Abstract
Lung cancer is the leading cause of cancer deaths with small cell lung cancer (SCLC) as the most aggressive subtype. Preferential occurrence of TP53 missense mutations rather than loss implicates a selective advantage for TP53 mutant expression in SCLC pathogenesis. We show that lung epithelial expression of R270H and R172H (R273H and R175H in humans), common Trp53 mutations in lung cancer, combined with Rb1 loss selectively results in two subtypes of neuroendocrine carcinoma, SCLC and large cell neuroendocrine carcinoma (LCNEC). Tumor initiation and progression occur in a remarkably consistent time frame with short latency and uniform progression to lethal metastatic disease by 7 months. R270H or R172H expression and Trp53 loss result in similar phenotypes demonstrating that Trp53 mutants promote lung carcinogenesis through loss-of-function and not gain-of-function mechanisms. Tumor responses to targeted and cytotoxic therapeutics were discordant in mice and corresponding tumor cell cultures demonstrating need to assess therapeutic response at the organismal level. Rapamycin did not have therapeutic efficacy in the mouse model despite inhibiting mTOR signaling and markedly suppressing tumor cell growth in culture. In contrast, cisplatin/etoposide treatment using a patient regimen prolonged survival with development of chemoresistance recapitulating human responses. R270H, but not R172H, expression conferred gain-of-function activity in attenuating chemotherapeutic efficacy. These data demonstrate a causative role for Trp53 mutants in development of chemoresistant lung cancer, and provide tractable preclinical models to test novel therapeutics for refractory disease.
Keywords: SCLC, Trp53 mutant proteins, Rb1, GEMM, chemotherapy
Introduction
Lung cancer is the leading cause of cancer deaths (1). Lung cancers are divided into small cell (SCLC) and non-small cell lung cancers (NSCLC) based upon distinct genetic, histologic and biologic features. SCLC account for 13% of lung cancers and are highly lethal malignancies with a dismal <5% 5-year survival (2, 3). Patients have an initial significant response to platinum based chemotherapy, but SCLC invariably relapses as treatment resistance disease (2, 3). Surgical resection and biopsy of posttreatment disease is rarely performed resulting in limited human tissue that represents a significant barrier to molecular understanding of disease progression and treatment resistance. Genetically engineered mouse models (GEMM) played a critical role in identifying molecular alterations that drive lung cancer (4, 5). Advanced GEMM that genetically mimic human disease, reflect the cellular heterogeneity of SCLC, and model both primary and resistant disease are now needed. These models should have consistent, short tumor latencies and progression with tools to measure tumor burden to serve as preclinical platforms to define mechanisms driving therapy resistance and test novel therapeutics.
Loss of TP53 and RB1 function is obligatory for development of human SCLC (6, 7). One allele of both genes is lost by deletion while alterations in the second allele are distinct for RB1 and TP53. RB1 genetic alterations result in loss of full length protein expression whereas the majority of TP53 alterations are missense mutations resulting in expression of a mutant protein (6). TP53 mutants can promote cancer through three mechanisms: 1) loss or diminution of wild type activity (loss-of-function), 2) forming heterotetrameric complexes with wild type TP53 that inhibit function (dominant negative function), and 3) acquisition of novel cancer promoting activities (gain-of-function) (8–10). Patients with germline TP53 missense mutations have significantly earlier cancer onset than patients with TP53 loss providing evidence for gain-of-function TP53 mutant activities (11, 12). Mice with germline Trp53 mutant expression have distinct tumor profiles and metastatic cancers not seen in Trp53 null mice providing direct evidence that Trp53 mutants drive carcinogenesis through gain-of-function mechanisms (13, 14).
TP53 mutant function is tumor type specific with unclear roles in lung cancer. TP53 codons 175 and 273 are among the most frequent sites of missense mutations in human lung cancer (15). Mice engineered to express Trp53 mutants R172H and R270H (R175H and R273H in humans) develop metastasizing adenocarcinomas that are not seen in Trp53 deficient mice supporting dominant negative and/or gain-of-function activities in NSCLC (13, 14). Similar metastatic lung adenocarcinomas occur in mice hemizygous for R172H and a latent oncogenic mutant K-ras allele that undergoes spontaneous activation to drive lung adenocarcinoma (16). In contrast, conditional expression of R172H and oncogenic K-ras induced by intranasal delivery of adenoviral Cre recombinase did not promote lung cancer (17). SCLC has not been reported in Trp53 mutant mice demonstrating that Trp53 mutants function differently in NSCLC and SCLC. In further support of this notion, codon distributions of TP53 mutations are distinct in NSCLC and SCLC (15). The function and mechanisms of action of Trp53 mutants in SCLC remain unknown.
Although significant progress has been made in NSCLC treatment, platinum based chemotherapy has remained the standard treatment for SCLC for >30 years. Despite initial tumor responses in 60–80% of patients, primary and acquired resistance limit chemotherapy efficacy (2). Mechanisms underlying chemoresistance in SCLC remain unknown. The significance of TP53 status in chemotherapy response is still ambiguous, and it is not yet clear if the enhanced resistance to cytotoxic agents conferred by TP53 mutants in cell lines in culture is operative at the organism level (18–20). The presence of “nondisruptive” TP53 missense mutations with conservative amino acid changes not predicted to markedly alter protein structure (including R175H and R273H) is an independent negative prognostic indicator of shorter survival in lung cancer patients providing evidence for a functional role for TP53 mutants in lung cancer (21). These clinical observations and findings in cultured cells suggest that specific TP53 mutants have gain-of-function activities that drive lung carcinogenesis and dictate treatment response. The goals of the current study were to determine the role of Trp53 mutants in lung cancer pathogenesis, and to develop a tractable preclinical model of chemoresistant disease to serve as a platform for identifying mechanisms of treatment resistance and testing novel therapeutics.
Materials and Methods
Mouse model
Mice were generated by mating previously described Sftpc-rtTA;tetO-Cre;Rbflox/flox or Scgb1a1-rtTA;tetO-Cre;Rbflox/flox mice with Trp53flox/flox, Trp53R172H/wt and Trp53R270H/wt mice obtained from NCI Mouse Repository (FVB.129P2-Trp53tm1Brn, 129S4-Trp53tm2Tyj, and 129S4-Trp53tm3Tyj, respectively) resulting in a mixed genetic background including FVB/N, 129 and C57Bl/6 (22, 23). Genotypes were determined by PCR analysis using established primers (14, 22, 23). Gene recombination was induced by feeding doxycycline food to pregnant dams throughout gestation. Mice were analyzed for lung tumors and metastases by gross examination of thoracic and abdominal organs with all tumors examined microscopically. Random histologic sections from livers lacking gross lesions were also examined microscopically for metastatic disease. Animals were maintained in a pathogen free facility with studies approved by Cincinnati Children’s Research Foundation Institutional Animal Care and Use Committee.
Histology, immunohistochemistry and immunofluorescence
Tissues were fixed in 10% formalin, paraffin embedded and analyzed as hematoxylin and eosin (H&E) stained sections. Methanol/hydrogen peroxide pretreatment, microwave 10mM citrate antigen retrieval, and serum blocking was performed. Antibodies were incubated at 4°C overnight: CALCA (Sigma-Aldrich, C8198, 1:8000), SCGB1A1 (Santa Cruz Biotechnology Inc., sc-9773, 1:2000), SFTPC (pro-SPC) (Seven Hill, RB458, 1:3000), NKX2-1 (Seven Hill, 1231, 1:1000), and SOX2 (Seven Hill, R1236, 1:200). For immunohistochemistry, slides were stained with Vectastain Elite ABC and DAB Substrate kits (Vector Laboratories) and counterstained with nuclear fast red. For immunofluorescence, slides were incubated with β-galactosidase (Abcam, ab9396, 1:1000) and CALCA (Sigma-Aldrich, C8198, 1:8000) followed by goat anti-chicken Alexa Fluor 484 (Invitrogen, A11039, 1:200) and goat anti-rabbit Alexa Fluor 594 (Invitrogen, R37117, 1:200) secondary antibodies for 1 hour at room temperature in the dark. Slides were coverslipped with Vectashield mounting medium with DAPI (Vector Laboratories) and analyzed using the Zeiss Apotome system (Zeiss, Jena, Germany).
Western blot analysis
Tissues were homogenized in lysis buffer supplemented with protease and phosphatase inhibitors (Thermo Scientific), and 25–50 μg total protein was resolved by SDS-polyacrylamide electrophoresis under reducing conditions. Proteins transferred to nitrocellulose membranes were probed with antibodies: phospho-S6K (Thr389) (Cell Signaling, 9234, 1:1000), S6K (Abcam, ab32529, 1:2000), phospho-S6 (Ser235/236) (Cell Signaling, 2211, 1:2000), S6 (Cell Signaling, 2217, 1:2000), phospho-4EBP1 (Thr37/46) (Cell Signaling, 2855, 1:2000), 4EBP1 (Cell Signaling, 9644, 1:2000), PTEN (Cell Signaling, 9188, 1:2000), ACTA1 (Sigma-Aldrich, A5060, 1:2000) and GAPDH (Cell Signaling, 8884, 1:20000). Detection was performed with horseradish peroxidase–conjugated secondary antibodies and ECL Prime Western Blotting Detection Reagent (GE Healthcare).
