ABSTRACT
Endophytic and rhizosphere actinobacteria isolated from the root system of 1-year-old grafted Vitis vinifera plants were evaluated for their activities against fungi that cause grapevine trunk diseases. A total of 58 endophytic and 94 rhizosphere isolates were tested. Based on an in vitro bioassay, 15.5% of the endophytic isolates and 30.8% of the rhizosphere isolates exhibited antifungal activity against the fungal pathogen Diplodia seriata, whereas 13.8% of the endophytic isolates and 16.0% of the rhizosphere isolates showed antifungal activity against Dactylonectria macrodidyma (formerly Ilyonectria macrodidyma). The strains which showed the greatest in vitro efficacy against both pathogens were further analyzed for their ability to inhibit the growth of Phaeomoniella chlamydospora and Phaeoacremonium minimum (formerly Phaeoacremonium aleophilum). Based on their antifungal activity, three rhizosphere isolates and three endophytic isolates were applied on grafts in an open-root field nursery in a 3-year trial. The field trial led to the identification of one endophytic strain, Streptomyces sp. VV/E1, and two rhizosphere isolates, Streptomyces sp. VV/R1 and Streptomyces sp. VV/R4, which significantly reduced the infection rates produced by the fungal pathogens Dactylonectria sp., Ilyonectria sp., P. chlamydospora, and P. minimum, all of which cause young grapevine decline. The VV/R1 and VV/R4 isolates also significantly reduced the mortality level of grafted plants in the nursery. This study shows that certain actinobacteria could represent a promising new tool for controlling fungal trunk pathogens that infect grapevine plants through the root system in nurseries.
IMPORTANCE Grapevine trunk diseases are a major threat to the wine and grape industry worldwide. They cause a significant reduction in yields as well as in grape quality, and they can even cause plant death. Trunk diseases are caused by fungal pathogens that enter through pruning wounds and/or the root system. Although different strategies have recently been developed to protect pruning wounds using antifungal compounds (natural or synthetic) or biocontrol agents, no tools are yet available for controlling soil pathogens that infect plants through their root system. This study shows that different actinobacterial isolates, when applied to grafts in a nursery, can significantly reduce the infection rate caused by fungal pathogens that enter through the root system. This is a new, promising, and green alternative for preventing the decline of young grapevines in nurseries and vineyards.
KEYWORDS: young grapevine decline, trunk diseases, actinobacteria, rhizosphere, endophytes
INTRODUCTION
Grapevines are one of the most important economic crops worldwide. Grapevine trunk diseases (GTD) are a major threat for the wine sector, causing serious economic losses to the wine industry (1, 2). GTD include different fungal diseases, the most relevant of which are Botryosphaeria dieback, esca, Eutypa dieback, Petri disease, and blackfoot. The incidence of GTD has increased over the last decades, primarily due to the lack of effective strategies for fighting these diseases (3, 4). Adult plant infections in mature vineyards can occur through the root system. This is the primary mode of colonization for pathogens belonging to the Dactylonectria or Ilyonectria genus (5), which are responsible for blackfoot disease. Other pathogens, such as species of the Botryosphaeriaceae family and Eutypa lata, mainly infect the plant through pruning wounds produced at the end of the growing season each year (6 – 8). Finally, other pathogens, like Phaeoacremonium minimum (formerly Phaeoacremonium aleophilum) or Phaeomoniella chlamydospora, can use both pathways to penetrate the plant, and these two microorganisms are the primary causal agents of Petri disease (9, 10). Young grapevine plants can be infected in the field by the same mechanisms described for adult plants. It is well known that planting material (grafted plants) produced in nurseries is frequently infected with fungal pathogens, especially those involved in blackfoot and Petri diseases. In fact, it has been widely reported that grafts can be infected at different stages of the propagation process that takes place in the nurseries (10, 11). GTD, particularly blackfoot disease and Petri disease, are also recognized as prevalent causes of young grapevine decline (YGD). Decline symptoms in young vineyards have dramatically increased all over the world since the early 1990s, and these plants are primarily infected through their root system (10).
Accordingly, in recent years, there have been many different studies aimed at controlling or diminishing fungal pathogen infection rates (6, 12 – 23). Most studies have focused on protecting pruning wounds by applying natural antifungal compounds (12) or different chemical fungicides. Benomyl and flusilazole are the most effective at controlling Eutypa lata infections (13, 14), thiophanate-methyl is used for treating both Petri disease and several Botryosphaeriaceae species (6) infections, and benzimidazole works effectively in protecting pruning wounds infected by Diplodia seriata, P. chlamydospora, or Inocutis sp. (15). The efficacy of several biological control agents (BCAs), including natural epiphytes and colonizers of pruning wounds (16 – 18), has been tested. The application of several strains of Trichoderma (19 – 22) has also been attempted.
Unfortunately, there is currently no treatment available to protect grafts in nurseries or grapevines in vineyards from phytopathogenic fungi that can infect plants through their root system. Nevertheless, some studies have been focused on the application of different Trichoderma strains to soils or the grapevine root system to prevent infection by fungi that cause GTD. Thus, in a nursery trial, Fourie and Hallen (23) reported that the application of Trichoderma strains had a growth-stimulating effect and reduced the infection rates of Cylindrocarpon sp., Phaeoacremonium sp., and P. chlamydospora. More recently, a study involving the application of Trichoderma harzianum at different vine growth stages in a nursery indicated that application during the rooting stage was the most effective in protecting against P. chlamydospora infections. Unfortunately, an increase in vine mortality was observed at the end of the growing season (19).
Actinobacteria, particularly streptomycetes, are a complex group of Gram-positive bacteria making up around 10% of the total soil microbiome (24). Although they are known as soil and rhizosphere bacteria, several reports indicate that they are also more intimately associated with plants, either as endophytic strains (25 – 27) or mycorrhiza-associated strains (28, 29). They are well known as important secondary metabolite producers, excreting antibiotic and antifungal compounds, and for their ability to control plant diseases (29 – 31).
Recently, bacterial communities associated with Vitis vinifera rhizospheres and their variations, including factors like geographical features (32) and vineyard management, have been characterized (33, 34). Although some reports indicate that grapevine rhizosphere-associated bacteria could have some beneficial effects on the plants, particularly in protecting them from reactive oxygen species (35), no special attention has been focused on actinobacteria and streptomycetes as a potential group from which to isolate strains to control phytopathogenic fungi that cause GTD. Loqman and colleagues (36) isolated 142 actinobacterial strains from rhizospheres of V. vinifera plants cultivated in Moroccan soils, and some of these isolates were effective against different fungal pathogens not involved in GTD. This study further characterizes the endophytic and rhizosphere microbiota associated with the grapevine root system, particularly as a potential source of strains of interest in controlling the incidence of GTD produced by fungi that penetrate plants through their root system and which are therefore involved in YGD. The best-performing isolates were tested in a 3-year field trial with grafted vines in a commercial nursery in Spain between 2013 and 2015.
RESULTS
Isolation and identification of culturable endophytic actinobacteria associated with the Vitis vinifera root system and their antifungal activity.
A total of 58 endophytic strains were obtained from four different young V. vinifera plants using various culture media (Table 1). 16S rRNA gene sequencing of all the strains revealed their genus levels. Streptomyces was the most abundant genus (26 out of 58 strains), representing 44.8% of the total isolates. Micromonospora (15 out of 58) and Saccharopolyspora strains (10 out of 58) represented 25.9% and 17.2% of the total isolates, respectively (see Fig. S1 in the supplemental material). Microorganisms from other taxa were also isolated in smaller numbers, including the rare actinomycete taxa Nonomuraea (37), Kribbella, Rathayibacter, and Corynebacterium (4 out of 58 strains [6.9%]). These strains, 1 out of 58 in each case, represented 1.7% of the total isolates. Since the primary goal of this work was to evaluate the putative application of selected actinobacteria (able to infect plants through their root system in order to control fungal pathogens involved in GTD and YGD), we screened all the endophytic strains for antifungal activity. A bioassay-based in vitro evaluation showed that 15.5% (9 out of 58) and 13.8% (8 out of 58) of the endophytic isolates, respectively, exhibited antifungal activity against D. seriata and Dactylonectria macrodidyma pathogens (Table 1). These pathogens were initially selected for their high growth rate. The five strains showing higher inhibition (I) index values were also tested for their antifungal activities against P. chlamydospora and P. minimum. Three endophytic isolates, Streptomyces sp. VV/E1, Streptomyces sp. VV/E2, and Streptomyces sp. VV/E5, showed antifungal activities against all the pathogens tested. These were selected for field assays.
TABLE 1.
