ABSTRACT
Histoplasma capsulatum is a pathogenic fungus that causes life-threatening lung infections. About 500,000 people are exposed to H. capsulatum each year in the United States, and over 60% of the U.S. population has been exposed to the fungus at some point in their life. We performed genome-wide population genetics and phylogenetic analyses with 30 Histoplasma isolates representing four recognized areas where histoplasmosis is endemic and show that the Histoplasma genus is composed of at least four species that are genetically isolated and rarely interbreed. Therefore, we propose a taxonomic rearrangement of the genus.
KEYWORDS: cryptic speciation, Histoplasma, phylogenetic species concept, genomic alignment, taxonomy
IMPORTANCE
The evolutionary processes that give rise to new pathogen lineages are critical to our understanding of how they adapt to new environments and how frequently they exchange genes with each other. The fungal pathogen Histoplasma capsulatum provides opportunities to precisely test hypotheses about the origin of new genetic variation. We find that H. capsulatum is composed of at least four different cryptic species that differ genetically and also in virulence. These results have implications for the epidemiology of histoplasmosis because not all Histoplasma species are equivalent in their geographic range and ability to cause disease.
INTRODUCTION
Histoplasma capsulatum is a dimorphic ascomycete fungus that causes histoplasmosis in humans. Originally discovered in Panama in 1906 by S. T. Darling (1), its morphology in infected tissues led to its original classification as a protozoan. After multiple reclassifications, this fungus was determined to be closely related to other pathogenic dimorphic fungi such as Coccidioides, Blastomyces, and Paracoccidioides (2, 3). The haploid nature of its genome, combined with a variety of plasmid-based molecular genetic tools, have made H. capsulatum the most developed genetic system among dimorphic fungi (4, 5).
Histoplasmosis, the disease caused by H. capsulatum, is arguably the most common fungal respiratory infection worldwide, with hundreds of thousands of new infections occurring annually in the United States alone (6, 7). The disease burden of pulmonary histoplasmosis is among the highest of any disease caused by a primary fungal pathogen (8–10). In 5 to 10% of cases, the infection progresses in the lung or disseminates to visceral organs and can be difficult to treat with antifungal drugs. Patients who appear to recover may suffer a recurrence months to years later, particularly if they are immunocompromised by chemotherapy (8), organ transplant (11), or AIDS (12, 13). In fact, up to 25% of AIDS patients living in regions where Histoplasma is endemic develop histoplasmosis and require prolonged antifungal therapy (14, 15). The potential for environmental exposure and individual reactivation ensures that histoplasmosis will persist as a public health problem: by age 20, more than 80% of individuals residing in areas of endemicity in the continental United States are skin test positive for a previous infection (7, 16).
Despite the importance of the disease and the development of Histoplasma as a genetic model system, many aspects regarding the natural history, evolution, and systematics of this pathogen remain largely unknown. The fungus is found in soil and is commonly associated with environments where guano and bird droppings are present (17–19). This association has led to the provocative hypothesis that flying animals might be involved in the dispersion of Histoplasma (17, 20). The fungus can also be found in soil (21–23) and mammalian reservoirs (24), but the extent of its natural occurrence remains largely unexplored (25, 26). Even though the fungus is known to produce a sexual stage under laboratory conditions (Emmonsiella capsulata [27]), this form has never been observed in nature. The magnitude of sexual recombination is thus still undetermined.
The number of species encompassed in the genus Histoplasma also remains unknown. Geographically, Histoplasma has the widest range of all the dimorphic fungi, being globally distributed (17, 28–30). There seems to be strong population structure across continents, which poses the possibility of undiscovered events of speciation (28, 31, 32); however, all populations of the genus are currently classified as H. capsulatum. Three distinct varieties, determined by geographic distribution, morphology, and clinical symptoms, have been historically recognized. This classification system subdivides H. capsulatum into New World human pathogens (H. capsulatum var. capsulatum), African human pathogens (H. capsulatum var. duboisii), and Old World horse pathogens (H. capsulatum var. farciminosum) (33, 34). Seminal studies revealed that these three classic groups are artificial. Instead, H. capsulatum is composed of multiple genetically distinct genetic groups (at least four) that until now have never been recognized as species (31). These genetically isolated groups have differences in virulence (35), and one of these groups (H. capsulatum var. duboisii, henceforth referred to as “Africa”) seems to cause a unique disease (36, 37). Notably, H. capsulatum var. farciminosum was found to be polyphyletic, which indicates the traditional methods to delimit Histoplasma varieties have significant shortcomings (31).
The identification of cryptic species and the delimitation of species boundaries in fungi have propelled the research required to identify differences in morphology, virulence strategies, drug susceptibility, and natural history that were initially not apparent (38, 39). In Histoplasma, some of these differences between genetic clusters have already been observed, ranging from extensive differences in virulence to drug susceptibility for multiple genetic clusters within the genus. The natural question is to assess whether these clusters are indeed isolated species or not. Disentangling the species limits within Histoplasma would be beneficial not only to understand the systematics and evolution of this group of pathogens. If the reported differences are ascribed to the existence of species within the group, then the identification of such species and of the genetic factors that make them unique might contribute to the identification of virulence strategies and the design of species-specific treatments.
In spite of the clear importance of defining species boundaries in pathogens, the task is not straightforward in fungi (reviewed in references 40 to 42). The biological species concept, the gold standard for animals and plants, states that species are groups of individuals that cannot interbreed in sexual crosses (38–43). However, delimiting species under this concept necessarily requires performing large-scale genetic crosses between individuals of potentially different species, which is challenging in eukaryotes such as fungi and almost impossible in Histoplasma, where sexual stages have not been observed in nature and crosses are formidably challenging to perform in vitro. In the absence of crosses, indirect measurements can be used to infer the occurrence of speciation in lieu of the ability to interbreed.
Genetic clusters that are discontinuous from each other can also be detected using DNA divergence. Several frameworks have been proposed to detect such discontinuities, but the phylogenetic species concept (PSC) is the most extensively used in fungi (reviewed in references 40, 41, and 43). The PSC has had several propositions over the years, but all of them are tailored to detect monophyletic and diagnosable groups that do not interbreed with each other (41, 44). The most practical and comprehensive test for the identification of phylogenetic species is to identify monophyletic groups with high levels of support, which might in turn reflect the existence of reproductively isolated groups (45–48). Dimorphic fungi are a natural group for the use of phylogenetic approaches to detect cryptic species because even though some of them can be crossed, the majority have no known sexual stage, which makes them unsuitable for the application of the biological species concept.
Here we test the hypothesis that Histoplasma capsulatum is composed of several monophyletic and identifiable species using whole-genome data. Population genetics and phylogenetic analyses indicate the existence of five isolated genetic lineages that are consistent with the existence of more than one species. Given the remarkable genealogical consistency across the whole genome, we propose that four of these genetic groups, all endemic to the Americas, should be elevated to species status. The fifth group, the African clade of Histoplasma (H. capsulatum var. duboisii, or “Africa”), is likely to be a separate species as well, but we refrain from modifying its taxonomic affinity without more sampling that allows us to reliably measure its within-group genetic diversity and quantify its level of differentiation from other Histoplasma species. Our approach provides a blueprint for the identification of species boundaries in fungi using whole-genome sequences. These results open the possibility of studying the biological barriers that lead to genetic discontinuities across fungal species but also to examine how much gene flow occurs between cryptic species of fungal pathogens.
RESULTS
Genomic data.
All genome sequences reported in this study have been deposited in the Short Read Archive, and intermediary processing files (.sam, and.bam) have been deposited in Dryad. The coverage for each reported genome is reported in Table S1 (at https://doi.org/10.5061/dryad.006bf). In spite of the differences in genome size (49), the coverage for all lines was over 40×, and in most cases, the levels of coverage were similar among species (one-way analysis of variance [ANOVA], F4,26 = 3.3822, P = 0.0235 [see Table S2 at https://doi.org/10.5061/dryad.006bf]). There were two exceptions. NAm 2 (i.e., North America 2) shows slightly lower coverage than NAm 1 (NAm 2 average coverage, 58.33; NAm 1 average coverage, 78.35; Tukey contrasts, t26 = −2.946, P = 0.0452) and LAm A (i.e., Latin America A) (LAm A average coverage, 83.18; Tukey contrasts, t26 = −2.917, P = 0.0482 [see Table S3 at https://doi.org/10.5061/dryad.006bf]). These results are consistent with NAm 2 having a larger genome than all other species (49).
Histoplasma harbors at least five discrete genetic clusters.
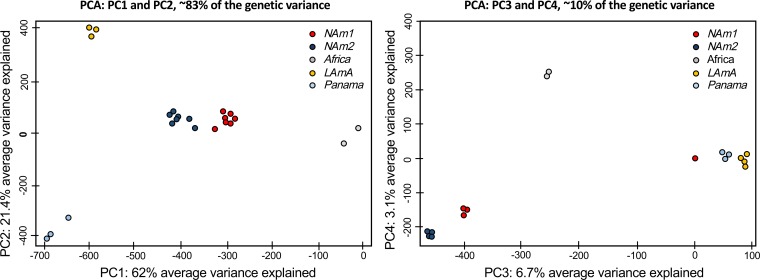
We employed a population genetics framework to quantitatively assess if the patterns of genetic variation were consistent with the existence of different species within Histoplasma. First, we calculated levels of within-species population variation. We did a principal-component analysis (PCA) to analyze allelic frequencies across the whole genome. The first five principal components (PC1 to -5) of the PCA explained about 95% of the genetic variance. The results suggested three genetic patterns (Fig. 1). The vast majority of variation is explained by the first four PCs. The first four PCs differentiate between species. PC1 (variance explained, 62%) clearly differentiates five genetic clusters within Histoplasma. These clusters are concordant with the groups reported by Kasuga et al. (31) and cluster according to geography. The differentiation between these genetic groups is large and complete as there is no overlap between isolates from different putative species. The NAm 1 and Africa groups are the most differentiated groups (in terms of their allele frequencies), and they appear to be highly differentiated along PC1. PC2 (variance explained, 21.4%) mainly differentiates between the Africa and Panama species, but all other species also appear differentiated. PC3 (~7%) separates the North American clade from the other three species, while PC4 (~3%) strongly differentiates between NAm 1 and NAm 2. No obvious patterns explained PC5 (explaining <2% of the variance), which might be related to variation within each species. These genotype analyses revealed that genetic variability in Histoplasma is portioned across genetic clusters and that such clusters seem to have little overlap in terms of allele frequencies. This is consistent with genetic differentiation and little gene flow between clusters. We thus explored whether these clusters fulfill the definition of a phylogenetic species and thus should be elevated in taxonomic status.
FIG 1 .
Principal-component analysis for Histoplasma using whole-genome allelic frequencies. The genetic variance in the genus Histoplasma indicates the existence of discrete genetic clusters. Red, NAm 1; blue, NAm 2; yellow, LAm A; gray, Africa; light blue, Panama. (Left panel) Principal components (PC) 1 and 2 are shown, and the percentage of variance explained by each eigenvalue is shown within parentheses on each axis. (Right panel) PC3 and PC4.
The genetic clusters within Histoplasma are cryptic species.
We next used whole-genome data to reconstruct the genealogical relationships between the sequenced individuals of Histoplasma. We rooted the tree with two closely related species: Emmonsia parva and Emmonsia crescens. The tree reveals two sets of results: whether the genetic clusters are phylogenetic species and the phylogenetic relationships between such species.
(i) Recognition of phylogenetic species.
