Genome-wide annotation and RNA-seq analysis identify small secreted peptides responsive to nodulation and macronutrient limitations regulating root and nodule development in Medicago truncatula.
Abstract
Growing evidence indicates that small, secreted peptides (SSPs) play critical roles in legume growth and development, yet the annotation of SSP-coding genes is far from complete. Systematic reannotation of the Medicago truncatula genome identified 1,970 homologs of established SSP gene families and an additional 2,455 genes that are potentially novel SSPs, previously unreported in the literature. The expression patterns of known and putative SSP genes based on 144 RNA sequencing data sets covering various stages of macronutrient deficiencies and symbiotic interactions with rhizobia and mycorrhiza were investigated. Focusing on those known or suspected to act via receptor-mediated signaling, 240 nutrient-responsive and 365 nodulation-responsive Signaling-SSPs were identified, greatly expanding the number of SSP gene families potentially involved in acclimation to nutrient deficiencies and nodulation. Synthetic peptide applications were shown to alter root growth and nodulation phenotypes, revealing additional regulators of legume nutrient acquisition. Our results constitute a powerful resource enabling further investigations of specific SSP functions via peptide treatment and reverse genetics.
Small, secreted peptides (SSPs) have emerged as critical regulators of a diverse array of growth and developmental processes in plants, including aspects of root growth, nutrient homeostasis, meristem maintenance, stress acclimation, pathogen defense, reproductive development, and symbiotic interactions (Marmiroli and Maestri, 2014; Murphy and De Smet, 2014; Djordjevic et al., 2015; Okamoto et al., 2016; de Bang et al., 2017). The biological roles demonstrated thus far suggest that SSPs hold great potential for improving diverse agronomic traits benefitting agricultural production. However, despite the clear importance of SSPs, the vast majority remain unstudied and, indeed, unannotated in plant genomes. Gene prediction algorithms are biased against smaller gene products, to avoid wrongful annotation of random or noncoding transcripts, thereby excluding most SSPs and other bona fide small proteins (Lease and Walker, 2006; Dinger et al., 2008; Andrews and Rothnagel, 2014). Moreover, many SSPs and other small proteins that are annotated have been identified merely as unknown proteins or hypothetical proteins, which, in practice, stalls their functional characterization.
SSPs, as their name implies, are small peptides of five to 50 amino acid residues that are secreted into the apoplast. A number of SSPs have been found to serve as ligands of cell surface receptor kinases (Tabata et al., 2014; Ou et al., 2016; Santiago et al., 2016; Shinohara et al., 2016; Doblas et al., 2017; Nakayama et al., 2017) and function in intercellular communication on spatial scales ranging from adjacent cells to distant organs (Okamoto et al., 2016). SSPs often are encoded within a longer protein sequence of about 100 to 200 amino acids called a preproprotein. These preproproteins contain an N-terminal signal peptide directing the proprotein out of the cell. Prediction of N-terminal signal peptides is done by specialized software programs, such as SignalP, which calculates the probability (D-value) that a given amino acid sequence comprises a signal peptide (Petersen et al., 2011). The proprotein resulting from signal peptide removal contains the final, bioactive peptide embedded within it, usually near the C terminus, which is released by endoproteolytic processing (Matsubayashi, 2014; Ghorbani et al., 2016; Schardon et al., 2016). A classification method for SSPs, based on the protein’s amino acid sequence, has been proposed (Tavormina et al., 2015). According to this classification, any SSP can be categorized into one of five classes: (1) posttranslationally modified (PTM), (2) Cys-rich, (3) non-Cys-rich, non-PTM, (4) functional precursor, and (5) short open reading frame (sORF). The first three classes consist of peptides derived from a larger precursor protein lacking a known function, the fourth class consists of peptides derived from a larger precursor protein with a separate known function (Chen et al., 2014), and peptides of the final class are expressed from sORFs and are not processed proteolytically (Lauressergues et al., 2015).
Recent efforts to assess the peptide-coding potential in plants have demonstrated that the complete set of plant peptides, or peptidome, is much larger and more diverse than anticipated previously (Tavormina et al., 2015; Hellens et al., 2016). In Arabidopsis (Arabidopsis thaliana), more than 1,000 predicted SSPs have been identified (Lease and Walker, 2006), and more than 8,000 sORFs with high coding potential have been found (Hanada et al., 2013). Pan et al. (2013) identified 101,048 potential SSP-encoding ORFs in rice (Oryza sativa ssp. japonica), of which two-thirds were located in intergenic regions. In the latest Medicago truncatula genome release (Mt4.0), SSP hidden Markov models (HMMs) were included specifically in the annotation pipeline, resulting in the prediction of hundreds of Cys-rich peptides (Zhou et al., 2013; Tang et al., 2014). Furthermore, the PlantSSP database (http://bioinformatics.psb.ugent.be/webtools/PlantSSP/), based on 32 plant species, has been released, containing 39,135 small proteins (less than 200 amino acids) with predicted N-terminal signal peptides grouped into 4,681 families based on sequence homology of the C-terminal 50 amino acids (Ghorbani et al., 2015).
Legumes represent a significant source of global food and feed and are a staple in many cropping systems. Due to their ability to fix nitrogen in cooperation with soil bacteria known as rhizobia, legumes play a crucial role in relation to nitrogen input into agricultural ecosystems (Downie, 2014). Growing evidence indicates that SSPs play a critical role in legume growth, development, and productivity, particularly in relation to nutrient acquisition and use (Funayama-Noguchi et al., 2011; Imin et al., 2013; Okamoto et al., 2013; Mohd-Radzman et al., 2015, 2016; Wang et al., 2015). The CLAVATA3/EMBRYO-SURROUNDING REGION (CLE) peptides are particularly well studied in legumes due to their role in the systemic negative feedback mechanism known as autoregulation of nodulation (Mortier et al., 2010; Okamoto et al., 2013; Kassaw et al., 2017) and in local regulation by nitrate-induced CLEs (Hastwell et al., 2015). Phosphate-induced CLEs also have been reported in Lotus japonicus (Funayama-Noguchi et al., 2011; Handa et al., 2015), while other legume CLEs are involved in root apical meristem maintenance (Oelkers et al., 2008). A member of the C-TERMINALLY-ENCODED PEPTIDE (CEP) family in M. truncatula, MtCEP1, inhibits lateral root growth and promotes nodulation via signaling pathways dependent on the Leu-rich repeat receptor-like kinase MtCRA2 (Imin et al., 2013; Huault et al., 2014; Mohd-Radzman et al., 2016). Likewise, RAPID-ALKALINIZATION FACTOR1 (MtRALF1) and ROTUNDIFOLIA/DEVIL1 (MtDVL1) expression levels increased upon nod-factor treatment, and it was shown that the two peptides are involved in the early stages of rhizobia infection (Combier et al., 2008). Recently, PHYTOSULFOKINES (PSKs), a family of Tyr-sulfated peptides, also were suggested to positively regulate nodulation in L. japonicus (Wang et al., 2015). Furthermore, the inverted repeat-lacking clade (IRLC) that includes the majority of agriculturally cultivated legumes is unique in that its members possess a family of NODULE CYSTEINE-RICH (NCR) SSPs (nearly 700 of which are found in M. truncatula), which act in host-rhizobia specificity (Wang et al., 2017; Yang et al., 2017) and are suspected to collectively orchestrate the terminal differentiation of bacteroids in nodule development (Horváth et al., 2015; Kim et al., 2015; Montiel et al., 2017). Taken together, the potential of SSPs to improve agricultural traits of legumes, in particular nutrient acquisition and use, is striking.
M. truncatula has emerged as an excellent model for studies of legume biology and symbiosis, yet the SSP-coding potential of M. truncatula, as for the majority of all other plant species, remains unclear. Moreover, only a tiny fraction of the known SSP-coding genes in M. truncatula have been connected to specific functions. There are 820 gene loci from 334 families in M. truncatula in the PlantSSP database (Ghorbani et al., 2015). However, the M. truncatula accessions are based on the previous Mt3.5 release, and no legumes were included in the initial HMM development, thus likely precluding legume-specific SSPs from identification. Several SSP genes are annotated in the most current M. truncatula genome assembly, including members of the CLE, EPIDERMAL PATTERNING FACTOR-LIKE (EPFL), PSK, RALF, S-LOCUS CYSTEINE RICH-LIKE (SCRL), and TAPETUM DETERMINANT (TPD) families. M. truncatula CLE genes were initially annotated by Mortier et al. (2010) using Mt2.0, and this was expanded recently to a total of 52 CLE genes by Hastwell et al. (2017) using Mt4.0 in their identification pipeline. Studies by Imin et al. (2013), Delay et al. (2013b), and Ogilvie et al. (2014) collectively identified 14 M. truncatula CEP (MtCEP) family members. Noteworthy, seven of the 14 MtCEPs identified in Mt3.5 are missing from Mt4.0, and conversely, a single MtCEP is present solely in Mt4.0. Taken together, the above studies highlight that a comprehensive bioinformatics strategy is required for an exhaustive genome-wide identification of M. truncatula SSPs.
The aims of this study were to generate a comprehensive data set of all potential M. truncatula SSPs and to utilize RNA sequencing (RNA-seq) data to identify SSP genes with macronutrient- and symbiosis-dependent changes and potential roles in nutrient use or acquisition. This approach led to the identification of hundreds of SSP genes regulated by macronutrient stress, rhizobial inoculation, and nodule development. Using synthetic peptides, we demonstrate that selected nutrient-responsive SSPs from the PLANT PEPTIDE CONTAINING SULFATED TYROSINE (PSY), PAMP-INDUCED SECRETED PEPTIDE (PIP), and INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) families enhance root growth characteristics, with potential relevance to nutrient acquisition. Additionally, a Focal List containing potential, novel SSP candidate genes within M. truncatula was developed based on shared characteristics with known SSP-coding genes. The utility of the Focal List was demonstrated by the identification of a new legume-specific SSP gene family that can suppress nodulation upon exogenous application of synthetic peptide. In summary, genome reannotation, SSP identification, and transcriptomic analyses have provided a comprehensive and valuable resource for further, detailed investigation of SSP regulation in legumes as well as in plants more broadly.
RESULTS
Reannotation of the M. truncatula Genome
While working with SSPs in M. truncatula, it has become clear that the annotation of ORFs in the genome is biased against those producing shorter gene products (i.e. the ORFs most likely to encode SSPs). To establish the full coding potential for SSPs within M. truncatula, it was essential to improve the identification of such sORFs. Thus, a reannotation of the M. truncatula genome using three bioinformatics software programs, MAKER (Cantarel et al., 2008), SPADA (Zhou et al., 2013), and sORF Finder (Hanada et al., 2010), was carried out (Fig. 1A). Of these software programs, SPADA and sORF Finder were developed to identify short gene products, and MAKER was fine-tuned for the same purpose.
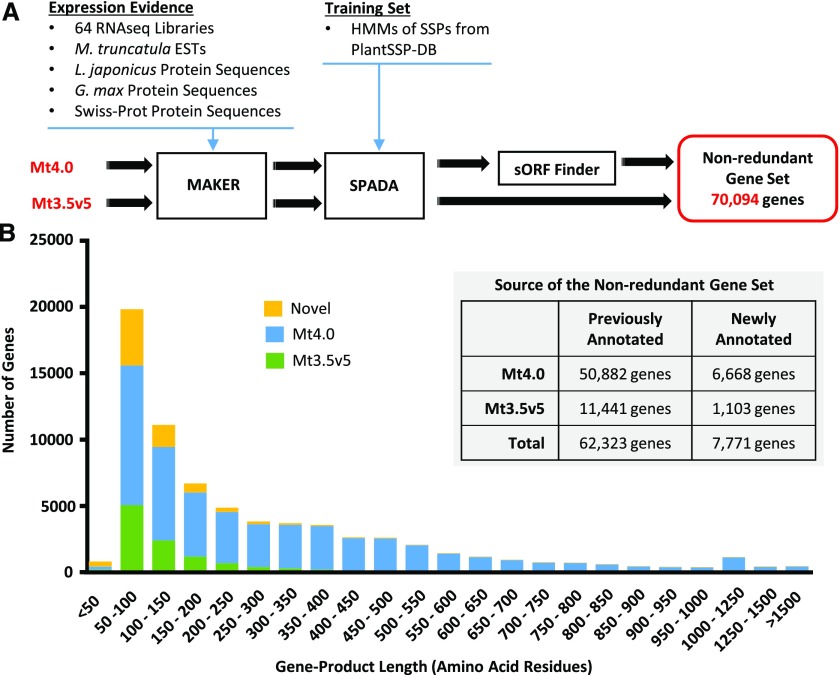
Figure 1.
Reannotation of the M. truncatula genome and identification of a nonredundant gene set. A, The reannotation pipeline was run in parallel on the current (Mt4.0) and most recent previous (Mt3.5v5) M. truncatula genome releases, resulting in a merged nonredundant gene set of 70,094 genes. The pipeline employed the MAKER software program with expression evidence from 64 in-house RNA-seq libraries of diverse tissue types and developmental stages, the M. truncatula EST Gene Index 11, and protein sequences from L. japonicus, Glycine max, and the Swiss-Prot manually curated protein database. SPADA was used to identify SSP gene models using HMMs from the PlantSSP database (Ghorbani et al., 2015) and the in-built HMMs of Cys-rich families, which facilitated the identification of sORFs. Finally, the sORF Finder program (Hanada et al., 2010) was run specifically on the Mt4.0 genome to identify sORF gene models from intergenic regions. B, Histogram illustrating the predicted protein size distribution (bins = 50 amino acids) of the nonredundant gene set, classified according to the database source of the sequence. Nearly 90% of the genes newly identified by our pipeline (Novel) encode products of 200 or fewer amino acid residues. The inset illustrates the sources of the nonredundant gene set, with a total of 7,771 newly annotated genes.
Comparing gene indices between the current M. truncatula genome release, Mt4.0, and the previous release, Mt3.5v5, many genes were found to be specific to one release, which may be caused by the distinct genome assembly methods used (Young et al., 2011; Tang et al., 2014). Hence, the annotation pipeline was performed on both genome releases in parallel, and the outputs were merged subsequently into a final nonredundant gene set, giving precedence to genes from Mt4.0 where redundancy was found. The reannotation pipeline yielded a set of 70,094 nonredundant genes that included 7,771 newly annotated gene loci (Fig. 1B; Supplemental Table S1). Size distribution analysis of the reannotated genome showed that 88% of the 7,771 newly annotated gene loci encoded proteins of less than 200 amino acids (Fig. 1B), of which 59% had clear experimental support from our RNA-seq data sets (see below). This supports the conclusion that the current annotation (Mt4.0) is biased against the prediction of shorter gene products and demonstrates the benefit of our reannotation.
