Corresponding lipidomic and transcriptomic data uncover the mechanism of P recycling and lipid class remodeling under P limitation in the alga Nannochloropsis oceanica.
Abstract
Molecular mechanisms of phosphorus (P) limitation are of great interest for understanding algal production in aquatic ecosystems. Previous studies point to P limitation-induced changes in lipid composition. As, in microalgae, the molecular mechanisms of this specific P stress adaptation remain unresolved, we reveal a detailed phospholipid-recycling scheme in Nannochloropsis oceanica and describe important P acquisition genes based on highly corresponding transcriptome and lipidome data. Initial responses to P limitation showed increased expression of genes involved in P uptake and an expansion of the P substrate spectrum based on purple acid phosphatases. Increase in P trafficking displayed a rearrangement between compartments by supplying P to the chloroplast and carbon to the cytosol for lipid synthesis. We propose a novel phospholipid-recycling scheme for algae that leads to the rapid reduction of phospholipids and synthesis of the P-free lipid classes. P mobilization through membrane lipid degradation is mediated mainly by two glycerophosphoryldiester phosphodiesterases and three patatin-like phospholipases A on the transcriptome level. To compensate for low phospholipids in exponential growth, N. oceanica synthesized sulfoquinovosyldiacylglycerol and diacylglyceroltrimethylhomoserine. In this study, it was shown that an N. oceanica strain has a unique repertoire of genes that facilitate P acquisition and the degradation of phospholipids compared with other stramenopiles. The novel phospholipid-recycling scheme opens new avenues for metabolic engineering of lipid composition in algae.
Phosphorus (P) availability is often a limiting factor in aquatic ecosystems (Van Mooy et al., 2009). In some microalgae, nutrient starvation limits cell division and leads to the accumulation of organic carbon (e.g. in the form of triacylglycerols [TAGs]). To cope with P limitation, the interplay of P recycling and changes in lipid classes is observed in the open sea and in culture studies of several stramenopiles and green algae (Van Mooy et al., 2009; Martin et al., 2011; Cañavate et al., 2017b). These studies showed a taxonomically diverse lipid response under P stress with unresolved questions related to the diversified mechanism behind the lipid responses. So far, only cursory insights into the lipid-remodeling process have been provided by either transcriptomic or lipidomic studies on P limitation (Riekhof et al., 2005; Cruz de Carvalho et al., 2016; Shemi et al., 2016; Cañavate et al., 2017b). To capture the taxonomically diverse response and niche partitioning, studies include ecological successful algae, such as Emiliania huxleyi and Thalassiosira pseudonana (Lessard et al., 2005; Van Mooy et al., 2009), and algae featuring high resilience and lipid content during low P, like Phaeodactylum tricornutum and Nannochloropsis spp. (Rodolfi et al., 2009; Yang et al., 2013; Mayers et al., 2014; Abida et al., 2015). During these studies, focus is often on changes of long-chain fatty acids (FAs) and their contribution to lipid classes in specific pathways such as TAG synthesis, leaving many unresolved questions. Therefore, it is still unclear by changes of lipid classes if there is a switch in lipid class synthesis or rather a shift of lipid classes (Brandsma, 2016). And what are their regulation patterns (Shemi et al., 2016)? Our study aims to answer parts of these questions through a combined analysis of lipidome and transcriptome data in the highly oleaginous alga Nannochloropsis oceanica. Our proposed novel phospholipid-recycling scheme in microalgae is shown to be a first response to P limitation, including pathways of phospholipid degradation and lipid biosynthesis. The acquired genetic information gives a better understanding of lipid metabolism, opening up possibilities for future genetic engineering approaches (Mühlroth et al., 2013).
P is an indispensable element for the structure and metabolic needs of the algal cell. It is required in biomolecules such as DNA, long-chain polyphosphates (polyPs), and phosphoproteins, all of which have long half-lives. In biomolecule systems that have shorter half-lives, P can be found in sugar molecules, RNAs, and cell membranes as phospholipids (Turner et al., 2005). Phosphate is used within the cell in the form of adenosine phosphates or NADPH. Thus, P limitation will affect a variety of energy-requiring processes, such as RNA transcription, the carbon cycle, and protein synthesis (Hu and Gao, 2003). Inorganic orthophosphate (Pi) is supplied and translocated into the cell by specific transporters, followed by incorporation into organic compounds or polyP storage through a plethora of biochemical reactions. Under low, external Pi, algae can assimilate dissolved organic phosphorus (DOP) by the enzymatic activity of alkaline phosphatases (APs) and purple acid phosphatases (PACPs; Theodorou et al., 1991; Lin et al., 2016; Cañavate et al., 2017a). The well-studied AP is a membrane-bound DOP-degrading enzyme found in a wide range of phytoplankton (Quisel et al., 1996; Lin et al., 2012). Very low or no enzymatic activity of AP was identified previously only in Nannochloropsis spp. and Picochlorum atomus, indicating unidentified enzymes for DOP uptake in this species (Lubian et al., 1992; Cañavate et al., 2017a). Additional P-scavenging enzymes are phosphodiesterases and nucleotidases that break phosphodiester bonds providing intracellular or extracellular P (Karl and Björkman, 2015; Dyhrman, 2016). The regulation of P acquisition and the involved enzymes or genes are currently studied intensively in Chlamydomonas reinhardtii and the higher plant Arabidopsis (Arabidopsis thaliana), as reviewed in previous work (Grossman and Aksoy, 2015; Karl and Björkman, 2015; Dyhrman, 2016).
Nannochloropsis is a genus of unicellular Eustigmatophyceae found in various habitats (Beacham et al., 2014). Currently, draft whole-genome sequences of N. oceanica CCMP1779 (Vieler et al., 2012) and Nannochloropsis gaditana CCMP526 (Radakovits et al., 2012) have been assembled and several genetic engineering tools established. Under P stress, Nannochloropsis spp. have been shown to accumulate TAGs, increasing their lipid content up to 50% of cell dry weight (Rodolfi et al., 2009; Mayers et al., 2014; Cañavate et al., 2017b). Low P content triggering TAG increase also is seen in the model algae C. reinhardtii and P. tricornutum, additionally replacing extraplastidial phosphatidylcholine (PC) by taxonomically specific betaine lipid (BL) classes such as diacylglyceroltrimethylhomoserine (DGTS; Iwai et al., 2014; Abida et al., 2015). This phenomenon of BL replacing PC under low P was shown to be common in freshwater and marine algae (Khozin-Goldberg and Cohen, 2006; Van Mooy et al., 2009; Martin et al., 2011; Iwai et al., 2015; Cañavate et al., 2017b). Recent studies comparing lipid class and P-uptake profiles of eight algae at low Pi showed replacement of phospholipid by BL with concomitant high P uptake in the green alga Tetraselmis suesica and the stramenopiles P. tricornutum, N. gaditana, and P. atomus (Cañavate et al., 2017a, 2017b). Phospholipid replacement and high P uptake under P stress indicate that these algae have a mechanism vital in temporal P limitation. Other stramenopiles were found to have no change in BL presence under low P but maximal P uptake, while other species had consistent (already in P-replete conditions) low phospholipid and high BL content, requiring little P uptake and being adapted to constant low P (Cañavate et al., 2017a, 2017b). Also seen was replacement of the essential lipid phosphatidylglycerol (PG) by sulfoquinovosyldiacylglycerol (SQDG) in C. reinhardtii to maintain the thylakoid membrane under P limitation (Iwai et al., 2014). Both SQDG and PG are important for the activity of PSII. Furthermore, land plants, green algae, and the freshwater Eustigmatophyceae Monodus subterraneus showed increased content of the photosynthetic membrane lipid digalactosyldiacylglycerol (DGDG) under P limitation, but the role of DGDG in this setup is unknown (Khozin-Goldberg and Cohen, 2006; Nakamura, 2013; Cañavate et al., 2017b). However, a lipid class study of the eight diverse species could not confirm a significant increase of SQDG or DGDG in any species except in P. atomus, making their role in lipid remodeling questionable (Cañavate et al., 2017b). Therefore, increased focus on lipid class changes on the transcriptome and proteome level will elucidate the importance of lipid classes in remodeling processes.
To our knowledge, P-induced changes in gene expression, with emphasis on lipid replacement, has not been investigated in Nannochloropsis spp. or other marine phytoplankton. In higher plants, however, the metabolic switch from phospholipids to galactolipids (PC is replaced by DGDG) and SQDG is well established (Nakamura, 2013). In this lipid class switch, P-containing head groups are hydrolyzed by phospholipase C (PLC) or phospholipase D (PLD) to diacylglycerol (DAG) or phosphatidic acid (PA), which then are converted into DGDG, monogalactosyldiacylglycerol (MGDG), and SQDG. Another pathway in Arabidopsis involves the activity of type A phospholipases (PLAs) and glycerophosphoryldiester phosphodiesterases (GDPDs), resulting in the release of free acyl-CoAs and glycerol-3-phosphate (G3P; Cheng et al., 2011; Nakamura, 2013). Acyl-CoA and G3P are precursors for the synthesis of P-free lipids in plastidial or cytosolic lipid synthesis (Mus et al., 2013; Li et al., 2014).
In this study, we focused on the lipid-remodeling mechanisms at the transcriptome level during the transition to P limitation. We report substantial agreement between transcriptomic and lipidomic data, as well as phenotypical changes under P limitation, in N. oceanica CCMP1779. Our results improved the understanding of adaptive P-acquiring mechanisms and a phospholipid-recycling scheme. Phospholipid-recycling genes, such as patatin-like PLAs and GDPDs, were assigned, showing expression as an immediate response to low Pi concentrations in exponential growth. We show that unique P transporters, as well as triose phosphate transporters (TPTs), play an important role for the P-carbon dynamic between cell compartments. Furthermore, we identify PACPs involved in DOP degradation and a lack of ubiquitous alkaline phosphatase. Former hypotheses for this drastic shift in lipid classes, such as DGTS and SQDG synthesis, due to P limitation were confirmed on both the transcriptomic and lipidomic levels. The phospholipid-recycling scheme reported here illustrates the flexible metabolism of Nannochloropsis spp. under P limitation.
RESULTS
Our experiments were designed to capture genes involved in P acquisition and the dynamic interplay between the phospholipid degradation and lipid synthesis in N. oceanica cells under transition to P limitation (Fig. 1). In order to detect the dynamic response more holistically, samples were taken for transcriptomic (DNA microarray), lipidomic, and physiological analysis during five time points of a 96-h cultivation period covering the transition from a P-replete to a P-depleted situation. Using tandem mass spectrometry (MS/MS) structural characterization, we were able to discern different molecular species of the 10 major glycerolipid classes in N. oceanica (see Fig. 3 below). Our analysis identified three phosphatidylinositol (PI) and five PG molecular species. Phospholipids PC and phosphatidylethanolamine (PE) exhibited nine and 11 identified species, respectively. The DAG lipid class contained six molecular species, whereas TAG showed 21 molecular species. DGTS was present in 16 different molecular species. Among lipids that are typically localized in plastids, our analysis distinguished nine MGDG, seven DGDG, and eight SQDG molecular species (Supplemental Tables S6–S9).
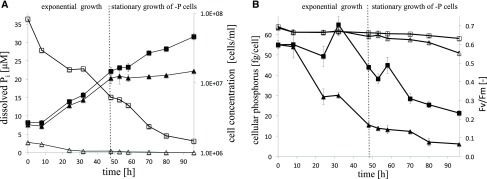
Figure 1.
