SUMO proteases regulate plant fertility through multiple pathways.
Abstract
In plants, the posttranslational modification small ubiquitin-like modifier (SUMO) is involved in regulating several important developmental and cellular processes, including flowering time control and responses to biotic and abiotic stresses. Here, we report two proteases, SUMO PROTEASE RELATED TO FERTILITY1 (SPF1) and SPF2, that regulate male and female gamete and embryo development and remove SUMO from proteins in vitro and in vivo. spf1 mutants exhibit abnormal floral structures and embryo development, while spf2 mutants exhibit largely a wild-type phenotype. However, spf1 spf2 double mutants exhibit severe abnormalities in microgametogenesis, megagametogenesis, and embryo development, suggesting that the two genes are functionally redundant. Mutation of SPF1 and SPF2 genes also results in misexpression of generative- and embryo-specific genes. In vitro, SPF1 and SPF2 process SUMO1 precursors into a mature form, and as expected in vivo, spf1 and spf2 mutants accumulate SUMO conjugates. Using a yeast two-hybrid screen, we identified EMBRYO SAC DEVELOPMENT ARREST9 (EDA9) as an SPF1-interacting protein. In vivo, we demonstrate that EDA9 is sumolyated and that, in spf1 mutants, EDA9-SUMO conjugates increase in abundance, demonstrating that EDA9 is a substrate of SPF1. Together, our results demonstrate that SPF1 and SPF2 are two SUMO proteases important for plant development in Arabidopsis (Arabidopsis thaliana).
Plant reproduction depends on a series of events, including flower formation, microgametogenesis, megagametogenesis, fertilization of the egg and central cells, and embryogenesis, that are regulated by many different factors, such as transcription factors, epigenetic complexes, small RNAs, and posttranslational protein modifications (for review, see Baroux and Autran, 2015; Gómez et al., 2015; ten Hove et al., 2015; Jiménez-Quesada et al., 2016; Li and Yang, 2016; Sampath and Ephrussi, 2016). A novel protein modifier, SUMO-E3 ligase, also is involved in gametophyte development in Arabidopsis (Arabidopsis thaliana; Ling et al., 2012; Liu et al., 2014) and rice (Oryza sativa; Thangasamy et al., 2011).
As a polypeptide tag, Small Ubiquitin-like Modifier (SUMO) was identified near the end of the last century (Meluh and Koshland, 1995; Matunis et al., 1998) and is covalently attached to diverse proteins and leads to various changes in protein activity, localization, or stability of the substrate proteins (Seeler and Dejean, 2003; Nabhan and Ribeiro, 2006). Deficiency in the SUMOylation system results in severe dysfunction and even lethality in most eukaryotes (Zhen et al., 1996; Huang et al., 2000; Fay et al., 2003; Nacerddine et al., 2005; Saracco et al., 2007).
Accumulating evidence is showing that, in plants, SUMO is involved in important developmental processes, such as flowering time regulation (Murtas et al., 2003; Jin et al., 2008; Budhiraja et al., 2009), meristem maintenance (Ishida et al., 2009), seed germination as well as root development (Huang et al., 2009; Miura et al., 2009), GA signaling pathway (Conti et al., 2014), gametophyte development (Thangasamy et al., 2011; Ling et al., 2012; Liu et al., 2014), nitrogen assimilation (Park et al., 2011), abiotic stress response (Kurepa et al., 2003; Lois et al., 2003; Catala et al., 2007; Miura et al., 2007, 2009; Conti et al., 2008; Cheong et al., 2009; Chen et al., 2011; Zheng et al., 2012), biotic stress response (Castillo et al., 2004; Lee et al., 2007; Bartetzko et al., 2009), and nutrient deficiency (Miura et al., 2005).
SUMOylation is a dynamic reversible process in which SUMO is covalently attached to its substrate protein and can be removed by a desumoylating enzyme (Meulmeester and Melchior, 2008). In Arabidopsis, at least eight genes encoding putative SUMO-specific enzymes have been identified (Colby et al., 2006). All tested Arabidopsis SUMO-specific enzymes, including EARLY IN SHORT DAYS4 (ESD4), ESD4-LIKE SUMO PROTEASE1 (ELS1), OVERLY TOLERANT TO SALT1 (OTS1), and OTS2, have peptidase activity that removes the C-terminal tail of the SUMO1/2 isoform precursors in vitro (Chosed et al., 2006; Colby et al., 2006). However, only ELS1 was able to cleave SUMO3 (Chosed et al., 2006; Colby et al., 2006). In terms of biological processes, both ESD4 and ELS1 regulate flowering time (Reeves et al., 2002; Murtas et al., 2003; Hermkes et al., 2011), whereas OTS1 and OTS2 increase salt tolerance (Conti et al., 2008). Recently, one additional SUMO protease (Arabidopsis SUMO Protease1 [ASP1]) was reported to be involved in flowering time regulation (Kong et al., 2017).
In this study, we describe the isolation and characterization of mutations in SUMO PROTEASE RELATED TO FERTILITY1 (SPF1) and SPF2 genes (previously called ULP2like2 and ULP2like1, respectively; Novatchkova et al., 2004) and analyze the biochemical functions of the proteins. Data from our experiments showed that SPF1 and SPF2 function as SUMO proteases to specifically regulate floral development, microgametogenesis, and megagametogenesis as well as embryogenesis. Finally, we identified a set of SPF1-interacting proteins, among which EDA9 may be a potential substrate of SPF1. Our results indicated that SPF1 and SPF2 are SUMO enzymes that play important roles during reproduction in Arabidopsis.
RESULTS
SPF1 and SPF2 Share the Conserved Domain with ESD4 Protease
In Arabidopsis, four SUMO proteases, ESD4, ELS1, OTS1, and OTS2, have been demonstrated to function in flowering time regulation and stress responses (Murtas et al., 2003; Conti et al., 2008; Hermkes et al., 2011). Two more ESD4-like genes, At1g09730 and At4g33620, exist in Arabidopsis and were previously named ULP2like2 and ULP2like1, respectively (Novatchkova et al., 2004). Recently, these two SUMO proteases were described as ASP1/2 and were described to have roles in flowering time regulation (Kong et al., 2017). Like ESD4, both proteins contain key amino acid residues in the ULP domain, including all the residues required for the Cys protease-like catalytic site (Supplemental Fig. S1, A and B), suggesting that these two proteins have similar biochemical functions to other SUMO proteases.
When T-DNA mutants in At1g09730 and At4g33620 were isolated in our laboratory, we identified clear defects in fertility and named the genes SPF1 and SPF2, respectively. To characterize the full-length mRNAs from both genes, we PCR amplified cDNAs of SPF1 and SPF2 based on the genomic sequences of At1g09730.1 and At4g33620.1 (TAIR8; http://www.arabidopsis.org/). For SPF1, a 2,796-bp clone was recovered that was shorter than the annotated At1g09730.1 and is predicted to result in a protein of 931 amino acids (Supplemental Fig. S2, A and B). For SPF2, a shorter cDNA also was amplified and is predicted to encode a putative protein of 774 amino acids, which was shorter than that of At4g33620.1 (Supplemental Fig. S2, C and D). Even though both SPF1 and SPF2 encode for predicted shorter proteins than the annotated genes At1g09730.1 and At4g33620.1, they both share the conserved catalytic domains with ESD4 (Supplemental Fig. S1A).
The full protein sequences of SPF1 and SPF2 are only 19% and 17% identical to ESD4, and the levels of amino acid similarity in their catalytic domains to ESD4 are 36% and 34%, respectively. The SPF1 and SPF2 catalytic domains are 63% identical to each other, and the catalytic active sites are identical to those of ESD4. However, unlike ESD4, where the ULP protease domain is located at the C terminus, the ULP domains of SPF1 and SPF2 are located in the middle region of the proteins (Supplemental Fig. S1B), similar to that of ULP2 in yeast (Li and Hochstrasser, 2000). Our phylogenetic analysis showed that SPF1 and SPF2 were grouped in the same clade (Supplemental Fig. S1C). The nucleotide and protein sequences of both SPF1 (EU877962) and SPF2 (EU877963) were deposited in GenBank.
Mutation of Both SPF1 and SPF2 Has Significant Effects on Fertility
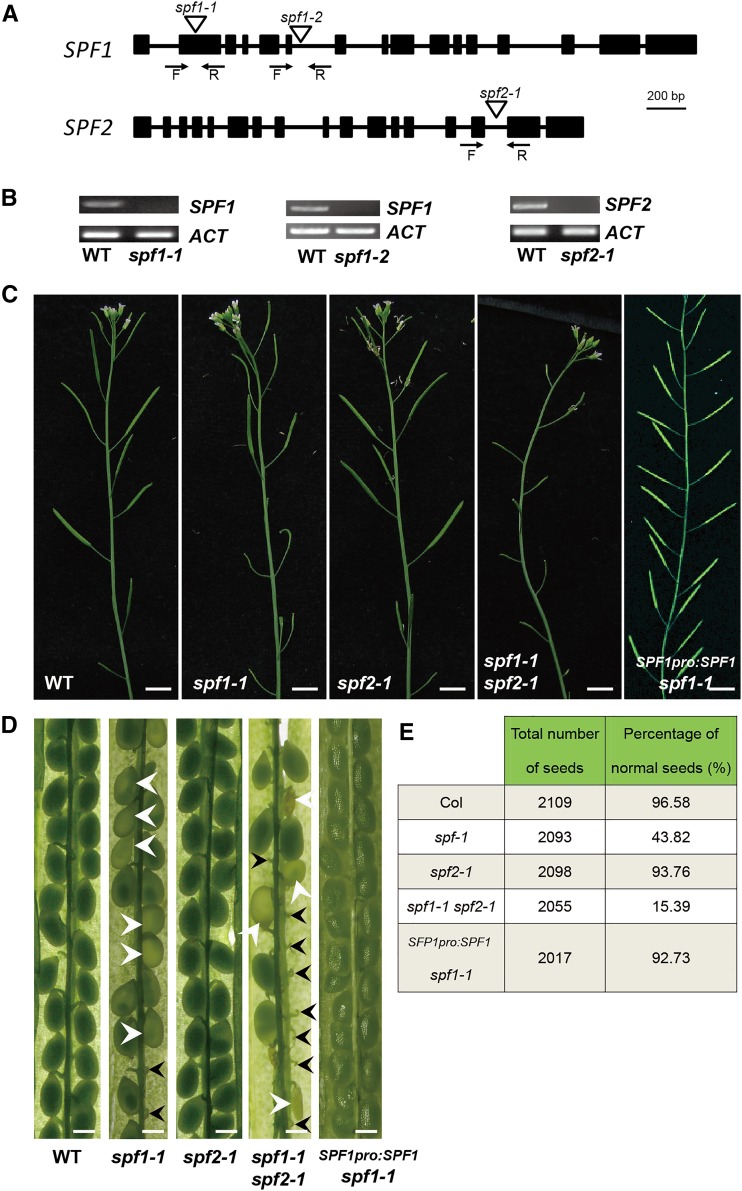
To elucidate the phenotypic and molecular effects of SPF1 and SPF2, several T-DNA insertion mutants in SPF1 and SPF2 genes were obtained from the Arabidopsis Biological Resource Center (ABRC; http://www.arabidopsis.org/). Only spf1-1 (Salk_040756), spf1-2 (Salk_049255), and spf2-1 (Salk_023493) were confirmed as null mutants (Fig. 1, A and B; see “Materials and Methods”); therefore, these mutants were used in this study. Double mutants of spf1 spf2 were produced from the crosses spf1-1 × spf2-1 and spf1-2 × spf2-1, resulting in either spf1-1 spf2-1 or spf1-2 spf2-1 double mutants. The spf1-1 single mutant had a clear phenotype on fertility (Fig. 1, C and D), as most of the siliques were shorter than those of the wild type (Supplemental Fig. S3) and nearly half of the seeds were abnormal (Fig. 1, D and E). In contrast, the spf2-1 mutant showed no obvious differences in fertility or seed development when compared with the wild type (Fig. 1, C–E; Supplemental Fig. S3). The spf1-1 spf2-1 double mutant had a more severe phenotype than the spf1-1 single mutant. The siliques of the double mutant were shorter than those of both the wild type and the spf1-1 single mutant (Supplemental Fig. S3), and most seeds developed abnormally (Fig. 1, D and E). A second mutant allele, spf1-2, had a much more severe phenotype than spf1-1, and the number of viable seeds in the spf1-2 spf2-1 double mutant was less than that in the spf1-1 spf2-1 double mutant (Supplemental Fig. S4). These observations led us to propose that SPF1 and SPF2 act partially redundantly in the regulation of fertility.
