Abstract
Genome stability ensures individual fitness and reliable transmission of genetic information. Hybridization between diverging lineages can trigger genome instability, highlighting its potential role in post-zygotic reproductive isolation. We argue that genome instability is not merely one of several types of hybrid incompatibility, but rather that genome stability is one of the very first and most fundamental traits that can break down when two diverged genomes are combined. Future work will reveal how frequent and predictable genome instability is in hybrids, how it affects hybrid fitness, and whether it is a direct cause or consequence of speciation.
Introduction
Speciation is the biological process by which populations diverge and become reproductively isolated. Much work has been devoted to identifying hybrid incompatibility genes that underlie reproductive isolation. Only a handful have been identified, highlighting firstly the confounding effect of other processes producing similar signatures of high divergence (e.g. drift and linked selection) and secondly the complex genetic basis of reproductive isolation [1,2]. Massively parallel sequencing has greatly improved our understanding of the genomic landscape of DNA sequence divergence and speciation [3]. One surprising finding is that there seems to be only rare instances where diverging populations carry differentially fixed alleles of genes involved in post-zygotic reproductive isolation [1,4]. This is because evolution and divergence seem to proceed mainly by soft sweeps from standing genetic variation and reproductive isolation tends to be highly polygenic. This also explains why so few “speciation genes” (more accurately termed “reproductive isolation genes”) have been identified outside of model systems [2,5]. The molecular basis of post-zygotic reproductive isolation thus remains difficult to identify using current strategies.
Here, we suggest that a focus on genome stability will be a fruitful strategy to tackle the molecular and genetic basis of post-zygotic reproductive isolation. Processes that maintain genome stability have frequently been observed to be “broken down” in hybrids, causing sterility and/or lethality through increased mutation rate or inaccurate chromosome segregation ([2,6–8] and references discussed below). This is because genomes are co-evolved units influenced by selective and neutral processes, and hybridization may disrupt this co-adaptation, leading to genome instability in multiple, additive and non-additive ways. We thus view genome stability as a trait, and genome instability as a hybrid incompatibility phenotype, with the underlying cause being sequence divergence and/or genome rearrangements between hybridizing populations or species. We suggest that genome instability is a widespread hybrid incompatibility phenotype because of the range of sequence classes whose divergence leads to genome instability. We present and discuss six mechanisms by which genome instability may arise in hybrids (Figure 1). Examples from the current literature illustrate how these mechanisms relate to divergence and how they may have genetic architectures of variable complexity. Finally we raise important questions to be addressed by future studies.
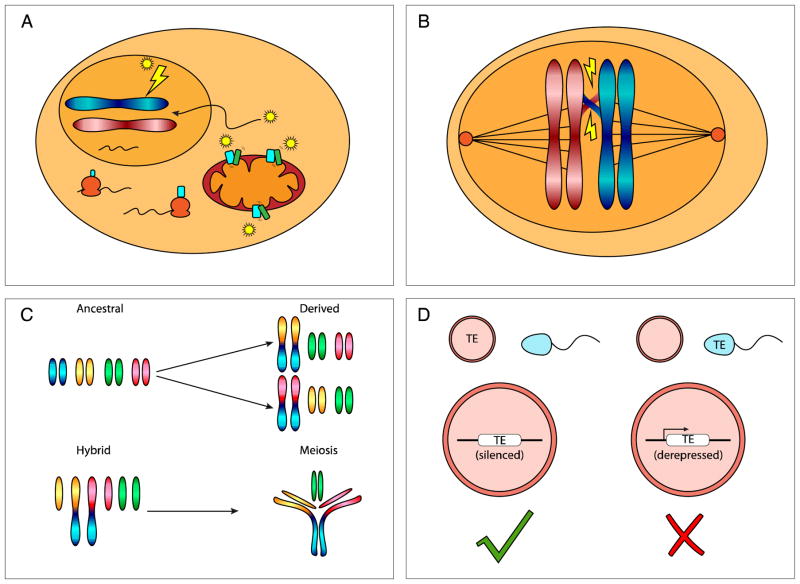
Figure 1. Schematic representation of genome instability mechanisms contributing to post-zygotic reproductive isolation.
A. Mito-nuclear incompatibilities may lead to increased reactive oxygen species production. A nucleus-encoded protein (light blue) is incompatible with a mitochondria-encoded protein (green). These two proteins interact together in the electron transport chain in sub-optimal ways due to amino acid changes between two lineages. This leads to inefficient oxidative phosphorylation and release of reactive oxygen species (yellow stars), which causes DNA damage. B. DNA mismatch proteins halt meiotic recombination between divergent sequences (here, between divergent homologous chromosomes in red and blue), reducing fertility of hybrids. C. Different Robertsonian chromosome rearrangements occur between diverging lineages (top panel). Upon secondary contact, these form complex and unstable arrangements during hybrid meiosis, reducing fertility (lower panel). D. Transposable element derepression in hybrids. Diverging lineages carry different transposable elements. Maternally loaded piRNAs initiate transposable element repression, maintaining silencing in the zygote (left panel). Paternally contributed transposable elements may be derepressed if the female fails to load repressive piRNAs. This leads to transposable element mobilization with potentially deleterious consequences for the progeny (right panel).
