Abstract
Members of the GroEL/HSP60 protein family have been studied for many years because of their critical roles as ATP-dependent molecular chaperones, so it might come as a surprise that some have important functions in ATP-poor conditions, for example, when secreted outside the cell. At least some members of each of the HSP10, HSP70, HSP90, HSP100 and HSP110 heat shock protein families are also ‘moonlighting proteins’. Moonlighting proteins exhibit more than one physiologically relevant biochemical or biophysical function within one polypeptide chain. In this class of multifunctional proteins, the multiple functions are not due to gene fusions or multiple proteolytic fragments. Several hundred moonlighting proteins have been identified, and they include a diverse set of proteins with a large variety of functions. Some participate in multiple biochemical processes by using an active site pocket for catalysis and a different part of the protein's surface to interact with other proteins. Moonlighting proteins play a central role in many diseases, and the development of novel treatments would be aided by more information addressing current questions, for example, how some are targeted to multiple cellular locations and how a single function can be targeted by therapeutics without targeting a function not involved in disease.
This article is part of the theme issue ‘Heat shock proteins as modulators and therapeutic targets of chronic disease: an integrated perspective’.
Keywords: moonlighting proteins, heat shock proteins, multifunctional proteins
1. Introduction
HSP60/GroEL heat shock proteins (HSPs) have been studied for many years because of their critical role as ATP-dependent molecular chaperones, but some have important functions in ATP-poor conditions, when secreted outside the cell. In fact, members of each of the HSP60/HSP10, HSP70, HSP90, HSP100 and HSP110 HSP/chaperone protein families have been found to be ‘moonlighting proteins’. Moonlighting proteins comprise a subset of multifunctional proteins in which one polypeptide chain exhibits more than one physiologically relevant biochemical or biophysical function [1]. In this class of multifunctional proteins, the multiple functions are not due to gene fusions or multiple proteolytic fragments. Several hundred moonlighting proteins have been identified, and they include a diverse set of proteins with a large variety of functions [2].
Among the first proteins to be recognized as performing two very different functions in the same organism were the taxon-specific crystallins. Crystallins make up a large part of the lens of the eye, and about a dozen are identical to catalytically active ubiquitious enzymes. For example, the epsilon crystalline found in birds (mallard duck, swans, geese and ostriches) and reptiles (crocodiles) [3–5] is the same protein as lactate dehydrogenase, which catalyses the interconversion of pyruvate and lactate and is found in many cell types in almost all species (figure 1). In other species, different enzymes were co-opted to serve as crystallins—quinone oxidoreductase is the zeta crystalline in camels, llamas, guinea pigs and frogs [6–8], and aldehyde dehydrogenase is the eta crystalline in elephant shrews [9].
Figure 1.
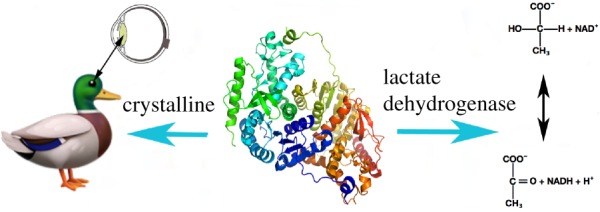
A moonlighting protein can have two very different functions in the same species. For example, in ducks, the epsilon crystalline found in the lens of the eye is the same protein as the ubiquitous enzyme lactate dehydrogenase, which catalyses the interconversion of pyruvate and lactate. (Online version in colour.)
Other soluble enzymes have evolved to serve a second function as transcriptional or translational regulators that bind DNA or RNA, respectively, in some cases as a feedback mechanism to regulate the level of expression of enzymes in the same biochemical pathway. Aconitase catalyses the isomerization of citrate to isocitrate in the citric acid cycle. When cellular iron concentrations decrease, the iron–sulfur cluster in the active site pocket is lost and the protein changes conformation. The new protein conformation binds RNA to regulate the expression of genes encoding proteins involved in iron uptake [10–12].
Moonlighting proteins are found throughout the evolutionary tree—bacteria, archaea, mammals, reptiles, birds, fish, worms, insects, plants, fungi, protozoans and even viruses. They include enzymes that serve as receptors, secreted cytokines, transcription factors, DNA stabilizers, components of the cytoskeleton or proteasome subunits. Other combinations of functions include a receptor and transcription factor, chaperone and cytokine, DNA-binding protein and component of the extracellular matrix, transmembrane channel and regulator of other channels, and components of the ribosome that are transcription factors.
2. Intracellular/extracellular moonlighting proteins
One of the largest subgroups of moonlighting proteins identified to date are intracellular chaperones and enzymes that play a different role outside the cell (figure 2). These are often ‘housekeeping proteins’ that are widespread in evolution and function in glycolysis, the citric acid cycle, the pentose phosphate pathway, and protein and DNA metabolism. The first to be identified was a glyceraldehyde 3-phosphate dehydrogenase (GAPDH) on the surface of pathogenic streptococci [13]. Many other intracellular/cell surface enzymes were later found, including other GAPDHs [14–28], phosphoglycerate kinase [29,30] and enolase [31–54]. Other intracellular/cell surface proteins (ICSPs) include chaperones (HSP60/GroEL, HSP70/DnaK) [54–58], a protein synthesis elongation factor (Ef-Tu) [59–61] and a histone (H1) [62].
Figure 2.

Several dozen moonlighting proteins have different functions inside and outside the cell. Many housekeeping proteins, including enzymes that convert a substrate (star) to a product (hexagon) or chaperones that assist in protein folding, have a second function when secreted or when attached to the cell surface. In most cases, how the intracellular/surface or intracellular/secreted moonlighting proteins are secreted and how some become attached to the cell surface are unknown. (Online version in colour.)
Some of these proteins function as cell surface receptors in humans and other mammals. GAPDH catalyses the conversion of D-glyceraldehyde 3-phosphate to 3-phospho-D-glyceroyl phosphate in glycolysis inside the cell in most cell types but in mammals also serves as a cell surface transferrin receptor to aid in iron uptake [63,64]. The HSP60 HSP is a chaperone assisting mitochondrial protein import in the cell, and is a cell surface receptor for high-density lipoproteins through its affinity for apolipoprotein apoA-II [65]. In humans, pyruvate kinase 3 (PK3) isoform 2, glutathione S-transferase Mu 3, triosephosphate isomerase and fructose-bisphosphate aldolase A play a second role on the sperm head membrane, where they are involved in interactions with zona pellucida proteins of egg [66–68].