PCR analysis for Trp53 and Rb1 gene recombination
Genomic DNA was subjected to 30 PCR cycles (30 sec at 94 °C, 30 sec at 58 °C, and 30 sec at 72 °C) using Trp53 primers Trp53-T008 (5′-CACAAAAACAGGTTAAACCCA-3′) and p53-T011 (5′-GAAGACAGAAAAGGG-3′), Trp53 mutant primers Trp53Rec-F (5′ - AGC CTG CCT AGC TTC CTC AGG-3′) and Trp53Rec-R (5′-CTT GGA GAC ATA GCC ACA CTG-3′), and Rb1 primers Rb212, Rb18 and Rb19E (24).
Establishment of tumor cell cultures and growth inhibition assay
Lung tumors were minced and cultured in HITES medium supplemented with 2% fetal bovine serum (FBS) and penicillin/streptomycin. Primary cells were established from the tumors (2010–2013), seeded in 96-well plates at 1×104 cells/well, treated with cisplatin and/or etoposide and assayed for cell growth by WST-1 assay. For 3D culture, cells were seeded in 96-well NanoCulture plates (Organogenix, Inc., Kawasaki, Japan) at 1×104 cells/well, cultured in 20% FBS-HITES medium until spheroids formed, and treated with cisplatin and etoposide.
Treatments with rapamycin and cisplatin-etoposide
Encapsulated rapamycin or empty eudragit vehicle control capsules (University of Texas Health Science Center, San Antonio, TX) were incorporated into Purina 5LG6 diet (Purina TestDiet, Richmond, IN). Mice were fed a diet containing 14ppm rapamycin or vehicle control. Cisplatin (Sigma-Aldrich) and etoposide (Sigma-Aldrich) were administrated by intraperitoneal injection.
Magnetic resonance imaging and tumor analysis
Magnetic Resonance Imaging (MRI) was performed ≤ 7 days prior to chemotherapy cycle 1 for baseline tumor measurements and then after cycle 4 and one week after cycle 6 to assess treatment response. Respiration during imaging was maintained at 80–160 breaths per minute using 1.25–2% isoflorane (Henry Schein) delivered with 0.4 L/min medical air supplemented with ketamine (Henry Schein) at 1–2.5 mg/25g body weight for erratic breathing to facilitate respiratory gating. Images were acquired on a 7T Bruker Avance system (Ettlingen, Germany) using a 38 mm linear Bruker coil. Isolated tumors ≥ 0.4mm in maximal dimension or arising during treatment were identified on coronal sections and followed during the course of treatment. Tumor volumes were determined by Region of Interest (ROI) measurements for each tumor slice using ImageJ software (National Institutes of Health, Bethesda, Maryland), converted to mm3 and summed to determine total tumor volumes. Tumor response was defined as: progressive disease (PD) - ≥ 20% increase in tumor volume; partial response (PR) - ≥ 30% reduction in tumor volume; stable disease (SD) - neither sufficient decrease to categorize as PR nor sufficient increase to categorize as PD; complete response (CR) - tumors no longer visible by imaging (25).
Statistical analysis
GraphPad Prism ver. 5.0d (GraphPad Software, La Jolla, CA) was used for statistical analyses. Symptom free survival was analyzed by Kaplan Meier log-rank test. Cell growth changes were determined using unpaired Student’s t-tests. One-way Anova followed by Tukey’s multiple comparison was used to analyze tumor size. Logistic regression was used for chemotherapy response. Statistical significance was defined as p < 0.05.
Results
Mutant Trp53 proteins promote development of lung neuroendocrine carcinomas through loss-of-function and not dominant-negative or gain-of-function mechanisms
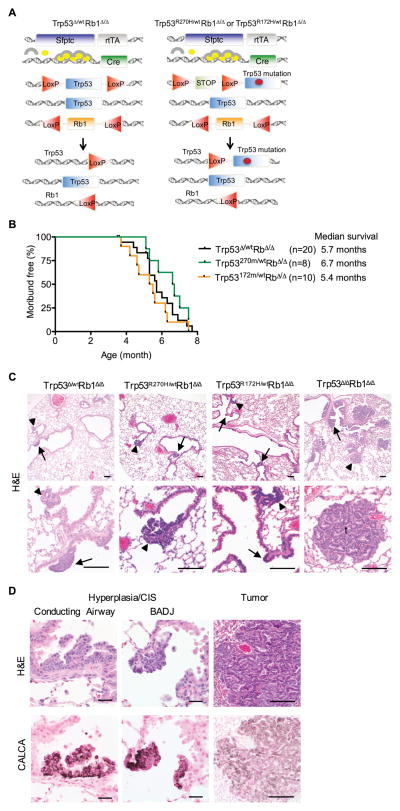
To determine the effects and mechanisms of action of Trp53 mutant proteins in Rb1 deficient lung epithelium, mice with heterozygous Trp53 mutant Rb1 deficient lung epithelium (Trp53R172H/wtRb1Δ/Δ and Trp53R270H/wtRb1Δ/Δ) were generated and phenotypes compared to heterozygous Trp53 deficient (Trp53Δ/wtRb1Δ/Δ) lungs (Fig. 1A). Trp53 and Rb1 gene recombination was targeted to the lung epithelium using doxycycline induced reverse tetracycline transactivator expression driven by the human surfactant protein C (Sftpc) gene promoter combined with a tetO-Cre transgene (Fig. 1A) (26). Using this system, gene expression is altered throughout the diverse cell types comprising the lung epithelium. Survival of Trp53R172H/wtRb1Δ/Δ, Trp53R270H/wtRb1Δ/Δ and Trp53Δ/wtRb1Δ/Δ mice were similar with indistinguishable lung phenotypes (Fig. 1B–C). Multifocal neuroendocrine hyperplasia and carcinomas-in-situ (CIS) involved the conducting airways and bronchoalveolar duct junctions (BADJ) in lungs of all three genotypes at 3.5–7.7 months of age (Fig 1C). A rare lymphoma and a 1 mm papillary lung adenoma were each detected in a single Trp53R172H/wtRb1Δ/Δ mouse. These data demonstrate that heterozygous Trp53 loss and R172H or R270H expression have the same biologic effects thus providing direct evidence that R172H and R270H promote lung tumorigenesis through loss-of-function and not dominant negative mechanisms.
Figure 1. R270H and R172H do not have dominant-negative effects in Rb1 deficient lung epithelium.
(A) Trp53Δ/wt Rb1Δ/Δ, Trp53R270H/wtRb1Δ/Δ and Trp53R172H/wtRb1Δ/Δ mice contain two transgenes: 1) the reverse tetracycline transactivator (rtTA) under control of the human Surfactant protein C (Sfptc) promoter, and 2) Cre recombinase under control of the tet operator. Treatment with doxycycline (circles) which combines with rtTA (arches) induces Cre recombinase expression resulting in recombination of floxed (LoxP) Trp53 and Rb1 alleles. Trp53Δ/wt Rb1Δ/Δ mice (left) contain one wild type and one floxed Trp53 allele leading to heterozygous Trp53 loss after recombination. Trp53R172H/wtRb1Δ/Δ and Trp53R270H/wtRb1Δ/Δ mice (right) contain one wild type and one mutant Trp53 allele containing a missense mutation in exon 5 (R172H) or exon 8 (R270H), that results in wild type Trp53 expression along with R172H or R270H expression after recombination of the floxed STOP cassette preceding the point mutation. All mice contain two floxed Rb1 alleles resulting in homozygous Rb1 loss after recombination. (B) Mice with Trp53Δ/wtRb1Δ/Δ, Trp53R270H/wtRb1Δ/Δ and Trp53R172H/wtRb1Δ/Δ lungs had similar survivals. Kaplan-Meier curves represent mice that died or were sacrificed when moribund or after losing > 10% body weight. (C) Trp53Δ/wtRb1Δ/Δ, Trp53R270H/wtRb1Δ/Δ and Trp53R172H/wtRb1Δ/Δ lungs had similar phenotypes consisting of multifocal neuroendocrine hyperplasia and carcinomas in situ (CIS) in conducting airways (arrows) and at bronchioalveolar duct junctions (BADJ, arrowheads) shown in H&E images. Trp53 Δ/ΔRb1Δ/Δ lungs with homozygous Trp53 and Rb1 loss had a more advanced neoplastic phenotype with extensive hyperplasia and CIS as well as tumors (t). Scale bars: 100 μm. (D) Cells comprising hyperplasia, CIS and tumors in all genotypes shown in H&E images (top row) were positive for calcitonin related polypeptide alpha (CALCA) staining by immunohistochemistry (bottom row). Scale bars: 20 μm (left and middle columns) and 100 μm (right column).
