ABSTRACT
All organisms must adapt to ever-changing environmental conditions and accordingly have evolved diverse signal transduction systems. In bacteria, the most abundant networks are built around the two-component signal transduction systems that include histidine kinases and receiver domains. In contrast, eukaryotic signal transduction is dominated by serine/threonine/tyrosine protein kinases. Both of these systems are also found in archaea, but they are not as common and diversified as their bacterial and eukaryotic counterparts, suggesting the possibility that archaea have evolved other, still uncharacterized signal transduction networks. Here we propose a role for KaiC family ATPases, known to be key components of the circadian clock in cyanobacteria, in archaeal signal transduction. The KaiC family is notably expanded in most archaeal genomes, and although most of these ATPases remain poorly characterized, members of the KaiC family have been shown to control archaellum assembly and have been found to be a stable component of the gas vesicle system in Halobacteria. Computational analyses described here suggest that KaiC-like ATPases and their homologues with inactivated ATPase domains are involved in many other archaeal signal transduction pathways and comprise major hubs of complex regulatory networks. We predict numerous input and output domains that are linked to KaiC-like proteins, including putative homologues of eukaryotic DEATH domains that could function as adapters in archaeal signaling networks. We further address the relationships of the archaeal family of KaiC homologues to the bona fide KaiC of cyanobacteria and implications for the existence of a KaiC-based circadian clock apparatus in archaea.
KEYWORDS: ATPase, Archaea, KaiC, circadian clock, signal transduction
IMPORTANCE
Little is currently known about signal transduction pathways in Archaea. Recent studies indicate that KaiC-like ATPases, known as key components of the circadian clock apparatus in cyanobacteria, are involved in the regulation of archaellum assembly and, likely, type IV pili and the gas vesicle system in Archaea. We performed comprehensive comparative genomic analyses of the KaiC family. A vast protein interaction network was revealed, with KaiC family proteins as hubs for numerous input and output components, many of which are shared with two-component signal transduction systems. Putative KaiC-based signal transduction systems are predicted to regulate the activities of membrane-associated complexes and individual proteins, such as signal recognition particle and membrane transporters, and also could be important for oxidative stress response regulation. KaiC-centered signal transduction networks are predicted to play major roles in archaeal physiology, and this work is expected to stimulate their experimental characterization.
INTRODUCTION
Signal transduction systems are essential components of all forms of life and serve as information channels between the organism and the environment. The general organization of a signal transduction system includes at least three components, namely, a sensor (input), a transmitter, and an effector (output). Bacterial signal transduction is dominated by two-component systems that typically transmit information through histidine kinases, whereas eukaryotic signal transduction systems are dominated by Ser/Thr/Tyr protein kinases (1–6).
Archaeal signal transduction systems have not been studied to a comparable extent (7), but studies support the existence of some archaea-encoded, complex signal transduction networks that mimic systems employed in bacteria and eukarya. Two-component systems homologous to bacterial counterparts have been experimentally characterized in Halobacteria that regulate complex protein-protein interaction networks that influence chemotaxis, phototaxis, and archaellum (archaeal rotary motor) activity (8). In the methanogenic archaeon Methanosaeta harundinacea, the histidine kinase FilI controls cell morphology and affects methane production (9). Phylogenetic analysis suggests that archaea acquired two-component signal transduction system components from bacteria on multiple occasions (10). This conclusion is compatible with the results of the reconstruction of the last archaeal common ancestor (LACA) on the basis of the arCOG (archaeal clusters of orthologous genes) database that estimated the probability of histidine kinases being present in the LACA at <0.2 (11). Notably, the two-component systems are mostly found in mesophilic archaea that appear to have captured numerous bacterial genes via horizontal gene transfer (HGT) (9, 10, 12–17) (Table 1). Protein phosphorylation, mostly attributed to Ser/Thr/Tyr (here, S/T) protein kinases, apparently plays an important role in archaea, but details about the specific roles of protein phosphorylation in signal transduction are scarce (7). Unlike histidine kinases, three S/T protein kinase families, RIO1, RIO2, and SPS1 (corresponding to COG1718, COG0478, and COG0515, respectively, in the Clusters of Orthologous Genes [COG] database [18]), have been traced back to the LACA (11). At least some of these kinases appear to be key regulators of the archaeal cell cycle, motility, and membrane remodeling (19). However, S/T kinases are not particularly prone to expansion in archaea (Table 1) and probably comprise only a limited part of the archaeal signal transduction networks.
TABLE 1 .
Three major protein superfamilies involved in signal transduction in selected archaea and bacteria
| Genome | No. of proteins in the familya |
||
|---|---|---|---|
| Serine/threonine protein kinasesb |
KaiC-like ATPasesc |
Sensor histidine kinasesd |
|
| Archaea | |||
| Aeropyrum pernix K1 | 3 | 3 | 0 |
| Desulfurococcus kamchatkensis 1221n | 3 | 3 | 0 |
| Ignicoccus hospitalis KIN4 I | 5 | 3 | 0 |
| Hyperthermus butylicus DSM 5456 | 4 | 5 | 0 |
| Pyrolobus fumarii 1A | 6 | 2 | 0 |
| Sulfolobus acidocaldarius DSM 639 | 9 | 2 | 0 |
| Pyrobaculum aerophilum IM2 | 5 | 12 | 0 |
| Archaeoglobus fulgidus DSM 4304 | 3 | 14 | 14 |
| Halobacterium salinarum R1 | 6 | 10 | 13 |
| Haloferax volcanii DS2 | 6 | 11 | 23 |
| Methanothermobacter thermautotrophicus Delta H | 2 | 6 | 16 |
| Methanocaldococcus jannaschii DSM 2661 | 3 | 2 | 0 |
| Methanococcus maripaludis S2 | 3 | 1 | 3 |
| Methanocella conradii HZ254 | 3 | 10 | 19 |
| Methanosarcina acetivorans C2A | 4 | 7 | 53 |
| Methanosarcina mazei Go1 | 3 | 7 | 34 |
| Pyrococcus furiosus COM1 | 4 | 21 | 0 |
| Thermococcus kodakarensis KOD1 | 3 | 32 | 1 |
| Thermoplasma acidophilum DSM 1728 | 3 | 3 | 0 |
| Candidatus Korarchaeum cryptofilum OPF8 | 2 | 3 | 0 |
| Nanoarchaeum equitans Kin4M | 2 | 1 | 0 |
| Nitrosoarchaeum koreensis MY1 | 3 | 2 | 12 |
| “Candidatus Caldiarchaeum subterraneum” | 2 | 5 | 0 |
| Bacteria | |||
| Escherichia coli K-12 MG1655 | 2 | 0 | 30 |
| Bacillus subtilis subsp. subtilis 168 | 5 | 0 | 36 |
| Nostoc sp. strain PCC 7120 | 54 | 2 | 139 |
| Thermus thermophilus HB8 | 5 | 0 | 11 |
The numbers of respective proteins were taken from previous publications (5, 73) and/or retrieved from recent updates of the COG (18) and arCOG (66) databases. The data were verified by using PSI-BLAST searches against the complete-genome database (as of March 2016).
COG0478, COG0515, COG0661, COG1718, COG2112, and COG2766.
COG0467.
COG0642, COG0643, COG2205, COG2972, COG3275, COG3290, and COG3920.
Given the relative paucity of identifiable signal transduction systems in Archaea, a search for new, perhaps, archaea-specific signal transduction systems is an important goal. Our previous analyses of type IV pili systems and the archaellum identified several KaiC-like ATPases (members of the COG0467 family) that appear to be involved in the regulation of these systems (20). These observations prompted us to undertake a comprehensive analysis of the KaiC family in archaea. Because of the high similarity to the eukaryotic recombinase component Rad55 (homologue of bacterial RecA and archaeal RadA), until recently, the COG0467 family (18) in archaea has been implicated in DNA recombination pathways (21). One of the archaeal proteins, namely, the SSO2452 protein of Sulfolobus solfataricus, has been experimentally studied in this context and was shown not to be an active recombinase but could bind single-stranded DNA and inhibit D-loop formation by RadA (22). However, outside the Archaea, the best-studied protein in this family is the cyanobacterial circadian clock ATPase KaiC, which does not appear to be involved in DNA recombination (23, 24).
