Abstract
Objectives:
Blood-based proteins might be an attractive option for early detection of colorectal cancer (CRC), but individually they are unlikely to achieve the diagnostic performance required for population based screening. We aimed at summarizing current evidence of diagnostic performance of signatures based on multiple proteins for early detection of CRC.
Methods:
A systematic literature review adhering to the PRISMA (preferred reporting items for systematic reviews and meta-analysis) guidelines was performed. PubMed and Web of Science databases were searched for potentially relevant studies published until 28th August, 2017. Relevant studies were identified by predefined eligibility criteria. Estimates of indicators of diagnostic performance such as sensitivity, specificity, and the area under the curve (AUC), along with information on validation and other key methodological procedures were extracted. Study quality was assessed by a QUADAS-2 (Quality Assessment of Diagnostic Accuracy Studies 2) instrument tool.
Results:
Thirty six eligible studies with numbers of CRC cases ranging from 23 to 512 and the number of proteins included in signatures ranged from 3 to 13 were identified. Reported Youden’s Index and AUC ranged from 0.19 to 0.95 and from 0.62 to 0.996, respectively. However most studies, especially those reporting better diagnostic performance, were conducted in clinical rather than screening setting and many studies lacked any internal or external validation of identified algorithm.
Conclusions:
Blood-based tests using signatures of multiple proteins may be a promising approach for non-invasive CRC screening. However, promising signatures identified in clinical settings still require rigorous evaluation in large studies conducted in true screening setting.
Introduction
With over 1.4 million incident cases accounting for 10% of all cancers and ~700,000 deaths per year, colorectal cancer (CRC) is the third most common cancer globally.1, 2 Most CRCs slowly develop from adenomas over many years, and opportunities for screening and early detection are much better than for most other types of cancers. Randomized trials have shown effective reduction of both CRC incidence and mortality by screening with stool tests or endoscopic examinations,3, 4, 5, 6 and screening programs are currently being implemented in an increasing number of countries.7
Screening colonoscopy and sigmoidoscopy are current gold standards for detection of CRC and its precursors in the total and distal colorectum, respectively. Although application of these screening procedures has been shown to effectively reduce CRC incidence and mortality,4 the implementation of and adherence to these invasive procedures in population-wide screening is limited on account of several disadvantages such as high costs, limited resources and low compliance.8, 9, 10 Compliance is also a major issue in CRC screening based on stool tests, such as the widely used guaiac-based or fecal immunochemical tests (FITs) for hemoglobin, or recently introduced DNA based stool tests.11, 12 Blood-based tests that could be easily integrated in routine exams, might therefore be a highly attractive alternative of minimally invasive CRC screening. It would therefore be of major interest to compare ability of blood-based tests and FITs with respect to their ability to detect early-stage CRC.
Over the years, several blood-based protein biomarkers such as CEA, CA19-9, or CA242 have been associated with diagnosis of CRC. However, none of these proteins individually is able to detect the majority of early-stage CRC manifestations.13, 14, 15, 16 A promising approach to increase early detection of CRC and its precursors could be to combine several different protein markers in a panel. The rationale behind this is that different protein markers may detect different morphological types of CRC and a combination might thus lead to increased overall accuracy. In this systematic review, we aim to provide a comprehensive overview of studies that have discovered, evaluated or validated blood-based protein signatures or panels for early detection of CRC, paying particular attention to the role of study design and study population characteristics. A particular focus of interest is diagnostic performance for detecting early-stage CRC or even adenomas, the precursors of most CRCs, which would be most relevant for potential application in CRC screening.
Methods
Data sources and search strategy
Observing the PRISMA (preferred reporting items for systematic reviews and meta-analysis) guidelines,17 the online scientific citation databases MEDLINE and Web of Science were searched from establishment until 28 August, 2017. Exact search terms which included keywords like “Tumor Marker”, “Protein”, “Colorectal Cancer”, or “Signature” are provided in Supplementary File 1.
Study selection
Our search was restricted to human research studies in English language with blood collected prior to any treatment. Studies reporting diagnostic performance of individual proteins markers were not included. We excluded studies that included not only primary CRC cases but also post-operative patients or patients with recurrent disease, or studies that had total number or cases <10. In addition, signatures where the diagnostic performance was reported for panels of unidentified peaks were left out. Studies with insufficient information on diagnostic indicators were omitted. Studies assessing the potential for predicting metastasis or prognosis of CRC were not considered. Moreover, studies that assessed proteins in combination with other types of biomarkers like DNA or RNA based markers were not considered. Furthermore, studies that exclusively reported TAA (tumor-associated autoantibody) signatures were excluded as a systematic review of such TAA signatures for CRC early detection has recently been reported elsewhere.18 A protein marker set was defined as a panel or signature if it was based on information from more than two proteins.
Data extraction and risk of bias in individual studies
Two authors (M.B. and A.G.) independently read and extracted data from the 36 studies. Information on key study characteristics like authors, year of publication, study population (country, numbers of cases and controls, age, and sex distribution), study design, type of protein signatures, type of protein detection technique, and diagnostic performance was retrieved. Data on the following diagnostic performance related indicators were extracted: overall and stage-specific (wherever reported) sensitivity and specificity, area under the receiver operating characteristics curve and P value. In addition, Youden’s Index (J) and 95% confidence interval (95% CI) of given sensitivities and specificities by Clopper–Pearson method using R 3.3.2 were calculated for each signature and are reported in Table 2.
To assess the risk of bias of individual studies, the QUADAS-2 (Quality Assessment of Diagnostic Accuracy Studies 2) instrument19 was applied to all the studies. Careful assessment was independently performed by the two above-mentioned authors by inserting and deleting standard signaling questions and rating risk of bias and applicability concerns as ‘high’, ‘low’, or ‘unclear’.
Results
Study selection
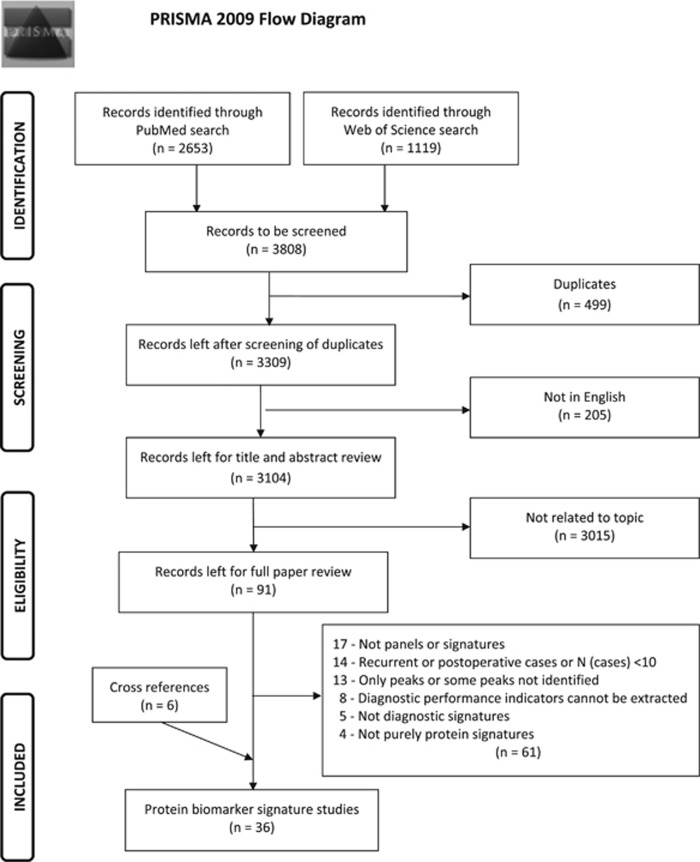
The search in above-mentioned databases using search terms (reported in Supplementary File 1) yielded 3,808 records. Details of the study selection are reported in Figure 1 observing the PRISMA 2009 Flow Diagram.20 As shown in Figure 1, upon application of inclusion and exclusion criteria and screening for relevance to the topic, 91 articles were selected for full review. Sixty-one articles had to be excluded based on aforementioned predefined criteria. Six other studies were identified by cross referencing. In the end, 36 protein signature studies were included in this review. Information regarding study populations and design is summarized in Table 1. Furthermore, information concerning number of proteins in the signature, type of detection method, diagnostic performance and measures for validation is reported in Table 2. The specific proteins included in the signatures are listed in Table 3. Evidence extracted with respect to stage specific results, risk of bias of individual studies, brief description of protein detection techniques and internal validation methods and the PRISMA checklist20 are reported in Supplementary Files 2–6.
