The deubiqitinase OTU5 coordinates Pi deficiency signals on DNA methylation, H3K4 and H3K27 trimethylation, and gene expression, resulting in Pi-dependent root phenotypes of otu5 mutants.
Abstract
Phosphate (Pi) starvation induces a suite of adaptive responses aimed at recalibrating cellular Pi homeostasis. Plants harboring a mutation in OVARIAN TUMOR DOMAIN-CONTAINING DEUBIQUITINATING ENZYME5 (OTU5) showed altered DNA methylation of root hair-related genes and altered Pi-responsive root traits. Unlike the wild type, homozygous otu5 mutants did not respond to Pi starvation by increased lateral root formation and increased root hair length but formed very short root hairs when grown on low-Pi media. Under Pi-replete conditions, otu5 plants developed more root hairs than the wild type due to attenuated primary root growth, a phenotype that resembled that of Pi-deficient plants. Growth of plants on low-Pi media altered both H3K4 and H3K27 trimethylation levels at the transcriptional start site of a subset of genes encoding key players in Pi homeostasis, which was correlated with mRNA abundance changes of these genes. Pi starvation had a minor impact on DNA methylation. Differentially methylated regions were enriched in transposable elements, suggesting that DNA methylation associated with low Pi supply is required for maintaining genome integrity. It is concluded that DNA methylation and histone methylation constitute critical, interdependent regulatory components that orchestrate the activity of a subset of Pi-responsive genes.
As a key component of nucleic acids, membrane phospholipids, signaling cascades, and energy metabolism, phosphorus (P) is essential for plant development and reproduction. Plants take up P in the form of inorganic phosphate (Pi). Due to numerous interactions with soil components, the phytoavailability of Pi can drop to levels well below the requirement of plants, impairing plant growth and productivity. Pi starvation triggers a suite of plant responses that improve the uptake, internal use, and recycling of Pi. This is achieved through a combination of reprogrammed energy and lipid metabolism, expression of Pi transporters, and altered root development (Ticconi and Abel, 2004; Desnos, 2008; Giehl et al., 2014). In response to low Pi availability, primary root growth is attenuated while the development of lateral roots is enhanced to improve foraging of Pi in topsoil layers enriched in Pi. Low-Pi conditions further increase the length and density of root hairs by expanding the duration of tip growth and reduced elongation of root epidermal cells, thereby enhancing the radial reach of the root (Bates and Lynch, 2000; Gilroy and Jones, 2000; Salazar-Henao et al., 2016).
Root hair morphogenesis and the expression of Pi starvation-responsive (PSR) genes are affected by alterations in chromatin structure. For example, compromised deposition of the histone variant H2A.Z in mutants defective in the expression of ARP6, a subunit of the SWR1 ATP-dependent chromatin-remodeling complex, was associated with constitutive activation of a suite of PSR genes (Smith et al., 2010). Also, arp6 plants formed longer and more abundant root hairs under Pi-replete conditions, a phenotype that is typical of Pi-deficient plants. Similarly, mutants defective in the expression of the histone deacetylase HDA19 showed alterations in Pi-responsive root traits and deregulation of a subset of PSR genes (Chen et al., 2015).
DNA cytosine methylation (mC) is an epigenetic modification that is important for transcriptional regulation, such as silencing of transposable elements (TEs; He et al., 2011). In plants, DNA methylation can respond rapidly and dynamically to environmental stimuli (Labra et al., 2002; Aina et al., 2004; Meaney and Szyf, 2005; Choi and Sano, 2007). In rice (Oryza sativa), Pi starvation increases DNA methylation in TEs close to highly induced genes (Secco et al., 2015). In Arabidopsis (Arabidopsis thaliana), different effects of Pi starvation on global DNA methylation were reported (Secco et al., 2015; Yong-Villalobos et al., 2015). While Secco et al. (2015) observed very limited effects of Pi starvation on the DNA methylation landscape in Arabidopsis, which were mainly located in TEs or in genes with undetectable expression levels, Yong-Villalobos et al. (2015) reported highly dynamic changes in DNA methylation levels upon Pi starvation in genic regions, which were associated with transcriptional regulation. Furthermore, mutants harboring defects in DNA methylation were found to be unable to regulate a subset of PSR genes, suggesting that some PSR components are epigenetically controlled by Pi-deficiency-induced changes in mC (Yong-Villalobos et al., 2015).
Deubiquitinating enzymes (DUBs) are specialized proteases that remove individual ubiquitin molecules or ubiquitin chains attached to target proteins. DUBs are involved in important biological processes such as proofreading ubiquitin-protein conjugates, chromatin remodeling, transcriptional regulation, cell cycle regulation, DNA repair, and signal transduction (Reyes-Turcu et al., 2009). Although clear roles in cellular processes have been defined for OVARIAN TUMOR DOMAIN-CONTAINING DEUBIQUITINATING ENZYME (OTU) deubiquitinases in Drosophila melanogaster, Saccharomyces cerevisiae, and Homo sapiens (King and Storto, 1988; Steinhauer et al., 1989; Lee et al., 2000; Stanisić et al., 2009; Nakada et al., 2010), in plants, a biological role has only been described for OTLD1, which functions as a direct interactor of the histone Lys demethylase complex KDM1C to suppress the expression of a subset of genes via deubiquitination of histone H2B (Krichevsky et al., 2011).
Phenotypic analysis of plants defective in the expression of members of the OTU family revealed that similar to arp6, Pi-replete otu5 plants display a root hair phenotype that resembles that of Pi-deficient plants. To understand the impact of Pi starvation on the epigenome and the role of OTU5 in root development, we performed comprehensive surveys of histone modifications, genome-wide DNA methylation, and transcription profiles of the otu5 mutant and its wild type under Pi-replete and Pi-deficient conditions. We found that gene regulation by OTU5 is conditionally dependent on the availability of Pi. Gene expression patterns validated significant interaction of root phenotypes with the environment. It is hypothesized that the function of OTU5 is associated with local alterations in DNA methylation, which directly or indirectly affect the activity of genes involved in root hair morphogenesis and primary root growth, traits that are also responsive to Pi starvation. Growth on low-Pi media altered transcriptional profiles, which was partly correlated with both H3K4 and H3K27 trimethylation at promoters. Intriguingly, differentially histone methylated genes encode key players in Pi acquisition and cellular Pi homeostasis. Our data suggest that DNA and histone methylation act interdependently to interpret environmental information.
