 bp DNA ladder. However, some larger byproducts existed in the crudes, which were shown as the smeared band behind the desired products in the electrophoresis. Therefore, further purification was applied to obtain pure circular structures.
Herein, SEC was introduced to purify the crude products (40,43). There are two main peaks on each chromatogram: a narrow peak with longer retention time and a broad peak with shorter retention time (Figure 2A). The narrower peak was identified by the agarose gel electrophoresis to be the target product with retention time of 16.2, 14.4, 13.2 and 12.4 min respectively from tetrahedron monomer to tetramer. The broad peaks were the large byproducts. After SEC separation, the target products were pure enough to show only one band on gel electrophoresis (Figure 2B). These results suggest the high purity of oligomeric nanostructures can be obtained.
The yields of each entity (60%, 57% and 40% for dimer, trimer and tetramer respectively) were calculated by integrating the peak area on SEC chromatogram. The slow decline in the assembly yields of these tetrahedron oligomers implied the assembly was a kinetic controlled process. Taken tetramer as an example, at the initial reaction state, the two linear dimers, T12 + T12’ and T1’2 + T1’2’, are supposed to firstly linked by one pair of overhang to form an open-state tetramer structure, which can further hybridize by another pair of overhang intramolecularly to form a circular structure, or intermolecularly with another linear dimer or open-state tetramer to form larger polymerized structures. The decrease of the yield from dimer to tetramer showed the competition between the intramolecular cyclization and intermolecular elongation. When the structure is small, like a dimer, the intramolecular hybridization is predominant because the distance between the two terminal tetrahedrons are close enough and the special orientations of vertex overhangs in the linear open structure favor ring closing assembly. In the case of larger circular assemblies, the distance between two terminal tetrahedrons in the linear open-state are extending spatially. As a result, intramolecular collision becomes less competitive versus intermolecular hybridization. To prove the hypothesis, the fraction of byproduct in tetramer was collected and characterized by AFM (Supplementary Figure S4). Larger aggregates composed of undefined high number of tetrahedrons were observed as expected. Based upon the kinetic control hypothesis, the intramolecular product (the circular structure) would be favored in a further diluted reaction solution. The idea was tested on tetramer structure. The yield of the circular tetramer can be increased to 49% from 40% once the concentration of two linear dimers were diluted for 2 folds (Supplementary Figure S5).
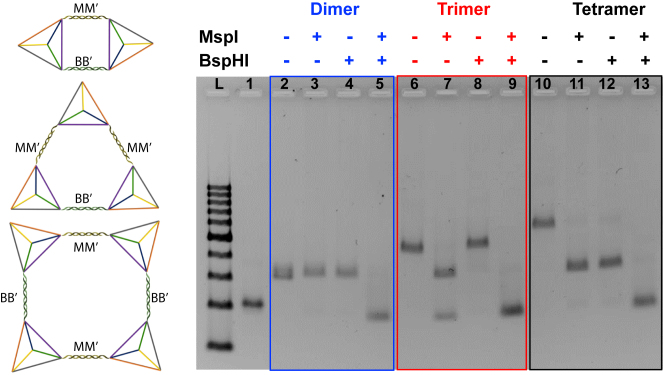
Although the assembly structures with desirable number of tetrahedron units were obtained, the circular assembly state needs to be further confirmed. Hence, endonuclease digestion was carried out in each entity to interrogate the complete hybridization of all the overhang pairs to close the ring structure. The overhangs of tetrahedron monomers were redesigned to incorporate the restriction sites of two endonucleases MspI (M) and BspHI (B). Five new tetrahedrons were prepared and named as TMB, TMB’, TM’B, TM’B’ and TM’M’, respectively. Herein, dimer was formed by mixing TMB and TM’B’ (Figure 3, left). The products after endonuclease digestion were analyzed by the agarose gel electrophoresis. The mobility of the structures remained almost the same as the starting dimer after the treatment of any endonuclease alone, indicating the dimer structure was remained after any one of the two linkages was already cleaved. The dimer can be digested into monomer only in the presence of both MspI and BspHI (blue frame in Figure 3). This result confirmed two linkages formed between TMB and TM’B’, and was consistent with our circular design. The trimer was made of (TMB + TMB’) + TM’M’ with the three linkages as two MM’ and one BB’ according to the design. After digesting by MspI, there were two bands on the electrophoresis gel which belonged to a dimer and a monomer, while only one band corresponding to a trimer structure appeared if only BspHI was used. In the presence of both MspI and BspHI, monomer band can be obtained exclusively because all linkages in the circular trimer were cleaved (red frame in Figure 3). The tetramer was composed of (TMB + TMB’) + (TM’B + TM’B’), and there should be two MM’ and two BB’ linkages in the parallel positions respectively. Only dimer structures were obtained after digested with either MspI or BspHI alone, while monomer was the only digestive product after both endonucleases were applied (black frame in Figure 3). All the observations above can be obtained only if the target assemblies are circular structure as designed. Hence, the enzyme digestion results confirm the circular assembly of the oligomers in solution.
