ABSTRACT
Microbial bioremediation is a promising approach for the removal of polycyclic aromatic hydrocarbon (PAH) contaminants. Many degraders of PAHs possess efflux pump genes in their genomes; however, their specific roles in the degradation of PAHs have not been clearly elucidated. In this study, two efflux pumps, TtgABC and SrpABC, were systematically investigated to determine their functions in a PAH-degrading Pseudomonas putida strain B6-2 (DSM 28064). The disruption of genes ttgABC or srpABC resulted in a defect in organic solvent tolerance. TtgABC was found to contribute to antibiotic resistance; SrpABC only contributed to antibiotic resistance under an artificial overproduced condition. Moreover, a mutant strain without srpABC did not maintain its activity in long-term biphenyl (BP) degradation, which correlated with the loss of cell viability. The expression of SrpABC was significantly upregulated in the course of BP degradation. BP, 2-hydroxybiphenyl, 3-hydroxybiphenyl, and 2,3-dihydroxybiphenyl (2,3-DHBP) were revealed to be the inducers of srpABC. 2,3-DHBP was verified to be a substrate of pump SrpABC; SrpABC can enhance the tolerance to 2,3-DHBP by pumping it out. The mutant strain B6-2ΔsrpS prolonged BP degradation with the increase of srpABC expression. These results suggest that the pump SrpABC of strain B6-2 plays a positive role in BP biodegradation by pumping out metabolized toxic substances such as 2,3-DHBP. This study provides insights into the versatile physiological functions of the widely distributed efflux pumps in the biodegradation of PAHs.
IMPORTANCE Polycyclic aromatic hydrocarbons (PAHs) are notorious for their recalcitrance to degradation in the environment. A high frequency of the occurrence of the efflux pump genes was observed in the genomes of effective PAH degraders; however, their specific roles in the degradation of PAHs are still obscure. The significance of our study is in the identification of the function and mechanism of the efflux pump SrpABC of Pseudomonas putida strain B6-2 (DSM 28064) in the biphenyl degradation process. SrpABC is crucial for releasing the toxicity caused by intermediates that are unavoidably produced in PAH degradation, which enables an understanding of how cells maintain the intracellular balance of materials. The findings from this study provide a new perspective on PAH recalcitrance and shed light on enhancing PAH degradation by genetic engineering.
KEYWORDS: biodegradation, polycyclic aromatic hydrocarbons, efflux pump, Pseudomonas putida
INTRODUCTION
Polycyclic aromatic hydrocarbons (PAHs) are some of the most serious organic pollutants, and they are commonly found in traffic exhaust and industrial emissions (1, 2). The recalcitrant characteristics of these compounds, especially their carcinogenicity or mutagenicity to living organisms, have made PAHs a concern in the field of environmental science. Bacterial remediation is the main method of natural removal of these pollutants, because pollutants can be completely mineralized; in addition, the process is relatively inexpensive. Some microorganisms that are capable of mineralizing a variety of PAH compounds have been isolated under laboratory conditions (3 – 6). Pseudomonas putida strain B6-2 was isolated and found to be a super degrader of PAHs (7, 8). It uses a wide spectrum of substrates and is able to cometabolically degrade a broad range of PAHs and dioxin-like compounds, such as fluorene, carbazole, dibenzothiophene, dibenzofuran, and benzothiophene (7, 8). Strain B6-2 cells can degrade 2 g · liter−1 biphenyl (BP) in 24 h. In addition, strain B6-2 is highly resistant to many substances in the environment, such as organic solvents, heavy metals, and antibiotics. Our previous studies have shown that strain B6-2 can grow in the presence of greater than 50% m-xylene or p-xylene. PAHs often coexist with many other toxic compounds, such as antimicrobial agents, heavy metals, and toxic derivatives produced by human industrial activities (9 – 11). The robustness of strain B6-2 makes it a promising tool for PAH degradation and bioremediation.
Resistance-nodulation-division (RND) efflux pumps are membrane proteins that exist in almost all microbial organisms and are assumed to play key roles in multidrug resistance, organic solvent tolerance, pathogenesis, microbial environmental adaptability, quorum sensing, and other important physiological processes (12, 13). A typical RND efflux pump consists of three components, namely, a transporter (efflux) protein trimer that is located in the inner (cytoplasmic) membrane of the bacterium (i.e., TtgB), an outer membrane protein trimer that penetrates the periplasmic space to form a channel (i.e., TtgC), and a lipoprotein trimer (a membrane fusion protein, i.e., TtgA) that is located in the periplasmic space and plays a role in stabilizing the interactions between the two other elements. RND efflux pumps of various Pseudomonas strains have been investigated, including P. putida strain S12 (14 – 16) and P. putida strain DOT-T1E (17 – 19). Three well-elucidated efflux pumps (toluene tolerance genes [Ttg]) are found in P. putida DOT-T1E (19). TtgABC is the first efflux system elucidated in P. putida DOT-T1E (17). It plays major roles in toluene tolerance and antibiotic resistance and is highly similar to ArpABC of P. putida S12 (16). TtgDEF is another efflux pump that can only transport styrene and toluene and is highly similar to SepABC in P. putida F1 (18, 20). It is crucial for toluene degradation and encoded by genes located near the toluene degradation gene cluster. The third efflux pump, TtgGHI, is involved in the exclusion of chloramphenicol, ampicillin, tetracycline, toluene, styrene, xylene, ethlylbenzene, and propylbenzene, and it is highly similar to SrpABC of P. putida S12 (15, 19, 21). The genome of P. putida strain B6-2 was sequenced in our previous work (8). The genome of strain B6-2 consists of a single circular chromosome that is 6,377,271 bp in length without plasmids, and more than 30 coding sequences were annotated efflux pump genes (8). Among them, two efflux pump systems, TtgABC and SrpABC, share more than 99% identities with the efflux pumps of P. putida DOT-T1E and P. putida S12, respectively. It is notable that RND efflux pumps have been repeatedly annotated in other PAH-degrading bacteria (22, 23), suggesting that RND efflux pumps might play a positive role in PAH degradation. It was reported that the plasmid pGRT1 harboring the TtgGHI efflux pump in P. putida DOT-T1E supported a superior toluene degradation process performance with unknown mechanisms (24). A similar phenomenon was observed for styrene degradation in Pseudomonas taiwanensis strain VLB120 (25). Meanwhile, efflux pumps, in some cases, decreased the efficiency of biodegradation by excreting substrates out of cells, such as the EmhABC pump of Pseudomonas fluorescens strain LP6α in phenanthrene degradation (26, 27). In general, the specific function of the efflux pumps on PAH degradation remains controversial.
In this study, ttgABC and srpABC of strain B6-2 were systematically investigated to determine their contributions to solvent tolerance, antibiotic resistance, and BP degradation. First, gene knockouts were performed to determine the effects of efflux pump activity on organic solvent tolerance, antibiotic resistance, and long-term BP degradation. Then, real-time quantitative PCR (RT-qPCR) was performed to examine the transcriptional response of the efflux pump genes in the presence of BP. The inducers of srpABC were also identified using a β-galactosidase assay. The substrates of the pump SrpABC were explored with resting cells. Finally, a fed-batch BP degradation assay was performed to analyze the effect of increasing the expression of srpABC on long-term BP degradation.
RESULTS
Gene clusters of ttgABC and srpABC in strain B6-2.
