Abstract
Cell invasion is a specialized cell behavior that likely co-evolved with the emergence of basement membranes in metazoans, as a mechanism to break down the barriers that separate tissues. A variety of conserved and lineage-specific biological processes that occur during development and homeostasis rely on cell invasive behavior. Recent innovations in genome editing and live-cell imaging have shed some light on the programs that mediate acquisition of an invasive phenotype; however, comparative approaches among species are necessary to understand how this cell behavior evolved. Here, we discuss the contexts of cell invasion, highlighting both established and emerging model systems, and underscore gaps in our understanding of the evolution of this key cellular behavior.
Introduction
The basement membrane (BM) is a metazoan innovation that arose at the dawn of multicellularity [1–3]. Comprised of basal and reticular laminae, this specialized extracellular matrix functions to separate epithelial, mesothelial, and endothelial tissues from adjacent connective tissue through a scaffold comprised mainly of collagen IV and laminin along with other structural proteins [4,5]. Recent genomic profiling of the earliest branching extant members of the Metazoa, the sponges and ctenophores, has identified that the evolution of collagen IV is likely to be correlated with the metazoan transition to multicellularity and epithelial organization [3,6]. Although evolution of the BM was pivotal to providing the mechanical structure and compartmentalization necessary to support multicellularity [3,7,8], it is likely that cells simultaneously needed to co-evolve the ability to cross these boundaries, to both migrate between, and anchor together, different tissues. Whether cell invasive behavior has a single evolutionary origin, or is a product of convergent evolution, remains unclear.
Invasive cellular behavior has been described in a variety of metazoan biological contexts and is critical for many aspects of development and homeostasis. As cell invasion is a dynamic process, often occurring deep within tissue layers where it is difficult to visualize using traditional microscopy, it is only in the last decade, with the advent of new imaging modalities [9–11], innovative genome engineering approaches such as CRISPR/Cas9 [12–15], and identification of new model systems to visualize invasion live [16,17], that we have begun to understand the basic principles that underlie acquisition of an invasive phenotype. Given the small, but growing, number of model systems available to study invasive cellular behavior, we are excited by the prospects to examine the evolution of this key cell biological process by making direct, functional comparisons among organisms. In this review, we provide a brief overview of the biological contexts where cell invasion occurs, the established and new model systems that are amenable to the study of cell invasive behavior, and our current understanding of the conserved and novel genetic mechanisms that regulate the acquisition of the invasive phenotype.
Contexts of cell invasion across the Metazoa
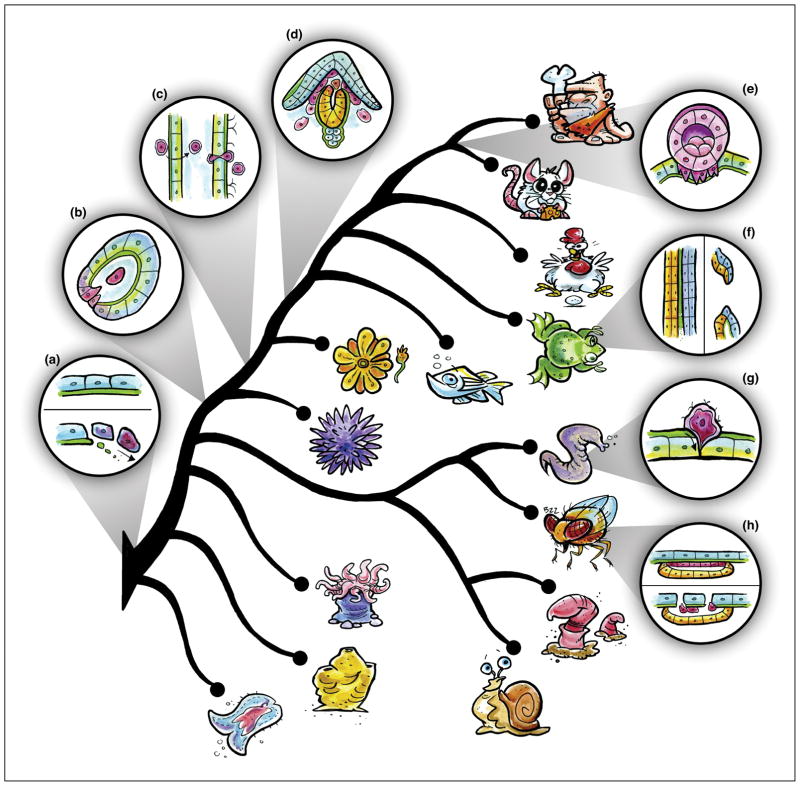
Cell invasion is a fundamental morphogenetic behavior that occurs at multiple times during the life history of many different taxa (Figure 1). Some of these contexts appear to be generally conserved across large phylogenetic distances, while others are unique to specific taxonomic groups. For example, in amphibian, avian, and mammalian embryos, a subset of mesendodermally-fated cells undergo an epithelial-to-mesenchymal transition (EMT), allowing them to penetrate the underlying BM and specify the site of gastrulation [18–21]. In sea urchin embryos, the earliest ingressing cells, the primary mesenchyme cells, undergo EMT and breach a BM before invading into the blastocoel [17] (Figure 1B). Many other EMT-dependent behaviors also coincide with invasive activity, including vertebrate cardiac valve development [22] and delamination of the neural crest from the roof plate of the neural tube [23,24] (Figure 1D). Thus, in many contexts where BM traversal is necessary, EMT and cell invasion function concomitantly. As EMT-like cellular behaviors have been observed in early branching phyla (i.e., sponges [25]), we posit here that EMT-driven invasive behaviors may be evolutionarily ancient in the Metazoa, coinciding with the evolution of the BM itself [3] (Figure 1A).
