Notwithstanding evidence of transgenerational drought-induced memory in one of six traits examined, the Arabidopsis methylome is relatively impervious to drought stress.
Abstract
Improving the responsiveness, acclimation, and memory of plants to abiotic stress holds substantive potential for improving agriculture. An unresolved question is the involvement of chromatin marks in the memory of agriculturally relevant stresses. Such potential has spurred numerous investigations yielding both promising and conflicting results. Consequently, it remains unclear to what extent robust stress-induced DNA methylation variation can underpin stress memory. Using a slow-onset water deprivation treatment in Arabidopsis (Arabidopsis thaliana), we investigated the malleability of the DNA methylome to drought stress within a generation and under repeated drought stress over five successive generations. While drought-associated epi-alleles in the methylome were detected within a generation, they did not correlate with drought-responsive gene expression. Six traits were analyzed for transgenerational stress memory, and the descendants of drought-stressed lineages showed one case of memory in the form of increased seed dormancy, and that persisted one generation removed from stress. With respect to transgenerational drought stress, there were negligible conserved differentially methylated regions in drought-exposed lineages compared with unstressed lineages. Instead, the majority of observed variation was tied to stochastic or preexisting differences in the epigenome occurring at repetitive regions of the Arabidopsis genome. Furthermore, the experience of repeated drought stress was not observed to influence transgenerational epi-allele accumulation. Our findings demonstrate that, while transgenerational memory is observed in one of six traits examined, they are not associated with causative changes in the DNA methylome, which appears relatively impervious to drought stress.
Plants, being sessile organisms, must be phenotypically plastic in a dynamic environment. This ability is relayed by various intricate intercellular and intracellular mechanisms, including hormone signaling and organelle-nuclear retrograde pathways that allow plants to perceive and respond to biotic and abiotic stresses (Karpinski et al., 1999; Fernández and Strand, 2008; Cutler et al., 2010; Goodger and Schachtman, 2010; Chan et al., 2016; Martín et al., 2016). Additionally, repeated exposure to stress can lead to plant stress priming, whereby prior stress exposure conveys an enhanced ability to respond to future events (Conrath et al., 2006; Ding et al., 2012; Gordon et al., 2013; Sani et al., 2013; Wang et al., 2014; Virlouvet and Fromm, 2015; Hilker et al., 2016; Wibowo et al., 2016). This notion has been extended to numerous considerations of the formation of plant stress memory, in which a state of altered stress responsivity is mitotically or meiotically transmissible (Bruce et al., 2007; Hauser et al., 2011b; Probst and Mittelsten Scheid, 2015; Crisp et al., 2016; van Loon, 2016). There is much interest in plant stress memory, including the underlying molecular mechanism(s) and its potential to impact crop yields, particularly in harsh and variable environments (Springer, 2013; Ji et al., 2015; Mickelbart et al., 2015).
Stable propagation of DNA methylation states has been suggested as a possible mechanism for the formation of plant stress memory (Boyko and Kovalchuk, 2010; Probst and Mittelsten Scheid, 2015; Crisp et al., 2016). DNA methylation, occurring in three sequence contexts in plants (mCG, mCHG, and mCHH, where H is anything but G), is largely considered to function in transposable element (TE) silencing, maintaining genome stability, and possible regulation of gene expression (Reinders et al., 2009; Law and Jacobsen, 2010; Jones, 2012; Eichten et al., 2014; Niederhuth and Schmitz, 2017). This potential effect on gene expression has raised the proposition that DNA methylation could complement genetic variation, as a mode for transferring heritable information, to contribute to phenotypic variation (Molinier et al., 2006; Heard and Martienssen, 2014; Quadrana and Colot, 2016). Indeed, DNA methylation states can be maintained faithfully over both mitotic and meiotic cell divisions by a suite of pathways and enzymes (Probst et al., 2009; Law and Jacobsen, 2010; Stroud et al., 2013). It is unclear the extent to which genome-wide patterns of DNA methylation (DNA methylome) in plants are reset; rather, it appears that the parental DNA methylome is reestablished and propagated during gametogenesis and spermatogenesis (Slotkin et al., 2009; Calarco et al., 2012). Since these processes occur within postembryonic growth in plants (Boavida et al., 2005), any accumulated variations in the DNA methylome (epi-allele), either environmentally induced or spontaneous, have the potential to be carried over generations. DNA methylation state has shown stable heritability (Li et al., 2014; Dubin et al., 2015; Hagmann et al., 2015), with the documented appearance of epi-alleles (in the form of differentially methylated regions [DMRs]) to occur at a frequency comparable to genetic polymorphisms (Becker et al., 2011; Schmitz et al., 2011, 2013), which possibly arise at elevated frequencies under abiotic stress (Jiang et al., 2014).
This potential has led to many investigations of transgenerational stress memory mediated by environmentally induced epi-alleles, which could open exciting possibilities for crop (epi)genomics (Hauser et al., 2011b; Springer, 2013; Ji et al., 2015). However, bona fide examples of transgenerational methylation changes leading to substantially altered plant behavior remain a rare observation (Pecinka et al., 2009; Becker et al., 2011; Jiang et al., 2014; Seymour et al., 2014; Crisp et al., 2016), with the majority of DNA methylome variation attributable to underlying genetic differences rather than being truly epigenetic (Eichten et al., 2013, 2014; Schmitz et al., 2013; Li et al., 2014; Seymour et al., 2014; Dubin et al., 2015; Hagmann et al., 2015). Furthermore, there are a growing number of conflicting reports of short-term adaptation to abiotic stresses, including salt or drought stress, that occur independently of DNA methylation changes (Cayuela et al., 1996; Jakab et al., 2005; Rossel et al., 2007; Ding et al., 2012, 2013, 2014; Gordon et al., 2013; Sani et al., 2013). This is in contrast to the potential for DNA methylation-mediated transgenerational memory (Luo et al., 1996; Tricker et al., 2013; Herman and Sultan, 2016; Nosalewicz et al., 2016; Wibowo et al., 2016; Zheng et al., 2017). Thus, the nature of DNA methylation and its contribution to short-term and transgenerational stress adaptation remain enigmatic, which is confounded by the limited identification of causative changes in the DNA methylome correlating with enhanced stress tolerance (Secco et al., 2015; Meng et al., 2016).
Given the conflicting nature of past studies, we sought to systematically investigate the potential for environmentally induced changes in the DNA methylome in the model species Arabidopsis (Arabidopsis thaliana) that could convey transgenerational stress memory. First, the potential for drought-induced formation of heritable epi-alleles that correlate with drought-responsive gene expression was tested. Subsequently, with an experiment designed to minimize genetic variation and stochastic DNA methylome variation, the ability of plants to form transgenerational stress memory using a repeated drought stress over consecutive generations was investigated and coupled with in-depth DNA methylome profiling.
RESULTS
Stress-Associated Variation in DNA Methylation Observed under a Slow-Onset Mild Drought Stress within a Generation
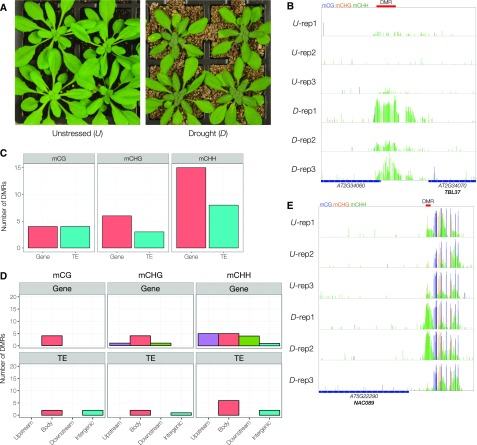
A slow-onset water deprivation treatment (drought stress) was imposed on soil-grown plants by withholding watering for 9 d to assess the potential for drought stress to induce epi-alleles in the DNA methylome. This stress led to a drop in relative water content to around ∼60% (measured in representative plants) and visible leaf wilting (Fig. 1A). Whole rosettes were harvested from unstressed plants (n = 3) and drought-treated plants (n = 3). The genome-wide DNA methylation patterns (DNA methylome) of these samples were assayed at single-base-pair resolution using whole-genome bisulfite sequencing (WGBS; Lister et al., 2008; Supplemental Table S1).
Figure 1.
A mild drought stress is associated with variations in DNA methylation. A, Representative plants that were either unstressed (U) or underwent a drought stress (D) involving 9 d of withheld watering. B, Representative drought stress-associated DMR identified by DSS. Rows represent individual samples. DNA methylation is shown at single C resolution for mCG, mCHG, and mCHH as blue, orange, and green bars, respectively. Underlying genomic elements are presented at the bottom, and the exact region identified as a DMR is shown on top (red bar). C, Numbers of filtered stress-associated DMRs occurring near annotated protein-coding genes and TEs for each methylation context. D, Detailed mapping of filtered stress-associated DMRs within, or near, annotated protein-coding genes and TEs for each context of methylation. Body refers to DMRs occurring within the genomic feature, Upstream refers to DMRs within 1 kb near the 5′ end of the feature, Downstream refers to DMRs within 1 kb of the 3′ end of the feature, and Intergenic refers to DMRs that are farther than 1 kb away from the nearest genomic feature. E, Drought stress-associated DMR identified upstream of a gene encoding NAC089, identified previously as a locus exhibiting transcriptional memory to repeated dehydration stress. Rows represent individual samples. DNA methylation levels are shown at single C resolution for mCG, mCHG, and mCHH as blue, orange, and green bars, respectively. Underlying genomic elements are presented at the bottom, and the region identified as a DMR is shown on top (red bar).
To explore variations in the methylome among our samples, pairwise comparisons of mean methylation levels, binned across 100-bp tiles, was performed to capture the full extent of variation between all samples (Eichten and Springer, 2015). This revealed 2,141 mCG, 1,039 mCHG, and 718 mCHH DMRs across all samples; however, hierarchical clustering of samples based on methylation levels at these regions did not cluster samples by treatment (Supplemental Fig. S1A). Instead, clustering revealed the existence of two to three putative preexisting DNA methylome states, herein referred to as epi-types. This suggests that the predominant source of variation in the DNA methylome between these samples arises from preexisting differences. As the seed stock for this experiment was derived from bulk seed harvestings, as opposed to single-seed descent, these differences are likely caused by distant relatedness between plants (Becker et al., 2011; Schmitz et al., 2011, 2013).
Notwithstanding the presence of epi-type DMRs, we hypothesized that, if the Arabidopsis DNA methylome is truly malleable to abiotic stress, then drought stress should induce conserved variations between control and treated plants among any epi-type variation. Despite the hierarchical clustering of 100-bp tile-based DMRs showing negligible evidence of conserved DMRs between treatments, we attempted to identify any evidence for statistically significant treatment-conserved changes through rank sum testing of DMRs (see “Materials and Methods”). However, upon correction for multiple testing, all of the observed changes were deemed to be insignificant. While tile-based DMRs are a powerful tool for exploring broad-scale methylome variation, it is limited in its ability to appropriately attribute biological and technical information at single C residues, thus losing statistical power to identify differential methylation. Thus, an alternative approach to evaluate differential methylation was performed using DSS (Feng et al., 2014). This method employs Bayesian hierarchical modeling to incorporate the variation that exists both within and between biological replicates, at single C resolution, to identify bona fide treatment-associated DMRs with greater statistical rigor, including posthoc P value adjustments.