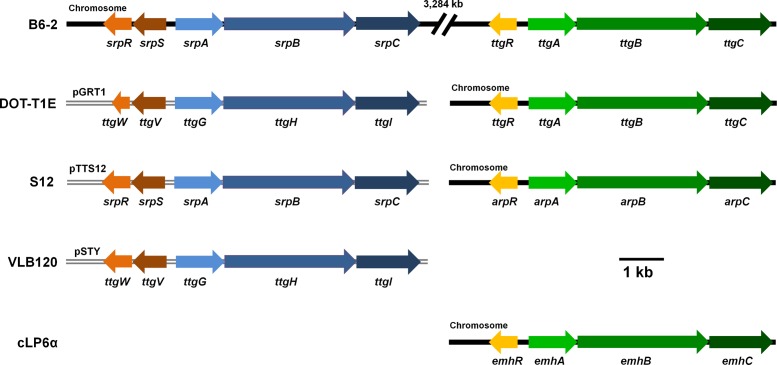
Two RND efflux pump systems, ttgABC and srpABC, were found in the genome of strain B6-2 by sequence alignment with other homologous efflux pump genes (Fig. 1). TtgABC is most related to TtgABC of P. putida DOT-T1E. Compared with those in P. putida DOT-T1E, TtgA has two amino acid substitutions, while TtgBC are identical in both strains. SrpABC is most related to SrpABC of P. putida S12. SrpA and SrpB have substitutions of one amino acid each compared with SrpA and SrpB from P. putida S12, respectively. SrpC is identical to SrpC of P. putida S12. In the opposite direction of ttgABC, ttgR encodes a putative transcriptional repressor that exhibits high amino acid sequence identity with TtgR (99%) of P. putida DOT-T1E. Two putative transcriptional repressor genes, srpS and srpR, are located upstream and in an opposite orientation to srpABC. SrpS, an IclR family regulator, shares the highest sequence identity with TtgV (99%) of P. putida DOT-T1E. SrpR belonging to the TetR family is most related to SrpR (96% identity) of P. putida S12. The two gene clusters ttgABCR and srpABCSR are located on the chromosome of strain B6-2. In contrast, the genes srpABC and their homologous sequences are usually harbored on plasmids in other strains (21, 28, 29).
FIG 1.
Genetic organization and locations of ttgABC and srpABC clusters. The ttgABC or srpABC genes are colored in the same pattern as for strain B6-2. The arrows indicate the size and direction of transcription of each gene. B6-2, P. putida B6-2; DOT-T1E, P. putida DOT-T1E (17, 19, 21); S12, P. putida S12 (15, 16, 28); VLB120, P. taiwanensis VLB120 (29); cLP6α, P. fluorescens cLP6α (22).
TtgABC and SrpABC are both involved in the solvent tolerance of strain B6-2.
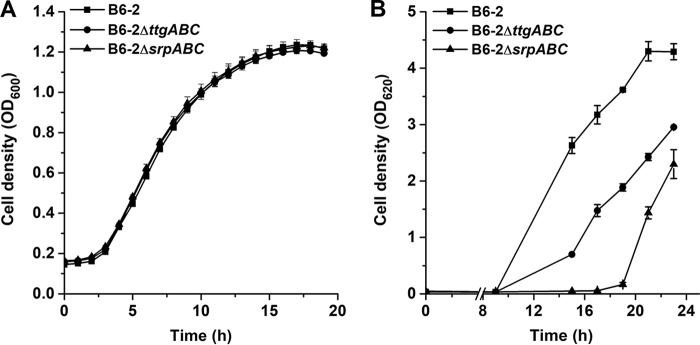
To investigate the roles of ttgABC and srpABC in organic solvent tolerance, we constructed two mutants, B6-2ΔttgABC and B6-2ΔsrpABC, by the homologous recombination method, and the resulting mutants are shown in Table 1. We first compared the growth of the mutants with that of parental strain B6-2 in citric acid (CA) medium by detecting their optical densities at 600 nm. As shown in Fig. 2A, there was no difference between the growth of the two mutants and that of strain B6-2 in CA medium, suggesting that the pumps TtgABC and SrpABC have no vital effect on cell growth in CA medium without organic solvent. To estimate the organic solvent tolerance of different mutants, p-xylene was selected as the model organic solvent. When 1% p-xylene was added to LB medium, each of the strains showed a lag phase of more than 9 h. In addition, strain B6-2ΔttgABC and strain B6-2ΔsrpABC showed longer lag phases (Fig. 2B). These results strongly suggest that both ttgABC and srpABC play critical roles in the solvent resistance of strain B6-2.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Reference or source |
|---|---|---|
| Strains (P. putida) | ||
| B6-2 | Wild type (DSM 28064) | 7, 8 |
| B6-2ΔsrpABC | srpABC-knockout mutant of strain B6-2 | This study |
| B6-2ΔttgABC | ttgABC-knockout mutant of strain B6-2 | This study |
| B6-2ΔsrpS | srpS-knockout mutant of strain B6-2 | This study |
| B6-2ΔttgABCΔsrpS | ttgABC- and srpS-null mutant of B6-2 | This study |
| B6-2ΔttgR | ttgR-null mutant of B6-2 | This study |
| B6-2ΔbphC | bphC-deficient mutant of strain B6-2 | This study |
| B6-2ΔbphD | bphD-deficient mutant of strain B6-2 | This study |
| B6-2ΔbphCΔsrpABC | srpABC-knockout and bphC-deficient mutant of strain B6-2 | This study |
| B6-2ΔbphA(pMEG) | bphA-deficient mutant containing plasmid pMEG | This study |
| B6-2ΔbphC(pMEG) | bphC-deficient mutant containing plasmid pMEG | This study |
| B6-2ΔbphD(pMEG) | bphD-deficient mutant containing plasmid pMEG | This study |
| B6-2(pMEG) | B6-2 containing plasmid pMEG | This study |
| B6-2(pMEA) | B6-2 containing plasmid pMEA | This study |
| B6-2ΔsrpS(pMEG) | B6-2ΔsrpS containing plasmid pMEG | This study |
| B6-2ΔttgR(pMEA) | B6-2ΔttgR containing plasmid pMEA | This study |
| Plasmids | ||
| pK18mobsacB | Mobilizable vector, mob+, sacB, Kmr | 35 |
| pK18G | pK18mobsacB::ΔsrpABC | This study |
| pK18A | pK18mobsacB::ΔttgABC | This study |
| pK18S | pK18mobsacB::ΔsrpS | This study |
| pK18R | pK18mobsacB::ΔttgR | This study |
| pK18bphA′ | pK18mobsacB::bphA′ | This study |
| pK18bphC′ | pK18mobsacB::bphC′ | This study |
| pK18bphD′ | pK18mobsacB::bphD′ | This study |
| pME6015 | promoterless lacZ vector, Tcr | This study |
| pMEG | Tcr, srpA promoter cloned in pME6015 | This study |
| pMEA | Tcr, ttgA promoter cloned in pME6015 | This study |
Kmr, resistance to kanamycin; Tcr, resistance to tetracycline.
FIG 2.
Growth curves of strain B6-2 and its two derivatives in citric acid (A) and LB medium containing 1% p-xylene (B). (A) The overnight culture samples (1%) were transferred into 100-well plates containing 300 μl mineral salts medium containing 0.5% citric acid as the sole carbon source. The growth of the strains was determined by the OD600 value using a Victor2 (PerkinElmer) spectrophotometer. (B) The strains were grown in LB medium in the presence of 1% p-xylene. Growth was monitored by measuring the absorbance at 620 nm. Data represent the averages from three parallel replicates ± standard deviations (SDs).
TtgABC is responsible for the antibiotic resistance of strain B6-2.
