Abstract
The coordinated control of Ca2+ signaling is essential for development in eukaryotes. Cyclic nucleotide-gated channel (CNGC) family members mediate Ca2+ influx from cellular stores in plants (Charpentier et al., 2016; Gao et al., 2016; Frietsch et al., 2007; Urquhart et al., 2007). Here, we report the unusual genetic behavior of a quantitative gain-of-function CNGC mutation (brush) in Lotus japonicus resulting in a leaky tetrameric channel. brush resides in a cluster of redundant CNGCs encoding subunits which resemble metazoan voltage-gated potassium (Kv1-Kv4) channels in assembly and gating properties. The recessive mongenic brush mutation impaired root development and infection by nitrogen-fixing rhizobia. The brush allele exhibited quantitative behavior since overexpression of the cluster subunits was required to suppress the brush phenotype. The results reveal a mechanism by which quantitative competition between channel subunits for tetramer assembly can impact the phenotype of the mutation carrier.
Research organism: A. thaliana, E. coli, S. cerevisiae, Xenopus
eLife digest
Plants constantly monitor and respond to changes in their environment. Central to this surveillance system is the movement of calcium ions into and out of cells. Calcium ions are normally kept at very low levels inside of cells and subtle changes in these levels relay information about the external environment. In the case of plant roots, changes in the concentration of calcium ions herald essential information about soil conditions and the presence of microorganisms, and in turn trigger appropriate responses.
Calcium ion signals are essential for peas, beans and other members of the legume family to form close relationships (known as symbioses) with soil bacteria called rhizobia. As such, many studies of calcium signalling have focused on root symbioses, particularly in a model legume called Lotus japonicus. Previous studies have identified one mutant version of this plant, called brush, which develops abnormal roots with brush-like arrays of root hairs near the tip. The brush mutant was also unable to form a symbiosis with rhizobia, and structures that allow the bacteria to enter the plant stopped developing before they were complete. However, the gene responsible had not been identified.
Chiasson, Haage et al. set out to identify the responsible mutation. At first the brush mutation escaped identification because a key experiment gave an unexpected result. The introduction of a normal, or wild type, copy of the proposed gene – referred to as BRUSH – into the brush mutant did not correct the problems with its roots. Further analysis revealed that it was actually the ratio between BRUSH and brush expression levels that was critical for determining how the plant’s roots developed.
The mutation in brush causes a small change in a protein belonging to the CNGC family. These proteins act as channels and allow ions to move across cell membranes. Further experiments found that the channel formed by the mutated CNGC protein is leaky and allows calcium ions to enter the cell in the absence of any cue from the environment. The leaky entry of calcium ions likely confuses the plant’s surveillance system, which disturbs the normal development of the root. It is also likely that the brush mutation’s effects on calcium signaling also interfere with the entry of rhizobia into the roots. These findings provide important insights into the function of CNGCs and reveal how a small change in a channel protein can have far reaching effects on an organism.
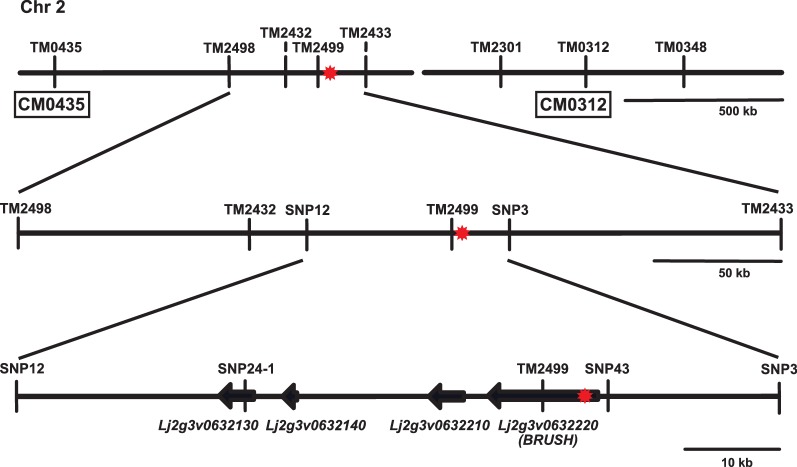
The legume-rhizobium symbiosis offers an excellent model system to study the role of Ca2+ signaling in eukaryotic cell development. Rhizobia produce lipochitooligosaccharides (LCOs) which stimulate signal transduction processes involving not only oscillations of [Ca2+] in the nucleus and perinuclear region but also rapid influx of calcium ions into the cytoplasm of legume root hairs (Felle et al., 1999; Cardenas et al., 1999; Ehrhardt et al., 1996; Harris et al., 2003; Miwa et al., 2006), preceeding rhizobial entry and organ development. The Lotus japonicus mutant brush was previously isolated in a screen of an ethyl-methanesulfonate (EMS)-mutagenised population for plants defective in symbiotic cell development (Maekawa-Yoshikawa et al., 2009). At 26°C, brush roots are stunted and root hair infection threads do not progress into the root cortex, resulting in the formation of non-infected (‘empty’) nodules. The evidence suggested that the recessive mutation was negatively interfering with infection thread progression and cell expansion in the root apical meristem. The brush mutation was mapped to the short arm of chromosome 2 at 8.8 cM (Maekawa-Yoshikawa et al., 2009), linked to the marker TM0312. Subsequently, a large-scale recombinant screen for fine-mapping was undertaken. In total, 20 of 1148 tested F2 individuals showed recombination events between the flanking markers TM2432 and TM0348 (Figure 1—figure supplement 1). F2 genotyping and subsequent F3 phenotyping refined the target region to 37 kb. One EMS-induced mutation was detected in the first exon of BRUSH, a predicted CNGC of unknown function. Because the brush mutant phenotype could not be complemented with the genomic region containing BRUSH including its native promoter (see below), we searched for additional possible missed mutations. The genome of brush was sequenced and aligned with the reference genome. Within the already delineated target interval the mutation in BRUSH was confirmed and no additional polymorphisms relative to the Gifu wild-type were detected.