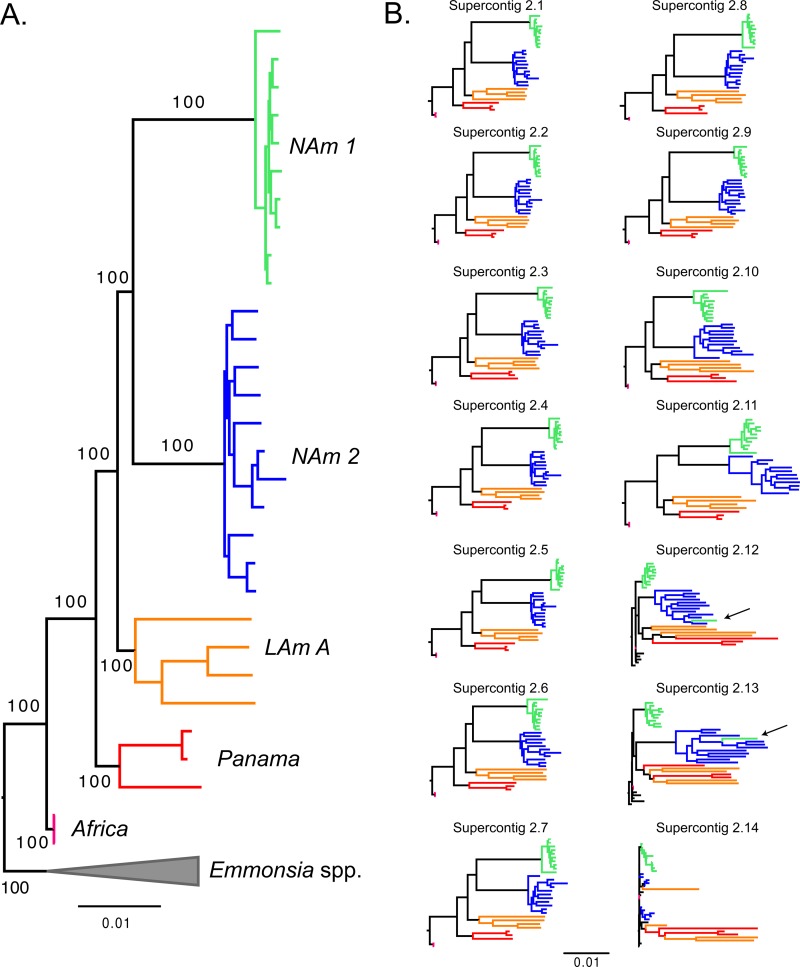
Our whole-genome concatenated phylogenetic reconstruction indicates Histoplasma is a highly genetically structured group. Congruent with the results from the PCA, five of the groups proposed by Kasuga et al. (the NAm 1, NAm 2, LAm A, Panama/H81 lineage, and Africa groups) (28, 31) form clades with high support (Fig. 2A). We next analyzed whether these groups fulfilled the requirements to be considered phylogenetic species (sensu [45, 50]). First, we found that all the genetic clusters are reciprocally monophyletic (i.e., all five groups coalesce within each other, before any coalescence events take place between groups), a primary requirement for species definition (51, 52). Second, we determined whether the five identifiable clades of Histoplasma had enough support to be classified as different phylogenetic species. In all five cases, the concordance factors were high enough to argue these clades are indeed different species (following reference 45). The support for the branches leading to each phylogenetic species is shown in Table S4 (at https://doi.org/10.5061/dryad.006bf). Single superscaffold analyses revealed a similar pattern. NAm 1 was monophyletic and strongly supported in all superscaffold trees, NAm 2 and H. capsulatum var. duboisii were supported by the 13 longest scaffolds, and all 5 groups were supported by the 11 longest scaffolds (Fig. 2B). The phylogenetic relationships between these groups in the superscaffold trees were identical to those observed in the concatenated tree in most cases, the exception being superscaffolds 10 to 13, where the Panama and LAm A groups were sister taxa, instead of sequential sister groups to the North American clade (NAm 1/NAm 2). It is worth noting these scaffolds are much smaller than the first 10 and thus contain fewer informative sites (see Table S5 at https://doi.org/10.5061/dryad.006bf). Collectively, the genetic differentiation, the reciprocal monophyly, and the high levels of support strongly suggest that the five genetic groups of Histoplasma are indeed phylogenetic species.
FIG 2 .
Rooted phylogram for the species of the Histoplasma genus using whole-genome data. The leftmost tree (A) shows a tree constructed using the variation from the whole genome. The trees to the right (B) were built with data from each of the 14 supercontigs. (The length of each supercontig is not shown). The length of each branch indicates the amount of genetic divergence. Arrows in the trees to the right show isolates with conflicting positioning for a given supercontig. All topologies are rooted with Emmonsia sp.
(ii) Species tree.
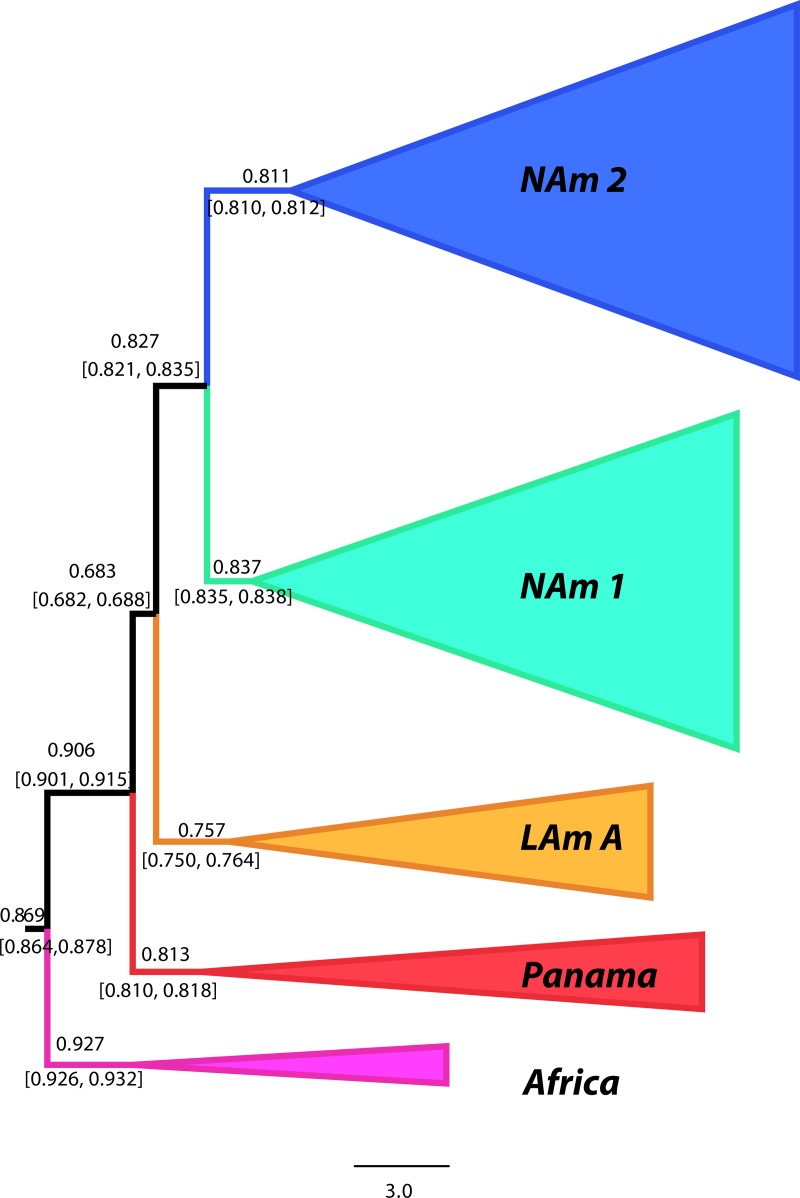
Our phylogenetic analyses indicate a large level of concordance among genomic windows and at the whole-genome level (Fig. 2). We quantified the magnitude of the concordance and inferred the likely Histoplasma species tree with gene tree analysis using BUCKy (53). This method inferred an identical species tree topology to the topology produced by the RAxML analysis of the whole genome (Fig. 2A and 3). We used 100-kb windows as the unit for analysis because of the high linkage disequilibrium (LD) in all species of Histoplasma (i.e., r2 is on the order of 0.2 after 100 kb [see Fig. S1 in the supplemental material]). The nodes grouping isolates for each species had concordance factor (CF) values larger than 0.75, and the other nodes in the tree had values above 0.68 which indicates that the vast majority of the genome recapitulates the species tree and is in concordance with our proposed phylogenetic species. These levels of concordance are higher than those shown by Neurospora species (54), which in turn provides further support that the phylogenetic species of Histoplasma are not simply the result of population structure but are indeed truly isolated genetic clusters.
FIG 3 .
Species tree of the genus Histoplasma shows that all species have high levels of genomic concordance. Shown is a genome-wide primary concordance tree produced by BUCKy (α = 5), based on 100-kb windows. Values above each branch show the concordance factor (CF) for each node, and values below branches show the 95% Bayesian credibility intervals for this statistic.
Linkage disequilibriums persist over large regions of the genome in Histoplasma. Average LD as measured by r2 between pairs of SNPs with distances binned every 100 bp. Download FIG S1, PDF file, 4.9 MB (5MB, pdf) .
Copyright © 2017 Sepúlveda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(iii) Phylogenetic relationships between groups.
We next assessed how the five phylogenetic species were related to each other. The tree topology is shown in Fig. 2A and 3. The 10 largest supercontigs, which account for 98.50% of the genome, also show the same genealogy (Fig. 2B). The relationship between these species was surprising because it strongly contradicts previously proposed relationships (20). First, and unlike previous reports, we find that all four American species are closely related and form a monophyletic group. The African specimens are an outgroup to the American group, suggesting a possible event of vicariance that led to the formation of the African and American clades. These results also indicate that several speciation events have occurred since Histoplasma’s arrival onto the American continent. These results seem to confirm the initial hypothesis of an American Histoplasma clade, initially dubbed H. capsulatum var. capsulatum, that is actually composed of at least four species (but see Discussion).
Notably, we find that the two North American species, NAm 1 and NAm 2, are sisters. This contradicts previous reports that had these two groups as highly divergent (20). A single isolate of the NAm 1 group (WU24) fell within the NAm 2 clade in supercontigs 10 to 13 (arrows in Fig. 2B), which might indicate these two species are still able to exchange genes. Alternatively, this may be due to low levels of incomplete lineage sorting (ILS). These alternatives are explored below. Collectively, these results constitute the first genome-wide approach to obtain resolution on the genealogical relationships between Histoplasma isolates.
The isolate MV3 collected in Colombia is part of the Panama lineage. This result expands the known range of the Panama species, initially thought to be restricted to Central America, to northern South America, which is part of the geographic range of LAm A (e.g., MZ5, also a Colombian isolate, is part of LAm A). This makes the Panama and LAm A groups sympatric. Just as it occurs with NAm 1 and NAm 2, this overlap is of particular importance to signal whether speciation has taken place because the two species pairs share a geographic range. This means that these two species have had the chance to interbreed; if no barriers to gene flow were in place, then their genomes would have undergone genome homogenization. Note that smaller supercontigs (2.12 to 2.14; less than 0.5% of the genome) show evidence of shared ancestry that might be caused by gene exchange or ILS circumscribed to these genomic regions. All these results are strongly suggestive of reproductive isolation between the Panama and LAm A groups and between NAm 1 and NAm 2.
(iv) Genetic distance.
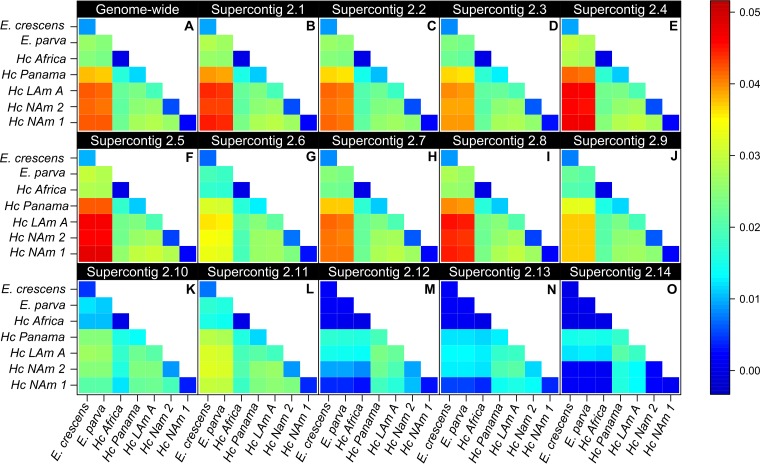
We measured the magnitude of genetic differentiation between species pairs. To this end, we next quantified πinter, a measurement of divergence among all putative species of Histoplasma, including the two sympatric pairs. By comparing the magnitude of between-species differentiation with within-species differentiation, one can assess the extent of differentiation and gene flow between putative species. We found that genetic variability is partitioned among species and that in all cases the magnitude of interspecific distances is at least 2× higher than that of intraspecific distances for all species pairs in genome-wide estimations (Fig. 4A; Table 1). In other words, the differentiation between species in all pairwise comparisons is larger than the amount of genetic variability within species. Notably, this pattern holds even though the sampling of some of the groups (e.g., the Africa and Panama groups) is rather modest. The 14 largest supercontigs show remarkably similar patterns, indicating that the genetic differentiation among species is pervasive across the genome (Fig. 4B to O). These results are in line with the PCA and the phylogenetic reconstruction and collectively suggest strong genetic differentiation among Histoplasma species.
FIG 4 .
Pairwise genetic distance indicates that Histoplasma species are genetically differentiated across the whole genome. Values in the diagonal are the estimate of πintra (intraspecific variability). All other values are πinter (genetic distance between species). As expected Emmonsia crescens is more distantly related to Histoplasma species than they are to themselves. In all cases, πinter is higher than πintra, indicating strong genetic differentiation (Table 1).
TABLE 1 .
