Phosphorylation of MKP1 is required for its function(s) in a subset of PAMP responses and for resistance to bacterial growth.
Abstract
Plants perceive potential pathogens via the recognition of pathogen-associated molecular patterns (PAMPs) by surface-localized pattern recognition receptors, which initiates a series of intracellular responses that ultimately limit bacterial growth. PAMP responses include changes in intracellular protein phosphorylation, including the activation of mitogen-activated protein kinase (MAPK) cascades. MAP kinase phosphatases (MKPs), such as Arabidopsis (Arabidopsis thaliana) MKP1, are important negative regulators of MAPKs and play a crucial role in controlling the intensity and duration of MAPK activation during innate immune signaling. As such, the mkp1 mutant lacking MKP1 displays enhanced PAMP responses and resistance against the virulent bacterium Pseudomonas syringae pv tomato DC3000. Previous in vitro studies showed that MKP1 can be phosphorylated and activated by MPK6, suggesting that phosphorylation may be an important mechanism for regulating MKP1. We found that MKP1 was phosphorylated during PAMP elicitation and that phosphorylation stabilized the protein, resulting in protein accumulation after elicitation. MKP1 also can be stabilized by the proteasome inhibitor MG132, suggesting that MKP1 is constitutively degraded through the proteasome in the resting state. In addition, we investigated the role of MKP1 posttranslational regulation in plant defense by testing whether phenotypes of the mkp1 Arabidopsis mutant could be complemented by expressing phosphorylation site mutations of MKP1. The phosphorylation of MKP1 was found to be required for some, but not all, of MKP1’s functions in PAMP responses and defense against bacteria. Together, our results provide insight into the roles of phosphorylation in the regulation of MKP1 during PAMP signaling and resistance to bacteria.
Plants have developed sophisticated basal immune systems to detect and fend off potential pathogens (Zipfel and Felix, 2005; Bittel and Robatzek, 2007). Basal defense responses are initiated from the recognition of conserved molecules from pathogens called pathogen-associated molecular patterns (PAMPs), such as flg22 (a conserved 22-amino acid flagellin peptide) and elf26 (a conserved 26-amino acid epitope from elongation factor Tu; Felix et al., 1999; Zipfel et al., 2006). This recognition occurs through plasma membrane-localized receptors and triggers diverse defense responses, such as the rapid phosphorylation of intracellular proteins and the transcription of defense-related genes (Boller and Felix, 2009; Zipfel, 2009). In later responses, PAMP treatment leads to the inhibition of seedling growth (Gómez-Gómez et al., 1999; Zipfel et al., 2006), and ultimately, it restricts the growth of bacterial invaders by what is called pattern-triggered immunity (Zipfel et al., 2004).
Mitogen-activated protein kinases (MAPKs) are activated in response to PAMP perception by the dual phosphorylation of Thr (T) and Tyr (Y) residues in the conserved T-X-Y motif of the MAPK activation loop by MAPK kinases, which are themselves phosphorylated and activated by upstream MAPK kinase kinases (Colcombet and Hirt, 2008). Active MAPKs are able to phosphorylate downstream substrates to generate appropriate physiological responses. Two Arabidopsis (Arabidopsis thaliana) MAPKs, MPK3 and MPK6, have been found to be activated by myriad stimuli, including PAMP elicitation (Nühse et al., 2000; Asai et al., 2002). MPK3 and MPK6 are partially redundant in their activities and function as positive regulators of defense responses (Pitzschke et al., 2009; Rodriguez et al., 2010; Tena et al., 2011; Meng and Zhang, 2013).
The magnitude and duration of MAPK activity are crucial determinants of correct biological outcomes. Therefore, once initiated, MAPK signaling needs to be deactivated properly to prevent the overstimulation of defense responses. In Arabidopsis, several phosphatases have been implicated in regulating PAMP responses and pathogen resistance through the dephosphorylation, and thus inactivation, of MAPKs. Two protein phosphatase type 2Cs (PP2Cs), AP2C1 and PP2C5, are phospho-Ser/Thr phosphatases that target a specific phospho-Thr in the MAPK activation loop (Meskiene et al., 2003; Fuchs et al., 2013). A mutant lacking both AP2C1 and PP2C5 displayed increased activation of MPK3 and MPK6 following abscisic acid treatment (Brock et al., 2010). Loss of AP2C1 also resulted in increased stimulus-induced callose deposition and enhanced resistance to bacterial infection (Shubchynskyy et al., 2017).
MAPK phosphatases (MKPs) are a different family of protein phosphatases. These dual-specificity phosphatases dephosphorylate both conserved Thr and Tyr residues of the MAPK activation loop, thereby fully inactivating the MAPKs (Camps et al., 2000; Luan, 2003; Bartels et al., 2010). MAPK PHOSPHATASE2 (MKP2) has been shown to interact functionally with MPK3 and MPK6, to positively regulate oxidative stress responses, and to exert differential functions in specific pathogen interactions (Lumbreras et al., 2010). MKP1 is an important negative regulator of plant immunity. Defense responses were hyperinduced in the mkp1 null mutant following PAMP treatment, including the activation of MPK6 and MPK3, accumulation of a subset of PAMP-regulated transcripts, and inhibition of seedling growth (Anderson et al., 2011). Consistent with enhanced PAMP responses, the mkp1 mutant also displayed enhanced resistance to the virulent pathogen Pseudomonas syringae pv tomato DC3000 (hereafter referred to as DC3000). The enhanced PAMP-induced growth inhibition and resistance in mkp1 are specifically dependent on MPK6, indicating a genetic interaction between MKP1 and MPK6 (Bartels et al., 2009; Anderson et al., 2011). Because of the importance of these negative regulators in modulating defense responses, it is also critical to understand how phosphatases such as MKP1 are regulated.
Studies in nonplant organisms have shown that the abundance and activity of phosphatases often are regulated by the activated MAPK pathways that they control, establishing an efficient negative feedback loop to attenuate responses (Millar et al., 1995; Brondello et al., 1999; Sohaskey and Ferrell, 2002; Li et al., 2007). However, the regulation of MKPs during PAMP responses in plants is not well understood. Both in vitro and in vivo studies have shown that Arabidopsis MKP1 interacts with three MAPKs (MPK3, MPK4, and MPK6), with the strongest interaction being with MPK6 (Ulm et al., 2002; Bartels et al., 2009). In addition, Arabidopsis MKP1 was shown to be phosphorylated in vitro by MPK6, and phosphorylation increased the phosphatase activity of MKP1, indicating that phosphorylation may be an important regulatory mechanism of MKP1 (Park et al., 2011). These results provide support for the idea that MKP1 may be regulated at least in part by a negative feedback loop during PAMP responses and bacterial resistance. However, in vivo studies have not been performed to investigate this possibility.
In this work, we investigated the role of MKP1 phosphorylation in response to PAMP elicitation. Here, we report that MKP1 is indeed phosphorylated in response to PAMP elicitation in vivo. We also show that MKP1 rapidly accumulates after PAMP treatment and that proper accumulation is dependent on protein phosphorylation. Importantly, the phosphorylation of MKP1 is required for only a subset of MKP1-dependent PAMP and resistance responses, indicating that phosphorylation is an important component for some, but not all, MKP1-mediated responses.
RESULTS
MKP1 Is Phosphorylated in Response to Bacterial PAMP elf26 in Vivo
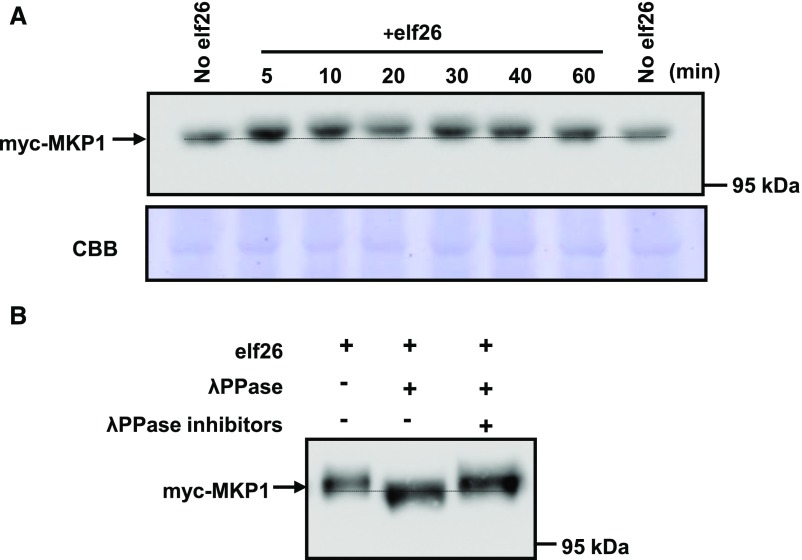
To determine if MKP1 is regulated in response to PAMP elicitation at the posttranslational level, we used Arabidopsis seedlings expressing myc-tagged MKP1 (myc-MKP1) driven by the constitutive cauliflower mosaic virus (CaMV) 35S promoter (González Besteiro and Ulm, 2013). Two-week-old seedlings were treated with elf26 and harvested at different times post elicitation. To achieve sufficient resolution, protein extracts were separated using large-format SDS-PAGE (16 × 16 cm) overnight. Immunoblot analysis showed that elf26 treatment resulted in a reduced-mobility form of the myc-MKP1 protein (Fig. 1A). The mobility shift occurred by 5 to 10 min after elicitation and gradually returned back to resting status within 60 min (Fig. 1A). Transient electrophoretic mobility shifts often are indicative of protein phosphorylation, so protein extracts from elf26-elicitated seedlings were treated with λ-phosphatase with or without inhibitors. Treatment of extracts with phosphatase without inhibitors eliminated the elf26-dependent high-Mr form of myc-MKP1 (Fig. 1B), while adding inhibitors during the phosphatase treatment maintained the band shift (Fig. 1B). These results indicate that MKP1 is phosphorylated during elf26 elicitation.
Figure 1.