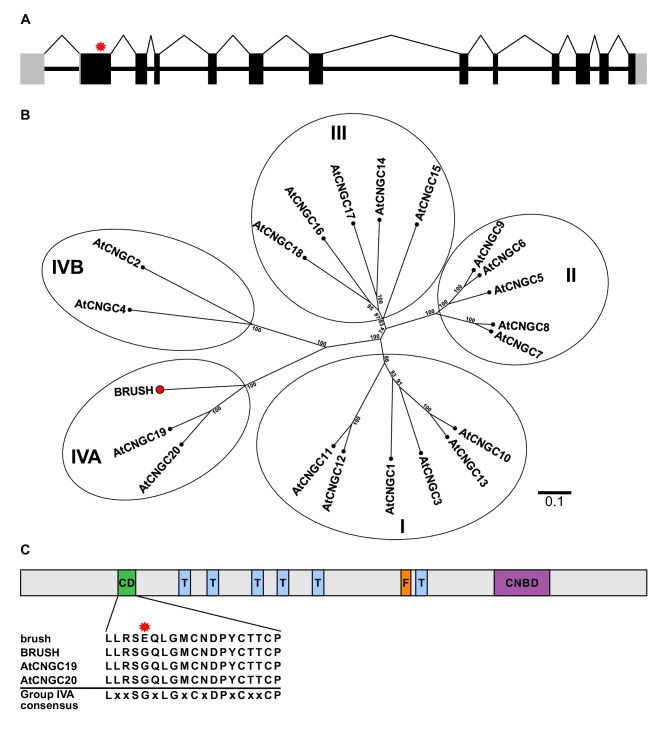
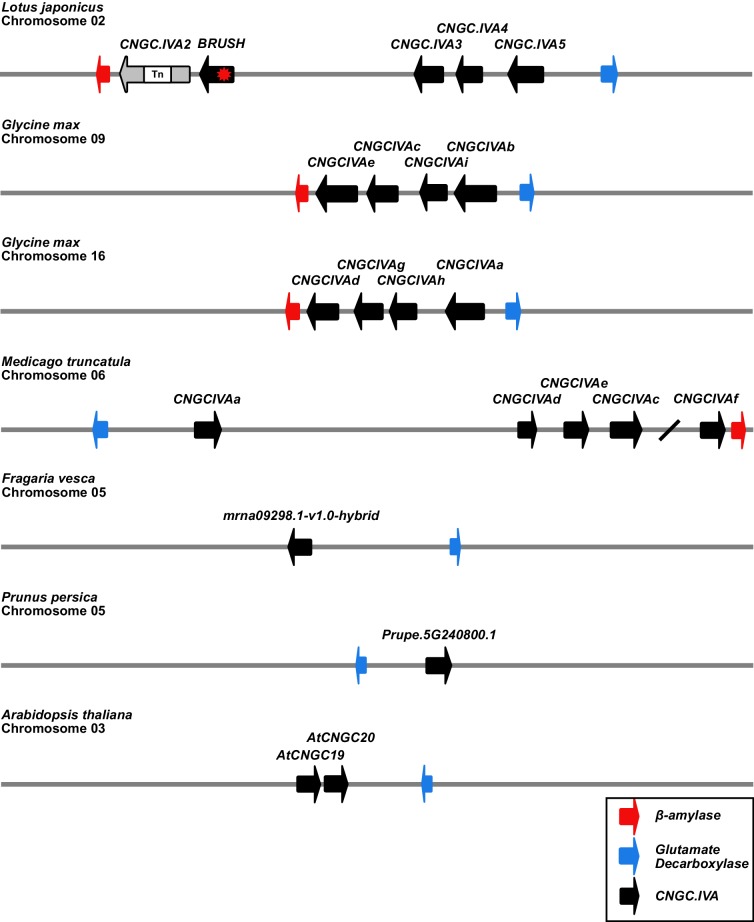
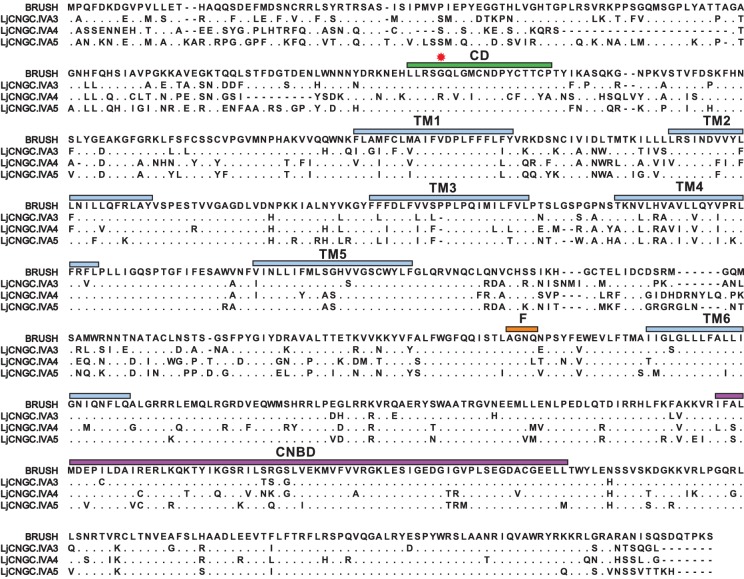
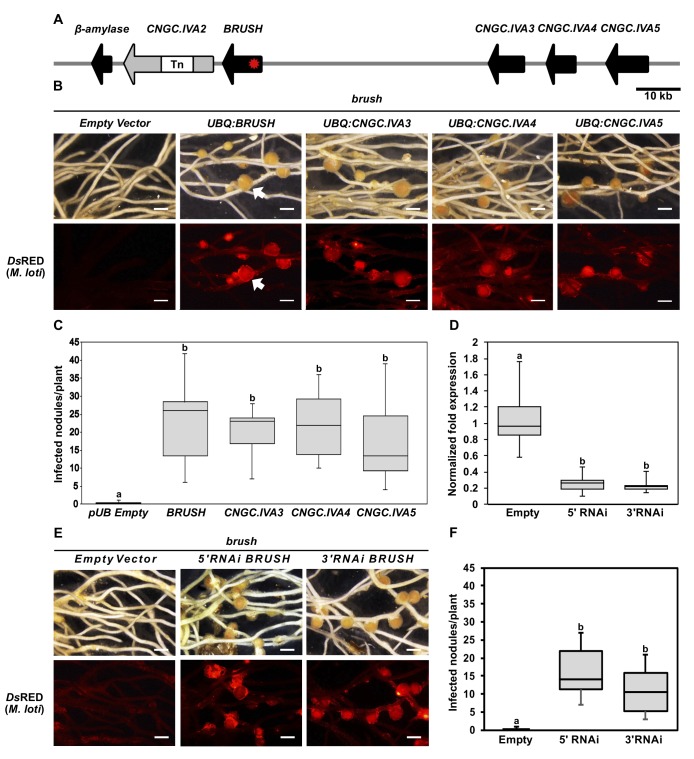
The predicted genomic sequence of BRUSH carried a guanine to adenine (G401A) transition in the first exon in brush (Figure 1A). Amplification of brush cDNA revealed an open-reading frame encoding a protein containing 773 amino acids with an amino acid exchange from glycine to glutamic acid (G134E). The genome of the model plant Arabidopsis thaliana contains 20 predicted CNGC genes, which can be classified into four distinct groups (I, II, III, IV) (Mäser et al., 2001). Phylogenetic and synteny analysis revealed that BRUSH (CNGC.IVA1) is orthologous to the Group IVA members AtCNGC19 and AtCNGC20 (Figure 1B). Similar to other Group IVA CNGCs, BRUSH contains a relatively long N-terminal extension followed by six predicted transmembrane domains and a cyclic nucleotide-binding domain (Figure 1C). The brush mutation is located in a conserved region previously identified as a putative sorting signal in Group IVA CNGCs (Yuen and Christopher, 2013). Sequencing coupled with gene prediction of the brush target region revealed that BRUSH resides in a cluster containing five CNGC loci (Figure 1—figure supplement 2). Analysis of syntenic genomic regions in legumes and non-legumes revealed that the CNGC.IVA cluster expansion occurred early in the legume lineage and was retained. Transcripts for three of the loci could be amplified (CNGC.IVA3, CNGC.IVA4, CNGC.IVA5) and encode proteins which are closely related to BRUSH (Figure 1—figure supplement 3). No transcript was detected for CNGC.IVA2 which contains a large transposon insertion in the seventh intron (Figure 2A).
Figure 1. brush contains a point mutation in CNGC.IVA1 (BRUSH).
(A) Schematic of the intron-exon structure of BRUSH highlighting the brush mutation in the first exon (red asterisk). (B) Phylogenetic tree of BRUSH (red node end) in relation to Arabidopsis thaliana CNGC proteins. BRUSH is orthologous to the Group IVA members AtCNGC19 and AtCNGC20. (C) Overview of the BRUSH protein domain structure highlighting the conserved Group IVA domain (CD, green), transmembrane domains (T, light blue), putative filter region (F, orange), and the predicted cyclic nucleotide-binding domain (CNBD, purple). Shown below is the CD sequence in brush (G134E mutation, asterisk) relative to BRUSH, AtCNGC19, and AtCNGC20 and the Group IVA CNGC consensus (Yuen and Christopher, 2013). Numbers at the branch points in (B) indicate the percentage bootstrap values (100 iterations) for the inferred tree. Scale bar in (B) indicates the number of amino acid substitutions per site.
Figure 1—figure supplement 1. Map-based cloning of the brush mutation on chromosome 2.
Figure 1—figure supplement 2. Syntenic chromosomal locations of CNGC.IVA genes in selected plant species.
Figure 1—figure supplement 3. Protein sequence comparison of Group IVA CNGCs from Lotus japonicus.
Figure 2. Genetic complementation of brush.
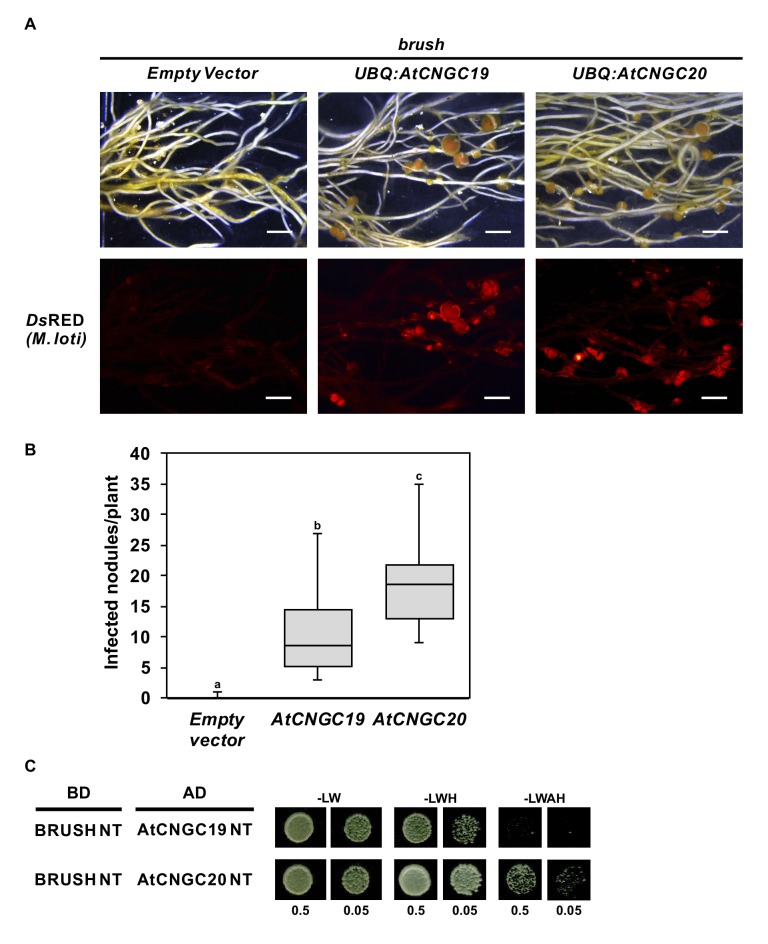
(A) Genomic region surrounding BRUSH on L. japonicus chromosome 2 showing a cluster of five CNGC.IVA genes (red asterisk, brush causative mutation). CNGC.IVA2 contains a large transposon (Tn) insertion. CNGC.IVA2 contains a large transposon insertion (shown as a gap). (B) Complementation assay of brush roots by overexpression (ubiquitin promoter, UBQ) of the four expressed CNGC.IVA cluster members (bright field, top panel). The presence of red fluorescent nodules (arrow) colonized by Mesorhizobium loti expressing the red fluorescent protein DsRED (lower panel) indicates successful bacterial infection and thus complementation. (C) Number of nodules per transformed plant from (B) (n = 10 for all constructs). (D) Quantitative reverse-transcription PCR analysis of brush transcript levels after RNAi targeting either the 5'UTR or 3' UTR of brush. The normalized fold expression of brush is shown relative to empty vector control roots (n = 6 for all constructs). (E) Complementation analysis of brush expressing RNAi fragments targeting either the 5'UTR or 3'UTR of brush in the brush mutant. Panels are the same as (B). (F) Number of nodules per transformed plant (n = 10 for all constructs) from (E). Roots for both complementation experiments were observed 6 weeks after inoculation with rhizobia. Scale bars in (B) and (E) represent 1 mm. Letters in (C), (D), and (F) indicate different statistical groups (ANOVA followed by Tukey’s HSD test). F(4, 45)=10.86, p < 0.001 (C), F(2, 15)=20.82, p < 0.001 (D), F(2, 27)=22.72, p < 0.001 (F).
