ABSTRACT
Pseudomonas aeruginosa and Aspergillus fumigatus are common opportunistic bacterial and fungal pathogens, respectively. They often coexist in airways of immunocompromised patients and individuals with cystic fibrosis, where they form biofilms and cause acute and chronic illnesses. Hence, the interactions between them have long been of interest and it is known that P. aeruginosa can inhibit A. fumigatus in vitro. We have approached the definition of the inhibitory P. aeruginosa molecules by studying 24 P. aeruginosa mutants with various virulence genes deleted for the ability to inhibit A. fumigatus biofilms. The ability of P. aeruginosa cells or their extracellular products produced during planktonic or biofilm growth to affect A. fumigatus biofilm metabolism or planktonic A. fumigatus growth was studied in agar and liquid assays using conidia or hyphae. Four mutants, the pvdD pchE, pvdD, lasR rhlR, and lasR mutants, were shown to be defective in various assays. This suggested the P. aeruginosa siderophore pyoverdine as the key inhibitory molecule, although additional quorum sensing-regulated factors likely contribute to the deficiency of the latter two mutants. Studies of pure pyoverdine substantiated these conclusions and included the restoration of inhibition by the pyoverdine deletion mutants. A correlation between the concentration of pyoverdine produced and antifungal activity was also observed in clinical P. aeruginosa isolates derived from lungs of cystic fibrosis patients. The key inhibitory mechanism of pyoverdine was chelation of iron and denial of iron to A. fumigatus.
IMPORTANCE Interactions between human pathogens found in the same body locale are of vast interest. These interactions could result in exacerbation or amelioration of diseases. The bacterium Pseudomonas aeruginosa affects the growth of the fungus Aspergillus fumigatus. Both pathogens form biofilms that are resistant to therapeutic drugs and host immunity. P. aeruginosa and A. fumigatus biofilms are found in vivo, e.g., in the lungs of cystic fibrosis patients. Studying 24 P. aeruginosa mutants, we identified pyoverdine as the major anti-A. fumigatus compound produced by P. aeruginosa. Pyoverdine captures iron from the environment, thus depriving A. fumigatus of a nutrient essential for its growth and metabolism. We show how microbes of different kingdoms compete for essential resources. Iron deprivation could be a therapeutic approach to the control of pathogen growth.
KEYWORDS: Aspergillus fumigatus, Pseudomonas aeruginosa, biofilms, intermicrobial interaction, iron, mutants, pyoverdine
INTRODUCTION
Pseudomonas aeruginosa and Aspergillus fumigatus are the most prominent bacterium and fungus, respectively, in the airways of cystic fibrosis (CF) patients (1–4). They have been associated with deterioration of lung function (1, 5–14), and it has been suggested that their combined presence in those airways has an even worse potential (15, 16). Both of these microbes are also prominent opportunists in immunocompromised patients, particularly neutropenic patients (17, 18).
In studies of P. aeruginosa-A. fumigatus interactions, it has long been noted in in vitro studies that P. aeruginosa products can be toxic to A. fumigatus. These P. aeruginosa products include phenazines such as pyocyanin (5-N-methyl-1-hydroxyphenazine) (19–22), 1-hydroxyphenazine (19, 21, 22), phenazine-1-carboxamide, phenazine-1-carboxylic acid (22), and dirhamnolipids (23). Other P. aeruginosa products have been noted to be toxic to other fungi, though their effects on A. fumigatus have not been studied, including 3,4-dihydroxy-2-heptylquinoline (PQS) and phenazin-1-ol (24), 3,4-dihydroxy-N-methyl-4-(4-oxochroman-2-yl)butanamide (25), and lipopolysaccharide (26). However, studying individual molecules for toxicity does not give insight into their relative contributions in a mixture, how the molecules would behave in the presence of other P. aeruginosa and/or A. fumigatus metabolites, or whether the concentrations studied are relevant to those that occur in intermicrobial interactions (27, 28). An alternative approach to begin to understand the intermicrobial factors involved is to study P. aeruginosa mutants and examine the effects of removing molecules, groups of them, or pathways by mutational analysis. We studied a series of mutants (Table 1) defective in the expression of most known P. aeruginosa virulence functions, including the production of bioactive molecules and secondary metabolites previously reported to interact with eukaryotic cells, and compared their inhibitory activity against A. fumigatus biofilms, as well as planktonic A. fumigatus growth, to that of the parent P. aeruginosa strain.
TABLE 1.
PA14 mutants used in this study
| Mutant no. | Mutation(s) | Result(s) of mutation(s)a | Reference(s) |
|---|---|---|---|
| 1 | pvdD pchE | Pyoverdine-pyochelin double-siderophore mutant | This study |
| 2 | pqsE | Gene mediates regulatory activity of MvfR system (see mvfR); HAQs similar to wild type but defective in pyocyanin, HCN | 87 |
| 3 | mvfR | Regulates transcription of pqsABCDE operon; underproduction of various QS-regulated factors related to loss of HAQs, phenazines, proteases, HCN, lectins, others (94) | 88 |
| 4 | pqsA | First step in HAQ synthesis; gene for anthranilate coenzyme A ligase lost; loss of extracellular quinolones, HAQ biosynthesis, including HHQ, PQS, DHQ; also decreased activity of MvfR (89) | 87 |
| 5 | pqsH | Product converts HHQ into PQS; loss of 2-heptyl-3-hydroxy-4(1H)-quinolone synthase; loss of PQS biosynthesis, thus decreased activity of MvfR; MvfR not as negatively affected as in pqsA mutant since pqsH mutant still produces HHQ and other HAQs that can act as ligands of MvfR | 90 |
| 6 | lasR rhlR | Double mutant defective for most QS-regulated metabolites, including phenazines, rhamnolipids, AHL, HAQs, proteases, HCN, chitinase, elastase, others | 91 |
| 7 | lasR | Lacks several QS-regulated factors, including proteases, oxo-C12-HSL; delayed activation of pqsABCDE and of RhlR QS pathway | 87 |
| 8 | rsmA | Global posttranscriptional regulator mutant; various effects, including less rhamnolipids, more phenazines, and HCN (92) | 93 |
| 9 | pqsA pqsH not polar | Same predicted outcome as pqsA mutant; in both, pqsA mutation is nonpolar on downstream genes in operon, thus not preventing their transcription | 90 |
| 10 | pvdD | Loss of pyoverdine (siderophore) | 93 |
| 11 | rhlR | Lacks several QS-regulated factors; loss of rhamnolipids, phenazines, HCN, lectins, C4-HSL | 94 |
| 12 | ΔHSI-I ΔHSI-II | Double-deletion mutant defective in 2 of 3 type VI secretion systems | 95 |
| 13 | pvcA | Loss of paerucumarin and pseudoverdin | 93 |
| 14 | rhlA | Loss of rhamnolipids (93) | 96 |
| 15 | phzC1 phzC2 | Double-phenazine mutant (completely abrogated); no pyocyanin | Unpublishedb |
| 16 | pchE | Loss of pyochelin (siderophore) | 93 |
| 17 | exoU | Loss of exotoxin U via type III secretion | 93 |
| 18 | rsmY rsmZ | Loss of genes for coding small regulatory RNAs; antagonistic to RsmA; decreased production of C4-HSL, phenazines, chitinase (92) | This study |
| 19 | ΔHSI-II ΔHSI-III | Double-deletion mutant defective in 2 of 3 type VI secretion systems | 95 |
| 20 | ΔHSI-I ΔHSI-III | Double-deletion mutant defective in 2 of 3 type VI secretion systems | 95 |
| 21 | pqsA pqsH polar | pqsA::TnPhoA, pqsH::Gm; mutation in pqsA is polar (thus theoretically preventing transcription of downstream genes in operon); Kanr Gmr | Unpublishedb |
| 22 | chiC | MrT7 transposon mutant; Gmr | 93 |
| 23 | lecA | MrT7 transposon mutant; Gmr | 93 |
| 24 | hcnA | MrT7 transposon mutant; Gmr | 93 |
| 25 | None | None (parental strain UBCPP-PA14) | 77, 78 |
Abbreviations: HAQ, 4-hydroxy-2-alkylquinolones; HCN, hydrogen cyanide; AHL, acylhomoserine lactones; HSL, homoserine lactone; HHQ, 4-hydroxy-2-heptylquinoline; PQS, 3,4-dihydroxy-2-heptylquinoline (97); HQNO, 4-hydroxy-2-heptylquinoline-N-oxide. Kan, kanamycin; Gm, gentamicin.
Unpublished laboratory strain (available from E. Déziel).
RESULTS
PA14 pvdD pchE, pvdD, lasR rhlR, and lasR mutants are most impaired in their antifungal activities.
Twenty-four PA14 mutants (Table 1) and wild-type PA14 were tested for the ability to interfere with A. fumigatus metabolism by using bacterial live cells (LC), P. aeruginosa planktonic culture filtrate (PCF), or P. aeruginosa biofilm culture filtrate (BCF) as the agents and A. fumigatus biofilm formation or preformed A. fumigatus biofilm as the target (resulting in six possible assay combinations for each assay method). The results will be considered in turn and then summarized at the end of Results.
Effects of LC, BCF, and PCF on A. fumigatus biofilm formation.
As A. fumigatus biofilm forms from conidia in agar, P. aeruginosa LC inhibition of growth can be manifested by clear zones around a well in which the LC are placed. Figure 1A shows a summary of four inhibition zone experiments. In these assays, 12 mutants produced zones of inhibition that were significantly smaller than the inhibition zones of wild-type PA14 LC. These are ranked by zone size in Table 2. An example of A. fumigatus inhibition zones generated by P. aeruginosa LC is shown in Fig. S1 in the supplemental material.
FIG 1.
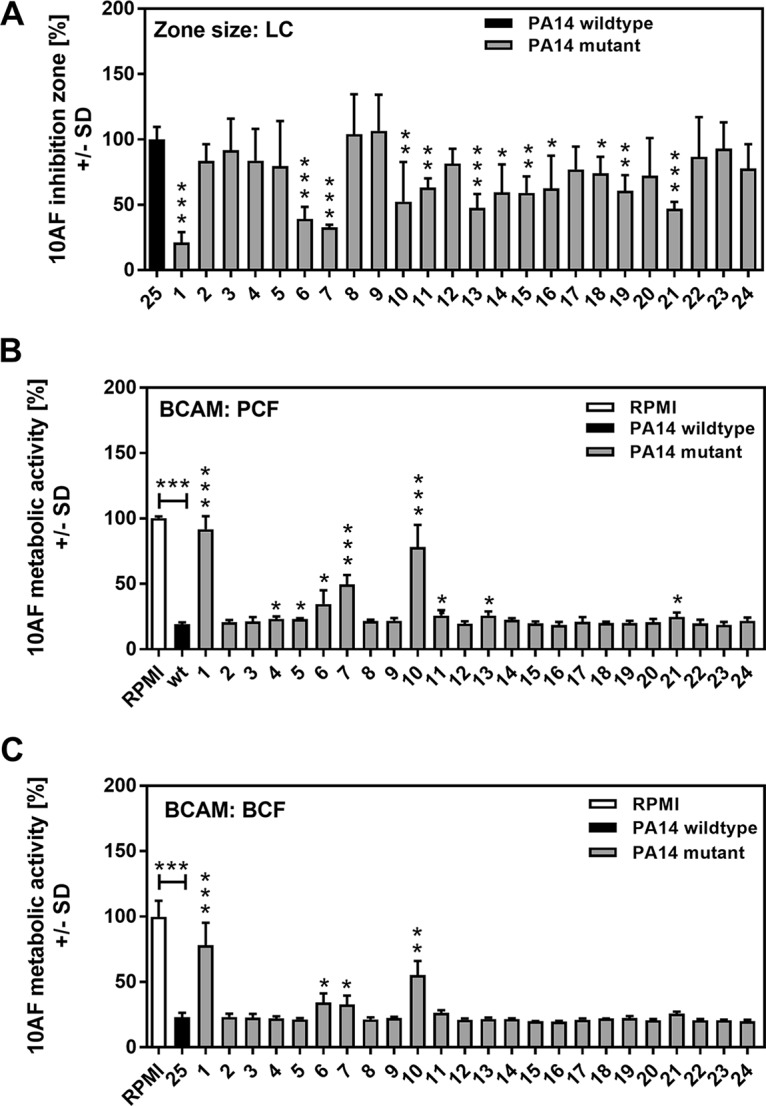
Effects of LC, PCF, and BCF on A. fumigatus strain 10AF biofilm formation and metabolism. (A) Wells in agar with A. fumigatus conidia were loaded with wild-type or mutant PA14 LC suspensions and incubated, and the area of the fungus-free (inhibition) zone around each well was measured. The inhibition zone created by wild-type PA14 LC was regarded as 100% inhibition, and inhibition zones created by mutants were compared to this. The results of four experiments are combined, with duplicates of each study article in each. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (mutant [no. 1 to 24] versus wild type [no. 25]). (B and C) Agar and A. fumigatus strain 10AF conidia were distributed into 96-well cell culture plates that were loaded with wild-type or mutant PA14 PCF (B) or wild-type or mutant PA14 BCF (C) and incubated at 37°C for 24 h. A. fumigatus strain 10AF metabolism was evaluated by XTT assay. Metabolism in the presence of RPMI medium alone was regarded as 100% A. fumigatus strain 10AF metabolic activity. The data shown are the mean ± SD of four independent experiments (with duplicates of each group studied in each experiment). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. Vertical asterisks, mutants (no. 1 to 24) versus wild-type PA14 (no. 25); horizontal asterisks, RPMI medium versus wild-type PA14. Bars: 1, pvdD pchE mutant; 2, pqsE mutant; 3, mvfR mutant; 4, pqsA mutant; 5, pqsH mutant; 6, lasR rhlR mutant; 7, lasR mutant; 8, rsmA mutant; 9, pqsA pqsH not polar mutant; 10, pvdD mutant; 11, rhlR mutant; 12, ΔHSI-I ΔHSI-II mutant; 13, pvcA mutant; 14, rhlA mutant; 15, phzC1 phzC2 mutant; 16, pchE mutant; 17, exoU mutant; 18, rsmY rsmZ mutant; 19, ΔHSI-II ΔHSI-III mutant; 20, ΔHSI-I ΔHSI-III mutant; 21, pqsA pqsH polar mutant; 22, chiC mutant; 23, lecA mutant; 24, hcnA mutant.
TABLE 2.
Ranking of PA14 mutant LC, PCF, and BCF activities against A. fumigatus strain 10AF forming biofilma
| Rank | Mutation(s) ranked on basis of: |
||
|---|---|---|---|
| Inhibition zone size, agar-LC | XTT conversion, agar-PCF | XTT conversion, agar-BCF | |
| 1 | pvdD pchE | pvdD pchE | pvdD pchE |
| 2 | lasR | pvdD | pvdD |
| 3 | lasR rhlR | lasR | lasR rhlR |
| 4 | pqsA pqsH polar | lasR rhlR | lasR |
| 5 | pvcA | rhlR | |
| 6 | phzC1 phzC2 | pvcA | |
| 7 | rhlA | pqsA pqsH polar | |
| 8 | ΔHSI-I ΔHSI-II | pqsA | |
| 9 | pchE | pqsH | |
| 10 | rhlR | ||
| 11 | rsmY rsmZ | ||
| 12 | pvdD | ||
All assays started from Aspergillus conidia. Ranking from 1 to n represents highest to lowest loss of antifungal activity. Ranking was performed as described in Materials and Methods (Fig. 1). Only isolates statistically significantly different from the parent strain are listed.