A total of 374 of the novel genes code for 50 amino acids or less (Supplemental Table S1). While the majority of these short gene products have no homology to known proteins, 78 encode for known protein families and another seven of these appear to be pseudogenes encoding truncated SSPs. In total, 87% of these short gene products had experimental support from the RNA-seq data sets, indicating that they may be novel SSPs or noncoding RNAs. In fact, 35% had SignalP D-values greater than 0.45, much higher than the fraction of all novel genes, underlining their SSP potential. Those genes without RNA-seq support may be expressed under highly specific conditions or represent spurious ORF identifications. There were 38 newly annotated genes coding for long products greater than 750 residues, all but two of which had strongly predicted conserved domains from PFAM and/or Uniprot, facilitating functional prediction. No conserved domain or functional category was enriched among these genes, and it is not clear why these genes were missed in previous genome annotations.
Known SSPs are typically embedded in gene products smaller than 200 amino acids. In our analysis, there were 38,193 genes encoding proteins shorter than 200 amino acids, of which 6,861 genes constituted novel gene models, with 60% having clear experimental RNA-seq support. This emphasizes the high number of additional, potential SSP-encoding genes that were gained by the genome reannotation (Fig. 1B). Taken together, the bioinformatics approach described here greatly extended the annotation of the M. truncatula genome and increased the potential to identify existing and novel SSP families.
Identification of M. truncatula Members of Established SSP Families
The number of recognized SSP gene families across all plant species has grown substantially in recent years, and the improved M. truncatula genome annotation was mined for homologs to these established SSP gene families. Mining was carried out with a three-pronged approach. First, a homology search using representative proteins from established SSP families was performed. Second, HMMs of known or putative plant SSPs from the PlantSSP database (Ghorbani et al., 2015) were searched for matches. Third, to identify additional SSPs, short (less than 200 amino acids) and secreted (SignalP D-value > 0.25) protein sequences were clustered based on shared sequence motifs using Markov Cluster Analysis (MCL; Enright et al., 2002; Van Dongen, 2008). Genes clustering with SSPs identified in the two initial steps were extracted as SSPs. For a list of all SSP families used for the searches along with functional and sequence annotations of each family, see Supplemental Table S2.
Based on amino acid sequences, the identified genes were grouped into individual, established SSP families, which included families of multiple peptidase inhibitors, antimicrobial peptides, known or putative signaling peptides, and peptides acting through unknown mechanisms. Multiple sequence alignments of each family were manually curated to confirm shared sequence motifs among family members. In a second search iteration, the genome was reinterrogated using HMMs constructed based on the compiled SSP families. Again, multiple sequence alignments of each family were manually curated to confirm proper inclusion of genes within families. This second search iteration resulted in an additional 312 SSP genes from 23 families, 155 of which were apparent pseudogenes that had lost one or more critical, conserved residues.
This exhaustive genome search resulted in a final, comprehensive list of 1,970 genes from 46 previously defined SSP families (Table I; Supplemental Tables S2 and S3). Some genes with highly conserved SSP motifs did not have predicted signal peptides. However, in a number of cases, manual curation of mapped reads identified alternative, upstream start codons that uncovered strongly predicted signal peptides (for examples, see Supplemental Figs. S1 and S2). Many of the families included presumptive pseudogene members, consistent with the high frequency of tandem duplications of SSP genes (Silverstein et al., 2007; Takeuchi and Higashiyama, 2012; Trujillo et al., 2014). There were 363 SSP genes, across 37 of the SSP families, that had not been identified previously as members of their respective families in either Uniprot or the Mt4.0 annotation, representing a 23% increase in the size of the M. truncatula SSP gene pool (Table I). Among those families known or suspected to act as receptor ligands (Signaling-SSPs), an additional 188 members were found, representing a 38% increase in size. Significantly, the Uniprot and Mt4.0 annotations of many of the families were found to be flawed, which was corrected during the extensive manual curation. Examples of these corrections from the NCR and RALF families are presented in Supplemental Figures S3 and S4.
Table I. Identified SSP families in M. truncatula.
| SSP Family | Full Name | Class | Mode of Action | No. of Genesa | No. Newly Identifiedb |
|---|---|---|---|---|---|
| ECL | Egg Cell1-Like | Cys-rich | Signal | 21 | 5 |
| EPFL | Epidermal Patterning Factor-Like | Cys-rich | Signal | 23 | 6 |
| GASA | Gibberellic Acid Stimulated in Arabidopsis | Cys-rich | Signal | 29 | 5 |
| Legin | Leginsulin | Cys-rich | Signal | 50 | 18 |
| MEG | Maternally Expressed Gene | Cys-rich | Signal | 2 | 1 |
| N26 | Nodulin26 | Cys-rich | Signal | 5 | 3 |
| nsLTP | Nonspecific Lipid Transfer Protein | Cys-rich | Signal | 134 | 7 |
| PCY | Plantacyanin/Chemocyanin | Cys-rich | Signal | 87 | 7 |
| LAT52/POE | LAT52/Pollen Ole e 1 Allergen | Cys-rich | Signal | 42 | 3 |
| RALF | Rapid Alkalinization Factor | Cys-rich | Signal | 15 | 3 |
| RC | Root Cap | Cys-rich | Signal | 9 | 2 |
| SCRL | S-Locus Cys-Rich-Like | Cys-rich | Signal | 17 | 1 |
| STIG/GRI | Stigma1/GRI | Cys-rich | Signal | 21 | 4 |
| TAX | Taximin | Cys-rich | Signal | 5 | 5 |
| TPD | Tapetum Determinant1 | Cys-rich | Signal | 23 | 20 |
| CAPE | CAP-Derived Peptide | Functional precursor | Signal | 21 | 0 |
| SUBPEP | Subtilisin-Embedded Plant Elicitor Peptide | Functional precursor | Signal | 1 | 1 |
| PEP | Plant Elicitor Peptide | Non-Cys, non-PTM | Signal | 1 | 1 |
| PNP | Plant Natriuretic Peptide | Non-Cys, non-PTM | Signal | 4 | 4 |
| CEP | C-Terminally Encoded Peptide | PTM | Signal | 17 | 17 |
| CIF | Casparian Strip Integrity Factor | PTM | Signal | 1 | 1 |
| CLE | Clavata/Embryo-Surrounding Region | PTM | Signal | 52 | 11 |
| GLV | Golven/Root Growth Factor | PTM | Signal | 15 | 14 |
| IDA | Inflorescence Deficient in Abscission | PTM | Signal | 42 | 21 |
| PIP | PAMP-Induced Secreted Peptide | PTM | Signal | 13 | 13 |
| PSK | Phytosulfokine | PTM | Signal | 11 | 2 |
| PSY | Plant Peptide Containing Sulfated Tyr | PTM | Signal | 10 | 10 |
| ENOD40 | Early Nodulin 40 | sORF | Signal | 2 | 0 |
| RTFL/DVL | Rotundifolia/Devil | sORF | Signal | 17 | 3 |
| Signaling-SSP total | 690 | 188 | |||
| BBPI | Bowman-Birk Peptidase Inhibitor | Cys-rich | Peptidase inhibitor | 17 | 1 |
| Kaz | Kazal Family Inhibitors | Cys-rich | Peptidase inhibitor | 2 | 0 |
| Kunitz | Kunitz-P Trypsin Inhibitor | Cys-rich | Peptidase inhibitor | 52 | 3 |
| T2SPI | Potato Type II Proteinase Inhibitor | Cys-rich | Peptidase inhibitor | 2 | 0 |
| CTLA | Cytotoxic T-Lymphocyte Antigen 2α | Non-Cys, non-PTM | Peptidase inhibitor | 15 | 0 |
| PhyCys | Phytocystatin | Non-Cys, non-PTM | Peptidase inhibitor | 47 | 2 |
| SubIn | Subtilisin Inhibitor | Non-Cys, non-PTM | Peptidase inhibitor | 12 | 8 |
| Peptidase inhibitor total | 147 | 14 | |||
| 2SA | 2S Albumin | Cys-rich | Antimicrobial | 3 | 0 |
| Plant Defensin | Cys-rich | Antimicrobial | 67 | 10 | |
| PDL | Plant Defensin-Like | Cys-rich | Antimicrobial | 34 | 3 |
| THL | Thionin-Like | Cys-rich | Antimicrobial | 35 | 8 |
| Antimicrobial total | 139 | 21 | |||
| LCR | Low-Mr Cys-Rich | Cys-rich | Unknown | 101 | 11 |
| NCR-A | Nodule-Specific Cys-Rich Group A | Cys-rich | Unknown | 361 | 48 |
| NCR-B | Nodule-Specific Cys-Rich Group B | Cys-rich | Unknown | 428 | 81 |
| LP | LEED..PEED | Non-Cys, non-PTM | Unknown | 22 | 0 |
| NodGRP | Nodule-Specific Gly-Rich Protein | Non-Cys, non-PTM | Unknown | 58 | 0 |
| PRP669 | Pro-Rich Protein Group 669 | Non-Cys, non-PTM | Unknown | 24 | 0 |
| Unknown total | 994 | 140 | |||
| Grand total |
1,970 | 363 | |||
The current, total number of gene members identified in the M. truncatula genome.
The number of newly identified gene members in this work (for details, see “Materials and Methods”).
The SSP families were categorized into one of the five SSP classes defined by Tavormina et al. (2015) and described above, which are based on shared structural properties. The greatest number of SSP genes in M. truncatula is from the Cys-rich class (1,585), which includes SSP families acting as signals, antimicrobials, and peptidase inhibitors (Table I; Supplemental Table S3).
Identification of Novel SSPs: The Focal List
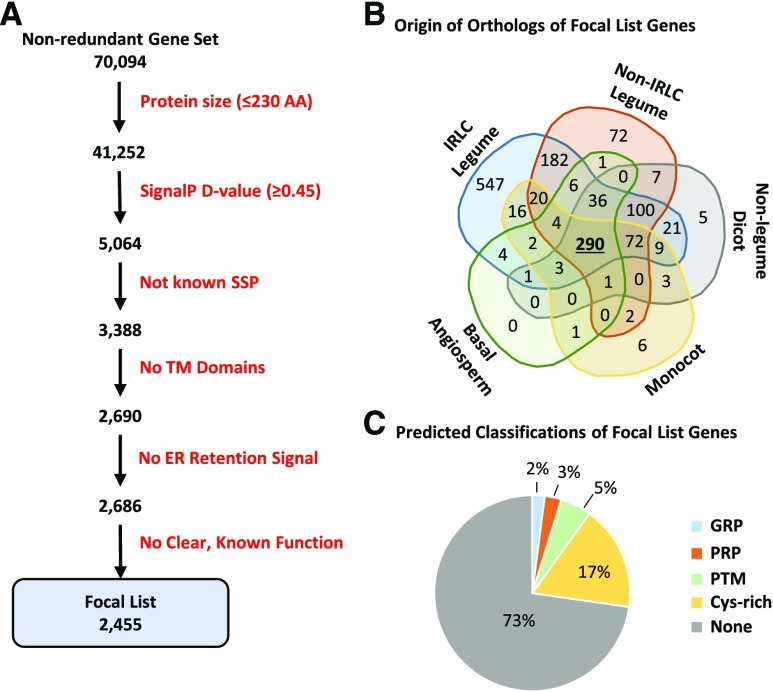
Although the systematic search of the reannotated M. truncatula genome identified nearly 2,000 SSP-encoding genes, this search necessarily relied on prior evidence of each SSP family. Thus, it was hypothesized that additional, novel SSP families remained to be discovered within the M. truncatula genome. Hence, the reannotated genome was mined to identify a list of genes that represent putative, novel SSPs based on a work flow of successive filtering steps. As this list represents excellent candidates for additional SSPs, likely including legume-specific SSPs, we dub this list the Focal List (Fig. 2A; Supplemental Table S4). The initial steps were based on a required short coding length and prediction of a signal peptide by the SignalP software (Petersen et al., 2011). The appropriate length and SignalP D-value criteria for filtering were determined empirically using all members of the known SSP families as a positive control (Supplemental Fig. S5). Additional filtering steps eliminated established SSP proteins, those with a predicted transmembrane helix, or those with an endoplasmic reticulum-retention signal. A final filtering step eliminated 232 proteins with known functions unrelated to signaling SSPs, such as photosystem subunits, transcription factors, or well-characterized metabolic enzymes (Fig. 2A). Filtering out these functional proteins may have increased the risk of discarding potential SSPs encoded within functional precursors. However, such SSPs would not necessarily be embedded within genes of less than 230 amino acids or lacking transmembrane domains. Accordingly, such potential SSP genes might have been discarded at earlier filtering steps. Therefore, all genes discarded at each filtering step were retained in Supplemental Table S4, allowing custom tailoring of candidate lists.
Figure 2.
Identification and characteristics of putative, novel SSP genes. A Focal List of putative, novel SSP genes was created based on shared characteristics of previously defined SSPs. This list represents an excellent starting point from which to identify novel SSP genes or families, some of which may be legume or IRLC specific. A, Six sequential filtering steps were used to develop our Focal List. B, A total of 1,371 Focal List members had orthologs identified from at least one of 16 other angiosperm species. These species are classified into five different phylogenetic groups. Indicated in the Venn diagram are the number of Focal List members that have orthologs in each of these groups. C, Focal List genes also were assigned to one of four predicted classifications, including a PTM group that is built on a PTM score calculation.
The final Focal List of candidate SSP genes comprised 2,455 genes, with over 74% having strong RNA-seq support based on in-house data sets. While a majority of these candidate genes had no prior annotation (71%), a number of them were incorrectly annotated as members of SSP families, including 15 NCRs and 30 RALFs. Although manual curation confirmed that these genes were not, in fact, members of any of the established SSP families, they are excellent candidates for novel SSP families.
To identify families of homologs among the Focal List members, MCL clustering was performed to group related genes. In this way, over half of the Focal List members clustered into one of 216 families. While most families were small, comprising only two or three genes, 21 families contained at least a dozen gene members, and eight of these appeared to be Cys-rich families based on the presence of conserved Cys pairs. To determine the degree of conservation of these genes among flowering plants and identify potential legume-specific SSPs, we searched for orthologs based on reciprocal best hits (Moreno-Hagelsieb and Latimer, 2008) in 16 other angiosperm species representing five different phylogenetic groups (Fig. 2B; Table II). A total of 1,371 Focal List genes were found to have an ortholog in at least one of the 16 selected species, 290 of which had orthologs in members of all five of the phylogenetic groups (Fig. 2B; Table II; Supplemental Table S4). Conversely, another 801 Focal List genes had orthologs only in other leguminous plants, 547 of which were only in the IRLC. These legume-specific Focal List members are excellent candidates for SSPs with nodulation-specific roles, as illustrated for Family 71 below.
