Abstract
Chromosome instability (CIN) is widely observed in both sporadic and hereditary colorectal cancer (CRC). Defects in APC and WNT signaling are primarily associated with CIN in hereditary CRC, but the genetic causes for CIN in sporadic CRC remain elusive. Using high-density SNP array and exome data from The Cancer Genome Atlas, we characterized loss of heterozygosity (LOH) and copy number variation (CNV) in the peripheral blood, normal colon and corresponding tumor tissue in 15 CRC patients with proficient mismatch repair (MMR) and 24 CRC patients with deficient MMR. We found a high frequency of 18q LOH in tumors and arm-specific enrichment of genetic aberrations on 18q in the normal colon (primarily copy neutral LOH) and blood (primarily copy gain). These aberrations were specific to the sporadic, pMMR CRC. Though in tumor samples genetic aberrations were observed for genes commonly mutated in hereditary CRC (e.g. APC, CTNNB1, SMAD4, BRAF), none of them showed LOH or CNV in the normal colon or blood. DCC located on 18q21.1 topped the list of genes with genetic aberrations in the tumor. In an independent cohort of 13 patients subjected to Whole Genome Sequencing, we found LOH and CNV on 18q in adenomatous polyp and tumor tissues. Our data suggests that patients with sporadic CRC may have genetic aberrations preferentially enriched on 18q in their blood, normal colon epithelium and non-malignant polyp lesions that may prove useful as a clinical marker for sporadic CRC detection and risk assessment.
Keywords: neoplastic transformation, colorectal cancer, genetic aberrations, LOH, CNV
Introduction
Colorectal cancer (CRC) is diagnosed in more than 140,000 people in the US each year, leading to nearly 51,000 deaths annually [1]. Although CRC is one of the best-understood neoplasms from a genetic perspective [2–4], this disease remains the second most common cause of cancer-related death. Decades of research have identified two major genetic classifications for CRC including microsatellite instability (MSI), and chromosomal instability (CIN) [5–7].
MSI is characterized by either germline mutations of DNA mismatch repair (MMR) genes associated with the hereditary CRC condition Lynch syndrome [8] or by hypermethylation-related inactivation of at least one of the four MMR genes (also known as CpG island methylator phenotype), which is responsible for 20% of sporadic CRC [7]. In both the hereditary and sporadic cases, deficient MMR (dMMR) allows small base pair mistakes to escape repair and leads to somatic alterations in nucleotide repeat sequences called microsatellites [9]. MSI may affect the function of tumor suppressor genes or oncogenes through mutations, and this hypermutator phenotype results in genomic rather than chromosomal instability [9,10].
However, the majority of CRC exhibit proficient MMR (pMMR) and are microsatellite stable (MSS), but exhibit chromosome instability (CIN) and have high amounts of loss of heterozygosity (LOH). Most sporadic CRC cases, in which there is no family history of CRC, are CIN [11–14]. The genetic reasons for association of sporadic CRC with CIN are complex, and may partly be attributable to somatic APC mutations [15]. However, aberrant crypt foci that are very early lesions in CRC carcinogenesis do exhibit LOH that is independent of somatic APC mutations [16].
The presence of some CIN features have been evaluated in sporadic CRC with LOH [11,14,16–19] or copy number variation (CNV) analyses [12,13]. The majority of the LOH studies compare the somatic allelotypes of CRC (or its putative precursors) to those of corresponding adjacent or distant normal (non-tumor) colon tissue, under the assumption that morphologically and histologically normal colon tissues are genetically normal [11,14,16–19]. This assumption may not hold, as normal colon mucosa from sporadic CRC patients exhibit monosomy, polysomy, and aneusomy on several chromosome arms shown by FISH [12]. In addition, a significant amount of genetic aberrations in CRC have been found in normal colon tissue [12]. Indeed, the method may underestimate the actual amount of genetic aberrations, given that Calhoun et al. demonstrated that copy number based methods (e.g. FISH, array-CGH) detect at maximum 47.2% of all forms of LOH [20].
The majority of CRC arises from a non-malignant precursor lesion called an adenomatous polyp [21–24]. It has been shown that CRC develops from an accumulation of genetic aberrations that can begin in the normal colon and progresses through an adenomatous polyp to cancer [2,25,26]. Currently, molecular features that characterize these precursor polyps as being at high risk for malignant transformation are currently limited, and LOH and CNV analyses are particularly limited in polyps that are associated with cancer [27–31].
We infer from these pioneering works that a broad spectrum of genetic aberrations may exist in morphologically and histologically non-malignant (either normal or polyp) colon tissue in sporadic CRC patients. Therefore, a more comprehensive evaluation of these aberrations is warranted. To this end, we used high-density SNP-array data from The Cancer Genome Atlas (TCGA) to detect genetic aberrations (both LOH and CNV) in matched peripheral blood and normal colon from sporadic CRC patients. The types of genetic aberrations we evaluated included classic LOH (copy loss LOH, CL-LOH), copy neutral LOH (CN-LOH), copy gain LOH (CG-LOH) and CNV (copy gain and homozygous deletion).
To validate the findings in the TCGA data, we examined specific LOH and CNV features in an independent set of 13 patients that were subjected to Whole Genome Sequencing (WGS). In addition to the blood, normal colon and CRC tissue, these patients also included the corresponding precursor adenomatous polyp tissue. We propose that a systematic analysis of blood, normal colon, polyp and corresponding tumor will help to identify potentially clinically valuable genetic markers for both sporadic CRC diagnosis and risk assessment. Importantly, identifying genetic markers that are aberrant and detectable in the patient’s blood, normal colon or polyp could advance screening and detection opportunities.
Materials and Methods
TCGA Patient information
A total of 15 patients with pMMR were studied. Each of these patients had contributed peripheral blood, colorectal adenocarcinoma and corresponding adjacent or distal normal colon tissue to TCGA. Histological images of the tissue samples are publicly available at http://www.cbioportal.org/public-portal/. The patients were five women and ten men, with age ranging from 53 to 90 years (mean=74, SD=11) old. Seven of the tumor samples were from the right colon, and five were from the sigmoid. The site of three tumors was unknown. Of the four patients with the distance of normal colon tissue to tumor recorded, all had distal (>2cm) normal colon tissue. Clinical information of the patients is available in Table S7.
These 15 patients exhibited normal expression of MMR genes in their tumor, and thus were pMMR. Patients were strictly screened so that they have no first degree relatives with cancer, no MSI, and have proficient MMR gene expression. Patients with dMMR tumors, having absent expression of any immunostains for MLH1, MSH2, PMS2, or MSH6 in their tumors, or patients who had one or more first-degree relative(s) with a history of any cancer were not included in the pMMR group. To strictly focus upon sporadic CRC, blood samples were evaluated for germline aberrations associated with CRC risk, and those cases excluded if known CRC risk related germline mutations were noted. None of the 15 patients had chemo or radiotherapy prior to the sample collection.
Blood samples from 24 dMMR patients (mean age=54, SD=14) were compared to the blood aberration profiles in the 15 pMMR patients. No limitation on the number of first-degree relative(s) with cancer was applied for these 24 dMMR patients.