Figure 3.
Schematic illustration (left) and 2.5% agarose gel electrophoresis (right) for endonuclease digestion of the circular dimer, trimer and tetramer with MspI and BspHI. Lane L: DNA ladder with 100–1000 base pairs. Lane 1: tetrahedron monomer as a control; Lane 2: dimer without any enzyme; lane 3: dimer digested with MspI; lane 4: dimer digested with BspHI; lane 5: dimer digested by both of MspI and BspHI; lane 6: trimer without any enzyme; lane 7: trimer digested with MspI; lane 8: trimer digested with BspHI; lane 9: trimer digested by both of MspI and BspHI; lane 10: tetramer without any enzyme; lane 11: tetramer digested with MspI; lane 12: tetramer digested with BspHI; lane 13: tetramer digested by both of MspI and BspHI.
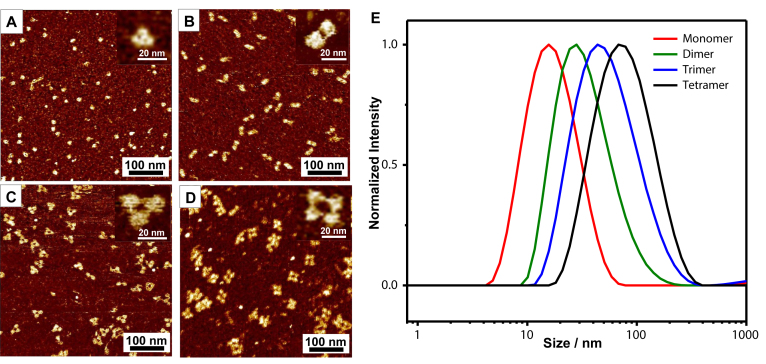
The morphologies of each assembly were further characterized by fluid mode AFM. The monomer was presented as small dots with a uniform size (Figure 4A). The high-resolution image in the inset showed a tetrahedron with the edge length of 11.9 ± 0.5 nm (Figure 4A and Supplementary Figure S6A). The size was larger than the theoretical value of 6.8 nm due to the broadening effect of the AFM tips (43). The dimers were shown as a short rod, and the two overhang linkages can be visualized in the amplified image. The lengths of the rod in short axial was 12.2 ± 1.1 nm, which was similar to the edge length of a single tetrahedron. The long axial was 28.4 ± 1.4 nm and was consistent with the length sum of two tetrahedral and one linkage (Figure 4B and Supplementary Figure S6B). An equilateral triangle structure can be clearly observed in AFM image of a circular trimer, with three DNA tetrahedrons distribute spatially equal at the vertex. The triangle was 26.8 ± 1.3 nm (Figure 4C and Supplementary Figure S6C) in edge length. In the image of the circular tetramer, the four tetrahedrons arranged into a square or rhombus shape as expected, with an edge length of 28.4 ± 1.5 nm (Figure 4D and Supplementary Figure S6D). The edge lengths of dimer, trimer and tetramer were similar as the tetrahedral building blocks used the universal core and the same lengths for all the overhangs. These AFM images further confirmed that circular structures were successfully assembled according to the design. The sizes of the assemblies in solution were also measured by dynamic light scattering (DLS). The diameters of monomer, dimer, trimer and tetramer were 18.6, 38.2, 62.3 and 87.2 nm respectively (Figure 4E). It was not surprising that the sizes measured by DLS was larger than those measured by AFM, because the hydration layer wrapped around the assemblies must be taken into consideration for the structural sizes in solution.
Figure 4.