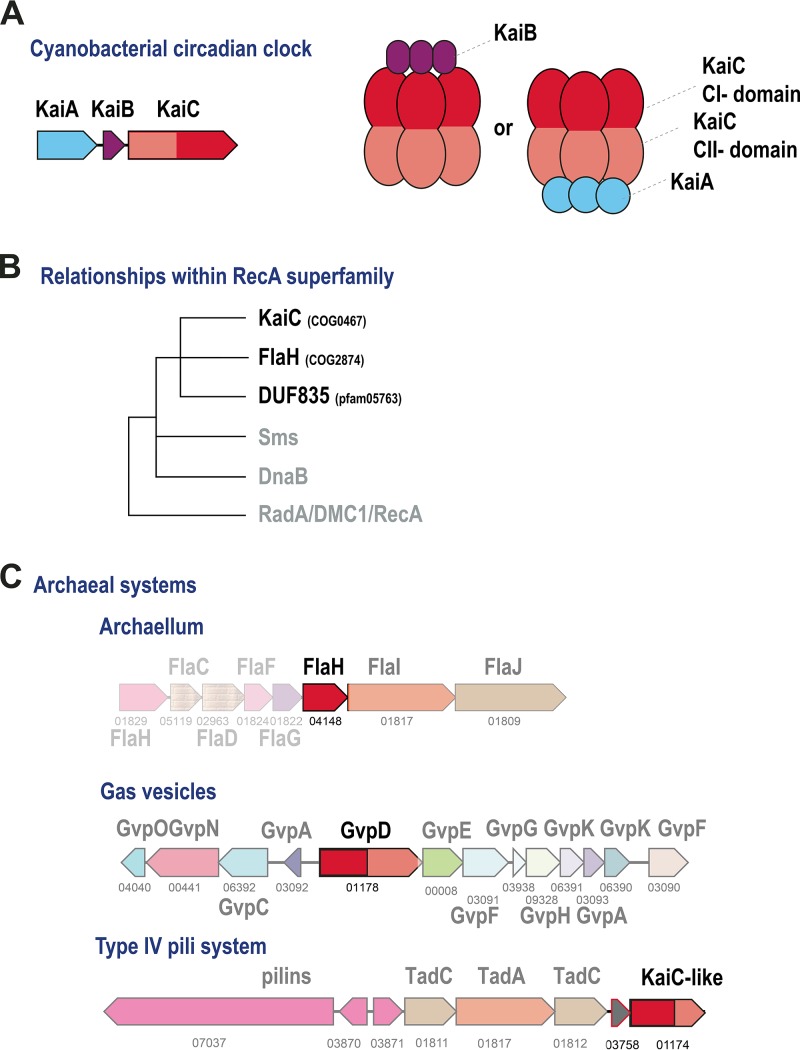
The cyanobacterial circadian clock system, an ATP-dependent, posttranslational molecular oscillator, has been thoroughly characterized biochemically, structurally, and functionally (25–31). Typically, the system consists of three protein components, KaiA, KaiB, and KaiC (Fig. 1A). The KaiA protein forms a homodimer that interacts directly with the C-terminal ATPase domain (CII) of KaiC and promotes its phosphorylation. Structurally, KaiA is a two-domain protein with an N-terminal four-helix bundle domain and a C-terminal OmpR-like winged helix-turn-helix (HTH) DNA-binding domain. KaiB has the thioredoxin fold and interacts with the N-terminal (CI) domain of KaiC, promoting dissociation of KaiA and dephosphorylation of KaiC. The cyanobacterium Prochlorococcus marinus encodes a minimal circadian system that lacks KaiA but nevertheless shows some features of an autonomous oscillator that, however, does not persist long under constant-light conditions, so that the system apparently requires a reset each diel cycle (26, 32). However, even when all three components are present, this is not always sufficient to reproduce all of the canonical properties of a circadian clock, as is the case in the purple nonsulfur bacterium Rhodopseudomonas palustris, which only poorly maintains rhythmicity under constant conditions (33). Multiple input and output components have been shown to interact with the cyanobacterial circadian clock system, forming a complex, interconnected network that includes transcriptional regulators, receiver (REC) domains, and sensory histidine kinases, as well as light-sensitive redox molecules such as quinones (26). Some of the input and output proteins contain KaiA- or KaiB-like domains and directly interact with KaiC.
FIG 1 .
Overview of the KaiC family. (A) Organization of the cyanobacterial circadian clock system. (B) Scheme of relationships of the KaiC family with other RecA-like ATPase families. (C) Known archaeal systems associated with KaiC-like proteins. KaiC family protein N- and C-terminal ATPase domains are red and pink, respectively. Genes are represented by arrows. For archaeal systems, arCOG numbers are shown below the arrows. Homologous genes are color coded. Models show interactions between subunits in the respective complexes (see the text for details and discussion).
Phylogenetic analysis indicates that the COG0467 family forms a separate clade within the RecA ATPase superfamily (34) (Fig. 1B), implying a separate function that does not involve DNA transactions. It has been hypothesized that, given the wide spread and major expansion of this family in archaea, which contrasts with its patchy distribution in bacteria, the KaiC component of the cyanobacterial circadian clock was acquired by HGT from archaea (34). Recently, the structure of the FlaH protein (COG2874 family), which is always encoded within the archaellum operon (Fig. 1C), has been solved and shown to be similar to the C-terminal domain of KaiC (35, 36). FlaH has been shown to form a hexamer and interact with the archaellum subunit FlaI, the motor ATPase, and in crenarchaea, with the FlaX ring (35, 36). KaiC family ATPase GvpD was found to be involved in the regulation of Halobacterium-specific gas vesicles (37) (Fig. 1C). Several halobacterial KaiC-like proteins have been studied with respect to their potential involvement in light-dependent gene expression (38). Very recently, KaiC proteins from the hyperthermophiles Thermococcus litoralis and Pyrococcus horikoshii were shown to be capable of KaiA-independent autophosphorylation at both 30°C and 75°C (34, 39). Finally, structural analysis of a distinct family of archaea-specific uncharacterized proteins (DUF835, PF05763 in the Pfam database [40]) has shown that these proteins are inactivated ATPases that are most closely related to KaiC (41). Thus, currently, at least two additional protein families can be included in the archaeal KaiC group (Fig. 1B). Evolutionary reconstructions suggest that KaiC-like ATPases from arCOG01171, arCOG001174, and arCOG04148 (FlaH) were likely present already in the LACA (11).
Prompted by the above observations and the extraordinary diversity of the KaiC ATPases in archaea, we performed a comprehensive phylogenomic analysis of this protein family. The results strongly suggest that the KaiC family ATPases and their homologues with inactivated ATPase domains are key components of the archaeal signaling network(s).
RESULTS
Genomic census of the KaiC ATPase family in archaea and bacteria.
To perform a comprehensive phylogenomic analysis of the KaiC ATPase superfamily, 2,635 sequences from the three KaiC subfamilies (COG0467, COG2874, and pfam05763) and related arCOGs (see Table S1 in the supplemental material) were extracted from the data set of complete archaeal and bacterial genomes. Genomic loci (five genes upstream and downstream from each kaiC-like gene) were retrieved for the genomic neighborhood analysis (Table S2). These loci were annotated by using PSI-BLAST and the CDD (Conserved Domain Database) collection of multiple sequence alignments, and the archaeal proteins were assigned to arCOGs (see Materials and Methods for details). Notably, members of the KaiC superfamily are present even in the archaea with the smallest genomes, such as Nanoarchaeota, and various KaiC families are expanded in many archaeal lineages, especially, in Thermococci and Thermoproteales (Table S1).
Phyletic patterns of KaiC-like and associated arCOGs. Download TABLE S1, XLSX file, 0.1 MB (63.6KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
KaiC-encoding genomic loci. Download TABLE S2, XLSX file, 2.5 MB (2.6MB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
From this collection of KaiC-like protein sequences, we selected a nonredundant set of proteins that could be expected to contain at least one full-sized ATPase domain (~200 amino acid residues). This nonredundant set was used to build a dendrogram by using a combination of the FastTree method and the unweighted pair group method using average linkages (UPGMA) (Text S1; see Materials and Methods for details). The resulting tree topology was largely consistent with results of previous phylogenetic analyses (34, 39).