Table II. Putative orthologs of Focal List members in 16 selected angiosperm species.
| Species | Classification | No. of Sequencesa | No. of Orthologsb | Percentage of Focal List |
|---|---|---|---|---|
| Medicago sativa (CADL)c | IRLC Legume | 84,484 | 1,230 | 50 |
| Cicer arietinum | IRLC Legume | 34,299 | 628 | 25 |
| Trifolium subterraneum | IRLC Legume | 42,186 | 615 | 25 |
| Nonredundant IRLC Legume orthologs | 1,318 | 53 | ||
| Glycine max | Non-IRLC Legume | 199,288 | 636 | 26 |
| Glycine soja | Non-IRLC Legume | 51,857 | 602 | 24 |
| Vigna angularis | Non-IRLC Legume | 103,454 | 579 | 23 |
| Phaseolus vulgaris | Non-IRLC Legume | 71,796 | 559 | 23 |
| Lotus japonicus | Non-IRLC Legume | 9,535 | 268 | 11 |
| Nonredundant Non-IRLC Legume orthologs | 798 | 32 | ||
| Vitis vinifera | Nonlegume Dicot | 95,739 | 450 | 18 |
| Solanum lycopersicum | Nonlegume Dicot | 43,173 | 426 | 17 |
| Arabidopsis thaliana | Nonlegume Dicot | 287,700 | 400 | 16 |
| Nicotiana benthamiana | Nonlegume Dicot | 787 | 19 | 1 |
| Nonredundant Nonlegume Dicot orthologs | 548 | 22 | ||
| Brachypodium distachyon | Monocot | 77,875 | 358 | 15 |
| Oryza sativa | Monocot | 346,600 | 349 | 14 |
| Zea mays | Monocot | 210,577 | 335 | 14 |
| Nonredundant Monocot orthologs | 430 | 17 | ||
| Amborella trichopoda | Basal Angiosperm | 48,088 | 349 | 14 |
| Total Focal List genes | 2,455 | |||
| Total Focal List genes with orthologs |
1,371 | |||
Total number of protein sequences used that were retrieved from the National Center for Biotechnology Information (NCBI) protein database (including GenBank, RefSeq, UniprotKB/Swiss-Prot, PIR, DDBJ, EMBL, and PDB).
Putative orthologs were identified based on the reciprocal-best-BLAST-hits approach using Protein-BLAST search.
The M. sativa protein sequences from the diploid alfalfa, Cultivated Alfalfa at the Diploid Level (CADL), were obtained from www.alfalfatoolbox.org.
Focal List genes also were assigned to SSP classes, such as PTM or Cys rich. A total of 68 genes were identified as Pro-rich proteins (PRPs), which contained at least 15% Pro residues outside of the predicted signal peptide sequence, and another 51 genes were identified as Gly-rich proteins (GRPs), again containing at least 15% Gly residues outside of the predicted signal peptide sequence. It has been noted that, while GRPs generally contain as much as 80% Gly content, nodule-specific GRPs are shorter and have a Gly content typically around 20% to 30% (Alunni et al., 2007). An additional 426 genes had at least one pair of conserved Cys residues, and these were classified as predicted Cys-rich SSPs. To identify putative PTM SSPs from the Focal List, a simple scoring method based on the unique amino acid frequencies of this SSP class was developed (Supplemental Fig. S6). In total, 128 Focal List members were predicted to be PTM SSPs (Fig. 2C; Supplemental Table S4).
The remainder of this work focused on SSPs known or suspected to act via receptor-mediated signaling (Signaling-SSPs), as opposed to antimicrobial or peptidase inhibitor SSPs, and more specifically on identifying Signaling-SSPs responding to nutrient stresses or symbiosis. In total, Signaling-SSPs represented 690 genes from all five SSP classes (Table I).
Signaling-SSP and Focal List Gene Response to Macronutrient Deficiency and Symbiosis
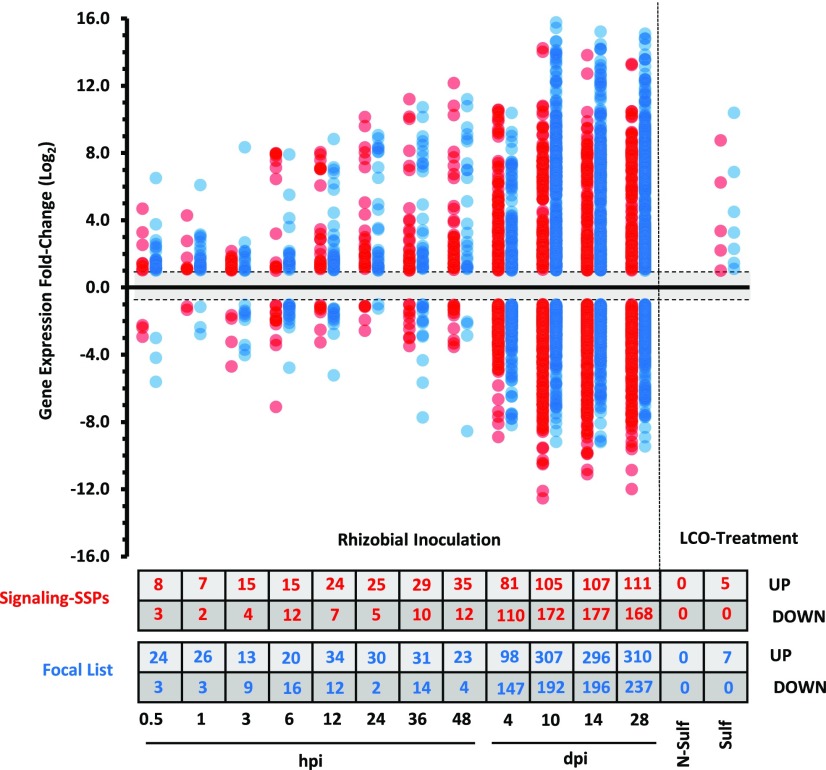
A small number of Signaling-SSPs from various species have been shown to impact traits related to plant nutrient homeostasis and acquisition, such as lateral root development and symbiotic interactions (Djordjevic et al., 2015; Okamoto et al., 2016; de Bang et al., 2017; Ohkubo et al., 2017). However, the full extent of Signaling-SSPs involved in acclimation to fluctuations in nutrient availability has not been explored. For a more comprehensive analysis of potential Signaling-SSP roles in acclimation to nutrient stress, changes in the expression of Signaling-SSPs and Focal List members were analyzed based on RNA-seq data of M. truncatula tissue from (1) four different macronutrient deficiencies, (2) a detailed time course covering early and late rhizobial signaling events and nodule development, and (3) mycorrhizal lipochitooligosaccharide (Myc-LCO) treatment, mimicking the early signaling events prior to mycorrhizal colonization (Fig. 3; Supplemental Tables S14 and S15). A total of 144 RNA-seq data sets were mapped to the reannotated M. truncatula genome, and gene expression was quantified as fragments per kilobase of transcript per million mapped reads (FPKM; Fig. 3). Differential expression (DE) analysis was carried out with the DEseq2 software program (Love et al., 2014), which employs dynamic filtering of low expressed genes to prevent spurious identifications of DE.
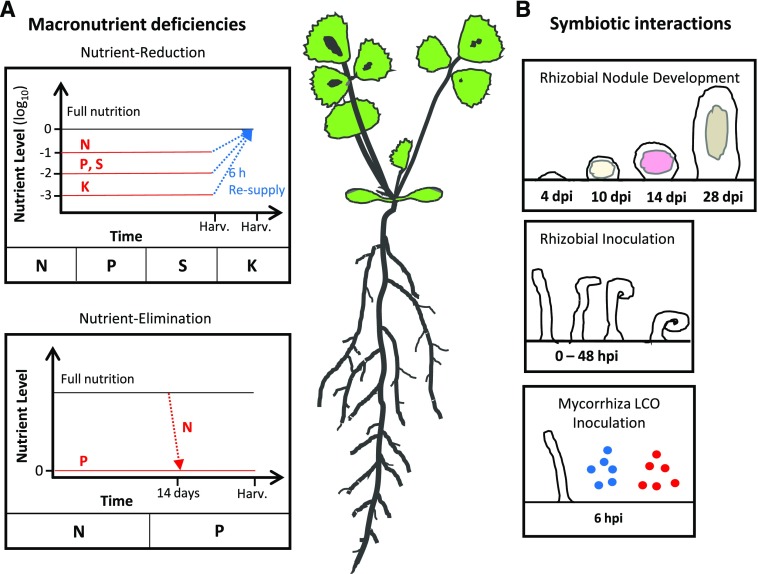
Figure 3.
Overview of RNA-seq experimental designs. A, Macronutrient deficiencies. Two different, complementary macronutrient experiments were conducted on plants grown over a time period of 3 weeks. RNA-seq was performed on both root and shoot tissues. In the nutrient-reduction experiment, plants were grown with a reduced external supply of one of four macronutrients, nitrogen (N), phosphorus (P), sulfur (S), or potassium (K), over the full 21 d. Six hours before harvest, a subset of deficient plants were resupplied with full nutrition (FN) solution. In the nutrient-elimination experiment, plants were either grown at FN for 14 d before complete elimination of external nutrient supply for a subsequent 7 d (N) or grown under complete elimination of external nutrient supply for the full 3-week time period. All experimental treatments were performed with appropriate control plants receiving full nutrition throughout the 3-week time period. The point of harvesting is indicated along the x axis as Harv. B, Symbiotic interactions. Three different experiments focusing on symbiotic interactions with M. truncatula roots were performed. RNA-seq data covering early rhizobial infection events at nine time points between 0 and 48 h post inoculation (hpi) were obtained from Larrainzar et al. (2015). RNA-seq data of roots inoculated with sulfated (blue) or nonsulfated (red) synthetic Myc-LCO molecules were obtained from Camps et al. (2015). RNA-seq data covering nodule development from 4 to 28 d post inoculation (dpi) were generated in house. The 4-dpi time point used nodule bumps, the portion of the root with nascent, developing nodules, while the latter three time points used isolated nodule tissue.
Macronutrient Deficiencies
To investigate macronutrient deficiency responses, RNA-seq data sets were produced from root and shoot tissues of plants grown under N, P, S, and K deficiency conditions (Fig. 3). Two distinct experimental setups were employed to identify a more complete set of responsive genes: (1) a nutrient-reduction experiment (N, P, K, or S) and (2) a nutrient-elimination experiment (N or P). To more specifically identify genes responding to changes in nutrient supply, samples from plants resupplied with the given nutrients 6 h before harvest were included. The experimental setups differed in the magnitude of nutrient limitation, as well as in the developmental stage of application, and thus formed complementary experiments. Nutrient depletion in the sampled tissue was confirmed by RNA-seq expression patterns and reverse transcription-quantitative PCR (RT-qPCR) data for published marker genes (Hirai et al., 2004; Scheible et al., 2004; Nussaume et al., 2011; Secco et al., 2012; Wipf et al., 2014; Supplemental Table S14). In addition, macronutrient deficiencies reduced shoot biomass production and lowered the concentrations of N, P, S, or K (Supplemental Fig. S7). Below, nutrient-responsive Signaling-SSPs and Focal List members are considered across the two experimental setups.
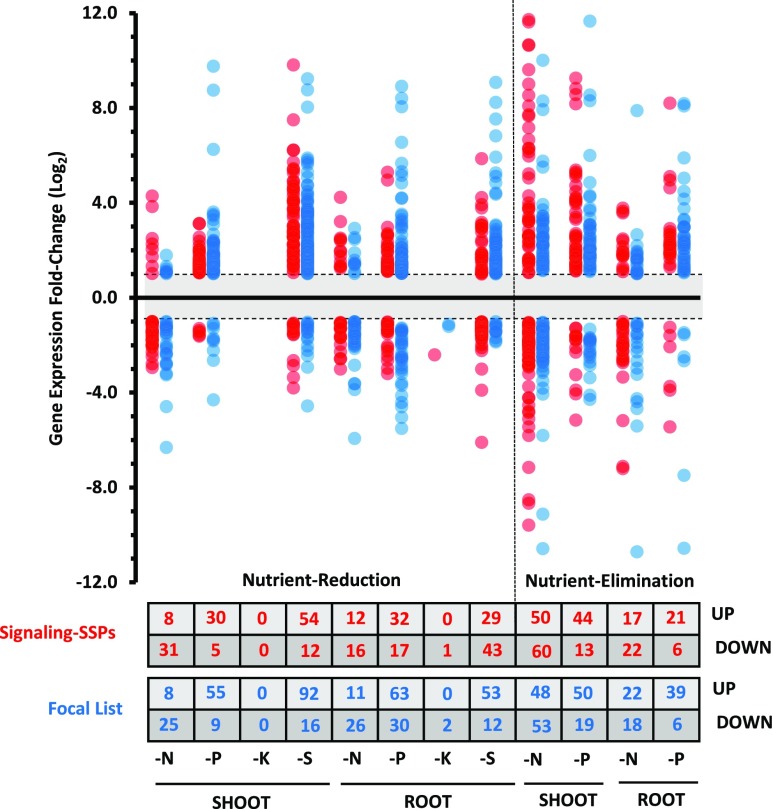
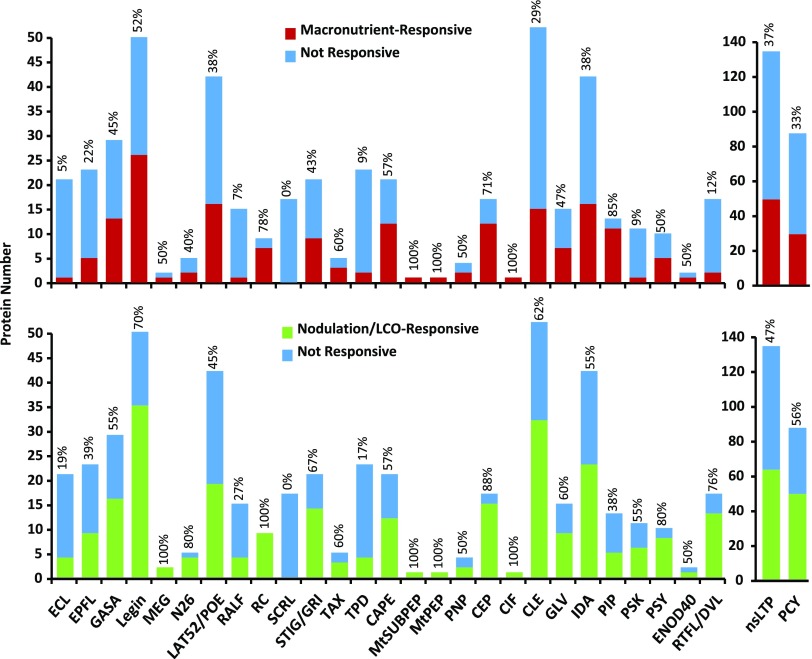
Transcript levels of 240 distinct Signaling-SSP genes were significantly altered under at least one macronutrient deficiency (i.e. an adjusted P ≤ 0.1 and a minimum 2-fold change), 138 genes in roots and 172 in shoots. Although only one gene, Leginsulin13 (MtLegin13) in the root, was responsive to K deficiency, numerous Signaling-SSP genes responded to the other three deficiencies, most dramatically in the N-deprived shoot tissue, which ranged from a 4,000-fold increase of MtCEP9 to a 730-fold decrease of MtLegin8 (Fig. 4; Supplemental Table S7). Comparing the Signaling-SSPs between the two experimental setups, shared responses (i.e. responding to the same nutrient in both experiments) were found for only one-third and one-sixth of the identified genes in shoots and roots, respectively (Supplemental Fig. S8). Significantly, macronutrient-responsive Signaling-SSPs represented all but one of the 27 Signaling-SSP families (Fig. 5). The family lacking macronutrient-responsive genes, the SCRL family, is believed to have a specific role in reproductive tissue (Vanoosthuyse et al., 2001). Individual Signaling-SSP gene responses were specific to the given macronutrients in 55% and 61% of the cases in shoots and roots, respectively (Supplemental Fig. S9). The remainder of the Signaling-SSP genes responded to two or three of the macronutrient deficiencies, while none responded to all four deficiencies. Sixteen genes in shoots and six genes in roots responded to N, P, and S deficiencies, including five PLANTACYANINs (PCYs) and three PIPs up-regulated in shoots and MtCLE34 and MtPSY3 down-regulated in roots (Supplemental Table S7). This small set of Signaling-SSP genes may represent general regulators of nutrient use or acquisition and, as such, are attractive targets for follow-up studies.