Figure 2—figure supplement 1. Expression of the BRUSH native genomic sequence in brush.
Figure 2—figure supplement 2. Complementation of brush by overexpression of BRUSH.
Figure 2—figure supplement 3. RNAi off-target controls and phenotypes associated with either overexpression of brush or a null BRUSH allele.
Figure 2—figure supplement 4. Complementation of brush by overexpressing AtCNGC19 or AtCNGC20.
Figure 2—figure supplement 5. CNGC.IVA gene expression in Lotus japonicus roots after rhizobial inoculation.
Figure 2—figure supplement 6. BRUSH expression in roots during nodulation.
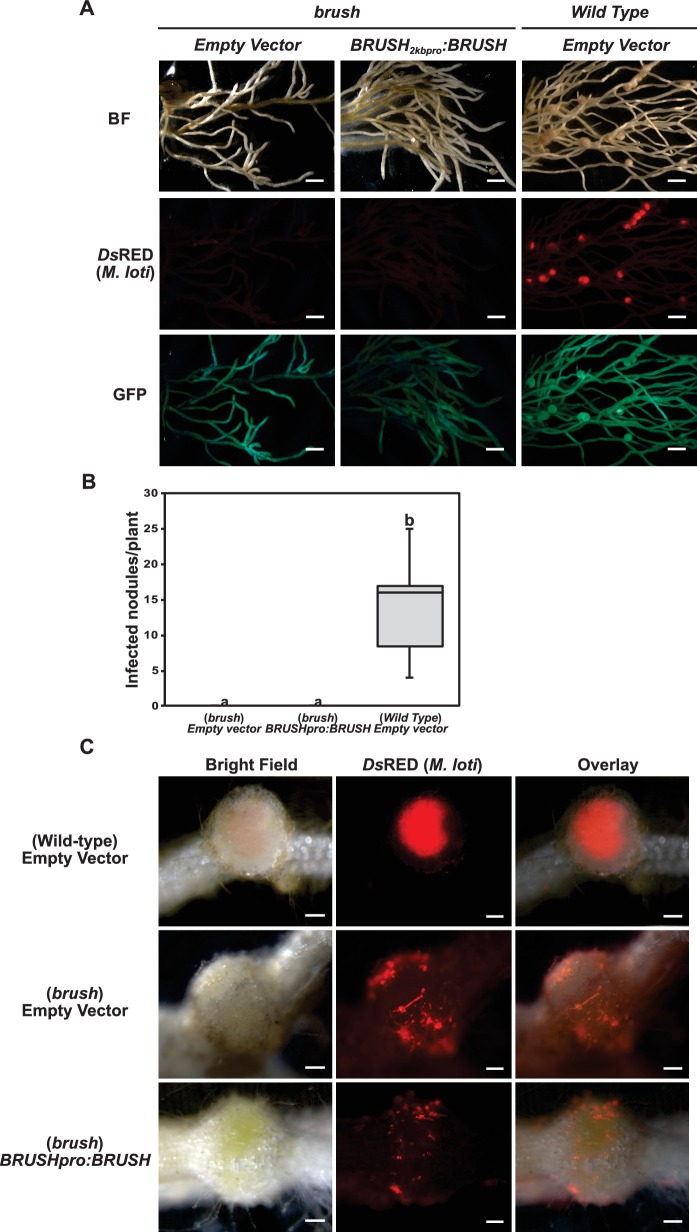
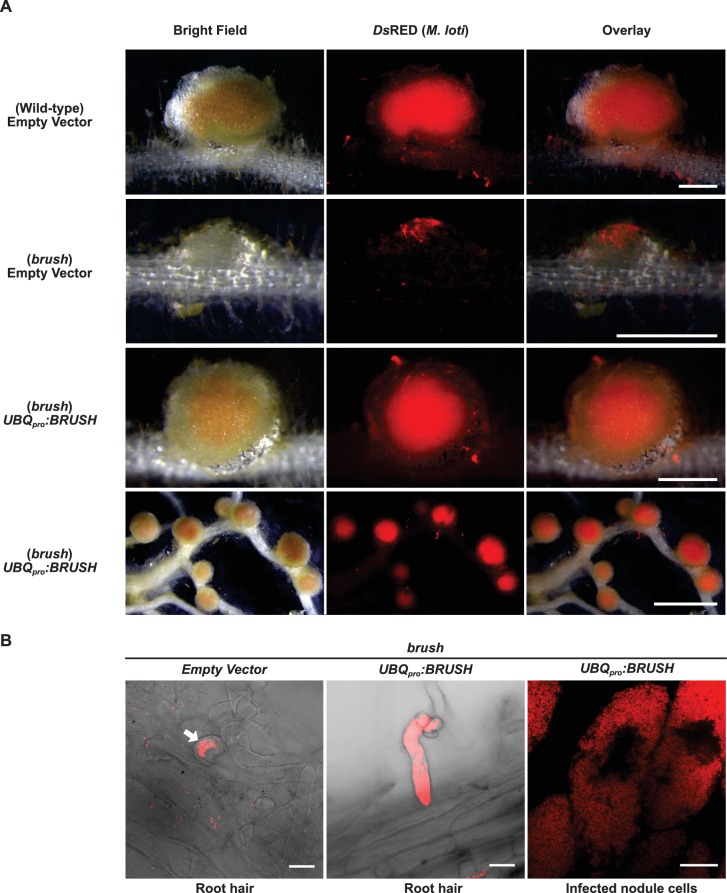
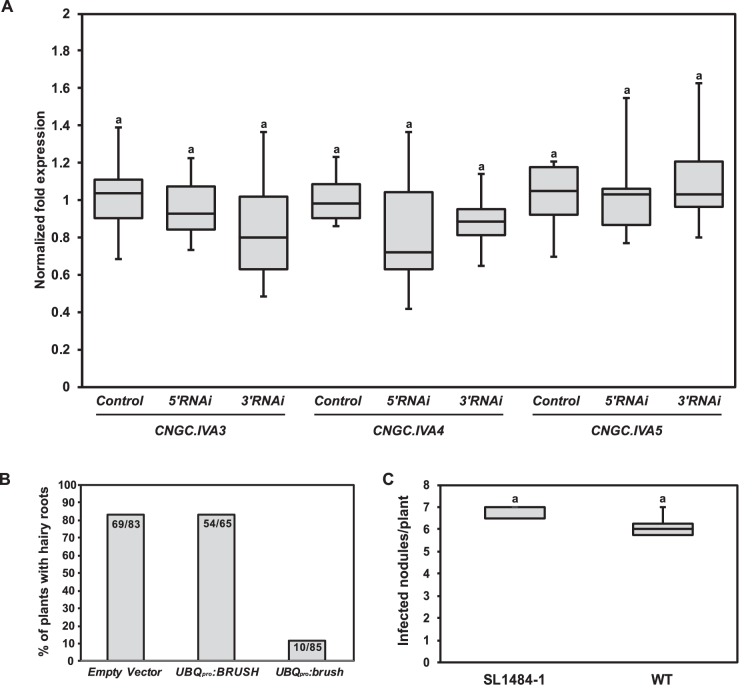
To confirm that brush carried the causative mutation, the BRUSH genomic sequence was expressed in brush transgenic hairy roots driven by its native 2 kb promoter. Surprisingly, we did not observe rescue of either the root or infection thread phenotype (Figure 2—figure supplement 1A–C). However, when brush was transformed with the BRUSH genomic sequence driven by the L. japonicus constitutive polyubiquitin promoter, we observed restoration of the root and infection thread phenotypes, including infected nodules (Figure 2B,C and Figure 2—figure supplement 2A,B). These results suggested that expression level of brush is critical for phenotype manifestation. To further analyze the relationship between brush expression levels and the observed phenotypes, we generated RNA interference (RNAi) constructs to target the untranslated regions (3’UTR or 5’UTR) of the brush transcript in the brush mutant. Transformation of each RNAi construct specifically silenced brush (Figure 2D, Figure 2—figure supplement 3A) and restored rhizobial infection of root cells (Figure 2E,F). We then overexpressed the brush allele in wild-type Gifu hairy roots to recapitulate the brush phenotype and observed that ectopic overexpression of brush impaired hairy root emergence (Figure 2—figure supplement 3B). Collectively, these results suggest that the expression level of brush is critical for the observed phenotypes and that the phenotypic penetrance of the allele appears to be dosage-dependent. An EMS mutant (SL1484-1) was then obtained by TILLING (Perry et al., 2009; Perry et al., 2003) containing a point mutation (W119stop) early in the BRUSH open-reading frame. Analysis of homozygous mutant plants did not reveal any phenotypic root or infection abnormalities after inoculation with rhizobia (Figure 2—figure supplement 3C). The finding that the null mutant of BRUSH is not recapitulating the brush phenotype indicates that brush is an interfering allele and that the loss of BRUSH is compensated by potential redundancy of other CNGCs within the cluster.