To minimize the chance that the normal colon tissue would be contaminated by neoplastic cells, we only studied patients with normal colon tissue samples recorded to have no tumor nuclei present. Only tumors with >60% tumor nuclei were included in the study to avoid dilution of the tumor signal with that of non-neoplastic DNA, which may result in ambiguous signals at LOH loci. The threshold was chosen in consideration that OncoSNP [32], which was adopted for downstream LOH and CNV analysis, can effectively handle this level of contamination in its statistical model.
LOH and CNV detection and somatic/germline mutation
The TCGA COAD (colorectal adenocarcinoma) gender annotation and level 2 copy number data (Affymetrix 6.0 platform) was downloaded from The Genomic Data Commons Data Portal legacy site (https://gdc-portal.nci.nih.gov/legacy-archive/search/f). The germline data from 378 patients was used to create a reference for the expected A & B intensity of the AA, AB, and BB genotype cluster. The B-Allele Frequency (BAF) was then computed using the formula from Wang et al., 2008 [33]. The BAF plots only include SNPs that are heterozygous in the germline sample. Furthermore, any SNP with minor allele frequency of less than 5% in the 378 patient reference was excluded (to exclude artifactual LOH stretches caused by the inclusion of rare variants in the Affymetrix 6.0 microarray). Tumor log2 ratio was computed from the level2 data, but recentered assuming that chromosomes 2,3,4p,5p,6,11,12p,13p,14p,15p,16,19p,21,22p had no copy number events (these chromosomes are rarely altered in CRC). BAF and log2 ratio was either plotted using R or was processed to be loadable into Affymetrix’s Chromosome Analysis Suite (Chas) for whole genome view plots and copy number state estimates.
According to the CNV state in corresponding tumor or normal colon tissue, LOH was further classified into copy-loss (CL-LOH), copy-neutral (CN-LOH), and copy-gain LOH (CG-LOH). LOH and CNV were analyzed in blood samples using single sample mode assuming no contamination and high homogeneity. In this case, CN-LOH calls were skipped and CG-LOH calls were converted to copy-gain since theoretically they are not detectable without a reference genotype to compare with.
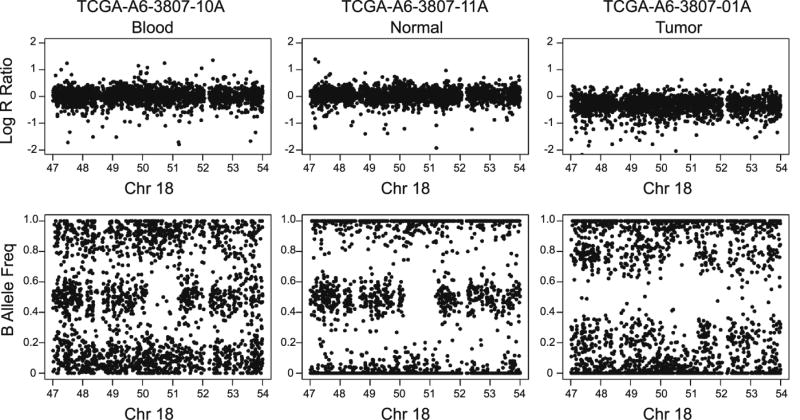
OncoSNP by default generates LOH and CNV calls using two different ploidy configurations (diploid or non-diploid) and attaches a likelihood score for each of them in the quality control report, and we used the highest likelihood score. We only kept the LOH and CNV calls that were covered by ≥30 probes. Visual inspection showed that this threshold can effectively exclude false positive calls. Additionally, the analysis in Chas for log2 ratios and BAF plots at the whole genome provided an additional level of confidence. The cutoff threshold left LOH with a minimum length of ~28.3Kb and CNV (copy-gain) with a minimum length of ~27.6Kb. Visualization of the log R ratio (LRR) and BAF of several LOH and CNV regions showed high concordance rate between the OncoSNP calls and manual calls (Figure 2, Figures S2–5).
Figure 2.
Example of CN-LOH in normal colon tissue and tumor that overlap with the DCC gene (49.8 ~ 51.0 Mb, hg19). From left to right: LRR and BAF of blood, normal colon tissue, and tumor.
For patient A6-5660, who showed massive CN-LOH in normal colon tissue, somatic mutations in normal colon tissue were obtained from exome sequencing results from TCGA. We used level2 data with controlled access (including both germline and somatic mutation).
Concurrent genetic aberration, fragile site database, and gene location analysis
Genetic aberrations among tumor, normal colon tissue, and blood are considered concurrent when they have genomic overlap of at least 1 base pair. Fragile site database was downloaded from HGNC http://www.genenames.org/cgi-bin/hgnc_stats. A total of 117 fragile sites were used for analysis. Genomic location (Version GRCh37/hg19) of genes in bed format was obtained from the UCSC website (http://genome.ucsc.edu/cgi-bin/hgTables). The UCSC gene identification was mapped to the HGNC approved gene symbol by using the mapping information from http://www.genenames.org/, and those without a match were dropped. For genes with multiple entries, the one with the longest basepair length was used.
Functional SNP analysis
Genotypes were called by using BirdSuite (version 1.5.5), which estimates allele specific genotype by considering both common and rare copy number variation regions. The Affymetrix annotation file (genome version NCBI37/hg19) was used to annotate the probe-set. All of the 45 samples from the 15 patients have SNP call rate > = 95% (mean=99.3%, min=97.7%).
For each SNP, we treated the major allele as reference and the minor allele as alternative (using allele frequency from CEU population). After uniformizing to the corresponding nucleotide on the positive strand, the effect of major to minor allele mutations were estimated by using SnpEff. For each variant, SnpEff predicts its functional consequence on corresponding transcripts as high, modifier, moderate, or low. From a total of 930k SNPs on Affymetrix SNP array 6.0, we identified 3,394 SNPs that, when mutated from the major to minor allele, have high or moderate effects on 4,624 transcripts from 2,644 genes. These variants cause changes including non-synonymous coding (n=4,516), stop gain/loss (n=53), splice site donor/acceptor (n=80), and start loss (n=9) to corresponding transcripts.
Independent validation set of PBL, normal, polyp and tumor tissues: sample characteristics, tissue preparation and nucleic acid extraction
A total of 13 patients, representing 44 tissues, were consented and collected at Mayo Clinic between 2000–2016 through our IRB approved Biobank for Gastrointestinal Health Research [BGHR] (IRB 622-00, PI LA Boardman). Patient IDs and tissue types are available in Table S8. Polyp and adjacent tumor tissues were harvested following surgical resection and snap frozen in liquid nitrogen and stored in a −80 freezer. Normal colon epithelium full thickness specimens that were at least 8 cm from the polyp/tumor margin were collected and stored as above. Peripheral blood leukocytes (PBL) were also collected when available. All polyps shown here are all matched for villous histology with low grade dysplasia. All cases exclude subjects with a prior history of any malignancy, a family history of Lynch syndrome or FAP, any other syndrome associated with hereditary CRC or inflammatory bowel disease. The patient medical history was collected and included lifetime polyp history, age, gender, body mass index, and other medical co-morbidities.
Tissues were macrodissected as described previously [28]. Briefly, a hematoxylin and eosin guide slide was reviewed and marked by a pathologist, and areas of normal epithelium, polyp or cancer identified. DNA was extracted by the PureGene method, and was quantified on the Qubit Fluorometer.