Genetic distances between species are at least two times larger than the amount of intraspecific variation in all pairwise comparisons among Histoplasma speciesa
| Species 1 | Species 2 | πintra1 | πintra2 | πinter | Z value | P value |
|---|---|---|---|---|---|---|
| NAm 1 | NAm 2 | 5.870 × 10−3 | 6.705 × 10−3 | 0.0243 | 11.767 | <1 × 10−4 |
| NAm 1 | LAm A | 5.870 × 10−3 | 1.246 × 10−2 | 0.0263 | 8.8614 | <1 × 10−4 |
| NAm 1 | Panama | 5.870 × 10−3 | 1.142 × 10−2 | 0.0254 | 8.5931 | <1 × 10−4 |
| NAm 1 | Africa | 5.870 × 10−3 | 3.290 × 10−6 | 0.0213 | 7.618 | <1 × 10−4 |
| NAm 2 | LAm A | 6.705 × 10−3 | 1.246 × 10−2 | 0.0282 | 8.1891 | <1 × 10−4 |
| NAm 2 | Panama | 6.705 × 10−3 | 1.142 × 10−2 | 0.0278 | 7.4744 | <1 × 10−4 |
| NAm 2 | Africa | 6.705 × 10−3 | 3.290 × 10−6 | 0.023 | 3.7805 | <1 × 10−4 |
| LAm A | Panama | 1.246 × 10−2 | 1.142 × 10−2 | 0.0235 | 2.6809 | 6.00 × 104 |
| LAm A | Africa | 1.246 × 10−2 | 3.290 × 10−6 | 0.0217 | 2.2539 | 0.0238 |
| Panama | Africa | 1.142 × 10−2 | 3.290 × 10−6 | 0.0163 | 1.8327 | 0.0404 |
πinter and πintra (i.e., πintra1 and πintra2 pooled) were compared using two-sample Fisher-Pitman permutation tests. The significance of the Z value was assessed using 9,999 Monte Carlo resamplings.
Approximate divergence between Histoplasma species pairs.
To assess whether any shared ancestry between species pairs (see immediately below) was most likely caused by incomplete lineage sorting, we calculated approximate divergence dates for four nodes in the tree: the node where NAm 1 and NAm 2 split, the node where (NAm 1/NAm 2) and LAm split, the node where [(NAm 1/NAm 2), LAm] and the Panama group split, and the node where {[(Nam 1/NAm 2), LAm A], Panama group} and the Africa group split. The approximate divergence times are shown in Table 2 and indicate that all speciation events are over 1.7 million years. We converted these divergence times to generations assuming that Histoplasma has 1, 10, or 100 generations per year and calculated the time required to lose an allele segregating in an ancestral population. These calculations are shown in Fig. S2 in the supplemental material. In all cases, and regardless of the assumptions regarding the number of generations per year or effective population size, the time to lose a neutral polymorphic allele in the ancestral species is much smaller than the divergence time between any of the species pairs of Histoplasma. (None of these calculations incorporates the possibility of trans-specific balancing selection.) These results suggest that any shared ancestry among Histoplasma species is most likely caused by introgression and not by incomplete lineage sorting.
TABLE 2 .
Approximate divergence times for four nodes in the tree shown in species pairs of Histoplasma
| Node/speciation event | MYA (CI)a | No. of generations since speciation (G)b: |
||
|---|---|---|---|---|
| 1 GPY | 10 GPY | 100 GPY | ||
| NAm 1/NAm 2 | 1.750 (1.37–2.12) | 1.75 × 106 | 1.75 × 105 | 1.75 × 104 |
| (NAm 1/NAm 2), LAm | 2.010 (1.76–2.26) | 2.01 × 106 | 2.01 × 105 | 2.01 × 104 |
| [(NAm 1/NAm 2), LAm], Panama | 2.110 (1.99–2.23) | 2.11 × 106 | 2.11 × 105 | 2.11 × 104 |
| {[(NAm 1/NAm 2), LAm], Panama}, Africa | 2.400 (2.19–2.61) | 2.40 × 106 | 2.40 × 105 | 2.40 × 104 |
MYA, million years ago; CI, confidence interval.
G, expected number of generations. We assumed three different numbers of generations per year (GPY) to convert from divergence time to number of generations.
Identified introgressions between Histoplasma species are unlikely to represent ancestral variation that is still segregating in the recipient species. Each panel shows the expected number of generations that an allele at a given frequency, p, would take to either be fixed (black line) or lost (red line) from the population. Horizontal lines denote the number of populations since the two species diverged, assuming 1 generation per year (solid blue line), 10 generations per year (dashed blue line), or 100 generations per year (dotted blue line). (A) Ne = 106. (B) Ne = 105. (C) Ne = 104. Note y axes are in log scale. Download FIG S2, PDF file, 0.03 MB (27.7KB, pdf) .
Copyright © 2017 Sepúlveda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The cryptic species of Histoplasma exchange genes rarely.
Species are genetic clusters discontinuous from each other. We tested whether the putative species of Histoplasma exchange genes with each other using three different approaches: (i) assessing the most likely number of admixture events in the tree using TreeMix, (ii) calculating the amount of introgression in different species pairs, and finally (iii) identifying potentially admixed individuals.
(i) Admixture events.
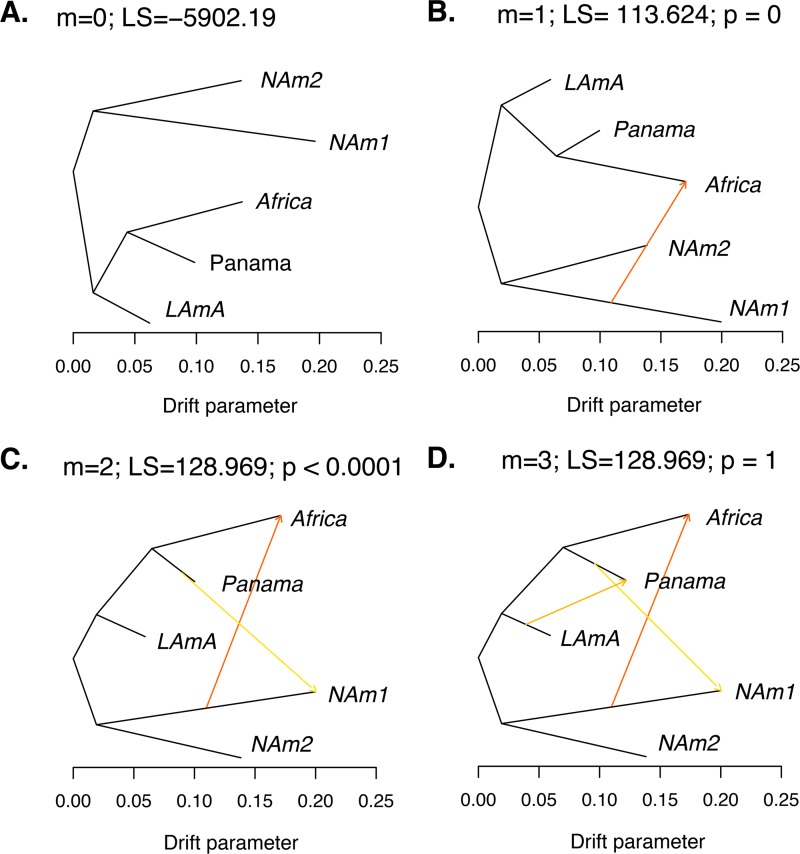
We measured the extent of gene exchange between phylogenetic species. To this end, we used the program TreeMix to identify instances of gene flow between Histoplasma species. TreeMix uses deviations from the genetic covariance predicted by a tree-like pattern of diversification to identify large events of gene exchange between species. This unrooted genealogy without accounting for gene flow (Fig. 5A) perfectly corresponds to our phylogenetic trees. We next sequentially added migration events and assessed whether they improved the likelihood of the tree. We found two migration edges were the most likely mixture scenario with both edges involving the NAm 1 branch (Fig. 5C). The most likely migration edge indicates the existence of a gene exchange event from NAm 1 to Africa (Fig. 5B and C; weight, 0.451). The second edge shows a milder migration event and indicates the migration of genetic material from Panama to NAm 1 (Fig. 5C; weight, 0.234). We inferred the same scenario using a likelihood ratio test (LRT) to assess the most likely number of migration edges or by weighted Akaike information criterion (wAIC [one migration edge versus two migration edges]) (Fig. 5B and C; Table 3). Addition of an additional migration event did not improve the log likelihood (two migration edges versus three migration edges) (Fig. 5D; Table 3). The drift parameter, which is proportional to the age of the migration, revealed that the two genetic exchange events are unlikely to be recent.
FIG 5 .
TreeMix results show genetic differentiation between species from the Histoplasma genus. (A) Best-fitting genealogy without admixture for Histoplasma species calculated from the variance-covariance matrix of genome-wide allele frequencies. We estimated the likelihood of three demographic scenarios with 1 to 3 migration events (m). The caption for each panel shows the number of migrations (m), the LS of the demographic scenario, and the P value (p) of the comparison between that model and the model with m−1 migration events (also shown in Table 3). (B) m = 1. (C) m = 2. (D) m = 3. To determine the best-fitting demographic scenario, we used LRTs to compare nested models (one migration versus two migrations and two migrations versus three migrations). We also used wAIC to find the best-supported migration scenario (see text). The caption for each panel contains the number of migrations, the likelihood, and the associated P value when comparing that demographic scenario with the previous one. The best-fitting demographic history involved two migration events: one from NAm 1 to Africa and a second one from Panama to NAm 1 (B).
TABLE 3 .
A demographic scenario that involves two migration events is the most likely explanation for the partition of allele frequencies across the genus Histoplasmaa
| m | LS | LRT | P value | AIC | wAIC |
|---|---|---|---|---|---|
| 0 | −5,902.19 | 11,804.38 | 2.431 × 10−49 | ||
| 1 | −5,845.378 | 113.624 | 0 | 11,692.76 | 4.205 × 10−25 |
| 2 | −5,788.566 | 128.969 | 2.17 × 10−7 | 11,581.13 | 0.731 |
| 3 | −5,788.566 | 0 | 1 | 11,583.13 | 0.269 |
As shown in Fig. 5C. All calculations were done with the model implemented in the package TreeMix. m, number of migration edges; LS, likelihood score; LRT, likelihood ratio test performed with the model that involved one fewer migration event; P value, level of significance of the LRT with the previous model (i.e., m versus m−1) using χ2 with 1 df; AIC, Akaike information criterion; wAIC, weighted AIC for each model.
(ii) Potential introgression.
We corroborated the results from TreeMix by assessing occurrence of ancestral admixture using the ABBA/BABA D statistic. This approach also rules out the possibility that shared ancestry is caused by incomplete lineage sorting. We tested three hypotheses: (i) that there has been introgression between NAm 1 and Africa, (ii) that there has been migration between NAm 2 and Panama, and (iii) that there has been introgression between NAm 1 and NAm 2. The reasoning for the first two migration events was the strong suggestion of gene exchange from TreeMix, while the third event was tested because these are the most closely related species in the genus and also the ones that cooccur. All these events were corroborated with the D statistic, which suggests introgression between these three species pairs (Table 4). Notably, the D statistic detects introgression between NAm 1 and NAm 2, which was not detected by TreeMix. The reasons for this incongruence remain unknown but might be related to the larger sample size of NAm 1 and NAm 2, which might reveal introgressions at low allele frequency, which are not detected by TreeMix.
TABLE 4 .
D statistic variations and fd show evidence for admixture between three species pairs of Histoplasmaa
We next used the fd statistic (55) to estimate the proportion of the donor genome that had introgressed from recipient in the three gene exchange scenarios described above. We found that on average, the proportion of the genome that has been introgressed across species boundaries is very small (less than 5% of the genome [Table 4]). This proportion was roughly equivalent regardless of the used outgroup. More work and a fine dissection of the precise allele frequency and haplotype size will be required to assess whether these low levels of introgression are caused by low allele frequencies (i.e., a few individuals are admixed), by small haplotypes (i.e., introgression might be common but small), or both.
(iii) Admixed individuals.