Figure 1.
PRISMA flow diagram for literature search process for records identified via PubMed and Web of Science database.20
Table 1. Participant characteristics of protein biomarker signature studies.
| First author,ref. | Case-finding Country | Compared groups | Population (gender) Cases N (female/males) Controls N (female/males) | AgeMean/median±s.d. or (range) Cases (years) Controls (years) | ||||
|---|---|---|---|---|---|---|---|---|
| Mercer,54 | SY USA | CRC Dz Controls | 23 (11/12) 10 (7/3) | (19–88) (38–81) | ||||
| Eskelinen,35 | CLIN Finland | CRC Dz Controls | 138 (76/62) 104 | 67.7±11.9 – | ||||
| Eskelinen,34 | CLIN Finland | CRC Dz Controls | 138 (76/62) 104 | 67.7±11.9 – | ||||
| Fernandez-Fernandez,29 | CLIN Spain | CRC Dz Controls | 127 (49/78) 70 (26/44) | 70.7±10.4 71.4±10.7 | ||||
| Ward,37 | UNCL UK | CRC Mi Controls | 62 (26/36) 31 (18/13) | 67.3±12.9 62.9±10.3 | ||||
| Zheng,46 | CLIN China | CRC Mi Controls | 111 40 | 56.7±7.3 54.2±3.5 | ||||
| Yoneda,51 | UNCL Japan | CRC Hy Controls | 159 (52/107) 40 (19/21) | (38–90) (29–85) | ||||
| Shimwell,36 | CLIN UK | CRC Hy Controls | Training Set 68 (26/42) 92 (48/44) | Test Set 38 (19/19) 41 (20/21) | Training Set 72(31–86) 67(31–89) | Test Set 73(32–89) 71(32–92) | ||
| Wild,41 | CLIN European population | CRC Controls AA Dz Controls Hy Controls | 301 (144/157) 266 (162/104) 143 (59/84) 141 (76/65) 176 (101/75) | 67(35–96) 62(18–89) 66(41–85) 62(23–91) 64(34–88) | ||||
| Hamelin,39 | UNCL France | CRC Hy Controls | Qualification Cohort 40 (15/25) 40 (13/27) | Verification Cohort 112 (51/61) 90 (36/54) | Qualification Cohort 71±11 55±5 | Verification Cohort 70±11 58±4 | ||
| Brock,31 | CLIN Russia | CRC Hy Controls | 172 (93/78) 259 (89/170) | 61.6(10.83) 56.8(4.75) | ||||
| Ladd,53 | TS USA | CRC Hy Controls | Discovery Set 90 (90/0) 90 (90/0) | Validation Set 32 (32/0) 32 (32/0) | Discovery Set 64.9 64.9 | Validation Set 68.1 67.8 | ||
| Lumachi,30 | UNCL Spain | CRC Dz Controls | 102 99 | 66(42–75) − | ||||
| Barderas,28 | UNCL Spain | CRC Hy Controls | 40 (17/23) 20 (8/12) | 67.5 60.5 | ||||
| Krzystek-Korpacka,40 | CLIN Poland | CRC Mi Controls | 99 (43/55)a 98 (39/60)a | 67.2(33–92) 60.7(41–87) | ||||
| Thorsen,23 | SY Denmark | CRC Adenoma Dz Controls Hy Controls | 70 (36/34) 70 (36/34) 70 (36/34) 70 (36/34) | (40–99) (40–99) (40–99) (40–99) | ||||
| Pengjun,43 | CLIN China | CRC Hy Controls | 149 (72/77) 69 (33/36) | 58(31–74) 57(34–71) | ||||
| Shin,52 | UNCL Korea | CRC Hy Controls | Verification Set 80 (37/43) 30 (15/15) | Validation Set 228 (90/137) 77 (44/33) | Verification Set 62.8±8.7 63.5±7.5 | Validation Set 62.6±8.7 68.2±7.0 | ||
| Bertuzzi,38 | TS Italy | CRC Hy Controls | Phase I 10 (3/7) 10 (1/9) | Phase II 10 (6/4) 10 (6/4) | Phase III 48 (28/20) 48 (28/20) | Phase I 61.6(11.1) 60.2(10.9) | Phase II 53.3(7.8) 53.3(7.6) | Phase III 55.1(6.2) 55.2(6.2) |
| Chen,25 | CLIN Germany | CRC Hy Controls | Training set 35 (10/25) 54 (27/27) | Validation Set 54 (22/32) 38 (22/16) | Training set 66.9±6.5 62.8±7.0 | Validation Set 66.0±10.8 63.8±9.5 | ||
| Fung,56 | CLIN Australia | CRC Hy Controls | Training Set 95 (50/45) 50 (25/25) | Test Set 98 (34/64) 99 (33/66) | Training Set 67(44–93) 70(50–85) | Test Set 67(25–89) 69(36–89) | ||
| Storm,22 | SY Denmark | Adenoma CRC Mi Controls | 196 (90/106) 99 (42/57) 696 (404/292) | 65(57–72) 69(62–79) 62(52–72) | ||||
| Surinova,26 | CLIN CZ, Germany | CRC Mi Controls | Training Cohort 100 (48/52) 100 (46/54) | Validation Cohort 202 (89/113) 67 (16/51) | Training Cohort 67(61–72) 64(58–69) | Validation Cohort 67(59–74.8) 49(52–65) | ||
| Taguchi,55 | SY USA | CRC Adenoma Hy Controls | 60 (36/24) 60 (29/31) 60 (16/44) | 61.9±10.2 55.7±9.9 | ||||
| Uchiyama,50 | UNCL Japan | CRC Controls | 72 (26/46) 63 (29/34) | 62.5 62.9±13.7 | ||||
| Xue,44 | CLIN China | CC Hy Controls | 120 (61/49) 40 | 66.8(26–93) 39.2(23–59) | ||||
| Blume,21 | SY Denmark | CRC Hy Controls | Discovery Set 75 (40/35) 75 (40/35) | Validation Set 75 (35/40) 75 (35/40) | Discovery Set 64.5±7.01 63.8±7.04 | Validation Set 65.6±6.09 64.8±5.76 | ||
| Butvilovskaya,32 | CLIN Russia | CRC Dz Controls Hy Controls | 33 (13/20) 27 (17/10) 67 (17/50) | (50–70) − 31; >70 − 2 (30–70) − 27 (50–70) − 64; >70 − 3 | ||||
| Hou,42 | UNCL China | CRC Dz Controls | 100 (44/56) 50 (29/21) | 58.5(25–80) 53.0(31–82) | ||||
| Ji,47 | UNCL China | RC Dz Controls | 100 (53/47) 100 (55/45) | 45(25–65) 52(31–73) | ||||
| Jones,33 | CLIN Russia | CRC Hy Controls | Discovery Set 69 (40/29) 69 (40/29) | Validation Set 68 (40/28) 68 (40/28) | Discovery Set 60.5 56.8 | Validation Set 62.0 58.0 | ||
| Werner,27 | TS Germany | CRC AA CIS Hy Controls | 36 (10/26) 420 (148/271) 4 (1/3) 1200 (652/545) | 66.0±6.2 63.6±6.7 63.0±5.3 62.0 ±6.1 | ||||
| Wilhelmsen,24 | SY Denmark | CRC Adenoma Dz controls Hy controls | 512 689 1342 1978 | – | ||||
| Xie,48 | UNCL China | CRC Hy controls | 30 (12/18) 30 (13/17) | (31–70) (31–70) | ||||
| Zhang,45 | UNCL China | CRC Dz controls | 120 (52/68) 86 (47/39) | (25–82) (32–80) | ||||
| Li,49 | UNCL China | CRC Dz Controls | 100 (44/56) 50 (29/21) | 59(25–82) 52.5(32–80) | ||||
AA, advanced adenomas; CC, colon cancer; CIS, carcinoma in situ; CLIN, recruitment of clinically diagnosed cases and collection of blood prior to any surgery or treatment; CRC, colorectal cancer; CZ, Czech Republic; Dz controls, Disease controls; Hy Controls, Healthy controls; Mi controls, mixed i.e. disease & healthy controls; N, number of participants; RC, rectal cancer; Ref, references; SY, recruitment and collection of blood of symptomatic individuals before establishment of clinical diagnosis; TS, recruitment of asymptomatic individuals and collection of blood prior to establishment of diagnosis in a true screening setting; UNCL, time of recruitment and collection of blood unclear; UK, United Kingdom; USA, United States of America.