AFM and DLS analysis of the tetrahedron assemblies. (A–D) The fluid mode AFM images of the tetrahedron monomer (A), dimer (B), trimer (C) and tetramer (D). The inset is the amplified single structure. (E) The DLS profile of the tetrahedron monomer (red), dimer (green), trimer (blue) and tetramer (black).
Construction and characterization of assemblies with more than four tetrahedron units
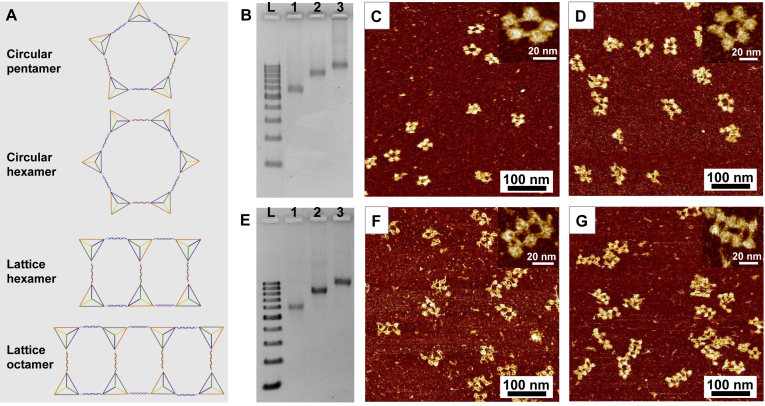
After constructing the circular dimer, trimer and tetramer successfully, much larger and more complex assemblies were further fabricated by using the DNA tetrahedron as building blocks. The assembly of circular pentamer and hexamer were attempted (Figure 5A). Following the stepwise assembly principle, the pentamer was constructed by hybridizing a linear dimer and trimer (Supplementary Figure S1A), and the hexamer was fabricated by using two linear trimers (Supplementary Figure S1B). On the agarose gel electrophoresis, the pentamer and hexamer showed reasonable mobility according to both circular tetramer and the DNA ladder (Figure 5B). AFM images indicated most of the pentamer stayed in a pentagonal shape, with DNA tetrahedron located at each vertex (Figure 5C). In case of the circular hexamer, although some hexagons can be observed in the AFM image (Figure 5D), hexamers with irregular shape were also detected. The various topologies of hexamer were due to the increasing structural flexibility as the circular structure possessed a larger ring size. Hence, circular assemblies with more than six units in a single ring and a defined uniform topology may not be able to achieve.
Figure 5.
AFM and 2.5% agarose gel electrophoresis of the circular pentamer, hexamer and the lattice hexamer and octamer. (A) The schematic illustration of the larger circular structures. (B) 2.5% agarose gel electrophoresis of circular pentamer and hexamer. Lane L: DNA ladder with 100–1000 base pairs. Lane 1: circular tetramer as a control; Lane 2 and 3: circular pentamer and hexamer respectively. (C and D) The fluid mode AFM images of circular pentamer and hexamer. The inset is the amplified single structure. (E) 2.5% agarose gel electrophoresis of lattice hexamer and octamer. Lane L: DNA ladder with 100–1000 base pairs. Lane 1: circular tetramer as a control; Lane 2 and 3: lattice hexamer and octamer respectively. (F and G) The fluid mode AFM images of lattice hexamer and octamer. The inset is the amplified single structure.
In order to assemble nanostructures with larger numbers of tetrahedron units and rigid 3D confirmations, we try to extend the circular tetramers in a lattice pattern. Therefore, the lattice structures with six and eight building blocks were fabricated, named as lattice hexamer and lattice octamer. The lattice hexamer was constructed by a circular tetramer and a linear dimer, while the lattice octamer was assembled by two circular tetramers (Figure 5A, Supplementary Figure S1C and S1D). One more overhang was introduced into the tetrahedron at the three-way vertex in the lattice structures. The band mobility confirmed the purity and the size of the nanolattice by comparing to the DNA ladder and the circular tetramer structure (Figure 5E). The morphology of 2 × 3 and 2 × 4 lattices can be visualized by AFM (Figure 5F and G). Thus, the lattice design with tetrameric ring as block unit can afford a more defined 3D topology of large assemblies than single ring design. These results suggest that larger and more complex structures can be constructed by self-assembly of DNA tetrahedron.