Genome stability is a complex trait
We consider genome stability as a trait with a complex genetic basis which requires a tight coordination of a complex network of signaling pathways [9]. Genome instability can arise from intrinsic factors such as transposable element derepression or mitochondrial metabolism, and from extrinsic factors such as environmental mutagens [9]. Therefore, genome instability may also involve gene-environment interactions. Molecular processes underlying genome stability include DNA damage sensors, DNA repair pathways and cell cycle checkpoints [9]. A major structural feature of genome stability is heterochromatin, a specialized type of chromatin that maintains repetitive DNA in a silent state. Heterochromatin is particularly important at telomeres and centromeres to stabilize chromosomes and maintain proper segregation [10].
The rate of sequence evolution of proteins involved in genome stability maintenance greatly varies. Many are highly conserved, sometimes from prokaryotes to eukaryotes (e.g. the recombinases recA and RAD51 [11]). Others, such as some genes required for centromere assembly, telomere capping and silencing of transposable elements evolve rapidly, sometimes under positive selection [12–14]. These rapidly evolving genes are good candidates when trying to identify the molecular basis of genome instability associated with reproductive isolation, as exemplified by genes such as PRDM9 in mammals (see below). Elucidating the forces driving rapid evolution of these genes is an important step to understanding why and how genome instability occurs in hybrids (Box 1).
Box 1. Relevant questions to genome stability and reproductive isolation.
General Questions
How frequently are hybrid genomes destabilized? Is this predictable from levels of DNA sequence divergence?
Why are certain genes involved in genome stability highly constrained (e.g. meiotic recombination machinery) while are others rapidly evolving (e.g. centromere definition, telomere capping)?
Questions related to molecular mechanisms
Do mito-nuclear incompatibilities lead to an increased mutation rate in hybrids?
Is there a “divergence threshold” disrupting meiosis through MMR that is strictly DNA sequence dependent?
Do hybrids show an increased mutation rate compared to parental lineages? Does this result from decreased replication fidelity?
Does DNA repair pathway choice during meiotic recombination impact divergence and reproductive isolation?
Are crossovers positioned differently in hybrids compared to parental lineages? Does this impact reproductive isolation?
How frequent is chromosome instability in hybrids?
Is transposable element derepression in hybrids a cause or a consequence of lineage divergence?
Mechanism 1: Mito-nuclear incompatibilities
Mitochondria produce ATP, the cell’s main source of energy, through oxidative phosphorylation. In animals, the mitochondrial genome is rapidly evolving compared to the nuclear genome [15], yet it codes for only a few genes and most proteins necessary for mitochondrial functions are encoded by the nuclear genome [16]. Therefore, the mitochondrial and nuclear genomes are thought to be tightly co-evolving within a species, and mito-nuclear incompatibilities have thus been predicted to underlie reproductive isolation [see 15 for a recent comprehensive review]. One way mito-nuclear incompatibilities could cause genome instability is by suboptimal electron transport resulting from the combination of maladapted alleles in a hybrid background. An increase in reactive oxygen species due to reduced respiratory activity occurs in mouse cybrid cell lines (cell lines which have different mtDNA haplotypes in an otherwise identical nuclear background) [17]. This may increase the amount of reactive oxygen species produced, a normal by-product of oxidative phosphorylation, but also potent DNA damage-inducing agents (Figure 1A) [15,18].
Geographically isolated populations of the marine copepod Tigriopus californicus exhibit a gradient of low to high levels of nuclear and mtDNA divergence (up to 15%), yet their hybrids are often at least partially viable. Barreto and Burton found that the fecundity of hybrids between increasingly divergent populations is correlated with 8-OH-dG levels, a marker of oxidative DNA damage induced by reactive oxygen species [19]. Hybrids from highly diverged populations also consistently show high 8-OH-dG levels, consistent with increased reactive oxygen species production due to sub-optimal electron transport. Similarly, Chang et al. reported reduced fertility in Caenorhabditis briggsae inter-population hybrids, which correlated with increased reactive oxygen species production [20]. Considering the number of systems where mito-nuclear incompatibilities have been reported (e.g. Drosophila [21], Nasonia wasp [22], Mimulus monkeyflower [23], and Anguilla eels [24]), we predict that increased reactive oxygen species production in hybrids may be found more widely. It would be interesting to test whether increased reactive oxygen species correlate with an increased mutation rate in hybrids, which would more directly indicate genome instability due to mitochondrial dysfunction (Box 1).