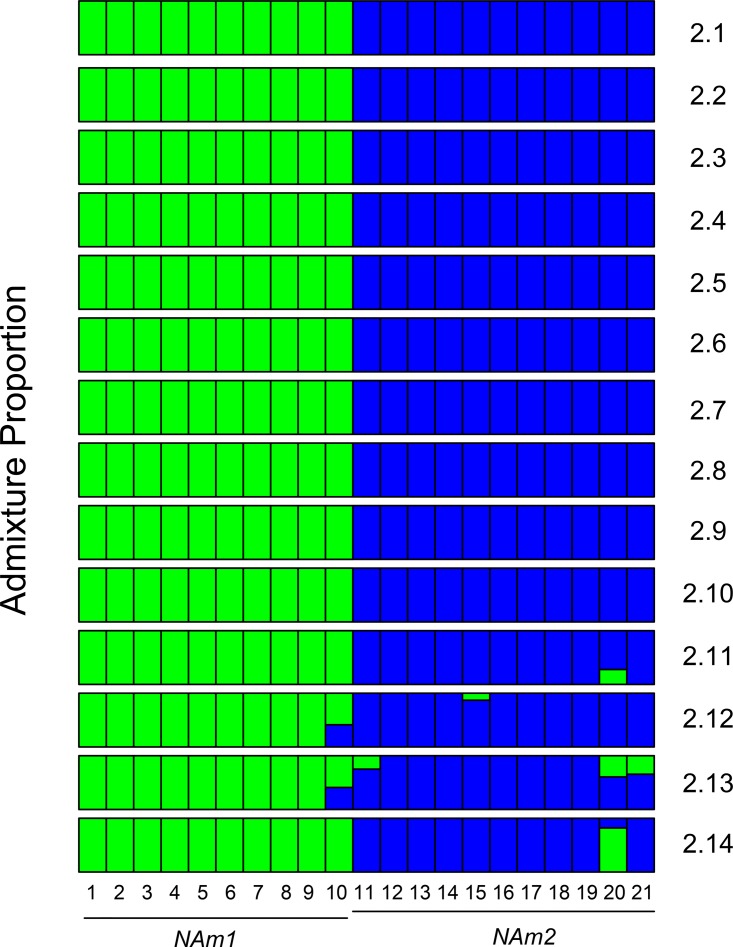
Finally, we assessed whether we could detect specific individuals with mixed ancestry. We focused on the most closely related species pair, which was also the pair with the largest population sample individuals: NAm 1/NAm 2. (The currently small sizes and deep divergence between conspecific isolates of the Africa, Panama, and LAmA samples preclude the possibility of this analysis.) We used ADMIXTURE to identify if any of the individuals from these two species showed evidence of being hybrids. The results from this analysis are largely congruent with the findings from our phylogenetic analyses. WU24, a NAm 1 isolate, showed conflicting positions in the genealogies from different supercontigs (supercontigs 2.12 and 2.13). This isolate also show evidence of being admixed in the ADMIXTURE analysis (Fig. 6). Notably, ADMIXTURE indicates the possibility of other admixed individuals in NAm 2. 1986, CI30, and G222B show possible admixture in one of the smaller supercontigs (2.11, 2.12, 2.13, or 2.14). G217B shows evidence of admixture in supercontigs 2.11, 2.13, and 2.14.
FIG 6 .
ADMIXTURE results for NAm 1 and NAm 2. ADMIXTURE proportions among NAm 1 and NAm 2 isolates (K = 2). Each of the two clusters is represented by a different color, and each isolate is represented by a vertical line divided into two colored segments with heights proportional to genotype memberships in the clusters. Thin black lines separate isolates. 1: 505; 2, CI_19; 3, CI_22; 4, CI_24; 5, CI_42; 6, CI_43; 7, CI_7; 8, Downs; 9, UCLA_531; 10, WU24; 11, 1986; 12, CI_10; 13, CI_17; 14, CI_18; 15, CI_30; 16, CI_35; 17, CI_4; 18, CI_6; 19, CI_9; 20, G217B; 21, G222B. We ran independent analyses for the largest 14 supercontigs, indicated by the number on the right. Results from the whole-genome analyses are concordant with the results from supercontigs 2.1 to 2.10.
Taken together, our results indicate that our putative species of Histoplasma represent genetically distinguishable, independently evolving lineages that seldom exchange genes, in some cases even while coexisting in sympatry. They should, therefore be elevated to species status.
Systematics.
Based on our results, we propose to restrict the name H. capsulatum to the Panamanian lineage, as this species was originally described in the Canal Zone (1), and describe three additional species, corresponding to the two North American and South American lineages. Our results also indicate that H. capsulatum var. duboisii (Africa) is probably a separate species from H. capsulatum. However, given that our sampling is limited to two clonal isolates, we refrain from modifying this variety’s taxonomic status for the sake of taxonomic stability. Wider and more systematic strain sampling and exploration of its genetic diversity need to be achieved before confidently defining its taxonomic status.
(i) Histoplasma capsulatum sensu stricto Darling 1906.
Histoplasma capsulatum sensu stricto Darling 1906. Mycobank accession number to be determined (TBD). Formerly known as the Panama or H81 lineage (31). Mycelial colonies on Histoplasma-macrophage medium (HMM) are white and cottony. Yeast colonies are variable in size, cream-colored (white to brown) on HMM agar. Yeast colonies are rough due to the presence of α-(1,3)-glucan in their cell wall. Histoplasma capsulatum can be discriminated from other Histoplasma species by PCR amplification, restriction fragment length polymorphism (RFLP) (mitochondrial DNA [mtDNA] and ysp3) (56), and randomly amplified polymorphic DNA (RAPD) (57, 58). The DNA fragment of α-tubulin amplified with the forward primer 5′-GGTGGCCAAATCGCAAACTC-3′ and reverse primer 5′-GGCAGCTTTCCGTTCCTCAGT-3′ (31) harbors 4 diagnostic single nucleotide polymorphisms (SNPs) of H. capsulatum (positions 1328, 1364, 1469, and 1575). Histoplasma capsulatum also has one diagnostic SNP at ADP-ribosylation factor (position 10; forward primer 5′-AGAATATGGGGCAAAAAGGA-3′ and reverse primer 5′-CGCAATTCATCTTCGTTGAG-3′), one at the H antigen precursor (position 710; forward primer 5′-CGCAGTCACCTCCATACTATC-3′ and reverse primer 5′-GCGCCGACATTAACCC-3′), and one at delta-9 fatty acid desaturase (position 1134; forward primer 5′ TTTAAACGAAGCCCCCACGG 3′ and reverse primer 5′-CACCACCTCCAACAGCAGCA-3′). Histoplasma capsulatum shows differential virulence in in vitro (macrophage) and in vivo (murine model [35]) assays compared to the two North American species.
Holotype: strain G186A. Isolated from a human patient in Panama in 1967 or before (31, 59, 60).
Etymology: We propose to restrict the use of H. capsulatum for the Panama group, as the organism was first discovered and described in Panama (1).
(ii) Histoplasma mississippiense sp. nov.
Histoplasma mississippiense sp. nov. (MycoBank accession number MB823360). Formerly known as NAm 1. Mycelial colonies on HMM are white and cottony. Yeast colonies are variable in size, cream-colored (white to brown) on HMM agar. Yeast colonies are rough due to the presence of α-(1,3)-glucan in their cell wall. No sexual stage is known. The DNA fragment of α-tubulin amplified with the forward primer 5′-GGTGGCCAAATCGCAAACTC-3′ and reverse primer 5′-GGCAGCTTTCCGTTCCTCAGT-3′ (28) harbors 7 diagnostic SNPs of H. mississippiense (positions 1385, 1415, 1426, 1447, 1506, 1532, and 1556). Histoplasma mississippiense also has three diagnostic SNPs at ADP-ribosylation factor (positions 126, 235, and 310; forward primer 5′-AGAATATGGGGCAAAAAGGA-3′ and reverse primer 5′-CGCAATTCATCTTCGTTGAG-3′), four at the H antigen precursor (positions 504, 620, 640, and 740; forward primer 5′-CGCAGTCACCT CCATACTATC-3′ and reverse primer 5′-GCGCCGACATTAACCC-3′), and five at delta-9 fatty acid desaturase (positions 943, 1080, 1193, 1195, and 1206; forward primer 5′-TTTAAACGAAGCCCCCACGG-3′ and reverse primer 5′-CACCACCTCCAACAGCAGCA-3′) (28). Additionally, H. mississippiense can be discriminated from other Histoplasma species by RFLP (mtDNA and ysp3 [58]) and RAPD (57). Histoplasma mississippiense also uses distinctive chymotrypsin-like serine proteases, which facilitates extracellular proteolytic activity (61). These enzymes are not present in any other Histoplasma species and are a diagnostic trait for H. mississippiense.
Holotype: Downs strain. Isolated in 1968 from an octogenarian woman with disseminated disease. Ex-type culture is preserved in the American Type Culture Collection (ATCC 38904).
Etymology: mississippiense; refers to the distribution of the species in the North American continent and the high density of its occurrence in the Mississippi Valley (7).
(iii) Histoplasma ohiense sp. nov.
Histoplasma ohiense sp. nov. (MycoBank accession number MB823361). Formerly known as NAm 2. Mycelial colonies on HMM are white and cottony. Yeast colonies are variable in size, cream-colored (white to brown) on HMM agar. Yeast colonies are smooth due to the lack of α-(1,3)-glucan in their cell wall. No sexual stage is known. The DNA fragment of H antigen precursor amplified with the forward primer 5′-CGCAGTCACCTCCATACTATC-3′ and reverse primer 5′-GCGCCGACATTAACCC-3′ harbors seven diagnostic SNPs of H. ohiense (positions 631, 648, 654, 758, 782, 814, and 837). Histoplasma ohiense also has four diagnostic SNPs at ADP-ribosylation factor (positions 917, 942, 1026, and 1200; forward primer 5′-AGAATATGGGGCAAAAAGGA-3′ and reverse primer 5′-CGCAATTCATCTTCGTTGAG-3′), three at α-tubulin (positions 1369, 1473, and 1493; forward primer 5′-GGTGGCCAAATCGCAAACTC-3′ and reverse primer 5′-GGCAGCTTTCCGTTCCTCAGT-3′), and four at delta-9 fatty acid desaturase (positions 917, 942, 1037, and 1206; forward primer 5′-TTTAAACGAAGCCCCCACGG 3′ and reverse primer 5′-CACCACCTCCAACAGCAGCA-3′) (28). Additionally, H. ohiense can be discriminated from other Histoplasma species by RFLP (mtDNA and ysp3 [56]) and RAPD (57, 58). Multiple studies have shown that Histoplasma ohiense is much more virulent than its sister species Histoplasma mississippiense (35). Histoplasma ohiense strains, unlike other Histoplasma species, have a smooth colony morphology and are virulent despite the lack of α-(1,3)-glucan in their cell wall. α-(1,3)-Glucan is a well-studied virulence factor in H. capsulatum that gives colonies a rough morphology. NAm 2 uses Yps3p, a homologue of the Blastomyces dermatitidis adhesin Bad1p, as a yeast-specific virulence factor. Notably, Yps3p is only essential for virulence in H. ohiense and not in the other species (62).
Holotype: strain G217B. Isolated in Louisiana, United States, in 1973 or before (31, 59, 60), from a human patient. Ex-type culture is preserved in the American Type Culture Collection (ATCC 26032).
Etymology: ohiense; refers to the distribution of the species in the North American continent and the high prevalence of the disease it causes in the Ohio Valley (8).
(iv) Histoplasma suramericanum sp. nov.
Histoplasma suramericanum sp. nov. (MycoBank accession number MB823362). Formerly known as LAm A.
Histoplasma suramericanum can be discriminated from other cryptic species of Histoplasma by PCR amplification. The DNA fragment of H antigen precursor amplified with the forward primer 5′-CGCAGTCACCTCCATACTATC 3′ and reverse primer 5′-GCGCCGACATTAACCC-3′ (28) harbors three diagnostic SNPs of H. suramericanum (positions 591, 622, and 716). Histoplasma suramericanum also has one diagnostic SNP at ADP-ribosylation factor (position 154; forward primer 5′-AGAATATGGGGCAAAAAGGA-3′ and reverse primer 5′-CGCAATTCATCTTCGTTGAG-3′), and one at delta-9 fatty acid desaturase (position 1276; forward primer 5′-TTTAAACGAAGCCCCCACGG-3′ and reverse primer 5′-CACCACCTCCAACAGCAGCA-3′). Unlike H. mississippiense and H. ohiense, which cause a chronic pulmonary disease, Histoplasma suramericanum causes an acute pulmonary disease that leads to a higher patient mortality (63). Histoplasma suramericanum also causes a more pronounced lung pathology than H. ohiense (63).
Holotype: strain MZ5. Strain isolated in the Department of Antioquia (Colombia) in 2010 (Corporación para Investigaciones Biologicas [CIB] no. 15297).
Etymology: suramericanum; refers to the distribution of the species on the South American continent (31).
DISCUSSION
Cryptic speciation in Histoplasma.
Previous studies have suggested extensive differentiation among genetic clusters of Histoplasma (28, 31). Up to 15 clusters have been proposed, but no taxonomic changes have been made, as these reports were based on a handful of loci. In particular, Kasuga et al. (31) reported the existence of strong genetic discontinuities in Histoplasma. To date, this has remained the most systematic effort to understand the partition of genetic variation in Histoplasma, proposing seven phylogenetic species. Such efforts were seminal to set the stage to eventually identify cryptic species in Histoplasma but fell short for two reasons. First, they identified genetic groups but left “orphan” isolates that did not belong to any phylogenetic species, thus making H. capsulatum a polyphyletic group. This is problematic because under a phylogenetic species recognition framework, all individuals must belong to monophyletic groups (40, 41). Second, the limited genetic sampling (4 coding loci) provided only a partial portrait of the amount of genealogical diversity and discordance across Histoplasma. Using whole-genome sequences of 30 Histoplasma isolates from five of the seven phylogenetic groups found by Kasuga et al. (31), we find enough phylogenetic structure and genetic differentiation between these clusters to consider them separate phylogenetic species. Moreover, our analyses suggest that some hybridization and gene flow has occurred between species, but the magnitude of gene exchange is too small to preclude species boundaries.