Circular tetramer as a stator in the rotary DNA motor
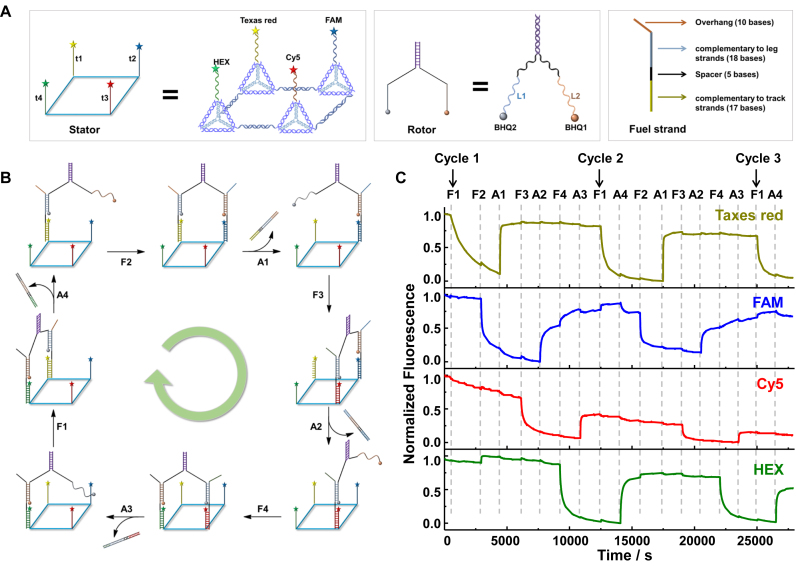
Inspired by the rotary protein motor, a dynamic rotary DNA motor system was designed with the circular tetramer as the stator as shown in Figure 6. There are four components in the rotary motor: a circular stator (CS), a rotor (R), fuel strands (F1-F4) and anti-fuel strands (A1-A4) (Figure 6A and B). The stator is a circular tetramer constructed by four different DNA tetrahedrons, T12-t1, TM’2’-t2, TMB-t3 and T1’B’-t4. Here, t1 to t4 were four track strands extend from the 5′ or 3′ end of S1 alternatively in T12, TM’2’, TMB and T1’B’. Four pairs of overhangs were used to fabricate the circular tetramer in order to assemble the four track strands in the order of t1→t2→t3→t4. To track the motion of DNA rotor via real-time fluorescence, the four track strands (t1–t4) were labeled with Texas red, FAM, Cy5 and HEX respectively at the strand terminal. To construct the circular stator, two tetrahedron dimers (T12-t1+TM’2’-t2) and (TMB-t3+T1’B’-t4) were assembled first, and then mixed together to assemble the target architecture. The stator was purified by SEC and further characterized by AFM (Supplementary Figure S7). The rotor, similar as the linear walker developed by Shin and Pierce (45), was composed of two partially hybridized DNA strands, with two single-stranded legs (L1 and L2). Two quencher BHQ2 and BHQ1 were labeled at the sing-stranded end of L1 and L2, respectively. The fuel strands (F1–F4) are 50-base single-stranded oligonucleotides with four functional parts: a 17-base fragment to hybridize with the track strand on the stator, an 18-base fragment to hybridize with the leg of the rotor, a 5-base spacer between the 17-base and 18-base sequence and a 10-base overhang for the strand exchange reaction with the anti-fuel strands. Anti-fuel strands are completely complementary to the corresponding fuel strands.
Figure 6.
The design and locomotion of the rotary DNA motor. (A) The schematic illustration of the components of the motor: stator, rotor and fuel strand. (B) The schematic locomotion of the rotary DNA motor and (C) the real-time fluorescent monitoring at four different channels. The excitation/ emission wavelengths for Texas red, FAM, Cy5 and HEX were 590/611, 490/520, 640/662 and 530/555 nm respectively.