Figure 4.
Macronutrient-responsive Signaling-SSP and Focal List genes. Using the DESeq2 software program (Love et al., 2014), DE genes of the Signaling-SSP and Focal List groups were identified from both the nutrient-reduction and nutrient-elimination experiments. DE genes required an adjusted cutoff value of P < 0.1 and a minimum log2 fold change of ±1. Log2 fold changes of each DE Signaling-SSP gene (red) or Focal List gene (blue) are plotted. The numbers of DE Signaling-SSP and Focal List genes up- or down-regulated are indicated in the table at bottom.
Figure 5.
Most Signaling-SSP families contain macronutrient- and nodulation-responsive genes. The numbers of genes from each family that are responsive (adjusted P < 0.1 and 2-fold or greater change) to at least one macronutrient deficiency (top) or one nodulation/LCO time point (bottom) are plotted for each Signaling-SSP family. The fraction of each family that is responsive is indicated above each column, as a percentage.
Nutrient resupply resulted in DE of 103 distinct Signaling-SSPs (relative to the end point of the deficiency): 54 responded in shoots and 68 responded in roots (Supplemental Fig. S10). The vast majority of these genes responded specifically to individual nutrients. However, seven Signaling-SSPs responded to resupply of both N and P, while four Signaling-SSPs responded to resupply of both P and S. The most dramatic responses to resupply were seen in P shoot and S root tissue, approaching 1,000-fold down-regulation in each case (Supplemental Fig. S10; Supplemental Table S7). The responses of three Signaling-SSPs to resupply were shared between P and K (MtCEP17, MtIDA10, and MtIDA31).
A total of 297 Focal List genes responded significantly to at least one macronutrient treatment (183 in shoots and 202 in roots; adjusted P ≤ 0.1 and a minimum 2-fold change; Supplemental Table S8). Several Focal List families showed consistent macronutrient responses across multiple members. Of particular note, members of Family 13 responded to deficiency of all four macronutrients and included both induced and repressed genes. Multiple members of the Cys-rich families, Family 101 and Family 189, also were broadly induced by N, P, and S deficiencies (Supplemental Table S9).
Further investigation of Focal List members that were not part of a family (i.e. singletons) identified Medtr1g043320.1, which was induced 69-fold in P-deficient shoots and 6-fold in P-deficient roots. The C-terminal sequence of this gene product resembles that of known PTM SSPs in its enrichment of Pro and His and the presence of a C-terminal Gln. Homology searches identified orthologs of this gene in other legume species, although it was not present in the PlantSSP database, consistent with this putative PTM SSP being legume specific (Supplemental Fig. S13A). Another singleton gene, Medtr2g095040.1, also had a C-terminal composition diagnostic of PTM SSPs. Homology searches revealed that this putative PTM SSP singleton gene is conserved throughout a diverse set of angiosperms as multigene families (Supplemental Fig. S13B). The expression of this gene was repressed specifically by N deficiency in shoots and roots (Supplemental Table S9).
Symbiosis
RNA-seq data sets representing varying stages of nodulation with Sinorhizobium meliloti were produced and supplemented with publicly available data sets from Larrainzar et al. (2015; only wild-type data) and Camps et al. (2015; Fig. 3). A total of 12 developmental stages of S. meliloti nodulation were investigated, ranging from the initial stages of interaction with the rhizobia (Larrainzar et al., 2015) to senescing nodule organs, as well as root tissue treated with sulfated and nonsulfated Myc-LCOs, representing a single early signaling step in mycorrhizal colonization (Camps et al., 2015).
In total, 365 differentially expressed Signaling-SSPs were identified ranging over all 12 time points of rhizobial inoculation, with the greatest numbers of responsive genes observed at later time points: 10, 14, and 28 dpi. However, as early as 24 hpi, there were Signaling-SSPs up-regulated near the same magnitude (approximately 700-fold) as the later time points (Fig. 6). Similar to macronutrient deficiencies, symbiosis-responsive Signaling-SSPs were derived from the majority of families; again, only the SCRL family was not regulated by nodulation (Fig. 5). The Myc-LCO experiment only considered changes at a single, early time point, perhaps explaining why only five Signaling-SSP genes were found to be responsive to the Myc-LCO experiment: MtPSK1, MtnsLTP62, MtPCY16, MtPCY29, and MtPCY33 were up-regulated by sulfated Myc-LCO (Fig. 6; Supplemental Table S7). The three PCYs and MtnsLTP62 also were induced by S. meliloti inoculation within 6 and 36 hpi, respectively. Hence, these four genes may play a role in the common, ancestral symbiotic pathway.
Figure 6.
Symbiosis-responsive Signaling-SSP and Focal List genes. Using the DESeq2 software program (Love et al., 2014), DE genes of the Signaling-SSPs (red) and Focal List members (blue) were determined relative to the preinoculated control specific to each experiment or to the untreated control treatment for the LCO experiment. DE genes required an adjusted cutoff value of P < 0.1 and a minimum log2 fold change of ±1. The numbers of DE Signaling-SSP and Focal List genes up- or down-regulated are indicated in the table at bottom.
To look at expression trends among the SSPs and Focal List members, hierarchical clustering was carried out on all SSP and Focal List members with mapped reads from at least one nodulation time point. The NCR genes, although not Signaling-SSPs, also were included in the hierarchical clustering for comparison. Clustering identified six groups, groups I to VI, with distinct expression patterns (Supplemental Fig. S11; Supplemental Table S10). Group I, comprising 115 genes, represented those genes highly up-regulated in nodule organs and included MtCLE12 and MtCLE13, implicated previously in nodulation (Mortier et al., 2010). Interestingly, a member of the EPFL family, MtEPFL14, had an expression pattern highly similar to that of MtCLE12, with strong induction beginning at 10 dpi (Supplemental Fig. S11B). The EPFL family includes well-known regulators of epidermal cell fate and stomatal patterning in leaf tissue, but its members have not been connected previously to any role in nodulation. Sixty-two Focal List members also clustered into group I. Of particular interest was Family 28, harboring six conserved Cys residues and annotated as an embryo-specific protein family by Mt4.0 (Supplemental Table S9). This family has nine members, two of which have orthologs throughout the angiosperm lineage, while the others appear to be legume specific. Five of the legume-specific members were clustered into group I and were induced in nodules as much as 1,000-fold relative to total root tissue (Supplemental Table S10). The expression and phylogenetic patterns indicate that this family has been coopted and expanded from an ancient embryo-specific function for a role in nodulation in legumes.
Genes strongly down-regulated within nodule organs are represented in group V. This group contains 131 genes, including three IDA genes, that are induced early in roots following inoculation but are strongly suppressed at all time points in nodules (Supplemental Table S10). The same expression pattern is seen in one to two members each from the families EPFL, PSY, PCY, PIP, nsLTP, Legin, and LAT52/POE. Specific induction of these SSPs at early time points indicates that they may represent components of root-rhizobia perception and nodule initiation.
A group of eight Focal List genes (five Cys rich and three Pro rich) were highly expressed early and persistently over the course of nodulation (Supplemental Fig. S12; Supplemental Table S11). Interestingly, three of these genes, two Pro-rich genes and the Cys-rich Medtr7g066110.1, a Proteinase-Activated Receptor1 gene, also were up-regulated in Myc-LCO-treated roots and, thus, may hold parallel roles in symbioses with bacteria and mycorrhizal fungi (Supplemental Table S11). Conversely, nine other Focal List genes were found to be down-regulated jointly in both Myc-LCO treatments and at early nodulation time points (Supplemental Table S11).
Systematic Investigation of Macronutrient- and Nodulation-Responsive CLE Genes
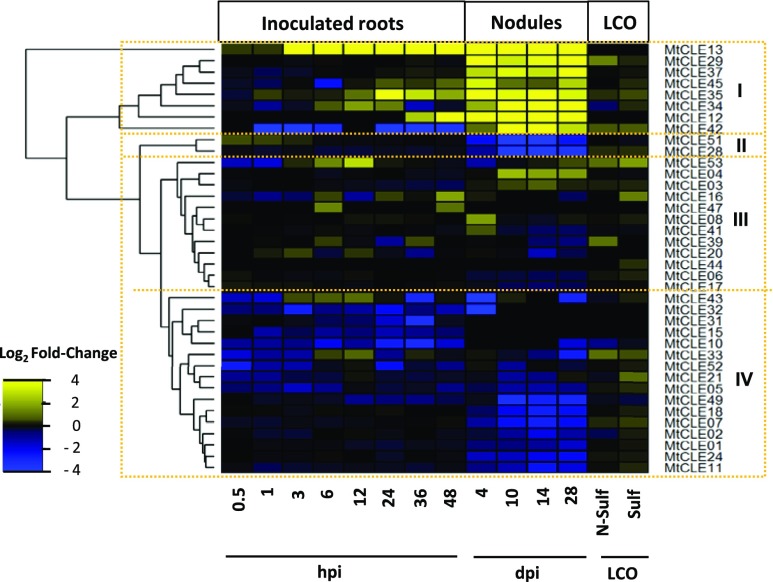
The CLE peptides are among the most studied SSPs in relation to macronutrient stresses and nodulation (Okamoto et al., 2016; de Bang et al., 2017). By combining an improved annotation (Hastwell et al., 2017) with our expression data, we were able to identify additional CLE genes with nodule-specific expression (Fig. 7). CLE gene expression during nodulation falls into three broad groups: (1) highly up-regulated in nodules (eight CLEs), (2) down-regulated by inoculation (18 CLEs), and (3) no clear effects of nodulation (12 CLEs). The remaining CLE genes were expressed in neither roots nor nodules (14 CLEs). Thus, six newly annotated CLEs with high expression in nodules emerge as attractive targets for future functional studies.
Figure 7.
CLE genes respond differentially to symbiotic interactions. M. truncatula CLE genes with detected expression in nodules and roots were collected. The heat map shows the differential expression (as log2 fold change) of inoculated roots (0.5 hpi–4 dpi) and nodules (10–28 dpi) compared with uninoculated roots as well as Myc-LCO-treated roots (sulfated and nonsulfated) compared with untreated controls. Hierarchical clustering has grouped the CLE genes into four groups (I–IV), labeled on the right.
In total, 18 different CLE genes responded to macronutrient deficiency (adjusted P < 0.1), 15 in roots and eight in shoots (Supplemental Table S12). Ten CLEs responded to N, five to P, and 10 to S deficiency, while no CLE genes responded to K deficiency. Two CLE genes, MtCLE05 and MtCLE34, responded to the N, P, and S treatments, with MtCLE05 up-regulated in both shoot and root and MtCLE34 down-regulated in the root. While MtCLE05 expression remained elevated upon resupply, MtCLE34 reverted quickly (within 6 h) to prestress levels when resupplied with each macronutrient. Interestingly, MtCLE34 also was found to be highly expressed in nodules (Fig. 7).
Synthetic Peptides of Nutrient-Responsive Signaling-SSPs Control Root Growth
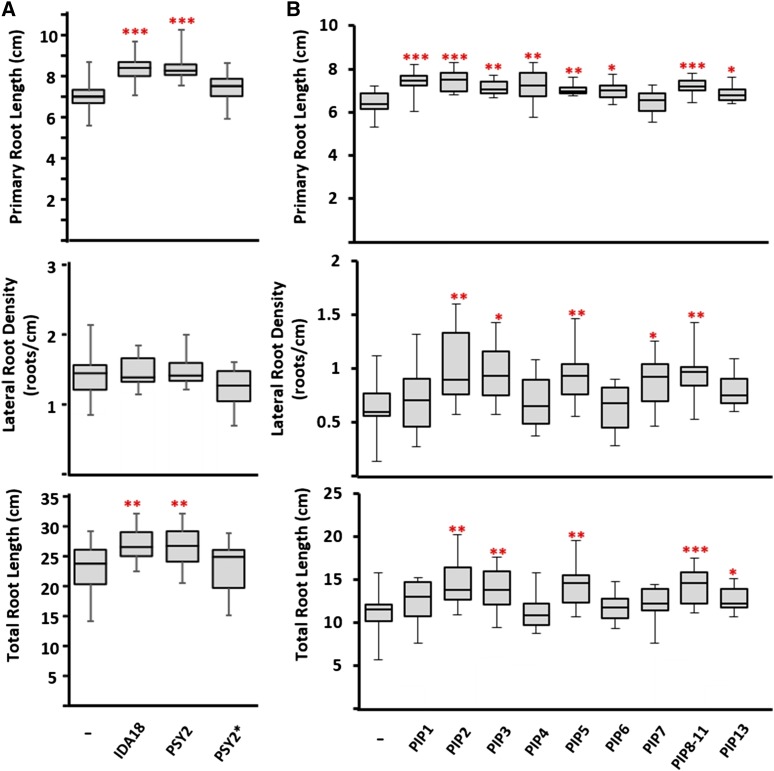
The high number of Signaling-SSP genes responsive to macronutrient deficiencies prompted us to test peptide effects on root traits related to nutrient homeostasis. First, two root-responsive genes were selected, MtIDA18 and MtPSY2, which were up-regulated 32- and 6-fold, respectively, under P deficiency in the nutrient-reduction experiment (Supplemental Table S7). Significantly, MtIDA18 represents a novel gene model resulting from our reannotation, while MtPSY2 was annotated as a putative transmembrane protein in Mt4.0. Synthetic peptides were created based on the mature sequences of AtIDA and AtPSY1, as determined by Santiago et al. (2016) and Amano et al. (2007), respectively, including the appropriate hydroxylated Pro and sulfated Tyr residues on the IDA and PSY peptides (Supplemental Table S13). Due to technical constraints, the glycosylation found on AtPSY1 was excluded from our synthetic peptide. Mature AtPSY1 contains a sulfated Tyr at the second residue, and to explore the relevance of the sulfation on bioactivity, an alternative MtPSY2 peptide was designed, which was phosphorylated instead of sulfated. M. truncatula seedlings were grown for 10 d on Fähræus medium in the presence of 1 µm peptide or in the absence of peptide and scored for root growth traits. Both primary root length and total root length were enhanced markedly in the presence of either the MtIDA18 or MtPSY2 peptide (Fig. 8A). Notably, the promotion of primary and total root length was lost in the presence of the alternative MtPSY2, containing a phosphorylated Tyr, indicating that sulfation is essential for the promotion of root growth under our assay conditions. While the MtIDA18 peptide may have been expected to promote lateral root growth based on the documented effect of AtIDA (Kumpf et al., 2013), none of the three peptides had a discernible effect on lateral root density under the tested conditions (Fig. 8A). Thus, the MtIDA18 and MtPSY2 peptides increased the total amount of root by stimulating root length but not lateral root emergence.