Figure 1. Cellular and phylogenetic contexts to study cell invasive behavior.
Cell invasion has been observed in a variety of contexts during embryonic development and homeostasis. (A–D) Invasive cell behaviors that may be more generally conserved between taxa include EMT (A), gastrulation (B), immune cell trafficking (C), and neural crest delamination (D). (E–H) Lineage-restricted cell invasive behaviors include deciduate placentation (E), mouth formation (F), AC invasion (G), wing disc eversion (H). Phylogeny based on [3,87–89].
Two critical mammalian embryological events also require BM invasion programs to be properly executed. The first occurs during endotheliochorial and hemochorial placentation in eutherian (placental) mammals (i.e., humans, primates, rodents, etc.). Syncytial trophoblast cells, which surround the embryonic placental villi, invade the BMs that line the endometrial layer of the maternal uterus and, in some instances, the spiral arteries that provide its blood supply, in order to establish pregnancy and nutrient circulation [26–28] (Figure 1E). A second key developmental event requiring cell invasive behavior has been observed in mouse embryonic development specifically, but is likely conserved among mammals. Following mouse embryo implantation, mechanical forces generated by maternal tissues promote BM rupture within the embryo, allowing the epiblast (embryonic ectoderm cells) to transmigrate and establish the anterior-posterior axis [29].
There are also specific examples of morphogenetic programs utilizing cell invasion during development. For example, during primary mouth formation in Xenopus, the BM between the ectoderm and endoderm is degraded, allowing for intercalation of cells from both tissues to form the buccopharyngeal membrane [30] (Figure 1F). Within the Ecdysozoa, cell invasion is required for eversion of specialized epithelial invaginations called imaginal discs during metamorphosis in Drosophila and other holometabolous insects. Specifically, squamous cells within the imaginal discs undergo a pseudo-EMT event and invade the BM lining the larval epidermis, ultimately giving rise to the adult exoskeleton [31–33] (Figure 1H). Cell invasion is also a critical aspect of organogenesis in the ecdysozoan nematodes. During uterine-vulval attachment in C. elegans and other rhabditid nematodes the uterine anchor cell (AC) invades the underlying BM to connect the gonad to the vulval epithelium, allowing for the future passage of eggs [34,35] (Figure 1G).
Outside of development, cell invasive behavior also plays an important role in immune system function, from the colonial urochordate ascidians, such as Botryllus schlosseri [36,37], to the chordates (Figure 1C). For example, leukocytes must exit the circulatory system via extravasation to reach sites of infection and injury [38–40]. Monocytes also extravasate during their differentiation into macrophages, and recent live cell imaging in zebrafish illustrates that hematopoietic stem cells must extravasate prior to their migration and formation of an endothelial stem cell niche [41]. Whether immune cells utilize the same molecular toolkit to escape the endothelial BM surrounding vasculature is poorly understood; although, at least in some contexts, it appears that extravasation occurs more frequently at sites that contain preformed BM gaps [39,42–44].
While invasion is critical to development and homeostasis, it also can elicit adverse consequences when aberrantly deployed, such as in the case of cancer metastasis. Following dissemination, the metastatic cascade entails migration, intravasation, and extravasation of cancer cells, which all require BM remodeling [45–48]. Given that cancer is known to hijack developmental regulatory programs [49–51], understanding the evolution of metazoan cell invasive programs will help us understand the mechanisms that cancer cells utilize to invade, providing potential alternative means for therapeutic alleviation of invasive behavior. As there are many excellent reviews that discuss cancer cell invasion [45–47], here, we focus on cell invasion events that occur during metazoan development, to best frame cell invasion in an evolutionary light.
A conserved genetic toolkit for invasive behavior?
Based on our current understanding of the genetic and molecular underpinnings that mediate acquisition of the invasive phenotype, there appear to be several conserved features common to an evolutionarily diverse group of organismal and developmental invasive contexts (Figure 2). In what follows, we summarize the current state of the field, focusing on both cell autonomous and non-autonomous control of invasion.
Figure 2. Conserved features of cellular invasion programs.
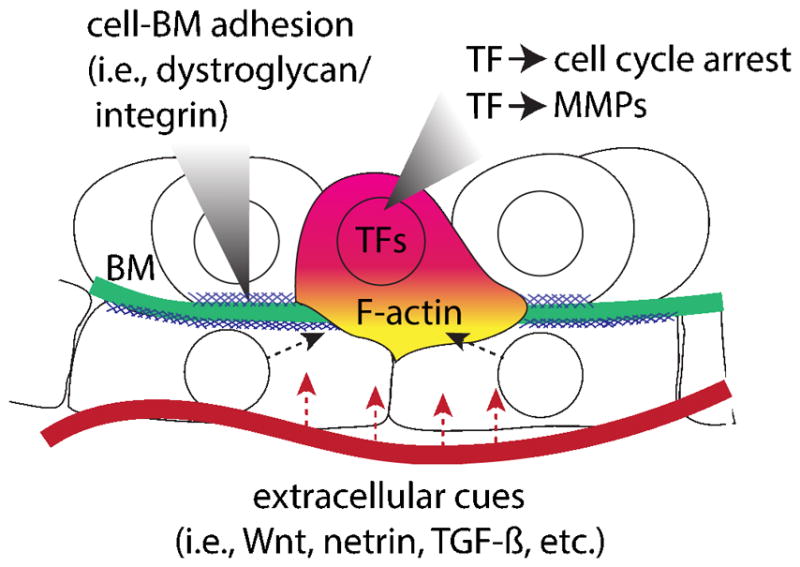
Schematic depicts key shared autonomous and non-autonomous genetic programs that appear to regulate cell invasive behavior.