The rotation of the motor system was triggered by the addition of fuel strands and anti-fuel strands successively (Figure 6B). The real-time locomotion of the motor system was monitored by the quenching of fluorochromes at different vertex positions of the stator (Figure 6C). One leg on the rotor (L1) was first anchored on the tract t1 of the stator by adding the fuel strand, F1. In this case, the quencher BHQ2 on the rotor was close enough to induce significant fluorescence attenuation of Texas red labeled on t1. Second step, F2 was added to walk L2 onto track t2. Similarly, a decrease on the fluorescent emission of FAM on track t2 was observed due to the quenching by BHQ1 on L2. Third step to free L1 from t1, A1 was added to pull off F1 by a strand displacement reaction, along with the production of an F1–A1 duplex as a waste. At the same time, the fluorescence of Texas red increased rapidly. Fourth step to walk rotor to position 3 on the stator, the addition of F3 trigger the movement of L1 to track t3, and the motor system rotated forward to the diagonal position. In the following steps, A2, F4 and A3 were added step by step to drive the rotor walk along the stator in a circular fashion, accompanied with the corresponding fluctuation on the fluorescent intensity of FAM, Cy5 and HEX. After F1 was added, the rotor entered the second cycle of rotation on the tetra-vertex stator (Figure 6C). The fluorescence of chromophores on the four vertexes were attenuated and regenerated in the same pattern as in the first cycle. The fluorescent intensity of all the xanthene dyes (Texas Red, FAM and HEX) remained without significant attenuation over several cycles. Even though Cy5 have experienced apparent photo bleaching, near full fluorescence recovery at the rest three tracks on stator indicated the successful operation of the rotary motor (Figure 6C). Here, two rotation cycles were achieved during the monitoring period. In principle, the motor can keep moving if the fuel strands and anti-fuel strands are continuously supplied.
DISCUSSION
The successful assembly of these circular structures lies in two aspects. First, the high purity and yield of the tetrahedron monomers must be guaranteed. The previous work showed that the tetrahedron building blocks need to be purified in advance to achieve a high yield in linear assembly (43). We found in this work that the tetrahedron can be used directly for further assembly. The key point lies in the concentration of the DNA stands applied for the tetrahedron formation. A relatively high concentration (such as 1 μM) could lead to higher amounts of byproducts than the desired tetrahedron. While in our work, a 100 nM annealing concentration made the tetrahedron pure enough (>95%) for direct higher-order structure assembling. While the stock solution with high concentration can be readily prepared via centrifuge filtration technique. Second, a stepwise assembly strategy was adopted. Hence, the number of overhang sequence was significantly reduced. Only the overhangs used in the same step were required to be different in the sequences, which allowed the recycle of overhang sequences and tetrahedral blocks in other assembly step and simplified the design of a complicate structure. For instance, only two pairs of overhang were used to achieve the nanostructure as large as a circular hexamer. One more pair overhang made the successful construction of the lattice structure possible.
The applicable property of the circular architectures was tested on a dynamic rotary motor system. The tetramer was taken as the stator in the present work, and two rotation circles were achieved. Due to the well-defined architectures of these circular assemblies, the relative distance and spatial arrangements between each tetrahedron are fixed. More remarkably, the sizes, steps and walking patterns of the rotary motor can be highly manipulated by the present assembly strategy because the number and position of the tetrahedron are readily controllable. In fact, by sophisticated design of the track sequences, different walking routes can be achieved on the same stator, such as a ‘Z’ path vs the ‘⋄’ path in the present work. The lattice structures can be also used as stators to achieve multiple walking routes, e.g. ‘S’ or ‘8’ shape. Furthermore, with adoption of functional DNA sequences to the tracks, besides the fuel strands, our stator could also be powered by and/or respond to multiple stimuli module, like those in a reported DNA stepper system with circular DNA as the stator (14). Whereas, the structural flexibility and the potential uncertainty on the distance and dynamic conformation of the walking route due to the single stranded circular DNA can be overcome in our stators made of DNA tetrahedron here. The circular rotating motion offered the promising potentials to mimic the rotary motions in many essential protein machineries. Though the central shaft was still missing in the present motor, the assembling manner with DNA tetrahedron as the subunit as well as the stepwise circular walking has shown the similarity as the proton channel subunits in ATP synthase and bacterial flagellar motor.
