ABSTRACT
We previously produced a heavy-chain-only antibody (Ab) VH domain (VHH)-displayed phage library from two alpacas that had been immunized with ricin toxoid and nontoxic mixtures of the enzymatic ricin toxin A subunit (RTA) and binding ricin toxin B subunit (RTB) (D. J. Vance, J. M. Tremblay, N. J. Mantis, and C. B. Shoemaker, J Biol Chem 288:36538–36547, 2013, https://doi.org/10.1074/jbc.M113.519207). Initial and subsequent screens of that library by direct enzyme-linked immunosorbent assay (ELISA) yielded more than two dozen unique RTA- and RTB-specific VHHs, including 10 whose structures were subsequently solved in complex with RTA. To generate a more complete antigenic map of ricin toxin and to define the epitopes associated with toxin-neutralizing activity, we subjected the VHH-displayed phage library to additional “pannings” on both receptor-bound ricin and antibody-captured ricin. We now report the full-length DNA sequences, binding affinities, and neutralizing activities of 68 unique VHHs: 31 against RTA, 33 against RTB, and 4 against ricin holotoxin. Epitope positioning was achieved through cross-competition ELISAs performed with a panel of monoclonal antibodies (MAbs) and verified, in some instances, with hydrogen-deuterium exchange mass spectrometry. The 68 VHHs grouped into more than 20 different competition bins. The RTA-specific VHHs with strong toxin-neutralizing activities were confined to bins that overlapped two previously identified neutralizing hot spots, termed clusters I and II. The four RTB-specific VHHs with potent toxin-neutralizing activity grouped within three adjacent bins situated at the RTA-RTB interface near cluster II. These results provide important insights into epitope interrelationships on the surface of ricin and delineate regions of vulnerability that can be exploited for the purpose of vaccine and therapeutic development.
KEYWORDS: antibody, biodefense, epitope, neutralizing, toxins, vaccines
INTRODUCTION
Ricin is the prototype of the type II ribosome-inactivating protein (RIP) family of toxins (1). Ricin toxin A subunit (RTA) is an RNA N-glycosidase that catalyzes the hydrolysis of a conserved adenine residue within the sarcin/ricin loop (SRL) of 28S rRNA, resulting in ribosome arrest and apoptosis (2–4). Ricin toxin B subunit (RTB) is a galactose- and N-acetylgalactosamine (Gal/GalNAc)-specific lectin that promotes toxin entry into mammalian cells, including the epithelial cells that line the respiratory tract (5, 6). Following endocytosis, RTB mediates the retrograde transport of ricin to the trans-Golgi network (TGN) and endoplasmic reticulum (ER). Once within the ER, the disulfide bond that links RTA to RTB is reduced by protein disulfide isomerase and RTA is retrotranslocated (dislocated) into the cell cytoplasm, where it triggers programmed cell death (7, 8). Ricin is classified as a category B biothreat agent by the Centers for Disease Control and Prevention (CDC) because of its potential, when delivered via aerosol, to induce severe lung inflammation and tissue damage (9, 10). Concerns over ricin's possible use as a biothreat agent stem from the fact that the toxin is relatively easy to procure from its natural source, castor beans (Ricinus communis), as well as from its extreme toxicity following inhalation or injection (11).
As part of a longstanding effort to engineer ricin antitoxins and immunotherapies, we previously produced a library of phage-displayed, heavy-chain-only antibodies (Abs; VHHs) from two alpacas that had been hyperimmunized with nontoxic mixtures of recombinant RTA, RTB, and ricin toxoid; the library is referred to here as “HobJo” in reference to the names of the two immunized alpacas, Hobbs and Johnnes (12). In our initial study, the phage-displayed VHH library was subjected to high- and low-stringency “panning” on purified native RTA or RTB that had been immobilized on plastic substrates. This initial screening strategy yielded 11 unique RTA-specific VHHs and 9 unique RTB-specific VHHs. Five (4 RTA-specific and 1 RTB-specific) VHHs among the original 20 had strong or moderate in vitro toxin-neutralizing activity (TNA), while the remaining 15 had weak TNA or no detectable TNA. A collection of homo- and heterodimeric (i.e., bispecific) VHHs were engineered with these original 20 VHHs and shown to have enhanced avidities for ricin and, in some cases, the capacity to passively protect mice from lethal-dose ricin challenge (12–15). As discussed below, the X-ray crystal structures of 10 different VHH-RTA complexes, including 7 VHHs from the original screen (16–18), have been solved to date.
The original strategy of panning the VHH library onto RTA- and RTB-coated immunotubes presented several drawbacks. First, there was a bias for VHHs directed against a single immunodominant region on RTA that we refer to as cluster I. Cluster I is defined by the murine monoclonal antibody (MAb) PB10, which recognizes a linear epitope encompassing α-helix B (residues 94 to 107) of RTA (see Fig. S1 in the supplemental material) (19–21). α-Helix B is a secondary motif conserved across the RIP family (22). Approximately half of the original RTA-specific VHHs recognized epitopes in cluster I, based on competition enzyme-linked immunosorbent assays (ELISAs) performed with PB10. The second shortcoming associated with the original screen was that it gave rise to VHHs that recognized cryptic epitopes that are exposed when RTA and RTB are adsorbed to polystyrene but that are not accessible to antibodies when ricin is in its native conformation. Finally, by performing screening with individual subunits, rather than with the holotoxin, we were likely missing antibodies directed against quaternary epitopes. With the long-term goal of generating a saturating B cell epitope map of ricin toxin using the HobJo library, which is estimated to contain ∼4 × 106 independent clones, we reasoned that it would be necessary to derive novel panning strategies.
To generate a more complete antigenic map of ricin toxin and to better define the epitopes associated with neutralizing activity, we have now subjected the HobJo VHH-displayed phage library to pannings on both receptor-bound and antibody-captured ricin holotoxins or subunits. We identified a total of 68 unique ricin-specific VHHs (31 against RTA, 33 against RTB, and 4 against ricin holotoxin) and present their full-length DNA sequences, binding affinities, and neutralizing activities. We used cross-competition ELISAs with a panel of well-characterized murine monoclonal antibodies (MAbs) as a strategy to localize the epitopes recognized by the new collection of VHHs. Epitope positioning was validated with available X-ray crystal structures and, in some instances, was followed up with hydrogen-deuterium mass spectrometry (HX-MS). We identified more than 20 different competition bins (a bin was comprised of all VHHs which shared the same competition profile) that cover >50% of RTA's surface area and an indefinite amount of RTB. Having this comprehensive collection of well-characterized, ricin-specific VHHs provides important insights into epitope interrelationships on the surface of ricin and delineates two or possibly three regions of vulnerability that can be exploited for the purpose of vaccine and therapeutic development.
RESULTS
In an effort to obtain a comprehensive map of toxin-neutralizing and nonneutralizing B cell epitopes on the surface of ricin, the HobJo VHH phage display library was subjected to nine different screens (Table 1). Screens 1 to 4 involved panning on immunotubes coated with RTA or RTB and yielded a total of 36 ricin-specific VHHs (Tables 1 and 2). The 20 VHHs resulting from screens 1 and 2 were described previously and included 11 against RTA (6 with TNA) and 9 against RTB (1 with TNA) (12). We successfully solved the X-ray crystal structures of seven of those VHHs in complex with RTA (16–18). Screens 3 and 4 yielded an additional 16 ricin-specific VHHs, including 7 against RTA, 1 of which was successfully crystallized (17), and 9 against RTB (Table 1).
TABLE 1.
Summary of HobJo screens
| Screenc | Target(s)d | No. of VHHs with indicated specificitya |
TNAb |
||||||
|---|---|---|---|---|---|---|---|---|---|
| A | B | R | Total | +++ | ++ | + | − | ||
| 1 | RTA | 11 | 0 | 0 | 11 | 2 | 2 | 2 | 5 |
| 2 | RTB | 0 | 9 | 0 | 9 | 1 | 0 | 0 | 8 |
| 3 | RTA | 7 | 0 | 0 | 7 | 0 | 2 | 2 | 3 |
| 4 | RTB | 0 | 9 | 0 | 9 | 0 | 0 | 0 | 9 |
| 5 | R-ASF | 5 | 0 | 1 | 6 | 0 | 1 | 1 | 4 |
| 6 | R-PB10 | 5 | 2 | 0 | 7 | 1 | 0 | 1 | 5 |
| 7 | Peptides | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | RTB-ASF | 0 | 5 | 0 | 5 | 0 | 0 | 1 | 4 |
| 9 | R-F5 + F6 | 4e | 8 | 3 | 15e | 3 | 2 | 0 | 9 |
| Total | 31 | 33 | 4 | 68e | 7 | 7 | 7 | 47 | |
A, RTA; B, RTB; R, ricin holotoxin.
Toxin neutralizing activity, based on IC50, indicated as follows: +++, IC50 < 10 nM; ++, IC50 < 100 nM; +, IC50 < 500 nM; −, IC50 > 500 nM or undetectable.
VHHs in screens 1 and 2 were reported previously (12).
R-ASF, ricin captured by ASF; R-PB10, ricin captured by PB10; RTB-ASF, RTB captured by ASF; R-F5 + F6, ricin captured with VHH JIV-F5 and then blocked with excess JIV-F6.
One anti-RTA VHH from screen 9 was not successfully expressed and thus was not characterized for TNA and also was not included in the final VHH total of 68.
TABLE 2.