Figure 8.
Synthetic peptides of nutrient-responsive Signaling-SSPs affect root growth traits. Synthetic peptides were tested for root growth and nodulation phenotypes in a Fähræus medium agar system with peptides embedded at a concentration of 1 µm. Plants were grown in the presence of the designated peptide or the absence of peptide for 10 d before measuring primary and lateral root lengths. Data are represented as box and whisker plots, n = 10 (peptide treatments) or n = 20 (no-peptide control treatments). Asterisks indicate statistically significant differences relative to the no-peptide control (*, P < 0.05; **, P < 0.01; and ***, P < 0.001, two-tailed, equal-variance Student’s t test). Data are representative of three independent experiments. A, Root growth measurements from MtIDA18 and MtPSY2 peptides. Both peptides promote root growth and increase total root length without impacting lateral root density. Peptides were designed according to the structure elucidated from AtIDA and AtPSY1, including the anticipated hydroxylation of Pro-7 of MtIDA18 and the sulfation of Tyr-2 and hydroxylations of Pro-13 and Pro-16 of MtPSY2. The PSY2* peptide replaced the sulfation with phosphorylation of Tyr-2, resulting in a loss of bioactivity. B, Root growth measurements from 12 of the 13 MtPIP family members. PIP peptides contained the predicted hydroxylation of Pro-6 in the sequence.
To further test the relationship between nutrient-responsive Signaling-SSPs and root growth, the PIP family was selected for further analysis, since a number of the M. truncatula PIP genes were regulated by macronutrient deficiencies (Supplemental Table S7). As for MtIDA18 and MtPSY2, none of the PIP genes were annotated previously as SSPs in the M. truncatula genome. Rather, most were annotated as hypothetical proteins or putative transmembrane proteins, while two PIP genes were novel gene models resulting from our reannotation (Supplemental Table S3). This Signaling-SSP family has been shown to be induced by pathogens as well as different abiotic factors such as cold and UV light (Hou et al., 2014; Vie et al., 2015); however, no reports have linked this family to macronutrient deficiency. Synthetic MtPIP peptides were designed based on C-terminal sequence homology and in accordance with previous studies in Arabidopsis that tested different Hyp-modified versions of AtPIP1, identifying a 12-residue peptide carrying Hyp modification at position 6 as most active (Hou et al., 2014). For five out of nine tested PIP peptides, M. truncatula seedlings exhibited significantly increased lateral root numbers and primary root lengths, which translated into increased total root lengths (Fig. 8B; Supplemental Table S13).
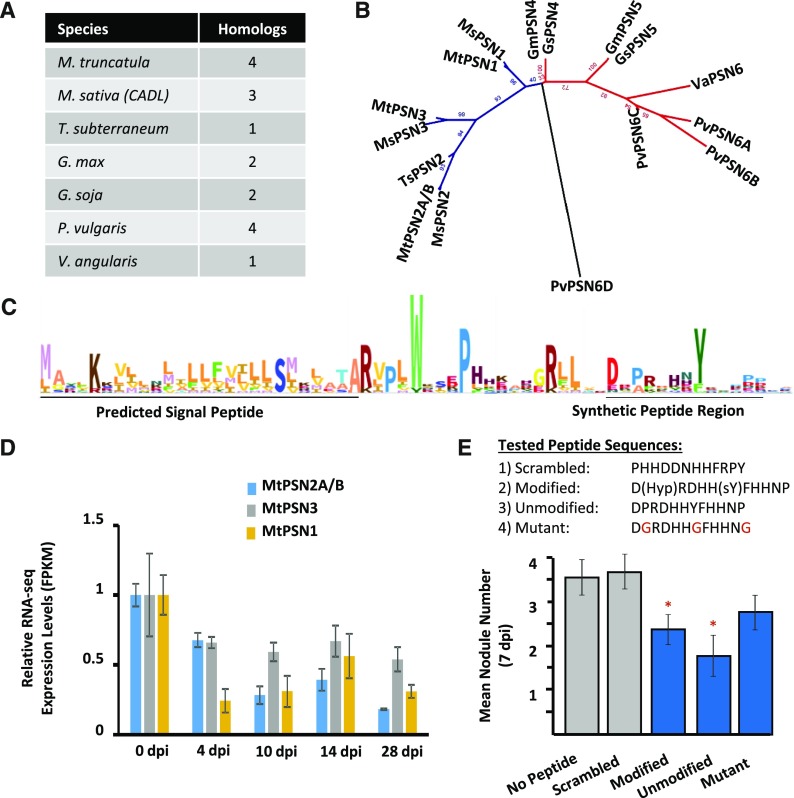
A Legume-Specific Focal List Family Affects Nodulation Number
Focal List Family 71, with four members in M. truncatula, is legume specific and displays reduced expression in nodule organs, which we call the PEPTIDE SUPPRESSING NODULATION (PSN) family (Fig. 9). To test the relevance of this family to legume nodules, we tested synthetic peptides for nodulation phenotypes. The bioactive peptide sequence of PSN members cannot be determined from their precursor sequences alone. However, other SSP families with known structures, such as the PSY family, have conserved upstream protease-cleavage motifs that also are found in PSN members: namely, two RxxL/RxLx motifs cleaved by subtilases and a conserved Asp forming the first residue of the processed peptide. In fact, it is possible that the same proteases are responsible for processing PSY and PSN proproteins. Additionally, the predicted PSN motif carries a highly conserved Tyr residue that is commonly modified by sulfation in other peptide families, although we point out that the Tyr position in PSN is not adjacent to Asp, which is the case for other known sulfated Tyr residues. Using these shared motifs as a guide, we synthesized a PTM-modified and a PTM-unmodified version of the predicted C-terminal peptide-coding region from PSN1 (Fig. 9C) along with two synthetic peptide controls: one mutant peptide modified at three conserved residues and one scrambled sequence peptide. Root inoculation with S. meliloti in the presence of either the PTM-modified or PTM-unmodified peptide provided a pronounced suppression of nodule numbers at 7 dpi (Fig. 9E). Importantly, the mutant peptide also appeared to moderately reduce the number of nodules, although not with statistical significance, while the scrambled control had no impact on nodule number (Fig. 9E). Based on the legume-specific occurrence and the effect on nodulation, we named this family the PSN family. Surprisingly, the MtPSN1 gene, but not the other members from M. truncatula, also was found to be induced in shoot and root tissue under nutrient deficiency (Supplemental Fig. S14A). Thus, we further tested our synthetic PSN peptides for effects on root growth or development. However, repeated testing failed to detect an impact compared with either the no-peptide or scrambled-peptide controls (Supplemental Fig. S14B).
Figure 9.
A synthetic peptide from the novel PSN family inhibits nodulation. The novel PSN family was selected for testing based on expression patterns and legume-specific occurrence. Modified and unmodified versions of the predicted SSP from MtPSN1 were synthesized and tested for effects on nodulation, along with two control peptides. A, A total of 16 homologs of the PSN family were identified in seven legume species. The number of homolog sequences is indicated per species. B, Phylogenetic tree of the 16 homologs calculated by maximum likelihood using RAxML. Homologs separate into an IRLC (blue) and a bean clade (red). Species are indicated by two-letter code at the beginning of each label: Gm, G. max; Gs, G. soja; Ms, M. sativa; Mt, M. truncatula; Pv, P. vulgaris; Ts, T. subterraneum; Va, V. angularis. C, An HMM from the members of the PSN family is illustrated as a WebLogo. The N-terminal predicted signal peptide and the predicted bioactive peptide regions are indicated. D, Normalized RNA-seq expression levels (as FPKM) demonstrate the down-regulated expression of PSN family members in nodules. Each gene is normalized to the 0-dpi control. Error bars represent se (n = 3). E, Nodulation was induced with S. meliloti on agar plates embedded with the designated peptide or without peptide. Nodule numbers were counted 7 dpi. PTM-modified and -unmodified forms of the peptide, and to a lesser extent the conserved residue mutant peptide, suppressed nodule numbers, while the scrambled peptide did not affect nodule numbers. sY, Sulfated Tyr residue. Mutated residues in the mutant sequence are indicated in red. Error bars represent se (n = 20). Asterisks indicate statistically significant differences relative to no peptide control (*, P < 0.05).
DISCUSSION
SSPs encompass many regulatory signaling molecules controlling a multitude of plant growth and development processes, including nodulation and acclimation to nutrient stress (Czyzewicz et al., 2013; Djordjevic et al., 2015; Ohkubo et al., 2017). The identification of such SSPs is of great interest for the improvement of agronomic traits and crop productivity (Bao et al., 2017), particularly in relation to nutrient acquisition. Coupling an improved genome annotation and comprehensive search of SSP homologs with extensive RNA-seq data sets, we have identified hundreds of SSPs responding to nutrient availability or symbiotic cues in M. truncatula. These results underscore the potential of SSPs in improving plant performance and provide a catalyst for follow-up studies. Furthermore, we have connected the nutrient-responsive expression of several SSP genes to beneficial root growth outcomes with synthetic peptide applications and identified a novel legume-specific SSP family that suppresses nodulation numbers in the presence of synthetic peptide.
Genome Reannotation and the Comprehensive Identification of SSPs
To improve the identification of SSP genes, typically derived from bona fide sORFs (fewer than 200 amino acids), we carried out a reannotation of the M. truncatula genome. This resulted in a 10% increase in the gene space, primarily of relatively short genes (fewer than 200 amino acids), which can be largely attributed to the use of extensive RNA-seq data sets that provided experimental evidence for a majority of the newly annotated, short genes (Fig. 1).
The improved annotation enabled a systematic and comprehensive search of homologs of established SSPs from the literature, leading to the identification of 1,970 SSP genes in M. truncatula from 46 different families (Table I). Previous SSP identification studies have typically focused on single families in many plant species, such as the CEP (Ogilvie et al., 2014), CLE (Goad et al., 2017), and RALF (Campbell and Turner, 2017) families. However, the extensive, manual curation performed in this study ensured high confidence in all known SSP families and the corresponding HMMs, which will enable high-quality annotation of a broad range of SSP families in other plant species.
Supplementing our identification of previously established SSP families, we also compiled a Focal List of 2,455 additional, putative SSP genes lacking homology to those described previously in the literature, many of which appear to be legume specific (Fig. 2B). Underscoring the value of our Focal List, the MtCIF gene was included originally in our Focal List as a predicted PTM-like SSP gene, until homologs of the CIF family were described recently in Arabidopsis and demonstrated to produce a bioactive SSP regulating Casparian strip formation (Doblas et al., 2017; Nakayama et al., 2017). The Focal List thus represents a rich source of leads for future research. Further illustrating this potential, we identified a novel PSN family from the Focal List, a legume-specific SSP family that contains a peptide sequence that suppresses nodule development upon exogenous application (Fig. 9).
The genome reannotation corrected many incorrectly predicted gene models and identified additional missed gene loci. In fact, more than 1,000 out of the 4,425 collective SSP and Focal List genes were not present in Mt4.0 prior to our reannotation. Further highlighting the improvement in the genome annotation, many incorrect gene models were corrected based on RNA-seq evidence (Supplemental Figs. S1 and S2). In addition, the wrongful assignments of many SSP genes, including members of the RALF and NCR families, also were corrected (Supplemental Figs. S3 and S4).
The PlantSSP database currently represents the most comprehensive list of known or putative SSP genes from plants, including 820 gene loci from M. truncatula, 265 of which are members of established SSP families (Ghorbani et al., 2015). Most of these 820 gene loci are included within our SSP and Focal List gene sets. However, only 709 Focal List genes (29%) and 612 established SSP genes (31%) could be associated with the HMMs constituting the PlantSSP database (Ghorbani et al., 2015; Supplemental Figs. S3 and S4). This is, in part, a reflection of the extensive manual curation and improved genome annotation employed in our searches, which has resulted in accurate and legume-specific HMM models. The 4,425 collective SSP and Focal List genes thus provide a valuable resource for identifying SSPs in both M. truncatula and other plants, and to date, they represent the most comprehensive and well-curated collection of SSPs for any plant species.
Hundreds of SSPs Are Regulated during Nodulation and by Macronutrient Availability
To exploit our identification of SSPs, we surveyed the expression responses of these genes under diverse experimental conditions relevant to nutrient acquisition. RNA-seq data sets derived from tissue covering early to late stages of nodulation, as well as tissue from plants grown at different availabilities of N, P, K, and S, were developed (Fig. 3). Previous studies connecting nutrient acquisition and SSP function in M. truncatula have focused largely on the roles of CLE and CEP genes in N status-dependent root development and nodulation (Mortier et al., 2010; Delay et al., 2013a; Imin et al., 2013; Okamoto et al., 2013; Araya et al., 2014; Huault et al., 2014; Tabata et al., 2014; Mohd-Radzman et al., 2016; Ohkubo et al., 2017). Demonstrating the validity of our data sets, we could confirm these previous reports of CLE and CEP regulation (Supplemental Table S7). However, our results extend these findings to identify additional CLE and CEP genes regulated by nodulation and nutrient deprivation as well as hundreds of other Signaling-SSP and Focal List genes, including members of all but one of the Signaling-SSP families (Fig. 7; Supplemental Tables S5 and S6). This greatly expands the number of SSP genes found to respond to nutrient availability and demonstrates the diversity of SSP families (and presumably developmental processes) regulated by nutrient deprivation. Interestingly, in a number of cases, some members of an SSP family will be up-regulated by a nutrient stress or nodulation time point while other members will be suppressed. One possible explanation is that competitive binding of related peptides is used to fine-tune signaling outputs. Such antagonism has been demonstrated, for example, within the EPFL family (Lee et al., 2015) and is suggested to be a common phenomenon among peptide hormones (Lee and De Smet, 2016).