Mechanism 2: DNA repair incompatibilities
Mismatch repair (MMR) proteins dramatically increase DNA replication fidelity by excising DNA mismatches that form as the result of DNA polymerase mis-incorporation errors. The MMR pathway also prevents recombination between divergent DNA sequences by recognizing and unwinding heteroduplex DNA that contains mismatches. This seems to occur during meiosis in hybrids between diverging yeast strains and species [25,26]. To our knowledge, the impact of sequence divergence has not been directly tested in other systems, but meiotic recombination may be inhibited in hybrids between diverged species, such as between the nematodes Caenorhabditis briggsae and C. nigoni [27]. These observations raise the possibility that sequence divergence itself may further promote divergence by inhibiting recombination through the activation of the MMR pathway (Box 1, Figure 1B). Species with exceptionally high levels of standing genetic variation, such as C. brenneri, may target the meiotic recombination machinery to loci with locally reduced polymorphism [28]. Another prediction is that incompatible MMR alleles could lead to a mutator phenotype, triggering genome instability in hybrids [29].
Recent studies have identified such incompatible alleles of MMR proteins. Demogines et al. [30] found that recombinant Saccharomyces cerevisiae strains carrying incompatible alleles of MMR proteins displayed increased mutation rates (although these strains are not reproductively isolated per se). Interestingly, this increased mutation rate accelerates adaptation to a stressful environment [31]. However, a mutator phenotype is predicted to be costly in the long term, due to the accumulation of deleterious mutations. Suppressor mutations within strains carrying incompatible alleles are thus likely to arise [32,but see 33 for a different interpretation]. This example illustrates how DNA repair pathways can be involved in reproductive isolation and potentially in adaptation. Considering the numerous factors influencing DNA replication fidelity [34], direct assessment of hybrid mutation rates compared to parental lineages is again likely to be an important step in deciphering such types of incompatibilities (Box 1). More detailed analyses of mutation type and context will help reveal the precise pathways involved.
Mechanism 3: Meiotic homologous recombination breakdown
Meiotic recombination directly influences eukaryotic evolution and is also generally a prerequisite to proper chromosome pairing and segregation [35]. Meiotic recombination requires the tight coordination of several steps, including homolog pairing, DNA double-strand break generation, homologous chromosome invasion, and DNA repair pathway choice and resolution. Reproductive isolation has been associated with the breakdown of several steps of meiotic recombination, including invasion and pathway choice (MMR incompatibility described above) and possibly double-strand break generation (see below), all of which may lead to genome instability.
PRDM9 is the only known mammalian hybrid incompatibility gene [36]. PRDM9 is a histone methyltransferase which directs the meiotic homologous recombination machinery to specific sites, determining recombination hotspot usage [37]. PRDM9 is particularly interesting because it is at the interface between recombination, chromatin modification, and chromosome segregation, emphasizing the multi-faceted aspect of genome stability. In male hybrids between Mus musculus musculus and M. m. domesticus, PRDM9-mediated meiotic arrest causes sterility by an unknown mechanism which relies on the DNA binding properties of PRDM9 [38], but may not involve hotspot determination [39,40]. Clearly, much remains to be discovered to understand the molecular mechanisms and genetic architecture underlying PRDM9-mediated hybrid infertility. More generally, there is considerable variation in how meiosis is coordinated and achieved across taxa [41,42]. For example, many organisms lack canonical hotspots (e.g. Drosophila [43]), and even within mammals, dogs and their relatives have lost PRDM9 [44,45]. A more general research avenue is to study how DNA repair pathway choice (homologous recombination versus gene conversion [46]) and crossover localization [3] influence divergence and speciation across a broad range of organisms (Box 1).
Mechanism 4: Chromosomal instability and aneuploidy in hybrids
Polymorphic chromosome rearrangements such as inversions and Robertsonian rearrangements (fusions and fissions involving an entire chromosome arm) are frequently found across eukaryotes [6,7]. Chromosome rearrangements have long been hypothesized to promote divergence by reducing fitness in hybrids, but such models suffer from a theoretical conundrum regarding the small probability of fixation of rearrangements that are deleterious in heterozygotes. More recently, inversions were proposed to bolster adaptation by limiting recombination, without necessitating reduced hybrid fitness [47,48].
The house mouse Mus musculus is perhaps the best-characterized model supporting a role for chromosome rearrangements in post-zygotic reproductive isolation. Extensive Robertsonian rearrangements are found in Mus musculus. Complex and unstable meiotic chains form in hybrids between chromosomal races, leading to meiotic nondisjunction and sterility (Figure 1C) [49]. Recent work combining genetic and simulation studies shows that hybrid meiotic breakdown itself explains reductions in gene flow between rearranged chromosomes and does not require recombination suppression mechanisms [50]. Robertsonian rearrangements can impact centromere strength [51], favoring the transmission of the rearranged chromosome during the asymmetrical female meiosis (i.e. meiotic drive), and thus spreading to entire populations [52].