RESULTS
The otu5 Mutant Shows Genotype-Specific Responses to Low-Pi Availability
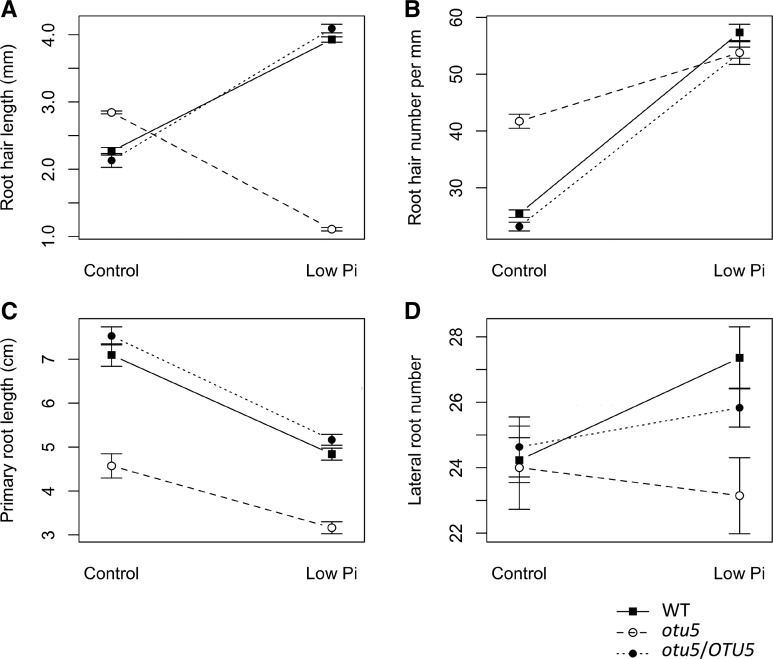
As anticipated from previous studies, wild-type plants responded to Pi starvation with an increase in root hair density and length, a decrease in primary root length, and an increase in the number of lateral roots (Fig. 1). By contrast, homozygous otu5 mutants formed longer and denser root hairs than the wild type under Pi-replete conditions but produced short root hairs with a density typical of Pi-deficient wild-type plants (Fig. 1, A and B). Similarly, under Pi-replete conditions, the primary root length of otu5 plants was comparable to that observed for Pi-deficient wild-type plants (Fig. 1C). In otu5, primary root length was further decreased upon exposure to low-Pi media, but to a lesser extent than in the wild type. No increase in the number of lateral roots was observed in otu5 plants (Fig. 1D). Interactions between genotype and environment (G × E; E = Pi conditions) were statistically significant for both the length and density of root hairs and for the number of lateral roots (two-way ANOVA, P < 0.05; Supplemental Table S1). Transgenic lines containing a wild-type copy of the OTU5 gene in the otu5 background showed a phenotype resembling that of the wild type in all the above-mentioned traits.
Figure 1.
Effect of growth on low-Pi media on root development of wild-type, otu5, and otu5 mutant plants complemented with a wild-type OTU5 copy (otu5/OTU5). A, Root hair length. B, Root hair number. C, Primary root length. D, Lateral root number.
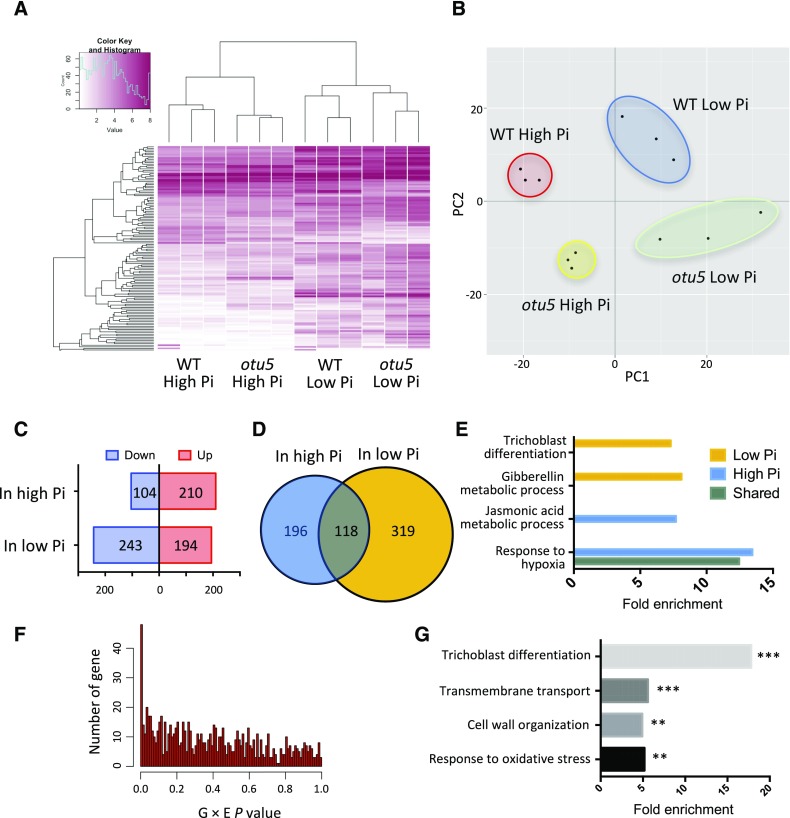
Next, we analyzed the gene expression profiles of wild-type and otu5 plants grown under Pi-replete and low-Pi conditions by RNA-seq (Supplemental Table S2). Differential gene expression was observed between all four groups (i.e. two growth conditions and two genotypes). Hierarchical clustering of these transcriptomes shows that both the Pi supply and the mutation in OTU5 impacted the transcriptome of the roots (Fig. 2A). It can also be inferred from the analysis that the Pi supply had a much broader impact on the transcriptional profile compared to that of the OTU5 mutation (Fig. 2A).
Figure 2.
RNA-seq analysis of wild-type and otu5 mutant plants under high-Pi (Pi-replete) and low-Pi conditions. A, Heat map of expression-variable genes. B, Principal component analysis of expression-variable genes. C, Numbers of otu5-associated DEGs (i.e. wild type versus otu5) under high-Pi and low-Pi conditions. D, Venn diagram showing otu5-associated DEGs under high-Pi and low-Pi conditions. E, Gene Ontology enrichment analysis of otu5-associated DEGs in Pi-replete, low-Pi conditions, or both conditions. F, Distribution of P values of two-way ANOVA using RNA-seq data of wild-type and otu5 plants under high-Pi and low-Pi conditions. A subset of 48 genes showed significant interaction between environment and genotypes (P < 0.01). G, Gene Ontology enrichment analysis of the 48 genes with significant interaction between environments and genotypes. P values are denoted by asterisks (**P < 0.01, ***P < 0.001).
Principal component analysis shows high variation between plants supplied with two different Pi concentrations (PC1) as well as between otu5 and the wild type under both growth conditions (low-Pi and Pi-replete), which is in line with the hierarchical clustering (Fig. 2B). In addition, principal component analysis also revealed higher variation among the triplicates from plants grown under low-Pi conditions than in those derived from plants grown on Pi-replete media, indicative of a higher level of transcriptional noise under the former conditions (Fig. 2B). This conclusion is confirmed by the fact that the mean expression standard deviations of plants grown in low-Pi conditions were higher when compared to those grown on Pi-replete media (Supplemental Table S3).
In order to understand the relationships between these groups, we performed a weighted gene coexpression network analysis (WGCNA; Langfelder and Horvath, 2008). This is an unbiased and unsupervised analysis that identifies modules corresponding to clusters of coexpressed transcripts in each group. This analysis identified three major modules containing tightly coexpressed genes (Supplemental Fig. S1; Supplemental Data Set 1). Two of the modules are specific to otu5 and low Pi, respectively, suggesting that despite producing similar root phenotypes, Pi deficiency and nonfunctional OTU5 affected the expression of different sets of genes.
