Dear Editor,
Campylobacter fetus is a zoonotic pathogen that rarely causes bacterial meningitis in humans.1 The majority (>90%) of intestinal campylobacter infections are caused by Campylobacter jejuni or Campylobacter coli, and only a small portion (~2.4%) are caused by C. fetus.2 However, C. fetus is the most common pathogen that causes Campylobacter bacteremia, and the clinical signs of human C. fetus infection vary from acute diarrheal illness to systemic illnesses, such as lung abscesses, arthritis and neurological infections.1 To date, only ~20 cases of C. fetus meningitis have been reported worldwide, mainly in North America and Western Europe.1, 3
Campylobacter fetus comprises 2 subspecies: C. fetus subspecies fetus (Cff) and C. fetus subspecies venerealis (Cfv). Cff has been isolated from the gastrointestinal tracts of sheep and cattle and causes infertility and abortion in these species. By contrast, Cfv is restricted to the genital tracts of cattle, where it causes bovine genital campylobacteriosis.4 Most of the C. fetus infections in humans are thought to be caused by Cff, but efforts to differentiate the subspecies of C. fetus are generally not made. Although Cfv has been isolated from humans in a few cases, its role in human disease is unknown.2 A newly proposed subspecies, C. fetus subspecies testudinum, has been recently isolated from sick humans;5 however, its clinical characteristics are largely unknown. Owing to the extremely small number of cases reported as C. fetus meningitis, efforts to differentiate the subspecies of C. fetus in meningitis patients are lacking. In this study, we demonstrate the first case of C. fetus meningitis diagnosed in South Korea, in which Cff was confirmed by 16S rRNA amplicon sequencing on a MinION nanopore sequencer (Oxford Nanopore Technologies, Oxford, UK).
In December 2016, a 64-year-old man with alcoholic liver cirrhosis and poorly controlled diabetes visited Seoul National University Hospital due to fever, nausea and seizure. He had undergone dental implant procedures 2 weeks prior to admission. He reported a 10-day history of upper respiratory infection and decreased oral intake due to gastrointestinal discomfort and recurrent diarrhea. A history of recent travel or animal contact was denied, and his food history was unremarkable.
On day 1, his temperature was 39.2 °C, blood pressure 140/100 mm Hg, heart rate 80 beats/min, and respiratory rate 20 breaths/min. Laboratory tests revealed borderline leukocytosis (10 050 cells/μL) with 79.8% neutrophils, hemoglobin 11.1 g/dL, and a platelet count within the reference range (148 × 103/μL). Oral moxifloxacin was given as an empirical antibiotic; however, on day 3, neck stiffness became pronounced, and a stuporous mental status was noted. Accordingly, a cerebrospinal fluid (CSF) analysis was performed on day 3 and showed pleocytosis (217 leukocytes/mm3; 73% neutrophils), a high protein level (188 (reference range 15–45) mg/dL), and a low glucose level (CSF/serum glucose ratio 36.4%). No focal lesions were observed on a brain MRI. Dexamethasone, ceftazidime, vancomycin and ampicillin were administered. C. fetus grew in the initial blood cultures but not in the CSF cultures, which were obtained 2 days after empirical antibiotic treatment. The results of other laboratory tests, including viral PCR and serological tests for a wide range of infectious causes of encephalitis, were all negative. Doripenem was administered for 8 days until the patient recovered from the meningitis symptoms without neurologic sequelae. However, the patient was diagnosed with gallbladder cancer during the systemic investigations and was referred to the surgical department for further management. For confirmation of the pathogen, the genomic DNA of the bacteria was extracted from subcultures of a single colony. The pathogen was confirmed as C. fetus by the sequence analysis of the full-length 16S rRNA gene using conventional Sanger sequencing.