A bacterial inhibition ring test was used to investigate the roles of ttgABC and srpABC in antibiotic resistance. Twenty structurally unrelated antibiotics were used in this assay. No significant differences were observed in inhibition ring diameters with rifampin, fosfomycin, teicoplanin, nystatin, lincomycin, or polymyxin B for any of the strains tested (Table 2). No significant differences were found in inhibition ring diameters for mutant strain B6-2ΔsrpABC and strain B6-2 with the remaining 14 antibiotics. Compared with that for strain B6-2, an inhibition of mutant strain B6-2ΔttgABC was detected with ampicillin, cefoxitin, novobiocin, chloramphenicol, erythromycin, streptomycin, and spectinomycin, and larger rings were observed with gentamicin, meropenem, nalidixic acid, cefepime, ceftazidime, cefotaxime, and tetracycline (Table 2). More recently, the overproduced TtgGHI of P. putida strain DOT-T1E was reported to contribute to the efflux of the antibiotics tetracycline, chloramphenicol, ampicillin, erythromycin, and norfloxacin (30). In strain B6-2, the deduced proteins SrpS and TtgR are also repressors of the SrpABC and TtgABC pumps, respectively (data not shown). We constructed a mutant strain B6-2ΔttgABCΔsrpS to overproduce SrpABC and investigate the roles of SrpABC in the background of TtgABC defeat. SrpABC of strain B6-2 can efflux ampicillin, tetracycline, novobiocin, cefoxitin, cefotaxime, and cefepime only under the condition in which SrpABC was overexpressed (Table 2). These results suggest that efflux pump TtgABC is the main efflux pump responsible for the multidrug resistance of strain B6-2.
TABLE 2.
Antibiotic susceptibility of P. putida B6-2 and its derivatives
| Antibiotic (μg/piece)a | Inhibition halo (cm) |
|||
|---|---|---|---|---|
| B6-2 | B6-2ΔsrpABC | B6-2ΔttgABC | B6-2ΔsrpSΔttgABC | |
| Ampicillin (10) | Nob | No | 1.9 ± 0.1 | No |
| Cefepime (30) | 2.6 ± 0.2 | 2.6 ± 0.2 | 3.6 ± 0.1 | 3.0 ± 0.2 |
| Cefotaxime (30) | 1.9 ± 0.1 | 1.8 ± 0.1 | 3.4 ± 0.1 | 2.5 ± 0.1 |
| Cefoxitin (30) | No | No | 2.8 ± 0.2 | No |
| Ceftazidime (30) | 2.5 ± 0.0 | 2.3 ± 0.2 | 2.9 ± 0.2 | 2.7 ± 0.1 |
| Nalidixic acid (30) | 1.8 ± 0.3 | 1.8 ± 0.2 | 3.1 ± 0.2 | 2.4 ± 0.1 |
| Novobiocin (30) | No | No | 1.5 ± 0.0 | 0.8 ± 0.1 |
| Tetracycline (30) | 1.3 ± 0.3 | 1.3 ± 0.1 | 2.3 ± 0.1 | 1.9 ± 0.0 |
| Chloramphenicol (30) | No | No | 1.1 ± 0.1 | 0.9 ± 0.1 |
| Erythromycin (15) | No | No | 1.5 ± 0.1 | 2.0 ± 0.0 |
| Gentamicin (10) | 1.5 ± 0.1 | 1.6 ± 0.0 | 2.2 ± 0.1 | 2.3 ± 0.1 |
| Meropenem (10) | 0.8 ± 0.1 | 1.0 ± 0.2 | 2.5 ± 0.2 | 2.6 ± 0.2 |
| Spectinomycin (100) | No | No | 1.5 ± 0.2 | 2.5 ± 0.1 |
| Streptomycin (10) | No | No | 2.0 ± 0.1 | 2.1 ± 0.0 |
Fourteen of the 20 antibiotics tested in this assay are shown. There were no differences found in susceptibility to rifampin, fosfomycin, teicoplanin, nystatin, lincomycin, or polymyxin B.
No, no bacteriostatic ring.
SrpABC promotes long-term BP degradation.
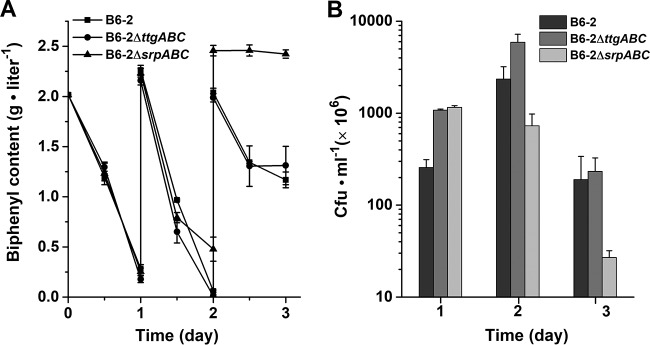
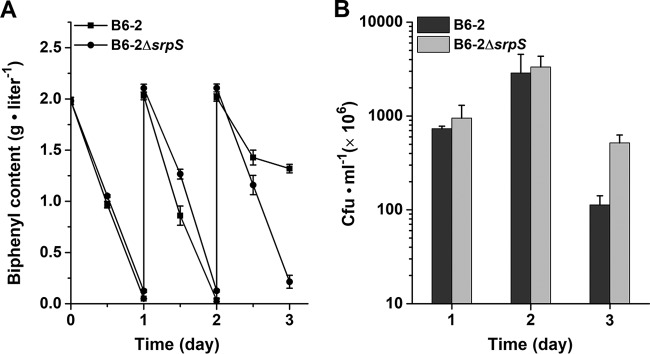
The sustainability of catabolic activity is important when considering the application of a degrader to a polluted environment (3). To investigate the roles of TtgABC and SrpABC in long-term BP degradation, the BP degradation performances of mutants B6-2ΔttgABC and B6-2 ΔsrpABC were compared with that of strain B6-2 in fed-batch BP degradation experiments. The initial BP content was 2 g · liter−1, and 2 g · liter−1 BP was added to the cultures every 24 h. The amount of BP was quantified by high-performance liquid chromatography (HPLC). As shown in Fig. 3A, on days 1 and 2, strains B6-2 and B6-2ΔttgABC almost completely degraded BP. However, strain B6-2ΔsrpABC did not completely degrade BP as did strains B6-2 and B6-2ΔttgABC on day 2. On day 3, the amounts of residual BP in the cultures of the two strains B6-2 and B6-2ΔttgABC were almost the same. However, no additional BP degradation was observed from day 2 to day 3 by strain B6-2ΔsrpABC (Fig. 3A). These results suggested that a defect of the pump SrpABC is detrimental to long-term BP degradation. Meanwhile, the viability of the strains was measured by plating serial dilutions of each culture on LB plates during the degradation process (Fig. 3B). On day 1, strains B6-2ΔttgABC and B6-2ΔsrpABC had higher viable counts than strain B6-2. This might be because BP, as the source of carbon and energy, must be consumed for pump expression and function. On day 2, the viability of strains B6-2 and B6-2ΔttgABC but not strain B6-2ΔsrpABC increased significantly. On day 3, severe drops in the viable counts were observed, and the viable count of strain B6-2ΔsrpABC was the least among the three strains. All of these results indicate that BP degradation ability is directly affected by a loss of cell viability which correlates with the loss of efflux pump SrpABC.
FIG 3.