This phenomenon of intracellular/surface moonlighting proteins has been observed widely in bacteria. Bacteria (and other pathogens) commonly use moonlighting cytosolic proteins on the cell surface for forming and maintaining interactions with the host species. Some of these proteins play important roles in infection, invasion, virulence and formation of biofilms. Colonization requires adhesion to the host, and many surface proteins bind to proteins in the extracellular matrix, including fibronectin, laminin, and/or collagen, or to mucin, a component of the mucosal epithelial lining. Other surface moonlighting proteins bind directly to proteins on host cell surfaces. These interactions enable a physical attachment to the host. Listeria makes use of alcohol acetaldehyde dehydrogenase/Listeria adhesion protein (LAP) to bind to intestinal epithelial cells [55]. Enolase in glycolysis has been found on the cell surface in many species of bacteria (Streptococcus, Mycoplasma and Plasmodium falciparum) where it plays a role in binding plasminogen, fibronectin, and other proteins and is important in infection of human, canine and avian hosts [69–72]. Translation Ef-Tu from Streptococcus gordonii binds to host mucin [73]. Streptococcus pneumoniae makes use of endopeptidase O to bind host plasminogen and fibronectin [74]. The GAPDH of Haemonchus contortus (barber pole worm), a nematode species that infects sheep and goat gastrointestinal tracts, binds the alternative complement pathway protein C3, inhibits the complement cascade, and helps the pathogen evade host immunity [75]. The HSP70/DnaK chaperone also serves as a cell surface receptor for plasminogen in many species— Neisseria [76], Mycobacterium tuberculosis [77], Bifidiobacterium lactis [78], etc.
Streptococcus pneumoniae and the many other pathogenic bacteria that use enolase GAPDH or other enzymes to bind plasminogen help the zymogen get converted to the active protease plasmin by using an endogenous protease or making use of the host's tissue-type plasminogen (tPA) activators and urokinase-type plasminogen activators [79,80]. The now active plasmin attached to the surface of the invading organism can be used as a general protease to digest host extracellular matrix and basement membrane, thereby assisting migration through tissues.
These interactions between cytoplasmic/cell surface moonlighting proteins with host tissue are not always the result of disease or infection. The commensual ‘probiotic’ bacterium Lactobacillus acidophilus uses GAPDH on its surface to help colonize the gut. In this case, the bacterial GAPDH binds to host mucin [81].
Moonlighting cytosolic proteins are also often used as secreted signalling molecules to regulate other cell types within an organism, or to modulate host responses in the case of a pathogen. Inside the cell, members of the HSP60/HSP10, HSP70, HSP90, HSP100 and HSP110 protein families are protein chaperones that prevent client proteins from misfolding and promote correct refolding and assembly of protein complexes, and members of each protein family have been identified that have additional functions outside the cell. The extracellular roles of mammalian members of these protein families are discussed further in other papers in this collection, so in this review the focus is on other examples of cytoplasmic proteins that perform other functions when secreted. Thymidine phosphorylase, an enzyme in pyrimidine metabolism, is the same protein as the secreted platelet-derived endothelial cell growth factor (PDGF) [82]. Lysyl-tRNA synthetase, which attaches lysine to tRNA for use in protein synthesis, acts extracellularly on macrophages and peripheral blood mononuclear cells to increase TNF-α production and target cell migration [83]. Thymosin β-4 is an intrinsically disordered protein involved in sequestering G-actin (monomeric actin) to prevent its polymerization to F-actin in polymorphonuclear leucocytes. Thymosin β-4 sulfoxide is generated in monocytes by the oxidation of a methionine (Met6) in the presence of glucocorticoids and is secreted to inhibit the anti-inflammatory response [84–87]. Phosphoglucose isomerase (PGI, also known as glucose-6-phosphate isomerase, autocrine motility factor, neuroleukin, differentiation and maturation mediator) catalyses the interconversion of glucose-6-phosphate and fructose-6-phosphate in glycolysis and gluconeogenesis, and is an extracellular cytokine/growth factor that binds to target cells and causes pre-B cells to mature into antibody secreting cells, supports the survival of embryonal neurons, and causes differentiation of several leukemia cell lines [88–94]. In the black footed ferret (Mustela nigripes), PGI was also found to be necessary for embryo implantation [95]. The growth factor effect of PGI can also play a role in disease. Extracellular PGI causes an increase in cell migration during breast cancer metastasis. Another cytoplasmic protein that also has roles in cancer development, threonyl aminoacyl-tRNA synthetase (TARS), is secreted from endothelial cells in response to TNF-α and VEGF and promotes vascular development. An association has been observed between TARS expression and advancing stage of ovarian cancer [96]. A mutation that prevents TARS synthetase activity is still active in promoting vascular development.
Some bacterial cytosolic enzymes are secreted to interfere with host defenses. GAPDH from A. vaginae is secreted to interfere with human C5a anaphylatoxin [97]. Leishmania donovani secretes another glycolytic enzyme, fructose-bisphosphate aldolase, as well as translation elongation factor EF-1 to cause activation of host macrophage protein tyrosine phosphatase-1 (SHP-1) and decreased activity of infected macrophages [98,99]. The GroEL/60 kDa chaperonin produced by Enterobacter aerogenes, the bacterial endosymbiont of an insect, antlions, is secreted and used as toxin to paralyze cockroaches [100]. Enolase from the nematode Steinernema glaseri, when on its cuticle surface or secreted in to the host hemolymph, suppresses its insect host's immune system [101].
Further examples of intracellular/surface/secreted moonlighting proteins are included in our list of moonlighting proteins in the MoonProt Database [2]. Most of the known ICSPs are from bacteria, although examples are found throughout the evolutionary tree. The bacterial species represented include typical Gram-positive and Gram-negative species, as well as mycobacteria, spirochetes and mycoplasma. In addition, many more cytoplasmic proteins have been found to be secreted or bound to the cell surface through proteomics studies. For most of the proteins found through proteomics studies, further experiments are needed to determine if the protein has a second function outside the cell, if it performs the same function as when inside the cell, or perhaps was found in the extracellular location due to experimental artefacts of the proteomics methods that were used.
3. Structural basis for intracellular/extracellular functions
One question that arises when a protein is found to be a moonlighting protein pertains to how one polypeptide chain can perform two different functions, because protein function is tied to protein structure and it might be presumed that it would be necessary to alter a protein structure a lot to gain a new function, which could result in loss of the original function. Some moonlighting proteins have been found to solve this problem of switching between functions by undergoing large conformational changes or transitions between intrinsically unfolded domains and multiple distinct folded structures so that different conformations of the protein structure can perform different functions. By contrast, for many of the intracellular/extracellular moonlighting proteins, large diversions from the structure or conformation performing a catalytic or chaperone function within the cell are not needed for performing the extracellular function. In many of the cases discussed above, the extracellular function involves binding to another molecule, often another protein. In addition to an active site pocket where catalysis occurs when the protein is inside the cell, the three-dimensional structure of an enzyme or chaperone includes a large amount of solvent exposed surface area. Through millions of years of evolution some portion of this surface can gain a pattern of amino acids needed for interacting with another molecule, whether a small molecule, a cell surface receptor, or another protein, without affecting the active site pocket [102]. This new binding site does not need to very large or complex. Ehinger and co-workers found that in the case of Streptococcus enolase, a small motif consisting of only nine amino acids (248FYDKERKVY256) containing lysines and negatively charged residues was sufficient for binding to plasminogen. The evolution of this binding site did not affect the ability of enolase to catalyse the reaction, and the overall subunit fold of the protein is identical to that of enolase proteins that are not known to bind plasminogen [103].