In conclusion, tetrahedron as a simple 3D DNA nanostructure built by oligonucleotide strands, was shown to be an ideal building block for constructing self-assembled architectures with well-defined 2D/3D configuration on nanometer-scale. A series of circular tetrahedron assemblies, from dimer to hexamer, and even complicate structures like lattice hexamer and octamer, were fabricated successfully. All of the architectures were well characterized by SEC, gel electrophoresis, enzyme digestion, AFM and DLS experiments. The highlight of using DNA tetrahedron as the building block lies in simple design, easy-preparation, facile assembly and high yields. Higher-order structures can be obtained easily by only reprogramming short overhang sequences, without the needs to redesign the sequences of the entire tetrahedron units. As a proof-of-concept, a rotary DNA motor system was successfully designed by using the circular tetramer as the stator. The DNA rotor could walk stepwise following a circular track. This work offered an indispensable missing piece in the library of DNA nanomotors and enriched the functionality of biomimetic nanomachinery. From this example, it can be envisioned that more complicate motions of biological machinery can be mimicked by DNA nanodevices with complex assembly architecture. This work could inspire a breakthrough in the construction and functions of the rotary DNA motor and extend the biomolecular motors to broad fields, such as templated chemical synthesis (46,47). Furthermore, the facile assembly of DNA tetrahedrons to well-defined architectures would show the promising potentials beyond DNA nanotechnology to drug delivery, bioimaging and other biomedical technologies.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
AcRF MOE Tier 1 [M4011554]. Funding for open access charge: AcRF MOE Tier 1 [M4011554].
Conflict of interest statement. None declared.
REFERENCES
- 1. Schliwa M., Woehlke G.. Molecular motors. Nature. 2003; 422:759–765. [DOI] [PubMed] [Google Scholar]
- 2. Erbas-Cakmak S., Leigh D.A., McTernan C.T., Nussbaumer A.L.. Artificial molecular machines. Chem. Rev. 2015; 115:10081–10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yoshida M., Muneyuki E., Hisabori T.. ATP synthase - a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2001; 2:669–677. [DOI] [PubMed] [Google Scholar]
- 4. Minamino T., Imada K., Namba K.. Molecular motors of the bacterial flagella. Curr. Opin. Struct. Biol. 2008; 18:693–701. [DOI] [PubMed] [Google Scholar]
- 5. Koumura N., Zijlstra R.W.J., van Delden R.A., Harada N., Feringa B.L.. Light-driven monodirectional molecular rotor. Nature. 1999; 401:152–155. [DOI] [PubMed] [Google Scholar]
- 6. Zhou C., Yang Z.Q., Liu D.S.. Reversible regulation of protein binding affinity by a DNA machine. J. Am. Chem. Soc. 2012; 134:1416–1418. [DOI] [PubMed] [Google Scholar]
- 7. Yurke B., Turberfield A.J., Mills A.P., Simmel F.C., Neumann J.L.. A DNA-fuelled molecular machine made of DNA. Nature. 2000; 406:605–608. [DOI] [PubMed] [Google Scholar]
- 8. Pan J., Li F.R., Cha T.G., Chen H.R., Choi J.H.. Recent progress on DNA based walkers. Curr. Opin. Biotechnol. 2015; 34:56–64. [DOI] [PubMed] [Google Scholar]
- 9. Mao C., Sun W., Shen Z., Seeman N.C.. A nanomechanical device based on the B-Z transition of DNA. Nature. 1999; 397:144–146. [DOI] [PubMed] [Google Scholar]
- 10. Lund K., Manzo A.J., Dabby N., Michelotti N., Johnson-Buck A., Nangreave J., Taylor S., Pei R., Stojanovic M.N., Walter N.G. et al. . Molecular robots guided by prescriptive landscapes. Nature. 2010; 465:206–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu D.S., Cheng E.J., Yang Z.Q.. DNA-based switchable devices and materials. NPG Asia Mater. 2011; 3:109–114. [Google Scholar]