The recognition of phylogenetic species is not devoid of caveats, as it does not explicitly assess the extent of reproductive incompatibility between species. It is possible that putative phylogenetic species actually represent genetic clusters formed by extrinsic (e.g., geographic) isolation, but which are still reproductively compatible. This issue might be the main limitation in the description of phylogenetic species; differentiation between cryptic speciation and local population structure is an issue especially pronounced when only a few loci are used (64, 65). It could be argued our approach might have a similar caveat because it is tailored to detect groups of isolated isolates using molecular markers. Yet, one would not expect congruent genealogies, reciprocal monophyly, and low levels of gene flow in locally structured populations at a genome-wide level (65, 66), and that is why our phylogenomic approach is superior.
Previous reports have evaluated the robustness of the criteria to identify phylogenetic species in fungi and have found that phylogenetic species recognition also predicts the boundaries dictated by the biological species concept. Yet, the origin of reproductive isolation is taxon specific, and a formal test of the suitability of the biological species concept in Histoplasma is still to be performed. However, we can unambiguously discard this scenario for at least one of the species pairs, H. mississippiense/H. ohiense, as their geographic distributions are largely overlapping, and they have been found in syntopy (28, 31). The existence of strong genetic differentiation despite the possibility to interbreed, facilitated by geographic and epidemiological overlap, indicates that these two lineages have speciated, since, in the absence of intrinsic reproductive isolation the observed genetic structure would have already broken down. This may also be the case for H. suramericanum and H. capsulatum, as their distributions overlap in central and possibly northwestern Colombia. However, no hybrid zones have been reported, although this could be due to sparse sampling of potential contact zones. The observation that most individual, unlinked, supercontigs, as well as 100-kb genomic windows, strongly support our proposed species delimitation further adds to the idea that the different genetic clusters within Histoplasma are not the result of early population structure but strongly imply that diversification in Histoplasma is at an advanced stage of divergence, probably past the point of speciation.
Shared ancestry between species of Histoplasma.
Speciation is an evolutionary process that facilitates the segregation of genetic variation between species and makes genetic clusters differentiated from others. We found evidence of small levels of shared ancestry in three species pairs of Histoplasma. Two possible scenarios might explain this shared genetic material. First, introgression (i.e., the genetic movement of alleles across species boundaries) through the formation of hybrids (through interspecific crossings or somatic fusion) might be a cause for the sharing of alleles. Meta-analyses have estimated that up to 10% of animal and plant species might be able to hybridize (67). The extent of hybridization and introgression remains unknown for fungi. A second possibility is that alleles that segregate in both species predate speciation (50). This phenomenon known as incomplete lineage sorting occurs because reciprocal monophyly across the genome is not an instantaneous process and ancestral alleles might remain postspeciation. Three of our results indicate that there is little chance of ancestral polymorphism across species of Histoplasma and strongly suggest the possibility of low rates of introgression. First, the age of divergence of each of the species pairs indicates that incomplete lineage sorting is an unlikely event. The most recent speciation event in Histoplasma is still old; regardless of the assumptions of the time between recombination events, NAm 1 and NAm 2 diverged 1.7 million years ago. This old divergence suggests there is little chance for segregating ancestral polymorphism in the genome of these two sister species. (The likelihood of segregating ancestral polymorphism between any of the other species is even lower.) The second line of evidence is that the vast majority of detectable shared ancestry colocalizes to the same regions in the genome, which might suggest that shared SNPs are clustered in haplotypes possibly caused by introgression. Finally, the results from our D statistic analyses argue against this possibility and are consistent with introgression. Nonetheless, the testing of this hypothesis will need to wait until shared alleles are finely mapped in the genome to identify the size of the shared haplotypes as potential introgressions should show larger haplotype sizes than ancestral polymorphisms. More widespread sampling is sorely needed before we can assess the possibility of shared ancestry and its causes among other Histoplasma species.
The diversification of the genus Histoplasma.
Our results also have important implications for the understanding of how fungal pathogens evolve. First, they contest the most recent available biogeographical hypothesis on the diversification of Histoplasma and the potential drivers of diversification in fungal pathogens. Teixeira et al. (20) argued that Histoplasma has experienced an accelerated rate of diversification in the last 4 million years, which might have been aided by dispersion of propagules by bats (also explored in references 17 and 68. Even though such a radiation—and the role of bats on it—remains a possibility that cannot be ruled out, our results lend weak evidence for this assertion. The phylogenetic relationships between Histoplasma spp. inferred from this multilocus sequence typing effort are not congruent with the phylogenomic information shown here. This type of inconsistency has been observed in other clades in which multilocus sequence typing can lead to incorrect phylogenetic relationships (65). Such a large overhaul of the understanding of the phylogenetic relationships among Histoplasma isolates renders all phylogenetic inference based on the erroneous phylogeny invalid.
The phylogenetic tree we produced also suggests the existence of a clade composed exclusively by American species of Histoplasma, with an African clade diverging early. It is premature to argue for (or against) the occurrence of an adaptive radiation after the colonization of the American continent. Before such hypothesis can be formally evaluated, it will be necessary to include isolates from a second Latin American group (LAm B) and evaluate the possibility of additional cryptic species within the clade. Moreover, samples of all the other localities where Histoplasma has been isolated (e.g., Eurasia, Australia, and Africa) are sorely needed to assess rates of diversification in Histoplasma. In general, a more extensive isolate collection will allow for testing the tempo and mode of evolution in the Histoplasma genus.
These results (and previous hypotheses) beg the question of what are the actual factors that have led to diversification in the Histoplasma genus. Dispersion by bats has been hypothesized to increase the geographic range of fungal pathogens and thus increase the likelihood of allopatric speciation. A robust analysis of coevolution between mammal reservoirs and different Histoplasma species would allow to quantify net diversification rates (speciation and extinction) and establish what have been the biological features leading to the existence of multiple species of Histoplasma and perhaps multiple virulence strategies in the genus. The current sampling in Histoplasma is too premature to perform this type of analyses, but our results provide a clear direction to identify the factors that underlie diversification in fungal human pathogens.
Health implications.
These results also have epidemiological implications, as genomic differentiation among Histoplasma species could lead to differences in important disease traits. The differences in virulence and drug resistance among species seem to confirm this notion. For example, one of the North American species (H. mississippiense) is less virulent than the others (35, 58) but is more resistant to regularly used antifungals (69) and shows unique extracellular proteolytic activity (61). Another example is α-(1,3)-glucan, a well-studied virulence factor in dimorphic fungi that allows Histoplasma to escape host immune responses (70, 71). Despite the fact that the other North American species, H. ohiense, lacks α-(1,3)-glucan (present in the rest of the Histoplasma species), it is still virulent and is optimized for greater virulence in high-dose infections (35). The absence of α-(1,3)-glucan in H. ohiense strains could have selected for additional adaptations, and coevolution of alternative virulence factors may have arisen to compensate for the lack of this polysaccharide; e.g., Yps3p, a homologue of the B. dermatitidis adhesin gene Bad1p (72), is a virulence factor restricted to H. ohiense (61, 73). Future studies should address the precise rate of gene exchange across species. Even if these events of gene exchange are rare, they deserve to be evaluated closely. Admixture might serve as a mechanism to generate pathogens with new trait combinations that could pose an additional threat to human health.
A special case is that of H. capsulatum var. duboisii, also known as Africa. This group is highly differentiated from all the American species of Histoplasma. Additionally, this clade has been associated not with a lung disease, but with granulomatous lesions predominantly in subcutaneous, cutaneous, and osseous tissue (36, 74). Even though this epidemiological assessment might be premature as there have been no controlled infection experiments to compare disease caused by all five species, the occurrence of a new type of disease would indicate the evolution of a new virulence strategy within the genus Histoplasma. Moreover, we present evidence that even though this group is highly differentiated from all American Histoplasma species, Africa (H. capsulatum var. duboisii) might rarely exchange genes with H. mississippiense. This strong genetic differentiation and the ability to rarely exchange genetic material might in turn facilitate the transmission of virulence syndromes across species boundaries and across large geographic ranges. We do not propose a formal taxonomic elevation of this group to species as we only collected data for two isolates. We do expect that a systematic sample collection of Africa (H. capsulatum var. duboisii) will lead to the formal description of a new species.
Conclusions and future directions.
Systematic explorations of phenotypic differences have revealed that Histoplasma encompasses a large phenotypic differentiation in terms of virulence, morphology, and pathogenicity strategies (35, 69) (described above). Here we have demonstrated that these phenotypic differences coincide with phylogeny and genetic differentiation and are probably the result of previously unidentified speciation events within Histoplasma. The identification of genetically isolated species represents a starting point to understand what genetic changes underlie medically important differences. Disease burden estimates as well as virulence studies are needed to recognize the incidence and phenotype of the diseases caused by each of the four formally described species here.
Unlike other dimorphic fungal pathogens, such as Paracoccidioides and Coccidioides, Histoplasma might hold the potential of producing sexual progeny in the lab. Even though this is not a trivial feat, Emmonsiella, the sexual stage of Histoplasma, can be produced in in vitro crosses (27, 75, 76). A possibility is to assess whether different species of Histoplasma can produce fertile progeny at the same rate as intraspecific crosses, which would also reveal what biological features might be involved in keeping species apart. This is of particular interest given that the species of Histoplasma show different levels of divergence, which in turn might represent different stages of diversification within the genus. This type of analysis might make Histoplasma a paradigm in the study of the evolution of reproductive isolation in ascomycetous pathogenic fungi. Of particular interest is the report that North American isolates of Histoplasma (likely to be NAm 1 or NAm 2) are able to produce sexual structures (cleistothecia) and mature ascospores when crossed to Africa (77). These groups are quite divergent, suggesting but not confirming that hybrid inviability might not occur readily in interspecific crosses of Histoplasma. This hypothesis needs to be tested with Histoplasma isolates of known taxonomic affinity and with recently collected isolates, as old isolates do not readily mate (75).
With the classification of different species showing strong genetic differentiation and displaying morphological and virulence differences, systematic analysis of worldwide isolate collections will be crucial to disentangle the ecology, evolution, and human incidence of each of these species. These efforts will help link how the molecular evolution of important virulence factors will ultimately impact the pathogenicity of different Histoplasma species and the evolution of other virulence factors.
MATERIALS AND METHODS
Isolate collection.
We gathered 30 Histoplasma isolates representing four recognized areas where histoplasmosis is endemic: 21 from North America (10 NAm 1 isolates, 11 NAm 2 isolates), 7 from Latin America (4 LAm A isolates, 3 Panama isolates), and 2 from the group Africa. Since all analyses reveal the existence of five clusters (see Results) congruent with the groups described by Kasuga et al. (31), we use the names proposed for each population by those authors: NAm 1 (North America 1), NAm 2 (North America 2), LAm A (Latin America A), Africa (Africa [H. capsulatum var. dubosii]), and Panama (Panama [H81 lineage]). Isolates were donated to W. Goldman during a span of 15 years. A list of donors and details for each strain is given in Table S6 at https://doi.org/10.5061/dryad.006bf. All isolates were kept in 15% glycerol at −80°C until they were ready to be subcultured and sequenced. Strains were grown in HMM (solid or liquid) at 37°C with 5% CO2 as described previously (5, 78). Solid medium contained 0.8% agarose (SeaKem, ME grade) and 25 mM FeSO4. All further analyses included only isolates for which we could obtain genomic sequences and which were available for further physiological, immunologic, and morphological experiments.
DNA extraction.
We extracted DNA from the 30 Histoplasma isolates (described above). Two hundred microliters of a solution containing 1 mM EDTA, 10 mM Tris (pH 8.0), 1% SDS, 2% Triton X-100, and 100 mM NaCl was added to the cell pellet, along with 200 μl of glass beads. Cells were broken by vortexing for 2 min (split into intervals of 30 s with 30 s of incubation on ice in between), followed by incubation at 65°C for 20 min. DNA was isolated using phenol-chloroform precipitation, followed by RNase treatment with 2 μl of 2 mg/ml RNase A (Sigma, St. Louis, MO) at 37°C for 1 h and a second phenol-chloroform precipitation. The integrity of the DNA was examined by running an aliquot in a 1.8% agarose gel with a DNA mass ladder and visualized with ethidium bromide and UV light.
Library construction.
Genomic libraries were built from high-molecular-weight DNA following the protocol described immediately above (section “DNA extraction”), using the Kappa protocol for TruSeq at the sequencing facility of the University of North Carolina, Chapel Hill. Approximately 10 µg of DNA was sonicated with a Covaris S220 to a mean fragment size of 160 bp (range, 120 to 200 bp) with the following program: 10% duty cycle at intensity 5, 100 cycles per burst, and 6 cycles of 60 s in frequency sweeping mode.
Sequencing.
We sequenced all libraries on Illumina HiSeq 2000 machines with v3.0 chemistry following the manufacturer’s instructions. The sequencing type (single end or paired end) and coverage for each library are not shown. Libraries were pooled prior to sequencing, and 6 libraries were sequenced per lane. If all genomes were of equivalent size, then we should expect homogenous coverage across lines. We assessed the quality of the individual reads using the HiSeq Control software 2.0.5 in combination with RTA 1.17.20.0 (real-time analysis). CASAVA-1.8.2 generated and reported run statistics of each of the final FASTQ files. Resulting reads ranged from 100 to 150 bp, and the target average coverage for each line was 30×. The coverage for each isolate is not shown.
