Abstract
Dissecting the genetic basis of natural phenotypic variation is a major goal in biology. We know that most traits are strongly heritable. However, their genetic architecture is a long-standing question, which is unfortunately confounded by the lack of complete knowledge of the genetic components as well as their phenotypic effect in a specific genetic background. Many genetic variants are known to affect phenotypes but the same functional variant can have a different effect on the phenotype in different individuals of the same species. Understanding the impact of genetic background on the expressivity of a given phenotype is essential because this effect complicates our ability to predict phenotype from genotype. Here, we briefly review recent progress on the exploration of the effect of genetic background and we discuss how a deeper characterization of the inheritance, expressivity and genetic interactions hidden behind the phenotypic landscape of natural variation could provide a better understanding of the relationship between genotype and phenotype.
Keywords: Genotypes, phenotypes, expressivity, penetrance, inheritance, monogenic mutations
Introduction
Highlighting the rules that govern trait variations in natural populations is a major goal in modern biology. And identifying the underlying genetic causes of such variation is very challenging. Despite the importance of understanding the genetic basis of complex traits, we currently lack complete knowledge of the relevant genetic components, even in scenarios where environment and other non-heritable contributing elements are well controlled [1]. The impact of genetic backgrounds, inter alia, on the phenotypic expression are still poorly understood to date. However, we argue here that a better understanding of background-specific effect on phenotypic expression variation would lead to a greater perception of the genotype-phenotype relationship.
Monogenic mutation, penetrance and expressivity
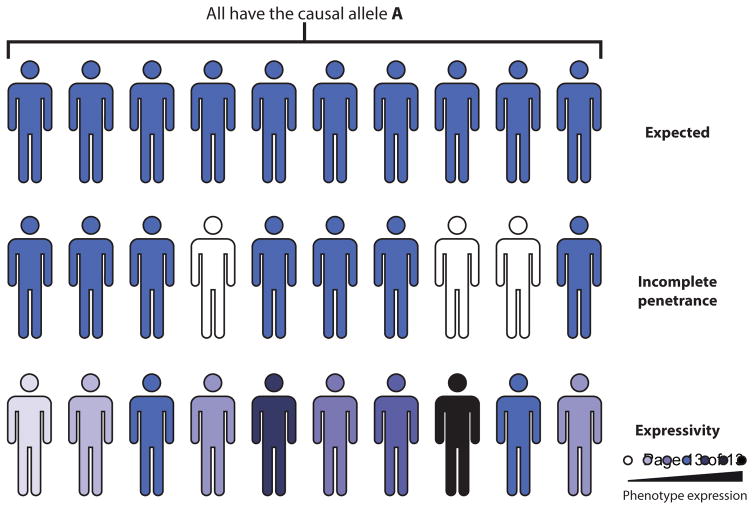
More than 150 years after Gregor Mendel laid the basis of genetics with his laws of heredity and experiments on hybrids [2], we still lack a general understanding of the genetic architecture of traits. For the past century, Mendelian and complex traits have been considered at the opposite ends of the phenotypic spectrum. The inheritance patterns of traits are usually classified as either monogenic, strongly influenced by variation within a single gene, or complex, resulting from variation within multiple genes and their interaction. While useful, this dichotomy is an overly simplistic and artificial view in most of the cases observed in natural populations. Almost from the beginning of modern genetics, the relevance of the genetic context or background was recognized when William Bateson coined the term epistasis to describe the departures from expected Mendelian ratios in his experimental crosses [3]. Behind the simplicity of a Mendelian inheritance, there is a clear hidden complexity of how variants exert a functional impact among individuals of the same species. Although this has been known for decades, the continuous level of the underlying phenotypic spectrum is overlooked. It is evident that most monogenic mutations do not always strictly follow Mendelian inheritance [4]. Many genetic disorders are referred as Mendelian i.e. caused by monogenic mutations. However, people inheriting the same mutation often display variation in phenotypic expression. This has come to be described by two words: “penetrance” and “expressivity” [5,6]. First, a mutation can exhibit incomplete penetrance, meaning that an individual may have this particular mutation but may not express the expected phenotype because of modifiers, epistatic interactions or suppressors present in the genome or because of the environment (Figure 1A). An example is the BRCA1 alleles, which predispose to breast and ovarian cancer in humans. Individuals with a mutation in the BRCA1 gene have a ~80% risk to develop this disease, therefore showing incomplete penetrance [7]. Second, the penetrance of a mutation is sometimes 100%, meaning that all the individuals present the expected trait (Figure 2A), but they exhibit different degrees of expressivity. Neurofibromatosis type I, a Mendelian disorder, is a notorious example of large variable expressivity. The disease is caused by dominant mutations in the NF1 gene [8] and individuals carrying a mutation show a significant phenotypic heterogeneity. In fact, this is the case of a large number of diseases referred as caused by mutations occurring in single genes such as cystic fibrosis, Huntington’s disease, and Fragile X [9–11]. In the case of cystic fibrosis, there is even evidence that modifiers, i.e. mutations in other genes, impact the phenotype [12,13]. Even for Down Syndrome, a whole chromosome disorder, there is evidence of phenotypic expression variation due to genetic background differences [14,15]. More broadly, the phenotypic expression can be modified by various factors with the two most reported being age [7] and sex [16]. However, phenotypic expression can also be impacted by genetic background with the presence of genetic interactions and modifiers as already mentioned, mutation type [17] and environment [18].