Phenotypes of Trp53R172H/wtRb1Δ/Δ and Trp53R270H/wtRb1Δ/Δ lungs were also compared to lungs with homozygous Trp53 loss (Trp53Δ/ΔRb1Δ/Δ). If R172H and R270H had dominant negative activity, phenotypes of Trp53R172H/wtRb1Δ/Δ, Trp53R270H/wtRb1Δ/Δ and Trp53Δ/ΔRb1Δ/Δ lungs would be expected to be similar; however, this was not the case. Trp53Δ/ΔRb1Δ/Δ lungs had a more severe phenotype with earlier and more extensive neuroendocrine hyperplasia, CIS and neuroendocrine tumors as compared to Trp53R172H/wtRb1Δ/Δ and Trp53R270H/wtRb1Δ/Δ lungs (Fig. 1C). Neuroendocrine tumors were detected in 83% (5/6) of Trp53Δ/ΔRb1Δ/Δ lungs at 3.3–4.1 months of age as compared to 10% (1/10) of Trp53R172H/wt lungs and no Trp53R270H/wt lungs despite analysis at an older average age of 5.9 months. Mice of all genotypes developed neuroendocrine medullary thyroid carcinomas as previously reported due to targeting a subpopulation of thyroid C-cells (22, 27). The neuroendocrine phenotype of the lung lesions was confirmed by expression of calcitonin/calcitonin-related polypeptide alpha (CALCA; calcitonin gene related peptide (CGRP)) (Fig. 1D). The median survival of mice with Trp53Δ/ΔRb1Δ/Δ lungs was 4.1 months (22), which was significantly shorter than the 5.4 and 6.7 month median survivals of mice with Trp53R172H/wtRb1Δ/Δ and Trp53R270H/wtRb1Δ/Δ lungs, respectively (p<0.004). The more severe phenotype in Trp53Δ/ΔRb1Δ/Δ lungs provide further evidence that R172H and R270H do not have dominant negative activity in the lung epithelium but rather promote lung carcinogenesis through loss-of-function mechanisms.
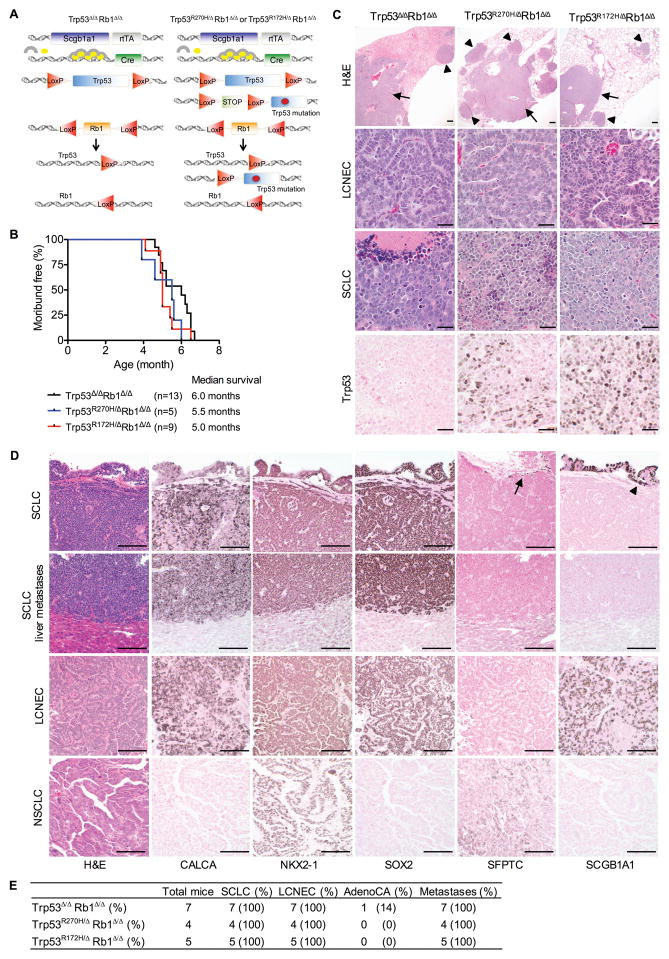
Potential gain-of-function activities were assessed by comparing effects of R172H and R270H in the absence of a wild type Trp53 allele (Trp53R172H/ΔRb1Δ/Δ and Trp53R270H/ΔRb1Δ/Δ) with homozygous Trp53 loss (Trp53Δ/ΔRb1Δ/Δ). If gain-of-function activities were present, additional phenotypes would be expected in Trp53R172H/ΔRb1Δ/Δ and/or Trp53R270H/ΔRb1Δ/Δ as compared to Trp53Δ/ΔRb1Δ/Δ lungs. A second mouse model was developed to avoid the uniform development of thyroid tumors seen in the Sftpc promoter driven model. Mice were generated with a similar strategy using the secretoglobin family 1A member 1 (Scgb1a1) gene promoter that targets the conducting airway and alveolar epithelium but rarely develop thyroid tumors (Fig. 2A) (24). The Scgb1a1driven model was used for the remaining experiments. Survival of mice with Trp53R172H/ΔRb1Δ/Δ, Trp53R270H/ΔRb1Δ/Δ and Trp53Δ/ΔRb1Δ/Δ lungs were identical (Fig. 2B) with no differences in lung phenotype or tumor spectra (Fig. 2C). Together, these data demonstrate that R172H and R270H do not have dominant negative or gain-of-function activity but rather promote lung carcinogenesis through loss-of-function mechanisms.
Figure 2. R270H and R172H promote lethal SCLC in Rb1 deficient lung epithelia through loss-of-function and not gain-of-function mechanisms.
(A) Mice with Trp53Δ/ΔRb1Δ/Δ, Trp53R270H/ΔRb1Δ/Δ or Trp53R172H/ΔRb1Δ/Δ lungs were generated with a similar strategy as in Fig. 1A using the rat secretoglobin family 1A member 1 (Scgb1a1) promoter rather than the Sfptc promoter to drive rtTA. The same floxed (LoxP) Trp53, Trp53 mutant and Rb1 alleles described in Fig. 1A were used to generate mice with homozygous Rb1 loss combined with homozyous Trp53 loss, or R270H or R172H expression targeted to the lung epithelium. (B) Mice with Trp53Δ/ΔRb1Δ/Δ, Trp53R270H/ΔRb1Δ/Δ and Trp53R172H/ΔRb1Δ/Δ lungs had similar median survivals of 5.0–6.0 months of age. Kaplan-Meier curves represent mice that died or were sacrificed when moribund or after losing > 10% body weight. (C) Trp53Δ/ΔRb1Δ/Δ, Trp53R270H/ΔRb1Δ/Δ and Trp53R172H/ΔRb1Δ/Δ lungs had similar phenotypes consisting of SCLC predominantly localized to hilar and proximal conducting airways (arrows), and LCNEC predominantly localized to distal conducting airways, BADJs and alveolar regions (arrowheads). Representative H&E images of Trp53Δ/ΔRb1Δ/Δ, Trp53R270H/ΔRb1Δ/Δ and Trp53R172H/ΔRb1Δ/Δ lungs from 5–7 month old mice are shown with higher magnification images showing differing morphologic features of LCNEC and SCLC. R270H and R172H protein was detected in Trp53R270H/ΔRb1Δ/Δ and Trp53R172H/ΔRb1Δ/Δ lungs, respectively by immunohistochemistry with no Trp53 detected in Trp53Δ/ΔRb1Δ/Δ lungs. Data is representative of tumors in 8 Trp53R270H/ΔRb1Δ/Δ, 6 Trp53R172H/ΔRb1Δ/Δ and 13 Trp53Δ/ΔRb1Δ/Δ mice. Scale bars: 200 μm (top row) and 20 μm (middle and bottom rows). (D) Mice with Trp53Δ/ΔRb1Δ/Δ, Trp53R270H/ΔRb1Δ/Δ and Trp53R172H/ΔRb1Δ/Δ lungs consistently developed metastatic SCLC and LCNEC with phenotypic characteristics of the corresponding human tumors. Primary SCLC and LCNEC lung tumors as well as SCLC metastases had a neuroendocrine phenotype, staining positively for the neuroendocrine marker, CALCA by immunohistochemistry. The tumors were also positive for the lung epithelial marker, NK2 homeobox 1 (NKX2-1) and the conducting airway marker, SRY-box 2 (SOX2), whereas they were negative for the parenchymal type II cell marker, SFTPC. Unlike SCLC, a subset of LCNEC had positive staining for the Club cell marker, SCGB1A1. Non-neoplastic type II cells were positive for SFTPC (arrow) and Club cells were positive for SCGB1a1 (arrowhead) serving as internal positive controls. One Trp53Δ/ΔRb1Δ/Δ mice developed a NSCLC papillary adenocarcinoma with a type II cell phenotype as indicated by positive staining for NKX2-1 and SFTPC, and absence of staining for CALCA, SOX2 and SCGB1A1. A second single adenocarcinoma developed in a Trp53Δ/ΔRb1Δ/Δ rapamycin treated mouse (see Fig. 4G). Scale bars: 100 μm. (E) Table summarizing pathologic findings in 5–7 month old mice with Trp53Δ/ΔRb1Δ/Δ, Trp53R270H/ΔRb1Δ/Δ and Trp53R172H/ΔRb1Δ/Δ lungs. Lethal metastatic SCLC consistently developed in mice of all genotypes with a single NSCLC adenocarcinoma (AdenoCA) being detected in a Trp53Δ/ΔRb1Δ/Δ mouse.