KaiC tree, Newick format. Download TEXT S1, TXT file, 0.1 MB (76.4KB, txt) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
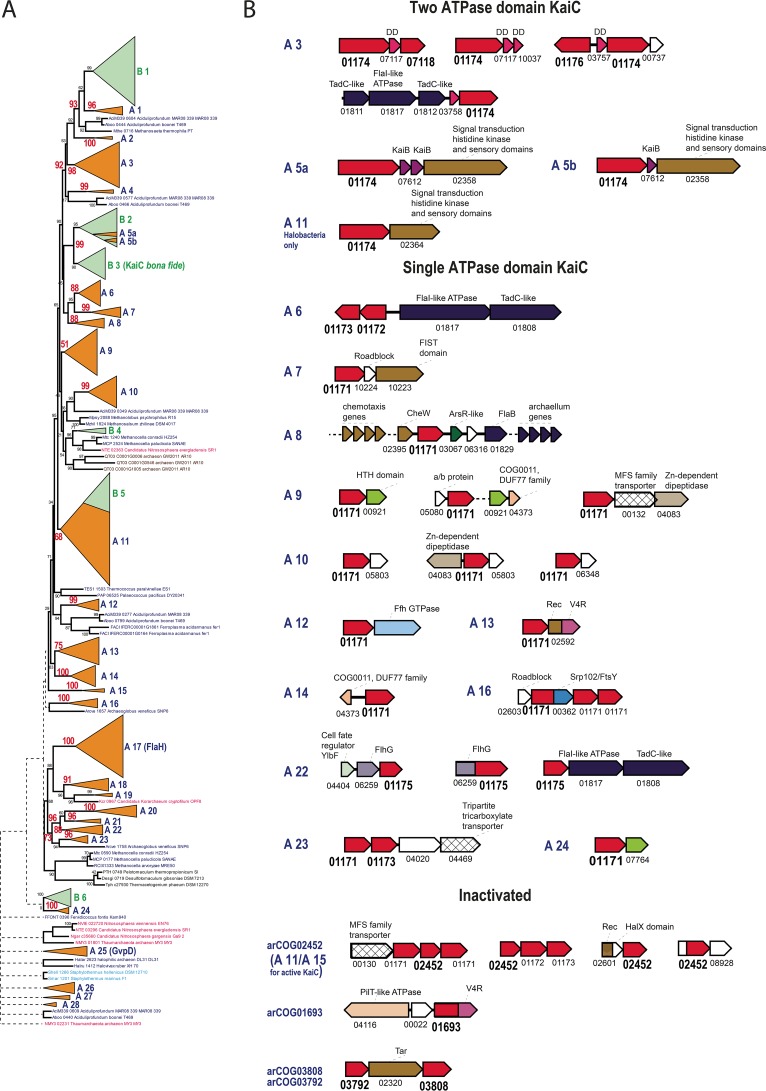
Despite the considerable overrepresentation of bacterial compared to archaeal genomes in the database, archaeal (and cyanobacterial) proteins dominate the KaiC family, in agreement with the previous conclusion that this family originated in Archaea (42). A phylogenetic tree was built for a nonredundant subset of KaiC family members (Fig. 2A). The tree contains 28 distinct strongly supported archaeal branches (A1 to A28) and 6 predominantly bacterial branches (B1 to B6). Bacterial sequences are mostly scattered over the tree, suggesting frequent HGT from archaea to bacteria. The large, mostly bacterial clade combining branches B2 and B3 corresponds to cyanobacterial KaiC components of the circadian clock (B3) and KaiC-like sequences (B2) including experimentally studied proteins of Rhodopseudomonas and Legionella (33, 43) (Fig. 2A). The strongly supported (95%) B2 clade contains several archaeal proteins, in addition to bacterial ones, all from different methanogens (branches A5a and A5b), which indicates likely HGT from bacteria to archaea. In Rhodopseudomonas (branch B2), involvement of the KaiC homologues in clock-like gene expression has been demonstrated, whereas in Legionella (branch B2), these proteins are implicated in oxidative and sodium stress resistance and do not appear to be components of an oscillator. This clade is deeply nested among diverse archaeal branches, in accord with the scenario in which the ancestral components of the circadian clock were transferred from archaea to bacteria (Fig. 2A). Proteins containing two ATPase domains and those with a single ATPase domain are interspersed in the tree, suggesting that multiple gene fusions and gene fissions occurred during the evolution of this family in Archaea. Furthermore, active and inactivated (as determined from the disruption of the Walker A and B signature motifs of the P-loop domain) ATPases are also interspersed, indicating multiple independent ATPase inactivations (Fig. 2A; Table 2). Here, we collectively refer to all groups of the KaiC homologues with inactivated ATPase domains as iKaiC; clearly, despite the abrogation of the ATPase activity, iKaiC could perform other functions, as discussed below. Archaeal branch A9 consists of KaiC-like proteins that are well represented in both Euryarchaeota and the TACK (Thaumarchaeota, Aigarchaeota, Crenarchaeota, Korarchaeota) superphylum, and thus appear to be ancestral (Table 2). Although the support of this branch is not very strong (44), all of these proteins belong to the same cluster, arCOG01171, and have a single ATPase domain, so two independent approaches to sequence clustering give similar results. The same considerations apply to branch A3, which includes KaiC-like proteins with two active ATPase domains. The third branch (A17) that appears to be ancestral consists of FlaH proteins, essential archaellum components (36, 45). The remaining tree branches are either lineage specific or include only a few archaeal lineages (Table 2; Table S3). Thus, this analysis supports the previous conclusions that at least three KaiC families could be represented in the LACA (11). The multiple long branches and inactivation of the ATPase domain imply frequent subfunctionalization of the KaiC family proteins, especially in Thermococci and Thermoproteaceae and to a lesser extent in Aciduliprofundum and Archaeoglobi. This evolutionary trend resulted in the appearance of numerous subfamilies of highly diverged iKaiC proteins (Table S1).
FIG 2 .
Phylogeny and conserved gene neighborhoods of the KaiC family. (A) The dendrogram reflecting the relationships between archaeal and bacterial representatives of the KaiC protein family was constructed as described in Materials and Methods. Major distinct branches are collapsed and shown as triangles numbered A1 to A28 for the archaeal branches and B1 to B6 for the bacterial branches. Bootstrap values calculated by the FastTree program are shown for several key nodes, and values for the major, well-supported branches are shown in red. Each sequence in the tree is described by the locus tag number and species name. Colors: green, bacterial genes; orange, archaeal genes. (B) For each branch, a conserved gene arrangement (if detected) is shown. Genes are shown as arrows. An arCOG number is shown for each gene. Functionally linked or homologous genes are represented as follows: KaiC-like genes, red; two-component signal transduction system genes, brown; type IV pili, dark blue; membrane transporters, angled grid; uncharacterized genes, white. Other domains are colored according to their descriptions provided above the domain icon. Abbreviations: V4R, V4R small-molecule-binding domain; FlhG, FlhG/MinD/FleN family ATPase, antiactivator of flagellar biosynthesis. For the complete tree, see Text S1.
TABLE 2 .
Descriptions of the major archaeal branches shown in Fig. 2
| Branch | Phyletic distribution | Commenta |
|---|---|---|
| A1 | Aciduliprofundum and several Methanomicrobia genomes | Mostly 2-domain ATPases; both domains are active |
| A2 | Methanocella only | 2-domain ATPase; second domain is inactivated and diverged |
| A3 | Patchy distribution in most archaeal lineages; three paralogs in Thermoproteales |
2 active ATPase domains; possibly an ancestral group |
| A4 | Few different archaea | Single active ATPase domain |
| A5 | Several methanomicrobia (A5a) and several Methanothermobacteriales (A5b) |
2 active ATPase domains, most closely related to bona fide cyanobacterial KaiC, likely lateral transfer from bacteria |
| A6 | Many euryarchaeal lineages but with patchy distribution | Single active ATPase domain; all belong to arCOG01173; ATPase is often fused to a large low-complexity N-terminal domain |
| A7 | Several euryarchaeal lineages but with patchy distribution | Single active ATPase domain |
| A8 | Patchy distribution in Halobacteria, Methanocella, and Nitrosopumilus, present in a small genome of archaeon_GW2011_AR10 |
Single active ATPase domain |
| A9 | Most archaeal lineages, including Nanoarchaeota | Single active ATPase domain; possibly an ancestral group |
| A10 | Most euryarchaeal lineages; duplication in Thermococci | Single active ATPase domain |
| A11 | Most archaeal lineages | Both a single active ATPase domain and 2 active ATPase domains (fused); includes bacterial branch B5, all with 2 ATPase domains |
| A12 | Thermoproteales only, 2 paralogs | Single active ATPase domain |
| A13 |
Methanomicrobia and Methanothermobacteriales, 2 paralogs in Methanosarcinales |
Single active ATPase domain |
| A14 | Most crenarcheal lineages and Koarchaeum | Single active ATPase domain |
| A15 | A few different archaea | Single active ATPase domain |
| A16 | Methanothermobacteriales | Single active ATPase domain |
| A17 | Most archaeal lineages | Single active ATPase domain; archaellum-associated protein FlaH; possibly an ancestral group |
| A18 | Thermoproteaceae only | Single active ATPase domain; arCOG05482 monophyletic |
| A19 | Several Halobacteria | Likely an active ATPase fused to metallochaperone-like domain (TRASH) |
| A20 | Patchy distribution in Archaeoglobi, Methanomicrobiales and Aciduliprofundum; present in most Halobacteria |
Single inactivated ATPase domain; arCOG01172 monophyletic |
| A21 | Archaeoglobi only | Single inactivated ATPase domain |
| A22 | Patchy distribution in Methanomicrobiales and Aciduliprofundum | Single inactivated ATPase domain; most sequences belong to arCOG01175 |
| A23 | Thermococci only | Single active ATPase domain |
| A24 | Archaeoglobi only | Single active ATPase domain; group with several bacteria |
| A25 | Several Halobacteria, all Methanosaeta and all Aciduliprofundum genomes |
2 ATPase domains; second domain is inactivated and diverged; in Halobacteria, it is GvpD, a component and regulator of a gas vesicle system |
| A26 | Thermoproteaceae only; 2 paralogs | Single active ATPase domain |
| A27 | Thermoproteaceae only | 2 ATPase domains; second domain is inactivated and diverged |
| A28 | Thermoproteaceae only | 2 ATPase domains; second domain is inactivated and diverged |
ATPase domains are denoted active if they have intact Walker A and B motifs.