Figure 1.
Mutation of SPF1 and SPF2 genes results in sterility. A, Structures of SPF1 and SPF2 genes and T-DNA positions in the spf1 and spf2 mutants. Black squares represent exons with matching sequences in TAIR, lines represent introns, triangles denote T-DNA, and arrows indicate the positions of forward primers (F) and reverse primers (R) for RT-PCR in B. B, No transcripts of SPF1 and SPF2 were detectable in the spf1-1, spf1-2, and spf2-1 mutants. ACTIN (ACT) served as a reference gene for RT-PCR. C, Primary stems bearing siliques of, from left to right, the wild type (WT), spf1-1, spf2-1, spf1-1 spf2-1, and a rescuing line of SPF1pro:SPF1 spf1-1. D, Opened siliques of, from left to right, the wild type, spf1-1, spf2-1, spf1-1 spf2-1, and a rescuing line of SPF1pro:SPF1 spf1-1. Ovules remaining unfertilized (black arrowheads) fail to initiate seed development, degenerate, and leave a void in the silique, whereas colorless ovules (white arrowheads) have been fertilized but then become either arrested in developmental stages or deformed. E, Percentage of normal seeds in the wild type (Columbia [Col]), spf1-1, spf2-1, spf1-1 spf2-1, and a rescuing line of SPF1pro:SPF1 spf1-1. Bars = 800 μm for C and 400 μm for D.
To confirm the above mutant phenotypes, we carried out a transgenetic complementation experiment by using a 2.5-kb fragment upstream of the start codon of the SPF1 gene fused to the SPF1 cDNA to produce an SPF1pro:SPF1 construct. The construct was transformed into the spf1-1 and spf1-2 mutants, and as expected, transgenic plants displayed a wild-type phenotype (Fig. 1, C–E). We also constructed a mutant version of SPF1 (SPF1C577S) driven by SPF1pro and transformed it into spf1-2, and as expected, the floral phenotypes were not complemented in transgenic plants (Supplemental Fig. S5). Together, these results demonstrated that the floral phenotype of spf1-1 mutants was caused by loss of function of SPF1.
To genetically analyze the contributions of SPF1 and SPF2 to microgametophyte and megagametophyte development, reciprocal crosses between wild-type plants and either the spf1 single or the spf1-1 spf2-1 double mutants were carried out. As shown in Table I, seed abortion was noticeable when the spf1-1 mutant was used as the female parent, but when the spf1-1 mutant was used as the male parent, the fertility was similar to that of the wild-type parent, suggesting that SPF1 is required during microgametophyte development. In contrast, in a set of hybrids between the double mutant and the wild type, the fertility between the reciprocal crosses was negligible (Table I), indicating that the fertility of both male and female gametes was severely defective in the spf1-1 spf2-1 double mutant.
Table I. Percentage of abnormal seeds from different cross combinations.
Abnormal seeds were counted as aborted seeds and undeveloped seeds, as shown in Figure 1. For more detail, see text. Col, Columbia wild type.
| Cross Combinations (♀ × ♂) | Abnormal Seeds |
Total Seeds | Percentage of Abnormal Seeds |
||||
|---|---|---|---|---|---|---|---|
| Aborted | Undeveloped | Total | Aborted | Undeveloped | Total | ||
| Col × Col | 0 | 27 | 27 | 954 | 0 | 2.83 | 2.83 |
| Col × spf1-1 | 9 | 21 | 30 | 1,074 | 0.84 | 1.95 | 2.79 |
| spf1-1 × Col | 13 | 64 | 77 | 795 | 1.64 | 8.05 | 9.69 |
| Col × spf1-1 spf2-1 | 153 | 728 | 881 | 1,059 | 14.44 | 68.75 | 83.19 |
| spf1-1 spf2-1 × Col | 42 | 789 | 831 | 973 | 4.32 | 81.09 | 85.41 |
Two distinct classes of abnormal seeds were observed in both the spf1-1 single and spf1-1 spf2-1 double mutants (Fig. 1D). In the first class, some ovules remained unfertilized and seed development was not initiated. These ovules subsequently degenerated, leaving a void in the silique. In the second class, embryo development initiated but was arrested at different stages. In this study, the first class of seeds are referred to as undeveloped seeds and the second class as aborted seeds. Most of the observed abnormal seeds belonged to the second class (Fig. 1D; Table I). These phenotypes suggested that both SPF1 and SPF2 are involved in embryo development.
The SPF1 Mutation Enhances the Growth of the Style
Two distinct types of flowers occurred in the spf1-1 and spf1-1 spf2-1 mutants (Fig. 2, A–D), while the spf2-1 mutant formed flowers that were indistinguishable from the wild type. In spf1-1 single or spf1-1 spf2-1 double mutants, about one-third (65 flowers from 10 individual plants) of the flowers appeared as wild type, but the remaining two-thirds produced abnormally long styles. When flowers were observed on the day the flowers opened, fewer pollen grains were present on the stigmas of spf1-1 mutant flowers than on the stigmas of wild-type flowers (Fig. 2, E and F).
Figure 2.
Elongated styles in spf1-1single mutant and spf1-1 spf2-1 double mutant flowers form a spatial barrier to pollination. A and E, Flowers of the wild type, showing the normal style and pollen on the stigma. B and D, Abnormal flowers of the spf1-1 and the spf1-1 spf2-1 mutants, showing the longer styles. C, A normal flower from the spf2-1 single mutant. F, Less pollen on the stigma of the double mutant. G and H, Siliques produced by natural (G) and manual (H) self-pollination of abnormal flowers (B) in the spf1-1 mutant. The numbers in G and H refer to the sterility ratios of 10 siliques. Bars = 50 μm for A to D and 100 μm for E to H.
To test whether spatial segregation of the pollen grains and stigmas in the spf1-1 mutant flowers prevented fertilization, manual self-pollination (carried out on the flower’s opening day) was compared with natural self-pollination. Interestingly, manual pollination was successful in alleviating the level of sterility (Fig. 2, G and H), suggesting that a longer style presented a spatial barrier to successful fertilization in the spf1-1 mutant.
The spf1-1 spf2-1 Double Mutants Show Pollen Abortion
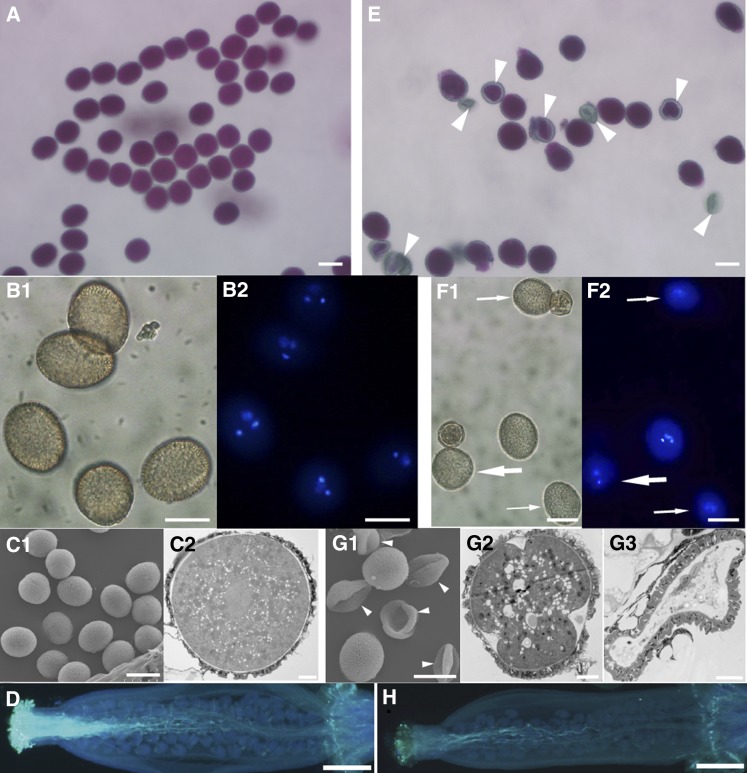
As shown in Table I, abnormal microgametophytes were partially responsible for reduced fertility in the spf1-1 spf2-1 double mutant. We investigated male fertility through different approaches. To monitor pollen viability, Alexander’s solution was applied to pollen grains from newly opened flowers. While most of the wild-type pollen appeared viable, many of the spf1-1 spf2-1 pollen grains remained unstained and, therefore, were considered to be nonviable (Fig. 3, A and E). 4′,6-Diamino-phenylindole (DAPI) staining of mature pollen showed that the spf1-1 spf2-1 pollen included a proportion of unicellular and bicellular stage grains as well as fully aborted ones (Fig. 3, B and F). While wild-type mature pollen grains generally progressed successfully through the tricellular stage, a significant proportion of spf1-1 spf2-1 pollen grains remained abnormal (23.4%, n = 454; Supplemental Table S1). Scanning electron microscopy indicated that many pollen grains of the double mutant had an abnormal shape even though they had a similar exine surface to wild-type pollen grains (Supplemental Fig. S3, C1 and G1), indicating that SPF1 and SPF2 did not affect the establishment of pollen surface structure. Further analysis by transmission electron microscopy showed that the spf1-1 spf2-1 pollen grains had reduced cytoplasmic contents (Supplemental Fig. S3, C2 and G2), while other pollen grains had a similar shape (sphericity) to wild-type pollen but their intracellular structure was abnormal (Supplemental Fig. S3G3). Together, these results demonstrate that SPF1 and SPF2 are involved in pollen grain development.