Growth and physiological parameters of N. oceanica in −P and +P cells. A, Time analysis of cell count of +P (black squares) and −P (black triangles) cells and dissolved Pi concentration in +P (white squares) and −P (white triangles) cultures. B, Photosynthetic activity (Fv/Fm) of +P (white squares) and −P (white triangles) cells and cellular P of +P (black squares) and −P (black triangles) cells as a function over time. Average values ± sd of three biological and two technical replicates from the first independent experiment are shown.
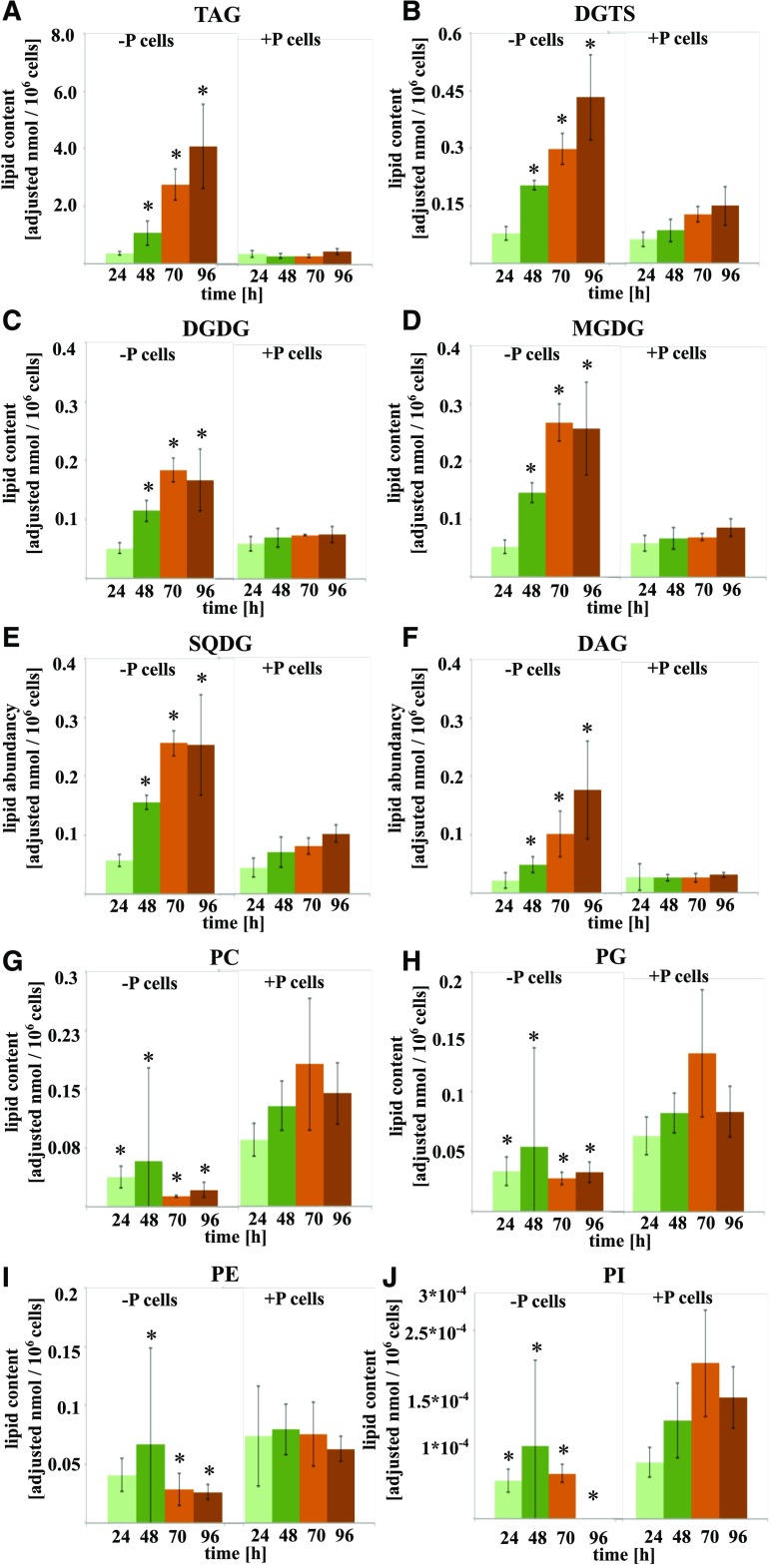
Figure 3.
Lipid class content per cell as a function of time in N. oceanica of −P and +P cells. Lipid class classification is based on linear trap quadrupole identification and liquid chromatography-MS/MS by target lipid profiling. A, TAG. B, DGTS. C, DGDG. D, MGDG. E, SQDG. F, DAG. G, PC. H, PG. I, PE. J, PI. Asterisks denote significant (P < 0.05) phosphate effects in –P cells compared with +P cells at each time point. Average values ± se of three biological and two technical replicates from the first independent experiment are shown.
Early Regulated Transcripts Related to P Acquisition
During the 96 h of cultivation, the −P culture (cells grown under P-limited conditions) grew exponentially until 48 h, whereas the +P cultures (cells grown under P-replete conditions) continued exponential growth until 92 h, when the PO43− concentration was reduced to less than 3.6 μm (Fig. 1A). Therefore, exponential and stationary growth refer only to the −P cultures in the following. The exponential growth of −P cells ended after 48 h at 0.4 μm PO43− and a cell concentration of 1.17 × 107 cells mL−1. In stationary growth, the PO43− concentration was lower than the detection limit (∼ 0.3 μm; Fig. 1A). The cellular P content, representing all cellular P-containing compounds, declined gradually from 0.055 ± 0.002 to 0.016 ± 0.001 pg cell−1 until 48 h, while +P cells showed only a minor reduction to 0.044 ± 0.002 pg cell−1 (Fig. 1B). The cellular P content per dry weight in −P cells also showed a drastic decline due to the changed dynamic in carbon-P ratio during the transition to P limitation (Supplemental Fig. S1). At 0 h, the carbon:P ratio was 63:1, but it increased to 1,193:1 at 96 h (Supplemental Table S1).
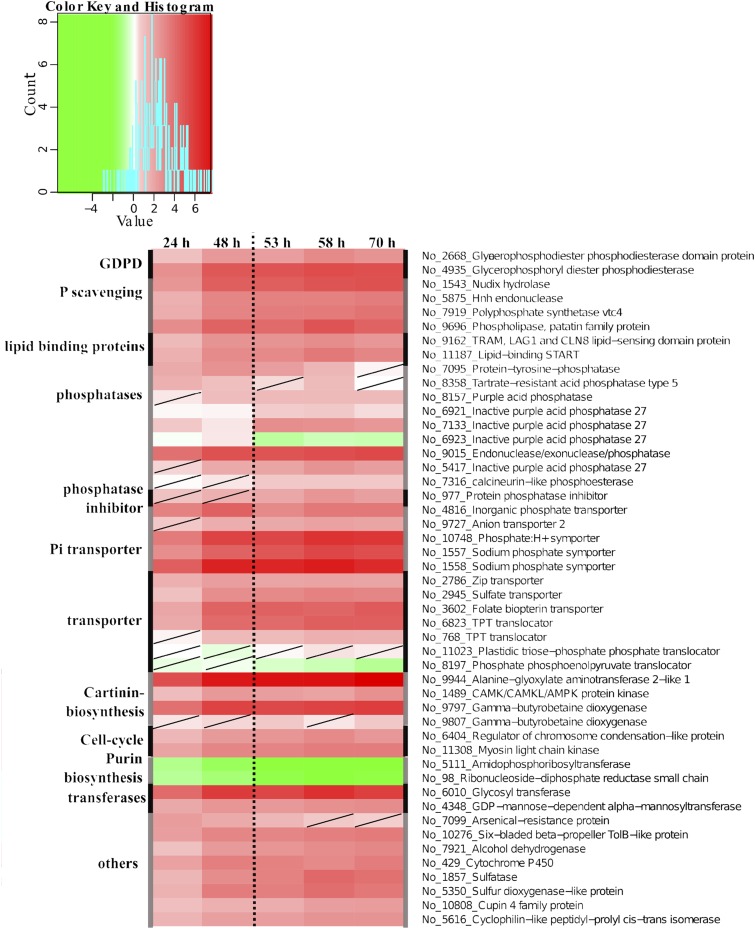
Global transcriptional regulation analyzed with oligonucleotide DNA microarrays showed significant (adjusted P/false discovery rate of 0.01) regulation of 60 genes after 24 h of P limitation. The initial response at 24 h showed that P limitation led to the up-regulation of genes related to Pi uptake (Fig. 2). Genes encoding for three putative Pi transporters (No_1558/No_1557, No_ 10748, and No_4816) and a sodium-phosphate symporter (No_9727) were up-regulated 2- to 16-fold. Additionally, two putative vacuolar transporter chaperone (VTC; No_663 and No_7919) transcripts showed 3-fold up-regulation at 24 and 48 h, respectively, compared with +P cells. In the exponential growth of −P cells, putative transcripts of PACPs (No_8358, No_7133, and No_9015) showed strong up-regulation out of nine identified PACPs (Fig. 2). These results indicate that P was low enough to trigger a cellular response already after 24 h of cultivation and significantly induced genes related to P acquisition and transportation.
Figure 2.
Transcriptional regulation of early responses and transporters under P limitation in N. oceanica. Regulation patterns (colored squares ranging from green to red) are derived from microarray data of putative genes encoding enzymes (black) that are involved in immediate responses to P limitation. All time points of the experiment are shown (24, 48, 53, 58, and 70 h). The scale at top represents gene expression ratio values, log2 transformed with P > 0.01 (nonsignificant regulation [diagonal line in a square]). No_ numbers indicate NannoCCMP1779 gene identifiers.
Early Phospholipid Degradation
Lipidomic data showed that the molecular species PE, PG, PI, and PC were ∼2 times less abundant in −P cells than in +P cells at 24 h (Fig. 3, G–J). The phospholipids PC and PE continued declining until 96 h, with reductions of 3.9 and 2.8 times, respectively, compared with +P cells at 24 h. The PI content remained constant from 24 to 48 h and dropped under the detection limit at 96 h. After the initial reduction, PG levels were stable in −P cells.
Changes in the expression of genes associated with phospholipid degradation in −P cells (Fig. 4A) revealed that 11 transcripts encoding PLAs of type A, D, or C were differently regulated compared with +P cells. The transcript of a patatin-like PLA (PLA_9696) was clearly up-regulated at 24 h. At later time points, two PLAs (PLA_4281 and PLA_11320) also were up-regulated. Together with the PLAs, two transcripts encoding for GDPD (GDPD_4935 and GDPD_2668) were up-regulated 2- to 4-fold throughout the experiment (Figs. 2 and 4A). Up-regulation of GDPD_4935 was confirmed by quantitative real-time (qRT)-PCR (Supplemental Table S2).