4. Regulation of multiple activities
Having the ability to perform multiple functions is often only part of the story for moonlighting proteins. The correct level of each of the multiple protein activities in the correct place and the correct time can also be important for maintaining health and homeostasis. Changes in the expression, level of activity and/or location of moonlighting proteins play a central role in many diseases. Mutations that result in altered activity of a moonlighting protein can lead to disease—whether it is a decrease in activity, or, in some cases, an increase in activity, for example, by disrupting the careful checks and balances of the immune system. In addition, mutations that change the amino acid sequence of a protein can on rare occasions result in an increase in function or a different function, sometimes referred to as ‘neomorphic moonlighting function’ if it adds a new catalytic function or results in aggregate formation [104]. The location and timing of each protein activity is also important. Sufficient amounts of protein need to be expressed by the appropriate cell types. The expression needs to occur at the correct time in development, and/or appropriately in response to signals in the environment. It is a common observation in libraries of gene knockouts that some gene knockouts result in death of the embryo even though the gene is expected to function only in the adult organism. It is likely that some of these genes encode moonlighting proteins that have additional functions during embryogenesis.
Once the correct level of protein is attained, each of the functions of a moonlighting protein needs to be performed at the appropriate level. A moonlighting protein might perform multiple functions simultaneously, each might be independently regulated, or it might alternately perform one or the other function and have a means of switching between them. A change in protein conformation as in aconitase, mentioned above, is one method of switching between functions. Post-translational modifications (PTMs) are commonly used to regulate protein function in general, and they can be used to prompt a moonlighting protein to switch to a different function. For example, several protein components of the ribosome become phosphorylated and leave the ribosome to enter the nucleus where they participate in other activities. Ribosomal protein S3 (rpS3) has additional functions in the nucleus in DNA damage repair and as a transcription factor [105,106]. L10a (rpL10a) can interfere with gemnivirus reproduction in plants [107,108]. L13a (rpL13a) joins a multiprotein transcription factor [109]. By contrast, the oestrogen receptor leaves the nucleus when it undergoes palmytoylation so that it can interact with a signalling pathway at the plasma membrane [110]. Whether because of PTMs or other means of targeting, many other moonlighting proteins also perform their different functions in different cellular locations, whether in the cytoplasm, in the nucleus, in or attached to another organelle, at the cell membrane, or outside the cell.
5. Secretion of intracellular/surface moonlighting proteins
While signals for switching between functions are known for some moonlighting proteins, less is known about how a portion of the cytosolic pool of an intracellular/surface moonlighting protein becomes targeted for secretion. There are several lines of evidence that the ICSPs are indeed secreted and do not get out of the cell due to cell leakage or cell death (reviewed in [111]). When secretion of intracellular/surface moonlighting proteins is observed, many other cytoplasmic proteins are not found in the supernatant, and the proteins found in the supernatant or on the cell surface do not correlate with the most abundant proteins in the cell. In Staphylococcus aureus, the highest level of secretion of enolase and aldolase is during exponential growth, when cell breakage is at a minimum [112]. In addition, for Escherichia coli enolase, single amino acid substitutions at K341 resulted in a loss of its secretion, even of a mutant protein that is still catalytically active [113]. Similarly, in Bacillus subtilis enolase, deletion of an internal α-helix also prevented secretion [114]. These results support the idea that there is likely to be a secretion system(s) for at least some intracellular/surface moonlighting proteins.
The system or mechanism by which the majority of cytoplasmic proteins with a second function outside the cell are secreted is not known (figure 2). The intracellular/surface moonlighting proteins do not contain an N-terminal signal sequence required for secretion by the canonical Sec secretion system. They also do not contain other sequence motifs, such as a twin arginine leader motif required for secretion by the bacterial TAT system. There are several additional non-canonical secretion systems, but the other secretion systems are generally used for transport of only a few specific proteins, for example, the type 1 secretion system (T1SS) in Gram-negative bacteria. In particular, a secretion system in which a large portion of the pool of each protein type remains inside the cell but some of the pool of the protein partitioned to the cell surface has not been identified. This may involve a novel version of a known secretion system or it may involve an as-yet-unknown secretion system. In mammals, some of the secreted moonlighting proteins are found in secretory lysozomes or exosomes, but how they are targeted there is not clear. How these intracellular/surface/secreted moonlighting proteins are selected for secretion, and why only a portion of the cytoplasmic pool of the protein is secreted are current questions in the field.
The mechanism(s) by which some moonlighting proteins become attached to the cell surface is also unknown. The mechanism of cell surface attachment could potentially be a part of a secretion mechanism, although some recombinantly expressed and purified intracellular/surface moonlighting proteins are capable of reattaching to the cell surface. With a few exceptions [115] most of the ICSPs do not contain known amino acid sequence or structural motifs for attaching to the cell surface. Cell surface attachment could involve a new version of a known mechanism or it may involve an as-yet-unknown mechanism.
6. New targets for therapeutics
The development of novel treatments of diseases involving HSPs or other moonlighting proteins would be aided by more information about their targeting and their functions. For many of the human intracellular/extracellular moonlighting proteins, there is still much to learn about their roles in disease, but many have been implicated in autoimmune disease, heart disease, obesity, diabetes and cancer. The current knowledge of the roles of HSPs in immunology and cancer is discussed in other articles in this collection. In general, for a moonlighting protein, it is important to identify and understand both functions as well as their regulation and targeting. It is necessary to clarify which of the functions of a moonlighting protein is involved in disease development so that the correct protein function can be targeted for the development of novel therapeutics without causing side effects due to targeting a function not involved in disease. This also extends to the roles of paralogs, and also splice variants in humans, because they can share all, some, one, or none of the functions of a moonlighting protein. Clarification of when and where each function is performed by each version of the protein is also important if the protein is to be used as a biomarker.
Elucidating how intracellular/cell surface/secreted moonlighting proteins are secreted might identify processes and proteins that are involved in the novel secretion systems (or additional versions of known secretion systems) or surface attachment mechanisms that could serve as novel targets for developing new strategies for controlling infection. Understanding how intracellular/cell surface moonlighting proteins are targeted to the surface of a pathogen might lead to a method to decrease the ability of bacteria to bind to host tissues and could provide new targets for developing therapeutics to treat infections. With the increasing problem of antibiotic resistance, new targets for inhibiting bacterial infection and virulence are needed.
7. Conclusion
Hundreds of proteins have been found to have multiple, apparently unrelated functions. A large portion of these, including many HSPs, are cytosolic proteins that have been found to have additional functions when targeted to the cell surface or secreted, and the results of cell surface proteomics studies suggest many more might join that group. There is still much to be learned about their roles in health and disease, especially the sometimes complex signalling functions of many secreted mammalian proteins. In the case of bacterial and other pathogens, elucidating the proteins needed for their secretion and membrane attachment could lead to novel treatments for infections.
Data accessibility
This article has no additional data.
Competing interests
I declare I have no competing interests.
Funding
I received no funding for this study.