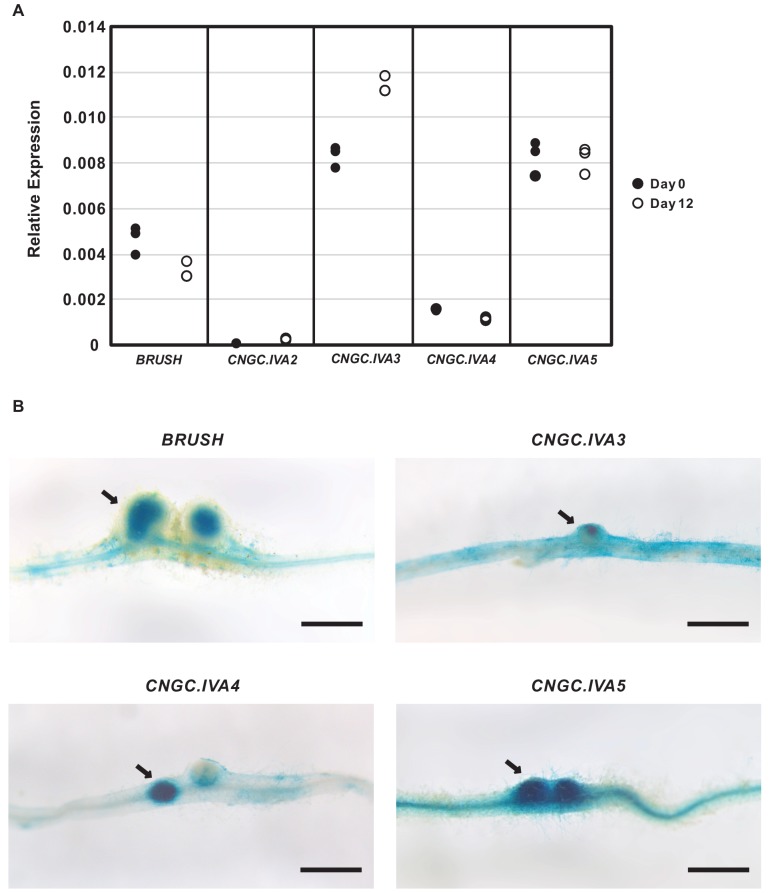
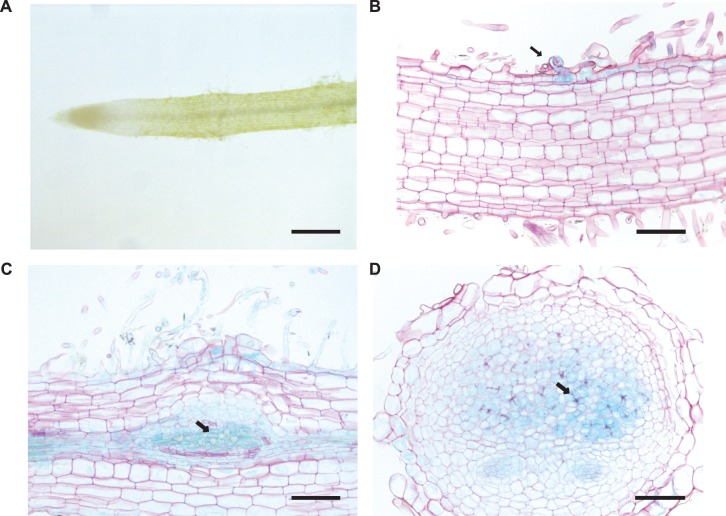
To determine if the other CNGC.IVA cluster genes are redundant with respect to BRUSH, we overexpressed CNGC.IVA3, CNGC.IVA4, CNGC.IVA5 in the brush mutant by hairy root transformation. Analysis of transgenic roots revealed that expression of each gene complemented brush, as evidenced by colonized root nodules (Figure 2B,C). Further, we found that overexpression of the predicted Arabidopsis orthologs AtCNGC19, or AtCNGC20 in brush also restored nodulation (Figure 2—figure supplement 4A,B). To assess if each L. japonicus gene is expressed in roots, RT-qPCR was performed before and after inoculation with rhizobia. Transcripts were detected for BRUSH, CNGC.IVA3, CNGC.IVA4, and CNGC.IVA5, the levels of which did not show significant changes (<2 fold) after inoculation with the rhizobial symbiont (Figure 2—figure supplement 5A). Spatial expression analysis of promoter:β-glucuronidase (GUS) fusions revealed BRUSH expression in root hairs and developing nodules after inoculation with rhizobia and that CNGC.IVA3, CNGC.IVA4, and CNGC.IVA5 are expressed in similar domains (Figure 2—figure supplement 5B). Closer investigation of the BRUSHpromoter:GUS activity revealed a lack of expression in roots prior to inoculation and subsequent activity associated with infected root hairs and nodule primordia after inoculation (Figure 2—figure supplement 6). The overlapping expression pattern of the Group IVA CNGCs together with their ability to dampen the brush phenotype indicate that these genes are redundant.
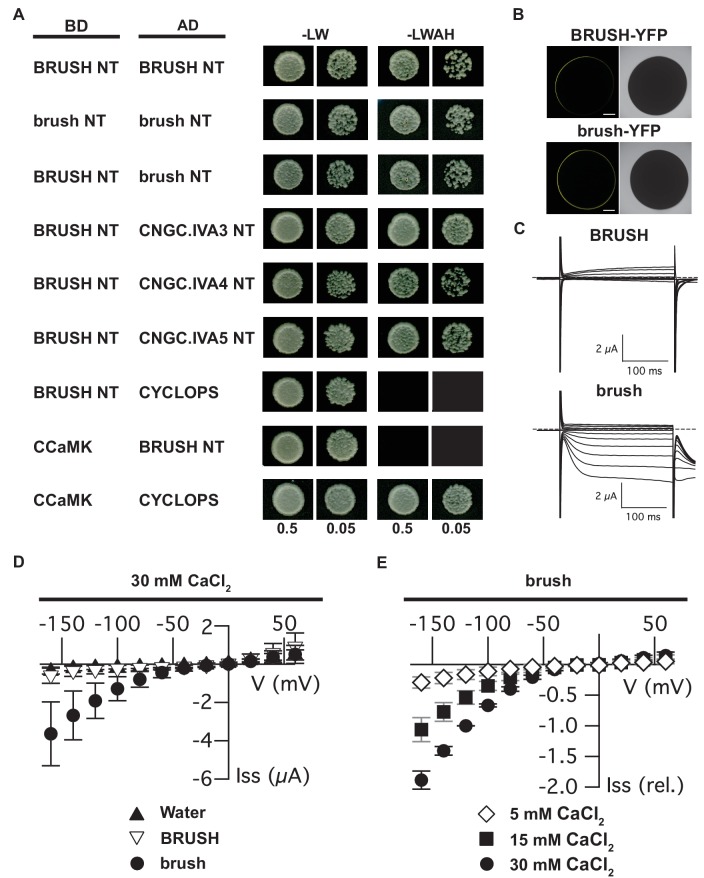
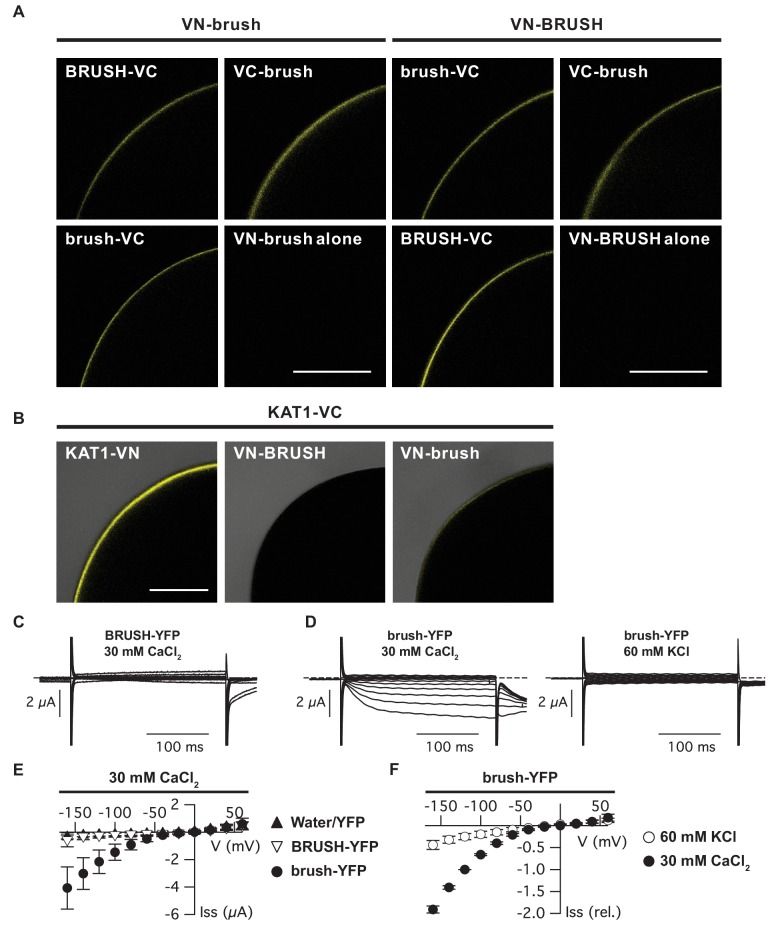
Given that plant CNGCs are anticipated to form both homomeric and heteromeric tetramers (Ma and Berkowitz, 2011), an interaction between BRUSH and redundant CNGCs is conceivable. We initially utilized the yeast split-ubiquitin interaction assay to determine the location of the BRUSH termini (Xing et al., 2016). The assay utilizes the N-terminal (Nub) and C-terminal (Cub) fragments of yeast ubiquitin (Ubi4) (Stagljar et al., 1998). Reconstitution of ubiquitin in the cytoplasm leads to proteolytic release of Cub (fused to LexA-VP16) and activation of genetic reporters. We observed that BRUSH-Cub interacted with both NubI-BRUSH and BRUSH-NubI fusions demonstrating that both BRUSH termini are located in the cytoplasm (Figure 3—figure supplement 1). Since voltage-gated ion channel subunits interact via their soluble domains (Barros et al., 2012), we focused on the CNGC.IVA soluble termini for yeast two-hybrid interaction assays. We observed a self-interaction for the BRUSH N-terminus (NT) as well as interaction with the NTs of brush, CNGC.IVA3, CNGC.IVA4, CNGC.IVA5 (Figure 3A) along with AtCNGC19 and AtCNGC20 (Figure 2—figure supplement 4C). To further substantiate the interaction observed in yeast, we co-injected full-length subunits into Xenopus laevis oocytes for bimolecular fluorescence complementation (BiFC) assays. Expression of BRUSH-BRUSH, BRUSH-brush, and brush-brush combinations resulted in successful complementation (Figure 3—figure supplement 2A,B). The yeast and oocyte interaction assays demonstrate that CNGC.IVA channels potentially form homo- and hetero-complexes in vivo, which may be mediated in part by their NT domains.