Whole Genome Sequencing (WGS) and analysis
Library construction, sequencing and initial processing of WGS data was performed as described previously [28]. All samples were sequenced on the Illumina HiSeq X instruments resulting in 150 base pair, paired-end reads to meet a goal of 30× average coverage. Using the Picard Informatics Pipeline, all data from a particular sample was aggregated into a single BAM file which included all reads, all bases from all reads, and original/vendor-assigned quality scores. The raw data in BAM file format for the WGS data analyzed in this manuscript are available in the dbGaP database with Study Accession number: phs001384.v1.p1. The study report page can be accessed at: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001384.v1.p1
Accession numbers for each individual WGS BAM file are located with the sample and tissue ID in Table S8.
Varscan 2.0 was run on pairs of samples: One test sample (Normal, Villous polyp, Tumor) compared to DNA from peripheral blood leukocytes (PBL). For each variant, the BAF was computed using the ratio of alternate read counts to total read counts and log2 ratio was computed using the log2 ratio of normalized count of each samples over that of the PBL. Only variants heterozygous in the PBL and called in all samples are included in plots. To reduce noise in plots: 1) only variants with coverage of >=40 × in PBL (and less than 80× in PBL) were included and 2) plots were limited to 10000 SNPs per chromosome to decrease noise. The log2 ratio and BAF plots for each chromosome in each patient evaluated by WGS is included as a Supplement.
Results
Copy neutral LOH characterizes the primary difference between blood and normal colon tissue
Normal colon tissue samples from the 15 patients showed large variances in the number of LOH and CNV events, compared to blood (Figure 1). This was exemplified by A6-3807 and A6-5660 which have genetic aberrations across whole autosomes, compared to AA-3529 that had only two aberrations (1 CL-LOH and 1 copy gain region). The normal colon tissue samples from A6-3807 and A6-5660 were distant normal (>2 cm away from the corresponding tumor) and had zero percentage of tumor nuclei, therefore their extensive genetic aberrations were unlikely a sign of contamination. Sorting of the patients by tumor TNM stage and gender indicated no association between the amount of genetic aberrations in normal colon tissue with either the tumor stage or sex of the CRC patients. Seventy percent of the CL-LOH and forty percent of the copy gain in the normal colon tissues had concurrent genetic aberrations in their matched blood sample (Table 1). The primary difference seen between normal colon tissue and blood was CN-LOH, which accounted for an average of 60% (median 63%) of all LOH and CNV in the normal colon tissues (SD=36%). The average length of CN-LOH in normal colon tissue was 97kbp and ranged from 46kbp in the 1st quantile to 122kbp in the 3rd quantile. Figure 2 illustrates a ~1Mb CN-LOH region on 18q21.1 of normal colon tissue from patient A6-3807.
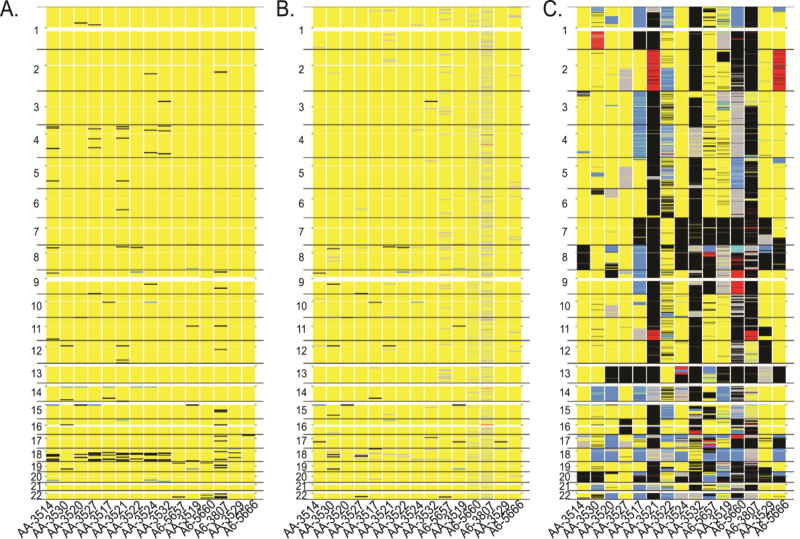
Figure 1.
LOH and CNV patterns of A) blood, B) normal colon tissue, and C) tumor from 15 pMMR patients ordered by gender and tumor TNM stages. Chromosome regions with CL-LOH (blue), CN-LOH (grey), CG-LOH (red), Copy Gain (black), and Homozygous Deletion (cyan) are shown. Green dots on the left and right sides mark the centromere location.
Table 1.
Count of LOH and CNV aberrations in normal colon tissue and concurrent aberrations in blood
| LOH/CNV Type | Count in normal colon tissue |
Count in blood |
No. and (%) of concurrency |
No. of patients with concurrency |
|---|---|---|---|---|
| Copy Gain | 39 | 130 | 17 (44%) | 11 |
| Copy-Loss (CL) LOH | 10 | 16 | 7 (70%) | 6 |
| Copy-Neutral (CN) LOH | 5651a | NAb | 31 (0.5%)c | 3 |
| Copy Gain (CG) LOH | 12 | NAb | 1 (8%)c | 1 |
Patients A6-3807 and A6-5660 had 3917 and 1637 CN-LOH, respectively. The remaining 97 CN-LOH were contributed by 10 patients. Three patients had no CN-LOH detected.
Copy-neutral and Copy-gain LOH are not detectable in blood.
Concurrent with copy gain in blood.
High frequency of genetic aberrations on 18q in blood normal colon and tumor tissue
At the chromosome arm level, the most frequent genetic aberrations in both normal colon tissue and blood were enriched on 18q. In blood, the aberration peaked at 18q12.3 (37.2 ~ 43.5MB) and 18q22, where six and nine patients had at least one copy gain region (mean=140KB, SD=59KB), respectively. Over the entire 18q region, 10 patients had at least two putative short regions of copy gain (Figure 1A, Table S1). These were considered putative since visualization of these regions indicated only slight increase in log R ratio [data not shown].
In normal colon tissue, the aberration peaked at 18q22 (61.6 ~ 73.1MB), where seven patients had at least one CN-LOH (mean=91KB, SD=71KB) detected. Over the span of the entire 18q arm, 10 patients had at least one CN-LOH, and three patients had at least one copy gain in their normal colon samples (Figure 1B, Table S1). The enrichment was retained under relatively stringent criteria, which showed that five patients had at least two CN-LOH, and two patients had at least two copy gain regions (Table S1). This was still higher than the frequency of genetic aberration observed on other arms.
To determine whether the recurrent copy gain signals on 18q were specific to pMMR patients, we further analyzed blood samples from 24 patients with dMMR tumors using the same parameters. The result indicated that only four patients (16.7%) had two or more aberrations on 18q, which was significantly lower than pMMR patients (66.7%, p < 0.05). Visual inspection (Figure S1) indicated that the blood sample of dMMR patients had more wide spread aberrations on chromosome 17, 19, and 22q than pMMR patients. In the pMMR cases, the amount of genetic aberration was much higher in tumor samples than that of normal colon tissue or blood. The highest frequency of LOH was observed on 18q, which had an average of 52% CL-LOH (SD=4.4%) and 17% CN-LOH (SD=4.6%) across the 15 patients (Figure 1C, Table S2). Furthermore, a high frequency of copy gain was observed, in decreasing order, on 20q, 13q, 7p/q, and 8q. These arms have copy gain in more than 50% of the patients (Table S2).