Figure 1. Penetrance and expressivity of traits.
In the case of a monogenic disease, all individual carrying the causal allele are expected to develop the same trait. However, in some cases, individuals with the causal allele do not express the expected phenotype, resulting in incomplete penetrance. For other traits, the phenotype will be expressed differentially in different individuals: some will develop more severe symptoms while others display milder symptoms thus representing phenotypic expressivity.
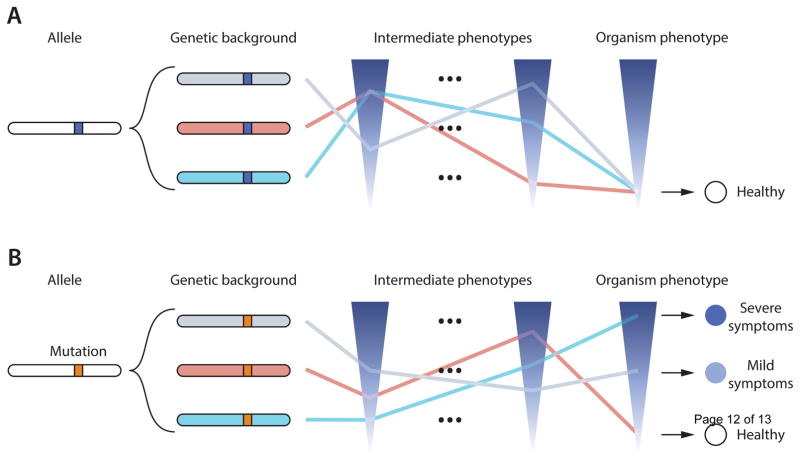
Figure 2. Phenotypic impact of the genetic background.
A: An allele present in different genetic backgrounds results in the same phenotypic outcome at the organismal level. However, this does not mean that intermediate phenotypes such as molecular traits (e.g. gene expression level) will be the same. Every layer of intermediate phenotype acts as a lens that can deflect the phenotype in a specific way with the organism phenotype as the focal point of all these superimposed lenses. B: Some mutations, however, alter intermediate phenotypes and change the final organism phenotype.
The distinction between penetrance and expressivity reflects an overly simplified view for several reasons. First, the full breadth of expression is not systematically characterized for any monogenic mutation in humans. Second, considerable uncertainty is introduced at the phenotypic level, because it is difficult to accurately characterize a trait measurement for most genetic disorders. Most diseases are obviously a complex layering of intermediate molecular traits, e.g. gene expression, methylation, protein and metabolite levels. Several layers of intermediate molecular traits account for the global phenotype at the individual level. Thus, two individuals can display the same trait at the organism level but exhibit completely different intermediate phenotypes at the molecular level, or vice-versa (Figure 2). To better understand the genetic basis of diseases, a more precise estimation of the phenotypic value as well as a more complete picture of the genetic architecture of the molecular traits are probably essential.
Genetic backgrounds, natural populations and model organisms
Variation among individuals of natural populations provides useful raw material to dissect the relationship between genetic variants and phenotypes [19–22]. Moreover, high-throughput genotyping and phenotyping technologies have greatly enhanced the power to dissect the genetic complexity hidden behind traits in model as well as in non-model organisms [23]. A focus on the effects of the genetic backgrounds in natural populations is timely given several recent technological developments. Besides classical examples in human diseases, variation of phenotypic expressivity of monogenic mutations were also observed in model organisms at a genome-wide scale such as in yeast [24,25], mouse [26–29] and worm [30,31].