MKP1 is phosphorylated in response to elf26 in vivo. A, Fourteen-day-old seedlings expressing a myc-tagged MKP1 expressed from the CaMV 35S promoter in mkp1 (Col-0) were treated with 1 µm elf26 for the times indicated. Protein extracts from treated seedlings were separated by 8% large-format (16 × 16 cm) SDS-PAGE overnight and immunoblotted with anti-myc antibody to detect myc-MKP1 (top). The membrane was stained with Coomassie Blue (CBB) as a loading control (bottom). The stippled line aligns the faster migrating forms in the untreated control lanes on each side of the loading. B, Immunoblot analysis with anti-myc antibody of protein extracts from 14-d-old transgenic seedlings treated for 20 min with 1 µm elf26. Extracts were treated with (+) or without (−) λ-phosphatase with or without the phosphatase inhibitors. Protein extracts were separated by 8% large-format (16 × 16 cm) SDS-PAGE overnight. Experiments were performed three times with similar results to those shown.
MKP1 Is Phosphorylated by elf26-Activated MPK6 on Thr-109
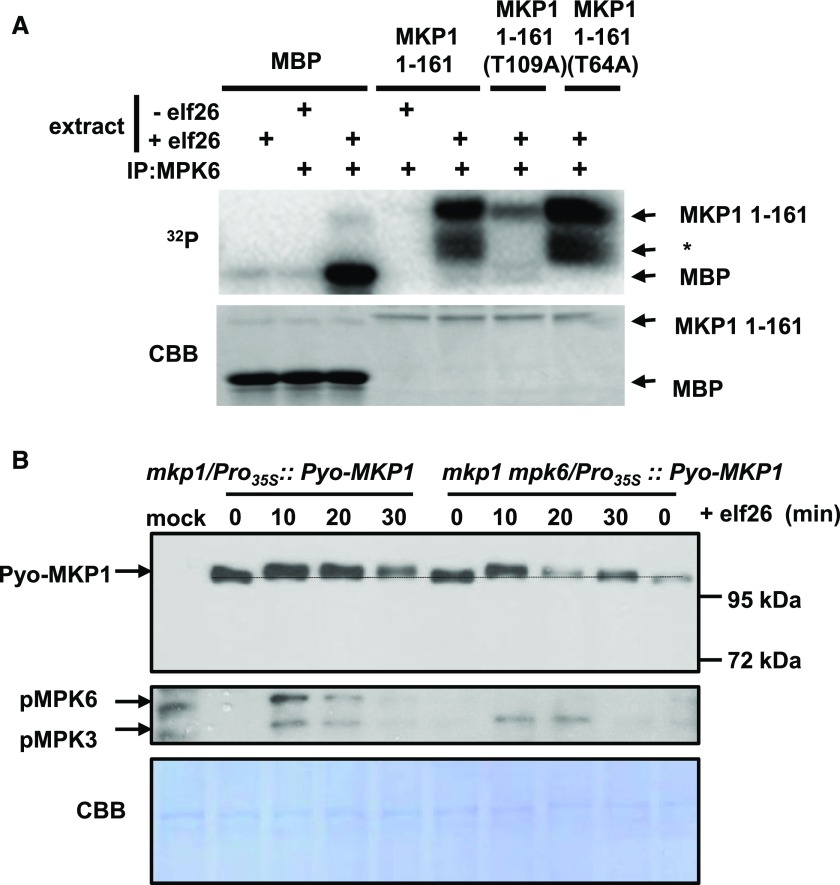
The phosphorylation-induced band shift of MKP1 occurs within the same time frame of elf26-induced activation of MPK6 (Anderson et al., 2011). Because defense-related phenotypes in the mkp1 mutant were shown to specifically require the presence of MPK6 (Anderson et al., 2011, 2014), we investigated whether MKP1 is phosphorylated by MPK6. MPK6 was immunoprecipitated from Arabidopsis cells before and after elicitation with elf26 and incubated with the N-terminal portion of the MKP1 protein (amino acids 1–161) containing two putative MAPK phosphorylation sites (S/T-P), Thr-64 and Thr-109. Myelin basic protein (MBP) also was included as a positive control. Both MBP and MKP1 were phosphorylated only in the presence of elf26-activated MPK6 (Fig. 2A), demonstrating that MKP1 is indeed a substrate of MPK6. To determine which residues may be targeted by MPK6, we mutated the putative MAPK phosphorylation sites to unphosphorylatable Ala. In the kinase assays, there was a strong preference of MPK6 for Thr-109, as mutating this residue (T109A) nearly abolished radioactive incorporation, whereas the other mutation (T64A) had little or no effect (Fig. 2A). These results were only in partial agreement with a previous study indicating that both Thr-64 and Thr-109 were in vitro MPK6 target residues (Park et al., 2011). However, these previous studies utilized autoactivated MPK6 from bacterial expression, which may display different specificity from the native protein immunoprecipitated from plants as used in our experiments here. However, both experiments indicate that MKP1 can be phosphorylated by MPK6 using in vitro kinase assays.
Figure 2.
MKP1 is phosphorylated by elf26-activated MPK6 on Thr-109 in vitro, but loss of MPK6 does not completely prevent elf26-induced phosphorylation in vivo. A, MPK6 was immunoprecipitated from Arabidopsis cells before and after elicitation with 100 nm elf26. The top gel is an autoradiograph of 32P incorporated in MBP, recombinant 6×His-tagged MKP1 1-161, MKP1 1-161 (T109A), or MKP1 1-161 (T64A). The bottom gel is a duplicate gel stained with Coomassie Blue (CBB) to verify equal loading. The asterisk indicates likely partially degraded MKP1. B, Pyo-MKP1 was transiently expressed from the CaMV 35S promoter in Arabidopsis protoplasts isolated from 5-week-old mkp1 (Ws) and mkp1 mpk6 (Ws) adult plants. The protoplasts were treated with 1 µm elf26 for the indicated times. Protein extracts were separated with 8% mini-format (8.3 × 7.3 cm) SDS-PAGE and immunoblotted with anti-Glu-Glu antibody to detect the Pyo-MKP1 protein (top) or the anti-phospho-p42/44 MAPKs to detect activated MAPKs (middle). The membrane was stained with Coomassie Blue as a loading control (bottom). Mock control samples were from protoplasts isolated from mkp1 (Ws) without transfection with Pyo-MKP1 plasmid. Experiments were performed at least three times with similar results to those shown.
MPK6 Is Not the Only Kinase Phosphorylating MKP1 during PAMP Responses
To examine whether MPK6 is the only kinase that phosphorylates MKP1, we transiently expressed Polyoma epitope-tagged MKP1 (Pyo-MKP1) in Arabidopsis protoplasts isolated from 5-week-old mkp1 (Wassilewskija [Ws]) and mkp1 mpk6 (Ws) adult plants. Treatment of protoplasts with elf26 consistently resulted in a gel mobility shift of Pyo-MKP1 in the mkp1 background (Fig. 2B). In the mkp1 mpk6 double mutant, where MPK6 is absent, Pyo-MKP1 was still phosphorylated early after elicitation (10 min), as indicated by the mobility shift. However, the duration of MKP1 phosphorylation appeared to be shorter in the absence of MPK6 based on the fact that the mobility shift returned to almost basal levels in the mkp1 mpk6 double mutant by 30 min, whereas the shift remained in the mkp1 single mutant (Fig. 2B). These results indicate that MPK6 contributes to the phosphorylation of MKP1 but that MPK6 is not the only kinase that phosphorylates MKP1.
Phosphorylation of MKP1 Stabilizes the Protein
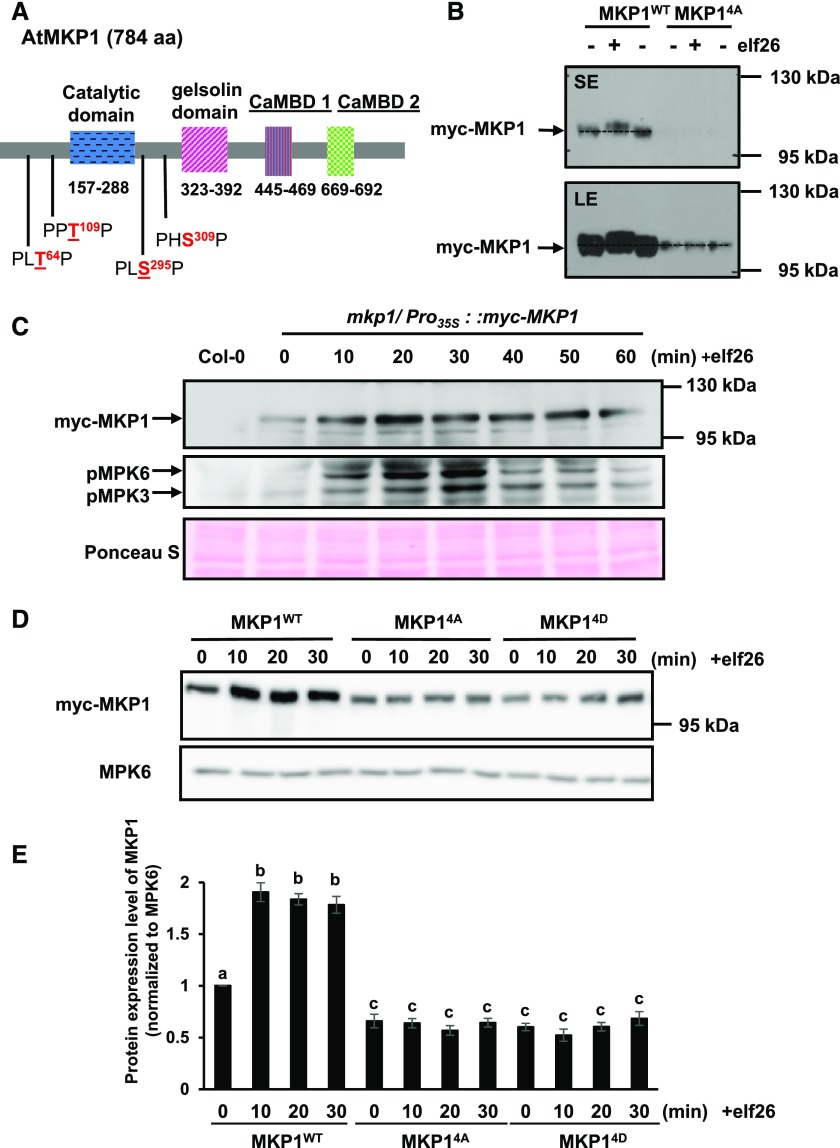
Our results indicated that MKP1 may be phosphorylated by kinases other than MPK6, and MKP1 contains four putative MAPK phosphorylation sites (Thr-64, Thr-109, Ser-295, and Ser-309) that are conserved across different plant species (González Besteiro and Ulm, 2013). Three of the sites (Thr-64, Thr-109, and Ser-295) have been found to be phosphorylated (Jones et al., 2009; Li et al., 2009; Park et al., 2011), and Ser-309 also is a potential MAPK phosphorylation site that is evolutionarily conserved. Therefore, to investigate if MAPK phosphorylation is required for any function(s) of MKP1, all four sites (Fig. 3A) were mutated to Ala (MKP14A) to prevent the phosphorylation or to Asp (MKP14D) to putatively mimic phosphorylation in order to investigate the potential role of the MAPK-dependent phosphorylation of MKP1. These mutant versions as well as the wild-type version of MKP1 were stably expressed with myc epitope tags in mkp1 (Columbia-0 [Col-0]) plants to investigate if they would complement phenotypes in the mutant.