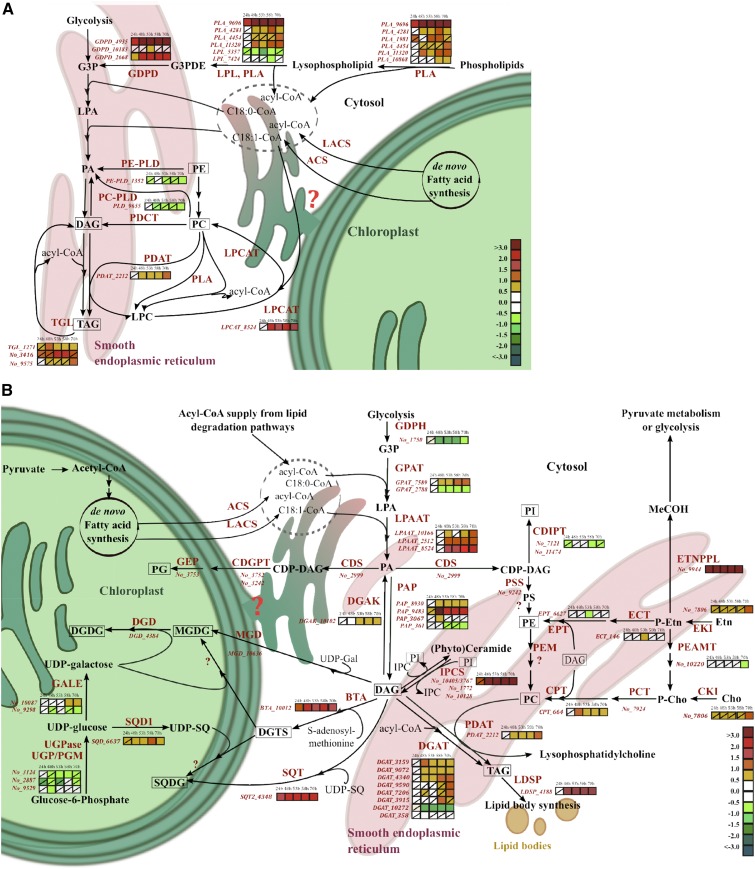
Figure 4.
Model of the phospholipid-recycling scheme in N. oceanica under P limitation. The gene expression (colored squares) derived from microarray data of putative genes encoding for enzymes (italic red) involved in phospholipid degradation (A) and lipid biosynthesis (B) is shown at different experimental time points (24, 48, 53, 58, and 70 h). Nonsignificant regulation was considered at P > 0.01 (diagonal line in a square). Gene identifiers are shown without regulation pattern if they were not significantly differently regulated at any time point. The scale on the bottom right represents gene expression ratio values, log2 transformed. No_ numbers indicate NannoCCMP1779 gene identifiers. Some putative genes encoding enzymes involved in the presented pathways are not annotated in the genome or are unknown (red question marks). Multiple enzyme localizations (cytosolic, plastidial [green], endoplasmic reticulum [red]) are not considered; therefore, the prokaryotic pathway of DAG formation is not visualized. Lipids identified by lipidomics are framed. Enzyme abbreviations are as follows: ACS, acyl-CoA synthetase; BTA, S-adenosyl-Met (AdoMet); DAG, 3-amino-3-carboxypropyltransferase; CDGPT, CDP-diacylglycerol-glycerol 3-phosphatidyltransferase; CDIPT, CDP-diacylglycerol-inositol 3-phosphatidyltransferase; CDS, cytidine diphosphate (CDP)-diacylglycerol synthase; Cho, choline; CKI, choline kinase; CPT, cholinephosphotransferase; DGD, digalactosyl diacylglycerol synthase; DGAK, diacylglyceride kinase; DGAT, acyl-CoA:diacylglycerol acyltransferase; ECT, ethanolamine phosphate cytidylyltransferase; EKI, ethanolamine kinase; EPT, ethanolaminephosphotransferase; Etn, ethanolamine; ETNPPL, phosphoethanolamine phospholyase; GALE, UDP-Glc 4-epimerase; GDPD, glycerophosphoryldiester phosphodiesterase; GEP, phosphatidylglycerophosphatase; GLA, α-galactosidase; GPAT, acyl-CoA:glycerol-3-phosphate acyltransferase; GPDH, glycerol-3-phosphate dehydrogenase; IPCS, inositol phosphorylceramide synthase; LACS, long-chain acyl-CoA synthetase; LDSP, lipid droplet surface protein; LPAAT, acyl-CoA:lysophosphatidic acyltransferase; LPCAT, lysophosphatidylcholine acyltransferase; LPL, lysophospholipase; MGD, monogalactosyldiacylglycerol synthase; PAP, phosphatidic acid phosphatase; P-Cho, phosphocholine; PCT, choline phosphate cytidyltransferase; PDAT, phospholipid/diacylglycerol acyltransferase; PDCT, phosphocholine/diacylglycerol transferase; P-Etn, phosphoethanolamine; PLA, phospholipase A; PLC, phospholipase C; PLD, phospholipase D; PEAMT, phosphoethanolamine N-methyltransferase; PEM, phosphatidylethanolamine N-methyltransferase; PSD, phosphatidylserine decarboxylase; PSS, phosphatidylserine synthase; SQD1, UDP-sulfoquinovose synthase; SQT, sulfolipid sulfoquinovosyldiacylglycerol synthase; UGPase, UDP-Glc-pyrophosphorylase; UGP/PGM, UDP-Glc pyrophosphorylase/phosphoglucomutase. Compound abbreviations are as follows: G3P, glycerol-3-phosphate; G3PDE, glycerol-3-phosphodiester (glycerophosphocholine, glycerophosphoethanolamine, glycerophosphoglycerol, etc.); IPC, inositolphosphorylceramide; LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; DAG, diacylglyceride; DGDG, digalactosyldiacylglycerol; DGTS, diacylglyceroltrimethylhomoserine; MeCOH, acetaldehyde; MGDG, monogalactosyldiacylglycerol; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; SQDG, sulfoquinovosyldiacylglycerol; TAG, triacylglyceride; UPD-Gal, UDP-galactose; UDP-SQ, UDP-sulfoquinovose.
Genes related to the Lands cycle, PLA2/patatin-like PLA (PLA_9696, PLA_4281, and PLA_11320) and one of three lysophosphatidylcholine acyltransferase (LPCAT; LPCAT_8524), were up-regulated (Fig. 4A). However, two PLD genes (PE-PLD and PC-PLD) and two lysophospholipase-encoding genes were suppressed in stationary growth in −P cells (Fig. 4A). Also, the putative phospholipid/diacylglycerol acyltransferase (PDAT; PDAT_2212) transcript showed significant up-regulation after 48 h in −P cells.
The phospholipid biosynthesis pathway is initiated by kinases coupled with several transferases that catalyze phosphorylation and the transfer of DAG to ethanolamine or choline, forming PE or PC, respectively. A putative choline/ethanolamine kinase (No_7806) showed up-regulation during the whole experiment but significantly only at 96 h in −P cells, while the putative cholinephosphotransferase (CPT; CPT_664) was up-regulated after 48 h during P limitation (Fig. 4B). These results indicate that phospholipids are synthesized continuously under transition to P limitation. Concomitant to phospholipid synthesis, phospholipid degradation and acyl-editing processes are boosted by increased mRNA levels of patatin-like PLAs, GDPDs, PDAT, and LPCAT.
Regulation of P-Free Lipids
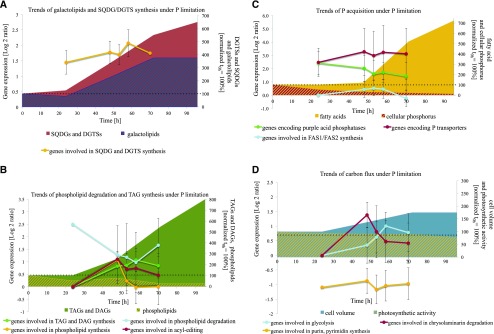
While the content of phospholipid classes showed a significant change already at 24 h in −P cells compared with +P cells, the other lipid classes exhibited changes from 48 h onward. A steady increase of TAG was observed, with a final 10.9 times increase in −P cells, shown to be the most abundant lipid, after 96 h of cultivation compared with 24-h +P cells. The dramatic TAG increase per cell is due to the cell size increase in −P cells. Calculation of the content of various lipid classes per dry weight showed divergence to those calculated per cell, due to the 60% cell volume expansion in stationary growth in −P cells, which was detected first at 70 h (Supplemental Fig. S2). It is worth noting that the TAG increase per dry weight showed only a 3.4 times increase in −P cells and no increase in +P cells (Supplemental Fig. S3A).
The content of DAGs increased significantly (6.6 times) in −P cells over the same period (Fig. 3A). The second most abundant lipid was DGTS, which increased (3.3 times at 48 h) and reached a 7 times change at 96 h compared with the content in 24-h +P cells (Fig. 3B). The galactolipids, MGDG and DGDG, showed 2.5 and 1.9 times increases at 48 h, respectively, and exhibited 4.5 and 3.1 times increases at 70 h in −P cells compared with the 24-h +P cells (Fig. 3, C and D). Comparing +P and −P cells at 70 h, P limitation increased SQDG level per cell 2.5 times. A significant increase in SQDG molecular species was observed in −P cells from 48 to 70 h, leading to a total 5.8 times change compared with 24-h +P cells (Fig. 3C).
The mRNA levels of the SQDG synthesis pathway (SQD1_6637 and SQT2_4348) and the putative gene BTA_10012 encoding an S-adenosyl-Met:DAG 3-amino-3-carboxypropyltransferase (BTA) showed constant and significant up-regulation from 24 h onward in −P cells (Fig. 4B). The results of qRT-PCR confirmed the increased expression level of BTA_10012 (Supplemental Table S3). The protein domain structure of the BTA_10012 gene in N. oceanica, and many other stramenopiles, showed a fusion of two domains (Supplemental Fig. S4), with a domain of unidentified function (DUF3419-domain)/BtaA-like located at the N terminus and a methyltransferase domain (MT)/BtaB-like at the C terminus.
Gene models used to design microarray probes for genes encoding synthases (MGD_10636 and DGD_4384) for MGDG and DGDG synthesis were deemed incorrect due to incorrect gene models and fused open reading frames in the draft sequences. During the P-limitation experiment, a novel, not described gene coding for inositol-phosphorylceramide synthase (IPCS) showed strong up-regulation from 48 h onward (Fig. 4B; IPCS_10405/3767 regulation also was confirmed by qRT-PCR; Supplemental Table S3). Increases in SQDG and DGTS were observed on both the transcriptome and lipidome levels at 24 h and later time points, and the BTA gene involved in DGTS synthesis was identified based on similarities to BTA1Cr from C. reinhardtii. Additionally, in stationary growth, the lipid classes TAGs and DAGs and galactolipid increased, which also was reflected by genes involved in the synthesis of these lipids at the transcriptome level.
Carbon and P Flux
During P limitation, the reduced P-carbon ratio dynamic leads to the stop of cell division and carbon accumulation in the storage compartment in the form of TAGs (Supplemental Table S1). Therefore, the down-regulation of genes linked to processes involved in purin and pyrimidine synthesis, generation of ribosomal proteins, and DNA replication was pronounced at the 48-h time point and later when cell growth was reduced in −P cells (Fig. 5D; Supplemental Fig. S5).
Figure 5.