Recent work from our own laboratory has highlighted a potentially deeply conserved aspect of the invasive cellular machinery - an inverse relationship between cell invasion and cell cycle progression. While this invasion/proliferation dichotomy was functionally identified in C. elegans AC invasion by our group [52], there exist many other examples where cells must stop cycling in order to invade, including deciduate placentation in which the extravillous trophoblast cells upregulate the cyclin-dependent kinase inhibitor p21 following their differentiation [53]. During sea urchin gastrulation, primary mesenchyme cells divide only after undergoing EMT and invading into the blastocoel [17,54], suggesting a similar dichotomy between proliferation and invasion. Indeed, in many contexts where cells undergo EMT and adopt an invasive mesenchymal phenotype, rates of proliferation decrease, often controlled by single zinc finger transcription factors (Zn-finger TFs) from the Snail/Slug family [55]. Finally, in many invasive cancers, correlative evidence suggests an inverse relationship between proliferation and invasion as well (see [56] for a comprehensive review). Together, these results suggest an ancient conservation between cell cycle arrest and invasive activity, although functional studies are needed in other model systems to test this intriguing hypothesis.
Transcriptionally, there also appear to be deeply conserved gene regulatory network interactions regulating invasive behavior. For example, it is well-documented that Zn-finger TFs from the Snail/Slug, Zeb and GATA families as well as the basic helix-loop-helix (bHLH) TF, Twist, promote EMT in many taxa (see [57] for a comprehensive review of developmental EMT). There is also a striking conservation of the regulatory circuit that activates matrix metalloproteinases (MMPs) during BM invasion programs across the Metazoa, as the AP-1 transcription factor (TF), FOS, regulates the expression of MMPs across a wide range of cell types and contexts [58], including C. elegans AC invasion [59] and placentation in deciduate mammals [60,61]. It is unclear if FOS is required for the MMP-mediated cell invasive contexts of Drosophila wing disc eversion [31], sea urchin EMT [17], vertebrate gastrulation [62,63] or neural crest delamination [23,64]. However, FOS family members are not always involved in activating MMP expression during invasion, suggesting that evolution has shaped the transcriptional networks mediating the deployment of proteases.
MMPs are not always required to breach BMs, suggesting that invasive cells can utilize different mechanisms to adopt an invasive phenotype. For example, non-proteolytic breakdown of the BM occurs during immune cell transmigration [39,42–44] and post-implantation mouse embryogenesis, where mechanical forces generated by maternal tissues have been implicated in the absence of MMPs to disrupt the BM [29]. A similar BM remodeling event may occur in hemimetaboluous insects (i.e., beetles, grasshoppers, dragonflies, etc.) during the reorganization of the extraembryonic amnion and serosa that occurs during embryonic development in the red flour beetle, Tribolium castaneum [65]. Researchers have yet to visualize the BM during this morphogenetic process, but similar to mouse epiblast migration [29], this process may be more reliant on physical forces rather than bulk proteolytic activity. Lastly, in C. elegans AC invasion, the initial BM breach requires the activity of fos-1a and potential MMP downstream targets [59]. However, the BM gap widens through a combination of sliding, facilitated by cell division of the underlying vulval precursor cells (VPCs) [34] and a tightly coordinated loss of cell-BM dystroglycan-mediated adhesion by the neighboring uterine cells [66] and integrin stabilization following cell cycle arrest of specific VPCs [34,67]. Finally, evidence from cancer biology supports the hypothesis that invasive cells can switch between MMP dependent and independent modes, as researchers have shown that based on matrix stiffness, cancer cells modulate their reliance on proteolytic activity during invasion[68]. Whether or not invasive cells in development or homeostasis can exhibit the same degree of plasticity as cancer cells remains an intriguing, but open question.
Other cell autonomous programs required for invasive behavior include the localization of the F-actin cytoskeleton during invasion, in the context of re-organization of apical/basal cell polarity and the formation of invadosomes, dynamic, punctate, F-actin-rich subcellular structures associated with BM degradation [16,69–72]. As this topic has been examined closely in the context of EMT and cell migration, we will refer readers to several recent reviews [73–75].
For many morphogenetic processes that require invasive cellular behavior, there are highly orchestrated interactions between the invading cell and the surrounding microenvironment, often requiring the cell(s) to receive input from multiple cell-cell signaling networks. Intriguingly, in two separate contexts, inhibition of the canonical Wnt signaling pathway has been utilized to promote BM degradation. Two secreted Wnt antagonists, Frzb-1 and Crescent, function during Xenopus mouth formation to promote the breakdown of BM separating the ectoderm and endoderm [76]. In both chick and frog embryos, DACT2, an intracellular inhibitor of nuclear β-catenin function, is required for the neural crest to delaminate [24]. Inversely, canonical Wnt can also activate invasion programs through EMT, as Wnt8 activity regulates the expression Twist and Snail at the vegetal pole of sea urchin embryos during EMT [77,78], as well as general roles for canonical Wnt, FGF, and BMP signaling during mouse and chick gastrulation and trunk neural crest delamination (reviewed in [57]). Together, these argue in favor of a model where pro-invasive extracellular cues are more likely to be evolutionarily malleable, though this has, to date, not been rigorously examined in any tractable system.