In contrast to what was observed in normal colon tissue and blood, which had relatively short regions with genetic aberrations, most of the genetic aberrations in tumors affected the whole arm, with a few samples exhibiting mosaic patterns of change. For example, AA-3524 had a mixed diploid and CL-LOH on 18q (Figure 1C, Figure S2). The p and q arm were observed to undergo different types of change; AA-3520 had CN-LOH on the whole 18p and CL-LOH on the whole 18q (Figure 1C, Figure S3). Both the whole arm and mosaic pattern aberrations were observed on other chromosome arms. A6-3807 featured interspersed LOH and copy gain on 10p (Figure 1C, Figure S4). AA-3517 displayed complete copy loss on 1p and complete copy gain on 1q (Figure 1C). Homozygous deletion was relatively rare and was observed only on 8p of A6-5660 (Figure 1C, S5).
Genes with aberrations in blood, normal colon and tumor
Analysis was conducted to determine how genetic aberrations in blood, normal colon, or tumor overlap between patients. In blood, 13 genes had genetic aberrations in ≥4 patients (Table 2, Table S3). The most frequent aberrations were copy gain at CCDC102B and KLHL14, both on 18q (close to the centromere) and affected six and five patients, respectively. CCDC102B also had a relatively high frequency of aberration in normal colon (Table 3) and topped the aberration list in tumor samples (Table 4). Frequent CL-LOH and copy gain were also observed at 15q11.2, encompassing 11 genes (Table S3).
Table 2.
Top 10 genes with genetic aberrations in blood
| Gene | Chr | Type of disordera | Count | No. of affected patients by type |
Total No. of affected patients |
|---|---|---|---|---|---|
| CCDC102B | 18q22.1 | 4 | 6 | 6 | 6 |
| KLHL14 | 18q12.1 | 4 | 5 | 5 | 5 |
| CXADRP2 | 15q11.2 | 1|4 | 2|2 | 2|2 | 4 |
| GOLGA6L6 | 15q11.2 | 1|4 | 2|2 | 2|2 | 4 |
| HERC2P3 | 15q11.2 | 1|4 | 2|2 | 2|2 | 4 |
| HERC2P7 | 15q11-q13 | 1|4 | 2|2 | 2|2 | 4 |
| NBEAP1 | 15q11.2 | 1|4 | 2|2 | 2|2 | 4 |
| NF1P2 | 15q11.2 | 1|4 | 2|2 | 2|2 | 4 |
| OR4M2 | 15q11.2 | 1|4 | 2|2 | 2|2 | 4 |
| OR4N3P | 15q11.2 | 1|4 | 2|2 | 2|2 | 4 |
1 is CL-LOH, 4 is Copy Gain.
Table 3.
Top 10 genes with genetic aberrations in normal colon tissue
| Gene | Chr | Type of disordera |
Count | No. of affected patients by type |
Total No. of affected patients |
|---|---|---|---|---|---|
| DCC | 18q21.1 | 2|3|4 | 5|1|1 | 3|1|1 | 4 |
| SGCZ | 8p22 | 2|4 | 4|1 | 3|1 | 4 |
| CCDC102B | 18q22.1 | 2|4 | 3|1 | 3|1 | 3 |
| CHEK2P2 | 15q11.1 | 1|4 | 1|2 | 1|2 | 3 |
| CXADRP2 | 15q11.2 | 1|4 | 1|2 | 1|2 | 3 |
| DAB1 | 1p32-p31 | 2 | 7 | 3 | 3 |
| DOK6 | 18q22.2 | 2 | 4 | 3 | 3 |
| DPP6 | 7q36.2 | 2 | 6 | 3 | 3 |
| FHIT | 3p14.2 | 2|4 | 7|1 | 2|1 | 3 |
| GOLGA6L6 | 15q11.2 | 1|4 | 1|2 | 1|2 | 3 |
1 is CL-LOH, 2 is CN-LOH, 3 is CG-LOH, 4 is Copy Gain, 5 is Homozygous Deletion
Table 4.
Top genes with genetic aberrations in tumor tissue
| Gene | Chr | Type of disordera |
Count | No. of affected patients by type |
Total No. of affected patients |
|---|---|---|---|---|---|
| CCDC102B | 18q22.1 | 1|2|3|4 | 7|3|1|3 | 7|3|1|3 | 14 |
| SGCZ | 8p22 | 1|2|4|5 | 6|6|7|1 | 5|3|7|1 | 14 |
| TNKS | 8p23.1 | 1|2|4|5 | 4|4|5|1 | 4|4|5|1 | 14 |
| ABHD16B | 20q13.33 | 1|4 | 2|11 | 2|11 | 13 |
| ADAM11 | 17q21.3 | 1|2|3|4 | 2|3|1|7 | 2|3|1|7 | 13 |
| ANGPT2 | 8p23 | 1|2|4|5 | 4|3|5|1 | 4|3|5|1 | 13 |
| APCDD1L | 20q13.32 | 1|4 | 2|11 | 2|11 | 13 |
| ARFRP1 | 20q13.3 | 1|4 | 2|11 | 2|11 | 13 |
| ARHGEF10 | 8p23 | 1|2|4|5 | 4|2|6|1 | 4|2|6|1 | 13 |
| ATP6V1B2 | 8p21.3 | 1|2|4|5 | 4|4|4|1 | 4|4|4|1 | 13 |
| Genes involved in non-sporadic (hereditary) CRC | |||||
| SMAD4 | 18q21.1 | 1|2 | 8|3 | 8|3 | 11 |
| TP53 | 17p13.1 | 1|2|3 | 7|3|1 | 7|3|1 | 11 |
| BRAF | 7q34 | 2|4 | 1|8 | 1|8 | 9 |
| PMS2 | 7p22.1 | 4 | 8 | 8 | 8 |
| POLD1 | 19q13.3 | 1|4 | 1|7 | 1|7 | 8 |
| APC | 5q21-q22 | 1|2|4 | 3|2|2 | 3|2|2 | 7 |
| STK11 | 19p13.3 | 1|2|4 | 1|2|4 | 1|2|4 | 7 |
| BMPR1A | 10q22.3 | 2|4 | 1|5 | 1|5 | 6 |
| EPCAM | 2p21 | 3|4 | 2|4 | 2|4 | 6 |
| MSH2 | 2p21 | 3|4 | 2|4 | 2|4 | 6 |
| MSH6 | 2p16 | 3|4 | 2|4 | 2|4 | 6 |
| MUTYH | 1p34.1 | 1|4 | 3|3 | 3|3 | 6 |
| PTEN | 10q23 | 2|4 | 1|5 | 1|5 | 6 |
| MLH1 | 3p22.3 | 1|2|4 | 1|2|2 | 1|2|2 | 5 |
| KRAS | 12p12.1 | 4 | 4 | 4 | 4 |
| POLE | 12q24.3 | 4 | 4 | 4 | 4 |
1 is CL-LOH, 2 is CN-LOH, 3 is CG-LOH, 4 is Copy Gain, 5 is Homozygous Deletion
In normal colon, the most frequent genetic aberration was observed in DCC, which is on 18q21.1 and affected four patients in the form of CN-LOH, CG-LOH, and copy gain (Table 3, Table S4). SGCZ, which is on 8p22, had CN-LOH and/or copy gain in the normal colon tissue of four patients. Fragile site gene FHIT (FRA3B), DOK6 (FRA18C), and DAB1 (FRA1L/1B) had aberrations in three patients. However, analysis of genes co-located with 117 fragile sites did not show significant enrichment of aberrations compared to genes in non-fragile site regions. Among tumors, the top affected genes were CCDC102B on 18q21.1, as well as SGC and TNKS on 8p22. These genes had LOH and/or CNV, and were observed in 14 patients (Table 4). Genetic aberrations in DCC were observed in 12 patient tumor samples.