When P. aeruginosa supernatants were used in the wells instead of live cells and A. fumigatus grown from conidia were used, no zones of inhibition were produced. However, there was inhibition of metabolism of A. fumigatus forming biofilm, and this could be measured by the 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide inner salt (XTT) assay (bioassay-conidium-agar-metabolic [BCAM] assays). Figure 1B shows the A. fumigatus inhibition results obtained with PCF of mutants 1 to 24 and wild-type PA14. In these assays, nine mutants demonstrated significantly impaired antifungal activity (ranked from most to least impaired Table 2). Figure 1C shows the results of this assay but with BCF. Four mutants showed significantly inhibited antifungal activity (ranked in Table 2).
In all three agar assays of A. fumigatus biofilm formation (Table 2), the pvdD pchE and pvdD mutants had impaired antifungal activity. The pvdD pchE mutant was the most impaired mutant in all three assays. The lasR rhlR and lasR mutants were also impaired in all three assays and among the four highest ranking mutants in all three assays. The rhlR, pvcA, and polar pqsA pqsH mutants were impaired in two of the three assays.
Effects of PCF and BCF on preformed A. fumigatus strain 10AF biofilm in agar (BHAM) assays.
The effects of supernatants on preformed A. fumigatus strain 10AF biofilm are summarized in Table 3. Figure 2A shows mutants 1 to 24, as well as wild-type PA14, tested in these bioassay-hypha-agar-metabolic (BHAM) assays with PCF. Five mutants showed significantly decreased antifungal activity, with again the two pyoverdine mutants, the pvdD and pvdD pchE mutants, and the two impaired las quorum-sensing (QS) system mutants, the lasR and lasR rhlR mutants, prominent (Table 3). With BCF, the picture was nearly identical to that obtained with PCF (Fig. 2B and Table 3). Thus, fewer mutants were impaired in the inhibition of preformed A. fumigatus biofilm than in the inhibition of A. fumigatus biofilm formation (compare Tables 2 and 3), although four mutants, the pvdD pchE, pvdD, lasR rhlR, and lasR mutants, appear prominently as impaired mutants on these lists against both forming and preformed A. fumigatus biofilms.
TABLE 3.
Ranking of PA14 mutant PCF and BCF activities against A. fumigatus strain 10AF preformed biofilma
| Rank | Mutation(s) tested on: |
|
|---|---|---|
| Agar-PCF | Agar-BCF | |
| 1 | pvdD | pvdD |
| 2 | pvdD pchE | pvdD pchE |
| 3 | lasR | lasR rhlR |
| 4 | rsmA | lasR |
| 5 | lasR rhlR | |
All assays started from Aspergillus hyphae. The readout in both assays was XTT conversion. Ranking from 1 to n represents the highest to lowest loss of antifungal activity. Ranking was performed as described in Materials and Methods. Only isolates statistically significantly different from the parent strain are listed.
FIG 2.
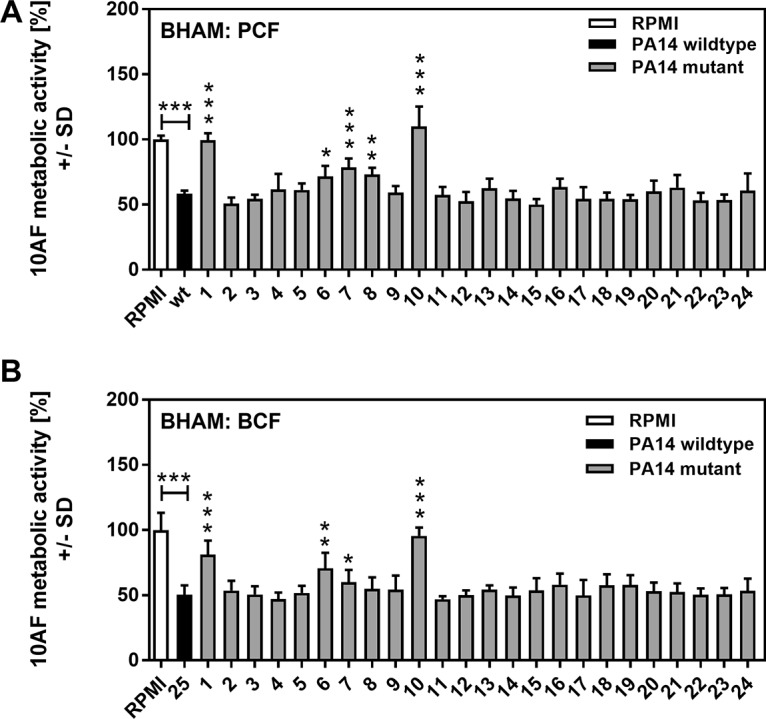
Effects of PCF or BCF on A. fumigatus strain 10AF preformed biofilm growth and metabolism. A. fumigatus strain 10AF conidia in agar were distributed into 96-well cell culture plates. Plates were incubated at 37°C for 24 h to allow hyphal growth and then loaded with wild-type or mutant PA14 PCF (A) or wild-type or mutant PA14 BCF (B) and incubated at 37°C for 24 h. A. fumigatus strain 10AF metabolism was evaluated by XTT assay. Metabolism in the presence of RPMI medium alone was regarded as 100% A. fumigatus strain 10AF metabolic activity. The data shown are the mean ± SD of four independent experiments (with duplicates of each group studied in each experiment). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (mutant [no. 1 to 24] versus wild type [no. 25]). Horizontal asterisks, RPMI medium versus wild-type PA14. Bars: 1, pvdD pchE mutant; 2, pqsE mutant; 3, mvfR mutant; 4, pqsA mutant; 5, pqsH mutant; 6, lasR rhlR mutant; 7, lasR mutant; 8, rsmA mutant; 9, pqsA pqsH not polar mutant; 10, pvdD mutant; 11, rhlR mutant; 12, ΔHSI-I ΔHSI-II mutant; 13, pvcA mutant; 14, rhlA mutant; 15, phzC1 phzC2 mutant; 16, pchE mutant; 17, exoU mutant; 18, rsmY rsmZ mutant; 19, ΔHSI-II ΔHSI-III mutant; 20, ΔHSI-I ΔHSI-III mutant; 21, pqsA pqsH polar mutant; 22, chiC mutant; 23, lecA mutant; 24, hcnA mutant.
Effects of BCF and PCF on forming and preformed A. fumigatus strain 10AF biofilms under hypoxic conditions.
To more closely mimic some focal CF lung conditions, we repeated part of our experiments under hypoxic conditions. PCF of wild-type PA14, the pvdD pchE and pvdD mutants, which had shown the lowest overall antifungal activity, and the pchE and phzC1 phzC2 mutants was tested in BCAM and BHAM assays under hypoxic conditions. Our results obtained under hypoxic conditions closely resemble those obtained under normoxic conditions (compare Fig. 3A to Fig. 1B and Fig. 3B to Fig. 2A), with the pvdD and pvdD pchE mutations affecting the antifungal activity of P. aeruginosa supernatants significantly.
FIG 3.
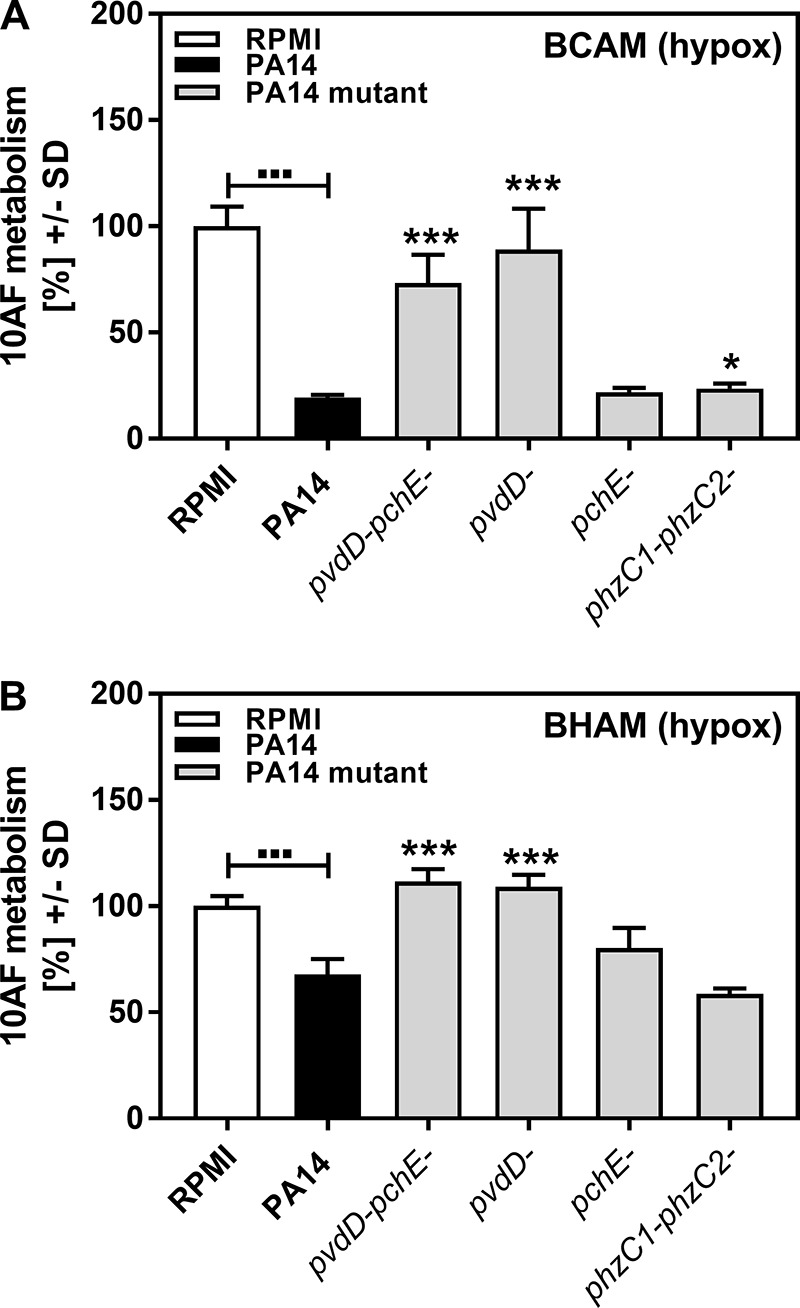
Effects of PCF on A. fumigatus strain 10AF forming and preformed biofilm metabolism under hypoxic conditions. A. fumigatus strain 10AF conidia in agar were distributed into 96-well cell culture plates. Plates were either loaded immediately after solidification (BCAM assay) (A) or incubated at 37°C for 24 h to allow hyphal growth (BHAM assay) (B). Plates were loaded with wild-type or mutant PA14 PCF and incubated under hypoxic conditions at 37°C for 24 h. A. fumigatus strain 10AF metabolism was evaluated by XTT assay. Metabolism in the presence of RPMI medium alone was regarded as 100% A. fumigatus strain 10AF metabolic activity. The data shown are the mean ± SD of four replicates. *, P ≤ 0.05; ***, P ≤ 0.001 (mutant versus wild-type PA14). Other comparisons are indicated by the ends of the brackets above the bars.
Effects of BCF and PCF on planktonic A. fumigatus strain 10AF growth (MIC).
Because there are demonstrated differences in P. aeruginosa activity against biofilm and planktonic A. fumigatus (29), we also investigated the activity of the mutants against planktonic A. fumigatus. Our results show that PCF and BCF derived from the pvdD pchE, pvdD, lasR rhlR, and lasR mutants interfered with planktonic fungal growth significantly less than wild-type supernatants did (Fig. 4). In addition, PCFs of the pqsA, pqsH, ΔHSI-I ΔHSI-II, and pvcA mutants, as well as BCF of the rsmY rsmZ mutant, showed significantly less antifungal activity. The reduction of antifungal activity by these mutants was much weaker than the reduction of antifungal activity by mutated inactivation of pvdD pchE, pvdD, lasR rhlR, or lasR (Table 4). Table S1 details the geometric mean values and ranges for all of the mutants shown in Table 4.
FIG 4.
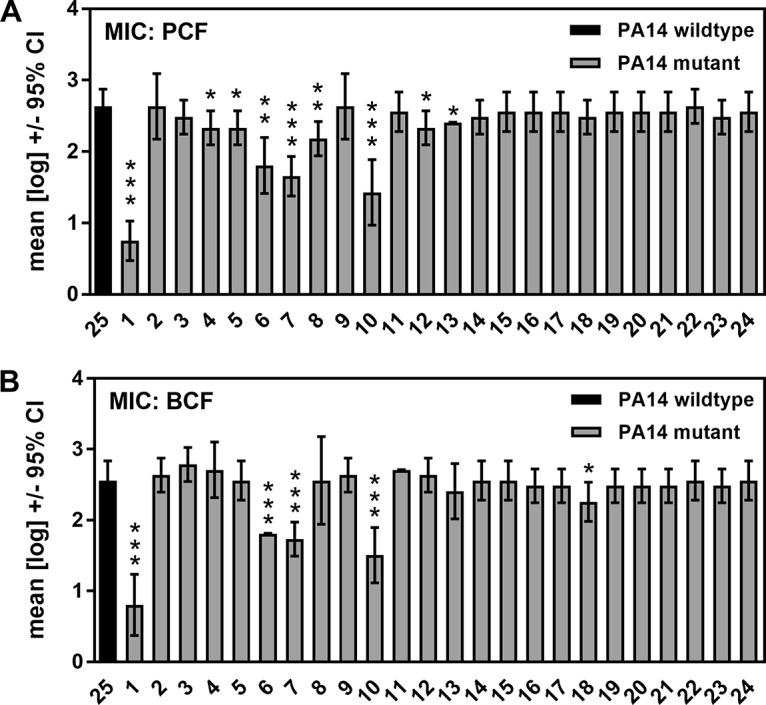
Effects of PCF or BCF on A. fumigatus strain 10AF planktonic growth (MIC). A. fumigatus strain 10AF conidia were incubated in the presence of RPMI medium and PCF dilutions (A) or RPMI medium and BCF dilutions (B) at 37°C for 48 h. Fungal growth was determined in 2-fold dilution steps for wild-type (no. 25) and mutant (no. 1 to 24) PA14 (dilutions, 1:2 to 1:1,024). For each supernatant, the last dilution showing fungal growth less than that in RPMI medium alone was determined. The means of four independent experiments were determined. The data shown are the mean ± SD. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (mutant [no. 1 to 24] versus wild type [no. 25]). Bars: 1, pvdD pchE mutant; 2, pqsE mutant; 3, mvfR mutant; 4, pqsA mutant; 5, pqsH mutant; 6, lasR rhlR mutant; 7, lasR mutant; 8, rsmA mutant; 9, pqsA pqsH not polar mutant; 10, pvdD mutant; 11, rhlR mutant; 12, ΔHSI-I ΔHSI-II mutant; 13, pvcA mutant; 14, rhlA mutant; 15, phzC1 phzC2 mutant; 16, pchE mutant; 17, exoU mutant; 18, rsmY rsmZ mutant; 19, ΔHSI-II ΔHSI-III mutant; 20, ΔHSI-I ΔHSI-III mutant; 21, pqsA pqsH polar mutant; 22, chiC mutant; 23, lecA mutant; 24, hcnA mutant. CI, confidence interval.
TABLE 4.