Note: age is mentioned as mean±standard deviation in years or mean/median (range or interquartile range) in years.
The gender distribution for this study does not match with total number of cases and controls.
Table 2. Diagnostic performance of protein biomarker signatures for all tumor stages for CRC.
| First author,ref. | Analytical method | No. of proteins | Sensitivity% (95% CI) | Specificity% (95% CI) | J | AUC (95% CI) | P value |
Validation |
|
|---|---|---|---|---|---|---|---|---|---|
| Internal | External | ||||||||
| Mercer,54 | IECC | 5 | 91 (71–99) | 70 (35–93) | 0.52 | – | – | – | – |
| Eskelinen,35 | IF | 3 | 47 (39–56)a | 88 (80–94)a | 0.35 | – | – | – | – |
| Eskelinen,34 | IF | 5 | 33 (25–42)b | 90 (83–95)b | 0.23 | – | – | – | – |
| Fernandez-Fernandez,29 | EIA | 3 | 62 (53–70) | 87 (77–94) | 0.49 | – | – | – | – |
| Ward,37 | SELDI | 4 | 95 (86–99) | 91 (75–98) | 0.86 | – | <0.05 | TFCV | – |
| Zheng,46 | SELDI | 3 | 62 (52–71) | 86 (71–95) | 0.48 | – | <0.005 | – | – |
| Yoneda,51 | SI | 3 | 63 (55–71) | 90 (76–97) | 0.53 | – | – | – | – |
| Shimwell,36 | ELISA | 4 | 43 (27–60) | 95 (83–99) | 0.38 | 0.85 (0.80–0.90) | – | LOOCV | – |
| Wild,41 | ELISA | 6 | 23 (16–31)c 70 (65–75) | 95 (92–97)c 95 (92–97) | 0.18 0.65 | – | – | MCCV | – |
| Hamelin,39 d | ELISA | 3 | 47 (38–57) | 90 (82–95) | 0.37 | 0.77 (0.70–0.84) | <0.0001 | SS | – |
| Brock,31 d | LC-MS/MS | 6 | 94 (89–97)e | 83 (78–87)e | 0.77 | 0.90 | – | SS | – |
| Ladd,53 d | LC-MS/MS | 4 | 41 (24–60) | 95 (81–100) | 0.36 | 0.72 | – | SS | – |
| Lumachi,30 | ELISA | 5 | 74 (64–82) | 94 (87–98) | 0.68 | – | – | – | – |
| Barderas,28 | ELISA | 3 | 60 (43–75) | 95 (75–100) | 0.55 | 0.88 | – | – | – |
| Krzystek-Korpacka,40 | FC EIA | 5 | 95 (89–98)f | 36 (27–46)f | 0.31 | 0.86 (0.79–0.81) | <0.001 | – | – |
| Thorsen,23 | PEA | 5 | 56 (44–68) | 90 (81–96) | 0.46 | – | – | CV | – |
| Pengjun,43 | SI | 3 | 96 (91–99) | 99 (93–100) | 0.95 | 0.996 (0.99–1.00) | – | – | – |
| Shin,52 d | ELISA | 3 | 68 (62–74) | 90 (81–96) | 0.58 | 0.82 (0.73–0.89) | <0.0001 | SS | – |
| Bertuzzi,38 d | LC-MS/MS | 3 | 90 (68–99)g | 32 (13–56)g | 0.22 | 0.62 (0.52–0.72) | <0.05 | SS | – |
| 3 | 88 (66–98)g | 32 (13–56)g | 0.20 | 0.62 (0.52–0.72) | – | ||||
| Chen,25 | PEA | 8 | 44 (31–58) | 90 (76–97) | 0.34 | 0.76 (0.65–0.85) | – | BS | – |
| Fung,56 | ELISA | 3 | 73 (63–81) | 95 (89–98) | 0.68 | 0.91 (0.87–0.95) | <0.0001 | BS | – |
| Storm,22 | TRIFMA | 3 | 36 (27–46) | 83 (80–86) | 0.19 | 0.68 | – | – | – |
| Surinova,26 | LC-MS/MS | 6 | 70 (63–76) | 79 (67–88) | 0.49 | 0.84 (0.75–0.92)h | – | TFCV | – |
| Taguchi,55 | AM, ELISA | 3 | 35 (23–48)i 43 (30–56) | 95 (86–99)i 95 (86–99) | 0.30 0.38 | – | – | BS | – |
| Uchiyama,50 | MALDI | 5 | 83 (72–91) | 92 (82–97) | 0.75 | 0.92 (0.88–0.96) | <0.05 | KFCV | – |
| Xue,44 | ELISA | 3 | 74 (65–82)j | 73 (56–85)j | 0.47 | 0.805 (0.74–0.87) | <0.001 | LOOCV | – |
| Blume,21 | ELISA | 8 | 80 (69–88) | 68 (56–78) | 0.48 | 0.82 (0.75–0.88) | 0.249 | TFCV | – |
| Butvilovskaya,32 | IA | 6 | 27 (13–45) | 98 (91–100) | 0.25 | 0.82 | – | – | – |
| Hou,42 | ELISA | 3 | 97 (92–99) | 91 (80–97) | 0.88 | 0.91 (0.85–0.96) | <0.01 | – | – |
| Ji,47 | CIA, BCA | 4 | 86 (78–92)k | 47 (37–57)k | 0.33 | – | – | – | – |
| Jones,33 | LC-MS, MRM | 13 | 87 (77–94) | 81 (70–90) | 0.68 | 0.91 | – | TFCV | – |
| Werner,27 | SI | 5 | 9 (6–12)c 42 (26–60) | 95 (94–96)c 95 (94–96) | 0.04 0.37 | 0.56 (0.53–0.59) 0.78 (0.68–0.87) | – | Yes | x |
| Wilhelmsen,24 | IA | 4 | 60 (56–64) | 89 (88–90) | 0.49 | 0.83 | <0.0001 | TFCV | – |
| Xie,48 | LC-MS, MRM | 4 | 87 (70–96) | 83 (65–94) | 0.70 | 0.90 (0.83–0.98) | <0.001 | – | – |
| Zhang,45 d | ELISA | 5 | 94 (88–98) | 98 (92–100) | 0.92 | 0.988 | – | SS | – |
| Li,49 d | ELISA | 6 | 85 (77–91) | 100 (93–100) | 0.85 | – | – | SS | – |
AM, antibody microarray; AUC, area under the receiver operating characteristic curve; BS, bootstrap; BCA, biochemical colorimetric assay; CIA, chemiluminescence immunoassay; CRC, colorectal cancer; CV, cross-validation; EIA, enzyme immunoassay; ELISA, enzyme-linked immunosorbent assay; FC, flow cytometry; IA, immunoassay; IECC, ion exchange column chromatography; IF, immunofluoroscence; J., Youden’s Index; KFCV, K-fold cross-validation; LC-MS/MS, liquid chromatography–mass spectrometry; LOOCV, leave-one-out cross-validation; MALDI, matrix-assisted laser desorption/ionization; MCCV, Monte Carlo Cross-Validation; MRM, multiple reaction monitoring; No., number; Ref, references; SI, Sandwich Immunoassay; SELDI, surface-enhanced laser desorption/ionization; SS, split-sample; TFCV, 10-fold cross-validation; TRIFMA, time resolved immunofluorometric assay.