It is noteworthy that genetic aberrations were observed for several genes that are frequently mutated in hereditary CRC. Eleven patients had somatic CL-LOH or CN-LOH in both SMAD4 and TP53, which when mutated in the germline may lead to juvenile polyposis syndrome (with or without Hereditary hemorrhagic telangiectasia overlap) or Li Fraumeni syndrome, respectively (Table 4). Seven patients had CL-LOH, CN-LOH or copy gain in APC, which is a marker of familial adenomatous polyposis (Table 4). However, it is remarkable that none of these genes exhibited any genetic aberrations in normal colon tissue or blood from the 15 patients. The normal colon tissue of patient A6-5660 had large amounts of CN-LOH, but exome sequencing showed no somatic or germline mutations in APC, CTNNB1 or any of the hereditary CRC risk genes shown in Table 4.
Concurrent gene aberrations in blood, normal colon and tumor tissue
We found gene aberrations that were concurrent in blood and normal colon tissue, or concurrent in normal colon tissue and tumor (Table S5). We defined concurrent gene aberrations as being present in two sample types from the same patient and that had genomic overlap of at least one base pair in known gene regions. For blood and normal colon tissue, the most frequent concurrency occurred at 15q11, where three patients had concurrent aberrations in 12 genes (Table S5). With the exception of patients A6-3807 and A6-5660, who contributed all the copy gain/CN-LOH concurrencies (160 events), all of the concurrent gene aberrations were copy gain/copy gain (68 events from 10 patients), or CL-LOH/CL-LOH (22 events from three patients) in both blood and matched normal colon tissue.
For normal colon tissue and tumor, we observed more complex concurrent gene aberrations including CN-LOH/copy gain (3322 events from eight patients), CN-LOH/CN-LOH (1470 events from 8 patients), and CN-LOH/CG-LOH (378 events from five patients). When sorted by the frequency per patient, DCC and SGCZ had seven and four concurrent gene aberrations, respectively, in four patients. CCDC102B, which had a high frequency of aberrations in blood and normal colon tissue, had five concurrent gene aberrations present in three patients. Fragile site gene FHIT had four concurrent gene aberrations in three patients (Table S6). However, none of the hereditary CRC risk genes (as shown in Table 4) had concurrent gene aberrations in blood and normal colon tissue, or in normal colon tissue and tumor.
The majority of concurrent LOH in normal colon and tumor tissues retained on the same allele
We further analyzed somatic mutations in normal colon tissue and tumor, and the germline mutations in blood of the 15 pMMR patients. To focus on the variants with functional consequence, we selected 3,394 SNPs (from Affymetrix SNP array 6.0) by restricting to those with high or moderate effects as predicted by SnpEff (see Materials and Methods for details). The analysis aimed to shed light on the direction of LOH (i.e. loss of reference or variant allele) in normal colon and tumor tissue, as well as important germline mutations in blood. We acknowledge that due to limited coverage, the analysis represents a small proportion of the whole mutation spectrum.
By comparing the genotypes across blood, normal colon and tumor tissue, we found that the majority (96.9%) of LOH, if present in both the normal colon and tumor, retained the same allele (Table 5). This was consistently observed in the two patients (A6-3807 and A6-5660) with a large amount of LOH in normal colon tissue as well as the remaining 13 patients, suggesting these LOH happened early in the common progenitor cell of normal colon and tumor. The primary differences were that tumor had significantly (p<0.05, Goodness-of-fit test) more independent LOH and retained more variant alleles than normal colon tissue. For example, for the 9,714 heterozygotes in blood, 918 (9.5%) were homozygotes in the corresponding normal colon while 1583 (16.3%) were homozygotes in tumor (Table 5). Among the homozygotes, 100 (10.9%) in normal colon tissue retained the variant allele, and 569 (35.9%) in tumor retained the variant allele.
Table 5.
LOH direction in normal colon and tumor tissue
| Genotype combinations a |
All 15 patients |
Patient A6-3807 b |
Patient A6-5660 b |
Other 13 patients |
|---|---|---|---|---|
| H_H_H | 7714 | 550 | 545 | 6619 |
| H_A_A | 551 | 328 | 111 | 112 |
| H_H_I | 511 | 18 | 62 | 431 |
| H_H_A | 456 | 27 | 52 | 377 |
| H_A_H | 238 | 12 | 188 | 38 |
| H_H_U | 107 | 16 | 10 | 81 |
| H_I_H | 59 | 2 | 12 | 45 |
| H_I_I | 39 | 10 | 8 | 21 |
| H_A_I | 17 | 0 | 15 | 2 |
| H_A_U | 12 | 5 | 5 | 2 |
| H_U_A | 5 | 4 | 0 | 1 |
| H_I_A | 2 | 0 | 0 | 2 |
| H_U_I | 2 | 1 | 0 | 1 |
| H_U_U | 1 | 0 | 0 | 1 |
| Total | 9714 c | 973 | 1008 | 7733 |
Genotype combinations are shown as Blood_normal colon tissue_tumor. H for heterozygote; A for homozygous reference allele; I for homozygous variant allele; U for unknown; All combinations were exhausted and only those with at least one count was shown.
Patients A6-3807 and A6-5660 were listed separately since they have much more LOH in normal colon tissue than other patients.
Only used SNPs that have variant alleles with predicted high or moderate effect on corresponding transcript.
We captured 39 unique regions of LOH that retained variant alleles in both normal colon tissue and tumor (Table 5). A6-3807 had CN-LOH that retained the abnormal stop codon (rs16883571, KHDC1, 6q13). AA-3521 had CN-LOH that retained the abnormal splice site (rs13240848, GALNTL5, 7q36.1). The rest of the 37 LOH were associated with retaining alleles that cause non-synonymous mutation. We captured 511 tumor-specific LOH that retained variant alleles (Table 5). Three genes on 18q topped this list of tumor-specific LOH, including include MRO (rs2849233, 18q21.2), DSG1 (rs1426310, 18q12.1), and FECH (rs1041951, 18q21.31) that had either CN-LOH or CL-LOH in five, three and three patients, respectively.
