Abstract
IMPORTANCE
Evidence is lacking regarding the consequences of antibiotic use in early life and the risk of certain autoimmune diseases.
OBJECTIVE
To test the association between early-life antibiotic use and islet or celiac disease (CD) autoimmunity in genetically at-risk children prospectively followed up for type 1 diabetes (T1D) or CD.
DESIGN, SETTING, AND PARTICIPANTS
HLA-genotyped newborns from Finland, Germany, Sweden, and the United States were enrolled in the prospective birth cohort of The Environmental Determinants of Diabetes in the Young (TEDDY) study between November 20, 2004, and July 8, 2010. The dates of analysis were November 20, 2004, to August 31, 2014. Individuals from the general population and those having a first-degree relative with T1D were enrolled if they had 1 of 9 HLA genotypes associated with a risk for T1D.
EXPOSURES
Parental reports of the most common antibiotics (cephalosporins, penicillins, and macrolides) used between age 3 months and age 4 years were recorded prospectively.
MAIN OUTCOMES AND MEASURES
Islet autoimmunity and CD autoimmunity were defined as being positive for islet or tissue transglutaminase autoantibodies at 2 consecutive clinic visits at least 3 months apart. Hazard ratios and 95% CIs calculated from Cox proportional hazards regression models were used to assess the relationship between antibiotic use in early life before seroconversion and the development of autoimmunity.
RESULTS
Participants were 8495 children (49.0% female) and 6558 children (48.7% female) enrolled in the TEDDY study who were tested for islet and tissue transglutaminase autoantibodies, respectively. Exposure to and frequency of use of any antibiotic assessed in this study in early life or before seroconversion did not influence the risk of developing islet autoimmunity or CD autoimmunity. Cumulative use of any antibiotic during the first 4 years of life was not associated with the appearance of any autoantibody (hazard ratio [HR], 0.98; 95% CI, 0.95–1.01), multiple islet autoantibodies (HR, 0.99; 95% CI, 0.95–1.03), or the transglutaminase autoantibody (HR, 1.00; 95% CI, 0.98–1.02).
CONCLUSIONS AND RELEVANCE
The use of the most prescribed antibiotics during the first 4 years of life, regardless of geographic region, was not associated with the development of autoimmunity for T1D or CD. These results suggest that a risk of islet or tissue transglutaminase autoimmunity need not influence the recommendations for clinical use of antibiotics in young children at risk for T1D or CD.
Since the introduction of penicillin in 1941, antibiotics have had a crucial role in combating infections, which has led to a sharp increase in the average life span in the industrialized world.1 However, the increasing use of antibiotics worldwide has been proposed as a cause for the growing incidence of autoimmune diseases in industrialized countries, particularly type 1 diabetes (T1D) and celiac disease (CD). The presence or absence of an association between antibiotic use and autoimmune diseases could have profound influences on future antibiotic use worldwide. Antibiotics administered to rodents predisposed to T1D have shown both protective and accelerating effects on disease development, mainly during the prenatal and neonatal periods.2–9 Yet, the antibiotics used in such rodent studies are not often prescribed for infections in children. In humans, maternal use before or during pregnancy did not increase the risk of childhood T1D, except in a few cases where proportional use by the cohort was so low that it could not explain the large increase in T1D incidence over the last 50 years.10 Increased CD risk was associated with antibiotic use in children11 and adults.12
Given the conflicting evidence on antibiotic use and autoimmunity risk, the aim herein was to test whether the use of oral β-lactam or macrolide antibiotics was associated with autoimmunity for T1D or CD during the first 4 years of life. Antibiotic use was investigated cumulatively from birth to assess any potential trigger associations before autoimmunity in The Environmental Determinants of Diabetes in the Young (TEDDY) cohort.
Methods
Study Design
The TEDDY is a large prospective cohort study that follows up children at high genetic risk for T1D or CD at 6 clinical centers in 4 countries (Finland, Germany, Sweden, and the United States).13 After screening 424 788 children at birth for HLA genes associated with T1D and CD between November 20, 2004, and July 8, 2010, the parents of 8676 genetically at-risk children gave written informed consent for enrollment in a 15-year follow-up study at age 3 months.14 The dates of analysis were November 20, 2004, to August 31, 2014. Individuals from the general population and those having a first-degree relative with T1D were enrolled if they had 1 of 9 HLA genotypes associated with a risk for T1D. Parental reports of the most common antibiotics (cephalosporins, penicillins, and macrolides) used between age 3 months and age 4 years were recorded prospectively. Factors associated with enrollment in the TEDDY study are described elsewhere15,16 and were adjusted for in the multivariable models. The TEDDY study was approved by the following ethical institutional review boards: the Colorado Multiple Institutional Review Board, the Hospital District of Southwest Finland Committee on Ethics, the University of Florida Health Center Institutional Review Board, the Augusta University Institutional Review Board (Georgia), the Ethik-Kommission der Bayerischen Landesarztekammer (Germany), the University of Pittsburgh Institutional Review Board, the Lund University Committee for Continuing Ethical Re view (Sweden), the Western Institutional Review Board (Washington), and the University of South Florida Institutional Review Board. The TEDDY study is also monitored by a National Institutes of Health external advisory board.
As of August 3, 2014, a total of 8495 children had reached age 4 years, had a confirmed HLA genotype at age 9 months, and were screened for T1D and CD autoantibodies during follow-up period (eFigure 1 in the Supplement). Blood samples used for antibody testing were collected at clinic visits conducted 4 times a year between age 3 months and age 4 years and biannually thereafter for those who had not developed islet autoimmunity (IA) by age 4 years.17
Islet autoantibodies, including insulin autoantibody (IAA), glutamic acid decarboxylase autoantibody (GADA), and tyrosine phosphatase IA-2 autoantibody (IA2A), were tested as previously described.18 Islet autoimmunity was defined as being positive for any one autoantibody at 2 consecutive clinic visits at least 3 months apart confirmed by 2 laboratories. The first primary outcome of this study was the risk of IA, but first-appearing IAA or GADA outcomes were also analyzed. Because young children who develop a second islet autoantibody are at high risk of developing T1D, the development of multiple persistent antibodies within 48 months was assessed (n = 276). Individuals who developed a single autoantibody (IAA, GADA, or IA2A) and did not progress to multiple autoantibodies before age 48 months (n = 36) were censored at 48 months as nonmultiple. The date of seroconversion was defined as the date of the first of 2 consecutive samples that tested positive for a specific islet autoantibody. For multiple islet autoantibodies, the date of seroconversion was defined as the date of the appearance of the second persistent autoantibody.
Screening for tissue transglutaminase autoantibodies (tTGAs) followed a modified schedule described previously,19 with children tested for tTGAs between age 12 months and age 48 months. After exclusion of children not tested for tTGAs between age 12 months and age 48 months (n = 1934), those who developed CD before tTGA testing (n = 1), and those who developed tTGAs before age 12 months (n = 2), the final cohort size for CD autoimmunity (CDA) outcome was 6558 children (eFigure 1 in the Supplement). The second primary outcome of this study was the risk of CDA, defined as being positive for tTGAs at 2 consecutive clinic visits at least 3 months apart. The date of seroconversion was defined as the date of the first of 2 consecutive samples that tested positive for tTGAs.