Trp53 mutant and deficient Rb1 ablated lungs uniformly develop metastasizing neuroendocrine tumors of two distinct subtypes
Primary neuroendocrine lung tumors in Trp53R172H/ΔRb1Δ/Δ, Trp53R270H/ΔRb1Δ/Δ and Trp53Δ/ΔRb1Δ/Δ lungs had two distinct histologic patterns: 1) tumors mimicking human SCLC comprised of sheets of small cells with high mitotic rates, frequent apoptotic cells, scant poorly defined cytoplasm and areas of geographic necrosis, and 2) tumors mimicking human large cell neuroendocrine carcinomas (LCNEC) comprised of nests of larger cells with more distinct cellular borders and abundant cytoplasm growing in a trabecular pattern with nuclear palisading (Fig. 2C–E). SCLC and LCNEC tumors were preferentially localized to distinct regions within the lung. SCLC predominated in the proximal conducting airways and central hilar lung regions similar to human SCLC. In contrast, LCNEC tumors were preferentially located in the lung parenchyma and distal conducting airways, most prominently at BADJ (Fig. 2C). Tumors with mixed morphology were also present. Extensive mediastinal spread was present along with metastases in lymph nodes, chest wall, liver, ovaries and uterus. Mutant Trp53 protein was detected in primary lung and tumor metastases arising in Trp53R172H/ΔRb1Δ/Δ and Trp53R270H/ΔRb1Δ/Δ mice with no Trp53 immunostaining in Trp53Δ/ΔRb1Δ/Δ tumors verifying the specificity of the assay (Fig. 2C and S1). Interestingly, all metastases were SCLC suggesting that the distinct tumor subtypes have differing biologic behavior.
Primary SCLC and LCNEC as well as metastatic SCLC were positive for expression of CALCA confirming the neuroendocrine phenotype (Fig. 2D). Primary and metastatic lung tumors irrespective of histologic type were also positive for the lung epithelial cell marker, NKX2-1, and the conducting airway epithelial cell marker, SOX2, while lacking expression of the parenchymal alveolar type II cell marker, SFTPC (Fig. 2D). SCLC and LCNEC tumors were however distinguished by expression of the Club cell marker, SCGB1A1, in a subset of LCNEC with no expression in primary or metastatic SCLC (Fig. 2D). These data raise the possibility that SCGB1A1 expression identifies a distinct subtype of neuroendocrine lung cancer.
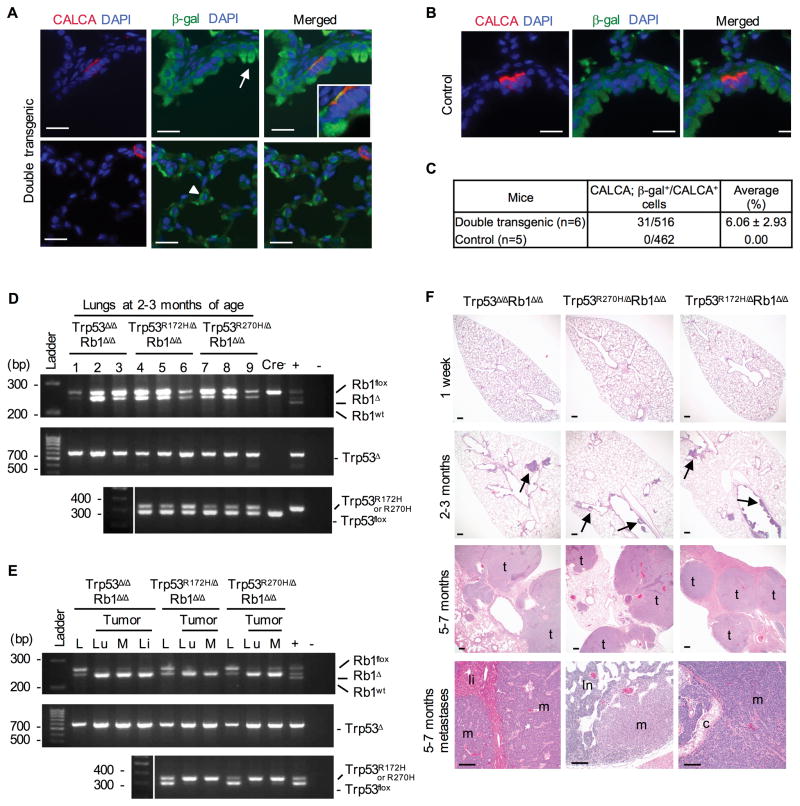
Despite targeting non-neuroendocrine cell lineages capable of giving rise to NSCLC, namely Club and alveolar type II cells, only two non-small cell adenocarcinomas were detected in a total of 65 analyzed Trp53R172H/ΔRb1Δ/Δ, Trp53R270H/ΔRb1Δ/Δ and Trp53Δ/ΔRb1Δ/Δ mice (Fig. 2D–E). In contrast to SCLC and LCNEC, the adenocarcinoma lacked expression of neuroendocrine markers and instead had a type II cell phenotype as indicated by expression of SFTPC and no expression of SCGB1A1 (Fig. 2D). Uniform targeting of the conducting airway epithelium including a subset of neuroendocrine cells was confirmed by crossing mice into the ROSA26 reporter stain (Fig. 3A–C) (24). Recombined Trp53 and Rb1 alleles were confirmed in lungs, primary tumors and metastases of all genotypes demonstrating tumor derivation from Trp53 and Rb1 modified epithelial cells (Fig. 3D–E). Thus loss of Trp53 function (either by allele loss or mutation) combined with Rb1 deletion preferentially drives pulmonary neuroendocrine carcinogenesis.
Figure 3. Targeting R270H, R172H or Trp53 loss to Rb1 deficient non-neuroendocrine and neuroendocrine cell lineages uniformly results in metastatic SCLC in a temporally consistent time course.
(A) Mice containing the Scgb1a1-rtTA+/−;tetO-Cre+/ transgenes described in Fig. 2A were crossed into the ROSA26 reporter strain, treated with doxycycline and adult lungs assessed for β-galactosidase (β-gal, green) expression by immunofluorescence to determine targeted cells. Club cells (arrow) and alveolar type II cells (arrowhead) as well as neuroendocrine cells were targeted as demonstrated by co-expression (yellow) of β-gal (green) and CALCA (red) in conducting airways in merged Zeiss Apotome acquired images. Scale bars: 20 μm. (B) No β-gal positive cells were detected in control lungs lacking one or both transgenes required for gene recombination. Scale bars: 20 μm. (C) Quantification of targeted neuroendocrine cells demonstrating an average 6% of CALCA positive neuroendocrine cells targeted with no CALCA/β-gal double positive cells detected in control lungs. (D) PCR analyses confirmed recombination of Trp53 (Trp53Δ, Trp53R172H or Trp53R270H) and Rb1 (Rb1Δ) floxed alleles in lungs from 2–3 month old mice. Wild type Rb1 (Rb1wt), floxed Rb1 (Rb1flox) and floxed Trp53 (Trp53flox) alleles are indicated. (+) = positive control, (−) = no DNA. (E) Tumors arose from Cre targeted cells as demonstrated by enrichment for recombined alleles in DNA isolated from lung (Lu) and metastatic mediastinal (M) and liver (Li) tumors as compared to nontumor lung (L) in all genotypes. (F) Tumor initiation and progression uniformly occurred within a consistent time course in Trp53Δ/ΔRb1Δ/Δ, Trp53R270H/ΔRb1Δ/Δ and Trp53R172H/ΔRb1Δ/Δ lungs. Lungs were normal at 1 week, neuroendocrine hyperplasia and CIS (arrows) developed by 2–3 months, and multifocal lung tumors (t) with SCLC metastases (m) to liver (li, bottom left), lymph nodes (ln, bottom middle) and mediastinum (c, tracheal cartilage, bottom right) were present at 5–7 months. Scale bars: 200 μm top and middle rows and 100 μm bottom row.
Tumor initiation and progression in Trp53 mutant or deficient Rb1 ablated lungs have a short latency and occur in a temporally consistent manner
Temporally uniform tumor initiation and progression in Trp53R172H/ΔRb1Δ/Δ, Trp53R270H/ΔRb1Δ/Δ and Trp53Δ/ΔRb1Δ/Δ lungs with consistent death by 6.7 months of age provides evidence that loss of Trp53 and Rb1 function is not only required, but sufficient, for SCLC development and progression. No gross or microscopic lung abnormalities were detected in mice of all genotypes at one week of age with consistent development of multifocal pulmonary neuroendocrine hyperplasia and CIS by 2–3 months (Fig. 3F). Tumor progression and metastases were present at 5 months resulting in uniform lethality between 3.9–6.7 months of age (Fig. 2B). Thus, in addition to providing mechanistic insights, these mice uniformly develop metastatic SCLC with short latency and temporally consistent progression providing a robust platform to study lung cancer evolution, identify mechanisms of treatment resistance, and test therapeutics.