References
- 1.Jeffery CJ. 1999. Moonlighting Proteins. Trends Biochem. Sci. 24, 8–11. ( 10.1016/S0968-0004(98)01335-8) [DOI] [PubMed] [Google Scholar]
- 2.Mani M, et al. 2015. MoonProt: a database for proteins that are known to moonlight. Nucleic Acids Res. 43, D277–D282. ( 10.1093/nar/gku954) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hendriks W, Mulders JWM, Bibby MA, Slingsby C, Bloemendal H, de Jong WW, Mulders JW, Bibby MA, de Jong WW. 1988. Duck lens epsilon-crystallin and lactate dehydrogenase B4 are identical: a single-copy gene product with two distinct functions. Proc. Natl Acad. Sci. USA 85, 7114–7118. ( 10.1073/pnas.85.19.7114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piatigorsky J. 1998. Multifunctional lens crystallins and corneal enzymes. More than meets the eye. Ann. N. Y. Acad. Sci. 842, 2–15. ( 10.1111/j.1749-6632.1998.tb09626.x) [DOI] [PubMed] [Google Scholar]
- 5.Wistow GJ, Mulders JWM, de Jong WW. 1987. The enzyme lactate dehydrogenase as a structural protein in avian and crocodilian lenses. Nature 326, 622–624. ( 10.1038/326622a0) [DOI] [PubMed] [Google Scholar]
- 6.Rao PV, Krishna CM, Zigler JS Jr. 1992. Identification and characterization of the enzymatic activity of zeta-crystallin from guinea pig lens. A novel NADPH:quinone oxidoreductase. J. Biol. Chem. 267, 96–102. [PubMed] [Google Scholar]
- 7.Rao PV, Zigler JS Jr. 1990. Zeta-crystallin from guinea pig lens is capable of functioning catalytically as an oxidoreductase. Biochem. Biophys. Res. Commun. 167, 1221–1228. ( 10.1016/0006-291X(90)90654-6) [DOI] [PubMed] [Google Scholar]
- 8.Huang QL, Russell P, Stone SH, Zigler JS Jr. 1987. Zeta-crystallin, a novel lens protein from the guinea pig. Curr. Eye Res. 6, 725–732. ( 10.3109/02713688709034836) [DOI] [PubMed] [Google Scholar]
- 9.Bateman OA, Purkiss AG, van Montfort, Slingsby C, Graham C, Wistow G. 2003. Crystal structure of eta-crystallin: adaptation of a class 1 aldehyde dehydrogenase for a new role in the eye lens. Biochemistry 42, 4349–4356. ( 10.1021/bi027367w) [DOI] [PubMed] [Google Scholar]
- 10.Banerjee S, Nandyala AK, Raviprasad P, Ahmed N, Hasnain SE. 2007. Iron-Dependent RNA-Binding Activity of Mycobacterium tuberculosis Aconitase. J. Bacteriol. 189, 4046–4052. ( 10.1128/JB.00026-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennedy MC, MendeMueller L, Blondin GA, Beinert H. 1992. Purification and characterization of cytosolic aconitase from beef liver and its relationship to the iron-responsive element binding protein. Proc. Natl Acad. Sci. USA 89, 11 730–11 734. ( 10.1073/pnas.89.24.11730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philpott CC, Klausner RD, Rouault TA. 1994. The bifunctional iron-responsive element binding protein/cytosolic aconitase: the role of active-site residues in ligand binding and regulation. Proc. Natl Acad. Sci. USA 91, 7321–7325. ( 10.1073/pnas.91.15.7321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pancholi V, Fischetti VA. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176, 415–426. ( 10.1084/jem.176.2.415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gozalbo D, Gil-Navarro I, Azorin I, Renau-Piqueras J, Martinez JP, Gil ML. 1998. The cell wall-associated glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is also a fibronectin and laminin binding protein. Infect. Immun. 66, 2052–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matta SK, Agarwal S, Bhatnagar R. 2010. Surface localized and extracellular Glyceraldehyde-3-phosphate dehydrogenase of Bacillus anthracis is a plasminogen binding protein. Biochim. Biophys. Acta 1804, 2111–2120. ( 10.1016/j.bbapap.2010.08.004) [DOI] [PubMed] [Google Scholar]
- 16.Aguilera L, Giménez R, Badia J, Aguilar J, Baldoma L. 2009. NAD+-dependent post-translational modification of Escherichia coli glyceraldehyde-3-phosphate dehydrogenase. Int. Microbiol. 12, 187–192. [PubMed] [Google Scholar]
- 17.Hurmalainen V, Edelman S, Antikainen J, Baumann M, Lähteenmäki K, Korhonen TK. 2007. Extracellular proteins of Lactobacillus crispatus enhance activation of human plasminogen. Microbiology 153, 1112–1122. ( 10.1099/mic.0.2006/000901-0) [DOI] [PubMed] [Google Scholar]
- 18.Kinoshita H, et al. 2008. Cell surface Lactobacillus plantarum LA 318 glyceraldehyde-3-phosphate dehydrogenase. GAPDH) adheres to human colonic mucin. J. Appl. Microbiol. 104, 1667–1674. ( 10.1111/j.1365-2672.2007.03679.x) [DOI] [PubMed] [Google Scholar]
- 19.Alvarez RA, Blaylock MW, Baseman JB. 2003. Surface localized glyceraldehyde-3-phosphate dehydrogenase of Mycoplasma genitalium binds mucin. Mol. Microbiol. 48, 1417–1425. ( 10.1046/j.1365-2958.2003.03518.x) [DOI] [PubMed] [Google Scholar]
- 20.Tunio SA, Oldfield NJ, Ala'Aldeen DA, Wooldridge KG, Turner DP. 2010. The role of glyceraldehyde 3-phosphate dehydrogenase. GapA-1) in Neisseria meningitidis adherence to human cells. BMC Microbiol. 10, 280 ( 10.1186/1471-2180-10-280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbosa MS, Bao SN, Andreotti PF, de Faria FP, Felipe MS, dos Santos Feitosa L, Mendes-Giannini MJ, Soares CMA. 2006. Glyceraldehyde 3-phosphate dehydrogenase of Paracoccidioides brasiliensis is a cell surface protein, involved in fungal adhesion to extracellular matrix proteins and interaction with cells. Infect. Immun. 74, 382–389. ( 10.1128/IAI.74.1.382-389.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Modun B, Williams P. 1999. The staphylococcal transferrin-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect. Immun. 67, 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winram SB, Lottenberg R. 1996. The plasmin-binding protein Plr of group A streptococci is identified as glyceraldehyde-3-phosphate dehydrogenase. Microbiology 142, 2311–2320. ( 10.1099/13500872-142-8-2311) [DOI] [PubMed] [Google Scholar]
