ABSTRACT
The ability to hydrolyze microcrystalline cellulose is an uncommon feature in the microbial world, but it can be exploited for conversion of lignocellulosic feedstocks into biobased fuels and chemicals. Understanding the physiological and biochemical mechanisms by which microorganisms deconstruct cellulosic material is key to achieving this objective. The glucan degradation locus (GDL) in the genomes of extremely thermophilic Caldicellulosiruptor species encodes polysaccharide lyases (PLs), unique cellulose binding proteins (tāpirins), and putative posttranslational modifying enzymes, in addition to multidomain, multifunctional glycoside hydrolases (GHs), thereby representing an alternative paradigm for plant biomass degradation compared to fungal or cellulosomal systems. To examine the individual and collective in vivo roles of the glycolytic enzymes, the six GH genes in the GDL of Caldicellulosiruptor bescii were systematically deleted, and the extents to which the resulting mutant strains could solubilize microcrystalline cellulose (Avicel) and plant biomass (switchgrass or poplar) were examined. Three of the GDL enzymes, Athe_1867 (CelA) (GH9-CBM3-CBM3-CBM3-GH48), Athe_1859 (GH5-CBM3-CBM3-GH44), and Athe_1857 (GH10-CBM3-CBM3-GH48), acted synergistically in vivo and accounted for 92% of naked microcrystalline cellulose (Avicel) degradation. However, the relative importance of the GDL GHs varied for the plant biomass substrates tested. Furthermore, mixed cultures of mutant strains showed that switchgrass solubilization depended on the secretome-bound enzymes collectively produced by the culture, not on the specific strain from which they came. These results demonstrate that certain GDL GHs are primarily responsible for the degradation of microcrystalline cellulose-containing substrates by C. bescii and provide new insights into the workings of a novel microbial mechanism for lignocellulose utilization.
IMPORTANCE The efficient and extensive degradation of complex polysaccharides in lignocellulosic biomass, particularly microcrystalline cellulose, remains a major barrier to its use as a renewable feedstock for the production of fuels and chemicals. Extremely thermophilic bacteria from the genus Caldicellulosiruptor rapidly degrade plant biomass to fermentable sugars at temperatures of 70 to 78°C, although the specific mechanism by which this occurs is not clear. Previous comparative genomic studies identified a genomic locus found only in certain Caldicellulosiruptor species that was hypothesized to be mainly responsible for microcrystalline cellulose degradation. By systematically deleting genes in this locus in Caldicellulosiruptor bescii, the nuanced, substrate-specific in vivo roles of glycolytic enzymes in deconstructing crystalline cellulose and plant biomasses could be discerned. The results here point to synergism of three multidomain cellulases in C. bescii, working in conjunction with the aggregate secreted enzyme inventory, as the key to the plant biomass degradation ability of this extreme thermophile.
KEYWORDS: Caldicellulosiruptor, extreme thermophile, cellulose degradation, lignocellulose, glycoside hydrolase, cellulase
INTRODUCTION
Anaerobic, Gram-positive bacteria from the genus Caldicellulosiruptor are the most thermophilic lignocellulosic biomass degraders currently identified (1, 2). These species can be isolated from globally distributed terrestrial hot springs and have optimal growth temperatures that range up to 78°C. Caldicellulosiruptor species can degrade unpretreated lignocellulosic substrates, driven by a variety of large, multidomain carbohydrate active enzymes (CAZymes), primarily glycoside hydrolases (GHs) (3). These large, single-polypeptide enzymes represent an alternative paradigm for plant biomass degradation compared to that of fungal or cellulosomal plant biomass degraders (1, 4, 5).
As crystalline cellulose is the primary recalcitrant carbohydrate component of the plant cell wall, its efficient degradation is essential for biomass conversion to biobased fuels and chemicals. However, the ability to degrade cellulose varies among the 13 Caldicellulosiruptor species that have been characterized (6). Comparative genomic and phenotypic analyses of these species have identified CAZyme domains that are common to the highly cellulolytic species, including carbohydrate binding module family 3 (CBM3) and GH families 9, 44, and 48 (7). The genes encoding these cellulolytic enzymes are concentrated in one genomic locus, which has been referred to as the glucan degradation locus (GDL) (6). In Caldicellulosiruptor bescii, the most well-studied member of the genus, this locus is approximately 50 kb of the 2.9-Mb genome and encodes six GHs (Athe_1867, -1866, -1865, -1860, -1859, and -1857), all of which contain repeated cellulose binding domains from CBM3 as well as Pro/Thr-rich linker regions between CAZyme domains. The repeated CBM3 domains and linkers, as well as repetitive GH5 and GH48 sequences, are characteristic of the architecture of the GDL in Caldicellulosiruptor species, which is hypothesized to be the result of recombination after gene duplication events (1, 8, 9). In fact, the large repetitive sequences within this locus led to errors in genome assembly of the GDL in the initial C. bescii genome sequence (10). This was recently corrected and resulted in the relocation of the Athe_1859 to -1857 genes between Athe_1867 and Athe_1866 (11). Transcriptomic and proteomic analyses of highly cellulolytic Caldicellulosiruptor species showed that the GDL is highly transcribed and expressed during growth on both crystalline cellulose and switchgrass (3, 12).
The most extensively studied enzyme from Caldicellulosiruptor species is that encoded by the first gene in the GDL, i.e., a GH9/GH48 cellulase referred to as CelA (Athe_1867 in C. bescii). CelA is highly active on crystalline cellulose and is one of the most highly expressed proteins in the extracellular proteome (12 – 14). The N-terminal GH9 domain of CelA functions as an endoglucanase, while the C-terminal GH48 domain is a cellobiohydrolase releasing primarily cellobiose from crystalline cellulose (15). Homologs of the CelA gene (seven full and one partial) are found in the genomes of eight Caldicellulosiruptor species which represent the most cellulolytic members of the genus. CelA exhibits a surface ablation mechanism common to that of processive cellulases. CelA also has a unique cavity-digging mechanism that allows it to burrow into cellulose microfibrils, making it one of the most effective cellulases characterized to date (14).
While CelA has been characterized most extensively, four of the other five enzymes from the GDL of C. bescii have been characterized to various extents (16 – 19). A summary of the biochemical data for these five enzymes is given in Table 1. The sixth enzyme, GH74/GH48 Athe_1860, has not been characterized. All of the characterized enzymes are optimally active at temperatures of 85 to 90°C and at pHs between 5.0 and 6.5. While CelA is particularly well suited for crystalline cellulose degradation, all of the other characterized enzymes from the GDL are also active on cellulosic substrates. This includes the GH10-containing enzyme Athe_1857, which exhibits a broader substrate specificity, including specificity for β-glucans, than that typical for enzymes with GH10 domains, which are typically active on xylans (19). In addition to the β-glucan activities of enzymes of the GDL, a highly repetitive GH5 domain, common to Athe_1859, Athe_1866, and Athe_1865, is active on mannan-containing substrates (18). Taken together, the GH domains of the GDL enzymes from families 5, 9, 10, 44, and 48 show activity on a broad array of polysaccharide substrates that make up plant biomass, including β-glucans, mannans, and xylans. This wide range of biocatalytic functions indicates that GDL enzymes play a critical role in enabling Caldicellulosiruptor spp. to scavenge growth substrates from a broad array of polysaccharides encountered in their globally diverse thermal environments.
TABLE 1.
Biochemical characteristics of glycoside hydrolases encoded within the glucan degradation locus
| Gene locus | Domains | No. of amino acids in protein | Predicted molecular mass (kDa) | Approx observed molecular massd (kDa) | Optimal temp (°C) | Optimal pH | Substratese | Specific activityc | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|
| Athe_1867 | GH9-CBM3-CBM3-CBM3-GH48 | 1,759 | 195 | 230 | 85 | 5–6.5 | Glucans (Avicel, barley beta-glucan, CMC), xylan | On Avicel, 23–55 U/μmol; on barley beta-glucan, 100,510 U/μmol | 13 – 15 |
| Athe_1859 | GH5-CBM3-CBM3-GH44 | 1,294 | 142 | 170 | 85 | 5 | Glucans (PASC, CMC, Avicel, lichenin), mannans (guar gum, 1,4-β-mannan), glucomannan (konjac), xylan (birchwood), xyloglucan | On Avicel, 5.3 U/μmol | 17 |
| Athe_1857a | GH10-CBM3-CBM3-GH48 | 1,478 | 165 | 200 | 85–90 | 6.5 | Xylan (beechwood), arabinoxylan (wheat), glucans (barley beta-glucan, lichenin, CMC, PASC, Avicel, filter paper), glucomannan (konjac), arabinans (arabic gum, debranched arabinan), pectin | On Avicel, 3.4 U/μmol; on beechwood xylan, 39,000 U/μmol; on barley beta-glucan, 3,400 U/μmol | 19 |
| Athe_1866b | GH5-CBM3-CBM3-CBM3-GH5 | 1,414 | 156 | 195 | Not available | 5.5–6.5 | For N-terminal GH5, mannans; for C-terminal GH5, glucans (PASC) | Not available | 17, 18 |
| Athe_1865 | GH9-CBM3-CBM3-CBM3-GH5 | 1,369 | 151 | 180 | 85–90 | 5.5–6.5 | Glucans (Avicel, filter paper, PASC, lichenin), mannans (locust bean gum, guar gum), glucomannan (konjac), arabinoxylan (wheat), xylan (oat spelt) | On Avicel, 10.2 U/μmol; on locust bean gum, 1,691 U/mg | 16, 18 |
GH10 only was characterized; the GH48 domain is 100% identical to that encoded by Athe_1867.
The Athe_1866-encoded N-terminal GH5 domain is 100% identical to the Athe_1859-encoded GH5 domain.
Enzyme activity units are defined as micromoles of reducing sugar per minute.
Observed via SDS-PAGE (34; this study).
CMC, carboxymethyl cellulose; PASC, phosphoric acid-swollen cellulose.
In the present study, the individual and collective roles of the GHs encoded in the GDL were examined, as these relate to the deconstruction of microcrystalline cellulose and plant biomasses by C. bescii. This study was made possible by the recent corrections in the GDL genome sequence and improvements in the C. bescii genetic system (11, 20) that facilitated gene deletions in this bacterium.
RESULTS
C. bescii GDL.
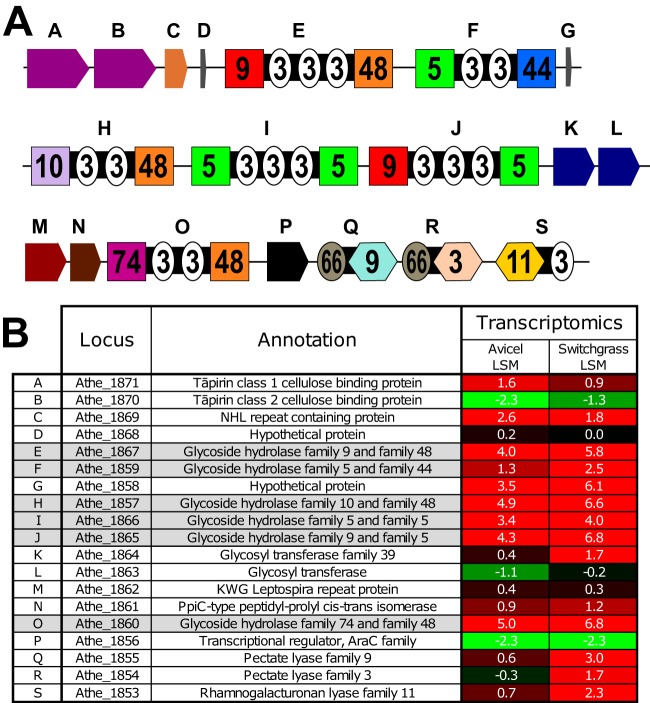
The glucan degradation locus (GDL) is highly conserved in the seven most cellulolytic Caldicellulosiruptor species characterized to date, including C. bescii. In C. bescii, this locus contains 19 genes, including genes encoding 6 GHs, 3 polysaccharide lyases (PLs), 2 cellulose binding proteins (tāpirins), and additional putative posttranslational protein-modifying enzymes. Figure 1A shows the C. bescii GDL with genes lettered A to S. Figure 1B provides locus tag and annotation information for these genes, along with transcriptomic data for C. bescii grown on microcrystalline cellulose (Avicel) and switchgrass. The 6 GH genes in the C. bescii GDL, each of which contains two catalytic GH domains, shown as boxes containing the GH family number in Fig. 1A, are shaded gray in Fig. 1B. Each enzyme contains either two or three cellulose binding modules from CBM family 3 (CBM3). While these GHs likely represent the core degradation ability of the GDL, other proteins and enzymes encoded in the locus likely contribute to C. bescii's plant biomass degradation ability.