Figure 3. Interaction of CNGC.IVA N-termini in yeast and channel activity in Xenopus oocytes.
(A) Yeast two-hybrid interaction of the soluble BRUSH N-terminus (NT) fused to the GAL4-binding domain (BD) and the NT of the indicated CNGC.IVA proteins fused to the GAL4 activation domain (AD). Yeast cells were resuspended in water (OD600= 0.5 and 0.05) and spotted onto -LW (leucine, tryptophan) and -LWAH (leucine, tryptophan, adenine, histidine) solid media. (B) Confocal fluorescence images of oocytes expressing either BRUSH-YFP or brush-YFP fusion proteins. (C) Plasma membrane currents of oocytes expressing BRUSH or brush in the presence of 30 mM CaCl2. Voltage steps ranged from +60 to −160 mV in 20 mV increments, starting from a holding potential of −40 mV. Dashed lines indicate 0 µA. (D) Current-voltage relations of oocytes injected with water or YFP (▲, n=15), BRUSH or BRUSH-YFP (▽, n=17), and brush or brush-YFP (●, n=26) . (E) Relative current-voltage relations for oocytes expressing brush and brush-YFP in the presence of 5 mM CaCl2 (◇ , n = 3), 15 mM CaCl2 (■, n = 3), and 30 mM CaCl2 (●, n = 6). Currents are shown relative to the current at −120 mV in the presence of 30 mM CaCl2. Data in (D) and (E) represent mean values ± standard deviations. Scale bars in images (B) represent 250 µm.
Figure 3—figure supplement 1. BRUSH topology determination in yeast.
Figure 3—figure supplement 2. Brush interaction in oocytes and permeability to Ca2+ and K+.
To characterize their channel properties, we injected either BRUSH or brush into Xenopus oocytes for two-electrode voltage clamping. Expression was confirmed for both BRUSH-YFP and brush-YFP by confocal microscopy (Figure 3B,C). Oocytes expressing BRUSH (Figure 3B,D) or BRUSH-YFP (Figure 3—figure supplement 2C,E) failed to yield significant inward currents at negative voltages in the presence of up to 30 mM CaCl2. In contrast, under the same experimental conditions, oocytes expressing brush (Figure 3C,D) or brush-YFP (Figure 3—figure supplement 2D,E) produced voltage- and time-dependent inward currents at negative voltages. The currents were evident starting from 15 mM external CaCl2 and increased in a dose-dependent manner with the external CaCl2 concentration (Figure 3E). Exchange of Ca2+ as a charge carrier to K+ abolished the voltage-dependent inward currents mediated by brush-YFP (Figure 3—figure supplement 2D,F), indicative of a hyperpolarization-activated Ca2+-permeable channel.
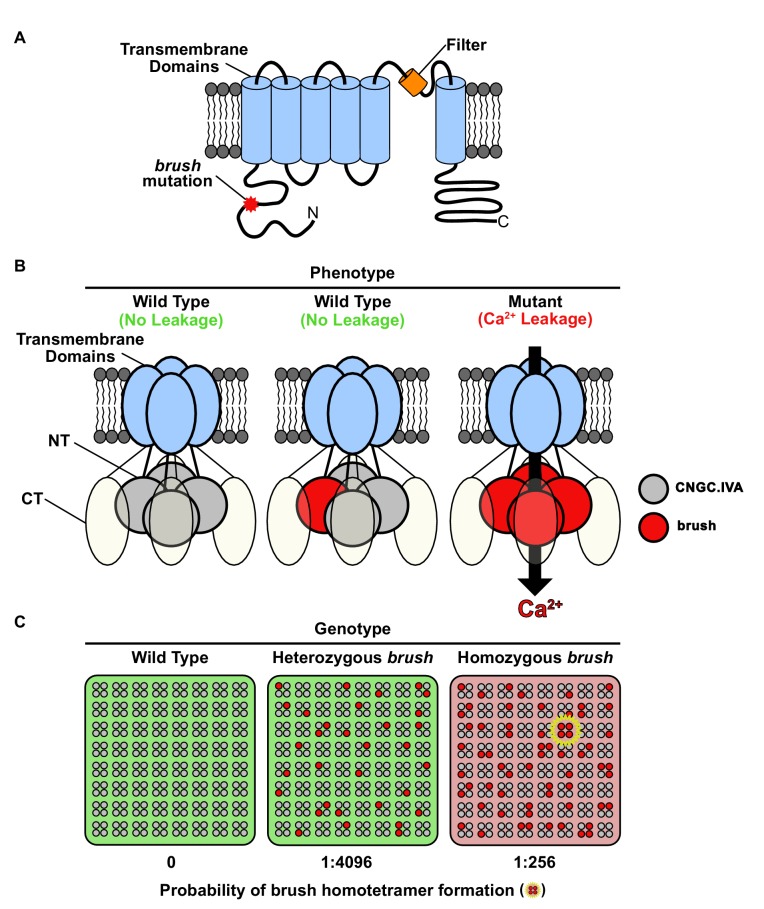
Since an N-terminal missense mutation leads to activation of brush, the conserved CNGC.IVA cytoplasmic domain may mediate channel gating (Figure 4A). The expression of brush alone in oocytes induces Ca2+ influx, therefore assembly of a brush homocomplex leads to deregulated activity (Figure 4B). As brush is recessive we speculate that brush is mainly positioned in silent heterotetrameric complexes in the heterozygous state, but assembles into a population of homocomplexes in homozygous plants triggering the phenotype (Figure 4C). Given that brush is expressed in root hairs and nodule primordia after inoculation with rhizobia and that Ca2+ spiking in brush is intact (Maekawa-Yoshikawa et al., 2009), the deregulated Ca2+ influx activity may impair rhizobial infection progression by interfering with downstream signaling events.
Figure 4. Model for brush activity in planta.
(A) The predicted two-dimensional topology of a brush monomer embedded in a lipid bilayer. (B) Model explaining the mechanistic connection between the relative abundance of the brush mutant channel with the unusual quantitative genetic behavior of the brush phenotype. Based on complementation and interaction studies, BRUSH engages in a tetrameric complex along with 3 other CNGC.IVA proteins. From the segregation analysis, plants heterozygous for brush exhibit a wild-type phenotype. Expression of brush in oocytes mediates leaky Ca2+ influx, indicating that a homotetramer is inappropriately active. (C) Probability-based overview of brush homotetramer formation assuming that each of the 4 CNGC.IVA subunits participates with equal likelihood in complex formation. Shown is a grid containing 8 × 8 tetramers with CNGC.IVA WT (grey dots) and brush (red dots) subunits. Both wild-type and heterozygous plants do not exhibit a phenotype (indicated by green background). The probability of forming brush homotetramer is 1:4096 for a heterozygous and 1:256 for a brush homozygous genotype. A brush homotetramer (shown inside yellow star) is required to trigger the phenotype (red background). This frequency-dependent assembly of the leaky brush tetramer explains the phenotypic differences observed in plants harboring different allele frequencies.
brush is a rare recessive gain-of-function missense mutation and exhibits an unusual quantitative genetic behavior. We pinpointed the CNGC tetrameric complex in combination with the expanded gene family as the causative factors for the unusual genetics. Although definitive evidence demonstrating that plant CNGCs form tetramers is required, their inclusion in the superfamily of voltage-gated ion channels predicts they will form such complexes. In support of this conclusion, the recent cryo-electron microscopy structures of the TAX-4 CNG channel from Caenorhabditis elegans (Li et al., 2017), the prokaryotic LliK CNG channel from Leptospira licerasiae (James et al., 2017), and the human hyperpolarization-activated cyclic nucleotide-gated (HCN1) channel (Lee and MacKinnon, 2017) all disclose a tetrameric assembly. Therefore, we speculate that quantitative competition amongst redundant subunits for tetramer inclusion clarifies why a BRUSH null is phenotypically wild type and why overexpression is required to suppress the brush phenotype.