In the germline variant analysis, one patient (AA-3527) had a homozygous variant allele on rs2228006, which causes gain of stop codon on gene PMS2, a gene involved in MMR. Also, the germline homozygous variant allele associated with stop gain/loss was observed on gene TAS2R46 (rs2708381, 12p13.2) in four patients, KHDC1 (rs16883571, 6q13) or SERPINB11 (rs4940595, 18q21.3) in three patients, and ZNF799 (rs2006651, 19p13.2) in one patient. These same genotypes were observed in the matched normal colon and tumor samples from these patients.
Validation of LOH and CNV in a model of neoplastic transformation
We sought to verify the LOH and CNV that we discovered in the above TCGA samples in an independent set of patients. Additionally, since we detected aberrations in the blood and normal colon epithelium, we were interested if adenomatous polyps would contain these genetic aberrations as well since they are an early, non-malignant and precursor legion to CRC. In these cases we have captured the colorectal cancer tissue along with the residual polyp of origin that is contiguous with the cancer, which enables a time lapse model of normal colon to non-malignant polyp and the contiguous cancer.
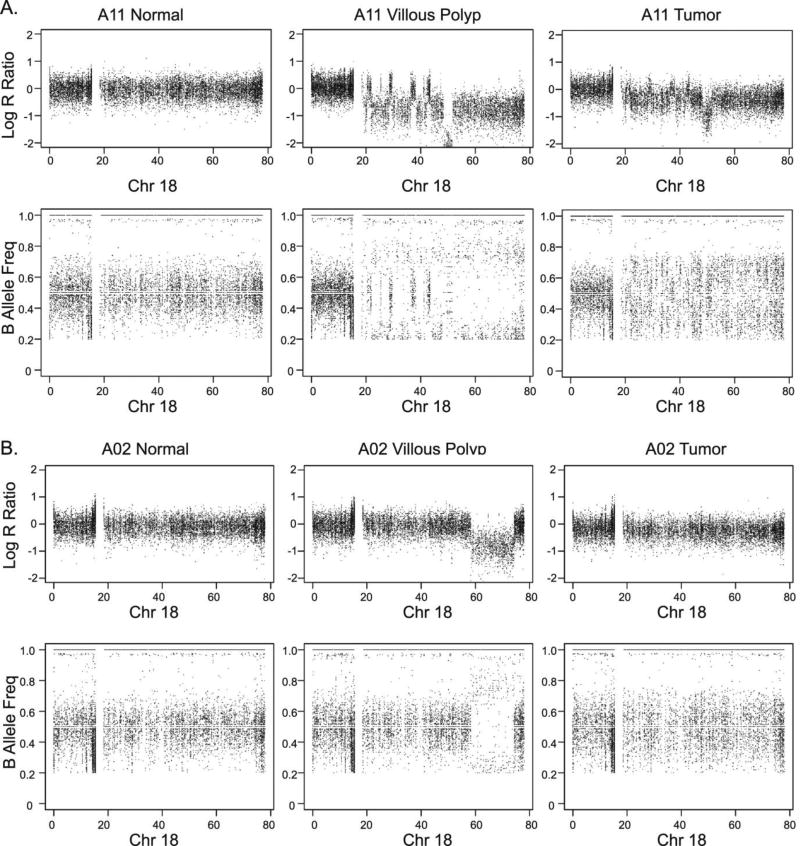
We have verified in a total of 13 independent patient samples representing 44 tissues that there are genetic aberrations on 18q as reported in the TCGA data. We found LOH and deletion events at 18q in the precursor polyp and tumor tissue, for patients A02 (Figure 3A) and A11 (Figure 3B). The interspersed LOH was found specifically at the 18q ~50–85MB and ~60–75MB regions in the polyp tissue of the two samples, respectively. In patient A11, the DCC gene is located at the region of LOH due to the deletion. In the remaining eight patients with either blood or normal colon, we were able to analyze LOH and CNV in the polyp and tumor tissue only. We discovered 18q LOH in both the polyp and tumor tissue in one patient, and 18q LOH due to deletion in the tumors of four of the patients (Figure S6).
Figure 3.
LOH and CNV patterns at chromosome 18 showing LOH at 18q. Patients A) A11 and B) A02 showing normal colon tissue, villous polyp tissue, and tumor of two patients from WGS data. From left to right: Log R Ratio and BAF of normal colon, villous polyp and tumor.
Discussion
In this study we have examined genetic aberrations in the form of LOH and CNV across the blood, normal colon and tumor tissues from CRC patients provided by the TCGA as well as in an independent cohort of patients that included adenomatous polyp tissue, the precursor lesion to CRC. By comparing genetic aberrations detected in the normal colon from patients with CRC with the genetic profile of their corresponding peripheral blood and polyp tissue, we documented the presence of genetic variants in normal colon, polyp and even the blood, which sometimes persisted into the CRC tumor itself. Our study highlights another advantage of utilizing peripheral blood as the reference standard rather than normal colon, since we identified non trivial regions of LOH that would have been missed if only normal colon to tumor comparisons had been performed.
In several cases, the identification of LOH was only possible because of the comparison between the peripheral blood and the normal colon, indicating that detection of LOH in CRC by comparison of tumor to corresponding normal colon’s allelic status without referencing the corresponding peripheral blood may yield significant underestimation. For example, a ~1MB LOH region that overlaps with the DCC gene on chromosome 18q21.1 was present in both normal colon and tumor. LOH in that tumor would not have been observed if normal colon rather than blood were used as the reference.
Since almost half of the CL-LOH and CNV (copy gain) aberrations in normal colon were also found in the blood, CN-LOH actually accounted for the primary difference between blood and normal colon. It is of note that CN-LOH can be deduced only by comparing to informative loci in the reference sample (i.e. heterozygous in blood for this study). Given that somatic changes accumulate in peripheral white blood cells during cell division [34], the actual amount of CN-LOH in normal colon tissue might be more than what was observed here. More “genuine” sources of germline information for detecting somatic aberrations would be tissues which undergo little to no cell division - such as nervous tissue or even occipital brain tissue [35]. However, these tissue types are not readily obtainable, and for the present, peripheral blood DNA is likely to remain the standard reference sample type.
Our data demonstrated high frequency loss of 18q in sporadic CRC tumors, which is consistent with previous reports [36]. The highest frequency of genetic aberration observed on 18q was in normal colon rather than tumor. This was due to the extent of genetic disruption present in the tumor in which 18q loss affected the whole q arm, while in normal colon, the genetic aberrations spanned only a few kilobases. Together with the observation that 96.9% of common LOH in normal colon and tumor retained the same allele, these findings suggest that the arm-specific aberrations in macroscopically and histologically normal colon predated the tumorigenic process and possibly promoted the acquisition of genetic changes in tumor. Similar observations were noted in another study [12], which found a similar aneusomy pattern between sporadic CRC tumor and distal normal colon mucosa.
Importantly, the genetic aberrations we found on 18q in normal colon most frequently overlapped the DCC gene and were also present in the corresponding tumors. DCC encodes a netrin receptor [37] that leads to proliferation and cell migration when bound with netrin-1 [38]. The genetic aberrations in DCC may cause a functional change in the DCC protein, which would contribute partly to CRC predisposition, as DCC mutant mice do not develop malignancies [39,40]. Unfortunately, due to limited SNP coverage here we were not able to cover DCC in the functional SNP analysis.