In the yeast S. paradoxus hybrids between diverging populations with rearranged chromosomes show chromosome instability [53]. Interestingly, hybridization also potentially led to speciation of a hybrid lineage in which chromosome rearrangements partially explain reproductive isolation with ancestral lineages [54]. Recently diverged populations of Lake Whitefish (<12–15,000 generations) show evidence of ongoing chromosomal divergence, and their hybrids suffer from extensive aneuploidy consistent with both meiotic and mitotic segregation errors [55,56]. Future efforts should aim in more systems to characterize genome organization comprehensively and to document the impact of chromosome rearrangements.
Mechanism 5: Heterochromatin divergence
Heterochromatin patterns and associated DNA sequences evolve rapidly and yet contribute to essential processes such as transposable element repression and chromosome segregation. This paradox led to the hypothesis that heterochromatin differences between diverging lineages may contribute to reproductive isolation [8]. Studies in hybrids from plants to mammals show aberrations in heterochromatin such as DNA methylation patterns [57,58] which may destabilize the genome [7].
Repetitive DNA sequences such as satellite repeats are prime candidates to trigger genome instability in hybrids. Heterochromatin differences directly cause reproductive isolation between D. simulans females and D. melanogaster males, where female F1 hybrids die during embryonic development. A large block of a 359bp satellite repeat on the X chromosome of D. melanogaster leads to mitotic instability in female hybrids [59]. This is thought to result from improper peri-centromeric heterochromatin formation, a hypothesis supported by recent work showing that peri-centromeric satellite repeat transcription is essential for proper chromosome segregation [60]. Heterochromatin-embedded sequences are challenging to study, but emerging long-read sequencing technologies combined with complementary approaches will help reveal whether these rapidly evolving structures are frequently disrupted in hybrids (Box 1 and 2).
Box 2. Technologies to detect genome instability in hybrids.
The combination of classical and state-of-the-art technologies helps is necessary to detect genome instability. Particularly challenging is the analysis of the non-coding fraction of the genome associated with heterochromatin, which is often involved in genome instability.
Long read sequencing: Repetitive sequences are challenging to resolve with short reads and may result in gaps and collapses of the assembly. Long read sequencing technologies at high coverage are starting to decipher the non-coding fraction of the genome and reveal its complex organization [e.g. 68].
Optical mapping: Draft genome assemblies are typically fragmented and incomplete. In optical mapping, long DNA molecules are digested with a site-specific nicking endonuclease and fluorescent probes are inserted. The resulting fluorescent molecule can be imaged and used to anchor and assemble contigs and scaffolds [e.g. 69].
Cytometry and “cytogenomics”: These methods allow the detection and characterization of genome size variation that may result from chromosome instability, repeat expansion or facultative chromosome polymorphism. These methods are generally inexpensive and can be integrated with genomic data [e.g. 55, 70]
RNA-sequencing: When properly designed, RNA-sequencing allows to test for differential gene expression in hybrids, to characterize genetic divergence between populations and to identify candidate genes that are rapidly evolving.
Mechanism 6: small RNAs and transposable elements
Small RNAs perform multiple functions that are fundamental to genome stability, from gene expression regulation to heterochromatin assembly and dosage compensation – many of which have been reported as “broken down” in hybrids (previous section and [61]). The diverse functions of small RNAs have been extensively reviewed elsewhere [e.g. 62]; here we briefly cover their role in transposable element repression and reproductive isolation.
Transposable elements (TEs) are selfish genetic parasites found in most (if not all) eukaryotic genomes. Transposable elements must be tightly controlled, as their mobilization may lead to highly mutagenic events by creating DNA double-strand breaks, interrupting open reading frames, deregulating gene expression, and causing ectopic recombination. Transposable element repression is initiated in part by maternal loading of PIWI interacting small RNAs (piRNAs) into the egg. When there is a mismatch between paternally transmitted transposable elements and maternally loaded piRNAs, this can lead to transposable element derepression, at least in Drosophila (Figure 1D). Transposable element derepression has frequently been reported in hybrids [e.g. 63–66].
There is an ever growing list of host-encoded transposable element repressors, many of which are rapidly evolving [67]. These are good candidates in the search for the molecular basis of reproductive isolation associated with genome instability, because divergent alleles may be incompatible, triggering transposable element derepression in hybrids. One test of this hypothesis, however, found surprisingly mild effects on TE expression when the D. melanogaster aubergine gene was replaced by its D. simulans ortholog [64].