The primary caregivers of children in the TEDDY study kept an ongoing record of the child’s exposures and medical history, starting at the first clinic visit at age 3 months.17 Exposures from the first 3 months of life were retrospectively collected at the first clinic visit. The parents recorded the use of any over-the-counter or prescription medication, including the brand name, reason for use, number of days of use, and age of the child at the time of use.17 A medical coding system was developed to combine TEDDY medication codes with standardized clinical drug nomenclature via RxNorm20 and pharmacological actions of substances. Pharmacists and TEDDY staff ratified this system to ensure proper classification of medications. Antibiotics were classified as any medication with bactericidal or bacteriostatic action. To reduce the number of comparisons during the data analysis stages, antibiotics were grouped into the following major categories: amoxicillins, penicillins, carbapenems, macrolides, and cephalosporins (eTable 1 in the Supplement). These antibiotic categories were further collapsed into β-lactam antibiotics, all penicillins, and “any antibiotic,” which was defined as the use of any of the antibiotics listed above (Figure1B). Carbapenems and other antibiotics were not analyzed as a separate category due to the low incidence of use. While the TEDDY study did not collect data on the route of administration, any known local or topical antibiotics belonging to the major antibiotic categories, as determined by TEDDY pediatric physicians, were removed. Antibiotic use was defined as a course of antibiotics prescribed by a physician.
Figure 1. Overview of Antibiotic Use in The Environmental Determinants of Diabetes in the Young (TEDDY) Study.
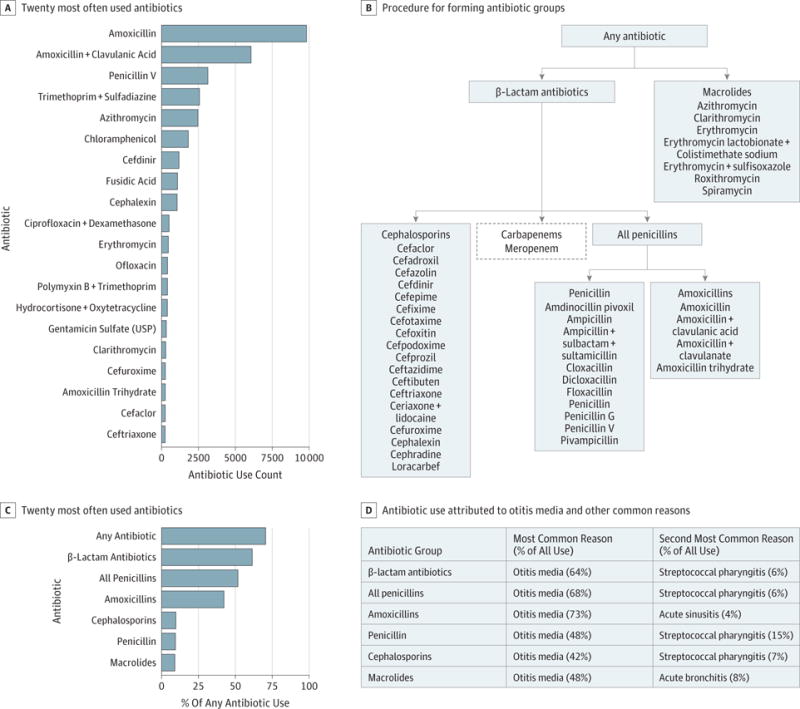
A, Count of the 20 most often used antibiotics in the first 48 months of life in the TEDDY study. B, Procedure for forming antibiotic groups. Medications listed under the headings were extracted from the TEDDY medication database. These medications were grouped into major antibiotic categories (cephalosporins, amoxicillins, penicillin, macrolides, and carbapenems). These categories were further collapsed into any antibiotic, β-lactam antibiotics, and all penicillins, as indicated by the flowchart. All antibiotic categories except carbapenems were considered for analysis. C, Percentage of any antibiotic use in the first 48 months of life for the chosen antibiotic groups. D, Table of the percentage of antibiotic use attributed to otitis media and the second most common reasons for use. USP indicates United States Pharmacopeia.
Maternal medication use during pregnancy was collected via a questionnaire mailed to the mothers of enrolled children and completed 3 to 4 months post partum. Probiotic exposure was defined as the timing of the first introduction of probiotics from a dietary supplement or infant formula. Duration of breast-feeding was collected from a questionnaire at study entry and was reviewed with TEDDY staff at all clinic visits.
Statistical Analysis
The objective was to examine whether early, cumulative, or recent antibiotic use was associated with instantaneous risk of IA or CDA. Each association was assessed by a hazard ratio (HR) and 95% CI calculated from Cox proportional hazards regression models that regressed antibiotic use as a time-varying covariate on the hazard of outcome (IA or CDA). In each model, children deemed negative for the outcome or missing 4 consecutive antibody samples were censored either at the age of their last autoantibody test or at age 48 months, whichever occurred first. Each risk set (the case and children still at risk) was formed on the day of the first positive sample. The between-visit period for that specific day was considered the seroconversion period for each child in the risk set. For the case, the actual day of seroconversion occurred between the last 3-month sample and the first positive sample (Figure 2). To ensure that antibiotic use was examined before autoantibodies developed, major antibiotics (categories shown in Figure 1B) were examined only before the seroconversion period. The number of antibiotics on the hazard of outcome was examined separately by the following 7 time frames: (1) cumulative count, (2) count to age 6 months, (3) count to age 12 months, and (4) count to age 48 months using sum and step functions, as well as the (5) count during 0 to 3 months before seroconversion, (6) count during 3 to 6 months before seroconversion, and (7) count during 0 to 12 months before seroconversion using sum and lag functions (Figure 2). In addition, antibiotic use within all exposure windows was also examined on first-appearing IAA or GADA outcomes to check for any age-specific association with IA.18 All models were adjusted for known risk factors, including country, sex, first-degree relative with T1D or CD, HLA-DR genotype, caesarian delivery, probiotic use before age 90 days, breastfeeding status at age 90 days, maternal antibiotic use during pregnancy, and season of birth.
Figure 2. Example of Sampling and Lag Periods in Relation to Islet Autoimmunity or Celiac Disease Autoimmunity Seroconversion.
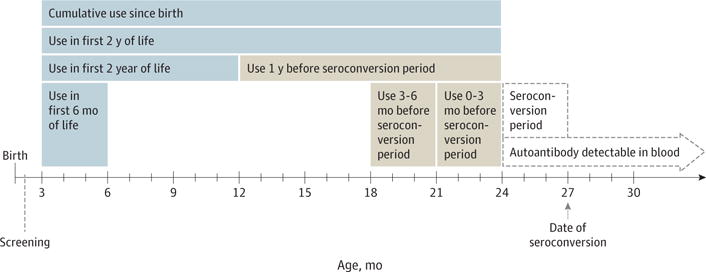
In this example, the 27-month and 30-month serum samples are confirmed positive for islet or tissue transglutaminase autoantibodies. Therefore, the date of seroconversion is set at 27 months. The seroconversion period is the 3-month interval between the last negative (24-month sample) and the first positive (27-month sample) autoantibody sample.