KaiC tree branches. Download TABLE S3, XLSX file, 0.1 MB (81.9KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Predicted interaction partners of KaiC proteins in Archaea.
Analysis of conserved gene neighborhoods (Fig. 2B) and domain fusions (Fig. 3) revealed a complex and diverse set of proteins and domains that can be predicted to interact with KaiC family members.
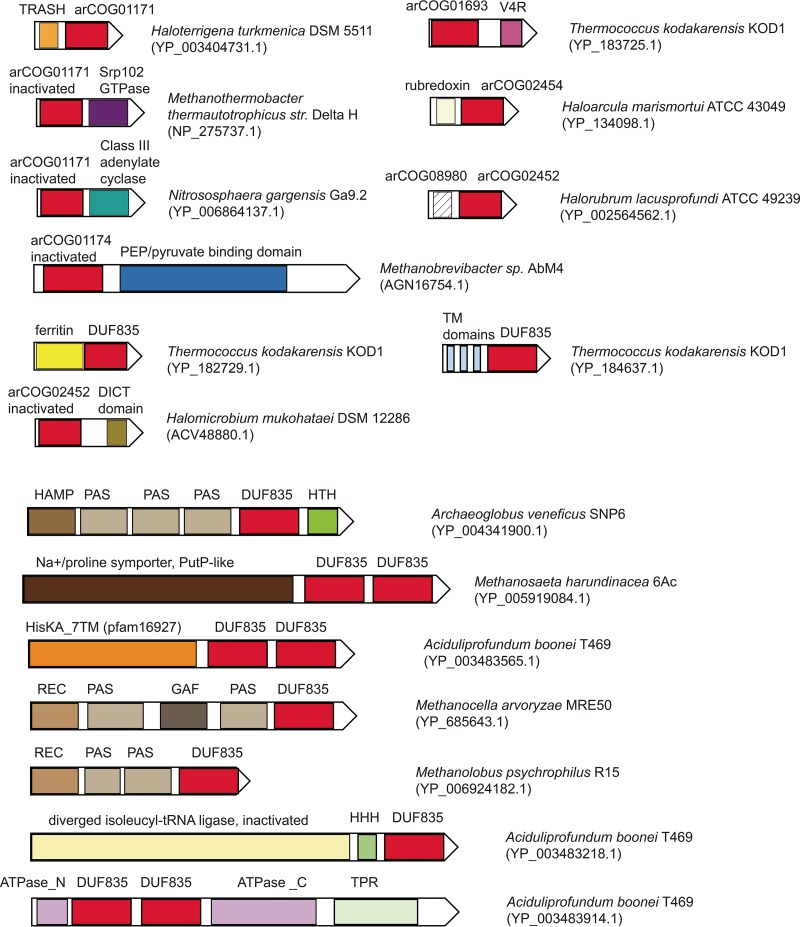
FIG 3 .
KaiC protein fusions. Individual domains are shown as rectangles. KaiC-related domains are designated by either arCOG numbers or Pfam identifiers. Species names and the respective protein IDs are show on the right. Homologous domains are color coded. Abbreviations: HAMP, PAS, REC, HisKA_7TM, and GAF, known domains shared with two-component signal transduction systems; HHH, triple-helix DNA-binding domain; TRASH, metal-binding domain predicted to be involved in heavy-metal sensing; ATPase_N, AAA ATPase N-terminal region; ATPase_C, AAA ATPase C-terminal region; TM, transmembrane segment; V4R, V4R small-molecule-binding domain.
The three most common contextual themes involving the KaiC family are (i) type IV pilus systems and other membrane-associated complexes such as the signal recognition particle (SRP) GTPase Ffh or a FlgN-like flagellar biosynthesis/secretory pathway chaperone (20), (ii) signal-transducing and sensory proteins that are typically associated with histidine kinases in bacteria, and (iii) membrane transporters (Fig. 2B).
Specifically, we can confidently predict the interacting partners for two ancestral KaiC families, in addition to FlaH, for which such partners are already known. Branch A9 KaiC proteins are predicted to interact with uncharacterized proteins from arCOG00921 (COG1318, predicted DNA-binding transcriptional regulators of the GlpR family) (Fig. 2B). The data in Table S1 indicate that arCOG00921 proteins and proteins from the A9 KaiC branch are always present in the same genome and often adjacently, even in the smallest archaeal genome of Nanoarchaeum equitans (NEQ174 and NEQ534, respectively). The coincidence of retention suggests that both components are involved in the same important cellular process(es). A third component also could be linked to this system, namely, a protein of the poorly characterized DUF77 family (pfam01910/COG0011) that is present in most archaea (arCOG04373) and appears to descend from the LACA (11). The structure of a protein from this family has been solved, revealing a ferredoxin fold, and it has been shown to form homotetramers and bind thiamine; in Thermotoga, the expression of the gene for this protein is upregulated under oxidizing conditions (46). Accordingly, it has been proposed that the protein is involved in an oxidative stress response mechanism (11). Additionally, arCOG007764 (a paralog of arCOG00921) is associated with KaiC-like ATPases of branch A24, whereas arCOG04373 is also associated with KaiC-like ATPases of branch A14, reinforcing the functional linkage of these three protein families (Fig. 2B).
Proteins with two ATPase domains, which most closely resemble the bona fide cyanobacterial KaiC protein, are typically associated with a small protein, either KaiB (branches A5a and A5b) or a member of an uncharacterized protein family (e.g., arCOG07117, arCOG03757, arCOG03758, arCOG11224, and arCOG10037) in ancestral branch A3 (Fig. 2B). The structure of one protein of this family has been solved (PDB code 2p9x), revealing a four-helix bundle fold. Structural comparison by using VAST (47) shows that the best match for this protein is the eukaryotic DEATH domain (a domain named for death, meaning its involvement in apoptosis, also often referred to as DD) with a root mean square deviation of 0.97 Å from the DD of the human RAIDD (DD-containing protein; the abbreviation is complex and is explained in reference 48) protein (PDB code 2O71) (49, 50) (Fig. S1). The DDs and related α-helical adapter domains are key components of eukaryotic signal transduction pathways, particularly those involved in programmed cell death (apoptosis), where these domains mediate connections between different components through homotypic interactions (i.e., different DD-related adapter domains interact with one another) (49, 50). Exceptions to this association are the two-domain ATPases from the halobacteria-specific clade of branch A11 and from mostly methanomicrobial branch A1, for which no small partner protein encoded in the same locus could be identified. Bacterial branch B1 lies within an archaeal subtree that includes branches A1 to A4; several internal branches within this subtree are strongly supported (>90%) (Fig. 2A), suggesting horizontal transfer from archaea to bacteria. The majority of the kaiC genes associated with branch B1 are located next to genes related to two-component systems, suggesting that archaeal KaiC of branch A1 could interact with the analogous components encoded in other loci in the respective archaeal genomes. DD-like domains are specifically expanded in the class Thermococci and several members of the phylum Thaumarchaeota (Fig. S1B; Table S1). Some of them are fused to a diverged iKaiC domain, REC domain, or ferritin domain, further linking these proteins to KaiC. Moreover, genes encoding DD-like domain proteins are found in several conserved neighborhoods together with other uncharacterized genes, suggesting that additional components could be linked to the KaiC-based signal transduction network (Fig. S1B). Taking these observations into account, we predict that the DD-like domains also serve as modulators of the autophosphorylation activity of KaiC.