Antifungal activity and culture media for the isolation of culturable endophytic actinobacteria isolated from the root system of grafted grapevines
| Isolate | I indexa |
Detection of antifungal activityb |
Isolation medium | 16S rRNA sequence accession no. | ||
|---|---|---|---|---|---|---|
| D. seriata | D. macrodidyma | P. chlamydospora | P. minimum | |||
| Streptomyces sp. VV/E1 | 53.47 | 33.91 | + | + | SC | KY978646 |
| Streptomyces sp. VV/E2 | 61.17 | 59.41 | + | + | ISP2 | KY978647 |
| Saccharopolyspora sp. VV/E3 | 31.97 | 35.87 | − | − | SC | KY978648 |
| Streptomyces sp. VV/E4 | 46.95 | 46.62 | − | + | SC | KY978649 |
| Streptomyces sp. VV/E5 | 52.50 | 47.15 | + | + | SC | KY978650 |
| Streptomyces sp. VV/E6 | 49.57 | 33.86 | ND | ND | DPA | KY978651 |
| Saccharopolyspora pathumthaniensis VV/E7 | 36.81 | — | ND | ND | ISP2 | KY978652 |
| Streptomyces sp. VV/E8 | 33.42 | — | ND | ND | DPA | KY978653 |
| Streptomyces sp. VV/E9 | — | 40.24 | ND | ND | DPA | KY978654 |
| Saccharopolyspora sp. VV/E10 | 29.97 | — | ND | ND | ISP2 | KY978655 |
| Streptomyces sp. VV/E11 | — | 40.24 | ND | ND | SAA | KY978656 |
| Streptomyces atrovirens VV/E12 | — | — | ND | ND | ISP2 | KY978657 |
| Streptomyces sp. VV/E13 | — | — | ND | ND | ISP2 | KY978658 |
| Streptomyces sp. VV/E14 | — | — | ND | ND | SAA | KY978659 |
| Streptomyces sp. VV/E15 | — | — | ND | ND | SC | KY978660 |
| Streptomyces sp. VV/E16 | — | — | ND | ND | SC | KY978661 |
| Streptomyces sp. VV/E17 | — | — | ND | ND | ISP2 | KY978662 |
| Streptomyces sp. VV/E18 | — | — | ND | ND | SAA | KY978663 |
| Micromonospora sp. VV/E19 | — | — | ND | ND | ISP2 | KY978664 |
| Micromonospora sp. VV/E20 | — | — | ND | ND | ISP2 | KY978665 |
| Micromonospora sp. VV/E21 | — | — | ND | ND | SC | KY978666 |
| Streptomyces sp. VV/E22 | — | — | ND | ND | SC | KY978667 |
| Streptomyces sp. VV/E23 | — | — | ND | ND | DPA | KY978668 |
| Streptomyces sp. VV/E24 | — | — | ND | ND | SC | KY978669 |
| Streptomyces sp. VV/E25 | — | — | ND | ND | SC | KY978670 |
| Streptomyces sp. VV/E26 | — | — | ND | ND | DPA | KY978671 |
| Micromonospora palomenae VV/E27 | — | — | ND | ND | ISP2 | KY978672 |
| S. pathumthaniensis VV/E28 | — | — | ND | ND | SC | KY978673 |
| Streptomyces sp. VV/E29 | — | — | ND | ND | ISP2 | KY978674 |
| Streptomyces sp. VV/E30 | — | — | ND | ND | SC | KY978675 |
| S. pathumthaniensis VV/E31 | — | — | ND | ND | ISP2 | KY978676 |
| Streptomyces sp. VV/32 | — | — | ND | ND | SC | KY978677 |
| S. pathumthaniensis VV/E33 | — | — | ND | ND | ISP2 | KY978678 |
| Rathayibacter caricis VV/E34 | — | — | ND | ND | ISP2 | KY978679 |
| Nonomuraea kuesteri VV/E35 | — | — | ND | ND | ISP2 | KY978680 |
| S. pathumthaniensis VV/E36 | — | — | ND | ND | SC | KY978681 |
| N. kuesteri VV/E37 | — | — | ND | ND | SC | KY978682 |
| S. pathumthaniensis VV/E38 | — | — | ND | ND | ISP2 | KY978683 |
| Micromonospora sp. VV/E39 | — | — | ND | ND | ISP2 | KY978684 |
| Micromonospora sp. VV/E40 | — | — | ND | ND | SC | KY978685 |
| Micromonospora sp. VV/E41 | — | — | ND | ND | ISP2 | KY978686 |
| Micromonospora sp. VV/E42 | — | — | ND | ND | ISP2 | KY978687 |
| Micromonospora sp. VV/E43 | — | — | ND | ND | ISP2 | KY978688 |
| Streptomyces sp. VV/E44 | — | — | ND | ND | SAA | KY978689 |
| Micromonospora sp. VV/E45 | — | — | ND | ND | SC | KY978690 |
| Micromonospora sp. VV/E46 | — | — | ND | ND | ISP2 | KY978691 |
| Micromonospora sp. VV/E47 | — | — | ND | ND | ISP2 | KY978692 |
| Micromonospora sp. VV/E48 | — | — | ND | ND | SC | KY978693 |
| S. pathumthaniensis VV/E49 | — | — | ND | ND | SC | KY978694 |
| Micromonospora sp. VV/E50 | — | — | ND | ND | ISP2 | KY978695 |
| Nonomuraea sp. VV/E51 | — | — | ND | ND | SC | KY978696 |
| S. pathumthaniensis VV/E52 | — | — | ND | ND | ISP2 | KY978697 |
| Streptomyces sp. VV/E53 | — | — | ND | ND | DPA | KY978698 |
| Nonomuraea sp. VV/E54 | — | — | ND | ND | ISP2 | KY978699 |
| Kribbella sp. VV/E55 | — | — | ND | ND | SAA | KY978700 |
| Corynebacterium sp. VV/E56 | — | — | ND | ND | ISP2 | KY978701 |
| Micromonospora sp. VV/E57 | — | — | ND | ND | ISP2 | KY978702 |
| Streptomyces sp. VV/E58 | — | — | ND | ND | SAA | KY978703 |
Inhibition index (I index) values shown are the average of three independent experiments. —, species not showing antifungal activity against D. seriata or D. macrodidyma.
ND, not determined.
Antifungal activities of rhizosphere actinobacteria.
A total of 94 rhizosphere strains were isolated. Bioassay-based in vitro screening showed that 30.8% (29 out of 94) and 16.0% (15 out of 94) of the isolates exhibited antifungal activity against D. seriata and D. macrodidyma pathogens, respectively (Table 2). Only those isolates exhibiting antifungal activity were identified by 16S rRNA sequencing, and all turned out to be members of the Streptomyces genus (Fig. S1). The five strains which showed the highest I index against D. macrodidyma were also tested for their antifungal activities against P. chlamydospora and P. minimum. Three rhizosphere isolates, Streptomyces sp. VV/R1, Streptomyces sp. VV/R4, and Streptomyces sp. VV/R5, showed antifungal activity against all the pathogens tested and accordingly were selected for field assays.
TABLE 2.
Antifungal activity and culture media for the isolation of culturable rhizospheric actinobacteria isolated from the root system of grafted grapevines
| Streptomyces isolate | I indexa |
Detection of antifungal activityb |
Isolation medium | 16S rRNA sequence accession no. | ||
|---|---|---|---|---|---|---|
| D. seriata | D. macrodidyma | P. chlamydospora | P. minimum | |||
| VV/R1 | 48.75 | 55.06 | + | + | ISP2 | KY978704 |
| VV/R2 | 45.52 | 53.20 | − | + | SC | KY978705 |
| VV/R3 | 42.58 | 49.13 | − | − | ISP2 | KY978706 |
| VV/R4 | 51.69 | 50.77 | + | + | SC | KY978707 |
| VV/R5 | 55.60 | 51.43 | + | + | SC | KY978708 |
| VV/R6 | 47.84 | 44.57 | ND | ND | SC | KY978709 |
| VV/R7 | 51.83 | 41.22 | ND | ND | DPA | KY978710 |
| VV/R8 | 51.04 | 44.40 | ND | ND | SAA | KY978711 |
| VV/R9 | 54.14 | 21.55 | ND | ND | ISP2 | KY978712 |
| VV/R10 | 45.65 | 48.44 | ND | ND | SC | KY978713 |
| VV/R11 | 45.52 | 35.23 | ND | ND | SC | KY978714 |
| VV/R12 | 44.61 | 49.07 | ND | ND | SC | KY978715 |
| VV/R13 | 52.03 | 46.41 | ND | ND | ISP2 | KY978716 |
| VV/R14 | 56.95 | 43.42 | ND | ND | SC | KY978717 |
| VV/R15 | 49.47 | 48.67 | ND | ND | ISP2 | KY978718 |
| VV/R16 | 45.65 | — | ND | ND | DPA | KY978719 |
| VV/R17 | 40.48 | — | ND | ND | SC | KY978720 |
| VV/R18 | 62.27 | — | ND | ND | SC | KY978721 |
| VV/R19 | 48.66 | — | ND | ND | SC | KY978722 |
| VV/R20 | 44.67 | — | ND | ND | SAA | KY978723 |
| VV/R21 | 40.61 | — | ND | ND | SAA | KY978724 |
| VV/R22 | 55.21 | — | ND | ND | ISP2 | KY978725 |
| VV/R23 | 48.27 | 47.02 | ND | ND | SC | KY978726 |
| VV/R24 | 40.71 | — | ND | ND | ISP2 | KY978727 |
| VV/R25 | 80.50 | — | ND | ND | ISP2 | KY978728 |
| VV/R26 | 62.87 | — | ND | ND | DPA | KY978729 |
| VV/R27 | 90.81 | — | ND | ND | SC | KY978730 |
| VV/R28 | 43.59 | — | ND | ND | SC | KY978731 |
| VV/R29 | 44.95 | — | ND | ND | ISP2 | KY978732 |
Inhibition index (I index) values shown are the average of three independent experiments. —, species not showing antifungal activity against D. seriata or D. macrodidyma.
ND, not determined.
Multilocus sequence analysis of actinobacterial strains selected for field assays.
As previously indicated, preliminary characterization of the selected endophytic and rhizosphere actinobacteria based on 16S rRNA sequencing led to their identification as species of the Streptomyces genus. To further characterize and more precisely identify them, we performed a multilocus sequence analysis (MLSA) study of five housekeeping genes (atpD, gyrB, recA, rpoB, and trpB).