To simplify presentation of the results, adjustment for multiple comparisons was performed first by examining each exposure window separately, followed by a Bonferroni correction (2-sided P < .007 was considered significant) in relation to the outcome. To further control for type I errors, a false discovery rate for all time-dependent associations with each outcome was also applied, with any change in significance of the association with antibiotic use reported for each outcome (Cochran Q<.05). Cumulative antibiotic use across time and by country was evaluated by generalized estimating equations with log link. All statistical analyses were performed using a software program (SAS, version 9.4; SAS Institute Inc).
Results
The baseline characteristics of the study participants are summarized in Table 1. There were 38 152 recorded instances of antibiotic use by TEDDY participants in the first 48 months of life, with 95 unique antibiotic compounds listed. Amoxicillins, in any combination, were the single most commonly used antibiotic compound, comprising 42.3% of all reported use (Figure 1A). All other antibiotic compounds accounted for less than 9% of the total recorded uses. Otitis media was the most common reason for antibiotic use, accounting for 62.0% of all instances (Figure 1D). The β-lactam and macrolides antibiotics that were chosen for further analysis accounted for most (70.5%) of all recorded use in the first 48 months of life (Figure 1C). The rest of the antibiotics were either used too in frequently to have sufficient power for the analysis (17.7%) or were applied topically (11.8%).
Table 1.
Population Characteristics of the Islet Autoimmunity (IA) and Celiac Disease Autoimmunity (CDA) Cohorts
| Variable | IA, % | CDA, % | |||||
|---|---|---|---|---|---|---|---|
| No Islet Autoantibodies (n = 8032) |
First-Appearing Islet Autoantibody | Multiple Islet Autoantibodies | |||||
| Any (n = 463) |
GADA (n = 157) |
IAA (n = 224) |
Second Islet Autoantibody (n = 276) |
No CDA (n = 5775) |
CDA (n = 783) |
||
| Country | |||||||
| Finland | 20.9 | 25.7 | 16.6 | 31.7 | 28.6 | 23.3 | 20.3 |
| Germany | 6.7 | 8.2 | 4.5 | 8.0 | 9.8 | 6.2 | 5.2 |
| Sweden | 29.2 | 33.3 | 35.7 | 32.6 | 30.8 | 29.8 | 40.9 |
| United States | 43.2 | 328 | 42.3 | 27.7 | 30.8 | 40.7 | 33.6 |
| Female sex | 49.5 | 44.9 | 49.7 | 40.2 | 44.6 | 47.4 | 57.9 |
| First-degree relative with T1D or CD | 10.2 | 22.0 | 20.4 | 22.8 | 26.5 | 2.5 | 8.0 |
| HLA-DR genotype | |||||||
| DR4/4, X | 40.0 | 36.7 | 26.8 | 43.7 | 37.0 | 42.8 | 16.0 |
| DR3/4 | 38.3 | 50.3 | 46.5 | 50.0 | 56.5 | 39.8 | 36.5 |
| DR3/3, X | 21.7 | 13.0 | 26.7 | 6.3 | 6.5 | 17.4 | 47.5 |
| Cesarean delivery | 26.2 | 21.8 | 24.2 | 20.5 | 21.7 | 26.6 | 21.5 |
| Probiotic use before age 90 d | 14.1 | 13.2 | 10.2 | 13.8 | 14.9 | 15.1 | 16.2 |
| Breastfeeding status at age 90 d | |||||||
| Still exclusive breastfeeding | 27.1 | 26.4 | 26.8 | 27.6 | 25.3 | 27.3 | 31.9 |
| Breastfeeding but not exclusively | 48.1 | 52.7 | 49.5 | 52.0 | 53.1 | 50.7 | 51.9 |
| Stopped breastfeeding | 24.8 | 21.0 | 23.7 | 20.4 | 21.6 | 21.9 | 16.2 |
| Maternal antibiotic use during pregnancy | 22.8 | 21.2 | 23.6 | 20.5 | 19.9 | 22.7 | 22.7 |
| Season of birth | |||||||
| Spring (March to May) | 23.4 | 24.0 | 28.0 | 23.2 | 26.5 | 23.2 | 25.7 |
| Summer (June to August) | 25.3 | 23.8 | 19.8 | 26.8 | 25.7 | 25.4 | 27.1 |
| Fall (September to November) | 25.3 | 28.5 | 28.7 | 29.5 | 26.1 | 25.4 | 23.2 |
| Winter (December to February) | 26.1 | 23.8 | 23.6 | 20.5 | 21.7 | 26.0 | 24.0 |
Abbreviations: CD, celiac disease; GADA, glutamic acid decarboxylase autoantibody; IAA, insulin autoantibody; T1D, type 1 diabetes.
Antibiotic use differed by country (eFigure 2 in the Supplement). In Sweden, the preferred penicillin is penicillin V, while amoxicillin is preferred in the United States and Finland (eFigure 2C in the Supplement). Cephalosporins and macrolides are used sparingly compared with their use in the United States, Germany, and Finland (eFigure 2E and F in the Supplement). Most children (72.2%) had received 0 to 2 doses of any antibiotic type by age 1 year; by age 4 years, 34.7% of children had received 2 to 3 doses of any antibiotic, and 37.6% had received 4 or more doses (Figure 3).
Figure 3. Frequency of Use by Antibiotic Type.
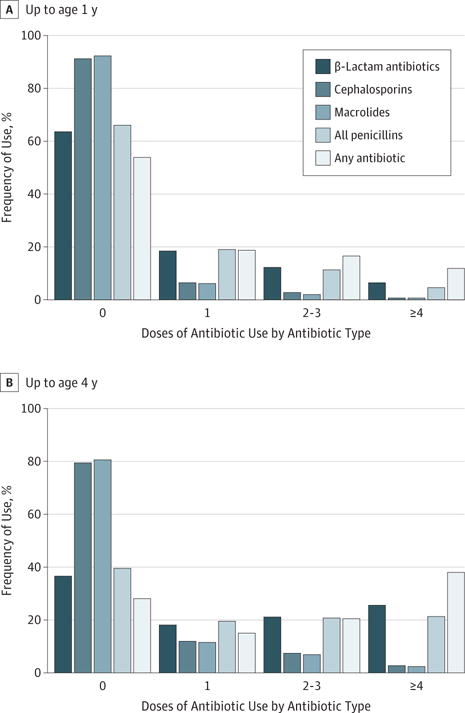
Frequency of use (in doses) is shown categorically for each specific antibiotic up to age 1 year (A) and up to age 4 years (B).