The number of study participants in this study is not same for each protein assay test and the analysis is based on probably 114 CRC cases.
The number of study participants in this study is not same for each protein assay test and the analysis is based on probably 73 CRC cases and 87 controls.
The diagnostic performance is reported for the outcome Advanced Adenoma.
Studies where correction for overoptimism is performed in form of Split-sampling method, diagnostic performance mentioned here is for validated set.
Diagnostic performance in this study is reported for 110 samples.
The diagnostic performance values were reported only for early stage and not all stages.
Here the signature was validated on a male-only cohort of 20 CRC cases and 20 controls. Two panels were reported with one protein out of three different as specified in Table 3.
The confidence interval for AUC in this study was reported for 99% CI
The diagnostic performance is reported for the outcome adenoma.
The diagnostic performance is reported for the outcome colon cancer.
The diagnostic performance is reported for the outcome rectal cancer.
Table 3. Proteins included into the biomarker signatures.
| First author, ref. | CEA | CA199 | CA242 | OPN | Separase | AAG | A1AT | CA125 | CA153 | CA724 | CYFRA21-1 | CO9 | Ferritin | HSP60 | Other proteins markers |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild,41 | x | x | x | x | x | anti-p53 | |||||||||
| Lumachi,30 | x | x | x | x | x | ||||||||||
| Zhang,45 | x | x | x | x | x | ||||||||||
| Jones,33 | x | x | x | x | x | CLUS, TIMP1, AMY2B, ECH1, FRIL, GELS, SBP1, SPON2 | |||||||||
| Werner,27 | x | x | x | x | anti-p53 | ||||||||||
| Butvilovskaya,32 | x | x | x | x | AFP, HCG | ||||||||||
| Li,49 | x | x | x | x | AFP, NSE | ||||||||||
| Fernandez-Fernandez,29 | x | x | x | ||||||||||||
| Zheng,46 | x | x | x | ||||||||||||
| Yoneda,51 | x | x | Cystatin SN | ||||||||||||
| Hamelin,39 | x | x | x | ||||||||||||
| Thorsen,23 | x | x | x | TFRC, MIF | |||||||||||
| Xue,44 | x | x | AZGP1 | ||||||||||||
| Blume,21 | x | x | x | CO3, CATD, AACT, MIF, PSGL | |||||||||||
| Ji,47 | x | x | x | TSGF | |||||||||||
| Hou,42 | x | x | x | ||||||||||||
| Wilhelmsen,24 | x | x | x | hs-CRP | |||||||||||
| Mercer,54 | x | x | CRP, MCK2, FHAP | ||||||||||||
| Eskelinen,35 | x | x | CA50 | ||||||||||||
| Eskelinen,34 | x | x | CA50, SIMA I, SIMA II | ||||||||||||
| Brock,31 | x | x | CO3, GSN, HABP2, SAA2 | ||||||||||||
| Ward,37 | x | complement C3a des-arg, transferrin, apolipoprotein C1 | |||||||||||||
| Shimwell,36 | x | DR-70, sCD26, MMP9 | |||||||||||||
| Ladd,53 | x | LRG1, MAPRE1, IGFBP2 | |||||||||||||
| Shin,52 | x | PAI-1, TRFM | |||||||||||||
| Chen,25 | x | IFNγ, EMMPRIN, ERBB4, PSA, CD69, AREG, HGF receptor | |||||||||||||
| Taguchi,55 | x | MAPRE1, AK1 | |||||||||||||
| Uchiyama,50 | x | fibrinogen alpha chain, AHSG S-cysteinylated form, F.XIIIa, VASP | |||||||||||||
| Barderas,28 | GDF15, S100A8/A9, SERPINI1 | ||||||||||||||
| Krzystek-Korpacka,40 | TNF-α, FGF2, IL-6, G-CSF, GM-CSF | ||||||||||||||
| Pengjun,43 | TNF-α, IL-8, MMP-2 | ||||||||||||||
| Fung,56 | IGFBP2, DKK3, PKM2 | ||||||||||||||
| Bertuzzi,38 | CLU, APOC2, CO4-B/MASP2 | ||||||||||||||
| Storm,22 | CL-L1, M-ficolin, MAp44 | ||||||||||||||
| Surinova,26 | LRG1, TIMP1, CP, PON1, SERPINA3 | ||||||||||||||
| Xie,48 | CELA1, CEL2A, CTRL, TRY2 |
A1AG1/AAG/ORM1, α1-acid glycoprotein1; A1AT, α1-antitrypsin; AACT, α1-antichymotrypsin; AFP, alpha-fetoprotein; AHSG S-cysteinylated form, α2-HS-glycoprotein; AK1-adenylate kinase 1; ALB, Albumin; AMY2B, α-amylase 2B; APOC2, apolipoprotein C-II; AREG, amphiregulin; AZGP1, zinc-α-2-glycoprotein; CA125, cancer antigen 125; CA153, cancer antigen 153; CA19-9/CA199, carbohydrate antigen 19-9; CA242, cancer antigen 242; CA50, cancer antigen 50; CA724, cancer antigen 72-4; CATD, Cathepsin D; CC, colon cancer; CD69, early activation antigen CD69; CEA, carcinoembryonic antigen; CELA1, chymotrypsin-like elastase 1; CEL2A, chymotrypsin-like elastase 2 A; CL-L1, collectin-liver 1; CLUS/CLU, clusterin; CO3/C3, complement component C3; CO4-B, complement C4-B; CO9/C9, Complement component C9; CP, ceruloplasmin; CRC, Colorectal cancer; CRP, C-reactive protein or hs-CRP, high sensitivity C-reactive protein; CTRL, chymopasin; CYFRA21-1, cytokeratin fragment 21-1; DKK3, dickkopf-3; DR-70-a commercially available test for fibrinogen degredation products; ECH1, delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial; EMMPRIN, extracellular matrix metalloproteinase inducer; ERBB4, receptor tyrosine-protein kinase ErbB-4; FGF2, basic fibroblast growth factor; F.XIIIa, coagulation factor XIII subunit a; FHAP, homoarginine-sensitive alkaline phosphatase; FRIL, ferritin light chain; G-CSF, granulocyte colony-stimulating factor; GDF15, growth differentiation factor 15; GM-CSF, granulocyte macrophage colony-stimulating factor; GSN/GELS, gelsolin; HABP2, hyaluronan binding protein 2; HCG, human chorionic gonadotropin; HGF receptor, hepatocyte growth factor receptor; HSP60, heat shock protein 60; IFNγ, interferon gamma; IGF-1, insulin-like growth factor-1; IGFBP2, insulin-like growth factor-binding protein 2; IGFBP-3, insulin-like growth factor-binding protein-3; IL-6, interleukin-6; IL-8, interleukin-8; LASS5, longevity assurance gene homologous 5; LRG1, leucine-rich alpha-2-glycoprotein; MAp44, mannose-binding lectin-associated protein 44; MAPRE1, microtubule-associated protein RP/EB family member 1; MASP2, mannan-binding lectin serine protease 2; MCK2, macrocreatine kinase type 2; MIF, macrophage inhibitory factor; MMP-2, matrix metallopeptidase 2; MMP9, matrix metallopeptidase 9; NSE, neuron-specific enolase; OSTP/OPN/SPP1/OPN, osteopontin; PAI-1, plasminogen activator inhibitor-1; PKM2, pyruvate kinase M2; PON1, serum paraoxonase/arylesterase 1; PSA, prostate-specific antigen; PSGL, P-selectin glycoprotein ligand 1; Ref, references; S100A8/A9, S100 calcium-binding protein A8/A9; SAA2, serum amyloid A protein; SBP1, selenium binding protein 1; sCD26, serum-soluble CD26; SEPR, seprase; SERPINA3, serpin peptidase inhibitor clade A; SERPINI1, neuroserpin; SIMA I, small intestinal mucin anitgen I; SIMA II, small intestinal mucin antigen II; SPON2, spondin-2; TFRC, transferrin receptor-1; TIMP1, tissue inhibitor of metalloproteinases 1; TNF-α, tumor necrosis factor alpha; TRFM, melanotransferrin; TRY2, trypsin 2; TSGF, tumor-specific growth factor; TSLC1, tumor supressor in lung cancer I; VASP, vasodilator-stimulated phosphoprotein.