DD-like family in archaea. Download FIG S1, PDF file, 1.2 MB (1.3MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Single-domain KaiC-like ATPases are often encoded as doublets of paralogs, of which some are active and others are inactivated, suggesting that they might form heterodimers, recapitulating the organization of the two-domain KaiC-like ATPases (Fig. 2B).
The fusions of KaiC with other domains are also informative, showing either the same trend as that observed for the conserved neighborhoods or suggesting the involvement of KaiC-like domains in more complex signal transduction pathways (Fig. 3). Many of these fusions (e.g., to the TRASH [trafficking, resistance, and sensing of heavy metals] sensory domain, rubredoxins, and ferritins) point to an involvement in oxidative stress. Most often, we observe ferritin domains both fused to KaiC or DUF835 and found in the respective neighborhoods (Fig. 3; Table S2). Ferritins are iron-binding proteins whose role in the oxidative stress response is well established (51).
The association with the SRP GTPase Ffh and the regulatory GTPase Srp102/FtsY suggests that KaiC-like proteins might regulate the targeting of nascent secreted or membrane proteins from the ribosome to the membrane through the SRP (44, 52).
The iKaiC domains of the DUF835 family are often found in multidomain proteins (Fig. 3). Many of these contain sensory and signal-transducing domains that have been thoroughly studied in the context of bacterial two-component signal transduction systems (53, 54). This connection suggests that DUF835 proteins are involved in signal transduction pathways. Many proteins of this family are membrane associated, presumably interacting with other membrane proteins, some of which are fused to the DUF835 domain (e.g., the Na+/proline symporter-like domain) (Fig. 3; Tables S1 and S2). Fusions with other regulatory and signal transduction proteins, such as AAA ATPases containing tetratricopeptide repeats and cyclases, in particular (Fig. 3), suggest that KaiC family proteins are involved in highly complex pathways, which include cross-talk with other signal transduction systems.
Finally, the previously described MEDS (methanogen/methylotroph DcmR sensory) domain (arCOG03567, pfam14417) shows a clear affinity for the KaiC family. PSI-BLAST searches initiated with any of the MEDS domain sequences against the arCOG database reveal significant sequence similarity of this domain with the members of KaiC-like arCOG01171 (E value of 4e-05 in the second iteration), although the MEDS domain is unlikely to be an active ATPase because of the lack of catalytic residues in the Walker A and B ATPase motifs. The MEDS domain has been described previously (55) both as a stand-alone domain, often encoded in the genomic neighborhoods of other components of signal transduction systems, and in a fusion with sensory histidine kinases along with other sensory domains (55). Here we also identified fusions of MEDS and DD-like domains (Fig. S1). Taken together, these observations suggest that the MEDS domain could be functionally similar to the DUF835 domain described above.
Many domains and genes linked to iKaiC proteins remain uncharacterized. There is an expansion of two protein families in Halobacteria that are associated with iKaiC of arCOG02452. One of these has been discussed previously in the context of signal transduction systems and is called HalX (arCOG02601/pfam08663) (56). The HalX domain is often fused to the REC domain and is found in the context of genes associated with a two-component signal transduction system (Fig. 2; Tables S1 and S2). The second expanded domain has not been previously described. In halobacterial genomes, it is represented by multiple paralogs that belong to five arCOGs, arCOG08928, arCOG08103, arCOG08989, arCOG09008, and arCOG08980. Among these, only arCOG08928 is often located next to an iKaiC of arCOG02452, and a few arCOG08980 members are fused to iKaiC (Fig. 2 and 3). Both domains might function as input domains for the respective KaiC-like proteins.
Models of KaiC-based signal transduction systems.
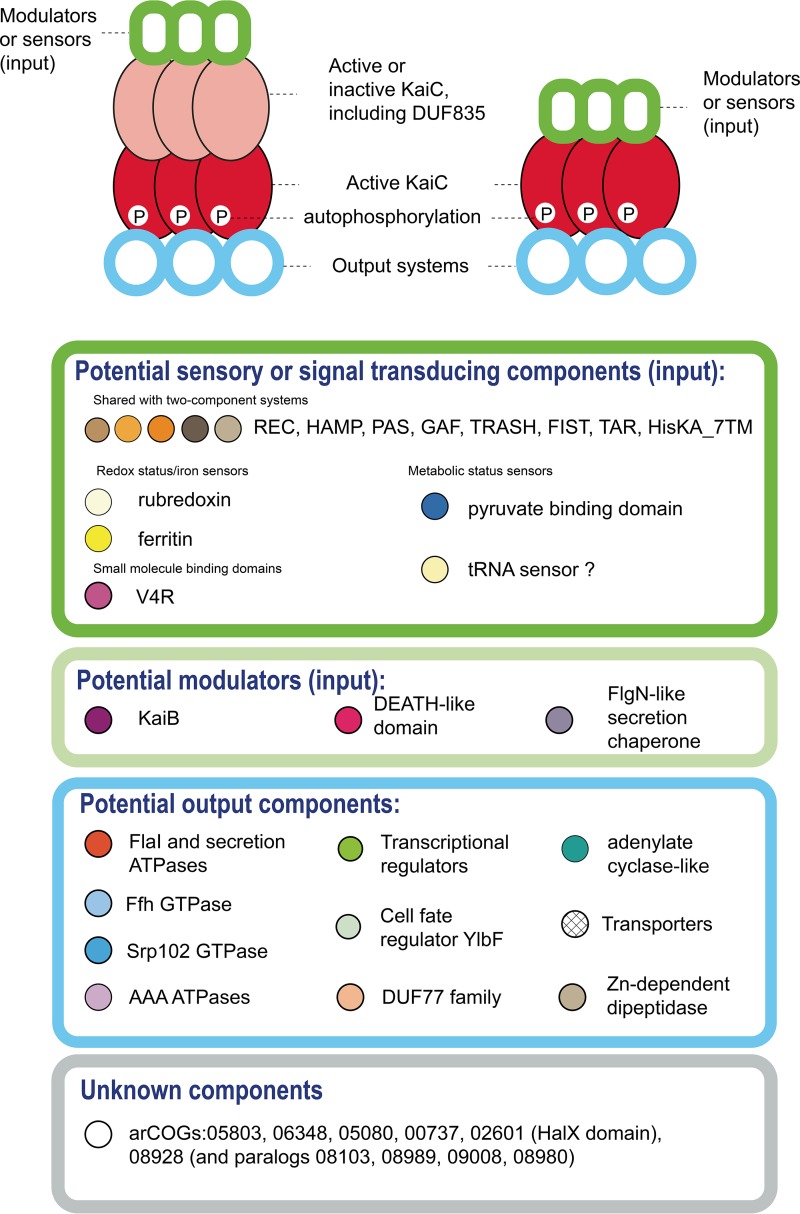
The multiple lines of evidence discussed above indicate that the KaiC family is likely a major hub of a versatile and complex archaeal signaling network that so far has largely escaped attention. Nevertheless, the available experimental data on a halobacterial circadian clock (24, 57) and the recent progress in the study of the functions of FlaH in the archaellum (35, 36, 45) allow us to propose two models of the roles of KaiC-like proteins in signal transduction (Fig. 4). The first model is essentially identical to the circadian clock mechanism and postulates the formation of either a homohexameric ring of KaiC proteins containing two ATPase domains or heterohexamers of interacting KaiC proteins, each containing a single ATPase domain. Both domains can be active ATPases, or alternatively, one of the domains could be inactivated, such as one of the multiple DUF835 domains, which would pass the signal from an input domain to the active KaiC-like domain (Fig. 4). Each of the hexameric ATPase rings would interact with multiple partners and, as with other signal transduction systems, such partners can be roughly classified into input and output components (Fig. 4). In addition, the KaiC rings could interact with modulators of the ATPase activity, such as KaiB in the circadian clock, which might compete for binding with other output proteins.