Expression of brush in oocytes revealed that the mutation renders the channel permeable to Ca2+ influx under hyperpolarizing conditions. Evidence obtained from Arabidopsis (Gao et al., 2016; Wang et al., 2017; Zhang et al., 2017), Medicago truncatula (Charpentier et al., 2016), and the moss Physcomitrella patens (Finka et al., 2012) CNGCs also supports the inward rectification of Ca2+ by plant CNGCs, while numerous physiological studies have implicated CNGCs as being intimately linked to Ca2+ (Wang et al., 2013; Guo et al., 2010; Chan et al., 2003; Urquhart et al., 2007). Although brush was impermeable to K+ in our assay, evidence exists that some plant CNGCs are permeable to other cations (K+ and Na+) in heterologous systems (Gobert et al., 2006; Ali et al., 2006; Leng et al., 2002; Leng et al., 1999). Collectively, the evidence demonstrates that plant CNGCs inwardly rectify cations.
Our results demonstrate that plant CNGC.IVAs may share more in common with metazoan Kv1-Kv4 K+ channels relative to typical mammalian CNGs. Similar to BRUSH, Kv1-Kv4 channel gating and subunit interactions are mediated by an N-terminal T1 domain (Barros et al., 2012; Zagotta et al., 1990). In contrast, human CNGs assemble via C-terminal interactions and are gated by binding of cyclic nucleotides (Barros et al., 2012; Giorgetti et al., 2005). In addition to CNGCs, plants contain a family of shaker-type K+ channels with cyclic nucleotide-binding domains. Similar to BRUSH, these channels are not gated by cyclic nucleotides, but instead are regulated by voltage and relative ion concentrations (Hedrich, 2012). Since plant CNGCs have been difficult to assess in heterologous systems (Ma and Berkowitz, 2011), the discovery that a single residue substitution in a conserved domain is sufficient for activation represents a significant advance towards understanding their regulation.
Materials and methods
Plant material and transformations
Lotus japonicus Gifu (wild-type, accession B-129) (Handberg and Stougaard, 1992), Miyakojima (accession MG-20) (Kawaguchi et al., 2001) and brush (EMS mutant SL0979-2, Gifu) (Perry et al., 2003) plants were used. The BRUSH TILLING line SL1484-1 was obtained from the L. japonicus TILLING facility (John Innes Centre, Norwich, UK). The seed bag numbers of critical lines are listed in Supplementary file 3. Seeds were scarified with sandpaper, sterilized for 10 min in 4% sodium hypochlorite, and imbibed overnight in sterile water at 4°C. Hairy roots were generated using the Agrobacterium rhizogenes strain AR1193 (Stougaard et al., 1987). Nodulation experiments were carried out by inoculating plants grown in pots or weck jars containing a sand-vermiculite mixture and Fåhraeus (Fahraeus, 1957) media with Mesorhizobium loti MAFF303099 expressing DsRed (Markmann et al., 2008). Transgenic roots were visualized with either a stereomicroscope (Leica M165FC) or confocal laser scanning microscope (Leica SP5). Hairy roots were stained for GUS and sectioned as described previously (Chiasson et al., 2014). Plants were cultivated in growth cabinets at 22°C (16 hr light/8 hr dark). All complementation and GUS experiments were carried out a minimum of three times and displayed similar results. Crossings were performed as described previously (Jiang and Gresshoff, 1997). Primers and plasmids used for all experiments are listed in Supplementary file 1 and Supplementary file 2, respectively.
Map-based cloning of the brush mutation
F2 plants from a cross between brush and MG-20 were used for fine mapping using SSR markers as described (Groth et al., 2013). Primer sequences were obtained from the Kazusa DNA Research Institute website (http://www.kazusa.or.jp/lotus/markerdb_index.html). The region was further refined using identified SNPs. The brush target interval between TM2432 and SNP3 (approximately 103 kb) was sequenced by Sanger sequencing. The brush genome was also reassembled after next-generation sequencing to identify mutant-specific polymorphisms. Nuclear DNA (see below) of brush seedlings was subjected to next-generation sequencing at Eurofins MWG, Germany, using an Illumina HiSeq 2000 (Illumina, USA) with a read length of 2 × 100 bp. Genes in the brush target region were annotated after sequencing using Genscan (Burge and Karlin, 1997) and Artemis (Rutherford et al., 2000). CLC Genomics Workbench (CLC bio, Denmark) was used to analyze the sequencing data.
Nuclear DNA extraction for next-generation sequencing
Four-week-old brush seedlings were transferred to the dark for 2 days before leaf material was harvested. Approximately 2 g of ground powder was resuspended in 20 ml ice-cold HB buffer (10 mM Tris, 80 mM KCl, 10 mM EDTA, 1 mM spermine, 1 mM spermidine, 0.5 M sucrose, 0.5% triton X-100, 0.15% β-mercaptoethanol, pH 9.4 with NaOH) by gentle shaking on ice. The solution was filtered through two layers of Miracloth (Calbiochem, Merck, Germany). The flow-through was transferred to a 15-ml Falcon tube and the nuclei were pelleted at 4°C by centrifugation (1800 x g) and washed two times by resuspension in HB buffer. The final pellet was resuspended in 500 μl CTAB buffer (55 mM cetyltrimethylammonium bromide, 1.4 M NaCl, 20 mM EDTA, 100 mM Tris, pH 8), and incubated at 60°C for 30 min. 500 μl chloroform:isoamylalcohol (24:1) was added and mixed by inverting the tube several times. After a centrifugation step at 8000 x g (4°C) for 10 min, the upper phase was transferred to a new tube. 5 μl of RNase (10 mg/ml stock concentration) was added and incubated at 37°C for 30 min. 0.6 volumes ice-cold isopropanol was added and mixed by inverting the tube several times. The nuclear DNA was then precipitated at −20°C overnight and centrifuged for 10 min at 16,000 x g and 4°C. The supernatant was discarded and the pellet was washed with 70% ethanol and resuspended in 55 μl TE buffer.
Yeast two-hybrid and split-ubiquitin assays
Yeast two-hybrid interaction assays were conducted with the haploid yeast strain AH109 (Clontech). Split ubiquitin interaction assays were carried out in the haploid strain THY.AP4 (Obrdlik et al., 2004). THY.AP4 and plasmids for split-ubiquitin were obtained from the Arabidopsis Biological Resource Center (http://abrc.osu.edu/). Plasmids used for both interaction assays are shown in Supplementary file 2. Bait and prey plasmids were introduced via double transformation using the lithium acetate method (Gietz and Schiestl, 2007) and selected on media lacking leucine and tryptophan (-LW). The interacting protein pair of CCaMK and CYCLOPS was used as a control for yeast two-hybrid (Yano et al., 2008). Positive transformants were restreaked on -LW, then used to inoculate overnight cultures in liquid -LW media. Overnight cultures were diluted to OD600 of 0.5 in sterile water and diluted 10-fold. 5 μl was spotted on –LW or solid media lacking leucine, tryptophan, adenine, and histidine (-LWAH). Yeast plates were incubated at 28°C for 3–5 days. All interaction assays were independently conducted a minimum of three times.
Clone preparation for Xenopus oocyte experiments
BRUSH and brush coding sequences were cloned for Xenopus expression with a custom Golden Gate cloning strategy using a modified backbone obtained from the Standard European Vector Architecture 2.0 database (Martinez-Garcia et al., 2015). The backbone (with flanking bacterial transcriptional terminators) was derived from pSEVA191 (http://wwwuser.cnb.csic.es/~seva/) and was chosen to alleviate toxicity issues uncovered while cloning CNGC.IVA sequences into pUC-based Golden Gate backbones and pGEMHE (Liman et al., 1992). A ccdB cassette compatible with Golden Gate cloning (Binder et al., 2014) was amplified and inserted into the AvrII/SacI sites of pSEVA191 to create the LII backbone pSEVA191 1–2. The coding sequences of BRUSH and brush were then combined in a BsaI cut-ligation with modules containing the T7 promoter as well as the 5’UTR and 3’UTR sequences of β-globin mRNA (amplified from pEMHE). The same backbone was used to express the constructs for BiFC analysis, where LI Golden Gate B-C or D-E parts encoding for the N-terminal (VN) or C-terminal (VC) portions of mVenus (Offenborn et al., 2015) were inserted. Plasmids were assembled in a 15 µl reaction containing 100 ng of each LI plasmid and backbone, 1.5 µl CutSmart buffer (NEB, Germany), 1.5 µl 10 mM ATP, 0.75 µl BsaI (NEB), 0.75 µl T4 ligase (NEB). The reaction was then cycled 6 times (10 min at 37°C, 10 min 16°C) in a PCR machine, followed by incubation at 37°C (10 min) and 65°C (20 min).
Functional analysis in Xenopus laevis oocytes
Capped RNA (cRNA) synthesis, oocyte injection, and voltage-clamp recordings were performed as described (Becker et al., 2004; Müller-Röber et al., 1995). cRNA was synthesized with a mMESSAGE mMACHINE T7 Transcription Kit (ThermoFisher, Germany) and oocytes were injected (General Valve Picospritzer III, Parker Hannifin Corp.) with approximately 25 ng cRNA or with RNase-free water as a control. Injected oocytes were stored at 18°C in ND96 solution (96 mM NaCl, 2 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 5 mM HEPES, 10 mM sorbitol, pH 7.4 with NaOH) adjusted to 220 mOsm/L with sorbitol and supplemented with 25 µg/ml gentamycin until use. Measurements were recorded 2 to 3 days after injection using the two-electrode voltage-clamp technique with a Turbo Tec-10Cx amplifier (NPI electronic GmbH). During two-electrode voltage clamp measurements, oocytes were constantly perfused with bath solution composed of 30 mM CaCl2, 10 mM MES-Tris pH 7.4, adjusted to 220 mOsm/L with mannitol and supplemented with either 100 µM 8-Bromo-cAMP (Sigma) or 100 µM 8-CPT-cAMP (BioLog). For analysis of channel permeabilities, CaCl2 was exchanged as indicated in the figure legends with 5 mM CaCl2, 15 mM CaCl2, or 60 mM KCl. Starting from a holding potential of −40 mV, voltage steps from +60 to −160 mV in 20 mV increments were applied (PatchMaster, HEKA Electronics Inc.). For localization, YFP was fused to the C-terminus of BRUSH or brush. Oocytes were imaged by confocal microscopy 2 to 3 days after injection with BRUSH-YFP and brush-YFP cRNA (Leica TCS SP5, excitation: 488 nm, detection: 525–575 nm) to confirm expression. The same protocol was used for BiFC experiments, except that cRNAs were mixed 1:1 prior to injection.