Figure 3.
Defective pollen grains in the spf1-1 spf2-1 double mutant. A to D, Wild-type pollen grains. E to H, spf1-1 spf2-1 pollen grains. A and E, Pollen visualized by transmission microscopy after staining with Alexander’s reagent. B and F, DAPI-stained pollen (B1 and F1 for bright field and B2 and F2 for UV light). C and G, Pollen visualized by scanning electron microscopy (C1 and G1) and transmission electron microscopy (C2, G2, and G3), showing abnormal shape and contents in the double mutant compared with the wild-type. D and H, Pollen from wild-type (D) and double mutant (H) plants were artificially pollinated on wild-type stigmas, and the growth of pollen tubes was visualized by Aniline Blue staining, showing that the growth of pollen of the double mutant was obviously affected. Arrowheads, Abnormal pollen grains; thin arrows, uninucleate pollen grains; thick arrows, binucleate pollen grains. Bars = 20 μm for A, B, E, and F, 5 μm for C and G, and 400 μm for D and H.
To further investigate pollen activity in vivo, manual pollination of mutant pollen onto wild-type stigmas was carried out. Pollen grains of both spf1-1 and spf2-1 single mutants germinated and grew similar to wild-type pollen (Fig. 3, D and H). However, double mutant pollen germination was reduced and pollen tubes grew slower than in the wild type. In vitro germination tests of pollen also showed very low growth of the double mutant pollen grains when compared with the wild-type (Supplemental Fig. S6). Together, these results demonstrate that SPF1 and SPF2 functioned redundantly in pollen development.
Development of the Embryo Sac Is Disrupted in the spf1-1 spf2-1 Double Mutant
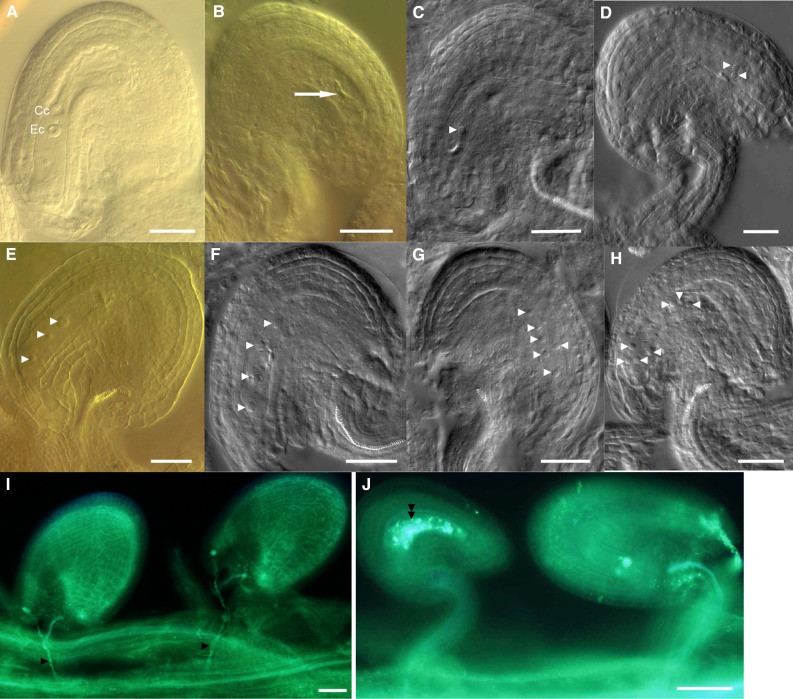
When we observed female reproductive development, we found that the morphology of ovules produced in the spf1-1 mutant was indistinguishable from that of the wild type, but many abnormal ovules were present in the spf1-1 spf2-1 double mutant (Fig. 4, B–H). In mature ovules of the double mutant, the defective phenotypes included arrested embryo sacs at different stages and degeneration of embryo sacs, suggesting that SPF1/SPF2 did not have a cell-specific effect but had a general effect on the development of embryo sacs.
Figure 4.
Defective embryo sacs in the spf1-1 spf2-1 double mutant. A, Wild-type embryo sac. Cc, Central cell; Ec, egg cell. B to H, Abnormal embryo sacs containing degenerated nuclei (B), one nucleus (C), two nuclei (D), three nuclei (E), four nuclei (F), six nuclei (G), and seven nuclei (H). Arrow, Degenerated nuclei; white arrowheads, nuclei. I and J, Siliques at the stage of the mature embryo sac of the wild type (I) and the spf1-1 spf2-1 mutant (J) stained by Aniline Blue for callose (in white, with double black arrowheads), indicating accumulating callose in the mature embryo sacs of the double mutant. Black arrowheads indicate growing pollen tubes. Bars = 50 μm for A to H and 100 μm for I and J.
Irregular callose clusters in ovules of the spf1-1 spf2-1 double mutant were obvious (Fig. 4, I and J) after staining with Aniline Blue (Pagnussat et al., 2005), and they accumulated at the position normally occupied by the embryo sac. Previous studies have shown that, if callose does not degrade in the late stage of megagametophyte development, callose is associated with degradation and/or abnormal behavior of ovule cell nuclei (Pimienta and Polito, 1982; Vishnyakova, 1991). Therefore, SPF1 and SPF2 may control embryo sac development partially through the regulation of callose degradation.
The spf1-1 spf2-1 Double Mutant Displays Arrested Embryos
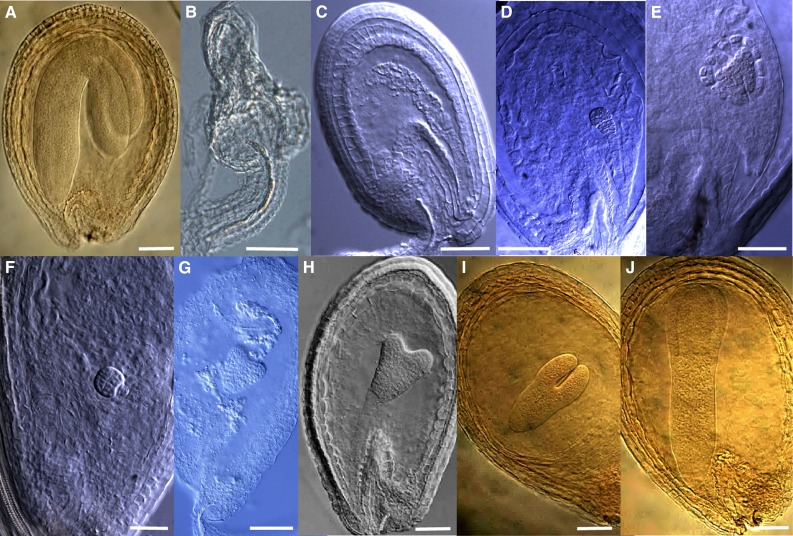
To follow the development of embryos, we collected siliques containing immature white and green seeds from the spf1-1 spf2-1 double mutant. These siliques contained a mixture of normal, undeveloped and aborted seeds (Fig. 5, B–J). The various stages of embryo development (globular, heart, torpedo, and cotyledon stages) as well as a number of irregular morphologies (abnormal structure, outgrowth suspensors, undeveloped embryos, and degenerated embryos) were identified in the spf1-1 spf2-1 double mutant, suggesting that the whole course of embryogenesis was affected in the double mutant. In contrast, the spf1-1 single mutant exhibited some arrested embryos, whereas embryos of the spf2-1 single mutant appeared like the wild type. These results suggested that SPF1 and SPF2 also function redundantly during embryo development.
Figure 5.
Defective phenotypes of embryos in the spf1-1 spf2-1 double mutant. A, The wild type. B, An embryo-free seed. C to J, Abnormal embryos that were arrested in different stages. Bars = 20 μm.
Marker Gene Misexpression in the spf1 spf2 Double Mutant
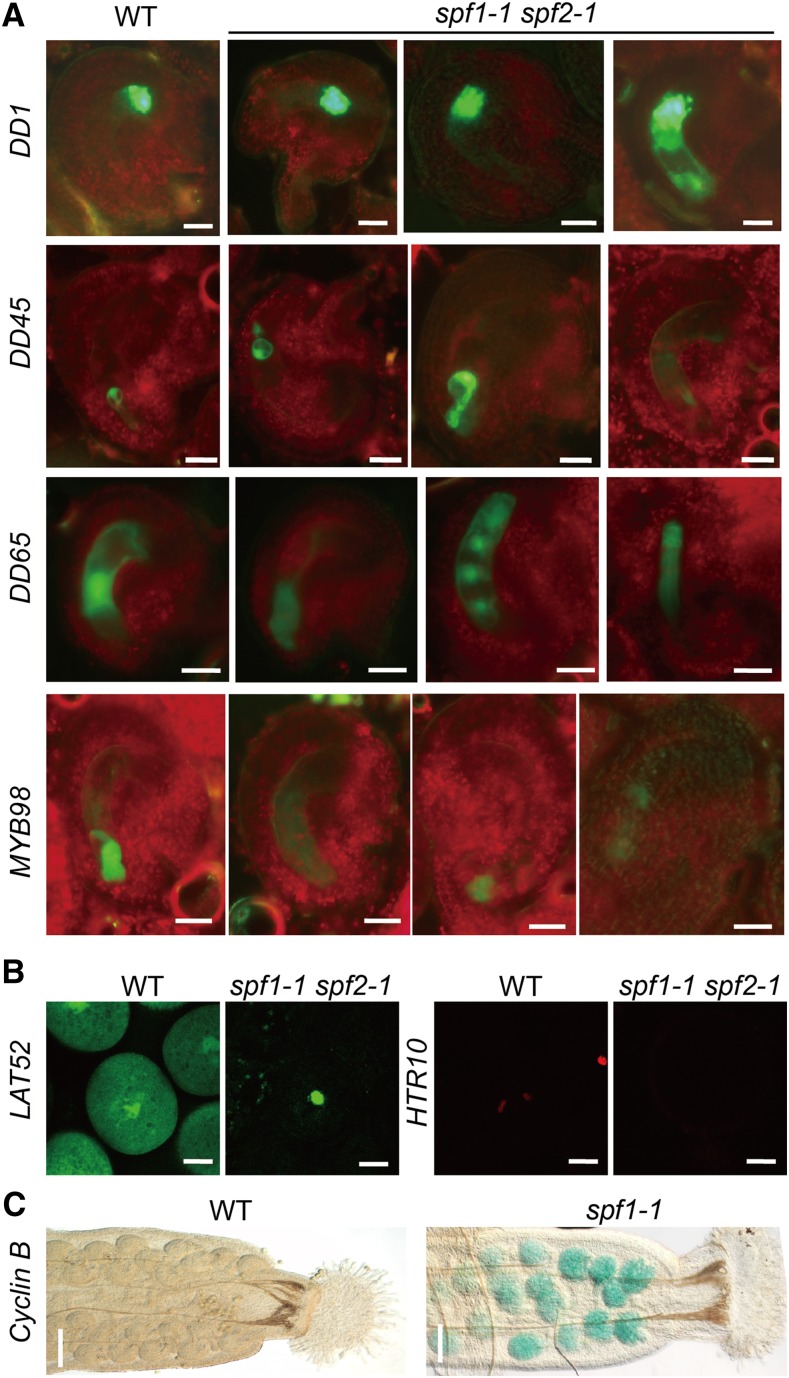
Benefiting from extensive studies of previous researchers, we checked the expression patterns of fertility marker genes in the spf1-1 spf2-1 double mutant by introducing marker lines into the mutant. These specific marker genes include DD1 (specific to the antipodal cell; Steffen et al., 2007), DD45 (specific to the egg cell; Steffen et al., 2007), MYB98 (specific to the synergid cell; Kasahara et al., 2005), DD65 (specific to the central cell; Steffen et al., 2007), LAT52 (specific to the vegetative cell of pollen; Eady et al., 1994; Berger, 2011), HTR10 (specific to the generation of pollen; Okada et al., 2005; Ingouff et al., 2007; Berger, 2011), and Cyclin B (specific to cell division of the ovule; Colón-Carmona et al., 1999; Wang et al., 2012). Figure 6 clearly shows that all these markers exhibited abnormal signals in the wrong positions or areas when compared with the wild type, providing more genetic evidence for defective development of pollen, embryo sacs, and embryos in the spf1-1 spf2-1 double mutant.