Metastatic SCLC develops in the absence of PTEN loss and is not sensitive to mTOR pathway inhibition
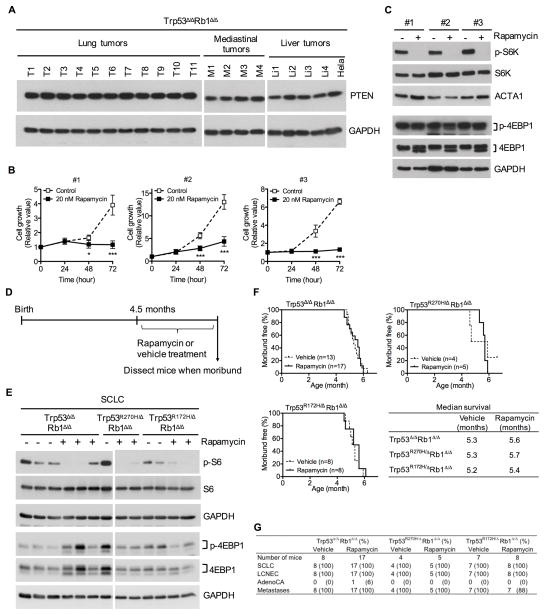
Recent studies in GEMMs identified PTEN as a potent suppressor of SCLC with complete loss of PTEN in 55% of high grade Trp53/Rb1 deficient tumors (28, 29). Surprisingly, PTEN expression was detected in all 29 analyzed lung and metastatic tumors representing all genotypes (Fig. 4A and S2). mTOR activity, a downstream effector in the PTEN pathway, was consistently detected at variable levels in primary and metastatic tumors as well as tumor cells grown in culture as determined by expression of phosphorylated S6 kinase and S6 (Fig. 4C and E). Blocking mTOR pathway activity with rapamycin markedly inhibited tumor cell growth in culture but was not sufficient to enhance survival of mice of any genotype despite inhibition of mTOR signaling in 67% of tumors analyzed from moribund mice (Fig. 4B–F and S3). Lung tumor phenotypes were similar in rapamycin and vehicle treated mice (Fig. 4G). These results demonstrate that Trp53/Rb1 deficient tumors in the current model retain PTEN, and directly demonstrate that mTOR pathway inhibition with rapamycin is not an effective therapy in vivo despite markedly suppressing growth of corresponding tumor cells in culture.
Figure 4. Primary lung and metastatic tumors retain PTEN expression and have activate mTOR signaling with rapamycin treatment being insufficient to alter survival despite marked suppression of mTOR signaling and inhibition of tumor cell growth in culture.
(A) PTEN expression was detected in all 11 primary lung (T1–T11), 4 metastatic mediastinal (M1–M4) and 4 metastatic liver (Li1–Li4) tumors derived from Trp53Δ/ΔRb1Δ/Δ mice by western blot analysis. Hela cell lysate was used as a positive control, and blots were reprobed with GAPDH as a loading control. (B) Growth of primary tumor cells established in culture from three independent Trp53Δ/ΔRb1Δ/Δ lung tumors arising in three different mice (#1–#3) was markedly inhibited by rapamycin (20 nM). Results are representative of three independent experiments with data represented as mean ± SD (n=4 wells/group). * p<0.05 and *** p<0.001. (C) Rapamycin (20 nM) inhibited mTOR pathway signaling in primary tumor cell cultures by 24 hours as demonstrated by reduced expression of phosphorylated ribosomal protein S6 kinase (p-S6K) by western blot analysis. Rapamycin did not alter expression of phosphoryated eukaryotic translation initiation factor 4E-binding protein 1 (p-4EBP1). Blots were reprobed for total S6K and 4EBP1 as well as alpha 1 actin (ACTA1) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as loading controls. (D) Mice with Trp53Δ/ΔRb1Δ/Δ, Trp53R270H/ΔRb1Δ/Δ or Trp53R172H/ΔRb1Δ/Δ lungs were treated with rapamycin or vehicle control at 4.5 months of age and dissected when moribund. (E) mTOR pathway signaling was downregulated by rapamycin treatment in 67% of lung tumors obtained from moribund mice as demonstrated by decreased phosphorylated S6 ribosomal protein (p-S6). Expression of p-4EBP1 was not consistently changed in tumors from rapamycin treated mice. Representative images are shown with decreased expression of p-S6 and no change in p-4EBP1 expression seen in 7/11 Trp53Δ/ΔRb1Δ/Δ, 3/3 Trp53R270H/ΔRb1Δ/Δ and 4/7 Trp53R172H/ΔRb1Δ/Δ tumors. Expression of p-S6 was detected in 14/15 tumors from vehicle control mice at variable levels. Blots were reprobed with total S6 and 4EBP1 as well as GAPDH as a loading control. (F) Rapmaycin did not alter survival of Trp53Δ/ΔRb1Δ/Δ, Trp53R270H/ΔRb1Δ/Δ or Trp53R172H/ΔRb1Δ/Δ mice as compared to vehicle controls. (G) Lung cancer phenotypes were the same in rapamycin and vehicle treated mice of all genotypes. All mice developed multifocal SCLC and LCNEC with only a single NSCLC adenocarcinoma (AdenoCA).
Trp53 mutant and Trp53 deficient Rb1 ablated tumors cohabitating the same lung have differential sensitivity to chemotherapy that is not recapitulated in tumor cells in culture
To determine whether the biologic response to chemotherapy seen in patients was mimicked in our model, mice with established tumors were treated with a standard cisplatin/etoposide regime used for SCLC patients (Fig. 5A). Chemotherapy treatment significantly prolonged survival of mice with Trp53Δ/ΔRb1Δ/Δ lungs as compared to vehicle treated mice demonstrating a significant chemotherapy response (Fig. 5B).
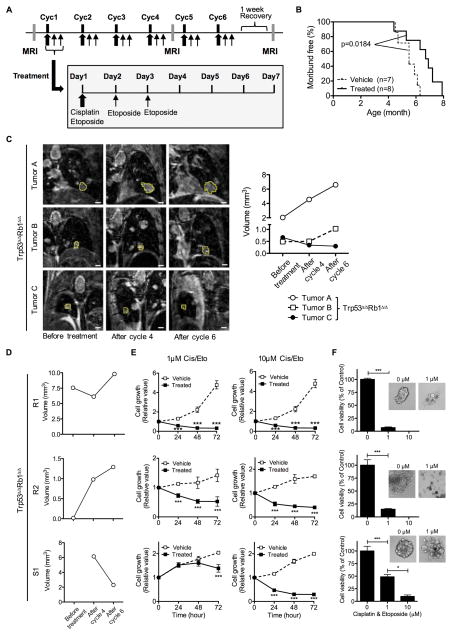
Figure 5. Chemotherapy treatment extends survival of tumor bearing mice with tumor response in vivo not retained in cell culture.
(A) Mice with Trp53Δ/ΔRb1Δ/Δ lungs were treated with a chemotherapy regime mimicking standard-of-care treatment for human SCLC patients with six weekly cycles (Cyc1–6) of chemotherapy consisting of cisplatin (6 mg/kg/day) and etoposide (12 mg/kg/day) intraperitoneally administered on the first day followed by additional administration of etoposide (12 mg/kg/day) on days 2 and 3. Three MRIs were performed during the course of treatment: before treatment to confirm tumor presence and serve as baseline, after cycle 4, and after cycle 6. (B) Chemotherapy treatment significantly prolonged survival (p=0.0184) of Trp53Δ/ΔRb1Δ/Δ mice with median survivals of 6.7 and 5.5 months for chemotherapy treated and vehicle control mice, respectively. Kaplan-Meier curves represent mice that died or were sacrificed when moribund or after losing > 10% body weight. (C) Trp53Δ/ΔRb1Δ/Δ tumors in an individual mouse have differing responses to chemotherapy. Tumors were imaged by MRI and volumes quantified before treatment, after cycle 4 and after cycle 6. Representative images of tumors (outlined in yellow) within a single mouse and graph showing corresponding tumor volumes over time are shown. Examples of a tumor refractory to chemotherapy (A), stable disease followed by tumor progression (chemoresistant tumor) (B) and a chemosensitive tumor (C) are depicted. (D) Trp53Δ/ΔRb1Δ/Δ primary cell cultures were established from chemoresistant (R1 and R2) and chemosensitive (S1) lung tumors. (E) Chemotherapeutic response of tumors in vivo is not retained in cell culture. Cells derived from R1, R2 and S1 tumors were all growth inhibited by cisplatin/etoposide (Cis/Eto) treatment at 1 μM and 10 μM doses as compared to vehicle treated controls in a 2D culture system as assessed by WST-1 assay. (F) Similar cisplatin/etoposide induced decreased cell viability was seen for cells from both chemoresistant and chemosensitive tumors grown in the 3D NanoCulture system. Cells were grown for 7 days and then treated with Cis/Eto (1 μM and 10 μM) for 5–7 days. Cell viability was significantly decreased at both concentrations of Cis/Eto irrespective of tumor response in vivo as assessed by CellTiter-Glo assay. Results in E–F are representative of three independent experiments with data represented as mean ± SD (n=4 (E) and 3 (F) wells/group). * p<0.05 and *** p<0.001.