OTU5 Modulates the Transcriptional Response to Low Pi
Growth on low Pi media increased the number of genes that were differentially expressed (DEGs) between otu5 mutant and wild-type plants (Fig. 2C; Supplemental Fig. S2). Of the 633 DEGs, only 118 (19%) were differentially expressed between otu5 and the wild type under both growth conditions, indicating that the effect of OTU5 on gene expression is dependent on the availability of Pi (Fig. 2D; Supplemental Data Set 2). Gene Ontology analysis showed that among the 319 OTU5-dependent genes that were only differentially expressed between otu5 and wild-type plants when plants were grown under low-Pi conditions, 25 genes are related to root hair development (Fig. 2E; Supplemental Table S4). This subset includes several genes that play key roles in root hair development, e.g. MRH6, XTH14, COW1, and several ROOT HAIR SPECIFIC genes that carry the Root Hair Element consensus sequence in their promoters (Kim et al., 2006). Of particular interest is the presence of ROOT HAIR DEFECTIVE6-LIKE2 in this subset, which encodes a functional homolog of ROOT HAIR DEFECTIVE6-LIKE4 that acts as an integrator of developmental and environmental signals and controls the elongation of root hairs (Datta et al., 2015). Consistent with the short root hair phenotype of Pi-deficient otu5 plants, all of the root hair genes were down-regulated in the mutant under Pi-deficient conditions. In addition to the low-Pi condition, we also identified 314 OTU5-dependent DEGs under Pi-replete conditions (Fig. 2C), of which two genes, IRON-REGULATED TRANSPORTER1 (IRT1) and AT5G62340, are related to root hair development. These OTU5-dependent DEGs specific to Pi-replete conditions are related to root hair initiation and root development and potentially resulted in the observed root phenotypes.
Two-way ANOVA identified 48 genes with statistically significant interaction in their expression in otu5 versus wild type and the two Pi conditions (Fig. 2F; Supplemental Table S5). Several genes from this subset have predicted or experimentally verified roles in root hair formation (Grierson et al., 1997; Velasquez et al., 2011; Lan et al., 2013). In particular, transcripts derived from genes encoding proteins involved in cell wall organization and cell wall modifications were highly (>500-fold) enriched in root hairs (Lan et al., 2013; Fig. 2G; Supplemental Table S6).
OTU5 Is Critical for the Establishment of DNA Methylation Patterns
To investigate whether the root phenotype of otu5 plants is caused by differential mC, we conducted whole-genome bisulfite sequencing of roots from wild-type and otu5 mutant plants grown under Pi-replete and low-Pi conditions (see Supplemental Table S7 for mapping statistics and methods for whole-genome bisulfite sequencing). Methylation of cytosines was categorized into CG, CHG, and CHH contexts (where H denotes A, T, or C) for subsequent analyses.
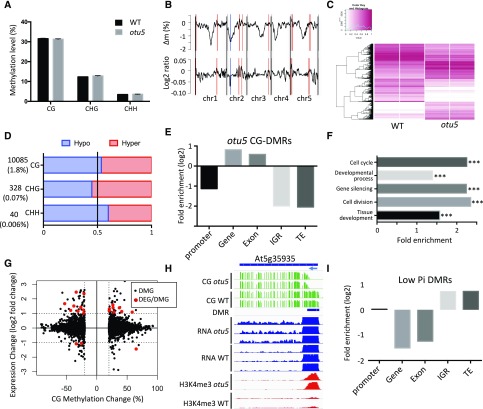
Global methylation levels were calculated from 3.7, 3.9, and 14.6 M of common sites in the CG, CHG, and CHH context, respectively, and found to be similar in wild-type and otu5 plants in all three contexts (Fig. 3A). On average, the methylation level of otu5 roots was approximately 31% for CG, 12% for CHG, and 3% for CHH sites. Although the global differences in methylation levels (∆ methylation) between otu5 and wild type were minor (0.35%, 0.4%, and 0.05% in CG, CHG, and CHH, respectively), highly variable mCs were observed locally in several chromosomal regions. Methylation levels of large genomic regions were compared using tiling windows of 100 kb. Figure 3B marks the top 10% of the regions with differential CG methylation between otu5 and the wild type (both ∆ methylation and log2 ratio of methylation levels) in more than three serial windows. The pericentromeric regions of chromosome 2 contain clusters of hypomethylated regions in otu5 relative to the wild type, and several hyper-methylated regions were observed throughout the otu5 genome. Hierarchical clustering revealed marked OTU5-driven differences in methylation patterns (Fig. 3C; Supplemental Fig. S3A). Together, the results suggest that OTU5 defines a distinct DNA methylation profile by targeting local genomic regions.
Figure 3.
DNA methylation patterns. A, Bulk DNA methylation. B, Chromosomal distribution of CG methylation between wild-type and otu5 plants. The upper line plot indicates the Δ CG methylation (otu5 – wild type); the lower line plot denotes the log2 CG methylation ratio (otu5/wild type). Red vertical lines indicate hypermethylated regions; the blue line depicts hypomethylated regions. C, Heat map of variable methylation regions of wild-type and otu5 mutant plants in the CG context. D, Total number of otu5-specific DMRs. Brackets indicate the percentage of DERs relative to the genome. E, Enrichment of otu5-specific CG DMRs. F, Gene Ontology enrichment analysis of DMR genes between wild type and the otu5 mutant. P values are denoted by asterisks (***P < 0.001). G, Scatterplot of otu5-associated DEGs/DMGs. The x axis indicates the Δ CG methylation (otu5 – wild type) and y axis indicates log2 expression changes (otu5/wild type). H, Genome browser snapshot showing CG methylation levels, DMRs, RNA expression, and abundance of H3K4me3 for the transposable element gene At5g35935 in both wild type and the otu5 mutant. I, Enrichment of low-Pi-associated DMRs.
A search for differentially methylated regions (DMRs; see “Materials and Methods” for DMR identification) identified 10,085, 328, and 40 DMRs in the CG, CHG, and CHH contexts, respectively, with an average size of 213 bp (Fig. 3D; Supplemental Data Set 3) and similar numbers of hyper- and hypo-DMRs. We found that 89% of the differentially methylated cytosines are CG sites (Supplemental Fig. S3B), suggesting that CG-specific DMRs (CG-DMRs) may contain the most pronounced methylation changes in otu5 plants. otu5-associated CG-DMRs were enriched in gene bodies, and exons and depleted from promoters, intergenic regions, and TEs (Fig. 3E). Non-CG-DMRs mostly distributed in intergenic regions and were depleted from coding regions (Supplemental Fig. S3C). In total, we identified 6,932 differentially methylated genes (DMGs) that contain at least one CG-DMR in either the gene body or promoter of the gene. These OTU5-associated DMGs were significantly enriched in biological functions, such as cell cycle, cell division, and tissue development (Fig. 3F; Supplemental Data Set 4). Several otu5-associated DMGs are related to ubiquitin-associated processes and also include the DUB UBP14, which was shown to be essential for the Pi-deficiency-induced elongation of root hairs (Supplemental Table S8; Li et al., 2010). otu5-associated methylation changes had very limited impact on transcription (Fig. 3G; Supplemental Fig. S4A); only 21 out of the 6,932 DMGs were differentially expressed between otu5 and the wild type, none of which is directly associated with PSR, suggesting the otu5-driven DMG has no direct impact on PSR. A total of four genes in which Δ methylation in the gene body exceeded 30% showed a clear anticorrelation of DNA methylation and gene expression (Supplemental Fig. S5). Such anticorrelation was also observed in some TEs (Fig. 3H).