Trends of cellular changes in −P cells of N. oceanica as a function of time. A, Changes of genes derived from microarray data involved in DGTS and SQDG synthesis (yellow line, one S-adenosyl-Met [AdoMet]:DAG 3-amino-3-carboxypropyltransferase [BTA], one UDP-sulfoquinovose synthase, one sulfolipid sulfoquinovosyldiacylglycerol synthase), shown with changes of galactolipid (blue striped area) and SQDG/DGTS change (red area) normalized to time point 0 h. B, Changes of TAGs and DAGs (green area), phospholipids (yellow striped area) normalized to time point 0 h, and the expression of genes involved in phospholipid degradation (blue line; three glycerophosphoryl diester phosphodiesterases, two patatin-like phospholipases A, four phospholipases A, two lysophospholipases), acyl editing (red line; one lysophosphatidylglycerol acyltransferase, one cholinephosphotransferase, one phospholipid/diacylglycerol acyltransferase, two phospholipases D), TAG and DAG biosynthesis (green line; two glycerol-3-phosphate acyltransferases, three lysophosphatidylglycerol acyltransferases, four phosphoesterase PA-phosphatases, eight diacylglycerol acyltransferases, one diacylglycerol kinase, one lipid droplet surface protein), and phospholipid biosynthesis (yellow line; one kinase, one ethanolamine phosphate cytidylyltransferase and phosphotransferase, one cholinephosphotransferase, one phosphoethanolamine N-methyltransferase, one CDP-diacylglycerol-inositol 3-phosphatidyltransferase). C, Changes of FA abundance (yellow area) and cellular P (red striped area) normalized to time point 0 h and of the expression of genes involved in P transporters (red line; five Pi transporters, two triose phosphate translocators, two vacuolar chaperone transporters), 10 purple acid phosphatases (green line), and FAS1/FAS2 synthesis (blue line; three acetyl-CoA carboxylases, two ketoacyl-synthases, three acyl-CoA synthetases, two long-chain acyl-synthetases, one thioesterase, two polyketide synthases type II, one peptide synthase, one hydroxyacyl-CoA synthase, six desaturases, two elongases). D, Changes of cell volume (blue area) and photosynthetic activity (yellow striped area) normalized to time point 0 h and the expression of genes involved in glycolysis (blue line; two Fru-2,6-bisphosphatases, two phosphoglycerate kinases, one glycerol-3-phosphate dehydrogenase, one dihydroxy-butanonkinase, one glyceraldehyde-3-phosphate dehydrogenase, four phosphoglycerate mutases, one enolase, three pyruvate kinases), chrysolaminarin degradation (red line; 11 endoglucanases, two exoglucanases), and purin, pyrimidine synthesis (yellow line). The presentation shows average log2 ratios of significantly expressed genes (P < 0.01) involved in metabolic clusters and changes of lipid classes (adjusted nmol 106 cells−1) or physiological parameters, such as cell volume (μm3) and FAs (pg cell−1) normalized to 0 h as 100%. For gene identifiers used for the calculation of average gene regulation and means, see Supplemental Excel Sheet S1. Average values ± se of the regulation of genes (P > 0.01 at each specific time point) involved in the specific pathway/catalysis are shown.
During the experiment, genes coding for proteins predicted to be involved in CO2 fixation and the relocalization of carbon via the glycolysis pathway were up-regulated slightly in stationary growth. The predicted cytosolic and plastid glycolysis genes, which convert Glc into phosphoenolpyruvate by phosphoglycerate kinase (No_3272 and No_4644), phosphoglycerate mutase (No_2186 and No_4572), and enolase (No_11908), showed significant up-regulation in −P cells from 48 h onward (Fig. 5D; Supplemental Excel Sheet S1). Also, two putative TPTs (No_6823 and No_768) showed significant up-regulation after 24 or 48 h in −P cells. The photosynthetic quantum yield started to differ between −P cells and +P cells at 70 h, where −P and +P cells had photosynthetic activities of 0.63 ± 0.01 and 0.66 ± 0.01, respectively (Fig. 1B). These data show that the cells’ reduced photosynthetic activity also was reflected on the transcriptome level with reduced expression of mRNA for proteins that are involved in the Calvin cycle and with a shift toward the accumulation of lipids, FAs, and carbon, resulting in a cell volume increase (Supplemental Fig. S2; Supplemental Excel Sheet S1; Supplemental Tables S4 and S5).
DISCUSSION
Whether lipid classes in algae are remodeled during the transition to P limitation or if there is a switch in the synthesis of lipid classes (Brandsma, 2016), and which genes trigger these responses (Shemi et al., 2016), are unresolved questions. Our study addresses these questions and provides a comprehensive scheme for phospholipid-recycling by employing complementary lipidomic and transcriptomic data. The novel assigned phospholipid-recycling scheme features the active breakdown of phospholipids, acyl-editing mechanisms, and the synthesis of P-free lipid classes. The phospholipid degradation occurs in both exponential and stationary growth of −P cells in N. oceanica CCMP1779 (Figs. 4A and 5A). Furthermore, our study described adaptive P-acquisition and -scavenging mechanisms that deviate from other stramenopiles due to their evolutionary history (Parker et al., 2008).
P Acquisition
Metabolite and mRNA transcript profiles during the onset of P limitation suggest a strong increase in the expression of the machinery that mediates P acquisition (Figs. 2 and 5B). The drastic physiological decline in cellular P in −P cells (down to ∼50% of P content per cell after 24 h compared with +P cells) reflects the reduced P uptake and availability. Studies of plankton communities showed that 50% of the P uptake in marine cyanobacteria was accounted for by the synthesis of RNA and, dependent on the marine microbe, 5% to 20% for phospholipid synthesis (Van Mooy and Devol, 2008; Van Mooy et al., 2008). Therefore, it is likely that the internal P in the form of the RNA pool is reduced immediately and has a higher turnover under low P availability, leading to a reduced growth rate and, finally, to stop of cell division (Dyhrman, 2016). This is reflected in the transcriptomic data, where genes coupled to the synthesis of RNA and cell division are down-regulated after 48 h in −P cells (Supplemental Fig. S5). Further P stress-specific response genes, such as Pi transporters and PACPs, were up-regulated in N. oceanica in the exponential growth in −P cells (Fig. 5C). The specific P stress response genes were shown to differ notably from other stramenopile P acquisition genes, as Nannochloropsis spp. lack APs and P acquisition genes have bacterial or green algal origins (Cruz de Carvalho et al., 2016; Lin et al., 2016).
Novel Assigned P Transporter and VTCs
The P transporters and VTCs of N. oceanica have not been described previously. Phylogenetic studies indicate that these transporters are sometimes not related to transporters in other stramenopiles but rather have a green algae origin (Supplemental Fig. S6). The amount of responsive P transporter to low Pi concentration varies significantly between algae species. While P. tricornutum cells up-regulated 12 of 24 putative P transporters, N. oceanica exhibited four of six identified P transporters up-regulated under low Pi concentration (Cruz de Carvalho et al., 2016). Also, C. reinhardtii exhibited 14 putative Pi transporters, indicating either gene loss in Nannochloropsis spp. or gene expansion in the other studied algae (Grossman and Aksoy, 2015; Cruz de Carvalho et al., 2016). The high-affinity P transporters encoded by No_1557/1558 show closer relation to Pho4-type P transporters in green algae, plants, and mosses, while five Na+/Pi cotransporters of P. tricornutum share sequence homology to mammalian renal transporters (Supplemental Fig. S6; Cruz de Carvalho et al., 2016). Closer relation to green algae, plants, and mosses also holds true for the phylogenetic affiliation of the VTC1-type protein (No_7919), which likely is involved in trafficking P to a potential polyP compartment (Aksoy et al., 2014). An indication that such polyP storage exists comes from the presence of vacuolar phosphatases and No_7919, which have a similar pattern of regulation to genes involved in the electron-dense acidocalcisomes-like compartment in the green alga C. reinhardtii (Docampo and Moreno, 2011). As in C. reinhardtii, this compartment may be a key component for polyP storage in Nannochloropsis spp. Another VTC transcript encoded by No_663 has a high degree of similarity to bacterial proteins containing an SPX domain that is known to be involved in regulating Pi homeostasis (Secco et al., 2012). A distinct role of the VTC proteins in polyP metabolism during P limitation is suggested not only in N. oceanica but also in the diatoms P. tricornutum and T. pseudonana (Dyhrman et al., 2012; Cruz de Carvalho et al., 2016). However, further investigations are needed for in-depth understanding of P uptake and storage in algae.
Nannochloropsis spp.-Specific Response to Organic Phosphate Uptake
Typically, aquatic microbes are equipped with one or more specific P-induced enzymes to utilize DOP (e.g. phosphoester or phosphonat) during P limitation (Dyhrman, 2016). The most widespread and studied of those DOP-degrading enzymes is the membrane-bound AP, being available mostly in multiple gene copy numbers in phytoplankton genomes (Quisel et al., 1996; Lin et al., 2012). In our study, no gene transcripts of such APs were identified, making Nannochloropsis spp. interesting study organisms regarding P acquisition and P niche partitioning. The absence of the AP gene in the genome of N. oceanica confirms earlier studies that reported a lack of or very low membrane-bound AP enzyme activities in N. oceanica and N. gaditana (Lubian et al., 1992; Cañavate et al., 2017a). The absence of AP genes in some Nannochloropsis spp. may originate from a species-specific gene-loss event. However, the mentioned enzymatic studies verified the degradation of cellular DOP as an alternative P source in Nannochloropsis spp. by intracellular and extracellular phosphatase activity within a pH range of 3 to 8.6 and a maximum of pH 5.5 (Lubian et al., 1992; Cañavate et al., 2017a). During the initial P limitation, three putative PACP genes showed strong up-regulation, suggesting that the corresponding gene products are responsible for degrading DOP compounds (Fig. 2). Few PACPs are common in N. oceanica and P. tricornutum. PAPCs such as the PACP-like phosphoesterase (No_9483) have an unusual origin in N. oceanica and are more closely related to bacterial proteins and suggest horizontal gene transfer.
The enzymatic activity of multiple APs has been a reliable indicator of P stress in plankton (Lin et al., 2016). In previous studies, surface-associated APs of low-P-tolerant species have been shown to have an important role in circumventing heterotrophic regeneration of DOP in seawater (with typical pH of 8) under P limitation (Kuenzler and Perras, 1965; Lin et al., 2016). Nannochloropsis spp. do not comprise any AP, still grow effectively in low-Pi seawater, and have a P uptake probably due to PACP activities (Mayers et al., 2014; Cañavate et al., 2017a). Why the evolutionary shift from membrane-bound AP to extracellular PACPs occurred with a pH maximum of 5.5 and a very weak activity at pH 8.6 remains unresolved and merits further investigation (Lubian et al., 1992). PACPs are the only phosphomonoester-degrading enzymes in N. oceanica, indicating an inherent difference in the mechanism of P acquisition compared with other phytoplankton species (Kuenzler and Perras, 1965; Dyhrman, 2016).
Phospholipid-Recycling Scheme
Our data suggest a novel assigned phospholipid-recycling scheme that is triggered by P limitation in N. oceanica. In this scheme, phospholipids are broken down to G3P, FAs, and DAG. The products are used to synthesize mainly DGTS and SQDG in exponential growth and, later, for the synthesis of TAG and galactolipid in stationary growth (Fig. 5, A and B). This scheme is consistent with lipidomic and transcriptomic data (Figs. 3 and 4). Characteristic of this scheme is the expression of genes encoding patatin-like PLAs, GDPDs, and genes encoding enzymes of the acyl-editing mechanism to degrade phospholipids as well as genes involved in the Kennedy pathway and P-free lipid synthesis, such as SQD1_6637, SQT2_4348, and BTA_10012 (Fig. 4B).