BP degradation curves (A) and cell viability (B) of strain B6-2 and its two mutants. Overnight culture samples of strain B6-2 and mutants grown in LB medium were inoculated (1:10) in BP (2 g · liter−1) medium. Additional 2 g · liter−1 BP was added to the culture every 24 h. At each data point, the amount of residual BP was analyzed by HPLC. On days 1, 2, and 3, the viable counts were measured by plating serial dilutions of each culture on LB plates. Data represent the averages from three parallel replicates ± SDs.
Expression of SrpABC was upregulated in the presence of BP.
To further investigate the physiological roles of srpABC and ttgABC, the transcriptional levels of ttgA and srpA were measured under BP and non-BP conditions. Strain B6-2 cells were cultured in the absence or presence of 2 mM BP and harvested when a turbidity of about 1.0 at 620 nm was reached, and then RNA was extracted. Quantitative RT-PCR analyses showed that the mRNA level of srpA increased 6.9-fold from the basal level, whereas the mRNA level of ttgA decreased 4.2-fold in response to BP (Fig. 4A). To confirm these results, a β-galactosidase assay was performed. We constructed fusions of the promoters for srpABC or ttgABC and a promoterless lacZ gene. The resulting plasmids were transformed into strain B6-2, and expression from the srpA and ttgA promoters was determined in the absence or presence of BP. The level of expression with the srpA promoter was increased 3.5-fold in the presence of BP (171.9 ± 8.8 U) relative to that in the absence of BP (49.0 ± 4.8 U) (Fig. 4B). The opposite tendency was observed with expression of the ttgA promoter (226.5 ± 5.3 U with BP and 207.0 ± 0.9 U without BP; P value < 0.05). These results suggest that strain B6-2 upregulates the expression of srpABC in response to biphenyl degradation. On the basis of the results of the transcription analysis and fed-batch BP degradation experiment, the TtgABC system did not appear to respond significantly during BP degradation; therefore, only the SrpABC system was examined in subsequent work.
FIG 4.
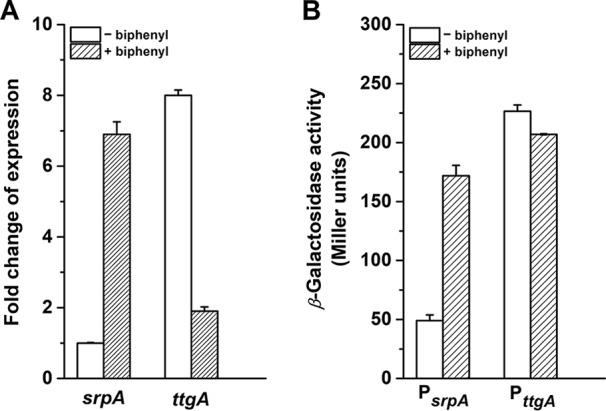
Transcription analysis of pump genes ttgA and srpA. (A) Quantitative RT-PCR analysis of the genes srpA and ttgA. The relative expression levels of the genes were measured using RNA extracted from P. putida B6-2 grown with or without BP. (B) Analysis of promoter activity. β-Galactosidase activity was measured with or without BP. Data represent the averages from three parallel replicates ± SDs.
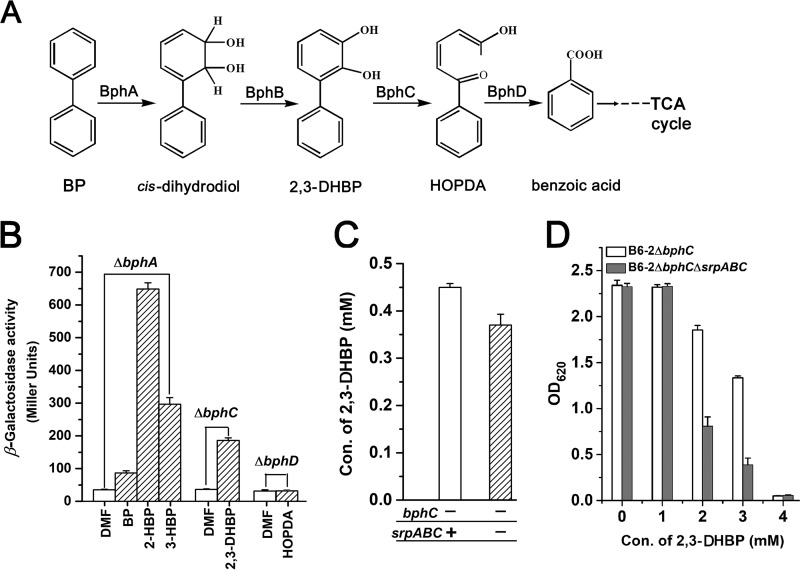
BP and BP metabolites serve as inducers of the srpABC promoter.
The fact that srpABC was upregulated in the course of BP degradation led us to search for the inducers of srpABC. To probe for inducers, we constructed several mutants in which BP degradation was interrupted; therefore, the investigated compound, BP or BP metabolites, would not be degraded and would remain stable (Fig. 5A). The reporter vector PsrpA::′lacZ fusion (pMEG) was transformed into strain B6-2 and its mutants, and the promoter activity of PsrpA was measured in a β-galactosidase assay in the presence of BP or BP metabolites. The β-galactosidase activity assay showed that BP, 2-hydroxybiphenyl (2-HBP), 3-hydroxybiphenyl (3-HBP), and 2,3-dihydroxybiphenyl (2,3-DHBP) are inducers of the srpA promoter (Fig. 5B). The effectors that caused the highest levels of induction (>5-fold increase) were 2-HBP, 3-HBP, and 2,3-DHBP. BP increased expression from the PsrpA promoter by approximately 2-fold. An intermediate, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid (HOPDA), did not induce the srpA promoter in strain B6-2. This suggested that strain B6-2 actively increases the expression of srpABC in response to the presence of BP and some metabolic intermediates.
FIG 5.
Analysis of inducers and substrates of pump SrpABC. (A) Proposed upper pathway for the conversion of biphenyl in strain B6-2. cis-Dihydrodiol can be spontaneously transformed into 2-hydroxybiphenyl (2-HBP) or 3-hydroxybiphenyl (3-HBP), which cannot be degraded by BphB (biphenyl dehydrogenase). In contrast, 2-HBP and 3-HBP can be catalyzed by BphA (biphenyl dioxygenase). BphC, 2,3-dihydroxybiphenyl-1,2-dioxygenase; BphD, hydrolase. (B) Analysis of PsrpA promoter activity. When the optical density of the cells was approximately 0.8, BP and intermediates dissolved in N,N-dimethylformamide (DMF) were added individually at a concentration of 2 mM, except for 0.5 mM 3-HBP. After 2 h, β-galactosidase activity was measured. (C) Mutants were grown in LB medium at 30°C for 14 h with 200 rpm shaking. The cells were washed and resuspended in BP medium with 3 mM BP at a final OD620 of 4.0 at 30°C with 150 rpm shaking. After 4 h, the samples were taken and the amounts of 2,3-DHBP were determined by HPLC. (D) The mutants were grown overnight, diluted 100-fold in LB medium, and incubated with different concentrations of 2,3-DHBP with 200 rpm shaking at 30°C. After 12 h, bacterial growth was monitored by measuring the optical density at 620 nm. All experiments were performed with triplicate samples.
2,3-DHBP is a substrate of efflux pump SrpABC.