- 24.Seifert KN, McArthur WP, Bleiweis AS, Brady LJ. 2003. Characterization of group B streptococcal glyceraldehyde-3-phosphate dehydrogenase: surface localization, enzymatic activity, and protein-protein interactions. J. Microbiol. 49, 350–356. ( 10.1139/w03-042) [DOI] [PubMed] [Google Scholar]
- 25.Bergmann S, Rohde M, Hammerschmidt S. 2004. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pneumoniae is a surface-displayed plasminogen-binding protein. Infect. Immun. 72, 2416–2419. ( 10.1128/IAI.72.4.2416-2419.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin H, Song YP, Boel G, Kochar J, Pancholi V. 2005. Group A streptococcal surface GAPDH, SDH, recognizes uPAR/CD87 as its receptor on the human pharyngeal cell and mediates bacterial adherence to host cells. J. Mol. Biol. 350, 27–41. ( 10.1016/j.jmb.2005.04.063) [DOI] [PubMed] [Google Scholar]
- 27.Jobin MC, Brassard J, Quessy S, Gottschalk M, Grenier D. 2004. Acquisition of host plasmin activity by the Swine pathogen Streptococcus suis serotype 2. Infect. Immun. 72, 606–610. ( 10.1128/IAI.72.1.606-610.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lama A, Kucknoor A, Mundodi V, Alderete JF. 2009. Glyceraldehyde-3-phosphate dehydrogenase is a surface-associated, fibronectin-binding protein of Trichomonas vaginalis. Infect. Immun. 77, 2703–2711. ( 10.1128/IAI.00157-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boone TJ, Burnham CA, Tyrrell GJ. 2011. Binding of group B streptococcal phosphoglycerate kinase to plasminogen and actin. Microb. Pathog. 51, 255–261. ( 10.1016/j.micpath.2011.06.005) [DOI] [PubMed] [Google Scholar]
- 30.Fulde M, et al. 2013. Pneumococcal phosphoglycerate kinase interacts with plasminogen and its tissue activator. Thromb. Haemost. 111, 401–416. ( 10.1160/TH13-05-0421) [DOI] [PubMed] [Google Scholar]
- 31.Bergmann S, Schoenen H, Hammerschmidt S. 2013. The interaction between bacterial enolase and plasminogen promotes adherence of Streptococcus pneumoniae to epithelial and endothelial cells. Int. J. Med. Microbiol. 303, 452–462. ( 10.1016/j.ijmm.2013.06.002) [DOI] [PubMed] [Google Scholar]
- 32.Bergmann S, Rohde M, Chhatwal GS, Hammerschmidt S. 2001. Alpha-enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40, 1273–1287. ( 10.1046/j.1365-2958.2001.02448.x) [DOI] [PubMed] [Google Scholar]
- 33.Bergmann S, Wild D, Diekmann O, Frank R, Bracht D, Chhatwal GS, Hammerschmidt S. 2003. Identification of a novel plasmin(ogen)-binding motif in surface displayed alpha-enolase of Streptococcus pneumoniae. Mol. Microbiol. 49, 411–423. ( 10.1046/j.1365-2958.2003.03557.x) [DOI] [PubMed] [Google Scholar]
- 34.Bergmann S, Rohde M, Chhatwal GS, Hammerschmidt S. 2004. Characterization of plasmin(ogen) binding to Streptococcus pneumoniae. Indian J. Med. Res. 119(Suppl), 29–32. [PubMed] [Google Scholar]
- 35.Sha J, Erova TE, Alyea RA, Wang S, Olano JP, Pancholi V, Chopra AK. 2009. Surface-expressed enolase contributes to the pathogenesis of clinical isolate SSU of Aeromonas hydrophila. J. Bacteriol. 191, 3095–3107. ( 10.1128/JB.00005-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agarwal S, Kulshreshtha P, Bambah Mukku D, Bhatnagar R. 2008. Alpha-enolase binds to human plasminogen on the surface of Bacillus anthracis. Biochim. Biophys. Acta 1784, 986–994. ( 10.1016/j.bbapap.2008.03.017) [DOI] [PubMed] [Google Scholar]
- 37.Candela M, et al. 2009. Bifidobacterial enolase, a cell surface receptor for human plasminogen involved in the interaction with the host. Microbiology 155, 3294–3303. ( 10.1099/mic.0.028795-0) [DOI] [PubMed] [Google Scholar]
- 38.Floden AM, Watt JA, Brissette CA. 2011. Borrelia burgdorferi enolase is a surface-exposed plasminogen binding protein. PLoS ONE 6, e27502 ( 10.1371/journal.pone.0027502) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jong AY, Chen SH, Stins MF, Kim KS, Tuan TL, Huang SH. 2003. Binding of Candida albicans enolase to plasmin(ogen) results in enhanced invasion of human brain microvascular endothelial cells. J. Med. Microbiol. 52, 615–622. ( 10.1099/jmm.0.05060-0) [DOI] [PubMed] [Google Scholar]
- 40.Miles LA, Dahlberg CM, Plescia J, Felez J, Kato K, Plow EF. 1991. Role of cell-surface lysines in plasmin(ogen)-binding to cells: identification of alpha-enolase as a candidate plasminogen receptor. Biochemistry 30, 1682–1691. ( 10.1021/bi00220a034) [DOI] [PubMed] [Google Scholar]
- 41.Redlitz A, Fowler BJ, Plow EF, Miles LA. 1985. The role of an enolase-related molecule in plasminogen binding to cells. Eur. J. Biochem. 227, 407–415. ( 10.1111/j.1432-1033.1995.tb20403.x) [DOI] [PubMed] [Google Scholar]
- 42.Vanegas G, Quiñones W, Carrasco-López C, Concepción JL, Albericio F, Avilán L. 2007. Enolase as a plasminogen binding protein in Leishmania mexicana. Parasitol. Res. 101, 1511–1516. ( 10.1007/s00436-007-0668-7) [DOI] [PubMed] [Google Scholar]
- 43.Jolodar A, Fischer P, Bergmann S, Buttner DW, Hammerschmidt S, Brattig NW. 2003. Molecular cloning of an a-enolase from the human filarial parasite Onchocerca volvulus that binds human plasminogen. Biochim. Biophys. Acta 1627, 111–120. ( 10.1016/S0167-4781(03)00083-6) [DOI] [PubMed] [Google Scholar]
- 44.Donofrio FC, Calil AC, Miranda ET, Almeida AM, Benard G, Soares CP, Veloso SN, Soares CM, Mendes Giannini MJ. 2009. Enolase from Paracoccidioides brasiliensis: isolation and identification as a fibronectin-binding protein. J. Med. Microbiol. 58, 706–713. ( 10.1099/jmm.0.003830-0) [DOI] [PubMed] [Google Scholar]
- 45.Nakajima K, Hamanoue M, Takemoto N, Hattori T, Kato K, Kohsaka S. 1994. Plasminogen binds specifically to a alpha-enolase on rat neuronal plasma membrane. J. Neurochem. 63, 2048–2057. ( 10.1046/j.1471-4159.1994.63062048.x) [DOI] [PubMed] [Google Scholar]
- 46.de la Torre-Escudero E, Manzano-Román R, Pérez-Sánchez R, Siles-Lucas M, Oleaga A. 2010. Cloning and characterization of a plasminogen-binding surface associated enolase from Schistosoma bovis. Vet. Parasitol. 173, 76–84. ( 10.1016/j.vetpar.2010.06.011) [DOI] [PubMed] [Google Scholar]