High-throughput experiments are very useful to quantify the prevalence of the genetic background effects on functional variants between individuals. As an example, model organisms allow for systematic testing of loss-of-function phenotypes. In this context, systematic gene deletion collections were obtained for two closely related yeast Saccharomyces cerevisiae laboratory isolates (S288c and Σ1278b) [26,32]. An extensive difference of gene essentiality was found by comparing those two gene-knockout libraries. In fact, nearly 5% of the genes identified as essential in one isolate are dispensable for survival in the other. In addition, rescue of the viable phenotype generally is of high order of complexity, requiring several modifier genes to counter the effect of a conditionally lethal deletion [24]. The genetic basis behind the disparity observed between these genetic backgrounds is still unknown.
A similar study has been conducted using the nematode Caenorhabditis elegans by knocking down ~1,400 genes with RNAi in the two canonical N2 Bristol and CB4856 Hawaiian isolates [30]. Reduced expression of ~20% of the tested genes led to a trait that varied considerably across the lines. In parallel, the same conclusion was reached by targeting 29 maternal-effect genes in 55 wild C. elegans strains from around the world [31]. By perturbing known embryonic genes, the variability of the embryonic lethality expressivity across genetic backgrounds was clearly highlighted.
Finally, the same mutation has also been recently expressed in a large number of Drosophila genetic backgrounds [33,34]. The Rh1G69D allele, which is a model for retinitis pigmentosa (RP), was crossed in multiple isolates of the Drosophila Genetic Reference Panel representing roughly 200 wild-derived strains [34,35]. It turns out that the retinal phenotype of Rh1G69D varies in a quantitative manner throughout the population, suggesting strong background effects. Using genome-wide association followed by functional validation with RNAi knock-down, the authors identified 10 modifier loci involved in the expressivity of RP [34]. Many of these modifiers have human orthologs and most have not yet been implicated in the onset of retinitis pigmentosa.
All together, these examples highlight that the phenotypic expression of a specific mutation varies tremendously and heritably, depending on the interacting alleles present in each genetic background.
The hidden complex inheritance of simple Mendelian cases is a continuum
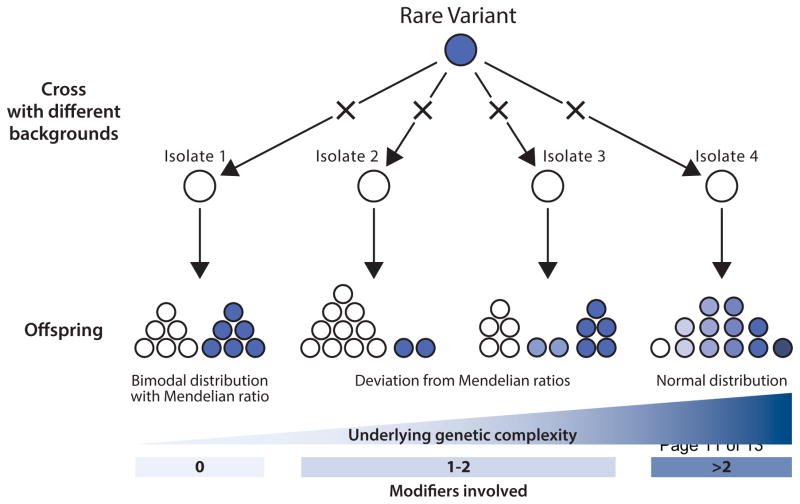
By performing a species-wide survey of monogenic variants in the yeast S. cerevisiae, it has been recently shown that genes and alleles underlying the onset of Mendelian traits are variable in terms of their type, frequency and genomic distribution at the population level [25]. The effect of a rare monogenic mutation of the PDR1 gene, which confers resistance to cycloheximide and anisomycin, was explored and highlighted a continuum of the phenotypic spectrum. The Pdr1p protein is a transcription factor regulating the expression of various multidrug resistance ATP-Binding Cassette (ABC) transporters. In a yeast clinical isolate (YJM326), the presence of a non-synonymous mutation in the sequence of the inhibitory domain of Pdr1p leads to constitutive expression of the downstream transporter coding genes, conferring the drug resistance trait. Twenty sensitive natural isolates were crossed with the resistant YJM326 isolate and the fitness distribution as well as the segregation of the drug resistance in the offspring were evaluated (Figure 3). Seventy percent of the cases displayed a classic Mendelian inheritance. But more interestingly, increased genetic complexity was observed in 30 % of the cases, with significant and continuous deviations from the Mendelian expectation (Figure 3). In five cases, a slight deviation from Mendelian inheritance was observed. The level of genetic complexity was low and the variation of expressivity observed in these cases was due to the presence of one or two modifiers and/or gene interactions. Finally, the fitness distribution appeared to be normal for one given cross, which is characteristic of a complex trait. This study clearly demonstrated that the genetic complexity of traits could be dynamic, transitioning from clear Mendelian to diverse complex inheritance patterns depending on various genetic backgrounds. The power of this study lies in the fact that assumptions regarding the number of modifiers involved can be made by looking at the phenotypic distribution and segregation patterns in the offspring (Figure 3). Consequently, it is possible to more accurately estimate the genetic complexity of traits. Deeper dissection of the transition between simple and complex traits in natural populations might therefore lead to new insights into the genetic architecture of traits.