To test whether the inducers serve as the substrates of SrpABC, the amount of metabolic intermediate 2,3-DHBP during biphenyl degradation was monitored using parent resting cells and mutant B6-2ΔsrpABC resting cells. To avoid the degradation of 2,3-DHBP, bphC was interrupted in strains B6-2 and B6-2ΔsrpABC. As shown in Fig. 5C, the content of 2,3-DHBP in the solution of B6-2ΔbphC cells was higher than that in the solution of B6-2ΔbphCΔsrpABC cells, indicating that SrpABC can pump out 2,3-DHBP. To confirm these results, we compared the growth of strains B6-2ΔbphC and B6-2ΔbphCΔsrpABC in the presence of 2,3-DHBP (Fig. 5D). At concentrations of up to 2 mM, 2,3-DHBP strongly inhibited cell growth. The toxic effect of 2,3-DHBP on strain B6-2ΔbphCΔsrpABC was more severe than that on strain B6-2ΔbphC. All of these results indicate that the pump SrpABC can pump out 2,3-DHBP and enhance 2,3-DHBP tolerance.
BP degradation was enhanced in the srpABC-overexpressing strain.
When SrpABC was knocked out BP degradation decreased. Therefore, we speculated that increasing the expression of srpABC would improve the BP degradation capability of this strain. The repressor gene srpS was knocked out in strain B6-2 to obtain strain B6-2ΔsrpS. Degradation experiments were then performed using strains B6-2 and B6-2ΔsrpS. As shown in Fig. 6A, on days 1 and 2 no significant difference in the extend of BP degradation was observed between strains B6-2 and B6-2ΔsrpS; both were almost completely degraded. On day 3 strain B6-2ΔsrpS still maintained its BP degradation capability, but strain B6-2 only degraded nearly half of BP. Viability was similar for the two strains on days 1 and 2, but on day 3 the viability of strain B6-2ΔsrpS decreased less severe than strain B6-2, which coincided with more BP degradation achieved by strain B6-2ΔsrpS (Fig. 6B). All of these results show that artificially increasing the expression of srpABC prolongs BP degradation.
FIG 6.
BP degradation curves (A) and cell viability (B) of strains B6-2 and B6-2ΔsrpS. Data represent the averages from three parallel replicates ± SDs.
DISCUSSION
In this study, TtgABC and SrpABC were systematically evaluated to determine their roles in BP degradation. In strain B6-2, TtgABC shows a higher level of basal expression than that of SrpABC and is responsible for the multidrug resistance (Fig. 4 and Table 2). In the presence of 1% p-xylene, all strains showed much longer lag phases, especially strains B6-2ΔttgABC and B6-2ΔsrpABC (Fig. 2). In previous studies, most bacterial cells died with the sudden addition of an organic solvent (17 – 19), which matches the longer lag phases shown here. These results strongly suggest that both ttgABC and srpABC of strain B6-2 play critical roles in solvent tolerance. When srpABC was knocked out, the mutant strain showed a defect in long-term BP degradation activity (Fig. 3A). In contrast, the mutant strain with srpABC overexpression showed prolonged fed-batch BP degradation compared with that of strain B6-2 (Fig. 6A). Further study indicated that strain B6-2 is capable of pumping the intermediate metabolite 2,3-DHBP, which is toxic to cells (Fig. 5). Meanwhile, a loss of efflux pump SrpABC correlates with a loss of cell viability, which directly affects BP degradation ability (Fig. 3). These results suggest strain B6-2 can respond positively to the toxicity caused by intermediates during BP degradation by upregulating the expression of efflux pump SrpABC.
RND efflux pumps are encoded on chromosomes of almost all organisms and participate in many important physiological processes. For example, in Escherichia coli, AcrAB-TolC is activated by metabolites that accumulate as a result of the interruption of the central biosynthetic pathway (31). In Pseudomonas aeruginosa, MexXY-OprM protects the cell from the adverse consequences of disrupted translation (32). These studies have shown that efflux pumps are closely associated with physiological processes within cells, including the elimination of toxic metabolites and the stabilization of the intracellular environment. The PAH degradation process usually consists of multiple steps and generates many intermediates that can accumulate in the lipid membranes of cells, disrupting the membrane integrity and leading to an abnormal permeability to protons and ions (33). In our study, strain B6-2 upregulated the expression of pump SrpABC to efflux toxic intermediates out of cells during BP degradation. The viable count of B6-2ΔsrpABC was less than that of the strain B6-2, which directly resulted in the dramatic drop of BP degradation activity. Therefore, cells of strain B6-2 might be protected from the damage caused by an overload of BP metabolites. This characteristic rendered strain B6-2 capable of solving the problem of metabolite toxicity.
Other RND efflux pumps also discharge aromatic chemicals that are structurally similar to PAHs or their derivatives. For instance, EmhABC in P. fluorescens strain LP6α can excrete phenanthrene, anthracene, and fluoranthene (26). TtgGHI in P. putida DOT-T1E can be induced by more than 30 aromatic hydrocarbons, including naphthalene, 1-naphthol, 2,3-dihydroxynaphthalene, catechol, and indole, suggesting they may be substrates for the efflux pump TtgGHI (34). On the basis of the high structural similarity among PAHs and the broad-spectrum of substrates for SrpABC, we speculate that other PAH degradation processes may benefit from the activity of the SrpABC efflux pump.
In summary, two RND efflux pumps, TtgABC and SrpABC, in strain B6-2 were investigated for their functions in strain robustness and BP degradation. TtgABC shows a high level of basal expression and is responsible for the multidrug resistance of strain B6-2. Both TtgABC and SrpABC can excrete organic solvents out of cells. SrpABC of strain B6-2 is crucial for releasing the toxicity caused by intermediates that are unavoidably produced during PAH degradation. The findings from this study provide a new perspective on the PAH recalcitrance and shed light on enhancing PAH degradation by genetic engineering.
MATERIALS AND METHODS
Chemicals.
BP, p-xylene, and ethyl acetate were purchased from Sinopharm Chemical Reagent Co., Ltd. (China). Methanol, 2-hydroxybiphenyl, 3-hydroxybiphenyl, 2,3-dihydroxybiphenyl, and o-nitrophenyl-β-d-galactopyranoside (ONPG) were obtained from Sigma-Aldrich Co. Antimicrobial susceptibility disks were purchased from Hangzhou Microbial Reagent Co., Ltd.
Preparation method of HOPDA.
A 1-liter overnight LB culture of strain B6-2ΔbphD induced by BP was washed twice and then suspended in 600 ml BP medium with 3 mM BP. After incubating for 4 h at 30°C with shaking, the supernatant was removed after centrifugation and 200 ml medium was then added for a further 2-h incubation. After centrifugation, the supernatants were merged and extraction was performed with an equivalent volume of ethyl acetate at pH 2.0. The obtained material was concentrated with a rotary evaporator, resolved in HPLC buffer (80% methanol, 20% H2O) and further purified by HPLC. HPLC was performed with a C18 column (Shimadzu PRC-ODS, 15 μm, 20 cm by 25 cm) at a flow rate of 15 ml · min−1 and monitored by absorbance at 342 nm. The fraction was collected, concentrated by evaporation, and resolved in N,N-dimethylformamide.
Bacterial strains, plasmids, and culture conditions.
P. putida B6-2, Escherichia coli strains, and plasmids used in this study are listed in Table 1. E. coli strains were routinely cultured in Luria-Bertani broth at 37°C. Strain P. putida B6-2 and its derivative strains were grown in LB broth or in minimal salts medium (3.7 g · liter−1 of KH2PO4, 5.2 g · liter−1 of K2HPO4·3H2O, 2.0 g · liter−1 of NH4Cl, 1.0 g · liter−1 of Na2SO4, 0.1 g · liter−1 of MgSO4, 1 ml of trace metal solution) containing either 2 g · liter−1 BP dissolved with N,N-dimethylformamide (BP medium) or 0.5% citric acid (CA medium) as a sole source of carbon at 30°C with shaking at 200 rpm (7). When needed, the culture medium was supplemented with tetracycline (15 μg · ml−1) or kanamycin (50 μg · ml−1).