MLSA (Fig. S2) confirmed that isolates VV/E1 and VV/R4 are the same species (sharing an MLSA evolutionary distance of 0.000; Table 3), although they exhibit macroscopic differences when grown in different culture media (Fig. S3). Both strains are closely related to Streptomyces albaduncus (showing an MLSA distance of 0.028 with this species; Table 3). Isolates VV/R1 and VV/E5 were located in a well-delineated subclade including Streptomyces peucetius and Streptomyces xantholiticus strains (Fig. S2). Both isolates share an MLSA distance of 0.108, indicating they are different species. Isolate VV/E5 was identified as S. peucetius, as it had MLSA distances smaller than 0.007 (Table 3) with the S. peucetius species. Strain VV/R1 matched between S. peucetius (0.108 MLSA distance) and S. xantholiticus (0.112 MLSA distance) (Fig. S2), suggesting that it could be a novel Streptomyces species. Isolate VV/E2 was located in a subclade including Streptomyces albidochromogenes, Streptomyces flavidovirens, and Streptomyces helvaticus (Fig. S2). MLSA distances with these strains were always higher than 0.007, indicating that isolate VV/E2 could be a novel species. Isolate VV/R5 is closely related to members of a subclade including Streptomyces phaeochromogenes and Streptomyces umbrinus (Fig. S2), although its MLSA distance with respect to both species clearly indicated that this isolate belongs to a different species.
TABLE 3.
MLSA distances for strains phylogenetically near to our isolates
| No. | Straina | MLSA (Kimura 2-parameter) distance |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | ||
| 1 | S. albaduncus NRRL B-3605T | ||||||||||||||||||
| 2 | S. albaduncus NRRL b-3605gT | 0.000 | |||||||||||||||||
| 3 | VV/E1 | 0.028 | 0.028 | ||||||||||||||||
| 4 | VV/R4 | 0.028 | 0.028 | 0.000 | |||||||||||||||
| 5 | S. umbrinus NRRL B-2572T | 0.094 | 0.094 | 0.092 | 0.092 | ||||||||||||||
| 6 | VV/R5 | 0.096 | 0.096 | 0.091 | 0.091 | 0.040 | |||||||||||||
| 7 | S. phaeochromogenes NRRL B-1248T | 0.094 | 0.094 | 0.091 | 0.091 | 0.034 | 0.013 | ||||||||||||
| 8 | S. phaeochromogenes subsp. phaeochromogenes NRRL B-3010T | 0.094 | 0.094 | 0.091 | 0.091 | 0.034 | 0.013 | 0.000 | |||||||||||
| 9 | S. helvaticus NRRL B-12365T | 0.124 | 0.124 | 0.130 | 0.130 | 0.121 | 0.124 | 0.126 | 0.126 | ||||||||||
| 10 | VV/E2 | 0.119 | 0.119 | 0.126 | 0.126 | 0.118 | 0.122 | 0.122 | 0.122 | 0.043 | |||||||||
| 11 | S. albidochromogenes NRRL B-24308T | 0.113 | 0.113 | 0.117 | 0.117 | 0.110 | 0.112 | 0.113 | 0.113 | 0.045 | 0.057 | ||||||||
| 12 | S. flavidovirens subsp. flavidovirens NRRL B-2708T | 0.113 | 0.113 | 0.113 | 0.113 | 0.106 | 0.113 | 0.111 | 0.111 | 0.067 | 0.055 | 0.048 | |||||||
| 13 | S. flavidovirens DSM 40150T | 0.113 | 0.113 | 0.113 | 0.113 | 0.106 | 0.115 | 0.112 | 0.112 | 0.067 | 0.055 | 0.050 | 0.001 | ||||||
| 14 | S. peucetius subsp. peucetius NRRL B-3826T | 0.104 | 0.104 | 0.105 | 0.105 | 0.111 | 0.113 | 0.108 | 0.108 | 0.131 | 0.128 | 0.131 | 0.124 | 0.124 | |||||
| 15 | VV/E5 | 0.106 | 0.106 | 0.107 | 0.107 | 0.112 | 0.114 | 0.110 | 0.110 | 0.132 | 0.130 | 0.132 | 0.126 | 0.126 | 0.002 | ||||
| 16 | S. peucetius CGMCC 4.1799T | 0.122 | 0.122 | 0.105 | 0.105 | 0.111 | 0.113 | 0.111 | 0.111 | 0.131 | 0.128 | 0.131 | 0.124 | 0.124 | 0.000 | 0.002 | |||
| 17 | VV/R1 | 0.219 | 0.219 | 0.187 | 0.187 | 0.201 | 0.200 | 0.196 | 0.196 | 0.218 | 0.212 | 0.214 | 0.210 | 0.210 | 0.108 | 0.108 | 0.108 | ||
| 18 | S. xantholiticus NRRL B-12153T | 0.106 | 0.106 | 0.108 | 0.108 | 0.113 | 0.114 | 0.111 | 0.111 | 0.130 | 0.125 | 0.128 | 0.124 | 0.124 | 0.012 | 0.014 | 0.112 | 0.112 | |
Our isolates are in bold.
Field assays. (i) Growth data and mortality rate.
The application of actinobacteria to grafted plants did not have a significant effect, either positive or negative, on the growth of the surviving plants, as determined by measurement of total plant height [F(6.413) = 0.37, P = 0.90] (Fig. 1A) or elongation of the seventh internode [F(6.413) = 0.12, P = 0.29] (Fig. 1B).
FIG 1.
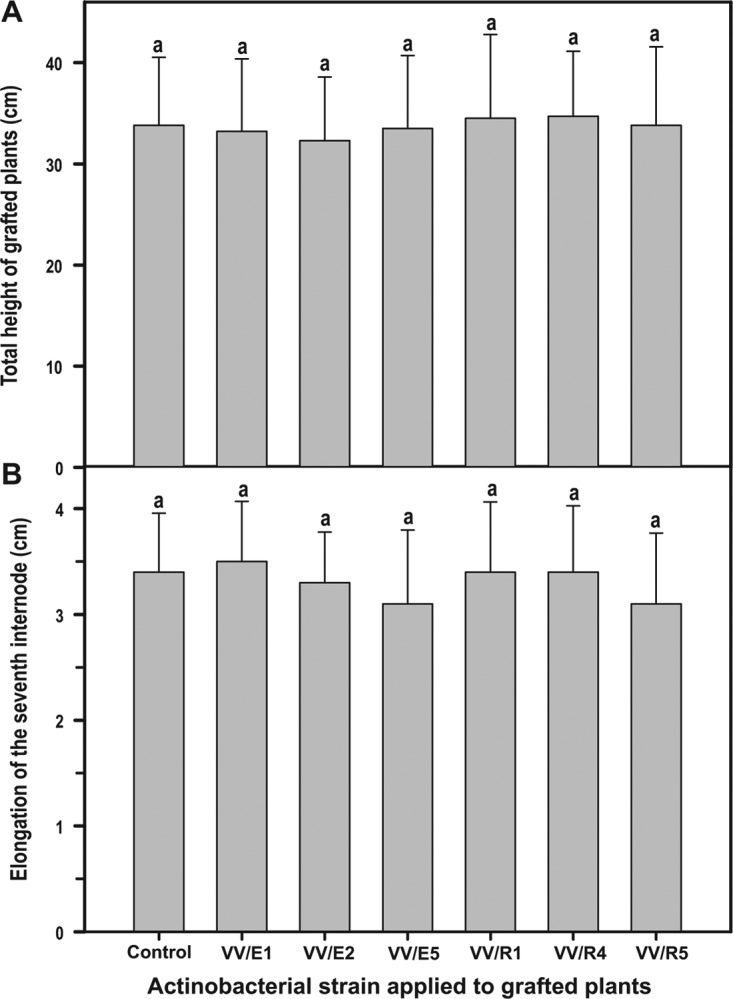
Plant growth in the experimental nurseries (2013, 2014, and 2015 growing seasons) as determined by total height (A) or length of the 7th internode (B) in grafted plants treated with different endophytic (VV/E1, VV/E2, or VV/E5) or rhizosphere (VV/R1, VV/R4, or VV/R5) actinobacteria compared to untreated control plants. Each bar corresponds to the arithmetic mean of the values determined for the total number of surviving plants in the three seasons tested and their corresponding standard error (SE). Bars marked with the same letter are not significantly different (P ≥ 0.05).
The average mortality rate of noninoculated untreated (negative control) plants in the 3-year field study was 28.4% (64 dead plants out of a total of 225 plants) (Fig. 2). Significant differences in mortality rates were observed for two of the treatments, according to the chi-square tests. The mortality rates of plants inoculated with VV/R1 (19.1% [43 dead plants out of 225 plants]) [χ2 (1) = 5.41, P < 0.05] or VV/R4 (17.8% [40 dead plants out of 225 plants]) [χ2 (1) = 7.20, P < 0.01] strains were significantly lower (Fig. 2). Thus, we could conclude that the odds of plant survival were 1.68 times higher on VV/R1 treatment than on untreated control plants, whereas the odds of plant survival were 1.84 times higher on VV/R4 treatment than on untreated control plants.
FIG 2.

Average mortality rates (%) and their corresponding SE values of grafted grapevine plants observed in the experimental nurseries (2013, 2014, and 2015 growing seasons) after treatment with different endophytic (VV/E1, VV/E2, or VV/E5) or rhizosphere (VV/R1, VV/R4, or VV/R5) strains compared to untreated control plants. Bars marked with the same letter are not significantly different (P ≥ 0.05).
(ii) Isolation of pathogenic fungi.
Two hundred days after planting (once the plants had completed their vegetative growth cycle and were in a dormant state), vines were carefully removed from the soil to obtain a root system that was as intact as possible. The incidence of fungal pathogens causing YGD (P. chlamydospora, P. minimum, and Dactylonectria sp.-Ilyonectria sp. group) and able to infect the plants through their root system was determined by collecting samples of wood taken from the root insertion point. All the selected actinobacteria led to a reduction in the number of fungal pathogens involved in YGD (Fig. 3) compared to the number of fungal pathogens isolated from control plants (raw data corresponding to this study are shown in Table S1).
FIG 3.