To identify stress-associated DMRs, we performed separate DMR calling between treatment groups accounting for within-group variation (Table I). First, to account for the contribution of preexisting methylome variation, DMRs were identified between epi-type groups (epi-type DMRs) using DSS (Table I; Supplemental Fig. S1B). The locations of epi-type DMRs were mapped relative to genomic features (Supplemental Table S2) based on the Araport11 genome reannotation (Cheng et al., 2017). Epi-type associated DMRs had comparable numbers mapping to annotated protein-coding genes and TEs; however, they were predominantly in the mCG context within gene and TE bodies (Supplemental Fig. S1, C and D).
Table I. Numbers of DMRs between epi-type groups and treatments identified by DSS.
The numbers of drought stress DMRs exclude those that were also identified between epi-type groups. U, Unstressed; D, drought stressed.
| DMR Class | Contrast | Sequence Context |
||
|---|---|---|---|---|
| mCG | mCHG | mCHH | ||
| Epi-type | U-1, U-3, D-2 versus U-2, D-1, D-3 | 144 | 41 | 33 |
| Drought stress | U-1, U-2, U-3 versus D-1, D-2, D-3 | 8 | 9 | 23 |
Second, 49 stress-associated DMRs were identified using DSS, nine of which also were identified as preexisting epi-type DMRs and filtered from further analysis to produce a final list of 40 drought-associated DMRs (Table I; Fig. 1B; Supplemental Table S3). Positional mapping of drought- and epi-type-associated DMRs relative to protein-coding genes and annotated TEs was compared to explore whether they exhibited similar characteristics. Drought-associated DMRs were more likely to be found within 1 kb of genes (24 of 40; 60%) compared with epi-type-associated DMRs (91 of 218; 42%). Interestingly, there were proportionally fewer mCG stress-associated DMRs (eight of 40 DMRs; 20%) than mCG epi-type-associated DMRs (144 of 218; 66%), with the majority in the mCHH context (Fig. 1C). Beyond this, stress-associated DMRs located near genes were predominantly non-mCG (20 of 24; 83%; Fig. 1D; Table II). These results potentiate the involvement of the RNA-directed DNA methylation (RdDM) pathway as a source of stress-induced methylome variation near genes (Matzke et al., 2009; Schmitz et al., 2013). The exact mechanism underpinning mCG-DMRs remains elusive; however, they have been suggested to be more likely truly epigenetic (i.e. unlinked from underlying genetic variation; Schmitz et al., 2013).
Table II. Numbers of drought stress DMRs mapping to protein-coding genes directly (gene body), within 1 kb (upstream/downstream) from the nearest gene, or greater than 1 kb from the nearest gene (intergenic).
| Location | Sequence Context |
||
|---|---|---|---|
| mCG | mCHG | mCHH | |
| Gene body | 4 | 4 | 5 |
| Upstream region (less than 1 kb from nearest gene) | 0 | 1 | 5 |
| Downstream region (less than 1 kb from nearest gene) | 0 | 1 | 4 |
| Intergenic (more than 1 kb from nearest gene) | 0 | 0 | 1 |
mRNA Sequencing and Promoter Methylation Profiling of a Single Drought
Next, the 24 stress-associated DMRs mapping near protein-coding genes were further investigated for effects on the expression of neighboring genes. There were 4,369 differentially expressed genes under this drought treatment compared with unstressed controls (Crisp et al., 2017). Our comparison with this mRNA sequencing data set revealed only four significant differentially regulated genes correlating with drought-associated DMRs (Supplemental Table S4). Not only is there a negligible relationship of drought DMRs to drought-responsive genes (hypergeometric test; P[X ≥ 4] = 0.54), but three of the correlating genes (ERD2, AT2G20920, and AT2G34060) exhibited only weak gene expression changes. Interestingly, NAC089, which showed the strongest transcriptional response (approximately 7-fold up-regulated) under these conditions, has been reported to demonstrate transcriptional memory in response to repeated dehydration stress (Ding et al., 2013). While there is an observable increase in mCHH, the hypermethylated state is not conserved across drought-stressed samples, and similar levels of methylation remain in the adjacent downstream region from the identified DMR (Fig. 1E). Therefore, while this methylation difference may have biological significance, it is unclear whether this is truly associated with drought stress.
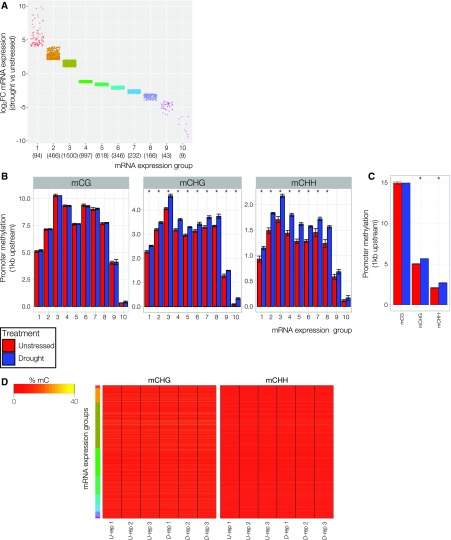
Our profiling of the DNA methylome suggests that it is relatively unresponsive to drought stress. Yet, this does not rule out an association; it is possible that the methylation profile of drought-responsive genes could distinguish them from other nonresponsive genes. For instance, given that most up-regulated genes do not display a change in DNA methylation, we would predict that their promoters are free from repressive DNA methylation to enable responsiveness. To investigate this possibility, the methylation state for all methylation contexts across the promoter region (considered as 1 kb upstream from gene annotation) of drought-responsive genes was profiled in unstressed and drought-treated plants. Methylation levels were averaged across genes clustered, using a k-means method, based on their log2 fold change in mRNA expression (Fig. 2A). There was no clear relationship between promoter methylation levels and the fold change in mRNA observed, although either strongly up- or down-regulated genes appeared to show lower levels of DNA methylation compared with other groups (Fig. 2B). There also appeared to be a slight, yet general, increase in promoter mCHG and mCHH levels in drought-treated samples. To test whether this increase reflected any characteristic of the promoters of drought-responsive loci, promoter methylation levels were averaged across 437 (mean group size from Fig. 2A) randomly selected loci that did not respond to drought stress (Fig. 2C). These loci, while generally having higher promoter methylation levels, also showed an increase in non-CG methylation, providing further evidence that this methylation difference was not reflective of gene expression changes. It was also apparent that some expression groups, with transcripts showing relatively small changes in expression, had higher levels of promoter methylation, possibly reflecting mRNA abundance under unstressed conditions. However, further inspection of transcripts in each expression group suggested that promoter methylation levels were not reflective of mRNA abundance under unstressed conditions (Supplemental Fig. S1E). We explored promoter non-CG methylation levels further to test whether there was a subset of drought-responsive genes driving this difference. However, these regions were found to be largely devoid of methylation, with the exception of a subset of loci (Fig. 2D). Despite the lack of association with drought-responsive mRNA expression, these findings implicate altered RdDM function under drought stress leading to elevated non-CG methylation upstream at a subset of genes (Matzke and Mosher, 2014).
Figure 2.
Promoter methylation levels at drought-responsive genes. A, Strip chart depicting log2 fold change in mRNA expression of all drought-responsive genes grouped based on k-means clustering. Dots represent individual drought-responsive loci. Numbers of genes in each cluster are presented in parentheses. B, Summarized methylation levels in the 1-kb region directly upstream of drought-responsive loci averaged across all genes in each expression group as defined in A. Bars denote mean, error bars denote standard error of the mean (n = 3), and asterisk denotes significant differences (P < 0.05) between treatments as determined by a Kruskal-Wallis rank sum test. C, Summarized methylation levels in the 1-kb region directly upstream of 437 randomly selected non-drought-responsive loci. Bars denote mean, error bars denote standard error of the mean (n = 3), and asterisk denotes significant differences (P < 0.05) between treatments as determined by a Kruskal-Wallis rank sum test. D, Heat maps of mCHG and mCHH levels summarized 1 kb directly upstream of drought-responsive loci for individual transcripts ordered by expression group (as defined in A) and subsequently by log2 fold change (top = highest; bottom = lowest).
Transgenerational Recurring Drought Stress
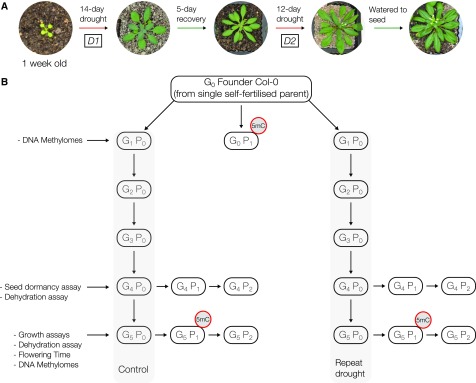
The experiment above highlights that DMRs do appear after the onset of a drought within a single generation. Although DMRs are present, the experimental design limits the ability to examine their biological relevance in a number of ways. First, the seed stock used contained existing epi-types that may interfere with stress-responsive changes to DNA methylation and/or prohibit detailed analyses by diluting any signal from stress-induced variation. Second, a single-generation experiment does not provide any insight into the heritable nature of methylation changes (Hauser et al., 2011b; Gutzat and Mittelsten Scheid, 2012). Third, biologically relevant DMRs may increase in number and persist over time if the stress is experienced repeatedly at different developmental stages both within a generation and across generations. Therefore, we performed a single-seed descent, recurring, and transgenerational drought stress experiment to directly address these experimental challenges.
Multiple independent lineages originating from a single inbred progenitor were propagated by single-seed descent, akin to previous mutant accumulation line experiments (Shaw et al., 2000; Becker et al., 2011; Schmitz et al., 2011). Plant lineages were subjected to either control growth conditions (unstressed lineages) or repeated drought stress composed of a 14-d drought, 5 d of recovery, followed by a second 12-d drought (Fig. 3A; Supplemental Fig. S2A). The first stress occurred during vegetative growth (D1) and the second during flowering (D2). This repeated drought treatment was performed through successive generations starting from founding plants (G0) through to fifth generation plants (G5; Fig. 3B). Direct progeny (P1) and progeny one generation removed (P2) of G4 and G5 plants, from independent lineages per treatment, were compared for altered growth and resilience. The DNA methylomes of six G5 P1 progeny per lineage condition, each from an independently propagated lineage, were assayed using WGBS. G0 P1 progeny also were assayed for a representation of initial DNA methylome patterns prior to generations of experimental treatment.
Figure 3.
Transgenerational repeated drought stress experiment. A, Scheme of the repeated drought stress treatment performed every generation. Initial water deprivation (D1) began on 1-week-old seedlings and lasted for 14 d. After a recovery period of 5 d, a second treatment was performed (D2) that lasted for 12 d. B, Multiple independent lineages were propagated by single-seed descent for five generations, from a founding inbred progenitor, with half of the lines being exposed to the repeated drought stress treatment every generation. Testing for transgenerational stress memory was performed on the P1 and P2 progeny of G4 and G5 plants with plants from each independent lineage. WGBS was performed on P1 progeny, each from an independent lineage, of G0 and G5 plants (red circles).