FIG 1.
GDL of C. bescii. (A) Genes (lettered A to S) comprising the GDL. GH domains are shown as boxes containing the GH family number. CBM domains are shown as circles containing the CBM family number. PL domains are shown as hexagons containing the PL family number. (B) Locus tag, annotation, and transcriptomic data for genes A to S. Transcriptomic values are shown as log squared means (LSM) for transcript levels for C. bescii grown on Avicel (microcrystalline cellulose) or switchgrass. An LSM value of 0 represents the average transcription level, positive values (highlighted in red) represent higher-than-average transcription, and negative values (highlighted in green) represent lower-than-average transcription. The rows for the six GDL GHs are shaded gray.
Downstream of the GH genes in the GDL are three polysaccharide lyase-encoding genes: Athe_1855 to -1853. The PL domains of these genes are shown as hexagons containing the PL family number in Fig. 1A. Athe_1854 was previously characterized, and a crystal structure for the PL3 domain has been solved (21, 22). These genes were also previously deleted in the genetically tractable C. bescii strain JWCB005, and the resulting strain (JWCB010) consequently showed a decreased ability to grow on pectin-based substrates (23). These three PL genes are not found in the GDLs of all seven highly cellulolytic species. For example, the GDL of C. saccharolyticus is missing all three PL genes, while the GDL of C. obsidiansis is missing the PL9 and PL11 genes, and in C. kronotskyensis the GDL is missing the PL9 gene. Located upstream of Athe_1867 (CelA) are genes for previously characterized cellulose binding proteins termed tāpirins (Athe_1871 and Athe_1870) (24). The location of these cellulose-binding proteins near highly cellulolytic enzymes is probably not coincidental. The tāpirins putatively play a role in cell attachment and anchoring to cellulose, which in turn aids in the localization of the secretome, including the GDL enzymes, such that it is in close proximity to the biomass substrate.
Between the GH genes Athe_1865 and Athe_1860 are four genes that are highly conserved in the cellulolytic Caldicellulosiruptor species. The Athe_1864 and Athe_1863 enzymes belong to glycosyltransferase protein families that are involved in O-mannosylation of proteins. Protein O-mannosyltransferases (PMTs) are found in both prokaryotes and eukaryotes and typically have several hydrophobic transmembrane domains (25). Because of their location in the cytoplasmic membrane in bacteria, glycosylation is likely coordinated with secretion (26). This was seen previously for Athe_1867 in C. bescii, where the extracellular protein was glycosylated but the intracellularly expressed protein lacking the signal peptide was not glycosylated (27). O-mannosylation of various proteins from Gram-positive Mycobacterium species was found on Thr residues, particularly in Pro-, Thr-, and Ala-rich sequences (28, 29). Glycosylation patterns for the GDL GHs are not known, but highly Pro/Thr-rich linker sequences are prevalent in these proteins and may be the sites of glycosylation.
Athe_1862 encodes a homolog of KWG Leptospira repeat proteins, so named because of their high abundance in Leptospira interrogans, although their function is unknown. The Athe_1861 protein is annotated as a peptidyl-prolyl cis-trans isomerase (PPIase), which may be involved in the interconversion of the cis and trans forms of proline during protein maturation. Several types of PPIases are found in bacteria. However, the Athe_1861 protein has sequence similarity to PrsA PPIases, which are thought to be anchored between the cell membrane and peptidoglycan and help to refold proteins as they are exported (30, 31). This enzyme may play a role in the export and proper extracellular folding of the GDL enzymes, particularly as this relates to their Pro/Thr-rich linkers.
Previously reported transcriptomic data comparing the growth of C. bescii on Avicel crystalline cellulose and a complex plant biomass substrate (switchgrass) (3) show that the GDL GH-encoding genes were very highly transcribed under both conditions (Fig. 1B). During C. bescii growth on switchgrass, Athe_1867, Athe_1857, Athe_1865, and Athe_1860 were among the top 15 transcripts (top 0.5%) among 2,780 total genes. One notable exception to this was GH5/GH44 Athe_1859, which was transcribed at approximately 6-fold lower levels than those of GH9/GH48 Athe_1867 (CelA). GH74/GH48 Athe_1860 is separated from the other GDL GH genes by the four genes Athe_1864 to -1861, which were transcribed at more moderate levels. Nevertheless, Athe_1860 was transcribed at levels comparable to those of other GH genes in the GDL of C. bescii.
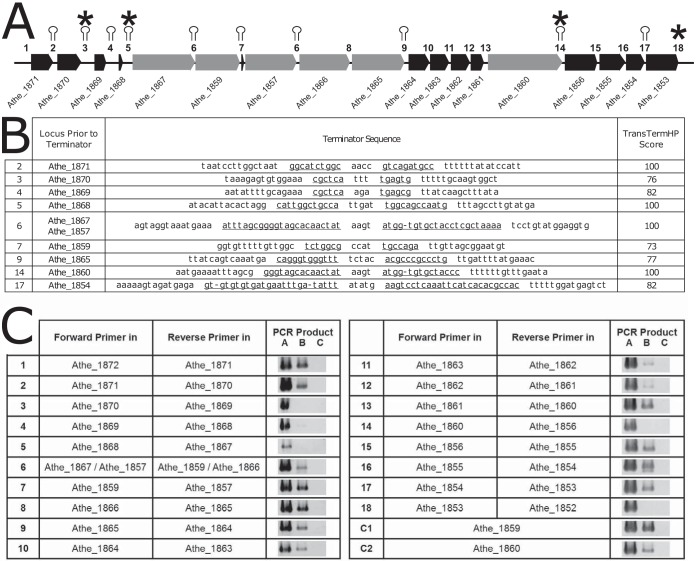
The structure of the operons in the GDL is interesting for considering the transcriptomic profiles of the very large GH enzyme genes (3.9 to 5.7 kb) and the accessory genes present here. To this end, rho-independent terminators were predicted in silico with TransTermHP (32) and are shown as stem-loops in Fig. 2A, with the intergenic regions between GDL genes numbered 1 to 18. The corresponding stem-loop sequences, with complementary stem sequences underlined, and the TransTermHP combined scores are shown in Fig. 2B. TransTermHP scores range from 0 for poor-quality terminators to 100 for high-quality terminators. In the GDL, predicted terminators with scores of 100 appear after Athe_1871, Athe_1868, GH9/GH48 Athe_1867, GH10/GH48 Athe_1857, and GH74/GH48 Athe_1860. Terminators are also predicted after the GH5/GH44 Athe_1859, GH9/GH5 Athe_1865, and PL3 Athe_1854 CAZyme genes, albeit with slightly lower scores (73, 77, and 82, respectively). No terminator is predicted between GH5/GH5 Athe_1866 and GH9/GH5 Athe_1865 or between peptidyl-prolyl cis-trans isomerase Athe_1861 and GH74/GH48 enzyme Athe_1860.
FIG 2.
In silico and experimental examination of the GDL operon structure. (A) Structure of the C. bescii GDL, with in silico terminator predictions shown as stem-loops and PCR-determined terminators marked with asterisks. Intergenic regions are numbered 1 to 18. Note that intergenic region 6 appears twice because the DNA sequence for Athe_1867 to Athe_1859 is identical to that for Athe_1857 to Athe_1866 (see the discussion of Fig. 3 in the text). (B) In silico terminator sequence predictions corresponding to intergenic regions 1 to 18 in the GDL of C. bescii, as predicted by TransTermHP (32). The complementary sequence of each stem-loop is underlined, and the TransTermHP score is shown. (C) Experimental verification of terminator sequences by PCRs targeting the regions between genes. Forward primers were designed to bind in one gene, with the reverse primer binding in the next sequential gene (see Table S2 in the supplemental material) to amplify through intergenic regions 1 to 18. PCR A was a positive control using C. bescii genomic DNA as the template. PCR B used reverse-transcribed RNA as the template. PCR C was a negative control on non-reverse-transcribed RNA to verify that there was no DNA contamination in the isolated RNA sample. A band for PCR B indicates that there is an mRNA stretching between the two genes to which the forward and reverse primers bind. Two positive-control reactions (C1 and C2) were designed, with both the forward and reverse primers targeting a single gene (Athe_1859 or Athe_1860).
As a complementary approach to this in silico terminator prediction, PCR was used to amplify the intergenic regions between GDL genes, with one primer in each of the adjacent genes using reverse-transcribed RNA as the template (PCR product B) (Fig. 2C). PCR products for intergenic regions should be produced when the two genes appear on the same mRNA, while intergenic regions containing a terminator that prevents transcription of the genes on the same mRNA should not produce a PCR product. Genomic DNA (PCR product A) and RNA that was not reverse transcribed (PCR product C) were used as positive and negative controls, respectively. PCR products for the intergenic regions of the GDL are shown in Fig. 2C. No PCR products were amplified for the regions of Athe_1870-Athe_1869, Athe_1868-Athe_1867, Athe_1860-Athe_1859, and Athe_1853-Athe_1852, showing that breaks in transcription occur at these locations (Fig. 2A, asterisks). While breaks in transcription are easily detected using this method, a terminator that is read through, even at a low frequency, may provide an mRNA template for PCR. Thus, additional terminators that do not always terminate transcription and allow some readthrough may exist in the operon, such as those predicted by TransTermHP. While these PCR results suggest that Athe_1867 to -1860 have the potential to be transcribed in a single operon, at least some of the time, the various transcription levels of the GDL shown in Fig. 1B suggest that internal promoters and/or transcriptional readthrough of terminators leads to a range of transcript levels for the GDL genes.
Repetitive nature of the GDL.