Patatin-Like PLAs: Breakdown Phospholipids
Significant increases in mRNA levels of genes involved in phospholipid degradation were seen in −P cells at 24 h. Two novel GDPDs (GDPD_4935 and GDPD_2668) and a patatin-like PLA (PLA_9696), later followed by PLA_4281 and PLA_11320, strongly point to the formation of G3P from phospholipids (Fig. 4A). Based on phylogenetic analysis, the most up-regulated lipases had no good ortholog in plants or diatoms, apart from the PLA_10868 transcript coding for a PLA2 Atgl-type gene from Arabidopsis. The single-copy genes (PLA_9696, PLA_4281, and PLA_11320) encode proteins of the multifunctional and diverse patatin-like PLA subtype (Fig. 4A). While PLA1 and PLA2 may hydrolyze the FA ester bond at position sn-1 or sn-2 of the glycerol backbone, respectively, the patatin-like PLA shows activity toward both positions (Chen et al., 2011). Therefore, the up-regulated patatin-like PLAs may consecutively deacylate the phospholipid and form glycerol-3-phosphodiester, which is catalyzed to G3P by GDPD. However, due to the complexity of patatin-like PLAs and lipases in general, enzymatic studies are required for elucidation (Scherer et al., 2010). In diatoms (P. tricornutum and T. pseudonana) and plants, several putative PLCs and PLDs show strong up-regulation on the mRNA level during P limitation, indicating contributions to phospholipid degradation and regulation on the transcript level (Dyhrman et al., 2012; Nakamura, 2013; Cruz de Carvalho et al., 2016). Different from plants and diatoms, the described PLCs and PLDs had a trend of down-regulation in N. oceanica during P limitation. This either indicates that the description of the phospholipid-recycling scheme is not completely deciphered (lacking described lipases in N. oceanica) or shows the taxonomically diverse degradation of phospholipids within stramenopiles. The latter hypothesis is more likely, given that a computational study showed algae possessing just a few putative eukaryote-type PLDs with the catalytic motif HKD, compared with higher plants containing 12 to 23 PLDs per genome (Beligni et al., 2015). Stramenopiles are shown to have two or three HKD PLDs, with the exception of Nannochloropsis spp., which contain only a single protein of the rare abundant PXPH-like PLDs (No_9655), of which the PLD activity has not been proven (Beligni et al., 2015). Based on the down-regulation of PLD_9655 and PE-PLD_3352 mRNA levels in our experiment, it seems as if PLD activity may not be acquired and PA is generated mainly via up-regulation of the acyl-CoA:lysophosphatide acyltransferase (LPAAT; LPAAT_10166, LPAAT_2512, and LPAAT_8524) mRNA level under low Pi in N. oceanica (Fig. 4).
Up-regulated mRNA transcripts of GDPDs also have been confirmed in diatoms under P limitation (Dyhrman et al., 2012). Protein sequence analyses of the highly expressed GDPDs predicted one cytosolic location (GDPD_2668), pointing to a cytosolic supply of G3P and a potential leader sequence for secretion (GDPG_4935). Secreted GDPDs also might contribute to external DOP degradation (Dyhrman, 2016). It is likely that the formed G3P by GDPDs was used as a precursor for the de novo Kennedy pathway. This hypothesis is supported by the down-regulation of genes related to G3P utilization and formation, such as glycerol-3-phosphate dehydrogenase (No_1750; Fig. 4B). During exponential growth, PLAs and GDPDs appear mainly responsible for phospholipid degradation and supplying precursors for the de novo Kennedy pathway. Even though there are numerous unidentified lipase candidates in Nannochloropsis spp., three patatin-like PLAs have been shown to play an important role in early phospholipid degradation, consistent with the recycling scheme in P-limited cells.
Acyl Editing of Newly Synthesized Phospholipids
In the stationary growth of −P cells, acyl-editing mechanisms showed increased activity of degrading PC. The genes PDAT (PDAT_2212) and LPCAT (LPCAT_8524) were up-regulated, giving further strength to the lipid-reshuffling mechanisms (Schneider and Roessler, 1995; Bates and Browse, 2011). In Arabidopsis, the Lands cycle, an evolutionarily conserved eukaryotic acyl-editing mechanism, stabilizes the acyl-CoA pool by the activity of PLA2 and LPCAT (Fig. 4B). The Lands cycle provides the Kennedy pathway with acyl-CoA and, thereby, prevents a bottleneck in the de novo synthesis of DAG (Wang et al., 2012). Released G3Ps, FAs, and DAGs from the phospholipid degradation serve as precursors for the synthesis of P-free lipids (DGTS, SQDG, galactolipids, and TAGs). Therefore, it is not surprising that the regulation of these genes seems to be a specific P stress response and is not found during TAG accumulation under nitrate limitation (Li et al., 2014).
For the synthesis of PC and PE, two pathways are known: (1) part of the Kennedy pathway (CDP-choline and CDP-ethanolamine pathway) and (2) the methylation pathway, in which PE is a precursor for PC (Gibellini and Smith, 2010). The sequential methylation performed by phosphatidylethanolamine N-methyltransferase (PEM) has not been identified in plants or in N. oceanica in this study (lack of the PEM gene; Fig. 4B; Liu et al., 2015). Therefore, it is suggested that, similar to plants, the synthesis from choline/ethanolamine to PC/PE, via the intermediate CDP-choline/ethanolamine, is essential for Nannochloropsis spp. during cell growth (Liu et al., 2015). Plant-based similarities of this pathway also are seen in the sequence homology between the putative CPT of N. oceanica (No_664) and the Arabidopsis genes AAPT1 and AAPT2 (Goode and Dewey, 1999). AAPT1 and AAPT2 encode a moderate activity toward CDP-ethanolamine and a high activity toward CDP-choline using DAG to form PE and PC, respectively (Goode and Dewey, 1999). Probably CPT of N. oceanica (No_664) can catalyze the formation of PE and PC. In N. oceanica, the kinase (No_7806) and CPT (No_664) were up-regulated in stationary growth of −P cells, pointing to an active phospholipid biosynthesis pathway (Fig. 4B). The formation of PC or PE in N. oceanica under P limitation will not be utilized in membrane lipid construction (seen in the 50% reduction of phospholipids per cell) but probably contributes to a high turnover of PC and PE through active acyl-editing mechanisms and the phospholipid degradation pathway supplying precursors to P-free lipid synthesis. Even though no down-regulated phospholipid synthesis was observed, a reduction of phospholipid biosynthesis intermediates seems to occur in order to reduce phospholipid synthesis (Fig. 4B). The intermediate phosphoethanolamine seems to be reduced to acetaldehyde by the up-regulated gene transcript of phosphoethanolamine phosphorylase (No_9944; regulation confirmed by qRT-PCR; Supplemental Table S3). Acetaldehyde can be fed into the pyruvate or glycolysis metabolism.
Specifically, induced patatin-like PLAs under P limitation show involvement in phospholipid breakdown in both growth phases, while acyl-editing mechanisms (the Lands cycle and PDAT) are activated mainly in stationary growth concomitant with higher gene expression of the whole Kennedy pathway, including PC, PE, and TAG synthesis. This regulation pattern leads to the conclusion that Nannochloropsis spp. favor lipid reshuffling rather than a switch between the down-regulation of phospholipid synthesis and the up-regulation of other lipid synthesis pathways on the transcriptome level (Hunter, 2015).
P-Free Lipid Synthesis during Exponential Growth
Our study revealed a dramatic shift of many lipid classes during the transition to P limitation. Cells were still dividing in P-limited conditions, although phospholipids declined by 50%, while the content of P-free DGTS and SQDG per cell increased (Fig. 3, B and E). The synthesis of DGTS and SQDG is consistent with transcriptomic data at 24 h. Substitution of P-containing lipid with P-free lipids such as extraplastidial DGTS and plastidial SQDG may enable the cells to grow under P limitation. Our findings, therefore, complement the hypotheses formulated in previous studies that assessed either the transcription or lipid composition in response to P limitation in T. pseudonana, P. tricornutum, P. atomus, and the freshwater stramenopile M. subterraneus (Khozin-Goldberg and Cohen, 2006; Van Mooy et al., 2009; Cruz de Carvalho et al., 2016). P stress-inducible SQDG genes also were identified in C. reinhardtii, and the SQD2 promoter of C. reinhardtii was transformed and confirmed to be P stress induced in Nannochloropsis spp. strain NIES-2145 (Iwai et al., 2015). The most recent study that comprises elaborated lipidomic data of eight algae species, including N. gaditana and P. tricornutum, under P limitation showed a rather constant SQDG content and no DGDG increase. Therefore, SQDG is not being considered as a major lipid-remodeling process (Cañavate et al., 2017b). This observation also is seen in our biomass-based calculation of lipid classes, indicating that the ratio of lipid classes relative to the overall cell biomass is not changing (Fig. 3; Supplemental Fig. S3). However, a physiological response can be seen in a cell-based lipid class calculation, where SQDG gene expression is up-regulated significantly at 24 h with concomitant SQDG and DGDG cell content increases. This net lipid class increase is accompanied by a cell size increase in the stationary phase (Supplemental Fig. S2). Whether the −P cells increase gene expression and the synthesis of P-free membrane lipids just to keep steady as cells grow in size or if the cells increase these lipid classes before P restriction is unclear. However, cell size increase as a P-stress response was observed in several other algae but not in P. tricornutum, which also showed increased SQDG synthesis and SQDG increase in cell-based calculations (Abdullahi et al., 2006; Abida et al., 2015; Peter and Sommer, 2015; Shemi et al., 2016). Cañavate et al. (2017b) also saw a decline of DGDG content in N. gaditana, whereas our study showed an increase of DGDG in N. oceanica. This could be a genus-specific response, although an increase in DGDG is consistent with other studies and the fact that the cell volume increases by 60% and, therefore, required more plastidial membrane lipids (Khozin-Goldberg and Cohen, 2006; Cruz de Carvalho et al., 2016).
The DGTS synthesis via BTA_10012 was confirmed by our phylogenetic studies and is likely the case in Nannochloropsis spp. (Supplemental Fig. S4). BTA_10012 has the same domains as those identified in BTA1Cr from C. reinhardtii (Riekhof et al., 2005). Even though the domain order between BTA1Cr and BTA_10012 is switched, it can be speculated that DGTS synthesis activity is not affected by this domain swapping. By expressing the fused MT-DUF3419 domain protein BTA1Cr from C. reinhardtii in Escherichia coli, the two-step reaction synthesized DGTS (Riekhof et al., 2005). Based on the strong up-regulation of BTA_10012 and the increase in the nitrate-rich lipid DGTS at 24 h, it is highly likely that DGTS compensates for the reduction of the structurally similar membrane lipids PC and PE in N. oceanica. This interpretation is supported by lipidomic studies in diatoms and other phytoplankton (Van Mooy et al., 2009; Abida et al., 2015). Furthermore, we show here that BTA proteins in prasinophytes such as Bathycoccus prasinos have an additional TRAM/TLC domain (Supplemental Fig. S4). Such a single gene (No_9162) containing the TRAM/TLC domain also was identified in N. oceanica with significant and high expression under P limitation. Whether the additional TRAM/TLC domain plays a significant role in DGTS formation remains to be seen. DGTS is an important substitution for phospholipids, especially PC, under P limitation. However, the 7 times increase of DGTS also is caused by its structural role as an acyl donor during increased FA and lipid synthesis (Eichenberger and Gribi, 1997; Cañavate et al., 2017b).
Indication of Phytoceramide Synthesis
In −P cells, two putative genes encoding for IPCSs (IPCS_10405/3767) showed strong expression in exponential growth, suggesting the induction of phytoceramide synthesis. The observed PI reduction and increased DAG content per cell during P limitation probably caused an imbalance between PI and DAG, leading to high gene expression of IPCS and the synthesis of phytoceramide from DAG. The hypothesis that PI reduction and DAG increase under P limitation can induce phytoceramide synthesis in N. oceanica is consistent with studies in yeast (Brice et al., 2009) and is supported by untargeted lipidomic studies in T. pseudonana in which diglycosylceramides increased under P limitation (Hunter, 2015). Therefore, the identification of genes involved in sphingolipid synthesis and understanding their function in N. oceanica are of great interest (Supplemental Fig. S7).
Carbon and P Flux in Stationary Growth
The expression levels of RNAs encoding proteins involved in the biosynthesis of nucleotides, mRNA, rRNA, and genome replication were reduced after 48 h in −P cells, when the cell number stabilized (Figs. 1 and 5D; Supplemental Fig. S5). This is likely to result in a reduced number of ribosomes and increased recycling of RNA pools, and in a reduced flux of P into catabolic processes such as metabolites, nucleotides, and, consequently, reduction in protein synthesis.
