ABSTRACT
Staphylococcus aureus is a common biofilm-forming pathogen. Low doses of disinfectants have previously been reported to promote biofilm formation and to increase virulence. The aim of this study was to use transcriptome sequencing (RNA-seq) analysis to investigate global transcriptional changes in S. aureus in response to sublethal concentrations of the commonly used food industry disinfectants ethanol (EtOH) and chloramine T (ChT) and their combination (EtOH_ChT) in order to better understand the effects of these agents on biofilm formation. Treatment with EtOH and EtOH_ChT resulted in more significantly altered expression profiles than treatment with ChT. Our results revealed that EtOH and EtOH_ChT treatments enhanced the expression of genes responsible for regulation of gene expression (sigB), cell surface factors (clfAB), adhesins (sdrDE), and capsular polysaccharides (cap8EFGL), resulting in more intact biofilm. In addition, in this study we were able to identify the pathways involved in the adaptation of S. aureus to the stress of ChT treatment. Further, EtOH suppressed the effect of ChT on gene expression when these agents were used together at sublethal concentrations. These data show that in the presence of sublethal concentrations of tested disinfectants, S. aureus cells trigger protective mechanisms and try to cope with them.
IMPORTANCE So far, the effect of disinfectants is not satisfactorily explained. The presented data will allow a better understanding of the mode of disinfectant action with regard to biofilm formation and the ability of bacteria to survive the treatment. Such an understanding could contribute to the effort to eliminate possible sources of bacteria, making disinfectant application as efficient as possible. Biofilm formation plays significant role in the spread and pathogenesis of bacterial species.
KEYWORDS: biofilm formation, gene expression, bacterial decontamination
INTRODUCTION
Staphylococcus aureus is a common biofilm-forming pathogen found in a wide variety of environments (1, 2). It frequently causes implant and catheter-associated infections and is considered to be one of the most common foodborne diseases worldwide (3). S. aureus strains have been isolated from foods, indicating a potential risk of their dissemination through the food production chain (4). Food contamination arises mainly because of inadequately sanitized food-processing equipment and the subsequent formation of biofilms on surfaces (5).
The ability of S. aureus to form biofilms can enhance the persistence of the microorganism at infection sites or contaminated surfaces (6). Bacteria within biofilms display increased tolerance of disinfectants, antibiotics, and phagocytosis (7, 8). Thus, the difficult eradication of bacteria within biofilms could potentially lead to substantial economic losses and health problems (7). The S. aureus biofilm mode of growth is regulated by complex genetic factors. It has been shown that intercellular signaling plays a significant role during biofilm development and dispersal (6, 9, 10). However, the mechanisms and processes of biofilm formation in S. aureus are poorly understood, and studies detailing the changes in gene expression during this process are still limited.
Cleaning agents containing ethanol (EtOH) are commonly used as disinfectants to sanitize or sterilize surfaces in health facilities and food-processing environments. Several studies have suggested that treatment with low concentrations of alcohols can enhance biofilm formation by Staphylococcus species (11, 12). Interestingly, it was reported that low concentrations (sub-MIC) of residual disinfectants may even provide an opportunity for pathogens to adapt and grow (13). It is generally thought that alcohols act by disrupting membranes, inhibiting protein synthesis, and interfering with cell division (14, 15). However, alcohols lack sporicidal action, and they inadequately penetrate protein-rich materials. For this reason, alcohols are not optimal as single-agent antiseptics for the disposal of biofilms.
One of the antimicrobial agents most commonly used to manage biofilm growth in the food industry is chloramine T (ChT). It belongs to the group of chlorine-releasing agents, and its mechanism of action is not fully known (16, 17).
The growth of S. aureus biofilms can be enhanced by some processing methods encountered in the food industry, such as suboptimal temperatures, improper disinfection, or a combination of salt and glucose (18). Sublethal concentrations of disinfectants were described to promote the expression of virulence factors involved in biofilm growth (13, 19). To investigate this issue in more depth, we formed biofilms from S. aureus strains treated with sublethal concentrations of commonly used food industry disinfectants (EtOH, ChT, and a combination of the two [EtOH_ChT]). The aim of this study was to investigate global transcriptional changes using transcriptome sequencing (RNA-seq) analysis in order to better understand the effects of these disinfectants on biofilm formation.
RESULTS AND DISCUSSION
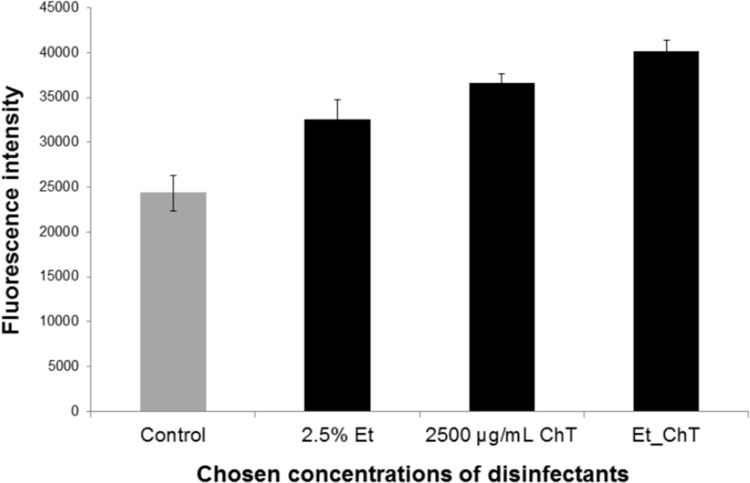
As reported previously, sublethal concentrations of some disinfectants can lead to enhanced biofilm formation, probably due to the adaptation of bacteria to these low-level stress conditions (19). Cincarova et al. determined the concentration ranges of EtOH and ChT that significantly enhanced biofilm formation of strong or weak S. aureus biofilm formers using Syto9 labeling (19). The effects of both tested disinfectants were higher in weak biofilm formers, and the sublethal concentrations with the strongest stimulating activity were approximately 2.5% EtOH and 2,500 μg/ml ChT (19). With reference to the work of Cincarova et al., we also chose to determine the effects of 2.5% EtOH and 2,500 μg/ml ChT on a weak S. aureus biofilm former. We further analyzed a combination of sublethal concentrations of EtOH (final concentration, 2.5%) and ChT (final concentration, 2,500 μg/ml) and observed a synergistic effect of this combination: the stimulation of biofilm formation was even higher than when each disinfectant was administered singly (Fig. 1).