Figure 3. Trait complexity acts as a continuum at the species level.
When crossing a rare variant into multiple genetic backgrounds, the underlying genetic complexity of a trait can range from Mendelian or monogenic trait to complex. Genetic complexity underlying the trait can be assessed by looking at the offspring phenotypic distribution. A bimodal distribution following Mendelian ratios (2:2 for haploids and 3:1 for diploids) suggests a monogenic trait. Deviations from these ratios are signs of higher but intermediate level of complexity. Ultimately, a normal phenotypic distribution depicts a complex phenotype.
Conclusion and perspectives
Understanding the phenotypic effects of natural genetic variants remains a major challenge in biology. This is obviously clear in the case of personalized medicine, with the hope to predict an individual’s disease risk from his genetic data. The advances of high-throughput sequencing technologies hold the promise that whole-genome sequencing will be routine in medical care and will enhance the power to determine the genetic basis of traits. Comprehensive dissection of the genetic mechanisms underlying natural phenotypic diversity seems to be within reach. Since the rise of next-generation sequencing, a lot of effort has been put into genome-wide association and linkage mapping strategies to dissect the genotype-phenotype relationship. Nevertheless, limitations have been clearly highlighted by all association studies in humans, where all causal variants found fail to explain the entirety of the observed phenotypic variance. This unexplained variance is better known as the missing heritability [36,37]. Because of this missing heritability, predictions about phenotypic variation remain limited. Possible reasons for this grey zone are the presence of a high number of rare variants, which are background specific, in natural populations and the intricate pattern of genetic interaction between all the genes that cannot be detected using these methods. Rare variants and genetic interactions clearly contribute to phenotypic expressivity variation. Deeper characterization of the inheritance, expressivity and genetic interactions hidden behind the phenotypic landscape of natural populations will bring further valuable insight into the conversion of genetic into phenotypic variation.
Highlights.
Phenotypic expression of a specific mutation can vary across individuals of the same species
Monogenic variants might display a continuous expressivity spectrum in natural population
Background specific effects lead to a continuum of genetic complexity of traits
Acknowledgments
The authors are grateful to David Stern for critical reading of the manuscript. TF is supported by a grant from the French “Ministère de l’Enseignement Supérieur et de la Recherche”. JS is funded by the National Institutes of Health (NIH Grant R01 GM101091-01). JS is a Fellow of the University of Strasbourg Institute for Advanced Study (USIAS) and a member of the Institut Universitaire de France.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mackay TFC, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nat Rev Genet. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- 2.Mendel G. Versuche über Plflanzenhybriden. Verhandlungen des naturforschenden Vereines in Brünn, Bd. IV für das Jahr 1865. 1866 [Google Scholar]
- 3.Bateson W. Facts limiting the theory of heredity. Science. 1907;26:649–660. doi: 10.1126/science.26.672.649. [DOI] [PubMed] [Google Scholar]
- 4.Antonarakis SE, Chakravarti A, Cohen JC, Hardy J. Mendelian disorders and multifactorial traits: the big divide or one for all? Nat Rev Genet. 2010;11:380–384. doi: 10.1038/nrg2793. [DOI] [PubMed] [Google Scholar]
- 5.Zlotogora J. Penetrance and expressivity in the molecular age. Genet Med. 2003;5:347–352. doi: 10.1097/01.gim.0000086478.87623.69. [DOI] [PubMed] [Google Scholar]