The synchronous development of multiple lung tumors in a single mouse allows for evaluation of individual tumor responses within the same tumor microenvironment to identify potential tumor specific chemotherapeutic responses. Despite widespread genetic alteration of Trp53 and Rb1 throughout the lung epithelium, distinct multifocal lung tumors developed that could be individually followed by MRI (Figs 2C, 3F, 5C and S4). Development of distinct tumors despite expected field effects of widespread Trp53 and Rb1 mutation likely reflects differential sensitivity of distinct epithelial cell types to Trp53/Rb1 dependent tumorigenesis. Chemosensitive and chemoresistent tumors were identified within individual lungs of all genotypes (Fig. 5C). Similar to human SCLC, some tumors were refractory to chemotherapy with tumor progression occurring during treatment while other tumors were initially chemosensitive and subsequently acquired chemoresistance typical of tumor relapse. To determine if the biologic response to cisplatin and etoposide evident in tumors in vivo was retained in culture, tumor cells were cultured from chemosensitive and chemoresistant tumors (Fig. 5D and S4). The tumor response to chemotherapy in vivo was not recapitulated in cells cultured in two or three dimensional systems (Fig. 5E–F and S5). Growth of all cell cultures was inhibited by cisplatin and etoposide treatment irrespective of whether cells were derived from chemoresistant or chemosensitive tumors. All tumor cell cultures derived from untreated mice were also growth inhibited by chemotherapy treatment (Fig. S6). Thus, tumor cells were uniformly sensitive to chemotherapy in cell culture systems despite showing differing chemotherapy response in vivo. These findings, along with the similar non-corresponding in vivo tumor and in vitro cell culture response to rapamycin, demonstrate discordant biologic responses to therapeutics in corresponding in vitro and in vivo SCLC models highlighting the need to test therapeutic effectiveness at the organismal level.
R270H has gain-of-function effects in chemotherapeutic response
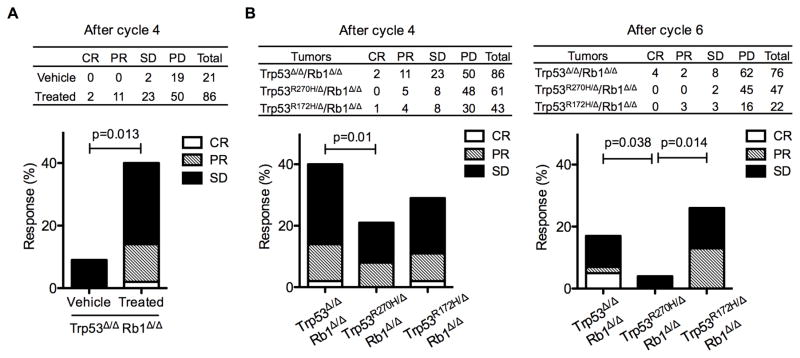
To identify potential biologic effects of Trp53 mutants in dictating chemotherapeutic response, Trp53R172H/ΔRb1Δ/Δ, Trp53R270H/ΔRb1Δ/Δ and Trp53Δ/ΔRb1Δ/Δ tumor growth was followed in vivo over 4–6 cycles of chemotherapy; the same duration of treatment given to human patients. Combined cisplatin/etoposide treatment resulted in a fourfold increase in the proportion of Trp53Δ/ΔRb1Δ/Δ tumors with stable or decreased growth as compared to vehicle treated controls (Fig. 6A). Importantly, there was a statistically significant reduction in chemotherapy sensitive tumors in Trp53R270H/ΔRb1Δ/Δ versus Trp53Δ/ΔRb1Δ/Δ lungs after both four and six cycles of treatment that was not seen in Trp53R172H/ΔRb1Δ/Δ lungs (Fig. 6B). These data indicate that R270H has mutant specific gain-of-function activities that negatively impact response of neuroendocrine lung cancers to chemotherapeutics.
Figure 6. R270H has mutant specific gain-of-function effects that decrease efficacy of cisplatin/etoposide treatment.
(A) An overall significant tumor response to cisplatin/etoposide treatment was present in mice with Trp53Δ/ΔRb1Δ/Δ tumors as compared to vehicle treated mice after 4 cycles of treatment. Table indicates number of tumors with complete response (CR), partial response (PR), stable disease (SD) and progressive disease (PD) in vehicle and chemotherapy treated mice with Trp53Δ/ΔRb1Δ/Δ lungs. Graph depicts overall response with 40% of tumors responding to chemotherapy treatment as CR, PR or SD vs 10% of tumors with SD in vehicle treated control mice. (B) Cisplatin/etoposide treatment was significantly less effective in Trp53R270H/ΔRb1Δ/Δ lung tumors as compared to Trp53Δ/ΔRb1Δ/Δ and Trp53R172H/ΔRb1Δ/Δ lung tumors. Tables and graphs indicate the number and percentage of tumors with CR, PR, SD and PD after cycle 4 (left) and after cycle 6 (right). Overall response of Trp53R270H/ΔRb1Δ/Δ lung tumors to chemotherapy treatment was statistically significantly reduced as compared to Trp53Δ/ΔRb1Δ/Δ lung tumors after cycle 4, and Trp53Δ/ΔRb1Δ/Δ and Trp53R172H/ΔRb1Δ/Δ lung tumors after cycle 6. Logistic regression was used to analyze tumor response with p values indicated in the graphs.
Discussion
Preferential occurrence of TP53 missense mutations rather than TP53 loss strongly implicates a selective advantage for TP53 mutant protein expression in the pathogenesis of SCLC. A direct functional role for TP53 mutant proteins in driving carcinogenesis, however, has not been demonstrated. The current studies directly demonstrate that Trp53 mutants commonly seen in human lung cancer drive lung carcinogenesis through loss-of-function, and not dominant negative or gain-of-function, mechanisms. Despite targeting Trp53 and Rb1 alterations to cells of origin for both NSCLC and SCLC, mice selectively developed neuroendocrine SCLC and LCNEC with only rare adenocarcinomas demonstrating that loss of Trp53 and Rb1 function preferentially promotes neuroendocrine carcinomas. Tumor initiation and progression occurred over a remarkably consistent time frame with short tumor latency and uniform progression to lethal metastatic disease by 7 months of age providing a tractable preclinical model to explore cancer evolution and chemotherapy resistance, and test drug efficacy. Interestingly, tumor progression was not associated with PTEN loss as seen in other genetically similar models, providing a complementary preclinical model for the >80% of human LCNEC and SCLC that retain PTEN expression (29). Given active mTOR signaling in lung tumors and frequent mutations in the PI3K/AKT/mTOR pathway in human SCLC and LCNEC, therapeutic efficacy of inhibiting mTOR signaling with rapamycin was tested. Despite marked rapamycin suppression of tumor cell growth in culture, rapamycin treatment did not alter lung tumor phenotypes or mouse survival. In contrast, treatment with a cisplatin/etoposide regimen that mimics current standard-of-care for human patients extended animal survival demonstrating efficacy of cytotoxic chemotherapy. Distinct tumors within an individual mouse showed differing sensitivity to chemotherapy providing evidence for a tumor autonomous mechanism dictating chemotherapeutic response that were not recapitulated in corresponding tumor cells in culture, highlighting the need to assess therapeutic efficacy in vivo. Importantly, R270H had a mutant specific gain-of-function activity that compromised efficacy of standard platinum based chemotherapy used to treat SCLC patients, identifying a novel in vivo function for Trp53 mutants in conferring chemoresistance.
These studies establish a direct casual role for TP53 mutants in SCLC and LCNEC tumorigenesis, and provide evidence that TP53 mutation and RB1 loss is sufficient for generation of these morphologically distinct malignancies. Metastatic adenocarcinomas were not detected in Trp53 mutant expressing Rb1 deficient lung epithelium which differs from germline Trp53R270H/+ and Trp53R172H/+ mice that developed lung adenocarcinomas with occasional metastasis in up to 19% of mice (13, 14). The lack of this gain-of-function phenotype could be due to concurrent inactivation of RB1-E2F pathway signaling and/or differences in genetic background. The rare occurrence of adenocarcinomas in the current model is also likely due to the short SCLC latency and rapid progression to death by 7 months which differs from the 15–16 month mean survival of germline Trp53R270H/+ and Trp53R172H/+ mice. In support of this notion, lung adenocarcinomas were also rarely detected in germline Trp53R270H/−, Trp53R172H/− and Trp53−/− mice that have shortened mean survivals of 4.4–4.6 months due to development of sarcomas and lymphomas.
Trp53 and Rb1 ablation by intranasal or intratracheal adenoviral Cre recombinase delivery results in similar SCLC but with an extended median latency of 7 months (30). NSCLC adenocarcinomas rarely developed and were associated with Trp53 loss alone whereas SCLC development required combined Trp53/Rb1 loss. Detailed analysis of Trp53/Rb1 and Trp53/Rb1/Pten deficient SCLC models provide evidence that the combination of genetic alterations and cell type targeted define lung cancer phenotypes (31). Trp53/Rb1 ablation by adenoviral Cre delivery resulted in predominantly late, centrally arising SCLC with widespread metastases after 1 year. Minor tumors types included LCNEC and NSCLC, each comprising ~10% of tumors. Additional PTEN ablation resulted in a transition from SCLC to NSCLC expressing neuroendocrine markers (NSCLC-NE). Conversely, PTEN ablation targeted to neuroendocrine cells with the CGRP promoter rather than the ubiquitous CMV promoter resulted in predominantly LCNEC rather than NSCLC-NE. The lack of NSCLC-NE tumors in the current model is consistent with retention of PTEN expression, and the co-occurrence of LCNEC with SCLC in all lungs that was not seen after adenoviral Cre induced Trp53/Rb1 ablation likely reflects more consistent gene ablation in distal smaller bronchioles wherein we show LCNEC preferentially arise. A uniform finding across all models is the SCLC phenotype of metastases irrespective of the predominant tumor type in the lung. Together, these murine models genetically and biologically mimic the entire spectrum of high grade neuroendocrine carcinomas and provide crucial models for studying multistage pathogenesis hindered by lack of human tissues for these seldom resected tumors.