- 12. Gu H.Z., Chao J., Xiao S.J., Seeman N.C.. A proximity-based programmable DNA nanoscale assembly line. Nature. 2010; 465:202–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yan H., Zhang X.P., Shen Z.Y., Seeman N.C.. A robust DNA mechanical device controlled by hybridization topology. Nature. 2002; 415:62–65. [DOI] [PubMed] [Google Scholar]
- 14. Wang Z.G., Elbaz J., Willner I.. DNA Machines: bipedal Walker and Stepper. Nano Lett. 2011; 11:304–309. [DOI] [PubMed] [Google Scholar]
- 15. Elbaz J., Wang Z.-G., Wang F., Willner I.. Programmed dynamic topologies in DNA catenanes. Angew. Chem. Int. Ed. 2012; 51:2349–2353. [DOI] [PubMed] [Google Scholar]
- 16. Lu C.H., Cecconello A., Elbaz J., Credi A., Willner I.. A three-station DNA catenane rotary motor with controlled directionality. Nano Lett. 2013; 13:2303–2308. [DOI] [PubMed] [Google Scholar]
- 17. Lu C.H., Cecconello A., Willner I.. Recent advances in the synthesis and functions of reconfigurable interlocked DNA nanostructures. J. Am. Chem. Soc. 2016; 138:5172–5185. [DOI] [PubMed] [Google Scholar]
- 18. Lohmann F., Ackermann D., Famulok M.. Reversible light switch for macrocycle mobility in a DNA rotaxane. J. Am. Chem. Soc. 2012; 134:11884–11887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rajendran A., Endo M., Hidaka K., Sugiyama H.. Direct and real-time observation of rotary movement of a DNA nanomechanical device. J. Am. Chem. Soc. 2013; 135:1117–1123. [DOI] [PubMed] [Google Scholar]
- 20. Ketterer P., Willner E.M., Dietz H.. Nanoscale rotary apparatus formed from tight-fitting 3D DNA components. Sci. Adv. 2016; 2:e1501209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tomaru T., Suzuki Y., Kawamata I., Nomura S.-I.M., Murata S.. Stepping operation of a rotary DNA origami device. Chem. Commun. 2017; 53:7716–7719. [DOI] [PubMed] [Google Scholar]
- 22. Yang Y., Tashiro R., Suzuki Y., Emura T., Hidaka K., Sugiyama H., Endo M.. A photoregulated DNA-bsed rotary system and direct observation of its rotational movement. Chem. Eur. J. 2017; 23:3979–3985. [DOI] [PubMed] [Google Scholar]
- 23. Goodman R.P., Berry R.M., Turberfield A.J.. The single-step synthesis of a DNA tetrahedron. Chem. Commun. 2004; 1372–1373. [DOI] [PubMed] [Google Scholar]
- 24. Goodman R.P., Schaap I.A.T., Tardin C.F., Erben C.M., Berry R.M., Schmidt C.F., Turberfield A.J.. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science. 2005; 310:1661–1665. [DOI] [PubMed] [Google Scholar]
- 25. Li J., Pei H., Zhu B., Liang L., Wei M., He Y., Chen N., Li D., Huang Q., Fan C.H.. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano. 2011; 5:8783–8789. [DOI] [PubMed] [Google Scholar]
- 26. Walsh A.S., Yin H.F., Erben C.M., Wood M.J.A., Turberfield A.J.. DNA cage dlivery to mammalian cells. ACS Nano. 2011; 5:5427–5432. [DOI] [PubMed] [Google Scholar]
- 27. Lee H., Lytton-Jean A.K.R., Chen Y., Love K.T., Park A.I., Karagiannis E.D., Sehgal A., Querbes W., Zurenko C.S., Jayaraman M. et al. . Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotech. 2012; 7:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim K.R., Kim D.R., Lee T., Yhee J.Y., Kim B.S., Kwon I.C., Ahn D.R.. Drug delivery by a self-assembled DNA tetrahedron for overcoming drug resistance in breast cancer cells. Chem. Commun. 2013; 49:2010–2012. [DOI] [PubMed] [Google Scholar]
- 29. Kim K.R., Lee Y.D., Lee T., Kim B.S., Kim S., Ahn D.R.. Sentinel lymph node imaging by a fluorescently labeled DNA tetrahedron. Biomaterials. 2013; 34:5226–5235. [DOI] [PubMed] [Google Scholar]
- 30. Tay C.Y., Yuan L., Leong D.T.. Nature-Inspired DNA Nanosensor for Real-Time in Situ Detection of mRNA in Living Cells. ACS Nano. 2015; 9:5609–5617. [DOI] [PubMed] [Google Scholar]
- 31. Peng Q., Shao X.R., Xie J., Shi S.R., Wei X.Q., Zhang T., Cai X.X., Lin Y.F.. Understanding the biomedical effects of the self-assembled tetrahedral DNA nanostructure on living cells. ACS Appl. Mater. Interfaces. 2016; 8:12733–12739. [DOI] [PubMed] [Google Scholar]