Figure 3.
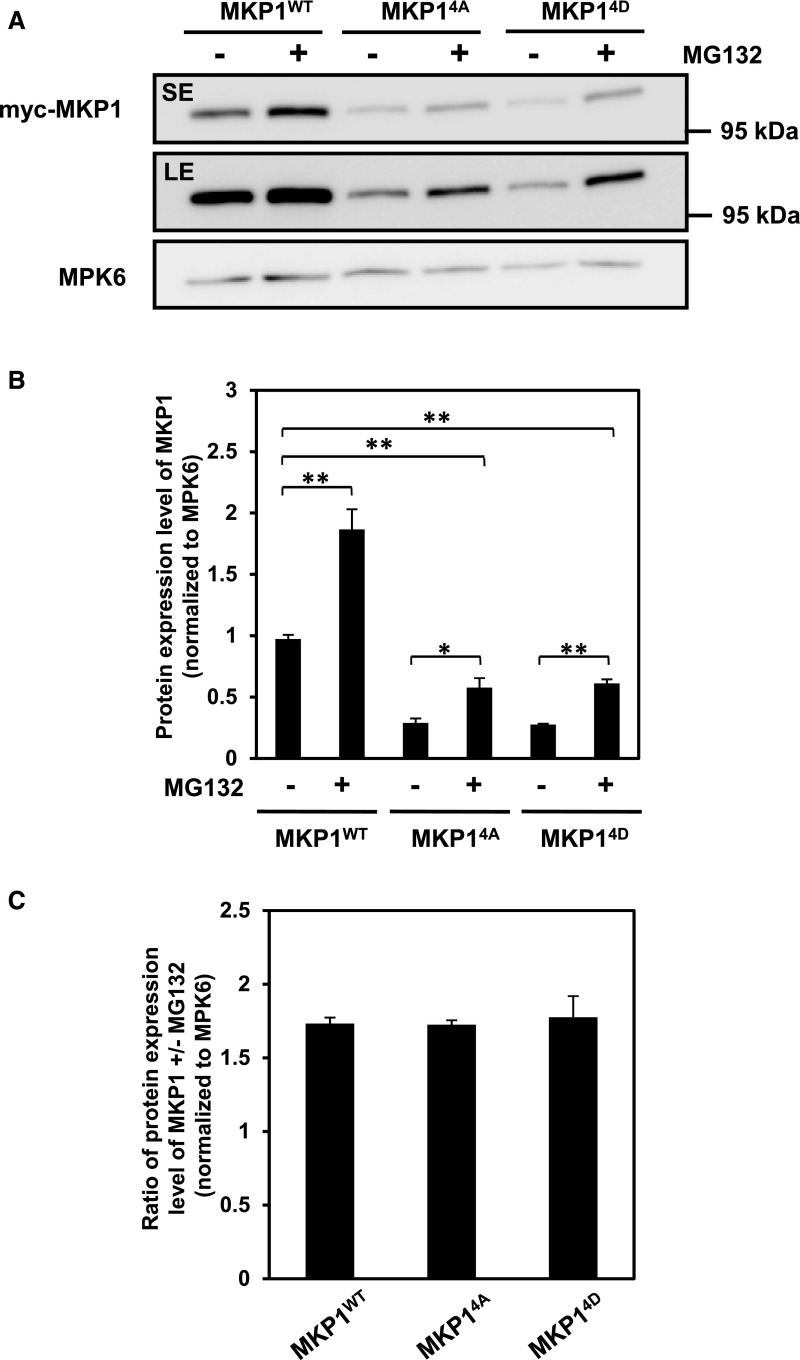
Phosphorylation of MKP1 stabilizes the protein. A, Domain structure of the Arabidopsis MKP1 protein, showing the positions of the conserved putative MAPK phosphorylation sites. Underlined residues have been experimentally shown to be phosphorylated (see refs. in the text). B, Immunoblot analysis with anti-myc antibody of protein extracts from 14-d-old myc-tagged MKP1 (MKP1WT) or phosphorylation site mutant (MKP14A) transgenic seedlings treated with or without 1 µm elf26 for 20 min. Protein extracts were separated with 8% large-format (16 × 16 cm) SDS-PAGE. LE, Long exposure; SE, short exposure. C, Fourteen-day-old myc-tagged MKP1 transgenic seedlings were treated with 1 μm elf26 for the times indicated. Protein extracts from treated seedlings were separated by 8% mini-format (8.3 × 7.3 cm) SDS-PAGE and immunoblotted with anti-myc antibody to detect myc-MKP1 (top) or anti-phospho-p42/44 MAPK antibody to detect phosphorylated MAPKs (middle). Ponceau S staining of the membrane was used as a loading control (bottom). D, Fourteen-day-old transgenic seedlings expressing MKP1 wild-type protein (MKP1WT) or phosphorylation site substitutions (MKP14A and MKP14D) were treated with or without 1 µm elf26 for the times indicated. Protein extracts from treated seedlings were separated by 8% mini-format (8.3 × 7.3 cm) SDS-PAGE and immunoblotted with anti-myc antibody to detect the myc-MKP1 protein (top) or anti-MPK6 antibody as a loading control (bottom). This experiment was performed three times with similar results to those shown. E, Quantification of the western-blot band intensity of MKP1 protein normalized to the intensity of MKP6 used as a loading control. Graphed are means ± se, representative of three independent biological replicates (n = 3). Lowercase letters indicate significant groupings (P < 0.05). The statistical test was performed using ANOVA with Tukey’s pairwise comparison.
First, we examined the transcript and protein accumulation in these transgenic lines. MKP1 transcript levels were higher in all transgenic lines than the native transcript levels in Col-0 (wild-type) plants (Supplemental Fig. S1), and transcript levels of the phosphorylation site mutants MKP14A and MKP14D were comparable to myc-tagged wild-type MKP1 (MKP1WT), all in mkp1. At the protein level, however, the myc-tagged MKP1WT consistently accumulated more than either of the phosphorylation site mutants (MKP14A or MKP14D), indicating that normal phosphorylation may be required for proper protein accumulation (Supplemental Fig. S1). For subsequent molecular and biological tests, results from the two phosphorylation mutant lines with comparatively higher protein levels (MKP14A 10-2 and MKP14D 12-3) were used for experiments shown in the main text (the other independent lines displayed similar phenotypes, which are shown in the supplemental figures).
To determine if these four phosphorylation sites could account for the elf26-induced phosphorylation band shift of MKP1, 2-week-old transgenic seedlings expressing the wild-type MKP1 protein (MKP1WT) and the phosphorylation site mutant protein (MKP14A) were treated with elf26 for 20 min. Protein extracts of treated seedlings were separated using the large-format SDS-PAGE to detect the phosphorylation-induced mobility shift followed by immunoblot analysis with anti-myc antibody. Although the elf26-induced mobility shift could be observed for MKP1WT, the mobility shift was completely abolished in the phosphorylation site mutant (MKP14A; Fig. 3B). This result indicates that mutation of these four sites is sufficient to eliminate the elf26-induced phosphorylation of MKP1, at least as detected by the mobility shift. In addition, mutation of these four sites also reduced the protein accumulation of MKP1 (Fig. 3B; Supplemental Fig. S1), indicating the elf26-induced phosphorylation may affect the protein stability of MKP1. Indeed, after elf26 elicitation, MKP1 began to accumulate within 10 min and remained at elevated levels until approximately 60 min after treatment (Fig. 3C), which is consistent with the timing of elf26-induced phosphorylation of MKP1 (Fig. 1A) and the activation of MPK3 and MPK6 (Fig. 3C). Comparing the protein accumulation of MKP1WT and phosphorylation site mutants MKP14A and MKP14D treated with elf26, we found that neither MKP14A nor MKP14D showed any significant increase (Fig. 3, D and E), indicating that phosphorylation is essential for elf26-induced protein accumulation. The failure of the putative phosphomimetic mutation to accumulate indicates that, in this case, substitution of a negative charge alone is not sufficient to replace the effects of the phosphorylated residue. Together, these results suggest that, in response to elf26 elicitation, MKP1 becomes phosphorylated and the phosphorylation of MKP1 stabilizes the protein, resulting in a rapid increase in protein levels.