FIG 1.
Syto9 quantification of S. aureus biofilm levels in response to treatment with disinfectants. Graph shows biofilm levels in samples cultivated with disinfectants versus controls (samples cultivated without disinfectants). S. aureus biofilm was cultivated for 24 h in medium (control) or in medium with selected sub-MICs of disinfectants that were previously proved to enhance S. aureus biofilm formation (19): 2.5% ethanol (Et) or 2,500 μg/ml chloramine T (ChT). Et_ChT, combination of both disinfectants at concentrations of 2.5% EtOH and 2,500 μg/ml ChT. Columns represent mean values of fluorescence, and vertical bars represent 95% confidence intervals regarding the means.
General changes in gene expression after treatment.
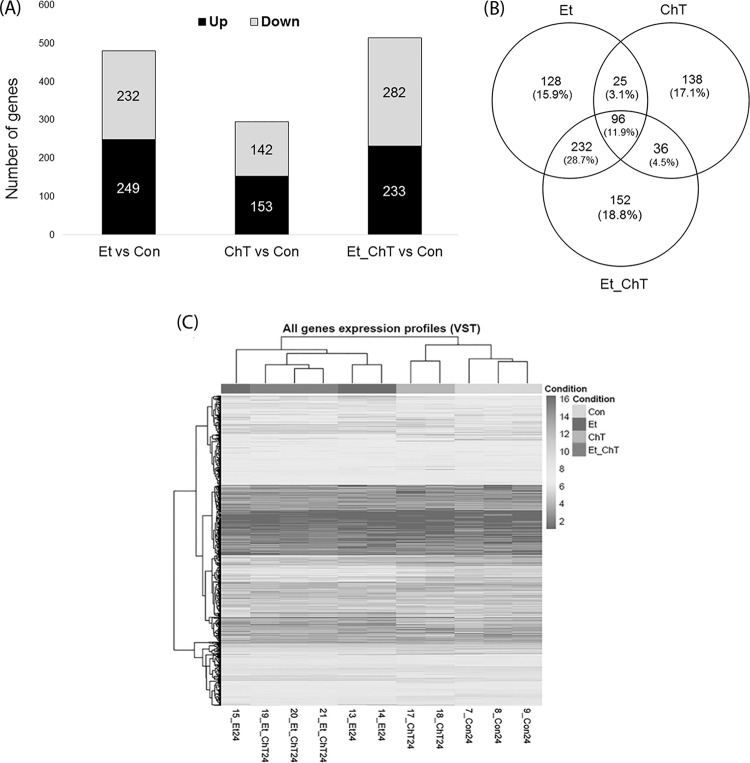
To determine the differences in gene expression between treatment groups and control groups, RNA samples from cells from each condition were sequenced (Fig. 2). The transcriptome of S. aureus biofilm changed significantly after incubation with all applied treatments (Fig. 2 and 3). Our analysis revealed an overlap of 81 upregulated genes in treated biofilms, including some genes related to biofilm formation, metabolism, transport, or membrane composition (Fig. 2B).
FIG 2.
Overview of the significant changes in the gene expression profiles of S. aureus biofilm formed after 24 h in the presence of sublethal concentrations of disinfectants. (A) Number of genes with significantly increased or decreased expression (adjusted P value < 0.05 and logFC ≥ 1). (B) Overlap of genes with significantly altered expression among the tested conditions compared to nontreated biofilm. (C) Heat map showing gene expression patterns and hierarchical clustering of all samples. Gene expression values are variance stabilizing transformation (VST) transformed. Con, untreated biofilm; Et, ethanol treatment; ChT, chloramine T treatment; Et_ChT, combination of both disinfectants.
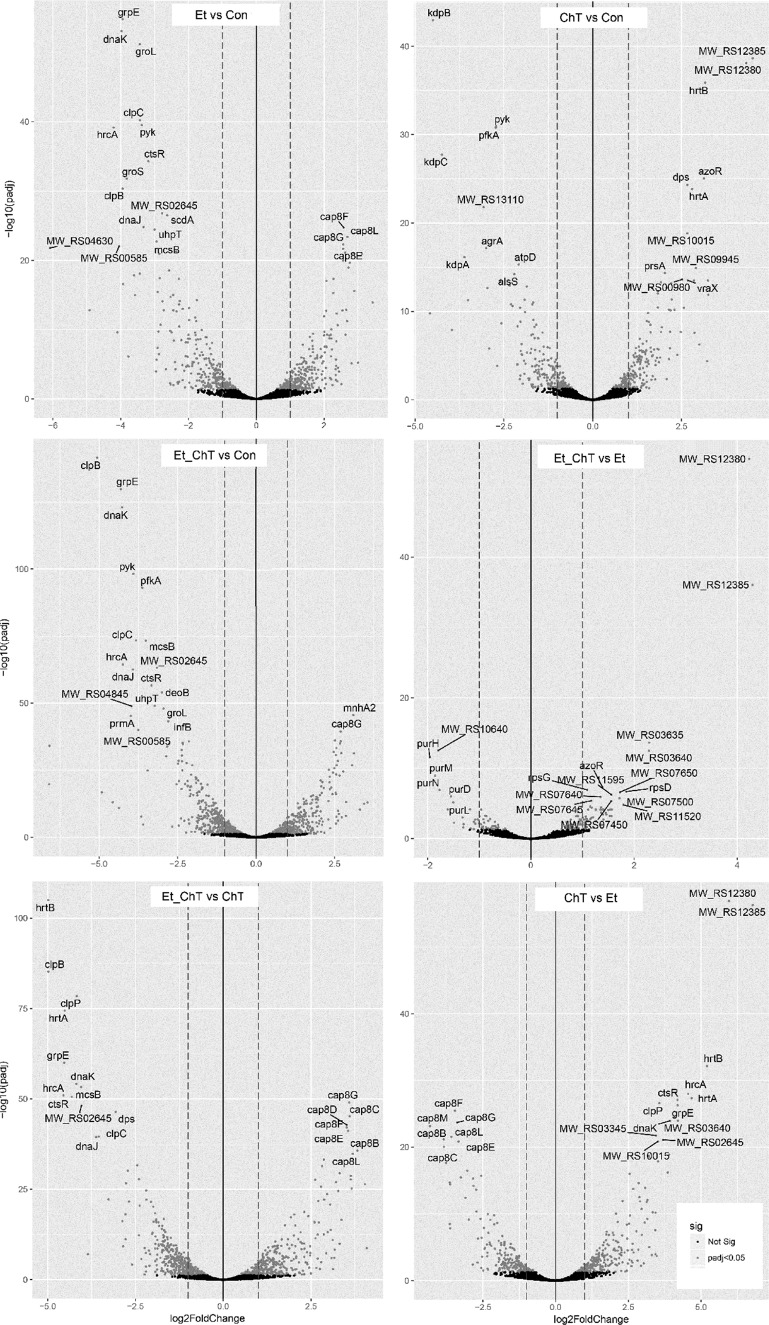
FIG 3.