The highest frequency of genetic aberrations found in blood samples were also on 18q, where a short region with weak copy gain signal was detected in at least two different loci in 10 of the 15 patients with pMMR CRC. In the dMMR patients, there were significantly fewer aberrations found on 18q, but instead had wide spread aberrations on 17, 19, and 22q. These findings confirm genetic aberration differences based on the MMR status of the CRC patient. In six pMMR patients the copy gain overlapped with CCDC102B, a protein-coding gene on 18q22.1 that was most likely to be mutated in the tumors. CCDC102B has not yet been reported as either a tumor suppressor or promoter gene, and does not overlap with known common copy number polymorphism loci [41]. This implies two possible explanations that require further experimental validation. First, this might be a sporadic pMMR CRC specific germline genetic aberration. Second, this might be a signal from circulating tumor cells. If the latter case is true, it can potentially be used as a “liquid biopsy” marker for sporadic CRC.
In the tumor samples, we found genetic aberrations on several genes that are commonly mutated in blood samples from hereditary CRC (e.g. APC, CTNNB1, SMAD4, and BRAF). Although most of them have been shown to be functionally involved in tumorigenesis, their causal relationships with CRC remain elusive [36]. In the current study, none of these genes showed LOH or CNV aberrations in normal colon or blood. Moreover, results from exome sequencing of the normal colon of a patient, neither somatic nor germline mutations were detected in APC or CTNNB1, suggesting the genomic instability seen in the corresponding normal colon was independent of APC and the associated WNT signaling pathway. This observation has two mutually exclusive implications. First, the aberrations in APC-like genes drive the normal-to-tumor cell transition. Second, their aberrations are a by-product of global CIN and reflect a selection advantage over other cells. Our finding of discernible arm-specific aberrations in both normal colon and blood supports the latter scenario and is also in line with the previously proposed mathematical model [42]. Nevertheless, we are aware that the current methodology captures only a limited fraction of the whole mutation spectrum. Further analysis involving epigenetics data is required to provide a comprehensive evaluation of the aberrations in normal colon or blood from these sporadic CRC patients. Viral infection is another confounding factor to consider, as existing studies have shown it may be associated with LOH in CRC [43].
From this study, we are aware of the potential heterogeneity in macroscopically normal colon tissue, which as of yet cannot be modeled by the statistical framework in the current software package. We utilized an independent set of patients, which included macrodissected normal colon epithelial tissues (although stromal and mucosal tissue still could be present) to enrich for the purity of the specimen, and did not observe aberrations in these normal tissues. However, we did observe alterations in the pre-malignant polyp tissues, which are also macrodissected and enriched for the polyp portion of the tissue. Single cell based techniques could be helpful in providing more insight into the current observations.
Our analysis has demonstrated arm-specific enrichment of genetic aberrations on 18q in both normal colon and blood from sporadic CRC patients. It is of interest to note that similar LOH enrichment was also observed in our previous study on breast infiltrating ductal carcinoma distant normal tissue, albeit on different arms [44]. Concurrent with our previous work on NCI-60 cancer cell lines [45], these findings suggest that these aberrant genetic patterns are tissue specific and may be the precursors for the larger genetic aberrations present in tumor.
We were able to validate our findings by WGS in an independent cohort of patients, which included the precursor polyp tissue in addition to the blood, normal colon, and tumor tissue. Collectively, we have identified genetic aberrations at 18q occurring in the non-malignant polyp lesion in three patients (23%), and in the tumor tissue of five patients (38%) from the 13 total patients. While we did not observe the 18q LOH in the normal colon tissue in the independent cohort, we did in the non-malignant polyp tissues. This validates our discovery of early genetic aberrations in the TCGA and shows that the genetic aberrations extend into the early lesion of the polyp. Our findings support the hypothesis that CIN may be an early event and therefore causative factor in the tumorigenesis of sporadic CRC. This finding indicates an important role for aberrations at 18q in the development and progression of CRC. The blood, normal colon and polyp are all readily attainable specimens through a blood-draw or colonoscopy. The ability to detect early molecular aberrations in these pre-malignant samples points to the potential for a blood-based or a normal colon and/or polyp collected at colonoscopy diagnostic marker for assessment of risk for transformation. It will be necessary to evaluate the functional significance and clinical utility of these aberrations in future studies. Our findings further highlight the existence of detectable aberrations in blood, normal colon and polyp tissue that maybe useful as a sporadic CRC risk marker in patient evaluation within the clinical setting.
Supplementary Material
Acknowledgments
Funding:
This work was supported by the National Cancer Institute at the National Institutes of Health (CA170357 and CA204013), and the Clinical Core of Mayo Clinic Center for Cell Signaling in Gastroenterology (P30DK084567). We thank the Center for Individualized Medicine of Mayo Clinic for partially funding this project.
Footnotes
Disclosures: The authors declare no conflicts of interest.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 3.Fearon ER, Jones PA. Progressing toward a molecular description of colorectal cancer development. FASEB J. 1992;6(10):2783–2790. doi: 10.1096/fasebj.6.10.1321771. [DOI] [PubMed] [Google Scholar]
- 4.Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005;307(5717):1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 5.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386(6625):623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 6.Rajagopalan H, Nowak MA, Vogelstein B, Lengauer C. The significance of unstable chromosomes in colorectal cancer. Nat Rev Cancer. 2003;3(9):695–701. doi: 10.1038/nrc1165. [DOI] [PubMed] [Google Scholar]
- 7.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96(15):8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terdiman JP, Gum JR, Jr, Conrad PG, et al. Efficient detection of hereditary nonpolyposis colorectal cancer gene carriers by screening for tumor microsatellite instability before germline genetic testing. Gastroenterology. 2001;120(1):21–30. doi: 10.1053/gast.2001.20874. [DOI] [PubMed] [Google Scholar]
- 9.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260(5109):816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 10.Armaghany T, Wilson JD, Chu Q, Mills G. Genetic alterations in colorectal cancer. Gastrointest Cancer Res. 2012;5(1):19–27. [PMC free article] [PubMed] [Google Scholar]