To test the connection between expression patterns under nutrient depletion and developmental outcomes, we tested the P-responsive genes MtPSY2, MtIDA18, and the MtPIP family for effects on root growth using synthetic peptide applications. Importantly, MtIDA18 and MtPSY2 significantly promoted primary root growth, while most of the MtPIPs were found to promote lateral root development. Both of these phenotypes led to enhanced capacity for nutrient acquisition via increased total root length (Fig. 8). Several root morphogenic SSPs have been reported in the literature, including members of the PSY and PIP families (de Bang et al., 2017). As in this study, primary root growth in Arabidopsis was found to be stimulated by AtPSY (Amano et al., 2007). However, the reported testing of Arabidopsis PIP peptides showed decreased primary and lateral root lengths (Hou et al., 2014; Ghorbani et al., 2015), contrary to what was found in this study, indicating that some SSPs have species-specific functions.
Our transcriptome analysis of nodule development indicates a surprisingly large number of Signaling-SSPs expressed in nodules, which includes members of all 29 Signaling-SSP families. A recently published M. truncatula proteome atlas, including nodules at 10, 14, and 28 dpi (Marx et al., 2016), verified 66 of these nodule-expressed Signaling-SSP genes. The magnitude and diversity of Signaling-SSPs far surpass previous reports in the literature and strongly indicate prominent roles for Signaling-SSPs in the orchestration of nodulation. These findings warrant follow-up studies to unravel the various roles of SSPs in the nodule. For example, we find that MtIDA18, MtIDA26, and MtPSK3 are induced 5- to 26-fold within the first 30 min after rhizobia inoculation, pointing to their involvement in the initial stages of host-rhizobia recognition. The Myc-LCO treatment only identified five Signaling-SSP and seven Focal List members expressed differentially (Fig. 6). In L. japonicus, Handa et al. (2015) performed a transcriptomic profiling of mycorrhiza-colonized roots and identified 27 differentially expressed Cys-rich SSPs as well as six CLE peptides. A Cys-rich SSP from the LTP family also was found to be up-regulated in L. japonicus nodules. Similarly, MtnsLTP62 induced by Myc-LCO also was induced by more than 100-fold from 6 hpi onward in this study (Supplemental Table S7).
CONCLUSION
To our knowledge, this study provides the most comprehensive set of SSPs ever reported for a plant species and will thus serve as an excellent platform for identifying SSPs in other plants. Transcriptomic analysis based on RNA-seq data of various stages of nodulation and different macronutrient stresses revealed that many more SSPs are likely to be involved in the signaling events controlling these processes. Taken together, this analysis provides novel information about SSPs and their roles in nodulation and acclimation to macronutrient stresses and highlights that the diversity of SSPs taking part in these processes is much greater than was thought previously. The data set constitutes an invaluable resource with regard to the identification of key regulatory SSPs and will be highly beneficial for subsequent mechanistic studies.
MATERIALS AND METHODS
Medicago truncatula Genome Reannotation
The Mt4.0 and Mt3.5v5 genome assemblies were first reannotated using MAKER (Cantarel et al., 2008). The input resource of MAKER comprised an in-house database consisting of 64 Illumina RNA-seq data sets representing different tissues, developmental stages, and stress conditions (Supplemental Table S16). In addition, the DFCI Medicago Gene Index version 11 (ftp://occams.dfci.harvard.edu/pub/bio/tgi/data/Medicago_truncatula/) and protein sequences from Glycine max and Lotus japonicus were included as input for MAKER. RepeatMask (http://repeatmasker.org) was applied to remove low-complexity regions in the genome during gene model prediction in MAKER. The pipeline was adjusted to allow any short gene products. Novel MAKER gene models with annotation edit distance values less than 0.5 were included in the further analysis.
Second, the SPADA pipeline (Zhou et al., 2013) was run to identify SSP genes in the genome assembly versions Mt4.0 and Mt3.5v5. In addition to the built-in HMM for Cys-rich protein families, SPADA was enhanced with the HMMs from the PlantSSP database (Ghorbani et al., 2015). The generated SSP gene models from SPADA were integrated into a novel gene model data set, provided that these genes did not overlap with any known gene models and/or newly identified MAKER gene models on coordinates. sORF Finder (Hanada et al., 2010) was run on Mt4.0 to identify additional gene models from intergenic regions.
After reannotation of both genome assemblies, the novel gene models from the Mt3.5v5 assembly were considered as distinct genes and merged into a final data set if their protein sequence similarities with Mt4.0 gene models were less than 50% based on NCBI BLAST search. The predicted transcripts were mapped onto the Mt4.0 genome assembly using HiSat2 (Pertea et al., 2016). Where appropriate, subsequent adjustments were made to SSP gene models by manual curation of SSP gene models based on transcript evidence from RNA-seq read mapping. Mapped reads were visualized with the Integrative Genomics Viewer program (Thorvaldsdóttir et al., 2013).
Identification of M. truncatula Genes of Established SSP Families
Homology Search
The reannotated M. truncatula genome was analyzed for sequence homology to SSP genes from previously described SSP families from the literature (for a list of SSP families searched, see Supplemental Table S2) using Ssearch (Ropelewski et al., 2002). Genuine homologs generally matched with an e-value ≤ 1 × 10−10. Additionally, homologous sequences were identified using HMMs from the PlantSSP database (http://bioinformatics.psb.ugent.be/webtools/PlantSSP/; Ghorbani et al., 2015) using HMMER with default settings (http://hmmer.org/). For the prediction of N-terminal signal peptides, all gene models were analyzed by SignalP 4.1 using default settings (Petersen et al., 2011).
Markov Cluster Analysis
MCL algorithm analysis (inflation value = 1.4) was performed using the last 70 amino acids of proteins smaller than 200 amino acids having a SignalP D-value above 0.25 as query (Enright et al., 2002). Identified genes of each family were aligned using the MUSCLE tool (https://toolkit.tuebingen.mpg.de/#/tools/muscle; Edgar, 2004; Alva et al., 2016) with a maximum number of 15 iterations, visualized in Jalview-2 (Waterhouse et al., 2009) and manually curated to produce the final list of M. truncatula SSP members of each family. In a second iteration, HMMs were produced for each of the SSP families. These HMMs were searched against the predicted protein sequences of the reannotated genome to identify additional members of each family. After merging genes from the first and second iterations, compiled family members were once more aligned using MUSCLE, visualized in Jalview-2, and manually curated. The previously established numbering for NCR peptides (Mergaert et al., 2003; Montiel et al., 2017) was mapped to gene accession identifiers by BLAST search of the NCR peptide sequence as query against the M. truncatula nonredundant gene list from our genome reannotation, requiring an e-value threshold of less than 1 × 10−6. In several cases, an NCR peptide sequence matched to two gene accession identifiers, in which case that with the higher bit score was used. To determine the number of novel SSPs identified in this work, previous identifications were determined from Mt4.0 annotations and/or Uniprot annotations, with the exception of the CEP family, for which previous identifications were made by Imin et al. (2013) and Delay et al. (2013b), and the CLE family, for which previous identifications were made by Mortier et al. (2010) and Hastwell et al. (2017). Where an SSP was not annotated as a member of its respective family by Mt4.0 or Uniprot, it was considered to be newly identified in this work.
Development and Analysis of the SSP Focal List
Gene models of the reannotated M. truncatula genome were filtered in six sequential steps, first according to size (230 or fewer amino acids) and then by SignalP D-value (0.45 or greater). The appropriate length and SignalP D-value criteria for filtering were determined empirically using all members of the established SSP families as a positive control (except those known to not be secreted [i.e. the RTFL/DVL and SubIn families]). The greatest proportion of established SSPs passing the filter, while limiting the number of negative controls, was achieved with a protein length criterion of 230 or fewer residues and a SignalP D-value of 0.45 or greater (Supplemental Fig. S5). This D-value is the recommended, default D-value for confident prediction of a signal peptide (Petersen et al., 2011). With these two sequential filtering criteria, 92.3% of established SSPs were retained but only 5.3% of the negative control set was retained. All members of established SSP families, identified as described above, were then removed. Subsequently, genes were filtered for the absence of transmembrane helices (TMs), which were predicted as follows: genes were input to the SignalP 4.1 browser (http://www.cbs.dtu.dk/services/SignalP/; Petersen et al., 2011), and signal peptides were predicted using the Input sequences do not include TM regions setting. Processed peptide sequences (i.e. with predicted signal peptides removed) were extracted and input to TM-HMM version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) to predict TMs. Genes predicted to harbor at least one TM were discarded. Next, filtered genes were searched for an endoplasmic reticulum-retention signal, defined as KDEL, HDEL, or KQEL, in the C-terminal four residues. Finally, genes were filtered for those strongly predicted, well-known functions unrelated to peptide hormones, such as transcription factors, photosynthetic subunits, or metabolic enzymes. Functional predictions were based primarily on PFAM predictions and required e-value matches of less than 1 × 10−10. To identify families of conserved genes among the Focal List, genes were clustered using the MCL tool (inflation value = 1.4) using the complete protein sequences. To identify the conservation of Focal List genes outside of M. truncatula, Focal List genes were BLAST searched against 16 selected angiosperm species, representing varying evolutionary distances to M. truncatula (Medicago sativa, Cicer arietinum, Trifolium subterraneum, Lotus japonicus, Glycine max, Glycine soja, Phaseolus vulgaris, Vigna angularis, Arabidopsis thaliana, Vitis vinifera, Solanum lycopersicum, Nicotiana benthamiana, Zea mays, Oryza sativa, Brachypodium distachyon, and Amborella trichopoda). Protein sequences for all species but M. sativa were retrieved from the NCBI protein database (https://www.ncbi.nlm.nih.gov/protein/). The M. sativa protein sequences of the genome annotation of diploid alfalfa, CADL, were obtained from www.alfalfatoolbox.org. Putative orthologs were identified based on the reciprocal best BLAST hit approach (Moreno-Hagelsieb and Latimer, 2008).
Plant Materials and Growth Conditions
Nutrient-Reduction Experiment
M. truncatula ‘Jemalong A17’ seeds (Young et al., 2011) were scarified in concentrated H2SO4 as described earlier (Garcia et al., 2006). After acid removal, seeds were washed seven times in cold and sterile Milli-Q water. Scarified seeds were then sterilized in a mixture containing 1.5% (v/v) sodium hypochlorite and 0.1% (v/v) Tween 20 for 5 min. After sterilization, seeds were washed seven times in sterile Milli-Q water and imbibed for 2 h. Imbibed seeds were transferred onto moist filter paper in petri dishes, covered with foil, and vernalized in the dark at 4°C for 3 d. Germination was initiated by moving the seeds to 22°C. Seeds were kept in the dark for 12 h to promote root growth.
Germinated seedlings were placed in a growth chamber at 22°C with a 16-h-day/8-h-night cycle for 2 d. Seedlings were transferred to Deepots (D60) containing a 3:1 mixture of sand and silica, with a porosity of 0.25, and grown under controlled greenhouse conditions. The response to persistent deficiency was tested by growing M. truncatula for 3 weeks under FN or under deficiency of N, P, K, or S. Appropriate fold reductions of each macronutrient were determined empirically in pilot experiments seeking to impair plant growth or induce visual macronutrient deficiency phenotypes. Plants were split into groups and watered every 2 d with 25 mL of the following nutrient solutions: FN, 10-fold reduced nitrogen, 100-fold reduced phosphate, 100-fold reduced sulfate, or 1,000-fold reduced potassium. For individual nutrient solution recipes, see Supplemental Table S15. Shoot and root tissues were separated upon harvest and quickly frozen in liquid nitrogen (N2). Frozen tissue was ground into a fine powder in liquid N2 and stored at –80°C until further analysis. On the day of harvest, additional samples were collected from plants resupplied with 25 mL of FN solution four times in 1.5-h intervals, and tissue was collected 6 h after beginning the resupply treatment.
Nutrient-Elimination Experiment
Germinated seedlings were placed in a growth chamber for 2 d and subsequently transferred to Deepots containing a sand:perlite mixture (1:3). Growth chamber settings were set to a 12-h-day/12-h-night cycle at 23°C/22°C with a light intensity of ∼125 µmol m−2 s−1 and a relative humidity of 50%. The plants were divided into two groups and watered with 5× nutrient solution for 3 d before continuing the watering regime with 1× nutrient solution (Supplemental Table S15). For the P-limitation experiment, plants were watered with 5× nutrient solution with or without P for 3 d (Supplemental Table S15). Subsequently, the nutrient solution was diluted to 1× and supplied to the plants on alternating days. The plants were grown for a total of 21 d until the typical P-limitation phenotype (dark-green leaves and smaller plants) became apparent in the P-limited group. The positions of the trays/plants were randomized every 2 d to minimize position effects. P limitation in plants was confirmed before harvesting by measuring inorganic phosphate in the leaves colorimetrically using the Malachite Green assay (Itaya and Ui, 1966) as described by Pant et al. (2015). Roots and shoots were washed with deionized water, harvested, flash frozen in liquid N2, and stored at −80°C until further analysis. For the N-deprivation treatment, a subset of 2-week-old plants grown in FN conditions were uprooted, rinsed with deionized water, transferred to N-limited conditions, and watered with 1× nutrient solution lacking N on alternate days. The plants were grown under N-deprivation conditions for an additional 8 d until the typical symptoms of N limitation (yellowish leaves and smaller plants) appeared.
Nodulation Experiment
M. truncatula ‘Jemalong A17’ seeds were scarified with concentrated H2SO4, rinsed, sterilized with 2% sodium hypochlorite, and vernalized at 4°C for 3 d on moist, sterile filter paper. For the early nodulation time points of 0, 4, 10, and 14 dpi, plants were grown in aeroponic chambers as described previously (Dickstein et al., 2002; Catalano et al., 2004) and misted in a nitrogen-free nutrient medium (Lullien et al., 1987) containing 5 mM NH4NO3 for 11 d. Plants were then placed in N-free media for 4 d to induce N starvation. Roots were collected and the remaining plants were inoculated with Sinorhizobium meliloti strain ABS7. At 4 dpi, root bumps were collected, and at 10 and 14 dpi, nodules were collected. A subset of plants received 10 mM KNO3 at 14 dpi, for 12 and 48 h. Plants were maintained on a regime of 16 h of light and 8 h of dark at 22°C. All experiments were performed with three biological replicates. All harvested material was frozen immediately in liquid nitrogen and stored at −80°C prior to RNA isolation.