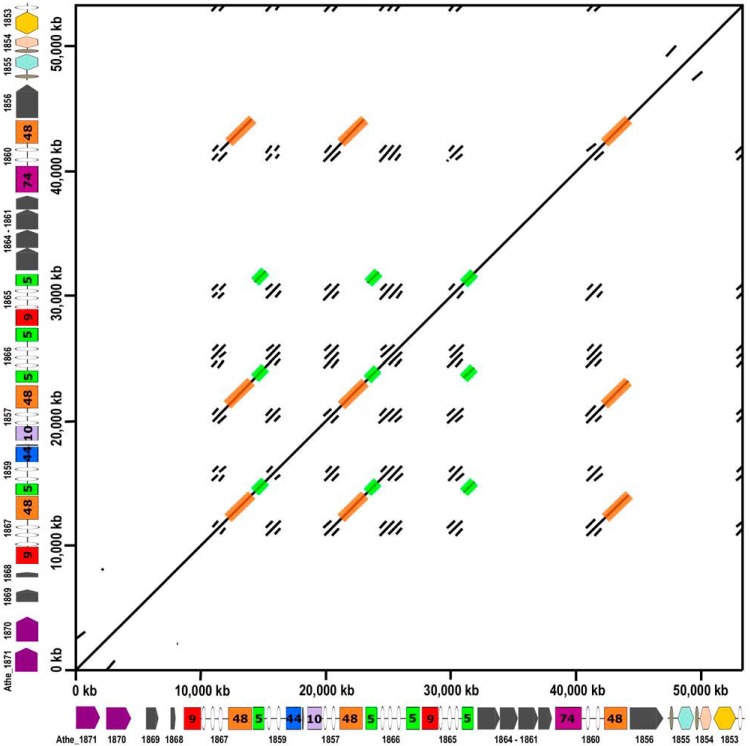
The GDL characteristically has repetitive CBM3, GH5, and GH48 domains, not only at the amino acid sequence level but also at the nucleotide sequence level. Recently, resequencing of the C. bescii DSMZ 6725 genome (11) by use of a PacBio platform revealed that the GDL was incorrectly assembled in the originally published sequence (10). In the C. bescii genome, Athe_1859 to -1857 are in fact between Athe_1867 and Athe_1866. This error occurred because the GH48 nucleotide sequences in Athe_1867, Athe_1857, and Athe_1860 are identical and the short reads of 454 sequencing technology were not able to read through these >1-kb repeats. Even manual curation of the sequence via Sanger sequencing led to mistakes in assembling this genomic locus. To visualize the repetitiveness of the nucleotide sequence of the GDL in C. bescii, the dot plot shown in Fig. 3 was constructed using EMBOSS dottup (http://www.bioinformatics.nl/cgi-bin/emboss/dottup), with a sequence word length of 50 bp, to compare the recently revised C. bescii sequence to itself. In Fig. 3, black pixels indicate where a 50-bp sequence at a position on the x axis is an exact match to 50 bp at a position on the y axis. The diagonal line from the bottom left to top right represents the direct match of the sequence to itself, but the large number of additional lines indicate many other areas where nucleotide sequences of 50 bp or larger are repeated. Many of these areas correspond to repeated CBM3 domains. To better visualize the repeated GH48 domains (1.8 kb) and GH5 domains (1 kb), these areas are shaded orange and green, respectively. In particular, note the 5.7-kb repeated region stretching from the CBM3 and GH48 domains of Athe_1867 through the GH5 and CBM3 domains of Athe_1859, which is repeated in Athe_1857 and Athe_1866 and partially repeated in the CBM3 and GH48 domains of Athe_1860. This extended large repeat is the reason that the genome was originally misassembled. It is important to consider these extended repeats in C. bescii to avoid issues with many standard biomolecular laboratory procedures, including PCR, sequencing, and targeted gene deletions. To this point, C. bescii strain JWCB029, in which CelA was deleted, showed a reduced ability to grow on microcrystalline cellulose as well as some lignocellulosic substrates, suggesting an essential role for CelA in the degradation of cellulosic substrates (33). Due to the repetitive nature of the GDL and the now corrected sequence, it is not clear whether this strain in fact contains a deletion of only Athe_1867 (CelA) or a larger deletion that includes Athe_1867, Athe_1859, and Athe_1857. The reverse screening primer used in the study of Young et al. binds in the repeated GH5 domain highlighted in green in Fig. 3. Thus, while CelA was certainly deleted in strain JWCB029 (33), it is not clear that only the CelA gene, and not a larger portion encompassing multiple GH enzyme genes, was deleted. Strain JWCB029 showed a severe growth phenotype on microcrystalline cellulose (∼80% reduction in cell count at 24, 48, and 96 h of growth compared to that of the genetic parent strain). But without knowing precisely which genes were deleted from the GDL, it is unclear that the absence of only the CelA gene was responsible for this phenotype.
FIG 3.
Dot plot showing the repeated DNA sequence of the C. bescii GDL. The sequence of the C. bescii GDL was used to construct a dot plot by using EMBOSS dottup (http://www.bioinformatics.nl/cgi-bin/emboss/dottup). A sequence word length of 50 bp was used to construct the plot. Sequences of 50 bp at positions on the x axis that match 50-bp sequences at positions on the y axis result in black pixels. The diagonal line from bottom left to top right show the direct match of the sequence to itself. All other markings represent repeated DNA sequences. The repeated GH48 domains of the GDL are shaded in orange. The repeated GH5 domains of the GDL are shaded in green.
In vivo analysis of GDL enzymes through targeted gene deletions.
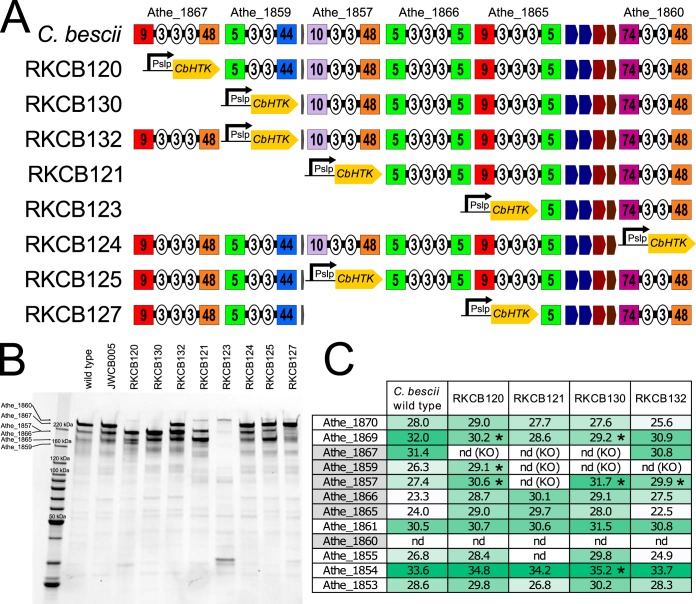
To further probe the role of CelA and the other GH-containing enzymes encoded by the GDL, a series of targeted gene deletions was constructed in C. bescii strain JWCB005, which was also recently sequenced (11). To enable clear selection of modified strains and prevent reversion to the parent strain during counterselection on 5-fluoroorotic acid (5-FOA), a codon-optimized, highly thermostable kanamycin resistance gene (Cbhtk) (20), driven by the promoter (Pslp) from S-layer protein (Athe_2303), was used to replace the genes that were deleted in these strains. The resulting genotypes of the knockout strains constructed using these methods are shown in Table 2, and the schemes for the GDL in these strains are shown in Fig. 4A. The fitness of these knockout strains did not appear to be affected by the genetic manipulations, as all strains grew equally well on cellobiose (data not shown). PCR screening was used as an initial verification of the strains (data not shown), but the repeated regions of the GDL led to primer binding in several locations, leading to multiple PCR products for each reaction. As a final verification of these strains, the whole genomes were reanalyzed using PacBio sequencing to obtain long reads allowing for assembly through the repeated areas of the GDL. The genome sequences of these strains verify that the knockout strains are indeed missing the genes targeted for deletion and that these are replaced by the PslpCbhtk construct. This approach replaced the deleted gene(s) with a strong promoter and a kanamycin resistance gene to enable genetic manipulation in this complex area of the genome. But a corresponding consequence of this strategy is that genes within the same operon as the deleted gene may be transcribed and expressed more highly than those in the wild-type or JWCB005 parent strain. In silico operon analysis (Fig. 2A and B) predicted terminator sequences after all of the GH deletion/PslpCbhtk insertion sites, but PCR analysis (Fig. 2C) suggested that these predicted terminators, except the one following GH74/GH48 Athe_1860, are read through at least some of the time.
TABLE 2.
C. bescii strains used and constructed for this study
| Strain | Parent | Transforming plasmid | Genotype | Reference |
|---|---|---|---|---|
| DSM 6725 | Wild type | 63 | ||
| JWCB005 | DSM 6725 | pDCW88 | ΔpyrFA | 64 |
| RKCB120 | JWCB005 | pJMC021 | ΔpyrFA ΔAthe_1867::PslpCbhtk | This study |
| RKCB121 | JWCB005 | pJMC021 | ΔpyrFA Δ(Athe_1867-Athe_1857)::PslpCbhtk | This study |
| RKCB123 | JWCB005 | pJMC021 | ΔpyrFA Δ(Athe_1867-Athe_1865′)::PslpCbhtk | This study |
| RKCB124 | JWCB005 | pJMC033 | ΔpyrFA ΔAthe_1860::PslpCbhtk | This study |
| RKCB125 | JWCB005 | pJMC034 | ΔpyrFA ΔAthe_1857::PslpCbhtk | This study |
| RKCB127 | JWCB005 | pJMC034 | ΔpyrFA Δ(Athe_1857-Athe_1865′)::PslpCbhtk | This study |
| RKCB130 | JWCB005 | pJMC069 | ΔpyrFA Δ(Athe_1867-Athe_1859)::PslpCbhtk | This study |
| RKCB132 | JWCB005 | pJMC070 | ΔpyrFA ΔAthe_1859::PslpCbhtk | This study |
FIG 4.
GDL knockout strains of C. bescii. (A) GDL layout for the eight knockout strains generated from C. bescii strain JWCB005. The location of the PslpCbhtk cassette insertion at the gene deletion location is shown. (B) SDS-PAGE gel containing secretome samples from the C. bescii wild-type strain, the parent strain JWCB005, and knockout strains RKCB120 to -132. Lanes were loaded with equal protein masses. The expected locations of the GDL enzymes are shown on the left. (C) Log2-transformed apex AUC values for GDL proteins from the C. bescii wild type and strains RKCB120, RKCB121, RKCB130, and RKCB132. Significant differences between the wild-type and mutant strains (two-tailed t test; P ≤ 0.05) are marked with asterisks. The full proteomics data set is available in Table S1 in the supplemental material. nd, not detected; KO, genes knocked out in mutant strains.
To determine the impacts of the gene knockouts on the extracellular proteins produced by these strains, cultures of the C. bescii wild type, the parent strain JWCB005, and all of the GDL knockout strains (RKCB120 to RKCB132) were grown on cellobiose. The supernatants from these cultures were concentrated and separated by SDS-PAGE (Fig. 4B). In addition to this analysis via SDS-PAGE, proteomic analysis was performed (Fig. 4C) on extracellular protein samples from triplicate cultures of the C. bescii wild type and strains RKCB120, RKCB121, RKCB130, and RKCB132. In Fig. 4B, the predicted locations of the GDL enzymes are shown on the left, based on a proteomic analysis performed previously on C. bescii extracellular proteins (34), and reflect the gene deletions in strains RKCB120 to -132. The GH9/GH48 Athe_1867 (CelA) band is clearly the predominant band in both the C. bescii wild-type and strain JWCB005 samples, suggesting that CelA is the most highly expressed protein (approximately 10% of the extracellular protein by densitometry). The enzymes deleted in the various RKCB knockout strains were not detected via SDS-PAGE or liquid chromatography-tandem mass spectrometry (LC-MS/MS) proteomic analysis. For example, strains RKCB120, RKCB130, RKCB121, and RKCB123 all lack Athe_1867 (CelA), and the expected band corresponding to Athe_1867 is absent in these lanes (Fig. 4B). Strain RKCB123, which is missing all but Athe_1860 and the GH5 domain of Athe_1865, shows no large extracellular proteins except for the Athe_1860 band at approximately 240 kDa. Log2-transformed apex area under the curve (AUC) data for the GDL proteins (Fig. 4C; see full data set in Table S1 in the supplemental material) show that the insertion of the PslpCbhtk construct did increase expression of GDL enzymes downstream of the resistance marker insertion to some extent, with the largest increase seen primarily for genes directly after the insertion. Significant changes (up or down) in protein level from the wild type to a modified strain, as determined by the two-tailed t test (P ≤ 0.05), are marked by asterisks in Fig. 4C. The increases in GH5/GH44 Athe_1859 expression (7-fold in RKCB120) and GH10/GH48 Athe_1857 expression (9-fold in RKCB120, 19-fold in RKCB130, and 6-fold in RKCB132) were all significantly (P ≤ 0.05) above the wild-type expression levels. Increases in GH5/GH5 Athe_1866 and GH9/GH5 Athe_1865 expression were also observed, but Athe_1866 and Athe_1865 were not consistently detected in all triplicate samples (Table S1), and these increases are not statistically significant. Interestingly, GH74/GH48 Athe_1860 was expressed at low enough levels that it was not detected in the proteomic samples from the wild type or any of the modified strains. The proteomics data set (Table S1) also suggests some compensatory effects of the genetic manipulations on the expression of extracellular proteins in these modified strains. A small number of proteins showed a significant difference (P ≤ 0.05) between the C. bescii wild type and each of the four modified strains, including Athe_0161 (hypothetical protein; 2- to 4-fold decrease), Athe_0594 (surface layer GH5-CBM28 protein; 4- to 8-fold decrease), Athe_1762 (hypothetical protein; 3- to 12-fold increase), Athe_1913 (ABC transporter solute binding protein; 4- to 10-fold increase), Athe_2303 (S-layer protein; 3- to 5-fold decrease), and Athe_2464 (hypothetical protein; 2-fold decrease). This relatively small set of significantly changing proteins, taken together with the unknown role of the hypothetical proteins identified by this method, indicates no substantial metabolic response or altered protein expression pattern outside the GDL in the mutant strains.