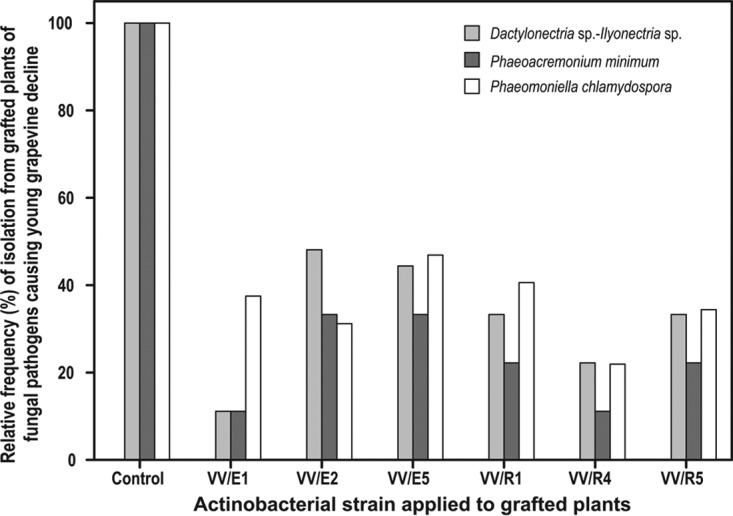
Relative isolation frequency (%) of fungal pathogens involved in young grapevine decline from grafted plants treated with different endophytic and rhizosphere actinobacteria with respect to untreated plants. A value of 100 was arbitrarily assigned to the total number of fungal pathogens of a particular species (P. chlamydospora or P. minimum) or Dactylonectria sp.-Ilyonectria sp. group analyzed once the plants had been removed from the field. The relative frequency for each type of pathogen analyzed in the plants treated with the different actinobacteria was calculated as a percentage (%) compared against the value of 100 assigned to the control plants. Data shown correspond to the average of 3 batches of 25 plants in 3 experimental nurseries (2013, 2014, and 2015 growing seasons).
In order to analyze the impact of every actinobacterial treatment on the number of each particular type of fungal pathogen involved in YGD, we used nested generalized linear models (Poisson distribution, with intercept versus intercept plus treatment as dependent variables into the model). The existence of significant differences between the number of fungal pathogens isolated from plants treated with the different actinobacteria and the number of pathogens isolated from control plants was determined using chi-square tests. Thus, in accordance with the nested generalized linear models, significant associations between the nature of the treatment and the number of fungal pathogens isolated were found in the Dactylonectria sp.-Ilyonectria sp. group and for P. chlamydospora (P < 0.05 in the chi-square tests conducted to compare the nested models) for all the actinobacteria assayed. On the other hand, no significant association (P > 0.05 in the chi-square tests) between the nature of the treatment and the number of fungal pathogens isolated was detected for P. minimum, probably due to the low number of individuals of this species isolated.
After that, chi-square and Fisher's tests were performed to establish the existence of significant differences between treatments. The results are shown as heatmaps of independence assumptions between the type of actinobacterial treatment and the number of fungal pathogens isolated (Fig. 4). Surprisingly, despite the data shown by nested generalized linear models, significant differences (P < 0.05) between the negative control and the three treatments (VV/E1, VV/R1, and VV/R4) were found with respect to P. minimum (Fig. 3). However, these significant associations were very close to the edge of the significance threshold (0.05). It may be that the nested model tests are more conservative than individual tests performed between treatments.
FIG 4.
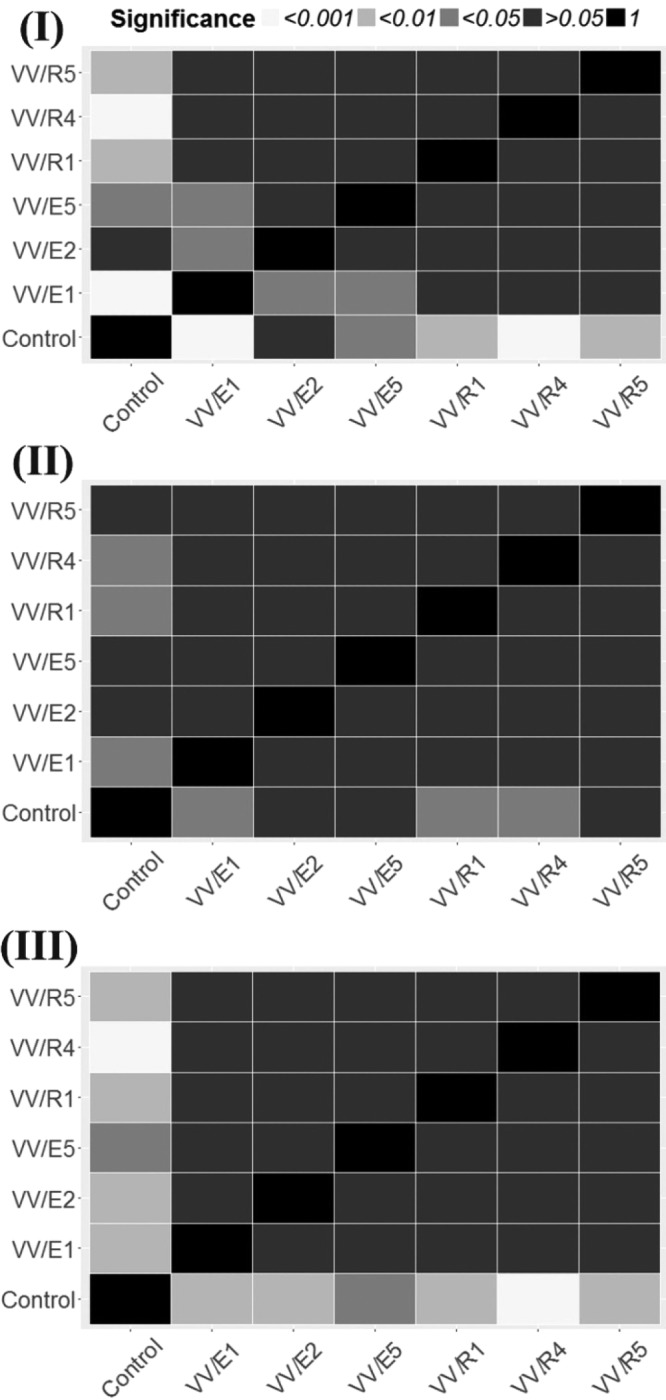
Analysis of significant differences between the number of fungal pathogens isolated from plants treated with the different actinobacteria versus the number of pathogens isolated from control plants. The analysis is shown as heatmaps of independence assumptions between the type of actinobacterial treatment and the number of fungal pathogens isolated. Color key corresponds to the significance of chi-square or Fisher's test results (according to expected cell values). (I) Data relative to Dactylonectria sp.-Ilyonectria sp. group; (II) data relative of P. minimum; (III) data relative to P. chlamydospora. Data shown correspond to 3 batches of 25 surviving plants in 3 experimental nurseries (2013, 2014, and 2015 growing seasons).
In the cases related to the Dactylonectria sp.-Ilyonectria sp. group isolation, and solely focusing on those highly significant associations between the nature of the treatment and the number of fungal pathogens isolated (P < 0.01 and <0.001), the following results should be highlighted: control and VV/E1 [χ2 (1) = 46.4, P < 0.001], control and VV/R4 [χ2 (1) = 12.0, P < 0.001], control and VV/R1 [χ2 (1) = 7.15, P < 0.01], and control and VV/R5 [χ2 (1) = 5.91, P < 0.01] (Fig. 4). If we focus on P. chlamydospora, the following highly significant associations were detected: control and VV/R4 [χ2 (1) = 15.8, P < 0.001], control and VV/E1 [χ2 (1) = 10.8, P < 0.01], control and VV/E2 [χ2 (1) = 10.8, P < 0.01], control and VV/R1 [χ2 (1) = 10.8, P < 0.01], and control and VV/R5 [χ2 (1) = 9.43, P < 0.01] (Fig. 4).
(iii) Rootstock colonization by selected actinobacteria.
The ability of the selected actinobacteria to colonize rootstocks at the root insertion point was analyzed 10 and 200 days after their application. As shown in Table 4, 10 days after application, all the actinobacteria were successfully recovered from rootstocks at percentages ranging between 11.43% (Streptomyces sp. VV/R5) and 36.43% (Streptomyces sp. VV/E1), indicating that the application method was effective enough to allow actinobacterial penetration into the rootstock. The differences were significant in all cases with respect to the isolation values for the same strain from control plants.
TABLE 4.
Rootstock colonization rates by selected actinobacteria at 10 and 200 days after their application
| Actinobacterium isolated by batch of plants | % of isolated actinobacteria aftera: |
|
|---|---|---|
| 10 days | 200 days | |
| Control plants | ||
| VV/E1 | 6.43 (9) | 4.29 (6) |
| VV/E2 | 1.43 (2) | 0.71 (1) |
| VV/E5 | 4.29 (6) | 2.14 (3) |
| VV/R1 | 3.57 (5) | ND (0) |
| VV/R4 | 2.86 (4) | 3.57 (5) |
| VV/R5 | ND (0) | 0.71 (1) |
| Treated plants | ||
| VV/E1 | 36.43 (51) | 13.57 (19) |
| VV/E2 | 27.14 (38) | 7.86 (11) |
| VV/E5 | 16.43 (23) | 4.29 (6) |
| VV/R1 | 30.00 (42) | ND (0) |
| VV/R4 | 20.71 (29) | 7.14 (10) |
| VV/R5 | 11.43 (16) | ND (0) |
ND, not detected. The percentage of each particular strain was calculated as the number of clones growing from wood chips and identified by sequencing of 16S rRNA against a number of 140 chips tested under each condition (7 chips isolated from two stubs per 10 plants analyzed). Values are the average of two independent experiments. Raw data (number of each particular actinobacterium isolated from 140 chips) are indicated in parentheses.
At the end of the trials (200 days after planting), only four strains could be reisolated from the plants, and these were always at lower levels than those detected 10 days after application. Significant differences compared to the control were observed for Streptomyces sp. VV/E1 and Streptomyces sp. VV/E2 strains. These data showed that with the passage of time, the degree of colonization tends to decrease.