Ranking of PA14 mutant BCF and PCF activities against planktonic A. fumigatus strain 10AF growtha
| Rank | Mutation(s) tested for effect on antifungal activity of: |
|
|---|---|---|
| PCF | BCF | |
| 1 | pvdD pchE | pvdD pchE |
| 2 | pvdD | pvdD |
| 3 | lasR | lasR |
| 4 | lasR rhlR | lasR rhlR |
| 5 | rsmA | rsmY rsmZ |
| 6 | ΔHSI-I ΔHSI-II | |
| 7 | pqsA | |
| 8 | pqsH | |
| 9 | pvcA | |
BCF and PCF were evaluated by four MIC tests each. Only mutants with statistically significant differences from wild-type PA14 are shown. Ranking from 1 to n represents the highest to lowest loss of antifungal activity.
Overview of different assays of mutants as inhibitors of A. fumigatus.
In summary, when the activities of all of the mutants tested in all seven normoxic assays were compared, the pvdD pchE, pvdD, lasR rhlR, and lasR mutants consistently showed the greatest magnitude of decreased antifungal activity (Table 5).
TABLE 5.
PA14 mutations producing significant loss of antifungal activity in at least one of seven assays
| Mutation(s) | No. of assays with significant loss/total |
|---|---|
| pvdD, pvdD pchE, lasR, lasR rhlR | 7/7 |
| pvcA | 3/7 |
| pqsA, pqsH, pqsA pqsH polar, rhlR, rsmA, ΔHSI-I ΔHSI-II | 2/7 |
| rhlA, phzC1 phzC2, pchE, rsmY rsmZ | 1/7 |
Pyoverdine is a crucial factor in P. aeruginosa antifungal activity.
One common feature of mutants we found impaired in antifungal activity is the loss or lack of pyoverdine production. Figure 5A visualizes pyoverdine production in all 24 PA14 mutants, as well as the wild type. The pvdD pchE and pvdD mutants show complete loss of pyoverdine production, while the lasR rhlR and lasR mutants show substantial reductions of pyoverdine production. Some mutants actually show slightly increased pyoverdine production, i.e., the pqsE, rsmA, rhlR, pchE, lecA, and hcnA mutants. The most significant overproducer of pyoverdine is the pchE mutant; this was not unexpected, since it cannot produce the other siderophore, pyochelin, thus corroborating our assays. Figure 5B shows that the ability of wild-type or mutant PA14 to grow in RPMI medium and pyoverdine production do not correlate. Specifically, the lack of pyoverdine production in the pvdD pchE, pvdD, lasR rhlR, and lasR mutants is not related to their inability to grow in the medium.
FIG 5.
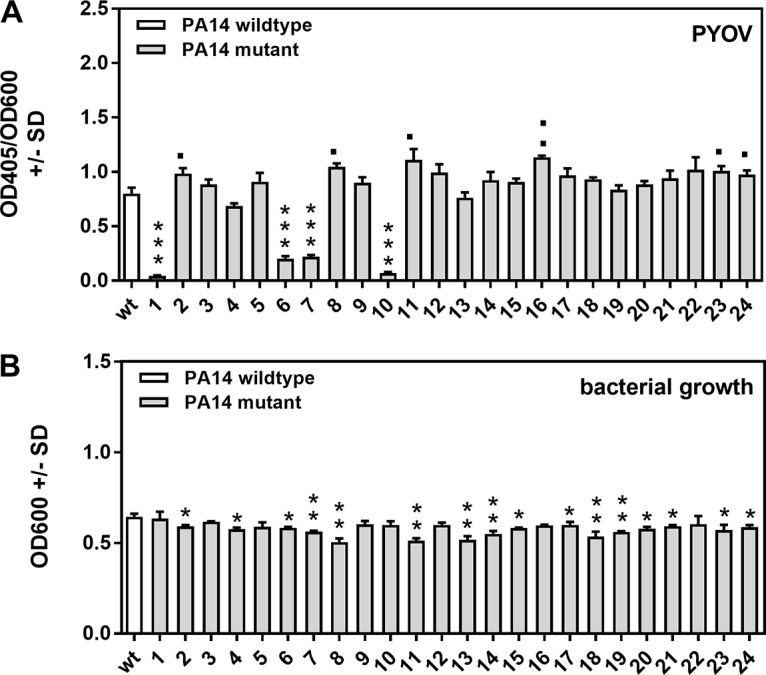
Pyoverdine production by wild-type and mutant PA14. Wild-type or mutant PA14 bacteria (5 × 107/ml) were incubated in RPMI medium overnight. Bacterial growth was measured spectrophotometrically at 600 nm. PCF was harvested, and pyoverdine (PYOV) production was measured at 405 nm (A). Measurements were normalized to bacterial growth (B) with the formula relative pyoverdine expression = OD405/OD600. Experiments in both panels were performed three times (four replicates of each group in each experiment), and the results were combined. ***, P ≤ 0.001 (decrease in pyoverdine production or bacterial growth in mutants compared to wild-type PA14); ■, P ≤ 0.05; ■■, P ≤ 0.01 (significant increase in pyoverdine production compared to wild-type PA14). Bars: 1, pvdD pchE mutant; 2, pqsE mutant; 3, mvfR mutant; 4, pqsA mutant; 5, pqsH mutant; 6, lasR rhlR mutant; 7, lasR mutant; 8, rsmA mutant; 9, pqsA pqsH not polar mutant; 10, pvdD mutant; 11, rhlR mutant; 12, ΔHSI-I ΔHSI-II mutant; 13, pvcA mutant; 14, rhlA mutant; 15, phzC1 phzC2 mutant; 16, pchE mutant; 17, exoU mutant; 18, rsmY rsmZ mutant; 19, ΔHSI-II ΔHSI-III mutant; 20, ΔHSI-I ΔHSI-III mutant; 21, pqsA pqsH polar mutant; 22, chiC mutant; 23, lecA mutant; 24, hcnA mutant.
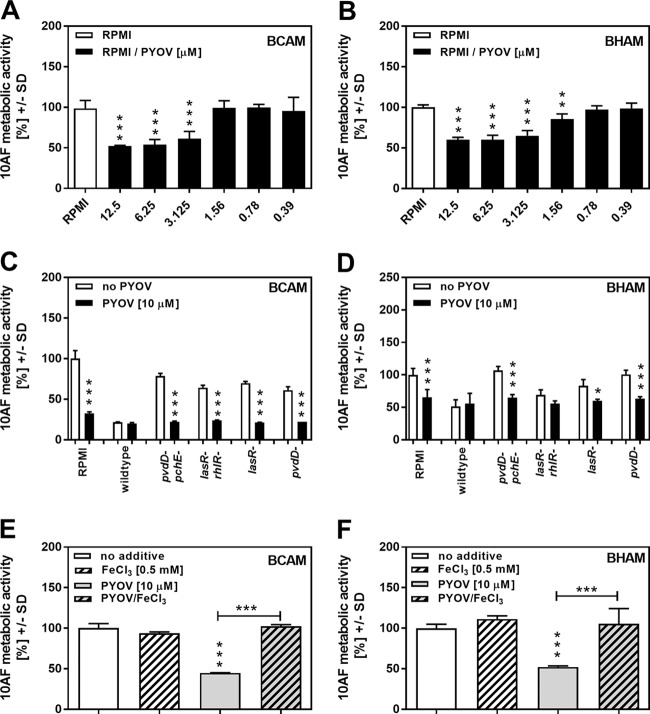
The correlation between mutants lacking pyoverdine production and their decreased antifungal activity demonstrated in our assays indicates that pyoverdine is a major P. aeruginosa factor conferring antifungal activity. To test this hypothesis, we diluted pure pyoverdine in RPMI medium and investigated its impact on A. fumigatus strain 10AF forming and preformed biofilm metabolism. Figure 6A shows that pyoverdine concentrations as low as 3.125 μM significantly and dose dependently interfered with the metabolism of A. fumigatus strain 10AF forming biofilm, whereas preformed A. fumigatus strain 10AF biofilm metabolism was affected by pyoverdine at ≥1.56 μM (Fig. 6B). Comparison to a pyoverdine standard curve revealed that wild-type PA14 PCF contained about 30 μM pyoverdine (data not shown). Supplementation with only 10 μM pyoverdine was able to restore antifungal activities of the pvdD pchE, pvdD, lasR rhlR, and lasR mutants against forming A. fumigatus strain 10AF biofilm (Fig. 6C) and of the pvdD pchE, pvdD, and lasR mutants against preformed A. fumigatus strain 10AF biofilm (Fig. 6D).
FIG 6.
Effects of pyoverdine on A. fumigatus strain 10AF biofilm metabolism and antifungal activities of PA14 mutants. (A and B) Increasing concentrations of pyoverdine (PYOV) in RPMI 1640 (0.39 to 12.5 μM) were assayed for activity against A. fumigatus strain 10AF biofilm formation (A) or preformed A. fumigatus strain 10AF biofilm (B). Fungal metabolism was measured by XTT assay. Statistical analysis was performed by one-way ANOVA of the RPMI 1640 control versus pyoverdine concentrations: **, P ≤ 0.01; ***, P ≤ 0.001. (C and D) Antifungal activities of wild-type PA14 and pyoverdine-lacking mutants with or without supplementation with 10 μM pyoverdine were measured against forming A. fumigatus strain 10AF biofilm (C) or preformed A. fumigatus strain 10AF biofilm (D). No pyoverdine supplementation versus pyoverdine supplementation: *, P ≤ 0.05; ***, P ≤ 0.001. (E and F) A. fumigatus strain 10AF conidia were incubated in the presence of RPMI 1640 (unstriped bars) or RPMI 1640 supplemented with 0.5 μM FeCl3 (striped bars) with (gray bars) or without (white bars) 10 μM pyoverdine. A. fumigatus strain 10AF metabolism was evaluated by XTT assay. Metabolism in the presence of RPMI 1640 without FeCl3 or pyoverdine was regarded as 100% A. fumigatus strain 10AF metabolic activity. Vertical asterisks, comparison with RPMI 1640; horizontal asterisks, RPMI 1640 containing pyoverdine versus RPMI 1640 containing pyoverdine and FeCl3. ***, P ≤ 0.001. Experiments in panel A were performed twice, those in panel B were performed three times, those in panel D were performed six times, and representative results are shown. Every experiment had four replicates per group studied.
Since pyoverdine is a siderophore produced by Pseudomonas that binds iron and facilitates iron uptake (30, 31), we investigated if complexing of iron during Aspergillus growth, thus decreasing iron bioavailability, might be the reason for its antifungal effects. We therefore compared the metabolism of A. fumigatus strain 10AF during biofilm formation and in preformed biofilm in the presence of pyoverdine with or without an excess of iron ions. Our results show that detrimental effects of pyoverdine on both forming A. fumigatus strain 10AF biofilm (Fig. 6E) and preformed A. fumigatus strain 10AF biofilm (Fig. 6F) were abolished by iron supplementation.
P. aeruginosa predominantly affects A. fumigatus by iron starvation.
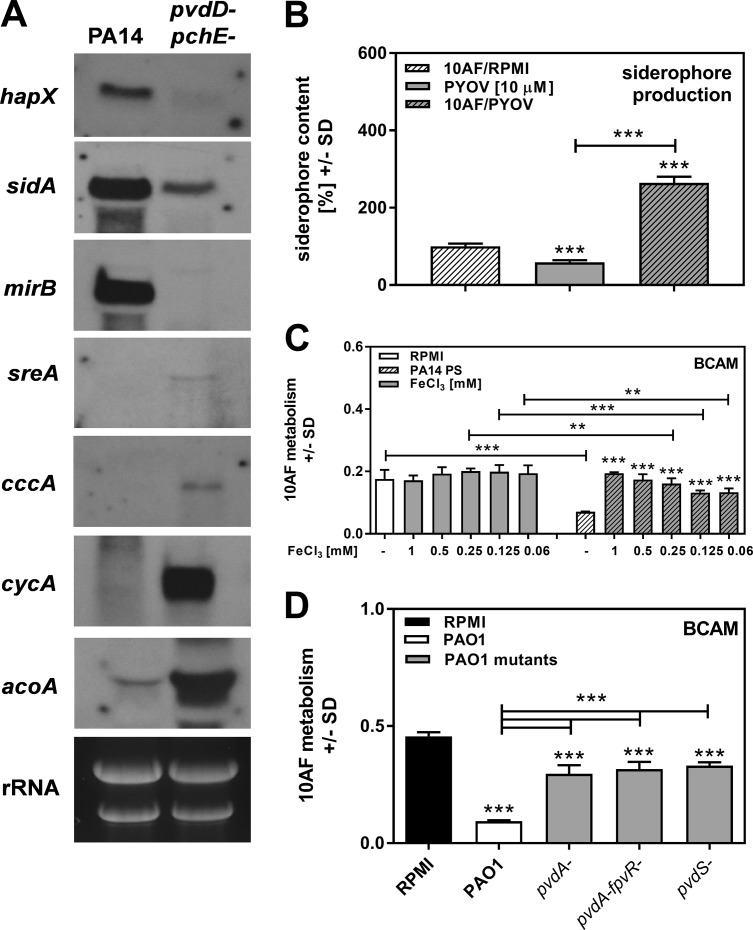
As shown in Fig. 6E and F, iron was able to interfere with the antifungal activity of PA14. To support the idea that iron starvation was the reason for the observed antifungal activity of PA14 supernatants, we performed Northern blot assays of A. fumigatus cultivated in the presence of wild-type or pvdD pchE mutant PA14 PCF. Our results show that wild-type PA14 PCF, in contrast to pvdD pchE mutant PCF, resulted in increased expression of A. fumigatus genes that are known to be responsive to iron starvation (hapX, sidA, and mirB), as opposed to the reduced expression of genes indicating iron availability (sreA, cccA, cycA, and acoA) (Fig. 7A). The hypothesis that iron denial affects A. fumigatus is further supported by the fact that pyoverdine increased A. fumigatus siderophore production by about 3-fold, as measured by chrome azurol S (CAS) assay (Fig. 7B). Induction of A. fumigatus siderophore production is a counteraction to lower iron levels in the fungal environment. Additionally, we found that iron concentrations as low as 60 μM were able to significantly interfere with the antifungal activity of PA14 PCF, supporting the relevance of iron for counteracting the effects of P. aeruginosa on A. fumigatus (Fig. 7C).
FIG 7.