Gene expression analysis
For analysis of gene expression after rhizobial inoculation, Lotus japonicus Gifu seeds were germinated and grown on half-strength B5 agar plates for 14 days. Six plants were planted per weck jar containing sand/vermiculite with Fåhraeus media. After 7 days, root tissue from a single jar was collected and pooled (represents a biological replicate) for the Day 0 time point. Mesorhizobium loti MAFF303099 expressing DsRed was added to the remaining jars and tissue was collected in the same manner after 12 days. To analyze gene expression after RNAi, positive hairy roots were isolated from individual plants 6 weeks after inoculation with Mesorhizobium loti MAFF303099 DsRed. For both experiments, root tissue was ground in liquid nitrogen and RNA was extracted with a Spectrum Plant Total RNA Kit (Sigma). Genomic DNA was removed using a Turbo DNA-free Kit (Ambion) and total RNA (1 μg for the time course and 200 ng for RNAi) was used for cDNA synthesis with Superscript III (ThermoFisher). cDNA was then checked for genomic DNA contamination by PCR. Expression of CNGC.IVA cluster genes after rhizobia inoculation was analyzed by qPCR using SYBR Select Master Mix (Applied Biosystems) with a CFX96 real-time PCR machine. brush expression after RNAi was analyzed by qPCR using mi-real-time EvaGreen Master Mix (Metabion) with a QuantStudio 5 Real-Time PCR System (ThermoFisher). In both cases, the plotted data point for each biological replicate represents the mean of three technical replicates. The relative expression was calculated with the 2-ΔΔCT method (Schmittgen and Livak, 2008) using eEF-1Aα (GenBank: BP045727) as the reference.
Bioinformatics and statistics
Arabidopsis thaliana protein sequences were obtained from The Arabidopsis Information Resource (TAIR). A multiple sequence alignment was generated using MUSCLE in CLC Main Workbench (CLC bio, Denmark). A Maximum Likelihood phylogenetic tree was calculated using UPGMA (100 bootstrap iterations were performed). One-way ANOVA statistical analysis of data followed by a post-hoc Tukey’s multiple comparisons test and t-tests were calculated using GraphPad Prism.
Acknowledgements
This work was supported by funding from European Research Council (MP; Project No:340904), Deutsche Forschungsgemeinschaft (MP, PD; Forschergruppe 964), and the Alexander von Humboldt Foundation (DC). We would like to thank C Korbmacher (FAU Erlangen) for providing Xenopus laevis oocytes, Gudrun Steingräber (FAU Erlangen) for cRNA synthesis, and Christopher Grefen (University of Tübingen) for advice regarding the split-ubiquitin system.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Martin Parniske, Email: parniske@lmu.de.
Maria J Harrison, Boyce Thompson Institute for Plant Research, United States.
Funding Information
This paper was supported by the following grants:
Deutsche Forschungsgemeinschaft FOR964 (Calcium) to Kristina Haage, Petra Dietrich, Martin Parniske.
FP7 Ideas: European Research Council 340904 (EvolvingNodules) to Martin Parniske.
Alexander von Humboldt-Stiftung to David M Chiasson.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Supervision, Funding acquisition, Investigation, Writing—original draft, Acquisition of Humboldt Fellowship, Supervision of Katharina Sollweck, Cloning, Gene expression analysis, Yeast interaction assays, Complementation, Promoter analysis, Microscopy.
Validation, Investigation, Writing—review and editing, Map-based cloning of brush, Complementation.
Investigation, Writing—review and editing, Yeast-two hybrid, Electrophysiology, Complementation and promoter analysis.
Resources, Formal analysis, Writing—review and editing, Sequencing and analysis of the CNGC cluster.
Resources, Formal analysis, Supervision, Investigation, Writing—review and editing, Supervision of electrophysiology experiments, Equipment, Conducting electrophysiology and BiFC experiments.
Conceptualization, Resources, Formal analysis, Supervision, Funding acquisition, Investigation, Project administration, Writing—review and editing, Supervision of doctoral student Kristina Haage.
Additional files
References
- Ali R, Zielinski RE, Berkowitz GA. Expression of plant cyclic nucleotide-gated cation channels in yeast. Journal of Experimental Botany. 2006;57:125–138. doi: 10.1093/jxb/erj012. [DOI] [PubMed] [Google Scholar]
- Barros F, Domínguez P, de la Peña P. Cytoplasmic domains and voltage-dependent potassium channel gating. Frontiers in Pharmacology. 2012;3:49. doi: 10.3389/fphar.2012.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D, Geiger D, Dunkel M, Roller A, Bertl A, Latz A, Carpaneto A, Dietrich P, Roelfsema MRG, Voelker C, Schmidt D, Mueller-Roeber B, Czempinski K, Hedrich R. AtTPK4, an Arabidopsis tandem-pore K+ channel, poised to control the pollen membrane voltage in a pH- and Ca2+-dependent manner. PNAS. 2004;101:15621–15626. doi: 10.1073/pnas.0401502101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder A, Lambert J, Morbitzer R, Popp C, Ott T, Lahaye T, Parniske M. A modular plasmid assembly kit for multigene expression, gene silencing and silencing rescue in plants. PLoS One. 2014;9:e88218. doi: 10.1371/journal.pone.0088218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. Journal of Molecular Biology. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Cardenas L, Feijo JA, Kunkel JG, Sanchez F, Holdaway-Clarke T, Hepler PK, Quinto C. Rhizobium nod factors induce increases in intracellular free calcium and extracellular calcium influxes in bean root hairs. The Plant Journal. 1999;19:347–352. doi: 10.1046/j.1365-313X.1999.00517.x. [DOI] [PubMed] [Google Scholar]
- Chan CW, Schorrak LM, Smith RK, Bent AF, Sussman MR. A cyclic nucleotide-gated ion channel, CNGC2, is crucial for plant development and adaptation to calcium stress. Plant Physiology. 2003;132:728–731. doi: 10.1104/pp.102.019216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M, Sun J, Vaz Martins T, Radhakrishnan GV, Findlay K, Soumpourou E, Thouin J, Véry AA, Sanders D, Morris RJ, Oldroyd GE. Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science. 2016;352:1102–1105. doi: 10.1126/science.aae0109. [DOI] [PubMed] [Google Scholar]
- Chiasson DM, Loughlin PC, Mazurkiewicz D, Mohammadidehcheshmeh M, Fedorova EE, Okamoto M, McLean E, Glass ADM, Smith SE, Bisseling T, Tyerman SD, Day DA, Kaiser BN. Soybean SAT1 ( Symbiotic Ammonium Transporter 1 ) encodes a bHLH transcription factor involved in nodule growth and NH 4+ transport. PNAS. 2014;111:4814–4819. doi: 10.1073/pnas.1312801111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt DW, Wais R, Long SR. Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell. 1996;85:673–681. doi: 10.1016/S0092-8674(00)81234-9. [DOI] [PubMed] [Google Scholar]
- Fahraeus G. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. Microbiology. 1957;16:374–381. doi: 10.1099/00221287-16-2-374. [DOI] [PubMed] [Google Scholar]
- Felle HH, Kondorosi E, Kondorosi A, Schultze M. Elevation of the cytosolic free [Ca2+] is indispensable for the transduction of the Nod factor signal in alfalfa. Plant Physiology. 1999;121:273–280. doi: 10.1104/pp.121.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finka A, Cuendet AF, Maathuis FJ, Saidi Y, Goloubinoff P. Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. The Plant Cell. 2012;24:3333–3348. doi: 10.1105/tpc.112.095844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frietsch S, Wang Y-F, Sladek C, Poulsen LR, Romanowsky SM, Schroeder JI, Harper JF. A cyclic nucleotide-gated channel is essential for polarized tip growth of pollen. PNAS. 2007;104:14531–14536. doi: 10.1073/pnas.0701781104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q-F, Gu L-L, Wang H-Q, Fei C-F, Fang X, Hussain J, Sun S-J, Dong J-Y, Liu H, Wang Y-F. Cyclic nucleotide-gated channel 18 is an essential Ca 2+ channel in pollen tube tips for pollen tube guidance to ovules in Arabidopsis. PNAS. 2016;113:3096–3101. doi: 10.1073/pnas.1524629113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature Protocols. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- Giorgetti A, Nair AV, Codega P, Torre V, Carloni P. Structural basis of gating of CNG channels. FEBS Letters. 2005;579:1968–1972. doi: 10.1016/j.febslet.2005.01.086. [DOI] [PubMed] [Google Scholar]
- Gobert A, Park G, Amtmann A, Sanders D, Maathuis FJ. Arabidopsis thaliana cyclic nucleotide gated channel 3 forms a non-selective ion transporter involved in germination and cation transport. Journal of Experimental Botany. 2006;57:791–800. doi: 10.1093/jxb/erj064. [DOI] [PubMed] [Google Scholar]