Read mapping and variant calling.
Reads were mapped to the Histoplasma capsulatum genome H. capsulatum var. duboisii H88 (https://www.ncbi.nlm.nih.gov/bioproject/29163; also deposited on Dryad [https://doi.org/10.5061/dryad.006bf]), currently assembled into 16 supercontigs using bwa version 0.7.12 (79). Histoplasma strains vary in their karyotype and are thought to have between five and seven chromosomes, depending on the strain (80, 81). The resulting bam files were merged using Samtools version 0.1.19. Indels were identified and reads locally remapped in the merged bam files using the GATK version 3.2-2 RealignerTargetCreator and IndelRealigner functions (82, 83). Subsequently, SNPs were called using the GATK UnifiedGenotyper function with the parameter “het” set to 0.01 and all others left as default. The following filters were applied to the resulting vcf (variant call format) file: QD = 2.0, FS_filter = 60.0, MQ_filter = 30.0, MQ_Rank_Sum_filter = −12.5, and Read_Pos_Rank_Sum_filter = −8.0. Sites were excluded if the coverage was less than 5 or greater than the 99th quantile of the genomic coverage distribution for the given line or if the SNP failed to pass one of the GATK filters. Read coverage among species was compared with a one-way ANOVA followed by post hoc Tukey tests.
Public data.
To root our trees, we obtained sequencing reads from two species of Emmonsia, E. crescens UAMH3008 and E. parva UAMH139, from the NCBI SRA (accession no. PRJNA178252 and PRJNA178178, respectively [84]). These species are among some of the closest relatives of Histoplasma (84).
PCA.
To assess the extent of between-isolate genetic differentiation, we used principal-component analysis (PCA), a general method for summarizing high-dimensional data, as is the case of genome-wide individual genotypes. The PCA transforms a set of possibly correlated variables into a reduced set of orthogonal variables. Sampled individuals are then projected in a two-dimensional space, where the axes are the new uncorrelated variables, or principal components. Closely related individuals will form clusters, thus revealing population relationships or within-population stratification. The allele frequencies were simply the counts of each allelic class inferred from the VCF files as Histoplasma is haploid (49). We next calculated the proportion of genetic variance explained by each principal component (PC) and plotted two PC combinations—PC1 and PC2/PC2 and PC3/PC4—that explain the majority of the genetic variation in Histoplasma. All PCAs were done with the R package adegenet (85).
LD.
We measured linkage disequilibrium (LD) within these five groups to explore the extent of recombination in Histoplasma. We used r2 as an LD metric, and estimated it using PLINK v.1.90b4.5. r2 ranges between 0 and 1, indicating completely independent segregation of alleles or complete linkage, respectively. For computational feasibility, we restricted our analysis to 1,000,000 randomly selected SNPs and only estimated LDs between those separated by 500 kb or less. PLINK was run with the following parameters: plink—allow-extra-chr—r2—ld-window-kb 1000—ld-window 99999—ld-window-r2 0.0. Finally we estimated the mean r2 between sites separated by physical distances, grouped in 10-bp bins (i.e., r2 between sites separated by 1 to 10 bp, 11 to 20 bp, 21 to 30 bp, etc., were averaged together).
Phylogenetic reconstructions.
Our goal was to determine whether the genetic clusters found in Histoplasma satisfy the requirements to be considered phylogenetic species. We followed a phylogenetic species concept (45, 86) to recognize species, defining species as genetic clusters that are reciprocally monophyletic and for which there was genealogical concordance across genome-wide unlinked loci. We used two parallel approaches: (i) maximum likelihood (ML) trees at the genome and at the supercontig level, and (ii) Bayesian concordance analysis of the genealogies of 100-kb genomic windows.
(i) ML phylogenetics.
To gain initial insight on the phylogenetic relationships among our sequenced isolates, and especially to explore the degree of monophyly of our putative species, we used traditional maximum likelihood (ML) phylogenetics. Reciprocal monophyly between two putative species is unlikely to be observed if they have had the chance to interbreed and exchanged genes extensively (50, 87). Given the low rates of recombination found in Histoplasma (31) and the high levels of linkage disequilibrium within all putative species (Fig. S1), we used whole supercontigs as unlinked loci. We focused exclusively on nuclear genomes as mitochondrial genomes have the propensity to be affected by horizontal gene transfer, thus obscuring species relationships (88, 89). We obtained whole supercontig sequences for each individual from the VCF file using the FastaAlternateReferenceMaker tool in GATK, realigned them using Mafft v.7 (90), and used them to build ML trees using RAxML version 8.2.9 (91). We inferred individual trees for each supercontig, except for 15 and 16, which were found to be completely monomorphic, as well as a “genome-wide” tree of a concatenated alignment of all supercontigs. Analyses were run under the GTR+Γ model, with 1,000 bootstrap pseudoreplicates to assess nodal support. The “genome-wide” analysis was partitioned by supercontig, with each partition having its own set of GTR+Γ model parameters, but sharing a common topology and branch lengths. All trees were rooted using Emmonsia parva and E. crescens, which are thought to have diverged from Histoplasma close to 100 million years ago (92).
(ii) Bayesian concordance analyses.
To shed light on the extent of genealogical concordance at a finer scale in the genome, we performed a Bayesian concordance analysis (BCA) using BUCKy v.1.4.4 (53, 93). BUCKy estimates the level of concordance among multiple genealogies and uses it to infer the tree with the most concordant nodes, which is considered an estimate of the species tree. The level of support for each node is expressed as a concordance factor (CF), which ranges between 0 (i.e., no concordance between genealogies) and 1 (complete concordance). Therefore, this algorithm allowed us to infer the phylogenetic relationships between our putative species, while estimating the genome-wide genealogical support for their monophyly.
This analysis has two steps. First, we split the supercontig alignments into nonoverlapping 100-kb windows, and used the parallel version of MrBayes v.3.2.6 (94, 95) to generate posterior tree distributions for each one. Supercontigs shorter than 100 kb were analyzed as a single locus, and supercontigs 15 and 16 were also left out of this analysis, given their lack of genetic variation. For each window, two independent Markov chain Monte Carlo (MCMC) analyses were run for 10 million generations, sampling every 1,000, and using four chains (three heated and one cold). Likelihood estimations were done under the GTR+Γ model. Convergence was evaluated using Tracer v.1.6 (96). Runs with split frequency standard deviations above 0.1 and/or effective sample size (ESS) scores below 200 for the likelihood or posterior traces (34 of 382) were excluded from further analyses. Posterior distributions were then summarized using the mbsum tool (part of BUCKy), skipping the first 1,000 trees (10%) as burn in, to generate input files for BUCKy. Finally, we ran two independent BUCKy chains for 2 million generations (200,000 additional burn in). BUCKy uses parameter α to describe the a priori level of gene tree incongruence. A larger value of α corresponds to greater gene tree incongruence—the probability that 2 genes share the same topology is approximately 1/(1 + α). At the two extremes, α = 0 indicates that all genes share the same tree, while α = ∞ indicates all genes have different topologies (93). We ran our analysis using several different α values (α = 0.1, 1, 3, 5, 7, 10, and infinity) to assess whether the choice of priors affected the results and obtained nearly identical results for all values of α. Therefore, we report the results obtained using α = 5. Output files generated with other values of α for this article are available in the Dryad repository.
Genetic distance.
To further assess the extent of genetic differentiation between phylogenetic species, we used the metric πinter, the average number of nucleotide differences between one sequence randomly chosen from a population and another sequence randomly chosen from a second population or species. πinter followed the form πinter ij = pairwise differences between individuals i and j/sequence length. Mean πinter was the mean of all pairwise comparisons between individuals from two species. We calculated 21 mean πinter values.
We also calculated πintra, the average pairwise genetic distance between individuals of the same species. πintra followed the same form as πinter, but instead of calculating the average number of differences between species, it calculates the average number of differences between two randomly selected individuals of the same species. Mean πintra was the mean within-species value for each of the five species. For both types of calculations, we used the function dna.dist (model=″raw") in the R package “ape” (97).
In cases where speciation is complete, πinter is expected to be much larger than πintra. For each species pair, πintra can take two values (i.e., from each of the two species), so for the pairwise comparisons πintra is the pooled set of the two intraspecific distances. To compare the values of πinter and πintra for each species pair (10 pairwise comparisons), we used two-sample Fisher-Pitman permutation tests as implemented in the “coin” library in R (function “oneway_test”; 9,999 Monte Carlo resamplings [98]). We also calculated πinter for each of the 10 pairwise comparisons and πintra for the five species for each of the largest 15 supercontigs. (Supercontig 2.16 had only 4,498 bp.)
Approximate divergence times and incomplete lineage sorting.
We calculated a rough estimate of the age of each of the phylogenetic species of Histoplasma, leveraging the information from our rooted phylogenetic tree. Due to the lack of an available calibration point for the Onygenales (i.e., fossils), we had to rely on genetic divergence and mutation rates alone. We assumed that the mutation and substitution rates in Histoplasma are equivalent to those measured in other fungi. The assumed substitution rate for nuclear DNA was 1 × 10−9 substitution/site/year (99, 100). (It is worth noting that no effort has been made to estimate the precise rate for Histoplasma or any other pathogenic fungus.) We converted the tree branch lengths to approximate node ages assuming a constant mutation and substitution rate.
Next, we calculated the likelihood of ancestral variation still segregating in the recipient species. We calculated the expected time for an allele segregating in the ancestral population to be either fixed (101):
or lost (99):
Where p is the allele frequency prespeciation and Ne is the ancestral effective population size. We used three different values of Ne: 104, 105, and 106.
Shared ancestry and gene exchange between species of Histoplasma. (i) TreeMix.
We looked for evidence of introgression in the nuclear genome among species of Histoplasma and its possible directions using the model implemented in TreeMix (102). TreeMix estimates the most likely evolutionary history of a group of populations by estimating the levels of genetic drift at a set of markers. In this case, we used genome-wide polymorphism. The analysis is done in two steps. First, it estimates the relationships between sampled populations and estimates the most likely dendrogram. Second, it compares the covariance structure modeled by this dendrogram to the actual observed covariance between populations. If a pair of populations is more closely related than expected by the strictly bifurcating tree, then the model suggests a preset number of admixture events in the history of those populations.
Since we had allele frequency data for five populations (NAm 1, NAm 2, LAm A, Panama, and Africa) and the number of inferred migrations must be lower than the number of populations, we could estimate between 0 and 4 migration events (m). TreeMix was run allowing for m = 0, 1, 2, 3, or 4. We found the most likely demographic scenario in terms of migration between species in two ways. First, we compared the log likelihood scores (LS) of each demographic event using a likelihood ratio test (LRT) with 2 df to account for changes in the number of migration edges and the direction of gene flow. The LRT follows a χ2 distribution and compares hierarchically nested models. Since addition of an extra migration event constitutes two additional parameters (i.e., the event itself and its direction), models with different numbers of migrations can be compared with LRTs. We did sequential LRTs to determine when addition of a new population improved the model likelihood. Second, we calculated the Akaike information criterion (AIC) for each migration scenario (103). To determine which migration scenario (i.e., number of migrations) best explained the data, we compared the AIC values for each migration model by ascribing weights to each model using weighted AIC (wAIC) with the following equation (78):
(ii) D statistic.
To confirm whether interspecific gene flow has indeed occurred between species of Histoplasma (as suggested by TreeMix [see Results]), we calculated historical levels of gene flow between three pairs of species: Africa and NAm 1, Panama and NAm 2 (both suggested by TreeMix [see below]), and NAm 1 and NAm 2, the only syntopic (i.e., cooccurring in the same geographical location) species pair of Histoplasma for which we had extensive sampling. We used the ABBA-BABA/D statistic (104). The ABBA-BABA test compares patterns of ancestral (A) and derived (B) alleles between four taxa. In the absence of gene flow, one expects to find equal numbers of sites for each pattern (ABBA versus BABA). However, gene flow from the third to the second population can lead to an excess of the ABBA pattern with respect to the BABA pattern; such departure from the expectation is the basis of the D statistic. As an outgroup we used Emmonsia parva (34, 84). Instead of sites, we used allele frequencies to obtain the ABBA and BABA counts (as described in references 55, 104, and 105). Significance was assessed using weighted block jackknifing with 50-kb windows (106). We also estimated the proportion of the genome that was introgressed with the fd statistic (55). fd compares the observed difference between the ABBA and BABA counts to the expected difference when the entire genome is introgressed. To test the potential influence of the choice of outgroup, we did the same calculations but instead of using E. parva, we used Paracoccidioides brasiliensis. All results were identical with this outgroup.
(iii) Admixture.
We assessed whether there was any evidence of admixed individuals in the most extensively sampled species pair, NAm 1/NAm 2. We estimated each isolate’s admixture proportion using ADMIXTURE (107). We assumed the existence of two ancestral populations from which all observed individuals descend (K = 2), since we were aiming to determine the extent of admixture between isolates of two putative species, and ran independent analyses for each supercontig, to obtain information on the genomic distribution of admixture.
Accession number(s).