Ion Chromatography
Finely ground M. truncatula tissue (∼50 mg) was resuspended in 500 µL of Milli-Q water and subjected to three freeze-thaw cycles: liquid N2 for 2 min and a 60°C water bath for 2 min. Samples were centrifuged at 14,000g for 1 min, and the supernatant (400 µL) was transferred to a separate tube. The pellet was resuspended in 500 µL of Milli-Q water and centrifuged at 14,000g for 1 min, and the supernatant (350 µL) was collected. Ion extracts were diluted 20 times before ion chromatography analysis. Chromatographic separation of 25 µL of diluted ion extract was achieved on a Dionex ICS-5000 IC system (Thermo Fisher Scientific) with a flow rate of 0.3 mL min−1 and a column temperature of 30°C. For cations, a Dionex CS12A Ion Pac (2 × 250 mm) analytical column with an AG12A guard column (2 × 50 mm) was used. The eluent used was Thermo Scientific Dionex EGC III methanesulfonic acid using the following gradient: from 12 to 20 mm during 7 min followed by 20 mm for 8 min. Anions were separated on a Dionex AS11 Ion Pac (2 × 250 mm) analytical column with an AG11 guard column (2 × 50 mm) with the following elution gradient: from 0 to 6 mm during 1 min, from 6 to 45 mm over 9 min, and from 45 to 55 mm in 2 min. Quantification was achieved using Chromeleon 7 version 7.1.2.1478 software. Standard curves were prepared using dilutions of the following standards: for anions, Thermo Scientific Dionex Seven Anion Standard II; and for cations, Thermo Scientific Dionex Six Cation Standard II.
RT-qPCR
RNA was extracted from ∼100 mg of tissue using the Spectrum Plant Total RNA Kit (Sigma-Aldrich), and subsequently, genomic DNA was removed using the TURBO DNA-free kit (Ambion). Both kits were used according to the manufacturer’s instructions. RNA yield and quality were assessed with a Nanodrop 1000 (A260/A280 > 1.8 and A260/A230 > 2). Reverse transcription was performed by mixing 5 µg of RNA with 1.5 µL of oligo(dT)20 (50 µm) and 1.5 µL of dNTP (10 µm each of dATP, dGTP, dCTP, and dTTP), followed by the addition of DEPC-treated water to a total volume of 36.5 µL. Samples were heated to 65°C for 5 min and cooled on ice for 2 min. Next, 10 µL of 5× first-strand buffer, 2 µL of DTT (0.1 mm), 0.5 µL of RNaseOUT, and 1 µL of SuperScript III (Invitrogen) were added, and samples were heated to 50°C for 60 min followed by 15 min at 70°C. All cDNA samples were diluted 8-fold, and the amplification of a stably expressed ubiquitin gene (MtUBQ, Medtr3g091400.1; Kakar et al., 2008) was analyzed to assess cDNA yield and quality (primer sequences can be found in Supplemental Table S14). Primers used to amplify macronutrient-responsive gene transcripts were designed using Primer3v4.0.0 (http://bioinfo.ut.ee/primer3/) based on the design criteria given by Udvardi et al. (2008). RT-qPCR was performed in 10-µL reactions using KiCqStart SYBR Green qPCR ReadyMix (Sigma-Aldrich) with final primer concentration of 200 nm each. Thermal cycling was carried out with the 7900 HT Real-Time PCR System (Applied Biosystems) using the following PCR protocol: denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 59°C for 45 s. A dissociation step was added to assess primer specificity. Differential gene expression was quantified based on the ΔΔCt method using MtUBQ as a reference gene.
RNA-Seq Library Preparation and Transcriptome Sequencing
Nodule Experiment
Total RNA was extracted using TRIzol reagent (Chomczynski and Mackey, 1995), treated with DNaseI (Ambion), and column purified with the RNeasy MinElute CleanUp Kit (Qiagen). RNA was quantified using a Nanodrop Spectrophotometer (ND-100; NanoDrop Technologies) and evaluated for purity with a Bioanalyzer 2100 (Agilent). RNA integrity number values were between 8 and 10. One microgram of total RNA was used to prepare the RNA-seq nodule data sets using the TruSeq RNA Sample Preparation Kit according to the standard protocol (Illumina). Library DNA size distribution was analyzed using an Agilent 2100 Bioanalyzer. Data sets were quantified before sequencing with the Illumina HiSeq 2000 system.
Nutrient-Reduction and Nutrient-Elimination Experiments
Total RNA was extracted from 100 mg of tissue using the Spectrum Plant Total RNA Kit (Sigma-Aldrich). RNA integrity and quality were assessed with an Agilent 2100 Bioanalyzer. RNA integrity number values were between 7 and 10. Total RNA (1 μg) was used to prepare RNA-seq data sets using the TruSeq Stranded mRNA LT Sample Prep Kit according to the standard protocol (Illumina). Library DNA size distribution was analyzed using an Agilent 2100 Bioanalyzer. Data sets were quantified using the Qubit 2.0 fluorometer before sequencing with the Illumina NextSeq 500 system using the NextSeq 500 High Output Reagent Cartridge version 2 (150 cycles) according to the manufacturer’s instructions (Illumina NextSeq 500 System Guide).
RNA-Seq Mapping and DE Gene Analysis
RNA-seq data sets were mapped against the in-house reannotated M. truncatula genome to estimate raw counts and effective read lengths using Salmon, an improved version of the Sailfish tool (Patro et al., 2014). Raw counts from Salmon were normalized across all samples by median normalization, and DE was estimated using the DEseq2 module from the Bioconductor R package (Love et al., 2014) and an in-house Perl script. The sequencing read depth was estimated to be 10×. Since only the longest transcript of each gene model was chosen for Illumina read mapping, the representative transcript was used directly to estimate the expression level of each gene model. Gene expression was quantified as FPKM, and identified DE genes were required to have a false discovery rate of less than 0.1. DESeq2 (Love et al., 2014) was used further to perform DE analysis, which included dynamic filtering of low expressed genes to prevent spurious identification of DE genes.
Hierarchical Clustering
Log2 fold changes were calculated based on average FPKM values from individual treatments, and hierarchical clustering was performed using heatmap.2 from the gplots package in the Bioconductor R packages (Warnes et al., 2016).
Synthetic Peptide Assays
Synthetic peptides (Pepscan) were diluted in Fähræus medium to a concentration of 1 µm and polymerized in 12-cm × 12-cm plates with 2% (w/v) Gelzan CM (Sigma-Aldrich).
Root Growth Assays
Germinated seedlings (two per plate) were transferred to the plates and placed between two filter papers (Whatman; grade 0858, reference no. 10 334 365) overlaying the gel medium. The lower three-quarters of the plates were wrapped in black plastic foil and placed upright in a growth chamber at 22°C with a 16-h-day/8-h-night cycle at approximately 150 µmol photons m−2 s−1 for 10 d. Prior to collecting images, plates were unwrapped and placed on a light box with the top filter paper sheet removed. Images were collected from a Canon EOS Rebel T3i. Images were subsequently converted to grayscale for analysis with SmartRoot (Lobet et al., 2011).
Nodulation Assays
Nodulation plate assays were adapted from Breakspear et al. (2014). Briefly, 50 µL of a 1 mm stock concentration of peptides was added to 50 mL of a water agarose medium for a final concentration of 1 μm. Ten overnight-germinated seedlings were transferred to these plates between two filter papers. Seedlings were allowed to grow for 24 h before inoculation with Rm 2011 dsRED at a final OD600 of 0.05. Nodules were counted with a stereomicroscope 7 dpi.
Accession Numbers
RNA-seq data sets produced in-house can be retrieved from the NCBI Short Read Archive at https://www.ncbi.nlm.nih.gov/sra with the study identifiers SRP110143, SRP110041, and SRP109847.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Manual curation of RNA-seq results identifies alternative gene models revealing strongly predicted signal peptides.
Supplemental Figure S2. Manual curation of RNA-seq results uncovers an additional PSY gene at the Medtr6g027320 locus.
Supplemental Figure S3. Improved annotation of the NCR family in M. truncatula.
Supplemental Figure S4. Improved annotation of the RALF family in M. truncatula.
Supplemental Figure S5. Optimization of the protein length and SignalP D-value filtering criteria.
Supplemental Figure S6. Amino acid enrichment in PTM SSP preproproteins among the C-terminal 25 residues.
Supplemental Figure S7. Biomass and ion contents of samples from the nutrient-reduction experiment.
Supplemental Figure S8. Overlap of nutrient-reduction and nutrient-elimination Signaling-SSP responses.
Supplemental Figure S9. Specificity of macronutrient-responsive Signaling-SSPs.
Supplemental Figure S10. Resupply-responsive Signaling-SSP and Focal List genes.
Supplemental Figure S11. Heat map and hierarchical clustering of Signaling-SSPs and Focal List members during nodulation.
Supplemental Figure S12. Heat map and hierarchical clustering of predicted PTM and Cys-rich Focal List members based on nodulation data.
Supplemental Figure S13. Alignments of selected Focal List SSP families.
Supplemental Figure S14. Nutrient deficiency expression patterns and root growth tests of MtPSN1.
Supplemental Table S1. Output from the reannotation pipeline.
Supplemental Table S2. Characteristics of established SSP families present in M. truncatula.
Supplemental Table S3. Curated list of M. truncatula SSP genes.
Supplemental Table S4. Creation of the Focal List by sequential filtering steps.
Supplemental Table S5. RNA-seq experimental meta data.
Supplemental Table S6. Normalized FPKM values of SSP and Focal List genes from RNA-seq experiments.
Supplemental Table S7. Macronutrient- and symbiosis-responsive Signaling-SSPs.
Supplemental Table S8. Macronutrient- and symbiosis-responsive Focal List members.
Supplemental Table S9. Analysis of Focal List members with interesting expression patterns during macronutrient deficiencies.
Supplemental Table S10. Hierarchical clustering of Signaling-SSPs and Focal List members expressed at minimum one time point during nodulation.
Supplemental Table S11. Analysis of Focal List members with interesting expression patterns during symbioses.
Supplemental Table S12. CLE peptide expression under macronutrient deficiency and resupply conditions.
Supplemental Table S13. Synthetic peptides used in plant growth experiments.
Supplemental Table S14. Expression of macronutrient deficiency marker genes analyzed by RT-qPCR.
Supplemental Table S15. Nutrient solution recipes.
Supplemental Table S16. RNA-seq data sets as input for genome reannotation by MAKER.
Acknowledgments
We thank Khem Kadel for assistance with plant growth and collection, Yongxiang Li and Shulan Zhang for assistance with synthetic peptide assays, Stacy Allen of the Genomics Core Facility at the Noble Research Institute for assistance with RNA-seq data production, David Huhman of the Chemistry Core Facility at the Noble Research Institute for assistance with ion chromatography analysis, Dr. Maria Monteros for providing the genome assemblies of CADL, and Drs. Eva Kondorosi and Jesus Montiel for discussions about annotation of NCR genes.
Footnotes
Funding was provided by the Noble Research Institute, the National Science Foundation (award no. 1444549 to W.R.S., P.X.Z., and M.K.U. and award no. 1127155 to M.K.U. and R.D.), and the EU Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 659251 to T.C.d.B.
Articles can be viewed without a subscription.
