Dear Editor,
Plant receptor kinases (RKs) can function as pattern recognition receptors (PRRs) for perception of pathogen-associated molecular patterns (PAMPs) to induce immune responses1,2. One of such PRRs is the leucine-rich repeat RK (LRR-RK) FLAGELLIN-SENSING 2 (FLS2) that recognizes bacteria-derived flagellin (flg22 epitope)3,4. The smaller LRR-RK BRI1-associated kinase 1 (BAK1) acts as a co-receptor with FLS25,6.
The Arabidopsis LRR-RK BAK1-interacting receptor-like kinase 1 (BIR1) was initially identified through a reverse genetic screen and the phenotypes of bir1-1 can be suppressed by the adapter LRR-RK SOBIR1 (suppressor of BIR1)7. SOBIR1 interacted with BAK1 in planta when the expression of BIR1 was silenced, suggesting that BIR1 sequesters BAK1 from SOBIR1 in resting cells to inhibit cell death and immune responses8. All four BIR members (BIR1-BIR4) in Arabidopsis interacted with BAK1 when expressed in Nicotiana benthamiana. Like bir1, BAK1-interacting receptor-like kinase 2 (bir2) mutants also display enhanced SA-dependent cell-death9. BIR2 also has a critical role in negative regulation of flg22-induced responses by controlling BAK1-FLS2 complex formation in a ligand-dependent manner9.
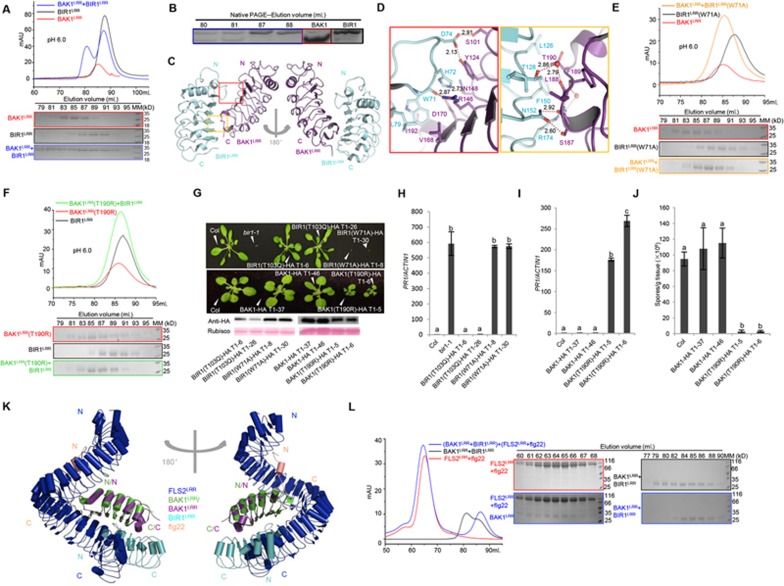
We first examined BAK1-BIR1 interaction using their extracellular LRR portions expressed in insect cells. BIR1LRR and BAK1LRR formed a stable heterodimeric complex at pH 6.0 in gel filtration (Figure 1A, Supplementary information, Figure S1), which was further confirmed by native gel analysis of the gel filtration fractions (Figure 1B). The BAK1LRR-BIR1LRR interaction was further supported by Isothermal Titration Calorimetry (ITC) and Sedimentation-Velocity Analytical UltraCentrifugation (SV-AUC) analyses (Supplementary information, Figure S2). At pH 4.0, BAK1LRR and BIR1LRR still interacted with each other (Supplementary information, Figure S3A) in gel filtration. However, BAK1LRR lost its activity of interacting with BIR1LRR at pH 8.0 (Supplementary information, Figure S3B). These results indicate that the BAK1LRR-BIR1LRR interaction in vitro is pH-dependent, which is further supported by the ITC data (Supplementary information, Figure S4). Similar to BIR1LRR, BIR3LRR and BIR4LRR also displayed interaction with BAK1LRR at pH 6.0 (Supplementary information, Figure S5A, S5C, S5D). But unlike BIR1LRR, BIR3LRR and BIR4LRR still interacted with BAK1LRR at pH 8.0 in both gel filtration and ITC assays (Supplementary information, Figure S6A, S6C, S6D). Compared to BIR1LRR, BIR3LRR and BIR4LRR, BIR2LRR exhibited a much weaker affinity towards BAK1LRR at pH 6.0 (Supplementary information, Figure S5A, S5B). Like BIR1LRR, BIR2LRR also had no detectable interaction with BAK1LRR at pH 8.0 in gel filtration and ITC assays (Supplementary information, Figure S6A, S6B). These biochemical data indicate that BIR1-4 and BAK1 directly interact with each other through their ecto-domains.