Volcano plots of differential gene expression for S. aureus biofilm formed in response to treatment with sublethal concentrations of disinfectants. DESeq2 gene expression analysis was performed, and the top 20 differentially expressed genes (adjusted P value < 0.05 and logFC ≥1, sorted by adjusted P value) between individual treatments are indicated. The dashed line shows the logFC 1 threshold for downregulated (left line) and upregulated (right line) genes. Highlighted points are differentially expressed genes (P value < 0.05). Con, untreated biofilm; Et, ethanol treatment; ChT, chloramine T treatment; Et_ChT, combination of both disinfectants.
Expression analysis revealed that biofilm formed in the presence of ChT showed the fewest differences in gene expression compared to other treatments (Fig. 2A and 3), i.e., the clustering of all analyzed samples showed that treatment with EtOH and EtOH_ChT resulted in more changes in expression profiles than treatment with ChT (Fig. 2C).
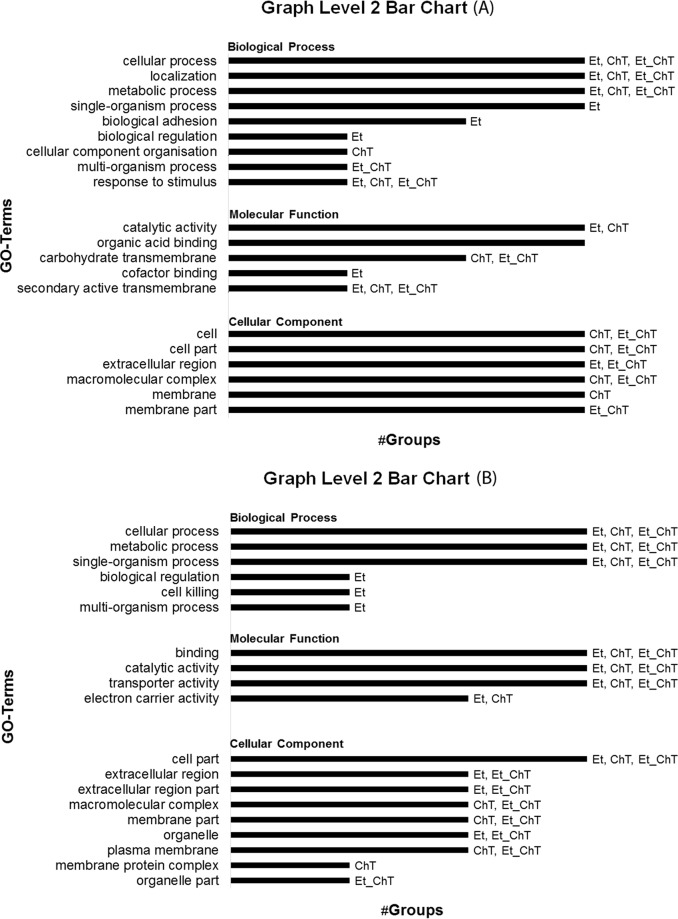
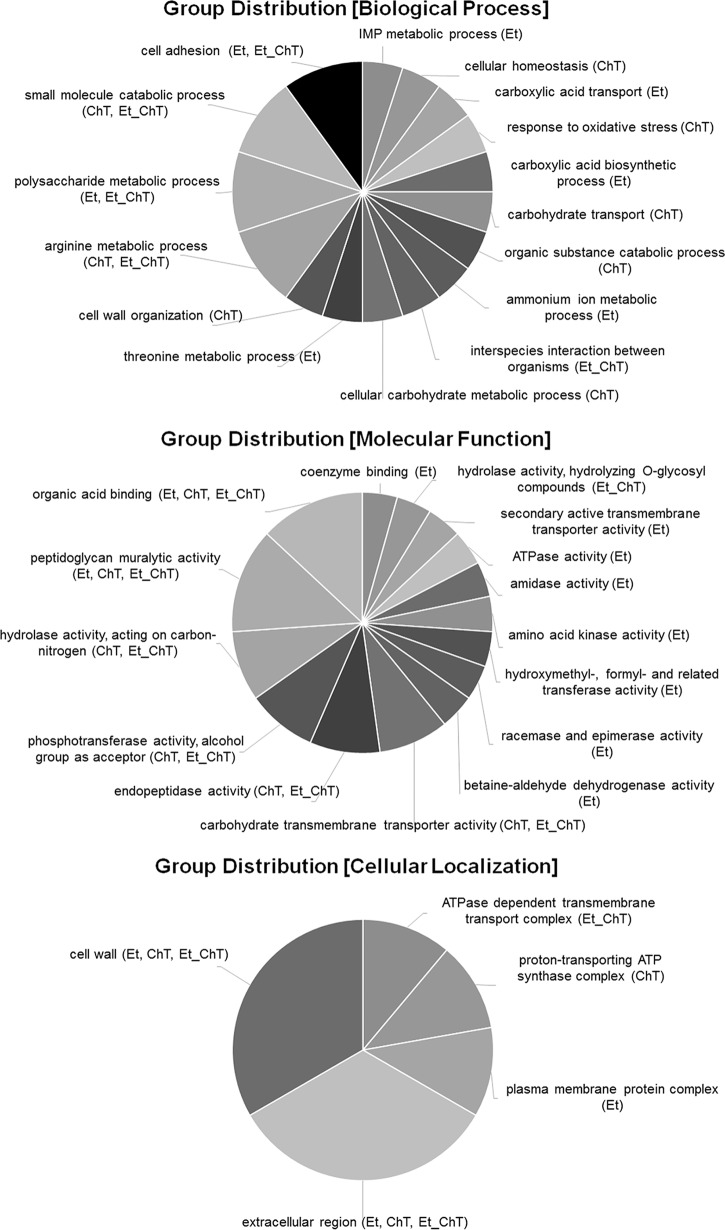
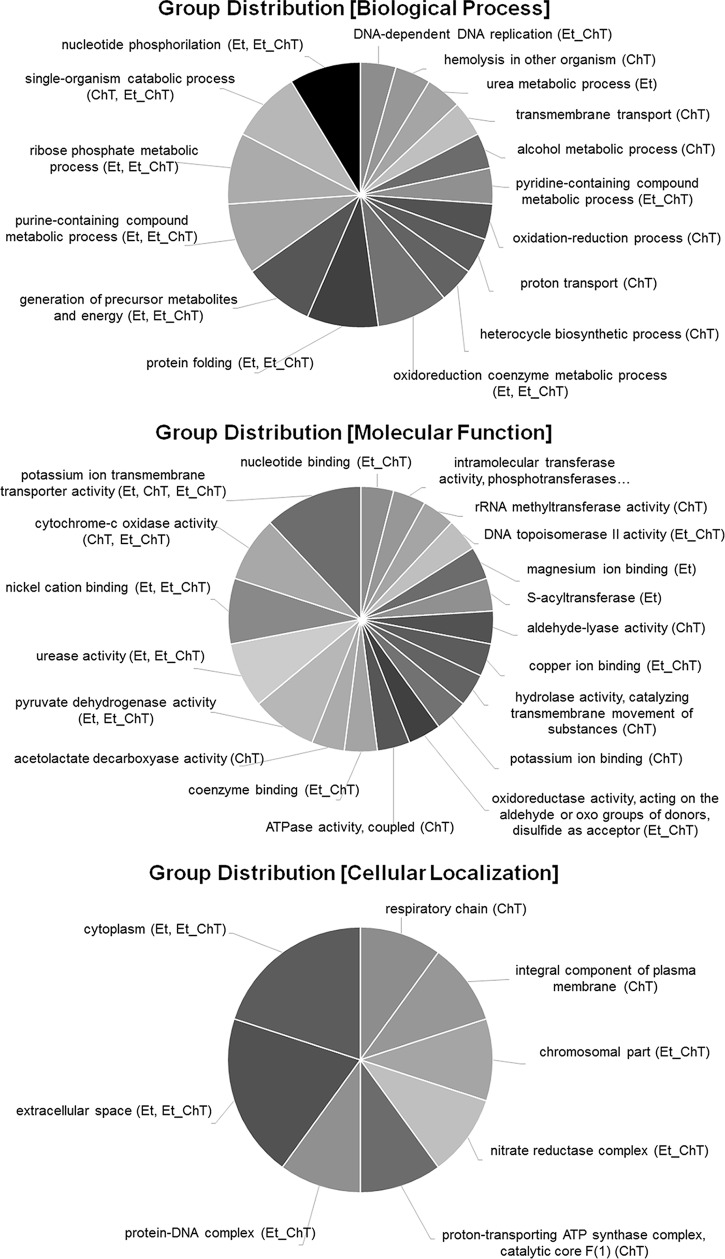
In addition, the comparison of gene ontology analysis results between individual treatments was carried out to identify significant alterations in biological processes, molecular functions, or cellular composition (Fig. 4, 5 and 6).
FIG 4.
Gene ontology (GO) analysis of upregulated (A) and downregulated (B) genes (adjusted P value < 0.05 and logFC ≥1) in S. aureus biofilms formed in response to treatment with the studied sublethal concentrations of disinfectants. Data were analyzed and visualized in Blast2go. Et, ethanol; ChT, chloramine T; Et_ChT, combination of both disinfectants.
FIG 5.
Multilevel visualization of group distribution for GO terms in response to all treatments. Group classification from differential expression analysis of genes identified as upregulated (adjusted P value < 0.05 and logFC ≥1) in biofilms formed in response to treatment with sublethal concentrations of the studied disinfectants versus untreated biofilm is shown. Data were analyzed and visualized in Blast2go.
FIG 6.