Properties of ricin-specific VHHs described in this study
| VHH | Screena | GenBank accession no. | PDB IDb | TNAc | KD (nM)d | Specificitye | Cluster(s)f | Familyg |
|---|---|---|---|---|---|---|---|---|
| JIV-F5 | 1 | KF746018 | 4Z9K | +++ | 0.019 | A | I | |
| JIV-F6 | 1 | KF746019 | − | 1.86 | A | III | JIV-F6 | |
| JIV-G12 | 1 | KF746020 | 4LGS | + | 0.129 | A | I | |
| JIY-A7 | 1 | KF746021 | 4LHQ | − | 4.3 | A | I | JIY-A7 |
| JIY-D9 | 1 | KF746022 | − | 0.174 | A | I + II | JIY-D9 | |
| JIY-D10 | 1 | KF746023 | 4LGR | ++ | 0.114 | A | I + II | |
| JIY-E1 | 1 | KF746024 | 5BOZ | + | 0.686 | A | II | |
| JIY-E3 | 1 | KF746025 | − | 0.296 | A | I | ||
| JIY-E5 | 1 | KF746026 | 4LGP | +++ | 0.191 | A | I + IV | |
| JIY-F10 | 1 | KF746027 | − | NB | A' | |||
| JIY-G11 | 1 | KF746028 | 4LHJ | ++ | 0.553 | A | I | |
| JIW-B1 | 2 | KF746029 | − | NB | B' | |||
| JIW-C12 | 2 | KF746030 | − | NB | B' | |||
| JIW-D12 | 2 | KF746031 | − | NB | B' | JIZ-B7 | ||
| JIW-G5 | 2 | KF746032 | − | NB | B' | |||
| JIW-G10 | 2 | KF746033 | − | NB | B' | JIW-G10 | ||
| JIZ-B7 | 2 | KF746034 | +++ | 0.031 | B | SII | JIZ-B7 | |
| JIZ-B9 | 2 | KF746035 | − | NB | B' | |||
| JIZ-D8 | 2 | KF746036 | − | NB | B' | |||
| JIZ-G4 | 2 | KF746037 | − | NB | B' | |||
| JNM-A11 | 3 | KY824576 | − | 0.200 | A | I | ||
| JNM-B5 | 3 | KY824577 | − | 0.060 | A | I | ||
| JNM-C12 | 3 | KY824578 | ++ | 0.099 | A | I + IV | ||
| JNM-D1 | 3 | KY824579 | − | 1.19 | A | II + III | ||
| JNM-E4 | 3 | KY824580 | ++ | 0.083 | A | I | ||
| JNM-F8 | 3 | KY824581 | 5E1H | + | 0.203 | A | I | |
| JNM-G4 | 3 | KY824582 | + | 0.128 | A | I | JIY-A7 | |
| JNN-A3 | 4 | KY824583 | − | NB | B' | JIW-G10 | ||
| JNN-B6 | 4 | KY824584 | − | NB | B' | |||
| JNN-B8 | 4 | KY824585 | − | NB | B' | JIZ-B7 | ||
| JNN-C5 | 4 | KY824586 | − | NB | B' | |||
| JNN-C8 | 4 | KY824587 | − | NB | B' | |||
| JNN-D1 | 4 | KY824588 | − | NB | B' | JIZ-B7 | ||
| JNN-D8 | 4 | KY824589 | − | NB | B' | |||
| JNN-E4 | 4 | KY824590 | − | NB | B' | |||
| JNN-H9 | 4 | KY824591 | − | NB | B' | |||
| V1B4 | 5 | KY824592 | − | 4.09 | R | SII | ||
| V1B10 | 5 | KY824593 | + | 0.917 | A | III | JIV-F6 | |
| V1B11 | 5 | KY824594 | − | 8.84 | A | I + III | ||
| V1C7 | 5 | KY824595 | 5J56 | − | 3.19 | A | II | V1C7 |
| V1D3 | 5 | KY824596 | ++ | 0.460 | A | III | JIV-F6 | |
| V1G6 | 5 | KY824597 | − | 5.34 | A | III | V2G10 | |
| V2A11 | 6 | KY824598 | − | 1.82 | A | III | ||
| V2B8 | 6 | KY824599 | − | 32.4 | A | I + II | JIY-D9 | |
| V2B9 | 6 | KY824600 | − | 1.03 | A | II | V1C7 | |
| V2C11 | 6 | KY824601 | +++ | 0.015 | B | SII | V2C11 | |
| V2D4 | 6 | KY824602 | + | 3.2 | B | SII | V2C11 | |
| V2E8 | 6 | KY824603 | − | 5.74 | A | II | V1C7 | |
| V2G10 | 6 | KY824604 | − | 1.16 | A | III | V2G10 | |
| V4A1 | 8 | KY824605 | + | 0.667 | B | SII | ||
| V4C6 | 8 | KY824606 | − | NB | B | V4C6 | ||
| V4C8 | 8 | KY824607 | − | NB | B | V4C6 | ||
| V4D12 | 8 | KY824608 | − | NB | B | |||
| V4G3 | 8 | KY824609 | − | NB | B | |||
| V5A2 | 9 | KY824610 | − | 1.46 | A | III | ||
| V5B1 | 9 | KY824611 | − | 2.17 | B | |||
| V5B6 | 9 | KY824612 | − | 0.668 | B | |||
| V5C1 | 9 | KY824613 | ++ | 0.301 | A | II | V1C7 | |
| V5C4 | 9 | KY824614 | − | 5.01 | B | |||
| V5D1 | 9 | KY824615 | ++ | 0.216 | R | SII | ||
| V5D5 | 9 | KY824616 | − | 5.54 | B | |||
| V5E1 | 9 | KY824617 | 5J57 | +++ | 0.019 | A | II | |
| V5E4 | 9 | KY824618 | +++ | 0.006 | B | SII | ||
| V5G1 | 9 | KY824619 | +++ | 0.106 | B | SII | ||
| V5G6 | 9 | KY824620 | − | 1.24 | R | SII | ||
| V5G12 | 9 | KY824621 | − | 0.994 | R | SII | ||
| V5H2 | 9 | KY824622 | − | 2.18 | B | SII | V2C11 | |
| V5H6 | 9 | KY824623 | − | 23.2 | B |
VHHs in screens 1 and 2 were reported previously (12).
Protein Data Bank IDs.
Toxin-neutralizing activity (TNA), as described in the legend of Table 1.
NB, no binding detected in SPR.
A, RTA; A′, RTA′; B, RTB; B′, RTB′; R, ricin holotoxin.
Data represent RTA epitope cluster(s) (I to IV) bound by a particular VHH. SII refers to “supercluster II” and is the designation given to RTB- or holotoxin-specific VHHs that are subject to competition from RTA binding cluster II MAb SyH7.
See Table 3 for descriptions of families.
Screens 5 and 6 were designed to circumvent two issues that we had encountered in screens 1 to 4. First, roughly half of the 36 VHHs from screens 1 to 4 failed to bind soluble ricin (R), even though they recognized plate-adsorbed ricin (R′), RTA (A′), or RTB (B′). This finding is consistent with the exposure of cryptic epitopes on protein antigens under conditions of binding to polystyrene surfaces (23, 24). Second, the majority of the RTA-specific VHHs were directed against cluster I, which is defined functionally by competition with MAb PB10 and is known to be immunodominant (12). To overcome these issues, the HobJo library was panned, in screen 5, on ricin captured with asialofetuin (R-ASF) to approximate the conformation of soluble ricin and, in screen 6, on ricin that was captured with PB10 (R-PB10) to mask immunodominant epitopes within cluster I. VHHs identified in screen 5 were cross-checked for binding to R-PB10, while VHHs identified in screen 6 were cross-checked for reactivity with R-ASF. Only VHHs that were positive in both assays were characterized further (Tables 1 and 2). This strategy yielded a total of 56 VHHs that were reactive with both R-ASF and R-PB10. After we eliminated siblings from the pool, a total of 13 VHHs remained: 10 against RTA, 2 against RTB, and 1 specific for ricin holotoxin.
Screen 7 was designed to identify VHHs that recognize two different linear epitopes on RTA defined by the murine toxin-neutralizing MAbs PB10 and SyH7 (19). Two biotin-tagged 18-mer peptides, E12 and G3, corresponding to the core epitopes of PB10 and SyH7, respectively, were captured on immunotubes via the use of avidin and then subjected to panning with the HobJo library. The screen proved unsuccessful, however, as none of the ∼100 clones that we examined bound to ricin or either of the E12 or G3 peptides (data not shown). The failure to obtain E12- or G3-specific VHHs was not completely surprising, considering that there are very few examples of single-domain antibodies that recognize linear epitopes, possibly because of the absence of a light chain to provide a cleft for peptide binding (25, 26).
Two final screens were conducted using the HobJo library. Screen 8, which was designed to enrich for VHHs that bind native (not cryptic) epitopes on RTB, involved panning the HobJo library on RTB captured via ASF (RTB-ASF). The screen gave rise to five unique RTB-specific VHHs. In screen 9, the VHH library was panned using two VHHs identified in screen 1; ricin holotoxin was captured with JIV-F5 to obstruct immunodominant epitopes in cluster I and then saturated with JIV-F6 to mask a subdominant epitope in cluster III (R-F5 + F6). The screen yielded a total of 15 unique VHHs, including 4 targeting RTA (3 of which were successfully expressed), 8 targeting RTB, and 3 targeting ricin holotoxin.
In total, the nine screens resulted in 68 unique anti-ricin VHHs: 31 specific for RTA, 33 specific for RTB, and 4 specific for ricin holotoxin (Tables 1 and 2). Fourteen additional VHHs (7 anti-RTA and 7 anti-RTB) found in various screens were dropped from further study because of their similarity to other VHHs (i.e., with very few, generally conservative mutations, usually within framework regions) or, in one case, because the VHH failed to be expressed at sufficiently high levels in Escherichia coli to be amenable to analysis (see Table S1 in the supplemental material). The complementarity determining region 3 (CDR3) nucleotide sequences of the entire collection of VHHs were aligned using BLAST and grouped into families based on length and nucleotide identity. There were nine different clonal families encompassing a total of 24 VHHs (Tables 2 and 3); the remaining 44 VHHs were unrelated to each other. The VHH sequences have been deposited in GenBank (Table 2). Within the group of 68 VHHs, CDR3 length ranged from 7 to 22 residues, with an average of 14.8 residues.
TABLE 3.
VHH families based on CDR3 similaritya
| Familyb | Member(s) | Specificityc |
|---|---|---|
| JIV-F6 | V1B10, V1D3 | A |
| JIY-A7 | JNM-G4 | A |
| JIY-D9 | V2B8 | A |
| V1C7 | V2B9, V2E8, V5C1 | A |
| V2G10 | V1G6 | A |
| JIZ-B7 | JIW-D12, JNN-B8, JNN-D1 | B |
| JIW-G10 | JNN-A3 | B′ |
| V2C11 | V2D4, V5H2 | B |
Bold and underlined data indicate a VHH with strong TNA; bold data indicate moderate TNA; underlined data indicate weak TNA.
A family was defined as composed of two or more VHHs having CDR3 sequences of equal lengths and >90% identity at the nucleotide level.
A, RTA; B, RTB; B′, plate-bound RTB.
Classification and characterization of ricin-specific VHHs.