FIG 4 .
Models of protein complex architectures and putative functions of the components of KaiC-based signal transduction pathways in archaea. KaiC pathway protein components are shown as colored shapes. Below the scheme of predicted protein-protein interaction, selected input, modulator, and output components are listed inside the oval borders, which are colored according to the predicted functions of these components. Each protein family name is shown next to a circle of the same color used for this component in Fig. 2 and 3.
The second model postulates interaction of a single-domain KaiC-like ATPase homohexamer directly with an output domain, similar to the potential interaction between FlaH and FlaI in the archaellum (36, 45) (Fig. 4). Many of the predicted components remain uncharacterized, and thus no specific functions can be predicted for them at this time. Furthermore, the KaiC-centered signaling systems could be interconnected with other signal transduction pathways, in particular, with two-components systems, via shared domains of input proteins (Fig. 4), and with Ras-like GTPases, either directly through the interaction with Srp102/FtsY or through Roadblock family proteins as described for both bacteria and eukaryotes (58, 59). The mode of signaling apparently can be modified with relative ease. For example, a two-domain KaiC-like ATPase and a DD-like protein are encoded in the type IV pilus loci in Thermoproteales, whereas in Euryarchaeota, these loci contain a gene coding for a single-domain KaiC (arCOG01175) linked to a FlhG-like secretion chaperone. Thus, two distinct models could apply to the same process of regulation of type IV pilus (or archaellum) assembly in different archaea (Fig. 4).
It can be predicted that many KaiC-like proteins lack autophosphorylation activity but could bind and/or hydrolyze ATP to transduce the signal. Indeed, many of these proteins lack the pair of serine/threonine residues that are conserved among the bona fide KaiC proteins and are autophosphorylated in the circadian clock system (30). However, several archaeal KaiC subfamilies, especially the two-domain ATPases, retain this motif or at least one of the two hydroxy amino acids and could be active autokinases.
Implications for the archaeal circadian clock.
Among archaea, diurnal gene expression has been demonstrated only in Halobacteria (24, 57, 60). It has been shown that KaiC-like proteins undergo cyclic expression, and deletion of most of them affected the expression of the others, suggesting that Halobacteria indeed might have a bona fide KaiC-based circadian mechanism (38). Similarly to cyanobacteria, Halobacteria adjust their metabolism to light conditions via rhodopsin-based proton pumps that generate a proton gradient and sensory rhodopsins that control phototaxis (61). Halobacteria encode two-domain KaiC-like ATPases (both within branch A11 in Fig. 2A), which do not group with KaiC from cyanobacteria. Furthermore, neither KaiB nor KaiA nor any potential analogue of these KaiC interactors could be identified in the genomic neighborhoods of the halobacterial KaiC-like ATPases. Moreover, there was a weak, if any, correlation between the presence of two-domain KaiC ATPases and rhodopsin-like proteins in halobacterial genomes (Table S1). Accordingly, the functions of these proteins in Halobacteria remain unclear. To the best of our knowledge, no evidence of a circadian clock in any other archaea has been reported and no rhodopsins have been identified.
A putative minimal circadian clock system consisting of KaiC from branch A5 and KaiB is present in some methanogens (Fig. 2; Table S3). However, as in the case in Legionella, this system could be involved in regulatory pathways distinct from the circadian clock. Apart from Halobacteria, there seems to be no indication that archaea can sense light and modulate their metabolism accordingly. It thus appears unlikely that most archaea possess circadian clocks similar to those of photosynthetic bacteria. However, if indications of clock mechanisms in archaea (other than Halobacteria) were found, the best candidates would be the systems containing the two-domain KaiC-like ATPases of branch A5, which is associated with KaiB, and those of branch A3, associated with a DEATH-like domain, a potential analogue of KaiB (Fig. 2 and 4).
Concluding remarks.
The striking proliferation and diversification of the KaiC-like ATPase family in archaea imply that these proteins comprise the core of diverse, unexplored, and apparently, archaea-specific signal transduction networks. These signal transduction systems are likely involved in the regulation of membrane-associated complexes and individual proteins, such as the archaellum, type IV pili, SRP, and membrane transporters. Additionally, the KaiC-centered signal transduction machinery can be predicted to regulate a response to oxidative stress. However, it appears unlikely that archaea, apart, maybe, from Halobacteria, possess cyanobacterial-type, KaiC-centered circadian clocks. The KaiC-based signaling mechanisms appear to be ancestral in Archaea, with at least three KaiC paralogs projected to the LACA. One ancestral KaiC subfamily that includes a protein containing an HTH domain (arCOG00921) might be involved in as-yet-uncharacterized global response pathways because it is encoded even in the minimal genome of the Nanoarchaeota. The predicted KaiC-based signal transduction system appears to be interconnected with two-component signal transduction systems through iKaiC of the DUF835 family and MEDS domains that are predicted to interact with active KaiC ATPases. In contrast, we could not identify any connections between the KaiC-centered network and genes involved in the S/T kinase pathway. Additionally, inspection of the available data on archaeal phosphoproteomes yielded no indications of extensive phosphorylation of KaiC pathway-related genes (62–64). Thus, the KaiC network appears to be largely disjointed from the S/T kinase-mediated regulatory pathways in Archaea. The phylogenomic analysis reported here can produce only crude models of archaeal signal transduction. Nevertheless, these observations expose multiple experimental directions that can be expected to shed light on key aspects of archaeal cell biology.
MATERIALS AND METHODS
Archaeal and bacterial complete genome sequences were downloaded from the NCBI FTP site (ftp://ftp.ncbi.nlm.nih.gov/genomes/all/) in March 2016. Altogether, the database includes 4,961 completely sequenced and assembled genomes. These genomes were assigned to COGs and Pfam families by using the PSI-BLAST program with an E-value cutoff of 1e-4 and low-complexity filtering turned off against a collection of multiple sequence alignments (profiles) from the CDD database (65) derived from COGs, Pfam, and CDD itself. The same approach was used to assign archaeal proteins to arCOGs as described previously (66).
All proteins that were assigned to any of the three groups (COG0467/pfam06745, COG2874, pfam05763) or to arCOGs associated with the KaiC family were retrieved. Genomic loci containing five genes upstream and downstream of all kaiC-like genes were extracted for neighborhood analysis. KaiC-like sequences were clustered by using BLASTCLUST (ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.html) with a length coverage of 90% and a sequence identity threshold of 90% to obtain a nonredundant set of sequences. Among those, readily alignable groups of predominantly active ATPase sequences were selected for phylogenetic analysis (several inactivated ATPases aligned poorly and were not included in this analysis; also, protein fragments in the nonredundant set were discarded). The final set used for tree reconstruction included 1,011 sequences. Tree reconstruction was performed by two approaches, (i) a combination of FastTree and UPGMA for full-length sequences and (ii) the default FastTree method for the N-terminal ATPase domain only. For the first approach, initial sequence clusters were obtained by using UCLUST (67) with a sequence similarity threshold of 0.5; the sequences were aligned within clusters by using MUSCLE (68). Cluster-to-cluster similarity scores were then obtained by using HHsearch (69) (including trivial clusters consisting of a single sequence each). A UPGMA dendrogram was constructed from the pairwise similarity scores. Highly similar clusters (pairwise-score to self-score ratio, >0.1) were aligned with each other by using HHALIGN (69). This procedure was repeated iteratively. At the last step, sequence-based trees were reconstructed from the cluster alignments by using FastTree (70) as described below and rooted by midpoint; these trees were grafted onto the tips of the profile similarity-based UPGMA dendrogram. Sites with gap character fraction values of >0.5 and homogeneity values of <0.1 were removed from the alignment (71). In both cases, the FastTree program (70) was executed with the WAG evolutionary model and the discrete gamma model with 20 rate categories. The same program was used to compute SH (Shimodaira-Hasegawa)-like node support values.
To identify remote sequence similarity, HHpred with default parameters (69) and CD search (72) with an E value cutoff of 10 and composition-based statistics adjustment turned off were used. In addition, web-based, manually curated PSI-BLAST searches were run with and without the composition-based statistics adjustment and with low-complexity filtering turned off. Inclusion E-value thresholds of 0.1 to 1e-8, depending on sequence length and content, were used, and some searches were run against the archaeal subset of the NCBI nonredundant protein database. The VAST program (47) was used with default parameters for structural comparison.