PSII performance was monitored using measures of chlorophyll fluorescence, allowing for nondestructive assaying of plant stress and vitality under drought, to maximize survival rate (Haitz and Lichtenthaler, 1988; Woo et al., 2008). Representative traces of various PSII parameters (see Table IV below) are shown for plants under control conditions, plants at the end of D1, and plants at the end of D2 (Supplemental Fig. S2, B–F). D1 and D2 plants demonstrated a corresponding reduction in both PSII quantum efficiency and photochemical quenching capacity, a reduction in the estimated fraction of open PSII centers, and some reduction in the maximal potential efficiency of PSII. For all these measures, D1 and D2 plants largely demonstrated similar trends, although the severity appeared greater after D2. For example, D2 plants showed a severely impaired nonphotochemical quenching profile, suggesting that plants after D2 were severely stressed to the point that they could not sufficiently activate photoprotective mechanisms. This suggests a greater impact of drought in mature plants undergoing the transition to reproduction.
Table IV. Photosynthetic parameters, and corresponding equations, to nondestructively assay the impact of the drought stress utilized in this study.
| Parameter | Equation | Interpretation |
|---|---|---|
| Fv | Fm – Fo | Variable fluorescence, the ability of PSII to perform photochemistry |
| Fv′ | Fm′ – Fo′ | |
| Fq′ | Fm′ – Ft′ | Photochemical quenching of fluorescence by open PSII centers |
| Fv/Fm | (Fm – Fo)/Fm | Maximum quantum efficiency of PSII |
| Fv′/Fm′ | (Fm′ – Fo′)/Fm′ | Estimate of the maximum quantum efficiency of PSII under actinic light |
| ΦPSII | Fq′/Fm′ | PSII quantum efficiency: the proportion of light absorbed by chlorophyll used for PSII photochemistry |
| qP | Fq′/Fv′ | Coefficient of photochemical quenching: relates PSII maximum efficiency to PSII operating efficiency |
| qL | (Fq′/Fv′)/(Fo′/Ft′) | Estimates the fraction of open PSII centers |
| NPQ | (Fm/Fm′) – 1 | Nonphotochemical quenching constant: estimates the rate constant for heat loss from PSII |
| Rfd | (Fp/Ft′) – 1 | Fluorescence decline ratio calculated using steady-state fluorescence: correlates with CO2 fixation rate, with Rfd > 3 indicative of highly efficient PSII and Rfd < 1 reflecting negligible net CO2 gain; this correlation allows this to be used as a vitality index; here, Fp is taken during the initial phase of the Kautsky effect, where the peak fluorescence is induced by actinic light |
Drought-Exposed Lineages Exhibit Enhanced Seed Dormancy
The progeny of G4 and G5 plants from unstressed and drought-treated lineages were compared to test whether sustained and repeated drought exposure over successive generations could lead to the formation of drought stress memory that might be evidenced as altered plant behavior or enhanced drought tolerance in drought-treated lineages.
The growth of 3-week-old descendants, from unstressed and drought-exposed lineages, was compared under control growth conditions for G5 P1 and P2 progeny. There were no intragenerational differences in plant size, using either green pixel count or fresh biomass, between descendants of watered and drought-treated parents (Supplemental Fig. S3, A and B). The growth rate in G5 P1 progeny, using green pixel counts of plant area over 3 weeks, showed that progeny from unstressed and drought lineages had equivalent growth rates (Supplemental Fig. S3C) and flowering times (Supplemental Fig. S3D). Thus, gross plant growth and development appears unaltered after experiencing repeated drought stress over previous generations.
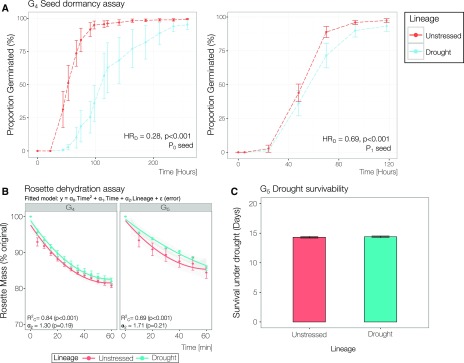
Seed provisioning is considered to be a significant mechanism for the transmission of adaptive transgenerational effects. For instance, seeds developed during periods of stress often have altered nutrient or hormone profiles, which holds important biological consequences such as propensity to germinate (Herman and Sultan, 2011). Indeed, previous transgenerational studies have reported phenotypes reliant on maternal exposure to stress (Murgia et al., 2015; Nosalewicz et al., 2016). Therefore, altered seed provisioning was tested by comparing dormancy in seed from P0 and P1 progeny from G4 plants of both lineages (Fig. 4A). Seed dormancy was compared by constructing a Cox proportional hazards model producing a comparative HR (McNair et al., 2012). Seeds from G5 P0 drought were 72% less likely to germinate (HRD = 0.28, P < 0.001) than seeds from unstressed lineages. It is possible that this was conveyed through maternal effects, such as increased abscisic acid (ABA) synthesis under drought stress, particularly since D2 occurred during early reproductive stages (Cutler et al., 2010). When seed dormancy was tested further in P1 seed, one generation removed from stress, the size of this effect was reduced but still statistically significant (HRD = 0.69, P < 0.001). While these observations are consistent with observations of maternal effects, in the form of altered seed provisioning, some dormancy is still retained in the seed of P1 progeny one generation removed from experimental drought.
Figure 4.
Progeny from drought-exposed lineages show enhanced seed dormancy. A, Independent dormancy assays performed on seeds from P0 (n = 6; more than 25 seeds per plate) and P1 (n = 9; more than 25 seeds per plate) progeny of G4 plants (P0 and P1 seeds, respectively). Points denote mean proportions of seeds germinated; error bars denote se. HR denotes the calculated hazard ratio from a fitted Cox proportional hazards model, representing the likelihood of germination between groups (HRD = drought versus unstressed lineage). B, Dehydration assay performed on detached rosettes of P1 progeny of G4 (n = 12) and G5 (n = 11) plants (independent experiments). A second-order polynomial regression, with a 95% confidence interval (shading), was performed to determine the coefficient for the lineage predictor term (α2). R2C denotes the conditional R2 calculated to assess model fit. C, Survival under the terminal drought experiment on transgenerational descendants (unstressed, n = 44; drought, n = 51). Bars denote means, and error bars denote se across two independent experiments.
It is possible that any form of transgenerational memory might only be observable in conditions of water limitation. One of the key responses to drought stress is stomatal closure (Verslues et al., 2006), and recent investigations have found that environmentally induced variation in stomatal development and index is regulated, at least in part, by DNA methylation, with some evidence for DNA methylation-mediated transgenerational transmission (Tricker et al., 2012, 2013). Greater stomatal control to prevent dehydration would be beneficial under water limitation; therefore, stomatal responsiveness was compared between lineages. A detached rosette dehydration experiment (see “Materials and Methods”) was performed on G4 and G5 P1 progeny. Independent experiments revealed that progeny from each lineage had very comparable rates of water loss, with lineage holding a very weak effect (α2 = 1.3–1.71, P > 0.05; Fig. 4B). Ultimately, if any form of drought stress memory was conveyed to the progeny of drought-stressed plants, then these progeny would be expected to exhibit improved survivability under drought. However, G5 P1 progeny from both lineages demonstrated near identical survivability under a longer term drought measured using the fluorescence decline ratio (Rfd) as a vitality index (Fig. 4C; Haitz and Lichtenthaler, 1988). In total, phenotypic assessment of transgenerational drought lineages revealed enhanced seed dormancy to be the only form of drought stress memory, which was partially retained in seeds one generation removed from stress.
Negligible DNA Methylome Epi-Alleles Associated with Transgenerational Drought Stress
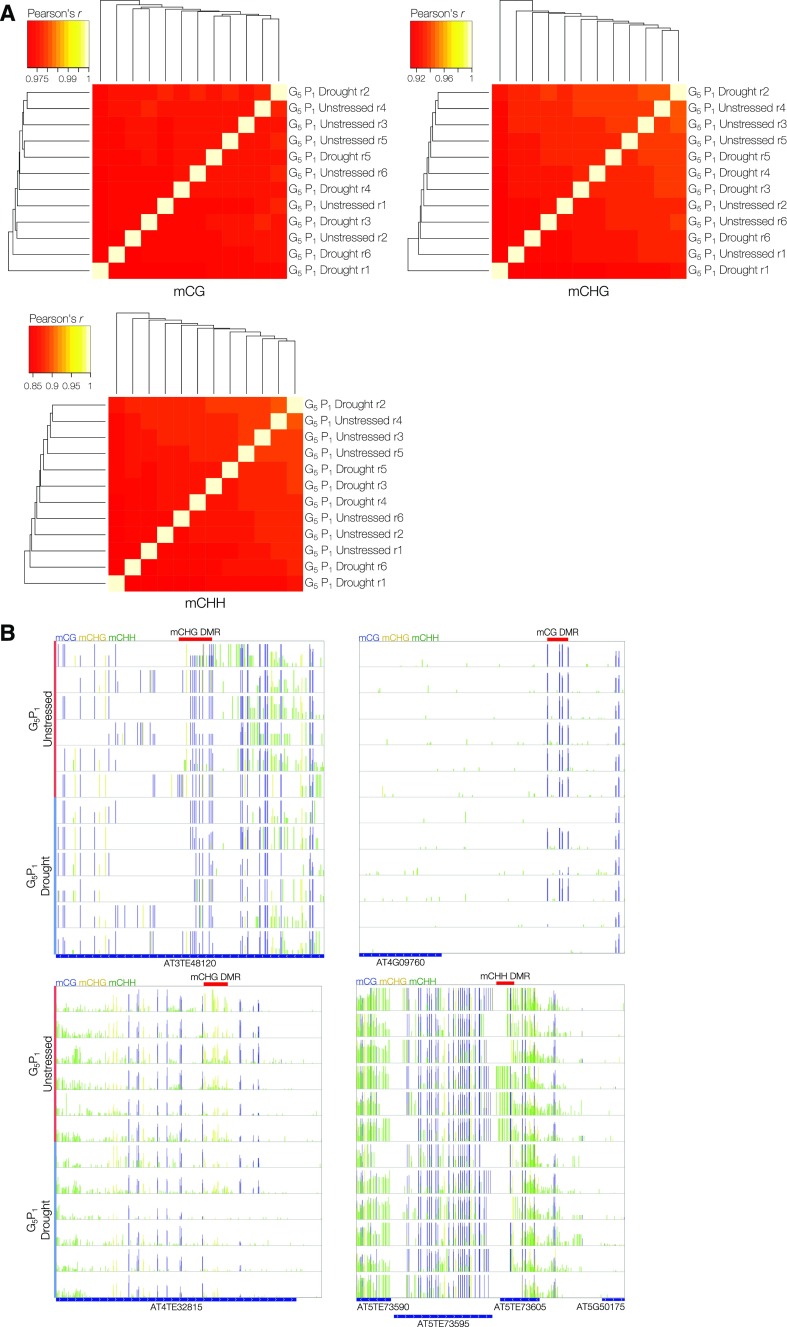
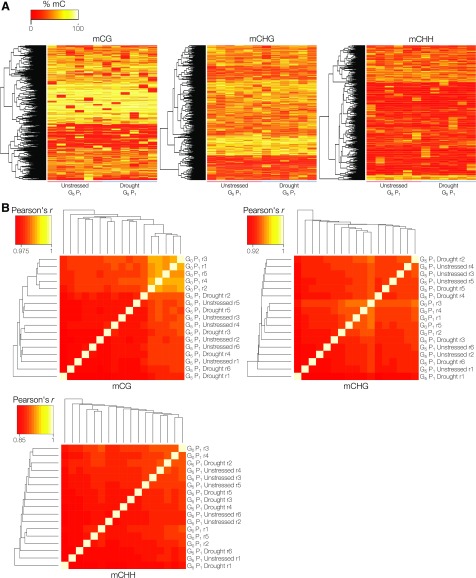
Beyond phenotypic measures of memory, we were interested in the extent of DNA methylation variation between these lineages associated with the transgenerational repeated drought stress. DNA methylomes were produced from whole rosettes of approximately 3-week-old G0 progeny (G0 P1) and G5 progeny from six independent lineages per condition (G5 P1 unstressed and G5 P1 drought), grown under control growth conditions (Supplemental Table S1). Each progeny plant that was sequenced came from an independently propagated lineage. Hierarchical clustering of all G5 P1 samples by genome-wide methylation levels, binned into 100-bp regions, confirmed that broad methylome patterns were highly similar among all progeny, excluding the possibility of genetic contamination, such as seed contamination or outcrossing, which could affect the DNA methylome patterns observed (Fig. 5A). In contrast to the previous experiment (Supplemental Fig. S1A), no clear epi-types were detected in the profiled G5 P1 progeny, despite being derived from independent lineages, confirming the importance of comparing relatively closely related plants. To identify conserved drought-induced heritable changes in the DNA methylome, we utilized DSS to call DMRs between progeny of G5 control and G5 drought lineages. This yielded just four transgenerational drought stress-associated DMRs (Table III; Supplemental Table S5). None of these overlapped with the epi-type or stress-associated DMRs that were identified in the previous within-generation drought stress experiment. This lack of variation was unexpected, since 40 DMRs were observed within a generation from a single drought stress; however, this reinforces the notion that heritable stress-induced variations in the DNA methylome are rare. Despite this conservative approach, none of the identified DMRs demonstrated complete conservation within treatment groups, and three of the DMRs mapped to repetitive regions of the genome (Fig. 5B). The fourth DMR was in intergenic space, 800 bp upstream of CEK3 (AT4G09760), and was present in only four of the six drought lineage progeny that were profiled.