- 6.Jarvik GP, Evans JP. Mastering genomic terminology. Genet Med. 2017;19:491–492. doi: 10.1038/gim.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mavaddat N, Peock S, Frost D, Ellis S, Platte R, Fineberg E, Evans DG, Izatt L, Eeles RA, Adlard J, et al. Cancer risks for BRCA1 and BRCA2 mutation carriers: Results from prospective analysis of EMBRACE. J Natl Cancer Inst. 2013;105:812–822. doi: 10.1093/jnci/djt095. [DOI] [PubMed] [Google Scholar]
- 8.Pasmant E, Vidaud M, Vidaud D, Wolkenstein P. Neurofibromatosis type 1: from genotype to phenotype. J Med Genet. 2012;49:483–489. doi: 10.1136/jmedgenet-2012-100978. [DOI] [PubMed] [Google Scholar]
- 9.Cutting GR. Modifier genes in Mendelian disorders: the example of cystic fibrosis. Ann N Y Acad Sci. 2010;1214:57–69. doi: 10.1111/j.1749-6632.2010.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arning L. The search for modifier genes in Huntington disease – Multifactorial aspects of a monogenic disorder. Mol Cell Probes. 2016;30:404–409. doi: 10.1016/j.mcp.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Garber KB, Visootsak J, Warren ST. Fragile X syndrome. Eur J Hum Genet. 2008;16:666–672. doi: 10.1038/ejhg.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emond MJ, Louie T, Emerson J, Zhao W, Mathias RA, Knowles MR, Wright FA, Rieder MJ, Tabor HK, Nickerson DA, et al. Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat Genet. 2012;44:886–889. doi: 10.1038/ng.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosendahl J, Landt O, Bernadova J, Kovacs P, Teich N, Bödeker H, Keim V, Ruffert C, Mössner J, Kage A, et al. CFTR, SPINK1, CTRC and PRSS1 variants in chronic pancreatitis: is the role of mutated CFTR overestimated? Gut. 2013;62:582–592. doi: 10.1136/gutjnl-2011-300645. [DOI] [PubMed] [Google Scholar]
- 14.Ackerman C, Locke AE, Feingold E, Reshey B, Espana K, Thusberg J, Mooney S, Bean LJH, Dooley KJ, Cua CL, et al. An Excess of Deleterious Variants in VEGF-A Pathway Genes in Down-Syndrome-Associated Atrioventricular Septal Defects. Am J Hum Genet. 2012;91:646–659. doi: 10.1016/j.ajhg.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Cherry S, Klinedinst D, DeLeon V, Redig J, Reshey B, Chin MT, Sherman SL, Maslen CL, Reeves RH. Genetic modifiers predisposing to congenital heart disease in the sensitized down syndrome population. Circ Cardiovasc Genet. 2012;5:301–308. doi: 10.1161/CIRCGENETICS.111.960872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tai YC, Domchek S, Parmigiani G, Chen S. Breast cancer risk among male BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst. 2007;99:1811–1814. doi: 10.1093/jnci/djm203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thauvin-Robinet C, Munck A, Huet F, Génin E, Bellis G, Gautier E, Audrézet M-P, Férec C, Lalau G, Des Georges M, et al. The very low penetrance of cystic fibrosis for the R117H mutation: a reappraisal for genetic counselling and newborn screening. J Med Genet. 2009;46:752–758. doi: 10.1136/jmg.2009.067215. [DOI] [PubMed] [Google Scholar]
- 18.Lachance J, Jung L, True JR. Genetic background and GxE interactions modulate the penetrance of a naturally occurring wing mutation in Drosophila melanogaster. G3. 2013;3:1893–901. doi: 10.1534/g3.113.007831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durbin RM, Altshuler DL, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, Clark AG, Collins FS, De La Vega FM, Donnelly P, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, Clark AG, Donnelly P, Eichler EE, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walter K, Min JL, Huang J, Crooks L, Memari Y, McCarthy S, Perry JRB, Xu C, Futema M, Lawson D, et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alonso-Blanco C, Andrade J, Becker C, Bemm F, Bergelson J, Borgwardt KM, Cao J, Chae E, Dezwaan TM, Ding W, et al. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell. 2016;166:481–491. doi: 10.1016/j.cell.2016.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellegren H. Genome sequencing and population genomics in non-model organisms. Trends Ecol Evol. 2014;29:51–63. doi: 10.1016/j.tree.