MKP1 Is Degraded through Proteasome-Mediated Pathways in Unelicited Plants
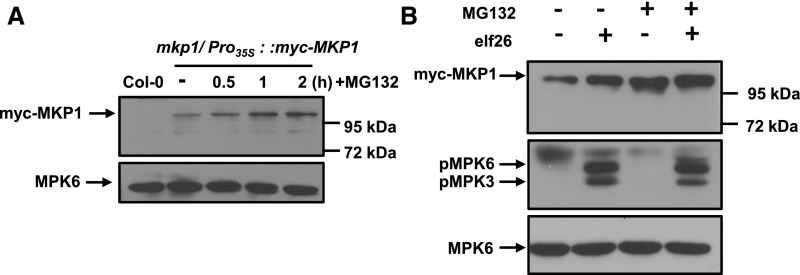
A possible explanation for the rapid protein accumulation of MKP1 after PAMP elicitation would be if phosphorylation affects the proteolysis of MKP1 that is constitutively occurring in naïve plants. To examine whether MKP1 may be degraded through the proteasome, transgenic myc-MKP1WT seedlings were treated with the proteasome inhibitor MG132. In comparison with the control (− in Fig. 4A), MG132-treated plants began to rapidly accumulate MKP1 protein within 30 min post treatment, and the accumulation of MKP1 continued for at least 2 h (Fig. 4A). This accumulation was not due to the myc tag, as MKP1 with an independent sequence tag (Pyo) showed the same MG132-induced accumulation (Supplemental Fig. S2). In contrast to MKP1, protein levels of MPK6 were not affected by the proteasome inhibitor (Fig. 4A), demonstrating that it was not a nonspecific effect of the treatment. These results are consistent with the previously proposed hypothesis that MKP1 is constitutively turned over through the proteasome-mediated protein degradation pathway in unelicited cells (González Besteiro and Ulm, 2013). In addition, MG132 treatment did not activate MPK3 or MPK6 (Fig. 4B), suggesting that the stabilizing effect of MG132 is not through the phosphorylation of MKP1 by active MAPKs. Therefore, it is likely that MAPK-induced phosphorylation precedes the suppression of proteasomal degradation.
Figure 4.
A portion of MKP1 is constitutively degraded by the proteasome in naïve plants. A, Fourteen-day-old transgenic seedlings expressing myc-MKP1 were treated with or without 40 µm MG132 for the times indicated. Protein extracts from treated seedlings were separated by 8% mini-format (8.3 × 7.3 cm) SDS-PAGE and immunoblotted with anti-myc antibody to detect the myc-MKP1 protein (top) or anti-MPK6 antibody used as a loading control (bottom). B, Fourteen-day-old transgenic seedlings were pretreated with or without 40 µm MG132 for 40 min and then treated with 1 µm elf26 for 20 min. Protein extracts from treated seedlings were separated by 8% mini-format (8.3 × 7.3 cm) SDS-PAGE and immunoblotted with anti-myc antibody to detect the myc-MKP1 protein (top), anti-phospho-p42/44 antibody to detect phosphorylated MAPKs (middle), or anti-MPK6 antibody as a loading control (bottom). This experiment was performed three times with similar results to those shown.
The Proteasome May Not Be the Only Pathway Affecting MKP1 Stability
To examine if MG132 treatment could restore protein levels in the MKP14A and MKP14D phosphorylation site mutants, seedlings were treated with MG132 for 1 h, which resulted in approximately maximal accumulation of MKP1 (Fig. 4A). As expected, MKP1 showed an increase in protein abundance as a result of the MG132 treatment for both the wild-type protein and the phosphorylation site mutants (Fig. 5A; Supplemental Fig. S3). However, inhibiting the proteasome alone could not fully restore protein levels in the phosphorylation site mutants even back to the basal levels found for MKP1WT (Fig. 5, A and B). In addition, the extent of the MG132-induced accumulation (i.e. the ratio between with and without MG132 treatment) in the phosphorylation site mutants was not significantly different from that in MKP1WT (Fig. 5C). This result indicates that phosphorylation is required for full accumulation and that this may involve a proteasome-independent mechanism.
Figure 5.
Phosphorylation of MKP1 is not required for MG132 stabilization. A, Fourteen-day-old transgenic seedlings were treated with or without 40 µm MG132 for 1 h. Protein extracts from treated seedlings were separated with 8% mini-format (8.3 × 7.3 cm) SDS-PAGE and immunoblotting with anti-myc antibody to detect the myc-MKP1 protein (top) and anti-MPK6 antibody as a loading control (bottom). LE, Long exposure; SE, short exposure. B and C, Quantification of the western-blot band intensity of MKP1 protein, which was normalized to that of MKP6 as a loading control. Graphed are means ± se, pooling from three independent biological replicates (n = 3). The asterisks indicate significant differences between pairwise groups (marked by parentheses: *, P < 0.05 and **, P < 0.01). The statistical test was performed using ANOVA with multiple pairwise comparisons under the protection of an overall F test.
Phosphorylation Site Mutants Complement the Growth Inhibition of Adult Plants and Some of the PAMP-Responsive Transcripts That Accumulate to Higher Levels in mkp1 (Col-0)
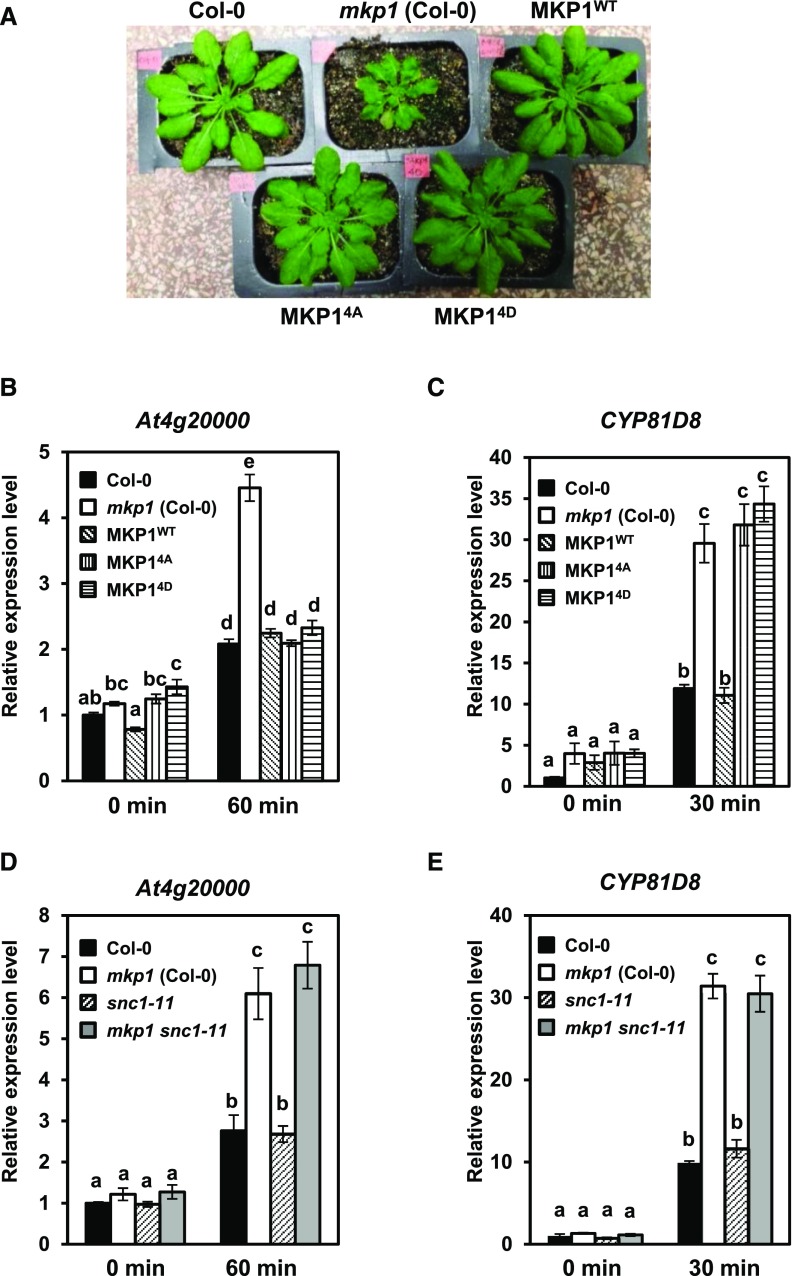
To examine whether the phosphorylation of MKP1 is required for its biological functions, we performed complementation tests using the phosphorylation site mutants on a series of mkp1-dependent phenotypes. When grown on soil, mkp1 (Col-0) displays a dwarf phenotype (Bartels et al., 2009). In agreement with previous observations (González Besteiro and Ulm, 2013), under our growth conditions, the expression of either MKP14A or MKP14D could complement the mkp1 (Col-0) dwarf phenotype of adult plants (Fig. 6A). This result indicates that, although the phosphorylation site mutants of MKP1 have lower levels of protein than MKP1WT, the proteins accumulate to sufficient levels to functionally suppress the growth defect in mkp1 (Col-0) to the same degree as MKP1WT. In addition, both phosphorylation site mutants also can suppress the hyperaccumulation of the At4g20000 transcript in mkp1 (Col-0) seedlings in response to elf26 elicitation (Fig. 6B). However, only MKP1WT, but not the phosphorylation site mutants, can suppress the increased accumulation of the PAMP-responsive transcript CYP81D8 in mkp1 (Col-0) (Fig. 6C). It was shown previously that the growth defect in mkp1 (Col-0) is dependent on the presence of the resistance gene homolog SUPPRESSOR OF npr1-1, CONSTITUTIVE1 (SNC1; Bartels et al., 2009). Because the phosphorylation site mutants could suppress both growth defects and the hyperaccumulation of the PAMP-responsive transcript At4g20000 in mkp1 (Col-0), it raised the possibility that the hyperaccumulation of At4g20000 also may be an mkp1-dependent phenotype that required SNC1. To test this hypothesis, we measured the transcript levels of At4g20000 and CYP81D8 in Col-0, mkp1 (Col-0), snc1-11 (Col-0), and the mkp1 snc1-11 (Col-0) double mutant after elf26 elicitation. We observed that both At4g20000 and CYP81D8 transcripts accumulated to similarly higher levels in mkp1 snc1-11 (Col-0) as in mkp1 (Col-0) (Fig. 6, D and E). These results demonstrate that the hyperaccumulation of At4g20000 transcripts is independent of SNC1. These results demonstrate that the phosphorylation site mutant proteins accumulate and can functionally complement some mkp1-dependent phenotypes with two different genetic requirements (SNC1 dependent and SNC1 independent) but that phosphorylation of the protein is required for other functions of MKP1 (e.g. transcript accumulation profile of CYP81D8).
Figure 6.