Pi Deficiency-Induced Changes in the Methylome
Environmental conditions such as Pi deficiency can alter both gene expression and DNA methylation patterns (Yong-Villalobos et al., 2015). In total, we identified 62, 31, and 76 DMRs between low-Pi and Pi-replete conditions in the CG, CHG, and CHH contexts, respectively (Supplemental Data Set 5). The distribution of differentially methylated cytosines did not enrich toward any specific sequence contexts (Supplemental Fig. S6A) when compared to the genome distribution of cytosines in CG, CHG, and CHH contexts of common cytosines among the samples (Supplemental Fig. S6B). CG-DMRs associated with Pi supply were enriched in intergenic regions and TEs, a pattern that is very different from what was observed for DMRs between otu5 and wild type, in which DMRs were mainly observed within genes (Fig. 3I). This suggests that genetic (otu5) and environmental (low-Pi) conditions differentially impact DNA methylation. Only 27 protein-coding genes were differentially methylated between Pi-replete and Pi-deficient plants (Supplemental Table S9). Both DMGs and genes in the vicinity of intergenic regions in which DMRs were observed showed little alterations in gene expression (Supplemental Data Set 6). No overlap of DEGs and DMGs was observed between Pi-replete and Pi-deficient plants (Supplemental Fig. S4B), suggesting that transcriptional regulation of Pi-responsive genes is not linked to differential methylation.
Pi Starvation Induces Differential Trimethylation of H3K4 of a Suite of Pi Starvation Genes
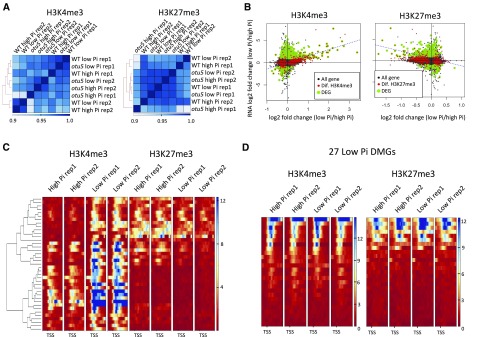
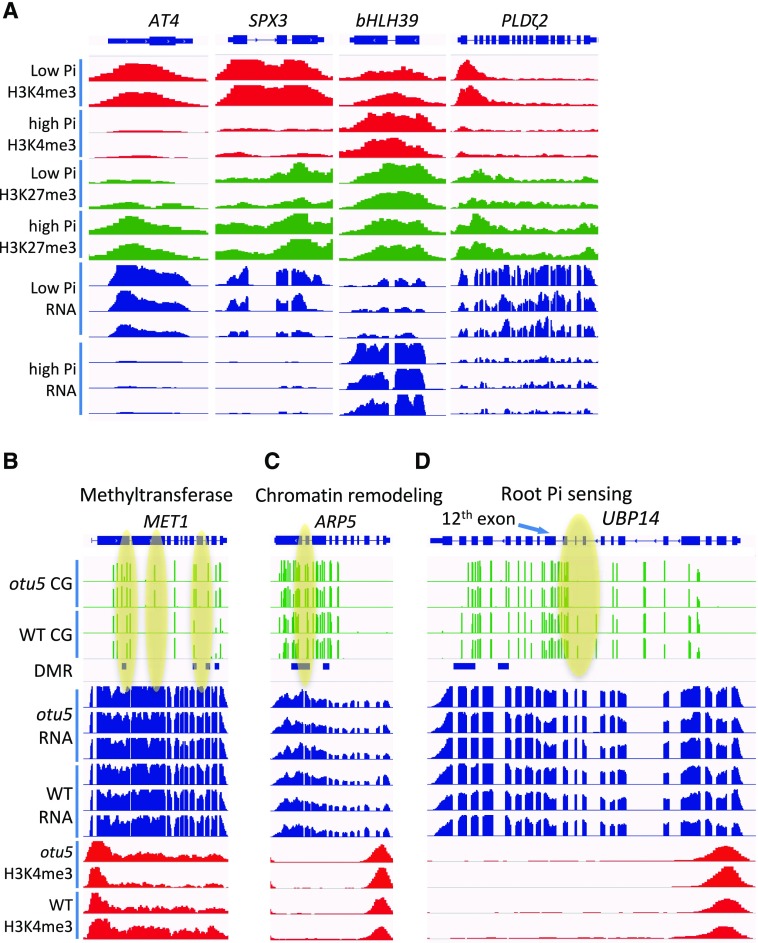
DNA and histone Lys methylation systems are highly interrelated, resulting in an epigenetic code that orchestrates gene activity. In particular, DNA methylation is correlated with the absence of methylation of Lys 4 of histone 3 (H3K4; Cedar and Bergman, 2009). To investigate whether the alterations in DNA methylation are correlated with changes in histone 3 methylation pattern, we determined the level of trimethylated H3K4 (H3K4me3), which is enriched in transcriptionally active promoters, and of trimethylated H3K27 (H3K27me3) as a repressive histone modification in wild type and otu5 plants under Pi-replete and Pi-deficient conditions using chromatin immunoprecipitation (ChIP)-seq. Globally, major differences were neither observed between plants grown with different Pi supply nor between the genotypes (Fig. 4A; Supplemental Fig. S7), indicating that both Pi supply and the otu5 mutation did not alter the genome-wide distribution of these two histone marks. Although no global changes of H3K4me3 and H3K27me3 abundance were observed, it is possible that local changes are associated with the expression or DNA methylation of individual genes. Plotting expression changes against alterations in H3K4me3 and H3K27me3 induced by growth on low Pi revealed a subset of DEGs in which H3K4me3 was positively and H3K27me3 negatively correlated with transcriptional changes in both wild-type and otu5 backgrounds (Fig. 4B; Supplemental Fig. S8). Among 39 low-Pi-induced DEGs that are all upregulated in low-Pi condition with clear change in histone marks, we observed at the transcriptional start site (TSS) a clear enrichment of H3K4me3 (active mark) in low-Pi conditions and a slightly less-clear enrichment of H3K27me3 (repressive mark) in high-Pi conditions (Fig. 4C; Supplemental Fig. S9). Interestingly, there is no clear change of H3K4me3, and H3K27me3 abundance was observed in low-Pi-induced DMGs, indicating that DNA methylation is not tightly associated with H3K4me3 and H3K27me3 (Fig. 4D).
Figure 4.
Analysis of H3K4 and H3K27 trimethylation in wild-type plants grown on low-Pi and high-Pi media. A, Correlation plots for H3K4me3 and H3K27me3 among the samples. B, Scatterplots of differential histone trimethylated genes and differentially expressed genes between Pi-deficient and Pi-replete wild-type plants. The y axis denotes changes in expression between low-Pi and Pi-replete conditions; x axis indicates change in H3K4me3/H3K27me3 abundance between low-Pi and Pi-replete conditions. Low-Pi-specific DEGs are denoted in yellow-green color; genes with differential TSS abundance of the histone mark are shown in red color; the blue dashed line represents the linear regression of DEGs with differential histone mark abundance. C, Heat maps of low-Pi-specific DEGs with differential histone marks at either H3K4me3 or H3K27me3 at TSSs. D, Heat map of H3K4me3 (left) and H3K27me3 (right) abundance at TSSs for low-Pi-specific DMGs.
Very few differentially methylated genes between otu5 mutant and the wild type did show difference in H3K4me3 and H3K27me3 abundance (Supplemental Fig. S10), but significant alterations in H3K4 and H3K27 trimethylation were observed for a small subset of DEGs (Supplemental Table S10). Growth on low-Pi media yielded the smallest difference between the two genotypes. Under both Pi-replete and Pi-deficient conditions, none of the genes relevant for root hair formation were differentially trimethylated at H3K4 between otu5 and the wild type. Only a small subset of genes that showed marked anticorrelation between DNA methylation and mRNA expression exhibited differential H3K4me3 abundance (Supplemental Fig. S11A).