DNA techniques.
Genomic DNA was isolated from P. putida B6-2 with a genomic DNA purification kit (Laifeng Bio Co., Ltd., Shanghai). Plasmid isolations were performed with a TIANprep mini plasmid kit (Tiangen Biotech Co., Ltd., Beijing). Amplified fragments were purified with a gel/PCR extraction kit (Generay Biotech Co., Ltd., China). Restriction enzyme digestions were performed according to the manufacturer's instructions (New England BioLabs). Ligations were performed with T4 DNA ligase (New England BioLabs). Chemically competent E. coli DH5α and S17-1 cells were transformed according to the standard procedure. The primers were obtained from Generay Biotech (China) and are listed in Table 3. DNA was sequenced by Biosun Biotech (China).
TABLE 3.
Oligonucleotides used in this study
| Primer | Sequence (5′→3′)a | Purpose |
|---|---|---|
| G-A1 | GCATGAATTCTCCCCGCGTGCATTACGGGT | Knock out srpABC |
| G-A2 | TGGGCTTGGTTCACCCATGCGGCAAGCGCATCGTC | |
| G-B1 | TGCGCTTGCCGCATGGGTGAACCAAGCCCAGGCCC | |
| G-B2 | GCATAAGCTTGGCTCGCAGCTTTCCTGCGT | |
| A-A1 | GCATGAATTCACACGCTCACTGGCCGAACG | Knock out ttgABC |
| A-A2 | GCCTTGAGCAGGTGCTGAACCGAGGCGGCGTTGTC | |
| A-B1 | CGCCGCCTCGGTTCAGCACCTGCTCAAGGCTGCCA | |
| A-B2 | GCATAAGCTTGCGTTCGGCCAAGCGGTAGT | |
| G-F | CTCTGAAGTTTTCCGAGGTG | Identify the ΔsrpABC mutant |
| G-R | AGCCGAGGCTTCTGTGGTAG | |
| A-F | GTCGGCGTCGTGACCATCCA | Identify the ΔttgABC mutant |
| A-R | CACCGATGCCTGCTGGTTGG | |
| S-A1 | GCATGAATTCGGGGATCGTATCTGTCTCAC | Knock out srpS |
| S-A2 | AGCTGGATCCTCTGGCGATGACCTGGATGC | |
| S-B1 | AGCTGGATCCCAGCATTACCTGACGAAACCCTA | |
| S-B2 | GCATAAGCTTCATCTTATCTAGGGAGCTTTCTTCGA | |
| S-F | GCATGAATTCCGCTCCACCGTTCAGAGAAT | Identify the ΔsrpS mutant |
| S-R | GCATAAGCTTGTTTGACAAGCGCCTTTCGT | |
| R-A1 | GCATGAATTCCTGCTGCAAGGGGGTTTCGAATTG | Knock out ttgR |
| R-A2 | AGCTGGATCCATATGCTGCGCTTGAGCCCG | |
| R-B1 | AGCTGGATCCCGCGCCAAATGGTCATGGGTCT | |
| R-B2 | GCATAAGCTTAGCTTATCCGAGAGGCCCCG | |
| R-F | CAGCCCGGTGTCGACCCATT | Identify the ΔttgR mutant |
| R-R | CTGGTGCTCGATGCCCGAAC | |
| PsrpA-F | GCATGAATTCCCCGGCCTTGCCAATATTTT | Clone the promoter of srpA |
| PsrpA-R | GCATCTGCAGTAATGCACGCGGGGATCGTA | |
| PttgA-F | GCATGAATTCGGGTTTCCTGGGCTTCTTCTTTGGT | Clone the promoter of ttgA |
| PttgA-R | GCATCTGCAGTGGCTTGAATTGCATGAGGAT | |
| q A-F | GCAGTGAGGTCAAGGAAGG | RT-qPCR for ttgA |
| q A-F | CGTGTAGCCAGCAGGTTG | |
| q G-F | CGTCACTCAGCCAATCAC | RT-qPCR for srpA |
| q G-F | GATAAGCACTTCCGTCATCC | |
| 16S-F | ACGCTAATACCGCATACG | RT-qPCR for 16S rRNA |
| 16S-R | CATCCTCTCAGACCAGTTAC | |
| pK-bphA-F | GCATGAATTCGGTTTTTGGTCGCTCTTGGC | Construct a bphA interruption |
| pK-bphA-R | GCATAAGCTTGAAGACCAGGCCCTTGTAGGTG | |
| pK-bphC-F | GCATGAATTCGACGGTTCCTGTGTACTTCCT | Construct a bphC interruption |
| pK-bphC-R | AGCTGGATCCCTCGGACTGTCGTGCCTCA | |
| pK-bphD-F | GCATGAATTCTAACGGCGAAACCGTCATCA | Construct a bphD interruption |
| pK-bphD-R | AGCTGGATCCAGCAGCTTGATGCCTTCCAT | |
| pK269 | GCTTCCCAACCTTACCAGA | Identify the single exchange mutant |
Engineered restriction sites are underlined.
Construction of mutant strains.
Gene deletion mutants were generated using the homologous recombination method. To construct a ttgABC-deleted mutant, we first amplified internal segments of ttgA and ttgC individually using primer pairs A1/A2 and B1/B2 and recombined these segments by PCR using primers A1 and B2 to obtain a recombination DNA fragment. Then, the amplified DNA fragment was inserted into the EcoRI-BamHI sites of plasmid pK18mobsacB to generate pK18A. Plasmid pK18A was transferred by intergeneric conjunction from E. coli S17-1 to B6-2 cells. Since pK18A is unable to replicate in P. putida B6-2, transconjugants only arise after the integration of pK18A into the chromosome by homologous recombination as described by Schäfer et al. (35). E. coli S17-1 and P. putida B6-2 were grown in 10 ml of LB medium for 3 h with shaking at 200 rpm. Cells were harvested by centrifugation at 4,500 rpm for 7 min at 4°C and washed three times with 0.9% NaCl. Cells from both of these strains were suspended and mixed (E. coli S17-1-P. putida B6-2, 5:1) in 100 μl 0.9% NaCl. The mixture was transferred to LB agar and incubated at 37°C for 6 h and then at 30°C for 18 h. The cells were harvested and washed three times, and then the cells were selected in modified M9 agar medium containing 50 mg · liter−1 kanamycin and confirmed by PCR using the oligonucleotide pair A1 and pK269 (shown in Table 3). The mutants, in which BP degradation was interrupted, were constructed by the method as described here. To select double crossover events, a single colony was grown for 24 h in LB medium. Approximately 2 × 106 cells were plated on LBS (LB medium containing 10% sucrose) agar and incubated for 48 h at 30°C. The mutant strain B6-2ΔttgABC was confirmed by PCR using the oligonucleotide pair A1 and B2. Other strains with pump gene mutations were derived by the same method, and bph mutants were made by interrupting bph via an insertion of plasmid pK18mobsacB by means of homologous single exchange.
Determination of organic solvent tolerance and antibiotic resistance.