All genome sequences reported in this study have been deposited in the Short Read Archive under accession numbers PRJNA416769 (samples SRX3350816–SRX3350845). Intermediary processing files (.bam and .bai) have been deposited in Dryad (https://doi.org/10.5061/dryad.006bf).
ACKNOWLEDGMENTS
We thank R. Brem, L. Noble, B. Barker, J. G. McEwen, S. Restrepo, and the members of the Matute lab for helpful scientific discussions and comments. We also thank J. G. McEwen, E. Arathoon, and B. Samayoa for donating isolates.
The authors have no conflicts of interest.
Footnotes
Citation Sepúlveda VE, Márquez R, Turissini DA, Goldman WE, Matute DR. 2017. Genome sequences reveal cryptic speciation in the human pathogen Histoplasma capsulatum. mBio 8:e01339-17. https://doi.org/10.1128/mBio.01339-17.
REFERENCES
- 1.Darling ST. 1906. A protozoön general infection producing pseudotubercles in the lungs and focal necroses in the liver, spleen and lymph nodes. JAMA 46:1283–1285. [Google Scholar]
- 2.Wang H, Xu Z, Gao L, Hao B. 2009. A fungal phylogeny based on 82 complete genomes using the composition vector method. BMC Evol Biol 9:195. doi: 10.1186/1471-2148-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fitzpatrick DA, Logue ME, Stajich JE, Butler G. 2006. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol 6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batanghari JW, Goldman WE. 1997. Calcium dependence and binding in cultures of Histoplasma capsulatum. Infect Immun 65:5257–5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eissenberg LG, Goldman WE. 1991. Histoplasma variation and adaptive strategies for parasitism: new perspectives on histoplasmosis. Clin Microbiol Rev 4:411–421. doi: 10.1128/CMR.4.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knox KS. 2014. Perspective on coccidioidomycosis and histoplasmosis. Am J Respir Crit Care Med 189:752–753. doi: 10.1164/rccm.201311-2024LE. [DOI] [PubMed] [Google Scholar]
- 7.Hage CA, Azar MM, Bahr N, Loyd J, Wheat LJ. 2015. Histoplasmosis: up-to-date evidence-based approach to diagnosis and management. Semin Respir Crit Care Med 36:729–745. doi: 10.1055/s-0035-1562899. [DOI] [PubMed] [Google Scholar]
- 8.Kauffman CA. 2007. Histoplasmosis: a clinical and laboratory update. Clin Microbiol Rev 20:115–132. doi: 10.1128/CMR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wheat LJ, Azar MM, Bahr NC, Spec A, Relich RF, Hage C. 2016. Histoplasmosis. Infect Dis Clin North Am 30:207–227. doi: 10.1016/j.idc.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Davies SF, Sarosi GA. 1997. Epidemiological and clinical features of pulmonary blastomycosis. Semin Respir Infect 12:206–218. [PubMed] [Google Scholar]
- 11.Freifeld AG, Iwen PC, Lesiak BL, Gilroy RK, Stevens RB, Kalil AC. 2005. Histoplasmosis in solid organ transplant recipients at a large Midwestern University transplant center. Transpl Infect Dis 7:109–115. doi: 10.1111/j.1467-8365.2005.00105.x. [DOI] [PubMed] [Google Scholar]
- 12.Graybill JR. 1988. Histoplasmosis and AIDS. J Infect Dis 158:623–626. doi: 10.1093/infdis/158.3.623. [DOI] [PubMed] [Google Scholar]
- 13.Johnson PC, Khardori N, Najjar AF, Butt F, Mansell PWA, Sarosi GA. 1988. Progressive disseminated histoplasmosis in patients with acquired immunodeficiency syndrome. Am J Med 85:152–158. doi: 10.1016/S0002-9343(88)80334-6. [DOI] [PubMed] [Google Scholar]
- 14.Sternberg S. 1994. The emerging fungal threat. Science 266:1632–1634. doi: 10.1126/science.7702654. [DOI] [PubMed] [Google Scholar]
- 15.Edwards LB, Acquaviva FA, Livesay VT, Cross FW, Palmer CE. 1969. An atlas of sensitivity to tuberculin, PPD-B, and histosplasmin in the United States. Am Rev Respir Dis 99:1–132. [PubMed] [Google Scholar]
- 16.Joseph Wheat L. 2003. Current diagnosis of histoplasmosis. Trends Microbiol 11:488–494. doi: 10.1016/j.tim.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Hoff GL, Bigler WJ. 1981. The role of bats in the propagation and spread of histoplasmosis: a review. J Wildl Dis 17:191–196. doi: 10.7589/0090-3558-17.2.191. [DOI] [PubMed] [Google Scholar]
- 18.Lyon GM, Bravo AV, Espino A, Lindsley MD, Gutierrez RE, Rodriguez I, Corella A, Carrillo F, McNeil MM, Warnock DW, Hajjeh RA. 2004. Histoplasmosis associated with exploring a bat-inhabited cave in Costa Rica, 1998–1999. Am J Trop Med Hyg 70:438–442. [PubMed] [Google Scholar]
- 19.Chick EW, Compton SB, Pass T, Mackey B, Hernandez C, Austin E Jr, Pitzer FR, Flanigan C. 1981. Hitchcock’s Birds, or the increased rate of exposure to Histoplasma from blackbird roost sites. Chest 80:434–438. doi: 10.1378/chest.80.4.434. [DOI] [PubMed] [Google Scholar]
- 20.Teixeira Mde M, Patané JS, Taylor ML, Gómez BL, Theodoro RC, de Hoog S, Engelthaler DM, Zancopé-Oliveira RM, Felipe MS, Barker BM. 2016. Worldwide phylogenetic distributions and population dynamics of the genus Histoplasma. PLoS Negl Trop Dis 10:e0004732. doi: 10.1371/journal.pntd.0004732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsh HW, Hinton A, Cozad GC. 1956. Natural reservoir of Histoplasma capsulatum. Am J Hyg 63:18–27. [DOI] [PubMed] [Google Scholar]
- 22.Zeidberg LD, Ajello L. 1954. Environmental factors influencing the occurrence of Histoplasma capsulatum and Microsporum gypseum in soil. J Bacteriol 68:156–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steenwyk JL, Soghigian JS, Perfect JR, Gibbons JG. 2016. Copy number variation contributes to cryptic genetic variation in outbreak lineages of Cryptococcus gattii from the North American Pacific Northwest. BMC Genomics 17:700. doi: 10.1186/s12864-016-3044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emmons CW. 1950. Histoplasmosis: animal reservoirs and other sources in nature of pathogenic fungus, Histoplasma. Am J Publ Health Nation’s Health 40:436–440. doi: 10.2105/AJPH.40.4.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emmons CW, Bell JA, Olson BJ. 1947. Naturally occurring histoplasmosis in Mus musculus and Rattus norvegicus. Public Health Rep 62:1642. [PubMed] [Google Scholar]
- 26.Naiff RD, Mok WY, Naiff MF. 1985. Distribution of Histoplasma capsulatum in Amazonian wildlife. Mycopathologia 89:165–168. [DOI] [PubMed] [Google Scholar]
- 27.Kwon-Chung KJ. 1972. Emmonsiella capsulata: perfect state of Histoplasma capsulatum. Science 177:368–369. doi: 10.1126/science.177.4046.368. [DOI] [PubMed] [Google Scholar]
- 28.Kasuga T, Taylor JW, White TJ. 1999. Phylogenetic relationships of varieties and geographical groups of the human pathogenic fungus Histoplasma capsulatum Darling. J Clin Microbiol 37:653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards PQ, Klaer JH. 1956. Worldwide geographic distribution of histoplasmosis and histoplasmin sensitivity. Am J Trop Med Hyg 5:235–257. doi: 10.4269/ajtmh.1956.5.235. [DOI] [PubMed] [Google Scholar]
- 30.Ajello L. 1971. Coccidioidomycosis and histoplasmosis. A review of their epidemiology and geographical distribution. Mycopathol Mycol Appl 45:221–230. doi: 10.1007/BF02051969. [DOI] [PubMed] [Google Scholar]
- 31.Kasuga T, White TJ, Koenig G, McEwen J, Restrepo A, Castañeda E, Da Silva Lacaz C, Heins-Vaccari EM, De Freitas RS, Zancopé-Oliveira RM, Qin Z, Negroni R, Carter DA, Mikami Y, Tamura M, Taylor ML, Miller GF, Poonwan N, Taylor JW. 2003. Phylogeography of the fungal pathogen Histoplasma capsulatum. Mol Ecol 12:3383–3401. doi: 10.1046/j.1365-294X.2003.01995.x. [DOI] [PubMed] [Google Scholar]
- 32.Arunmozhi Balajee SA, Hurst SF, Chang LS, Miles M, Beeler E, Hale C, Kasuga T, Benedict K, Chiller T, Lindsley MD. 2013. Multilocus sequence typing of Histoplasma capsulatum in formalin-fixed paraffin-embedded tissues from cats living in non-endemic regions reveals a new phylogenetic clade. Med Mycol 51:345–351. doi: 10.3109/13693786.2012.733430. [DOI] [PubMed] [Google Scholar]
- 33.Ajello L. 1968. Comparative morphology and immunology of members of the genus Histoplasma. A review. Mykosen 11:507–514. doi: 10.1111/j.1439-0507.1968.tb03370.x. [DOI] [PubMed] [Google Scholar]
- 34.Guého E, Leclerc MC, de Hoog GS, Dupont B. 1997. Molecular taxonomy and epidemiology of Blastomyces and Histoplasma species. Mycoses 40:69–81. doi: 10.1111/j.1439-0507.1997.tb00191.x. [DOI] [PubMed] [Google Scholar]
- 35.Sepúlveda VE, Williams CL, Goldman WE. 2014. Comparison of phylogenetically distinct Histoplasma strains reveals evolutionarily divergent virulence strategies. mBio 5:e01376-14. doi: 10.1128/mBio.01376-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gugnani HC, Muotoe-Okafor F. 1997. African histoplasmosis: a review. Rev Iberoam Micol 14:155–159. [PubMed] [Google Scholar]
- 37.Vanbreuseghem R. 1953. Histoplasma duboisii and African histoplasmosis. Mycologia 45:803–816. [Google Scholar]
- 38.Fisher MC, Koenig GL, White TJ, Taylor JW. 2002. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia 94:73–84. doi: 10.1080/15572536.2003.11833250. [DOI] [PubMed] [Google Scholar]
- 39.Turissini DA, Gomez OM, Teixeira MM, McEwen JG, Matute DR. 2017. Species boundaries in the human pathogen Paracoccidioides. Fungal Genet Biol 106:9–25. doi: 10.1016/j.fgb.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor JW, Turner E, Townsend JP, Dettman JR, Jacobson D. 2006. Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom Fungi. Philos Trans R Soc Lond B Biol Sci 361:1947–1963. doi: 10.1098/rstb.2006.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- 42.Taylor JW, Jacobson DJ, Fisher MC. 1999. The evolution of asexual fungi: reproduction, speciation and classification. Annu Rev Phytopathol 37:197–246. doi: 10.1146/annurev.phyto.37.1.197. [DOI] [PubMed] [Google Scholar]
- 43.Kohn LM. 2005. Mechanisms of fungal speciation. Annu Rev Phytopathol 43:279–308. doi: 10.1146/annurev.phyto.43.040204.135958. [DOI] [PubMed] [Google Scholar]
- 44.Baum DA, Donoghue MJ. 1995. Choosing among alternative “phylogenetic” species concepts. Syst Bot 20:560. doi: 10.2307/2419810. [DOI] [Google Scholar]
- 45.Dettman JR, Jacobson DJ, Taylor JW. 2003. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57:2703–2720. doi: 10.1111/j.0014-3820.2003.tb01514.x. [DOI] [PubMed] [Google Scholar]
- 46.Dettman JR, Jacobson DJ, Turner E, Pringle A, Taylor JW. 2003. Reproductive isolation and phylogenetic divergence in Neurospora: comparing methods of species recognition in a model eukaryote. Evolution 57:2721–2741. doi: 10.1111/j.0014-3820.2003.tb01515.x. [DOI] [PubMed] [Google Scholar]
- 47.Liti G, Barton DBH, Louis EJ. 2006. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics 174:839–850. doi: 10.1534/genetics.106.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hughes KW, Petersen RH, Lickey EB. 2009. Using heterozygosity to estimate a percentage DNA sequence similarity for environmental species’ delimitation across basidiomycete fungi. New Phytol 182:795–798. doi: 10.1111/j.1469-8137.2009.02802.x. [DOI] [PubMed] [Google Scholar]
- 49.Carr J, Shearer G. 1998. Genome size, complexity, and ploidy of the pathogenic fungus Histoplasma capsulatum. J Bacteriol 180:6697–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baum DA. 2007. Concordance trees, concordance factors, and the exploration of reticulate genealogy. Taxon 56:417–426. [Google Scholar]
- 51.De Queiroz K. 2007. Species concepts and species delimitation. Syst Biol 56:879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- 52.Knowles LL, Carstens BC. 2007. Delimiting species without monophyletic gene trees. Syst Biol 56:887–895. doi: 10.1080/10635150701701091. [DOI] [PubMed] [Google Scholar]
- 53.Larget BR, Kotha SK, Dewey CN, Ané C. 2010. BUCKy: gene tree/species tree reconciliation with Bayesian concordance analysis. Bioinformatics 26:2910–2911. doi: 10.1093/bioinformatics/btq539. [DOI] [PubMed] [Google Scholar]