Cellulose and plant biomass solubilization by GDL GH knockout strains.
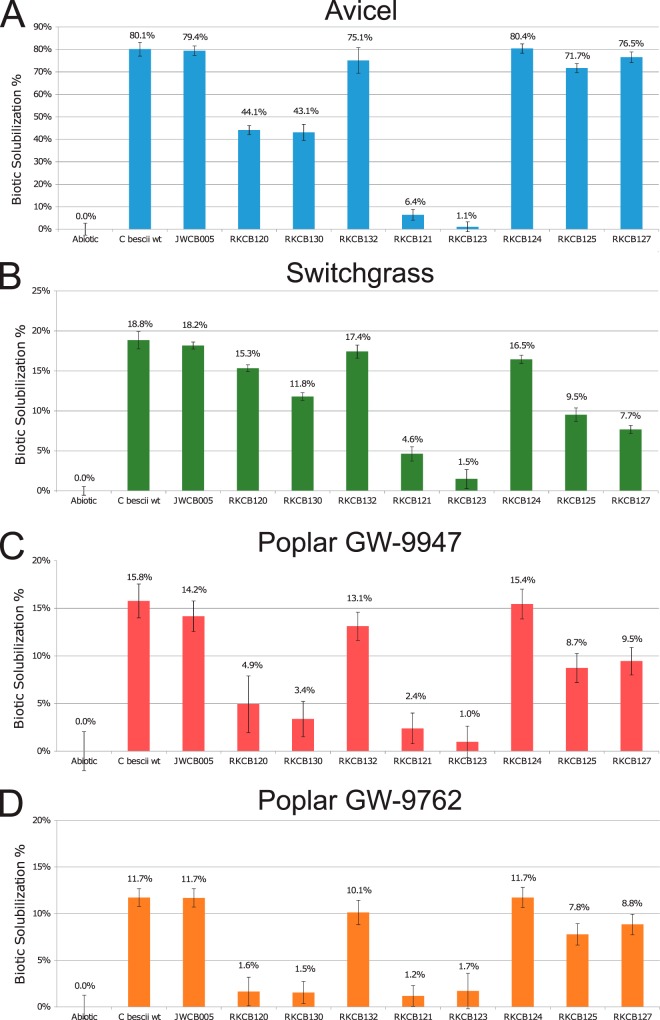
The extent to which Caldicellulosiruptor species solubilize plant biomass substrates is a reflection of the extracellular enzyme inventory. Thus, the C. bescii wild type, the genetic parent strain JWCB005, and the GDL enzyme knockout strains RKCB120 to -132 were grown on microcrystalline cellulose (Avicel), switchgrass, and two lines of natural variant poplar to assess the extents to which these substrates could be solubilized. Solubilization of these substrates was determined at 70°C in static cultures with defined medium in which the plant biomass substrate was the sole carbon source. Figure 5 shows the biotic solubilization for each strain, which is directly related to the associated secretome enzyme inventory and the ability to degrade and grow on the substrate. Biotic solubilization does not include the thermal contribution to solubilization determined by an abiotic control, which was 2.1% for Avicel, 19.6% for switchgrass, 10.6% for poplar GW-9947, and 10.7% for poplar GW-9762. In these experiments, the C. bescii wild type and the parent strain JWCB005 performed nearly identically, showing that the modifications to strain JWCB005 that made it genetically tractable and were identified in its newly sequenced genome (11) did not affect its ability to degrade these substrates. C. bescii strains RKCB120, RKCB121, RKCB130, and RKCB132, which have the PslpCbhtk cassette insertion, showed higher expression of the genes downstream in the locus (Fig. 4C). Overexpression of GDL enzymes would be expected to increase solubilization; thus, the observation of a reduction in plant biomass solubilization by a given strain can be interpreted as a demonstrable effect of the deletion of GDL genes and the lack of that enzyme's activity in the secretome enzyme mixture.
FIG 5.
Biotic solubilization of plant biomass substrates by GDL knockout strains. (A) Avicel microcrystalline cellulose; (B) Cave-in-Rock switchgrass; (C) poplar natural variant GW-9947; (D) poplar natural variant GW-9762. Biotic solubilization was calculated by subtracting the solubilization in the thermal abiotic control to reflect only the biotic contribution to solubilization. The abiotic thermal solubilization values were as follows: 2.1% for Avicel, 19.6% for switchgrass, 10.6% for poplar GW-9947, and 10.7% for poplar GW-9762. Error bars represent standard deviations (n = 3).
On Avicel (Fig. 5A), strains RKCB120, RKCB130, RKCB121, and RKCB123, which have knockouts of one, two, three, and five genes, respectively, starting at GH9/GH48 Athe_1867 (CelA) and continuing downstream, solubilized Avicel significantly less than the wild type did. The lack of Athe_1867 alone in strain RKCB120 resulted in a 45% reduction in the biotic solubilization of the wild type. The double knockout of Athe_1867 and GH5/GH44 Athe_1859 showed a similar reduction (46%) in solubilization. Strain RKCB121, which lacks Athe_1867, Athe_1859, and GH10/GH48 Athe_1857, had a severe growth phenotype, with a 92% reduction in biotic solubilization, suggesting that the three encoded enzymes are the primary cellulases used by C. bescii. The individual deletion of Athe_1857 in strain RKCB125 showed only a 10% solubilization reduction, and individual deletion of Athe_1859 in strain RKCB132 showed only a 6% reduction relative to the solubilization by the wild type. It is interesting that the three strains lacking only individual genes encoding the three main cellulases (Athe_1867, Athe_1859, and Athe_1857) had, at worst, a 45% reduction in cellulose solubilization (RKCB120). However, the combined deletion of all three genes in RKCB121 reduced cellulose degradation by 92%. This indicates that the actions of Athe_1867, Athe_1859, and Athe_1857 are synergistic or coordinated in some way. Additionally, strains RKCB124 (0% reduction) and RKCB127 (4% reduction) showed no significant reduction in biomass solubilization compared to that of the wild type, suggesting that the other three GDL GHs contribute little to the degradation of microcrystalline cellulose.
On switchgrass (Fig. 5B), a complex herbaceous biomass substrate containing hemicelluloses in addition to cellulose, a different set of GDL GH enzymes appear to be most important for degradation. In this case, strain RKCB120, lacking GH9/GH48 Athe_1867 (CelA), had only a 20% reduction in the total solubilization of the wild type. Successive deletions of more genes downstream of Athe_1867 in strains RKCB130, RKCB121, and RKCB123 further reduced switchgrass solubilization. Strain RKCB125, which lacks only GH10/GH48 Athe_1857, showed a 50% reduction in biotic solubilization compared to that of the wild type. This was the largest biotic solubilization decrease for a single GH gene deletion on switchgrass, indicating the importance of GH10/GH48 Athe_1857 on this substrate. The GH10 domain of the Athe_1857 enzyme is active on both xylan and glucan substrates (19), so the importance of this enzyme on a complex substrate containing xylan and cellulose is consistent with its broad activity. GH5/GH5 Athe_1866 and GH9/GH5 Athe_1865 may contribute modestly to switchgrass solubilization, as their deletion from RKCB121 to RKCB123 and from RKCB125 to RKCB127 resulted in small decreases in biotic solubilization (4.6% to 1.5% for RKCB121 to RKCB123 and 9.5% to 7.7% for RKCB125 to RKCB127). Additionally, strain RKCB124, with GH74/GH48 Athe_1860 deleted, had a slight reduction in solubilization from that of the wild type (16.5% for RKCB124 versus 18.8% for the wild type), indicating that the enzyme may also contribute modestly to switchgrass deconstruction.
Two natural variant lines of Poplar trichocarpa (GW-9947 and GW-9762) with reduced lignin content were used to assess biotic solubilization of a complex hardwood substrate (Fig. 5C and D). Poplar GW-9947 contains 22.7% lignin, with a 1.7 syringyl-to-guaiacyl (S/G) ratio, and GW-9762 contains 21.7% lignin, with a 1.5 S/G ratio. These were the lowest-level lignin variants and the most easily solubilized poplar variants in a previous solubilization study of five natural variant poplar lines with three cellulolytic Caldicellulosiruptor species, including C. bescii (35). In a different study of sugar release during enzymatic treatment, poplar GW-9947 ranked as the second best and GW-9762 as sixth best among 22 poplar variants for combined glucan and xylan release (36). In the present study, the biotic solubilization of poplar GW-9762 was 11.7% for the C. bescii wild type, with that of GW-9947 being about a third higher, at 15.8%. Unlike its role on Avicel and switchgrass, GH9/GH48 Athe_1867 (CelA) appears to play a very critical role in poplar degradation. Strain RKCB120, with only Athe_1867 deleted, had a biotic solubilization of 4.9% on poplar GW-9947, a reduction of 70% from the wild-type level, and on poplar GW-9762 this strain had almost all biotic solubilization eliminated (1.6% biotic solubilization, an 86% reduction). The strains with knockouts of successively larger portions of the GDL (RKCB130, RKCB121, and RKCB123) performed just as poorly on the more recalcitrant poplar variant, GW-9762. On the less recalcitrant poplar variant, GW-9947, these successively larger knockouts showed slightly decreased solubilization relative to that of the single GH9/GH48 Athe_1867 knockout strain (RKCB120). Similar to the case on switchgrass, strain RKCB125, lacking GH10/GH48 Athe_1857, achieved about half the biotic solubilization of the wild type, again indicating the importance of the encoded enzyme on complex biomasses. The deletion of GH5/GH5 Athe_1866 and GH9/GH5 Athe_1865 from strains RKCB125 to RKCB127 did not affect poplar solubilization, suggesting that the encoded enzymes are not critical for poplar degradation. Additionally, the deletion of GH74/GH48 Athe_1860 in strain RKCB124 did not reduce poplar solubilization at all for either natural variant.
Total solubilization reflects the mass loss of all components from the biomass substrate, including carbohydrate, protein, and lignin. To assess the carbohydrate solubilization performed by the C. bescii wild type, the genetic parent strain JWCB005, and mutant strains RKCB120 to -132, measurements of the glucan and xylan contents in the raw and spent switchgrass samples were performed, and the solubilization of these components was determined (Fig. 6). These data reflect a trend similar to that seen for switchgrass biotic total solubilization (Fig. 5B): as sequentially more GDL genes were deleted from strains RKCB120 to RKCB130 to RKCB121 to RKCB123, glucan and xylan solubilization levels also decreased sequentially. For the wild type and the majority of the knockout strains, glucan and xylan solubilization levels were about equal; however, for strains RKCB130, RKCB121, and RKCB123, glucan solubilization levels were significantly lower than xylan solubilization levels. With the deletion in strain RKCB121 of Athe_1867, Athe_1859, and Athe_1857, encoding the three enzymes most critical for Avicel degradation (Fig. 5A), glucan solubilization was reduced to 20% that of the wild type, only slightly above that of the abiotic control. For strain RKCB123, in which the most GDL genes are deleted, glucan solubilization was not higher than that of the abiotic control, suggesting that glucans are not efficiently solubilized by strain RKCB123's secreted enzymes and that the majority of its biotic solubilization consists of xylan and noncarbohydrate components. Strain RKCB123's poor solubilization of the glucan components of switchgrass mirrored the poor biotic solubilization of microcrystalline cellulose (Avicel) (Fig. 5A).