Figure 1.
Interaction between BAK1LRR and BIR1LRR is required for BIR1 inhibition of BAK1-mediated plant immunity. (A) Gel filtration profiles of BAK1LRR and BIR1LRR at pH 6.0. The vertical and horizontal axes represent ultraviolet absorbance (λ = 280 nm) and elution volume (mL), respectively. Bottom panel, coomassie blue staining of the peak fractions following SDS-PAGE. The numbers shown on the top of the SDS-PAGE gels indicate elution volumes (mL). Frame colors of the SDS-PAGE gels are equivalents to those of the gel filtration profiles for proteins indicated. MM: molecular weight maker. Hiload 200 was used for the gel filtration assays. (B) Native-PAGE coomassie blue staining of the peak fractions for the gel filtration at pH 6.0 in (A). (C) Overall structure of the BAK1LRR-BIR1LRR complex. “N” and “C” represent the N- and C-terminus, respectively. (D) Detail interactions between BAK1LRR and BIR1LRR. The left panel, the interface between the N-terminal side of BIR1LRR and BAK1LRR. The right panel, the interface between the C-terminal portion of BAK1LRR and BIR1LRR. Red dashed lines indicate polar interactions and their distances are labeled. T, Thr; L, Leu; N, Asn; S, Ser; Y, Tyr; D, Asp; H, His; R, Arg; W, Trp; V, Val; I, Ile; F, Phe. (E) Mutagenesis analysis of the BAK1LRR-BIR1LRR (W71A) complex using gel filtration. (F) Mutagenesis analysis of the BAK1LRR (T190R)-BIR1LRR complex using gel filtration. (E and F) The assays were performed as described in (A). (G) Morphological phenotypes of transgenic plants expressing the BIR1 (T103Q) -HA or BIR1 (W71A)-HA protein in bir1-1, and the BAK1-HA or BAK1 (T190R)-HA protein under its native promoter in wild type background (Col-0). The photograph shows four-week-old soil-grown plants. Expression of proteins was detected by western blot using an anti-HA antibody. (H and I) Expression levels of PR1 in the indicated genotypes as determined by quantitative RT-PCR. Two-week-old seedlings grown on ½ MS plates were used for the assays. Values were normalized to the expression levels of ACTIN1. The data are shown as means ± SD (n = 3) with one-way ANOVA and Tukey's test. Different letters indicate significant differences (P < 0.01). The experiments were repeated three times with similar results. (J) Growth of H. a. Noco2 on seedlings of the indicated genotypes. The data are shown as mean ± SD (n = 3) with one-way ANOVA and Tukey's test. Different letters indicate significant differences (P < 0.01). The experiments were repeated three times with similar results. (K) Structural superimposition of the BAK1LRR-BIR1LRR complex with that of FLS2LRR-flg22-BAK1LRR using BAK1LRR as the template. “N” and “C” represent the N- and C-terminus, respectively. Color codes are indicated. This alignment was performed by the program COOT. (L) FLS2LRR-flg22 releases BAK1LRR from the BAK1LRR-BIR1LRR complex in gel filtration at pH 6.0. BIR1LRR and BAK1LRR with a molar ratio of about 2.5:1 were mixed together and incubated at 4 °C for 30 min. The FLS2LRR-flg22 complex was then added to the mixture for gel filtration. The molar ratio between FLS2LRR and BAK1LRR was about 2:1. Shown in the left panel are gel filtration profiles of proteins indicated. The vertical and horizontal axes represent ultraviolet absorbance (λ = 280 nm) and elution volumes (mL), respectively. Right panel, coomassie blue staining of the peak fractions following SDS-PAGE. The numbers shown on the top of the gels indicate elution volumes (mL). MM: molecular weight maker.