Antibiotic Use and the Appearance of Islet Autoantibodies
A total of 463 of 8495 individuals (5.5%) developed any islet autoantibodies before age 49.5 months (Table 1 and eFigure 1 in the Supplement). The median age at seroconversion to any islet autoantibodies was 21.4 months (range, 12.2–33.0 months), with IAA-first seroconversion occurring earlier (median, 16.2 months) than GADA-first seroconversion (median, 27.0 months). The median age at seroconversion to multiple islet autoantibodies was 24.3 months (range, 16.5–33.4 months). Cumulative, first-year, and second-year exposure, as well as exposure to any antibiotic category in the year before the seroconversion period, was not associated with IA or multiple islet auto-antibodies (Table 2). Further examination during the more vulnerable periods showed that antibiotic use soon after birth and just before seroconversion was also not associated with IA or multiple islet autoantibodies (eTable 2 in the Supplement).
Table 2.
Antibiotic Use Over Time Relative to the Risk of Islet Autoimmunity (IA), Multiple Islet Autoantibodies, and Celiac Disease Autoimmunity (CDA) Examined in Separate Cox Proportional Hazards Regression Models
| Variable | Hazard Ratio (95% CI)a | ||
|---|---|---|---|
| IA | Multiple Islet Autoantibodies | CDA | |
| Cumulative Use Since Birth | |||
| Any antibiotic | 0.98 (0.95–1.01) | 0.99 (0.95–1.03) | 1.00 (0.98–1.02) |
| β-Lactam antibiotics | 0.98 (0.95–1.02) | 1.00 (0.96–1.04) | 1.00 (0.97–1.02) |
| Cephalosporins | 0.89 (0.77–1.02) | 0.90 (0.76–1.06) | 1.01 (0.94–1.09) |
| All penicillins | 0.99 (0.95–1.03) | 1.01 (0.96–1.06) | 0.99 (0.97–1.02) |
| Amoxicillins | 0.98 (0.94–1.03) | 1.00 (0.95–1.03) | 0.99 (0.96–1.02) |
| Penicillin | 1.04 (0.92–1.18) | 1.02 (0.87–1.20) | 1.03 (0.92–1.09) |
| Macrolides | 0.90 (0.79–1.04) | 0.85 (0.72–1.02) | 0.99 (0.91–1.08) |
| Use in First 2 y of Life | |||
| Any antibiotic | 0.99 (0.95–1.02) | 0.99 (0.95–1.04) | 1.01 (0.99–1.04) |
| β-Lactam antibiotics | 0.99 (0.95–1.03) | 1.00 (0.95–1.05) | 1.01 (0.98–1.04) |
| Cephalosporins | 0.91 (0.78–1.06) | 0.93 (0.78–1.11) | 1.04 (0.96–1.13) |
| All penicillins | 1.00 (0.95–1.05) | 1.01 (0.96–1.07) | 1.01 (0.98–1.04) |
| Amoxicillins | 0.99 (0.94–1.04) | 1.01 (0.95–1.07) | 1.01 (0.97–1.04) |
| Penicillin | 1.09 (0.94–1.26) | 1.05 (0.87–1.27) | 1.00 (0.91–1.11) |
| Macrolides | 0.92 (0.79–1.07) | 0.86 (0.71–1.04) | 1.04 (0.95–1.14) |
| Use in First Year of Life | |||
| Any antibiotic | 1.01 (0.95–1.07) | 1.00 (0.93–1.07) | 1.03 (0.99–1.07) |
| β-Lactam antibiotics | 1.01 (0.95–1.07) | 1.00 (0.93–1.08) | 1.03 (0.99–1.07) |
| Cephalosporins | 0.87 (0.69–1.09) | 0.95 (0.74–1.24) | 1.03 (0.90–1.17) |
| All penicillins | 1.03 (0.96–1.10) | 1.01 (0.92–1.10) | 1.04 (0.99–1.09) |
| Amoxicillins | 1.03 (0.95–1.10) | 1.01 (0.92–1.11) | 1.03 (0.98–1.09) |
| Penicillin | 1.09 (0.87–1.36) | 1.03 (0.76–1.40) | 1.10 (0.94–1.30) |
| Macrolides | 0.92 (0.74–1.14) | 0.92 (0.70–1.19) | 1.12 (0.96–1.31) |
| Use 1 y Before Seroconversion Period | |||
| Any antibiotic | 1.00 (0.95–1.06) | 0.99 (0.93–1.06) | 1.04 (0.99–1.08) |
| β-Lactam antibiotics | 1.01 (0.95–1.07) | 1.00 (0.93–1.08) | 1.04 (0.99–1.09) |
| Cephalosporins | 0.89 (0.72–1.11) | 0.71 (0.50–1.00) | 1.08 (0.93–1.26) |
| All penicillins | 1.02 (0.96–1.09) | 1.03 (0.96–1.12) | 1.04 (0.99–1.09) |
| Amoxicillins | 1.02 (0.96–1.10) | 1.03 (0.95–1.12) | 1.04 (0.98–1.10) |
| Penicillin | 0.99 (0.80–1.23) | 1.06 (0.82–1.37) | 1.06 (0.93–1.21) |
| Macrolides | 0.93 (0.75–1.15) | 0.87 (0.67–1.14) | 1.05 (0.90–1.22) |
Adjusted for country of residence, sex, first-degree relative with type 1 diabetes or celiac disease, HLA genotype, caesarian delivery, probiotic use before age 90 days, breastfeeding status at age 90 days, maternal antibiotic use during pregnancy, and season of birth.
Antibiotic Use and the Risk of CDA
Celiac disease autoimmunity developed in 783 of 6558 individuals (11.9%) before age 49.5 months (eFigure 1 in the Supplement). The median age at CDA seroconversion was 30.6 months (range, 23.7–37.9). Exposure to antibiotics cumulatively, up to 6 months, during the first or second year of life, and within 6 months before the seroconversion period was not associated with CDA (Table 2 and eTable 2 in the Supplement). There was a weak association between any antibiotic use 0 to 3 months before the seroconversion period (HR, 1.11; 95% CI, 1.00–1.22; P = .05), which was not statistically significant after correction for multiple testing (eTable 2 in the Supplement).
Sensitivity Analysis
Sensitivity analyses were performed to evaluate the consistency of the primary results across the TEDDY countries. No significant associations with IA or CDA were found. There were no significant nonlinear associations with frequency of use (0, 1–2, 2–3, or ≥4 doses) up to ages 1, 2, and 4 years for antibiotic use on the risk of developing IA. No significant associations were found with the interaction of season of antibiotic use and specific antibiotic type assessed for the intervals 0 to 3 months and 3 to 6 months before the seroconversion period and the risk of developing IA or CDA. Although not significant after adjustment for multiple comparisons, a small subgroup of children who received 2 or more doses of macrolides within the first year of life (157 of 6558 [2.4%]) had elevated CDA risk (HR, 1.77; 95% CI, 1.18–2.66; P = .006 before adjustment) compared with children who received fewer than 2 doses of macrolides (6401 of 6558 [97.6%]). No other associations were observed with CDA risk. Finally, maternal use of antibiotics during pregnancy was evaluated as a risk factor but did not significantly contribute to IA or CDA risk (P > .05).