Study characteristics
An overview on the characteristics of the studies and the participants is given in Table 1. The majority of studies were conducted in Europe (21/36). Of these, four studies were carried out in Denmark,21, 22, 23, 24 three each in Germany,25, 26, 27 Spain,28, 29, 30 and Russia,31, 32, 33 two each in Finland34, 35 and the United Kingdom,36, 37 one study each in Italy,38 France,39 Poland,40 and Czech Republic,26 and at last one study was multicenter.41 Eleven studies that were carried out in Asia, included eight studies from China,42, 43, 44, 45, 46, 47, 48, 49 two studies from Japan,50, 51 and one study from Korea.52 Three studies were carried out in the United States of America53, 54, 55 and one in Australia.56 Blood was collected prior to colonoscopy or any other clinical diagnostic procedure in a true screening setting from asymptomatic individuals only in three studies.27, 38, 53 Numbers of CRC cases ranged from 2354 to 512.24 Six studies21, 23, 30, 33, 53, 56 reported having matched cases and controls by age (n=6), gender (n=6), or ethnicity (n=1);53 two studies 22, 36 used age- and sex-matched cases and controls in the pilot phase but did not match the population for the later validation phase. Only healthy controls were used in 18 studies, nine studies used only disease controls, whereas nine studies used a mix of both types of controls. One study each was carried out in male-only (n=20)38 and female only (n=32)53 populations. In 23 studies, the number of male patients was higher than the number of females, whereas eight studies did not specify gender distribution of study participants. Six studies included (as a separate case group) adenoma cases.22, 23, 24, 27, 41, 55 Twenty-four studies reported stage-specific distribution of cases (Supplementary File 2).
Protein detection techniques
Table 2 provides an overview on the protein detection and quantitation methods applied by the studies and among the included studies Immunoassay-based platforms like ELISA (enzyme-linked immunosorbent assay), EIA (enzyme immunoassay) and so on. were used by 21 studies and mass spectrometry-based platforms were used by nine studies, respectively. A brief description of all methods used is reported in Supplementary File 4. All in all, 93 different proteins were examined in 36 different protein signatures (Table 3) in this review. The number of proteins in the signatures ranged from 3 to 13, with almost half (15/36) of the signatures using three proteins. Several markers were found in more than one signature, including tumor markers like CEA (24 studies), CA19-9 (10 studies), CA242 (5 studies) and CA125, CA153, CA724, CYFRA21-1 (in 3 studies each). Other frequently used markers like Osteopontin and Separase were included in five and four studies, respectively, and AAG, A1AT, CO9, ferritin, and HSP60 were included in three different signatures each. Inflammation markers like TNF-alpha, interleukins, and CRP and tumor suppressor markers like anti-p53 were included in only two signatures each.
Correcting for overoptimistic prediction of statistical model
To avoid overoptimistic results, a signature derived in a specific study needs to be validated in independent samples. Initial internal evaluation of model performance was performed by 20 out of 36 different studies as shown in Table 2. Split-sampling method, the simplest form of internal validation, was utilized in seven different studies.31, 38, 39, 45, 49, 52, 53 More advanced techniques including various variants of cross-validation21, 23, 24, 26, 33, 36, 37, 41, 44, 50 and.632 bootstrap25, 55, 56 were used by 10 and 3 studies, respectively. External validation of results was not performed by any study except Werner et al.,27 which validated results from a previous clinical setting study (i.e., Wild et al.41) in a true screening setting. Supplementary File 5 contains a brief description of each validation method.
Diagnostic performance of protein signatures
The diagnostic indicators of all the selected studies as shown in Table 2 varied greatly, with sensitivities and specificities ranging from 27–97% and 32–99%, respectively. The P value specifying the level of significance in test performance between cases and controls was stated in only 13 studies and ranged from 0.249 to <0.001. The area under the curve (AUC) was reported by 19 out of 23 studies published in the last five years and ranged from 0.6238 to 0.996.43 Notably, AUCs for CRC were much lower for studies conducted in true screening settings (range 0.62–0.78) than in studies conducted in clinical settings (range 0.68–0.996), and the AUCs of studies conducted in true screening settings were much lower than the summary AUC for FITs derived from such studies (0.95, 95% CI 0.93–0.97) in a recent meta-analysis.57
Among 21 studies that performed some form of validation measure, best diagnostic performance (94% sensitivity, 98% specificity, and AUC of 0.988) was reported by Zhang et al.45 for a panel of tumor markers (CA199, CA242, CA125, CA153, and CEA). However, this study was conducted in a clinical setting and the time of recruitment and blood collection was unclear. Among 15 studies that did not employ any form of validation, Pengjun et al.43 reported best diagnostic performance (96% sensitivity, 99% specificity, and AUC of 0.996) for a combination of inflammatory markers (IL-8, MMP-2, and TNF-α). Again, this study was conducted in a clinical setting with recruitment of clinically detected rather than screening-detected cases.
All of the protein signatures were identified or evaluated in just one study, except one signature constituting of three proteins HSP60, CA19-9, and CEA which was identical in two studies.39, 42 Hamelin et al.39 who conducted their study in a French population reported 47% sensitivity at 90% specificity (J=0.37) and an AUC of 0.77 in a split-sample validation, whereas Hou et al.42 reported a sensitivity of 97% at 91% specificity (J=0.88) and an AUC of 0.906 in a study in a Chinese population without any internal or external validation. Both studies were conducted in clinical settings, however, with unclear information on the time of recruitment and blood collection.
A notable difference was observed when a protein signature first evaluated in a clinical setting was later validated in a true screening setting. Wild et al.41 had reported a diagnostic sensitivity of 70% at 95% specificity for differentiating between CRC cases and controls for a signature with CYFRA21-1, ferritin, Osteopontin, anti-p53, seprase, and CEA. However, when these candidate proteins combined to a five biomarker blood test (excluding CYFRA21-1 as it was dispensable for the reoptimized algorithm) was later validated in a true screening setting, the diagnostic sensitivity decreased to 42% at 95% specificity.27
Stage distribution of cases reported by 24 studies and stage-specific diagnostic indicators of signatures reported by 11 studies are summarized in the Supplementary File 2. Stage-specific numbers of CRC cases were often small and several studies included larger proportion of late stage cancers compared with early-stage cancers with a tendency towards higher sensitivity for the more advanced cancers.