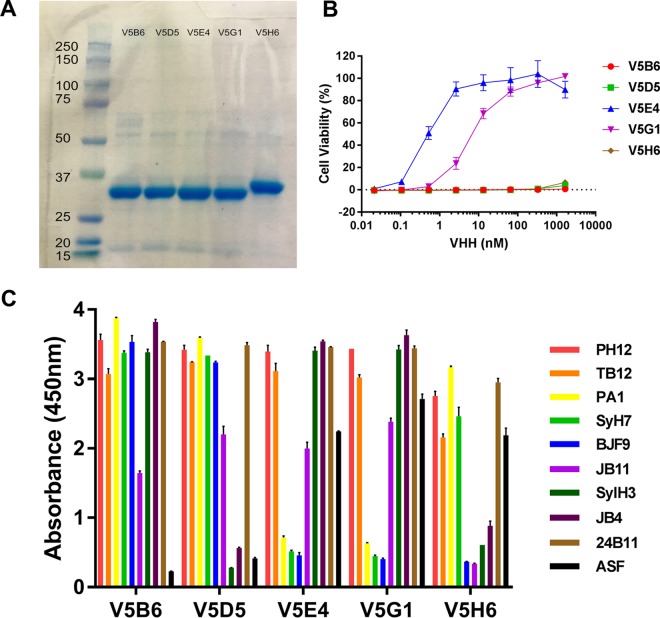
All 68 VHHs were expressed in E. coli as 33- to 35-kDa E-tagged thioredoxin fusion proteins, as described in the supplemental methods and in previous publications (12). The relative purity and integrity of the VHH preparations were assessed by SDS-PAGE (Fig. 1A). Each of the 68 ricin-specific VHHs was also assessed for the ability to neutralize ricin toxin in a Vero cell cytotoxicity assay (Fig. 1B and Table 2) (27, 28). Relative 50% inhibitory concentrations (IC50s) were determined by mixing each VHH at a range of concentrations (0.01 to 10,000 nM) with a fixed amount of ricin before applying it to Vero cells, as described in Materials and Methods. The VHHs were assigned to one of four groups based on their relative IC50s as follows: <10 nM, strong toxin-neutralizing activity; <100 nM, moderate toxin-neutralizing activity; <500 nM, weak toxin-neutralizing activity; >500 nM, no neutralizing activity. The majority (∼80%) had weak (n = 7) or no detectable (n = 47) TNA, while the remainder (∼20%) were evenly divided between strong (n = 7) and moderate (n = 7) toxin-neutralizing activities (Tables 1 and 2). The results of the toxin-neutralizing assays performed for the 14 VHHs with strong or moderate TNA are shown in Fig. S2 in the supplemental material. Among the seven VHHs with the strongest TNA, three were specific for RTA and four were specific for RTB.
FIG 1.
Expression and characterization of ricin-specific VHHs. All 68 VHHs in this study (Table 2) were subjected to analysis by SDS-PAGE, toxin-neutralizing assays, and competition ELISA. Data shown here represent results of analysis of five representative RTB-specific VHHs: V5B6, V5D5, V5E4, V5G1, and V5H6. (A) VHHs were subjected to SDS-PAGE, and the resulting gel was stained with Coomassie blue. Molecular mass standards (with kilodalton notations) are shown in the far left lane. The major band (∼33 to ∼35 kDa) in each lane corresponds to the expected molecular mass of the recombinant VHHs, with an N-terminal thioredoxin fusion partner, a hexa-His tag, and a C-terminal epitope E-tag, as described in the supplemental methods. (B) Toxin-neutralizing activity, as determined in a Vero cell-based microtiter plate assay. The representative set of VHHs consists of two VHHs with strong TNA (IC50 = <10 nM) and three with no TNA. (C) Competition ELISA, as described in Materials and Methods. Microtiter plates were coated with the indicated MAbs or ASF (legend on right) and then overlaid with ricin. The plates were probed with VHHs, as indicated along the x axis, and were developed with anti-E-tag secondary antibodies. As such, the sandwich ELISA data provided relative competition values against a panel of RTA- and RTB-specific MAbs. The failure of the VHHs to recognize ricin toxin when bound to ASF is indicative of the antibodies recognizing epitopes near one of the RTB Gal/GalNAc binding sites. The full array of competition ELISAs is presented in Data Set S1.
VHH association (Ka) and dissociation (Kd) rates and equilibrium dissociation constants (KD) for ricin were determined by surface plasmon resonance (SPR) using sensor chips coated with native toxin via amine coupling, as described in Materials and Methods. Kinetic constants were fitted using the Langmuir 1:1 binding model (29). We found that KD values ranged from 6 pM to ∼32 nM (Table 2; see also Table S2 and Fig. S3 in the supplemental material). In general, the VHHs with the highest binding affinity (KD) generally had the strongest TNA (R2 = 0.6966; Fig. 2A). For example, all 14 VHHs with strong and moderate TNA had KD values of <1 nM, while VHHs with KD values of ≥1 nM lacked TNA, with one exception. Furthermore, the VHHs with the strongest TNA also tended to have the slowest off-rates (Kd) (R2 = 0.6018; Fig. 2B), a relationship that has been noted by other investigators (30). The relationship between binding affinity and TNA was most apparent within VHH clonal families (Fig. 2C), the members of which would be expected to have epitopes similar to each other (31). The failure of antibodies such as JNM-B5 to neutralize ricin despite its relatively high binding affinity (e.g., 60 pM) demonstrates once again that other factors such as epitope specificity contribute to antibody functionality.
FIG 2.
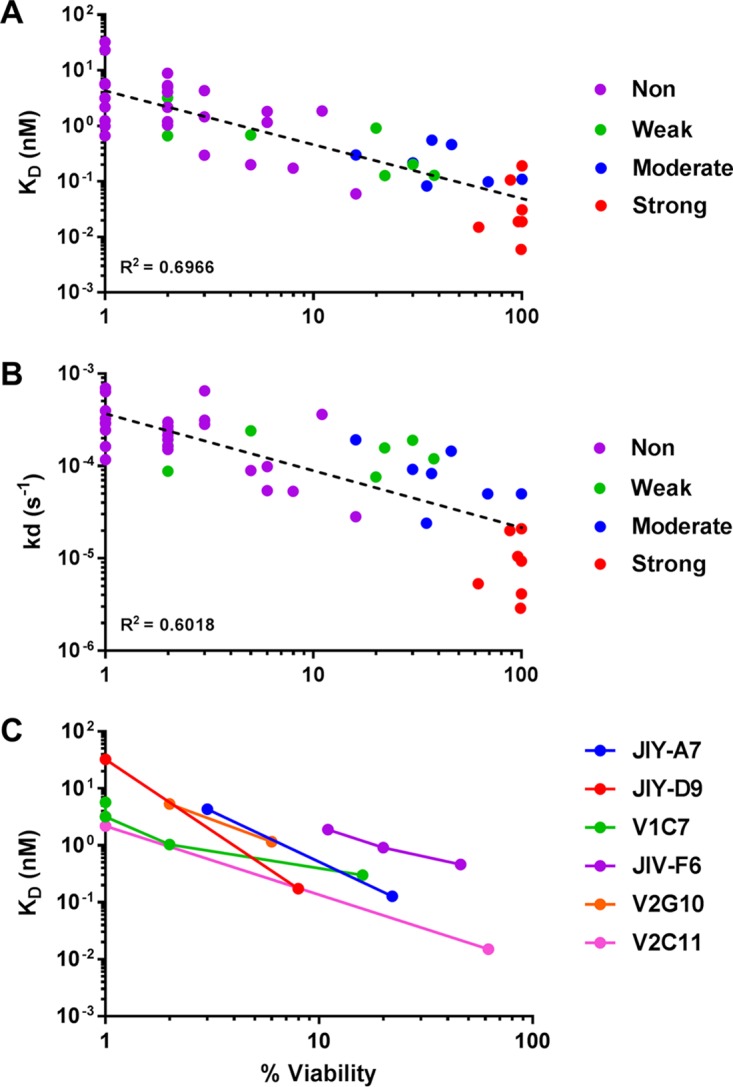
Relationships between VHH binding affinities and toxin-neutralizing activities. (A and B) Using values presented in Table 2, toxin-neutralizing activity (percent viability at 66 nM VHH) of representative VHHs was plotted versus its respective (A) equilibrium dissociation constant (KD) or (B) off-rate (Kd) value. For simplicity's sake, individual VHHs are color coded based on their neutralizing activity (i.e., strong, moderate, weak, or no neutralizing [Non] activity). To enable the use of a logarithmic axis, VHHs that lacked TNA were arbitrarily assigned a value of 1 for percent viability (x axis). R2 values were calculated using the Power Trendline function in Microsoft Excel. (C) TNA as a function of equilibrium dissociation constant (KD) for the six VHH clonal families represented in Table 3. Each color represents a family; each dot represents an individual VHH within that family. The downward slope of the lines suggests a relationship between KD and TNA within a clonal family (i.e., VHHs with the highest affinity for ricin also have the strongest TNA). The percentages of cell viability as plotted are from a single representative experiment done in triplicate.
Localization of VHH epitopes on RTA.
We next sought to locate the relative binding sites of all VHHs that recognized soluble ricin, with the expectation that toxin-neutralizing antibodies would segregate into specific bins or clusters of epitopes on RTA and RTB.
We addressed RTA first. In collaboration with Michael Rudolph (New York Structural Biology Center, New York, NY), all 30 RTA-specific VHHs (excepting JIY-F10, which did not recognize soluble ricin) have been subjected to X-ray crystallography trials. To date, we have reported 10 VHH-RTA cocrystal structures: 6 that involve epitope cluster I, 3 that involve epitope cluster II, and 1 that straddles clusters I and II (Table 2) (16–18). The two cluster I VHHs with the strongest TNA, JIY-E5 and JIV-F5, each make considerable contact with RTA's α-helix B (i.e., ∼472 to ∼477 Å2), an interaction that has been proposed to be critical for toxin inactivation (17–22, 26, 32). The other cluster I VHHs, and the cluster I/II VHH, interacted with β-strand h and α-helix D to various degrees. Within cluster II, the two VHHs with weak TNA, V1C7 and JIY-E1, made primary contact with α-helices D and E, while V5E1, which has strong TNA, associated with α-helices A and F and the F-G loop. For perspective, we provide a PyMol surface representation of all 10 solved VHH structures superimposed onto RTA (see Fig. S4 in the supplemental material).