The biologic relationship of LCNEC to SCLC versus NSCLC is unclear resulting in uncertainty regarding optimal clinical management. LCNEC shares many clinicopathologic features with SCLC including aggressive clinical behavior, association with smoking, extremely high proliferation rates, and a neuroendocrine phenotype (32). However, LCNEC have morphologic features of NSCLC and thus are classified as a NSCLC subtype. Most important clinically, there is lack of consensus on whether LCNEC should be managed with platinum/etoposide based chemotherapy as for SCLC or be managed for local, regional and metastatic disease similar to NSCLC. This clinical uncertainty is fueled by the highly variable chemosensitivity of LCNEC to regimes used to treat SCLC (33–35). Genomic profiling of human LCNEC revealed distinct genetic subsets that may explain the variable biologic behavior of LCNEC. Three LCNEC subsets were identified with distinct genomic signatures resembling: 1) SCLC with TP53 and RB1 co-mutation/loss, 2) NSCLC lacking co-altered TP53 and RB1 with mutations in NSCLC associated genes like KRAS, and 3) carcinoid tumors with MEN1 mutations (36). The current studies provide direct evidence that combined TP53/RB1 loss drives development of LCNEC, and reveals previously undescribed phenotypic heterogeneity in this subset exemplified by expression of the non-neuroendocrine Club cell marker, SCGB1A1, typically seen in NSCLC. SCGB1A1 was restricted to LCNEC which had had a different spatial distribution and metastatic potential than SCLC that could result from distinct cells of origin (37–39). Neuroendocrine cells are the predominant cell of origin for SCLC with previous studies concluding that SCGB1A1 expressing Club cells are largely resistant to transformation by conditional Trp53 and Rb1 ablation (40, 41). The distinct LCNEC and SCLC tumor phenotypes could also result from regional variations in neuroendocrine cell subsets and/or cellular niches with distinct signaling programs known to exist within the lung (42). Regardless of the cell of origin, however, a question of clinical importance is whether the distinct LCNEC and SCLC tumor phenotypes dictate clinical behavior and/or predict treatment response. The finding that all metastatic tumors are SCLC suggests that LCNEC have a less aggressive biologic behavior. The rarity of LCNEC that comprise ~3% of all human lung cancers has resulted in a paucity of tumor specimens as well as clinical trials to guide therapeutic decision making and test drug efficacy (33). The current model now offers a valuable tool to explore LCNEC evolution, and gain further insights into the biologic relationship between SCLC and LCNEC. The consistent, short tumor latency and progression to metastatic lethal disease by 7 months of age in the current GEMM as compared to tumor latencies of 7–18 months in other models also provides a tractable preclinical platform to efficiently test drug efficacy to guide patient management.
PTEN mutations and/or loss are detected in 4–9% of human SCLC and LCNEC (6, 36, 43, 44). Loss of PTEN expression in 55% of murine SCLC despite expression in precursor lesions suggests that PTEN loss promotes SCLC progression (28, 29). Interestingly, PTEN has oncogenic properties in R273H and R175H expressing human cancer cell lines that differ from tumor suppressive effects in cells with wild type TP53, predicting a potential selective advantage for retaining PTEN in TP53 mutant cancers (45). We demonstrate that PTEN expression was uniformly maintained in primary and metastatic Trp53 mutant murine SCLC serving as a model for the majority of human SCLC that retain PTEN. Loss of PTEN in Trp53/Rb1 deficient tumors most frequently resulted from chromosome 19 loss which contains the PTEN locus (29). The extended animal survivals of 10–20 months versus 7 months in the current model may allow for acquisition of these additional genetic alterations (29, 30). Despite PTEN expression, mTOR signaling was active in the lung tumors. Loss of Trp53 function can promote mTOR activation through downregulation of PTEN/mTOR pathway inhibitors that function downstream of PTEN including tuberous sclerosis 2 (Tsc2), polo-like kinase 2 (Plk2) and sestrin 2 (Sesn2) (22). Indeed, Trp53 suppressed progression of Rb1 deficient medullary thyroid cancer, an aggressive neuroendocrine carcinoma, by inhibiting mTOR signaling (22). Inhibition of mTORC1 by rapamycin was not sufficient to enhance survival of mice with SCLC and LCNEC despite marked suppression of tumor cell growth in culture. Targeting other components within the pathway or combination therapy may be required to gain a durable response. In support of this notion, inhibition of mTOR signaling restored cisplatin sensitivity in lung cancer cell lines (46).
Development of chemotherapy resistance is a major problem in SCLC patients. Maintenance of cancer-initiating cells and transformation from SCLC to NSCLC have been implicated as mechanisms of chemotherapy resistance (47, 48). Expansion of tumor propagating cells with inherent chemoresistance does not contribute to chemotherapy resistance in murine Trp53/Rb1 deficient SCLC (49). Interesting, R273H conferred doxorubicin resistance to human colon cancer cell lines by promoting epithelial-mesenchymal transition and cancer stem cell phenotypes through gain-of-function mechanisms prompting need to test the role of cancer initiating stem cells in conferring chemoresistance in the context of Trp53 mutant expression (50). Importantly, we identify R270H expression as a mechanism to confer chemotherapy resistance in vivo. We demonstrate that R270H lung tumors have increased resistance to cisplatin/etoposide treatment as compared to R172H and Trp53 deficient tumors demonstrating mutant specific gain-of-function activities that decrease cytotoxic chemotherapeutic efficacy. Gain-of-function Trp53 mutants commonly exert biologic effects by interacting with other transcription factors, altering chromatin structure and inducing epigenetic modifications resulting in novel gene expression (8–10). Studies in pancreatic adenocarcinoma cell lines provide evidence that R273H confers resistance to gemcitabine induced apoptosis and induces expression of cell cycle genes Cdk1 and CCNB1 resulting in hyperproliferation (20). In NSCLC cell lines, R273H or R175H overexpression confers resistance to chemotherapy induced apoptosis in a mutant and drug specific manner (18). Both R273H and R175H inhibited cisplatin induced apoptosis whereas only R175H conferred resistance to etoposide. R273H also conferred resistance to combined treatment with cisplatin and panobinostat, a histone deacetylase inhibitor, by inhibiting apoptosis (19). Herein we show that R270H, but not R172H, confers cisplatin/etoposide resistance in vivo demonstrating a Trp53 mutant specific gain-of-function chemoresistance mechanism that is not faithfully recapitulated in cells in culture. The ability to track individual tumor responses in vivo in the current model provides a valuable system to now identify downstream molecular signals that mediate R270H dependent chemoresistance; an important step toward identifying predictive biomarkers for treatment response, improving durability of chemotherapeutic response, and for rational design of novel SCLC therapies.
Supplementary Material
Acknowledgments
Grant Support
This work was supported by National Heart Lung and Blood Institute HL079193 (to K.A.W-B) and the American Cancer Society RSG-10-194-01-TBG (to K.A.W-B.).