Effects of P. aeruginosa siderophores on A. fumigatus iron metabolism. (A) A. fumigatus strain AfS77 conidia were grown at 37°C in 15 ml of liquid 2YT medium for 15 h. Subsequently, 10 ml of the culture supernatant was replaced with 10 ml of wild-type or pvdA pvdE mutant PA14 PCF and incubated for 3 h. A Northern blot analysis was performed for genes inducible by iron starvation (hapX, sidA, and mirB) and genes repressed by iron starvation (sreA, cccA, cycA, and acoA). Measurement of rRNA served as a loading control. (B) A. fumigatus strain 10AF conidia were incubated in the presence of RPMI 1640 (striped bar) or RPMI 1640 supplemented with 10 μM pyoverdine (gray striped bar) for 24 h. Supernatants were sterile filtered. As a control, 10 μM pyoverdine (PYOV) solution in RPMI 1640 was prepared (solid gray bar). A CAS assay measuring siderophore content in A. fumigatus supernatants was performed, and the results were normalized to those of an XTT assay prepared in parallel with the CAS assay measuring fungal metabolism. Siderophore production by A. fumigatus was regarded as 100% and compared to all other bars. Other comparisons are indicated by the ends of the brackets above the bars. ***, P ≤ 0.001. Pyoverdine itself as a siderophore was measurable by CAS assay. (C) RPMI 1640 or PA14 PCF with or without increasing amounts of FeCl3 was subjected to a BCAM assay measuring effects on the metabolism of A. fumigatus strain 10AF forming biofilm. A. fumigatus strain 10AF metabolism was evaluated by XTT assay. Metabolism in the presence of RPMI 1640 alone was regarded as 100% A. fumigatus strain 10AF metabolic activity. The data shown are the mean ± SD from four replicates. **, P ≤ 0.01; ***, P ≤ 0.001 (RPMI 1640 versus RPMI 1640 containing FeCl3 [left side] or PCF versus PCF containing FeCl3 [right side]). Other comparisons are indicated by the ends of the brackets above the bars. (D) Agar and A. fumigatus strain 10AF conidia were distributed into 96-well cell culture plates that were loaded with wild-type or mutant PAO1 PCF and incubated at 37°C for 24 h. A. fumigatus strain 10AF metabolism was evaluated by XTT assay. Metabolism in the presence of RPMI medium alone was compared to metabolism in the presence of PCF. Other comparisons are indicated by the ends of the brackets above the bars. ***, P ≤ 0.001.
Iron concentrations and pyoverdine affect signaling pathways in P. aeruginosa leading to the expression of virulence factors, i.e., exotoxin A (ExoA), PrpL protease, and pyoverdine itself (32). Using a P. aeruginosa mutant that is unable to express pyoverdine but is still able to produce PvdS-dependent virulence factors (pvdA fpvR mutant) in comparison with a P. aeruginosa mutant that does not express pyoverdine but expresses small amounts of ExoA and PrpL protease (pvdA mutant) and a P. aeruginosa mutant that is unable to produce pyoverdine, as well as ExoA and PrpL protease (pvdS mutant), we found no significant differences in the low antifungal activities of these mutants (Fig. 7D). Hence, PvdS-dependent expression of ExoA or PrpL protease does not contribute to the antifungal activity of P. aeruginosa. The predominant mode of action of pyoverdine appears to be deprivation of A. fumigatus of iron, resulting in fungal inhibition.
It has to be noted that although pyoverdine appears to have primacy over other potential P. aeruginosa microbial inhibitors (Table 5), other mutations are likely to affect the antifungal activity of P. aeruginosa to a lesser extent, since pyoverdine mutants were not constantly found completely impaired in their antifungal activity (Fig. 1 to 4 and 7D).
Effects of pharmacologically decreased pyoverdine levels on the antifungal activities of P. aeruginosa supernatants.
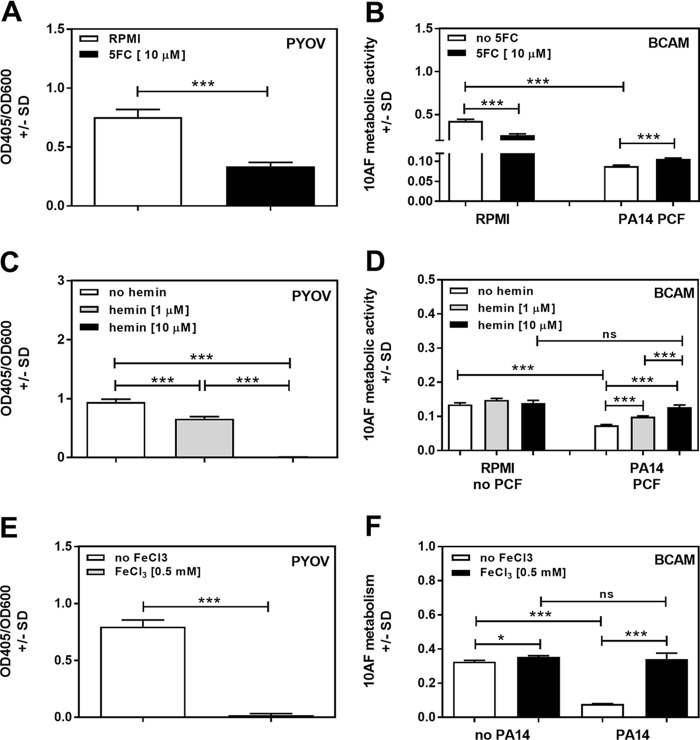
It has been shown previously that 5-fluorocytosine (5FC) is able to decrease pyoverdine production by P. aeruginosa strain PAO1 (33). Using 10 μM 5FC, we found a similar effect on PA14 (Fig. 8A). PCF produced in the presence of 10 μM 5FC, i.e., containing less pyoverdine, was found to be significantly less active against A. fumigatus biofilm formation than supernatant produced without 5FC (Fig. 8B, right side). It has to be noted that 10 μM 5FC by itself showed significant antifungal effects (Fig. 8B, left side) (34), which might partially mask the loss of antifungal activity in 5FC-containing P. aeruginosa supernatants.
FIG 8.
Effects of pyoverdine reduction on A. fumigatus strain 10AF biofilm metabolism. RPMI 1640 with or without 10 μM 5FC (A, B), 1 or 10 μM hemin (C, D), or 0.5 mM FeCl3 (E, F) was inoculated with 5 × 107 PA14 bacteria/ml. Pyoverdine (PYOV) production was measured after 24 h of incubation (A, C, E). PCFs derived from panels A, C, and E were subjected to BCAM assays measuring effects on the metabolism of A. fumigatus strain 10AF forming biofilm (B, D, F). Groups being compared are indicated by the ends of the brackets above the bars. The experiments shown were performed three times (four replicates of each group in each experiment), and the results were combined. *, P ≤ 0.05; ***, P ≤ 0.001.
Because of the perfusion of lungs with blood, exacerbated by the presence of blood components in airways as a result of chronic inflammation in the lungs of individuals with CF, both P. aeruginosa and A. fumigatus are exposed to blood components in CF lungs. Blood, owing to its heme component, can be a vast source of iron for organisms expressing heme oxygenases (35, 36). P. aeruginosa possesses two heme uptake systems, encoded by has and phu (37), and expresses the heme oxygenase HemO (38) and thus is able to use hemin as an iron source. We investigated whether addition of the heme oxygenase inducer hemin, which also serves as a heme oxygenase substrate, would affect pyoverdine production by PA14, as well as the antifungal activity of PA14 supernatant produced in the presence of hemin. Our results show that 10 μM hemin abolished pyoverdine production by PA14 (Fig. 8C), whereas the bacteria grew better in the presence of hemin than in its absence, showing the dissociation between growth and pyoverdine production, as well as lack of hemin toxicity for P. aeruginosa. Hemin alone had no significant effect on A. fumigatus metabolism (Fig. 8D, left side). Planktonic PA14 supernatants produced in the presence of hemin showed significantly less antifungal activity than control supernatants produced without hemin (Fig. 8D, right side). Supernatants produced in the presence of 10 μM hemin had no significant antifungal effects on A. fumigatus strain 10AF forming biofilm (Fig. 8D), similar to the effects of PA14 mutants lacking pyoverdine (Fig. 1B and 4A). Thus, iron from blood in patients' lungs might suppress pyoverdine production by P. aeruginosa and aggravate the detrimental effects of A. fumigatus in, e.g., CF. These data are supported by Fig. 8E and F, showing that the presence of iron indeed interferes with the ability of P. aeruginosa to produce pyoverdine (Fig. 6E). Iron did not exert toxic effects on P. aeruginosa and significantly increased bacterial growth. Similar to our observations regarding hemin, iron abolished the antifungal properties of PA14 PCF (Fig. 8F). In the experiments presented in Fig. 8E and F, we used 0.5 mM iron but observed similar effects over a range of iron concentrations (1 to 0.1 mM; data not shown). The results support our finding that the ability to produce pyoverdine via iron denial is a major factor determining the antifungal activity of P. aeruginosa.
Corroboration of the studies with PA14.
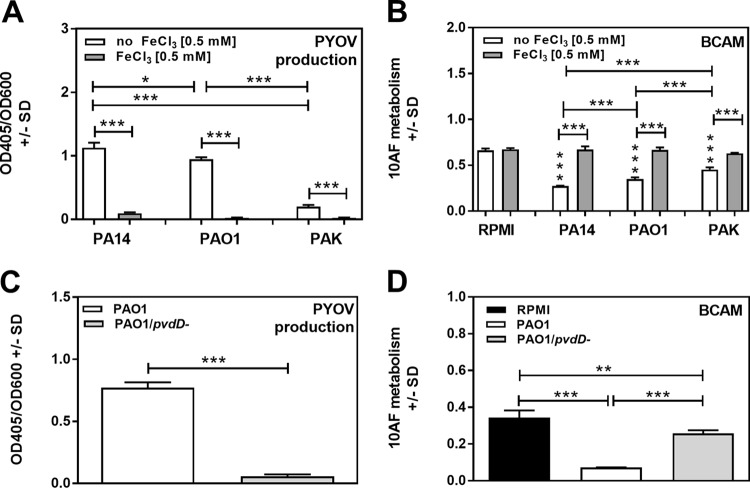
In addition to wild-type PA14, we also studied P. aeruginosa isolates PAO1 (producer of a type I pyoverdine, as is PA14) and PAK. PCFs of PAO1 and PAK also inhibited A. fumigatus strain 10AF biofilm formation (P ≤ 0.01 to 0.001 in four experiments) (Fig. 9B). Pyoverdine production (Fig. 9A) and inhibition of A. fumigatus biofilm formation by their PCFs were also abolished by iron (P ≤ 0.01 to 0.001) (Fig. 9B). PAK produced less pyoverdine than PA14 or PAO1 (P ≤ 0.001) (Fig. 9A), and also in three experiments, its PCF produced the least inhibition of A. fumigatus biofilm formation (Fig. 9B). We also studied a pvdD mutant in the PAO1 background; compared to the parent, its ability to inhibit A. fumigatus forming biofilm was reduced (P ≤ 0.01 to 0.001 in four experiments) (Fig. 9D), its pyoverdine production was negligible (Fig. 9C), and added iron did not affect either inhibition of A. fumigatus or pyoverdine production. Representative experiments for each of these findings are shown in Fig. 9.
FIG 9.
Pyoverdine production by P. aeruginosa PAO1, PAK, and PA14 and effects on A. fumigatus strain 10AF forming biofilm. (A) RPMI 1640 with or without 0.5 mM FeCl3 was inoculated with P. aeruginosa strains PAO1, PAK and PA14 at 5 × 107 bacteria/ml. Pyoverdine (PYOV) production was measured after 24 h of incubation. (B) PCFs derived from panel A were studied in assays measuring effects on the metabolism of A. fumigatus strain 10AF forming biofilm (BCAM assay). Vertical asterisks, comparison with the negative control (RPMI 1640 alone). (C) Pyoverdine production by wild-type or pvdD mutant PAO1 bacteria (5 × 107/ml) was measured after 24 h of incubation. (D) PCFs derived from panel C were studied in assays measuring effects on the metabolism of A. fumigatus strain 10AF forming biofilm (BCAM). Groups being compared are indicated by the ends of the brackets above the bars. Representative results are shown. Each group in each experiment contained at least four replicates. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Compared to the effects of PCF from the PAO1 parent strain on preformed A. fumigatus biofilm, those of pvdD mutant-derived PCF were also reduced (P ≤ 0.05 to 0.001 in three experiments). PAK-derived PCF showed less inhibition of A. fumigatus strain 10AF preformed biofilm than PA14- or PAO1-derived PCF (P ≤ 0.05 to 0.001 in four experiments).
Study of pyoverdine types.
Pseudomonas species generate three, possibly four, classes of pyoverdines, and P. aeruginosa strains produce one representative of each principal class (30, 31). For P. aeruginosa, these representatives are termed types, with every P. aeruginosa strain producing only one type. We measured the activity of each pyoverdine type to assess the global applicability of our observations on antifungal activity. Dose titrations of 2-fold dilutions of purified pyoverdine types ranging from 20 to 2.5 μM were tested against A. fumigatus strain 10AF biofilm formation. Significant inhibition of A. fumigatus biofilm formation by types I and III was observed at concentrations as low as 5 μM. However, 5 μM type II was not significantly inhibitory. At 2.5 μM, no type was different from the control (Fig. 10A). These results suggest that all three pyoverdine types are similarly active against A. fumigatus biofilm formation and type II is slightly less active.
FIG 10.
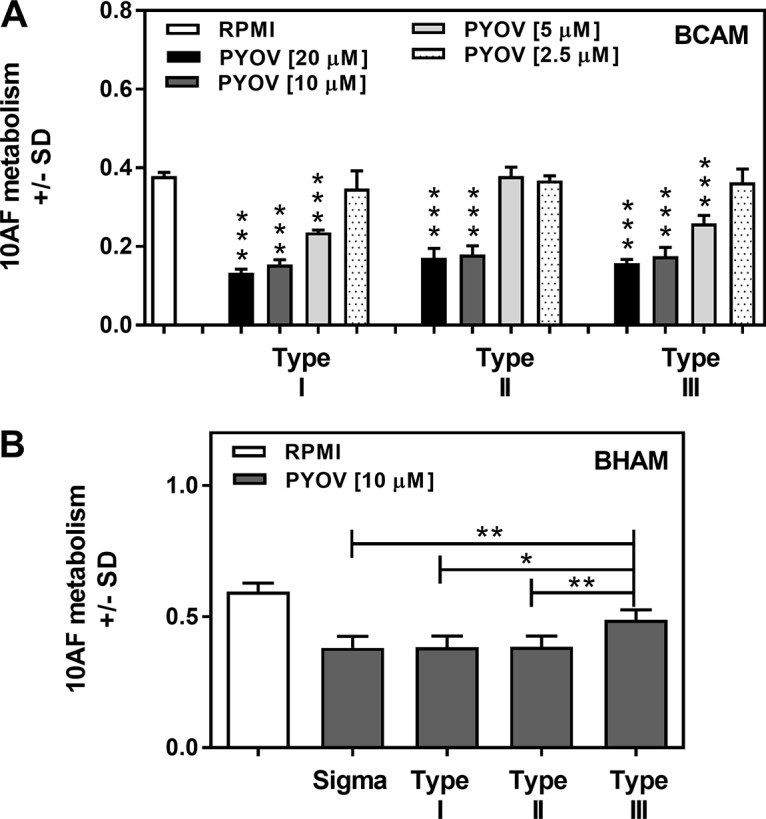
Titration of purified pyoverdines. (A) Pyoverdine types were titrated 2-fold from 20 to 2.5 μM against A. fumigatus strain 10AF biofilm formation. Mean results of four replicates of each group are shown. ***, P ≤ 0.001 (versus RPMI 1640 control). (B) Part of a titration series against preformed A. fumigatus strain 10AF biofilm (10 μM comparison) is shown. Sigma is the commercial pyoverdine (PYOV; type I) used in the experiments whose results are shown in the preceding figures. Mean results of four replicates of each group are shown. Groups being compared are indicated by the ends of the brackets above the bars. *, P ≤ 0.05; **, P ≤ 0.01. All three types of pyoverdine inhibited preformed biofilm metabolism compared to the RPMI 1640 control. Type III, P ≤ 0.01; type I or II, P ≤ 0.001.