ACKNOWLEDGMENTS
We thank Thomas Santangelo of Colorado State University and Sonja Albers of the University of Freiburg for helpful discussions and critical reading of the manuscript. We are also grateful to Yuri Wolf of the National Center for Biotechnology Information for technical assistance and for in-house scripts that we used during this project.
Our research is supported by the NIH Intramural Research Program at the National Library of Medicine, U.S. Department of Health and Human Services.
Footnotes
Citation Makarova KS, Galperin MY, Koonin EV. 2017. Proposed role for KaiC-like ATPases as major signal transduction hubs in archaea. mBio 8:e01959-17. https://doi.org/10.1128/mBio.01959-17.
REFERENCES
- 1.Zschiedrich CP, Keidel V, Szurmant H. 2016. Molecular mechanisms of two-component signal transduction. J Mol Biol 428:3752–3775. doi: 10.1016/j.jmb.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin J, Pawson T. 2012. Modular evolution of phosphorylation-based signalling systems. Philos Trans R Soc Lond B Biol Sci 367:2540–2555. doi: 10.1098/rstb.2012.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor SS, Kornev AP. 2011. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem Sci 36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wuichet K, Zhulin IB. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal 3:ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galperin MY. 2010. Diversity of structure and function of response regulator output domains. Curr Opin Microbiol 13:150–159. doi: 10.1016/j.mib.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitworth DE, Cock PJ. 2009. Evolution of prokaryotic two-component systems: insights from comparative genomics. Amino Acids 37:459–466. doi: 10.1007/s00726-009-0259-2. [DOI] [PubMed] [Google Scholar]
- 7.Esser D, Hoffmann L, Pham TK, Bräsen C, Qiu W, Wright PC, Albers SV, Siebers B. 2016. Protein phosphorylation and its role in archaeal signal transduction. FEMS Microbiol Rev 40:625–647. doi: 10.1093/femsre/fuw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlesner M, Miller A, Besir H, Aivaliotis M, Streif J, Scheffer B, Siedler F, Oesterhelt D. 2012. The protein interaction network of a taxis signal transduction system in a halophilic archaeon. BMC Microbiol 12:272. doi: 10.1186/1471-2180-12-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Zheng X, Guo X, Qi L, Dong X. 2014. Characterization of an archaeal two-component system that regulates methanogenesis in Methanosaeta harundinacea. PLoS One 9:e95502. doi: 10.1371/journal.pone.0095502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koretke KK, Lupas AN, Warren PV, Rosenberg M, Brown JR. 2000. Evolution of two-component signal transduction. Mol Biol Evol 17:1956–1970. doi: 10.1093/oxfordjournals.molbev.a026297. [DOI] [PubMed] [Google Scholar]
- 11.Wolf YI, Makarova KS, Yutin N, Koonin EV. 2012. Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer. Biol Direct 7:46. doi: 10.1186/1745-6150-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koonin EV, Makarova KS, Aravind L. 2001. Horizontal gene transfer in prokaryotes—quantification and classification. Annu Rev Microbiol 55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson-Sathi S, Sousa FL, Roettger M, Lozada-Chávez N, Thiergart T, Janssen A, Bryant D, Landan G, Schönheit P, Siebers B, McInerney JO, Martin WF. 2015. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517:77–80. doi: 10.1038/nature13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.López-García P, Zivanovic Y, Deschamps P, Moreira D. 2015. Bacterial gene import and mesophilic adaptation in archaea. Nat Rev Microbiol 13:447–456. doi: 10.1038/nrmicro3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garushyants SK, Kazanov MD, Gelfand MS. 2015. Horizontal gene transfer and genome evolution in Methanosarcina. BMC Evol Biol 15:102. doi: 10.1186/s12862-015-0393-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papke RT, Corral P, Ram-Mohan N, Haba RR, Sánchez-Porro C, Makkay A, Ventosa A. 2015. Horizontal gene transfer, dispersal and haloarchaeal speciation. Life 5:1405–1426. doi: 10.3390/life5021405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deschamps P, Zivanovic Y, Moreira D, Rodriguez-Valera F, López-García P. 2014. Pangenome evidence for extensive interdomain horizontal transfer affecting lineage core and shell genes in uncultured planktonic Thaumarchaeota and Euryarchaeota. Genome Biol Evol 6:1549–1563. doi: 10.1093/gbe/evu127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galperin MY, Makarova KS, Wolf YI, Koonin EV. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makarova KS, Yutin N, Bell SD, Koonin EV. 2010. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat Rev Microbiol 8:731–741. doi: 10.1038/nrmicro2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarova KS, Koonin EV, Albers SV. 2016. Diversity and evolution of type IV pili systems in Archaea. Front Microbiol 7:667. doi: 10.3389/fmicb.2016.00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haldenby S, White MF, Allers T. 2009. RecA family proteins in archaea: RadA and its cousins. Biochem Soc Trans 37:102–107. doi: 10.1042/BST0370102. [DOI] [PubMed] [Google Scholar]
- 22.McRobbie AM, Carter LG, Kerou M, Liu H, McMahon SA, Johnson KA, Oke M, Naismith JH, White MF. 2009. Structural and functional characterisation of a conserved archaeal RadA paralog with antirecombinase activity. J Mol Biol 389:661–673. doi: 10.1016/j.jmb.2009.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Axmann IM, Hertel S, Wiegard A, Dörrich AK, Wilde A. 2014. Diversity of KaiC-based timing systems in marine cyanobacteria. Mar Genomics 14:3–16. doi: 10.1016/j.margen.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Johnson CH, Zhao C, Xu Y, Mori T. 2017. Timing the day: what makes bacterial clocks tick? Nat Rev Microbiol 15:232–242. doi: 10.1038/nrmicro.2016.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egli M. 2017. Architecture and mechanism of the central gear in an ancient molecular timer. J R Soc Interface 14:20161065. doi: 10.1098/rsif.2016.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shultzaberger RK, Boyd JS, Diamond S, Greenspan RJ, Golden SS. 2015. Giving time purpose: the Synechococcus elongatus clock in a broader network context. Annu Rev Genet 49:485–505. doi: 10.1146/annurev-genet-111212-133227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen SE, Golden SS. 2015. Circadian rhythms in cyanobacteria. Microbiol Mol Biol Rev 79:373–385. doi: 10.1128/MMBR.00036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pattanayek R, Egli M. 2015. Protein-protein interactions in the cyanobacterial circadian clock: structure of KaiA dimer in complex with C-terminal KaiC peptides at 2.8 Å resolution. Biochemistry 54:4575–4578. doi: 10.1021/acs.biochem.5b00694. [DOI] [PubMed] [Google Scholar]
- 29.Egli M, Johnson CH. 2013. A circadian clock nanomachine that runs without transcription or translation. Curr Opin Neurobiol 23:732–740. doi: 10.1016/j.conb.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishiwaki T, Satomi Y, Kitayama Y, Terauchi K, Kiyohara R, Takao T, Kondo T. 2007. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J 26:4029–4037. doi: 10.1038/sj.emboj.7601832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dvornyk V, Vinogradova O, Nevo E. 2003. Origin and evolution of circadian clock genes in prokaryotes. Proc Natl Acad Sci U S A 100:2495–2500. doi: 10.1073/pnas.0130099100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zinser ER, Lindell D, Johnson ZI, Futschik ME, Steglich C, Coleman ML, Wright MA, Rector T, Steen R, McNulty N, Thompson LR, Chisholm SW. 2009. Choreography of the transcriptome, photophysiology, and cell cycle of a minimal photoautotroph, Prochlorococcus. PLoS One 4:e5135. doi: 10.1371/journal.pone.0005135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma P, Mori T, Zhao C, Thiel T, Johnson CH. 2016. Evolution of KaiC-dependent timekeepers: a proto-circadian timing mechanism confers adaptive fitness in the purple bacterium Rhodopseudomonas palustris. PLoS Genet 12:e1005922. doi: 10.1371/journal.pgen.1005922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leipe DD, Aravind L, Grishin NV, Koonin EV. 2000. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res 10:5–16. [PubMed] [Google Scholar]