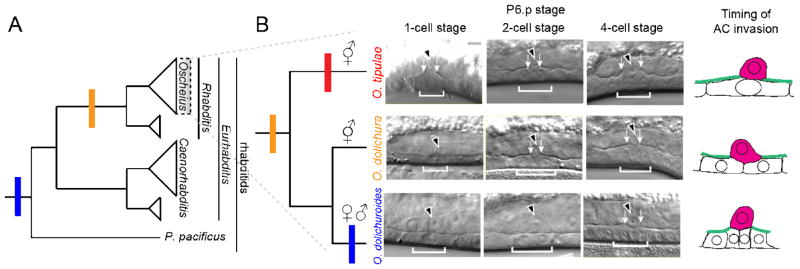
One such system to examine this hypothesis is rhabditid nematode AC invasion into the vulval epithelium. Research from the Sherwood lab has identified a role for netrin signaling from the underlying ventral nerve cord and an as-of-yet still unknown, secreted cue from the primary-fated vulval precursor cells [35,79] in providing the spatial and temporal controls guiding AC invasion. Whether these same signaling pathways are conserved in promoting invasion outside of C. elegans is currently unknown, but examination of the timing of AC invasion between related rhabditid nematodes suggests that, at the very least, the deployment of these signals can vary among species, as we have observed species-specific heterochrony in the timing of AC invasion in relation to the division of the underlying VPCs (Figure 3) [34]. Additionally, netrin signaling has been shown to function as a pro-invasive cue in certain cancers [80–83] and in leukocyte migration [84], suggesting that invasive cells may broadly coopt axon-guidance cues to provide spatiotemporal polarity.
Figure 3. Heterochrony in AC invasion timing observed in rhabditid nematodes.
Changes in the timing of nematode AC invasion in relation to vulval precursor cell division. A) Simplified rhabditid nematode phylogeny (left) depicts timing when AC invasion is complete based on DIC imaging [34]. B) Changes in timing of AC invasion have occurred in the evolution of the Oscheius genus (yellow). Red bar denotes invasion at the P6.p 1-cell stage, orange at the 2-cell stage, and blue at the 4-cell stage, summarized in the schematic (right). Micrographs modified from [34].
Conclusions & future directions/prospects
Moving forward, we envision parallel approaches to better understand how cell invasive behaviors evolved within the Metazoa. One potential approach is to dissect invasive behaviors that occur in organisms closely related to current classical genetic model systems such as C. elegans or Drosophila. Suitable systems would include holometabolous imaginal disc eversion or hemimetabolous extraembryonic rupture to compare to Drosophila melanogaster [31] or Tribolium castaneum [65], respectively. Our own work comparing AC invasion during rhabditid nematode vulval development also has the potential for adding insight into how a single conserved cell, the AC, modulates an invasion program, particularly as we can examine the intrinsic and extrinsic factors regulating AC invasion across large evolutionary distances [34]. Lastly, examining EMT during echinoderm gastrulation, where the transcriptional control of BM removal has been well-characterized [17], and comparing invasive programs with those of distantly related species will complement existing gene regulatory network analyses between lineages [85,86] and shed insight into the evolution of invasive gene batteries.
In parallel, it would be important to examine the earliest branching extant taxa that evolved BMs – the sponges and ctenophores. Recent genomic profiling identifies these taxa as having true BMs, suggesting that the BM was a key innovation leading to multicellularity [3]. Thus, it stands to reason that there are specific cells that must cross BMs during embryogenesis or homeostasis in these phyla. For these potential emerging model systems, the ability to generate transgenic animals will be key to visualizing invasion live. Hopefully, the ease of CRISPR/Cas9-mediated genome engineering will allow for functional testing of candidate pro-invasive genes and signaling pathways, facilitating the identification of conserved and novel features of cellular invasion programs, thus shedding insight into the number of evolutionary solutions there are to the question of how to adopt an invasive phenotype.
Highlights.
Cell invasion through basement membranes is a conserved cellular behavior that is critical to development and homeostasis in metazoans.
There appears to be a conserved genetic toolkit for the cell autonomous and non-autonomous programs mediating acquisition of the invasive phenotype.
Technological advances combined with the emergence of new model systems in which to study cell invasion have the potential to shed light on how this key cellular behavior evolved.
Acknowledgments
We apologize to the authors whose work could not be cited due to space limitations. We thank Abraham Kohrman and Ben Martin for helpful comments and Travis Hill for providing the illustrations used in Figure 1. We also thank Eric Haag, whose constructive feedback greatly improved the manuscript. This work was supported by NIH National Cancer Institute (5R00CA154870-05) and National Institute of General Medical Sciences (1R01GM121597-01) awards (to D.Q.M.), as well as by the Carol M. Baldwin Foundation. D.Q.M is a Damon Runyon-Rachleff Innovator supported (in part) by the Damon Runyon Cancer Research Foundation (DRR-47-17). In addition, some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Hynes RO, Naba A. Overview of the matrisome--an inventory of extracellular matrix constituents and functions. Cold Spring Harb Perspect Biol. 2012;4:a004903. doi: 10.1101/cshperspect.a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ozbek S, Balasubramanian PG, Chiquet-Ehrismann R, Tucker RP, Adams JC. The evolution of extracellular matrix. Mol Biol Cell. 2010;21:4300–4305. doi: 10.1091/mbc.E10-03-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3**.Fidler AL, Darris CE, Chetyrkin SV, Pedchenko VK, Boudko SP, Brown KL, Gray Jerome W, Hudson JK, Rokas A, Hudson BG. Collagen IV and basement membrane at the evolutionary dawn of metazoan tissues. Elife. 2017:6. doi: 10.7554/eLife.24176. This recent paper examines the evolution of key BM components in the earliest branching metazoan taxa, the sponges and ctenophores, and concludes that the evolution of type IV collagen was critical to the acquisition of multicellularity within the Metazoa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halfter W, Oertle P, Monnier CA, Camenzind L, Reyes-Lua M, Hu H, Candiello J, Labilloy A, Balasubramani M, Henrich PB, et al. New concepts in basement membrane biology. FEBS J. 2015;282:4466–4479. doi: 10.1111/febs.13495. [DOI] [PubMed] [Google Scholar]