The dose titration against preformed A. fumigatus biofilm gave similar results. At 5 to 20 μM, inhibition by all three types was different from that of the control (P ≤ 0.01), as was the case for 2.5 μM type I, whereas at that apparently limiting dilution, the other two types showed nonsignificant inhibition (data not shown). At 10 μM, pyoverdine types I and II produced significantly more inhibition of preformed A. fumigatus biofilm (P < 0.001) than the control, similar to the commercial pyoverdine (class I) used in all of the preceding parts of this study, as did P. aeruginosa type III (P < 0.01) (Fig. 10B). However, type III was significantly (P ≤ 0.05) less active than the other three pyoverdine comparators. These results suggest that type I may be slightly more active against preformed A. fumigatus biofilm and type III may be slightly less active. These data were confirmed in a second experiment.
Pyoverdine production by clinical P. aeruginosa isolates correlates with their antifungal activity.
To verify the clinical relevance of our observation that P. aeruginosa antifungal activity is related primarily to pyoverdine production, we compared the pyoverdine production (Fig. 11A) and antifungal activity (Fig. 11B) levels of 10 P. aeruginosa isolates derived from the lungs of CF patients. Our results showed a correlation between pyoverdine production and antifungal activity (Fig. 11C). Isolates producing small amounts of pyoverdine displayed low antifungal activity (isolates a, c, d, e, g, and k), while isolates with higher pyoverdine production showed increased antifungal activity (isolates b and i).
FIG 11.
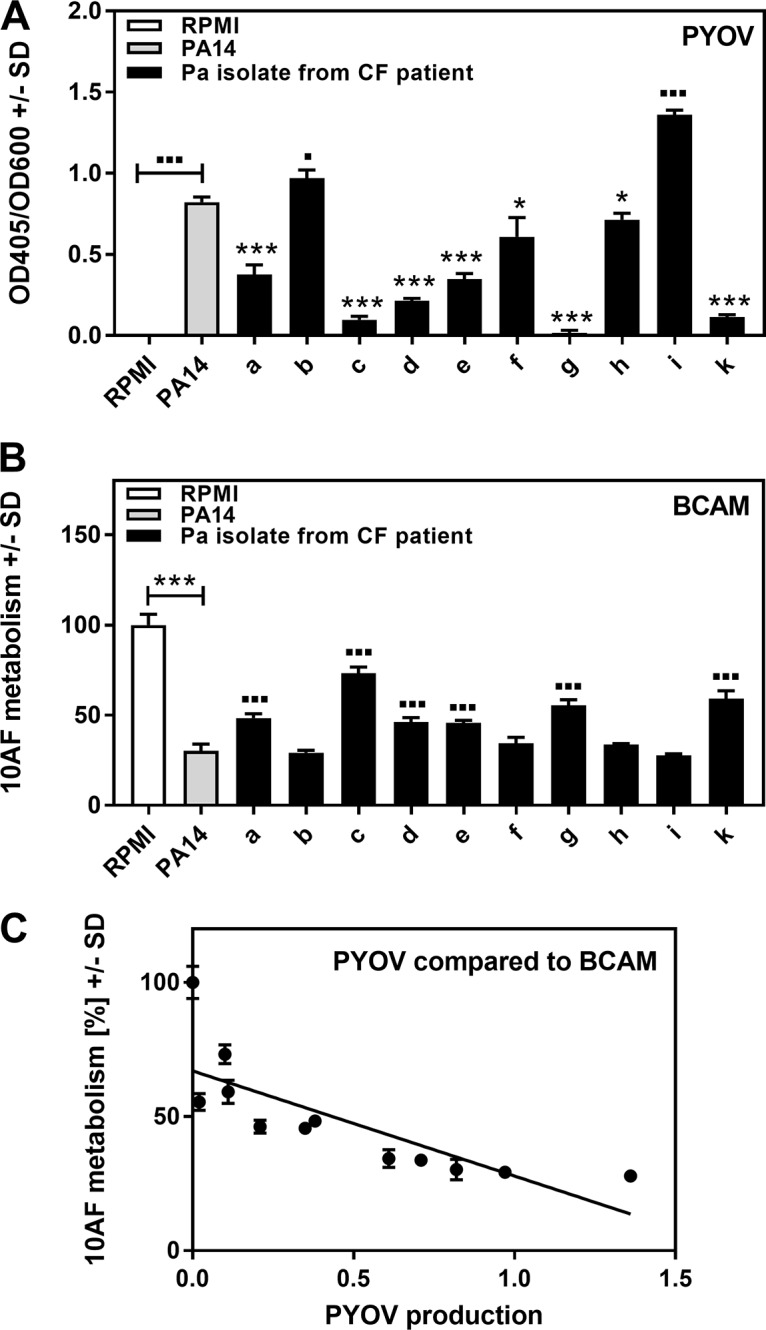
Correlation between pyoverdine production and antifungal activity of clinical P. aeruginosa (Pa) isolates. Ten CF isolates designated a to i and k, as well as PA14, were tested for pyoverdine (PYOV) production (A), as well as for activity against the metabolism of A. fumigatus strain 10AF forming biofilm (BCAM assay) (B). RPMI medium served as a control. Results of panels A and B were compared by linear regression curve analysis (panel C). Symbols: ■ or *, P ≤ 0.01; ■■■ or ***, P ≤ 0.001. Each comparison without brackets in panels A and B is PA14 versus 1 of the 10 CF isolates. Comparisons with brackets are as indicated. *, significant decreases; ■, significant increases. n = 3 for pyoverdine production. n = 5 for A. fumigatus strain 10AF metabolic activity.
Pyoverdine does not act as a xenosiderophore for A. fumigatus.
P. aeruginosa has been shown to be able to use several xenosiderophores of bacterial origin, as well as the fungal siderophore ferrichrome (reviewed in reference 39). To investigate whether pyoverdine or other P. aeruginosa siderophores could act as xenosiderophores for A. fumigatus, we performed a growth bioassay using the A. fumigatus sidA ftrA mutant (40) lacking both high-affinity iron uptake systems (siderophore biosynthesis and reductive iron assimilation). We incubated the mutant fungus in the presence of a large amount of iron (5 mM FeSO4), the A. fumigatus endogenous siderophore ferricrocin (FC), or the fusarinine-type siderophore triacetylfusarinine C (TAFC) (40), enabling growth of the sidA ftrA mutant (Fig. 12, rows 1 to 3). Incubation of the A. fumigatus sidA ftrA mutant with pyoverdine (Fig. 12, row 4) or PA14 PCF containing all PA14 siderophores or chelators (Fig. 12, row 5) did not enable fungal growth. Our results show that PA14 PCF does not contain xenosiderophores usable by A. fumigatus at concentrations necessary to overcome the antifungal effects of pyoverdine.
FIG 12.
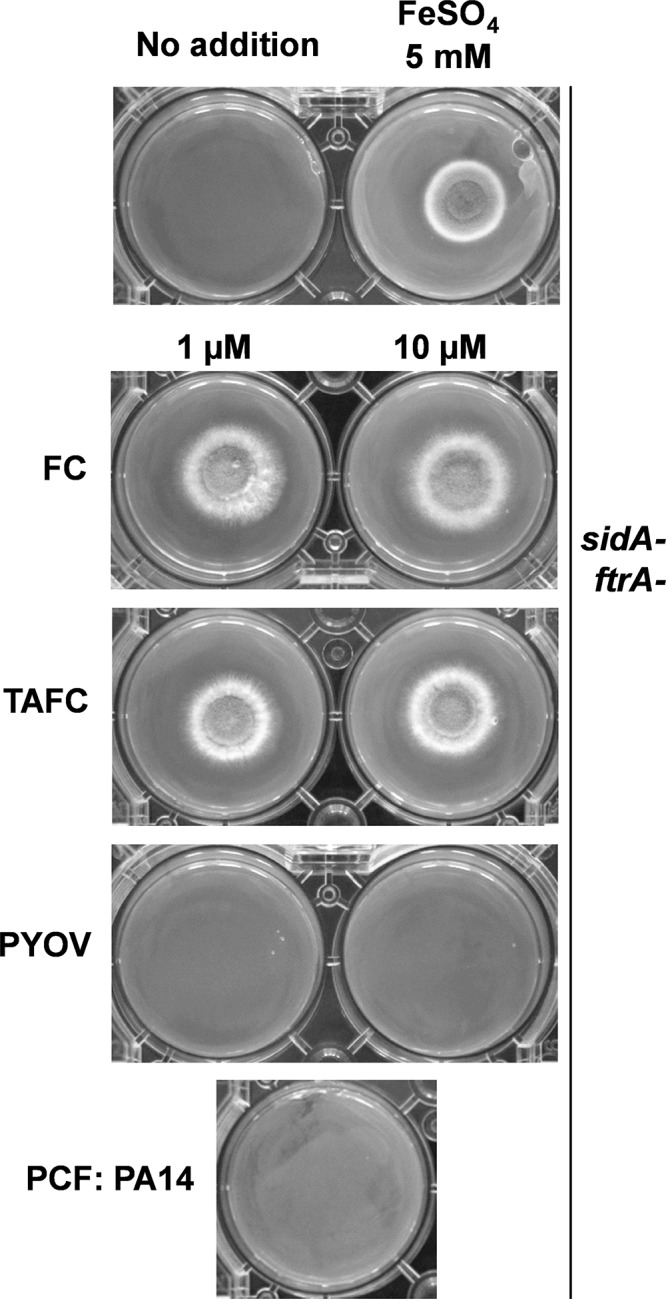
Measurement of xenosiderophores in PCF. A. fumigatus sidA ftrA mutant conidia (104) were point inoculated onto solid minimal medium agar with or without supplementation with FeSO4 to a final concentration of 5 mM, pyoverdine (PYOV) to final concentrations of 1 and 10 μM, TAFC or FC to a final concentration of 1 or 10 μM, or 600 μl of wild-type PA14 PCF. The plates were incubated for 48 h at 37°C.
DISCUSSION
P. aeruginosa has a relatively large genome compared to those of other bacteria and produces numerous extracellular products that are able to interfere with bacteria and fungi that might compete with P. aeruginosa in its ecological niches. Those niches include water and soil, as well as the human microbiome. Elaboration of these products explains the ubiquitous nature of P. aeruginosa and its success as a colonizer and pathogen. These products include toxins, as well as molecules that enable P. aeruginosa to compete with other microbes, such as molecules that can seize the crucial element iron from the environment or from the host and deny iron to these competitors (22, 39, 41, 42).
For decades (19), it has been known that P. aeruginosa can inhibit fungi, particularly A. fumigatus, a fungus that P. aeruginosa would encounter in human sinuses and airways, as well as in soil and water. A number of P. aeruginosa-produced molecules have been studied for the ability to inhibit A. fumigatus, as cited in the introduction, and the strengths and shortcomings of that approach have been discussed (such as the difficulty in knowing the relevance of the concentrations of materials studied in vitro to physiologic conditions). We have taken a different approach to investigate these questions, namely, to look at the effects of deletion mutations, to assess the importance of the subtraction of a gene on the intermicrobial activity against A. fumigatus. Because P. aeruginosa can modulate its metabolism and offensive weapons depending on the environment and the competitors (39, 43–46), we have studied the P. aeruginosa-A. fumigatus interaction in several different ways. We have studied the mutants in direct cellular interactions, as well the effects of extracellular P. aeruginosa products, and studied products of P. aeruginosa produced during planktonic growth, as well as from growth as P. aeruginosa biofilms. Initially, this was pursued in liquid in wells (47) and here in agar too. We have studied the effects on planktonic A. fumigatus and on A. fumigatus trying to form biofilm, as well as on preformed A. fumigatus biofilm. Biofilm formation by A. fumigatus leads to morphological and physiologic changes that produce a protected environment for A. fumigatus, with special importance for diseases such as CF (47–56).
A complexity of our approach is that some genes of P. aeruginosa regulate downstream pathways, where the elaboration of a number of products could be affected at once. Despite this complexity and despite mutations that affect many downstream pathways, a clear pattern emerged, where a limited number of mutants consistently appeared deficient in A. fumigatus inhibition in multiple settings. These were the pvdD, pvdD pchE, lasR, and lasR rhlR mutants (Table 5). The first two of these mutants pointed to the importance of P. aeruginosa siderophores, and especially a single such molecule, pyoverdine, in the interplay with A. fumigatus. The three classes of pyoverdines appear to have similar iron binding properties and appear to have similar levels of activity. Type II pyoverdine is the one prevalent in CF strains (57).
Some studies have indicated that pyoverdine may have antifungal activity independent of its activity as a P. aeruginosa siderophore (58), but our studies, demonstrating reversal of pyoverdine inhibition in the presence of small amounts of iron and A. fumigatus siderophore induction in the presence of pyoverdine, indicate that iron denial is the main mechanism by which pyoverdine inhibits A. fumigatus. Although pyoverdine regulates the production of virulence factors (32), our studies indicate the importance of iron chelation by pyoverdine itself as key. Residual antifungal activity of pyoverdine mutants was independent of the mutant‘s ability to produce PvdS-dependent virulence factors. We could also demonstrate that wild-type PA14 PCF, but not P. aeruginosa siderophore-deficient PCF, affected A. fumigatus iron metabolism at a molecular level, inducing genes that are known to be upregulated during iron denial, e.g., hapX (59, 60), and suppressing the expression of genes that are expressed under high-iron conditions, e.g., sreA (61).
The prominent appearance of the other two mutants, the lasR rhlR and lasR mutants, suggests that QS-regulated metabolites also play an important role in A. fumigatus inhibition. Many of these products have potential as intermicrobial inhibitors, but the advantage of studying downstream mutants as we did enables us to suggest that it is possible that the effect of the lasR rhlR and lasR mutations is a result of the loss of the combined activity of many of the downstream products, since few of the single downstream mutants appear as consistently impaired (Table 5). However, we note that both the lasR rhlR and lasR mutants had diminished pyoverdine production as well, which may help explain their reduced inhibitory activity. This was rather unexpected, since the initial observation (62) of reduced production of pyoverdine in a lasR mutant has never since been confirmed at the regulatory level, likely because the QS transcriptome of P. aeruginosa is also dependent on culture conditions (63).
For completeness, inhibition assays in 12-well plates had first shown the dramatic impairment of the pvdD pchE mutant (47). In the more extensive studies using the 96-well format, this mutant also had impaired activity against biofilm formation in all three assays (LC, PCF, and BF) (47). The lasR rhlR, pvdD, and rhlR mutants also appear to be impaired in some 96-well biofilm formation assays, as also described for the agar assays here. Also, for comparison, only in wells was assay of inhibition of preformed biofilm possible with LC, and those assays and the well assays with PCF and BCF revealed several impaired mutants, including the lasR and pvdD pchE mutants (47). The only other study of the interaction of P. aeruginosa mutants with A. fumigatus that we are aware of is a study of only the lasR and rhlR mutants (64), which were deficient in inhibition compared to the wild type. Our results are consistent with that report.
Perhaps the most striking absence of effects on antifungal activity was observed regarding the double phenazine mutant. This mutant was similar to the wild type in activity both initially in wells (47) and in the present studies. This observation is striking, because several in vitro studies had emphasized the inhibitory power of phenazines, including pyocyanin, on A. fumigatus (19–22), whereas complete subtraction of these molecules via mutation appeared to make little difference. This may suggest that the concentrations that have been previously studied in vitro are irrelevant, although we cannot exclude the upregulation of other factors in the mutant that could compensate for the loss of inhibiting activity. However, a recent study with another CF fungal pathogen has indicated that neither pyocyanin nor phenazine has a fungus inhibitory effect (65), and others have found pyocyanin to only be fungistatic (66).