Emerging conclusions and moving forward
The examples discussed herein illustrate three key points. First, integrative studies are needed to tackle the molecular basis of post-zygotic reproductive isolation and speciation. Post-zygotic reproductive isolation is difficult to observe in the wild, and understanding its mechanisms requires the integration of data from natural populations with carefully controlled laboratory studies. Best-understood examples come from model organisms, but pioneering work has also been done in non-classical genetic systems such as sticklebacks, the monkey flower Mimulus, Helianthus sunflowers, and the Lake Whitefish. Further integrative work in a broad taxonomic range should be encouraged. Second, the genetic changes contributing to lineage divergence and post-zygotic reproductive isolation can be associated with non-coding DNA, which is a substantial and rapidly evolving part of the genome. Emerging technologies will help reveal which changes destabilize hybrid genomes and how they do so (Box 2). Third, genome stability is a complex trait influenced by hundreds of genes in addition to the genome structure itself and the environment. This has two consequences: 1)first, it may be difficult to identify its complete genetic basis; and 2)second, a certain level of genome instability may be tolerated, until a “threshold” is reached. The best-characterized examples, such as some presented here, have a simple genetic basis, but these may be the exception rather than the rule.
Finally, one may ask whether genome instability is merely one of many types of genetic incompatibility, or is instead a fundamental characteristic of the hybrid genome. We argue that because genomes are highly co-adapted evolutionary units, genome stability can break down at the earliest stages of development following hybridization. As such, genome instability has a central role in consolidating divergence between nascent species. More work in diverse organisms will provide the data to determine the frequency of genome instability in hybrids, to test if it directly contributes to reproductive isolation and speciation, and to address other outstanding questions (Box 1).
Genome stability is intimately linked to speciation and evolution.
Genome stability is a complex trait.
Genome instability may be a widespread cause of hybrid incompatibility.
Hybridization can trigger genome instability in multiple ways.
Acknowledgments
We thank Eric Alani, Guillaume Charron, Anne Dalziel, Eric Haag, Christian Landry, Souhir Marsit, Alexander Suh and one anonymous reviewer for stimulating discussions and helpful comments on an earlier version of this article. This work was supported by the National Institutes of Health R01GM074737 to DAB and a postdoctoral training award to AMDC from the Fonds de Recherche du Québec – Santé.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Recommended reading
* of special interest
** of outstanding interest
- 1.Wolf JBW, Ellegren H. Making sense of genomic islands of differentiation in light of speciation. Nature Reviews Genetics. 2017;18:87–100. doi: 10.1038/nrg.2016.133. [DOI] [PubMed] [Google Scholar]
- 2.Maheshwari S, Barbash DA. The genetics of hybrid incompatibilities. Annual Review of Genetics. 2011;45:331–355. doi: 10.1146/annurev-genet-110410-132514. [DOI] [PubMed] [Google Scholar]
- 3.Payseur BA, Rieseberg LH. A genomic perspective on hybridization and speciation. Molecular Ecology. 2016;25:2337–2360. doi: 10.1111/mec.13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cutter AD. The polymorphic prelude to Bateson–Dobzhansky–Muller incompatibilities. Trends Ecol Evol. 2012;27:210–219. doi: 10.1016/j.tree.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Presgraves DC. The molecular evolutionary basis of species formation. Nature Reviews Genetics. 2010;11:175–180. doi: 10.1038/nrg2718. [DOI] [PubMed] [Google Scholar]
- 6.Faria R, Navarro A. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol Evol. 2010;25:660–669. doi: 10.1016/j.tree.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Brown JD, O’Neill RJ. Chromosomes, conflict, and epigenetics: chromosomal speciation revisited. Annu Rev Genom Human Genet. 2010;11:291–316. doi: 10.1146/annurev-genom-082509-141554. [DOI] [PubMed] [Google Scholar]
- 8.Michalak P. Epigenetic, transposon and small RNA determinants of hybrid dysfunctions. Heredity. 2009;102:45–50. doi: 10.1038/hdy.2008.48. [DOI] [PubMed] [Google Scholar]