Multilevel visualization of group distribution for GO terms in response to all treatments. Group classification from differential expression analysis of genes identified as downregulated (adjusted P value < 0.05 and logFC ≥1) in biofilm formed in response to treatment with sublethal concentrations of the studied disinfectants versus untreated biofilm is shown. Data were analyzed and visualized in Blast2go.
Differentially expressed genes related to biofilm formation and virulence.
Notably, genes involved in biofilm formation and the virulence of S. aureus, including genes encoding transcription regulators, virulence factors, surface proteins, proteases, capsular polysaccharides, and others, were significantly upregulated/downregulated under all conditions (Fig. 3; see also Tables S1 and S2 in the supplemental material).
Two different pathways resulting in S. aureus biofilm have been described: polysaccharide intercellular adhesin (PIA)-dependent biofilm promoted by the ica operon and PIA-independent biofilm (4). The expression pattern (under all treatments) of genes previously described as being associated with biofilm development was suggestive of the PIA-independent pathway. All applied treatments (EtOH, ChT, EtOH_ChT) resulted in upregulation of the major autolysin (atl) and downregulation of the staphylococcal respiratory response regulator (srrAB) responsible for PIA induction under anaerobic conditions (20). Atl promotes primary attachment to surfaces, following which proteolytic cleavage of Atl leads to some cell lysis, extracellular DNA (eDNA) release, and cell accumulation as described previously (21, 22). More importantly, no significant upregulation of icaADBC was observed.
The expression of genes encoding the major global regulators of S. aureus gene expression, staphylococcal accessory regulator (sarA), RNA polymerase sigma factor (sigB), and accessory gene regulator (agr), was also altered in treated biofilms.
Our data revealed that EtOH/EtOH_ChT treatment induced the expression of genes responsible for gene regulation (sigB), cell surface factors (clfAB), adhesins (sdrDE), and capsular polysaccharides (cap8EFGL), resulting in increased biological adhesion (gene ontology [GO] category present in Fig. 4 and 5); treatment with ChT alone led to no significant upregulation of the genes listed above (Table S1).