In the present assays, we noted that there was some residual inhibition left for pyoverdine mutants. This suggests the presence of other inhibitors, which may contribute to the total fungal inhibition by wild-type P. aeruginosa. Some of these residuals in the pvdD pchE and pvdD mutants may correspond to the roster of absent molecules resulting from loss of QS-regulated metabolites in the lasR rhlR and lasR mutants, such as rhamnolipids (23). P. aeruginosa also produces iron chelators other than pyoverdine, such as pyochelin, PQS, and paerucumarin, but their absence appears to have a much lesser effect (Tables 1 and 5).
In lungs of CF patients, the microenvironment frequently contains A. fumigatus, as well as P. aeruginosa (15, 16). In our experiments, we used the well-studied A. fumigatus reference strain 10AF (29, 67–69). However, a recent survey found no differences among non-CF and CF A. fumigatus strains and included A. fumigatus strain 10AF, with respect to inhibition by P. aeruginosa products (70), indicating that the present findings are broadly applicable to A. fumigatus.
Using P. aeruginosa isolates from the lungs of 10 CF patients, we were able to demonstrate that the correlation between pyoverdine production and antifungal activity observed in PA14, PAO1, and PAK was also present in clinical samples. A prospective study would likely show that the ability to produce pyoverdine is correlated with clinical events and changes in the microbiome, contributing to a long-term prognosis for P. aeruginosa-A. fumigatus lung infections.
Although we were able to show in many assay settings that PA14 PCF has antifungal activity, there was the possibility that P. aeruginosa could also produce some profungal compounds, thus blunting the antifungal activity measured. A candidate class of such compounds is P. aeruginosa siderophores, which potentially could be used as xenosiderophores by A. fumigatus. Using bioassays measuring A. fumigatus growth, we did not find evidence of the use of P. aeruginosa siderophores by A. fumigatus.
The assays we used are a model system for a much more complex interplay in the in vivo setting. P. aeruginosa and A. fumigatus are competitors for iron in their microenvironment, and high pyoverdine expression antagonizes A. fumigatus metabolism and growth, which might support antifungal treatment. Most of the work presented here was performed under normoxic conditions. In CF progression, however, hypoxia may exist in focal lung sites (71–73). When experiments were repeated under hypoxic conditions, we observed the same outcomes, with pyoverdine being the principal mediator of antifungal activity on both forming and preformed A. fumigatus biofilms. This is consistent with our previous observations with nonmutant P. aeruginosa cells and filtrates under hypoxic conditions (74). As CF progresses, P. aeruginosa mutants frequently appear, and commonly these mutants are hypoproducers of pyoverdine (39). In the areas of hypoxia, the ratio of Fe3+ to Fe2+ decreases and P. aeruginosa depends more on the less avid Fe2+ transport into the bacterium for iron uptake via production of phenazines and a permease (39, 41, 75). The majority of P. aeruginosa isolates from CF patients we tested showed low pyoverdine expression. The resulting low antifungal activity of P. aeruginosa represents an unwanted advantage for A. fumigatus. Also, iron derived from blood, e.g., present in inflamed lungs, would decrease pyoverdine production and provide beneficial conditions for coexisting A. fumigatus. These conditions would be more favorable to A. fumigatus in the P. aeruginosa-A. fumigatus struggles and may help explain why A. fumigatus appears in CF airways later in the course of the disease (5, 6, 9, 10, 13, 14, 16). Evolution of P. aeruginosa may act in concert with other factors favoring A. fumigatus colonization, including repeated antibacterial courses and inhaled corticosteroids (76).
The present studies have focused on the effects of P. aeruginosa on A. fumigatus. As shown above, iron sequestering by pyoverdine leads to an increase in A. fumigatus siderophore excretion, providing further evidence that A. fumigatus is iron starved by pyoverdine. In subsequent studies, we are examining the countering effects of A. fumigatus. Adding A. fumigatus supernatants, presumably containing A. fumigatus siderophores, at the time of a pyoverdine challenge of A. fumigatus blunts the inhibitory effect of pyoverdine. An A. fumigatus sidA mutant, which fails to produce A. fumigatus siderophores, was found to be hypersusceptible to P. aeruginosa products and to pyoverdine. This is consistent with the findings of the importance of siderophores and pyoverdine in the P. aeruginosa mutants in the present study, as a siderophoreless A. fumigatus mutant that has a decreased ability to defend itself in the contest for iron would be expected to be hypersusceptible to P. aeruginosa's weapons. Our observations suggest that the competition between A. fumigatus and P. aeruginosa seems to rely on the relative amounts of siderophores produced by them, the speed at which they are produced, and their relative affinity for Fe.
MATERIALS AND METHODS
Materials.
Pyoverdine, 5FC, FeCl3, hemin, XTT, N-dodecyl-N,N-dimethyl-3-ammonio-1-propanesulfonate (DDAPS), and menadione were purchased from Sigma-Aldrich (St. Louis, MO). CAS was purchased from MP Biomedicals (Solon, OH). The type I (derived from P. aeruginosa ATCC 15692), type II (derived from P. aeruginosa ATCC 27853), and type III (derived from the P. aeruginosa strain Pa6) pyoverdines were purchased from EMC Microcollections GmbH, Tuebingen, Germany. The Sigma-Aldrich pyoverdine is of type I and was used throughout, except in experiments comparing the three types (obtained from EMC Microcollections GmbH).
Isolates.
The A. fumigatus strain used in these studies, 10AF, is a virulent patient isolate (67, 68) that can be obtained from the ATCC (ATCC 90240). The ATCC 46645 sidA ftrA mutant (40) lacks the high-affinity iron permease (FtrA) essential for reductive iron assimilation, as well as l-ornithine-N5-mono-oxygenase (SidA), which is essential for siderophore production. The use of all microbes in our lab was approved by the CIMR Biological Use Committee (approval no. 001-03Yr.11). P. aeruginosa isolates from CF patient respiratory cultures were obtained after written informed consent for biobanking and subsequent use of the patients' specimens, approved by the Stanford Institutional Review Board, was obtained (29). No patient names are associated with any of the organisms. PA14, a widely studied P. aeruginosa strain (77, 78), is the parental strain of all of the PA14 mutants studied here. The PA14 mutants investigated here, their descriptions and phenotypes, and references for production are shown in Table 1. P. aeruginosa strains PAO1 (ATCC 15692) and PAK (ATCC 25102) and the PAO1 pvdD mutant were kindly provided by P. R. Secor, Departments of Microbiology and Medicine, University of Washington, Seattle, WA. The PAO1 pvdA, pvdA fpvR, and pvdS mutants were generated and described previously (79). None of these PAO1 mutants is able to produce pyoverdine, whereas they differ in the production of ExoA and PrpL protease virulence factors. Briefly, the pvdA mutant shows reduced production of PvdS-dependent ExoA and PrpL protease virulence factors, the pvdA fpvR mutant is fully able to direct PvdS-dependent virulence factor production, and the pvdS mutant does not produce ExoA and PrpL protease.
PA14 mutants. (i) Construction of the markerless PA14 ΔrsmY ΔrsmZ (rsmY rsmZ) mutant.
Overnight cultures of the single PA14 ΔrsmY (receiver strain) mutant and the donor strain SM10 containing the pEXG2-rsmZ suicide plasmid (80) grown at 37°C were diluted to an optical density at 600 nm (OD600) of 0.05 and subcultured until the mid-exponential phase was reached (an OD600 of 0.5). Once the desired OD was achieved, 1 ml of each culture was washed twice with 1 ml of phosphate-buffered saline (PBS). A 50-μl volume of both the donor and receiver strains was plated on lysis broth (LB) agar and incubated overnight at 37°C. On the following day, the biomass was scraped off the plate and resuspended in 1 ml of PBS. Serial dilutions (100 to 10−6) were made, plated on LB agar supplemented with 100 μg/ml gentamicin and 25 μg/ml triclosan, and incubated overnight at 37°C. The colonies obtained were inoculated and incubated overnight at 37°C in LB supplemented with 100 μg/ml gentamicin for first recombinant screening. On the next day, a colony PCR assay of the gentamicin resistance cassette was performed to confirm the integration of the suicide plasmid. A second recombination was initiated by inoculating the first recombinants into LB with no salt (LBNS) and incubating them at 37°C overnight. On the following day, the cultures were serially diluted (100 to 10−7) and a 100-μl volume was plated on LBNS agar containing 10% sucrose. The plates were incubated overnight at 30°C. Once colonies were visible, they were replated on both LBNS supplemented with 10% sucrose and LBNS supplemented with 100 μg/ml gentamicin before overnight incubation at 37°C to screen for second recombinants. Colonies that grew only on LBNS containing 10% sucrose and not on LBNS plus 100 μg/ml gentamicin were selected and verified by colony PCR for the presence of a truncated gene and loss of the sacB gene.
(ii) Construction of the pvdD pchE double mutant.
A mutant fragment of pvdD containing a kanamycin resistance cassette flanked by flippase recognition sites (FRT) from pUC18-miniTn7T-kan-FRT was created by PCR overlap extension as described previously (81). The primers used to create the pvdD::kan-FRT fragment contained HindIII restriction sites, which allowed it to be digested and ligated into HindIII-digested pEX18Ap (82). The ligation product was then transformed into Escherichia coli DH5α, and positive clones were selected on 35 μg/ml kanamycin and 100 μg/ml carbenicillin. Confirmation was done by plasmid extraction (miniprep) and digestions with HindIII visualized by agarose gel electrophoresis. The confirmed pEX18Ap-pvdD::kan-FRT construct was then transformed into E. coli SM10 λpir with 100 μg/ml carbenicillin for selection. Replacement of the pvdD gene in the pchE::MrT7 mutant (83) was performed by a sacB-mediated strategy as described previously (82). Merodiploid selection was performed with 400 μg/ml kanamycin and 250 μg/ml carbenicillin. Ten percent sucrose was then used for counterselection of double recombinants on LBNS agar, and resistant colonies were screened for kanamycin resistance and carbenicillin sensitivity. Carbenicillin-sensitive clones were confirmed as pvdD pchE mutants by colony PCR. Absence of pyoverdine production was verified by cultivating selected clones in King's B medium and measuring fluorescence at 398 and 455 nm with a Cytation plate reader (BioTek, Winooski, VT).
Pseudomonas culture filtrate and cell production.
PA14 wild-type and mutant LC, PCF, and BCF were prepared as detailed previously (29). In brief, RPMI 1640 medium (Lonza, Walkersville, MD) was used and quantitated P. aeruginosa suspensions were grown for 24 h at 37°C. For BCF, a 2-h adhesion step and wash preceded 22 h of culture. PCF and BCF were filtered (0.22 μm) after the growth period.
Assays used to determine effects of Pseudomonas on Aspergillus.
In our initial studies, we studied A. fumigatus biofilm formation and preformed A. fumigatus biofilm and the mutants in plastic wells, initially 12-well plates, as previously described (29). This platform has advantages and disadvantages; the advantages include large volumes that enable more studies of the supernatants, and the disadvantages include greater consumption of materials per study and fewer replicates possible for each variable studied. In further developments, a 96-well plate assay was used as described previously (69), which had the advantage of an increased number of possible replicates and decreased consumption of reagents but required more precision in the delivery of smaller quantities of materials per well. Results of studies of the mutants in wells were presented in a previously published abstract and poster (47). However, even the 96-well format does not allow tests that would include a study of all of the mutants on the same plate, allowing the same controls for all; thus, the well studies are based on eight replicates per test condition studied but studied one to three times for each condition and not all on the same plate with the same controls. Because of these limitations, the assays on agar plates described below were developed, which overcome the above-described limitations and also are less cumbersome to perform. For that reason, although the well assay results are briefly summarized in the Discussion, for comparative purposes and completeness, the focus of the present paper is on the newer assays, which we regard as more robust for the concurrent screening of multiple isolates.
Forming (BCAM) and preformed (BHAM) A. fumigatus strain 10AF biofilm plate assays on agar.
A 1.25-g sample of Bacto agar (final experimental concentration, 1.25%; BD Biosciences, Durham, NC) was added to 25 ml of distilled water and autoclaved. After cooling to 56°C, 75 ml of RPMI 1640 medium and 2.5 × 104 A. fumigatus (strain 10AF) conidia/ml of agar were added. The conidium-containing agar was distributed into the inner 60 wells of sterile flat-bottom 96-well cell culture plates (Costar, Corning Inc., Corning, NY) at 100 μl/well. Upon agar solidification, plates were either loaded immediately with test articles (BCAM assays) or incubated at 37°C for 24 h before loading (BHAM assays). Immediately before loading, all of the empty peripheral wells of each 96-well plate were filled with 200 μl of sterile water to limit evaporation from the test wells. Test wells were loaded with 100 μl of test substances, e.g., P. aeruginosa wild-type or mutant planktonic or biofilm supernatant. On each supernatant test plate, all PA14 mutant and wild-type supernatants were present in duplicate. Control wells on each test plate contained 100 μl of RPMI 1640 medium, allowing the conversion of test results from each plate to percentages of the individual RPMI medium control (as 100%) for better comparison of results between plates. Loaded plates were incubated at 37°C for 24 h. On the agar, hyphal mats with biofilm form (48), as verified by optical microscopy, showing the same arrangement as has been studied in liquid wells, studied by optical, confocal, and electron microscopy (29, 83). Studies were also conducted in a hypoxic environment, as detailed previously (74). In brief, the assay plates were incubated in the Gas-Pak EZ Campy pouch system (Becton Dickinson, Franklin Lakes, NJ), generating an atmosphere of 10% oxygen (74). Plates were evaluated by XTT metabolic assay (29). Briefly, 100 μl of an XTT-menadione mixture (150 μg/ml XTT, 30 μM menadione) was added to each test well and incubated at 37°C for 30 min (BHAM assays) to 1 h (BCAM assays). Supernatant from each well (100 μl) was assayed with a plate reader (Opsys MR; DYNEX Technologies, Chantilly, VA).
Agar-based bioassay to measure effects of PA14 LC on A. fumigatus strain 10AF growth.
Bacto agar (3 g) was dissolved in 50 ml of distilled water and autoclaved. After cooling to 56°C, 150 ml of RPMI 1640 medium and 2.5 × 104 A. fumigatus strain 10AF conidia/ml of agar were added. The conidium-containing agar was distributed into a plastic bioassay dish (Thermo Scientific, Roskilde, Denmark). The agar-conidium mixture described above was sufficient for one plate. Upon agar solidification, wells were cut into the agar with a vacuum pump attached to a metal suction tool. Each plate was loaded with 40 μl (OD610 of 0.4) of wild-type or mutant PA14 LC suspension per well in duplicate. Loaded plates were incubated at 37°C for 24 h. Plates were evaluated by measuring the area of the fungus-free zone around each well; the area of the well was subtracted. A mean was calculated for each duplicate value on the same plate. Each plate contained all of the mutants, wild-type PA14, and RPMI medium negative controls. The experiment was performed four times, resulting in four inhibition zone area values per wild type/mutant. The mean of the wild-type inhibition zones was regarded as 100% inhibition, and the inhibition zones of the mutants were scored into relation to that.
For visualization of inhibition zones (Fig. S1), we used 60- by 15-mm petri dishes (Corning Inc., Corning, NY) containing 2 ml of RPMI 1640 agar and 2.5 × 104 A. fumigatus (strain 10AF) conidia/ml of agar. Upon solidification, a single well was punched into the middle of each plate and filled with a wild-type or mutant PA14 LC suspension. After 24 h of incubation at 37°C, inhibition zones were measured, plates were washed with PBS, and photographs were taken.