- 9.Aguilera A, Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nature Reviews Genetics. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 10.Grewal SIS, Jia S. Heterochromatin revisited. Nature Reviews Genetics. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 11.Chintapalli SV, Bhardwaj G, Babu J, Hadjiyianni L, Hong Y, Todd GK, Boosalis CA, Zhang Z, Zhou X, Ma H, et al. Reevaluation of the evolutionary events within recA/RAD51 phylogeny. BMC Genomics. 2013;14:240. doi: 10.1186/1471-2164-14-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kursel LE, Malik HS. Recurrent gene duplication leads to diverse repertoires of centromeric histones in Drosophila species. Molecular Biology and Evolution. 2017 doi: 10.1093/molbev/msx091. [no volume] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henikoff S, Ahmad K, Malik HS. The Centromere Paradox: Stable Inheritance with Rapidly Evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 14.Lee YCG, Leek C, Levine MT. Recurrent Innovation at Genes Required for Telomere Integrity in Drosophila. Molecular Biology and Evolution. 2016 doi: 10.1093/molbev/msw248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burton RS, Pereira RJ, Barreto FS. Cytonuclear Genomic Interactions and Hybrid Breakdown. Annu Rev Ecol Evol Syst. 2013;44:281–302. [Google Scholar]
- 16.Chou J-Y, Leu J-Y. The Red Queen in mitochondria: cyto-nuclear co-evolution, hybrid breakdown and human disease. Front Gene. 2015;6:3354. doi: 10.3389/fgene.2015.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreno-Loshuertos R, Acín-Pérez R, Fernández-Silva P, Movilla N, Pérez-Martos A, de Cordoba SR, Gallardo ME, Enríquez JA. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat Genet. 2006;38:1261–1268. doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]
- 18.Sabharwal SS, Schumacker PT. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nature Publishing Group. 2014;14:709–721. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barreto FS, Burton RS. Elevated oxidative damage is correlated with reduced fitness in interpopulation hybrids of a marine copepod. Proceedings of the Royal Society B Biological Sciences. 2013;280:20131521–20131521. doi: 10.1098/rspb.2013.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Chang CC, Rodriguez J, Ross J. Mitochondrial–nuclear epistasis impacts fitness and mitochondrial physiology of interpopulation Caenorhabditis briggsae hybrids. G3: Genes| Genomes| Genetics. 2016;6:209–219. doi: 10.1534/g3.115.022970. The authors test the hypothesis that mitonuclear incompatibilities underlie reproductive isolation between diverging populations. They show that reduced hybrid fecundity correlates with increased reactive oxygen species levels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montooth KL, Meiklejohn CD, Abt DN, Rand DM. Mitochondrial-Nuclear Epistasis Affects Fitness Within Species but Does Not Contribute to Fixed Incompatibilities Between Species of Drosophila. Evolution. 2010;64:3364–3379. doi: 10.1111/j.1558-5646.2010.01077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellison CK, Burton RS. Interpopulation hybrid breakdown maps to the mitochondrial genome. Evolution. 2008;62:631–638. doi: 10.1111/j.1558-5646.2007.00305.x. [DOI] [PubMed] [Google Scholar]
- 23.Fishman L, Willis JH. A Cytonuclear Incompatibility Causes Anther Sterility in Mimulus Hybrids. Evolution. 2006;60:1372. doi: 10.1554/05-708.1. [DOI] [PubMed] [Google Scholar]
- 24.Gagnaire P-A, Normandeau E, Bernatchez L. Comparative genomics reveals adaptive protein evolution and a possible cytonuclear incompatibility between European and American Eels. Molecular Biology and Evolution. 2012;29:2909–2919. doi: 10.1093/molbev/mss076. [DOI] [PubMed] [Google Scholar]
- 25.Hunter N, Chambers SR, Louis EJ, Borts RH. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. The EMBO Journal. 1996;15:1726–1733. [PMC free article] [PubMed] [Google Scholar]
- 26.Greig D, Travisano M, Louis EJ, Borts RH. A role for the mismatch repair system during incipient speciation in Saccharomyces. J Evol Biol. 2003;16:429–437. doi: 10.1046/j.1420-9101.2003.00546.x. [DOI] [PubMed] [Google Scholar]
- 27.Bi Y, Ren X, Yan C, Shao J, Xie D, Zhao Z. A Genome-Wide Hybrid Incompatibility Landscape between Caenorhabditis briggsae and C. nigoni. PLoS Genet. 2015;11:e1004993. doi: 10.1371/journal.pgen.1004993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dey A, Chan CKW, Thomas CG, Cutter AD. Molecular hyperdiversity defines populations of the nematode Caenorhabditis brenneri. P Natl Acad Sci USA. 2013;110:11056–11060. doi: 10.1073/pnas.1303057110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heck JA, Argueso JL, Gemici Z, Reeves RG, Bernard A, Aquadro CF, Alani E. Negative Epistasis between Natural Variants of the Saccharomyces cerevisiae MLH1 and PMS1 Genes Results in a Defect in Mismatch Repair. P Natl Acad Sci USA. 2006;103:3256–3261. doi: 10.1073/pnas.0510998103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demogines A, Wong A, Aquadro C, Alani E. Incompatibilities Involving Yeast Mismatch Repair Genes: A Role for Genetic Modifiers and Implications for Disease Penetrance and Variation in Genomic Mutation Rates. PLoS Genet. 2008;4:e1000103. doi: 10.1371/journal.pgen.1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31*.Bui DT, Dine E, Anderson JB, Aquadro CF, Alani EE. A Genetic Incompatibility Accelerates Adaptation in Yeast. PLoS Genet. 