- 5.Morrissey MA, Sherwood DR. An active role for basement membrane assembly and modification in tissue sculpting. J Cell Sci. 2015;128:1661–1668. doi: 10.1242/jcs.168021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leys SP, Riesgo A. Epithelia, an evolutionary novelty of metazoans. J Exp Zool B Mol Dev Evol. 2012;318:438–447. doi: 10.1002/jez.b.21442. [DOI] [PubMed] [Google Scholar]
- 7.Fidler AL, Vanacore RM, Chetyrkin SV, Pedchenko VK, Bhave G, Yin VP, Stothers CL, Rose KL, McDonald WH, Clark TA, et al. A unique covalent bond in basement membrane is a primordial innovation for tissue evolution. Proc Natl Acad Sci U S A. 2014;111:331–336. doi: 10.1073/pnas.1318499111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halfter W, Candiello J, Hu H, Zhang P, Schreiber E, Balasubramani M. Protein composition and biomechanical properties of in vivo-derived basement membranes. Cell Adh Migr. 2013;7:64–71. doi: 10.4161/cam.22479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu Q, Martin BL, Matus DQ, Gao L. Imaging multicellular specimens with real-time optimized tiling light-sheet selective plane illumination microscopy. Nature Communications. 2016:7. doi: 10.1038/ncomms11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao L, Shao L, Chen BC, Betzig E. 3D live fluorescence imaging of cellular dynamics using Bessel beam plane illumination microscopy. Nat Protoc. 2014;9:1083–1101. doi: 10.1038/nprot.2014.087. [DOI] [PubMed] [Google Scholar]
- 11.Royer LA, Lemon WC, Chhetri RK, Wan Y, Coleman M, Myers EW, Keller PJ. Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat Biotechnol. 2016;34:1267–1278. doi: 10.1038/nbt.3708. [DOI] [PubMed] [Google Scholar]
- 12.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo TW, Pickle CS, Lin S, Ralston EJ, Gurling M, Schartner CM, Bian Q, Doudna JA, Meyer BJ. Precise and heritable genome editing in evolutionarily diverse nematodes using TALENs and CRISPR/Cas9 to engineer insertions and deletions. Genetics. 2013;195:331–348. doi: 10.1534/genetics.113.155382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickinson DJ, Pani AM, Heppert JK, Higgins CD, Goldstein B. Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics. 2015 doi: 10.1534/genetics.115.178335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013 doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagedorn EJ, Ziel JW, Morrissey MA, Linden LM, Wang Z, Chi Q, Johnson SA, Sherwood DR. The netrin receptor DCC focuses invadopodia-driven basement membrane transmigration in vivo. J Cell Biol. 2013;201:903–913. doi: 10.1083/jcb.201301091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saunders LR, McClay DR. Sub-circuits of a gene regulatory network control a developmental epithelial-mesenchymal transition. Development. 2014;141:1503–1513. doi: 10.1242/dev.101436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders EJ, Prasad S. Invasion of a basement membrane matrix by chick embryo primitive streak cells in vitro. J Cell Sci. 1989;92(Pt 3):497–504. doi: 10.1242/jcs.92.3.497. [DOI] [PubMed] [Google Scholar]
- 19.Nakaya Y, Sukowati EW, Sheng G. Epiblast integrity requires CLASP and Dystroglycan-mediated microtubule anchoring to the basal cortex. J Cell Biol. 2013;202:637–651. doi: 10.1083/jcb.201302075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakaya Y, Sukowati EW, Wu Y, Sheng G. RhoA and microtubule dynamics control cell–basement membrane interaction in EMT during gastrulation. Nature Cell Biology. 2008;10:765–775. doi: 10.1038/ncb1739. [DOI] [PubMed] [Google Scholar]
- 21.Williams M, Burdsal C, Periasamy A, Lewandoski M, Sutherland A. Mouse primitive streak forms in situ by initiation of epithelial to mesenchymal transition without migration of a cell population. Dev Dyn. 2012;241:270–283. doi: 10.1002/dvdy.23711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tao G, Levay AK, Gridley T, Lincoln J. Mmp15 is a direct target of Snai1 during endothelial to mesenchymal transformation and endocardial cushion development. Dev Biol. 2011;359:209–221. doi: 10.1016/j.ydbio.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monsonego-Ornan E, Kosonovsky J, Bar A, Roth L, Fraggi-Rankis V, Simsa S, Kohl A, Sela-Donenfeld D. Matrix metalloproteinase 9/gelatinase B is required for neural crest cell migration. Dev Biol. 2012;364:162–177. doi: 10.1016/j.ydbio.2012.01.028. [DOI] [PubMed] [Google Scholar]