- 11.Shih IM, Zhou W, Goodman SN, Lengauer C, Kinzler KW, Vogelstein B. Evidence that genetic instability occurs at an early stage of colorectal tumorigenesis. Cancer Res. 2001;61(3):818–822. [PubMed] [Google Scholar]
- 12.Cianciulli A, Cosimelli M, Marzano R, et al. Genetic and pathologic significance of 1p, 17p, and 18q aneusomy and the ERBB2 gene in colorectal cancer and related normal colonic mucosa. Cancer Genet Cytogenet. 2004;151(1):52–59. doi: 10.1016/j.cancergencyto.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Lin CH, Lin JK, Chang SC, et al. Molecular profile and copy number analysis of sporadic colorectal cancer in Taiwan. J Biomed Sci. 2011;18:36. doi: 10.1186/1423-0127-18-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melcher R, Hartmann E, Zopf W, et al. LOH and copy neutral LOH (cnLOH) act as alternative mechanism in sporadic colorectal cancers with chromosomal and microsatellite instability. Carcinogenesis. 2011;32(4):636–642. doi: 10.1093/carcin/bgr011. [DOI] [PubMed] [Google Scholar]
- 15.Rowan AJ, Lamlum H, Ilyas M, et al. APC mutations in sporadic colorectal tumors: A mutational "hotspot" and interdependence of the "two hits". Proc Natl Acad Sci U S A. 2000;97(7):3352–3357. doi: 10.1073/pnas.97.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo L, Shen GQ, Stiffler KA, Wang QK, Pretlow TG, Pretlow TP. Loss of heterozygosity in human aberrant crypt foci (ACF), a putative precursor of colon cancer. Carcinogenesis. 2006;27(6):1153–1159. doi: 10.1093/carcin/bgi354. [DOI] [PubMed] [Google Scholar]
- 17.Ruivenkamp CA, van Wezel T, Zanon C, et al. Ptprj is a candidate for the mouse colon-cancer susceptibility locus Scc1 and is frequently deleted in human cancers. Nat Genet. 2002;31(3):295–300. doi: 10.1038/ng903. [DOI] [PubMed] [Google Scholar]
- 18.Vogelstein B, Fearon ER, Kern SE, et al. Allelotype of colorectal carcinomas. Science. 1989;244(4901):207–211. doi: 10.1126/science.2565047. [DOI] [PubMed] [Google Scholar]
- 19.Ruivenkamp C, Hermsen M, Postma C, et al. LOH of PTPRJ occurs early in colorectal cancer and is associated with chromosomal loss of 18q12-21. Oncogene. 2003;22(22):3472–3474. doi: 10.1038/sj.onc.1206246. [DOI] [PubMed] [Google Scholar]
- 20.Calhoun ES, Gallmeier E, Cunningham SC, Eshleman JR, Hruban RH, Kern SE. Copy-number methods dramatically underestimate loss of heterozygosity in cancer. Genes Chromosomes Cancer. 2006;45(11):1070–1071. doi: 10.1002/gcc.20365. [DOI] [PubMed] [Google Scholar]
- 21.Church JM. Clinical significance of small colorectal polyps. Dis Colon Rectum. 2004;47(4):481–485. doi: 10.1007/s10350-003-0078-6. [DOI] [PubMed] [Google Scholar]
- 22.Heitman SJ, Ronksley PE, Hilsden RJ, Manns BJ, Rostom A, Hemmelgarn BR. Prevalence of adenomas and colorectal cancer in average risk individuals: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2009;7(12):1272–1278. doi: 10.1016/j.cgh.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 23.Martinez ME, Sampliner R, Marshall JR, Bhattacharyya AK, Reid ME, Alberts DS. Adenoma characteristics as risk factors for recurrence of advanced adenomas. Gastroenterology. 2001;120(5):1077–1083. doi: 10.1053/gast.2001.23247. [DOI] [PubMed] [Google Scholar]
- 24.Winawer SJ, Zauber AG, O'Brien MJ, et al. Randomized comparison of surveillance intervals after colonoscopic removal of newly diagnosed adenomatous polyps. The National Polyp Study Workgroup. N Engl J Med. 1993;328(13):901–906. doi: 10.1056/NEJM199304013281301. [DOI] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 27.Chakradhar S. Colorectal cancer: 5 big questions. Nature. 2015;521(7551):S16. doi: 10.1038/521S16a. [DOI] [PubMed] [Google Scholar]
- 28.Druliner BR, Rashtak S, Ruan X, et al. Colorectal Cancer with Residual Polyp of Origin: A Model of Malignant Transformation. Transl Oncol. 2016;9(4):280–286. doi: 10.1016/j.tranon.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hershkovitz D, Simon E, Bick T, et al. Adenoma and carcinoma components in colonic tumors show discordance for KRAS mutation. Hum Pathol. 2014;45(9):1866–1871. doi: 10.1016/j.humpath.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Jones S, Chen WD, Parmigiani G, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105(11):4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sedivy R, Wolf B, Kalipciyan M, Steger GG, Karner-Hanusch J, Mader RM. Genetic analysis of multiple synchronous lesions of the colon adenoma-carcinoma sequence. Br J Cancer. 2000;82(7):1276–1282. doi: 10.1054/bjoc.1999.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yau C, Mouradov D, Jorissen RN, et al. A statistical approach for detecting genomic aberrations in heterogeneous tumor samples from single nucleotide polymorphism genotyping data. Genome Biol. 2010;11(9):R92. doi: 10.1186/gb-2010-11-9-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K, Bucan M. Copy Number Variation Detection via High-Density SNP Genotyping. CSH Protoc. 2008;2008 doi: 10.1101/pdb.top46. pdb top46. [DOI] [PubMed] [Google Scholar]
- 34.Holstege H, Pfeiffer W, Sie D, et al. Somatic mutations found in the healthy blood compartment of a 115-yr-old woman demonstrate oligoclonal hematopoiesis. Genome Res. 2014;24(5):733–742. doi: 10.1101/gr.162131.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spalding KL, Bhardwaj RD, Buchholz BA, Druid H, Frisen J. Retrospective birth dating of cells in humans. Cell. 2005;122(1):133–143. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 36.Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138(6):2059–2072. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keino-Masu K, Masu M, Hinck L, et al. Deleted in Colorectal Cancer (DCC) encodes a netrin receptor. Cell. 1996;87(2):175–185. doi: 10.1016/s0092-8674(00)81336-7. [DOI] [PubMed] [Google Scholar]
- 38.Arakawa H. Netrin-1 and its receptors in tumorigenesis. Nat Rev Cancer. 2004;4(12):978–987. doi: 10.1038/nrc1504. [DOI] [PubMed] [Google Scholar]
- 39.Fazeli A, Dickinson SL, Hermiston ML, et al. Phenotype of mice lacking functional Deleted in colorectal cancer (Dcc) gene. Nature. 1997;386(6627):796–804. doi: 10.1038/386796a0. [DOI] [PubMed] [Google Scholar]
- 40.Cho KR, Oliner JD, Simons JW, et al. The DCC gene: structural analysis and mutations in colorectal carcinomas. Genomics. 1994;19(3):525–531. doi: 10.1006/geno.1994.1102. [DOI] [PubMed] [Google Scholar]
- 41.McCarroll SA, Kuruvilla FG, Korn JM, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40(10):1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 42.Nowak MA, Komarova NL, Sengupta A, et al. The role of chromosomal instability in tumor initiation. Proc Natl Acad Sci U S A. 2002;99(25):16226–16231. doi: 10.1073/pnas.202617399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rollison DE. JC virus infection: a cause of colorectal cancer? J Clin Gastroenterol. 2010;44(7):466–468. doi: 10.1097/MCG.0b013e3181e0084b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruan X, Liu H, Boardman L, Kocher JP. Genome-wide analysis of loss of heterozygosity in breast infiltrating ductal carcinoma distant normal tissue highlights arm specific enrichment and expansion across tumor stages. PLoS One. 2014;9(4):e95783. doi: 10.1371/journal.pone.0095783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruan X, Kocher JP, Pommier Y, Liu H, Reinhold WC. Mass homozygotes accumulation in the NCI-60 cancer cell lines as compared to HapMap Trios, and relation to fragile site location. PLoS One. 2012;7(2):e31628. doi: 10.1371/journal.pone.0031628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.