2015;11:e1005407. doi: 10.1371/journal.pgen.1005407. This article demonstrates that an increased mutation rate caused by incompatible alleles of mismatch repair genes accelerates adaptation to a stressful environment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32**.Bui DT, Friedrich A, Al-Sweel N, Liti G, Schacherer J, Aquadro CF, Alani E. Mismatch Repair Incompatibilities in Diverse Yeast Populations. Genetics. 2017 doi: 10.1534/genetics.116.199513. Bui et al. show that the mutator phenotype conferred by incompatible mismatch repair alleles is buffered by suppressor mutations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skelly DA, Magwene PM, Meeks B, Murphy HA. Known mutator alleles do not markedly increase mutation rate in clinical Saccharomyces cerevisiaestrains. Proceedings of the Royal Society B Biological Sciences. 2017;284:20162672. doi: 10.1098/rspb.2016.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganai RA, Johansson E. DNA Replication—A Matter of Fidelity. Molecular Cell. 2016;62:745–755. doi: 10.1016/j.molcel.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 35.Zickler D, Kleckner N. Recombination, Pairing, and Synapsis of Homologs during Meiosis. Cold Spring Harbor Perspectives in Biology. 2015;7:a016626. doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. A Mouse Speciation Gene Encodes a Meiotic Histone H3 Methyltransferase. Science. 2009;323:373–375. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- 37.Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. PRDM9 Is a Major Determinant of Meiotic Recombination Hotspots in Humans and Mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38**.Davies B, Hatton E, Altemose N, Hussin JG, Pratto F, Zhang G, Hinch AG, Moralli D, Biggs D, Diaz R, et al. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature. 2016 doi: 10.1038/nature16931. This study leveraged a humanized allele of PRDM9 in mice to show that binding asymmetry conferred by different alleles of PRDM9 played a key role is hybrid infertility. This suggests that hotspot erosion (i.e. the sequence itself) rather than hotspot position may be involved in hybrid infertility. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flachs P, Mihola O, Simecek P, Gregorova S, Schimenti JC, Matsui Y, Baudat F, de Massy B, Pialek J, Forejt J, et al. Interallelic and Intergenic Incompatibilities of the Prdm9 (Hst1) Gene in Mouse Hybrid Sterility. PLoS Genet. 2012;8:e1003044. doi: 10.1371/journal.pgen.1003044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flachs P, Bhattacharyya T, Mihola O, Pialek J, Forejt J, Trachtulec Z. Prdm9 Incompatibility Controls Oligospermia and Delayed Fertility but No Selfish Transmission in Mouse Intersubspecific Hybrids. PLoS ONE. 2014;9:e95806. doi: 10.1371/journal.pone.0095806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lenormand T, Engelstädter J, Johnston SE, Wijnker E, Haag CR. Evolutionary mysteries in meiosis. Philosophical Transactions of the Royal Society B Biological Sciences. 2016;371:20160001. doi: 10.1098/rstb.2016.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loidl J. Conservation and Variability of Meiosis Across the Eukaryotes. Annual Review of Genetics. 2016;50:293–316. doi: 10.1146/annurev-genet-120215-035100. [DOI] [PubMed] [Google Scholar]
- 43.Comeron JM, Ratnappan R, Bailin S. The Many Landscapes of Recombination in Drosophila melanogaster. PLoS Genet. 2012;8:e1002905. doi: 10.1371/journal.pgen.1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muñoz-Fuentes V, Di Rienzo A, Vilà C. Prdm9, a Major Determinant of Meiotic Recombination Hotspots, Is Not Functional in Dogs and Their Wild Relatives, Wolves and Coyotes. PLoS ONE. 2011;6:e25498. doi: 10.1371/journal.pone.0025498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Axelsson E, Webster MT, Ratnakumar A, Ponting CP, Lindblad-Toh K The LUPA Consortium. Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Research. 2012;22:51–63. doi: 10.1101/gr.124123.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korunes KL, Noor MAF. Gene Conversion and Linkage: Effects on Genome Evolution and Speciation. Molecular Ecology. 2016 doi: 10.1111/mec.13736. [DOI] [PubMed] [Google Scholar]
- 47.Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol Evol. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- 48.Noor MA, Grams KL, Bertucci LA, Reiland J. Chromosomal inversions and the reproductive isolation of species. P Natl Acad Sci USA. 2001;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pialek J, Hauffe HC, Searle JB. Chromosomal variation in the house mouse. Biol J Linn Soc. 2005;84:535–563. [Google Scholar]
- 50.Gimenez MD, White TA, Hauffe HC, Panithanarak T, Searle JB. Understanding the Basis of Diminished Gene Flow Between Hybridizing Chromosome Races of the House Mouse. Evolution. 2013 doi: 10.1111/evo.12054. [DOI] [PubMed] [Google Scholar]
- 51.Chmátal L, Gabriel SI, Mitsainas GP, Martínez-Vargas J, Ventura J, Searle JB, Schultz RM, Lampson MA. Centromere Strength Provides the Cell Biological Basis for Meiotic Drive and Karyotype Evolution in Mice. Curr Biol. 2014 doi: 10.1016/j.cub.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henikoff S, Malik HS. Centromeres: selfish drivers. Nature. 2002;417:227. doi: 10.1038/417227a. [DOI] [PubMed] [Google Scholar]