Figure 5.
Limited methylome variation associated with transgenerational drought stress. A, Heat maps representing two-dimensional hierarchical clustering of correlations (Pearson’s r) in genome-wide DNA methylation levels, in all sequence contexts, averaged across 100-bp bins confirms similar broad DNA methylome patterns between all G5 descendants. B, Integrative Genomics Viewer (IGV) visualization of lineage-associated DMRs identified by DSS (red bars). Vertical blue, yellow, and green bars denote mean mCG, mCHG, and mCHH, respectively, at single C resolution.
Table III. Lineage-associated and spontaneous DSS-based DMRs identified in the transgenerational drought experiment.
| DMR Class | Contrast | Methylation Context |
|||
|---|---|---|---|---|---|
| mCG | mCHG | mCHH | Total | ||
| Lineage | G5 P1 unstressed versus G5 P1 drought | 1 | 2 | 1 | 4 |
| Spontaneous | G0 P1 versus G5 P1 unstressed | 1 | 10 | 12 | 23 |
| G0 P1 versus G5 P1 drought | 1 | 6 | 8 | 15 | |
Core ABA Signaling and Documented Memory Loci Remain Stable under Transgenerational Recurring Drought Stress
Given the negligible detection of transgenerational drought-associated DMRs using unbiased approaches, we undertook targeted analyses of subsets of the genome. The rationale for this strategy relates to the hypothesized biological relevance of methylation as a regulatory mechanism near, or within, annotated genes related to drought response and tolerance (Gutzat and Mittelsten Scheid, 2012). The directed approaches were used to examine DNA methylation levels at loci encoding the core signaling components in the ABA signaling pathway crucial for drought response and at previously characterized loci described to have stress-induced, transgenerational DNA methylome variation.
ABA induces a signaling cascade, involving both transcriptional and posttranscriptional changes, that activates drought-response mechanisms (Verslues et al., 2006; Cutler et al., 2010). Given the importance of this pathway and the observation of enhanced dormancy in progeny of drought-exposed lineages, it was postulated that loci encoding key components of the ABA signaling pathway (Hauser et al., 2011a) could be targets for memory formation. However, when differences in DNA methylation levels were assayed at these loci in G5 P1 progeny, between unstressed and treated lineages, they were found to be nearly identical, with the largest difference being a 4.45% decrease in mCG (Supplemental Table S6).
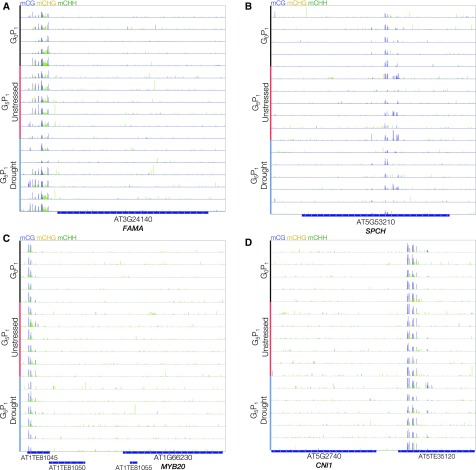
Low humidity-induced hypermethylation at the FAMA and SPCH loci was described to be transmitted to progeny in Landsberg erecta (Tricker et al., 2012, 2013). These loci were profiled across P1 progeny of G0 and G5 plants to look for evidence of hypermethylation (Fig. 6, A and B). Interestingly, both loci were found to be largely devoid of DNA methylation across all experimental samples, comparable to the unmethylated nonstressed plants reported previously (Tricker et al., 2012). However, there was no evidence of any transmissible hypermethylation at these loci, neither lineage dependent nor drought dependent. Notably, there was a region of stochastic differences, in all three sequence contexts of DNA methylation, downstream from the protein-coding region of FAMA. This observation raises the following possibilities: (1) regulation of the DNA methylome can be stress type specific; and (2) different ecotypes within a species may have altered stress-induced regulation of the DNA methylome.
Figure 6.
DNA methylation levels at loci reported to exhibit transgenerational stress-induced methylation variation. IGV visualization is shown for DNA methylation, in G0 P1 control plants and G5 P1 plants of both watered and drought-stressed lineages, across loci documented to exhibit transgenerational memory of stress-induced changes in DNA methylation: A, FAMA (low humidity-induced hypermethylation); B, SPCH (low humidity-induced hypermethylation); C, MYB20 (upstream HS-induced hypermethylation); and D, CNI1 (downstream HS-induced hypomethylation). Blue, orange, and green bars denote mean mCG, mCHG, and mCHH, respectively, at single C resolution.
Transgenerational hyperosmotic stress (HS) was reported recently to induce enhanced salt tolerance in P1 progeny of lineages exposed to salt stress for at least two generations (Wibowo et al., 2016). This enhanced tolerance was correlated with HS-DMRs, two of which occurred at TEs adjacent to MYB20 and CNI1. In the case of MYB20, HS-induced hypermethylation across an upstream TE correlated with persistent down-regulated MYB20 expression. In the case of CNI1, HS-induced hypomethylation across a downstream TE correlated with increased stress-responsive expression in the P1. Thus, we investigated these loci in the context of transgenerational drought stress to see if any hypermethylation or hypomethylation was evident in G5 P1 progeny. The DNA methylation pattern across and upstream region of MYB20, in all samples assayed in this study, was similar to that of unstressed G0 P1 progeny (Fig. 6C). Interestingly, select drought lineages did appear to show hypomethylation in an upstream TE akin to the P2 progeny, one generation removed from 75 mm salt stress, which did not exhibit enhanced salt tolerance (Wibowo et al., 2016). The CNI1 locus also was largely devoid of DNA methylation; however, the downstream TE was partly methylated in all sequence contexts (Fig. 6D). However, there was no transgenerational drought-induced hypomethylation at this downstream TE, as was observed across P0, P1, and P2 progeny of 75 mm salt-stressed parents. This supports the observed stochastic nature of DNA methylome variation in that methylome changes are not always universal and/or stable. Single studies may only capture a portion of this and potentiate the possibility that different abiotic stresses induce changes in the DNA methylome to different efficacies.
Greater Stochastic Variation and the Appearance of Spontaneous DNA Methylome Epi-Alleles in Transgenerational Lineages
Having observed a limited number of transgenerational drought stress-associated DMRs, we explored the extent of stochastic variation in the DNA methylome. Using the aforementioned 100-bp tile-based analysis revealed extensive variation using pairwise comparisons of all G5 P1 progeny across lineages (2,871 mCG, 2,284 mCHG, and 1,292 mCHH DMRs; Supplemental Table S7). Almost all changes appeared to be unique to individual lineages, with negligible conservation within treatment groups (Fig. 7A). Rank sum testing was repeated on these 100-bp tile-based DMRs to test for association with treatment; however, none of the DMRs were significant after P value correction for multiple comparisons. Collectively, this suggests that the Arabidopsis DNA methylome is relatively impervious to transgenerational drought stress, with the predominant source of variation being due to stochastic differences between lineages.
Figure 7.
Exploring stochastic and spontaneous methylome variation across transgenerational lineages. A, Heat maps of average methylation across 100-bp tile-based DMRs identified from pairwise comparisons, with one-dimensional hierarchical clustering of rows, between all samples. B, Heat maps representing two-dimensional hierarchical clustering of correlations (Pearson’s r) of genome-wide DNA methylation, in all sequence contexts, averaged across 100-bp tiles for all G0 and G5 progeny.
The spontaneous nature of epi-allele appearance in the Arabidopsis DNA methylome is well characterized and also has been documented to increase in frequency under environmental stress (Becker et al., 2011; Schmitz et al., 2011; Jiang et al., 2014). The appearance of spontaneous DMRs in the lineages generated in this study was explored by comparing P1 progeny from G0 and G5 plants using DSS (Table III; Supplemental Tables S8 and S9). Interestingly, more DSS-based DMRs were identified between G0 and G5 progeny regardless of lineage (Table III) at a magnitude comparable to previous observations of epi-allele accumulation (in the form of DMRs; Becker et al., 2011). Indeed, G0 siblings were found to have more similar genome-wide DNA methylation patterns to each other than to G5 descendants, particularly in the mCG context (Fig. 7B). Exposure to repeated drought stress for five successive generations did not lead to a greater number of DMRs; in fact, progeny from stressed lineages had fewer DMRs, than unstressed lineages, when compared with G0 P1 (Table III). Nine regions were in common between variations accumulated in unstressed and drought-exposed lineages, which may reflect truly labile DNA methylation sites from this data set. None of these nine regions were in common with previously identified labile regions. From the total 38 spontaneous DMRs identified here, only three were found to overlap with regions associated previously with spontaneous variation (Becker et al., 2011; Schmitz et al., 2011). Curiously, a handful of the overlapping sites occur across a hypothetical protein surrounded by TEs on chromosome 4 (AT4G19270), where there has been extensive non-mCG hypomethylation, yet unaffected mCG, in G5 progeny (Supplemental Fig. S4A).
Labile regions of the Arabidopsis DNA methylome have been identified previously, whether spontaneous, stress induced, or driven by genetic divergence across diverse environments (Becker et al., 2011; Schmitz et al., 2011; Jiang et al., 2014; Hagmann et al., 2015; Wibowo et al., 2016). The four transgenerational drought stress-associated DMRs, identified in this study, were overlapped with regions identified across the aforementioned data sets to test whether any of the four regions were in common with previously reported stress-induced regions. One of these transgenerational drought-associated DMRs overlapped; however, this region was not associated with a stress-induced change (Supplemental Fig. S4B; Supplemental Table S10).