- 35.Meshcheryakov VA, Wolf M. 2016. Crystal structure of the flagellar accessory protein FlaH of Methanocaldococcus jannaschii suggests a regulatory role in archaeal flagellum assembly. Protein Sci 25:1147–1155. doi: 10.1002/pro.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaudhury P, Neiner T, D’Imprima E, Banerjee A, Reindl S, Ghosh A, Arvai AS, Mills DJ, van der Does C, Tainer JA, Vonck J, Albers SV. 2016. The nucleotide-dependent interaction of FlaH and FlaI is essential for assembly and function of the archaellum motor. Mol Microbiol 99:674–685. doi: 10.1111/mmi.13260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfeifer F. 2015. Haloarchaea and the formation of gas vesicles. Life 5:385–402. doi: 10.3390/life5010385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maniscalco M, Nannen J, Sodi V, Silver G, Lowrey PL, Bidle KA. 2014. Light-dependent expression of four cryptic archaeal circadian gene homologs. Front Microbiol 5:79. doi: 10.3389/fmicb.2014.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmelling NM, Lehmann R, Chaudhury P, Beck C, Albers SV, Axmann IM, Wiegard A. 2017. Minimal tool set for a prokaryotic circadian clock. BMC Evol Biol 17:169. doi: 10.1186/s12862-017-0999-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang HJ, Kubota K, Ming H, Miyazono K, Tanokura M. 2009. Crystal structure of KaiC-like protein PH0186 from hyperthermophilic archaea Pyrococcus horikoshii OT3. Proteins 75:1035–1039. doi: 10.1002/prot.22367. [DOI] [PubMed] [Google Scholar]
- 42.Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL, Wolf YI, Koonin EV. 1999. Comparative genomics of the Archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell. Genome Res 9:608–628. [PubMed] [Google Scholar]
- 43.Loza-Correa M, Sahr T, Rolando M, Daniels C, Petit P, Skarina T, Gomez Valero L, Dervins-Ravault D, Honoré N, Savchenko A, Buchrieser C. 2014. The Legionella pneumophila kai operon is implicated in stress response and confers fitness in competitive environments. Environ Microbiol 16:359–381. doi: 10.1111/1462-2920.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Egea PF, Tsuruta H, de Leon GP, Napetschnig J, Walter P, Stroud RM. 2008. Structures of the signal recognition particle receptor from the archaeon Pyrococcus furiosus: implications for the targeting step at the membrane. PLoS One 3:e3619. doi: 10.1371/journal.pone.0003619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banerjee A, Neiner T, Tripp P, Albers SV. 2013. Insights into subunit interactions in the Sulfolobus acidocaldarius archaellum cytoplasmic complex. FEBS J 280:6141–6149. doi: 10.1111/febs.12534. [DOI] [PubMed] [Google Scholar]
- 46.Dermoun Z, Foulon A, Miller MD, Harrington DJ, Deacon AM, Sebban-Kreuzer C, Roche P, Lafitte D, Bornet O, Wilson IA, Dolla A. 2010. TM0486 from the hyperthermophilic anaerobe Thermotoga maritima is a thiamin-binding protein involved in response of the cell to oxidative conditions. J Mol Biol 400:463–476. doi: 10.1016/j.jmb.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Madej T, Lanczycki CJ, Zhang D, Thiessen PA, Geer RC, Marchler-Bauer A, Bryant SH. 2014. MMDB and VAST+: tracking structural similarities between macromolecular complexes. Nucleic Acids Res 42:D297–D303. doi: 10.1093/nar/gkt1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duan H, Dixit VM. 1997. RAIDD is a new “death” adaptor molecule. Nature 385:86–89. doi: 10.1038/385086a0. [DOI] [PubMed] [Google Scholar]
- 49.Kersse K, Verspurten J, Vanden Berghe T, Vandenabeele P. 2011. The death-fold superfamily of homotypic interaction motifs. Trends Biochem Sci 36:541–552. doi: 10.1016/j.tibs.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 50.Park HH, Wu H. 2007. Crystallization and preliminary X-ray crystallographic studies of the oligomeric death-domain complex between PIDD and RAIDD. Acta Crystallogr Sect F Struct Biol Cryst Commun 63:229–232. doi: 10.1107/S1744309107007889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arosio P, Levi S. 2002. Ferritin, iron homeostasis, and oxidative damage. Free Radic Biol Med 33:457–463. doi: 10.1016/S0891-5849(02)00842-0. [DOI] [PubMed] [Google Scholar]
- 52.Peluso P, Shan SO, Nock S, Herschlag D, Walter P. 2001. Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry 40:15224–15233. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- 53.Galperin MY. 2004. Bacterial signal transduction network in a genomic perspective. Environ Microbiol 6:552–567. doi: 10.1111/j.1462-2920.2004.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Airola MV, Huh D, Sukomon N, Widom J, Sircar R, Borbat PP, Freed JH, Watts KJ, Crane BR. 2013. Architecture of the soluble receptor Aer2 indicates an in-line mechanism for PAS and HAMP domain signaling. J Mol Biol 425:886–901. doi: 10.1016/j.jmb.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anantharaman V, Aravind L. 2005. MEDS and PocR are novel domains with a predicted role in sensing simple hydrocarbon derivatives in prokaryotic signal transduction systems. Bioinformatics 21:2805–2811. doi: 10.1093/bioinformatics/bti418. [DOI] [PubMed] [Google Scholar]
- 56.Galperin MY. 2006. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol 188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitehead K, Pan M, Masumura K, Bonneau R, Baliga NS. 2009. Diurnally entrained anticipatory behavior in archaea. PLoS One 4:e5485. doi: 10.1371/journal.pone.0005485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levine TP, Daniels RD, Wong LH, Gatta AT, Gerondopoulos A, Barr FA. 2013. Discovery of new Longin and Roadblock domains that form platforms for small GTPases in Ragulator and TRAPP-II. Small GTPases 4:62–69. doi: 10.4161/sgtp.24262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miertzschke M, Koerner C, Vetter IR, Keilberg D, Hot E, Leonardy S, Søgaard-Andersen L, Wittinghofer A. 2011. Structural analysis of the Ras-like G protein MglA and its cognate GAP MglB and implications for bacterial polarity. EMBO J 30:4185–4197. doi: 10.1038/emboj.2011.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, Maywood ES, Hastings MH, Baliga NS, Merrow M, Millar AJ, Johnson CH, Kyriacou CP, O’Neill JS, Reddy AB. 2012. Peroxiredoxins are conserved markers of circadian rhythms. Nature 485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharma AK, Walsh DA, Bapteste E, Rodriguez-Valera F, Ford Doolittle W, Papke RT. 2007. Evolution of rhodopsin ion pumps in haloarchaea. BMC Evol Biol 7:79. doi: 10.1186/1471-2148-7-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu WL, Lai SJ, Yang JT, Chern J, Liang SY, Chou CC, Kuo CH, Lai MC, Wu SH. 2016. Phosphoproteomic analysis of Methanohalophilus portucalensis FDF1(T) identified the role of protein phosphorylation in methanogenesis and osmoregulation. Sci Rep 6:29013. doi: 10.1038/srep29013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aivaliotis M, Macek B, Gnad F, Reichelt P, Mann M, Oesterhelt D. 2009. Ser/Thr/Tyr protein phosphorylation in the archaeon Halobacterium salinarum—a representative of the third domain of life. PLoS One 4:e4777. doi: 10.1371/journal.pone.0004777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reimann J, Esser D, Orell A, Amman F, Pham TK, Noirel J, Lindås AC, Bernander R, Wright PC, Siebers B, Albers SV. 2013. Archaeal signal transduction: impact of protein phosphatase deletions on cell size, motility, and energy metabolism in Sulfolobus acidocaldarius. Mol Cell Proteomics 12:3908–3923. doi: 10.1074/mcp.M113.027375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Makarova KS, Wolf YI, Koonin EV. 2015. Archaeal clusters of orthologous genes (arCOGs): an update and application for analysis of shared features between Thermococcales, Methanococcales, and Methanobacteriales. Life 5:818–840. doi: 10.3390/life5010818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 68.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV. 2008. The deep archaeal roots of eukaryotes. Mol Biol Evol 25:1619–1630. doi: 10.1093/molbev/msn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marchler-Bauer A, Bryant SH. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Galperin MY. 2005. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol 5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phyletic patterns of KaiC-like and associated arCOGs. Download TABLE S1, XLSX file, 0.1 MB (63.6KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
KaiC-encoding genomic loci. Download TABLE S2, XLSX file, 2.5 MB (2.6MB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
KaiC tree, Newick format. Download TEXT S1, TXT file, 0.1 MB (76.4KB, txt) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
KaiC tree branches. Download TABLE S3, XLSX file, 0.1 MB (81.9KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DD-like family in archaea. Download FIG S1, PDF file, 1.2 MB (1.3MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.