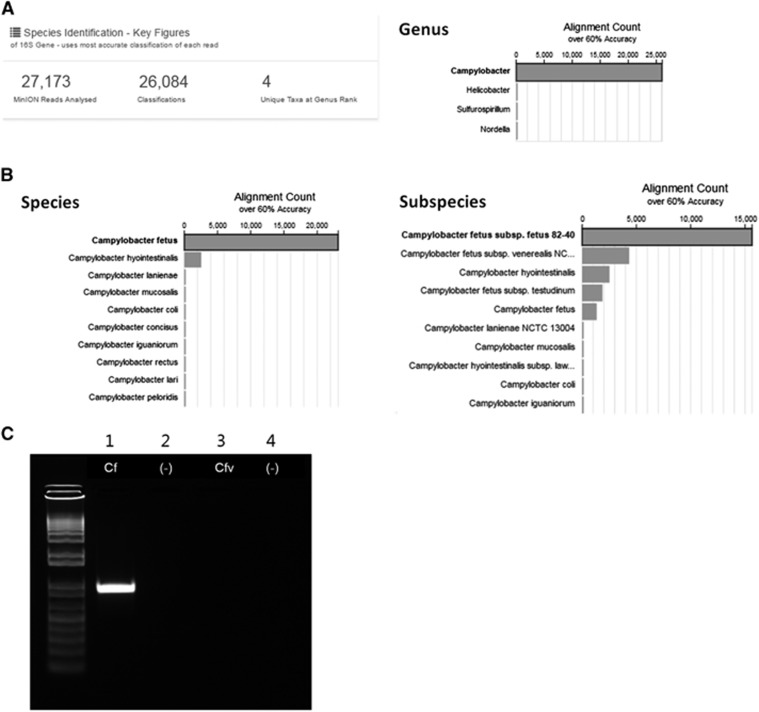
Additionally, 16S rRNA amplicon sequencing was performed on the MinION nanopore sequencer (Oxford Nanopore Technologies, Oxford, UK). The 16S rRNA genes were PCR amplified from the genomic DNA of the pathogen using the universal bacterial primers 27F 5′-AGA GTT TGA TCM TGG CTC AG-3′ and 1492R 5′-GGT TAC CTT GTT ACG ACT T-3′. Nanopore sequencing libraries were constructed using these amplicons. A total of 43 044 reads were generated during the 51 min sequencing run time. The cloud-based Metrichor 16S-BLAST workflow was applied. Of the 27 173 successfully base-called reads, 26 084 reads were aligned to one of the bacterial 16S rRNA gene sequences. Of the total aligned reads, 26 066 (99.9%) were aligned to the genus Campylobacter (Figure 1A). C. fetus (23 172 reads, 88.8%) and Cff (15 643 reads, 60.0%) were the top listed species and subspecies, respectively (Figure 1B). The standard methods for the differentiation of C. fetus subspecies are tolerance to 1% glycine and H2S production,6 which were unavailable because the causative strain of this meningitis case had not been stored for further analysis. Instead, the remaining genomic DNA was tested with an additional PCR using previously described primers,7 to differentiate the C. fetus subspecies. Finally, the pathogen was specified as Cff (Figure 1C).
Figure 1.
Identification of the pathogen at the species and subspecies level. (A) After the MinION 16S rRNA amplicon sequencing, 99.9% of the reads were aligned to the genus Campylobacter. (B) C. fetus (23 172 reads, 88.8%) and C. fetus subspecies fetus (15 643 reads, 60.0%) were the top listed species and subspecies, respectively. (C) Agarose gel electrophoresis of PCR products obtained using primers specific to C. fetus (Cf) and C. fetus subspecies venerealis (Cfv). Lanes 2 and 4 were loaded with a negative control. The band present in lane 1 but absent from lane 3 indicates C. fetus subspecies fetus.
Only 22 cases of C. fetus meningitis have been reported worldwide in adults.1, 3 The median age was 48 years (range 23–84 years), and the majority of the patients were men (73%). Sixteen out of the 22 patients were in an immunocompromised state, mostly due to alcoholism (41%) and diabetes (27%). Most of the patients were reported from North America or Western Europe, except for 2 patients reported from Japan.8, 9 Although C. fetus is a zoonotic pathogen, contact with animals or animal products was identified in <70% of patients.3 Food products from cattle and sheep are the most likely routes of transmission in human C. fetus infections.2 Some studies proposed dental procedures as a possible invasion route for a C. fetus infection, and cancer has been suggested as a risk factor for C. fetus bacteremia.10 The treatment outcome of C. fetus meningitis has been favorable, with 17 patients showing full recovery, 3 patients showing a neurologic deficit and 2 deaths.3 Several reports have indicated that human C. fetus isolates are resistant to ceftriaxone, cefotaxime, penicillin and erythromycin,3, 11 while carbapenems are effective.1, 3
The MinION is a portable nanopore sequencer that is a third-generation sequencing method. Because it performs low cost, real-time, long-read sequencing, it is increasingly being used for rapid metagenomics analyses of disease/pathogen surveillance.12, 13 Real-time surveillance of Ebola was carried out using the MinION.14 Recently, remarkable performances of the MinION for rapid bacterial identification by full-length 16S rRNA amplicon sequencing have been reported.15 Since deep sequencing of the 16S rRNA gene is possible with MinION, multiple bacterial infections can be detected and their relative abundance can be analyzed by a single sequencing run. However, only a limited number of studies have reported its application in human cases.