To probe the molecular mechanism underlying BIRLRR-BAK1LRR interaction, we solved the crystal structure of the complex (Figure 1C). Interaction between the two proteins is mediated by packing of one lateral side of BIR1LRR against the C-terminal inner surface and the C-terminal capping domain of BAK1LRR(Figure 1C). Specifically, a loop region from the N-terminal capping domain of BIR1LRR makes extensive contacts with BAK1LRR(Figure 1C). While the overall structure of BIR1LRR remarkably resembles that of BAK1LRR (Supplementary information, Figure S7A), the loop regions from these two proteins are strikingly different in their conformations and primary sequences (Supplementary information, Figure S7).
The BIR1LRR-BAK1LRR interaction is mediated by both polar and hydrophobic contacts, and can be divided into two interfaces (Figure 1C). One is mainly mediated by packing of the loop region from the N-terminal capping domain of BIR1LRR against the C-terminal inner surface of BAK1LRR. Interaction between one lateral side of BIR1LRR and one short helix of BAK1LRR forms the other interface (Figure 1D). Trp71 of BIR1LRR from the first interface forms extensive interactions with BAK1LRR by being sandwiched by Val168, Asp170 and Ile192 of BAK1LRR(Figure 1D, Supplementary information, Figure S8). Several hydrogen bonds also contribute to the interaction around this interface. At the other interface, Thr190 positioned immediately underneath the C-terminal capping domain of BAK1LRR tightly stacks against Phe150 of BIR1LRR, and Thr128 of BIR1LRR and Leu188 of BAK1LRR forms a water-mediated hydrogen bond (Figure 1D and Supplementary information, Figure S8).
To confirm our structural observations, we chose two residues from the centers and two from the peripheries of the BAK1LRR-BIR1LRR interfaces for mutagenesis analyses. In support of the structure, mutating Trp71 in BIR1 to the smaller alanine residue resulted in loss of interaction with BAK1LRR (Figure 1E, Supplementary information, Figure S9A). Mutations in the equivalent residues in BIR2LRR (W73), BIR3LRR (W67) and BIR4LRR (W60) caused similar effects on their interaction with BAK1LRR (Supplementary information, Figure S10). Similarly, mutating Thr190 in BAK1 to arginine (Figure 1F, Supplementary information, Figure S9B) and His72 in BIR1 to asparagine (Supplementary information, Figure S11) also resulted in no detectable BAK1LRR-BIR1LRR interaction. In contrast, mutating Val168 in BAK1 to arginine compromised but did not abolish the BAK1LRR-BIR1LRR interaction as indicated by the results of gel filtration and ITC assays (Supplementary information, Figure S12).
We then used the luciferase (LUC) complementation assay to test the effect of the above mutations on BIR1-BAK1 interaction in N. benthamiana. Co-infiltration of Agrobacteria containing BAK1-CLuc and BIR1-NLuc, resulted in strong LUC activity (Supplementary information, Figure S13A). In support of our biochemical data, the BIR1 W71A and BAK1 T190R mutations greatly reduced the LUC activity generation (Supplementary information, Figure S13B, S13C). In contrast, the BAK1 V168R and BIR1 H72N mutations only modestly affected the LUC activity (Supplementary information, Figure S13D, S13E). The positive control BIR1 T103Q mutation, found in other BIR proteins (Supplementary information, Figure S14) and located between the two BAK1-BIR1 interfaces (Supplementary information, Figure S8B), did not impact BIR1 interaction with BAK1 in the assay (Supplementary information, Figure S13A). The reduced interaction was not caused by difference in protein levels as the wild type and mutant BAK1-CLuc and BIR1-NLuc proteins were expressed at comparable levels (Supplementary information, Figure S13F).
We then generated transgenic plants and examined their defense responses. The bir1-1::BIR1 (W71A) but not the bir1-1::BIR1 (T103Q) transgenic plants displayed seeding lethality phenotype, phenocopying the bir1-1 mutant (Figure 1G). Furthermore, expression levels of pathogenesis-related genes PR1 and PR2 were upregulated in bir1-1::BIR1 (W71A) but not in bir1-1::BIR1 (T103Q) plants (Figure 1H, Supplementary information, Figure S15A). Supporting our biochemical and cell-based assays, expression of BAK1 (T190R) but not wild type BAK1 or BAK1 (V168R) led to plant dwarfism, constitutive expression of PR1 and PR2 and increased resistance to Hyaloperonospora parasitica Noco2 (Figure 1G, 1I, 1J, Supplementary information, Figures S15B and S16). These results suggest that defense responses were constitutively activated in the BAK1 (T190R) transgenic plants. Different from the bir1-1::BIR1 (W71A) plants, the bir1-1::BIR1 (H72N) plants had wild type morphology, did not constitutively express PR genes, and were fully susceptible to the virulent oomycete pathogen Hyaloperonospora parasitica Noco2 (Supplementary information, Figure S17). The precise reason for the discrepancy between these functional data and the in vitro biochemical data remains unclear.