MKP1 protein lacking phosphorylation sites complements some of the mkp1(Col-0) phenotypes. A, Constitutive defense-related phenotypes of adult mkp1(Col-0) plants compared with wild-type (Col-0) and transgenic mkp1 (Col-0) plants expressing myc-tagged wild-type MKP1 or phosphorylation site mutants MKP14A and MKP14D. The photograph shows 5-week-old adult plants grown on soil. B to E, mRNA levels of PAMP-responsive transcripts of At4g2000 (B and D) or CYP81D8 (C and E) measured by quantitative real-time PCR from 12-d-old seedlings treated with and without 1 μm elf26 for the indicated times. Experiments were performed to test for the requirements of MKP1 phosphorylation (B and C) or for the presence of SNC1 (D and E) in altering the accumulation pattern in mkp1 (Col-0). Transcript levels were normalized to the amount of At2g28390 transcript detected in each sample, then to the transcript level at time 0 in Col-0. Data were pooled from three independent biological experiments with an additional technical replicate for each sample (n = 6). Lowercase letters indicate significant groupings (P < 0.01). The statistical test was performed using ANOVA with Tukey’s pairwise comparison.
Phosphorylation of MKP1 Is Required for Elicitor-Induced Seedling Growth Inhibition Phenotypes in mkp1 (Col-0)
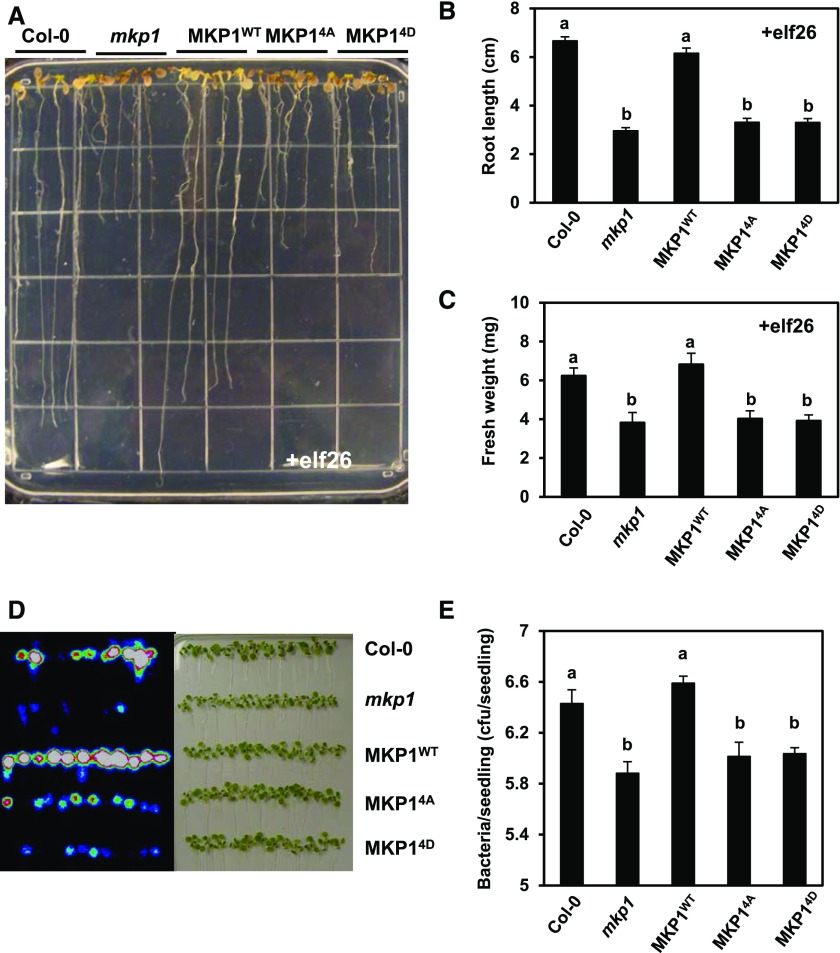
We reported previously that the mkp1 seedlings displayed enhanced growth inhibition in response to elf26 (Anderson et al., 2011). To examine whether the phosphorylation sites are required for MKP1 in regulating the elf26-mediated inhibition of seedling growth, 6-d-old seedlings of Col-0 (wild type), mkp1 (Col-0), or mkp1 (Col-0), expressing MKP1WT, MKP14A, or MKP14D, were transferred from agar plates to liquid growth medium supplemented with or without 1 µm elf26. After a 2-week incubation, we observed that the phosphorylation site mutants MKP14A and MKP14D were not able to rescue the decreased primary root length (Fig. 7, A and B; Supplemental Fig. S4A) or seedling fresh weight (Fig. 7C; Supplemental Fig. S4B) observed in the elf26-treated mkp1 (Col-0) seedlings. No significant differences in the root length or fresh weight of untreated seedlings were observed in any genotypes (Supplemental Fig. S5), indicating that all growth differences were PAMP dependent. These results indicate that phosphorylation is required for MKP1 function in regulating the seedling growth inhibition triggered by elf26.
Figure 7.
Phosphorylation of MKP1 is required for regulating elf26-induced growth inhibition and resistance to bacteria. A, Six-old-day seedlings of Col-0, mkp1 (Col-0), MKP1WT, MKP14A, and MKP14D were aseptically transferred from MS agar to the wells of a 24-well microtiter plate containing 1 mL of liquid MS medium with 1 µm elf26 for 14 d. After 14 d, seedlings were placed on an agar surface to photograph the plants, and the image is representative of at least three independent experiments. B and C, Primary root length and fresh weight were measured for the experiment described in A. Graphed are means ± se (n = 24), pooled from three independent experiments. Lowercase letters indicate significant groupings (P < 0.05). The statistical test was performed using ANOVA with Tukey’s pairwise comparison. D, Fourteen-day-old seedlings of Col-0, mkp1 (Col-0), MKP1WT, MKP14A, and MKP14D were immersed in 1 × 107 cfu mL−1 DC3000 LuxCDABE. Three days post infection, seedlings were removed, rinsed with water, and placed on an agar surface. A heat map image of bacterial luminescence in DC3000-infected seedlings detected using a photon-detection camera (left) and a bright-field image of the same seedlings (right) are shown. Results are representative of three independent experiments. E, Bacterial levels in DC3000-infected seedlings 3 d post infection were measured by serial dilution plating of seedling extracts. Graphed are means ± se (n = 6). Lowercase letters indicate significant groupings (P < 0.05). The statistical test was performed using ANOVA with Tukey’s pairwise comparison. Experiments were performed three times with similar results to those shown.
Phosphorylation of MKP1 Is Required for Enhanced Resistance to DC3000 in mkp1 (Col-0)
The mkp1 mutant is more resistant to the virulent bacterial pathogen DC3000 (Anderson et al., 2011, 2014). To investigate whether the phosphorylation of MKP1 is required to complement the resistance phenotype, we assessed the growth of DC3000 in the seedlings. To facilitate the detection of DC3000 growth in seedlings, we used an autoluminescent strain of DC3000 expressing the LuxCDABE operon (Fan et al., 2008). Plants were immersed in a bacterial solution of 1 × 107 colony-forming units (cfu) mL−1 DC3000 LuxCDABE, and 3 d post infection, seedlings were removed, rinsed with water, and placed on an agar surface for imaging. Consistent with previous results (Anderson et al., 2011, 2014), the DC3000-infected mkp1 (Col-0) seedlings displayed significantly decreased bacterial growth, as determined by luminescence (Fig. 7D) and by serial dilution plating (Fig. 7E; Supplemental Fig. S6). Similar to the result from the growth inhibition assay, the phosphorylation site mutants failed to complement the enhanced resistance in mkp1 (Col-0), displaying a decrease in bacterial growth compared with the wild type that was indistinguishable from that in mkp1 (Col-0) (Fig. 7, D and E). From these results, we conclude that phosphorylation is required for MKP1 function in regulating resistance to bacterial infection.
DISCUSSION
Uncontrolled activation of MAPKs can have detrimental effects on the cell. For instance, constitutive activation of MPK3 and MPK6 by expressing the activated form of the upstream MAPKK, AtMKK4, results in cell death (Ren et al., 2002; Lassowskat et al., 2014). Therefore, MAPK activity must be controlled tightly to ensure the proper coordination of physiological events. MKP1 is a dual-specificity phosphatase that appears to dephosphorylate and inactivate MAPKs after PAMP elicitation, thus resetting MAPK activity to basal levels after the initiation of defense responses (Bartels et al., 2009; Anderson et al., 2011). Arabidopsis MKP1 interacts with the MAPKs MPK3, MPK4, and MPK6 in yeast two-hybrid assays, in vitro pull-down experiments, and transient bimolecular fluorescence complementation in planta, and it deactivates MPK6 in protoplasts (Ulm et al., 2002; Bartels et al., 2009). In addition, genetic knockouts have demonstrated that MKP1 is an important negative regulator of MPK6-mediated PAMP responses and bacterial resistance (Anderson et al., 2011). However, little is known regarding the mechanisms by which cells regulate the functions of MKP1 during immune responses. Here, we reveal the importance of phosphorylation in regulating some, but not all, PAMP responses regulated by MKP1.