The main carbon flux in this study was in agreement with N limitation studies in N. oceanica (Li et al., 2014) but showed differences from P starvation in diatoms (Dyhrman et al., 2012; Cruz de Carvalho et al., 2016). In N. oceanica, the flux has a clear direction from the Calvin cycle via glycolysis toward FA and TAG accumulation, while in diatoms, the activation of cytosolic gluconeogenesis and a stress-induced cytosolic oxidative pentose phosphate pathway seems to occur (Cruz de Carvalho et al., 2016). However, due to incomplete protein sequences, the localization of pathways is somewhat difficult to assign in N. oceanica, restricting our conclusions here.
In contrast to P limitation in diatoms, the up-regulation of TPT antiporters (No_6823 and No_768) in N. oceanica suggest that the chloroplast is a prioritized compartment for Pi supply in stationary growth. This is likely to support anabolic processes in the plastid, such as the Calvin cycle and ATP generation, which seemed to stay active until 70 h, after which photosynthetic activity declined (Fig. 1B). Simultaneously, TPTs promote cytosolic glycolysis by exporting triose phosphate into the cytosol. By this, the carbon flux toward the cytosol is increased and probably channeled toward lipid synthesis. An opposite transcriptional regulation was observed in Nannochloropsis spp. under N starvation (Li et al., 2014) and high light (Alboresi et al., 2016), which showed down-regulation of TPT isoforms. Thus, intermediates of glycolysis are utilized differently by plastid and cytosol under different environmental stresses. Reduction of the TPT antiporter transcripts and up-regulation of the expression of genes involved in cytosolic glycolysis suggested that the cytosolic FA synthesis operated independently and is the only supplier of cytosolic FAs for TAG and DGTS accumulation (Alboresi et al., 2016). This finding is supported by the recently discovered Δ0-elongase1, where a cytosolic, middle-chain FA synthesis system may direct FAs to specific glycerolipid classes in N. gaditana (Dolch et al., 2017). In both high light and N starvation, genes involved in the synthesis of plastidial lipids showed no increased expression, and no increased content of plastidial lipids was observed (Li et al., 2014; Alboresi et al., 2016). These findings indicate that a strict separation of the metabolism in plastid and cytosol occurs in Nannochloropsis spp. However, under P limitation, the synthesis of FA and lipid classes was promoted in both compartments.
CONCLUSION
Previous studies showed taxonomically diverse lipid class responses and DOP degradation under P limitation within stramenopiles, but with P. tricornutum and N. oceanica having common physiological P stress-specific responses, such as phospholipid degradation, DGTS and SQDG increase, and high P uptake (Cañavate et al., 2017a, 2017b). On the gene expression level, however, our study suggests that adaptive strategies to cope with P stress differ significantly between P. tricornutum and N. oceanica and probably stramenopiles in general. N. oceanica responds only through the expression of PACPs, GDPDs, PLAs, and genes related to new lipid class synthesis to recruit Pi throughout stationary growth. Interestingly, diatoms additionally use AP and phosphodiesterase genes and show no/low expression of genes for TAG biosynthesis. These findings indicate different trades in lipid metabolism under P stress between stramenopiles. The phospholipid-recycling scheme, which is regulated largely on the gene expression level and seen on the lipidome level, includes both the degradation mechanism of phospholipids and the synthesis of new non-P lipids. In the exponential growth of −P cells, patatin-like PLAs together with GDPDs degrade the phospholipids. Simultaneously, extraplastidial DGTS and plastidial SQDG are synthesized as membrane replacements. In stationary growth during P limitation, genes involved in the Kennedy pathways, PC/PE, and TAG synthesis were up-regulated. PC and PE were immediately turned into non-P lipids (TAG, DGTS, SQDG, and galactolipids) via up-regulated acyl-editing pathways (the Lands cycle and PDAT). This recycling scheme provides promising targets for molecular studies of switches in lipid classes and for increasing lipid production in Nannochloropsis spp. In particular, genes such as BTA_1002, IPCS (IPCS_10405/3767), and PLA_9696 may be involved in lipid class reshuffling. The phospholipid-recycling scheme and assignment of gene functionalities reported in this study will contribute to controlling and engineering lipid composition in Nannochloropsis spp.
MATERIALS AND METHODS
Growth Conditions
The CCMP1779 strain of Nannochloropsis oceanica was obtained from the National Center for Marine Algae and Microbiota (https://ncma.bigelow.org/). N. oceanica was grown in axenic cultures in f/2 medium under constant light (350 µmol photons m−2 s−1) at 23°C. The flasks were aerated with sterile-filtered air, and the medium from filtered (1 μm; Whatman GF/C filter) and autoclaved seawater was supplemented with macronutrients and micronutrients and vitamins (Guillard, 1975). For the axenic growth cultures, 100 μg mL−1 ampicillin was added. The precultures were grown to exponential growth (OD750nm = 0.4, ∼8 × 106 cells mL−1) in 6 L of f/2 medium in three 2-L flasks (Thermo Fisher Nalgene). The precultures were harvested and washed with sterile seawater by centrifugation (4,700g, 20 min). The experimental cultures were inoculated to an OD750nm of 0.06 (∼2.5 × 106 cells mL−1) in either f/2 medium (36.38 μm PO43−; +P cells) or f/2 medium with a reduced phosphate concentration (2.79 μm PO43−; −P cells). Three biological replicates of the algal cultures under each of the mentioned conditions were analyzed. For most of the analyses, two technical samples of the three biological replicates were analyzed every 12 h, unless stated. Two independent experiments were performed, and physiological variables such as photosynthetic activity, Nile Red assay, and cell count were analyzed. The first experiment was used for transcriptomic and P footprint studies, and the second experiment was used for lipidomic studies. Axenic growth was analyzed by plating 200-µL culture suspensions on f/2 medium agar containing 1 mg mL−1 peptone and incubating the plates in darkness for 1 week at 23°C.
Physiological Parameters and P Footprint
The OD750nm was measured using 2 mL of culture in a Hitachi U-5100 spectrophotometer; culture was diluted if necessary (Hitachi High Technologies America). The photosynthetic efficiency was measured with the FluorPen fluorometer (AquaPen-C AP-C 100; Photon Systems Instruments). The cell size was determined from 20 cells in two parallel runs of the replicates with a fluorescence microscope (Zeiss Axio Imager Z.2) and the software AxioVision Rel.4.8. Volumes were calculated from linear dimensions assuming a spherical shape. For Nile Red, 204 g L−1 glycerol was added to the sample along with 12.5 μg of Nile Red solution (stock solution, 0.25 mg mL−1 Nile Red dissolved in acetone). After 10 to 15 min of dark incubation, fluorescence was measured at excitation/emission wavelengths of 525/575 nm, respectively (Aquafluor spectrofluorometer; Turner Designs). Relative fluorescence intensity was calculated by subtracting the autofluorescence of the cells and the self-fluorescence of the Nile Red. The fluorescence was normalized to the cell counts. A BD Accuri C6 flow cytometer (BD Bioscience) was used for cell counting. A 5-mL culture was fixed with 0.1% (v/v) glutaraldehyde, then snap frozen for 30 min with liquid nitrogen, and stored at −80°C. After slowly thawing the cell suspension, the algal cells were counted using excitation light of 488 nm, a forward angle light scatter (0° ± 13°), and the fluorescence filter FL3 with absorption at 670 nm.
Dissolved Pi was determined according to the standard method (Strickland and Parsons, 1968). Defined volumes of the cell cultures (30–40 mL) were filtered on a 47-mm GF/C Whatman filter and stored at −20°C. Two 9-mm pieces of filter were excised from the 47-mm filter and used to determine the cellular P after Norwegian standard NS4725 (Grasshoff, 1967).
Culture suspensions of time points 24 and 96 h (120 and 144 h were not used for other analysis) were washed by vacuum filtration on a Durapore membrane filter (DVPP; 0.65 µm; Merck Millipore) with 30 mL of 0.5 m ammonium formate (AF). After resuspension in 0.5 m AF and OD750 determination, a defined volume of culture was filtered on a preweighed 25-mm GF/C filter (Whatman, Schleicher & Schuell). The filters had been washed with 100 mL of distilled water and dried overnight at 60°C and 3 h at 120°C. The filtered cells were lyophilized for 24 h, and the relative biomass dry weight was determined after adapting to room temperature in a desiccator. The relationship between OD750nm and freeze dry weight (mg L−1) was used to determine together with the measured parameters cell count (cells mL−1), lipid composition (lipid nmol−1 mg−1 dry weight), and lipid content per cell (lipid nmol−1 106 cells−1).
Lipid Content and Composition
To determine the lipid classes, duplicate aliquots between 120- and 200-mL cultures were harvested by vacuum filtration on Durapore membrane filters (DVPP; 0.65 µm; Merck Millipore). The harvested culture was washed with 100 mL of 0.5 m AF, and the filtered samples were resuspended in 1 mL of 0.5 m AF. After centrifugation (1 min, 20,000g), the pellet was flushed with nitrogen gas and quickly frozen in liquid nitrogen. Before lyophilization, the samples were stored at −80°C. The defined cell number (∼8 × 108–2 × 109 cells mL−1) of freeze-dried algae was extracted as described previously (Simionato et al., 2013). The nitrogen-air dried lipid extract was stored at −80°C. After resuspension in 1 mL of chloroform, total FA content was determined after methylation and the addition of hexane. The hexane phase was analyzed by gas-liquid chromatography (PerkinElmer) on a BPX70 (SGE) column (Simionato et al., 2013).
To determine the lipid molecules that were present in each extract, 2D thin-layer chromatography with lipid extract from cells grown with or without Pi was run as described previously (Abida et al., 2015). Each lipid spot was extracted from the silica, and each lipid molecule was identified by mass spectrometry according to Jouhet et al. (2017).
The lipid extracts, corresponding to 25 nmol of total FAs, were dissolved in 100 µL of chloroform:methanol (2:1, v/v) containing 500 pmol of each internal standard. The internal standards used were PE 18:0-18:0, DAG 18:0-22:6 (Avanti Polar Lipid), and SQDG 16:0-18:0 extracted from spinach (Spinacia oleracea) thylakoid and hydrogenated as described in other studies (Buseman et al., 2006; Demé et al., 2014). Lipids were separated using HPLC and quantified by MS/MS. The HPLC separation method was adapted from Rainteau et al. (2012). Lipid classes were separated using an Agilent 1200 HPLC system using a 150-mm × 3-mm (length × i.d.), 5-µm diol column (Macherey-Nagel) at 40°C. The mobile phases consisted of 1 m hexane:isopropanol:water:ammonium acetate, pH 5.3 (625:350:24:1, v/v/v/v [A]) and 1 m isopropanol:water:ammonium acetate, pH 5.3 (850:149:1, v/v/v [B]). The injection volume was 20 µL. After 5 min, the percentage of B was increased linearly from 0% to 100% during a period of 30 min and maintained at 100% for 15 min. This elution sequence was followed by a return to 100% A during a period of 5 min and an equilibration for 20 min with 100% A before the next injection, leading to a total run time of 70 min. The flow rate of the mobile phase was 200 µL min−1. The distinct phospholipid classes were eluted successively as a function of the polar head group. Mass spectrometric analysis was done on a 6460 triple quadrupole (Q1Q2Q3) mass spectrometer (Agilent) equipped with a jet stream electrospray ion source under the following settings: drying gas heater, 260°C; drying gas flow, 13 L min−1; sheath gas heater, 300°C; sheath gas flow, 11 L min−1; nebulizer pressure, 25 p.s.i.; capillary voltage, ±5,000 V; and nozzle voltage, ±1,000 V. Nitrogen was used as the collision gas. The quadrupoles Q1 and Q3 were operated at widest and unit resolution, respectively. The PC and DGTS analysis was carried out in positive ion mode by scanning for precursors of m/z 184 and 236, respectively, at collision energy of 34 and 52 eV. The SQDG analysis was carried out in negative ion mode by scanning for precursors of m/z −225 at a collision energy of −56 eV. For PE, PI, PG, MGDG, and DGDG, measurements were performed in positive ion mode by scanning for neutral losses of 141, 277, 189, 179, and 341 D at collision energies of 20, 12, 16, 8, and 8 eV, respectively. Quantification was done by multiple reaction monitoring with a 50-ms dwell time. DAG and TAG species were identified and quantified by multiple reaction monitoring as singly charged ions [M+NH4]+, at collision energies of 16 and 22 eV, respectively, with a 50-ms dwell time. Mass spectra were processed by MassHunter Workstation software (Agilent) for the identification and quantification of lipids. Lipid amounts (pmol) were corrected for response differences between internal standards and endogenous lipids and by comparison with a known N. gaditana lipid extract qualified and quantified by thin-layer chromatography and gas chromatography-flame ionization detection as described previously (Abida et al., 2015; Dolch et al., 2017).