Structural comparison between BAK1LRR-BIR1LRR and FLS2LRR-flg22-BAK1LRR showed that the C-terminal portion of FLS2LRR, which interacts with BAK1LRR, completely overlaps with BIR1LRR(Figure 1K), suggesting that the flg22-bound FLS2 may compete with BIR1LRR to release BAK1LRR from the BIR1LRR-BAK1LRR complex. Supporting this hypothesis, BAK1LRR from the pre-incubated BAK1LRR-BIR1LRR complex formed a stable interaction with FLS2LRR-flg22 at pH 6.0 (Figure 1L), indicating that the BAK1LRR-BIR1LRR interaction had been outcompeted by FLS2LRR-flg22.
Our study shows that the ecto-domains of BIR1 and BAK1 are sufficient for them to interact with each other in vitro and the interaction is critical to the inhibition of BAK1 function by BIR1. Consistently, over-expression of the ecto-domain together with the trans-membrane segment of BAK1 in plants phenocopies the BAK1-overexpressing phenotypes, presumably through sequestering BIR110. Our primary sequence analysis (Supplementary information, Figure S14) suggests that all 4 BIR members may share a common mechanism of interacting with BAK1. We also showed that the flg22-bound FLS2 outcompeted BIR1LRR for binding to BAK1LRR, even with an excess of BIR1LRR, explaining the observation that BIR1 has no effect on FLS2-mediated immune responses7. Release of the BIR1-sequestered BAK1 by the flg22-bound FLS2 could become easier by an increase in pH during flg22-induced plant immunity. BIR1 has likely evolved to keep BAK1-mediated cell death signaling under tight control, thus preventing undesired autoimmunity. The observation that the ecto-domain of BIR1 is sufficient for inhibition of BAK1 suggests that the signal relieving BIR1-mediated inhibition of BAK1, if present, comes from the extracellular space.
Acknowledgments
We thank Huang S and He J at Shanghai Synchrotron Radiation Facility (SSRF). We acknowledge the China National Center for Protein Sciences Beijing for providing the facility support. This research was funded by grants from Projects of International Cooperation and Exchanges NSFC (31420103906), the Chinese Ministry of Science and Technology (2015CB910200), the National Natural Science Foundation of China (31421001) to JC and Natural Sciences and Engineering Research Council (Discovery grant 249922 to YZ).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
Supplementary information, Figure S1–S17 and Data S1 Materials and Methods
References
- Macho AP, Zipfel C. Mol Cell 2014; 54:263–272. [DOI] [PubMed]
- Ryan CA, Huffaker A, Yamaguchi Y. Cell Microbiol 2007; 9:1902–1908. [DOI] [PubMed]
- Zipfel C, Robatzek S, Navarro L, et al. Nature 2004; 428:764–767. [DOI] [PubMed]
- Gomez-Gomez L, Boller T. Mol Cell 2000; 5:1003–1011. [DOI] [PubMed]
- Chinchilla D, Shan L, He P, et al. Trends Plant Sci 2009; 14:535–541. [DOI] [PMC free article] [PubMed]
- Sun Y, Li L, Macho AP, et al. Science 2013; 342:624–628. [DOI] [PubMed]
- Gao M, Wang X, Wang D, et al. Cell Host Microbe 2009; 6:34–44. [DOI] [PubMed]
- Liu Y, Huang X, Li M, et al. New Phytol 2016; 212:637–645. [DOI] [PubMed]
- Halter T, Imkampe J, Mazzotta S, et al. Curr Biol 2014; 24:134–143. [DOI] [PubMed]
- Dominguez-Ferreras A, Kiss-Papp M, Jehle AK, et al. Plant Physiol 2015; 168:1106–1121. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information, Figure S1–S17 and Data S1 Materials and Methods