- 24*.Rabadan MA, Herrera A, Fanlo L, Usieto S, Carmona-Fontaine C, Barriga EH, Mayor R, Pons S, Marti E. Delamination of neural crest cells requires transient and reversible Wnt inhibition mediated by Dact1/2. Development. 2016;143:2194–2205. doi: 10.1242/dev.134981. This paper reports that Wnt/β-catenin signaling is autonomously inhibited through the activity of the scaffolding proteins Dact1/2 during neural crest delamination during Xenopus and chick embryogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakanishi N, Sogabe S, Degnan BM. Evolutionary origin of gastrulation: insights from sponge development. BMC Biol. 2014;12:26. doi: 10.1186/1741-7007-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carter AM, Enders AC, Pijnenborg R. The role of invasive trophoblast in implantation and placentation of primates. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140070. doi: 10.1098/rstb.2014.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Red-Horse K, Zhou Y, Genbacev O, Prakobphol A, Foulk R, McMaster M, Fisher SJ. Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J Clin Invest. 2004;114:744–754. doi: 10.1172/JCI22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Genbacev O, Miller RK. Post-implantation differentiation and proliferation of cytotrophoblast cells: in vitro models--a review. Placenta. 2000;21(Suppl A):S45–49. doi: 10.1053/plac.1999.0523. [DOI] [PubMed] [Google Scholar]
- 29.Hiramatsu R, Matsuoka T, Kimura-Yoshida C, Han S, Mochida K, Adachi T, Takayama S, Matsuo I. External Mechanical Cues Trigger the Establishment of the Anterior-Posterior Axis in Early Mouse Embryos. Dev Cell. 2013;27:131–144. doi: 10.1016/j.devcel.2013.09.026. [DOI] [PubMed] [Google Scholar]
- 30.Dickinson AJ, Sive H. Development of the primary mouth in Xenopus laevis. Dev Biol. 2006;295:700–713. doi: 10.1016/j.ydbio.2006.03.054. [DOI] [PubMed] [Google Scholar]
- 31.Srivastava A, Pastor-Pareja JC, Igaki T, Pagliarini R, Xu T. Basement membrane remodeling is essential for Drosophila disc eversion and tumor invasion. Proc Natl Acad Sci U S A. 2007;104:2721–2726. doi: 10.1073/pnas.0611666104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fessler LI, Condic ML, Nelson RE, Fessler JH, Fristrom JW. Site-specific cleavage of basement membrane collagen IV during Drosophila metamorphosis. Development. 1993;117:1061–1069. doi: 10.1242/dev.117.3.1061. [DOI] [PubMed] [Google Scholar]
- 33.Pastor-Pareja JC, Grawe F, Martin-Blanco E, Garcia-Bellido A. Invasive cell behavior during Drosophila imaginal disc eversion is mediated by the JNK signaling cascade. Dev Cell. 2004;7:387–399. doi: 10.1016/j.devcel.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 34.Matus DQ, Chang E, Makohon-Moore SC, Hagedorn MA, Chi QY, Sherwood DR. Cell division and targeted cell cycle arrest opens and stabilizes basement membrane gaps. Nature Communications. 2014;5:13. doi: 10.1038/ncomms5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherwood DR, Sternberg PW. Anchor cell invasion into the vulval epithelium in C. elegans. Dev Cell. 2003;5:21–31. doi: 10.1016/s1534-5807(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 36.Weissman IL. Stem cells are units of natural selection for tissue formation, for germline development, and in cancer development. Proc Natl Acad Sci U S A. 2015;112:8922–8928. doi: 10.1073/pnas.1505464112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rinkevich Y, Voskoboynik A, Rosner A, Rabinowitz C, Paz G, Oren M, Douek J, Alfassi G, Moiseeva E, Ishizuka KJ, et al. Repeated, long-term cycling of putative stem cells between niches in a basal chordate. Dev Cell. 2013;24:76–88. doi: 10.1016/j.devcel.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madsen CD, Sahai E. Cancer dissemination--lessons from leukocytes. Dev Cell. 2010;19:13–26. doi: 10.1016/j.devcel.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 39.Nourshargh S, Hordijk PL, Sixt M. Breaching multiple barriers: leukocyte motility through venular walls and the interstitium. Nat Rev Mol Cell Biol. 2010;11:366–378. doi: 10.1038/nrm2889. [DOI] [PubMed] [Google Scholar]
- 40.Dominguez F, Yanez-Mo M, Sanchez-Madrid F, Simon C. Embryonic implantation and leukocyte transendothelial migration: different processes with similar players? Faseb J. 2005;19:1056–1060. doi: 10.1096/fj.05-3781hyp. [DOI] [PubMed] [Google Scholar]
- 41.Tamplin OJ, Durand EM, Carr LA, Childs SJ, Hagedorn EJ, Li P, Yzaguirre AD, Speck NA, Zon LI. Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell. 2015;160:241–252. doi: 10.1016/j.cell.2014.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pflicke H, Sixt M. Preformed portals facilitate dendritic cell entry into afferent lymphatic vessels. J Exp Med. 2009;206:2925–2935. doi: 10.1084/jem.20091739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sixt M, Engelhardt B, Pausch F, Hallmann R, Wendler O, Sorokin LM. Endothelial cell laminin isoforms, laminins 8 and 10, play decisive roles in T cell recruitment across the blood-brain barrier in experimental autoimmune encephalomyelitis. J Cell Biol. 2001;153:933–946. doi: 10.1083/jcb.153.5.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu C, Ivars F, Anderson P, Hallmann R, Vestweber D, Nilsson P, Robenek H, Tryggvason K, Song J, Korpos E, et al. Endothelial basement membrane laminin alpha5 selectively inhibits T lymphocyte extravasation into the brain. Nat Med. 2009;15:519–527. doi: 10.1038/nm.1957. [DOI] [PubMed] [Google Scholar]
- 45.Rowe R, Weiss S. Breaching the basement membrane: who, when and how? Trends in Cell Biology. 2008;18:560–574. doi: 10.1016/j.tcb.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 46.Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 47.Sahai E. Illuminating the metastatic process. Nat Rev Cancer. 2007;7:737–749. doi: 10.1038/nrc2229. [DOI] [PubMed] [Google Scholar]