Phosphorylation of MKP1 Is Required for a Subset of Biological Functions
Ala substitutions of putative MAPK phosphorylation sites (Thr-64, Thr-109, Ser-295, and Ser-309) in MKP1 prevented the complementation of most mkp1-dependent phenotypes, including the enhanced elf26-induced seedling growth inhibition (Fig. 7, A–C; Supplemental Fig. S4, A and B), the elevated resistance to the virulent bacterial pathogen DC3000 (Fig. 7, D and E; Supplemental Fig. S6), and some of the hyperaccumulation of PAMP-induced transcripts such as CYP81D8 (Fig. 6C). These results demonstrate the functional importance of phosphorylating these residues. One possibility with mutations, of course, is that the amino acid changes may affect the structure and/or accumulation of the protein, rendering it a nonfunctional protein. However, the phosphorylation site mutants were able to suppress the dwarf phenotype in adult plants of mkp1 (Col-0) (Fig. 6A). The dwarf phenotype in mkp1 (Col-0) is dependent on SNC1, a Col-specific TIR-NB-LRR protein (Bartels et al., 2009). This result can be explained by the guard hypothesis (Jones and Dangl, 2006), whereby MKP1 (or its activity) is monitored by the resistance (R) gene-like SNC1 (or another R gene that indirectly activates SNC1) such that the absence of MKP1 in the knockout mutant results in the activation of SNC1 signaling pathways leading to constitutive defense responses. The fact that the phosphorylation site mutants can complement the dwarf phenotype indicates that the mutant form of MKP1 both can accumulate to sufficient levels and adopt the proper structure to be recognized by SNC1, indicating that mutations do not grossly affect the protein. The phosphorylation site mutants also suppressed a molecular PAMP response in the form of the mkp1-dependent hyperaccumulation of At4g20000-encoded transcripts (Fig. 6B). This aberrant accumulation in mkp1 was not suppressed by the loss of SNC1 (Fig. 6D), indicating that this response is not a secondary effect of SNC1 signaling in the absence of MKP1. These results indicate that the function of MKP1 in regulating some of the PAMP responses does not require phosphorylation. However, we cannot rule out the possibility that different responses may have different sensitivities in terms of the requirement for MKP1 protein levels, as the phosphorylation site mutants do not accumulate to the same levels as the wild-type protein (see below).
Both nonphosphorylatable (MKP14A) and putative phosphomimetic (MKP14D) mutations were equally unable to complement the same subset of PAMP and/or defense responses. The most likely explanation is that MKP14D does not actually mimic phosphorylation in this case. Although often used, substitutions with Asp or Glu do not always act like a phosphorylated residue (Peck, 2006), especially if the physical presence of the phosphate group is involved in regulating a conformational change in the protein or specific protein-protein interactions (Corbit et al., 2003; Paleologou et al., 2008). We should also note that, although three of the four sites have been found to be phosphorylated in planta on MKP1 and the other is evolutionarily conserved, it is not known if combinatorial complexity may exist between positive and negative actions of the different sites. Therefore, it is possible that, for these responses, the phosphomimetic mutations of some residues may act positively while others act negatively, thus canceling out positive effects in the MKP14D mutants. Our analyses of MKP14A and MKP14D have established the importance of phosphorylation among the four targeted residues, but a more detailed dissection of each single residue (or combinations) will be required to test this more complex possibility.
Stabilization of MKP1 Follows the PAMP-Induced Phosphorylation
At least one of the actions of phosphorylation involves the apparent stabilization of MKP1. The amount of MKP1 protein increased significantly rapidly after PAMP elicitation and coincided with the timing of MKP1 phosphorylation (Fig. 1A). Moreover, mutation of the four phosphorylation sites that eliminated the PAMP-induced phosphorylation of MKP1 based on mobility shift assays (Fig. 3B) also eliminated the PAMP-induced accumulation of MKP1 (Fig. 3, D and E). These results indicate that one of the functions of phosphorylation is to increase the levels of MKP1. A likely scenario for this result is that a portion of MKP1 may be constantly degraded via the ubiquitin-proteasome pathway, a hypothesis that is supported by the fact that MKP1 can be stabilized by addition of the proteasome inhibitor, MG132 (Fig. 4, A and B). However, the proteasome-mediated protein degradation pathway may not be the only mechanism affecting MKP1 stability. Although the transcript level of MKP1 phosphorylation site mutants is not significantly different from that in the wild-type MKP1 transgenic line, the protein level compared with that of MKP1WT is reduced significantly in MKP14A (Supplemental Fig. S1). If the difference was completely dependent on the proteasome, MG132 treatment should fully bypass the requirement for phosphorylation to revert the protein level of MKP14A back to the wild-type level. However, although MG132 did cause an increase in MKP1 protein, MG132 treatment could not restore the amount of MKP14A to even basal levels of MKP1WT (Fig. 5, A and B). In addition, the ratio of MKP1 protein with and without MG132 was the same in both MKP1WT and MKP14A types (Fig. 5C). These results indicate that, although a portion of the control of MKP1 levels may be through the proteasome, phosphorylation of MKP1 appears to have an added effect on MKP1 stability that is proteasome independent. For instance, the phosphorylation of MKP1 may induce a conformational change or interfere with protein-protein interactions that affect protein stability. Alternatively, there is evidence from mammalian pathways for ubiquitin-mediated degradation that occurs through zinc-dependent but proteasome-independent pathways (Wan et al., 2014), but it is unknown if similar mechanisms may exist in plants.
MKP1 Stabilization Likely Confers a Negative Feedback Loop upon the MAPK Cascade during PAMP Elicitation
The phosphorylation-induced stabilization of MKP1 would be consistent with models from mammalian systems in which processes leading to the activation of MAPKs also result in the accumulation of MAPK phosphatases as part of a negative feedback loop to turn off the signaling (Martín et al., 2005). For example, activation of the p42/p44 MAPK cascade in hamster fibroblasts results in MKP1 phosphorylation followed by reduced proteasomal degradation and, thus, protein stabilization (Brondello et al., 1999). A similar mechanism also was reported in Xenopus laevis oocytes (Sohaskey and Ferrell, 2002). In Arabidopsis, MPK6 was found to phosphorylate MPK1 in vitro (Park et al., 2011), and we report in this study that PAMP-activated MPK6 can phosphorylate MKP1 primarily on Thr-109 (Fig. 2A). Moreover, the PAMP-induced phosphorylation and accumulation of MKP1 is closely correlated with the timing of MAPK activation, supporting the possibility of a negative feedback loop in planta during PAMP elicitation. Our data, however, suggest that MPK6 may not be the only kinase phosphorylating MKP1 in vivo, as MKP1 still undergoes a mobility shift in protoplasts of mkp1 mpk6 mutants, although the duration of phosphorylation does appear to be affected (Fig. 2B). Due to the coregulation and at least partial functional redundancy of MPK3 and MPK6 (Colcombet and Hirt, 2008; Ren et al., 2008; Wang et al., 2008), we hypothesize that MPK3 and MPK6 both may be involved in MKP1 phosphorylation.
A consideration of the timing of MKP1 regulation and MAPK activation when comparing biotic and abiotic stress responses indicates that MKP1 may play a greater role in attenuating the activity of MAPKs rather than fully inactivating them. After both UV-B stress (González Besteiro et al., 2011) and PAMP treatment (Anderson et al., 2011), MPK3 and MPK6 are hyperactivated in plants lacking a functional MKP1. However, in the case of UV-B treatment, MAPK activation is maintained for a longer time (e.g. greater than 6 h; González Besteiro et al., 2011), whereas MAPKs show a more transient activation, reaching a maximal point approximately 20 to 30 min after PAMP treatment (Anderson et al., 2011). In response to both treatments, activation of MAPKs is concomitant with phosphorylation and accumulation of MKP1 (González Besteiro and Ulm, 2013; this work). If the function of MKP1 was to completely inactivate the MAPKs, it is difficult to reconcile how the protein can accumulate rapidly in both cases yet have very different outcomes with regard to the duration of MAPK activation. One possible explanation is if the duration of MAPK activation may be more a function of the signal input from the receptors. It is known that FLS2, the receptor for flagellin, is removed via endocytosis between 30 and 60 min after PAMP treatment, which has been proposed as a mechanism for attenuating PAMP signaling (Smith et al., 2014). It is not clear how UV-B activates MAPKs, as the MKP1-dependent misregulation of the MAPKs is independent of the UVR8 photoreceptor (González Besteiro et al., 2011), but is has been proposed that the activation results from cellular damage when UV-B protection is not sufficient. Therefore, it is plausible that the UV-B damage signal is longer lived than that from PAMPs. In both cases, however, MAPK activation is higher in mkp1 mutants but shows similar temporal profiles for inactivation compared with wild-type responses, clearly indicating an important role for other phosphatases that also must control these processes.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds were sterilized with 1% sodium hypochlorite and 0.01% Tween 20 for 20 min, rinsed with water, and plated aseptically on 0.5% agar containing 2.1 g L−1 Murashige and Skoog (MS) salts (PhytoTechnology Laboratories), pH 5.7, 1% Suc, and 6.4 g mL−1 MS salts vitamin powder (PhytoTechnology Laboratories). After stratification for 2 d at 4°C, seeds were grown in a Sanyo MLR-351H growth chamber at 21°C with a 9/15-h light/dark cycle. Seedlings were maintained in the same growth chamber under the same conditions during elf26, MG132, and Pseudomonas syringae pv tomato DC3000 treatments. The mutants mkp1 (Col-0), mkp1/Pro35s::myc-MKP1, mkp1/Pro35s::myc-MKP14A, mkp1/Pro35s::myc-MKP14D, snc1-11 (Col-0), and mkp1 snc1-11 (Col-0) are in the Col-0 background (Bartels et al., 2009; González Besteiro and Ulm, 2013). mkp1-1 (Ws) and mkp1-1 mpk6-1 (Ws) are in the Ws background (Ulm et al., 2001; Liu and Zhang, 2004; Bartels et al., 2009; Anderson et al., 2011, 2014).
Protein Extraction, λ-Phosphatase Treatment, and Immunoblot Analysis
For each treatment condition, four 14-d-old seedlings were transferred from MS agar plates to a single well of a 12-well microtiter plate with 2 mL of sterile water per well and incubated overnight (24 h). For elicitor or MG132 treatment, water from the overnight incubation was removed and replaced with sterile water containing 1 µm elf26 or 40 µm MG132. At the corresponding time points, the seedlings were frozen in liquid nitrogen, and proteins were isolated by grinding tissue with a pestle in 100 µL of SDS-PAGE sample buffer (5% SDS, 10% glycerol, and 0.1 m Tris-HCl, pH 6.8), and 15 µL of the extract per lane was separated by SDS-PAGE for immunoblot analyses. For phosphatase treatment, the proteins were isolated by grinding in protein extraction buffer (100 mm HEPES-KOH, pH 7.5, 5% glycerol, 0.5% PVP, 1 mm PMSF], and 10 µm leupeptin) and incubated with λ-phosphatase (New England Biolabs) at 30°C for 10 min, in the presence or absence of a phosphatase inhibitor mix (50 mm NaF, 20 mm NaVO3, and 5 mm EDTA). For detection of the band shift caused by phosphorylation, total cellular proteins or λ-phosphatase-treated extracts were separated using 8% large-format (16 × 16 cm) SDS-PAGE overnight. Otherwise, protein extracts were separated using 8% mini-format (8.3 × 7.3 cm) SDS-PAGE. Proteins were transferred to PVDF, and immunoblotting was performed using primary mouse anti-myc antibody (Cell Signaling Technologies, Fisher) for detection of myc-tagged MKP1 protein, rabbit anti-phospho-p44/42 MAPK antibody (Cell Signaling Technologies) for detection of active MPK3/6, rabbit anti-Glu-Glu (EMD, Millipore) antibody for detection of Pyo-tagged MKP1, or previously described rabbit anti-MPK6 antibody for detection of MPK6 (Merkouropoulos et al., 2008). Chemiluminescence-based detection (Pierce) was performed using horseradish peroxidase-conjugated goat anti-rabbit antibody (Cell Signaling Technologies) or goat anti-mouse antibody (Sigma). The protein band intensity was quantified using the software ImageJ (https://imagej.net/).