We thank Xiaolan Ma for generating and genotyping mice, Diana Lindquist, Scott Dunn and the Imaging Research Center at Cincinnati Children’s Hospital Medical Center (CCHMC) for expertise in radiographic imaging, Susan Wert at CCHMC for assistance with histologic imaging and analysis, Veterinary Services at CCHMC for excellent animal care, the Pathology Research Core at CCHMC for expertise and assistance with tissue processing, and Organogenix, Inc. for providing NanoCulture plates.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Pillai RN, Owonikoko TK. Small cell lung cancer: therapies and targets. Semin Oncol. 2014;41(1):133–42. doi: 10.1053/j.seminoncol.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Meerbeeck JP, Fennell DA, De Ruysscher DK. Small-cell lung cancer. Lancet. 2011;378(9804):1741–55. doi: 10.1016/S0140-6736(11)60165-7. [DOI] [PubMed] [Google Scholar]
- 4.Gazdar AF, Hirsch FR, Minna JD. From Mice to Men and Back: An Assessment of Preclinical Model Systems for the Study of Lung Cancers. J Thorac Oncol. 2016;11(3):287–99. doi: 10.1016/j.jtho.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev. 2015;29(14):1447–62. doi: 10.1101/gad.263145.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44(10):1104–10. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26(12):1268–86. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim MP, Zhang Y, Lozano G. Mutant p53: Multiple Mechanisms Define Biologic Activity in Cancer. Front Oncol. 2015;5:249. doi: 10.3389/fonc.2015.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25(3):304–17. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birch JM, Blair V, Kelsey AM, Evans DG, Harris M, Tricker KJ, et al. Cancer phenotype correlates with constitutional TP53 genotype in families with the Li-Fraumeni syndrome. Oncogene. 1998;17(9):1061–8. doi: 10.1038/sj.onc.1202033. [DOI] [PubMed] [Google Scholar]
- 12.Bougeard G, Sesboue R, Baert-Desurmont S, Vasseur S, Martin C, Tinat J, et al. Molecular basis of the Li-Fraumeni syndrome: an update from the French LFS families. J Med Genet. 2008;45(8):535–8. doi: 10.1136/jmg.2008.057570. [DOI] [PubMed] [Google Scholar]
- 13.Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119(6):861–72. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119(6):847–60. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, et al. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum Mutat. 2016;37(9):865–76. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 16.Zheng S, El-Naggar AK, Kim ES, Kurie JM, Lozano G. A genetic mouse model for metastatic lung cancer with gender differences in survival. Oncogene. 2007;26(48):6896–904. doi: 10.1038/sj.onc.1210493. [DOI] [PubMed] [Google Scholar]
- 17.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65(22):10280–8. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 18.Blandino G, Levine AJ, Oren M. Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene. 1999;18(2):477–85. doi: 10.1038/sj.onc.1202314. [DOI] [PubMed] [Google Scholar]
- 19.Cai Y, Yan X, Zhang G, Zhao W, Jiao S. The predictive value of ERCC1 and p53 for the effect of panobinostat and cisplatin combination treatment in NSCLC. Oncotarget. 2015;6(22):18997–9005. doi: 10.18632/oncotarget.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiorini C, Cordani M, Padroni C, Blandino G, Di Agostino S, Donadelli M. Mutant p53 stimulates chemoresistance of pancreatic adenocarcinoma cells to gemcitabine. Biochim Biophys Acta. 2015;1853(1):89–100. doi: 10.1016/j.bbamcr.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Molina-Vila MA, Bertran-Alamillo J, Gasco A, Mayo-de-las-Casas C, Sanchez-Ronco M, Pujantell-Pastor L, et al. Nondisruptive p53 mutations are associated with shorter survival in patients with advanced non-small cell lung cancer. Clin Cancer Res. 2014;20(17):4647–59. doi: 10.1158/1078-0432.CCR-13-2391. [DOI] [PubMed] [Google Scholar]
- 22.Akeno N, Miller AL, Ma X, Wikenheiser-Brokamp KA. p53 suppresses carcinoma progression by inhibiting mTOR pathway activation. Oncogene. 2015;34(5):589–99. doi: 10.1038/onc.2013.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mason-Richie NA, Mistry MJ, Gettler CA, Elayyadi A, Wikenheiser-Brokamp KA. Retinoblastoma function is essential for establishing lung epithelial quiescence after injury. Cancer Res. 2008;68(11):4068–76. doi: 10.1158/0008-5472.CAN-07-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wikenheiser-Brokamp KA. Rb family proteins differentially regulate distinct cell lineages during epithelial development. Development. 2004;131(17):4299–310. doi: 10.1242/dev.01232. [DOI] [PubMed] [Google Scholar]
- 25.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45(2):228–47. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 26.Simpson DS, Mason-Richie NA, Gettler CA, Wikenheiser-Brokamp KA. Retinoblastoma family proteins have distinct functions in pulmonary epithelial cells in vivo critical for suppressing cell growth and tumorigenesis. Cancer Res. 2009;69(22):8733–41. doi: 10.1158/0008-5472.CAN-09-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perl AK, Wert SE, Nagy A, Lobe CG, Whitsett JA. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc Natl Acad Sci U S A. 2002;99(16):10482–7. doi: 10.1073/pnas.152238499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui M, Augert A, Rongione M, Conkrite K, Parazzoli S, Nikitin AY, et al. PTEN is a potent suppressor of small cell lung cancer. Mol Cancer Res. 2014;12(5):654–9. doi: 10.1158/1541-7786.MCR-13-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McFadden DG, Papagiannakopoulos T, Taylor-Weiner A, Stewart C, Carter SL, Cibulskis K, et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell. 2014;156(6):1298–311. doi: 10.1016/j.cell.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003;4(3):181–9. doi: 10.1016/s1535-6108(03)00220-4. [DOI] [PubMed] [Google Scholar]
- 31.Gazdar AF, Savage TK, Johnson JE, Berns A, Sage J, Linnoila RI, et al. The comparative pathology of genetically engineered mouse models for neuroendocrine carcinomas of the lung. J Thorac Oncol. 2015;10(4):553–64. doi: 10.1097/JTO.0000000000000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.William D, Travis EB, Burke Allen P, Marx Alexander, Nicholson Andrew G. WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart. 4. Lyon, France: IARC; 2015. [DOI] [PubMed] [Google Scholar]
- 33.Naidoo J, Santos-Zabala ML, Iyriboz T, Woo KM, Sima CS, Fiore JJ, et al. Large Cell Neuroendocrine Carcinoma of the Lung: Clinico-Pathologic Features, Treatment, and Outcomes. Clin Lung Cancer. 2016;17(5):e121–e9. doi: 10.1016/j.cllc.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossi G, Cavazza A, Marchioni A, Longo L, Migaldi M, Sartori G, et al. Role of chemotherapy and the receptor tyrosine kinases KIT, PDGFRalpha, PDGFRbeta, and Met in large-cell neuroendocrine carcinoma of the lung. J Clin Oncol. 2005;23(34):8774–85. doi: 10.1200/JCO.2005.02.8233. [DOI] [PubMed] [Google Scholar]
- 35.Varlotto JM, Medford-Davis LN, Recht A, Flickinger JC, Schaefer E, Zander DS, et al. Should large cell neuroendocrine lung carcinoma be classified and treated as a small cell lung cancer or with other large cell carcinomas? J Thorac Oncol. 2011;6(6):1050–8. doi: 10.1097/JTO.0b013e318217b6f8. [DOI] [PubMed] [Google Scholar]
- 36.Rekhtman N, Pietanza MC, Hellmann MD, Naidoo J, Arora A, Won H, et al. Next-Generation Sequencing of Pulmonary Large Cell Neuroendocrine Carcinoma Reveals Small Cell Carcinoma-like and Non-Small Cell Carcinoma-like Subsets. Clin Cancer Res. 2016;22(14):3618–29. doi: 10.1158/1078-0432.CCR-15-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121(6):823–35. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 38.Mainardi S, Mijimolle N, Francoz S, Vicente-Duenas C, Sanchez-Garcia I, Barbacid M. Identification of cancer initiating cells in K-Ras driven lung adenocarcinoma. Proc Natl Acad Sci U S A. 2014;111(1):255–60. doi: 10.1073/pnas.1320383110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutherland KD, Song JY, Kwon MC, Proost N, Zevenhoven J, Berns A. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc Natl Acad Sci U S A. 2014;111(13):4952–7. doi: 10.1073/pnas.1319963111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park KS, Liang MC, Raiser DM, Zamponi R, Roach RR, Curtis SJ, et al. Characterization of the cell of origin for small cell lung cancer. Cell Cycle. 2011;10(16):2806–15. doi: 10.4161/cc.10.16.17012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19(6):754–64. doi: 10.1016/j.ccr.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 42.Hogan BL, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CC, et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15(2):123–38. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyoshi T, Umemura S, Matsumura Y, Mimaki S, Tada S, Makinoshima H, et al. Genomic Profiling of Large-Cell Neuroendocrine Carcinoma of the Lung. Clin Cancer Res. 2017;23(3):757–65. doi: 10.1158/1078-0432.CCR-16-0355. [DOI] [PubMed] [Google Scholar]
- 44.Umemura S, Mimaki S, Makinoshima H, Tada S, Ishii G, Ohmatsu H, et al. Therapeutic priority of the PI3K/AKT/mTOR pathway in small cell lung cancers as revealed by a comprehensive genomic analysis. J Thorac Oncol. 2014;9(9):1324–31. doi: 10.1097/JTO.0000000000000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, Guessous F, Kwon S, Kumar M, Ibidapo O, Fuller L, et al. PTEN has tumor-promoting properties in the setting of gain-of-function p53 mutations. Cancer Res. 2008;68(6):1723–31. doi: 10.1158/0008-5472.CAN-07-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wangpaichitr M, Wu C, You M, Kuo MT, Feun L, Lampidis T, et al. Inhibition of mTOR restores cisplatin sensitivity through down-regulation of growth and anti-apoptotic proteins. Eur J Pharmacol. 2008;591(1–3):124–7. doi: 10.1016/j.ejphar.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brambilla E, Moro D, Gazzeri S, Brichon PY, Nagy-Mignotte H, Morel F, et al. Cytotoxic chemotherapy induces cell differentiation in small-cell lung carcinoma. J Clin Oncol. 1991;9(1):50–61. doi: 10.1200/JCO.1991.9.1.50. [DOI] [PubMed] [Google Scholar]
- 48.Sehested M, Hirsch FR, Osterlind K, Olsen JE. Morphologic variations of small cell lung cancer. A histopathologic study of pretreatment and posttreatment specimens in 104 patients. Cancer. 1986;57(4):804–7. doi: 10.1002/1097-0142(19860215)57:4<804::aid-cncr2820570420>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 49.Jahchan NS, Lim JS, Bola B, Morris K, Seitz G, Tran KQ, et al. Identification and Targeting of Long-Term Tumor-Propagating Cells in Small Cell Lung Cancer. Cell Rep. 2016;16(3):644–56. doi: 10.1016/j.celrep.2016.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hosain SB, Khiste SK, Uddin MB, Vorubindi V, Ingram C, Zhang S, et al. Inhibition of glucosylceramide synthase eliminates the oncogenic function of p53 R273H mutant in the epithelial-mesenchymal transition and induced pluripotency of colon cancer cells. Oncotarget. 2016;7(37):60575–92. doi: 10.18632/oncotarget.11169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.