- 47.Carneiro CR, Postol E, Nomizo R, Reis LF, Brentani RR. 2004. Identification of enolase as a laminin-binding protein on the surface of Staphylococcus aureus. Microbes Infect. 6, 604–608. ( 10.1016/j.micinf.2004.02.003) [DOI] [PubMed] [Google Scholar]
- 48.Kinnby B, Booth NA, Svensater G. 2008. Plasminogen binding by oral streptococci from dental plaque and inflammatory lesions. Microbiology 154, 924–931. ( 10.1099/mic.0.2007/013235-0) [DOI] [PubMed] [Google Scholar]
- 49.Jones MN, Holt RG. 2007. Cloning and characterization of an alpha-enolase of the oral pathogen Streptococcus mutans that binds human plasminogen. Biochem. Biophys. Res. Commun. 364, 924–929. ( 10.1016/j.bbrc.2007.10.098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kolberg J, Aase A, Bergmann S, Herstad TK, Rødal G, Frank R, Rohde M, Hammerschmidt S. 2006. Streptococcus pneumoniae enolase is important for plasminogen binding despite low abundance of enolase protein on the bacterial cell surface. Microbiology 152, 1307–1317. ( 10.1099/mic.0.28747-0) [DOI] [PubMed] [Google Scholar]
- 51.Antikainen J, Kuparinen V, Lähteenmäki K, Korhonen TK. 2007. Enolases from Gram-positive bacterial pathogens and commensal lactobacilli share functional similarity in virulence-associated traits. FEMS Immunol. Med. Microbiol. 51, 526–534. ( 10.1111/j.1574-695X.2007.00330.x) [DOI] [PubMed] [Google Scholar]
- 52.Castaldo C, Vastano V, Siciliano RA, Candela M, Vici M, Muscariello L, Marasco R, Sacco M. 2009. Surface displaced alfa-enolase of Lactobacillus plantarum is a fibronectin binding protein. Microb. Cell Fact. 8, 14 ( 10.1186/1475-2859-8-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pancholi V, Fischetti VA. 1998. Alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273, 14 503–14 515. ( 10.1074/jbc.273.23.14503) [DOI] [PubMed] [Google Scholar]
- 54.Esgleas M, Li Y, Hancock MA, Harel J, Dubreuil JD, Gottschalk M. 2008. Isolation and characterization of alpha-enolase, a novel fibronectin-binding protein from Streptococcus suis. Microbiology 154, 2668–2679. ( 10.1099/mic.0.2008/017145-0) [DOI] [PubMed] [Google Scholar]
- 55.Kim KP, Jagadeesan B, Burkholder KM, Jaradat ZW, Wampler JL, Lathrop AA, Morgan MT, Bhunia AK. 2006. Adhesion characteristics of Listeria adhesion protein. (LAP)-expressing Escherichia coli to Caco-2 cells and of recombinant LAP to eukaryotic receptor Hsp60 as examined in a surface plasmon resonance sensor. FEMS Microbiol. Lett. 256, 324–332. ( 10.1111/j.1574-6968.2006.00140.x) [DOI] [PubMed] [Google Scholar]
- 56.Wampler JL, Kim KP, Jaradat Z, Bhunia AK. 2004. Heat shock protein 60 acts as a receptor for the Listeria adhesion protein in Caco-2 cells. Infect. Immun. 72, 931–936. ( 10.1128/IAI.72.2.931-936.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burkholder KM, Bhunia AK. 2010. Listeria monocytogenes uses Listeria adhesion protein (LAP) to promote bacterial transepithelial translocation and induces expression of LAP receptor Hsp60. Infect. Immun. 78, 5062–5073. ( 10.1128/IAI.00516-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Candela M, Centanni M, Fiori J, Biagi E, Turroni S, Orrico C, Bergmann S, Hammerschmidt S, Brigidi P. 2010. DnaK from Bifidobacterium animalis subsp. Lactis is a surface-exposed human plasminogen receptor upregulated in response to bile salts. Microbiology 156, 1609–1618. ( 10.1099/mic.0.038307-0) [DOI] [PubMed] [Google Scholar]
- 59.Dallo SF, Kannan TR, Blaylock MW, Baseman JB. 2002. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 46, 1041–1051. ( 10.1046/j.1365-2958.2002.03207.x) [DOI] [PubMed] [Google Scholar]
- 60.Kunert A, et al. 2007. Immune evasion of the human pathogen Pseudomonas aeruginosa: elongation factor Tuf is a factor H and plasminogen binding protein. J. Immunol. 179, 2979–2988. ( 10.4049/jimmunol.179.5.2979) [DOI] [PubMed] [Google Scholar]
- 61.Granato D, Bergonzelli GE, Pridmore RD, Marvin L, Rouvet M, Corthésy-Theulaz IE. 2004. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect. Immun. 72, 2160–2169. ( 10.1128/IAI.72.4.2160-2169.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brix K, Summa W, Lottspeich F, Herzog V. 1998. Extracellularly occurring histone H1 mediates the binding of thyroglobulin to the cell surface of mouse macrophages. J. Clin. Invest. 102, 283–293. ( 10.1172/JCI1614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vincent S, Fort P. 1990. Nucleotide sequence of hamster glyceraldehyde-3-phosphate dehydrogenase mRNA. Nucleic Acids Res. 18, 3054 ( 10.1093/nar/18.10.3054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar S, Sheokand N, Mhadeshwar MA, Raje CI, Raje M. 2012. Characterization of glyceraldehyde-3-phosphate dehydrogenase as a novel transferrin receptor. Int. J. Biochem. Cell Biol. 44, 189–199. ( 10.1016/j.biocel.2011.10.016) [DOI] [PubMed] [Google Scholar]
- 65.Bocharov AV, Vishnyakova TG, Baranova IN, Remaley AT, Patterson AP, Eggerman TL. 2000. Heat shock protein 60 is a high-affinity high-density lipoprotein binding protein. Biochem. Biophys. Res. Commun. 277, 228–235. ( 10.1006/bbrc.2000.3663) [DOI] [PubMed] [Google Scholar]
- 66.Petit FM, Serres C, Bourgeon F, Pineau C, Auer J. 2013. Identification of sperm head proteins involved in zona pellucida binding. Hum. Reprod. 28, 852–865. ( 10.1093/humrep/des452) [DOI] [PubMed] [Google Scholar]
- 67.Gopalakrishnan B, Aravinda S, Pawshe CH, Totey SM, Nagpal S, Salunke DM, Shaha C. 1998. Studies on glutathione S-transferases important for sperm function: evidence of catalytic activity-independent functions. Biochem. J. 329, 231–241. ( 10.1042/bj3290231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Auer J, Camoin L, Courtot AM, Hotellier F, De Almeida M. 2004. Evidence that P36, a human sperm acrosomal antigen involved in the fertilization process is triosephosphate isomerase. Mol. Reprod. Dev. 68, 515–552. ( 10.1002/mrd.20107) [DOI] [PubMed] [Google Scholar]