Upregulation of sigB was described as downregulating protease production but additionally promoting expression of adherence factors that aid in initial biofilm formation (4, 23). Staphylococcal capsules are important in the pathogenesis of S. aureus infections. The capsule enhances staphylococcal virulence by hindering phagocytosis, resulting in bacterial persistence in the bloodstream of infected hosts (24). Characterized serotypes of S. aureus include those with 1, 2, 5, and 8 capsules, with more than 80% of S. aureus isolates expressing a serotype 5 or 8 capsular polysaccharide (cap) located in cap5 or cap8 gene clusters (25). Upregulation of the cap8 gene cluster is consistent with the fact that ClpC (whose expression was repressed in response to EtOH and EtOH_ChT treatment) strongly activates transcription of the sae operon, whose products are known to negatively regulate capsule synthesis (26). The polysaccharide metabolic process shown as a GO category in Fig. 5 supports our suggestion that expression of the cap8 gene cluster is induced by the presence of EtOH during biofilm treatment.
Transcripts of sarA are upregulated in biofilms compared with planktonic cultures (27). Expression of sarA acts to downregulate proteases and thermostable nuclease, allowing for development of an immature biofilm (28). sarA was upregulated in all treated samples, but only ChT treatment resulted in significant changes in expression (Cht log fold change [logFCChT] = 1.12, logFCEt = 0.71, and logFCEt_ChT = 0.65).
Our results show downregulation of agr in all treated samples. Agr is the key component of quorum sensing in S. aureus. Repression of the agr operon has been shown to be necessary for immature biofilm formation contradictory to its upregulation in developed (mature) biofilm, where it induces the production of detergent-like peptides, proteases, and thermostable nucleases responsible for biofilm dispersal (29, 30). There also appears to be a correlation between the ability of S. aureus to adhere to polystyrene and the agr quorum-sensing system phenotype, as 78% of agr-negative, but only 6% of agr-positive, strains tested form biofilms (6).
Many virulence factors in S. aureus are controlled by Agr and Sar (31). This regulation may be affected by the environment in which the organisms are grown. Agr and Sar have pleiotropic effects on the surface expression of molecules responsible for binding to different substrates (32). The analysis of biofilm treated with ChT showed the lowest number of expressed genes associated with production of virulence factors. This is possibly due to the effect of the stress conditions discussed below. More importantly, the combination of EtOH with ChT enhanced the expression of the gene for staphylococcal serine protease (sspA).
The downregulation of the agr operon and the absence of expression of the genes responsible for production of extracellularly released virulence factors (hemolysins hlgA, MW_RS10655, and MW_RS05690) in response to all treatments suggest the formation of an immature stage of biofilm after 24 h.
Stress response.
The ability to combat oxidative stress is important for survival in the environment, for protection against disinfectant treatments in the food industry, and for defense against host phagocytes when bacteria are attacked during infection (33). Gene ontology analysis (biological process) revealed a pronounced stress response for ChT treatment (response to stimulus shown in Fig. 4A; response to oxidative stress shown in Fig. 5). ChT, as an active chlorine compound and well-known biocide with strong oxidative activity, induced the expression of several genes that are involved in detoxification and that are under the control of the general stress regulator PerR (34). These included the genes for catalase (katA), superoxide dismutase (sodA), and alkyl hydroperoxide reductase (ahpC) (Table S1 in the supplemental material). The role of these enzymes in oxidative stress protection was previously inferred from studies performed in deficient S. aureus strains which exhibited lower resistance to oxidative stress and protection from DNA damage and reduced virulence (35, 36). This finding is supported by ChT-associated upregulation of dps (DNA starvation/stationary-phase protection protein). Dps family proteins possess ferroxidase activity that contributes to oxidative stress resistance (37).
EtOH and EtOH_ChT treatments did not result in any stress response as downregulation of genes that are part of the stress-induced multichaperone system (clpB, clpC, dnaJ, dnaK, groL, grpE) was observed, while ChT treatment resulted in no significant change of gene expression compared to untreated biofilm. GroL prevents misfolding and promotes the refolding and proper assembly of unfolded polypeptides generated under stress conditions (38). Clp homologues are important for a variety of stress conditions, virulence, growth recovery (recovery from the stationary phase), and death (39). Clp ATPase and CLP proteolytic complexes control several key processes that contribute to the success of S. aureus as a pathogen (39, 40).
Transport and metabolism.
The applied treatments altered the expression of genes associated with multiple cellular processes, including transport, localization, or metabolic pathways (amino acids, lipids, and carbohydrates). GO analysis focused on molecular function revealed a significant decrease in transporter activity (Fig. 3). We detected a downregulation of K+ transport-associated operon (kdpABC) in all treated samples. The role of the kdpABC operon in many bacteria still remains unclear. More importantly, we observed ChT-associated downregulation of the kdpDE operon previously shown to regulate transcription for a series of virulence factors through sensing external K+ concentrations (41). Repression of kdpDE seems to be associated with agrB levels, because agrB was significantly downregulated in response to ChT treatment (logFC = −1,68), while changes in response to EtOH (logFC = −0.42) and EtOH_ChT (logFC = −0.62) treatments were below the applied cutoffs. Previously published work revealed that kdpDE is upregulated by the Agr/RNAIII system and may be an important virulence regulator during pathogenesis in S. aureus.
Interestingly, treatment with ChT (alone or in combination with EtOH) resulted in the activation of the anaerobic alternative energy-generating arginine deamination (ADI) pathway composed of three enzymes (arginine deiminase [ArcA]; ornithine transcarbomoylase [ArcB]; carbamate kinase [ArcC]), that enable the utilization of arginine as a source of energy for growth under anaerobic conditions (42, 43). Recent transcriptional profiling of staphylococci growing in biofilms has suggested that the bacteria grow anaerobically (40). Our data demonstrate that ChT upregulates expression of the ADI pathway, leading us to speculate that ChT enhances biofilm formation, in part, through shunting metabolic flux toward the ADI pathway.
Conclusions.
Several studies documented the effect of sublethal concentrations of individual disinfectants on biofilm growth and the expression of virulence factors. So far, the effect of disinfectants is not satisfactorily explained. To our best knowledge there is no study describing transcriptomic changes in S. aureus biofilm treated with ChT (alone or combined with EtOH). The data presented here enhance the knowledge necessary to better understand the mode of action of disinfectants with regard to their effectivity and the possibility of bacteria surviving the treatment. This knowledge could contribute to the effort to eliminate possible sources of bacteria, making disinfectant application as efficient as possible.