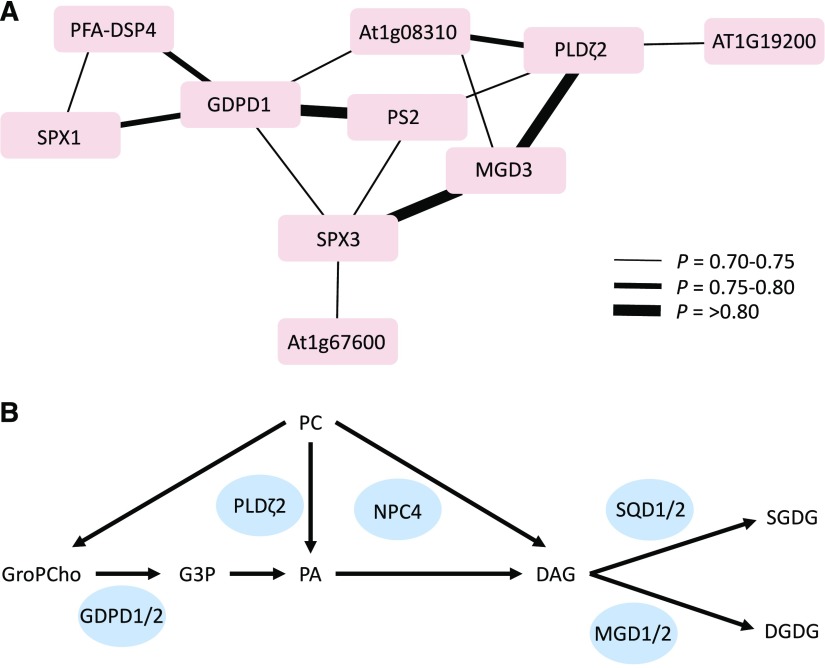
Within the 450 low-Pi-induced DEGs in wild-type plants, 47 genes showed differential H3K4me3 marks at the TSS between Pi-replete and Pi-deficient wild-type plants, the majority of which (37 genes) showed increased abundance of H3K4 trimethylation (Supplemental Data Set 7). This group comprises several highly Pi-deficiency-induced key regulators of Pi homeostasis, such as the noncoding RNAs AT4 and IPS1 as well as the SPX (SYG1/Pho81/XPR1)-domain proteins SPX1 and SPX3. Root-specific coexpression analysis of genes with both differential expression and differential H3K4me3 in low Pi (Supplemental Data Set 7) revealed a cluster of putatively coregulated genes encoding proteins involved in lipid metabolism such as MGD3, GDPD1, PLDζ2, the phosphatase PFA-DSP4, and three proteins of unknown functions, which may represent novel players in this process (Fig. 5A; Supplemental Fig. S11B). A subset of these genes have proven roles in membrane lipid remodeling (Fig. 5B), a process that recycles Pi from phospholipids, which is strongly induced upon Pi starvation (Supplemental Fig. S11B; Nakamura, 2013). Among the genes that showed both decreased expression and H3K4me3 upon Pi starvation, three genes, IRON-RESPONSIVE PROTEIN1 (IRP1), IRP6, and bHLH39, have been shown to be strongly induced by iron starvation (Rodríguez-Celma et al., 2013) and may constitute a regulatory unit that prevents the accumulation of toxic iron levels under conditions of Pi starvation (Fig. 6A; Supplemental Fig. S11C). A subset of nine low-Pi DEGs was differentially trimethylated at H3K27 (Supplemental Data Set 7). Most interestingly, a suite of seven low-Pi DEGs showed anticorrelated H3K4 and H3K27 trimethylation at their promoters (Supplemental Data Set 7), including NPC4, PFA-DSP2, PFA-DSP4, IPS1, and the Pi transporter Pht1;4. It can thus be concluded that differential trimethylation of H3K4 and H3K27 is involved in the transcriptional regulation of genes with critical function in Pi homeostasis.
Figure 5.
Differentially H3K4 trimethylated genes with putative functions in membrane lipid remodeling. A, Coexpression network. The network was constructed with the MACCU toolbox (http://maccu.sourceforge.net) against a database derived from root-related experiments. B, Scheme for membrane lipid remodeling upon Pi deficiency. Genes that are differentially trimethylated at H3K4 are framed in boxes. GroPCho, Glycerophosphocholine; PC, phosphatidylcholine; G3P, glycerol-3-phosphate; PA, phosphatidic acid; DAG, diacylglycerol; DGDG, galactolipid digalactosyldiacylglycerol; SQDG, sulfoquinovosyldiacylglycerol.
Figure 6.
A, Genome browser snapshots showing the abundance of H3K4me3, H3K27me3, and transcript levels of genes that are differentially expressed between plants grown under high-Pi and low-Pi conditions. B to D, Snapshots showing CG methylation, gene expression, and abundance of H3K4me3 of genes with differentially methylated regions between wild-type and otu5 mutant plants: MET1 (B), ARP5 (C), and UBP14 (D).
DISCUSSION
Homozygous otu5 plants exhibit a pleiotropic phenotype that comprises alterations in root architecture and root hair development. Chromatin-level regulation of root traits, particularly those responsive to Pi-deficient conditions, has been inferred from previous studies. A phenotype similar to that of otu5 was observed for arp6, a mutant which harbors a defect in a subunit of the chromatin remodeler SWR1 that mediates eviction of the canonical histone H2A-H2B dimer and deposition of the H2A.Z-H2B pair (Smith et al., 2010). Furthermore, the histone deacetylase HDA19 was identified as regulator of epidermal cell length, root hair frequency, and several Pi-responsive genes (Chen et al., 2015).
A suite of genes that are likely to contribute to the root hair phenotype was found to have significant G × E interactions (Supplemental Table S6). This group comprised several proteins that are essential for the assembly, modification, and growth of cell walls. Notably, several genes involved in DNA methylation such as MET1, NRPD2A, DCL3, and DCL4 (Chan et al., 2005; Matzke and Mosher, 2014) were differentially methylated in otu5 (Fig. 6B). This may introduce a feed-forward loop that amplifies the methylation of target genes. Similarly, several genes involved in chromatin remodeling (INO80, ARP5, CHR3, CHR4, CHR6, CHR20, and SDG3) were differentially methylated and may alter the activity of downstream genes at the transcriptional level or indirectly via impacting the expression of regulatory proteins (Fig. 6C). It can thus be speculated that effects on chromatin structure are amplified and diversified in the otu5 mutant by altering methylation levels of genes that mediate such changes.
Notably, the expression of the genes that showed significant G × E interactions does not appear to be regulated by mC or histone Lys methylation. Inspection of GC meta plots shows that GC methylation is similar among all four groups with minor differences between wild-type and mutant plants downstream of the TSS (Supplemental Fig. S3B). Meta plots of H3K4me3 also show similarity between the four sample groups with a slightly higher trimethylation abundance under low-Pi conditions (Supplemental Fig. S3C). H3K27me3 meta plots reveal a slightly higher level of trimethylation in otu5 samples, which was, however, not negatively correlated with gene expression. It can thus be inferred that the observed G × E interactions are not directly caused by changes in the DNA or histone Lys methylation patterns.