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Dowell RD, Ryan O, Jansen A, Cheung D, Agarwala S, Danford T, Bernstein DA, Rolfe PA, Heisler LE, Chin B, et al. Genotype to phenotype: a complex problem. Sci (New York, NY) 2010;328:469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **25.Hou J, Sigwalt A, Fournier T, Pflieger D, Peter J, de Montigny J, Dunham MJ, Schacherer J. The hidden complexity of Mendelian traits across natural yeast populations. Cell Rep. 2016;16:1106–1114. doi: 10.1016/j.celrep.2016.06.048. By crossing a strain carrying a rare monogenic variant displaying an extreme phenotype with 20 genetic backgrounds, the authors enlightened the continuum of genetic complexity underlying a trait: from a monogenic trait rises more complex phenotypes governed by multiple genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montagutelli X. Effect of the genetic background on the phenotype of mouse mutations. J Am Soc Nephrol. 2000;11(Suppl 1):S101–S105. [PubMed] [Google Scholar]
- 27.Yoshiki A, Moriwaki K. Mouse phenome research: implications of genetic background. ILAR J. 2006;47:94–102. doi: 10.1093/ilar.47.2.94. [DOI] [PubMed] [Google Scholar]
- 28.Doetschman T. Influence of genetic background on genetically engineered mouse phenotypes. Methods Mol Biol. 2009;530:423–433. doi: 10.1007/978-1-59745-471-1_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *29.Percival CJ, Marangoni P, Tapaltsyan V, Klein O, Hallgrímsson B. The interaction of genetic background and mutational effects in regulation of mouse craniofacial shape. G3. 2017;7:1439–1450. doi: 10.1534/g3.117.040659. In this paper, authors studied mice craniofacial size and shape in three genetic backgrounds knocked-out for three genes. They aimed to map genetic interactions in a given pathway resulting in a difference in phenotype expressivity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **30.Vu V, Verster AJ, Schertzberg M, Chuluunbaatar T, Spensley M, Pajkic D, Hart GT, Moffat J, Fraser AG. Natural variation in gene expression modulates the severity of mutant phenotypes. Cell. 2015;162:391–402. doi: 10.1016/j.cell.2015.06.037. The authors knocked-down 1,400 genes in two Caenorhabditis elegans isolates and showed that around 20 % of the phenotypes differed between them. Differences in expressivity seem to be predictable from expression. [DOI] [PubMed] [Google Scholar]
- **31.Paaby AB, White AG, Riccardi DD, Gunsalus KC, Piano F, Rockman MV. Wild worm embryogenesis harbors ubiquitous polygenic modifier variation. Elife. 2015:4. doi: 10.7554/eLife.09178. By performing knock-down with RNAi of 29 embryogenesis related genes in 55 Caenorhabditis elegans strains, they elegantly revealed the astonishing trait expressivity across different backgrounds at the species level. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 33.He BZ, Ludwig MZ, Dickerson DA, Barse L, Arun B, Vilhjálmsson BJ, Jiang P, Park S-Y, Tamarina NA, Selleck SB, et al. Effect of genetic variation in a Drosophila model of diabetes-associated misfolded human proinsulin. Genetics. 2014;196:557–567. doi: 10.1534/genetics.113.157800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **34.Chow CY, Kelsey KJP, Wolfner MF, Clark AG. Candidate genetic modifiers of retinitis pigmentosa identified by exploiting natural variation in Drosophila. Hum Mol Genet. 2016;25:651–659. doi: 10.1093/hmg/ddv502. The authors used a Drosophila model strain of retinitis pigmentosa (RP) carrying the RH1G69D allele and crossed it with 176 isolates from the Drosophila Genetic Reference Panel. After performing a genome-wide association study and functional verification with RNAi, they highlighted 10 candidate modifiers genes that modulate the phenotype expressivity of RP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. The Drosophila melanogaster genetic reference panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuk O, Schaffner SF, Samocha K, Do R, Hechter E, Kathiresan S, Daly MJ, Neale BM, Sunyaev SR, Lander ES. Searching for missing heritability: Designing rare variant association studies. Proc Natl Acad Sci. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]