- 32. Erben C.M., Goodman R.P., Turberfield A.J.. Single-molecule protein encapsulation in a rigid DNA cage. Angew. Chem. Int. Ed. 2006; 45:7414–7417. [DOI] [PubMed] [Google Scholar]
- 33. Mastroianni A.J., Claridge S.A., Alivisatos A.P.. Pyramidal and chiral groupings of gold nanocrystals assembled using DNA scaffolds. J. Am. Chem. Soc. 2009; 131:8455–8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Crawford R., Erben C.M., Periz J., Hall L.M., Brown T., Turberfield A.J., Kapanidis A.N.. Non-covalent single transcription factor encapsulation inside a DNA cage. Angew. Chem. Int. Ed. 2013; 52:2284–2288. [DOI] [PubMed] [Google Scholar]
- 35. Mitchell N., Schlapak R., Kastner M., Armitage D., Chrzanowski W., Riener J., Hinterdorfer P., Ebner A., Howorka S.. A DNA nanostructure for the functional assembly of chemical groups with tunable stoichiometry and defined nanoscale geometry. Angew. Chem. Int. Ed. 2009; 48:525–527. [DOI] [PubMed] [Google Scholar]
- 36. Pei H., Lu N., Wen Y.L., Song S.P., Liu Y., Yan H., Fan C.H.. A DNA nanostructure-based biomolecular probe carrier platform for electrochemical biosensing. Adv. Mater. 2010; 22:4754–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Z.G., Xue Q.W., Tian W.Z., Wang L., Jiang W.. Quantitative detection of single DNA molecules on DNA tetrahedron decorated substrates. Chem. Commun. 2012; 48:9661–9663. [DOI] [PubMed] [Google Scholar]
- 38. Lin M.H., Wang J.J., Zhou G.B., Wang J.B., Wu N., Lu J.X., Gao J.M., Chen X.Q., Shi J.Y., Zuo X.L. et al. . Programmable engineering of a biosensing interface with tetrahedral DNA nanostructures for ultrasensitive DNA detection. Angew. Chem. Int. Ed. 2015; 54:2151–2155. [DOI] [PubMed] [Google Scholar]
- 39. Liu Y.J., Wei M., Liu X., Wei W., Zhao H., Zhang Y.J., Liu S.Q.. Label-free ultrasensitive detection of telomerase activity via multiple telomeric hemin/G-quadruplex triggered polyaniline deposition and a DNA tetrahedron-structure regulated signal. Chem. Commun. 2016; 52:1796–1799. [DOI] [PubMed] [Google Scholar]
- 40. Rendek K.N., Fromme R., Grotjohann I., Fromme P.. Crystallization of a self-assembled three-dimensional DNA nanostructure. Acta Cryst. 2013; F69:141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilks T.R., Bath J., de Vries J.W., Raymond J.E., Herrmann A., Turberfield A.J., O’Reilly R.K.. “Giant Surfactants” created by the fast and efficient functionalization of a DNA tetrahedron with a temperature-responsive polymer. ACS Nano. 2013; 7:8561–8572. [DOI] [PubMed] [Google Scholar]
- 42. Li J., Hong C.Y., Wu S.X., Liang H., Wang L.P., Huang G.M., Chen X., Yang H.H., Shangguan D.H., Tan W.H.. Facile phase transfer and surface biofunctionalization of hydrophobic nanoparticles using janus DNA tetrahedron nanostructures. J. Am. Chem. Soc. 2015; 137:11210–11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xing S., Jiang D.W., Li F., Li J., Li Q., Huang Q., Guo L.J., Xia J.Y., Shi J.Y., Fan C.H. et al. . Constructing higher-order DNA nanoarchitectures with highly purified DNA nanocages. ACS Appl. Mater. Interfaces. 2015; 7:13174–13179. [DOI] [PubMed] [Google Scholar]
- 44. Barth A., Kobbe D., Focke M.. DNA-DNA kissing complexes as a new tool for the assembly of DNA nanostructures. Nucleic Acids Res. 2016; 44:1502–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shin J.S., Pierce N.A.. A synthetic DNA walker for molecular transport. J. Am. Chem. Soc. 2004; 126:10834–10835. [DOI] [PubMed] [Google Scholar]
- 46. He Y., Liu D.R.. Autonomous multistep organic synthesis in a single isothermal solution mediated by a DNA walker. Nat. Nanotech. 2010; 5:778–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Meng W.J., Muscat R.A., McKee M.L., Milnes P.J., El-Sagheer A.H., Bath J., Davis B.G., Brown T., O’Reilly R.K., Turberfield A.J.. An autonomous molecular assembler for programmable chemical synthesis. Nat. Chem. 2016; 8:542–548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.