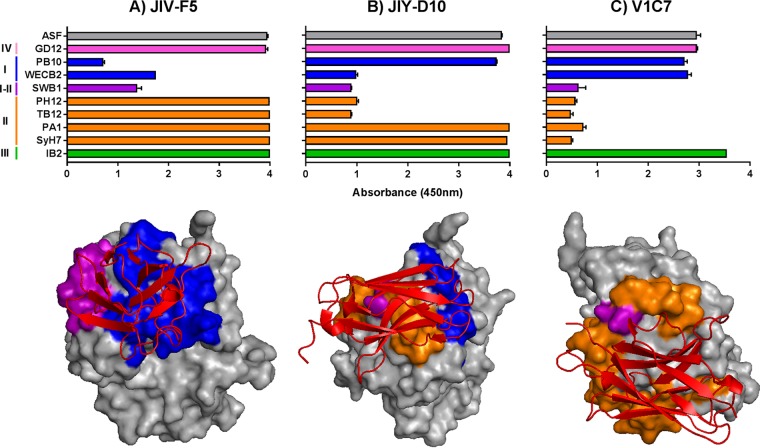
In the absence of additional X-ray crystal structures, epitope localization on RTA was done by competitive sandwich ELISA with a collection of nine different MAbs against epitopes in clusters I (PB10 and WECB2), I and II (SWB1), II (SyH7, PA1, TB12, and PH12), III (IB2), and IV (GD12) (Fig. 3; see also Table S3 in the supplemental material) (19, 20, 53). As described in Materials and Methods, each of the 30 anti-RTA VHHs was tested for the ability to recognize ricin that had been captured by each of the nine cluster I to IV MAbs. The validity of the sandwich ELISA for epitope mapping was evaluated using the following three VHHs whose epitopes have been resolved by X-ray crystallography (Fig. 4): JIV-F5 (PDB identifier [ID]: 4Z9K), JIY-D10 (PDB ID: 4LGR), and V1C7 (PDB ID: 5J56) (16–18). In the sandwich ELISA, the binding of JIV-F5 to ricin was affected by PB10, WECB2, and SWB1 but not by SyH7, PA1, PH12, TB12, IB2, or GD12 (Fig. 4A). This competition profile is consistent with the structural epitope of JIV-F5 encompassing α-helix B, β-strand h, and the C-terminal portion of α-helix D (17). By the same token, JIY-D10 was able to bind to ricin that had been captured by GD12, PB10, PA1, SyH7, and IB2 but not WECB2, SWB1, TB12, or PH12, a profile congruent with JIY-D10's structural epitope being localized to α-helices D and E and β-strand h (Fig. 4B). Finally, V1C7 was able to bind ricin that had been captured by GD12, PB10, WECB2, and IB2 but not the cluster II-specific MAbs (SyH7, PA1, PH12, and TB12) or the I-II overlap MAb SWB1. This profile is consistent with V1C7 making contact with RTA's α-helix D and α-helix E, along with several additional interactions with loops e-d, C-D, and D-E (16) (Fig. 4C). The sandwich ELISA also successfully positioned the epitopes of the seven additional VHHs (JIV-G12, JIY-A7, JIY-E1, JIY-E5, JIY-G11, JNM-F8, and V5E1) whose X-ray crystal structures in complex with RTA have been solved (16–18) (see Data Set S1 in the supplemental material). However, it is important to note that the epitope localization determined by the sandwich ELISA is only approximate, as the assay does not discriminate between direct epitope overlap, steric hindrance, and allostery. Indeed, we noted that the sandwich ELISA overestimated V1C7's contact on RTA compared to its known structural epitope as defined by X-ray crystallography (see Fig. S5).
FIG 3.
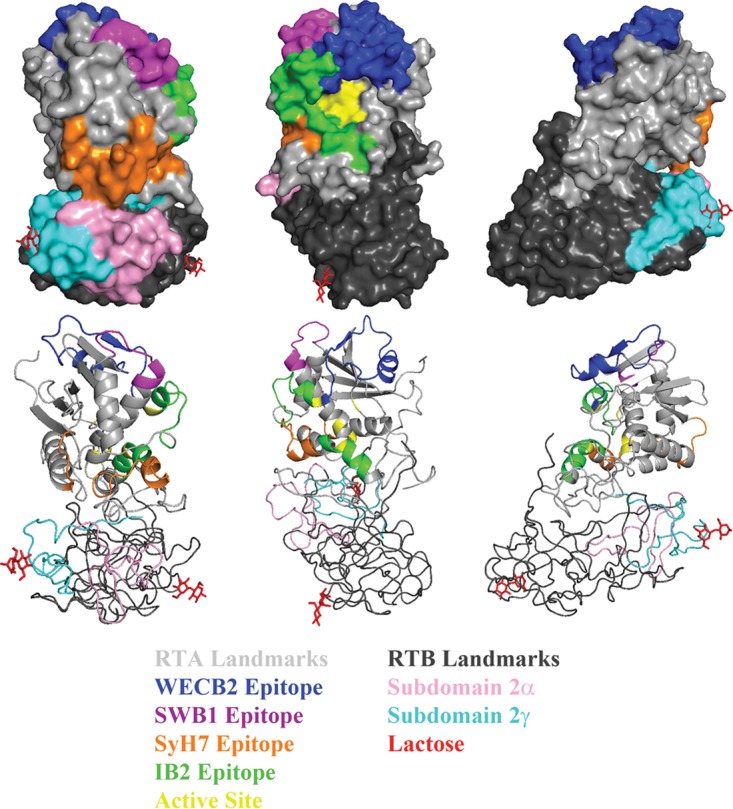
Representative landmarks and functional elements on ricin toxin. Surface (top) and ribbon (bottom) PyMol representations of ricin toxin (PDB ID: 2AAI) at 120° rotations. Landmarks on RTA (gray) include the epitopes recognized by WECB2 (blue), SWB1 (magenta), SyH7 (orange), IB2 (green), and the active site (yellow). Landmarks on RTB (black) include the two galactose binding sites (lactose moieties in red) and subdomains 2α (pink) and 2γ (cyan).
FIG 4.
Validation of cross-competition ELISA as a strategy for epitope localization on RTA. (Top panels) VHHs (A) JIV-F5, (B) JIY-D10, and (C) V1C7 were subjected to competition ELISAs with a panel of RTA-specific MAbs representing neutralizing clusters I to IV, as described in Materials and Methods. As a point of reference, VHH binding to R-ASF is shown in each panel (gray). (Bottom panels) The known X-ray crystal structures of VHHs (colored red) (A) JIV-F5, (B) JIY-D10, and (C) V1C7 bound to RTA with relevant murine MAb epitopes (clusters I to IV), color coded as in the top panel. PDB IDs are 4Z9K (JIV-F5), 4LGR (JIY-D10), and 5J56 (V1C7).
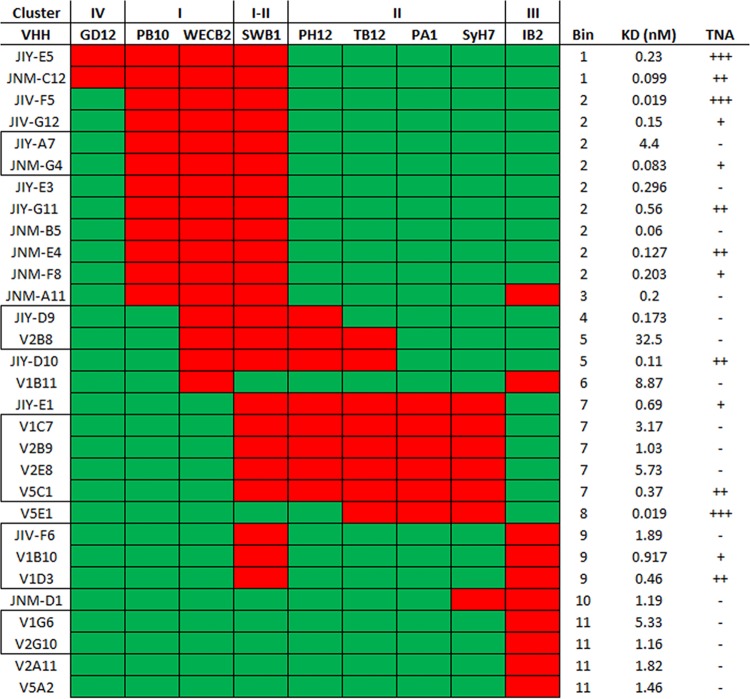
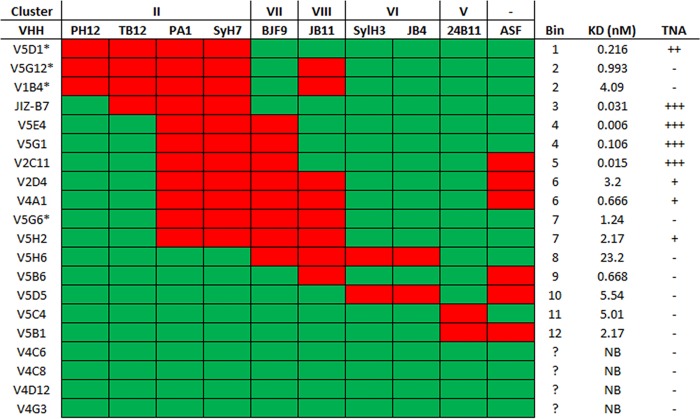
The sandwich ELISA was next used to approximate the binding sites of the remaining 20 RTA-specific VHHs on the surface of RTA relative to clusters I to IV. All 20 VHHs were inhibited from binding to ricin by at least one MAb, and the majority (16/20) were inhibited by two or more MAbs (Fig. 5; see also Fig. S6 and Data Set S1 in the supplemental material). In total, the 30 anti-RTA VHHs fell into 11 different competition profiles, or “bins” (a bin was comprised of all VHHs which shared the same competition profile). When the bins were aligned vertically and the epitope clusters horizontally, as shown in Fig. 5, it was evident that the putative VHH binding sites were spatially contiguous across the four previously identified epitope clusters. Moreover, each of the 11 competition bins overlapped WECB2 (cluster I; bins 1 to 6), SyH7 (cluster II; bins 7 and 8), or IB2 (cluster III; bins 9 to 11), suggesting that these three MAbs constitute the immunodominant regions on RTA.
FIG 5.
Relative locations of the epitopes recognized by 30 different RTA-specific VHHs. Thirty RTA-specific VHHs (left column) were subjected to cross-competition sandwich ELISA with a panel of nine murine MAbs (top row) representing the four different epitope clusters on RTA. Shown here is a binary representation of the actual sandwich ELISA (Fig. S6) in which individual cells are colored red for >50% competition (VHH versus MAb) and green for <50% competition. VHH clonal families are boxed (left column). VHHs with the same competition profile were grouped into bins (right column). For reference, VHH binding affinities and TNA (from Table 2) are shown in the far right columns.