MIC assay.
MIC testing for wild-type and mutant PA14 PCF and BCF against planktonic A. fumigatus growth was performed by using the CLSI M38-A2 protocol for broth macrodilution (84). Briefly, an inoculum of 2 × 103 conidia/ml was added to 500 μl of fresh RPMI 1640 medium and combined with 500 μl of wild-type or mutant PA14 supernatant dilutions (1:2 to 1:2,048) in 5-ml polystyrene round-bottom tubes (BD Biosciences). Controls containing dilutions of 500 μl of RPMI 1640 instead of supernatants were added. A positive control contained 103 conidia in 1 ml of fresh RPMI 1640. The tubes were incubated at 37°C with shaking (100 rpm) for 48 h, and A. fumigatus strain 10AF growth was determined visually. The highest dilution with fungal growth diminished from the growth in the RPMI 1640 control was determined for wild-type and mutant supernatants. The experiment was performed four times.
Ranking of PA14 mutants.
For each of four experiments per assay, wild-type and mutant PA14 effects on A. fumigatus strain 10AF were expressed as percentages of the RPMI 1640 control (in BCAM and BHAM assays, the RPMI 1640 control = 100% = undisturbed metabolism or growth) or percentages of wild-type PA14 activity (for zone sizes; the MIC is wild-type PA14 activity = 100% = maximal antifungal effect). The results of the four experiments per assay were combined and statistically evaluated with Student's t test. Mutants with antifungal effects significantly different from that of wild-type PA14 were ranked by their mean values, with a rank of 1 assigned to the mutant with the lowest antifungal activity.
Pyoverdine measurement.
RPMI 1640 medium was incubated with PA14 (5 × 107 bacteria/ml) at 37°C and 100 rpm overnight. Bacterial growth was measured at 600 nm in a spectrophotometer (Genesys 20; Thermo Fisher Scientific Inc., Waltham, MA). All cultures were centrifuged at 200 × g for 30 min at room temperature. Supernatants were filtered (0.22 μm) to obtain sterile PCFs. For all PCFs, pyoverdine production was measured at 405 nm. Measurements were normalized to bacterial growth with the formula Relative PYOV production = OD405/OD600.
Northern blotting.
An A. fumigatus wild-type strain (AfS77; 106 conidia/ml) was grown in 15 ml of liquid 2YT medium at 37°C for 15 h (22). Subsequently, 10 ml of the culture supernatant was removed and replaced with 10 ml of culture supernatant of wild type or pvdA pvdE mutant P. aeruginosa PA14, respectively, and culturing was continued for another 3 h. For Northern blot analysis, RNA was isolated with TRI Reagent (Sigma); 10 μg of total RNA was separated in formaldehyde-containing agarose gels, blotted onto Hybond-N+ membranes (Amersham Biosciences), and hybridized with digoxigenin-labeled probes. The primers used for generation of the hybridization probes were described previously (22).
Liquid CAS assay.
We prepared 10× CAS assay reagent as described previously (85). One part 10× CAS reagent was combined with 9 parts siderophore-containing liquid, and the mixture was incubated at 37°C for 6 h. Mixtures were measured with a plate reader (Opsys MR; DYNEX Technologies) and compared to RPMI medium –1× CAS reagent as reference values.
Aspergillus growth assays.
The A. fumigatus sidA ftrA mutant strain (104 conidia) was point inoculated onto 2 ml of solid minimal medium (86) with or without supplementation with FeSO4 to a final concentration of 5 mM, pyoverdine to final concentrations of 1 and 10 μM, TAFC to final concentrations of 1 and 10 μM, or 600 μl of wild-type PA14 PCF. The plates were incubated for 48 h at 37°C.
Statistical analysis.
Results were analyzed with Student's t test if two groups were compared and one-way analysis of variance (ANOVA) combined with Tukey's posttest for multiple comparisons. All data in this study are expressed as the mean ± the standard deviation (SD). Data reported as percentages of the control value were compared with Student's t test after arcsin transformation of the proportions; these data are presented as percentages.
Supplementary Material
ACKNOWLEDGMENTS
We thank Marife Martinez for excellent technical support.
These studies were partially supported by a gift from John Flatley (CIMR no. 3770) and by a grant from the Child Health Research Institute, Stanford Transdisciplinary Initiatives Program (CIMR no. 3777). This work was partially supported by the Austrian Science Fund/Infect-ERA program (FWF grant I1616/Infect-ERA project AspMetNet to H.H.). A.-M.D. is an associate student of the HOROS doctoral program (W1253).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00345-17.
REFERENCES
- 1.Williams HD, Davies JC. 2012. Basic science for the chest physician: Pseudomonas aeruginosa and the cystic fibrosis airway. Thorax 67:465–467. doi: 10.1136/thoraxjnl-2011-201498. [DOI] [PubMed] [Google Scholar]
- 2.Smyth AR, Hurley MN. 2010. Targeting the Pseudomonas aeruginosa biofilm to combat infections in patients with cystic fibrosis. Drugs Future 35:1007–1014. doi: 10.1358/dof.2010.035.012.1537937. [DOI] [Google Scholar]
- 3.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Høiby N, Molin S. 2012. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol 10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 4.Sabino R, Ferreira JA, Moss RB, Valente J, Veríssimo C, Carolino E, Clemons KV, Everson C, Banaei N, Penner J, Stevens DA. 2015. Molecular epidemiology of Aspergillus collected from cystic fibrosis patients. J Cyst Fibros 14:474–481. doi: 10.1016/j.jcf.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Fillaux J, Brémont F, Murris M, Cassaing S, Rittié JL, Tétu L, Segonds C, Abbal M, Bieth E, Berry A, Pipy B, Magnaval JF. 2012. Assessment of Aspergillus sensitization or persistent carriage as a factor in lung function impairment in cystic fibrosis patients. Scand J Infect Dis 44:842–847. doi: 10.3109/00365548.2012.695454. [DOI] [PubMed] [Google Scholar]
- 6.Speirs JJ, van der Ent CK, Beekman JM. 2012. Effects of Aspergillus fumigatus colonization on lung function in cystic fibrosis. Curr Opin Pulm Med 18:632–638. doi: 10.1097/MCP.0b013e328358d50b. [DOI] [PubMed] [Google Scholar]
- 7.Ramsey KA, Ranganathan S, Park J, Skoric B, Adams AM, Simpson SJ, Robins-Browne RM, Franklin PJ, de Klerk NH, Sly PD, Stick SM, Hall GL, AREST CF. 2014. Early respiratory infection is associated with reduced spirometry in children with cystic fibrosis. Am J Respir Crit Care Med 190:1111–1116. doi: 10.1164/rccm.201407-1277OC. [DOI] [PubMed] [Google Scholar]
- 8.de Boer K, Vandemheen KL, Tullis E, Doucette S, Fergusson D, Freitag A, Paterson N, Jackson M, Lougheed MD, Kumar V, Aaron SD. 2011. Exacerbation frequency and clinical outcomes in adult patients with cystic fibrosis. Thorax 66:680–685. doi: 10.1136/thx.2011.161117. [DOI] [PubMed] [Google Scholar]
- 9.Nicolai T, Arleth S, Spaeth A, Bertele-Harms RM, Harms HK. 1990. Correlation of IgE antibody titer to Aspergillus fumigatus with decreased lung function in cystic fibrosis. Pediatr Pulmonol 8:12–15. doi: 10.1002/ppul.1950080106. [DOI] [PubMed] [Google Scholar]
- 10.Forsyth KD, Hohmann AW, Martin AJ, Bradley J. 1988. IgG antibodies to Aspergillus fumigatus in cystic fibrosis: a laboratory correlate of disease activity. Arch Dis Child 63:953–957. doi: 10.1136/adc.63.8.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schønheyder H, Jensen T, Høiby N, Andersen P, Koch C. 1985. Frequency of Aspergillus fumigatus isolates and antibodies to Aspergillus antigens in cystic fibrosis. Acta Pathol Microbiol Immunol Scand B 93:105–112. [DOI] [PubMed] [Google Scholar]
- 12.Coughlan CA, Chotirmall SH, Renwick J, Hassan T, Low TB, Bergsson G, Eshwika A, Bennett K, Dunne K, Greene CM, Gunaratnam C, Kavanagh K, Logan PM, Murphy P, Reeves EP, McElvaney NG. 2012. The effect of Aspergillus fumigatus infection on vitamin D receptor expression in cystic fibrosis. Am J Respir Crit Care Med 186:999–1007. doi: 10.1164/rccm.201203-0478OC. [DOI] [PubMed] [Google Scholar]
- 13.Mirković B, Lavelle GM, Azim AA, Helma K, Gargoum FS, Molloy K, Gernez Y, Dunne K, Renwick J, Murphy P, Moss RB, Greene CM, Gunaratnam C, Chotirmall SH, McElvaney NG. 2016. The basophil surface marker CD203c identifies Aspergillus species sensitization in patients with cystic fibrosis. J Allergy Clin Immunol 137:436–443. doi: 10.1016/j.jaci.2015.07.045. [DOI] [PubMed] [Google Scholar]
- 14.Baxter CG, Moore CB, Jones AM, Webb AK, Denning DW. 2013. IgE-mediated immune responses and airway detection of Aspergillus and Candida in adult cystic fibrosis. Chest 143:1351–1357. doi: 10.1378/chest.12-1363. [DOI] [PubMed] [Google Scholar]
- 15.Shoseyov D, Brownlee KG, Conway SP, Kerem E. 2006. Aspergillus bronchitis in cystic fibrosis. Chest 130:222–226. doi: 10.1378/chest.130.1.222. [DOI] [PubMed] [Google Scholar]
- 16.Amin R, Dupuis A, Aaron SD, Ratjen F. 2010. The effect of chronic infection with Aspergillus fumigatus on lung function and hospitalization in patients with cystic fibrosis. Chest 137:171–176. doi: 10.1378/chest.09-1103. [DOI] [PubMed] [Google Scholar]
- 17.de Bentzmann S, Plésiat P. 2011. The Pseudomonas aeruginosa opportunistic pathogen and human infections. Environ Microbiol 13:1655–1665. doi: 10.1111/j.1462-2920.2011.02469.x. [DOI] [PubMed] [Google Scholar]
- 18.Walsh TJ, Stevens DA. 2011. Aspergillosis, chapter 347. In Goldman L, Schafer A (ed), Goldman's Cecil medicine, 24th ed Elsevier, New York, NY. [Google Scholar]
- 19.Mangan A. 1969. Interactions between some aural Aspergillus species and bacteria. J Gen Microbiol 58:261–266. doi: 10.1099/00221287-58-2-261. [DOI] [PubMed] [Google Scholar]
- 20.Blyth W, Forey A. 1971. The influence of respiratory bacteria and their biochemical fractions on Aspergillus fumigatus. Sabouraudia 9:273–282. doi: 10.1080/00362177185190531. [DOI] [PubMed] [Google Scholar]
- 21.Kerr JR, Taylor GW, Rutman A, Høiby N, Cole PJ, Wilson R. 1999. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J Clin Pathol 52:385–387. doi: 10.1136/jcp.52.5.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Briard B, Bomme P, Lechner BE, Mislin GL, Lair V, Prévost MC, Latgé JP, Haas H, Beauvais A. 2015. Pseudomonas aeruginosa manipulates redox and iron homeostasis of its microbiota partner Aspergillus fumigatus via phenazines. Sci Rep 5:8220. doi: 10.1038/srep08220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Briard B, Rasoldier V, Bomme P, ElAouad N, Guerreiro C, Chassagne P, Muszkieta L, Latgé JP, Mulard L, Beauvais A. 2017. Dirhamnolipids secreted from Pseudomonas aeruginosa modify antifungal susceptibility of Aspergillus fumigatus by inhibiting β1,3 glucan synthase activity. ISME J 11:1578–1591. doi: 10.1038/ismej.2017.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorantla JN, Kumar SN, Nisha GV, Sumandu AS, Dileep C, Sudaresan A, Kumar MM, Lankalapalli RS, Kumar BS. 2014. Purification and characterization of antifungal phenazines from a fluorescent Pseudomonas strain FPO4 against medically important fungi. J Mycol Med 24:185–192. doi: 10.1016/j.mycmed.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Illakkiam D, Ponraj P, Shankar M, Muthusubramanian S, Rajendhran J, Gunasekaran P. 2013. Identification and structure elucidation of a novel antifungal compound produced by Pseudomonas aeruginosa PGPR2 against Macrophomina phaseolina. Appl Biochem Biotechnol 171:2176–2185. doi: 10.1007/s12010-013-0469-7. [DOI] [PubMed] [Google Scholar]
- 26.Bandara HM, K Cheung BP, Watt RM, Jin LJ, Samaranayake LP. 2013. Pseudomonas aeruginosa lipopolysaccharide inhibits Candida albicans hyphae formation and alters gene expression during biofilm development. Mol Oral Microbiol 28:54–69. doi: 10.1111/omi.12006. [DOI] [PubMed] [Google Scholar]
- 27.Moree WJ, Phelan VV, Wu CH, Bandeira N, Cornett DS, Duggan BM, Dorrestein PC. 2012. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc Natl Acad Sci U S A 109:13811–13816. doi: 10.1073/pnas.1206855109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith K, Rajendran R, Kerr S, Lappin DF, Mackay WG, Williams C, Ramage G. 2015. Aspergillus fumigatus enhances elastase production in Pseudomonas aeruginosa co-cultures. Med Mycol 53:645–655. doi: 10.1093/mmy/myv048. [DOI] [PubMed] [Google Scholar]
- 29.Ferreira JA, Penner JC, Moss RB, Haagensen JA, Clemons KV, Spormann AM, Nazik H, Cohen K, Banaei N, Carolino E, Stevens DA. 2015. Inhibition of Aspergillus fumigatus and its biofilm by Pseudomonas aeruginosa is dependent on the source, phenotype and growth conditions of the bacterium. PLoS One 10:e0134692. doi: 10.1371/journal.pone.0134692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cézard C, Farvacques N, Sonnet P. 2015. Chemistry and biology of pyoverdines, Pseudomonas primary siderophores. Curr Med Chem 22:165–186. doi: 10.2174/0929867321666141011194624. [DOI] [PubMed] [Google Scholar]
- 31.Meyer JM, Stintzi A, De Vos D, Cornelis P, Tappe R, Taraz K, Budzikiewicz H. 1997. Use of siderophores to type pseudomonads: the three Pseudomonas aeruginosa pyoverdine systems. Microbiology 143:35–43. doi: 10.1099/00221287-143-1-35. [DOI] [PubMed] [Google Scholar]
- 32.Lamont IL, Beare PA, Ochsner U, Vasil AI, Vasil ML. 2002. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 99:7072–7077. doi: 10.1073/pnas.092016999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imperi F, Massai F, Facchini M, Frangipani E, Visaggio D, Leoni L, Bragonzi A, Visca P. 2013. Repurposing the antimycotic drug flucytosine for suppression of Pseudomonas aeruginosa pathogenicity. Proc Natl Acad Sci U S A 110:7458–7463. doi: 10.1073/pnas.1222706110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nobre G. 1977. Sensitivity to 5-fluorocytosine and virulence for mice of some human isolates of Aspergillus. Mycopathologia 62:57–60. doi: 10.1007/BF00491997. [DOI] [PubMed] [Google Scholar]
- 35.Maines MD, Trakshel GM, Kutty RK. 1986. Characterization of two constitutive forms of rat liver microsomal heme oxygenase. Only one molecular species of the enzyme is inducible. J Biol Chem 261:411–419. [PubMed] [Google Scholar]
- 36.Maines MD. 1988. Heme oxygenase: multifunction, multiplicity, regulatory mechanisms and clinical application. FASEB J 2:2557–2568. [PubMed] [Google Scholar]