DISCUSSION
It is well established that grapevine plants are mainly infected by fungal pathogens causing GTD through pruning wounds or the root system, with a reduced number of fungal strains that can use both routes of entry. There have been many different studies to test pruning wound protection with chemical antifungals (6, 13 – 15), natural antifungal compounds (12), or BCAs (16 – 18). Given the many years that a plant remains in the ground and the consequent extent and depth of root development and soil penetration, it is clear that the protection of grapevines from fungi that penetrate through the root system is much more complicated. Current studies have not gone much beyond the application of BCAs to the root system, mainly different strains of Trichoderma sp. (19, 23). Thus, it seems evident that development of technologies that can limit the infection of vine plants through their root systems is of great interest for the grape and wine industry. Thus, we decided to investigate the putative application of certain actinobacteria that might help mitigate this problem.
Actinobacteria are an integral part of soil microbial communities, making up around 10% of the total soil microbiome (24), and they stand out, particularly streptomycetes, for their ability to produce a wide range of secondary metabolites, including antifungal compounds (29 – 31). Several studies indicate that actinobacteria can enter root tissues and establish an endophytic lifestyle with plants (26 – 30). As reported in other plants, the grapevine root system contains many different endophytic actinobacteria. Streptomyces and Micromonospora were the most frequently isolated genera, representing up to 70.7% of the total isolates. Similarly, when the population of endophytic actinobacteria in Artemisia annua was analyzed, Streptomyces and Micromonospora represented 57.0% of the total identified isolates (26). Other minority genera found in grapevine plants, like Nonomuraea and Kribbella, were also detected in A. annua (26). However, Corynebacterium and Saccharopolyspora, genera isolated from V. vinifera, were not detected in A. annua (26). These data confirm that actinobacteria are frequent endophytes in the root systems of both plants.
Streptomyces species identification is a great challenge, further complicated by factors, such as the fact that there are a great number of species, many of which are poorly defined. The insufficient resolution of 16S rRNA as a phylogenetic marker, difficulties in effectively addressing phenotypic differences, and the absence of rapid molecular identification methods applicable to a high number of species also hinder the accurate identification of species of Streptomyces (38, 39). Since 16S rRNA sequencing does not provide sufficient resolution for species-level identification of streptomycetes, we performed an MLSA study of the six isolates used in field trials by partially sequencing five housekeeping genes. MLSA has proven to be an efficient molecular tool for the improved taxonomic resolution of Streptomycetaceae family (39), as well as for the right identification of Streptomyces species for the S. albidoflavus (40), S. griseus (41), and S. hygroscopicus (38) clades. These clades include some of the more widely studied species in the genera, many of which produce antibiotics and other industrially and agronomically important secondary metabolites (38). In our case, the MLSA allowed us to identify the isolate VV/E5 as belonging to the S. peucetius species. Unfortunately, this study did not lead to an accurate identification of the rest of the strains at the species level, probably due to a poor characterization of those closely related phylogenetic strains. This setback makes it difficult to establish whether these strains are unique to the root system of grapevines or to assess their ability to produce known antifungal compounds. We cannot even rule out that some of them are new species. The case of Streptomyces sp. VV/E1 and VV/R4 is particularly intriguing, since the MLSA study indicated that the two isolates belong to the same species. In fact, these strains share 99.5% homology at the 16S rRNA level. However, they exhibit quite different macroscopic traits when growing on various culture media, as shown in Fig. S3. Remember that Streptomyces sp. VV/E1 was isolated as an endophytic strain, whereas Streptomyces sp. VV/R4 was isolated from the rhizosphere environment. We can speculate that the observed macroscopic differences could be a putative adaptation to two different lifestyles. However, currently, we do not know for certain whether a particular strain present in the rhizosphere might also develop an endophytic lifestyle. Works under way for the development of molecular markers for the specific detection of these strains, as well as genome sequencing of the VV/E1 strain, should enable us in the future to address many of these issues that remain unanswered for now.
Recently, there have been several attempts to characterize the endophytic bacterial microbiota of grapevine plants. Andreolli et al. (42) analyzed the diversity of bacterial endophytes in 3- and 15-year-old plants. In 3-year-old plants, Actinobacteria was the second dominant class (26%), and the genera detected were Nocardioides, Curtobacterium, Microbacterium, Brachybacterium, Kocuria, and Micrococcus. None of these genera were detected in our study, and surprisingly, Andreolli and colleagues did not detect any Streptomyces sp. strains. In 15-year-old plants, the population of endophytic actinobacteria was reduced to 5%, with only 2 strains detected, belonging to the Microbacterium and Curtobacterium genera. Also recently, Rezgui et al. (43) analyzed the population of endophytic bacteria that inhabit the wood tissues from grapevines of Tunisian vineyards. Actinobacteria were hardly detected, and Curtobacterium was the only genus found. These differences can be attributed to many different factors. First, in both studies, endophytes were isolated from vine stems and mature arms, which are aerial habitats far from the ground. In fact, it has been reported that the population of actinobacteria decreases along the plant axis from 20% in the root, to 5% in the graft union, and just 2% in the cane samples (44). Second, the observed differences may be attributed to some extent to the different isolation culture media used, and even to the different molecular techniques used in the two works (43, 44).
Recently, different studies have focused on the characterization of the grapevine microbiome (45, 46) and vineyard soils (32, 34). After Proteobacteria, Actinobacteria are the second most abundant microorganisms in vineyard soils, representing averages of 9.72% and 20.48% of the total microorganisms detected in vineyards of Argentina (34) and California (32), respectively. In New York vineyards, Actinobacteria were present at 5.1% in grapevine roots, whereas their relative abundance was lower in all aboveground samples (grapes, leaves and flowers) tested (45). Actinobacteria are also a prominent group detected in both rhizosphere (around 44%), and roots (around 36%) of plants from Austrian vineyards (46). Unfortunately, most of these studies dealing with soil and root microbiomes do not delve deeply into characterization of the actinobacteria detected beyond the taxonomic class level. Future studies, focused more specifically on the population of actinobacteria detected in the root system of grapevine plants at the genus or even species level, should shed light on many issues that remain unresolved.
Actinobacteria were also easily isolated from young grapevine plant rhizospheres, and a screening based on antifungal activity against fungi causing GTD and YGD resulted in the identification of a total of 29 isolates, all belonging to the genus Streptomyces. Furthermore, Loqman and colleagues (36) have isolated strains of Streptomyces with antifungal activity against Botrytis cinerea from a grapevine rhizosphere in Morocco. Together, these data indicate that both the rhizosphere and the interior of the grapevine root system are highly interesting sources for actinobacteria which may be potential BCAs for the treatment of different vineyard pathologies.
Blackfoot and Petri diseases, recognized as prominent causes of YGD, along with failure of planting material, are some of the most worrisome problems affecting the wine grape industry since the 1990s (10). Evaluations of declining young vineyards has revealed that many factors are involved. Among all the diseases, the grafted plant root system infections by fungal trunk pathogens in nurseries are the most relevant (10). The main fungal trunk pathologies associated with YGD are Petri disease, primarily produced by P. chlamydospora and different Phaeoacremonium species, and blackfoot disease, mainly produced by species of Campylocarpon, Dactylonectria, and Ilyonectria. These pathogens can infect a significant portion of all the grafted plants produced every year in nurseries all around the world. They are frequently found infecting the rootstock mother vines (10, 23). They can also infect grafts once they are planted in an open-root field nursery.
This step seems to be particularly critical, since it is supposed that the pathogens present in the soil can infect grafts through the incipient root system that the vines must produce to survive and develop in soil. Thus, when Halleen and colleagues analyzed the presence of fungal pathogens involved in YGD in nurseries during the different steps of the propagation process, they found that less than 1% of the plants were infected with Cylindrocarpon spp. before planting in the nursery, whereas 50% or more of the plants were infected at the end of the season (47). This result is highly significant, since it demonstrates that most of the grafted plants in the nurseries can be infected through the roots once they are planted in an open-root field nursery. Our study shows that the application of actinobacteria, selected for their in vitro antifungal properties against fungal pathogens which cause YGD, is highly effective in controlling the infection at this step. Application of all the selected actinobacteria resulted in a clear reduction in the isolation of fungal pathogens involved in YGD from surviving plants. Furthermore, the application of actinobacterial strains VV/E1, VV/R1, and VV/R4 led to a statistically significant decrease (P < 0.05) in the presence of three types of pathogenic fungi analyzed, Dactylonectria sp.-Ilyonectria group, P. chlamydospora, and P. minimum. None of the actinobacteria had a significant effect, either positive or negative, on the growth of the surviving plants. However, whereas the average mortality rate of untreated (negative control) plants in the 3-year field study was 28.4%, the mortality rate was significantly reduced when the actinobacteria VV/R1 and VV/R4 were applied to the grafts. Graft failure is a serious problem for nurseries since, on average, it results in the failure of 30 to 60% of the grafts taken to the field. Many different factors can contribute to graft failure, including abiotic causes, such as improperly healed rootstock, disbudding sites and graft unions, or improper storage and management of the plant material used for obtaining grafts (10).
However, despite the large number of studies showing the presence of fungal pathogens in nursery plants at the end of the propagation process, to date, no clear data are available that correlate the presence of pathogens with mortality levels. In fact, after analyzing the presence of fungal pathogens in plants with failed graft unions, Rumbos and Rumbou concluded that those pathogens were not the cause of YGD (48). Our data suggest a putative relationship between the number of fungal pathogens and the mortality rate, since the application of actinobacterial strains VV/R1 and VV/R4 led to both a significant reduction in the number of fungal pathogens detected and a significant reduction in the mortality rate. Interestingly, the dead plants at the end of the field trials showed a very small, almost nonexistent root system. We can speculate that the early infection with fungi causing YGD may interfere with the development of a potent root system, resulting in a premature graft death. Clearly, graft failure is a complex problem with many different factors, but in view of these results, it seems that the presence of fungal pathogens could significantly contribute to this problem.