Overall, the competition profiles associated with each of the bins enabled us to assign their location on the surface of RTA with confidence, especially if a bin contained one or more VHHs whose X-ray crystal structure in complex with RTA has been solved. Bin 2, for example, is defined by competition with PB10, WECB2, and SWB1. These three MAbs have overlapping epitopes on RTA's α-helix B, β-strand h, and α-helix D and the D-E loop (53). Moreover, the structural epitopes of five of the nine VHHs in bin 2 have been reported (17, 18). Therefore, we can assume that the remaining four VHHs in bin 2, JIY-E3, JNM-B5, JNM-E4, and JNM-G4, contact α-helix B, β-strand h, and/or α-helix D (Fig. 3; see also Fig. S1).
Similarly, bin 7 is defined by competition with SWB1 and all four cluster II MAbs (PH12, TB12, PA1, and SyH7). PA1 and SyH7 contact the loop before α-helix A and α-helices F and G, while PH12, TB12, and SWB1 contact β-strand d and α-helices D and E. Moreover, as noted above, the structural epitopes of two VHHs in bin 7, JIY-E1 and V1C7, have recently been reported (16). The remaining three VHHs in bin 7, V2B9, V2E8, and V5C1, are members of the V1C7 family (Table 3). Rosetta-based homology modeling suggests that the V1C7 family members, despite their minor differences in CDR sequences, interact with RTA in similar fashions (31). Thus, the epitopes of all five VHHs in bin 7 can be positioned on RTA with relative accuracy.
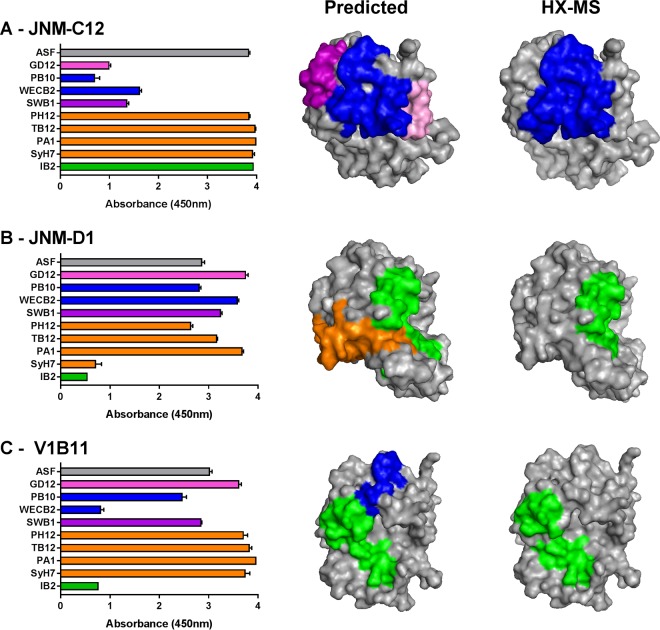
In some instances, the results of the sandwich ELISA were more difficult to reconcile. In the case of V1B11 and JNM-D1, representing bins 6 and 10, respectively, considerable cross-cluster competition was involved. V1B11 was competed with by WECB2 (cluster I) and IB2 (cluster III), while JNM-D1 was affected by SyH7 (cluster II) and IB2 (cluster III). When the cross-cluster competition results were taken at face value, the predicted epitopes for V1B11 and JNM-D1 encompassed too large a surface on RTA to localize actual antibody-antigen contact points (Fig. 6). To refine these ambiguous antibody binding sites, we subjected V1B11 and JNM-D1 to epitope mapping by hydrogen-deuterium exchange mass spectrometry (HX-MS), as described recently (31). HX-MS affords peptide-level resolution of antibody binding sites and is therefore an extremely useful methodology for epitope localization (33). As part of this analysis, we also included JNM-C12 (bin 1) since it also demonstrated a degree of cross-cluster competition.
FIG 6.
Confirmation of epitope positioning by HX-MS. (Left column) VHHs (A) JNM-C12, (B) JNM-D1, and (C) V1B11 were subjected to cross-competition ELISAs, as described in the legend to Fig. 4. (Middle column) Based on the competition profiles, the predicted epitopes recognized by JNM-C12, JNM-D1, and V1B11 were positioned on the surface of RTA (PDB ID: 1RTC). (Right column) Results of HX-MS analysis of JNM-C12, JNM-D1, and V1B11 (Fig. S7), mapped onto RTA. The regions of strong protection, shown in the same orientation as in the middle column for comparison, are indicated using cluster-specific colors.
JNM-C12's binding to ricin was inhibited by cluster I (PB10 and WECB2), cluster I/II (SWB1), and cluster IV (GD12) MAbs (Fig. 6A). This profile differs from that seen with bin 2, for which we have several crystal structures, determined solely by competition with cluster IV (GD12). On the basis of this profile, we predicted that JNM-C12's core epitope would encompass RTA's α-helix B and β-strand h and might also potentially include β-strands b to d. This prediction was partially borne out when JNM-C12 was subjected to epitope mapping by HX-MS. Specifically, JNM-C12 strongly protected RTA peptides corresponding to α-helix B (residues 92 to 109) and β-strand h (residues 108 to 122) but did not protect β-strands b to d (see Fig. S7A and Table S5 in the supplemental material). Thus, competition with GD12 may be steric in nature. In this respect, JNM-C12's core epitope involves contact with α-helix B and β-strand h and resembles VHHs in bin 2.
The sandwich ELISA predicted that JNM-D1 would recognize α-helices C and G and the F-G loop (Fig. 6B). Again, HX-MS analysis partially confirmed this prediction, as JNM-D1 strongly protected peptides encompassing RTA's α-helices C (residues 123 to 135) and G (residues 205 to 210) but not the F-G loop (see Fig. S7B and Table S5 in the supplemental material). On the basis of the sandwich ELISA, V1B11 was predicted to recognize an epitope situated between β-strand h and α-helix C (Fig. 6C). HX-MS analysis of V1B11 revealed that it strongly protected RTA's α-helices C (residues 123 to 135) and G (residues 205 to 210), consistent with the observed IB2 results (see Fig. S7C and Table S5). Thus, HX-MS enabled us to predict with greater confidence the epitopes of V1B11 and JNM-D1 on RTA. Interestingly, despite the differential competition profiles of these two VHHs, their binding epitopes as determined by HX-MS are almost identical. Thus, the sandwich ELISA may provide additional information regarding the orientation of binding of these VHHs to that footprint on RTA, which results in differential steric hindrance with WECB2 and SyH7.
HX-MS also revealed another interesting difference between JNM-D1 and V1B11 in residues 249 to 255, which span a 3/10-helix and a β-bridge in RTA's C terminus. This region is on the opposite side of RTA from the residues in helices C and G that are strongly protected by both VHHs and would not be exposed to solvent in the context of ricin holotoxin. While JNM-D1 induced strong deprotection (i.e., increased flexibility leading to faster hydrogen exchange) in this region, V1B11 induced strong protection (see Fig. S7B and C in the supplemental material). We hesitate to speculate about the significance of the observed deprotection/protection associated with RTA's C terminus, considering that this region of RTA is prone to unfolding in the absence of RTB. Nonetheless, the observation that two VHHs (JNM-D1 and V1B11) with seemingly identical binding sites (and similar affinities) have opposite effects (protection versus deprotection) on RTA's C terminus raises interesting questions about potential allosteric changes that can occur within the context of ricin toxin.
Spatial location and secondary elements on RTA associated with neutralizing epitopes.
We had originally proposed that toxin-neutralizing antibodies would be confined to a very limited number of sites on RTA, a hypothesis that proved to be somewhat accurate. As shown in Fig. 5, although VHHs with strong and moderate TNA were distributed across six different bins (namely, bins 1, 2, 5, 7, 8, and 9), the six bins likely constitute two hot spots. One hot spot, defined by competition with WECB2 and SWB1 and represented by JNM-C12, JIV-F5, and JIY-D10, among others, involves α-helix B and possibly the C-terminal portion of α-helix D (Fig. 3; see also Fig. S1) (17, 18). Although β-strand h sits between these structural elements, evidence provided by X-ray crystal structure analysis suggests that the interaction with strand h alone is not sufficient to confer TNA (17, 18). The other hot spot, defined by competition with SyH7 and PA1 and represented by V5E1, involves contact with the loop preceding α-helix A, α-helix F itself, and the F-G loop, with the F-G loop likely the critical determinant of strong neutralizing activity (16).
With that in mind, we can account, at the structural level, for strong and moderate TNA associated with other RTA-specific VHHs. VHHs in bins 1 to 6 with detectable TNA (e.g., JNM-E4) would be predicted to engage with α-helix B and/or α-helix D, while VHHs in bins 7 and 8 would be expected to contact α-helix F and the F-G loop. V5C1 (bin 7) sits between these two hot spots, being competed with by all three of SWB1 and SyH7/PA1, and thus may neutralize via contact with either or both hot spots. V1D3 (bin 9) is interesting because it is competitively inhibited by both IB2 and SWB1. On the basis of this profile, V1D3 would be expected to make contact with α-helices C and G and/or the C-terminal portion of α-helix D. Its neutralizing activity may therefore be due to contact with α-helix D and/or proximity to the F-G loop. Alternatively, it may represent a third neutralizing hot spot associated with α-helix C.
The story is more complicated, however, because even among VHHs with identical competition profiles there were often disparities in TNA that are explained only partially by binding affinities (KD). In bin 2, for example, JIV-F5, JNM-B5, and JNM-G4 have identical competition profiles and similar binding affinities (0.02 nM, 0.06 nM, and 0.128 nM, respectively) but very different neutralizing activities; JNM-B5 lacks TNA, JNM-G4 has weak TNA, and JIV-F5 has strong TNA. On the basis of the six reported structural epitopes in this area of RTA (17, 18), we hypothesize that JNM-B5 engages solely with β-strand h and not with α-helix D or α-helix B. CDR3 of JIV-F5 is relatively short (16 residues) and contains a disulfide bond between Cys50 and Cys97 that renders it spatially constrained; we previously showed that its short length and constrained conformation prevent the formation of an intermolecular β-sheet with β-strand h (17). In contrast, JNM-B5 and JNM-G4 have longer CDR3 sequences (19 and 22 residues, respectively) that are devoid of the cysteine residues necessary for disulfide bond formation. In this respect, JNM-B5 and JNM-G4 resemble JIY-G11 (PDB ID: 4LHJ), JIY-A7 (PDB ID: 4LHQ), and JIV-G12 (PDB ID: 4LGS), whose unconstrained CDR3 elements form strand-strand interactions with β-strand h. However, definitive conclusions about the actual contact points for JNM-B5 and JNM-G4 on RTA await HX-MS and/or X-ray crystal structure analysis.
