Abstract
Circular RNAs (circRNAs) are a novel class of RNA whose physiological function has yet to be investigated. A recent publication in Science provides the first evidence of the biological relevance of a circRNA in an in vivo model and unveils an unexpected twist on their crosstalk with miRNAs.
In 1993, Cocquerelle et al.1, inspired by the discovery of scrambled exons, investigated the existence of two nonpolyadenylated circular RNA (circRNA) isoforms generated from the ets-1 locus in human cells. In a concurrent study, Capel et al.2 identified a circRNA arising from the Sry locus. Two decades later, tremendous advances in RNA sequencing technologies and bioinformatic analysis allowed three groups to validate the data of Cocquerelle and Capel3,4,5,6. So far thousands of circRNAs have been uncovered. They appear to be generated as the product of a proactive form of backsplicing of pre-mRNAs.
Potential mechanisms of action have been investigated for a number of circRNAs. The vast majority appear to be non-coding RNAs that play a role in transcriptional and translational regulation, similar to the better-characterized long non-coding RNAs (lncRNAs). The role of circRNAs as “miRNA-sponges” has arguably been extensively explored. miRNAs are short regulatory RNAs that recognize target RNAs through their binding sites (miRNA recognition elements, MRE) and can negatively mediate their abundance and translational potential.
A few years ago, our group proposed a competing endogenous RNA (ceRNA) hypothesis7, in which different RNAs can communicate with and regulate each other by competing for miRNAs through MREs. This model implies intricate crosstalk by which the modulation of a RNA with multiple MREs for specific miRNAs results in the concomitant regulation of other targets of these miRNAs. This hypothesis was subsequently validated for both coding and non-coding linear RNAs by our group and others. The Rajewsky and Kjems laboratories focused on a circRNA, Cdr1as, and its ceRNA activity4,6. Specifically expressed in the mammalian nervous systems, the Cdr1as circRNA is the only RNA isoform arising from a non-coding locus. Notably, it harbors ∼70 conserved MREs for miR-7 and one MRE for miR-671, and it is extensively bound by AGO2, a critical effector of miRNA-mediated gene silencing.
Since it is highly expressed, stable, cytoplasmic and contains an extraordinary number of miR-7 MREs, Cdr1as represents the perfect archetype of a ceRNA. Accordingly, the Rajewsky lab and the Kjems lab provided evidence supporting a functional association between Cdr1as, miR-7 and miR-7 targets. Morpholino knockdown of miR-7 in zebrafish induced defects of brain development which were phenocopied by injection of plasmid expressing CDR1as into embryos. The effect was partially rescued by injecting a miR-7 precursor4. Similarly, Cdr1as could inhibit miR-7 in HeLa cells. A further layer of regulation is offered by miR-671, which is able to trigger Cdr1as degradation, and as an indirect consequence, to affect miR-7 levels6. Knockdown of CDR1as in colorectal cancer and hepatocellular carcinoma cell lines resulted in decreased tumor proliferation, increased miR-7 levels, and downregulation of miR-7 targets8,9. It is important to note that all these studies exploited shRNAs and siRNAs to target circRNAs. Thus, the physiological role of a circRNA in a genetically engineered in vivo model was yet to be elucidated.
Rajewsky and colleagues, in collaboration with the Birchmeier lab, recently published results from a Cdr1as knockout (KO) mouse model in Science10. To date, this is the first and only KO model of a circRNA, and the first full deletion of a circRNA transcript involved in ceRNA crosstalk. Cdr1as KO mice are viable and fertile without gross abnormality. However, Piwecka et al.10 observed that these mice suffer from defects in sensorimotor gating, possibly due to altered excitatory synaptic transmission in the absence of Cdr1as. The direct interaction between Cdr1as and both miR-7 and miR-671 was confirmed in vivo by a modified CLIP assay in which miRNAs that co-precipitate with AGO2 are ligated to their RNA target sites and the chimeric RNAs can be sequenced. Consistently, miR-sequencing of four major brain regions from Cdr1as KO mice and their wild-type counterpart showed that miR-7 and miR-671 were among the most differentially distributed miRNAs. Surprisingly, miR-7 was consistently and markedly post-transcriptionally downregulated, and its target genes were specifically upregulated in the brain.