Discussion
This multinational prospective cohort study found no evidence of antibiotic use influencing the risk of T1D or CD–related autoimmunity in the first 4 years of life in a genetically at-risk population. This finding was true for cephalosporins, penicillins, and macrolides, which composed 70.5% of all antibiotic use in the first 48 months of life among children in the TEDDY study. While the use of specific antibiotics differed across the TEDDY countries (eFigure 2 in the Supplement), there was no association between antibiotic use and either autoimmune state (IA or CDA) in any of the 4 countries studied.
The results of previously published rodent studies2–9 suggest that diabetes outcomes are dependent on both the type of antibiotic and the timing of exposure. However, 9 of 10 antibiotics used in those studies did not have significant use in the TEDDY study (<3% of use before age 48 months). Trimethoprim was the only antibiotic used in a rodent study3 that was commonly used in the TEDDY study (10.9% of use before age 48 months) but was not investigated herein because the level of use was insufficient to power the analysis. In the TEDDY study, commonly prescribed antibiotics were not associated with IA or CDA risk.
Previous cohort studies10,21–24 have provided conflicting results on an association between antibiotic use and T1D. However, even when an association was observed, it could not be determined in those studies if the association between increased antibiotic use before T1D was caused by preclinical T1D and impaired glycemic control,10,25 which may increase susceptibility to infections.25–27 In contrast, prospective recording of antibiotic use over a 4-year period in the TEDDY study allowed for determination of exposures before the onset of IA and preclinical T1D. Therefore, the antibiotics analyzed in this study were taken before the development of any autoantibodies and were shown not to be a risk factor for IA or CDA. To date, one other relevant prospective study22 exists, and it limited examination of antibiotic use to the first week of life. Screening for islet autoantibodies also allowed for stratified analyses based on the type of first-appearing autoantibody because IAA-first and GADA-first seroconversion may have different primary triggers.18
Two previous studies11,12 have shown a link between antibiotic use and CD risk in humans, but neither could delineate whether antibiotic use was simply a response to the manifestation of undiagnosed disease.28 A study29 in Sweden found no evidence of increased CD risk with antibiotic use in the first 6 months of life. To our knowledge, the present TEDDY study is the first tTGA screening study to examine associations between antibiotic use and CDA.
Recently, no association between reported bacterial infectious episodes and IA or CDA risk was demonstrated in the first 4 years of life in the TEDDY cohort,30,31 consistent with the observation herein that antibiotic use also did not influence the risk of autoimmunity. Meanwhile, evidence is mounting for a role of viral infections in the etiology of T1D and CD in the TEDDY study, with gastrointestinal and respiratory infections associated with an increased risk of CDA and IA, respectively.30,31
However, both T1D and CD have also been linked to alterations of the human gut microbiome,32–36 which is necessary for proper development of the immune system and establishment of oral tolerance in early life.37,38 Antibiotic influences on the gut microbiome during gestation and immediately after birth may have a detrimental effect on proper development of the microbiome and thus priming of the immune system.39–44 However, previous cohort studies have not found any significant associations between antibiotic use in the first week of life and the risk of T1D autoimmunity32,44 or between antibiotic use during pregnancy and increased risk of either T1D or CD.44 Similarly, in the Finnish study,32 in which Bacteroides dorei was found to have increased abundance before the onset of T1D autoimmunity, no effect of antibiotic use was observed, and each B dorei genome sequenced from this cohort comprised more than 50 antibiotic resistance genes.45 Therefore, B dorei in those children would likely survive after treatment with antibiotics. Because the use of β-lactam and macrolides antibiotics was not a risk factor for IA or CDA in the TEDDY study, the gut bacteria influenced by these antibiotics likely do not have a role in autoimmunity.
Limitations
A limitation of this study was that antibiotic use was not captured by medical records or pharmacy-based registries but was reported by the parents. To reduce error in parental record keeping, the TEDDY protocol required the parents to record any medication use in real time. This method captured the use of medications beyond those prescribed by a physician. The present study was also limited by retrospective collection of maternal antibiotic use during pregnancy and lacked detailed records of the specific antibiotic used by the mother during pregnancy. Therefore, although recollection of medication use during pregnancy may not be optimal, maternal antibiotic use during pregnancy was a binary variable, which limits the potential for overestimating or underestimating use.
Conclusions
The use of antibiotics in early childhood does not increase the risk of IA or CDA in genetically at-risk individuals. This finding suggests that autoimmunity for these 2 diseases is not associated with bacterial infections but does not exclude an association with viral infections or gut bacteria resistant to commonly prescribed antibiotics. Therefore, there is no evidence herein to suggest that current clinical practice in the administration of the major classes of antibiotics should be revised to avoid an increased risk of T1D or CDA in young children.
Supplementary Material
Key Points.
Question
Is the use of β-lactam or macrolide antibiotics in early life associated with autoimmunity for type 1 diabetes or celiac disease?
Findings
Included in this cohort study were 8495 children and 6558 children enrolled in The Environmental Determinants of Diabetes in the Young (TEDDY) study who were tested for islet and tissue transglutaminase autoantibodies, respectively. Among these children at genetic risk for type 1 diabetes or celiac disease, the use of β-lactam or macrolide antibiotics in early life and before seroconversion was not associated with a risk of autoimmunity for either disease.
Meaning
These results suggest that the use of the most common antibiotics in early life does not increase the risk of autoimmunity in children at increased genetic risk.
Acknowledgments
Funding/Support: This study was funded by grants U01 DK63829, U01 6 DK63861, U01 DK63821, U01 DK63865, U01 DK63863, U01 DK63836, 7 U01 DK63790, UC4 DK63829, UC4 DK63861, UC4 DK63821, UC4 8 DK63865, UC4 DK63863, UC4 DK63836, UC4 DK95300, UC4 DK100238, and UC4 DK106955 and by contract HHSN267200700014C from the National Institute of Diabetes and Digestive and Kidney Diseases. The TEDDY study is also funded by grants from the National Institute of Allergy and Infectious Diseases, the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the National Institute of Environmental Health Sciences, the Juvenile Diabetes Research Foundation, and the Centers for Disease Control and Prevention.
Role of the Funder/Sponsor: The funders of this study were represented in The Environmental Determinants of Diabetes in the Young (TEDDY) Steering Committee. The funders had a role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. The corresponding author had the final say in submitting the manuscript for publication.
Footnotes
Author Contributions: Dr Triplett had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Kemppainen, Elding Larsson, V. Simell, Liu, O. G. Simell, Toppari, Ziegler, Rewers, Lernmark, Hagopian, She, Akolkar, Schatz, Atkinson, Blaser, Krischer, Hyöty, Agardh, Triplett.
Acquisition, analysis, or interpretation of data: Kemppainen, Vehik, Lynch, Elding Larsson, Canepa, Koletzko, Liu, Toppari, Ziegler, Hagopian, She, Schatz, Atkinson, Blaser, Krischer, Hyöty, Agardh, Triplett.
Drafting of the manuscript: Kemppainen, Vehik, Schatz, Agardh, Triplett.
Critical revision of the manuscript for important intellectual content: All authors.