FIG 6.
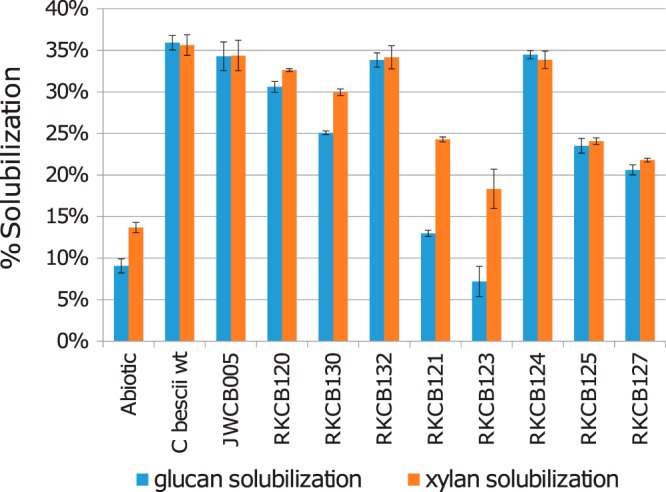
Glucan and xylan solubilization in switchgrass for C. bescii wild-type and GDL knockout strains. Glucan and xylan solubilization levels for switchgrass were calculated as the percentages of the glucan or xylan lost from untreated switchgrass for the switchgrass solubilized by C. bescii strains. Raw switchgrass contained 30.7% glucan, 22.2% xylan, 3.2% arabinan, and 43.9% inert components. Error bars represent standard deviations (n = 3).
Switchgrass solubilization by mixtures of GDL knockout strains.
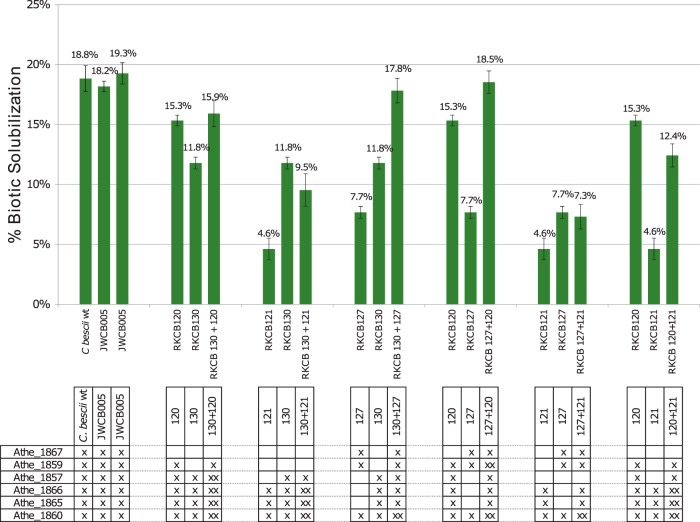
To further confirm that the decreases in biotic solubilization were a result of a decreased secretome enzyme inventory, switchgrass solubilization with a mixture of two knockout strains was examined. For these experiments, RKCB120, RKCB121, RKCB127, and RKCB130 were selected because these strains exhibited biotic switchgrass solubilization at levels (15.3%, 4.6%, 7.7%, and 11.8%, respectively) intermediate to the maximum (18.8%) of the wild type and the minimum (0.0%) of the abiotic control. All possible combinations of two strains from this group were examined. The biotic solubilization results for the mixed cultures are shown alongside the biotic solubilization levels of the two individual strains cultured separately in Fig. 7. At the bottom of Fig. 7, the six GDL GH enzymes are listed, and those present in the specific strains are indicated with “×” symbols. For the mixed cultures, the enzymes produced by one of the two strains are marked once, and enzymes produced by both of the strains in the mixtures are marked twice. Two of the combinations, RKCB130 + RKCB127 and RKCB127 + RKCB120, restored the full complement of GDL GH enzymes to the culture between the two combined strains. For the RKCB130 + RKCB127 combination, all of the enzymes were produced by one of the two strains, except the Athe_1860 enzyme, which was produced by both. The combination of RKCB127 and RKCB120 was identical, except that Athe_1859 was also produced by both strains. In each of these mixed-culture cases, restoring the full complement of GDL GH enzymes in the culture, even produced by different strains, restored the biotic solubilization to levels similar to that of the wild type. Likewise, the RKCB127 + RKCB121 mix, while not restoring the full GDL GH complement, provided a secreted enzyme mixture similar to that of strain RKCB125, which lacks only Athe_1857. The mixture of RKCB127 and RKCB121 had a biotic solubilization level of 7.3%, while that of strain RKCB125 individually was 9.5%. These mixtures also generated cases where the strains each produced the same set of enzymes and then one of the two strains in the mixture produced an additional one or two enzymes, as in the mixtures of RKCB130 and RKCB120, RKCB130 and RKCB121, and RKCB120 and RKCB121. For example, in the mix of RKCB130 and RKCB120, strain RKCB130 duplicates the enzymes produced by RKCB120, RKCB120 additionally produces Athe_1859, and neither produces Athe_1867. In this case, the mixture showed 15.9% solubilization, which is the same as that of the better-solubilizing strain, RKCB120, alone (15.3%). A similar situation occurred for the RKCB130 + RKCB121 and RKCB120 + RKCB121 mixtures, where solubilization was close to that of the better-solubilizing strain alone, i.e., RKCB130 and RKCB120, respectively. Taken together, data from these mixed cultures show that the GDL can be separated between two different strains of the same species and perform as well as if the genes were all produced by a single strain. This shows that the most important factor in solubilization is the cocktail of extracellular enzymes produced in the culture, independent of the source organism producing the individual enzymes. Thus, in natural settings with mixed communities, individual species need not contain the full GDL to benefit from fermentable sugars liberated from biomass by the collective secreted enzymes of the community.
FIG 7.
Switchgrass solubilization by mixed cultures of GDL knockout strains. Biotic solubilization values on switchgrass were determined for GDL knockout strains (RKCB120, RKCB121, RKCB127, and RKCB130) alone and combined in mixed cultures inoculated with equal proportions of two strains. Data for three replicates of C. bescii containing the full GDL (C. bescii wild type and C. bescii strain JWCB005) are shown at the left for comparison. The genes contained in each strain are marked in the table (×). Genes marked “××” for a mixed culture represent enzymes produced by each of the two strains in the mixture. Error bars represent standard deviations (n = 3).
DISCUSSION
From a genomics perspective, the presence of genes encoding GH9, GH44, and GH48 enzymes in the genomes of Caldicellulosiruptor species is the major differentiating factor between cellulolytic and noncellulolytic phenotypes of these extremely thermophilic bacteria (6, 7). These particular GH families are all encoded in the GDL of cellulolytic Caldicellulosiruptor species. The GDL encodes six enzymes in C. bescii, containing four GH5, two GH9, one GH10, one GH44, three GH48, and one GH74 domains. Genomic loci with similar sets of GH domains are also found in many other cellulolytic bacterial species, including Clostridium cellulolyticum (two GH5, GH8, five GH9, PL11, and GH48 domains) (37, 38) and Acidothermus cellulolyticus (two GH5, GH12, GH6, GH48, and GH74 domains) (39). However, other bacteria do not have a concentrated locus of cellulase genes, and instead these genes are distributed throughout the genome (40). Nevertheless, all cellulolytic bacteria produce complementary GHs from the major cellulase families, i.e., GH5, GH6, GH8, GH9, GH12, GH44, GH45, GH48, and GH124 (5).
While there are many bacteria that produce cellulases, the multidomain structure of the cellulase enzymes encoded in the GDL is unique to Caldicellulosiruptor species. The architectures of these genes are highly similar, encoding GH domains at the N terminus and C terminus and two or three CBM3 domains between. This multidomain architecture, combined with the highly repetitive nature on the nucleotide sequence level (Fig. 3), suggests that this locus arose by domain shuffling (8, 9). This is further supported by comparing GDLs across Caldicellulosiruptor species (1). For example, the GDL of C. saccharolyticus is missing the repeated CBM3-GH48 and GH5-CBM3 segments found between Athe_1867-Athe_1859 and Athe_1857-Athe_1866. Instead, C. saccharolyticus has one enzyme, which has been characterized and termed CelB (41), made up of a GH10 domain homologous to that of the Athe_1857 protein and a C-terminal GH5 domain homologous to that of the Athe_1866 protein. It is unclear whether this difference is the result of gene duplication and recombination to expand the GDL repertoire from C. saccharolyticus to C. bescii or if the repeated domains in C. bescii were lost at some point by C. saccharolyticus. The GDLs of recently sequenced Caldicellulosiruptor species (42) contain GH12 domain sequences located among the typical GH5, GH9, GH44, and GH48 domain sequences, also likely the result of domain shuffling. The repeated domain shuffling of the GDL sequence in different species represents the evolution of the cellulolytic function in Caldicellulosiruptor and offers a suite of multidomain enzymes with a mixture of different activities and domains for degrading plant polysaccharide substrates.
Not only are these unique multidomain enzymes encoded in the genomes of Caldicellulosiruptor species, but their genes are also highly transcribed and translated (Fig. 1) (3, 12) under most growth conditions. The large proportion of the secretome associated with the GDL can be seen in the protein gels shown in Fig. 4B. Because of the large size of these enzymes, the cell would expend significant energy producing these very large proteins (1,414 to 1,904 amino acids) at relatively high levels. While the secretome of C. bescii (43) and GDL enzymes or their domains (14, 17, 19) are highly effective at degrading crystalline cellulose and plant biomass substrates, herein, using recently improved genetic tools for C. bescii (20) and a corrected genome (11), their roles in vivo could be examined more closely.
Various knockout strains with different combinations of GDL enzyme deletions (Fig. 3A) were constructed, and the impacts of these deletions on the ability to degrade microcrystalline cellulose (Avicel), switchgrass, and poplar revealed that the relative importance of the GDL enzymes is biomass dependent. In the case of switchgrass solubilization (Fig. 5B), the deletion of GH10/GH48 Athe_1857 in strain RKCB125 reduced the biotic solubilization by 50%, more than the case for any other single-gene knockout. While for poplar degradation (Fig. 5C and D) the contribution of GH10/GH48 Athe_1857 appears to be important (strain RKCB125), GH9/GH48 Athe_1867 (CelA) appears to be the most critical gene, as knockout strain RKCB120, lacking only Athe_1867, performs very little biotic solubilization. On Avicel (Fig. 5A), no one enzyme appears to be critical, as the single-gene knockout strains degraded >55% as well as the wild type, but the combined deletion of GH9/GH48 Athe_1867, GH5/GH44 Athe_1859, and GH10/GH48 Athe_1857 in strain RKCB121 abolished 92% of the ability to degrade crystalline cellulose. This strain demonstrates that it is the combination of the Athe_1867, Athe_1859, and Athe_1857 enzymes that is critical for cellulose degradation, and further, the criticality of the three enzymes suggests that their action is synergistic. Previously, an attempt was made to knock out the GH9/GH48 Athe_1867 enzyme (CelA) from C. bescii to demonstrate the importance of this enzyme (33). However, upon closer examination of the repetitive genome and the phenotypes reported for strain JWCB029, this strain likely was missing Athe_1867, Athe_1859, and Athe_1857 and therefore was a deletion mutant similar to strain RKCB121. For the biomasses tested, the deletion of GH5/GH5 Athe_1866 and GH9/GH5 Athe_1865 (from RKCB121 to RKCB123 or from RKCB125 to RKCB127) had only a minor effect. The deletion of GH74/GH48 Athe_1860 in strain RKCB124 showed no measurable effect on Avicel or poplar solubilization and only a slight effect for switchgrass. These observations indicate that these three genes are not essential for degrading these biomass substrates. The GH5 domains encoded in these genes have significant activity on mannans (Table 1), so a substrate not tested here that contains higher mannan levels, such as pine or spruce (44), might require the encoded enzymes for utilization.