- 37.Ochsner UA, Johnson Z, Vasil ML. 2000. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146:185–198. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- 38.Barker KD, Barkovits K, Wilks A. 2012. Metabolic flux of extracellular heme uptake in Pseudomonas aeruginosa is driven by the iron-regulated heme oxygenase (HemO). J Biol Chem 287:18342–18350. doi: 10.1074/jbc.M112.359265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cornelis P, Dingemans J. 2013. Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front Cell Infect Microbiol 3:75. doi: 10.3389/fcimb.2013.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schrettl M, Bignell E, Kragl C, Joechl C, Rogers T, Arst HN Jr, Haynes K, Haas H. 2004. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J Exp Med 200:1213–1219. doi: 10.1084/jem.20041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen AT, Oglesby-Sherrouse AG. 2015. Spoils of war: iron at the crux of clinical and ecological fitness of Pseudomonas aeruginosa. Biometals 28:433–443. doi: 10.1007/s10534-015-9848-6. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen AT, Jones JW, Ruge MA, Kane MA, Oglesby-Sherrouse AG. 2015. Iron depletion enhances production of antimicrobials by Pseudomonas aeruginosa. J Bacteriol 197:2265–2275. doi: 10.1128/JB.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dixon EF, Hall RA. 2015. Noisy neighbourhoods: quorum sensing in fungal-polymicrobial infections. Cell Microbiol 17:1431–1441. doi: 10.1111/cmi.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toyofuku M, Uchiyama H, Nomura N. 2012. Social behaviours under anaerobic conditions in Pseudomonas aeruginosa. Int J Microbiol 2012:405191. doi: 10.1155/2012/405191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eichner A, Günther N, Arnold M, Schobert M, Heesemann J, Hogardt M. 2014. Marker genes for the metabolic adaptation of Pseudomonas aeruginosa to the hypoxic cystic fibrosis lung environment. Int J Med Microbiol 304:1050–1061. doi: 10.1016/j.ijmm.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 46.Tashiro Y, Yawata Y, Toyofuku M, Uchiyama H, Nomura N. 2013. Interspecies interaction between Pseudomonas aeruginosa and other microorganisms. Microbes Environ 28:13–24. doi: 10.1264/jsme2.ME12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nazik H, Penner J, Ferreira JAG, Clemons KV, Groleau M-C, Déziel E, Stevens DA. 2016. Study of Pseudomonas aeruginosa (Pa) mutants to indicate factors in Pseudomonas inhibition of Aspergillus fumigatus (Af) biofilm, abstr 39. 7th Advances Against Aspergillosis Conference, Manchester, United Kingdom, 3 to 5 March 2016 https://www.aspergillus.org.uk/content/study-pseudomonas-aeruginosa-pa-mutants-indicate-factors-pseudomonas-inhibition-aspergillus. [Google Scholar]
- 48.Beauvais A, Latgé J-P. 2015. Aspergillus biofilm in vitro and in vivo. Microbiol Spectr 3. doi: 10.1128/microbiolspec.MB-0017-2015. [DOI] [PubMed] [Google Scholar]
- 49.Seidler MJ, Salvenmoser S, Müller FM. 2008. Aspergillus fumigatus forms biofilms with reduced antifungal drug susceptibility on bronchial epithelial cells. Antimicrob Agents Chemother 52:4130–4136. doi: 10.1128/AAC.00234-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beauvais A, Schmidt C, Guadagnini S, Roux P, Perret E, Henry C, Paris S, Mallet A, Prévost MC, Latgé JP. 2007. An extracellular matrix glues together the aerial-grown hyphae of Aspergillus fumigatus. Cell Microbiol 9:1588–1600. doi: 10.1111/j.1462-5822.2007.00895.x. [DOI] [PubMed] [Google Scholar]
- 51.Beauvais A, Fontaine T, Aimanianda V, Latge JP. 2014. Aspergillus cell wall and biofilm. Mycopathologia 178:371–377. doi: 10.1007/s11046-014-9766-0. [DOI] [PubMed] [Google Scholar]
- 52.Kaur S, Singh S. 2014. Biofilm formation by Aspergillus fumigatus. Med Mycol 52:2–9. doi: 10.3109/13693786.2013.819592. [DOI] [PubMed] [Google Scholar]
- 53.Joubert LM, Ferreira JA, Stevens DA, Nazik H, McDonald KL, Cegelski L. 2017. Visualization of Aspergillus fumigatus biofilms with scanning electron microscopy and variable pressure-scanning electron microscopy: a comparison of processing techniques. J Microbiol Methods 132:46–55. doi: 10.1016/j.mimet.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 54.Loussert C, Schmitt C, Prevost MC, Balloy V, Fadel E, Philippe B, Kauffmann-Lacroix C, Latgé JP, Beauvais A. 2010. In vivo biofilm composition of Aspergillus fumigatus. Cell Microbiol 12:405–410. doi: 10.1111/j.1462-5822.2009.01409.x. [DOI] [PubMed] [Google Scholar]
- 55.Müller FM, Seidler M, Beauvais A. 2011. Aspergillus fumigatus biofilms in the clinical setting. Med Mycol 49(Suppl 1):S96–S100. doi: 10.3109/13693786.2010.502190. [DOI] [PubMed] [Google Scholar]
- 56.Reichhardt C, Stevens DA, Cegelski L. 2016. Fungal biofilm composition and opportunities in drug discovery. Future Med Chem 8:1455–1468. doi: 10.4155/fmc-2016-0049. [DOI] [PubMed] [Google Scholar]
- 57.De Vos D, De Chial M, Cochez C, Jansen S, Tümmler B, Meyer JM, Cornelis P. 2001. Study of pyoverdine type and production by Pseudomonas aeruginosa isolated from cystic fibrosis patients: prevalence of type II pyoverdine isolates and accumulation of pyoverdine-negative mutations. Arch Microbiol 175:384–388. doi: 10.1007/s002030100278. [DOI] [PubMed] [Google Scholar]
- 58.Lopez-Medina E, Fan D, Coughlin LA, Ho EX, Lamont IL, Reimmann C, Hooper LV, Koh AY. 2015. Candida albicans inhibits Pseudomonas aeruginosa virulence through suppression of pyochelin and pyoverdine biosynthesis. PLoS Pathog 11:e1005129. doi: 10.1371/journal.ppat.1005129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schrettl M, Beckmann N, Varga J, Heinekamp T, Jacobsen ID, Jöchl C, Moussa TA, Wang S, Gsaller Blatzer FM, Werner ER, Niermann WC, Brakhage AA, Haas H. 2010. HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog 6:e1001124. doi: 10.1371/journal.ppat.1001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gsaller F, Hortschansky P, Beattie SR, Klammer V, Tuppatsch K, Lechner BE, Rietzschel N, Werner ER, Vogan AA, Chung D, Mühlenhoff U, Kato M, Cramer RA, Brakhage AA, Haas H. 2014. The Janus transcription factor HapX controls fungal adaptation to both iron starvation and iron excess. EMBO J 33:2261–2276. doi: 10.15252/embj.201489468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schrettl M, Kim HS, Eisendle M, Kragl C, Nierman WC, Heinekamp T, Werner ER, Jacobsen I, Illmer P, Yi H, Brakhage AA, Haas H. 2008. SreA-mediated iron regulation in Aspergillus fumigatus. Mol Microbiol 70:27–43. doi: 10.1111/j.1365-2958.2008.06376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stintzi A, Evans K, Meyer JM, Poole K. 1998. Quorum-sensing and siderophore biosynthesis in Pseudomonas aeruginosa: lasR/lasI mutants exhibit reduced pyoverdine biosynthesis. FEMS Microbiol Lett 166:341–345. doi: 10.1111/j.1574-6968.1998.tb13910.x. [DOI] [PubMed] [Google Scholar]
- 63.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. 2003. Analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol 185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mowat E, Rajendran R, Williams C, McCulloch E, Jones B, Lang S, Ramage G. 2010. Pseudomonas aeruginosa and their small diffusible extracellular molecules inhibit Aspergillus fumigatus biofilm formation. FEMS Microbiol Lett 313:96–102. doi: 10.1111/j.1574-6968.2010.02130.x. [DOI] [PubMed] [Google Scholar]
- 65.Kaur J, Pethani BP, Kumar S, Kim M, Sunna A, Kautto L, Penesyan A, Paulsen IT, Nevalainen H. 2015. Pseudomonas aeruginosa inhibits the growth of Scedosporium aurantiacum, an opportunistic fungal pathogen isolated from the lungs of cystic fibrosis patients. Front Microbiol 6:866. doi: 10.3389/fmicb.2015.00866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rella A, Yang MW, Gruber J, Montagna MT, Luberto C, Zhang YM, Del Poeta M. 2012. Pseudomonas aeruginosa inhibits the growth of Cryptococcus species. Mycopathologia 173:451–461. doi: 10.1007/s11046-011-9494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Denning DW, Clemons KV, Hanson LH, Stevens DA. 1990. Restriction endonuclease analysis of total cellular DNA of Aspergillus fumigatus isolates of geographically and epidemiologically diverse origin. J Infect Dis 162:1151–1158. doi: 10.1093/infdis/162.5.1151. [DOI] [PubMed] [Google Scholar]
- 68.Denning DW, Stevens DA. 1991. Efficacy of cilofungin alone and in combination with amphotericin B in a murine model of disseminated aspergillosis. Antimicrob Agents Chemother 35:1329–1333. doi: 10.1128/AAC.35.7.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nazik H, Penner JC, Ferreira JA, Haagensen JA, Cohen K, Spormann AM, Martinez M, Chen V, Hsu JL, Clemons KV, Stevens DA. 2015. Effects of iron chelators on the formation and development of Aspergillus fumigatus biofilm. Antimicrob Agents Chemother 59:6514–6520. doi: 10.1128/AAC.01684-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nazik H, Moss RB, Karna V, Clemons KV, Banaei N, Cohen K, Choudhary V, Stevens DA. 2017. Are cystic fibrosis Aspergillus fumigatus isolates different? Intermicrobial interactions with Pseudomonas. Mycopathologia 182:315–318. doi: 10.1007/s11046-016-0087-3. [DOI] [PubMed] [Google Scholar]
- 71.Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Weiss T, Botzenhart K, Yankaskas JR, Randell S, Boucher RC, Döring GJ. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest 109:317–325. doi: 10.1172/JCI0213870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cowley ES, Kopf SH, LaRiviere A, Ziebis W, Newman DK. 2015. Pediatric cystic fibrosis sputum can be chemically dynamic, anoxic, and extremely reduced due to hydrogen sulfide formation. mBio 6:e00767. doi: 10.1128/mBio.00767-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lambiase A, Catania MR, Rossano F. 2010. Anaerobic bacteria infection in cystic fibrosis airway disease. New Microbiol 33:185–194. [PubMed] [Google Scholar]
- 74.Anand R, Clemons KV, Stevens DA. 2017. Effect of anaerobiasis or hypoxia on Pseudomonas aeruginosa inhibition of Aspergillus fumigatus biofilm. Arch Microbiol 199:881–890. doi: 10.1007/s00203-017-1362-5. [DOI] [PubMed] [Google Scholar]
- 75.Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. 2006. Feo—transport of ferrous iron into bacteria. Biometals 19:143–157. doi: 10.1007/s10534-006-0003-2. [DOI] [PubMed] [Google Scholar]
- 76.Noni M, Katelari A, Dimopoulos G, Kourlaba G, Spoulou V, Alexandrou-Athanassoulis H, Doudounakis SE, Tzoumaka-Bakoula C. 2014. Inhaled corticosteroids and Aspergillus fumigatus isolation in cystic fibrosis. Med Mycol 52:715–722. doi: 10.1093/mmy/myu038. [DOI] [PubMed] [Google Scholar]
- 77.O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 78.Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S, Diggins LT, He J, Saucier M, Déziel E, Friedman L, Li L, Grills G, Montgomery K, Kucherlapati R, Rahme LG, Ausubel FM. 2006. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol 7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Minandri F, Imperi F, Frangipani E, Bonchi C, Visaggio D, Facchini M, Pasquali P, Bragonzi A, Visca P. 2016. Role of iron uptake systems in Pseudomonas aeruginosa virulence and airway infection. Infect Immun 84:2324–2335. doi: 10.1128/IAI.00098-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S. 2009. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol 73:434–445. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choi KH, Schweizer HP. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol 5:30. doi: 10.1186/1471-2180-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 83.Reichhardt C, Ferreira JA, Joubert LM, Clemons KV, Stevens DA, Cegelski L. 2015. Analysis of the Aspergillus fumigatus biofilm extracellular matrix by solid-state nuclear magnetic resonance spectroscopy. Eukaryot Cell 14:1064–1072. doi: 10.1128/EC.00050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard, 2nd ed CLSI document M38-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 85.Andrews MY, Santelli CM, Duckworth OW. 2016. Layer plate CAS assay for the quantitation of siderophore production and determination of exudation patterns for fungi. J Microbiol Methods 121:41–43. doi: 10.1016/j.mimet.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 86.Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AW. 1953. The genetics of Aspergillus nidulans. Adv Genet 5:141–238. [DOI] [PubMed] [Google Scholar]
- 87.Déziel E, Lépine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A 101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cao H, Krishnan G, Goumnerov B, Tsongalis J, Tompkins R, Rahme LG. 2001. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc Natl Acad Sci U S A 98:14613–14618. doi: 10.1073/pnas.251465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wade DS, Calfee MW, Rocha ER, Ling EA, Engstrom E, Coleman JP, Pesci EC. 2005. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J Bacteriol 187:4372–4380. doi: 10.1128/JB.187.13.4372-4380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiao G, Déziel E, He J, Lépine F, Lesic B, Castonguay MH, Milot S, Tampakaki AP, Stachel SE, Rahme LG. 2006. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol Microbiol 62:1689–1699. doi: 10.1111/j.1365-2958.2006.05462.x. [DOI] [PubMed] [Google Scholar]
- 91.Dekimpe V, Déziel E. 2009. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 155:712–723. doi: 10.1099/mic.0.022764-0. [DOI] [PubMed] [Google Scholar]
- 92.Petrova OE, Sauer K. 2010. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J Bacteriol 192:5275–5288. doi: 10.1128/JB.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Déziel E, Gopalan S, Tampakaki AP, Lépine F, Padfield KE, Saucier M, Xiao G, Rahme LG. 2005. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-l-homoserine lactones. Mol Microbiol 55:998–1014. [DOI] [PubMed] [Google Scholar]
- 95.Lesic B, Starkey M, He J, Hazan R, Rahme LG. 2009. Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology 155:2845–2855. doi: 10.1099/mic.0.029082-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ochsner UA, Fiechter A, Reiser J. 1994. Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J Biol Chem 269:19787–19795. [PubMed] [Google Scholar]
- 97.Lépine F, Déziel E, Milot S, Rahme LG. 2003. A stable isotope dilution assay for the quantification of the Pseudomonas quinolone signal in Pseudomonas aeruginosa cultures. Biochim Biophys Acta 1622:36–41. doi: 10.1016/S0304-4165(03)00103-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.