Figure 6.
Marker genes misexpress in the spf1-1 spf2-1 double mutant. The marker lines were introduced into the spf1-1 spf2-1 double mutant (A and B) or the spf1-1 single mutant (C) by crossing, and the signal of markers was observed with a confocal microscope. A, Marker lines for the embryo sac. B, Marker lines for pollen. C, Marker line for embryos. WT, The wild type. Bars = 20 μm in DD1-GFP, DD45-GFP, DD65-GFP, MYB98-GFP, LAT52-GFP, and HTR10-RFP lines and 100 μm in pCYCB1-CYCB1-GUS lines.
SPF1 and SPF2 Regulate the Expression of Genes Related to Gametogenesis and Embryogenesis
To elucidate the possible mechanism of SPF1 and SPF2 regulating reproduction, we analyzed the expression profiles of 89 genes related to fertility by quantitative real-time RT-PCR in RNA isolated from inflorescence tissues of the spf1 spf2 double mutant and wild-type plants. Previously, these genes were demonstrated to be related to the development of microgametophytes, megagametophytes, or embryos (Supplemental Fig. S7; Supplemental Table S2). The RT-PCR results showed that the expression level of most embryo-related genes in the spf1-1 spf2-1 double mutant was about 20% higher or lower than that in wild-type plants, whereas less than 30% of the gametogenesis genes displayed such a difference (Supplemental Fig. S7; Supplemental Table S2). These differentially expressed genes are involved in a range of cellular processes, indicating that SPF1 and SPF2 influenced many molecular processes impacting on fertility.
SPF1 and SPF2 Are Expressed Mainly in Reproductive Organs
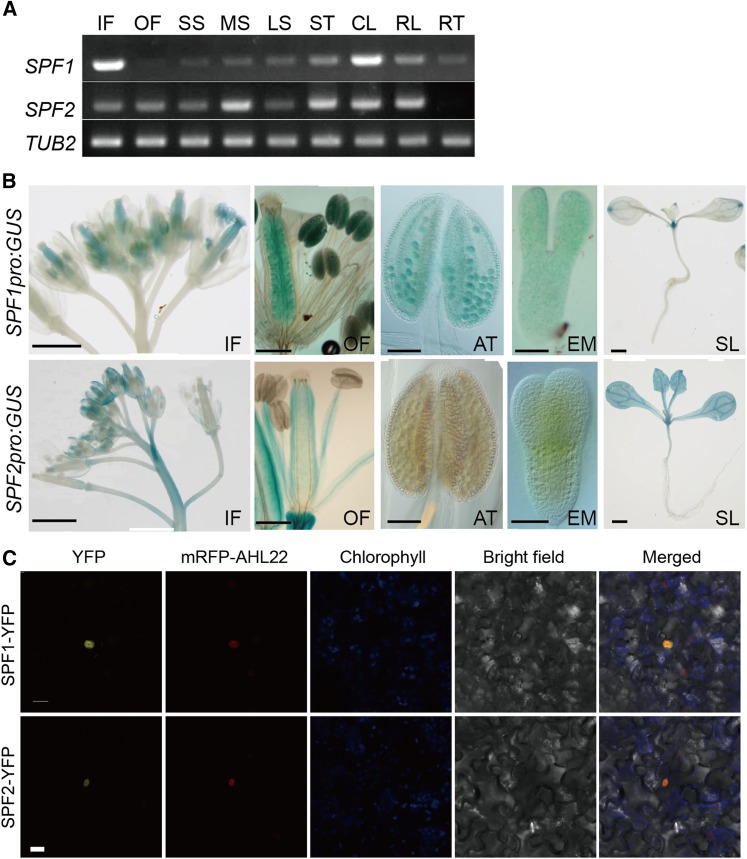
Next, we investigated the expression patterns of SPF1 and SPF2 genes in planta with two approaches. Semiquantitative RT-PCR demonstrated that both SPF1 and SPF2 mRNAs were present in most organs, with higher levels in developing reproductive organs (flower buds and siliques) and cauline leaves (Fig. 7A). SPF2 generally had a higher mRNA level when compared with that of SPF1. In parallel, GUS fusions to the SPF1 and SPF2 promoters were constructed and introduced into wild-type plants. In these transgenic plants, GUS activity was detected mainly in the floral organs and developing embryos (Fig. 7B). Significant spatial differences also were observed between SPF1:GUS and SPF2:GUS plants. In flowers, SPF1 was expressed mainly in the anthers and embryo sacs, whereas SPF2 was expressed mainly in maternal tissues, indicating that these two genes diverged in expression patterns. In the case of leaves, SPF1 was expressed mainly in the tip, while SPF2 was expressed in all areas. These observations supported the notion that SPF1 and SPF2 are involved primarily in reproductive growth with a partially overlapping expression pattern.
Figure 7.
Expression patterns and subcellular localization of SPF1 and SPF2. A, RT-PCR analysis of SPF1 and SPF2 expression in various tissues at the flowering stage (ACT was used as a reference gene). IF, Inflorescence; OF, open flower; SS, short siliques; MS, middle siliques; LS, long siliques; ST, stems; CL, cauline leaves; RL, rosette leaves; RT, roots. B, GUS staining for SPF1 and SPF2 promoter analysis. IF, Inflorescences; OF, flowers with strong signals in pistils and stamens; AT, anthers; EM, embryos; SL, seedlings. Bars = 500 μm in IF and SL, 250 μm in OF, 100 μm in AT, and 50 μm in EM. C, Subcellular localization of SPF1- and SPF2-YFP proteins. YFP was fused to the C terminus of SPF1 and SPF2, and the fusion genes were transformed into N. benthamiana epidermal cells and then observed with a confocal microscope. YFP, YFP fluorescence; Chlorophyll, chlorophyll fluorescence; BF, bright field; Merge, YFP fluorescence images merged with chlorophyll fluorescence and bright field. Bar = 10 μm.
To study the subcellular localization of SPF1 and SPF2 proteins, SPF1:YFP and SPF2:YFP translational fusions driven by the 35S promoter were separately introduced into Nicotiana benthamiana epidermal cells (Yoo et al., 2007), and the YFP signal was observed by epifluorescence microscopy. The results showed clearly that both proteins were localized exclusively in nuclei (Fig. 7C).
SPF1 and SPF2 Have SUMO-Specific Peptidase Activity
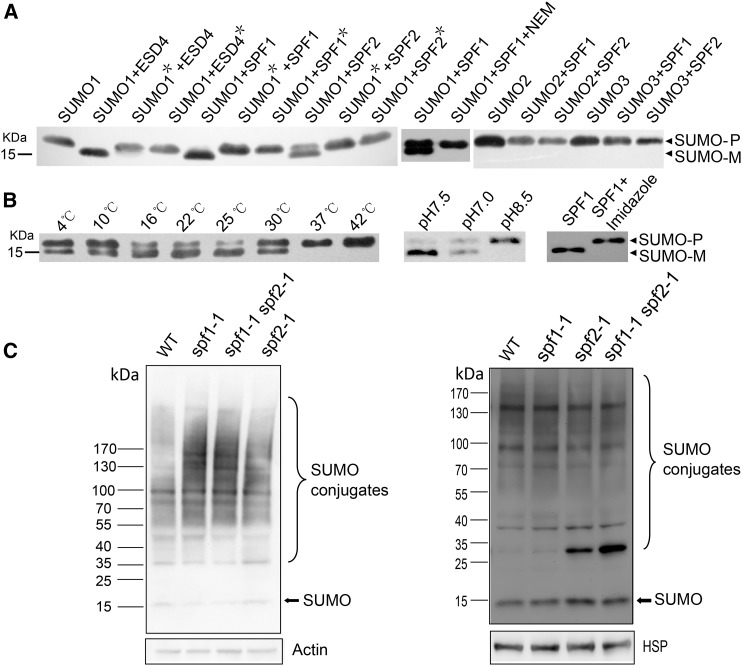
As both SPF1 and SPF2 share a ULP1 domain that is present in the SUMO-processing enzyme ESD4 (Supplemental Fig. S1), we investigated whether SPF1 and SPF2 showed in vitro SUMO-processing activity (Murtas et al., 2003). First, we carried out yeast two-hybrid (Y2H) analysis to detect the interaction between SPF1/2 and three SUMO isoforms. As expected, SPF1 and SPF2 had strong interaction patterns with SUMO isoforms in yeast (Supplemental Fig. S8). Second, we expressed various versions of SPF1, SPF2, ESD4, and SUMOs in Escherichia coli cells for the analysis of in vitro enzyme activity. Then, we purified the His-SPF1, His-SPF1C577S, His-SPF2, His-SPF2C485S, His-ESD4, His-ESD4C448S, and His-SUMO1 proteins from E. coli by nickel-exchange chromatography as reported previously (Murtas et al., 2003). The protease activities of SPF1 and SPF2 were then tested compared with ESD4, using His-SUMO1 precursor as the substrate. As Figure 8A shows, His-SPF1 and His-SPF2 were able to process the SUMO1 precursor into its mature form, while His-SPF1C577S and His-SPF2C485S, in which the key site Cys had been replaced with Ser, had no such activity. The activity of His:SPF1 was blocked by the thiol reagent N-ethylmaleimide, a Cys protease inhibitor (Li and Hochstrasser, 1999; Murtas et al., 2003; Fig. 8A). These results suggested that SPF1 and SPF2 were potent SUMO-specific proteases and had common characteristics of SUMO proteases.