The solubilization experiments with the individual knockout strains showed that the enzymes required for degradation vary with the substrate. Using mixed cultures containing pairwise combinations of four knockout strains (Fig. 7), solubilization was found to be dependent on the mix of enzymes produced in the culture and independent of the strain that produces them. The two mixed cultures (RKCB130 + RKCB127 and RKCB127 + RKCB120) both collectively contained all six GDL enzymes, and the combination in the culture achieved solubilization similar to that achieved by the wild type. Because it appears that the most important factor is the extracellular enzyme mixture, strategies based on coculture of different species producing different enzymes or addition of industrially produced enzymes to alter the enzyme mixture in the culture can be considered. Not discussed here, however, is the contribution of the attachment of the cells to the substrate. Caldicellulosiruptor species are known to attach to biomass substrates while they degrade them (24, 45), and this may significantly enhance the action of the enzymes by releasing them in close proximity to their substrates. If this is an important factor in how these enzymes function, addition of exogenously produced enzymes may not significantly improve solubilization.
Conclusions.
Using in vivo analyses of the glucan degradation locus (GDL) in C. bescii, we showed here, for the first time, the nuanced roles of large multidomain GHs in the degradation of plant biomass substrates. While GH9/GH48 Athe_1867 (CelA) has been highly studied and encodes a prolific cellulase in its own right, we showed that its combined action with the other GHs of the GDL, particularly those encoded by GH5/GH44 Athe_1859 and GH10/GH48 Athe_1857, is key to cellulose and plant biomass degradation. We demonstrated that the essentiality of individual C. bescii GDL GHs depends on the plant biomass substrate, with certain GDL enzymes being required to different extents depending on the substrate. Through mixed cultures of knockout mutant strains, we also showed that the degradation ability is tied to the secreted enzyme mixture produced by the community. Ultimately, these findings advance how we understand and inform efforts to deploy these enzymes responsible for the cellulolytic phenotype in Caldicellulosiruptor species.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
An axenic strain of C. bescii was obtained from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures. C. bescii strain JWCB005 (46) and the nonreplicating vector pDCW121 (47) were obtained from Janet Westpheling (University of Georgia, Athens, GA). Caldicellulosiruptor genomic DNA (gDNA) for cloning, vector construction, and genome sequencing was extracted as described previously (48). Genes of interest were amplified by PCR and inserted into vectors by Gibson assembly (49), using Gibson assembly master mix (New England BioLabs). For vector construction, Escherichia coli NEB 5-alpha (New England BioLabs) was used. Plasmids were isolated using Zymo Research plasmid miniprep classic and ZymoPURE midiprep kits (Zymo Research); sequences were confirmed by Sanger sequencing (Genewiz). Carbohydrates and biomass used in this study were as follows: Avicel PH-101 crystalline cellulose; Cave-in-Rock switchgrass (Panicum virgatum L.) cv. Cave-in-Rock, field grown in Monroe County, IA, obtained from the National Renewable Energy Laboratory (NREL), ground using a Wiley mill (Thomas Scientific), and sieved using 40/80 mesh; and poplars from 4-year-old Poplar trichocarpa variants GW-9947 and GW-9762, grown in Clatskanie, OR, milled using a 20-mesh screen on a Wiley mill (Thomas Scientific), and obtained as such from Gerald A. Tuskan (Oak Ridge National Laboratory). The growth conditions for the natural variant poplars were described previously (50, 51).
Media and culture conditions.
All E. coli cultures were maintained in either Luria-Bertani (LB) medium (5 g/liter yeast extract, 10 g/liter tryptone, 10 g/liter NaCl) or LB medium with 1.5% (wt/vol) agar. Media were supplemented with 50 μg/ml apramycin (Fisher Scientific) as appropriate.
C. bescii strains were grown anaerobically with a 20% CO2-80% N2 headspace at 70°C without agitation in the following growth media. For routine cultivation, a modified version of DSM516 medium (DSMZ) was used, containing 0.33 g/liter NH4Cl, 0.33 g/liter KCl, 0.33 g/liter MgCl2·6H2O, 0.14 g/liter CaCl2·2H2O, 0.16 mM sodium tungstate, 1 ml/liter trace element solution SL-10, 1 ml/liter vitamin solution, 0.25 mg/liter resazurin, 1 g/liter l-cysteine-HCl·H2O, 1 g/liter sodium bicarbonate, 1 mM potassium phosphate, and 5 g/liter glucose or cellobiose. The medium as described above is defined and is designated D516 medium. A complex version of this medium, designated C516 medium, is supplemented with 0.5 g/liter yeast extract. To grow C. bescii for competent cell preparation, LOD medium (52), containing either cellobiose or glucose as the carbon source, was supplemented with 1 × 19 amino acid solution (53). C. bescii was plated by embedding in LOD medium, D516 medium, or C516 medium with 1.5% agar and grown at 65°C under anaerobic conditions with 2% hydrogen-98% nitrogen. For the growth of all uracil-auxotrophic mutants, the medium was supplemented with 40 mM uracil. Kanamycin (50 μg/ml) was added to the cultures for strains resistant to kanamycin.
For solubilization experiments, Caldicellulosiruptor strains were grown on a modified defined version of DSM671 medium, containing 1 g/liter NH4Cl, 0.02 g/liter NaCl, 0.1 g/liter MgCl2·6H2O, 0.05 g/liter CaCl2·2H2O, 0.06 g/liter K2HPO4, 1 ml/liter trace element solution SL-10, 1 ml/liter vitamin solution, 0.25 mg/liter resazurin, 1 g/liter l-cysteine-HCl·H2O, and 2.6 g/liter sodium bicarbonate. Solubilization cultures contained 5 g/liter plant biomass substrate as the carbon source.
The vitamin solution for all medium recipes contained the following: 20 mg/liter biotin, 20 mg/liter folic acid, 100 mg/liter pyridoxine-HCl, 50 mg/liter thiamine-HCl·2H2O, 50 mg/liter riboflavin, 50 mg/liter nicotinic acid, 50 mg/liter d-calcium pantothenate, 50 mg/liter vitamin B12, 50 mg/liter p-aminobenzoic acid, and 50 mg/liter lipoic acid.
Construction of genetic knockout strains of C. bescii.
The following knockout vectors were constructed via Gibson assembly in the pDCW121 vector backbone (47), using 1-kb regions flanking the desired gene deletion location: pJMC021 for RKCB120, RKCB121, and RKCB123; pJMC033 for RKCB124; pJMC034 for RKCB125 and RKCB127; pJMC069 for RKCB130; and pJMC070 for RKCB132. For all knockout vectors, the PslpCbhtk high-temperature resistance cassette was placed between the flanking regions to allow for direct selection of second crossovers by use of kanamycin, as described previously (20). Purified vectors were methylated using recombinantly expressed M.CbeI methylase, as described previously (54).
Competent cells were prepared as described previously (20). Cells (50 μl) were mixed with 1 μg of plasmid DNA at room temperature and electroporated using 1-mm-gap cuvettes (USA Scientific) and a Gene Pulser II system with a Pulse Controller Plus module (Bio-Rad) under the following conditions: 2.0 kV, 200 Ω, and 25 μF. Immediately following electroporation, cells were transferred to 10 ml of prewarmed complex medium and incubated at 70°C. At 15 min and 1 h postelectroporation, 1 to 5 ml of the recovery culture was transferred into selective complex medium supplemented with 50 to 100 μg/ml kanamycin. Cultures were incubated at 70°C for 1 to 4 days, until growth was observed. These cultures were passaged on liquid selective medium once and then plated on selective medium lacking uracil to select individual colonies. For integrating vectors, a successful first-crossover mutant strain was plated on medium supplemented with 40 mM uracil, 4 mM 5-fluoroorotic acid (5-FOA), and 50 μg/ml kanamycin to select for second-crossover mutants. Strains were screened by PCR, and successful second-crossover knockout strains were plated on medium supplemented with uracil and kanamycin two additional times to ensure the purity of the strains. All PCR screening of modified strains was performed on genomic DNA, which was isolated using a ZymoBead genomic DNA kit (Zymo Research).
Genomic sequencing of modified C. bescii strains.
Assembled draft genome sequences were generated by the U.S. Department of Energy's Joint Genome Institute (JGI) (55) for C. bescii strains RKCB120, RKCB121, RKCB123, RKCB124, RKCB125, RKCB127, RKCB130, and RKCB132. Sequencing was performed on a PacBio RS/RS II platform with SMRTbell libraries (56).
Secretome analysis.
C. bescii strains were grown in 300-ml D516 medium cultures containing cellobiose as the substrate at 70°C. Growth was monitored, and cultures were harvested during early stationary phase (1 × 108 to 3 × 108 cells/ml) by rapidly cooling the culture in a dry ice bath and centrifuging it at 8,000 × g for 10 min. The supernatant was centrifuged at 18,000 × g for 20 min to remove any residual cell debris and then concentrated and buffer exchanged into 50 mM sodium acetate buffer, pH 5.5, containing 10 mM calcium chloride and 100 mM sodium chloride by using 50,000-molecular-weight-cutoff 20-ml Vivaspin concentrators, to a final volume of 5 ml. Total protein concentrations were determined using a bicinchoninic acid (BCA) assay (Thermo Fisher Scientific). A Mini-PROTEAN TXG stain-free 4 to 15% gel (Bio-Rad) was run using these supernatant samples, with equal total protein mass loading across all strains. Benchmark protein ladder (ThermoFisher) was used as the molecular weight standard. Gel imaging was performed using a Syngene G:BOX gel imaging system.
For proteomics analysis, each culture was grown in 100 ml of D516 medium containing cellobiose at 70°C to early stationary phase. The supernatant was harvested by centrifugation at 8,000 × g for 10 min. The resulting supernatant was filtered using a 5-kDa filter and concentrated to 1 ml. The concentrated protein solution was transferred to a 2-ml 10-kDa filter, washed and filtered several times using 100 mM ammonium bicarbonate (ABC), denatured and reduced with 4% sodium deoxycholate (SDC) prepared in 100 mM ABC and 10 mM dithiothreitol (DTT), and alkylated using 30 mM iodoacetamide (IAA), followed by digestion with two applications of sequencing-grade trypsin (Sigma-Aldrich) at an enzyme-to-protein ratio of 1:50 (wt/wt). The filter tube was then centrifuged at 10,000 × g for 15 to 30 min, and the flowthrough was collected in a new Eppendorf tube. SDC was removed via precipitation by adding formic acid to 0.5%, followed by centrifugation at 16,000 × g for 15 min. Cleared supernatant peptide concentrations were then measured using a BCA assay (Pierce). Subsequently, 25-μg samples of peptides were desalted with C18 StageTips (Thermo Fisher) as recommended by the manufacturer.
To measure the extracellular proteomes of different strains, 5 μg of each peptide sample was analyzed by two-dimensional LC-MS/MS, using a Vanquish UHPLC instrument plumbed directly in line with a Q Exactive Plus mass spectrometer (QE+; Thermo Scientific) outfitted with a nanospray emitter (75-μm internal diameter [ID]; fused silica) packed with 25 cm of 5-μm Kinetex C18 reversed-phase (RP) resin (Phenomenex). Autosampled peptides were loaded onto a 100-μm-ID MudPIT (57, 58) precolumn packed with 5-μm Luna strong-cation-exchange (SCX; Phenomenex) and Kinetex C18 RP resins. Bound peptides were then washed, separated, and analyzed by data-dependent MS/MS over 2 successive salt cuts of ammonium acetate (50 mM and 500 mM), each followed by a 90-min, split-flow (300 nl/min) organic gradient (0 to 3% B over 1 min, 3 to 25% B over 80 min, 25 to 50% B over 5 min, and 50 to 0% B over 4 min).
MS data analysis and evaluation.