RNA Isolation and Microarray Analysis
For the RNA isolation and following microarray analysis, 50- to 100-mL samples of the biological replicates were quickly harvested through vacuum filtration on Durapore membrane filters, resuspended in seawater, centrifuged, and then flash frozen in liquid nitrogen and stored at −80°C. Three biological replicates were used for the 24-h time point, while two biological samples were used for the 48, 53, 58, and 70-h samples. RNA was isolated after Chauton et al. (2013). A total of 200 ng of RNA was reverse transcribed, amplified, and labeled according to the Low Input Quick Amp Labeling Kit, One-Color (Agilent; 5190-2305). The cRNA hybridization and fragmentation as well as the image processing were performed according to Chauton et al. (2013). A custom microarray chip for N. oceanica CCMP1779 was produced by Agilent Technologies using an 8- × 15-K format (design identifier, Agilent-048674; Gene Expression Omnibus platform identifier, GPL23012). A total of 15,208 60-mer oligonucleotide probes were designed after draft gene models from GSE36959 (http://www.ncbi.nlm.nih.gov/geo/). The identification of significant differentially expressed genes and normalization of the data were done according to the method of Chauton et al. (2013). A design matrix was created that included the sample time points (24, 48, 53, 58, and 70 h) for −P cells and the 24-h time point of +P cells. Pairwise comparisons were processed for all time points using the +P cells at 24 h as a reference. The +P cells at 24 h were chosen as a reference because the cells showed maximal growth rate with no limitation (e.g. by light shading), ensuring no additional undetermined impact affecting the global gene regulation profile. It is worth noting that the DNA microarray data were generated for −P cells at times points 24, 48, 53, 58, and 70 h and for +P cells at 24 and 58 h to evaluate the effect of cell aging and growth inhibition. The +P cells at 58 h showed a slight induction of P limitation by minimal up-regulation of P stress indicator genes such as PACPs and P transporters. However, the regulation was minor compared with the gene regulation of −P cells (Supplemental Excel Sheet S1). The false discovery rate was estimated using the Benjamini-Hochberg method, and genes with an adjusted P < 0.01 were considered to be significantly differently expressed. Gene and protein models were generated from the genome assembly of N. oceanica CCMP1779 (Vieler et al., 2012). For validation of the microarray data, qRT-PCR was carried out on total RNA from the three biological replicates of both conditions and experiments.
qRT-PCR
For validation of the microarray data, qRT-PCR was carried out on total RNA from the three biological replicates of both conditions and experiments. cDNA synthesis was performed with the PrimeScript First-Strand cDNA Synthesis Kit (TaKaRa) on 500 ng of total RNA that was isolated for the microarray analysis. Oligo(T) and random primers were used for cDNA synthesis. The standard method, with a preincubation for 5 min at 95°C followed by 50 cycles of amplification consisting of 10 s at 95°C, 10 s at 55°C, and 10 s at 72°C, was used with the LightCycler 480 SYBR Green I Master kit for the LightCycler 96 (Roche Applied Science). Genes of interest and housekeeping genes (exportin1 [No_11171], nuclear transporter [No_4337], and 14-3-3-like protein [No_6482]) were analyzed. The reference genes were evaluated with geNorm (http://medgen.ugent.be/genorm). Both genes of interest and housekeeping genes were picked based on the microarray data to analyze their expression at time points 24, 32, 58, and 72 h. The forward and reverse primers listed in Supplemental Table S2 resulted in amplification products of 100 to 300 bp. LinRegPCR 11.1 software was used to determine the means of primer efficiency, and the LightCycler 96 software was used to identify the threshold cycle value (crossing points) of each sample (Ramakers et al., 2003; Ruijter et al., 2009). Means of the individual biological samples (n = 3) were calculated and used to determine the ratio and the respective log2 value by the qBasePLUS software (Biogazelle).
Accession Numbers
This study is MIAME compliant, and the microarray data have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE95774.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Comparison of the physiological parameters of two independent experiments in N. oceanica CCMP1779.
Supplemental Figure S2. Cell volume increase and cellular P per dry weight in N. oceanica CCMP1779.
Supplemental Figure S3. Lipid class content per dry weight as a function of time in N. oceanica of −P cells and +P cells.
Supplemental Figure S4. Phylogenetic tree of BTA sequences in the kingdom of eukaryotes.
Supplemental Figure S5. Transcriptional regulation of cell replication stop in N. oceanica CCMP1779.
Supplemental Figure S6. Phylogenetic tree of phosphate transporter (No_8918) sequences in the kingdom of eukaryotes.
Supplemental Figure S7. Model of sphingolipid biosynthesis in N. oceanica CCMP1779 under P limitation.
Supplemental Table S1. Carbon, P, and nitrogen ratio during cultivation.
Supplemental Table S2. Primer pairs used for qRT-PCR for experiments 1 and 2.
Supplemental Table S3. Gene regulation of lipid-related genes obtained by qRT-PCR experiments 1 and 2 compared with microarray results.
Supplemental Table S4. FA profile of −P cells.
Supplemental Table S5. FA profile of +P cells.
Supplemental Table S6. Increased FA moieties of all lipid classes in −P cells.
Supplemental Table S7. Decreased FA moieties of all lipid classes in −P cells.
Supplemental Table S8. FA moieties of lipid classes with no change in −P cells.
Supplemental Table S9. Comparison of marker genes of P limitation regulated in +P cells and −P cells at 58 h.
Supplemental Excel Sheet S1. Significantly regulated genes involved in clusters of lipid and carbon pathways.
Acknowledgments
We thank Torfinn Sparstad and Kjersti Andresen for excellent support in experimental analysis as well as Nora Gigli Bisceglia, Julie Despres, Daniel Felten, and Jacob Lamb for their kind and amazing technical support.
Footnotes
This work was supported by the Research Council of Norway through their funding of the project Microbially Produced Raw Materials for Aquafeed (MIRA; project nos. 239001 and 234229) and a scholarship from Norwegian University of Science and Technology to A.M. cofounded by the Faculty of Science and Technology and the strategic research area NTNU Oceans. J.J. and E.M. were supported by an Investissement d’Avenir grant (Océanomics).
Articles can be viewed without a subscription.
References
- Abdullahi AS, Underwood GJ, Gretz MR (2006) Extracellular matrix assembly in diatoms (Bacillariophyceae). J Phycol 42: 363–378 [Google Scholar]
- Abida H, Dolch LJ, Meï C, Villanova V, Conte M, Block MA, Finazzi G, Bastien O, Tirichine L, Bowler C, et al. (2015) Membrane glycerolipid remodeling triggered by nitrogen and phosphorus starvation in Phaeodactylum tricornutum. Plant Physiol 167: 118–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksoy M, Pootakham W, Grossman AR (2014) Critical function of a Chlamydomonas reinhardtii putative polyphosphate polymerase subunit during nutrient deprivation. Plant Cell 26: 4214–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alboresi A, Perin G, Vitulo N, Diretto G, Block M, Jouhet J, Meneghesso A, Valle G, Giuliano G, Maréchal E, et al. (2016) Light remodels lipid biosynthesis in Nannochloropsis gaditana by modulating carbon partitioning between organelles. Plant Physiol 171: 2468–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Browse J (2011) The pathway of triacylglycerol synthesis through phosphatidylcholine in Arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. Plant J 68: 387–399 [DOI] [PubMed] [Google Scholar]
- Beacham TA, Bradley C, White DA, Bond P, Ali ST (2014) Lipid productivity and cell wall ultrastructure of six strains of Nannochloropsis: implications for biofuel production and downstream processing. Algal Res 6: 64–69 [Google Scholar]
- Beligni MV, Bagnato C, Prados MB, Bondino H, Laxalt AM, Munnik T, Ten Have A (2015) The diversity of algal phospholipase D homologs revealed by biocomputational analysis. J Phycol 51: 943–962 [DOI] [PubMed] [Google Scholar]
- Brandsma J. (2016) Phytoplankton phenotype plasticity induced by phosphorus starvation may play a significant role in marine microbial ecology and biogeochemistry. New Phytol 211: 765–766 [DOI] [PubMed] [Google Scholar]
- Brice SE, Alford CW, Cowart LA (2009) Modulation of sphingolipid metabolism by the phosphatidylinositol-4-phosphate phosphatase Sac1p through regulation of phosphatidylinositol in Saccharomyces cerevisiae. J Biol Chem 284: 7588–7596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buseman CM, Tamura P, Sparks AA, Baughman EJ, Maatta S, Zhao J, Roth MR, Esch SW, Shah J, Williams TD, et al. (2006) Wounding stimulates the accumulation of glycerolipids containing oxophytodienoic acid and dinor-oxophytodienoic acid in Arabidopsis leaves. Plant Physiol 142: 28–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañavate JP, Armada I, Hachero-Cruzado I (2017a) Aspects of phosphorus physiology associated with phosphate-induced polar lipid remodelling in marine microalgae. J Plant Physiol 214: 28–38 [DOI] [PubMed] [Google Scholar]
- Cañavate JP, Armada I, Hachero-Cruzado I (2017b) Interspecific variability in phosphorus-induced lipid remodelling among marine eukaryotic phytoplankton. New Phytol 213: 700–713 [DOI] [PubMed] [Google Scholar]
- Chauton MS, Winge P, Brembu T, Vadstein O, Bones AM (2013) Gene regulation of carbon fixation, storage, and utilization in the diatom Phaeodactylum tricornutum acclimated to light/dark cycles. Plant Physiol 161: 1034–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GQ, Snyder CL, Greer MS, Weselake RJ (2011) Biology and biochemistry of plant phospholipases. Crit Rev Plant Sci 30: 239–258 [Google Scholar]
- Cheng Y, Zhou W, El Sheery NI, Peters C, Li M, Wang X, Huang J (2011) Characterization of the Arabidopsis glycerophosphodiester phosphodiesterase (GDPD) family reveals a role of the plastid-localized AtGDPD1 in maintaining cellular phosphate homeostasis under phosphate starvation. Plant J 66: 781–795 [DOI] [PubMed] [Google Scholar]
- Cruz de Carvalho MH, Sun HX, Bowler C, Chua NH (2016) Noncoding and coding transcriptome responses of a marine diatom to phosphate fluctuations. New Phytol 210: 497–510 [DOI] [PubMed] [Google Scholar]
- Demé B, Cataye C, Block MA, Maréchal E, Jouhet J (2014) Contribution of galactoglycerolipids to the 3-dimensional architecture of thylakoids. FASEB J 28: 3373–3383 [DOI] [PubMed] [Google Scholar]