Figure 8.
SUMO-specific protease activity of SPF1 and SPF2. A, Both SPF1 and SPF2 can process SUMO1 precursor (SUMO-P) into its mature form (SUMO-M) in vitro. SUMO protease activity can be blocked by N-ethylmaleimide or mutation of the key active site. The western blot was probed by His antibody. A total of 100 μg of total purified protein was loaded for each lane. Stars indicate the mutant proteins. B, High temperature, pH value, and the eluent imidazole can suppress the activity of SPF1 protease. A total of 100 μg of total purified protein was loaded for each lane. C, Activity analysis of SPF1/2 in vivo. The SUMO conjugate profiles in inflorescences (left) and seedlings (right) are shown for wild-type (WT) plants and mutants. The western blot was probed by SUMO antibody. SPF1C577S/SPF2C485S, Mutated SPF1/SPF2 with changes of Cys-577/485 to Ser (see Supplemental Fig. S1); HSP/Actin, loading controls.
As plant fertility is influenced by environmental conditions and our data above showed that SPF1/SPF2 contributed to Arabidopsis fertility, we further checked SPF1 activity at different temperatures and pH values. We found that high temperatures and pH values suppressed the SUMO protease activity of SPF1 (Fig. 8B). Additionally, we found that the purified SPF1 eluted by imidazole had no SUMO protease activity. But when imidazole was removed from the eluent by dialysis, SPF1 activity on SUMO processing could be detected (Fig. 8B), confirming that imidazole inhibited SPF1 protease.
When the in vivo protease activity of SPF1 and SPF2 was investigated immunologically, it was apparent that greater amounts of SUMO conjugates were present in the floral extracts, but not in seedling extracts, of all mutants when compared with the wild type, especially in the double mutant (Fig. 8C). However, in seedlings, there was one band with a size around 30 kD that accumulated significantly in the spf2-1 single and spf1-1 spf2-1 double mutants. We further investigated SUMO conjugates in another allele of the SPF1 gene mutant spf1-2. As expected, the spf1-2 mutant also accumulated SUMO conjugates and the SPF1 gene rescued the spf1-2 phenotype. However, the mutated SPF1 gene (SPF1C577S) did not recover the profile of SUMO conjugates in the spf1-2 mutant (Supplemental Fig. S9; Kong et al., 2017). Taken together, our results indicate that SPF1 and SPF2 were required to desumoylate substrates in vivo.
Identification of SPF1 Substrates
To identify candidate SPF1 substrates, an Arabidopsis Y2H library (Invitrogen) was screened with SPF1 as a bait, and 57 full-length positive targets were identified, which are involved in a wide range of biological functions, including fertility formation, such as EDA9, LUMINAL BINDING PROTEIN, and DEFECTIVE KERNEL1 proteins (Supplemental Table S3). To confirm the relationship between these candidate interacting proteins with the SUMO system, a Y2H screen was carried out between these proteins and SPF1, SPF2, E2, E3, SUMO1, SUMO2, or SUMO3. Our results showed that these proteins had differential specificity (Supplemental Table S4) and could be grouped into two classes: SUMO substrates and non-SUMO substrates. The non-SUMO substrates may be SPF1-interacting proteins, which are related to its function or metabolism. For SUMO substrates, they also showed different specificity to SUMO isoforms, suggesting their specificities to SUMO1/2/3. Some SUMO substrates did not interact with E2 or E3, indicating that some SPF1 substrates may employ another unidentified E2 or E3 for their sumoylation. Supplemental Table S4 shows some substrates that were common to both SPF1 and SPF2, while other substrates were only specific to SPF1. It was clear that more detailed evidence was required to elucidate their relationship with SUMOs.
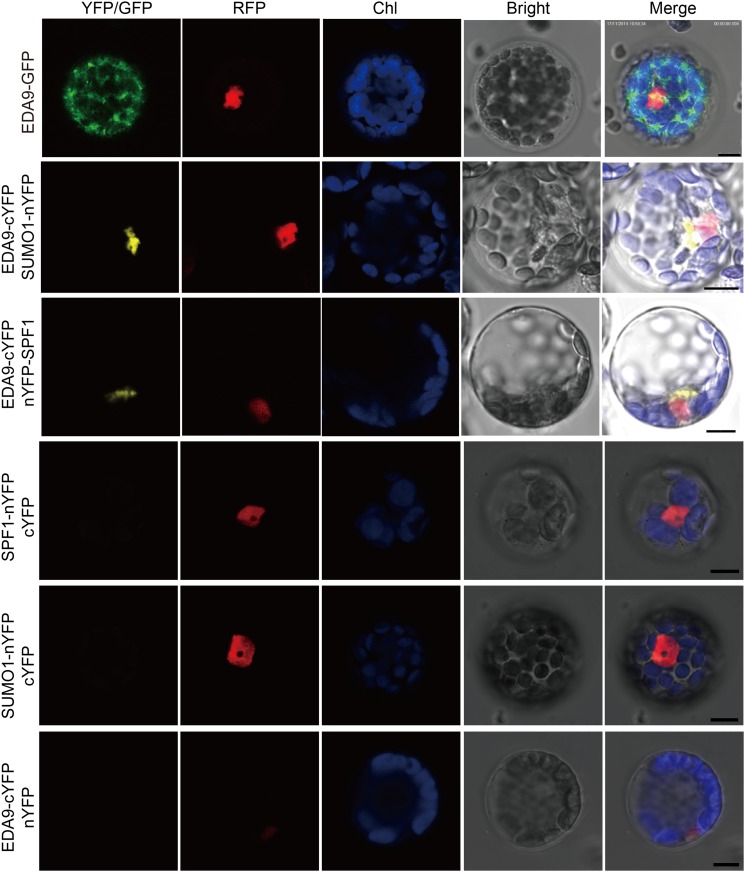
The bimolecular fluorescence complementation (BiFC) approach was employed to confirm SPF substrates. EDA9 is a gene coding one of the 3-phosphoglycerate dehydrogenases in Ser biosynthesis. The EDA9:GFP protein localized in chloroplasts (Fig. 9), which is consistent with the previous report (Toujani et al., 2013). Not surprisingly, EDA9 interacted with both SPF1 and SUMO1 in planta. However, the interacting location was in the cytoplasm and near the nucleus (Fig. 9), not where EDA9 and SPF1 proteins localize. SUMOylated proteins are distributed throughout the whole cell (Elrouby and Coupland, 2010). Therefore, free proteins of SUMO, SPF1, and EDA9 have distinct subcellular localizations, but their interaction happens at specific sites. However, the exact mechanism and biological significance remain to be uncovered.
Figure 9.
EDA9 interacts with both SPF1 and SUMO1. EDA9 proteins localize in the chloroplast but interact with both SUMO1 and SPF1 proteins at specific sites in the cytoplasm and adjacent to the nuclei. GFP, EDA9:GFP; YFP, interaction between EDA9 (marked with cYFP at the C terminus) and SPF1 (marked with nYFP at the N terminus) or SUMO1 (marked with nYFP at the C terminus just before the double Gly residues); RFP, nuclear marker AHL22-mRFP (Xiao et al., 2009); Chl, chlorophyll; Bright, white light; Merge, merged images. Bars = 10 μm.
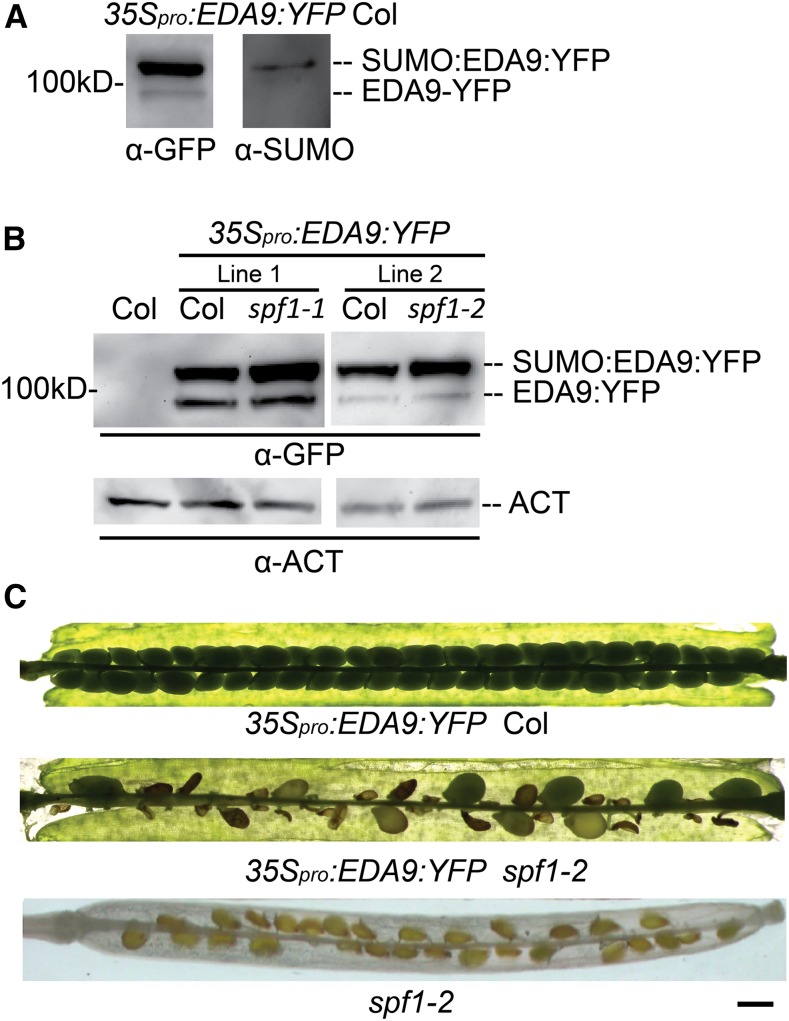
To further elucidate the relationship between SPF1 and EDA9, we checked whether SPF1 could desumoylate SUMO-EDA9 proteins in vitro. First, we overexpressed EDA9-YFP in wild-type plants, purified EDA9-YFP proteins from 35Spro:EDA9-YFP seedlings by immunoprecipitation, and then probed by GFP and SUMO antibodies. Western-blot analysis showed that EDA9-YFP proteins could be sumoylated in vivo (Fig. 10A). However, we failed to desumoylate these SUMO-EDA9-YFP proteins by using SPF1 purified from E. coli cells, due to self-desumoylation of SUMO-EDA9-YFP in vitro. So, we introduced 35Spro:EDA9-YFP into spf1-1 and spf1-2 mutants from wild-type plants by crossing. As expected, the resulting plants (35Spro:EDA9-YFP spf1-1 and 35Spro:EDA9-YFP spf1-2) accumulated much higher levels of SUMO-EDA9-YFP proteins (Fig. 10B). Moreover, the 35Spro:EDA9-YFP spf1 plants exhibited sterility phenotypes, which were more severe than that of the spf1-1 mutant (Fig. 10C). These results indicated that EDA9 could be sumoylated in vivo and then desumoylated by SPF1 and that sumoylated EDA9 affects fertility.