Finally, in recent years, it has become clear that endophytic microorganisms, particularly actinobacteria, can be a significant reservoir of genetic diversity and an important source for the discovery of novel bioactive secondary metabolites (27) that can help control many different crop pathologies. This work represents one of the first reports on the characterization of the endophytic actinobacterial population in the root system of young grapevines at the levels of genus and species. Several of the isolates tested had a beneficial impact when applied to grafts in nurseries, resulting in a significant decrease in both the mortality and infection rates by fungal pathogens involved in YGD. Therefore, the application of endophytic and/or rhizosphere actinobacteria to grafts in nurseries is a novel, green, and promising technology that can reduce the incidence of grapevine plant fungal infections through the root system.
MATERIALS AND METHODS
Isolation of culturable endophytic and rhizosphere actinobacteria from the grapevine root system.
A total of four 7-month-old Vitis vinifera cv. Tempranillo plants grafted on Richter 110 (110R) rootstock were collected from an open-root field nursery (Viveros Villanueva Vides S.L.) located in Larraga (Spain) at 350 m above sea level (a.s.l.) (01°48′45.8″W and 42°32′12.9″N). Actinobacteria associated with the root system were isolated from root-adjacent soil (rhizosphere) and from interior root tissue (endophytic strains). Roots and soil were placed in sterile containers to be taken to the lab. Two different approaches were used to isolate the culturable actinobacterial population associated with the root system. Rhizosphere strains were isolated from 1 g of root-adjacent soil samples obtained from every plant collected. Soil samples were suspended in a final volume of 10 ml of sterile water and homogenized using a vortex (1 min). Tenfold serial dilutions were made in sterile water, and 0.1-ml aliquots of each dilution were inoculated on starch-casein (SC) (49), ducitol-proline agar (DPA) (26), International Streptomyces Project 2 (ISP2) (50), and sodium succinate-asparagine agar (SAA) media (26), supplemented with cycloheximide (150 μg · ml−1) and nalidixic acid (25 μg · ml−1) (Sigma-Aldrich, Madrid, Spain) in order to prevent fungal and Gram-negative bacterial growth, respectively. Plates were incubated at 28°C for 3 to 7 days.
Endophytic strains were isolated from root tissue samples. Two different sections of root tissue were used: 2-cm stubs corresponding to the root insertion point, and root fragments corresponding to the 6-cm apical ends. Wood from both samples was mixed, and microorganisms were extracted as indicated below. Roots were washed in sterile phosphate-buffered saline (PBS) and sonicated (160 W; 2 min) to dislodge soil and organic matter from the sample surface. After drying at room temperature (RT), roots were cut into 2.0-cm-long fragments. Root fragments were surface-sterilized by immersing in 20 ml of Tween 20 (0.1%) (Sigma-Aldrich) for 30 s, followed by sodium hypochlorite (1%) for 6 min and then Na2S2O3 (2.5%) (Panreac, Barcelona, Spain) for 10 min to remove the residual chlorine. After that, samples were washed three times with sterile water. Next, the root fragments were submerged in 70% (vol/vol, in sterile water) ethanol for 6 min, followed by three washes with sterile water and air-dried (in petri plates) in a laminar flow hood (Telstar AV-100; Telstar, Terrassa, Spain). To confirm the effectiveness of the surface disinfection process, 0.2 ml of liquid from the final washing step was spread onto ISP2 medium and incubated at 28°C. Two different methods were used to isolate endophytic actinobacteria from the disinfected root fragments. The first method involved the disruption of 5 g of root material using a mortar and pestle. A volume of 20 ml of 0.9% (wt/vol) NaCl was added to the samples, and they were incubated at 28°C with strong agitation (220 rpm) to favor microorganism extraction. The second method entailed disruption of the samples (0.1 g) in 2-ml tubes containing glass beads (0.5 mm diameter) with a FastPrep-24 homogenizer (MP Biomedicals, Santa Ana, CA) at 4,000 oscillations per min for 8 s, for a total of 10 times. Samples from both methods were diluted 10-fold, and 0.1-ml aliquots were plated on SC, DPA, ISP2, and SAA media. Plates were incubated at 28°C for 3 to 7 days.
Preliminary identification of actinobacteria by 16S rRNA sequencing.
Different isolates were selected according to their morphological and cultural characteristics, including colony properties, presence/absence of aerial mycelia, spore mass color, distinctive reverse colony color, and production of diffusible pigments. Isolates were routinely cultivated and maintained on ISP2 medium at 4°C. Spore-producing isolates were maintained as spore suspensions at −20°C in glycerol (40%). Endophytic and rhizosphere strains exhibiting antifungal activity were identified by 16S rRNA sequencing. Briefly, genomic DNA extraction was performed as described by Hopwood et al. (51). 16S rRNA genes were amplified using the oligonucleotides 27F and 1492R (52). Isolates were identified by comparing them to corresponding sequences of the type strain found on the EzTaxon-e database (53) (http://www.ezbiocloud.net/eztaxon/identify). Sequence alignment and generation of phylogenetic trees were performed using the MEGA 6.0 software (http://www.megasoftware.net/) using a neighbor-joining (NJ) algorithm. Evolutionary distance was calculated using the Kimura two-parameter (K2P) model for nucleotide sequences (54).
Identification of the actinobacteria selected by field assays using multilocus sequence analysis.
Multilocus sequence analysis (MLSA) was carried out using five housekeeping genes, namely, atpD (ATP synthase F1, beta subunit), gyrB (DNA gyrase B subunit), recA (recombinase A), rpoB (RNA polymerase, beta subunit), and trpB (tryptophan synthase, beta subunit) (38). PCR amplification of housekeeping genes was carried out using the primers and under the amplification conditions previously described by Guo et al. (41) and Rong et al. (40). The GenBank accession numbers of DNA sequences from the housekeeping genes are listed in Table 5. Phylogenetic trees were constructed from a concatenation of the five housekeeping genes. All the sequences were concatenated by joining them head to tail. DNA sequences were manually trimmed at the same position before being aligned using MEGA 6.0 software with sequences from type strains obtained from the ARS Microbial Genomic Sequence Database server (http://199.133.98.43). Phylogenetic trees were constructed using the maximum likelihood method with the Kimura two-parameter model (54). MLSA evolutionary distances were calculated using MEGA 6.0 by calculating the K2P distance. Strain pairs having ≤0.007 MLSA evolutionary distance were considered conspecific based on the guideline empirically determined by Rong and Huang (38).
TABLE 5.
GenBank accession numbers of DNA sequences from the endophytic and rhizosphere Streptomyces sp. strains isolated from the root system of grapevine plants used in the MLSA
| GenBank accession no. |
||||||
|---|---|---|---|---|---|---|
| Genus/species | Isolate | atpD | gyrB | recA | rpoB | trpB |
| Streptomyces sp. | VV/E1 | MF437320 | MF437326 | MF437332 | MF437338 | MF437344 |
| Streptomyces sp. | VV/E2 | MF437321 | MF437327 | MF437333 | MF437339 | MF437345 |
| Streptomyces peucetius | VV/E5 | MF437322 | MF437328 | MF437334 | MF437340 | MF437346 |
| Streptomyces sp. | VV/R1 | MF437323 | MF437329 | MF437335 | MF437341 | MF437347 |
| Streptomyces sp. | VV/R4 | MF437324 | MF437330 | MF437336 | MF437342 | MF437348 |
| Streptomyces sp. | VV/R5 | MF437325 | MF437331 | MF437337 | MF437343 | MF437349 |
Evaluation of antifungal activity.
All isolates were tested for antifungal activity using an in vitro antifungal assay, as described by Lamsal et al. (55), with minor modifications. Briefly, isolates were inoculated on potato dextrose agar (PDA) plates (Sigma-Aldrich) in a 1.0-cm2 area (4 isolates per plate inoculated at 1 cm from the edge of the plates). An agar plug (0.7 mm) containing D. seriata CBS 112555 or Ilyonectria macrodidyma CBS 120170 (now reclassified as Dactylonectria macrodidyma) was placed in the middle of the plates. The plates were incubated up to 12 days at 25°C to detect growth inhibition areas. Growth was quantified by calculating the inhibition index (I index) according to the following formula: I index (%) = [1 − (Ra − R)/(Rc − R)] × 100, where Ra is the radius of the fungal colony opposite the bacterial colony, Rc is the maximum radius of the fungal colony (farther away from the bacterial effect), and R is the radius of the agar plug containing the fungi (3.5 mm). This assay was performed in triplicate for each actinobacterium tested (Fig. S4A).
Strains showing antifungal activity against both pathogens were also screened for their ability to inhibit the growth of P. chlamydospora and P. minimum in PDA, Czapek Dox (Sigma-Aldrich), or ISP5 (50) agar plates as follows (Fig. S4B): fungal pathogens were inoculated on an agar plate forming a circle 2 cm from the periphery of the petri dish. Actinobacterial strains were inoculated in 2.25-cm2 patches located 5 mm from the edge of the fungal strain. Plates were incubated at 25°C for up to 20 days to detect the effect of growth inhibition.
Field evaluation in nurseries.