- 69.Fulde M, Rohde M, Polok A, Preissner KT, Chhatwal GS, Bergmann S. 2013. Cooperative plasminogen recruitment to the surface of Streptococcus canis via M protein and enolase enhances bacterial survival. mBio 4, e00629-12 ( 10.1128/mBio.00629-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bao S, et al. 2014. Mycoplasma synoviae enolase is a plasminogen/fibronectin binding protein. BMC Vet. Res. 10, 223 ( 10.1186/s12917-014-0223-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Read M, Hicks KE, Sims PFG, Hyde JE. 1994. Molecular characterization of the enolase gene from the human malaria parasite Plasmodium falciparum. Evidence for ancestry within a photosynthetic lineage. Eur. J. Biochem. 220, 513–520. ( 10.1111/j.1432-1033.1994.tb18650.x) [DOI] [PubMed] [Google Scholar]
- 72.Ghosh AK, Coppens I, Gårdsvoll H, Ploug M, Jacobs-Lorena M. 2011. Plasmodium ookinetes coopt mammalian plasminogen to invade the mosquito midgut. Proc. Natl Acad. Sci. USA 108, 17 153–17 158. ( 10.1073/pnas.1103657108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kesimer M, Kilic N, Mehrotra R, Thornton DJ, Sheehan JK. 2009. Identification of salivary mucin MUC7 binding proteins from Streptococcus gordonii. BMC Microbiol. 9, 163 ( 10.1186/1471-2180-9-163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Agarwal V, Kuchipudi A, Fulde M, Riesbeck K, Bergmann S, Blom AM. 2013. Streptococcus pneumoniae endopeptidase O (PepO) is a multifunctional plasminogen- and fibronectin-binding protein, facilitating evasion of innate immunity and invasion of host cells. J. Biol. Chem. 288, 6849–6863. ( 10.1074/jbc.M112.405530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sahoo S, Murugavel S, Devi IK, Vedamurthy GV, Gupta SC, Singh BP, Joshi P. 2013. Glyceraldehyde-3-phosphate dehydrogenase of the parasitic nematode Haemonchus contortus binds to complement C3 and inhibits its activity. Parasite Immunol. 35, 457–467. ( 10.1111/pim.12058) [DOI] [PubMed] [Google Scholar]
- 76.Knaust A, Weber MV, Hammerschmidt S, Bergmann S, Frosch M, Kurzai O. 2007. Cytosolic proteins contribute to surface plasminogen recruitment of Neisseria meningitidis. J. Bacteriol. 189, 5404 ( 10.1128/JB.01966-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xolalpa W, Vallecillo AJ, Lara M, Mendoza-Hernandez G, Comini M, Spallek R, Singh M, Espitia C. 2007. Identification of novel bacterial plasminogen-binding proteins in the human pathogen Mycobacterium tuberculosis. Proteomics 7, 3332–3341. ( 10.1002/pmic.200600876) [DOI] [PubMed] [Google Scholar]
- 78.Candela M, Bergmann S, Vici M, Vitali B, Turroni S, Eikmanns BJ, Hammerschmidt S, Brigidi P. 2007. Binding of Human Plasminogen to Bifidobacterium. J. Bacteriol. 189, 5929–5936. ( 10.1128/JB.00159-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Collen D, Verstraete M. 1975. Molecular biology of human plasminogen. II. Metabolism in physiological and some pathological conditions in man. Thromb. Diath. Haemorrh. 34, 403–408. [PubMed] [Google Scholar]
- 80.Dano K, Andreasen PA, Grondahl-Hansen J, Kristensen P, Nielsen .S, Skriver L. 1985. Plasminogen activators, tissue degradation, and cancer. Adv. Cancer Res. 44, 139–266. ( 10.1016/S0065-230X(08)60028-7) [DOI] [PubMed] [Google Scholar]
- 81.Patel DK, Shah KR, Pappachan A, Gupta S, Singh DD. 2016. Cloning, expression and characterization of a mucin-binding GAPDH from Lactobacillus acidophilus. Int. J. Biol. Macromol. 91, 338–346. ( 10.1016/j.ijbiomac.2016.04.041) [DOI] [PubMed] [Google Scholar]
- 82.Furukawa T, Yoshimura A, Sumizawa T, Haraguchi M, Akiyama S, Fukui K, Ishizawa M, Yamada Y. 1992. Angiogenic factor. Nature 356, 668 ( 10.1038/356668a0) [DOI] [PubMed] [Google Scholar]
- 83.Park SG, Kim HJ, Min YH, Choi EC, Shin YK, Park BJ, Lee SW, Kim S. 2005. Human lysyl-tRNA synthetase is secreted to trigger proinflammatory response. Proc. Natl Acad. Sci. USA 102, 6356–6361. ( 10.1073/pnas.0500226102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Safer D, Elzinga M, Nachmias VT. 1991. Thymosin beta 4 and Fx, an actin-sequestering peptide, are indistinguishable. J. Biol. Chem. 266, 4029–4032. [PubMed] [Google Scholar]
- 85.Cassimeris L, Safer D, Nachmias VT, Zigmond SH. 1992. Thymosin beta-4 sequesters the majority of G-actin in resting human polymorphonuclear leukocytes. J. Cell Biol. 119, 1261–1270. ( 10.1083/jcb.119.5.1261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Safer D, Sosnick TR, Elzinga M. 1997. Thymosin beta 4 binds actin in an extended conformation and contacts both the barbed and pointed ends. Biochemistry 36, 5806–5816. ( 10.1021/bi970185v) [DOI] [PubMed] [Google Scholar]
- 87.Young JD, Lawrence AJ, MacLean AG, Leung BP, McInnes IB, Canas B, Pappin DJ, Stevenson RD. 1999. Thymosin 4 sulfoxide is an anti-inflammatory agent generated by monocytes in the presence of glucocorticoids. Nat. Med. 5, 1424–1427. ( 10.1038/71002) [DOI] [PubMed] [Google Scholar]
- 88.Chaput M, Claes V, Portetelle D, Cludts I, Cravador A, Burny A, Gras H, Tartar A. 1988. The neurotrophic factor neuroleukin is 90% homologous with phosphohexose isomerase. Nature 332, 454–455. ( 10.1038/332454a0) [DOI] [PubMed] [Google Scholar]
- 89.Faik P, Walker JI, Redmill AA, Morgan MJ. 1988. Mouse glucose-6-phosphate isomerase and neuroleukin have identical 3′ sequences. Nature 332, 455–457. ( 10.1038/332455a0) [DOI] [PubMed] [Google Scholar]
- 90.Watanabe H, Takehana K, Date M, Shinozaki T, Raz A. 1996. Tumor cell autocrine motility factor is the neuroleukin/phosphohexose isomerase polypeptide. Cancer Res. 56, 2960–2963. [PubMed] [Google Scholar]
- 91.Xu W, Seiter K, Feldman E, Ahmed T, Chiao JW. 1996. The differentiation and maturation mediator for human myeloid leukemia cells shares homology with neuroleukin or phosphoglucose isomerase. Blood 87, 4502–4506. [PubMed] [Google Scholar]
- 92.Gurney ME, Heinrich SP, Lee MR, Yin HS. 1986. Molecular cloning and expression of neuroleukin, a neurotrophic factor for spinal and sensory neurons. Science 234, 566–574. ( 10.1126/science.3764429) [DOI] [PubMed] [Google Scholar]