Epitope localization of toxin-neutralizing and nonneutralizing RTB- and ricin holotoxin-specific VHHs.
We next turned our attention to the 20 VHHs isolated from the HobJo library that were RTB or holotoxin specific and that could bind to soluble ricin. Using a sandwich ELISA with five benchmark anti-RTB MAbs (see Table S4 in the supplemental material), four RTA-specific cluster II MAbs (SyH7, PA12, PH12, and TB12), and the surrogate receptor, ASF, we tentatively localized the epitopes recognized by 16 of the RTB- and holotoxin-specific VHHs (Fig. 1C and 7; see also Fig. S8 and Data Set S1). Four VHHs were unaffected by the panel of MAbs used in the sandwich ELISA, indicating that they recognize epitopes situated outside our existing panel of MAbs.
FIG 7.
Relative locations of the epitopes recognized by holotoxin- or RTB-specific VHHs. Twenty holotoxin (*)- or RTB-specific VHHs (left column) were subjected to cross-competition sandwich ELISA with a panel of nine murine MAbs and ASF (top row). Shown here is a binary representation of the actual sandwich ELISA (Fig. S8) in which individual cells are colored red for >50% competition (VHH versus MAb) and green for <50% competition. VHHs with the same competition profile were grouped together into bins (right column). For reference, VHH binding affinity data and TNA data (from Table 2) are shown in the far right columns.
The 16 RTB- and holotoxin-specific VHHs constituted 12 different bins, based on their competition profiles in the sandwich ELISA. The 11 VHHs in bins 1 to 7 shared the common trait, in the sandwich ELISA, of being inhibited by PA1 and SyH7, a finding that has important implications for epitope localization on RTB. SyH7 and PA1 each make contact with RTA near the interface with RTB's subdomains 2α (residues 148 to 183) and 2γ (residues 228 to 262) (Fig. 3) (53). We would predict, therefore, that all 11 VHHs in bins 1 to 7 recognize epitopes within RTB's subdomains 2α and/or 2γ and that the observed competition with SyH7 and PA1 in the sandwich ELISA is due to steric hindrance. VHHs in bins 1 to 3 would be predicted to bind epitopes abutting the RTA-RTB interface, based on the observed competition with TB12 and PH12, whose epitopes are considerably “north” of the border. In fact, V5D1, V5G12, and V1B4 (comprising all of bins 1 and 2) bind ricin holotoxin, but not the individual subunits, indicating that they recognize quaternary epitopes that straddle the RTA-RTB interface. Finally, based on the observed competition with ASF, VHHs in bins 5 and 6 are predicted to have epitopes situated in close proximity to (or even overlapping) the Gal/GalNAc recognition element in subdomain 2γ (Fig. 3). Elsewhere, we report results of extensive cross-competition studies performed to refine the epitopes on RTB's domain 2 recognized by the 11 VHHs in bins 1 to 7 (A. Y. Poon, D. Vance, Y. Rong, D. Ehrbar, and N. Mantis, unpublished data).
Bins 8 to 12 each contained a single VHH. On the basis of competition with SylH3 and JB4, we would propose that V5H6 (bin 8) and V5D5 (bin 10) also recognize epitopes in RTB domain 2. Moreover, the fact that V5D5 is unable to recognize R-ASF suggests that its epitope is occluded upon RTB-receptor interaction and is therefore in close proximity to the Gal/GalNAc recognition domain but on the opposite side from VHHs in bins 5 and 6. In contrast, V5C4 (bin 11) and V5B1 (bin 12) were each competitively inhibited from binding to ricin by 24B11, which is postulated to recognize RTB subdomain 1 (34–36). V5B1 was also unable to recognize R-ASF, presumably because its epitope is near the domain 1 Gal-specific binding element (5). We recently identified three additional MAbs specific for RTB's domain 1, although they were not included in this study (37).
Among the 20 RTB- and holotoxin-specific VHHs isolated from the HobJo library, four were classified as having strong TNA, one moderate TNA, and three weak TNA (Fig. 7 and Table 2). The four VHHs with strong TNA had KD values of <110 pM and were located in bins 3 (JIZ-B7), 4 (V5E4 and V5G1), and 5 (V2C11). They are also defined by competition with the two anti-RTA MAbs, SyH7 and PA1. On the basis of their competition profiles, we hypothesize that JIZ-B7, V5E4, V5G1, and V2C11 recognize a neutralizing hot spot at the RTA-RTB interface. Two V2C11 family members, V2D4 and V5H2, are also predicted to target this hot spot but have only weak TNA, probably because of their relatively low binding affinities.
Along those same lines, it should be underscored that V5E1 and JIZ-B7 have identical competition profiles, even though V5E1 is classified as an RTA-specific antibody and JIZ-B7 as an RTB-specific antibody (Fig. 5 and 7). Had we conducted the epitope binning studies with ricin holotoxin rather than the individual RTA and RTB subunits, V5E1 and JIZ-B7 would theoretically have been in the same bin. We speculate that JIZ-B7 recognizes a quaternary epitope that straddles the RTA-RTB border, possibly even in a configuration that mirrors V5E1 (16).
DISCUSSION
In this study, we sought to advance our longstanding efforts to generate a comprehensive B cell epitope map of ricin toxin with a focus on localizing the epitopes recognized by toxin-neutralizing antibodies. Ricin's enzymatic subunit (RTA) is of particular interest to the biodefense research community as two recombinant RTA-based vaccines, RiVax and RVEc, are being evaluated in preclinical and clinical studies (38–40). Predicting the efficacy of these two vaccine candidates in humans is challenging, because neither the anti-ricin serum IgG titer nor the TNA is necessarily a reliable predictor of protective immunity (41). However, there is evidence that epitope-specific antibody responses may be more useful measures in this regard (42). The availability of a high-resolution epitope map of RTA would enable a targeted analysis of specific antibody responses following vaccination, possibly even resulting in the identification of a serum antibody profile that correlates with immunity. Understanding the antibody response to RTA is also relevant to the development of RTA-based immunotoxins, whose efficacy (in some instances) has been limited by the onset of one or more toxin-neutralizing antibodies that render the immunotoxin inactive (43, 44).
We estimate that the current collection of 68 VHHs described in this study covers slightly more than 50% of the surface area of RTA, with a particular bias toward two immunodominant regions of RTA, referred to as epitope clusters I and II (20). Areas not covered in the current VHH repertoire include the entire “backside” of RTA (Fig. 3, right panel; see also Fig. S1 in the supplemental material). In terms of RTB, we identified an aggregate of neutralizing epitopes localized at the RTA-RTB interface positioned in close proximity to the Gal/GalNAc recognition element of RTB's domain 2γ (5). In a separate report, we refer to this region of RTB as “supercluster II” and demonstrate that toxin-neutralizing activity is associated with antibody binding in the vicinity of the RTA-RTB interface (as manifested through competition ELISA performed with SyH7 and V5E1) rather than with obstruction of the Gal/GalNAc recognition element (A. Y. Poon, D. Vance, Y. Rong, D. Ehrbar, and N. Mantis, unpublished data). This interfacial region of ricin toxin is also apparently the target of a number of holotoxin-specific VHHs (e.g., V5D1) whose epitopes likely involve residues on both sides of the border. Unfortunately, our current report does not address the mechanism(s) by which the supercluster II antibodies neutralize ricin. We can only speculate that they interfere with a region of the toxin that is required for retrograde transport from the plasma membrane to the ER and/or retrotranslocation from the ER to the cytoplasm (45–48).
Nonetheless, our results are consistent with there being two putative neutralizing hot spots on RTA, with “hot spot” being defined as a cluster of closely spaced epitopes recognized by antibodies with potent toxin-neutralizing activity. The first hot spot encompasses RTA's α-helix B and α-helix D. The importance of α-helix B as a target of neutralizing antibodies has been recognized for more than 2 decades (22, 49), although it is only recently that the critical holdfasts have been identified (16, 18, 21, 26, 32). The second hot spot (as noted above) is located at the RTA-RTB interface, defined by RTA's α-helix F and the F-G loop and (possibly) RTB's subdomains 2α and/or 2γ (16). At present, it is not clear why these regions render ricin vulnerable to antibody attack, although evidence is mounting that neutralization occurs intracellularly rather than extracellularly (13, 45–47).
The HobJo VHHs described here should be a valuable resource for the ricin toxin research community and may have important applications not only for vaccine development but also for diagnostics and therapeutics. For example, Turner et al. recently showed that pairing a llama-derived anti-ricin VHH (D12fneg) with a modified version of alpaca-derived VHH JIV-F6 (F6m+) yielded a highly sensitive assay that could detect less than 1 ng/ml of ricin (12, 50). Here we have described two new JIV-F6 family members, V1B10 and V1D3, each with higher affinities for ricin than JIV-F6. In another application, Legler et al. used JIV-F6, JIY-D10, and JIY-E1, along with several of their own anti-ricin VHHs, to show that the ability of a VHH to increase the melting temperature of RTA correlated with its neutralization ability (26). The availability of dozens of additional VHH coding sequences, affinities, and TNAs (Table 2) enables those types of analyses to be extended. Finally, the VHHs are amenable to recombinant engineering and may provide a platform for therapeutic development. We demonstrated, for example, that select VHH heterodimers effectively aggregate ricin in solution and inactive the toxin in vitro and in vivo (12–14). With the available epitope maps derived from the current study, it is now possible to rationally design different VHH bispecific antibodies with appropriate linkers intended to target notable hot spots on RTA and RTB.
MATERIALS AND METHODS
Ricin-specific VHH phage display library.