Figure 10.
SPF1 desumoylates sumoylated EDA9 proteins in vivo. 35Spro:EDA9:YFP was expressed in Columbia (Col) wild-type plants and introduced into spf1-1 and spf1-2 mutants by crossing, and then homozygous plants were used for western blots. A, EDA9:YFP proteins were purified from 35Spro:EDA9:YFP seedlings by GFP antibody affinity beads and then applied to western blots, which were probed by GFP and SUMO antibodies (Murtas et al., 2003). Each lane was loaded with 20 µL of the eluate from the GFP beads. B, Whole proteins were extracted from the transgenic seedlings and then applied to western blots, which were probed by GFP and ACT antibodies. ACT was used as a loading control. The top band is predicted as sumoylated EDA9:GFP (SUMO:EDA9:GFP), as shown in A. C, Siliques of 35Spro:EDA9:YFP Col and 35Spro:EDA9:YFP spf1-2. The experiments were repeated at least three times. Bar = 200 μm.
DISCUSSION
Successful production of viable seeds requires the formation of normal flowers capable of pollination, production of living pollen grains and their compatible egg cells in embryo sacs, successful pollination and fertilization, as well as healthy development of embryos. Sumoylation appears to be implicated in nearly all developmental processes in eukaryotes, including reproductive growth in different organisms (Budhiraja et al., 2009; Hashiyama et al., 2009). Here, we exhibit a set of data showing that two SUMO proteases, SPF1 and SPF2, are involved in Arabidopsis fertility.
SPF1/2 Are Two Novel SUMO Proteases in Arabidopsis
SPF1 and SPF2 share conserved active domains of SUMO proteases, in particular their His/Asp/Glu/Cys active sites (Li and Hochstrasser, 1999), and mutation of Cys in these domains results in the loss of SUMO protease activity (Fig. 8). However, global peptide sequences of SPF1 and SPF2 share low similarity with other Arabidopsis SUMO proteases. Different from ESD4, the ULP1 protease domains of SPF1 and SPF2 are flanked by an extensive amino acid stretch at both ends (Supplemental Fig. S1). ExPASy analysis showed that the extending sequences of SPF1 and SPF2 contain predicted sites for phosphorylation, glycosylation, and myristylation. These results suggest that the function and regulation of SPF1 and SPF2 may be quite different from those of other SUMO proteases in plants.
Even though the in vitro analysis of SPF1/2 activity supports the sequence-based notion that SPF1/2 act as SUMO-specific peptidases for the maturation of SUMO precursors, the in vivo analysis indicates that they may function as SUMO isopeptidases for removing SUMO from their substrates, because SUMO conjugates accumulate in the inflorescence of the spf1-1 spf2-1 double mutant (Fig. 8). Furthermore, the abundance of SUMO-EDA9-GFP is higher in the spf1-1 mutant than in the wild type (Fig. 10).
In yeast, SPF1 and SPF2 can interact with all three SUMO isoforms and share some common substrates, but they also have individual substrates (Supplemental Table S4). Therefore, SPF1 and SPF2 may function both individually and synergistically, dependent on the substrates. SPF1 activity is dependent on environmental conditions (Fig. 8). Both high-temperature and low-pH conditions inhibit its SUMO protease activity. Therefore, environmental stresses may reduce SPF1 activity and affect plant fertility, and this may explain the reduction in fertility observed at high temperature (Jagadish et al., 2007, 2015).
SPF1 and SPF2 Appear to Function in Different Subcellular Sections
Subcellular localization is a spatially significant constraint on SUMO isopeptidase specificity and their precise functions. SUSP1, a human SUMO protease in reproductive organs, localizes within the nucleoplasm (Mukhopadhyay et al., 2006), while human ULP1 (Li and Hochstrasser, 2003), Drosophila spp. Ulp1 (Smith et al., 2004), and Arabidopsis ESD4 proteins localize specifically to the nuclear periphery (Murtas et al., 2003; Xu et al., 2007). Our data indicate that SPF1 and SPF2 are nucleoplasm proteins (Fig. 7). Therefore, SPF1 and SPF2 function in different subcellular sites from that of ESD4. A potential SPF1 substrate, EDA9, localizes in chloroplasts (Fig. 9; Toujani et al., 2013), but it interacts with SPF1 or SUMO1 in a distinct site adjacent to the nucleus in the cytoplasm (Fig. 9). This may suggest that sumoylation and desumoylation of EDA9 happen at specialized locations.
Both SPF1 and SPF2 Are Involved in Multiple Aspects of Reproductive Growth
The spf1-1 spf2-1 double mutant showed several abnormal developmental phenotypes. The spf1-1 single and spf1-1 spf2-1 double mutants (Figs. 2–5) had similar phenotypes to delayed dehiscence1 (Sanders et al., 2000) and sterile apetala mutants (Byzova et al., 1999) with long-style flowers. However, previous reports did not mention whether long styles were a barrier to pollination or not. We found here that the long styles in the spf1-1 and spf1-1 spf2-1 mutants created a spatial obstacle for pollination; however, artificial pollination can overcome the barrier (Fig. 2).
The spf1-1 spf2-1 double mutant shows distinctly aberrant phenotypes in microgametogenesis, megagametogenesis, and embryogenesis. The spf1-1 spf2-1 double mutant included abnormal pollen in aspects of morphology, physiology, activity, and cell division, abnormal embryo sacs with different numbers of nuclei and spatial distribution, and abnormal embryos arrested at different stages. All marker lines presented abnormal signals in the mutant when compared with that in wild-type plants. The abnormal expression of these genes and potential interacting proteins supports our speculation that these genes are involved in many different cellular processes (Supplemental Fig. S9). For example, TAPETAL DEVELOPMENT AND FUNCTION1 (Zhu et al., 2008) and DUO POLLEN1 (Durbarry et al., 2005) are related to mitosis, and SUCCINATE DEHYDROGENASE controls cell structure and the accumulation of cell contents in pollen (León et al., 2007). RING-H2 Group F1a (Liu et al., 2008), LACHESIS (Gross-Hardt et al., 2007), and GAMETOPHYTIC FACTOR1 (Moll et al., 2008) are key genes for embryo sac development. Several genes involved in metabolic pathways were misexpressed in the double mutant and led to abnormal embryos, including phosphatidylethanolamine biosynthesis (CTP:PHOSPHORYLETHANOLAMINE CYTIDYLYLTRANSFERASE; Mizoi et al., 2006), nitrate transport (NITRATE TRANSPORTER; Almagro et al., 2008), mRNA adenosine methyltransferase (METHYLASE; Zhong et al., 2008), DNA glycosylase (DEMETER; Choi et al., 2002), and vitamin B6 biosynthesis (PYRIDOXAL PHOSPHATE SYNTHASE; Titiz et al., 2006).
Our results also suggest that SPF1 and SPF2 are partially redundant. The spf1 spf2 double mutant had a much more severe phenotype than the spf1 single mutant. SPF1 is expressed mainly in microgametophytes, megagametophytes, and embryo cells, while SPF2 is expressed in embryos and maternal cells/tissues, such as filaments, styles, and inflorescence axes (Fig. 7).
Obviously, SPF1 and SPF2 may not be the only SUMO proteases related to fertility in Arabidopsis, because mutation in the ESD4 gene also results in sterility and the phenotypes of mutants that are stronger than the spf1-1 spf2-1 double mutant (Reeves et al., 2002; Murtas et al., 2003). However, the esd4 mutant shows a general effect on plant development (Reeves et al., 2002; Murtas et al., 2003), while vegetative growth in the spf1-1 spf2-1 double mutant is similar to that in the wild type. Therefore, SPF1 and SPF2 may regulate fertility more specifically, perhaps due to tissue-specific expression in reproductive organs.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia and various of its derived mutants were grown under either long-day (16 h/8 h, light/dark) or short-day (8 h/16 h, light/dark) conditions with 100 µmol m−2 s−1 lighting provided by fluorescent lamps. Seeds of the T-DNA insertions into spf1-1 (Salk_040756), spf1-2 (Salk_049255), and spf2-1 (Salk_023493) were obtained from the ABRC. Homozygous screening was according to the protocol provided by SALK (http://signal.salk.edu/).
The effect on fertility was measured on the first 10 siliques of the primary inflorescence stem. The ratio of normal to abnormal flowers on the primary inflorescence stem of at least 20 individual plants per genotype per replicate was assessed 15 d after bolting. Generally, at least 20 plants per treatment were assessed to calculate the fertility percentage.
Gene and Promoter Cloning, and Plasmid Construction
Standard Gateway (Invitrogen) methods were employed for cloning and plasmid construction. The full-length SPF1 and SPF2 open reading frames were PCR amplified with primers (Supplemental Table S2) that contained the recombination sites of attB1/2 sequences. BP and LR reactions were performed according to the Invitrogen protocols, and the entry clones were sequenced for confirmatory purposes. The destination vectors included pLeela (for overexpression), p35S-GW-YFP (for protein localization), pGW-GUS (for promoter analysis), and pGreen-GW-MCS (for mutant complementation). A 2.5-kb fragment upstream of each of the SPF1 and SPF2 coding sequences was cloned from the genomic DNA of the Columbia ecotype. For the complementation experiment, the SPF1 gene was ligated into pGreen-GW-MCS, and then the resulting vector was LR reacted with either the SPF1 or SPF2 promoter to produce pGreen-SPF1pro:SPF1 or pGreen-SPF2pro:SPF2, respectively. For GUS fusion expression, SPF1 and SPF2 promoters were fused to GUS through an LR reaction (Invitrogen) between SPF1 or SPF2 promoter entry clones and the pGW-GUS destination vector, resulting in SPF1:GUS and SPF2:GUS, respectively. The resulting binary vectors were introduced into Agrobacterium tumefaciens strain GV3101 pMP90RK and then transformed into Arabidopsis using the floral dipping method (Clough and Bent, 1998). Sequence alignment and phylogenetic analysis were performed using MEGA version 3.1. Protein analysis was achieved with ExPASy (http://www.expasy.org/tools/; MotifScan).
Semiquantitative PCR and Quantitative Real-Time RT-PCR
RNA purification, cDNA synthesis, and both quantitative real-time and semiquantitative RT-PCR were carried out following Xiao et al. (2009), except for the use of At4g34270 as a reference gene (Czechowski et al., 2005; Gutierrez et al., 2008). All relevant primer sequences are listed in Supplemental Table S2. The primers for real-time PCR were designed by Beacon Designer 7.0. All the samples were analyzed by real-time PCR using StepOne (ABI) and RealMasterMix (SYBR Green) with At4g34270 as the reference gene (Czechowski et al., 2005; Gutierrez et al., 2008). Total RNA was extracted from flower buds and siliques of wide-type and mutant plants using standard procedures. The RNA samples were treated with DNase I and then used to synthesize the first-strand cDNA using the cDNA synthesis kit (Takara). Reaction mixtures contained 1 μL of cDNA, 7.5 μL of 2×SYBR Primix Ex Taq (TakaRa), and 0.3 μL of each of 10 μm primers in a total volume of 15 μL. PCR was performed using a thermal cycling program comprising an initial denaturizing step of 95°C for 30 s followed by 40 cycles of 95°C for 5 s and finally 60°C for 30 s. The specificity of PCR products was determined by melting-curve analysis and electrophoresis on a 2% agarose gel to ensure that PCRs were free of primer dimers. For each RNA extraction, measurements of gene expression were obtained in triplicate, and the mean of these values was used for further analysis.