- Groth M, Kosuta S, Gutjahr C, Haage K, Hardel SL, Schaub M, Brachmann A, Sato S, Tabata S, Findlay K, Wang TL, Parniske M. Two Lotus japonicus symbiosis mutants impaired at distinct steps of arbuscule development. The Plant Journal. 2013;75:117–129. doi: 10.1111/tpj.12220. [DOI] [PubMed] [Google Scholar]
- Guo KM, Babourina O, Christopher DA, Borsic T, Rengel Z. The cyclic nucleotide-gated channel AtCNGC10 transports Ca2+ and Mg2+ in Arabidopsis. Physiologia Plantarum. 2010;139:303–312. doi: 10.1111/j.1399-3054.2010.01366.x. [DOI] [PubMed] [Google Scholar]
- Handberg K, Stougaard J. Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. The Plant Journal. 1992;2:487–496. doi: 10.1111/j.1365-313X.1992.00487.x. [DOI] [Google Scholar]
- Harris JM, Wais R, Long SR. Rhizobium-Induced Calcium Spiking in Lotus japonicus. Molecular Plant-Microbe Interactions. 2003;16:335–341. doi: 10.1094/MPMI.2003.16.4.335. [DOI] [PubMed] [Google Scholar]
- Hedrich R. Ion channels in plants. Physiological Reviews. 2012;92:1777–1811. doi: 10.1152/physrev.00038.2011. [DOI] [PubMed] [Google Scholar]
- James ZM, Borst AJ, Haitin Y, Frenz B, DiMaio F, Zagotta WN, Veesler D. CryoEM structure of a prokaryotic cyclic nucleotide-gated ion channel. PNAS. 2017;114:4430–4435. doi: 10.1073/pnas.1700248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Gresshoff PM. Classical and Molecular Genetics of the Model Legume Lotus japonicus. Molecular Plant-Microbe Interactions. 1997;10:59–68. doi: 10.1094/MPMI.1997.10.1.59. [DOI] [PubMed] [Google Scholar]
- Kawaguchi M, Motomura T, Imaizumi-Anraku H, Akao S, Kawasaki S. Providing the basis for genomics in Lotus japonicus: the accessions Miyakojima and Gifu are appropriate crossing partners for genetic analyses. Molecular Genetics and Genomics : MGG. 2001;266:157–166. doi: 10.1007/s004380100540. [DOI] [PubMed] [Google Scholar]
- Lee CH, MacKinnon R. Structures of the Human HCN1 Hyperpolarization-Activated Channel. Cell. 2017;168:111–120. doi: 10.1016/j.cell.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Q, Mercier RW, Yao W, Berkowitz GA. Cloning and first functional characterization of a plant cyclic nucleotide-gated cation channel. Plant Physiology. 1999;121:753–761. doi: 10.1104/pp.121.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Q, Mercier RW, Hua BG, Fromm H, Berkowitz GA. Electrophysiological analysis of cloned cyclic nucleotide-gated ion channels. Plant Physiology. 2002;128:400–410. doi: 10.1104/pp.010832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhou X, Wang S, Michailidis I, Gong Y, Su D, Li H, Li X, Yang J. Structure of a eukaryotic cyclic-nucleotide-gated channel. Nature. 2017;542:60–65. doi: 10.1038/nature20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-A. [DOI] [PubMed] [Google Scholar]
- Ma W, Berkowitz GA. Coding and Decoding of Calcium Signals in Plants. Springer; 2011. Cyclic nucleotide gated channels (CNGCs) and the generation of Ca2+ signals; pp. 93–110. [Google Scholar]
- Maekawa-Yoshikawa M, Müller J, Takeda N, Maekawa T, Sato S, Tabata S, Perry J, Wang TL, Groth M, Brachmann A, Parniske M. The temperature-sensitive brush mutant of the legume Lotus japonicus reveals a link between root development and nodule infection by rhizobia. Plant Physiology. 2009;149:1785–1796. doi: 10.1104/pp.108.135160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmann K, Giczey G, Parniske M. Functional adaptation of a plant receptor-kinase paved the way for the evolution of intracellular root symbioses with bacteria. PLoS Biology. 2008;6:e68. doi: 10.1371/journal.pbio.0060068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia E, Aparicio T, Goni-Moreno A, Fraile S, de Lorenzo SEVA 2.0: an update of the Standard European Vector Architecture for de-/re-construction of bacterial functionalities. Nucleic Acids Research. 2015;43:D1183–D1189. doi: 10.1093/nar/gku1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H, Sun J, Oldroyd GE, Downie JA, Allan Downie J. Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. The Plant Journal. 2006;48:883–894. doi: 10.1111/j.1365-313X.2006.02926.x. [DOI] [PubMed] [Google Scholar]
- Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJ, Sanders D, Harper JF, Tchieu J, Gribskov M, Persans MW, Salt DE, Kim SA, Guerinot ML. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiology. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Röber B, Ellenberg J, Provart N, Willmitzer L, Busch H, Becker D, Dietrich P, Hoth S, Hedrich R. Cloning and electrophysiological analysis of KST1, an inward rectifying K+ channel expressed in potato guard cells. The EMBO Journal. 1995;14:2409–2416. doi: 10.1002/j.1460-2075.1995.tb07238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrdlik P, El-Bakkoury M, Hamacher T, Cappellaro C, Vilarino C, Fleischer C, Ellerbrok H, Kamuzinzi R, Ledent V, Blaudez D, Sanders D, Revuelta JL, Boles E, Andre B, Frommer WB. K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. PNAS. 2004;101:12242–12247. doi: 10.1073/pnas.0404467101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offenborn JN, Waadt R, Kudla J. Visualization and translocation of ternary Calcineurin-A/Calcineurin-B/Calmodulin-2 protein complexes by dual-color trimolecular fluorescence complementation. New Phytologist. 2015;208:269–279. doi: 10.1111/nph.13439. [DOI] [PubMed] [Google Scholar]
- Perry JA, Wang TL, Welham TJ, Gardner S, Pike JM, Yoshida S, Parniske M. A TILLING reverse genetics tool and a web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiology. 2003;131:866–871. doi: 10.1104/pp.102.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J, Brachmann A, Welham T, Binder A, Charpentier M, Groth M, Haage K, Markmann K, Wang TL, Parniske M. TILLING in Lotus japonicus identified large allelic series for symbiosis genes and revealed a bias in functionally defective ethyl methanesulfonate alleles toward glycine replacements. Plant Physiology. 2009;151:1281–1291. doi: 10.1104/pp.109.142190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature Protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Stagljar I, Korostensky C, Johnsson N, te Heesen S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. PNAS. 1998;95:5187–5192. doi: 10.1073/pnas.95.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stougaard J, Abildsten D, Marcker KA. The Agrobacterium rhizogenes pRi TL-DNA segment as a gene vector system for transformation of plants. MGG Molecular & General Genetics. 1987;207:251–255. doi: 10.1007/BF00331586. [DOI] [Google Scholar]
- Urquhart W, Gunawardena AH, Moeder W, Ali R, Berkowitz GA, Yoshioka K. The chimeric cyclic nucleotide-gated ion channel ATCNGC11/12 constitutively induces programmed cell death in a Ca2+ dependent manner. Plant Molecular Biology. 2007;65:747–761. doi: 10.1007/s11103-007-9239-7. [DOI] [PubMed] [Google Scholar]
- Wang YF, Munemasa S, Nishimura N, Ren HM, Robert N, Han M, Puzõrjova I, Kollist H, Lee S, Mori I, Schroeder JI. Identification of cyclic GMP-activated nonselective Ca2+-permeable cation channels and associated CNGC5 and CNGC6 genes in Arabidopsis guard cells. Plant Physiology. 2013;163:578–590. doi: 10.1104/pp.113.225045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kang Y, Ma C, Miao R, Wu C, Long Y, Ge T, Wu Z, Hou X, Zhang J, Qi Z. CNGC2 Is a Ca2+ Influx Channel That Prevents Accumulation of Apoplastic Ca2+ in the Leaf. Plant Physiology. 2017;173:1342–1354. doi: 10.1104/pp.16.01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Wallmeroth N, Berendzen KW, Grefen C. Techniques for the Analysis of Protein-Protein Interactions in Vivo. Plant Physiology. 2016;171:pp.00470.2016–.00470.2758. doi: 10.1104/pp.16.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Yoshida S, Muller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, Asamizu E, Tabata S, Murooka Y, Perry J, Wang TL, Kawaguchi M, Imaizumi-Anraku H, Hayashi M, Parniske M. CYCLOPS, a mediator of symbiotic intracellular accommodation. PNAS. 2008;105:20540–20545. doi: 10.1073/pnas.0806858105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen CCY, Christopher DA. The group IV-A cyclic nucleotide-gated channels, CNGC19 and CNGC20, localize to the vacuole membrane in Arabidopsis thaliana. AoB Plants. 2013;5:plt012. doi: 10.1093/aobpla/plt012. [DOI] [Google Scholar]
- Zagotta WN, Hoshi T, Aldrich RW. Restoration of inactivation in mutants of Shaker potassium channels by a peptide derived from ShB. Science. 1990;250:568–571. doi: 10.1126/science.2122520. [DOI] [PubMed] [Google Scholar]
- Zhang S, Pan Y, Tian W, Dong M, Zhu H, Luan S, Li L. Arabidopsis CNGC14 Mediates Calcium Influx Required for Tip Growth in Root Hairs. Molecular Plant. 2017;10:1004–1006. doi: 10.1016/j.molp.2017.02.007. [DOI] [PubMed] [Google Scholar]