Assessment of risk of bias across studies
Supplementary File 3 summarizes the results of our assessment of risk of bias using the QUADAS-2 tool. In the QUADAS-2 assessment for four domains, only Werner et al.27 out of 36 studies presented low risk of bias and low applicability concerns in all domains as this was the only study that collected blood from asymptomatic participants in a true screening setting, analyzed the index test without knowledge of the results of colonoscopy and accounted for analytical, internal, and external validity. Risk of bias was highest for the domain ‘Patient selection’ given that most studies were conducted in a clinical rather than a screening setting. For more than two-third of the studies risk of bias was also high or unclear for the domain ‘Reference standard’, because it was not clearly reported whether all CRC patients and controls received a reference standard colonoscopy. Applicability concerns were common for the ‘Patient selection’ domain because blood was collected in most of the studies either from symptomatic patients or after establishment of clinical diagnosis. Risk of bias was low for all ‘Index Test’ criteria as none of the included studies conducted or interpreted the test in a way that would conflict the review question.
Discussion
This review provides an overview of protein signature studies associated with diagnosis of CRC. Overall, 36 studies published from 1985 to 2017 and investigating 93 different proteins in multimarker signatures were identified. Great discrepancies in estimates of diagnostic performance were observed with sensitivities ranging from 27 to 99%. However, study setting and study quality were very heterogeneous and require careful consideration in the interpretation of the results.
In recent years, there have been studies from major prospective cohorts like the European Prospective Investigation into Cancer and Nutrition (EPIC)58 and Women’s Health Initiative (WHI).59, 60 In addition, studies were also carried out in screening cohorts like BLiTz (Begleitende Evaluierung innovativer Testverfahren zur Darmkrebs-Früherkennung)61, 62, 63 where both cases and controls were selected from participants of screening colonoscopy. Such studies have major advantages as they represent the target population for CRC screening, ensure fully comparable recruitment conditions for cases and controls and confirm absence of pre-clinical CRC from controls. However, they are costly, time consuming and the typically low prevalence and number of CRC cases restricts the possibility of subgroup-specific analysis. Thus, almost all previous studies recruited study participants in clinical settings which may give rise to various potential biases. For example, blood protein marker levels might be affected by diagnostic or early therapeutic interventions or lifestyle and diet modifications following diagnosis. Furthermore, patients recruited in clinical settings typically include higher proportions of advanced stage cases than screening-detected cases, which may often lead to overestimation of sensitivity13 and limit applicability to the target screening population due to spectrum bias.64, 65
For the analysis of multiple protein markers, it is essential to fit statistical models to the data. Because models are meant to offer best fit for the specific data analyzed, they will typically provide less-accurate prediction when applied to other study populations or data sets. Hence, validation is crucial to prevent overfitting and overoptimistic estimates of diagnostic performance.66 Internal validation of some form was performed for 20 out of the 36 studies included in this review and seven out of these used the split-sampling method, which may be subject to major variability of results owing to specific split of the sample.67 Complementary to internal validity, external validation on participants that differ in place, setting, time, and so on from the subjects used to develop a model is crucial for comprehensive evaluation of diagnostic signatures. In the current review signatures with AUC as high as 0.996,43 0.988,45 and 0.92450 have been identified but as these signatures were not externally validated it would be interesting to find the extent to which the results reported in one study would hold accurate for a separate study population from a different point of time and setting.
Protein signatures and their performance in particular may vary between populations from different parts of the world. One protein signature identified by Hamelin et al.39 in a French study population was identical to that of Hou et al.42 evaluated in a Chinese population. Despite the use of identical panels including commonly assessed proteins such as HSP60, CA19-9, and CEA reported diagnostic performances were far from identical. Sensitivity and specificity were 47% and 90% in the French study compared with 97% and 90% in the Chinese study. Given that the Chinese study, in contrast to the French study, did not employ any internal validation, the apparently much better performance might partly reflect overfitting and overoptimism, which underlines once more the importance of both internal and external validation in the evaluation of biomarker signatures. Apart from this no other signature was identical in Asian or Western population. Also as the proteins in all signatures in the review are functionally different, it is challenging to suggest if any particular class of proteins like tumor markers or tumor suppressor or inflammatory markers possess a higher diagnostic potential. Nevertheless, validation of all the identified proteins on same study population would yield interesting and generalizable results.
Out of the several methods used in the studies included in this review, immunoassay, and mass spectrometry-based methods were the most commonly used. Both mass spectrometry and immunoassay have a limited dynamic range with limit of detections in nanogram range and so several low abundance proteins are often missed. Whereas mass spectrometry can identify >1,000 proteins per run, immunoassays work for only one target at a time. Nevertheless, as the sample handling and collection procedure was very heterogeneous and the analytical procedure was also different across studies, the influence of the method upon result cannot be clearly established. In addition, several studies in this review used a candidate approach when selecting which proteins to analyze, whereas others used approaches without a priori defined targets. Although it is therefore not possible from this review to derive firm conclusions on which signatures would work best in practice, the overview of identified signatures provided by this review should help designing studies that directly compare promising signatures in the same study population and under comparable preanalytical conditions.
To our knowledge, this is the first systematic review summarizing the existing literature on protein panels or signatures rather than individual proteins. This is a rapidly emerging field, with 23 out of 36 identified articles published in the past five years only. Given the numerous differences between studies in terms of study populations, sample handling, number, composition, and overlap of proteins assessed, analytic strategies, and proneness to the various types of biases outlined above it is difficult if not impossible to identify one or more specific protein panels to be the most promising. Rather, our review may serve as a basis to identify potentially promising candidates of both individual proteins and protein signatures that ideally should be evaluated in parallel in the same study population in a study conducted in a true screening setting, avoiding or at least minimizing the various types of biases outlined above. Only this way will it be possible to disentangle to what extent the apparent large variations in diagnostic performance may result from differences in study populations, signature compositions or various degrees of bias control.
A number of limitations of our review should be kept in mind. Our search was restricted to English language articles, which could be a source of language bias. Even though a comprehensive literature search was performed in two databases and careful cross referencing was done, it cannot be excluded that relevant articles, especially those published in the “gray literature” were missed. Several studies had to be excluded in full text review because they did not report the diagnostic sensitivity/specificity of the protein signatures. This selective reporting could be a potential source of outcome reporting bias.68, 69 Furthermore, this systematic review only provides an overview and summary of individual studies, whereas a meta-analysis of results was not meaningful on account of multiple differences across the studies.
Although the number of studies assessing diagnostic performance of protein signatures for early detection of CRC has been rapidly increasing in recent years, evidence on diagnostic performance for detection of CRC precursors, such as advanced adenomas, is still very limited. The few studies that addressed this issue generally found diagnostic performance to be rather poor and much worse than diagnostic performance for CRC. This property is shared with other, established screening approaches, such as fecal occult blood testing. For the time being, thoroughly validated blood protein signatures cannot compete with FITs in terms of diagnostic performance in detecting CRC. There might be a large potential for protein signatures to enhance CRC screening in the future if sensitive analytical techniques could be developed that would more reliably allow detecting proneness to and presence of advanced adenomas in addition to CRC.