The single-domain-antibody (VHH) library has been described previously (12). The library is referred to as “HobJo” here in reference to the names of the two immunized alpacas, Hobbs and Johnnes. The VHH-pIII phagemid library was maintained in E. coli TG1 at −80°C. The library was subjected to nine different screens, as described in Table 1 and in Results. Each screen consisted of low-stringency (round 1) and high-stringency (round 2) panning procedures in which target antigen was present at 10 μg/ml and 1 μg/ml, respectively. To begin each screen, an aliquot of the library was used to inoculate Super Broth (SB) medium supplemented with 100 μg/ml ampicillin and 2% glucose. The culture was grown at 37°C to an absorbance at 600 nm (OD600) of ∼0.6 to ∼0.8 and then infected with VCSM13 helper phage carrying kanamycin resistance (Agilent Technologies, Santa Clara, CA). After a 2-h infection at 37°C, the bacteria were pelleted at 8,000 rpm, reconstituted in SB medium supplemented with ampicillin (100 μg/ml), kanamycin (70 μg/ml), and 0.1% glucose, and incubated overnight at 30°C. The next morning, the bacteria were pelleted by centrifugation and discarded. The resulting supernatants containing VHH-displaying phages were precipitated with 1:5 (vol/vol) 20% polyethylene glycol (PEG 8000)–2.5 M NaCl at 4°C for 2 h. The supernatant was subjected to centrifugation at 8,000 rpm for 20 min to pellet the phages, which were then resuspended in phosphate-buffered saline (PBS) and used for round 1 of panning. Further phage panning protocols are presented in detail in the supplemental material. Chosen VHHs were expressed as fusion proteins with an N-terminal thioredoxin tag, a linker containing a hexa-His purification tag, and a C-terminal epitopic E-tag (GAPVPYPDPLEPR) in E. coli Rosetta-gami 2(DE3)pLacI (EMD Millipore, Billerica, MA) as previously described (14, 15) and presented in detail in the supplemental material.
Peptides.
Biotin-tagged peptides E12 (HPDNQEDAEAITHLFTDV) and G3 (IEGEMRTRIRYNRRSAPD) used in screen 7 were synthesized by Genemed Synthesis, Inc. (San Antonio, TX).
ELISA.
For cross-competition sandwich ELISAs, Nunc microtiter plates were coated with competitor capture MAbs (1 μg/ml in PBS) or ASF (4 μg/ml in PBS) overnight at 4°C and then blocked for 2 h with 2% goat serum–0.1% Tween 20 phosphate-buffered saline (PBST). Ricin (1 μg/ml), RTA (0.5 μg/ml), or RTB (0.5 μg/ml) was then captured by the MAbs or ASF and probed with VHH analytes at 330 nM. Bound VHHs were detected with an anti-E tag–horseradish peroxidase (HRP) secondary antibody (Bethyl Labs, Montgomery, TX) and developed with SureBlue TMB (3,3′,5,5′-tetramethylbenzidine) substrate (KPL). After quenching was performed with 1 M phosphoric acid, absorbance was read at 450 nm on a VersaMax microplate reader (Molecular Devices, Sunnyvale, CA).
Vero cell cytotoxicity assays.
Relative IC50s were determined in Vero cell cytotoxicity assays, as previously described (28). Vero cells were detached from culture dishes with trypsin, adjusted to ∼5 ×104 cells per ml, seeded (100 μl/well) into white 96-well plates (Corning Life Sciences, Corning, NY), and allowed to adhere overnight. The cells were then treated with ricin (0.01 μg/ml; 154 pM), ricin-VHH mixtures, or medium alone (negative control) for 2 h at 37°C. The cells were washed and then incubated for 48 h at 37°C, after which cell viability was assessed using CellTiter-GLO (Promega, Madison, WI). All treatments were performed in triplicate. Relative IC50s were determined following published guidelines in which 100% viability was derived from cells treated with medium only, while 0% viability was defined as the average value obtained from ricin-treated cells (51). Reported relative IC50s for each VHH were derived from at least three independent toxin-neutralizing assays (each done in triplicate).
SPR and affinity determinations.
VHH association and dissociation rates for ricin toxin were determined by surface plasmon resonance (SPR) using a ProteOn XPR36 system (Bio-Rad Inc., Hercules, CA), as described previously (18, 37). For ricin immobilization, general layer compact (GLC) chips were equilibrated in running buffer (PBS–0.005% Tween) at a flow rate of 30 μl/min. Following EDAC [N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide hydrochloride] (200 mM)–sulfo-NHS (N-hydroxysulfosuccinimide) (50 mM) activation (3 min), ricin was diluted in 10 mM sodium acetate (pH 5.0) at either 4 μg/ml or 2 μg/ml and coupled for 2 min. A third vertical channel received only acetate buffer and served as a reference channel. The surfaces were deactivated using 1 M ethanolamine for 5 min. A ProteOn array system multichannel module (MCM) was rotated to the horizontal orientation for affinity determination experiments. Each VHH was serially diluted in running buffer and then injected at 50 μl/min for 180 s, followed by 1 to 3 h of dissociation. After each experiment, the chip was regenerated with 10 mM glycine (pH 1.5) at 100 μl/min for 18 s, until the response unit (RU) values had returned to baseline. All kinetic experiments were performed at 25°C. Kinetic constants for the antibody/ricin interactions were obtained with ProteOn Manager software 3.1.0 (Bio-Rad Inc.) using the Langmuir fit model.
Epitope mapping by hydrogen deuterium exchange-mass spectrometry (HX-MS).
The HX-MS method for epitope mapping VHH antibodies is described in more detail elsewhere (R. Toth IV and D. D. Weis, unpublished results) (31). For safety reasons, epitope mapping studies were conducted with a nontoxic variant of RTA known as RiVax (52). Briefly, regions of RiVax where the rate of HX was significantly lower (protection) or higher (deprotection) in the presence of bound VHH were identified using k-means clustering and significance testing based on time-averaged HX measurements (ΔHX). However, unlike that previous work, here the results were not filtered for the solvent accessibility of the RiVax residues. Using k-means clustering, the regions that were assigned as showing strong and intermediate protection were used to define the epitope. The RiVax peptides are numbered sequentially from the N terminus to the C terminus and described in Table S5 in the supplemental material.
Molecular graphics.
Images of ricin holotoxin (PBD ID: 2AAI), RTA (PDB ID: 1RTC), and RTA-VHH complexes (see Table 2 for PDB IDs) were generated using PyMOL (The PyMOL Molecular Graphics System; Schrodinger LLC, San Diego, CA).
Accession number(s).
All VHH nucleotide sequences were deposited into GenBank (see Table 2) and the Immune Epitope Database (IEDB) (pending).
Supplementary Material
ACKNOWLEDGMENTS
We thank Amanda Poon (University at Albany), Greta Van Slyke (Wadsworth Center), and Michael Rudolph (New York Structural Biology Center) for technical assistance with epitope mapping studies. We thank the Wadsworth Center's Applied Genomics Technology Core for DNA sequencing and the Biochemistry/Immunology Core facility for access to SPR instrumentation.
Research reported in this work was supported by contract no. HHSN272201400021C from the National Institutes of Allergy and Infectious Diseases, National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/CVI.00237-17.
For a commentary on this article, see https://doi.org/10.1128/CVI.00275-17.
Supplemental material for this article may be found at https://doi.org/10.1128/CVI.00236-17.
REFERENCES
- 1.Schrot J, Weng A, Melzig MF. 2015. Ribosome-inactivating and related proteins. Toxins (Basel) 7:1556–1615. doi: 10.3390/toxins7051556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Endo Y, Mitsui K, Motizuki M, Tsurugi K. 1987. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. J Biol Chem 262:5908–5912. [PubMed] [Google Scholar]
- 3.Endo Y, Tsurugi K. 1987. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J Biol Chem 262:8128–8130. [PubMed] [Google Scholar]
- 4.Tesh VL. 2012. The induction of apoptosis by Shiga toxins and ricin. Curr Top Microbiol Immunol 357:137–178. [DOI] [PubMed] [Google Scholar]
- 5.Rutenber E, Ready M, Robertus JD. 1987. Structure and evolution of ricin B chain. Nature 326:624–626. doi: 10.1038/326624a0. [DOI] [PubMed] [Google Scholar]
- 6.Sandvig K, Olsnes S, Pihl A. 1976. Kinetics of binding of the toxic lectins abrin and ricin to surface receptors of human cells. J Biol Chem 251:3977–3984. [PubMed] [Google Scholar]
- 7.Sandvig K, Skotland T, van Deurs B, Klokk TI. 2013. Retrograde transport of protein toxins through the Golgi apparatus. Histochem Cell Biol 140:317–326. doi: 10.1007/s00418-013-1111-z. [DOI] [PubMed] [Google Scholar]
- 8.Spooner RA, Lord JM. 2012. How ricin and Shiga toxin reach the cytosol of target cells: retrotranslocation from the endoplasmic reticulum. Curr Top Microbiol Immunol 357:19–40. doi: 10.1007/82_2011_154. [DOI] [PubMed] [Google Scholar]
- 9.Reisler RB, Smith LA. 2012. The need for continued development of ricin countermeasures. Adv Prev Med 2012:149737. doi: 10.1155/2012/149737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfe DN, Florence W, Bryant P. 21 February 2013. Current biodefense vaccine programs and challenges. Hum Vaccin Immunother doi: 10.4161/hv.24063. [DOI] [PubMed] [Google Scholar]