Subcellular Localization and GUS Staining
To determine the subcellular localization of SPF1 and SPF2, SPF1:YFP and SPF2:YFP translational fusions driven by the cauliflower mosaic virus 35S promoter were transiently expressed in Arabidopsis protoplasts and observed by confocal microscopy (Wang et al., 2013). SPF1:GUS and SPF2:GUS plants were used as materials for GUS activity analysis. The staining time for GUS activity was 16 h. Detecting YFP fluorescence and GUS activity followed the method described by Xiao et al. (2009). For BiFC, the N-terminal part of YFP (nYFP) was fused to SPF1, SPF2, or SUMO1 (immediately before the double Gly residues [GG], as SUMO proteases can recognize GG and remove the peptide after GG); the C-terminal part of YFP (cYFP) was fused to potential substrates of SPF1 or SPF2. BiFC was carried out in protoplasts of Arabidopsis or Nicotiana benthamiana leaves, and fluorescent signals were visualized by confocal microscopy (Wang et al., 2013).
Microscopy
Microgametophyte
The viability of mature pollen grains was assayed using Alexander’s reagent (Alexander, 1969), which differentially stains the cytosol of viable pollen. DAPI staining followed the protocol of Park et al. (1998). Pollen grain morphology was observed by differential interference contrast (DIC) microscopy (Leica), after initial cell clearing using Hoyer’s solution (Liu and Meinke, 1998; Gingerich et al., 2005). The assessment of in vivo pollen germination followed the methods described by Pagnussat et al. (2005).
Megagametophyte
Pistils were treated following the method described by Boisnard-Lorig et al. (2001). Cleared ovules were mounted on slides, covered slightly with a cover slide, and observed using a Leica microscope with DIC optics (Leica). Callose was stained by Aniline Blue (Pagnussat et al., 2005).
Embryo
Immature seeds were removed from siliques at different stages and cleared in Hoyer’s solution (Liu and Meinke, 1998). Preparations were assessed by DIC microscopy (Leica).
In Vitro and in Vivo Analyses of Protease Activity
The induction of all proteins in Escherichia coli, purification, enzymatic reaction, preparation of plant tissues, and western blotting were performed according to the approaches of Murtas et al. (2003). HSP and Actin were used as loading controls (Kuras et al., 2007). SUMO antibody was raised as described by Murtas et al. (2003) and can recognize all SUMO isoforms, including SUMO1, SUMO2, and SUMO3. HSP and Actin antibodies were purchased from Sigma.
Y2H and Screening Library
The binding domain vector constructs (pDEST22; Invitrogen) and the activation domain vector constructs (pDEST32; Invitrogen) with genes of SPF1, SPF2, and different SUMO isoforms were used for Y2H analysis in appropriate pairs as described in the Supplemental Figure S8 legend. To introduce activation domain and binding domain plasmids into yeast strain AH109, a lithium acetate-mediated transformation method was used. Transformants were selected on synthetic complete agar (SC)-Trp-Leu medium and then transferred to the interaction selection SC-Trp-Leu-His and SC-Trp-Leu-His-adenine media to score growth as an indicator of protein-protein interaction (Li et al., 2006). To confirm the interaction between SPF1, SPF2, and SUMOs, LacZ colony-lift assays were used (Shi et al., 2010). Positive interactions were detected by the appearance of blue clones.
Statistical Analysis
Student’s t test was used to statistically analyze our data. Each experiment had at least three biological replicates from separate plants. Data in all bar graphs represent means ± sd. For digital statistical analysis, all statistical analyses were determined using the SPSS software package. At least 20 plants per line per experiment were assessed to analyze fertility, and each experiment was repeated at least three times. More than 2,000 seeds were evaluated for their fertility. Other statistics are indicated in each figure or table.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: SPF1 (At1g09730 in ABRC; EU877962 in this study), SPF2 (At4g33620 in ABRC; EU877963 in this study), SUMO1 (At4g26840), SUMO2 (At5g55160), SUMO3 (At5g55170), ESD4 (At4g15880), E2 (At3g57870), E3 (At5g60410), EDA9 (At4g34200), DD1 (At1g36340), DD45 (At2g21740), DD65 (At3g10890), HTR10 (At1g19890), MYB98 (At4g18770), and Cyclin B (At4g37490).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Domain sequence analysis of main SUMO proteases in Arabidopsis.
Supplemental Figure S2. Sequence alignment.
Supplemental Figure S3. Silique morphology of single and double mutants.
Supplemental Figure S4. Phenotypes of the spf1-2 single and spf1-2 spf2-1 double mutants.
Supplemental Figure S5. The mutated SPF1 gene is nonfunctional.
Supplemental Figure S6. In vitro analysis of pollen activity.
Supplemental Figure S7. Expression analysis of genes related to fertility in the spf1-1 spf2-1 double mutant compared with the wild type.
Supplemental Figure S8. SPF1 and SPF2 interact with SUMO1, SUMO2, or SUMO3 in Y2H analysis.
Supplemental Figure S9. The mutated SPF1 gene does not rescue the SUMO conjugate pattern.
Supplemental Table S1. Pollen viability of mutants and the wild type.
Supplemental Table S2. List of genes and their primers used in this study.
Supplemental Table S3. Results from Y2H screening with SPF1 as bait.
Supplemental Table S4. Classification of SPF-interacting proteins in Arabidopsis.
Acknowledgments
We thank Dr. Gary N. Drews for providing seeds of DD1pro:GFP, DD45pro:GFP, DD65pro:GFP, and MYB98pro:GFP, Dr. Frederic Berger for seeds of LAT52pro:GFP HTR10pro:HTR:RFP, and Dr. Li-Jia Qu for seeds of CYCB1pro:CYCB1-GUS. We also thank Drs. Chunming Liu, De Ye, Lijia Qu, Xiangdong Fu, Ming Yuan, Chunyi Zhang, and Chenyang Huang for helpful discussions and technical support and Dr. Qingzhu Zhang for helping to clone promoters. SALK T-DNA insertion lines were obtained from the Ohio State Stock Center.
Footnotes
This research was supported by the Chinese National Science Foundation (no. 30670189 and 31370324), the Beijing National Science Foundation (5132031), start-up funds from CAAS, the CAAS Special Funding of National Non-Profit Institutes, and the Agricultural Science and Technology Innovation Program. I.S. was supported by Australian Research Council Fellowship FT130100525.
References
- Alexander MP. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Almagro A, Lin SH, Tsay YF (2008) Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development. Plant Cell 20: 3289–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C, Autran D (2015) Chromatin dynamics during cellular differentiation in the female reproductive lineage of flowering plants. Plant J 83: 160–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartetzko V, Sonnewald S, Vogel F, Hartner K, Stadler R, Hammes UZ, Börnke F (2009) The Xanthomonas campestris pv. vesicatoria type III effector protein XopJ inhibits protein secretion: evidence for interference with cell wall-associated defense responses. Mol Plant Microbe Interact 22: 655–664 [DOI] [PubMed] [Google Scholar]
- Berger F. (2011) Imaging fertilization in flowering plants, not so abominable after all. J Exp Bot 62: 1651–1658 [DOI] [PubMed] [Google Scholar]
- Boisnard-Lorig C, Colon-Carmona A, Bauch M, Hodge S, Doerner P, Bancharel E, Dumas C, Haseloff J, Berger F (2001) Dynamic analyses of the expression of the HISTONE:YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13: 495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhiraja R, Hermkes R, Müller S, Schmidt J, Colby T, Panigrahi K, Coupland G, Bachmair A (2009) Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation. Plant Physiol 149: 1529–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byzova MV, Franken J, Aarts MG, de Almeida-Engler J, Engler G, Mariani C, Van Lookeren Campagne MM, Angenent GC (1999) Arabidopsis STERILE APETALA, a multifunctional gene regulating inflorescence, flower, and ovule development. Genes Dev 13: 1002–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo AG, Kong LJ, Hanley-Bowdoin L, Bejarano ER (2004) Interaction between a geminivirus replication protein and the plant sumoylation system. J Virol 78: 2758–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, Chua NH (2007) The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19: 2952–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Chen YY, Tang IC, Liang HM, Lai CC, Chiou JM, Yeh KC (2011) Arabidopsis SUMO E3 ligase SIZ1 is involved in excess copper tolerance. Plant Physiol 156: 2225–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong MS, Park HC, Hong MJ, Choi W, Jin JB, Bohnert HJ, Lee SY, Bressan RA, Yun DJ (2009) Specific domain structures control ABA, SA, and stress mediated SIZ1 phenotypes. Plant Physiol 151: 1930–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL (2002) DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110: 33–42 [DOI] [PubMed] [Google Scholar]
- Chosed R, Mukherjee S, Lois LM, Orth K (2006) Evolution of a signalling system that incorporates both redundancy and diversity: Arabidopsis SUMOylation. Biochem J 398: 521–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colby T, Matthäi A, Boeckelmann A, Stuible HP (2006) SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol 142: 318–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P (1999) Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Conti L, Nelis S, Zhang C, Woodcock A, Swarup R, Galbiati M, Tonelli C, Napier R, Hedden P, Bennett M, et al. (2014) Small ubiquitin-like modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin. Dev Cell 28: 102–110 [DOI] [PubMed] [Google Scholar]
- Conti L, Price G, O’Donnell E, Schwessinger B, Dominy P, Sadanandom A (2008) Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20: 2894–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbarry A, Vizir I, Twell D (2005) Male germ line development in Arabidopsis: duo pollen mutants reveal gametophytic regulators of generative cell cycle progression. Plant Physiol 137: 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eady C, Lindsey K, Twell D (1994) Differential activation and conserved vegetative cell-specific activity of a late pollen promoter in species with bicellular and tricellular pollen. Plant J 5: 543–550 [Google Scholar]
- Elrouby N, Coupland G (2010) Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc Natl Acad Sci USA 107: 17415–17420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS, Large E, Han M, Darland M (2003) lin-35/Rb and ubc-18, an E2 ubiquitin-conjugating enzyme, function redundantly to control pharyngeal morphogenesis in C. elegans. Development 130: 3319–3330 [DOI] [PubMed] [Google Scholar]
- Gingerich DJ, Gagne JM, Salter DW, Hellmann H, Estelle M, Ma L, Vierstra RD (2005) Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J Biol Chem 280: 18810–18821 [DOI] [PubMed] [Google Scholar]
- Gómez JF, Talle B, Wilson ZA (2015) Anther and pollen development: a conserved developmental pathway. J Integr Plant Biol 57: 876–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Hardt R, Kägi C, Baumann N, Moore JM, Baskar R, Gagliano WB, Jürgens G, Grossniklaus U (2007) LACHESIS restricts gametic cell fate in the female gametophyte of Arabidopsis. PLoS Biol 5: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, et al. (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6: 609–618 [DOI] [PubMed] [Google Scholar]