- 53.Charron G, Leducq J-B, Landry CR. Chromosomal variation segregates within incipient species and correlates with reproductive isolation. Molecular Ecology. 2014;23:4362–4372. doi: 10.1111/mec.12864. [DOI] [PubMed] [Google Scholar]
- 54**.Leducq J-B, Nielly-Thibault L, Charron G, Eberlein C, Verta J-P, Samani P, Sylvester K, Hittinger CT, Bell G, Landry CR. Speciation driven by hybridization and chromosomal plasticity in a wild yeast. Nature Microbiology. 2016;1:1–10. doi: 10.1038/nmicrobiol.2015.3. This study builds on earlier work showing that hybrid inviability correlates with chromosomal instability and chromosome rearrangements between parental lineages and in hybrids. Here the authors show that these rearrangements underlie speciation of a hybrid lineage. [DOI] [PubMed] [Google Scholar]
- 55.Dion-Côté A-M, Symonová R, Ráb P, Bernatchez L. Reproductive isolation in a nascent species pair is associated with aneuploidy in hybrid offspring. Proceedings of the Royal Society B Biological Sciences. 2015;282:20142862–20142862. doi: 10.1098/rspb.2014.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dion-Côté A-M, Symonová R, Lamaze FC, Pelikánová Š, Ráb P, Bernatchez L. Standing chromosomal variation in Lake Whitefish species pairs: the role of historical contingency and relevance for speciation. Molecular Ecology. 2016 doi: 10.1111/mec.13816. [DOI] [PubMed] [Google Scholar]
- 57.Comai L, Madlung A, Josefsson C, Tyagi A. Do the different parental “heteromes” cause genomic shock in newly formed allopolyploids? Philosophical Transactions of the Royal Society B Biological Sciences. 2003;358:1149–1155. doi: 10.1098/rstb.2003.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waugh O’Neill RJ, O’Neill MJ, Marshall Graves JA. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature. 1998;393:68–72. doi: 10.1038/29985. [DOI] [PubMed] [Google Scholar]
- 59.Ferree PM, Barbash DA. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 2009;7:e1000234. doi: 10.1371/journal.pbio.1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rošić S, Köhler F, Erhardt S. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. The Journal of Cell Biology. 2014;207:335–349. doi: 10.1083/jcb.201404097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li R, Ren X, Bi Y, Ho V, Hsieh CL, Young A. Specific down-regulation of spermatogenesis genes targeted by 22G RNAs in hybrid sterile males associated with an X-Chromosome introgression. Genome. 2016 doi: 10.1101/gr.204479.116. [no volume] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sabin LR, Delás MJ, Hannon GJ. Dogma Derailed: The Many Influences of RNA on the Genome. Molecular Cell. 2013;49:783–794. doi: 10.1016/j.molcel.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dion-Côté A-M, Renaut S, Normandeau E, Bernatchez L. RNA-seq reveals transcriptomic shock involving transposable elements reactivation in hybrids of young lake whitefish species. Molecular Biology and Evolution. 2014;31:1188–1199. doi: 10.1093/molbev/msu069. [DOI] [PubMed] [Google Scholar]
- 64.Kelleher ES, Edelman NB, Barbash DA. Drosophila interspecific hybrids phenocopy piRNA-pathway mutants. PLoS Biol. 2012;10:e1001428. doi: 10.1371/journal.pbio.1001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ungerer MC, Strakosh SC, Zhen Y. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr Biol. 2006;16:R872–3. doi: 10.1016/j.cub.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 66.Vela D, Fontdevila A, Vieira C, García Guerreiro MP. A Genome-Wide Survey of Genetic Instability by Transposition in Drosophila Hybrids. PLoS ONE. 2014;9:e88992. doi: 10.1371/journal.pone.0088992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodier JL. Restricting retrotransposons: a review. Mobile DNA. 2016 doi: 10.1186/s13100-016-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68*.Khost DE, Eickbush DG, Larracuente AM. Single-molecule sequencing resolves the detailed structure of complex satellite DNA loci in Drosophila melanogaster. Genome Research. 2017 doi: 10.1101/gr.213512.116. Khost et al. apply single-molecule sequencing long reads from PacBio to assemble and resolve the structure of complex satellites, providing unprecedented insights on the evolution of these structures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weissensteiner MH, Pang AWC, Bunikis I, Höijer I, Vinnere-Petterson O, Suh A, Wolf JBW. Combination of short-read, long-read, and optical mapping assemblies reveals large-scale tandem repeat arrays with population genetic implications. Genome Research. 2017 doi: 10.1101/gr.215095.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Valente GT, Conte MA, Fantinatti BEA, Cabral-de-Mello DC, Carvalho RF, Vicari MR, Kocher TD, Martins C. Origin and Evolution of B Chromosomes in the Cichlid Fish Astatotilapia latifasciata Based on Integrated Genomic Analyses. Molecular Biology and Evolution. 2014;31:2061–2072. doi: 10.1093/molbev/msu148. [DOI] [PubMed] [Google Scholar]