Different treatments applied to shunting during biofilm formation resulted in distinct transcriptomic profiles in this study. All applied treatments induced biofilm formation via the PIA-independent pathway. Growth in response to treatments lasting 24 h resulted in the formation of immature biofilm, possibly due to adaptation to the applied treatment and short incubation time. Nevertheless, treatment with EtOH and EtOH_ChT resulted in more significantly altered expression profiles than treatment with ChT. Treatment with EtOH and EtOH_ChT enhanced the expression of genes responsible for gene regulation (sigB), cell surface factors (clfAB), adhesins (sdrDE), and capsular polysaccharides (cap8EFGL), resulting in more intact biofilm. This study enabled us to identify the pathways involved in the adaptation of the bacterium to the stressful conditions encountered during ChT treatment, which contributes to its survival and persistence. Finally, considering the general expression profile we assume that the effect of ChT is antagonized by the effect of EtOH when both are used together at sublethal concentrations. This raises the possibility that this combination of disinfectants may not be effective for sanitation, but further research should be performed.
MATERIALS AND METHODS
Growth conditions.
S. aureus samples were collected from food contact equipment in a meat processing plant located in the South Moravian region of the Czech Republic. Briefly, sampling was carried out in 2 visits. Samples were taken aseptically from a surface covering approximately 100 cm2 by using a sterile sampling sponge moistened with LPT neutralizing broth (P-LAB, Prague, Czech Republic) and then transported at 4°C to the laboratory for immediate processing. First, the strength of biofilm formation was determined according to a procedure described previously (44). For further analysis, the weak biofilm former B+ (internal collection number 1053; isolated from knives used in slaughtering) was chosen, because according to previous measurements the highest induction of biofilm formation caused by ethanol or chloramine T was observed in weak isolates (19). The S. aureus biofilm was grown in Trypticase soy broth (TSB) (Oxoid Ltd., Hampshire, England) supplemented with 1% NaCl (Penta, Chrudim, Czech Republic) and 1% d-glucose (Penta). To prepare biofilm, S. aureus inoculum (5 h cultivation in shaking water bath at 37°C) was grown to reach 1.5 × 109 CFU/ml, diluted 1:100 in TSB, and dispensed (in 10-ml aliquots) into 6-well polystyrene flat-bottomed, non-growth-enhanced tissue culture plates (Falcon, NY) to obtain sufficient amounts for RNA isolation; or in 200-μl aliquots into 96-well plates (Falcon) for biofilm quantification. Bacteria were incubated in their respective media (control medium, 2.5% EtOH in medium, 2,500 μg/ml ChT in medium, or a combination of 2.5% EtOH and 2,500 μg/ml ChT at their final concentrations) in an incubator (Sanyo, Tokyo, Japan). Biofilm was cultivated at 37°C for 24 h. Medium was discarded from 6-well plates, and biofilm was washed twice (2 ml ice-cold PBS, pH = 7.2) and then centrifuged at 5,000 × g for 10 min to harvest cells. Samples were stored at −80°C. Biofilms in 96-well plates were quantified using Syto9 staining.
Biofilm quantification.
S. aureus biofilm in 96-well tissue culture plates was quantified after 24 h of growth using the green fluorescent nucleic acid stain Syto9 (Life Technologies, Eugene, OR) as described previously (19). Briefly, the medium was discarded, and biofilms were washed with phosphate-buffered saline (PBS) (pH = 7.2) at room temperature for 15 min at 250 rpm in a TS-100 thermo-shaker (BioSan, MI). Syto9 was diluted in PBS (1:3,600). One hundred μl of PBS/well and 100 μl of a diluted solution of Syto 9/well were added consecutively to the washed biofilm. After 1 h of incubation in a thermo-shaker at 37°C at 250 rpm in the dark, fluorescence was measured using a Synergy H1 hybrid reader (BioTek, VT) (excitation, 478 nm; emission, 510 nm; gain, 60%).
Total RNA isolation.
After the biofilms were rinsed, the compound-free and treated biofilm cells were harvested by a washing step and placed in the RNAprotect bacteria reagent (Qiagen GmbH, Germany). The sessile cell suspension was then transferred to a microcentrifuge tube and incubated for 5 min at room temperature to stabilize the mRNA. Next, the suspension was centrifuged at 5,000 × g for 5 min at 4°C to harvest the cells. Total RNA was purified using a combination of two different kits. Briefly, cell disruption (only zirconium beads with no enzymatic treatment) followed by phenol-chloroform extraction and ethanol precipitation was achieved using the RiboPure-Bacteria kit (ThermoFisher Scientific, Waltham, MA) according to the manufacturer's protocol. Precipitated RNA was then loaded on columns of the RNeasy minikit (Qiagen) and purified according to the manufacturer's instructions. On-column DNase digestion using RNase-Free DNase (Qiagen) was applied to remove residual contaminating DNA. Each total RNA sample was suspended in 30 μl of RNase-free water, and the quality of total RNA obtained was determined using an Agilent 2100 bioanalyzer (Agilent, Santa Clara, CA).
RNA-seq library preparation and sequencing.
Three independent biological replicates of each 6-well plate experiment were used for further analysis. A ScriptSeq complete bacteria kit (Epicentre, Madison, WI) was used to deplete rRNA from total RNA samples (2 μg of DNA-free RNA) with Ribo-Zero magnetic beads and to generate an RNA-seq library according to the manufacturer's specifications. During library preparation, individual samples were indexed with ScriptSeq index PCR primers (set 1 and set 2) to enable library pooling. The construction, quality, and quantity of the libraries were estimated using the KAPA library quantification kit (Kapa Biosystems), Qubit dsDNA HS assay kit (ThermoFisher Scientific), and Agilent DNA 1000 kit (Agilent). Sequencing data were generated from paired-end reads (2 × 75 bp) on a MiSeq system (Illumina, San Diego, CA) using an RNA-seq library of 10 nM.
Analysis pipeline.