- 54.Sun Y, Corcoran P, Menkis A, Whittle CA, Andersson SGE, Johannesson H. 2012. Large-scale introgression shapes the evolution of the mating-type chromosomes of the filamentous ascomycete Neurospora tetrasperma. PLoS Genet 8:e1002820. doi: 10.1371/journal.pgen.1002820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin SH, Davey JW, Jiggins CD. 2015. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol Biol Evol 32:244–257. doi: 10.1093/molbev/msu269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keath EJ, Kobayashi GS, Medoff G. 1992. Typing of Histoplasma capsulatum by restriction fragment length polymorphisms in a nuclear gene. J Clin Microbiol 30:2104–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kersulyte D, Woods JP, Keath EJ, Goldman WE, Berg DE. 1992. Diversity among clinical isolates of Histoplasma capsulatum detected by polymerase chain reaction with arbitrary primers. J Bacteriol 174:7075–7079. doi: 10.1128/jb.174.22.7075-7079.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eissenberg LG, West JL, Woods JP, Goldman WE. 1991. Infection of P388D1 macrophages and respiratory epithelial cells by Histoplasma capsulatum: selection of avirulent variants and their potential role in persistent histoplasmosis. Infect Immun 59:1639–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berliner MD, Biundo N. 1973. Effects of continuous light and total darkness on cultures of Histoplasma capsulatum. Sabouraudia 11:48–51. doi: 10.1080/00362177385190101. [DOI] [PubMed] [Google Scholar]
- 60.Berliner MD. 1968. Primary subcultures of Histoplasma capsulatum. I. Macro and micro-morphology of the mycelial phase. Sabouraudia 6:111–118. [PubMed] [Google Scholar]
- 61.Zarnowski R, Connolly PA, Wheat LJ, Woods JP. 2007. Production of extracellular proteolytic activity by Histoplasma capsulatum grown in Histoplasma-macrophage medium is limited to restriction fragment length polymorphism class 1 isolates. Diagn Microbiol Infect Dis 59:39–47. doi: 10.1016/j.diagmicrobio.2007.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bohse ML, Woods JP. 2007. RNA interference-mediated silencing of the YPS3 gene of Histoplasma capsulatum reveals virulence defects. Infect Immun 75:2811–2817. doi: 10.1128/IAI.00304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Durkin MM, Connolly PA, Karimi K, Wheat E, Schnizlein-Bick C, Allen SD, Alves K, Tewari RP, Keath E. 2004. Pathogenic differences between North American and Latin American strains of Histoplasma capsulatum var. capsulatum in experimentally infected mice. J Clin Microbiol 42:4370–4373. doi: 10.1128/JCM.42.9.4370-4373.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nichols R. 2001. Gene trees and species trees are not the same. Trends Ecol Evol 16:358–364. doi: 10.1016/S0169-5347(01)02203-0. [DOI] [PubMed] [Google Scholar]
- 65.Shen XX, Hittinger CT, Rokas A. 2017. Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat Ecol Evol 1:126. doi: 10.1038/s41559-017-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sukumaran J, Knowles LL. 2017. Multispecies coalescent delimits structure, not species. Proc Natl Acad Sci U S A 114:1607–1612. doi: 10.1073/pnas.1607921114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mallet J. 2005. Hybridization as an invasion of the genome. Trends Ecol Evol 20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 68.Emmons CW. 1962. Soil reservoirs of pathogenic fungi. J Wash Acad Sci 52:3–9. [Google Scholar]
- 69.Goughenour KD, Balada-Llasat JM, Rappleye CA. 2015. Quantitative microplate-based growth assay for determination of antifungal susceptibility of Histoplasma capsulatum yeasts. J Clin Microbiol 53:3286–3295. doi: 10.1128/JCM.00795-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rappleye CA, Engle JT, Goldman WE. 2004. RNA interference in Histoplasma capsulatum demonstrates a role for alpha-(1,3)-glucan in virulence. Mol Microbiol 53:153–165. doi: 10.1111/j.1365-2958.2004.04131.x. [DOI] [PubMed] [Google Scholar]
- 71.Marion CL, Rappleye CA, Engle JT, Goldman WE. 2006. An alpha-(1,4)-amylase is essential for alpha-(1,3)-glucan production and virulence in Histoplasma capsulatum. Mol Microbiol 62:970–983. doi: 10.1111/j.1365-2958.2006.05436.x. [DOI] [PubMed] [Google Scholar]
- 72.Keath EJ, Painter AA, Kobayashi GS, Medoff G. 1989. Variable expression of a yeast-phase-specific gene in Histoplasma capsulatum strains differing in thermotolerance and virulence. Infect Immun 57:1384–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bohse ML, Woods JP. 2007. Expression and interstrain variability of the YPS3 gene of Histoplasma capsulatum. Eukaryot Cell 6:609–615. doi: 10.1128/EC.00010-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Loulergue P, Bastides F, Baudouin V, Chandenier J, Mariani-Kurkdjian P, Dupont B, Viard JP, Dromer F, Lortholary O. 2007. Literature review and case histories of Histoplasma capsulatum var. duboisii infections in HIV-infected patients. Emerg Infect Dis 13:1647–1652. doi: 10.3201/eid1311.070665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kwon-Chung KJ. 1973. Studies on Emmonsiella capsulata. I. Heterothallism and development of the ascocarp. Mycologia 65:109–121. doi: 10.2307/3757791. [DOI] [PubMed] [Google Scholar]
- 76.Kwon-Chung KJ, Weeks RJ, Larsh HW. 1974. Studies on Emmonsiella capsulata (Histoplasma capsulatum). II. Distribution of the two mating types in 13 endemic states of the United States. Am J Epidemiol 99:44–49. doi: 10.1093/oxfordjournals.aje.a121583. [DOI] [PubMed] [Google Scholar]
- 77.Kwon-Chung KJ. 1975. Perfect state (Emmonsiella capsulata) of the fungus causing large-form African histoplasmosis. Mycologia 67:980–990. doi: 10.2307/3758589. [DOI] [PubMed] [Google Scholar]
- 78.Worsham PL, Goldman WE. 2009. Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. J Med Vet Mycol 26:137–143. [PubMed] [Google Scholar]
- 79.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Steele PE, Carle GF, Kobayashi GS, Medoff G. 1989. Electrophoretic analysis of Histoplasma capsulatum chromosomal DNA. Mol Cell Biol 9:983–987. doi: 10.1128/MCB.9.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Canteros CE, Zuiani MF, Ritacco V, Perrotta DE, Reyes-Montes MR, Granados J, Zúñiga G, Taylor ML, Davel G. 2005. Electrophoresis karyotype and chromosome-length polymorphism of Histoplasma capsulatum clinical isolates from Latin America. FEMS Immunol Med Microbiol 45:423–428. doi: 10.1016/j.femsim.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 82.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Muñoz JF, Gauthier GM, Desjardins CA, Gallo JE, Holder J, Sullivan TD, Marty AJ, Carmen JC, Chen Z, Ding L, Gujja S, Magrini V, Misas E, Mitreva M, Priest M, Saif S, Whiston EA, Young S, Zeng Q, Goldman WE, Mardis ER, Taylor JW, McEwen JG, Clay OK, Klein BS, Cuomo CA. 2015. The dynamic genome and transcriptome of the human fungal pathogen Blastomyces and close relative Emmonsia. PLoS Genet 11:e1005493. doi: 10.1371/journal.pgen.1005493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 86.Koufopanou V, Burt A, Taylor JW. 1997. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc Natl Acad Sci U S A 94:5478–5482. doi: 10.1073/pnas.94.10.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hudson RR, Coyne JA. 2002. Mathematical consequences of the genealogical species concept. Evolution 56:1557–1565. [DOI] [PubMed] [Google Scholar]
- 88.Moore WS. 1995. Inferring phylogenies from mtDNA variation: mitochondrial-gene trees versus nuclear-gene trees. Evolution 49:718–726. doi: 10.1111/j.1558-5646.1995.tb02308.x. [DOI] [PubMed] [Google Scholar]
- 89.Hoelzer GA. 1997. Inferring phylogenies from mtDNA variation: mitochondrial-gene trees versus nuclear-gene trees revisited. Evolution 51:622–626. doi: 10.1111/j.1558-5646.1997.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 90.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Taylor JW. 2006. Evolution of human-pathogenic fungi: phylogenies and species, p 113–132. In Heitman J, Filler SG, Edwards JE, Mitchell AP (ed), Molecular principles of fungal pathogenesis. American Society of Microbiology, Washington, DC. [Google Scholar]
- 93.Ané C, Larget B, Baum DA, Smith SD, Rokas A. 2007. Bayesian estimation of concordance among gene trees. Mol Biol Evol 24:412–426. doi: 10.1093/molbev/msl170. [DOI] [PubMed] [Google Scholar]
- 94.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. 2004. Parallel Metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20:407–415. doi: 10.1093/bioinformatics/btg427. [DOI] [PubMed] [Google Scholar]
- 96.Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014. Tracer v1.6. http://tree.bio.ed.ac.uk/software/tracer.
- 97.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 98.Hothorn T, Hornik K, Van De Wiel MA, Zeileis A. 2008. Implementing a class of permutation pests: the coin package. J Stat Software 28:1–23. [Google Scholar]
- 99.Kasuga T, White TJ, Taylor JW. 2002. Estimation of nucleotide substitution rates in Eurotiomycete fungi. Mol Biol Evol 19:2318–2324. doi: 10.1093/oxfordjournals.molbev.a004056. [DOI] [PubMed] [Google Scholar]
- 100.Torriani SFF, Penselin D, Knogge W, Felder M, Taudien S, Platzer M, McDonald BA, Brunner PC. 2014. Comparative analysis of mitochondrial genomes from closely related Rhynchosporium species reveals extensive intron invasion. Fungal Genet Biol 62:34–42. doi: 10.1016/j.fgb.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 101.Kimura M, Ohta T. 1969. The average number of generations until fixation of a mutant gene in a finite population. Genetics 61:763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pickrell JK, Pritchard JK. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet 8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Akaike H. 1974. A new look at the statistical model identification. IEEE Trans Automat Contr 19:716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- 104.Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, Li H, Mallick S, Dannemann M, Fu Q, Kircher M, Kuhlwilm M, Lachmann M, Meyer M, Ongyerth M, Siebauer M, Theunert C, Tandon A, Moorjani P, Pickrell J, Mullikin JC, Vohr SH, Green RE, Hellmann I, Johnson PLF, Blanche H, Cann H, Kitzman JO, Shendure J, Eichler EE, Lein ES, Bakken TE, Golovanova LV, Doronichev VB, Shunkov MV, Derevianko AP, Viola B, Slatkin M, Reich D, Kelso J, Pääbo S. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, Hansen NF, Durand EY, Malaspinas A-S, Jensen JD, Marques-Bonet T, Alkan C, Prüfer K, Meyer M, Burbano HA, Good JM, Schultz R, Aximu-Petri A, Butthof A, Höber B, Höffner B, Siegemund M, Weihmann A, Nusbaum C, Lander ES, Russ C, Novod N, Affourtit J, Egholm M, Verna C, Rudan P, Brajkovic D, Kucan Ž, Gušic I, Doronichev VB, Golovanova LV, Lalueza-Fox C, de la Rasilla M, Fortea J, Rosas A, Schmitz RW, Johnson PLF, Eichler EE, Falush D, Birney E, Mullikin JC, Slatkin M, Nielsen R, Kelso J, Lachmann M, Reich D, Pääbo S. 2010. A draft sequence of the Neandertal genome. Science 328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Busing FMTA, Meijer E, Van Der Leeden RVD. 1999. Delete-m JackKnife for Unequal m. Stat Comput 9:3–8. doi: 10.1023/A:1008800423698. [DOI] [Google Scholar]
- 107.Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Linkage disequilibriums persist over large regions of the genome in Histoplasma. Average LD as measured by r2 between pairs of SNPs with distances binned every 100 bp. Download FIG S1, PDF file, 4.9 MB (5MB, pdf) .
Copyright © 2017 Sepúlveda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Identified introgressions between Histoplasma species are unlikely to represent ancestral variation that is still segregating in the recipient species. Each panel shows the expected number of generations that an allele at a given frequency, p, would take to either be fixed (black line) or lost (red line) from the population. Horizontal lines denote the number of populations since the two species diverged, assuming 1 generation per year (solid blue line), 10 generations per year (dashed blue line), or 100 generations per year (dotted blue line). (A) Ne = 106. (B) Ne = 105. (C) Ne = 104. Note y axes are in log scale. Download FIG S2, PDF file, 0.03 MB (27.7KB, pdf) .
Copyright © 2017 Sepúlveda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.