Strain B6-2 and its derivatives were cultured in LB medium overnight. The following day, culture suspensions (1%) were transferred to 100-well plates containing 300 μl CA medium per well. The growth of the strains was monitored by measuring the optical densities at 600 nm (OD600) using a Victor2 (PerkinElmer) spectrophotometer. The solvent tolerance of strain B6-2 and its mutants was determined by growing the cells for 24 h in 50 ml liquid LB medium in 250-ml flasks supplemented with 1% p-xylene. Antibiotic resistance was determined on LB agar plates with individual thin wafers containing an antibiotic. Overnight cultures were inoculated (1:500) in fresh LB medium and then applied to LB agar plates with antibiotic wafers. The diameters of the inhibition zones were determined after 18 h. All data represent averages from at least three independent samples.
Fed-batch BP degradation experiments.
To examine BP-degrading activity, strain B6-2 and its mutants were grown in 50 ml LB medium overnight and then transferred to BP medium. Bacterial cultures were established in 250-ml conical flasks containing 50 ml BP medium and incubated with shaking at 200 rpm. For fed-batch degradation, 2 g · liter−1 BP was fed directly into the flasks every 24 h. Bacterial growth was verified by measuring the turbidity of the cultures. Residual BP concentrations were monitored by a high-performance liquid chromatography-diode array detector system at 254 nm using an Agilent 1200 system equipped with a reverse-phase C18 column (Agilent Eclipse XDB-C18, 5 μm, 4.6 mm by 150 mm) and were resolved with methanol-H2O (80:20 [vol/vol]) at a flow rate of 1.0 ml · min−1. All assays were performed using at least three biologically independent samples.
Quantitative RT-PCR.
The relative expression levels of ttgA and srpA were measured using RNA extracted from strain B6-2 grown in the presence or absence of BP. Strain B6-2 was grown overnight in LB medium. Cells were then diluted 100-fold in fresh medium, and aliquots were incubated in the absence or presence of 2 mM BP until the culture reached a turbidity of approximately 1.0 at 620 nm. Cells were harvested and RNA was extracted. Total cDNA was synthesized in a 20-μl reverse transcription reaction mixture containing 1.5 μg total RNA, 0.5 mM deoxynucleoside triphosphate (dNTP) mix, 200 U SuperScript III reverse transcriptase (Invitrogen, USA), and 12.5 ng random primers. Reverse transcription reactions were performed according to the method described by the manufacturer. The RT-qPCR was performed using a CFX96 real-time PCR detection system (Bio-Rad) with SYBR green I RealMasterMix (Tiangen Bio. Co., Ltd., Beijing) under the manufacturer's recommended reaction conditions. The primers used for RT-qPCR are listed in Table 3. All of the data were normalized to the expression levels of 16S rRNA and are presented as relative to the srpA expression level in cells grown in the absence of BP. All assays were performed at least three times from independent RNA preparations.
β-Galactosidase assay.
A fusion of the srpABC promoter and a promoterless lacZ gene was constructed in the low-copy-number vector pME6015. The srpS-srpA intergenic region (289 bp) was amplified by PCR with primers that incorporated restriction sites (an EcoRI site in the primer targeting the 5′ end, and a PstI site in the primer targeting the 3′ end). The DNA obtained was digested with EcoRI and PstI and linked to EcoRI- and PstI-digested pME6015 to yield pMEG. Plasmid pMEG was sequenced to ensure that no mutations were introduced in the corresponding promoter regions. The plasmid was transformed into the wild-type strain B6-2 and its derivatives at 15 kV · cm−1, 200 Ω, and 25 μF with a Bio-Rad Gene Pulser Xcell (Bio-Rad, USA) using a previously described method (36). The corresponding transformants were grown overnight in LB medium with tetracycline. The cultures were diluted 100 times in the same medium and grown until they reached an OD620 of 0.8, at which time chemical compound supplements were or were not added. After 2 h, β-galactosidase activity was assayed in permeabilized whole cells according to Miller's method (37). The promoter activity of PttgA was determined using the same method as for PsrpA. All experiments were repeated at least three times.
Accession number(s).
The genome sequence of strain B6-2 was submitted to the GenBank database under accession number NZ_CP015202. The accession numbers of genes srpABCS are kkk_RS03965, kkk_RS03970, kkk_RS03975, and kkk_RS03960, respectively. The accession numbers of genes ttgABCR are kkk_RS21310, kkk_RS21315, kkk_RS21320, and kkk_RS21305, respectively.
ACKNOWLEDGMENTS
This work was supported by grants from the Chinese National Natural Science Foundation (31570101, 31422004, and 21777098).
REFERENCES
- 1.Viguri J, Verde J, Irabien A. 2002. Environmental assessment of polycyclic aromatic hydrocarbons (PAHs) in surface sediments of the Santander Bay, Northern Spain. Chemosphere 48:157–165. doi: 10.1016/S0045-6535(02)00105-4. [DOI] [PubMed] [Google Scholar]
- 2.Chang K-F, Fang G-C, Chen J-C, Wu Y-S. 2006. Atmospheric polycyclic aromatic hydrocarbons (PAHs) in Asia: a review from 1999 to 2004. Environ Pollut 142:388–396. doi: 10.1016/j.envpol.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 3.Valenzuela J, Bumann U, Cespedes R, Padilla L, Gonzalez B. 1997. Degradation of chlorophenols by Alcaligenes eutrophus JMP134 (pJP4) in bleached Kraft mill effluent. Appl Environ Microbiol 63:227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chae J-C, Kim E, Park S-H, Kim C-K. 2000. Catabolic degradation of 4-chlorobiphenyl by Pseudomonas sp. DJ-12 via consecutive reaction of meta-cleavage and hydrolytic dechlorination. Biotechnol Bioprocess Eng 5:449–455. [Google Scholar]
- 5.L'Abbée JB, Barriault D, Sylvestre M. 2005. Metabolism of dibenzofuran and dibenzo-p-dioxin by the biphenyl dioxygenase of Burkholderia xenovorans LB400 and Comamonas testosteroni B-356. Appl Microbiol Biotechnol 67:506–514. doi: 10.1007/s00253-004-1791-3. [DOI] [PubMed] [Google Scholar]
- 6.Murphy CD, Quirke S, Balogun O. 2008. Degradation of fluorobiphenyl by Pseudomonas pseudoalcaligenes KF707. FEMS Microbiol Lett 286:45–49. doi: 10.1111/j.1574-6968.2008.01243.x. [DOI] [PubMed] [Google Scholar]
- 7.Li Q, Wang X, Yin G, Gai Z, Tang H, Ma C, Deng Z, Xu P. 2009. New metabolites in dibenzofuran cometabolic degradation by a biphenyl-cultivated Pseudomonas putida strain B6-2. Environ Sci Technol 43:8635–8642. doi: 10.1021/es901991d. [DOI] [PubMed] [Google Scholar]
- 8.Tang H, Yu H, Li Q, Wang X, Gai Z, Yin G, Su F, Tao F, Ma C, Xu P. 2011. Genome sequence of Pseudomonas putida strain B6-2, a superdegrader of polycyclic aromatic hydrocarbons and dioxin-like compounds. J Bacteriol 193:6789–6790. doi: 10.1128/JB.06201-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong P, Sun T-H, Beudert G, Hahn HH. 1997. Ecological effects of combined organic or inorganic pollution on soil microbial activities. Water Air Soil Pollut 96:133–143. doi: 10.1023/A:1026409628540. [DOI] [Google Scholar]