- 48.Harney AS, Arwert EN, Entenberg D, Wang Y, Guo P, Qian BZ, Oktay MH, Pollard JW, Jones JG, Condeelis JS. Real-Time Imaging Reveals Local, Transient Vascular Permeability, and Tumor Cell Intravasation Stimulated by TIE2hi Macrophage-Derived VEGFA. Cancer Discov. 2015;5:932–943. doi: 10.1158/2159-8290.CD-15-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aiello NM, Stanger BZ. Echoes of the embryo: using the developmental biology toolkit to study cancer. Dis Model Mech. 2016;9:105–114. doi: 10.1242/dmm.023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma Y, Zhang P, Wang F, Yang J, Yang Z, Qin H. The relationship between early embryo development and tumourigenesis. J Cell Mol Med. 2010;14:2697–2701. doi: 10.1111/j.1582-4934.2010.01191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holtan SG, Creedon DJ, Haluska P, Markovic SN. Cancer and pregnancy: parallels in growth, invasion, and immune modulation and implications for cancer therapeutic agents. Mayo Clin Proc. 2009;84:985–1000. doi: 10.1016/S0025-6196(11)60669-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52*.Matus DQ, Lohmer LL, Kelley LC, Schindler AJ, Kohrman AQ, Barkoulas M, Zhang W, Chi Q, Sherwood DR. Invasive Cell Fate Requires G1 Cell-Cycle Arrest and Histone Deacetylase-Mediated Changes in Gene Expression. Developmental Cell. 2015;35:162–174. doi: 10.1016/j.devcel.2015.10.002. This paper, published by our group, presents functional evidence that cell invasion requires a G1/G0 cell cycle arrest in order to be properly executed, during C. elegans AC invasion into the vulval epithelium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Genbacev O, McMaster MT, Fisher SJ. A repertoire of cell cycle regulators whose expression is coordinated with human cytotrophoblast differentiation. Am J Pathol. 2000;157:1337–1351. doi: 10.1016/S0002-9440(10)64648-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lyons DC, Kaltenbach SL, McClay DR. Morphogenesis in sea urchin embryos: linking cellular events to gene regulatory network states. Wiley Interdiscip Rev Dev Biol. 2012;1:231–252. doi: 10.1002/wdev.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56*.Kohrman AQ, Matus DQ. Divide or Conquer: Cell Cycle Regulation of Invasive Behavior. Trends Cell Biol. 2017;27:12–25. doi: 10.1016/j.tcb.2016.08.003. This review, published by our group, presents evidence that cell cycle arrest appears to be a prerequisite for invasiveness in many contexts of development and disease, supporting the hypothesized dichotomy between cell proliferation and invasion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lim J, Thiery JP. Epithelial-mesenchymal transitions: insights from development. Development. 2012;139:3471–3486. doi: 10.1242/dev.071209. [DOI] [PubMed] [Google Scholar]
- 58.Lin CW, Georgescu HI, Evans CH. The role of AP-1 in matrix metalloproteinase gene expression. Agents Actions. 1993;39(Spec No):C215–218. doi: 10.1007/BF01972770. [DOI] [PubMed] [Google Scholar]
- 59.Sherwood DR, Butler J, Kramer J, Sternberg P. FOS-1 Promotes Basement-Membrane Removal during Anchor-Cell Invasion in. Cell. 2005;121:951–962. doi: 10.1016/j.cell.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 60.Qiu Q, Yang M, Tsang BK, Gruslin A. EGF-induced trophoblast secretion of MMP-9 and TIMP-1 involves activation of both PI3K and MAPK signalling pathways. Reproduction. 2004;128:355–363. doi: 10.1530/rep.1.00234. [DOI] [PubMed] [Google Scholar]
- 61.Kent LN, Rumi MA, Kubota K, Lee DS, Soares MJ. FOSL1 is integral to establishing the maternal-fetal interface. Mol Cell Biol. 2011;31:4801–4813. doi: 10.1128/MCB.05780-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mogi K, Toyoizumi R. Invasion by matrix metalloproteinase-expressing cells is important for primitive streak formation in early chick blastoderm. Cells Tissues Organs. 2010;192:1–16. doi: 10.1159/000286231. [DOI] [PubMed] [Google Scholar]
- 63.Coyle RC, Latimer A, Jessen JR. Membrane-type 1 matrix metalloproteinase regulates cell migration during zebrafish gastrulation: evidence for an interaction with non-canonical Wnt signaling. Exp Cell Res. 2008;314:2150–2162. doi: 10.1016/j.yexcr.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 64.Duong TD, Erickson CA. MMP-2 plays an essential role in producing epithelial-mesenchymal transformations in the avian embryo. Dev Dyn. 2004;229:42–53. doi: 10.1002/dvdy.10465. [DOI] [PubMed] [Google Scholar]
- 65*.Hilbrant M, Horn T, Koelzer S, Panfilio KA. The beetle amnion and serosa functionally interact as apposed epithelia. Elife. 2016:5. doi: 10.7554/eLife.13834. This paper uses light sheet microscopy and new transgenic lines in the model beetle, Tribolium castaneum, to examine the rupturing of the extraembryonic tissues during embryogenesis, highlighting the potential for this new model system to examine basement membrane remodeling during morphogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66**.McClatchey ST, Wang Z, Linden LM, Hastie EL, Wang L, Shen W, Chen A, Chi Q, Sherwood DR. Boundary cells restrict dystroglycan trafficking to control basement membrane sliding during tissue remodeling. Elife. 2016:5. doi: 10.7554/eLife.17218. This paper uses the power of C. elegans forward genetics approaches paired with functional genomics and live-cell imaging to identify a dystroglycan-based mechanism for precisely regulating BM gap expansion during AC invasion into the vulval epithelium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ihara S, Hagedorn EJ, Morrissey MA, Chi Q, Motegi F, Kramer JM, Sherwood DR. Basement membrane sliding and targeted adhesion remodels tissue boundaries during uterine-vulval attachment in Caenorhabditis elegans. Nature cell biology. 