Acquired MS/MS spectra were assigned to specific peptide spectral matches (PSMs) by using the database search engine Tide (59), with a protein database specific to C. bescii concatenated with reversed/decoy sequence entries (to assess the false-discovery rate [FDR]) and common contaminant proteins. The scored PSMs were filtered using Percolator (60) for q values of 0.02 or less, and the resulting identified peptides were quantified by extracting their apex areas under the curve (AUCs), using moFF (61). The resulting peptide reports were adjusted to ∼1% FDR, and peptides were assembled into proteins, using the peptide summed AUC to calculate protein abundances. Protein abundance distributions were normalized and adjusted for protein length by using an in-house Python script. The final protein report was further filtered by keeping only proteins which were identified in at least two of three biological replicates.
Operon structure analysis.
RNAs were extracted from C. bescii cells harvested from early-stationary-phase cultures grown in D516 medium with cellobiose as the carbon source by using an RNeasy minikit (Qiagen). The harvested RNA was reverse transcribed using iScript reverse transcription (RT) supermix for RT-quantitative PCR (RT-qPCR) (Bio-Rad) and used as the template for PCR with primers to amplify from gene to gene (see primer sequences in Table S2 in the supplemental material). PCR controls were also performed with C. bescii genomic DNA and no-RT iScript control supermix (Bio-Rad) templates. PCR was performed using OneTaq Quick Load 2× master mix with standard buffer (New England BioLabs).
Solubilization of lignocellulosic biomass substrates.
Prior to starting the solubilization experiment, Caldicellulosiruptor strains were passaged on modified defined DSM671 medium with each of the biomass substrates 3 times to ensure growth on the biomass substrate as the sole carbon source. Fifty-milliliter solubilization cultures were prepared in triplicate for each substrate and strain combination, loaded with 5 g/liter substrate in 125-ml serum bottles. Caldicellulosiruptor strains were inoculated into these cultures at a density of 1 × 106 cells/ml. For mixtures of two strains in the same culture, each strain was added at a density of 5 × 105 cells/ml. After growth for 7 days at 70°C, cultures were harvested by centrifugation at 6,000 × g for 10 min and washed 3 times with 70°C sterile water. The residual substrate was dried at 70°C until it had a constant mass. The percent solubilization was determined from the difference in mass between the biomass used to prepare each culture and the residual remaining after harvest and drying.
Determination of switchgrass composition.
Analysis of the carbohydrate contents of switchgrass before and after solubilization by C. bescii strains was performed using a modified version of the National Renewable Energy Laboratory (NREL) procedure for determination of structural carbohydrates and lignin in biomass (62). Briefly, 40 mg of biomass was mixed with 600 μl 72% sulfuric acid and incubated at 30°C for 70 min, with agitation with a glass rod every 5 to 10 min. Next, 16.8 ml deionized water was added to dilute the sulfuric acid to 4%, and tubes were capped and autoclaved for 1 h to hydrolyze oligosaccharide sugars to monosaccharides. Using a high-pressure liquid chromatograph (HPLC) (Empire 1515 separation module; Waters) with a refractive index detector (model 2414; Waters), glucose and xylose in the hydrolyzed samples were quantified on a Rezex-ROA column (300 mm by 7.8 mm; Phenomenex). The column was operated with a 5 mM H2SO4 mobile phase at 0.6 ml/min and 60°C. The mass of initial switchgrass input into the procedure not accounted for by carbohydrate content represents the inert components (lignin and ash).
Accession number(s).
Genome sequence data for the modified C. bescii strains described here are archived in the National Center for Biotechnology Information database (NCBI; www.ncbi.nlm.nih.gov) under the following BioProject accession numbers: RKCB120, PRJNA371177; RKCB121, PRJNA371178; RKCB123, PRJNA371180; RKCB124, PRJNA371181; RKCB125, PRJNA371182; RKCB127, PRJNA371184; RKCB130, PRJNA402429; and RKCB132, PRJNA416008.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the help of Christa Pennacchio, Joel Martin, and Matthew Blow at the DOE Joint Genome Institute for sequencing of the recombinant Caldicellulosiruptor strains.
This work was funded in part by the BioEnergy Science Center, a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01828-17.
REFERENCES
- 1.Blumer-Schuette SE, Brown SD, Sander KB, Bayer EA, Kataeva I, Zurawski JV, Conway JM, Adams MWW, Kelly RM. 2014. Thermophilic lignocellulose deconstruction. FEMS Microbiol Rev 38:393–448. doi: 10.1111/1574-6976.12044. [DOI] [PubMed] [Google Scholar]
- 2.Yang S-J, Kataeva I, Hamilton-Brehm SD, Engle NL, Tschaplinski TJ, Doeppke C, Davis M, Westpheling J, Adams MWW. 2009. Efficient degradation of lignocellulosic plant biomass, without pretreatment, by the thermophilic anaerobe “Anaerocellum thermophilum” DSM 6725. Appl Environ Microbiol 75:4762–4769. doi: 10.1128/AEM.00236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zurawski JV, Conway JM, Lee LL, Simpson H, Izquierdo JA, Blumer-Schuette S, Nookaew I, Adams MWW, Kelly RM. 2015. Comparative analysis of extremely thermophilic Caldicellulosiruptor species reveals common and differentiating cellular strategies for plant biomass utilization. Appl Environ Microbiol 81:7159–7170. doi: 10.1128/AEM.01622-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conway JM, Zurawski JV, Lee LL, Blumer-Schuette SE, Kelly RM. 2015. Lignocellulosic biomass deconstruction by the extremely thermophilic genus Caldicellulosiruptor, p 91–120. In Li F. (ed), Thermophilic microorganisms, vol 1 Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 5.Bayer EA, Shoham Y, Lamed R. 2013. Lignocellulose-decomposing bacteria and their enzyme systems, p 215–266. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes: prokaryotic physiology and biochemistry. Springer, Berlin, Germany. [Google Scholar]
- 6.Ozdemir I, Blumer-Schuette SE, Kelly RM. 2012. S-layer homology domain proteins Csac_0678 and Csac_2722 are implicated in plant polysaccharide deconstruction by the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl Environ Microbiol 78:768–777. doi: 10.1128/AEM.07031-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumer-Schuette SE, Lewis DL, Kelly RM. 2010. Phylogenetic, microbiological, and glycoside hydrolase diversities within the extremely thermophilic, plant biomass-degrading genus Caldicellulosiruptor. Appl Environ Microbiol 76:8084–8092. doi: 10.1128/AEM.01400-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergquist PL, Gibbs MD, Morris DD, Te'o VSJ, Saul DJ, Morgan HW. 1999. Molecular diversity of thermophilic cellulolytic and hemicellulolytic bacteria. FEMS Microbiol Ecol 28:99–110. doi: 10.1111/j.1574-6941.1999.tb00565.x. [DOI] [Google Scholar]
- 9.Gibbs MD, Reeves RA, Farrington GK, Anderson P, Williams DP, Bergquist PL. 2000. Multidomain and multifunctional glycosyl hydrolases from the extreme thermophile Caldicellulosiruptor isolate Tok7B.1. Curr Microbiol 40:333–340. doi: 10.1007/s002849910066. [DOI] [PubMed] [Google Scholar]
- 10.Kataeva IA, Yang SJ, Dam P, Poole FL II, Yin Y, Zhou F, Chou WC, Xu Y, Goodwin L, Sims DR, Detter JC, Hauser LJ, Westpheling J, Adams MW. 2009. Genome sequence of the anaerobic, thermophilic, and cellulolytic bacterium “Anaerocellum thermophilum” DSM 6725. J Bacteriol 191:3760–3761. doi: 10.1128/JB.00256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams-Rhaesa AM, Poole FL II, Dinsmore J, Lipscomb GL, Rubinstein GM, Scott IM, Conway JM, Lee LL, Khatibi PA, Kelly RM, Adams MWW. 2017. Genome stability in engineered strains of the extremely thermophilic, lignocellulose-degrading bacterium Caldicellulosiruptor bescii. Appl Environ Microbiol 83:e00444-17. doi: 10.1128/AEM.00444-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lochner A, Giannone RJ, Rodriguez M Jr, Shah MB, Mielenz JR, Keller M, Antranikian G, Graham DE, Hettich RL. 2011. Use of label-free quantitative proteomics to distinguish the secreted cellulolytic systems of Caldicellulosiruptor bescii and Caldicellulosiruptor obsidiansis. Appl Environ Microbiol 77:4042–4054. doi: 10.1128/AEM.02811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zverlov V, Mahr S, Riedel K, Bronnenmeier K. 1998. Properties and gene structure of a bifunctional cellulolytic enzyme (CelA) from the extreme thermophile ‘Anaerocellum thermophilum’ with separate glycosyl hydrolase family 9 and 48 catalytic domains. Microbiology 144:457–465. doi: 10.1099/00221287-144-2-457. [DOI] [PubMed] [Google Scholar]
- 14.Brunecky R, Alahuhta M, Xu Q, Donohoe BS, Crowley MF, Kataeva IA, Yang S-J, Resch MG, Adams MWW, Lunin VV, Himmel ME, Bomble YJ. 2013. Revealing nature's cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA. Science 342:1513–1516. doi: 10.1126/science.1244273. [DOI] [PubMed] [Google Scholar]
- 15.Yi Z, Su X, Revindran V, Mackie RI, Cann I. 2013. Molecular and biochemical analyses of CbCel9A/Cel48A, a highly secreted multi-modular cellulase by Caldicellulosiruptor bescii during growth on crystalline cellulose. PLoS One 8:e84172. doi: 10.1371/journal.pone.0084172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su X, Mackie RI, Cann IK. 2012. Biochemical and mutational analyses of a multidomain cellulase/mannanase from Caldicellulosiruptor bescii. Appl Environ Microbiol 78:2230–2240. doi: 10.1128/AEM.06814-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye L, Su X, Schmitz GE, Moon YH, Zhang J, Mackie RI, Cann IKO. 2012. Molecular and biochemical analyses of the GH44 module of CbMan5B/Cel44A, a bifunctional enzyme from the hyperthermophilic bacterium Caldicellulosiruptor bescii. Appl Environ Microbiol 78:7048–7059. doi: 10.1128/AEM.02009-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang R, Gong L, Xue X, Qin X, Ma R, Luo H, Zhang Y, Yao B, Su X. 2016. Identification of the C-terminal GH5 domain from CbCel9B/Man5A as the first glycoside hydrolase with thermal activation property from a multimodular bifunctional enzyme. PLoS One 11:e0156802. doi: 10.1371/journal.pone.0156802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue X, Wang R, Tu T, Shi P, Ma R, Luo H, Yao B, Su X. 2015. The N-terminal GH10 domain of a multimodular protein from Caldicellulosiruptor bescii is a versatile xylanase/beta-glucanase that can degrade crystalline cellulose. Appl Environ Microbiol 81:3823–3833. doi: 10.1128/AEM.00432-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipscomb GL, Conway JM, Blumer-Schuette SE, Kelly RM, Adams MWW. 2016. Highly thermostable kanamycin resistance marker expands the toolkit for genetic manipulation of Caldicellulosiruptor bescii. Appl Environ Microbiol 82:4421–4428. doi: 10.1128/AEM.00570-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alahuhta M, Brunecky R, Chandrayan P, Kataeva I, Adams MW, Himmel ME, Lunin VV. 2013. The structure and mode of action of Caldicellulosiruptor bescii family 3 pectate lyase in biomass deconstruction. Acta Crystallogr D Biol Crystallogr 69:534–539. doi: 10.1107/S0907444912050512. [DOI] [PubMed] [Google Scholar]