Statistical analysis: Vehik, Lynch, Krischer, Hyöty, Triplett.
Obtained funding: O. G. Simell, Toppari, Ziegler, Rewers, Lernmark, Hagopian, She, Schatz, Atkinson, Krischer, Hyöty, Triplett
Administrative, technical, or material support: O. G. Simell, Toppari, Lernmark, Hagopian, Schatz, Krischer.
Study supervision: Liu, O. G. Simell, Toppari, Ziegler, Rewers, Hagopian, Atkinson, Krischer, Agardh, Triplett.
Conflict of Interest Disclosures: None reported.
Group Information: The Environmental Determinants of Diabetes in the Young (TEDDY) Study Group members are the following: Colorado Clinical Center: University of Colorado, Anschutz Medical Campus, Barbara Davis Center for Childhood Diabetes: Marian Rewers (principal investigator), Kimberly Bautista, Judith Baxter, Ruth Bedoy, Daniel Felipe-Morales, Kimberly Driscoll, Brigitte I. Frohnert, Patricia Gesualdo, Michelle Hoffman, Rachel Karban, Edwin Liu, Jill Norris, Adela Samper-Imaz, Andrea Steck, Kathleen Waugh, and Hali Wright. Finland Clinical Center: University of Turku, University of Tampere, University of Oulu, Turku University Hospital, Hospital District of Southwest Finland, Tampere University Hospital, Oulu University Hospital, National Institute for Health and Welfare, Finland, and University of Kuopio: Jorma Toppari (principal investigator), Olli G. Simell, Annika Adamsson, Suvi Ahonen, Heikki Hyöty, Jorma Ilonen, Sanna Jokipuu, Tiina Kallio, Leena Karlsson, Miia Kähönen, Mikael Knip, Lea Kovanen, Mirva Koreasalo, Kalle Kurppa, Tiina Latva-aho, Maria Lönnrot, Elina Mäntymäki, Katja Multasuo, Juha Mykkänen, Tiina Niininen, Sari Niinistö, Mia Nyblom, Petra Rajala, Jenna Rautanen, Anne Riikonen, Mika Riikonen, Jenni Rouhiainen, Minna Romo, Tuula Simell, Ville Simell, Maija Sjöberg, Aino Stenius, Maria Leppänen, Sini Vainionpää, Eeva Varjonen, Riitta Veijola, Suvi M. Virtanen, Mari Vähä-Mäkilä, Mari Åkerlund, and Katri Lindfors. Georgia/Florida Clinical Center: Center for Biotechnology and Genomic Medicine, Augusta University, University of Florida, and Pediatric Endocrine Associates, Atlanta: Jin-Xiong She (principal investigator), Desmond Schatz, Diane Hopkins, Leigh Steed, Jamie Thomas, Janey Adams, Katherine Silvis, Michael Haller, Melissa Gardiner, Richard McIndoe, Ashok Sharma, Joshua Williams, Gabriela Young, Stephen W. Anderson, and Laura Jacobsen. Germany Clinical Center: Forschergruppe Diabetes e.V. and Institute of Diabetes Research, Helmholtz Zentrum München, and Klinikum rechts der Isar, Technische Universität München, Center for Regenerative Therapies, Technische Universität Dresden, Dr von Hauner Children’s Hospital, Department of Gastroenterology, Ludwig Maximillians University München, and Research Institute for Child Nutrition, Dortmund: Anette G. Ziegler (principal investigator), Andreas Beyerlein, Ezio Bonifacio, Michael Hummel, Sandra Hummel, Kristina Foterek, Nicole Janz, Mathilde Kersting, Annette Knopff, Sibylle Koletzko, Claudia Peplow, Roswith Roth, Marlon Scholz, Joanna Stock, Katharina Warncke, Lorena Wendel, and Christiane Winkler. Sweden Clinical Center: Lund University: Åke Lernmark (principal investigator), Daniel Agardh, Carin Andrén Aronsson, Maria Ask, Jenny Bremer, Ulla-Marie Carlsson, Corrado Cilio, Emelie Ericson-Hallström, Lina Fransson, Thomas Gard, Joanna Gerardsson, Rasmus Bennet, Monica Hansen, Gertie Hansson, Susanne Hyberg, Fredrik Johansen, Berglind Jonsdottir, Helena Elding Larsson, Marielle Lindström, Markus Lundgren, Maria Månsson-Martinez, Maria Markan, Jessica Melin, Zeliha Mestan, Karin Ottosson, Kobra Rahmati, Anita Ramelius, Falastin Salami, Sara Sibthorpe, Birgitta Sjöberg, Ulrica Swartling, Evelyn Tekum Amboh, Carina Törn, Anne Wallin, Åsa Wimar, and Sofie Åberg. Washington Clinical Center: Pacific Northwest Diabetes Research Institute: William A. Hagopian (principal investigator), Michael Killian, Claire Cowen Crouch, Jennifer Skidmore, Josephine Carson, Maria Dalzell, Kayleen Dunson, Rachel Hervey, Corbin Johnson, Rachel Lyons, Arlene Meyer, Denise Mulenga, Alexander Tarr, Morgan Uland, and John Willis. Pennsylvania Satellite Center: Children’s Hospital of Pittsburgh of University of Pittsburgh Medical Center: Dorothy Becker, Margaret Franciscus, MaryEllen Dalmagro-Elias Smith, Ashi Daftary, Mary Beth Klein, and Chrystal Yates. Data Coordinating Center: University of South Florida: Jeffrey P. Krischer (principal investigator), Michael Abbondondolo, Sarah Austin-Gonzalez, Maryouri Avendano, Sandra Baethke, Rasheedah Brown, Brant Burkhardt, Martha Butterworth, Joanna Clasen, David Cuthbertson, Christopher Eberhard, Steven Fiske, Dena Garcia, Jennifer Garmeson, Veena Gowda, Kathleen Heyman, Francisco Perez Laras, Hye-Seung Lee, Shu Liu, Xiang Liu, Kristian Lynch, Jamie Malloy, Cristina McCarthy, Steven Meulemans, Hemang Parikh, Chris Shaffer, Laura Smith, Susan Smith, Noah Sulman, Roy Tamura, Ulla Uusitalo, Kendra Vehik, Ponni Vijayakandipan, Keith Wood, and Jimin Yang. Past staff are Lori Ballard, David Hadley, and Wendy McLeod. Project Scientist: National Institute of Diabetes and Digestive and Kidney Diseases: Beena Akolkar. Other Contributors: National Institute of Allergy and Infectious Diseases: Kasia Bourcier. Columbia University: Thomas Briese. Florida State University: Suzanne Bennett Johnson. University of Florida: Eric W. Triplett. Autoantibody Reference Laboratories: Barbara Davis Center for Childhood Diabetes, University of Colorado Denver, School of Clinical Sciences, University of Bristol, United Kingdom: Liping Yu, Dongmei Miao, Polly Bingley, Alistair Williams, Kyla Chandler, Saba Rokni, Claire Williams, Rebecca Wyatt, Gifty George, and Sian Grace. Repository: National Institute of Diabetes and Digestive and Kidney Diseases Biosample Repository at Fisher BioServices: Sandra Ke and Niveen Mulholland.