Stochastic DMRs, using the 100-bp tile-based method in this study, also were overlapped with previously published DMRs to look for conservation across methylation-labile regions. An overlap of the stochastic DMRs identified in this study showed that 617 of 6,447 (9.5%) occurred at regions identified previously. Overlaps with specific studies remained low, ranging from 0.2% to 4.4% of regions from this study being detected previously. Of particular interest was to compare the stochastic transgenerational DMRs identified here with previously identified transgenerational spontaneous DMRs across 30 generations of single-seed descent (Becker et al., 2011; Schmitz et al., 2011). Twenty-four of 72 regions characterized as a site hosting a spontaneous transgenerational epi-allele (Schmitz et al., 2011), unlinked from cis-genetic variation, were identified out of 6,447 stochastic transgenerational DMRs (Supplemental Table S10). These were predominantly changes in mCG, occurring largely at intragenic or repetitive regions, including TEs and pseudogenes.
DISCUSSION
Notions of transgenerational plant stress memory are often discussed alongside DNA and chromatin alterations, as a potential mechanism underpinning their storage and transmission (Herman and Sultan, 2011; Tricker, 2015; Crisp et al., 2016). In particular, DNA methylation is considered a key epigenetic mechanism for which there is now growing evidence (Luo et al., 1996; Boyko and Kovalchuk, 2010; Tricker et al., 2013; Herman and Sultan, 2016; Wibowo et al., 2016; Zheng et al., 2017). The specific contribution of DNA methylation at, or near, protein-coding genes toward basal plant growth and endurance remains unknown, albeit essential for proper development (Finnegan et al., 1996; Henderson and Jacobsen, 2008; Zemach et al., 2013; Yamamuro et al., 2014). Despite documentation of the stable inheritance of spontaneously occurring epi-alleles (Becker et al., 2011; Schmitz et al., 2011), there still remains uncertainty regarding the malleability of the DNA methylome to stress-induced variation (Seymour et al., 2014; Eichten and Springer, 2015; Secco et al., 2015, 2017). Whether such DNA methylation changes are necessary for transgenerational stress memory is still unclear, with various memory traits not always aligning with changes in the DNA methylome (Ding et al., 2012; Sani et al., 2013; Murgia et al., 2015; Nosalewicz et al., 2016). In this investigation, we examined and compared the effect of drought stress on the Arabidopsis methylome both within and between generations.
Within-Generation Methylation Profiles in Response to Drought Stress
Within a generation, plants experiencing a mild drought stress that induced a substantial transcriptional response exhibited 40 stress-associated DNA methylation epi-alleles. However, these did not appear to correlate with drought-responsive gene expression changes. Further investigation of promoter methylation status at drought-responsive genes did not reveal any methylation features that distinguish drought-responsive genes. This did, however, reveal widespread non-CG hypermethylation in gene promoter regions in drought-treated plants, implicating altered RdDM performance under drought stress. Such observations are comparable to the non-CG hypermethylation, predominantly in the mCHH context, that occurred in the root tissue of rice (Oryza sativa) under phosphate starvation (Secco et al., 2015). The use of a DCL3 knockdown line suggested that the phosphate-induced non-CG DMRs were largely RdDM independent, and a similar approach would be beneficial to address the putative involvement of RdDM here. In both cases, however, there was minimal evidence of such methylation changes affecting gene expression. It is worth noting that the effects of DNA methylation changes could be confounded by the complexity of interactions between all the chemical marks that contribute to chromatin state (Eichten et al., 2014; Crisp et al., 2016). It is also possible that preexisting epi-type differences could influence stress-inducible transcriptional changes. To systematically uncouple such effects would require a much larger scale sequencing effort, which may become a viable option in the future.
The identification of multiple epi-types within a seed stock derived from bulk harvesting further demonstrates the importance of appropriate experimental design when testing for DNA methylation-mediated stress memory. Furthermore, as the epi-type DMRs were predominantly mCG-DMRs, it is possible that these epi-type DMRs represent true epigenetic differences arising between distantly related plants rather than being reflective of genetic differences (Schmitz et al., 2013). Regardless, the relative lack of drought stress-associated DNA methylome epi-alleles observed within a generation aligns with other studies using phosphate, temperature, or UV radiation that all present a stoic DNA methylome unperturbed by abiotic stress (Seymour et al., 2014; Eichten and Springer, 2015; Secco et al., 2015; Meng et al., 2016).
Transgenerational Inheritance and Methylome Profiling
Between generations, descendants of lineages exposed to successive generations of recurring drought exhibited only four transgenerational drought stress-associated DMRs compared with unstressed lineages. Significantly, none of these were in common with the 40 drought-associated epi-alleles detected within a generation, reinforcing the notion that transgenerational adaptive DNA methylation is a rare occurrence, even under conditions of abiotic stress (Pecinka et al., 2009; Seymour et al., 2014; Eichten and Springer, 2015; Hagmann et al., 2015; Secco et al., 2015; Crisp et al., 2016). Three of the four mapped to repetitive and already heavily methylated regions of the genome. The fourth DMR also was in intergenic space, albeit 800 bp upstream from CEK3. CEK3 encodes a protein, most highly transcribed in the hypocotyl, that is a part of the Choline/Ethanolamine Kinase family, for which CEK4 has been implicated in phospholipid biosynthesis and embryo development; however, mutation of CEK3 did not lead to the same phenotypes (Lin et al., 2015). It is unclear whether this DMR upstream of CEK3 would be of biological significance; however, it does not appear to be required for transgenerational drought stress, as it was only evident in four of the six drought lineage progeny profiled. One possibility, since each progeny plant was derived from an independent lineage, is that this DMR is only weakly induced by drought stress; however, this would require further elucidation.
Targeted analyses of specific ABA-related loci were undertaken, as ABA-responsive genes are critical for drought responses. A recent study also reported that key ABA signaling kinases regulate the activity of a chromatin-remodeling ATPase (Peirats-Llobet et al., 2016). This regulation allowed for the fine-tuning of downstream components of the ABA pathway, in particular ABI5, further potentiating not only ABA-mediated chromatin variation but variation that feeds back onto the ABA signaling pathway itself. However, the targeted analysis of genes encoding ABA signaling components did not reveal treatment-specific methylation changes at any corresponding loci.
When compared with published data sets studying methylome variation, one transgenerational drought stress DMR overlapped with a previously identified spontaneous locus (Schmitz et al., 2011). Certainly, the nature of DNA methylome variation at all identified DMRs (stress associated and stochastic) is reminiscent of the spontaneous changes characterized previously comparing plants separated by approximately 30 generations (Becker et al., 2011; Schmitz et al., 2011). Whether those stochastic variations in the DNA methylome are tied to a particular lineage with biological consequence may warrant further investigation, despite not being tied to the experimental treatment. Furthermore, a vast majority of DMRs identified in this study mapped to TEs or unannotated genomic regions. This is unsurprising, given the expected relationship between DNA methylation and TEs. TE movement is considered to be a driving force in the appearance of epi-alleles (facilitated or obligatory epi-alleles; Richards, 2006), and, indeed, documented environmentally induced epigenetic changes correlate with, although are not always necessary for, TE activity (Ito et al., 2011, 2016; Eichten et al., 2013; Ong-Abdullah et al., 2015; Stuart et al., 2016). Future studies should take into consideration the impact of TE regulation, under conditions of abiotic stress particularly in species with greater TE content, which possibly underpins at least a subset of the stochastic or spontaneous epi-alleles observed in this study.
There is evidence building for the possibility of transgenerational plant stress memory irrespective of chromatin variation (Agrawal, 2002; Rasmann et al., 2012; Murgia et al., 2015; Nosalewicz et al., 2016; Zheng et al., 2017). Indeed, a distinction has been made between transgenerational epigenetic effects, referring to nongenetically determined transgenerational phenotypes, and transgenerational epigenetic inheritance, referring to nongenetically determined transgenerational phenotypes attributable to heritable chromatin modifications (Youngson and Whitelaw, 2008). Thus, evidence for the formation of transgenerational drought stress memory was investigated in drought-exposed Arabidopsis lineages propagated by single-seed descent. Despite successive generations of repeated drought stress, during both vegetative and reproductive growth stages, no altered aboveground morphological growth phenotypes were observed. This also was surprising given the recent reports of transgenerational memory phenotypes observed in Arabidopsis for salinity and low humidity (Sani et al., 2013; Tricker et al., 2013; Wibowo et al., 2016). A caveat of this study was that root phenotypes were not investigated, as previously Polygonum persicaria and barley (Hordeum vulgare) roots demonstrated transgenerational memory phenotypes in response to drought (Herman and Sultan, 2016; Nosalewicz et al., 2016). However, root memory phenotypes are not a general occurrence, as demonstrated in studies of phosphate starvation in rice (Secco et al., 2015). Recently, propagation of rice under drought stress led to aboveground differences in generation 11 plants compared with the first generation (Zheng et al., 2017). However, in that study, critically, there were no unstressed lineages incorporated to enable analysis of the phenotypic changes to be considered alongside associated DMRs, as opposed to the stochastic methylome variability observed herein that can arise over such a long-term experiment.
Here, the only evidence of transgenerational memory was in the form of increased seed dormancy (72% enhanced dormancy), which persisted, to some extent, beyond a generation of drought stress exposure (31% enhanced dormancy). This seed-specific memory might be expected of a rapid-cycling annual species whose success is dependent on seed behavior (Grime et al., 1981; Thompson, 1994; Springthorpe and Penfield, 2015). Any effect of enhanced seed dormancy on other developmental phenotypes, in this study, would have been masked by the seed stratification treatment performed prior to experimentation. Although the potential adaptive advantage of increased seed dormancy was not tested directly in this study, it would not be inconsequential, as seed dormancy dictates the environment that progeny plants would germinate in, thus having a potentially critical impact on early growth (Finch-Savage and Leubner-Metzger, 2006; Shu et al., 2016). Such a trait also has been suggested to be an advantage for progeny whose parents were affected by herbivory (Agrawal, 2002; Rasmann et al., 2012).
Increased seed dormancy is a classic form of maternal imprinting, whereby environmental conditions experienced by the maternal plant can influence seed development, altering seed properties including propensity to germinate. For example, seeds that develop under conditions of stress induce maternal ABA production, which can increase seed ABA content, thus enhancing dormancy (Finch-Savage and Leubner-Metzger, 2006). This altered seed provisioning would be the simplest explanation for the enhanced seed dormancy observed, especially since the D2 treatment occurred during reproductive development. Indeed, independent transgenerational studies of Fe deficiency also have shown memory phenotypes to be carried through altered seed provisioning that were lost in the absence of stress (Murgia et al., 2015). Here, however, the enhanced seed dormancy persisted in seeds developed in the absence of stress (P1 seed), albeit to a weaker magnitude. This persistent memory is more consistent with the notion of transgenerational memory. The mechanism conveying this memory is not resolved; however, it appears to be DNA methylation independent. Histone modifications were not assayed in this study, but variations also may have been induced. Indeed, osmotic stress-induced variation in histone methylation has been reported previously to mediate stress priming to HS within a generation, lending support to this hypothesis (Sani et al., 2013).