- 22.Alahuhta M, Taylor LE II, Brunecky R, Sammond DW, Michener W, Adams MW, Himmel ME, Bomble YJ, Lunin V. 2015. The catalytic mechanism and unique low pH optimum of Caldicellulosiruptor bescii family 3 pectate lyase. Acta Crystallogr D Biol Crystallogr 71:1946–1954. doi: 10.1107/S1399004715013760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung D, Pattathil S, Biswal AK, Hahn MG, Mohnen D, Westpheling J. 2014. Deletion of a gene cluster encoding pectin degrading enzymes in Caldicellulosiruptor bescii reveals an important role for pectin in plant biomass recalcitrance. Biotechnol Biofuels 7:147. doi: 10.1186/s13068-014-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blumer-Schuette SE, Alahuhta M, Conway JM, Lee LL, Zurawski JV, Giannone RJ, Hettich RL, Lunin VV, Himmel ME, Kelly RM. 2015. Discrete and structurally unique proteins (tāpirins) mediate attachment of extremely thermophilic Caldicellulosiruptor species to cellulose. J Biol Chem 290:10645–10656. doi: 10.1074/jbc.M115.641480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lommel M, Strahl S. 2009. Protein O-mannosylation: conserved from bacteria to humans. Glycobiology 19:816–828. doi: 10.1093/glycob/cwp066. [DOI] [PubMed] [Google Scholar]
- 26.Iwashkiw JA, Vozza NF, Kinsella RL, Feldman MF. 2013. Pour some sugar on it: the expanding world of bacterial protein O-linked glycosylation. Mol Microbiol 89:14–28. doi: 10.1111/mmi.12265. [DOI] [PubMed] [Google Scholar]
- 27.Chung D, Young J, Bomble YJ, Vander Wall TA, Groom J, Himmel ME, Westpheling J. 2015. Homologous expression of the Caldicellulosiruptor bescii CelA reveals that the extracellular protein is glycosylated. PLoS One 10:e0119508. doi: 10.1371/journal.pone.0119508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobos KM, Khoo KH, Swiderek KM, Brennan PJ, Belisle JT. 1996. Definition of the full extent of glycosylation of the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. J Bacteriol 178:2498–2506. doi: 10.1128/jb.178.9.2498-2506.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michell SL, Whelan AO, Wheeler PR, Panico M, Easton RL, Etienne AT, Haslam SM, Dell A, Morris HR, Reason AJ, Herrmann JL, Young DB, Hewinson RG. 2003. The MPB83 antigen from Mycobacterium bovis contains O-linked mannose and (1→3)-mannobiose moieties. J Biol Chem 278:16423–16432. doi: 10.1074/jbc.M207959200. [DOI] [PubMed] [Google Scholar]
- 30.Wiemels RE, Cech SM, Meyer NM, Burke CA, Weiss A, Parks AR, Shaw LN, Carroll RK. 2017. An intracellular peptidyl-prolyl cis/trans isomerase is required for folding and activity of the Staphylococcus aureus secreted virulence factor nuclease. J Bacteriol 199:e00453-16. doi: 10.1128/JB.00453-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veeraraghavan S, Nall BT, Fink AL. 1997. Effect of prolyl isomerase on the folding reactions of staphylococcal nuclease. Biochemistry 36:15134–15139. doi: 10.1021/bi971357r. [DOI] [PubMed] [Google Scholar]
- 32.Kingsford CL, Ayanbule K, Salzberg SL. 2007. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol 8:R22. doi: 10.1186/gb-2007-8-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young J, Chung D, Bomble YJ, Himmel ME, Westpheling J. 2014. Deletion of Caldicellulosiruptor bescii CelA reveals its crucial role in the deconstruction of lignocellulosic biomass. Biotechnol Biofuels 7:142. doi: 10.1186/s13068-014-0142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokoyama H, Yamashita T, Morioka R, Ohmori H. 2014. Extracellular secretion of noncatalytic plant cell wall-binding proteins by the cellulolytic thermophile Caldicellulosiruptor bescii. J Bacteriol 196:3784–3792. doi: 10.1128/JB.01897-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zurawski JV. 2016. Microbiological and engineering studies of extremely thermophilic Caldicellulosiruptor species for lignocellulose deconstruction and conversion. PhD thesis North Carolina State University, Raleigh, NC. [Google Scholar]
- 36.Bhagia S, Muchero W, Kumar R, Tuskan GA, Wyman CE. 2016. Natural genetic variability reduces recalcitrance in poplar. Biotechnol Biofuels 9:106. doi: 10.1186/s13068-016-0521-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belaich JP, Tardif C, Belaich A, Gaudin C. 1997. The cellulolytic system of Clostridium cellulolyticum. J Biotechnol 57:3–14. doi: 10.1016/S0168-1656(97)00085-0. [DOI] [PubMed] [Google Scholar]
- 38.Desvaux M. 2005. Clostridium cellulolyticum: model organism of mesophilic cellulolytic clostridia. FEMS Microbiol Rev 29:741–764. doi: 10.1016/j.femsre.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 39.Ding S-Y, Adney WS, Vinzant TB, Decker SR, Baker JO, Thomas SR, Himmel ME. 2003. Glycoside hydrolase gene cluster of Acidothermus cellulolyticus. ACS Symp Ser 855:332–360. doi: 10.1021/bk-2003-0855.ch020. [DOI] [Google Scholar]
- 40.Guglielmi G, Beguin P. 1998. Cellulase and hemicellulase genes of Clostridium thermocellum from five independent collections contain few overlaps and are widely scattered across the chromosome. FEMS Microbiol Lett 161:209–215. doi: 10.1111/j.1574-6968.1998.tb12950.x. [DOI] [PubMed] [Google Scholar]
- 41.VanFossen AL, Ozdemir I, Zelin SL, Kelly RM. 2011. Glycoside hydrolase inventory drives plant polysaccharide deconstruction by the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Biotechnol Bioeng 108:1559–1569. doi: 10.1002/bit.23093. [DOI] [PubMed] [Google Scholar]
- 42.Lee LL, Izquierdo JA, Blumer-Schuette SE, Zurawski JV, Conway JM, Cottingham RW, Huntemann M, Copeland A, Chen IM, Kyrpides N, Markowitz V, Palaniappan K, Ivanova N, Mikhailova N, Ovchinnikova G, Andersen E, Pati A, Stamatis D, Reddy TB, Shapiro N, Nordberg HP, Cantor MN, Hua SX, Woyke T, Kelly RM. 2015. Complete genome sequences of Caldicellulosiruptor sp. strain Rt8.B8, Caldicellulosiruptor sp. strain Wai35.B1, and “Thermoanaerobacter cellulolyticus.” Genome Announc 3:e00440-15. doi: 10.1128/genomeA.00440-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanafusa-Shinkai S, Wakayama Ji Tsukamoto K, Hayashi N, Miyazaki Y, Ohmori H, Tajima K, Yokoyama H. 2013. Degradation of microcrystalline cellulose and non-pretreated plant biomass by a cell-free extracellular cellulase/hemicellulase system from the extreme thermophilic bacterium Caldicellulosiruptor bescii. J Biosci Bioeng 115:64–70. doi: 10.1016/j.jbiosc.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 44.Girio FM, Fonseca C, Carvalheiro F, Duarte LC, Marques S, Bogel-Lukasik R. 2010. Hemicelluloses for fuel ethanol: a review. Bioresour Technol 101:4775–4800. doi: 10.1016/j.biortech.2010.01.088. [DOI] [PubMed] [Google Scholar]
- 45.Conway JM, Pierce WS, Le JH, Harper GW, Wright JH, Tucker AL, Zurawski JV, Lee LL, Blumer-Schuette SE, Kelly RM. 2016. Multidomain, surface layer-associated glycoside hydrolases contribute to plant polysaccharide degradation by Caldicellulosiruptor species. J Biol Chem 291:6732–6747. doi: 10.1074/jbc.M115.707810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chung D, Farkas J, Westpheling J. 2013. Overcoming restriction as a barrier to DNA transformation in Caldicellulosiruptor species results in efficient marker replacement. Biotechnol Biofuels 6:82. doi: 10.1186/1754-6834-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cha M, Chung D, Elkins JG, Guss AM, Westpheling J. 2013. Metabolic engineering of Caldicellulosiruptor bescii yields increased hydrogen production from lignocellulosic biomass. Biotechnol Biofuels 6:85. doi: 10.1186/1754-6834-6-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geslin C, Le Romancer M, Erauso G, Gaillard M, Perrot G, Prieur D. 2003. PAV1, the first virus-like particle isolated from a hyperthermophilic euryarchaeote, “Pyrococcus abyssi”. J Bacteriol 185:3888–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gibson DG. 2011. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol 498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans LM, Slavov GT, Rodgers-Melnick E, Martin J, Ranjan P, Muchero W, Brunner AM, Schackwitz W, Gunter L, Chen JG, Tuskan GA, DiFazio SP. 2014. Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations. Nat Genet 46:1089–1096. doi: 10.1038/ng.3075. [DOI] [PubMed] [Google Scholar]
- 51.Muchero W, Guo J, DiFazio SP, Chen JG, Ranjan P, Slavov GT, Gunter LE, Jawdy S, Bryan AC, Sykes R, Ziebell A, Klapste J, Porth I, Skyba O, Unda F, El-Kassaby YA, Douglas CJ, Mansfield SD, Martin J, Schackwitz W, Evans LM, Czarnecki O, Tuskan GA. 2015. High-resolution genetic mapping of allelic variants associated with cell wall chemistry in Populus. BMC Genomics 16:24. doi: 10.1186/s12864-015-1215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farkas J, Chung D, Cha M, Copeland J, Grayeski P, Westpheling J. 2013. Improved growth media and culture techniques for genetic analysis and assessment of biomass utilization by Caldicellulosiruptor bescii. J Ind Microbiol Biotechnol 40:41–49. doi: 10.1007/s10295-012-1202-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lipscomb GL, Stirrett K, Schut GJ, Yang F, Jenney FE Jr, Scott RA, Adams MW, Westpheling J. 2011. Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl Environ Microbiol 77:2232–2238. doi: 10.1128/AEM.02624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chung D, Farkas J, Huddleston JR, Olivar E, Westpheling J. 2012. Methylation by a unique alpha-class N4-cytosine methyltransferase is required for DNA transformation of Caldicellulosiruptor bescii DSM6725. PLoS One 7:e43844. doi: 10.1371/journal.pone.0043844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nordberg H, Cantor M, Dusheyko S, Hua S, Poliakov A, Shabalov I, Smirnova T, Grigoriev IV, Dubchak I. 2014. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res 42:D26–D31. doi: 10.1093/nar/gkt1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. 2009. Real-time DNA sequencing from single polymerase molecules. Science 323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 57.Washburn MP, Wolters D, Yates JR. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 58.McDonald WH, Ohi R, Miyamoto DT, Mitchison TJ, Yates JR. 2002. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int J Mass Spectrom 219:245–251. doi: 10.1016/S1387-3806(02)00563-8. [DOI] [Google Scholar]
- 59.Diament BJ, Noble WS. 2011. Faster SEQUEST searching for peptide identification from tandem mass spectra. J Proteome Res 10:3871–3879. doi: 10.1021/pr101196n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. 2007. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods 4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 61.Argentini A, Goeminne LJ, Verheggen K, Hulstaert N, Staes A, Clement L, Martens L. 2016. moFF: a robust and automated approach to extract peptide ion intensities. Nat Methods 13:964–966. doi: 10.1038/nmeth.4075. [DOI] [PubMed] [Google Scholar]
- 62.Sluiter A, Hames B, Ruiz R, Scarlata C, Sluiter J, Templeton D, Crocker D. 2012. Determination of structural carbohydrates and lignin in biomass, NREL/TP-510-42618. National Renewable Energy Laboratory, Golden, CO. [Google Scholar]
- 63.Yang SJ, Kataeva I, Wiegel J, Yin Y, Dam P, Xu Y, Westpheling J, Adams MW. 2010. Classification of ‘Anaerocellum thermophilum’ strain DSM 6725 as Caldicellulosiruptor bescii sp. nov. Int J Syst Evol Microbiol 60:2011–2015. doi: 10.1099/ijs.0.017731-0. [DOI] [PubMed] [Google Scholar]
- 64.Chung D, Cha M, Farkas J, Westpheling J. 2013. Construction of a stable replicating shuttle vector for Caldicellulosiruptor species: use for extending genetic methodologies to other members of this genus. PLoS One 8:e62881. doi: 10.1371/journal.pone.0062881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.