This is the first case of C. fetus meningitis reported in South Korea, and it is the first to apply MinION-based, full-length 16S rRNA amplicon sequencing in a patient with bacterial meningitis. The patient was in an immunocompromised state with alcoholic liver cirrhosis, uncontrolled diabetes and underlying gallbladder cancer. Although direct animal contact was denied and his food history was unremarkable, he had undergone dental procedures two weeks before admission; this could be the possible invasion route of the pathogen. The patient recovered fairly well after antimicrobial treatment, which included doripenem. The subspecies of C. fetus was successfully identified as Cff by 16S rRNA amplicon sequencing using the MinION. We believe this was achievable because of deep sequencing, which enables the identification of small sequence differences in the 16S rRNA gene between two subspecies. Evaluating the efficacy of 16S rRNA amplicon sequencing directly from clinical samples (blood or CSF) is extremely valuable, because it can significantly reduce the turnaround time by omitting the time required for bacterial growth in culture tests. Unfortunately, we could not evaluate this in the current patient because we did not obtain enough specimens before the antimicrobial treatment was initiated.
In conclusion, C. fetus should be considered a possible cause of bacterial meningitis, especially in immunocompromised patients with accompanying gastrointestinal symptoms. Nanopore sequencing of the 16S rRNA gene allowed the identification of C. fetus at the subspecies level. The capability of differentiating bacterial subspecies makes MinION extremely useful in the epidemiology and surveillance of bacterial infections; however, it should be verified for diverse bacteria obtained from clinical samples in the near future. Nevertheless, the nanopore sequencer would be very useful for pathogen detection in patients with bacterial infections because it enables full-length 16S rRNA amplicon sequencing and the real-time analysis of the reads.
Acknowledgments
This study was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2016R1C1B2016275) and by the SNUH Research Fund (03-2015-0430). Jangsup Moon was supported by a SK Young Investigator Award by SK chemicals, Korean Neurological Association.
References
- van Samkar A, Brouwer MC, van der Ende A et al. Zoonotic bacterial meningitis in human adults. Neurology 2016; 87: 1171–1179. [DOI] [PubMed] [Google Scholar]
- Wagenaar JA, Bergen V, van Bergen MA et al. Campylobacter fetus infections in humans: exposure and disease. Clin Infect Dis 2014; 58: 1579–1586. [DOI] [PMC free article] [PubMed]
- van Samkar A, Brouwer MC, van der Ende A et al. Campylobacter fetus meningitis in adults. Medicine 2016; 95: e2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mshelia G, Amin J, Woldehiwet Z et al. Epidemiology of bovine venereal campylobacteriosis: geographic distribution and recent advances in molecular diagnostic techniques. Reprod Domest Anim 2010; 45: e221–e230. [DOI] [PubMed] [Google Scholar]
- Patrick ME, Gilbert MJ, Blaser MJ et al. Human infections with new subspecies of Campylobacter fetus. Emerg Infect Dis 2013; 19: 1678–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett TJ, Patton CM, Morris GK. Differentiation of Campylobacter species using phenotypic characterization. Lab Med 1988; 19: 96–102. [Google Scholar]
- Schulze F, Bagon A, Müller W et al. Identification of Campylobacter fetus subspecies by phenotypic differentiation and PCR. J Clin Microbiol 2006; 44: 2019–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Wakasugi H, Mukuta T et al. Campylobacter fetus subspecies fetus meningitis with chronic alcoholism and diabetes mellitus. Jpn J Med 1990; 29: 542–544. [DOI] [PubMed] [Google Scholar]
- Umehara Y, Kudo M, Kawasaki M. Campylobacter fetus meningitis in a patient with Crohn's disease. Inflamm Bowel Dis 2009; 15: 645–646. [DOI] [PubMed] [Google Scholar]
- Pacanowski J, Lalande V, Lacombe K et al. Campylobacter bacteremia: clinical features and factors associated with fatal outcome. Clin Infect Dis 2008; 47: 790–796. [DOI] [PubMed] [Google Scholar]
- Herve J, Aissa N, Legrand P et al. Campylobacter fetus meningitis in a diabetic adult cured by imipenem. Eur J Clin Microbiol Infect Dis 2004; 23: 722–724. [DOI] [PubMed] [Google Scholar]
- Greninger AL, Naccache SN, Federman S et al. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med 2015; 7: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen T, Groseth A, Rosenke K et al. Nanopore sequencing as a rapidly deployable Ebola outbreak tool. Emerg Infect Dis 2016; 22: 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick J, Loman NJ, Duraffour S et al. Real-time, portable genome sequencing for Ebola surveillance. Nature 2016; 530: 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Lee S, Go M-J et al. Analysis of the mouse gut microbiome using full-length 16S rRNA amplicon sequencing. Sci Rep 2016; 6: 29681. [DOI] [PMC free article] [PubMed] [Google Scholar]