The quality of the sequencing data was checked using FastQC v0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) and MultiQC v0.8 (45). The presence of adapters was scanned for using Minion, Kraken package v15-065 (46). Preprocessing of raw reads was done using Trimmomatic v0.36 (47) in the following steps. (i) N and very-low-quality bases (Phred score <3) from both 5′ and 3′ ends were removed. (ii) Low-quality ends with average Phred scores of <5 of 4 consecutive bases were trimmed using a sliding window approach. (iii) Reads shorter than 20 bp and without proper pairing after the preprocessing were removed. Preprocessed reads were then mapped to the S. aureus subsp. aureus MW2 reference genome (GenBank accession number GCF_000011265.1) using STAR v2.5.2b (48). The quality of the mapping was scanned and evaluated using RSeQC v2.6.1 (49) and Picard tools v2.5.0 (http://broadinstitute.github.io/picard/). Only uniquely mapped reads were considered in subsequent analyses. Raw gene expression quantification was performed using STAR v2.5.2b, and stranded gene counts were selected. Differential gene expression was analyzed using DESeq2 v1.14.1 (50) and edgeR v3.16.5 (51) bioconductor packages (52). Genes with a P value < 0.05 and fold change ≥2 (mean value from all biological replicates) were considered being significantly differentially expressed.
Gene ontology (GO) enrichment analysis was performed using the BiNGO plugin in the Cytoscape software platform v3.4.0 (53). Blast2go was used for the mapping of gene ontology terms and the description of biological processes, molecular function, and cellular components associated with the biofilm expression profiles (54).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Czech projects RO0517, COST LD 14015, and NPUI LO1218. We acknowledge the CF New Generation Sequencing Bioinformatics supported by the CIISB research infrastructure (LM2015043 funded by MEYS CR) for their support with obtaining scientific data presented in this paper.
Access to computing and storage facilities owned by parties and projects contributing to the National Grid Infrastructure MetaCentrum, provided under the program “Projects of Large Research, Development, and Innovations Infrastructures” (CESNET LM2015042), is greatly appreciated.
Neysan Donnelly (Clear Science Editing Services) is thanked for grammatical correction of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01643-17.
REFERENCES
- 1.Mermel LA, Allon M, Bouza E, Flynn P, O'Grady NP, Raad II, Rijnders BJ, Sheretz RJ, Warren DK. 2010. Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis 49:1–45. doi: 10.1086/599376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graveland H, Duim B, van Duijkeren E, Heederik D, Wagenaar JA. 2011. Livestock-associated methicillin-resistant Staphylococcus aureus in animals and humans. Int J Med Microbiol 301:630–634. doi: 10.1016/j.ijmm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Crago B, Ferrato C, Drews SJ, Svenson LW, Tyrrell G, Louie M. 2012. Prevalence of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) in food samples associated with foodborne illness in Alberta, Canada from 2007 to 2010. Food Microbiol 32:202–205. doi: 10.1016/j.fm.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Doulgeraki AI, Di Ciccio P, Ianieri A, Nychas GJE. 2017. Methicillin-resistant food-related Staphylococcus aureus: a review of current knowledge and biofilm formation for future studies and applications. Res Microbiol 168:1–15. doi: 10.1016/j.resmic.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Gutierrez D, Delgado S, Vazquez-Sanchez D, Martinez B, Cabo ML, Rodriguez A, Herrera JJ, García P. 2012. Incidence of Staphylococcus aureus and analysis of associated bacterial communities on food industry surfaces. Appl Environ Microbiol 78:8547–8554. doi: 10.1128/AEM.02045-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gotz F. 2002. Staphylococcus and biofilms. Mol Microbiol 43:1367–1378. [DOI] [PubMed] [Google Scholar]
- 7.Bridier A, Briandet R, Thomas V, Dubois-Brissonnet F. 2011. Resistance of bacterial biofilms to disinfectants: a review. Biofouling 27:1017–1032. doi: 10.1080/08927014.2011.626899. [DOI] [PubMed] [Google Scholar]
- 8.Halliman DG, Ahearn DG. 2004. Relative susceptibilities to vancomycin and quinupristin-dalfopristin of adhered and planktonic vancomycin-resistant and vancomycin-susceptible coagulase-negative staphylococci. Curr Microbiol 48:214–218. doi: 10.1007/s00284-003-4091-8. [DOI] [PubMed] [Google Scholar]
- 9.Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 10.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. 2004. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol 186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knobloch JKM, Horstkotte MA, Rohde H, Kaulfers PM, Mack D. 2002. Alcoholic ingredients in skin disinfectants increase biofilm expression of Staphylococcus epidermidis. J Antimicrob Chemother 49:683–687. doi: 10.1093/jac/49.4.683. [DOI] [PubMed] [Google Scholar]
- 12.Korem M, Gov Y, Rosenberg M. 2010. Global gene expression in Staphylococcus aureus following exposure to alcohol. Microb Pathog 48:74–84. doi: 10.1016/j.micpath.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Kastbjerg VG, Larsen MH, Gram L, Ingmer H. 2010. Influence of sublethal concentrations of common disinfectants on expression of virulence genes in Listeria monocytogenes. Appl Environ Microbiol 76:303–309. doi: 10.1128/AEM.00925-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silveira MG, Baumgartner M, Rombouts FM, Abee T. 2004. Effect of adaptation to ethanol on cytoplasmic and membrane protein profiles of Oenococcus oeni. Appl Environ Microbiol 70:2748–2755. doi: 10.1128/AEM.70.5.2748-2755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert P, McBain AJ. 2003. Potential impact of increased use of biocides in consumer products on prevalence of antibiotic resistance. Clin Microbiol Rev 16:189–208. doi: 10.1128/CMR.16.2.189-208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rolland SL, Carrick TE, Walls AW, McCabe JF. 2007. Dentin decontamination using chloramine T prior to experiments involving bacteria. Dent Mater 23:1468–1472. doi: 10.1016/j.dental.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Bal Krishna KC, Sathasivan A, Ginige MP. 2013. Microbial community changes with decaying chloramine residuals in a lab-scale system. Water Res 47:4666–4679. doi: 10.1016/j.watres.2013.04.035. [DOI] [PubMed] [Google Scholar]
- 18.Rode TM, Langsrud S, Holck A, Moretro T. 2007. Different patterns of biofilm formation in Staphylococcus aureus under food-related stress conditions. Int J Food Microbiol 116:372–383. doi: 10.1016/j.ijfoodmicro.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 19.Cincarova L, Polansky O, Babak V, Kulich P, Kralik P. 2016. Changes in the expression of biofilm-associated surface proteins in Staphylococcus aureus food-environmental isolates subjected to sublethal concentrations of disinfectants. Biomed Res Int 2016:4034517. doi: 10.1155/2016/4034517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulrich M, Bastian M, Cramton SE, Ziegler K, Pragman AA, Bragonzi A, Memmi G, Wolz C, Schlievert PM, Cheung A, Döring G. 2007. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol Microbiol 65:1276–1287. doi: 10.1111/j.1365-2958.2007.05863.x. [DOI] [PubMed] [Google Scholar]