Additional Contributions: Steven Fiske, BS, and Rachel Richesson, BS, at The Environmental Determinants of Diabetes in the Young (TEDDY) Data Coordinating Center worked on the medical coding system and integration of RxNorm and TEDDY medication codes. Mike Haller, MD (College of Medicine, University of Florida) reviewed the antibiotic data to identify parenteral compounds for removal. No compensation was received by these individuals.
Contributor Information
Kaisa M. Kemppainen, Department of Microbiology and Cell Science, Institute of Food and Agricultural Sciences, University of Florida, Gainesville.
Kendra Vehik, Health Informatics Institute, Morsani College of Medicine, University of South Florida, Tampa.
Kristian F. Lynch, Health Informatics Institute, Morsani College of Medicine, University of South Florida, Tampa.
Helena Elding Larsson, Department of Clinical Sciences, Lund University Clinical Research Center, Skåne University Hospital, Malmö, Sweden.
Ronald J. Canepa, Department of Microbiology and Cell Science, Institute of Food and Agricultural Sciences, University of Florida, Gainesville.
Ville Simell, MediCity Laboratory, University of Turku, Turku, Finland.
Sibylle Koletzko, Division of Paediatric Gastroenterology and Hepatology, Dr von Hauner Children’s Hospital, Ludwig Maximilian University, München, Germany.
Edwin Liu, Digestive Health Institute, Children’s Hospital Colorado, Anschutz Medical Campus, University of Colorado Denver, Aurora.
Olli G. Simell, Research Centre of Applied and Preventive Cardiovascular Medicine, University of Turku, Turku, Finland.
Jorma Toppari, Department of Pediatrics, University of Turku, Turku University Hospital, Turku, FinlandDepartment of Physiology, Institute of Biomedicine, University of Turku, Turku, Finland.
Anette G. Ziegler, Institute of Diabetes Research, Helmholtz Zentrum München, München, GermanyKlinikum Rechts der Isar, Technische Universität München, München, GermanyForschergruppe Diabetes e.V., Neuherberg, Germany.
Marian J. Rewers, Barbara Davis Center for Childhood Diabetes, University of Colorado Denver, Aurora.
Åke Lernmark, Department of Clinical Sciences, Lund University Clinical Research Center, Skåne University Hospital, Malmö, Sweden.
William A. Hagopian, Pacific Northwest Diabetes Research Institute, Seattle, Washington.
Jin-Xiong She, Center for Biotechnology and Genomic Medicine, Medical College of Georgia, Augusta University, Augusta.
Beena Akolkar, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, Maryland.
Desmond A. Schatz, Department of Pediatrics, College of Medicine, University of Florida, Gainesville.
Mark A. Atkinson, Department of Pathology, Immunology, and Laboratory Medicine, College of Medicine, University of Florida, Gainesville.
Martin J. Blaser, Department of Medicine and Microbiology, New York School of Medicine, New York.
Jeffrey P. Krischer, Health Informatics Institute, Morsani College of Medicine, University of South Florida, Tampa.
Heikki Hyöty, Department of Virology, Faculty of Medicine and Life Sciences, University of Tampere, Tampere, FinlandFimlab Laboratories, Pirkanmaa Hospital District, Tampere, Finland.
Daniel Agardh, Department of Clinical Sciences, Lund University Clinical Research Center, Skåne University Hospital, Malmö, Sweden.
Eric W. Triplett, Department of Microbiology and Cell Science, Institute of Food and Agricultural Sciences, University of Florida, Gainesville.
References
- 1.Fletcher C. First clinical use of penicillin. BMJ (Clin Res Ed) 1984;289(6460):1721–1723. doi: 10.1136/bmj.289.6460.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown K, Godovannyi A, Ma C, et al. Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice. ISME J. 2016;10(2):321–332. doi: 10.1038/ismej.2015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brugman S, Klatter FA, Visser JT, et al. Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat: is the gut flora involved in the development of type 1 diabetes? Diabetologia. 2006;49(9):2105–2108. doi: 10.1007/s00125-006-0334-0. [DOI] [PubMed] [Google Scholar]
- 4.Candon S, Perez-Arroyo A, Marquet C, et al. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS One. 2015;10(5):e0125448. doi: 10.1371/journal.pone.0125448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen CH, Krych L, Nielsen DS, et al. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia. 2012;55(8):2285–2294. doi: 10.1007/s00125-012-2564-7. [DOI] [PubMed] [Google Scholar]
- 6.Hu Y, Peng J, Tai N, et al. Maternal antibiotic treatment protects offspring from diabetes development in nonobese diabetic mice by generation of tolerogenic antigen-presenting cells. J Immunol. 2015;195(9):4176–4184. doi: 10.4049/jimmunol.1500884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Y, Jin P, Peng J, Zhang X, Wong FS, Wen L. Different immunological responses to early-life antibiotic exposure affecting autoimmune diabetes development in NOD mice. J Autoimmun. 2016;72(8):47–56. doi: 10.1016/j.jaut.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz RF, Neu J, Schatz D, Atkinson MA, Wasserfall C. Comment on: Brugman S et al. (2006) Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat: Is the gut flora involved in the development of type 1 diabetes? Diabetologia 49:2105–2108. Diabetologia. 2007;50(1):220–221. doi: 10.1007/s00125-006-0526-7. [DOI] [PubMed] [Google Scholar]
- 9.Livanos AE, Greiner TU, Vangay P, et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat Microbiol. 2016;1(11):16140. doi: 10.1038/nmicrobiol.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kilkkinen A, Virtanen SM, Klaukka T, et al. Use of antimicrobials and risk of type 1 diabetes in a population-based mother-child cohort. Diabetologia. 2006;49(1):66–70. doi: 10.1007/s00125-005-0078-2. [DOI] [PubMed] [Google Scholar]
- 11.Canova C, Zabeo V, Pitter G, et al. Association of maternal education, early infections, and antibiotic use with celiac disease: a population-based birth cohort study in northeastern Italy. Am J Epidemiol. 2014;180(1):76–85. doi: 10.1093/aje/kwu101. [DOI] [PubMed] [Google Scholar]
- 12.Mårild K, Ye W, Lebwohl B, et al. Antibiotic exposure and the development of coeliac disease: a nationwide case-control study. BMC Gastroenterol. 2013;13:109. doi: 10.1186/1471-230X-13-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagopian WA, Lernmark A, Rewers MJ, et al. TEDDY: The Environmental Determinants of Diabetes in the Young: an observational clinical trial. Ann N Y Acad Sci. 2006;1079:320–326. doi: 10.1196/annals.1375.049. [DOI] [PubMed] [Google Scholar]