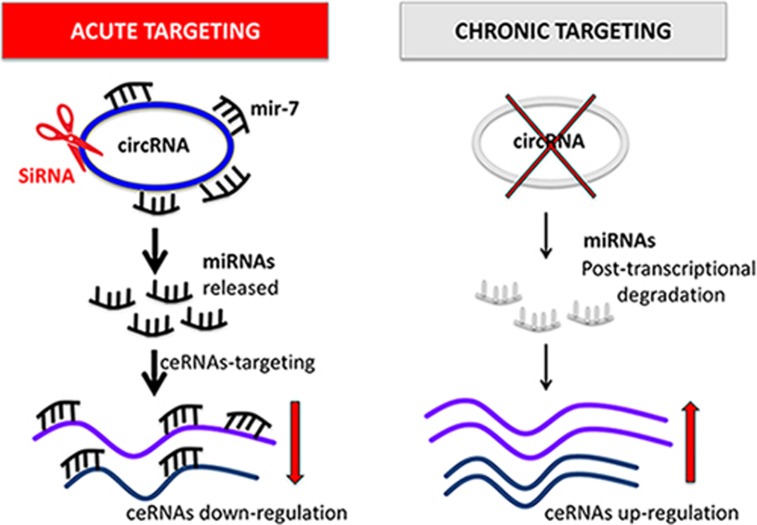
How can such an unexpected result that is in contrast with previous reports and with the canonical mechanism of action of a ceRNA be explained? The authors propose that the key to understanding the balance between these different transcripts is the Cdr1as-miR-7 binding site architecture. miR-7 MREs on Cdr1as lack complementarity beyond the seed region and therefore do not allow AGO2-mediated slicing. As a result, Cdr1as competes with other transcripts to stabilize and transport, rather than to inhibit, miR-7. On the contrary, the miR-671 binding site shows perfect complementarity with Cdr1as and consequently the level of miR-671 is increased in the brain of the Cdr1as KO mouse. In this scenario, as Piwecka et al. suggested, it is conceivable that the discrepancy with previous studies could be a consequence of the differences between an acute vs a chronic depletion of Cdr1as. In siRNA studies, the sudden targeting of Cdr1as releases stable miR-7:AGO2 complexes, which can rapidly bind and act on other miR-7 targets causing their repression in the short term. On the other hand, the constitutive, prolonged lack of Cdr1as deprived miR-7 in cytoplasm, reducing its overall level (Figure 1).
Figure 1.
CircRNAs can function as ceRNAs thus sponging for miRNAs. Once the circRNA is destabilized through acute treatments (e.g., with siRNAs, shRNAs, ASOs or LNA GapmeRs), the attached miRNAs are released and free to bind and repress ceRNA targets. As a final result of an acute intervention, ceRNAs are down-regulated. In a different scenario, the prolonged absence of a sponge circRNA, due to chronic treatments or knockout, leads to destabilization of its binding miRNAs through a post-transcriptional mechanism. As a consequence, other ceRNAs are no longer targeted and their expression increases.
The full dissection of this intricate balance will require further efforts. However, the hypothesis by Piwecka et al. is plausible and may have major implications. First, the data further enhance our understanding of ceRNA crosstalk. Currently, miRNA binding affinity, expression levels of both ceRNA and miRNA, their localization, and the actual binding by AGO2 are possible critical parameters in the modulation of ceRNA networks, and therefore in predicting whether targeting a specific ceRNA may result in the concomitant repression of a variety of other RNAs that share MREs for the same miRNA. We now need to systematically assess the target binding site architecture, considering that an imperfect match could result in a stabilizing competition rather than inhibitory sponging. Moreover, it would be interesting to understand whether linear ceRNA function like Cdr1as in this respect. Second, many circRNAs have been linked to cancer development and progression. Notably, aberrant circRNAs have been also identified in cancer as proto-tumorigenic products of chromosomal translocations11. Thus, the opposite outcome, in terms of gene expression, that is observed in acute vs chronic targeting of Cdr1as could have tremendous therapeutic implications, and it will be critical to test multiple treatment strategies in pre-clinical settings (e.g., metronomic vs continuous administration of the RNA medicine of interest) (Figure 1).
In conclusion, this study proves, for the first time, the functional importance of a circRNA in vivo and paves the way for further mechanistic studies. It is notable that Piwecka et al. took advantage of the CRISPR/Cas9 approach and removed the entire Cdr1as locus (2.9 kb). This strategy can have several unwanted consequences12. Nevertheless, the authors convincingly linked the observed phenotype to alteration of the Cdr1as binding partner miR-7 and confirmed that the Cdr1 locus exclusively expresses Cdr1as. Most circRNAs are generated by backsplicing RNAs from coding or non-coding genes, thus the CRISPR/Cas9 strategy used for Cdr1as would result in the concomitant deletion of protein-coding exons or the lincRNA/pseudogene. It is crucial to further uncover mechanisms of circRNA biogenesis and to find alternative approaches to delete and mutagenize them in vivo.
References
- Cocquerelle C, Mascrez B, Hétuin D, et al. FASEB J 1993; 7:155–160. [DOI] [PubMed]
- Capel B, Swain A, Nicolis S, et al. Cell 1993; 73:1019–1030. [DOI] [PubMed]
- Salzman J, Gawad C, Wang PL, et al. PLoS One 2012; 7:e30733. [DOI] [PMC free article] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Nature 2013; 495:333–338. [DOI] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. RNA 2013; 19:141–157. [DOI] [PMC free article] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Nature 2013; 495:384–388. [DOI] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. Cell 2011; 146:353–358. [DOI] [PMC free article] [PubMed]
- Tang W, Ji M, He G, et al. Onco Targets Ther 2017; 10:2045–2056. [DOI] [PMC free article] [PubMed]
- Yu L, Gong X, Sun L, et al. PLoS One 2016; 11:e0158347. [DOI] [PMC free article] [PubMed] [Retracted]
- Piwecka M, Glažar P, Hernandez-Miranda LR, et al. Science 2017; 357 pii: eaam8526. [DOI] [PubMed]
- Guarnerio J, Bezzi M, Jeong JC, et al. Cell 2016; 165:289–302. [DOI] [PubMed]
- Bassett AR, Akhtar A, Barlow DP, et al. eLife 2014; 3:e03058. [DOI] [PMC free article] [PubMed]