CONCLUSION
Herein, we present a systematic and comprehensive investigation of the possibility for DNA methylation variants to act as heritable stress-induced epi-alleles to convey transgenerational drought stress memory for multiple physiological traits that could be associated with drought responsiveness. Overall, Arabidopsis showed one specific memory trait: elevated seed dormancy in both the direct seed of drought-stressed parents (72% enhanced dormancy) and in seed produced from P1 progeny, from drought-exposed lineages, grown in the absence of stress, albeit to a lesser magnitude of dormancy (31% enhanced dormancy). Whether this conveys an adaptive advantage remains unclear, as seed stratification was done prior to experimentation for aboveground memory traits. Furthermore, there are likely to be cell type-specific responses that contribute to the complexity of plant stress memory, which will be important to consider in future investigations. Despite the appearance of 40 drought-associated DMRs within a generation, transgenerational drought stress-induced epi-alleles were rare and are unlikely to act as a mechanism to convey any form of transgenerational stress memory. Rather, the majority of DNA methylation states are highly stable, and the variation observed in this investigation, within and across generations, appears to occur stochastically. In conclusion, despite evidence of transgenerational drought stress memory for one of the six traits examined, the DNA methylome was relatively impervious to stress-induced changes.
MATERIALS AND METHODS
Plant Germplasm, Growth, and Drought Stress Treatments
All Arabidopsis (Arabidopsis thaliana) plants used in this study were in the Columbia background. Plants were cultivated on soil under a 12-h photoperiod (8 am to 8 pm) of 100 to 150 μmol photons m−2 s−1, 20°C ± 0.5°C, and 55% ± 5% relative humidity. The desired light intensity was achieved using 250-W metal halide lamps (Venture Lighting; MH 250W/U). Prior to light, seeds were sown onto moist soil and kept at 4°C for three nights to allow for seed stratification. A slow-onset water deprivation treatment (drought stress) was imposed, after saturating soil moisture, by withholding watering for 9 d for the within-generation drought treatment.
The growth conditions for propagation of lineages by single-seed descent were identical to those above, with the exception of a 16-h photoperiod (8 am to 12 am) to promote rapid cycling. All lineages were initiated from a common inbred G0 progenitor to minimize genetic difference and stochastic DNA methylome variations. An extended version of the initial water deprivation treatment was applied twice every generation to lineages propagated under drought stress (Fig. 3A). The first treatment was applied at 1 week of age, which involved saturating soil moisture and subsequently withholding water for 2 weeks. Plants were then watered and allowed to recover for 5 d. The second treatment was repeated following recovery, although this time for only 12 d to minimize plant death. Plants were then watered until rosette leaf senescence and the appearance of dried, mature siliques for seed harvesting following the guidelines set by the Arabidopsis Biological Resource Centre.
High-Throughput Phenotyping Using PlantScreen
PlantScreen (Photon Systems Instruments), a platform for high-throughput phenotyping, was used to monitor plant growth and photosynthetic performance by measuring plant area and chlorophyll fluorescence (Humplík et al., 2015; Rungrat et al., 2016). An ANOVA with subsequent Tukey’s honestly significant difference (HSD) method was utilized to test for size differences at single time points. A second-order mixed-effect polynomial model was constructed to test for differences in growth rate between lineages.
Monitoring PSII Performance Using Chlorophyll Fluorescence
To monitor PSII performance under drought stress, measures of chlorophyll fluorescence were obtained using a PSI FluorCam (Photon System Instruments). Images were analyzed using the accompanying FluorCam7 imaging software. Chlorophyll fluorescence was measured across the adaxial side of dark-adapted (30 min) plants for 7 min under actinic light (approximately 800 μmol photons m−2 s−1) followed by 3 min in the dark, with regular measures of chlorophyll fluorescence induced by a saturating pulse (approximately 3,000 μmol photons m−2 s−1) as well as minimal fluorescence in the presence of measuring light only (Humplík et al., 2015; Rungrat et al., 2016). Measurements on 30-min dark-adapted plants allowed measurements of base fluorescence (Fo), fluorescence immediately prior to a saturating pulse (Ft), maximal fluorescence after a saturating pulse (Fm), and variable fluorescence (Fv). Subsequently, the Kautsky effect was induced, with the initial signal giving peak fluorescence (Fp) as PSII activity engages. Regular saturating pulses occur under actinic light, allowing measurement of the light-adapted counterparts: Fo′, Fm′, Fv′, and Ft′. The parameters shown in Table IV were calculated using these obtained values and the corresponding equations (Haitz and Lichtenthaler, 1988; Lichtenthaler and Miehé, 1997; Maxwell and Johnson, 2000; Lichtenthaler et al., 2005; Baker, 2008; Brestic and Zivcak, 2013; Murchie and Lawson, 2013).
Plant Biomass and Rosette Dehydration Assay
The rosette dehydration assay was performed as reported previously (Wilson et al., 2009). Briefly, rosettes of approximately 4-week-old plants, grown under control growth conditions as described above, were excised at the base and weighed on a five-digit fine balance (Mettler Toledo). This was used as the measurement of fresh biomass. An ANOVA with Tukey’s HSD posthoc analysis was used to determine significant differences (P < 0.05) in rosette biomass. The mass of excised rosettes was then monitored at regular intervals for 1 h. A mixed-effect second-order polynomial model was constructed to test for significantly reduced rate of water loss between lineages (P < 0.05).
Flowering Time
Flowering time was measured on mature plants grown under the growth conditions described above. This was compared using two metrics upon emergence of the floral bud: (1) number of days since emergence; and (2) number of true leaves. Differences in flowering time were analyzed using ANOVA with Tukey’s HSD posthoc for each metric.
Seed Dormancy Assay
Seed dormancy was tested on seeds collected from G4 P0 and G4 P1 plants using recommended methods (McNair et al., 2012). Dried and matured siliques were collected from senescing plants. Each silique was taken from an individual plant and considered as a single biological replicate, with six biological replicates per lineage. Seeds from an individual silique were released onto a 0.8% agar plate and kept immediately in the growth cabinet under the growth conditions described above. For the first 5 d, photographs of the plates were taken twice daily; thereafter, they were taken only once daily. At each time point, all seeds per plate were scored as either germinated or ungerminated. To statistically compare seed dormancy, a Cox proportional hazards model was produced. From this model, calculation of the HR provides a comparative value between treatment groups. The HRD was calculated relative to unstressed lineages.
Assaying Survival under Prolonged Drought
The length of survival under drought was tested by performing a terminal drought experiment. G5 P1 progeny from multiple independent lineages, per treatment, were grown under control growth conditions, as described above, to approximately 3 weeks of age. Subsequently, soil was watered to saturation and excess water was drained. Watering was withheld thereafter, and plant vitality was monitored nondestructively with chlorophyll fluorescence measurements. The parameter Rfd was utilized as a vitality index, where plants that demonstrated Rfd < 1 are considered to be dead (Table IV; Haitz and Lichtenthaler,1988).
WGBS Library Preparation
WGBS was performed from snap-frozen leaf tissue of harvested whole rosettes. Details for all samples are provided in Supplemental Table S1. Genomic DNA was extracted using the Qiagen DNeasy Plant Mini Kit, as per the manufacturer’s instructions, and quantified using the ND-1000 Spectrophotometer (NanoDrop Technologies). One hundred to 200 ng of fragmented (Covaris) and purified genomic DNA was bisulfite converted using the Zymo DNA-Gold bisulfite conversion kit (Zymo Research). WGBS libraries were constructed using the Accel-NGS Methyl-Seq DNA Library Kit (Swift Biosciences) as per the manufacturer’s instructions. All purification steps were performed using Sera-mag SpeedBeads (GE Healthcare). The concentration and size distribution of bead-purified libraries were quantified on the Perkin-Elmer GXII using a DNA High Sensitivity kit. Libraries were subsequently pooled equimolar, in six-sample pools, and sequenced across a HiSeq2500 (100-bp single end; Illumina), with a 5% to 10% spike in PhiX DNA, depending on sample complexity, at the ACRF Biomolecular Research Facility (Australian National University, Canberra).
WGBS Analysis
Raw sequencing reads were quality controlled and trimmed using Trim Galore! (version 0.3.7), Cutadapt (version 1.9), and FastQC (version 0.11.2). Reads were then aligned to the TAIR10 reference genome using Bismark (version 0.14.5; Krueger and Andrews, 2011) with the flags-bowtie1 -n 2, -l 20 (Bowtie1 version 1.1.2; Langmead et al., 2009). Methylated Cs were extracted from aligned reads using the Bismark methylation extractor with default parameters. Bisulfite conversion efficiency was calculated from the proportion of unconverted Cs in the mCHH context from the chloroplast genome and ranged from 99.3% to 99.7% per sample. The proportion of mCG, mCHG, and mCHH was determined as weighted methylation (Schultz et al., 2012) across reads at single C resolution and across 100-bp tiles for genome-wide comparisons. Pearson’s correlation coefficient (r) of methylation levels, between samples, was performed on the 100-bp bin mean methylation levels in all sequence contexts. Methylation levels were assigned to annotated genes and TE features of the TAIR10 assembly using Bedtools (version 2.21.0; Quinlan and Hall, 2010) and the Araport11 genome reannotation (Cheng et al., 2017). Sequencing summary statistics are provided in Supplemental Table S1.
Identifying DMRs
This study employed two unbiased approaches to identify both stochastic and treatment-associated DMRs based on a previous combinatorial approach (Eichten et al., 2016).
First, to look at stochastic variation in the DNA methylome, regardless of treatment, DMRs were identified using pairwise comparisons employing a method based on average methylation binned to 100-bp tiles across the genome. In brief, pairwise comparisons were performed between corresponding 100-bp tiles in all samples. For each pairwise sample comparison, all 100-bp tiles were called differentially methylated if the absolute difference in methylation levels met a given threshold (mCG, 70%; mCHG, 50%; mCHH, 40%) alongside a minimum coverage and number of Cs (10× coverage, three Cs). Adjacent tiles identified as DMRs were collapsed into a single tile. All results were compared, and the largest region was kept for any overlapping DMRs between pairwise comparisons. A Kruskal-Wallis rank-sum test was performed to identify significant differences between treatment groups using 100-bp tile-based DMRs with a Bonferroni posthoc P value correction for multiple comparisons.
Second, a more conservative approach was used to identify statistically significant treatment-associated DMRs using a Bayesian hierarchical modeling approach, incorporating technical and biological variation at the individual C level, with the R package DSS (version 2.10.0; Feng et al., 2014). This was performed using the recommended default settings (with smoothing to allow for imputation of missing data) except for a reduced smoothing tile size (smoothing.span = 100). The threshold methylation difference for DMRs in each sequence context (delta) also was adjusted to 40% for mCG, 20% for mCHG, and 20% for mCHH. DSS calculates an adjusted P value (q value) based on the posterior probability that the difference specified (delta) is significant; DMRs were considered significant at q < 0.05.