- 10.Dua M, Singh A, Sethunathan N, Johri A. 2002. Biotechnology and bioremediation: successes and limitations. Appl Microbiol Biotechnol 59:143–152. doi: 10.1007/s00253-002-1024-6. [DOI] [PubMed] [Google Scholar]
- 11.Xie F, Koziar SA, Lampi MA, Dixon DG, Norwood WP, Borgmann U, Huang XD, Greenberg BM. 2006. Assessment of the toxicity of mixtures of copper, 9,10-phenanthrenequinone, and phenanthrene to Daphnia magna: evidence for a reactive oxygen mechanism. Environ Toxicol Chem 25:613–622. doi: 10.1897/05-256R.1. [DOI] [PubMed] [Google Scholar]
- 12.Piddock LJV. 2006. Multidrug-resistance efflux pumps-not just for resistance. Nat Rev Microbiol 4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 13.Martinez JL, Sánchez MB, Martínez-Solano L, Hernandez A, Garmendia L, Fajardo A, Alvarez-Ortega C. 2009. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol Rev 33:430–449. doi: 10.1111/j.1574-6976.2008.00157.x. [DOI] [PubMed] [Google Scholar]
- 14.Kieboom J, Dennis JJ, Zylstra GJ, de Bont JAM. 1998. Active efflux of organic solvents by Pseudomonas putida S12 is induced by solvents. J Bacteriol 180:6769–6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieboom J, Dennis JJ, de Bont JAM, Zylstra GJ. 1998. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J Biol Chem 273:85–91. doi: 10.1074/jbc.273.1.85. [DOI] [PubMed] [Google Scholar]
- 16.Kieboom J, de Bont JAM. 2001. Identification and molecular characterization of an efflux system involved in Pseudomonas putida S12 multidrug resistance. Microbiology 147:43–51. doi: 10.1099/00221287-147-1-43. [DOI] [PubMed] [Google Scholar]
- 17.Ramos JL, Duque E, Godoy P, Segura A. 1998. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-T1E. J Bacteriol 180:3323–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosqueda G, Ramos JL. 2000. A set of genes encoding a second toluene efflux system in Pseudomonas putida DOT-T1E is linked to the tod genes for toluene metabolism. J Bacteriol 182:937–943. doi: 10.1128/JB.182.4.937-943.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rojas A, Duque E, Mosqueda G, Golden G, Hurtado A, Ramos JL, Segura A. 2001. Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT-T1E. J Bacteriol 183:3967–3973. doi: 10.1128/JB.183.13.3967-3973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phoenix P, Keane A, Patel A, Bergeron H, Ghoshal S, Lau P. 2003. Characterization of a new solvent-responsive gene locus in Pseudomonas putida F1 and its functionalization as a versatile biosensor. Environ Microbiol 5:1309–1327. doi: 10.1111/j.1462-2920.2003.00426.x. [DOI] [PubMed] [Google Scholar]
- 21.Rodríguez-Herva JJ, García V, Hurtado A, Segura A, Ramos JL. 2007. The ttgGHI solvent efflux pump operon of Pseudomonas putida DOT-T1E is located on a large self-transmissible plasmid. Environ Microbiol 9:1550–1561. doi: 10.1111/j.1462-2920.2007.01276.x. [DOI] [PubMed] [Google Scholar]
- 22.Hearn EM, Dennis JJ, Gray MR, Foght JM. 2003. Identification and characterization of the emhABC efflux system for polycyclic aromatic hydrocarbons in Pseudomonas fluorescens cLP6a. J Bacteriol 185:6233–6240. doi: 10.1128/JB.185.21.6233-6240.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chain PSG, Denef VJ, Konstantinidis KT, Vergez LM, Agulló L, Reyes VL, Hauser L, Córdova M, Gómez L, González M. 2006. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc Natl Acad Sci U S A 103:15280–15287. doi: 10.1073/pnas.0606924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez M, Gouveia J, Segura A, Muñoz R, Villaverde S. 2009. Addressing the role of the extrusion pump-bearing pGRT1 plasmid in toluene biodegradation by Pseudomonas putida DOT-T1E under real case scenarios. Water Sci Technol 60:2391–2398. doi: 10.2166/wst.2009.681. [DOI] [PubMed] [Google Scholar]
- 25.Volmer J, Neumann C, Buhler B, Schmid A. 2014. Engineering of Pseudomonas taiwanensis VLB120 for constitutive solvent tolerance and increased specific styrene epoxidation activity. Appl Environ Microbiol 80:6539–6548. doi: 10.1128/AEM.01940-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bugg T, Foght JM, Pickard MA, Gray MR. 2000. Uptake and active efflux of polycyclic aromatic hydrocarbons by Pseudomonas fluorescens LP6a. Appl Environ Microbiol 66:5387–5392. doi: 10.1128/AEM.66.12.5387-5392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adebusuyi AA, Smith AY, Gray MR, Foght JM. 2012. The EmhABC efflux pump decreases the efficiency of phenanthrene biodegradation by Pseudomonas fluorescens strain LP6a. Appl Microbiol Biotechnol 95:757–766. doi: 10.1007/s00253-012-3932-4. [DOI] [PubMed] [Google Scholar]
- 28.Kuepper J, Ruijssenaars HJ, Blank LM, de Winde JH, Wierckx N. 2015. Complete genome sequence of solvent-tolerant Pseudomonas putida S12 including megaplasmid pTTS12. J Biotechnol 200:17–18. doi: 10.1016/j.jbiotec.2015.02.027. [DOI] [PubMed] [Google Scholar]
- 29.Köhler AK, Rückert C, Schatschneider S, Vorhölter F-J, Szczepanowski R, Blank LM, Niehaus K, Goesmann A, Pühler A, Kalinowski J, Schmid A. 2013. Complete genome sequence of Pseudomonas sp. strain VLB120 a solvent tolerant, styrene degrading bacterium, isolated from forest soil. J Biotechnol 168:729–730. doi: 10.1016/j.jbiotec.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 30.Molina-Santiago C, Daddaoua A, Fillet S, Duque E, Ramos JL. 2014. Interspecies signalling: Pseudomonas putida efflux pump TtgGHI is activated by indole to increase antibiotic resistance. Environ Microbiol 16:1267–1281. doi: 10.1111/1462-2920.12368. [DOI] [PubMed] [Google Scholar]
- 31.Helling RB, Janes BK, Kimball H, Tran T, Bundesmann M, Check P, Phelan D, Miller C. 2002. Toxic waste disposal in Escherichia coli. J Bacteriol 184:3699–3703. doi: 10.1128/JB.184.13.3699-3703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morita Y, Sobel ML, Poole K. 2006. Antibiotic inducibility of the MexXY multidrug efflux system of Pseudomonas aeruginosa: involvement of the antibiotic-inducible PA5471 gene product. J Bacteriol 188:1847–1855. doi: 10.1128/JB.188.5.1847-1855.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sikkema J, De Bont J, Poolman B. 1995. Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev 59:201–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guazzaroni ME, Krell T, Felipe A, Ruiz R, Meng C, Zhang X, Gallegos MT, Ramos JL. 2005. The multidrug efflux regulator TtgV recognizes a wide range of structurally different effectors in solution and complexed with target DNA: evidence from isothermal titration calorimetry. J Biol Chem 280:20887–20893. doi: 10.1074/jbc.M500783200. [DOI] [PubMed] [Google Scholar]
- 35.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 36.Trevors J, Starodub M. 1990. Electroporation of pKK1 silver-resistance plasmid from Pseudomonas stutzeri AG259 into Pseudomonas putida CYM318. Curr Microbiol 21:103–107. doi: 10.1007/BF02091827. [DOI] [Google Scholar]
- 37.Joseph S, David W. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]