- 93.Gurney ME, Apatoff BR, Spear GT, Baumel MJ, Antel JP, Bania MB, Reder AT. 1986. Neuroleukin: a lymphokine product of lectin-stimulated T cells. Science 234, 574–581. ( 10.1126/science.3020690) [DOI] [PubMed] [Google Scholar]
- 94.Watanabe H, Carmi P, Hogan V, Raz T, Silletti S, Nabi IR, Raz A. 1991. Purification of human tumor cell autocrine motility factor and molecular cloning of its receptor. J. Biol. Chem. 266, 13 442–13 448. [PubMed] [Google Scholar]
- 95.Schulz LC, Bahr JM. 2004. Potential endocrine function of the glycolytic enzyme glucose-6-phosphate isomerase during implantation. Gen. Comp. Endocrinol. 137, 283–287. ( 10.1016/j.ygcen.2004.04.003) [DOI] [PubMed] [Google Scholar]
- 96.Mirando AC, et al. 2015. Aminoacyl-tRNA synthetase dependent angiogenesis revealed by a bioengineered macrolide inhibitor. Sci. Rep. 5, 13160 ( 10.1038/srep13160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Querol-García J, et al. 2017. Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from the gram-positive bacterial pathogen A. vaginae, an Immunoevasive Factor that Interacts with the Human C5a Anaphylatoxin. Front. Microbiol. 8, 541 ( 10.3389/fmicb.2017.00541) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nandan D, Tran T, Trinh E, Silverman JM, Lopez M. 2007. Identification of leishmania fructose-1,6-bisphosphate aldolase as a novel activator of host macrophage Src homology 2 domain containing protein tyrosine phosphatase SHP-1 . Biochem. Biophys. Res. Commun. 364, 601–607. (doi:0.1016/j.bbrc.2007.10.065) [DOI] [PubMed] [Google Scholar]
- 99.Nandan D, Yi T, Lopez M, Lai C, Reiner NE. 2002. Leishmania EF-1alpha activates the Src homology 2 domain containing tyrosine phosphatase SHP-1 leading to macrophage deactivation. J. Biol. Chem. 277, 50 190–50 197. ( 10.1074/jbc.M209210200) [DOI] [PubMed] [Google Scholar]
- 100.Yoshida N, Oeda K, Watanabe E, Mikami T, Fukita Y, Nishimura K, Komai K, Matsuda K. 2001. Protein function. Chaperonin turned insect toxin. Nature 411, 44 ( 10.1038/35075148) [DOI] [PubMed] [Google Scholar]
- 101.Liu H, Zeng H, Yao Q, Yuan J, Zhang Y, Qiu D, Yang X, Yang H, Liu Z. 2012. Steinernema glaseri surface enolase: molecular cloning, biological characterization, and role in host immune suppression. Mol. Biochem. Parasitol. 185, 89–98. ( 10.1016/j.molbiopara.2012.06.006) [DOI] [PubMed] [Google Scholar]
- 102.Jeffery CJ, Bahnson BJ, Chien W, Ringe D, Petsko GA. 2000. Crystal structure of rabbit phosphoglucose isomerase, a glycolytic enzyme that moonlights as neuroleukin, autocrine motility factor, and differentiation mediator. Biochemistry 39, 955–964. ( 10.1021/bi991604m) [DOI] [PubMed] [Google Scholar]
- 103.Ehinger S, Schubert WD, Bergmann S, Hammerschmidt S, Heinz DW. 2004. Plasmin(ogen)-binding alpha-enolase from Streptococcus pneumoniae: crystal structure and evaluation of plasmin(ogen)-binding sites. J. Mol. Biol. 343, 997–1005. ( 10.1016/j.jmb.2004.08.088) [DOI] [PubMed] [Google Scholar]
- 104.Jeffery CJ. 2011. Proteins with neomorphic moonlighting functions in disease. IUBMB Life 63, 489–494. ( 10.1002/iub.504) [DOI] [PubMed] [Google Scholar]
- 105.Kim TS, Kim HD, Kim J. 2009. PKCdelta-dependent functional switch of rpS3 between translation and DNA repair. Biochim. Biophys. Acta 1793, 395–405. ( 10.1016/j.bbamcr.2008.10.017) [DOI] [PubMed] [Google Scholar]
- 106.Wan F, Weaver A, Gao X, Bern M, Hardwidge PR, Lenardo MJ. 2011. IKKbeta phosphorylation regulates RPS3 nuclear translocation and NF-kappaB function during infection with Escherichia coli strain O157:H7. Nat. Immunol. 12, 335–343. ( 10.1038/ni.2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Carvalho CM, Santos AA, Pires SR, Rocha CS, Saraiva DI, Machado JP, Mattos EC, Fietto LG, Fontes EP. 2008. Regulated nuclear trafficking of rpL10A mediated by NIK1 represents a defense strategy of plant cells against virus. PLoS Pathog. 4, e1000247 ( 10.1371/journal.ppat.1000247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rocha CS, Santos AA, Machado JP, Fontes EP. 2008. The ribosomal protein L10/QM-like protein is a component of the NIK-mediated antiviral signaling. Virology 380, 165–169. ( 10.1016/j.virol.2008.08.005) [DOI] [PubMed] [Google Scholar]
- 109.Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL. 2003. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 115, 187–198. ( 10.1016/S0092-8674(03)00773-6) [DOI] [PubMed] [Google Scholar]
- 110.Adlanmerini M, et al. 2014. Mutation of the palmitoylation site of estrogen receptor α in vivo reveals tissue-specific roles for membrane versus nuclear actions. Proc. Natl. Acad. Sci. USA 111, E283–E290. ( 10.1073/pnas.1322057111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ebner P, Rinker J, Götz F. 2016. Excretion of cytoplasmic proteins in Staphylococcus is most likely not due to cell lysis. Curr. Genet. 62, 19–23. ( 10.1007/s00294-015-0504-z) [DOI] [PubMed] [Google Scholar]
- 112.Ebner P, et al. 2015. Excretion of cytoplasmic proteins (ECP) in Staphylococcus aureus. Mol. Microbiol. 97, 775–789. ( 10.1111/mmi.13065) [DOI] [PubMed] [Google Scholar]
- 113.Boël G, et al. 2004. Is 2-phosphoglycerate-dependent automodification of bacterial enolases implicated in their export? J. Mol. Biol. 337, 485–496. ( 10.1016/j.jmb.2003.12.082) [DOI] [PubMed] [Google Scholar]
- 114.Yang CK, Zhang XZ, Lu CD, Tai PC. 2014. An internal hydrophobic helical domain of Bacillus subtilis enolase is essential but not sufficient as a non-cleavable signal for its secretion. Biochem. Biophys. Res. Commun. 446, 901–905. ( 10.1016/j.bbrc.2014.03.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Daubenspeck JM, Liu R, Dybvig K. 2016. Rhamnose Links Moonlighting Proteins to Membrane Phospholipid in Mycoplasmas. PLoS ONE 11, e0162505 ( 10.1371/journal.pone.0162505) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.