Conclusion
This review identified a large number of recent studies exploring protein panels or signatures (rather than single proteins) for diagnosis of CRC. Despite some apparently very promising results, the blood-based protein signatures are not ready to replace established screening and surveillance methods for CRC, such as fecal FITs, flexible sigmoidoscopy or colonoscopy. Even though a number of studies have reported promising diagnostic performance comparable to performance of FITs, thorough validation in studies conducted in true screening settings ideally in comparison with FITs and using screening colonoscopy as gold standard reference test in all participants would be essential. If the apparent good performance of promising signatures can be confirmed in such settings, they might become a promising alternative for CRC screening, given that higher adherence rates might be achieved in blood-based compared to stool-based screening. In addition, rapid progress in technologies for both targeted and untargeted proteomics screening may open new arenas for substantial future development of highly informative protein signatures.
Footnotes
Guarantor of the article: Megha Bhardwaj.
Specific author contributions: Hermann Brenner designed and supervised the study. Megha Bhardwaj carried out the literature search and drafted the manuscript. Megha Bhardwaj and Anton Gies extracted the data from the eligible studies. All authors critically reviewed, contributed to and approved the final manuscript.
Financial support: None.
Potential competing interests: The German Cancer Research Center has received industrial grants related to blood markers for early detection of colorectal cancer from Epigenomics, Applied Proteomics, Roche Diagnostics, and Volition. Megha Bhardwaj had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Supplementary Material
References
- Ferlay J, Soerjomataram I, Dikshit R et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136: E359–E386. [DOI] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL et al. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- Shroff J, Thosani N, Batra S et al. Reduced incidence and mortality from colorectal cancer with flexible-sigmoidoscopy screening: a meta-analysis. World J Gastroenterol 2014; 20: 18466–18476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner H, Stock C, Hoffmeister M. Effect of screening sigmoidoscopy and screening colonoscopy on colorectal cancer incidence and mortality: systematic review and meta-analysis of randomised controlled trials and observational studies. BMJ 2014; 348: g2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauber AG. The impact of screening on colorectal cancer mortality and incidence: has it really made a difference? Dig Dis Sci 2015; 60: 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JS, Piper MA, Perdue LA et al. Screening for colorectal cancer: updated evidence report and systematic review for the us preventive services task force. JAMA 2016; 315: 2576–2594. [DOI] [PubMed] [Google Scholar]
- Schreuders EH, Ruco A, Rabeneck L et al. Colorectal cancer screening: a global overview of existing programmes. Gut 2015; 64: 1637–1649. [DOI] [PubMed] [Google Scholar]
- Lansdorp-Vogelaar I, Knudsen AB, Brenner H. Cost-effectiveness of colorectal cancer screening. Epidemiol Rev 2011; 33: 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Efron JE. Screening for colorectal cancer. Adv Surg 2011; 45: 31–44. [DOI] [PubMed] [Google Scholar]
- Hassan C, Giorgi Rossi P, Camilloni L et al. Meta-analysis: adherence to colorectal cancer screening and the detection rate for advanced neoplasia, according to the type of screening test. Aliment Pharmacol Ther 2012; 36: 929–940. [DOI] [PubMed] [Google Scholar]
- Adler A, Geiger S, Keil A et al. Improving compliance to colorectal cancer screening using blood and stool based tests in patients refusing screening colonoscopy in Germany. BMC Gastroenterol 2014; 14: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracci F, Zorzi M, Grazzini G. Colorectal cancer screening: tests, strategies, and perspectives. Front Public Health 2014; 2: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao S, Hundt S, Haug U et al. Sensitivity estimates of blood-based tests for colorectal cancer detection: impact of overrepresentation of advanced stage disease. Am J Gastroenterol 2011; 106: 242–253. [DOI] [PubMed] [Google Scholar]
- Thomas DS, Fourkala EO, Apostolidou S et al. Evaluation of serum CEA, CYFRA21-1 and CA125 for the early detection of colorectal cancer using longitudinal preclinical samples. Br J Cancer 2015; 113: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatandoost N, Ghanbari J, Mojaver M et al. Early detection of colorectal cancer: from conventional methods to novel biomarkers. J Cancer Res Clin Oncol 2016; 142: 341–351. [DOI] [PubMed] [Google Scholar]
- Borrebaeck CA. Precision diagnostics: moving towards protein biomarker signatures of clinical utility in cancer. Nat Rev Cancer 2017; 17: 199–204. [DOI] [PubMed] [Google Scholar]
- Liberati A, Altman DG, Tetzlaff J et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ 2009; 339: b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Werner S, Tao S et al. Blood autoantibodies against tumor-associated antigens as biomarkers in early detection of colorectal cancer. Cancer Lett 2014; 346: 178–187. [DOI] [PubMed] [Google Scholar]
- Whiting PF, Rutjes AW, Westwood ME et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011; 155: 529–536. [DOI] [PubMed] [Google Scholar]
- Moher D, Liberati A, Tetzlaff J et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009; 6: e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume JE, Wilhelmsen M, Benz RW et al. Discovery and validation of plasma-protein biomarker panels for the detection of colorectal cancer and advanced adenoma in a danish collection of samples from patients referred for diagnostic colonoscopy. The JALM 2016; 1: 181–193. [DOI] [PubMed] [Google Scholar]
- Storm L, Christensen IJ, Jensenius JC et al. Evaluation of complement proteins as screening markers for colorectal cancer. Cancer Immunol Immunother 2015; 64: 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen SB, Lundberg M, Villablanca A et al. Detection of serological biomarkers by proximity extension assay for detection of colorectal neoplasias in symptomatic individuals. J Transl Med 2013; 11: 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsen M, Christensen IJ, Rasmussen L et al. Detection of colorectal neoplasia: combination of eight blood-based, cancer-associated protein biomarkers. Int J Cancer 2016; 140: 1436–1446. [DOI] [PubMed] [Google Scholar]
- Chen H, Zucknick M, Werner S et al. Head-to-head comparison and evaluation of 92 plasma protein biomarkers for early detection of colorectal cancer in a true screening setting. Clin Cancer Res 2015; 21: 3318–3326. [DOI] [PubMed] [Google Scholar]
- Surinova S, Choi M, Tao S et al. Prediction of colorectal cancer diagnosis based oncirculating plasma proteins. EMBO Mol Med 2015; 7: 1166–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Krause F, Rolny V et al. Evaluation of a 5-marker blood test for colorectal cancer early detection in a colorectal cancer screening setting. Clin Cancer Res 2016; 22: 1725–1733. [DOI] [PubMed] [Google Scholar]
- Barderas R, Mendes M, Torres S et al. In-depthcharacterization of the secretome of colorectal cancer metastatic cells identifies key proteins in cell adhesion, migration, and invasion. Mol Cell Proteomics 2013; 12: 1602–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Fernandez L, Tejero E, Tieso A. Significance of CA 72-4 in colorectal carcinoma. Comparison with CEA and CA 19-9. Eur J Surg Oncol 1995; 21: 388–390. [DOI] [PubMed] [Google Scholar]
- Lumachi F, Marino F, Orlando R et al. Simultaneous multianalyte immunoassay measurement of five serum tumor markers in the detection of colorectal cancer. Anticancer Res 2012; 32: 985–988. [PubMed] [Google Scholar]