- 14.Hagopian WA, Erlich H, Lernmark A, et al. TEDDY Study Group The Environmental Determinants of Diabetes in the Young (TEDDY): genetic criteria and international diabetes risk screening of 421 000 infants. Pediatr Diabetes. 2011;12(8):733–743. doi: 10.1111/j.1399-5448.2011.00774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baxter J, Vehik K, Johnson SB, Lernmark B, Roth R, Simell T, TEDDY Study Group Differences in recruitment and early retention among ethnic minority participants in a large pediatric cohort: the TEDDY study. Contemp Clin Trials. 2012;33(4):633–640. doi: 10.1016/j.cct.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lernmark B, Johnson SB, Vehik K, et al. Enrollment experiences in a pediatric longitudinal observational study: The Environmental Determinants of Diabetes in the Young (TEDDY) study. Contemp Clin Trials. 2011;32(4):517–523. doi: 10.1016/j.cct.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.TEDDY Study Group. The Environmental Determinants of Diabetes in the Young (TEDDY) study: study design. Pediatr Diabetes. 2007;8(5):286–298. doi: 10.1111/j.1399-5448.2007.00269.x. [DOI] [PubMed] [Google Scholar]
- 18.Krischer JP, Lynch KF, Schatz DA, et al. TEDDY Study Group The 6 year incidence of diabetes-associated autoantibodies in genetically at-risk children: the TEDDY study. Diabetologia. 2015;58(5):980–987. doi: 10.1007/s00125-015-3514-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu E, Lee HS, Aronsson CA, et al. TEDDY Study Group Risk of pediatric celiac disease according to HLA haplotype and country. N Engl J Med. 2014;371(1):42–49. doi: 10.1056/NEJMoa1313977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc. 2011;18(4):441–448. doi: 10.1136/amiajnl-2011-000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hviid A, Svanström H. Antibiotic use and type 1 diabetes in childhood. Am J Epidemiol. 2009;169(9):1079–1084. doi: 10.1093/aje/kwp038. [DOI] [PubMed] [Google Scholar]
- 22.Virtanen SM, Takkinen HM, Nwaru BI, et al. Microbial exposure in infancy and subsequent appearance of type 1 diabetes mellitus–associated autoantibodies: a cohort study. JAMA Pediatr. 2014;168(8):755–763. doi: 10.1001/jamapediatrics.2014.296. [DOI] [PubMed] [Google Scholar]
- 23.Boursi B, Mamtani R, Haynes K, Yang YX. The effect of past antibiotic exposure on diabetes risk. Eur J Endocrinol. 2015;172(6):639–648. doi: 10.1530/EJE-14-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fazeli Farsani S, Souverein PC, van der Vorst MM, Knibbe CA, de Boer A, Mantel-Teeuwisse AK. Population-based cohort study of anti-infective medication use before and after the onset of type 1 diabetes in children and adolescents. Antimicrob Agents Chemother. 2014;58(8):4666–4674. doi: 10.1128/AAC.03080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casqueiro J, Casqueiro J, Alves C. Infections in patients with diabetes mellitus: a review of pathogenesis. Indian J Endocrinol Metab. 2012;16(suppl 1):S27–S36. doi: 10.4103/2230-8210.94253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller LM, Gorter KJ, Hak E, et al. Increased risk of common infections in patients with type 1 and type 2 diabetes mellitus. Clin Infect Dis. 2005;41(3):281–288. doi: 10.1086/431587. [DOI] [PubMed] [Google Scholar]
- 27.Simonsen JR, Harjutsalo V, Järvinen A, et al. FinnDiane Study Group Bacterial infections in patients with type 1 diabetes: a 14-year follow-up study. BMJ Open Diabetes Res Care. 2015;3(1):e000067. doi: 10.1136/bmjdrc-2014-000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mårild K, Kahrs CR, Tapia G, Stene LC, Størdal K. Infections and risk of celiac disease in childhood: a prospective nationwide cohort study. Am J Gastroenterol. 2015;110(10):1475–1484. doi: 10.1038/ajg.2015.287. [DOI] [PubMed] [Google Scholar]
- 29.Myléus A, Hernell O, Gothefors L, et al. Early infections are associated with increased risk for celiac disease: an incident case-referent study. BMC Pediatr. 2012;12:194. doi: 10.1186/1471-2431-12-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kemppainen KM, Lynch KF, Liu E, et al. TEDDY Study Group Factors that increase risk of celiac disease autoimmunity after a gastrointestinal infection in early life. Clin Gastroenterol Hepatol. 2017;15(5):694–702.e5. doi: 10.1016/j.cgh.2016.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lönnrot M, Lynch KF, Elding Larsson H, et al. TEDDY Study Group Respiratory infections are temporally associated with initiation of type 1 diabetes autoimmunity: the TEDDY study. [published online August 2, 2017] Diabetologia. 2017 doi: 10.1007/s00125-017-4365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis-Richardson AG, Ardissone AN, Dias R, et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front Microbiol. 2014;5:678. doi: 10.3389/fmicb.2014.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis-Richardson AG, Triplett EW. A model for the role of gut bacteria in the development of autoimmunity for type 1 diabetes. Diabetologia. 2015;58(7):1386–1393. doi: 10.1007/s00125-015-3614-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knip M, Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol. 2016;12(3):154–167. doi: 10.1038/nrendo.2015.218. [DOI] [PubMed] [Google Scholar]
- 35.Marasco G, Di Biase AR, Schiumerini R, et al. Gut microbiota and celiac disease. Dig Dis Sci. 2016;61(6):1461–1472. doi: 10.1007/s10620-015-4020-2. [DOI] [PubMed] [Google Scholar]
- 36.McLean MH, Dieguez D, Jr, Miller LM, Young HA. Does the microbiota play a role in the pathogenesis of autoimmune diseases? Gut. 2015;64(2):332–341. doi: 10.1136/gutjnl-2014-308514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107(27):12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker WA. Initial intestinal colonization in the human infant and immune homeostasis. Ann Nutr Metab. 2013;63(suppl 2):8–15. doi: 10.1159/000354907. [DOI] [PubMed] [Google Scholar]
- 39.Becattini S, Taur Y, Pamer EG. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med. 2016;22(6):458–478. doi: 10.1016/j.molmed.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bokulich NA, Chung J, Battaglia T, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8(343):343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fouhy F, Guinane CM, Hussey S, et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother. 2012;56(11):5811–5820. doi: 10.1128/AAC.00789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Francino MP. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front Microbiol. 2016;6:1543. doi: 10.3389/fmicb.2015.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pérez-Cobas AE, Gosalbes MJ, Friedrichs A, et al. Gut microbiota disturbance during antibiotic therapy: a multiomic approach. Gut. 2013;62(11):1591–1601. doi: 10.1136/gutjnl-2012-303184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mårild K, Ludvigsson J, Sanz Y, Ludvigsson JF. Antibiotic exposure in pregnancy and risk of coeliac disease in offspring: a cohort study. BMC Gastroenterol. 2014;14:75. doi: 10.1186/1471-230X-14-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leonard MT, Davis-Richardson AG, Ardissone AN, et al. The methylome of the gut microbiome: disparate Dam methylation patterns in intestinal Bacteroides dorei. Front Microbiol. 2014;5:361. doi: 10.3389/fmicb.2014.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