In Vitro Kinase Assay
The MKP1 DNA sequence corresponding to the N-terminal 161 amino acids of MKP1 was PCR amplified from Arabidopsis Col-0 genomic DNA using primers 5′-ATGGTGGGAAGAGAGGATGC-3′ and 5′-TCATCCACCCACATATATATGATCAGC-3′ and subcloned into expression vector pET100D (Invitrogen). Quick-change mutagenesis (Stratagene) was used to introduce codon changes in the MKP1 (1-161)::pET100D plasmid, resulting in Ala substitutions at either Thr-64 or Thr-109. Primers used for mutagenesis were T64AF (5′-CCAGCTGCTCCTTTGGCACCTCGTTCACATC-3′), T64AR (5′-GATGTGAACGAGGTGCCAAAGGAGCAGCTGG-3′), T109AF (5′-GGCCTCATCCACCAGCACCTAGCGGGAAC-3′), and T109AR (5′-GTTCCCGCTAGGTGCTGGTGGATGAGGCC-3′). DNA mutations were confirmed by Sanger sequencing. For protein expression, MKP1::pET100D plasmids were transformed into Escherichia coli BL21 cells, and the transformation mixture was used to inoculate 5 mL of Luria-Bertani broth containing 50 μg mL−1 carbenicillin. After overnight culture at 37°C, 5 mL of culture was used to inoculate 100 mL of Luria-Bertani broth with 50 μg mL−1 carbenicillin. After 2 h of growth at 37°C, 1 mm of IPTG was added to induce protein expression. After 4 h at 37°C, the bacteria were collected by centrifugation and stored at −20°C. To purify proteins, the frozen pellets were resuspended in native lysis buffer (Qiagen; 50 mm NaH2PO4, 300 mm NaCl, and 10 mm imidazole, pH 8, with 1 mm PMSF and 10 µm leupeptin added to inhibit protease activity). The resuspended bacteria were sonicated to disrupt cells and centrifuged at 10,000g for 10 min to remove debris. Two milliliters of the clarified supernatant was mixed with 50 µL of Ni-NTA resin (Qiagen) and incubated at 4°C for 30 min. The resin was then washed three times with wash buffer (50 mm NaH2PO4, 300 mm NaCl, and 20 mm imidazole, pH 8), and proteins were eluted into 100 µL of elution buffer (50 mm NaH2PO4, 300 mm NaCl, and 250 mm imidazole, pH 7.5). MPK6 was immunoprecipitated from lysates prepared from 12 mL of a 7-d-old Arabidopsis suspension cell culture treated with 100 nm elf26 or DMSO-only mock control for 10 min. Treated Arabidopsis cells were collected by filtration, frozen in liquid nitrogen, ground to a powder using a mortar and pestle, and resuspended in 4 mL of cold immunoprecipitation (IP) buffer (100 mm Tris-HCl, pH 7.5, 5% glycerol, 50 mm NaPP, 1 mm NaMo, 25 mm NaF, 15 mm EGTA, 5 mm EDTA, 0.5% PVP, 1% Triton X-100, 150 mm NaCl, 1 mm PMSF, 10 µm leupeptin, 1 nm calyculin A, and 1 mm DTT). All subsequent steps for IP were performed at 4°C. Homogenates were centrifuged at 16,000g for 15 min, and 1 mL of the resulting supernatant was mixed with 10 µL of MPK6-protein A Sepharose beads (Sigma) prepared by preincubating the beads with 1.5 µL of a polyclonal α-MPK6 antibody (Merkouropoulos et al., 2008) in IP buffer for 1.5 h. After 2 h, the beads were washed three times with IP buffer and once with kinase buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm MnCl2, 10 mm MgCl2, 10% glycerol, and 1 mm DTT). For kinase assays, purified MKP1 protein or MBP (Sigma) was mixed with a 10-μL aliquot of MPK6-bound protein A Sepharose beads, 0.3 μL of [32P]γ-ATP, and 1 μL of 250 µm ATP in a total reaction volume of 25 µL of kinase buffer. Kinase reactions were incubated at 30°C for 10 min, then stopped by the addition of 6 µL of 5× SDS-PAGE loading buffer and incubating at 80°C for 10 min. Protein gels were stained with Coomassie Blue R-250 and dried, and 32P signal was detected by phosphor screen imaging.
Arabidopsis Protoplast Isolation and Transformation
Protoplasts from 5-week-old mkp1 (Ws) and mkp1 mpk6 (Ws) adult plant were isolated as described previously (Yoo et al., 2007). For transient protein expression, protoplasts (100 µL with 3 × 105 cells) were transfected with 10 µg of plasmid. After overnight incubation (14–16 h) in the dark at room temperature, the transfected protoplasts were treated with 1 µm elf26 or 40 µm MG132 for the times indicated in the figures. The treated protoplasts were frozen in liquid nitrogen and resuspended in 40 µL of SDS sample buffer. Twenty microliters of the sample per lane was separated by SDS-PAGE and used for immunoblot analyses.
Inhibition of Seedling Growth by elf26 Treatment
Six-day-old seedlings were aseptically transferred from MS agar to wells of a 24-well microtiter plate (one seedling per well) containing 1 mL of liquid MS medium (2.1 g L−1 MS salts, pH 5.7, and 1% Suc) with or without 1 µm elf26. After 14 d, seedlings were weighed and placed on an agar surface for photography and measurement of primary root length.
DC3000 Growth Measurements and Luciferase Imaging
DC3000 expressing the LuxCDABE operon has been described previously (Fan et al., 2008). For inoculation, 2 d prior to infection, a glycerol stock of DC3000 LuxCDABE stored at −80°C was streaked onto King’s B medium agar plates containing 50 μg mL−1 kanamycin and 60 μg mL−1 rifampicin and then incubated for 2 d at room temperature. Prior to infection, two 14-d-old seedlings were transferred from MS agar plates to 1 mL of sterile water in a single well of a 24-well microtiter plate. After transferring seedlings, the plate was returned to the growth chamber (Sanyo MLR-351H) at 21°C overnight for 16 to 20 h. Immediately before infection, an inoculum was prepared by scraping DC3000 bacteria from the agar plate and resuspending it in sterile water to a final OD600 of 0.01 (1 × 107 cfu mL−1). Seedlings were inoculated in microtiter plate wells by replacing the water used for overnight incubation with 2 mL of DC3000 inoculum, and the microtiter plates were returned to the growth chamber. Prior to luciferase imaging and serial dilution plating, infected seedlings were removed from microtiter plate wells, rinsed thoroughly by immersing in water, blotted dry, and placed on an agar surface for imaging. Luciferase signal from DC3000 bacteria in seedlings was detected using an HRPCS4 photon-detection camera and IFS32 software (Photek). For measurements of DC3000 by serial dilution plating, two seedlings from the same well of the microtiter plate were blotted dry, rinsed in water, blotted dry, and then homogenized in 400 µL of 10 mm MgCl2, and 10-fold serial dilutions were performed. A 20-µL aliquot of each dilution was spotted on King’s B medium agar plates containing appropriate antibiotics, and colonies were counted after 2 d of incubation at room temperature.
Transcript Analysis Using Quantitative Real-Time PCR
For each treatment condition, four 12-d-old seedlings were transferred from MS agar plates to 1 mL of sterile water in a single well of a 24-well microtiter plate and incubated overnight. For elicitor treatment, water from the overnight incubation was removed and replaced with sterile water containing 1 μm elf26.
Total RNA was isolated using TRI reagent (Sigma) and treated with DNase I (Fermentas), and 1µg of RNA was reverse transcribed in 25-µL reactions containing 5 mm DTT, 0.5 µL of RnaseOUT (Invitrogen), 2 μm oligo(dT), 1 mm each of dNTPs, and 0.5 μL M-MLV reverse transcriptase (Promega) for 1 h at 42°C, followed by 5 min at 85°C. Reverse transcription reactions were diluted to 100 µL using diethylpyrocarbonate-treated water. Real-time PCR was performed essentially as described previously (Libault et al., 2007) using the primers listed in Supplemental Table S1. In brief, 10-µL real-time PCRs containing 5 µL of SYBR Green PCR master mix (Applied Biosystems), 1 µL of cDNA, and 0.2 µm of each primer were performed using an ABI7500 real-time thermal cycler (Applied Biosystems). Three independent experiments were performed. Expression levels were calculated using the following equation: expression level = (PCR efficiency)−∆Ct, where ∆Ct = Ct (sample) – Ct (control), and PCR efficiencies and Ct for each reaction were obtained using the software program LinRegPCR (Ramakers et al., 2003). At2g28390 (SAND family protein) was used as the reference gene for normalization (Czechowski et al., 2005).
Statistical Analysis
Statistical analyses were performed using either ANOVA with multiple pairwise Student’s t test in Microsoft Excel or ANOVA with Tukey’s pairwise comparisons in Minitab software (Minitab 18).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At3g55270 (MKP1), At2g43790 (MPK6), At4g16890 (SNC1), At4g37370 (CYP81D8), and At4g20000.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. MKP1 mRNA levels and protein amounts of myc-MKP1 in different transgenic lines.