Data Visualization and Statistical Analyses
Data visualization and statistical analyses, as described above and throughout the article, were conducted in R (version 3.3.2) using the appropriate packages (Wickham, 2007, 2009, 2011; Bache and Wickham, 2014; R Core Team, 2016; Warnes et al., 2016). DNA methylome patterns, including identified DMRs, were viewed using the IGV (Robinson et al., 2011). The R lme4 package (version 1.1; Bates et al., 2015) was used for generalized mixed-effects modeling with fixed (e.g. lineage treatment) and random (e.g. blocking design) effects. Model fit was assessed using the conditional R2 value, calculated using piecewiseSEM (Lefcheck, 2016), and the Akaike information criterion to compare relative fit. Pearson’s r was calculated using the R cor function (method = “pearson”). A hypergeometric test was performed using the R phyper function. Expression-based clustering of drought-responsive transcripts was achieved using the R kmeans function (centers = 10). Kruskal-Wallis rank-sum tests were performed using the R kruskal.test function. The R p.adjust function (method = “bonferroni”) allowed for posthoc P value correction for multiple pairwise comparisons. Survival analyses to compare seed dormancy between lineages, using a Cox proportional hazards model, were performed using the R survival package (version 2.41; Therneau, 2015). All statistical analyses, including modeling, were produced on single data points. Biological replication, unless described otherwise, was considered to be on independent whole plants. The R DiGGer package (version 0.2.31; Coombes, 2011) was used to produce spatially optimized complete randomized experimental designs.
Accession Numbers
The next-generation sequencing data sets utilized for this article are available at the following National Center for Biotechnology Information Gene Expression Omnibus and Sequence Read Archive data repositories: GSE94075 (WGBS; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE94075) and PRJNA391262 (mRNA sequencing; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA391262/). Bioinformatic pipelines are freely available on github (https://github.com/dtrain16/NGS-scripts). Other accession numbers are as follows: ABI5 (AT2G36270), ERD2 (AT1G29330), MYB20 (AT1G66230), AT2G20920, AT2G34060, FAMA (AT3G24140), CEK3 (AT4G09760), AT4G19270, NAC089 (AT5G22290), CNI1 (AT5G27420), and SPCH (AT5G53210).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Preexisting differences in the methylome define multiple epi-types.
Supplemental Figure S2. Representative plants and chlorophyll fluorescence profiles characterizing the impacts of D1 and D2.
Supplemental Figure S3. Characterizing the growth of G4 and G5 progeny from unstressed and drought-exposed lineages.
Supplemental Figure S4. DNA methylation-labile regions identified in the transgenerational drought stress experiment.
Supplemental Table S1. WGBS summary statistics.
Supplemental Table S2. Epi-type-associated DMRs.
Supplemental Table S3. Drought stress-associated DMRs.
Supplemental Table S4. Gene-mapping DMRs associated with drought-responsive genes.
Supplemental Table S5. Transgenerational drought stress-associated DMRs.
Supplemental Table S6. DNA methylation differences at core ABA signaling genes.
Supplemental Table S7. Stochastic DMRs between all G5 progeny.
Supplemental Table S8. DSS-identified DMRs identified between G0 P1 progeny and G5 P1 progeny from lineages propagated under control conditions.
Supplemental Table S9. DSS-identified DMRs identified between G0 P1 progeny and G5 P1 progeny from lineages propagated with repeated drought stress.
Supplemental Table S10. Transgenerational drought-DMRs identified in public DMR data sets.
Supplemental Table S11. Overlap of stochastic DMRs with identifie epi-alleles.
Acknowledgments
We thank Ryan Lister, Dr. Ryan McQuinn, Dr. Andrew Bowerman, and Estee Tee for discussion and critical feedback on experiments and interpretations. We acknowledge the Biomolecular Resource Facility at the Australian National University for performing Illumina sequencing, the Australian Plant Phenomics Facility at the Australian National University for phenotyping and growth facilities, and the Genome Discovery Unit and the Statistical Consulting Unit at the Australian National University, particularly Dr. Terry Neeman, for extensive and knowledgeable assistance and insight with experimental design and analyses. We also make special mention of the contributions and wisdom of the late Dr. Sylvain Foret.
Footnotes
This project was supported by the ARC Centre of Excellence in Plant Energy Biology (CE140100008). D.R.G. and P.A.C. were supported by the Grains Research and Development Corporation (GRS10683 and GRS184) and Australian Postgraduate Awards. S.R.E. was funded by a Discovery Early Career Researcher Award (DE150101206).
Articles can be viewed without a subscription.
References
- Agrawal AA. (2002) Herbivory and maternal effects: mechanisms and consequences of transgenerational induced plant resistance. Ecology 83: 3408 [Google Scholar]
- Bache SM, Wickham H (2014) magrittr: a forward-pipe operator for R. https://cran.r-project.org/package=magrittr
- Baker NR. (2008) Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu Rev Plant Biol 59: 89–113 [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67: 1–48 [Google Scholar]
- Becker C, Hagmann J, Müller J, Koenig D, Stegle O, Borgwardt K, Weigel D (2011) Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480: 245–249 [DOI] [PubMed] [Google Scholar]
- Boavida LC, Becker JD, Feijó JA (2005) The making of gametes in higher plants. Int J Dev Biol 49: 595–614 [DOI] [PubMed] [Google Scholar]
- Boyko A, Kovalchuk I (2010) Transgenerational response to stress in Arabidopsis thaliana. Plant Signal Behav 5: 995–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brestic M, Zivcak M (2013) PSII fluorescence techniques for measurement of drought and high temperature stress signal in crop plants: protocols and applications. In G Rout and A Das, eds, Molecular Stress Physiology of Plants. Springer, India, pp 87–131 [Google Scholar]
- Bruce TJA, Matthes MC, Napier JA, Pickett JA (2007) Stressful “memories” of plants: evidence and possible mechanisms. Plant Sci 173: 603–608 [Google Scholar]
- Calarco JP, Borges F, Donoghue MTA, Van Ex F, Jullien PE, Lopes T, Gardner R, Berger F, Feijó JA, Becker JD, et al. (2012) Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayuela E, Perez-Alfocea F, Caro M, Bolarin MC (1996) Priming of seeds with NaCl induces physiological changes in tomato plants grown under salt stress. Physiol Plant 96: 231–236 [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2016) Learning the languages of the chloroplast: retrograde signaling and beyond. Annu Rev Plant Biol 67: 25–53 [DOI] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD (2017) Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J 89: 789–804 [DOI] [PubMed] [Google Scholar]
- Conrath U, Beckers GJM, Flors V, García-Agustín P, Jakab G, Mauch F, Newman MA, Pieterse CMJ, Poinssot B, Pozo MJ, et al. (2006) Priming: getting ready for battle. Mol Plant Microbe Interact 19: 1062–1071 [DOI] [PubMed] [Google Scholar]
- Coombes N. (2011) DiGGer: DiGGer design generator under correlation and blocking. http://www.austatgen.org/files/software/downloads
- Crisp PA, Ganguly D, Eichten SR, Borevitz JO, Pogson BJ (2016) Reconsidering plant memory: intersections between stress recovery, RNA turnover, and epigenetics. Sci Adv 2: e1501340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp PA, Ganguly DR, Smith AB, Murray KD, Estavillo GM, Searle I, Ford E, Bogdanović O, Lister R, Borevitz JO, et al. (2017) Rapid recovery gene downregulation during excess-light stress and recovery in Arabidopsis. Plant Cell 29: 1836–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Ding Y, Fromm M, Avramova Z (2012) Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat Commun 3: 740. [DOI] [PubMed] [Google Scholar]
- Ding Y, Liu N, Virlouvet L, Riethoven JJ, Fromm M, Avramova Z (2013) Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol 13: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Virlouvet L, Liu N, Riethoven JJ, Fromm M, Avramova Z (2014) Dehydration stress memory genes of Zea mays: comparison with Arabidopsis thaliana. BMC Plant Biol 14: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin MJ, Zhang P, Meng D, Remigereau MS, Osborne EJ, Casale FP, Drewe P, Kahles A, Jean G, Vilhjálmsson B, et al. (2015) DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. eLife 4: e05255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten SR, Briskine R, Song J, Li Q, Swanson-Wagner R, Hermanson PJ, Waters AJ, Starr E, West PT, Tiffin P, et al. (2013) Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell 25: 2783–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten SR, Schmitz RJ, Springer NM (2014) Epigenetics: beyond chromatin modifications and complex genetic regulation. Plant Physiol 165: 933–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten SR, Springer NM (2015) Minimal evidence for consistent changes in maize DNA methylation patterns following environmental stress. Front Plant Sci 6: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten SR, Stuart T, Srivastava A, Lister R, Borevitz JO (2016) DNA methylation profiles of diverse Brachypodium distachyon align with underlying genetic diversity. Genome Res 26: 1520–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Conneely KN, Wu H (2014) A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res 42: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández AP, Strand A (2008) Retrograde signaling and plant stress: plastid signals initiate cellular stress responses. Curr Opin Plant Biol 11: 509–513 [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G (2006) Seed dormancy and the control of germination. New Phytol 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA 93: 8449–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodger JQD, Schachtman DP (2010) Re-examining the role of ABA as the primary long-distance signal produced by water-stressed roots. Plant Signal Behav 5: 1298–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MJ, Carmody M, Albrecht V, Pogson B (2013) Systemic and local responses to repeated HL stress-induced retrograde signaling in Arabidopsis. Front Plant Sci 3: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grime JP, Mason G, Curtis AV, Rodman J, Band SR (1981) A comparative study of germination characteristics in a local flora. J Ecol 69: 1017 [Google Scholar]
- Gutzat R, Mittelsten Scheid O (2012) Epigenetic responses to stress: triple defense? Curr Opin Plant Biol 15: 568–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann J, Becker C, Müller J, Stegle O, Meyer RC, Wang G, Schneeberger K, Fitz J, Altmann T, Bergelson J, et al. (2015) Century-scale methylome stability in a recently diverged Arabidopsis thaliana lineage. PLoS Genet 11: e1004920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haitz M, Lichtenthaler HK (1988) The measurement of Rfd-values as plant vitality indices with the portable field chlorophyll fluorometer and the Pam-fluorometer. In Lichtenthaler HK, ed, Applications of Chlorophyll Fluorescence in Photosynthesis Research, Stress Physiology, Hydrobiolgy, and Remote Sensing. Springer, Dordrecht, The Netherlands, pp 249–254 [Google Scholar]
- Hauser F, Waadt R, Schroeder JI (2011a) Evolution of abscisic acid synthesis and signaling mechanisms. Curr Biol 21: R346–R355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser MT, Aufsatz W, Jonak C, Luschnig C (2011b) Transgenerational epigenetic inheritance in plants. Biochim Biophys Acta 1809: 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Martienssen RA (2014) Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157: 95–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE (2008) Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes Dev 22: 1597–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JJ, Sultan SE (2011) Adaptive transgenerational plasticity in plants: case studies, mechanisms, and implications for natural populations. Front Plant Sci 2: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JJ, Sultan SE (2016) DNA methylation mediates genetic variation for adaptive transgenerational plasticity. Proc Biol Sci 283: 20160988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilker M, Schwachtje J, Baier M, Balazadeh S, Bäurle I, Geiselhardt S, Hincha DK, Kunze R, Mueller-Roeber B, Rillig MC, et al. (2016) Priming and memory of stress responses in organisms lacking a nervous system. Biol Rev Camb Philos Soc 91: 1118–1133 [DOI] [PubMed] [Google Scholar]