Field assays were conducted in three experimental open-root field nurseries of grafted Vitis vinifera cv. Tempranillo plants that were grafted on Richter 110 (110R) rootstock in 2013, 2014, and 2015. Every experimental nursery consisted of 525 plants. Grafts were subdivided into 7 batches of 75 units (3 replicas of 25 units per batch), each of which was treated with one of the 6 selected actinobacteria, 3 endophytic (VV/E1, VV/E2, and VV/E5) and 3 rhizosphere (VV/R1, VV/R4, and VV/R5) isolates. A negative-control batch (grafts not treated with actinobacteria) was also included in the study. The biomass production of each strain was obtained from 200-ml liquid cultures of tryptic soy broth (TSB) medium (Sigma-Aldrich) incubated at 220 rpm and 28°C for 3 days in 500-ml Erlenmeyer flasks that were inoculated with 8 × 108 spores. Cells were centrifuged (20 min, 12,000 rpm), washed with saline solution (0.9% [wt/vol]), and resuspended in a final volume of 50.0 ml of saline solution. Actinobacterial isolates were applied to a batch of 75 grafts by partially immersing the grafts (up to a depth of 10 cm) in a rooting hormone solution (indole-3-butyric acid; 0.2 mg · ml−1) containing the selected actinobacteria (107 CFU · ml−1) for 24 h at RT. Whole sets of plants were planted (during the month of May in 2013, 2014, and 2015) in an open-root field nursery. After 60 days in the field (month of July), several parameters were measured: mortality rate, total plant height, and elongation of the 7th internode in surviving plants. Two hundred days after planting (month of December), once the plants had completed their cycle of vegetative growth and were in a dormant state, they were carefully removed from the soil to obtain a root system as intact as possible to be analyzed for the presence of fungal pathogens as described below. A total of 75 live plants per batch (corresponding to three replicates of 25 plants) were analyzed.
Analysis of rootstock colonization by selected actinobacteria.
In order to check rootstock colonization by the selected actinobacteria, we analyzed the graft interiors. A total of 10 grafts inoculated with each actinobacteria were analyzed 10 days and 200 days (at the time of uprooting the plants) after their application. The wood was surface-sterilized by immersion in 70% ethanol (1 min), 4% sodium hypochlorite (2 min), and then again in 70% ethanol (1 min). Two 1-cm-long stubs (isolated from 2 and 5 cm from the lower end of the rootstock) were split longitudinally, and samples were taken from 7 tissue sections (wood chips of 3 by 2 by 2 mm obtained using a sterile scalpel). Wood samples were plated on ISP2 agar medium containing cycloheximide (200 μg · ml−1) (Sigma-Aldrich) to prevent fungal growth and nalidixic acid (25 μg · ml−1) to inhibit the growth of Gram-negative bacteria. Plates were incubated at 28°C, and actinobacteria growing from the chips were identified by amplification and sequencing of 16S rRNA genes, as described above.
Fungal isolation from wood and pathogen identification.
Plants were removed from the soil and analyzed for the presence of pathogenic fungi. A 1-cm stub was cut 1 cm above the root insertion point. The wood was surface-sterilized as described above, and inner tissue sections were obtained as previously indicated and placed on PDA plates supplemented with chloramphenicol (150 μg · ml−1) to prevent bacterial growth. Plates were incubated at 25°C for up to 4 weeks, subculturing where necessary. Pathogenic fungi were identified by their cultural-morphological traits as P. chlamydospora, P. minimum, Dactylonectria sp., and Ilyonectria species. Subsequently, their identities were confirmed by sequencing their internal transcribed spacer 1 (ITS1)-5.8S-ITS2 regions using ITS1 and ITS4 primers under the PCR conditions described by White et al. (56). Other isolates were identified by sequencing the same region. DNA from wood samples was isolated using the REDExtract-N-Amp kit (XNAP) (Sigma-Aldrich).
Statistical data analysis.
Plant growth data were tested for univariate normality using the Shapiro-Wilk test. They were subjected to univariate analysis of variance using the general linear means procedure to determine if there were significant differences between treated and untreated plants. Previously, total height and length of the 7th internode data were tested for univariate normality using the Shapiro-Wilk test and then subjected to univariate analysis of variance using the general linear means procedure. In the case of grafted grapevine mortality data, we used a chi-square to test the null hypothesis of no differences in mortality rates against a two-tailed alternative hypothesis that the difference in mortality rates was not zero. When the expected frequencies were less than 5, Fisher's exact test was used. The chances of plant survival versus untreated control plants were calculated as the ratio between the odds of vine survival between a particular treatment and the control (the odds ratio for a particular treatment is the quotient between the number of live and dead plants). To research the treatment efficacy on the number of fungal pathogens isolated, we used the Poisson distribution (comparing the results achieved with nested models) to construct generalized linear models. The null hypothesis of the independence assumption between the frequency of fungal pathogen isolation and treatments was checked using either the chi-square or Fisher's exact test, depending on the nature of the data. Statistical analyses were performed using R Core Team (3.3.0) software (http://www.R-project.org/).
Accession number(s).
The accession numbers for the 16S RNA sequences in this article are presented in Tables 1, 2, and 5.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Viveros Villanueva Vides S.L. (Larraga, Spain), which was financed by the Centro para el Desarrollo Tecnológico Industrial (CDTI; Madrid, Spain) and the Government of Comunidad Foral de Navarra (Spain). S. González-García was supported by a FPU fellowship of the Ministerio de Educación, Cultura y Deporte (Madrid, Spain).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01564-17.
REFERENCES
- 1.Siebert JB. 2001. Eutypa: the economic toll in vineyards. Wines Vines 82:50–56. [Google Scholar]
- 2.Gubler WD, Rolshausen PE, Trouillas FP, Urbez JR, Voegel T, Leavitt GM, Weber EA. 2005. Grapevine trunk diseases in California. Pract Winery Vineyard Mag Jan-Feb):6–25. [Google Scholar]
- 3.Chiarappa L. 2000. Esca (black measles) of grapevine. An overview. Phytopathol Mediterr 39:11–15. [Google Scholar]
- 4.Graniti A, Surico G, Mugnai L. 2000. Esca of grapevine: a disease complex or a complex of diseases? Phytopathol Mediterr 39:16–20. [Google Scholar]
- 5.Halleen F, Fourie PH, Crous PW. 2006. A review of black foot disease of grapevine. Phytopathol Mediterr 45:S55–S67. [Google Scholar]
- 6.Rolshausen PE, Úrbez-Torres JR, Rooney-Latham S, Eskalen A, Smith RJ, Gubler WD. 2010. Evaluation of pruning wounds susceptibility and protection against fungi associated with grapevine trunk diseases. Am J Enol Vitic 61:113–119. [Google Scholar]
- 7.Van Niekerk JM, Halleen, Fourie PH. 2011. Temporal susceptibility of grapevine pruning wounds to trunk pathogen infections in South African grapevines. Phytopathol Mediterr 50:139–150. [Google Scholar]
- 8.Luque J, Elena G, García-Figueres F, Reyes J, Barrios G, Legorburu FJ. 2014. Natural infections of pruning wounds by fungal trunk pathogens in mature grapevines in Catalonia (Northeast Spain). Aust J Grape Wine Res 20:134–143. doi: 10.1111/ajgw.12046. [DOI] [Google Scholar]
- 9.Bertsch C, Ramírez-Suero M, Magnin-Robert M, Larignon P, Chong J, Abou-Mansour E, Spagnolo A, Clément C, Fontaine F. 2013. Grapevine trunk diseases: complex and still poorly understood. Plant Pathol 62:243–265. doi: 10.1111/j.1365-3059.2012.02674.x. [DOI] [Google Scholar]
- 10.Gramaje D, Armengol J. 2011. Fungal trunk pathogens in the grapevine propagation process: potential inoculum sources, detection, identification and management strategies. Plant Dis 95:1040–1055. doi: 10.1094/PDIS-01-11-0025. [DOI] [PubMed] [Google Scholar]
- 11.Agustí-Brisach C, Gramaje D, García-Jiménez J, Armengol J. 2013. Detection of black-foot disease pathogens in the grapevine nursery propagation process in Spain. Eur J Plant Pathol 137:103–112. doi: 10.1007/s10658-013-0221-8. [DOI] [Google Scholar]
- 12.Cobos R, Mateos RM, Álvarez-Pérez JM, Olego MA, Sevillano S, González-García S, Garzón-Jimeno E, Coque JJR. 2015. Effectiveness of natural antifungal compounds in controlling infection by grapevine trunk disease pathogens through pruning wounds. Appl Environ Microbiol 81:6474–6483. doi: 10.1128/AEM.01818-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sosnowski MR, Creaser ML, Wicks TJ, Lardner R, Scott ES. 2008. Protection of grapevine pruning wounds from infection by Eutypa lata. Aust J Grape Wine Res 14:134–142. doi: 10.1111/j.1755-0238.2008.00015.x. [DOI] [Google Scholar]
- 14.Sosnowski MR, Loschiavo AP, Wicks TJ, Scott ES. 2013. Evaluating treatments and spray application for the protection of grapevine pruning wounds from infection by Eutypa lata. Plant Dis 97:1599–1604. doi: 10.1094/PDIS-02-13-0201-RE. [DOI] [PubMed] [Google Scholar]
- 15.Díaz GA, Latorre BA. 2013. Efficacy of paste and liquid fungicide formulation to protect pruning wounds against pathogens associated with grapevine trunk diseases in Chile. Crop Prot 46:106–112. doi: 10.1016/j.cropro.2013.01.001. [DOI] [Google Scholar]