Supplemental Figure S2. MKP1 can be stabilized by MG132.
Supplemental Figure S3. Phosphorylation sites are not required for MG132 stabilization.
Supplemental Figure S4. Phosphorylation site mutants cannot complement the enhanced growth inhibition in mkp1 (Col-0).
Supplemental Figure S5. No difference in the fresh weight and primary root length of Col-0, mkp1 (Col-0), MKP1WT, MKP14A, and MKP14D seedlings without elf26 treatment.
Supplemental Figure S6. Phosphorylation site mutants cannot complement the enhanced bacteria resistance in mkp1 (Col-0).
Supplemental Table S1. Quantitative PCR primers used in this study.
Footnotes
The work was supported by National Science Foundation Grants IOS-1051286 and IOS-1456256 to S.C.P. The work of M.A.G.B. was supported by the University of Geneva and the Swiss National Science Foundation Grant 31003A_132902 to Roman Ulm.
Articles can be viewed without a subscription.
References
- Anderson JC, Bartels S, González Besteiro MA, Shahollari B, Ulm R, Peck SC (2011) Arabidopsis MAP Kinase Phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. Plant J 67: 258–268 [DOI] [PubMed] [Google Scholar]
- Anderson JC, Wan Y, Kim YM, Pasa-Tolic L, Metz TO, Peck SC (2014) Decreased abundance of type III secretion system-inducing signals in Arabidopsis mkp1 enhances resistance against Pseudomonas syringae. Proc Natl Acad Sci USA 111: 6846–6851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gómez-Gómez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Bartels S, Anderson JC, González Besteiro MA, Carreri A, Hirt H, Buchala A, Métraux JP, Peck SC, Ulm R (2009) MAP KINASE PHOSPHATASE1 and PROTEIN TYROSINE PHOSPHATASE1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 21: 2884–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels S, González Besteiro MA, Lang D, Ulm R (2010) Emerging functions for plant MAP kinase phosphatases. Trends Plant Sci 15: 322–329 [DOI] [PubMed] [Google Scholar]
- Bittel P, Robatzek S (2007) Microbe-associated molecular patterns (MAMPs) probe plant immunity. Curr Opin Plant Biol 10: 335–341 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Brock AK, Willmann R, Kolb D, Grefen L, Lajunen HM, Bethke G, Lee J, Nürnberger T, Gust AA (2010) The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiol 153: 1098–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondello JM, Pouysségur J, McKenzie FR (1999) Reduced MAP kinase phosphatase-1 degradation after p42/p44MAPK-dependent phosphorylation. Science 286: 2514–2517 [DOI] [PubMed] [Google Scholar]
- Camps M, Nichols A, Arkinstall S (2000) Dual specificity phosphatases: a gene family for control of MAP kinase function. FASEB J 14: 6–16 [PubMed] [Google Scholar]
- Colcombet J, Hirt H (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413: 217–226 [DOI] [PubMed] [Google Scholar]
- Corbit KC, Trakul N, Eves EM, Diaz B, Marshall M, Rosner MR (2003) Activation of Raf-1 signaling by protein kinase C through a mechanism involving Raf kinase inhibitory protein. J Biol Chem 278: 13061–13068 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Crooks C, Lamb C (2008) High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J 53: 393–399 [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Fuchs S, Grill E, Meskiene I, Schweighofer A (2013) Type 2C protein phosphatases in plants. FEBS J 280: 681–693 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Felix G, Boller T (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18: 277–284 [DOI] [PubMed] [Google Scholar]
- González Besteiro MA, Bartels S, Albert A, Ulm R (2011) Arabidopsis MAP kinase phosphatase 1 and its target MAP kinases 3 and 6 antagonistically determine UV-B stress tolerance, independent of the UVR8 photoreceptor pathway. Plant J 68: 727–737 [DOI] [PubMed] [Google Scholar]
- González Besteiro MA, Ulm R (2013) Phosphorylation and stabilization of Arabidopsis MAP kinase phosphatase 1 in response to UV-B stress. J Biol Chem 288: 480–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, MacLean D, Studholme DJ, Serna-Sanz A, Andreasson E, Rathjen JP, Peck SC (2009) Phosphoproteomic analysis of nuclei-enriched fractions from Arabidopsis thaliana. J Proteomics 72: 439–451 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Lassowskat I, Böttcher C, Eschen-Lippold L, Scheel D, Lee J (2014) Sustained mitogen-activated protein kinase activation reprograms defense metabolism and phosphoprotein profile in Arabidopsis thaliana. Front Plant Sci 5: 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Scott DA, Hatch E, Tian X, Mansour SL (2007) Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development 134: 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wong WS, Zhu L, Guo HW, Ecker J, Li N (2009) Phosphoproteomic analysis of ethylene-regulated protein phosphorylation in etiolated seedlings of Arabidopsis mutant ein2 using two-dimensional separations coupled with a hybrid quadrupole time-of-flight mass spectrometer. Proteomics 9: 1646–1661 [DOI] [PubMed] [Google Scholar]
- Libault M, Wan J, Czechowski T, Udvardi M, Stacey G (2007) Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol Plant Microbe Interact 20: 900–911 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S. (2003) Protein phosphatases in plants. Annu Rev Plant Biol 54: 63–92 [DOI] [PubMed] [Google Scholar]
- Lumbreras V, Vilela B, Irar S, Solé M, Capellades M, Valls M, Coca M, Pagès M (2010) MAPK phosphatase MKP2 mediates disease responses in Arabidopsis and functionally interacts with MPK3 and MPK6. Plant J 63: 1017–1030 [DOI] [PubMed] [Google Scholar]
- Martín H, Flández M, Nombela C, Molina M (2005) Protein phosphatases in MAPK signalling: we keep learning from yeast. Mol Microbiol 58: 6–16 [DOI] [PubMed] [Google Scholar]
- Meng X, Zhang S (2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51: 245–266 [DOI] [PubMed] [Google Scholar]
- Merkouropoulos G, Andreasson E, Hess D, Boller T, Peck SC (2008) An Arabidopsis protein phosphorylated in response to microbial elicitation, AtPHOS32, is a substrate of MAP kinases 3 and 6. J Biol Chem 283: 10493–10499 [DOI] [PubMed] [Google Scholar]
- Meskiene I, Baudouin E, Schweighofer A, Liwosz A, Jonak C, Rodriguez PL, Jelinek H, Hirt H (2003) Stress-induced protein phosphatase 2C is a negative regulator of a mitogen-activated protein kinase. J Biol Chem 278: 18945–18952 [DOI] [PubMed] [Google Scholar]
- Millar JB, Buck V, Wilkinson MG (1995) Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev 9: 2117–2130 [DOI] [PubMed] [Google Scholar]
- Nühse TS, Peck SC, Hirt H, Boller T (2000) Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J Biol Chem 275: 7521–7526 [DOI] [PubMed] [Google Scholar]
- Paleologou KE, Schmid AW, Rospigliosi CC, Kim HY, Lamberto GR, Fredenburg RA, Lansbury PT Jr, Fernandez CO, Eliezer D, Zweckstetter M, et al. (2008) Phosphorylation at Ser-129 but not the phosphomimics S129E/D inhibits the fibrillation of alpha-synuclein. J Biol Chem 283: 16895–16905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Song EH, Nguyen XC, Lee K, Kim KE, Kim HS, Lee SM, Kim SH, Bae DW, Yun DJ, et al. (2011) Arabidopsis MAP kinase phosphatase 1 is phosphorylated and activated by its substrate AtMPK6. Plant Cell Rep 30: 1523–1531 [DOI] [PubMed] [Google Scholar]
- Peck SC. (2006) Analysis of protein phosphorylation: methods and strategies for studying kinases and substrates. Plant J 45: 512–522 [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Schikora A, Hirt H (2009) MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Ren D, Liu Y, Yang KY, Han L, Mao G, Glazebrook J, Zhang S (2008) A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 105: 5638–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Yang H, Zhang S (2002) Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J Biol Chem 277: 559–565 [DOI] [PubMed] [Google Scholar]
- Rodriguez MC, Petersen M, Mundy J (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61: 621–649 [DOI] [PubMed] [Google Scholar]
- Shubchynskyy V, Boniecka J, Schweighofer A, Simulis J, Kvederaviciute K, Stumpe M, Mauch F, Balazadeh S, Mueller-Roeber B, Boutrot F, et al. (2017) Protein phosphatase AP2C1 negatively regulates basal resistance and defense responses to Pseudomonas syringae. J Exp Bot 68: 1169–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Salamango DJ, Leslie ME, Collins CA, Heese A (2014) Sensitivity to Flg22 is modulated by ligand-induced degradation and de novo synthesis of the endogenous flagellin-receptor FLAGELLIN-SENSING2. Plant Physiol 164: 440–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohaskey ML, Ferrell JE Jr (2002) Activation of p42 mitogen-activated protein kinase (MAPK), but not c-Jun NH(2)-terminal kinase, induces phosphorylation and stabilization of MAPK phosphatase XCL100 in Xenopus oocytes. Mol Biol Cell 13: 454–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena G, Boudsocq M, Sheen J (2011) Protein kinase signaling networks in plant innate immunity. Curr Opin Plant Biol 14: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Ichimura K, Mizoguchi T, Peck SC, Zhu T, Wang X, Shinozaki K, Paszkowski J (2002) Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J 21: 6483–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Revenkova E, di Sansebastiano GP, Bechtold N, Paszkowski J (2001) Mitogen-activated protein kinase phosphatase is required for genotoxic stress relief in Arabidopsis. Genes Dev 15: 699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Petris MJ, Peck SC (2014) Separation of zinc-dependent and zinc-independent events during early LPS-stimulated TLR4 signaling in macrophage cells. FEBS Lett 588: 2928–2935 [DOI] [PubMed] [Google Scholar]
- Wang H, Liu Y, Bruffett K, Lee J, Hause G, Walker JC, Zhang S (2008) Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell 20: 602–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zipfel C. (2009) Early molecular events in PAMP-triggered immunity. Curr Opin Plant Biol 12: 414–420 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Felix G (2005) Plants and animals: a different taste for microbes? Curr Opin Plant Biol 8: 353–360 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]