References
- Alunni B, Kevei Z, Redondo-Nieto M, Kondorosi A, Mergaert P, Kondorosi E (2007) Genomic organization and evolutionary insights on GRP and NCR genes, two large nodule-specific gene families in Medicago truncatula. Mol Plant Microbe Interact 20: 1138–1148 [DOI] [PubMed] [Google Scholar]
- Alva V, Nam SZ, Söding J, Lupas AN (2016) The MPI bioinformatics toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res 44: W410–W415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y (2007) Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc Natl Acad Sci USA 104: 18333–18338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SJ, Rothnagel JA (2014) Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet 15: 193–204 [DOI] [PubMed] [Google Scholar]
- Araya T, Miyamoto M, Wibowo J, Suzuki A, Kojima S, Tsuchiya YN, Sawa S, Fukuda H, von Wirén N, Takahashi H (2014) CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc Natl Acad Sci USA 111: 2029–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Clancy MA, Carvalho RF, Elliott K, Folta KM (2017) Identification of novel growth regulators in plant populations expressing random peptides. Plant Physiol 175: 619–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakspear A, Liu C, Roy S, Stacey N, Rogers C, Trick M, Morieri G, Mysore KS, Wen J, Oldroyd GE, et al. (2014) The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 26: 4680–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton WJ, Dilworth MJ (1971) Control of leghaemoglobin synthesis in snake beans. Biochem J 125: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell L, Turner SR (2017) A comprehensive analysis of RALF proteins in green plants suggests there are two distinct functional groups. Front Plant Sci 8: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps C, Jardinaud MF, Rengel D, Carrère S, Hervé C, Debellé F, Gamas P, Bensmihen S, Gough C (2015) Combined genetic and transcriptomic analysis reveals three major signalling pathways activated by Myc-LCOs in Medicago truncatula. New Phytol 208: 224–240 [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Sánchez Alvarado A, Yandell M (2008) MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18: 188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano CM, Lane WS, Sherrier DJ (2004) Biochemical characterization of symbiosome membrane proteins from Medicago truncatula root nodules. Electrophoresis 25: 519–531 [DOI] [PubMed] [Google Scholar]
- Chen YL, Lee CY, Cheng KT, Chang WH, Huang RN, Nam HG, Chen YR (2014) Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. Plant Cell 26: 4135–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Mackey K (1995) Modification of the TRI reagent procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. Biotechniques 19: 942–945 [PubMed] [Google Scholar]
- Combier JP, Küster H, Journet EP, Hohnjec N, Gamas P, Niebel A (2008) Evidence for the involvement in nodulation of the two small putative regulatory peptide-encoding genes MtRALFL1 and MtDVL1. Mol Plant Microbe Interact 21: 1118–1127 [DOI] [PubMed] [Google Scholar]
- Czyzewicz N, Yue K, Beeckman T, De Smet I (2013) Message in a bottle: small signalling peptide outputs during growth and development. J Exp Bot 64: 5281–5296 [DOI] [PubMed] [Google Scholar]
- de Bang TC, Lay KS, Scheible WR, Takahashi H (2017) Small peptide signaling pathways modulating macronutrient utilization in plants. Curr Opin Plant Biol 39: 31–39 [DOI] [PubMed] [Google Scholar]
- Delay C, Imin N, Djordjevic MA (2013a) Regulation of Arabidopsis root development by small signaling peptides. Front Plant Sci 4: 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delay C, Imin N, Djordjevic MA (2013b) CEP genes regulate root and shoot development in response to environmental cues and are specific to seed plants. J Exp Bot 64: 5383–5394 [DOI] [PubMed] [Google Scholar]
- Dickstein R, Hu X, Yang J, Ba L, Coque L, Kim DJ, Cook DR, Yeung AT (2002) Differential expression of tandemly duplicated Enod8 genes in Medicago. Plant Sci 163: 333–343 [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Mattick JS (2008) Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLOS Comput Biol 4: e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic MA, Mohd-Radzman NA, Imin N (2015) Small-peptide signals that control root nodule number, development, and symbiosis. J Exp Bot 66: 5171–5181 [DOI] [PubMed] [Google Scholar]
- Doblas VG, Smakowska-Luzan E, Fujita S, Alassimone J, Barberon M, Madalinski M, Belkhadir Y, Geldner N (2017) Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 355: 280–284 [DOI] [PubMed] [Google Scholar]
- Downie JA. (2014) Legume nodulation. Curr Biol 24: R184–R190 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30: 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funayama-Noguchi S, Noguchi K, Yoshida C, Kawaguchi M (2011) Two CLE genes are induced by phosphate in roots of Lotus japonicus. J Plant Res 124: 155–163 [DOI] [PubMed] [Google Scholar]
- Garcia J, Barker DG, Journet EP (2006) Seed storage and germination. In Mathesius UJ, Sumner LW, eds, Medicago truncatula Handbook. Noble Research Institute, https://www.noble.org/medicago-handbook/ [Google Scholar]
- Ghorbani S, Hoogewijs K, Pečenková T, Fernandez A, Inzé A, Eeckhout D, Kawa D, De Jaeger G, Beeckman T, Madder A, et al. (2016) The SBT6.1 subtilase processes the GOLVEN1 peptide controlling cell elongation. J Exp Bot 67: 4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbani S, Lin YC, Parizot B, Fernandez A, Njo MF, Van de Peer Y, Beeckman T, Hilson P (2015) Expanding the repertoire of secretory peptides controlling root development with comparative genome analysis and functional assays. J Exp Bot 66: 5257–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goad DM, Zhu C, Kellogg EA (2017) Comprehensive identification and clustering of CLV3/ESR-related (CLE) genes in plants finds groups with potentially shared function. New Phytol 216: 605–616 [DOI] [PubMed] [Google Scholar]
- Hanada K, Akiyama K, Sakurai T, Toyoda T, Shinozaki K, Shiu SH (2010) sORF finder: a program package to identify small open reading frames with high coding potential. Bioinformatics 26: 399–400 [DOI] [PubMed] [Google Scholar]
- Hanada K, Higuchi-Takeuchi M, Okamoto M, Yoshizumi T, Shimizu M, Nakaminami K, Nishi R, Ohashi C, Iida K, Tanaka M, et al. (2013) Small open reading frames associated with morphogenesis are hidden in plant genomes. Proc Natl Acad Sci USA 110: 2395–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa Y, Nishide H, Takeda N, Suzuki Y, Kawaguchi M, Saito K (2015) RNA-seq transcriptional profiling of an arbuscular mycorrhiza provides insights into regulated and coordinated gene expression in Lotus japonicus and Rhizophagus irregularis. Plant Cell Physiol 56: 1490–1511 [DOI] [PubMed] [Google Scholar]
- Hastwell AH, de Bang TC, Gresshoff PM, Ferguson BJ (2017) CLE peptide-encoding gene families in Medicago truncatula and Lotus japonicus, compared with those of soybean, common bean and Arabidopsis. Sci Rep 7: 9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastwell AH, Gresshoff PM, Ferguson BJ (2015) The structure and activity of nodulation-suppressing CLE peptide hormones of legumes. Funct Plant Biol 42: 229–238 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Brown CM, Chisnall MA, Waterhouse PM, Macknight RC (2016) The emerging world of small ORFs. Trends Plant Sci 21: 317–328 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K (2004) Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 10205–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth B, Domonkos Á, Kereszt A, Szűcs A, Ábrahám E, Ayaydin F, Bóka K, Chen Y, Chen R, Murray JD, et al. (2015) Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. Proc Natl Acad Sci USA 112: 15232–15237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, Wang X, Chen D, Yang X, Wang M, Turrà D, Di Pietro A, Zhang W (2014) The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PLoS Pathog 10: e1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huault E, Laffont C, Wen J, Mysore KS, Ratet P, Duc G, Frugier F (2014) Local and systemic regulation of plant root system architecture and symbiotic nodulation by a receptor-like kinase. PLoS Genet 10: e1004891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imin N, Mohd-Radzman NA, Ogilvie HA, Djordjevic MA (2013) The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. J Exp Bot 64: 5395–5409 [DOI] [PubMed] [Google Scholar]
- Itaya K, Ui M (1966) A new micromethod for the colorimetric determination of inorganic phosphate. Clin Chim Acta 14: 361–366 [DOI] [PubMed] [Google Scholar]
- Kakar K, Wandrey M, Czechowski T, Gaertner T, Scheible WR, Stitt M, Torres-Jerez I, Xiao Y, Redman JC, Wu HC, et al. (2008) A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula. Plant Methods 4: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassaw T, Nowak S, Schnabel E, Frugoli J (2017) ROOT DETERMINED NODULATION1 is required for M. truncatula CLE12, but not CLE13, peptide signaling through the SUNN receptor kinase. Plant Physiol 174: 2445–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Chen Y, Xi J, Waters C, Chen R, Wang D (2015) An antimicrobial peptide essential for bacterial survival in the nitrogen-fixing symbiosis. Proc Natl Acad Sci USA 112: 15238–15243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpf RP, Shi CL, Larrieu A, Stø IM, Butenko MA, Péret B, Riiser ES, Bennett MJ, Aalen RB (2013) Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc Natl Acad Sci USA 110: 5235–5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrainzar E, Riely BK, Kim SC, Carrasquilla-Garcia N, Yu HJ, Hwang HJ, Oh M, Kim GB, Surendrarao AK, Chasman D, et al. (2015) Deep sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between nodulation factor and ethylene signals. Plant Physiol 169: 233–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauressergues D, Couzigou JM, Clemente HS, Martinez Y, Dunand C, Bécard G, Combier JP (2015) Primary transcripts of microRNAs encode regulatory peptides. Nature 520: 90–93 [DOI] [PubMed] [Google Scholar]
- Lease KA, Walker JC (2006) The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol 142: 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, De Smet I (2016) Fine-tuning development through antagonistic peptides: an emerging theme. Trends Plant Sci 21: 991–993 [DOI] [PubMed] [Google Scholar]
- Lee JS, Hnilova M, Maes M, Lin YCL, Putarjunan A, Han SK, Avila J, Torii KU (2015) Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522: 439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobet G, Pagès L, Draye X (2011) A novel image-analysis toolbox enabling quantitative analysis of root system architecture. Plant Physiol 157: 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lullien V, Barker DG, de Lajudie P, Huguet T (1987) Plant gene expression in effective and ineffective root nodules of alfalfa (Medicago sativa). Plant Mol Biol 9: 469–478 [DOI] [PubMed] [Google Scholar]
- Marmiroli N, Maestri E (2014) Plant peptides in defense and signaling. Peptides 56: 30–44 [DOI] [PubMed] [Google Scholar]
- Marx H, Minogue CE, Jayaraman D, Richards AL, Kwiecien NW, Siahpirani AF, Rajasekar S, Maeda J, Garcia K, Del Valle-Echevarria AR, et al. (2016) A proteomic atlas of the legume Medicago truncatula and its nitrogen-fixing endosymbiont Sinorhizobium meliloti. Nat Biotechnol 34: 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y. (2014) Posttranslationally modified small-peptide signals in plants. Annu Rev Plant Biol 65: 385–413 [DOI] [PubMed] [Google Scholar]
- Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E (2003) A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol 132: 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Radzman NA, Binos S, Truong TT, Imin N, Mariani M, Djordjevic MA (2015) Novel MtCEP1 peptides produced in vivo differentially regulate root development in Medicago truncatula. J Exp Bot 66: 5289–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Radzman NA, Laffont C, Ivanovici A, Patel N, Reid D, Stougaard J, Frugier F, Imin N, Djordjevic MA (2016) Different pathways act downstream of the CEP peptide receptor CRA2 to regulate lateral root and nodule development. Plant Physiol 171: 2536–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel J, Downie JA, Farkas A, Bihari P, Herczeg R, Bálint B, Mergaert P, Kereszt A, Kondorosi É (2017) Morphotype of bacteroids in different legumes correlates with the number and type of symbiotic NCR peptides. Proc Natl Acad Sci USA 114: 5041–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Hagelsieb G, Latimer K (2008) Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics 24: 319–324 [DOI] [PubMed] [Google Scholar]
- Mortier V, Den Herder G, Whitford R, Van de Velde W, Rombauts S, D’Haeseleer K, Holsters M, Goormachtig S (2010) CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol 153: 222–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, De Smet I (2014) Understanding the RALF family: a tale of many species. Trends Plant Sci 19: 664–671 [DOI] [PubMed] [Google Scholar]
- Nakayama T, Shinohara H, Tanaka M, Baba K, Ogawa-Ohnishi M, Matsubayashi Y (2017) A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science 355: 284–286 [DOI] [PubMed] [Google Scholar]
- Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC (2011) Phosphate import in plants: focus on the PHT1 transporters. Front Plant Sci 2: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelkers K, Goffard N, Weiller GF, Gresshoff PM, Mathesius U, Frickey T (2008) Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol 8: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie HA, Imin N, Djordjevic MA (2014) Diversification of the C-TERMINALLY ENCODED PEPTIDE (CEP) gene family in angiosperms, and evolution of plant-family specific CEP genes. BMC Genomics 15: 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo Y, Tanaka M, Tabata R, Ogawa-Ohnishi M, Matsubayashi Y (2017) Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat Plants 3: 17029. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Shinohara H, Mori T, Matsubayashi Y, Kawaguchi M (2013) Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat Commun 4: 2191. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Tabata R, Matsubayashi Y (2016) Long-distance peptide signaling essential for nutrient homeostasis in plants. Curr Opin Plant Biol 34: 35–40 [DOI] [PubMed] [Google Scholar]
- Ou Y, Lu X, Zi Q, Xun Q, Zhang J, Wu Y, Shi H, Wei Z, Zhao B, Zhang X, et al. (2016) RGF1 INSENSITIVE 1 to 5, a group of LRR receptor-like kinases, are essential for the perception of root meristem growth factor 1 in Arabidopsis thaliana. Cell Res 26: 686–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B, Sheng J, Sun W, Zhao Y, Hao P, Li X (2013) OrysPSSP: a comparative platform for small secreted proteins from rice and other plants. Nucleic Acids Res 41: D1192–D1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Pant P, Erban A, Huhman D, Kopka J, Scheible WR (2015) Identification of primary and secondary metabolites with phosphorus status-dependent abundance in Arabidopsis, and of the transcription factor PHR1 as a major regulator of metabolic changes during phosphorus limitation. Plant Cell Environ 38: 172–187 [DOI] [PubMed] [Google Scholar]
- Patro R, Mount SM, Kingsford C (2014) Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol 32: 462–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11: 1650–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Ropelewski AJ, Nicholas HB, Deerfield DW (2002) Mathematically complete nucleotide and protein sequence searching using search. Curr Protoc Bioinformatics 3: Unit 3.10 [DOI] [PubMed] [Google Scholar]
- Santiago J, Brandt B, Wildhagen M, Hohmann U, Hothorn LA, Butenko MA, Hothorn M (2016) Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardon K, Hohl M, Graff L, Pfannstiel J, Schulze W, Stintzi A, Schaller A (2016) Precursor processing for plant peptide hormone maturation by subtilisin-like serine proteinases. Science 354: 1594–1597 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136: 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Wang C, Arpat BA, Wang Z, Poirier Y, Tyerman SD, Wu P, Shou H, Whelan J (2012) The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol 193: 842–851 [DOI] [PubMed] [Google Scholar]
- Shinohara H, Mori A, Yasue N, Sumida K, Matsubayashi Y (2016) Identification of three LRR-RKs involved in perception of root meristem growth factor in Arabidopsis. Proc Natl Acad Sci USA 113: 3897–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein KAT, Moskal WA Jr, Wu HC, Underwood BA, Graham MA, Town CD, VandenBosch KA (2007) Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J 51: 262–280 [DOI] [PubMed] [Google Scholar]
- Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H, Matsubayashi Y (2014) Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346: 343–346 [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Higashiyama T (2012) A species-specific cluster of defensin-like genes encodes diffusible pollen tube attractants in Arabidopsis. PLoS Biol 10: e1001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Krishnakumar V, Bidwell S, Rosen B, Chan A, Zhou S, Gentzbittel L, Childs KL, Yandell M, Gundlach H, et al. (2014) An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina P, De Coninck B, Nikonorova N, De Smet I, Cammue BPA (2015) The plant peptidome: an expanding repertoire of structural features and biological functions. Plant Cell 27: 2095–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo DI, Silverstein KAT, Young ND (2014) Genomic characterization of the LEED..PEEDs, a gene family unique to the Medicago lineage. G3 4: 2003–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi MK, Czechowski T, Scheible WR (2008) Eleven golden rules of quantitative RT-PCR. Plant Cell 20: 1736–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dongen S. (2008) Graph clustering via a discrete uncoupling process. SIAM J Matrix Anal Appl 30: 121–141 [Google Scholar]
- Vanoosthuyse V, Miege C, Dumas C, Cock JM (2001) Two large Arabidopsis thaliana gene families are homologous to the Brassica gene superfamily that encodes pollen coat proteins and the male component of the self-incompatibility response. Plant Mol Biol 46: 17–34 [DOI] [PubMed] [Google Scholar]
- Vie AK, Najafi J, Liu B, Winge P, Butenko MA, Hornslien KS, Kumpf R, Aalen RB, Bones AM, Brembu T (2015) The IDA/IDA-LIKE and PIP/PIP-LIKE gene families in Arabidopsis: phylogenetic relationship, expression patterns, and transcriptional effect of the PIPL3 peptide. J Exp Bot 66: 5351–5365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yu H, Zhang Z, Yu L, Xu X, Hong Z, Luo L (2015) Phytosulfokine is involved in positive regulation of Lotus japonicus nodulation. Mol Plant Microbe Interact 28: 847–855 [DOI] [PubMed] [Google Scholar]
- Wang Q, Yang S, Liu J, Terecskei K, Ábrahám E, Gombár A, Domonkos Á, Szűcs A, Körmöczi P, Wang T, et al. (2017) Host-secreted antimicrobial peptide enforces symbiotic selectivity in Medicago truncatula. Proc Natl Acad Sci USA 114: 6854–6859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw WHA, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, et al. (2016) Various R Programming Tools for Plotting Data. https://cran.r-project.org/web/packages/gplots/index.html [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipf D, Mongelard G, van Tuinen D, Gutierrez L, Casieri L (2014) Transcriptional responses of Medicago truncatula upon sulfur deficiency stress and arbuscular mycorrhizal symbiosis. Front Plant Sci 5: 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang Q, Fedorova E, Liu J, Qin Q, Zheng Q, Price PA, Pan H, Wang D, Griffitts JS, et al. (2017) Microsymbiont discrimination mediated by a host-secreted peptide in Medicago truncatula. Proc Natl Acad Sci USA 114: 6848–6853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GED, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KFX, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Silverstein KAT, Gao L, Walton JD, Nallu S, Guhlin J, Young ND (2013) Detecting small plant peptides using SPADA (Small Peptide Alignment Discovery Application). BMC Bioinformatics 14: 335. [DOI] [PMC free article] [PubMed] [Google Scholar]