- 16.Munkvold GP, Marois JJ. 1993. Efficacy of natural epiphytes and colonizers of grapevine pruning wounds for biological control of Eutypa dieback. Phytopathology 83:624–629. doi: 10.1094/Phyto-83-624. [DOI] [Google Scholar]
- 17.McMahan G, Yeh W, Marshall MN, Olsen M, Sananikone S, Wu JY, Block DE, VanderGheysnt JS. 2001. Characterizing the production of a wild-type and benomyl-resistant Fusarium lateritium for biocontrol of Eutypa lata on grapevine. J Ind Microbiol Biotech 26:151–155. doi: 10.1038/sj.jim.7000099. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt CS, Lorenz D, Wolf GA. 2001. Biological control of the grapevine dieback fungus Eutypa lata I: screening of bacterial antagonists. J Phytopathol 149:427–435. doi: 10.1111/j.1439-0434.2001.tb03874.x. [DOI] [Google Scholar]
- 19.Di Marco S, Osti F. 2007. Applications of Trichoderma to prevent Phaeomoniella chlamydospora infections in organic nurseries. Phytopathol Mediterr 46:73–83. [Google Scholar]
- 20.Halleen F, Fourie PH, Lombard PJ. 2010. Protection of grapevine pruning wounds against Eutypa lata by biological and chemical methods. S Afr J Enol Vitic 31:125–132. [Google Scholar]
- 21.Kotze Ç, Van Niekerk J, Mostert L, Halleen F, Fourie P. 2011. Evaluation of biocontrol agents for grapevine pruning wound protection against trunk pathogen infection. Phytopathol Mediterr 50:S247–S263. [Google Scholar]
- 22.Mutawila C, Fourie PH, Halleen F, Mostert L. 2011. Grapevine cultivar variation to pruning wounds protection by Trichoderma species against trunk pathogens. Phytopathol Mediterr 50:S264–S276. [Google Scholar]
- 23.Fourie PH, Halleen F. 2004. Proactive control of Petri disease trough treatment of propagation material. Plant Dis 88:1241–1245. doi: 10.1094/PDIS.2004.88.11.1241. [DOI] [PubMed] [Google Scholar]
- 24.Janssen PH. 2006. Identifying the dominant soil bacteria taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao L, Qiu Z, Dai X, Tan H, Lin Y, Zhou S. 2004. Isolation of endophytic actinomycetes from roots and leaves of banana (Musa acuminata) and their activities against Fusarium oxysporum f. sp. cubense. World J Microbiol Biotechnol 20:501–504. doi: 10.1023/B:WIBI.0000040406.30495.48. [DOI] [Google Scholar]
- 26.Li J, Zhao G-Z, Huang H-Y, Qin S, Zhu W-Y, Zhao L-X, Xu l-H, Zhang S, Li W-J, Strobel G. 2012. Isolation and characterization of culturable endophytic actinobacteria associated with Artemisia annua L. Antonie Van Leeuwenhoek 101:515–527. doi: 10.1007/s10482-011-9661-3. [DOI] [PubMed] [Google Scholar]
- 27.Joseph B, Sankarganesh P, Edwin BT, Raj SJ. 2012. Endophytic streptomycetes from plants with novel green chemistry: review. Int J Biolog Chem 6:42–45. doi: 10.3923/ijbc.2012.42.52. [DOI] [Google Scholar]
- 28.Scherey SD, Erkenbrack E, Früh E, Fengler S, Hommel K, Horlacher N, Schulz D, Ecke M, Kulik A, Fiedler H-P, Hampp R, Tarkka MT. 2012. Production of fungal and bacterial growth modulating secondary metabolites is widespread among mycorrhiza-associated streptomycetes. BMC Microbiol 12:164. doi: 10.1186/1471-2180-12-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrey SD, Tarkka MT. 2008. Friends and foes: streptomycetes as modulators of plant disease and symbiosis. Antonie Van Leeuwenhoek 94:11–19. doi: 10.1007/s10482-008-9241-3. [DOI] [PubMed] [Google Scholar]
- 30.Doumbou CL, Hamby-Salove MK, Crawford DL, Beaulieu C. 2001. Actinomycetes, promising tools to control plant diseases and to promote plant growth. Phytoprotection 82:85–102. doi: 10.7202/706219ar. [DOI] [Google Scholar]
- 31.Davelos AL, Kinkel LL, Samac DA. 2004. Spatial variations in frequency and intensity of antibiotic interactions among Streptomycetes from prairie soil. Appl Environ Microbiol 70:1051–1058. doi: 10.1128/AEM.70.2.1051-1058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burns KN, Kluepfel DA, Strauss SL, Bokulich NA, Cantu D, Steenwerth KL. 2015. Vineyard soil bacterial diversity and composition revealed by 16S rRNA genes: differentiation by geographic features. Soil Biol Biochem 91:232–247. doi: 10.1016/j.soilbio.2015.09.002. [DOI] [Google Scholar]
- 33.Burns KN, Bokulich NA, Cantu D, Greenhut RF, Kluepfel DA, O'Geen AT, Strauss SL, Steenwerth KL. 2016. Vineyard soil bacterial diversity and composition revealed by 16S rRNA genes: differentiation by vineyard management. Soil Biol Biochem 103:337–348. doi: 10.1016/j.soilbio.2016.09.007. [DOI] [Google Scholar]
- 34.Vega-Avila AD, Gumiere T, Andrade PAM, Lima-Perim JE, Durrer A, Baigori M, Vazquez F, Andreote FD. 2015. Bacterial communities in the rhizosphere of Vitis vinífera L. cultivated under distinct agricultural practices in Argentina. Antonie Van Leeuwenhoek 107:575–588. doi: 10.1007/s10482-014-0353-7. [DOI] [PubMed] [Google Scholar]
- 35.Salomon MV, Purpora R, Bottini R, Piccoli P. 2016. Rhizosphere associated bacteria trigger accumulation of terpenes in leaves of Vitis vinifera L. cv. Malbec that protect cells against reactive oxygen species. Plant Physiol Biochem 106:295–304. doi: 10.1016/j.plaphy.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Loqman S, Barka EA, Clément C, Ouhdouch Y. 2009. Antagonistic actinomycetes from Moroccan soil to control the grapevine gray mold. World J Microbiol Biotechnol 25:81–91. doi: 10.1007/s11274-008-9864-6. [DOI] [Google Scholar]
- 37.Sungthong R, Nakaew N. 2015. The genus Nonomuraea: a review of a rare actinomycete taxon for novel metabolites. J Basic Microbiol 55:554–565. doi: 10.1002/jobm.201300691. [DOI] [PubMed] [Google Scholar]
- 38.Rong X, Huang Y. 2012. Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA-DNA hybridization, validating the MLSA scheme for the systematic of whole genus. Syst Appl Microbiol 35:7–18. doi: 10.1016/j.syapm.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Labeda DP, Dunlap CA, Rong X, Huang Y, Doroghazi K-S, Metcalf WW. 2017. Phylogenetic relationships in the family Streptomycetaceae using multi-locus sequence analysis. Antonie Van Leeuwenhoek 110:563–583. doi: 10.1007/s10482-016-0824-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rong X, Guo Y, Huang Y. 2009. Proposal to reclassify the Streptomyces albidoflavus clade on the basis of multilocus sequence analysis and DNA-DNA hybridization, and taxonomic elucidation of Streptomyces griseus subsp. solvifaciens. Syst Appl Microbiol 32:314–322. doi: 10.1016/j.syapm.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 41.Guo Y, Zheng W, Rong X, Huang Y. 2008. A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: use of multilocus sequence analysis for streptomycete systematics. Int J Syst Evol Microbiol 58:149–159. doi: 10.1099/ijs.0.65224-0. [DOI] [PubMed] [Google Scholar]
- 42.Andreolli M, Lampis S, Zapparoli G, Angelini E, Vallini G. 2016. Diversity of bacterial endophytes in 3 and 15 year-old grapevines of Vitis vinifera cv. Corvina and their potential for plant growth promotion and phytopathogen control. Microbiol Res 183:42–52. [DOI] [PubMed] [Google Scholar]
- 43.Rezgui A, Ben Ghnaya-Chakroun A, Vallance J, Bruez E, Hajlaoui MR, Sadfi-Zouaoui N, Rey P. 2016. Endophytic bacteria with antagonistic traits inhabit the wood tissues of grapevines from Tunisian vineyards. Biol Control 99:28–37. doi: 10.1016/j.biocontrol.2016.04.005. [DOI] [Google Scholar]
- 44.Faist H, Keller A, Hentschel U, Deeken R. 2016. Grapevine (Vitis vinifera) crown galls host distinct microbiota. Appl Environ Microbiol 82:5542–5552. doi: 10.1128/AEM.01131-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zarraonaindia I, Owens SM, Weisenhorn P, West K, Hampton-Marcell J, Lax S, Bokulich NA, Mills DA, Martin G, Taghavi S, van der Lelie D, Gilbert JA. 2015. The soil microbiome influences grapevine-associated microbiota. mBio 6(2):e02527-14. doi: 10.1128/mBio.02527-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samad A, Trognitz F, Compant S, Antonielli L, Sessitsch A. 2017. Shared and host-specific microbiome diversity and functioning of grapevine and accompanying weed plants. Environ Microbiol 19:1407–1424. doi: 10.1111/1462-2920.13618. [DOI] [PubMed] [Google Scholar]
- 47.Halleen F, Crous PW, Petrini O. 2003. Fungi associated with healthy grapevine cuttings in nurseries, with special reference to pathogens involved in the decline of young vines. Aust Plant Pathol 32:47–52. doi: 10.1071/AP02062. [DOI] [Google Scholar]
- 48.Rumbos I, Rumbou A. 2001. Fungi associated with esca and young grapevine decline in Greece. Phytopathol Mediterr 40S:330–335. [Google Scholar]
- 49.Küster E, Williams ST. 1964. Selection of media for isolation of streptomycetes. Nature 202:928–929. doi: 10.1038/202928a0. [DOI] [PubMed] [Google Scholar]
- 50.Shirling EB, Gottlieb D. 1966. Methods for the characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340. doi: 10.1099/00207713-16-3-313. [DOI] [Google Scholar]
- 51.Hopwood DA, Bidd MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate DJ, Smith CP, Ward JM, Schrempf H. 1985. Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, United Kingdom. [Google Scholar]
- 52.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY. [Google Scholar]
- 53.Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J. 2012. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- 54.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 55.Lamsal K, Kim SW, Kim YS, Lee YS. 2012. Application of rhizobacteria for plant growth promotion effect and biocontrol of anthracnose caused by Colletotrichum acutatum on pepper. Mycobiology 40:244–251. doi: 10.5941/MYCO.2012.40.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