- 21.Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. 2011. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence 2:445–459. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houston P, Rowe SE, Pozzi C, Waters EM, O'Gara JP. 2011. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect Immun 79:1153–1165. doi: 10.1128/IAI.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholas RO, Li T, McDevitt D, Marra A, Sucoloski S, Demarsh PL, Gentry DR. 1999. Isolation and characterization of a sigB deletion mutant of Staphylococcus aureus. Infect Immun 67:3667–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Riordan K, Lee JC. 2004. Staphylococcus aureus capsular polysaccharides. Clin Microbiol Rev 17:218–234. doi: 10.1128/CMR.17.1.218-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cocchiaro JL, Gomez MI, Risley A, Solinga R, Sordelli DO, Lee JC. 2006. Molecular characterization of the capsule locus from non-typeable Staphylococcus aureus. Mol Microbiol 59:948–960. doi: 10.1111/j.1365-2958.2005.04978.x. [DOI] [PubMed] [Google Scholar]
- 26.Luong TT, Sau K, Roux C, Sau S, Dunman PM, Lee CY. 2011. Staphylococcus aureus ClpC divergently regulates capsule via sae and codY in strain Newman but activates capsule via codY in strain UAMS-1 and in strain Newman with repaired saeS. J Bacteriol 193:686–694. doi: 10.1128/JB.00987-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valle J, Toledo-Arana A, Berasain C, Ghigo JM, Amorena B, Penades JR, Lasa I. 2003. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol 48:1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 28.Beenken KE, Mrak LN, Griffin LM, Zielinska AK, Shaw LN, Rice KC, Horswill AR, Bayles KW, Smeltzer MS. 2010. Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS One 5:e10790. doi: 10.1371/journal.pone.0010790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan WC, Coyle BJ, Williams P. 2004. Virulence regulation and quorum sensing in staphylococcal infections: competitive AgrC antagonists as quorum sensing inhibitors. J Med Chem 47:4633–4641. doi: 10.1021/jm0400754. [DOI] [PubMed] [Google Scholar]
- 30.Boles BR, Horswill AR. 2008. agr-Mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog 4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arya R, Princy SA. 2013. An insight into pleiotropic regulators Agr and Sar: molecular probes paving the new way for antivirulent therapy. Future Microbiol 8:1339–1353. doi: 10.2217/fmb.13.92. [DOI] [PubMed] [Google Scholar]
- 32.Pratten J, Foster SJ, Chan PF, Wilson M, Nair SP. 2001. Staphylococcus aureus accessory regulators: expression within biofilms and effect on adhesion. Microb Infect 3:633–637. doi: 10.1016/S1286-4579(01)01418-6. [DOI] [PubMed] [Google Scholar]
- 33.Flannagan RS, Cosio G, Grinstein S. 2009. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol 7:355–366. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- 34.Horsburgh MJ, Ingham E, Foster SJ. 2001. In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J Bacteriol 183:468–475. doi: 10.1128/JB.183.2.468-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cosgrove K, Coutts G, Jonsson IM, Tarkowski A, Kokai-Kun JF, Mond JJ, Foster SJ. 2007. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J Bacteriol 189:1025–1035. doi: 10.1128/JB.01524-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolf C, Hochgrafe F, Kusch H, Albrecht D, Hecker M, Engelmann S. 2008. Proteomic analysis of antioxidant strategies of Staphylococcus aureus: diverse responses to different oxidants. Proteomics 8:3139–3153. doi: 10.1002/pmic.200701062. [DOI] [PubMed] [Google Scholar]
- 37.Ushijima Y, Yoshida O, Villanueva MJ, Ohniwa RL, Morikawa K. 2016. Nucleoid clumping is dispensable for the Dps-dependent hydrogen peroxide resistance in Staphylococcus aureus. Microbiology 162:1822–1828. doi: 10.1099/mic.0.000353. [DOI] [PubMed] [Google Scholar]
- 38.Michta E, Ding W, Zhu SC, Blin K, Ruan HQ, Wang R, Wohlleben W, Mast Y. 2014. Proteomic approach to reveal the regulatory function of aconitase AcnA in oxidative stress response in the antibiotic producer Streptomyces viridochromogenes Tu494. PLoS One 9:e87905. doi: 10.1371/journal.pone.0087905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graham JW, Lei MG, Lee CY. 2013. Trapping and identification of cellular substrates of the Staphylococcus aureus ClpC chaperone. J Bacteriol 195:4506–4516. doi: 10.1128/JB.00758-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chatterjee I, Becker P, Grundmeier M, Bischoff M, Somerville GA, Peters G, Sinha B, Harraghy N, Proctor RA, Herrmann M. 2005. Staphylococcus aureus ClpC is required for stress resistance, aconitase activity, growth recovery, and death. J Bacteriol 187:4488–4496. doi: 10.1128/JB.187.13.4488-4496.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue T, You Y, Hong D, Sun H, Sun B. 2011. The Staphylococcus aureus KdpDE two-component system couples extracellular K+ sensing and Agr signaling to infection programming. Infect Immun 79:2154–2167. doi: 10.1128/IAI.01180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Resch A, Rosenstein R, Nerz C, Gotz F. 2005. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl Environ Microbiol 71:2663–2676. doi: 10.1128/AEM.71.5.2663-2676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makhlin J, Kofman T, Borovok I, Kohler C, Engelmann S, Cohen G, Aharonowitz Y. 2007. Staphylococcus aureus ArcR controls expression of the arginine deiminase operon. J Bacteriol 189:5976–5986. doi: 10.1128/JB.00592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stepanovic S, Vukovic D, Hola V, Di Bonaventura G, Djukic S, Cirkovic I, Ruzicka F. 2007. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 115:891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 45.Ewels P, Magnusson M, Lundin S, Kaller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis MPA, van Dongen S, Abreu-Goodger C, Bartonicek N, Enright AJ. 2013. Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods 63:41–49. doi: 10.1016/j.ymeth.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang LG, Wang SQ, Li W. 2012. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 50.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:e550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oleœ AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. 2015. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maere S, Heymans K, Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 54.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.