- Brock R, Xiong B, Li L et al. A multiplex serum protein assay for determining the probability of colorectal cancer. Am J Cancer Res 2012; 2: 598–605. [PMC free article] [PubMed] [Google Scholar]
- Butvilovskaya VI, Popletaeva SB, Chechetkin VR et al. Multiplex determination of serological signatures in the sera of colorectal cancer patients using hydrogel biochips. Cancer Med 2016; 5: 1361–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JJ, Wilcox BE, Benz RW et al. A plasma-based protein marker panel for colorectal cancer detection identified by multiplex targeted mass spectrometry. Clinical Colorectal Cancer 2016; 15: 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen M, Pasanen P, Janatuinen E et al. Small intestinal mucin antigen (SIMA); a novel tumour marker in colorectal cancer? Anticancer Res 1995; 15: 2351–2356. [PubMed] [Google Scholar]
- Eskelinen M, Pasanen P, Kulju A et al. Clinical evaluation of serum tumour markers CEA, CA 50 and CA 242 in colorectal cancer. Anticancer Res 1994; 14: 1427–1432. [PubMed] [Google Scholar]
- Shimwell NJ, Wei W, Wilson S et al. Assessment of novel combinations of biomarkers for the detection of colorectal cancer. Cancer Biomark 2010; 7: 123–132. [DOI] [PubMed] [Google Scholar]
- Ward DG, Suggett N, Cheng Y et al. Identification of serum biomarkers for colon cancer by proteomic analysis. Br J Cancer 2006; 94: 1898–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuzzi M, Marelli C, Bagnati R et al. Plasma clusterin as a candidate pre-diagnosis marker of colorectal cancer risk in the Florence cohort of the European Prospective Investigation into Cancer and Nutrition: a pilot study. BMC Cancer 2015; 15: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelin C, Cornut E, Poirier F et al. Identification and verification of heat shock protein 60 as a potential serum marker for colorectal cancer. FEBS J 2011; 278: 4845–4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzystek-Korpacka M, Diakowska D, Kapturkiewicz B et al. Profiles of circulating inflammatory cytokines in colorectal cancer (CRC), high cancer risk conditions, and health are distinct. Possible implications for CRC screening and surveillance. Cancer Lett 2013; 337: 107–114. [DOI] [PubMed] [Google Scholar]
- Wild N, Andres H, Rollinger W et al. A combination of serum markers for the early detection of colorectal cancer. Clin Cancer Res 2010; 16: 6111–6121. [DOI] [PubMed] [Google Scholar]
- Hou PZ, Luo JC, Zhang JH. A study of optimized model of serum tumor markers of colorectal cancer based on intelligent algorithms. Int J Clin Exp Med 2016; 9: 17153–17164. [Google Scholar]
- Pengjun Z, Xinyu W, Feng G et al. Multiplexed cytokine profiling of serum for detection of colorectal cancer. Future Oncol 2013; 9: 1017–1027. [DOI] [PubMed] [Google Scholar]
- Xue Y, Yu F, Yan D et al. Zinc-alpha-2-glycoprotein: a candidate biomarker for colon cancer diagnosis in Chinese population. Int J Mol Sci 2014; 16: 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Liang XL, Gao HY et al. Models of logistic regression analysis, support vector machine, and back-propagation neural network based on serum tumor markers in colorectal cancer diagnosis. Genet Mol Res 2016; 15; doi: 10.4238/gmr.15028643. [DOI] [PubMed] [Google Scholar]
- Zheng GX, Wang CX, Qu X et al. Establishment of serum protein pattern for screening colorectal cancer using SELDI-TOF-MS. Exp Oncol 2006; 28: 282–287. [PubMed] [Google Scholar]
- Ji WB, Chen J, Mi YC et al. CEA, CA125, CA153 and TSGF act as diagnostic tumor biomarkers for patients with rectal carcinoma. Int J Clin Exp Med 2016; 9: 20109–20113. [Google Scholar]
- Xie YY, Chen LH, Lv XL et al. The levels of serine proteases in colon tissue interstitial fluid and serum serve as an indicator of colorectal cancer progression. Oncotarget 2016; 7: 32592–32606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ma XH. Study on specificity of colon carcinoma-associated serum markers and establishment of SVM prediction model. Saudi J Biol Sci 2017; 24: 644–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K, Naito Y, Yagi N et al. Peptidomic analysis via one-step direct transfer technology for colorectal cancer biomarker discovery. J Proteomics Bioinform 2015; S5. [Google Scholar]
- Yoneda K, Iida H, Endo H et al. Identification of Cystatin SN as a novel tumor marker for colorectal cancer. Int J Oncol 2009; 35: 33–40. [PubMed] [Google Scholar]
- Shin J, Kim HJ, Kim G et al. Discovery of melanotransferrin as a serological marker of colorectal cancer by secretome analysis and quantitative proteomics. J Proteome Res 2014; 13: 4919–4931. [DOI] [PubMed] [Google Scholar]
- Ladd JJ, Busald T, Johnson MM et al. Increased plasma levels of the APC-interacting protein MAPRE1, LRG1, and IGFBP2 preceding a diagnosis of colorectal cancer in women. Cancer Prev Res (Phila) 2012; 5: 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer DW, Talamo TS. Multiple markers of malignancy in sera of patients with colorectal carcinoma: preliminary clinical studies. Clin Chem 1985; 31: 1824–1828. [PubMed] [Google Scholar]
- Taguchi A, Rho JH, Yan QX et al. MAPRE1 as a plasma biomarker for early-stage colorectal cancer and adenomas. Cancer Prev Res 2015; 8: 1112–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung KY, Tabor B, Buckley MJ et al. Blood-based protein biomarker panel for the detection of colorectal cancer. PLoS ONE 2015; 10: e0120425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Liles EG, Bent S et al. Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta-analysis. Ann Intern Med 2014; 160: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riboli E, Kaaks R. The EPIC Project: rationale and study design. European Prospective Investigation into Cancer and Nutrition. Int J Epidemiol 1997; 26: S6–14. [DOI] [PubMed] [Google Scholar]
- Hays J, Hunt JR, Hubbell FA et al. The Women's Health Initiative recruitment methods and results. Ann Epidemiol 2003; 13: S18–S77. [DOI] [PubMed] [Google Scholar]
- Design of the Women's Health Initiative clinical trial and observational study.. The Women's Health Initiative Study Group. Control Clin Trials 1998; 19: 61–109. [DOI] [PubMed] [Google Scholar]
- Haug U, Hundt S, Brenner H. Quantitative immunochemical fecal occult blood testing for colorectal adenoma detection: evaluation in the target population of screening and comparison with qualitative tests. Am J Gastroenterol 2010; 105: 682–690. [DOI] [PubMed] [Google Scholar]
- Brenner H, Tao S, Haug U. Low-dose aspirin use and performance of immunochemical fecal occult blood tests. JAMA 2010; 304: 2513–2520. [DOI] [PubMed] [Google Scholar]
- Hundt S, Haug U, Brenner H. Comparative evaluation of immunochemical fecal occult blood tests for colorectal adenoma detection. Ann Intern Med 2009; 150: 162–169. [DOI] [PubMed] [Google Scholar]
- Kohn MA, Carpenter CR, Newman TB. Understanding the direction of bias in studies of diagnostic test accuracy. Acad Emerg Med 2013; 20: 1194–1206. [DOI] [PubMed] [Google Scholar]
- Whiting P, Rutjes AW, Reitsma JB et al. Sources of variation and bias in studies of diagnostic accuracy: a systematic review. Ann Intern Med 2004; 140: 189–202. [DOI] [PubMed] [Google Scholar]
- Smith GC, Seaman SR, Wood AM et al. Correcting for optimistic prediction in small data sets. Am J Epidemiol 2014; 180: 318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyerberg EW, Harrell FE Jr., Borsboom GJ et al. Internal validation of predictive models: efficiency of some procedures for logistic regression analysis. J Clin Epidemiol 2001; 54: 774–781. [DOI] [PubMed] [Google Scholar]
- McGauran N, Wieseler B, Kreis J et al. Reporting bias in medical research - a narrative review. Trials 2010; 11: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan M, Ansari MT, Berkman ND et alAssessing the risk of bias of individual studies in systematic reviews of health care interventions. In: Methods Guide for Effectiveness and Comparative Effectiveness Reviews: Rockville (MD), 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.