Most interestingly, several genes encoding proteins with validated functions in cellular Pi homeostasis and/or root hair morphogenesis were differentially methylated between otu5 and the wild type. This group includes the SUMO E3 ligase SIZ1 (Miura et al., 2011), the phospholipases PLDζ1 and PLDζ2 (Fig. 6A; Li et al., 2006; Miura et al., 2011), the phosphatidylinositol-4-phosphate 5-kinase At1g60890 (Stenzel et al., 2008; Wada et al., 2015), the phosphatidylinositol-4-phosphate phosphatase RHD4 (Stenzel et al., 2008; Thole et al., 2008; Wada et al., 2015), the ubiquitin-specific protease UBP14 (Li et al., 2010), the patterning protein TRN1 (Kwak et al., 2015), and the P5-type ATPase PDR2, a key player in root Pi sensing (Wang et al., 2010; Müller et al., 2015).
Our study showed a very limited impact of Pi starvation on DNA methylation levels that did not correlate with changes in transcriptional profiles, indicating that DNA methylation has a minor effect on gene activity at the transcriptional level under these conditions. Similar to what has been observed by Secco et al. (2015) for rice, Pi deficiency-induced DMRs were mainly located in TEs or intergenic regions. However, subtle differences in the abundance of proteins caused by differential methylation and/or other changes at the chromatin level may still affect the phenotypic readout. For example, a synonymous substitution in the third position of a conserved Pro in the 12th exon of UBP14 led to a minor decrease in protein abundance, which compromised root hair elongation under Pi-deficient but not under Pi-replete conditions (Fig. 6D; Li et al., 2010). In both mammals and plants, DNA methylation is closely interlinked with the positioning of nucleosomes (Chodavarapu et al., 2010; Zhu et al., 2013). A plausible mechanism for the action of OTU5 in roots could thus lie in the positioning of nucleosomes, probably as a component of a chromatin remodeler complex, which may affect posttranscriptional processes that alter the activity of root-hair-related genes.
Whereas changes in mC were largely incongruent with alterations in gene expression between genotypes and growth conditions, Pi starvation induced differential H3K4me3 profiles that corresponded to changes in the abundance of transcripts of a subset of Pi-responsive genes. Alterations in H3K4me3 abundance and expression level were homodirectional, indicating that these changes could be functionally linked. Moreover, for a subset of low-Pi DMRs, such homodirectional changes in H3K4me3 and transcription were associated with anticorrelated changes in H3K27me3. This subset comprised several regulatory factors of crucial importance for Pi homeostasis, suggesting that H3K4 methylation is a critical component of the regulatory machinery of the Pi-deficiency response. It should also be noted that the majority of transcriptionally Pi-responsive genes did not show alteration in histone Lys methylation, indicating that Pi starvation-induced histone modification is not necessarily associated with changes in transcription.
Differential H3K4 and H3K27 trimethylation affected disparate components of the PSR. The recycling of Pi via membrane lipid remodeling plays a central role in the recalibration of Pi homeostasis. This is of particular interest, as it has been shown that mutations in several genes involved in this process, i.e. NPC4, PS2, or SQD2, resulted in Pi-specific root hair defects (Chandrika et al., 2013). Expression of these genes was partially compromised in mutants with defects in the plant homeodomain protein ALFIN-LIKE6 (AL6). AL6 was identified in a forward genetic screen for Pi-specific defects in root hair development and shown to bind to di- and trimethylated H3K4 with its plant homeodomain (Chandrika et al., 2013; Lee et al., 2009). Similar to Pi-deficient otu5 plants, al6 mutants develop very short root hairs when grown on low-Pi media. It was hypothesized that AL6 acts by stabilizing transcripts encoding proteins required for the rapid tip growth of root hairs from Pi-deficient plants (Chandrika et al., 2013). Together, these results underline a close linkage of Pi-deficiency-induced H3K4/H3K27 trimethylation, reprogramming of lipid metabolism, and root hair morphogenesis.
Differential H3K4me3 abundance was not restricted to up-regulated genes. Three of the down-regulated genes with reduced H3K4me3 levels upon Pi deficiency, i.e. the transcription factor bHLH39 and two genes from a recently identified subset of highly iron-responsive small proteins designated as IRP1 and IRP6, were reportedly strongly induced in response to iron deficiency (Rodríguez-Celma et al., 2013). For bHLH39, also differential H3K27 trimethylation was observed that was anticorrelated with H3K4me3. In Pi-deficient plants, iron acquisition genes are generally down-regulated to compensate for increased iron levels (Misson et al., 2005; Lan et al., 2012). Down-regulation of iron acquisition genes is a consequence of oxidatively precipitated iron, which is deposited in the meristematic and elongation zones of the root as a prerequisite of Pi deficiency-induced determinacy of primary root growth (Müller et al., 2015; Hoehenwarter et al., 2016, Mora-Macías et al., 2017). While a molecular function has yet to be assigned to the two IRPs, bHLH39 was shown to form a heterodimer with the master regulator of iron homeostasis FIT (bHLH29), which controls the expression of a large subset of genes involved in the acquisition and homeostatic control of iron (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2008). It can thus be assumed that the small iron-related cluster identified here is involved in repressing iron acquisition genes in response to Pi starvation.
In summary, it can be deduced from the present data that OTU5 is required for the coordinated expression of genes involved in root hair morphogenesis and critical for the orchestration of Pi signaling on gene expression, thereby impacting the root phenotype of both Pi-replete and Pi-deficient plants. Whole-genome bisulfite sequencing revealed that OTU5 defines a distinct DNA methylation profile by targeting local genomic regions, altering the methylation of genes with functions in root hair morphogenesis and Pi signaling. The methylation patterns and gene expression profiles are not directly associated with each other, suggesting that subtle changes in posttranscriptional processes such as mRNA maturation or stability may contribute to the phenotype. Several, but not all, Pi-responsive genes showed anticorrelated H3K4 and H3K27 methylation marks, demonstrating that histone modifications are important for the activity of a subset of PSR genes. For Pi-dependent DNA methylation, no such correlation with gene expression was observed. In addition, DMRs were enriched in transposable elements and depleted from coding regions, indicating that mC has a minor or indirect effect on the activity of PSR genes and is likely to be critical for genome stability and integrity.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis (Arabidopsis thaliana; Col-0) was used as the wild type. otu5-1 is a T-DNA-inserted knockout mutant (Salk_032407) requested from the Arabidopsis Biological Resource Center at The Ohio State University (Columbus, OH). No full-length transcript or protein expression could be detected in the mutant. To complement the otu5 mutant, a construct encoding a GS-tagged OTU5 was made by PCR amplification and mobilization of the OTU5 coding region from a pET28a expression vector into pKNGSTAP (VIB, Belgium) using the gateway cloning method (Radjacommare et al., 2014). The Arabidopsis transformation was performed as previously described, and homozygous complementation lines were selected from T3 plants (Clough and Bent, 1998). Seeds were surface sterilized and germinated on media containing KNO3 (5 mm), MgSO4 (2 mm), Ca(NO3)2 (2 mm), KH2PO4 (2.5 mm), H3BO3 (70 µm), MnCl2 (14 µm), ZnSO4 (1 µm) CuSO4 (0.5 µm), CoCl2 (0.01 µm), Na2MoO4 (0.2 µm) and FeEDTA (40 µm), solidified with 0.4% Gelrite pure (CP Kelco), 1.5% Suc, and 1 g/L MES hydrate. The pH was adjusted to 5.5. Seeds were sown on petri plates and stratified for 2 d at 4°C in the dark and then transferred to a growth chamber at 21°C under continuous illumination (50 µmol m−2 s−1). Low-Pi conditions were obtained by growing plants on media containing 2.5 μm KH2PO4, while the Pi concentration of Pi-replete medium was 2.5 mm KH2PO4. The lower concentration of potassium due to the reduced KH2PO4 concentration was compensated for by the addition of KCl. Plants were grown on Pi-sufficient (2.5 mm) media for 7 d and transferred to Pi-sufficient and low (2.5 μm) Pi media, respectively, for another 7 d.