- Hashiyama K, Shigenobu S, Kobayashi S (2009) Expression of genes involved in sumoylation in the Drosophila germline. Gene Expr Patterns 9: 50–53 [DOI] [PubMed] [Google Scholar]
- Hermkes R, Fu YF, Nürrenberg K, Budhiraja R, Schmelzer E, Elrouby N, Dohmen RJ, Bachmair A, Coupland G (2011) Distinct roles for Arabidopsis SUMO protease ESD4 and its closest homolog ELS1. Planta 233: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Johnson KD, Petcherski AG, Vandergon T, Mosser EA, Copeland NG, Jenkins NA, Kimble J, Bresnick EH (2000) A HECT domain ubiquitin ligase closely related to the mammalian protein WWP1 is essential for Caenorhabditis elegans embryogenesis. Gene 252: 137–145 [DOI] [PubMed] [Google Scholar]
- Huang L, Yang S, Zhang S, Liu M, Lai J, Qi Y, Shi S, Wang J, Wang Y, Xie Q, et al. (2009) The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J 60: 666–678 [DOI] [PubMed] [Google Scholar]
- Ingouff M, Hamamura Y, Gourgues M, Higashiyama T, Berger F (2007) Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr Biol 17: 1032–1037 [DOI] [PubMed] [Google Scholar]
- Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, Adachi S, Minamisawa K, Umeda M, Sugimoto K (2009) SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21: 2284–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadish SV, Craufurd PQ, Wheeler TR (2007) High temperature stress and spikelet fertility in rice (Oryza sativa L.). J Exp Bot 58: 1627–1635 [DOI] [PubMed] [Google Scholar]
- Jagadish SV, Murty MV, Quick WP (2015) Rice responses to rising temperatures: challenges, perspectives and future directions. Plant Cell Environ 38: 1686–1698 [DOI] [PubMed] [Google Scholar]
- Jiménez-Quesada MJ, Traverso JA, Alché JdeD (2016) NADPH oxidase-dependent superoxide production in plant reproductive tissues. Front Plant Sci 7: 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim WY, Van Oosten M, Hyun Y, Somers DE, Lee I, et al. (2008) The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J 53: 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara RD, Portereiko MF, Sandaklie-Nikolova L, Rabiger DS, Drews GN (2005) MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 17: 2981–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Luo X, Qu GP, Liu P, Jin JB (2017) Arabidopsis SUMO protease ASP1 positively regulates flowering time partially through regulating FLC stability. J Integr Plant Biol 59: 15–29 [DOI] [PubMed] [Google Scholar]
- Kuras R, Saint-Marcoux D, Wollman FA, de Vitry C (2007) A specific c-type cytochrome maturation system is required for oxygenic photosynthesis. Proc Natl Acad Sci USA 104: 9906–9910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Vierstra RD (2003) The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis: accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem 278: 6862–6872 [DOI] [PubMed] [Google Scholar]
- Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, et al. (2007) Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49: 79–90 [DOI] [PubMed] [Google Scholar]
- León G, Holuigue L, Jordana X (2007) Mitochondrial complex II is essential for gametophyte development in Arabidopsis. Plant Physiol 143: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yang WC (2016) RLKs orchestrate the signaling in plant male-female interaction. Sci China Life Sci 59: 867–877 [DOI] [PubMed] [Google Scholar]
- Li L, Kim BG, Cheong YH, Pandey GK, Luan S (2006) A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc Natl Acad Sci USA 103: 12625–12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M (1999) A new protease required for cell-cycle progression in yeast. Nature 398: 246–251 [DOI] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M (2000) The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol 20: 2367–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M (2003) The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J Cell Biol 160: 1069–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y, Zhang C, Chen T, Hao H, Liu P, Bressan RA, Hasegawa PM, Jin JB, Lin J (2012) Mutation in SUMO E3 ligase, SIZ1, disrupts the mature female gametophyte in Arabidopsis. PLoS ONE 7: e29470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, Meinke DW (1998) The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J 16: 21–31 [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang Y, Qin G, Tsuge T, Sakaguchi N, Luo G, Sun K, Shi D, Aki S, Zheng N, et al. (2008) Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. Plant Cell 20: 1538–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Shi S, Zhang S, Xu P, Lai J, Liu Y, Yuan D, Wang Y, Du J, Yang C (2014) SUMO E3 ligase AtMMS21 is required for normal meiosis and gametophyte development in Arabidopsis. BMC Plant Biol 14: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois LM, Lima CD, Chua NH (2003) Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell 15: 1347–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis MJ, Wu J, Blobel G (1998) SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol 140: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Koshland D (1995) Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell 6: 793–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeester E, Melchior F (2008) Cell biology: SUMO. Nature 452: 709–711 [DOI] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM (2009) Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA 106: 5418–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoi J, Nakamura M, Nishida I (2006) Defects in CTP:PHOSPHORYLETHANOLAMINE CYTIDYLYLTRANSFERASE affect embryonic and postembryonic development in Arabidopsis. Plant Cell 18: 3370–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll C, von Lyncker L, Zimmermann S, Kägi C, Baumann N, Twell D, Grossniklaus U, Gross-Hardt R (2008) CLO/GFA1 and ATO are novel regulators of gametic cell fate in plants. Plant J 56: 913–921 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Ayaydin F, Kolli N, Tan SH, Anan T, Kametaka A, Azuma Y, Wilkinson KD, Dasso M (2006) SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. J Cell Biol 174: 939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtas G, Reeves PH, Fu YF, Bancroft I, Dean C, Coupland G (2003) A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell 15: 2308–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan JF, Ribeiro P (2006) The 19 S proteasomal subunit POH1 contributes to the regulation of c-Jun ubiquitination, stability, and subcellular localization. J Biol Chem 281: 16099–16107 [DOI] [PubMed] [Google Scholar]
- Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A (2005) The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell 9: 769–779 [DOI] [PubMed] [Google Scholar]
- Novatchkova M, Budhiraja R, Coupland G, Eisenhaber F, Bachmair A (2004) SUMO conjugation in plants. Planta 220: 1–8 [DOI] [PubMed] [Google Scholar]
- Okada T, Endo M, Singh MB, Bhalla PL (2005) Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J 44: 557–568 [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Park BS, Song JT, Seo HS (2011) Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat Commun 2: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Howden R, Twell D (1998) The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125: 3789–3799 [DOI] [PubMed] [Google Scholar]
- Pimienta E, Polito VS (1982) Ovule abortion in ‘Nonpareil’ almond (Prunus dulcis [Mill.] D.A. Webb). Am J Bot 69: 913–920 [Google Scholar]
- Reeves PH, Murtas G, Dash S, Coupland G (2002) early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development 129: 5349–5361 [DOI] [PubMed] [Google Scholar]
- Sampath K, Ephrussi A (2016) CncRNAs: RNAs with both coding and non-coding roles in development. Development 143: 1234–1241 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB (2000) The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco SA, Miller MJ, Kurepa J, Vierstra RD (2007) Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 145: 119–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler JS, Dejean A (2003) Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol 4: 690–699 [DOI] [PubMed] [Google Scholar]
- Shi H, Ren J, Xu H, Pan J, Hao X, Barlow LL, Dong W (2010) Upregulated expression of hITF in Crohn’s disease and screening of hITF interactant by a yeast two-hybrid system. Dig Dis Sci 55: 2929–2939 [DOI] [PubMed] [Google Scholar]
- Smith M, Bhaskar V, Fernandez J, Courey AJ (2004) Drosophila Ulp1, a nuclear pore-associated SUMO protease, prevents accumulation of cytoplasmic SUMO conjugates. J Biol Chem 279: 43805–43814 [DOI] [PubMed] [Google Scholar]
- Steffen JG, Kang IH, Macfarlane J, Drews GN (2007) Identification of genes expressed in the Arabidopsis female gametophyte. Plant J 51: 281–292 [DOI] [PubMed] [Google Scholar]
- ten Hove CA, Lu KJ, Weijers D (2015) Building a plant: cell fate specification in the early Arabidopsis embryo. Development 142: 420–430 [DOI] [PubMed] [Google Scholar]
- Thangasamy S, Guo CL, Chuang MH, Lai MH, Chen J, Jauh GY (2011) Rice SIZ1, a SUMO E3 ligase, controls spikelet fertility through regulation of anther dehiscence. New Phytol 189: 869–882 [DOI] [PubMed] [Google Scholar]
- Titiz O, Tambasco-Studart M, Warzych E, Apel K, Amrhein N, Laloi C, Fitzpatrick TB (2006) PDX1 is essential for vitamin B6 biosynthesis, development and stress tolerance in Arabidopsis. Plant J 48: 933–946 [DOI] [PubMed] [Google Scholar]
- Toujani W, Muñoz-Bertomeu J, Flores-Tornero M, Rosa-Téllez S, Anoman AD, Alseekh S, Fernie AR, Ros R (2013) Functional characterization of the plastidial 3-phosphoglycerate dehydrogenase family in Arabidopsis. Plant Physiol 163: 1164–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnyakova MA. (1991) Callose as an indicator of sterile ovules. Phytomorphology 41: 245–252 [Google Scholar]
- Wang X, Fan C, Zhang X, Zhu J, Fu YF (2013) BioVector, a flexible system for gene specific-expression in plants. BMC Plant Biol 13: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hou Y, Gu H, Kang D, Chen Z, Liu J, Qu LJ (2012) The Arabidopsis APC4 subunit of the anaphase-promoting complex/cyclosome (APC/C) is critical for both female gametogenesis and embryogenesis. Plant J 69: 227–240 [DOI] [PubMed] [Google Scholar]
- Xiao C, Chen F, Yu X, Lin C, Fu YF (2009) Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol Biol 71: 39–50 [DOI] [PubMed] [Google Scholar]
- Xu XM, Rose A, Muthuswamy S, Jeong SY, Venkatakrishnan S, Zhao Q, Meier I (2007) NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell 19: 1537–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhen M, Schein JE, Baillie DL, Candido EP (1996) An essential ubiquitin-conjugating enzyme with tissue and developmental specificity in the nematode Caenorhabditis elegans. EMBO J 15: 3229–3237 [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Schumaker KS, Guo Y (2012) Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc Natl Acad Sci USA 109: 12822–12827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG (2008) MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20: 1278–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang ZN (2008) Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J 55: 266–277 [DOI] [PubMed] [Google Scholar]