- Docampo R, Moreno SNJ (2011) Acidocalcisomes. Cell Calcium 50: 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolch LJ, Rak C, Perin G, Tourcier G, Broughton R, Leterrier M, Morosinotto T, Tellier F, Faure JD, Falconet D, et al. (2017) A palmitic acid elongase affects eicosapentaenoic acid and plastidial monogalactosyldiacylglycerol levels in Nannochloropsis. Plant Physiol 173: 742–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyhrman ST. (2016) Nutrients and their acquisition: phosphorus physiology in microalgae. In Borowitzka M, Beardall J, Raven J, eds, The Physiology of Microalgae: Developments in Applied Phycology, Vol 6 Springer International Publishing, Switzerland, pp 155–183 [Google Scholar]
- Dyhrman ST, Jenkins BD, Rynearson TA, Saito MA, Mercier ML, Alexander H, Whitney LP, Drzewianowski A, Bulygin VV, Bertrand EM, et al. (2012) The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response. PLoS ONE 7: e33768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberger W, Gribi C (1997) Lipids of Pavlova lutheri: cellular site and metabolic role of DGCC. Phytochemistry 45: 1561–1567 [Google Scholar]
- Gibellini F, Smith TK (2010) The Kennedy pathway: de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 62: 414–428 [DOI] [PubMed] [Google Scholar]
- Goode JH, Dewey RE (1999) Characterization of aminoalcoholphosphotransferases from Arabidopsis thaliana and soybean. Plant Physiol Biochem 37: 445–457 [Google Scholar]
- Grasshoff K. (1967) Methods of Seawater Analysis. Verlag Chemie, Weinheim, Germany [Google Scholar]
- Grossman AR, Aksoy M (2015) Algae in a phosphorus-limited landscape. In Plaxton WC, Lambers H, eds, Annual Plant Reviews, Vol 48 John Wiley & Sons, Hoboken, NJ, pp 163–210 [Google Scholar]
- Guillard RRL. (1975) Culture of Marine Invertebrate Animals. Plenum Press, New York [Google Scholar]
- Hu H, Gao K (2003) Optimization of growth and fatty acid composition of a unicellular marine picoplankton, Nannochloropsis sp., with enriched carbon sources. Biotechnol Lett 25: 421–425 [DOI] [PubMed] [Google Scholar]
- Hunter JE. (2015) Phytoplankton lipidomics: lipid dynamics in response to microalgal stressors. PhD thesis. University of Southampton, UK [Google Scholar]
- Iwai M, Hori K, Sasaki-Sekimoto Y, Shimojima M, Ohta H (2015) Manipulation of oil synthesis in Nannochloropsis strain NIES-2145 with a phosphorus starvation-inducible promoter from Chlamydomonas reinhardtii. Front Microbiol 6: 912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai M, Ikeda K, Shimojima M, Ohta H (2014) Enhancement of extraplastidic oil synthesis in Chlamydomonas reinhardtii using a type-2 diacylglycerol acyltransferase with a phosphorus starvation-inducible promoter. Plant Biotechnol J 12: 808–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet J, Lupette J, Clerc O, Magneschi L, Bedhomme M, Collin S, Roy S, Maréchal E, Rébeillé F (2017) LC-MS/MS versus TLC plus GC methods: consistency of glycerolipid and fatty acid profiles in microalgae and higher plant cells and effect of a nitrogen starvation. PLoS ONE 12: e0182423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl DM, Björkman KM (2015) Dynamics of dissolved organic phosphorus. In Hansell DA, Carlson CA, eds, Biogeochemistry of Marine Dissolved Organic Matter. Academic Press, Burlington, MA, pp 233–334 [Google Scholar]
- Khozin-Goldberg I, Cohen Z (2006) The effect of phosphate starvation on the lipid and fatty acid composition of the fresh water eustigmatophyte Monodus subterraneus. Phytochemistry 67: 696–701 [DOI] [PubMed] [Google Scholar]
- Kuenzler EJ, Perras JP (1965) Phosphatases of marine algae. Biol Bull 128: 271–276 [Google Scholar]
- Lessard EJ, Merico A, Tyrrell T (2005) Nitrate:phosphate ratios and Emiliania huxleyi blooms. Limnol Oceanogr 50: 1020–1024 [Google Scholar]
- Li J, Han D, Wang D, Ning K, Jia J, Wei L, Jing X, Huang S, Chen J, Li Y, et al. (2014) Choreography of transcriptomes and lipidomes of Nannochloropsis reveals the mechanisms of oil synthesis in microalgae. Plant Cell 26: 1645–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Litaker RW, Sunda WG (2016) Phosphorus physiological ecology and molecular mechanisms in marine phytoplankton. J Phycol 52: 10–36 [DOI] [PubMed] [Google Scholar]
- Lin X, Zhang H, Cui Y, Lin S (2012) High sequence variability, diverse subcellular localizations, and ecological implications of alkaline phosphatase in dinoflagellates and other eukaryotic phytoplankton. Front Microbiol 3: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang G, Wang X (2015) Role of aminoalcoholphosphotransferases 1 and 2 in phospholipid homeostasis in Arabidopsis. Plant Cell 27: 1512–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubian LM, Blasco J, Establier R (1992) A comparative-study of acid and alkaline-phosphatase activities in several strains of Nannochloris (Chlorophyceae) and Nannochloropsis (Eustigmatophyceae). Br Phycol J 27: 119–130 [Google Scholar]
- Martin P, Van Mooy BAS, Heithoff A, Dyhrman ST (2011) Phosphorus supply drives rapid turnover of membrane phospholipids in the diatom Thalassiosira pseudonana. ISME J 5: 1057–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JJ, Flynn KJ, Shields RJ (2014) Influence of the N:P supply ratio on biomass productivity and time-resolved changes in elemental and bulk biochemical composition of Nannochloropsis sp. Bioresour Technol 169: 588–595 [DOI] [PubMed] [Google Scholar]
- Mühlroth A, Li K, Røkke G, Winge P, Olsen Y, Hohmann-Marriott MF, Vadstein O, Bones AM (2013) Pathways of lipid metabolism in marine algae, co-expression network, bottlenecks and candidate genes for enhanced production of EPA and DHA in species of Chromista. Mar Drugs 11: 4662–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mus F, Toussaint JP, Cooksey KE, Fields MW, Gerlach R, Peyton BM, Carlson RP (2013) Physiological and molecular analysis of carbon source supplementation and pH stress-induced lipid accumulation in the marine diatom Phaeodactylum tricornutum. Appl Microbiol Biotechnol 97: 3625–3642 [DOI] [PubMed] [Google Scholar]
- Nakamura Y. (2013) Phosphate starvation and membrane lipid remodeling in seed plants. Prog Lipid Res 52: 43–50 [DOI] [PubMed] [Google Scholar]
- Parker MS, Mock T, Armbrust EV (2008) Genomic insights into marine microalgae. Annu Rev Genet 42: 619–645 [DOI] [PubMed] [Google Scholar]
- Peter KH, Sommer U (2015) Interactive effect of warming, nitrogen and phosphorus limitation on phytoplankton cell size. Ecol Evol 5: 1011–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quisel JD, Wykoff DD, Grossman AR (1996) Biochemical characterization of the extracellular phosphatases produced by phosphorus-deprived Chlamydomonas reinhardtii. Plant Physiol 111: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radakovits R, Jinkerson RE, Fuerstenberg SI, Tae H, Settlage RE, Boore JL, Posewitz MC (2012) Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropsis gaditana. Nat Commun 3: 686–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainteau D, Humbert L, Delage E, Vergnolle C, Cantrel C, Maubert MA, Lanfranchi S, Maldiney R, Collin S, Wolf C, et al. (2012) Acyl chains of phospholipase D transphosphatidylation products in Arabidopsis cells: a study using multiple reaction monitoring mass spectrometry. PLoS ONE 7: e41985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Riekhof WR, Sears BB, Benning C (2005) Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii: discovery of the betaine lipid synthase BTA1Cr. Eukaryot Cell 4: 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodolfi L, Chini Zittelli G, Bassi N, Padovani G, Biondi N, Bonini G, Tredici MR (2009) Microalgae for oil: strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol Bioeng 102: 100–112 [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer GFE, Ryu SB, Wang X, Matos AR, Heitz T (2010) Patatin-related phospholipase A: nomenclature, subfamilies and functions in plants. Trends Plant Sci 15: 693–700 [DOI] [PubMed] [Google Scholar]
- Schneider JC, Roessler PG (1995) A novel acyltransferase activity in an oleaginous alga. In Kader JC, Mazliak P, eds, Plant Lipid Metabolism. Springer, Dordrecht, The Netherlands, pp 105–107 [Google Scholar]
- Secco D, Wang C, Arpat BA, Wang Z, Poirier Y, Tyerman SD, Wu P, Shou H, Whelan J (2012) The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol 193: 842–851 [DOI] [PubMed] [Google Scholar]
- Shemi A, Schatz D, Fredricks HF, Van Mooy BAS, Porat Z, Vardi A (2016) Phosphorus starvation induces membrane remodeling and recycling in Emiliania huxleyi. New Phytol 211: 886–898 [DOI] [PubMed] [Google Scholar]
- Simionato D, Block MA, La Rocca N, Jouhet J, Maréchal E, Finazzi G, Morosinotto T (2013) The response of Nannochloropsis gaditana to nitrogen starvation includes de novo biosynthesis of triacylglycerols, a decrease of chloroplast galactolipids, and reorganization of the photosynthetic apparatus. Eukaryot Cell 12: 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland JDH, Parsons TR (1968) Practical Handbook of Seawater Analysis. Fisheries Research Board of Canada, Ottawa [Google Scholar]
- Theodorou ME, Elrifi IR, Turpin DH, Plaxton WC (1991) Effects of phosphorus limitation on respiratory metabolism in the green alga Selenastrum minutum. Plant Physiol 95: 1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BL, Frossard E, Baldwin DS (2005) Organic Phosphorus in the Environment. CAB International, Wallingford, UK [Google Scholar]
- Van Mooy BAS, Devol AH (2008) Assessing nutrient limitation of Prochlorococcus in the North Pacific subtropical gyre by using an RNA capture method. Limnol Oceanogr 53: 78–88 [Google Scholar]
- Van Mooy BAS, Fredricks HF, Pedler BE, Dyhrman ST, Karl DM, Koblízek M, Lomas MW, Mincer TJ, Moore LR, Moutin T, et al. (2009) Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature 458: 69–72 [DOI] [PubMed] [Google Scholar]
- Van Mooy BAS, Moutin T, Duhamel S, Rimmelin P, Van Wambeke F (2008) Phospholipid synthesis rates in the eastern subtropical South Pacific Ocean. Biogeoscience 5: 133–139 [Google Scholar]
- Vieler A, Wu G, Tsai CH, Bullard B, Cornish AJ, Harvey C, Reca IB, Thornburg C, Achawanantakun R, Buehl CJ, et al. (2012) Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genet 8: e1003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Shen W, Kazachkov M, Chen G, Chen Q, Carlsson AS, Stymne S, Weselake RJ, Zou J (2012) Metabolic interactions between the Lands cycle and the Kennedy pathway of glycerolipid synthesis in Arabidopsis developing seeds. Plant Cell 24: 4652–4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZK, Niu YF, Ma YH, Xue J, Zhang MH, Yang WD, Liu JS, Lu SH, Guan Y, Li HY (2013) Molecular and cellular mechanisms of neutral lipid accumulation in diatom following nitrogen deprivation. Biotechnol Biofuels 6: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]