Root Hair Length and Density
Confocal images with a scale bar of 100 µm at 10× resolution were used for measuring the root hair length. A Zeiss Discovery V12 microscope equipped with an ocular scale bar was used for measuring root hair density at 2 to 6 mm from the tip of primary root.
Interaction between Genotype and Pi Availability
Two-way ANOVA analysis with two levels of categories (E, Pi availability, and G, presence of functional OTU5) was performed for gene expression data. ANOVA analysis was conducted in R using the ANOVA function in the stats package (R Core Team, 2015). In order to focus on genes showing substantial differences between samples, only genes with at least a 2-fold change in expression level among all samples were included in the analysis. Genes with P < 0.01 in the interaction between genotype and Pi availability were deemed to have robust G × E interaction.
WGCNA
WGCNA was performed using the R package on transcriptome data following the standard method described on the authors’ website (Langfelder and Horvath, 2008). This unsupervised and unbiased analysis identifies distinct coexpression gene modules by clustering transcripts with similar expression patterns across samples. Transcriptionally variable genes with at least 2-fold changes between samples in the analysis were selected for the analysis. The genes were hierarchically clustered based on a dissimilarity measure of topological overlap matrix. The resulting dendrogram was used for module detection (minimum size 40; height cutoff 0.8). Gene modules were labeled in unique colors. Unassigned genes were labeled in gray.
RNA-Seq Library Preparation and Data Analysis
Total RNA was isolated from roots of 14-d-old plants grown under Pi-replete and Pi-deficient conditions using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. Equal amounts of total RNA were collected from three independent experiments for library construction. cDNA libraries for sequencing were prepared from 5 μg of purified RNA using Illumina TruSeq RNA Sample Preparation kit following the manufacturer's instructions. cDNA libraries were sequenced with Illumina HiSeq 2000 system using 101-bp single-end reads. Transcript abundance was calculated by mapping reads to the Arabidopsis TAIR10 genome using Tophat2 (Kim et al., 2013). The expression value of a gene was computed as RPKM (reads per kilobase of exons per million mapped reads). Differentially expressed genes between samples were identified using the Cuffdiff program (Trapnell et al., 2013), with at least 2-fold change in expression and a false discovery rate (FDR) <5% as criteria.
Profiling Genome-Wide DNA Methylation
Genomic DNA was extracted by grinding root tissue in liquid nitrogen followed by purification with the DNeasy Plant Mini Kit (Qiagen). For whole-genome bisulfite sequencing, 1 µg of fragmented DNA was first ligated with premethylated TruSeq DNA adapters (Illumina). The DNA fragments were then converted using the EZ DNA Methylation-Gold kit (Zymo Research) using the manufacturer’s protocol. The resulting libraries were sequenced using the Illumina/Solexa sequencing technology (HiSeq 2000 sequencer; Illumina). Bisulfite converted reads were aligned to the Arabidopsis reference genome (TAIR10) using BS-Seeker2 (Guo et al., 2013). Genome-wide DNA methylation profiles were generated by determining methylation levels for each cytosine in the genome. Because bisulfite treatment converts unmethylated cytosines (Cs) to thymines (Ts) after PCR amplification, the methylation level at each cytosine was estimated as #C/(#C + #T), where #C is the number of methylated reads and #T is the number of unmethylated reads. The methylation level per cytosine serves as an estimate of the percentage of cells that have a methylated cytosine at this locus. We only included cytosines that are covered by at least four reads. By spike in Lambda Phage DNA, we estimated the bisulfite conversion rate to be ≥99.6% in our libraries (see Supplemental Table S7).
Identifying DMRs
To identify putative DMRs, a nonoverlapping tiling window approach was used for genome-wide screening. Regions (200 bp) containing at least five cytosines, each of which covered by least four reads in all compared samples, were included in the analysis. To be considered as DMRs, the region had to (1) show a difference in average methylation level of ≥20% between the two groups, and (2) an FDR of less than 10%. To estimate the FDR, we constructed simulated methylomes with the same coverage as the real samples (Chen et al., 2013). For each cytosine site in each simulated sample, we simulated the reads (C as methylated and T as unmethylated) based on the average methylation level (Pm) of this cytosine site and coverage (n) from all real samples. The number of methylated reads Cs is a random number based on binomial distribution B (n, Pm). Since the simulated methylomes are constructed to have no methylation difference between two comparison groups, DMRs identified from the comparison of the simulated methylomes are considered false positives. FDR was estimated as the number of DMRs found in simulated methylomes (false positives) divided by that from the real samples (positive prediction). See Supplemental Table S11 for the cutoffs of Δ methylation level corresponding to FDR ≤ 10%.
ChIP-Seq Library Preparation and Data Analysis
ChIP was performed using root (∼1.5 g) of Arabidopsis (Col-0) following the published protocols (Langmead and Salzberg, 2012). For immunoprecipitation, 5 μg of antibodies against H3-core (Ab1791; Abcam), H3K4me3 (Ab8580; Abcam), and H3K27me3 (07-499; Upstate) were added to a 600-μL solution containing diluted chromatin extracts (∼25 μg) and incubated overnight at 4°C with gentle rotation. Immunoprecipitated chromatin-DNA (IP-DNA) and ∼5 μg of input chromatin-DNA (without IP) were subjected to reverse cross link and protease K treatment. DNA was purified using the MinElute PCR purification kit (Qiagen) for sequencing libraries preparation. DNA was quantified by Qubit and analyzed using the Agilent High Sensitivity DNA analysis chip (5067-4626). Libraries for ChIP-seq were prepared following KAPA Library Preparation Kits Illumina series (#KK8201; KAPA Biosystems) according to the manufacturer’s protocol. To summarize, 2 to 5 ng ChIPed DNA was end-repaired, “A” base was added to the 3′ end of DNA fragment and then ligated to the barcoded adapters. The products are purified and enriched with 12 cycles of PCR to create the final double-stranded cDNA library. Final libraries were analyzed using the Agilent High Sensitivity DNA analysis chip to estimate the quality and were then quantified by qPCR using the KAPA Library Quantification Kit (KK4824; KAPA Biosystems). The ChIP-seq libraries were sequenced with the Illumina HiSeq 2500 system. The reads were aligned to the TAIR10 Arabidopsis genome using Bowtie2 version 2.2.4 (Langmead and Salzberg, 2012; see Supplemental Table S12 for mapping statistics). Alignments were converted into coverage files (bigWig format) using the bamCoverage function in deepTools (Ramírez et al., 2014) and normalized to 1X. Correlation plots of ChIP-seq were generated using the multiBigwigSummary function followed by plotCorrelation. To generate heat maps of reads abundance associated with TSS, the reads density around TSS (−500 to +1,500 bp) was computed using the computeMatrix function, followed by the plotHeatmap. Genes with differentially binding of histone marks were detected by t test P value of the average density. A cutoff of P < 0.05 with differential abundance ≥2-fold and abundance ≥1 RPKM in one or more samples were used to select genes showing differential histone modification binding.
Coexpression Analysis
Gene clustering was performed using the MACCU software package (http://maccu.sourceforge.net/) to build coexpression clusters based on pairwise coexpression relationships of genes with Pearson coefficients greater than or equal to 0.70. In order to capture the coexpression relationships specifically in roots, Pearson coefficients were computed based on robust multiarray averaged array data derived from publically available root-specific experiments downloaded from NASCArrays (http://affymetrix.arabidopsis.info/). Visualization of the networks was performed with the Cytoscape software version 3.