- Humplík JF, Lazár D, Husičková A, Spíchal L (2015) Automated phenotyping of plant shoots using imaging methods for analysis of plant stress responses: a review. Plant Methods 11: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J (2011) An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472: 115–119 [DOI] [PubMed] [Google Scholar]
- Ito H, Kim JM, Matsunaga W, Saze H, Matsui A, Endo TA, Harukawa Y, Takagi H, Yaegashi H, Masuta Y, et al. (2016) A stress-activated transposon in Arabidopsis induces transgenerational abscisic acid insensitivity. Sci Rep 6: 23181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab G, Ton J, Flors V, Zimmerli L, Métraux JP, Mauch-Mani B (2005) Enhancing Arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol 139: 267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L, Neumann DA, Schmitz RJ (2015) Crop epigenomics: identifying, unlocking, and harnessing cryptic variation in crop genomes. Mol Plant 8: 860–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Mithani A, Belfield EJ, Mott R, Hurst LD, Harberd NP (2014) Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Res 24: 1821–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492 [DOI] [PubMed] [Google Scholar]
- Karpinski S, Reynolds H, Karpinska B, Wingsle G, Creissen G, Mullineaux P (1999) Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 284: 654–657 [DOI] [PubMed] [Google Scholar]
- Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefcheck JS. (2016) piecewiseSEM: piecewise structural equation modelling in R for ecology, evolution, and systematics. Methods Ecol Evol 7: 573–579 [Google Scholar]
- Li Q, Eichten SR, Hermanson PJ, Springer NM (2014) Inheritance patterns and stability of DNA methylation variation in maize near-isogenic lines. Genetics 196: 667–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK, Buschmann C, Knapp M (2005) How to correctly determine the different chlorophyll fluorescence parameters and the chlorophyll fluorescence decrease ratio RFd of leaves with the PAM fluorometer. Photosynthetica 43: 379–393 [Google Scholar]
- Lichtenthaler HK, Miehé JA (1997) Fluorescence imaging as a diagnostic tool for plant stress. Trends Plant Sci 2: 316–320 [Google Scholar]
- Lin YC, Liu YC, Nakamura Y (2015) The choline/ethanolamine kinase family in Arabidopsis: essential role of CEK4 in phospholipid biosynthesis and embryo development. Plant Cell 27: 1497–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E (1996) Origin of floral asymmetry in Antirrhinum. Nature 383: 794–799 [DOI] [PubMed] [Google Scholar]
- Martín G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E (2016) Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat Commun 7: 11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJM (2009) RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 21: 367–376 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394–408 [DOI] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN (2000) Chlorophyll fluorescence: a practical guide. J Exp Bot 51: 659–668 [DOI] [PubMed] [Google Scholar]
- McNair JN, Sunkara A, Frobish D (2012) How to analyse seed germination data using statistical time-to-event analysis: non-parametric and semi-parametric methods. Seed Sci Res 22: 77–95 [Google Scholar]
- Meng D, Dubin M, Zhang P, Osborne EJ, Stegle O, Clark RM, Nordborg M (2016) Limited contribution of DNA methylation variation to expression regulation in Arabidopsis thaliana. PLoS Genet 12: e1006141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelbart MV, Hasegawa PM, Bailey-Serres J (2015) Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat Rev Genet 16: 237–251 [DOI] [PubMed] [Google Scholar]
- Molinier J, Ries G, Zipfel C, Hohn B (2006) Transgeneration memory of stress in plants. Nature 442: 1046–1049 [DOI] [PubMed] [Google Scholar]
- Murchie EH, Lawson T (2013) Chlorophyll fluorescence analysis: a guide to good practice and understanding some new applications. J Exp Bot 64: 3983–3998 [DOI] [PubMed] [Google Scholar]
- Murgia I, Giacometti S, Balestrazzi A, Paparella S, Pagliano C, Morandini P (2015) Analysis of the transgenerational iron deficiency stress memory in Arabidopsis thaliana plants. Front Plant Sci 6: 745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederhuth CE, Schmitz RJ (2017) Putting DNA methylation in context: from genomes to gene expression in plants. Biochim Biophys Acta 1860: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosalewicz A, Siecińska J, Śmiech M, Nosalewicz M, Wiącek D, Pecio A, Wach D (2016) Transgenerational effects of temporal drought stress on spring barley morphology and functioning. Environ Exp Bot 131: 120–127 [Google Scholar]
- Ong-Abdullah M, Ordway JM, Jiang N, Ooi SE, Kok SY, Sarpan N, Azimi N, Hashim AT, Ishak Z, Rosli SK, et al. (2015) Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 525: 533–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A, Rosa M, Schikora A, Berlinger M, Hirt H, Luschnig C, Mittelsten Scheid O (2009) Transgenerational stress memory is not a general response in Arabidopsis. PLoS ONE 4: e5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirats-Llobet M, Han SK, Gonzalez-Guzman M, Jeong CW, Rodriguez L, Belda-Palazon B, Wagner D, Rodriguez PL (2016) A direct link between abscisic acid sensing and the chromatin-remodeling ATPase BRAHMA via core ABA signaling pathway components. Mol Plant 9: 136–147 [DOI] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G (2009) Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 10: 192–206 [DOI] [PubMed] [Google Scholar]
- Probst AV, Mittelsten Scheid O (2015) Stress-induced structural changes in plant chromatin. Curr Opin Plant Biol 27: 8–16 [DOI] [PubMed] [Google Scholar]
- Quadrana L, Colot V (2016) Plant transgenerational epigenetics. Annu Rev Genet 50: 467–491 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2016) R: A Language and Environment for Statistical Computing. The R Foundation, Vienna, Austria
- Rasmann S, De Vos M, Casteel CL, Tian D, Halitschke R, Sun JY, Agrawal AA, Felton GW, Jander G (2012) Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol 158: 854–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J, Wulff BBH, Mirouze M, Marí-Ordóñez A, Dapp M, Rozhon W, Bucher E, Theiler G, Paszkowski J (2009) Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev 23: 939–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ. (2006) Inherited epigenetic variation: revisiting soft inheritance. Nat Rev Genet 7: 395–401 [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossel JB, Wilson PB, Hussain D, Woo NS, Gordon MJ, Mewett OP, Howell KA, Whelan J, Kazan K, Pogson BJ (2007) Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell 19: 4091–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rungrat T, Awlia M, Brown T, Cheng R, Sirault X, Fajkus J, Trtilek M, Furbank B, Badger M, Tester M, et al. (2016) Using phenomic analysis of photosynthetic function for abiotic stress response gene discovery. The Arabidopsis Book 14: e0185, doi/10.1199/tab.0185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sani E, Herzyk P, Perrella G, Colot V, Amtmann A (2013) Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol 14: R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Lewsey MG, O’Malley RC, Urich MA, Libiger O, Schork NJ, Ecker JR (2011) Transgenerational epigenetic instability is a source of novel methylation variants. Science 334: 369–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Urich MA, Nery JR, Pelizzola M, Libiger O, Alix A, McCosh RB, Chen H, Schork NJ, et al. (2013) Patterns of population epigenomic diversity. Nature 495: 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MD, Schmitz RJ, Ecker JR (2012) ‘Leveling’ the playing field for analyses of single-base resolution DNA methylomes. Trends Genet 28: 583–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Wang C, Shou H, Schultz MD, Chiarenza S, Nussaume L, Ecker JR, Whelan J, Lister R (2015) Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife 4: 1–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Whelan J, Rouached H, Lister R (2017) Nutrient stress-induced chromatin changes in plants. Curr Opin Plant Biol 39: 1–7 [DOI] [PubMed] [Google Scholar]
- Seymour DK, Koenig D, Hagmann J, Becker C, Weigel D (2014) Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genet 10: e1004785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RGG, Byers DLL, Darmo E (2000) Spontaneous mutational effects on reproductive traits of Arabidopsis thaliana. Genetics 155: 369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Liu XD, Xie Q, He ZH (2016) Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant 9: 34–45 [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzić M, Becker JD, Feijó JA, Martienssen RA (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM. (2013) Epigenetics and crop improvement. Trends Genet 29: 241–247 [DOI] [PubMed] [Google Scholar]
- Springthorpe V, Penfield S (2015) Flowering time and seed dormancy control use external coincidence to generate life history strategy. eLife 4: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MVC, Feng S, Bernatavichute YV, Jacobsen SE (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Eichten SR, Cahn J, Karpievitch YV, Borevitz JO, Lister R (2016) Population scale mapping of transposable element diversity reveals links to gene regulation and epigenomic variation. eLife 5: 1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau TM. (2015) A package for survival analysis in S. https://CRAN.R-project.org/package=survival
- Thompson L. (1994) The spatiotemporal effects of nitrogen and litter on the population dynamics of Arabidopsis thaliana. J Ecol 82: 63–68 [Google Scholar]
- Tricker PJ. (2015) Transgenerational inheritance or resetting of stress-induced epigenetic modifications: two sides of the same coin. Front Plant Sci 6: 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricker PJ, Gibbings JG, Rodríguez López CM, Hadley P, Wilkinson MJ (2012) Low relative humidity triggers RNA-directed de novo DNA methylation and suppression of genes controlling stomatal development. J Exp Bot 63: 3799–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricker PJ, López CM, Gibbings G, Hadley P, Wilkinson MJ (2013) Transgenerational, dynamic methylation of stomata genes in response to low relative humidity. Int J Mol Sci 14: 6674–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC. (2016) The intelligent behavior of plants. Trends Plant Sci 21: 286–294 [DOI] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45: 523–539 [DOI] [PubMed] [Google Scholar]
- Virlouvet L, Fromm M (2015) Physiological and transcriptional memory in guard cells during repetitive dehydration stress. New Phytol 205: 596–607 [DOI] [PubMed] [Google Scholar]
- Wang X, Vignjevic M, Jiang D, Jacobsen S, Wollenweber B (2014) Improved tolerance to drought stress after anthesis due to priming before anthesis in wheat (Triticum aestivum L.) var. Vinjett. J Exp Bot 65: 6441–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw WHA, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, et al. (2016) gplots: various R programming tools for plotting data. https://cran.r-project.org/package=gplots
- Wibowo A, Becker C, Marconi G, Durr J, Price J, Hagmann J, Papareddy R, Putra H, Kageyama J, Becker J, et al. (2016) Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. eLife 5: 1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2007) reshaping data with the reshape package. J Stat Softw 21: 1–20 [Google Scholar]
- Wickham H. (2009) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York [Google Scholar]
- Wickham H. (2011) The split-apply-combine strategy for data analysis. J Stat Softw 40: 1–29 [Google Scholar]
- Wilson PB, Estavillo GM, Field KJ, Pornsiriwong W, Carroll AJ, Howell KA, Woo NS, Lake JA, Smith SM, Millar AH, et al. (2009) The nucleotidase/phosphatase SAL1 is a negative regulator of drought tolerance in Arabidopsis. Plant J 58: 299–317 [DOI] [PubMed] [Google Scholar]
- Woo NS, Badger MR, Pogson BJ (2008) A rapid, non-invasive procedure for quantitative assessment of drought survival using chlorophyll fluorescence. Plant Methods 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Miki D, Zheng Z, Ma J, Wang J, Yang Z, Dong J, Zhu JK (2014) Overproduction of stomatal lineage cells in Arabidopsis mutants defective in active DNA demethylation. Nat Commun 5: 4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngson NA, Whitelaw E (2008) Transgenerational epigenetic effects. Annu Rev Genomics Hum Genet 9: 233–257 [DOI] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D (2013) The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Chen L, Xia H, Wei H, Lou Q, Li M, Li T, Luo L (2017) Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant’s adaptation to drought condition. Sci Rep 7: 39843. [DOI] [PMC free article] [PubMed] [Google Scholar]