- 11.Pincus SH, Bhaskaran M, Brey RN III, Didier PJ, Doyle-Meyers LA, Roy CJ. 2015. Clinical and pathological findings associated with aerosol exposure of macaques to ricin toxin. Toxins (Basel) 7:2121–2133. doi: 10.3390/toxins7062121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vance DJ, Tremblay JM, Mantis NJ, Shoemaker CB. 2013. Stepwise engineering of heterodimeric single domain camelid VHH antibodies that passively protect mice from ricin toxin. J Biol Chem 288:36538–36547. doi: 10.1074/jbc.M113.519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrera C, Klokk TI, Cole R, Sandvig K, Mantis NJ. 2016. A bispecific antibody promotes aggregation of ricin toxin on cell surfaces and alters dynamics of toxin internalization and trafficking. PLoS One 11:e0156893. doi: 10.1371/journal.pone.0156893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrera C, Tremblay JM, Shoemaker CB, Mantis NJ. 2015. Mechanisms of ricin toxin neutralization revealed through engineered homodimeric and heterodimeric camelid antibodies. J Biol Chem 290:27880–27889. doi: 10.1074/jbc.M115.658070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrera C, Vance DJ, Eisele LE, Shoemaker CB, Mantis NJ. 2014. Differential neutralizing activities of a single domain camelid antibody (VHH) specific for ricin toxin's binding subunit (RTB). PLoS One 9:e99788. doi: 10.1371/journal.pone.0099788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudolph MJ, Vance DJ, Cassidy MS, Rong Y, Mantis NJ. 2017. Structural analysis of single domain antibodies bound to a second neutralizing hot spot on ricin toxin's enzymatic subunit. J Biol Chem 292:872–883. doi: 10.1074/jbc.M116.758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudolph MJ, Vance DJ, Cassidy MS, Rong Y, Shoemaker CB, Mantis NJ. 2016. Structural analysis of nested neutralizing and non-neutralizing B cell epitopes on ricin toxin's enzymatic subunit. Proteins 84:1162–1172. doi: 10.1002/prot.25062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudolph MJ, Vance DJ, Cheung J, Franklin MC, Burshteyn F, Cassidy MS, Gary EN, Herrera C, Shoemaker CB, Mantis NJ. 4 June 2014. Crystal structures of ricin toxin's enzymatic subunit (RTA) in complex with neutralizing and non-neutralizing single-chain antibodies. J Mol Biol doi: 10.1016/j.jmb.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Hara JM, Neal LM, McCarthy EA, Kasten-Jolly JA, Brey RN III, Mantis NJ. 2010. Folding domains within the ricin toxin A subunit as targets of protective antibodies. Vaccine 28:7035–7046. doi: 10.1016/j.vaccine.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Hara JM, Kasten-Jolly JC, Reynolds CE, Mantis NJ. 2014. Localization of non-linear neutralizing B cell epitopes on ricin toxin's enzymatic subunit (RTA). Immunol Lett 158:7–13. doi: 10.1016/j.imlet.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vance DJ, Mantis NJ. 2012. Resolution of two overlapping neutralizing B cell epitopes within a solvent exposed, immunodominant alpha-helix in ricin toxin's enzymatic subunit. Toxicon 60:874–877. doi: 10.1016/j.toxicon.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lebeda FJ, Olson MA. 1999. Prediction of a conserved, neutralizing epitope in ribosome-inactivating proteins. Int J Biol Macromol 24:19–26. doi: 10.1016/S0141-8130(98)00059-2. [DOI] [PubMed] [Google Scholar]
- 23.Friguet B, Djavadi-Ohaniance L, Goldberg ME. 1984. Some monoclonal antibodies raised with a native protein bind preferentially to the denatured antigen. Mol Immunol 21:673–677. doi: 10.1016/0161-5890(84)90053-1. [DOI] [PubMed] [Google Scholar]
- 24.Jemmerson R. 1987. Antigenicity and native structure of globular proteins: low frequency of peptide reactive antibodies. Proc Natl Acad Sci U S A 84:9180–9184. doi: 10.1073/pnas.84.24.9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J, Muyldermans S, Wyns L. 2006. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci U S A 103:4586–4591. doi: 10.1073/pnas.0505379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legler PM, Compton JR, Hale ML, Anderson GP, Olson MA, Millard CB, Goldman ER. 2017. Stability of isolated antibody-antigen complexes as a predictive tool for selecting toxin neutralizing antibodies. MAbs 9:43–57. doi: 10.1080/19420862.2016.1236882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konowalchuk J, Speirs JI, Stavric S. 1977. Vero response to a cytotoxin of Escherichia coli. Infect Immun 18:775–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wahome PG, Mantis NJ. 2013. High-throughput, cell-based screens to identify small-molecule inhibitors of ricin toxin and related category B ribosome inactivating proteins (RIPs). Curr Protoc Toxicol 2013:Chapter 2:Unit 2.23. doi: 10.1002/0471140856.tx0223s55. [DOI] [PubMed] [Google Scholar]
- 29.O'Shannessy DJ, Brigham-Burke M, Soneson KK, Hensley P, Brooks I. 1993. Determination of rate and equilibrium binding constants for macromolecular interactions using surface plasmon resonance: use of nonlinear least squares analysis methods. Anal Biochem 212:457–468. doi: 10.1006/abio.1993.1355. [DOI] [PubMed] [Google Scholar]
- 30.Rosenfeld R, Alcalay R, Mechaly A, Lapidoth G, Epstein E, Kronman C, S JF, Mazor O. 3 May 2017. Improved antibody-based ricin neutralization by affinity maturation is correlated with slower off-rate values. Protein Eng Des Sel doi: 10.1093/protein/gzx028. [DOI] [PubMed] [Google Scholar]
- 31.Bazzoli A, Vance DJ, Rudolph MJ, Rong Y, Angalakurthi SK, Toth RT IV, Middaugh CR, Volkin DB, Weis DD, Karanicolas J, Mantis NJ. 4 August 2017. Using homology modeling to interrogate binding affinity in neutralization of ricin toxin by a family of single domain antibodies. Proteins doi: 10.1002/prot.25353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai J, Zhao L, Yang H, Guo H, Fan K, Wang H, Qian W, Zhang D, Li B, Wang H, Guo Y. 2011. Identification of a novel functional domain of ricin responsible for its potent toxicity. J Biol Chem 286:12166–12171. doi: 10.1074/jbc.M110.196584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown KA, Wilson DJ. 13 July 2017. Bottom-up hydrogen deuterium exchange mass spectrometry: data analysis and interpretation. Analyst doi: 10.1039/c7an00662d. [DOI] [PubMed] [Google Scholar]
- 34.McGuinness CR, Mantis NJ. 2006. Characterization of a novel high-affinity monoclonal immunoglobulin G antibody against the ricin B subunit. Infect Immun 74:3463–3470. doi: 10.1128/IAI.00324-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yermakova A, Mantis NJ. 2011. Protective immunity to ricin toxin conferred by antibodies against the toxin's binding subunit (RTB). Vaccine 29:7925–7935. doi: 10.1016/j.vaccine.2011.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yermakova A, Vance DJ, Mantis NJ. 2012. Sub-domains of ricin's B subunit as targets of toxin neutralizing and non-neutralizing monoclonal antibodies. PLoS One 7:e44317. doi: 10.1371/journal.pone.0044317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rong Y, Van Slyke G, Vance DJ, Westfall J, Ehrbar D, Mantis NJ. 2017. Spatial location of neutralizing and non-neutralizing B cell epitopes on domain 1 of ricin toxin's binding subunit. PLoS One 12:e0180999. doi: 10.1371/journal.pone.0180999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pittman PR, Reisler RB, Lindsey CY, Guerena F, Rivard R, Clizbe DP, Chambers M, Norris S, Smith LA. 2015. Safety and immunogenicity of ricin vaccine, RVEc, in a phase 1 clinical trial. Vaccine 33:7299–7306. doi: 10.1016/j.vaccine.2015.10.094. [DOI] [PubMed] [Google Scholar]
- 39.Vitetta ES, Smallshaw JE, Coleman E, Jafri H, Foster C, Munford R, Schindler J. 2006. A pilot clinical trial of a recombinant ricin vaccine in normal humans. Proc Natl Acad Sci U S A 103:2268–2273. doi: 10.1073/pnas.0510893103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vitetta ES, Smallshaw JE, Schindler J. 2012. Pilot phase IB clinical trial of an alhydrogel-adsorbed recombinant ricin vaccine. Clin Vaccine Immunol 19:1697–1699. doi: 10.1128/CVI.00381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vance DJ, Mantis NJ. 2016. Progress and challenges associated with the development of ricin toxin subunit vaccines. Expert Rev Vaccines 15:1213–1222. doi: 10.1586/14760584.2016.1168701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy CJ, Brey RN, Mantis NJ, Mapes K, Pop IV, Pop LM, Ruback S, Killeen SZ, Doyle-Meyers L, Vinet-Oliphant HS, Didier PJ, Vitetta ES. 2015. Thermostable ricin vaccine protects rhesus macaques against aerosolized ricin: epitope-specific neutralizing antibodies correlate with protection. Proc Natl Acad Sci U S A 112:3782–3787. doi: 10.1073/pnas.1502585112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castelletti D, Fracasso G, Righetti S, Tridente G, Schnell R, Engert A, Colombatti M. 2004. A dominant linear B-cell epitope of ricin A-chain is the target of a neutralizing antibody response in Hodgkin's lymphoma patients treated with an anti-CD25 immunotoxin. Clin Exp Immunol 136:365–372. doi: 10.1111/j.1365-2249.2004.02442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vitetta ES, Stone M, Amlot P, Fay J, May R, Till M, Newman J, Clark P, Collins R, Cunningham D, Ghetie V, Uhr JW, Thorpe PE. 1991. Phase I immunotoxin trial in patients with B-cell lymphoma. Cancer Res 51:4052–4058. [PubMed] [Google Scholar]
- 45.Song K, Mize RR, Marrero L, Corti M, Kirk JM, Pincus SH. 2013. Antibody to ricin a chain hinders intracellular routing of toxin and protects cells even after toxin has been internalized. PLoS One 8:e62417. doi: 10.1371/journal.pone.0062417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yermakova A, Klokk TI, Cole R, Sandvig K, Mantis NJ. 2014. Antibody-mediated inhibition of ricin toxin retrograde transport. mBio 5:e00995-13. doi: 10.1128/mBio.00995-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yermakova A, Klokk TI, O'Hara JM, Cole R, Sandvig K, Mantis NJ. 2016. Neutralizing monoclonal antibodies against disparate epitopes on ricin toxin's enzymatic subunit interfere with intracellular toxin transport. Sci Rep 6:22721. doi: 10.1038/srep22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Hara JM, Mantis NJ. 2013. Neutralizing monoclonal antibodies against ricin's enzymatic subunit interfere with protein disulfide isomerase-mediated reduction of ricin holotoxin in vitro. J Immunol Methods 395:71–78. doi: 10.1016/j.jim.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lemley PV, Amanatides P, Wright DC. 1994. Identification and characterization of a monoclonal antibody that neutralizes ricin toxicity in vitro and in vivo. Hybridoma 13:417–421. doi: 10.1089/hyb.1994.13.417. [DOI] [PubMed] [Google Scholar]
- 50.Turner KB, Hardy S, Liu JL, Zabetakis D, Lee PAB, Goldman ER, Anderson GP. 2017. Pairing alpaca and llama-derived single domain antibodies to enhance immunoassays for ricin. Antibodies 6:3. doi: 10.3390/antib6010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sebaugh JL. 2011. Guidelines for accurate EC50/IC50 estimation. Pharm Stat 10:128–134. doi: 10.1002/pst.426. [DOI] [PubMed] [Google Scholar]
- 52.Legler PM, Brey RN, Smallshaw JE, Vitetta ES, Millard CB. 2011. Structure of RiVax: a recombinant ricin vaccine. Acta Crystallogr D Biol Crystallogr 67:826–830. doi: 10.1107/S0907444911026771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toth RT IV, Angalakurthi SK, Van Slyke G, Vance DJ, Hickey JM, Joshi SB, Middaugh CR, Volkin DB, Weis DD, Mantis NJ. 2017. High-definition mapping of four spatially distinct neutralizing epitope clusters on RiVax, a candidate ricin toxin subunit vaccine. Clin Vaccine Immunol 24:e00237-17. doi: 10.1128/CVI.00237-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.