2011;13:641–651. doi: 10.1038/ncb2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolf K, Te Lindert M, Krause M, Alexander S, Te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol. 2013;201:1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hagedorn EJ, Kelley LC, Naegeli KM, Wang Z, Chi Q, Sherwood DR. ADF/cofilin promotes invadopodial membrane recycling during cell invasion in vivo. J Cell Biol. 2014;204:1209–1218. doi: 10.1083/jcb.201312098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lohmer LL, Clay MR, Naegeli KM, Chi Q, Ziel JW, Hagedorn EJ, Park JE, Jayadev R, Sherwood DR. A Sensitized Screen for Genes Promoting Invadopodia Function In Vivo: CDC-42 and Rab GDI-1 Direct Distinct Aspects of Invadopodia Formation. PLoS Genet. 2016;12:e1005786. doi: 10.1371/journal.pgen.1005786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Z, Xu Y, Zhang C, Liu X, Jiang L, Chen F. Mammalian diaphanous-related formin 1 is required for motility and invadopodia formation in human U87 glioblastoma cells. Int J Mol Med. 2014;33:383–391. doi: 10.3892/ijmm.2013.1577. [DOI] [PubMed] [Google Scholar]
- 72.Morrissey MA, Hagedorn EJ, Sherwood DR. Cell invasion through basement membrane: The netrin receptor DCC guides the way. Worm. 2013;2:e26169. doi: 10.4161/worm.26169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lohmer LL, Kelley LC, Hagedorn EJ, Sherwood DR. Invadopodia and basement membrane invasion in vivo. Cell Adh Migr. 2014:8. doi: 10.4161/cam.28406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Glentis A, Gurchenkov V, Matic Vignjevic D. Assembly, heterogeneity, and breaching of the basement membranes. Cell Adh Migr. 2014;8:236–245. doi: 10.4161/cam.28733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murphy DA, Courtneidge SA. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nat Rev Mol Cell Biol. 2011;12:413–426. doi: 10.1038/nrm3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dickinson AJ, Sive HL. The Wnt antagonists Frzb-1 and Crescent locally regulate basement membrane dissolution in the developing primary mouth. Development. 2009;136:1071–1081. doi: 10.1242/dev.032912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wikramanayake AH, Huang L, Klein WH. beta-Catenin is essential for patterning the maternally specified animal-vegetal axis in the sea urchin embryo. Proc Natl Acad Sci U S A. 1998;95:9343–9348. doi: 10.1073/pnas.95.16.9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wikramanayake AH, Peterson R, Chen J, Huang L, Bince JM, McClay DR, Klein WH. Nuclear beta-catenin-dependent Wnt8 signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis. 2004;39:194–205. doi: 10.1002/gene.20045. [DOI] [PubMed] [Google Scholar]
- 79.Ziel JW, Hagedorn EJ, Audhya A, Sherwood DR. UNC-6 (netrin) orients the invasive membrane of the anchor cell in C. elegans. Nature Cell Biology. 2009;11:183–189. doi: 10.1038/ncb1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mehlen P, Guenebeaud C. Netrin-1 and its dependence receptors as original targets for cancer therapy. Curr Opin Oncol. 2010;22:46–54. doi: 10.1097/CCO.0b013e328333dcd1. [DOI] [PubMed] [Google Scholar]
- 81.Dumartin L, Quemener C, Laklai H, Herbert J, Bicknell R, Bousquet C, Pyronnet S, Castronovo V, Schilling MK, Bikfalvi A. Netrin-1 Mediates Early Events in Pancreatic Adenocarcinoma Progression, Acting on Tumor and Endothelial Cells. Gastroenterology. 2010;138:1595–1606. e1598. doi: 10.1053/j.gastro.2009.12.061. [DOI] [PubMed] [Google Scholar]
- 82.Chen H, Wei Q, Zhang J, Xu C, Tan T, Ji W. Netrin-1 signaling mediates NO-induced glial precursor migration and accumulation. Cell Res. 2010;20:238–241. doi: 10.1038/cr.2010.7. [DOI] [PubMed] [Google Scholar]
- 83.Rodrigues S, De Wever O, Bruyneel E, Rooney RJ, Gespach C. Opposing roles of netrin-1 and the dependence receptor DCC in cancer cell invasion, tumor growth and metastasis. Oncogene. 2007;26:5615–5625. doi: 10.1038/sj.onc.1210347. [DOI] [PubMed] [Google Scholar]
- 84.Ly NP, Komatsuzaki K, Fraser IP, Tseng AA, Prodhan P, Moore KJ, Kinane TB. Netrin-1 inhibits leukocyte migration in vitro and in vivo. Proc Natl Acad Sci U S A. 2005;102:14729–14734. doi: 10.1073/pnas.0506233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McCauley BS, Weideman EP, Hinman VF. A conserved gene regulatory network subcircuit drives different developmental fates in the vegetal pole of highly divergent echinoderm embryos☆. Dev Biol. 2010;340:200–208. doi: 10.1016/j.ydbio.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 86.Hinman VF, Nguyen AT, Cameron RA, Davidson EH. Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc Natl Acad Sci U S A. 2003;100:13356–13361. doi: 10.1073/pnas.2235868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ryan JF, Pang K, Schnitzler CE, Nguyen AD, Moreland RT, Simmons DK, Koch BJ, Francis WR, Havlak P, Program NCS, et al. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 2013;342:1242592. doi: 10.1126/science.1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hejnol A, Obst M, Stamatakis A, Ott M, Rouse GW, Edgecombe GD, Martinez P, Baguna J, Bailly X, Jondelius U, et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc Biol Sci. 2009;276:4261–4270. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dunn C, Hejnol A, Matus D, Pang K, Browne W, Smith S, Seaver E, Rouse G, Obst M, Edgecombe G, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–U745. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]