Accession Numbers
All sequence data can be downloaded from NCBI Gene Expression Omnibus under accession number GSE81407 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE81407).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Weighted gene coexpression network analysis groups genes into distinct modules using all samples with the y axis corresponding to coexpression distance between genes.
Supplemental Figure S2. Genes that are differentially expressed in otu5 plants.
Supplemental Figure S3. The expression, CG methylation, H3K4me3 abundance, and H3K27 abundance of 48 genes that showed significant interaction between environments and genotypes.
Supplemental Figure S4. Association between differentially methylated genes and differentially expressed genes.
Supplemental Figure S5. Heat map of differentially methylated and differentially expressed genes.
Supplemental Figure S6. Pie chart of differentially methylated cytosines in low-Pi conditions and common cytosine sites among samples.
Supplemental Figure S7. Correlation plots (using a Pearson correlation measure) for histone modification abundance around the TSS.
Supplemental Figure S8. Scatterplots of differentially histone marked genes and differentially expressed genes in otu5 plants under low-Pi conditions.
Supplemental Figure S9. ChIP-seq analysis on low-Pi and control conditions in wild-type background.
Supplemental Figure S10. ChIP-seq analysis of wild type and otu5 mutant in control condition.
Supplemental Figure S11. Snapshot of genome browser showing CG methylation, gene expression, and abundance of H3K4me3 for genes with differential methylation in otu5.
Supplemental Table S1. Two-way ANOVA analysis of genotype × environment interactions for various root phenotypes.
Supplemental Table S2. Mapping statistics of RNA-seq libraries.
Supplemental Table S3. Mean expression standard deviations of all four groups.
Supplemental Table S4. OTU5-dependent root development-related DEGs specific to low-Pi conditions.
Supplemental Table S5. Genes that showed significant between environment × phenotype interactions.
Supplemental Table S6. Genes with significant genotype × environment interactions that are preferentially expressed in root hairs.
Supplemental Table S7. Mapping statistics of whole-genome bisulfite sequencing libraries.
Supplemental Table S8. Ubiquitin-related DMRs (otu5/wild type).
Supplemental Table S9. List of low Pi-dependent DMGs.
Supplemental Table S10. Gene counts for DEGs and DMGs with differential histone marks abundant in different comparisons.
Supplemental Table S11. Δ Methylation cutoff for DMR identification.
Supplemental Table S12. Mapping statistics of ChIP-seq libraries.
Supplemental Data Set 1. Gene list for WGCNA modules.
Supplemental Data Set 2. Genes that are differentially expressed between otu5 and the wild type.
Supplemental Data Set 3. List of OTU5-specific DMRs.
Supplemental Data Set 4. Gene list of OTU5-dependent DMGs.
Supplemental Data Set 5. List of low Pi-specific DMRs.
Supplemental Data Set 6. List of genes adjacent to the low-Pi DMRs.
Supplemental Data Set 7. H3K4me3 and H3K27me3 TSS abundances of genes with differential mRNA expression of low Pi.
Acknowledgments
We thank Drs. Marjori Matzke and Yuki Nakamura (IPMB, Taiwan) for critical comments on the manuscript.
Footnotes
This work was supported by a grant from Academia Sinica to H.F. and W.S. (grant no. AS-102-TP-B05) and by grants from the Ministry of Science and Technology, Taiwan (MOST-103-2313-B-001-003-MY3 and MOST-103-2633-B-001-002) to P.-Y.C.
Articles can be viewed without a subscription.
References
- Aina R, Sgorbati S, Santagostino A, Labra M, Ghiani A, Citterio S (2004) Specific hypomethylation of DNA is induced by heavy metals in white clover and industrial hemp. Physiol Plant 121: 472–480 [Google Scholar]
- Bates TR, Lynch JP (2000) The efficiency of Arabidopsis thaliana (Brassicaceae) root hairs in phosphorus acquisition. Am J Bot 87: 964–970 [PubMed] [Google Scholar]
- Cedar H, Bergman Y (2009) Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet 10: 295–304 [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6: 351–360 [DOI] [PubMed] [Google Scholar]
- Chandrika NNP, Sundaravelpandian K, Yu SM, Schmidt W (2013) ALFIN-LIKE 6 is involved in root hair elongation during phosphate deficiency in Arabidopsis. New Phytol 198: 709–720 [DOI] [PubMed] [Google Scholar]
- Chen PY, Ganguly A, Rubbi L, Orozco LD, Morselli M, Ashraf D, Jaroszewicz A, Feng S, Jacobsen SE, Nakano A, et al. (2013) Intrauterine calorie restriction affects placental DNA methylation and gene expression. Physiol Genomics 45: 565–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Wu K, Schmidt W (2015) The histone deacetylase HDA19 controls root cell elongation and modulates a subset of phosphate starvation responses in Arabidopsis. Sci Rep 5: 15708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. (2010) Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CS, Sano H (2007) Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol Genet Genomics 277: 589–600 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Prescott H, Dolan L (2015) Intensity of a pulse of RSL4 transcription factor synthesis determines Arabidopsis root hair cell size. Nat Plants 1: 15138. [DOI] [PubMed] [Google Scholar]
- Desnos T. (2008) Root branching responses to phosphate and nitrate. Curr Opin Plant Biol 11: 82–87 [DOI] [PubMed] [Google Scholar]
- Giehl RF, Gruber BD, von Wirén N (2014) It’s time to make changes: Modulation of root system architecture by nutrient signals. J Exp Bot 65: 769–778 [DOI] [PubMed] [Google Scholar]
- Gilroy S, Jones DL (2000) Through form to function: Root hair development and nutrient uptake. Trends Plant Sci 5: 56–60 [DOI] [PubMed] [Google Scholar]
- Grierson CS, Roberts K, Feldmann KA, Dolan L (1997) The COW1 locus of Arabidopsis acts after RHD2, and in parallel with RHD3 and TIP1, to determine the shape, rate of elongation, and number of root hairs produced from each site of hair formation. Plant Physiol 115: 981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Fiziev P, Yan W, Cokus S, Sun X, Zhang MQ, Chen PY, Pellegrini M (2013) BS-Seeker2: A versatile aligning pipeline for bisulfite sequencing data. BMC Genomics 14: 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Chen T, Zhu JK (2011) Regulation and function of DNA methylation in plants and animals. Cell Res 21: 442–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehenwarter W, Mönchgesang S, Neumann S, Majovsky P, Abel S, Müller J (2016) Comparative expression profiling reveals a role of the root apoplast in local phosphate response. BMC Plant Biol 16: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P (2004) FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett 577: 528–534 [DOI] [PubMed] [Google Scholar]
- Kim DW, Lee SH, Choi SB, Won SK, Heo YK, Cho M, Park YI, Cho HT (2006) Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18: 2958–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RC, Storto PD (1988) The role of the otu gene in Drosophila oogenesis. BioEssays 8: 18–24 [DOI] [PubMed] [Google Scholar]
- Krichevsky A, Zaltsman A, Lacroix B, Citovsky V (2011) Involvement of KDM1C histone demethylase-OTLD1 otubain-like histone deubiquitinase complexes in plant gene repression. Proc Natl Acad Sci USA 108: 11157–11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak SH, Song SK, Lee MM, Schiefelbein J (2015) TORNADO1 regulates root epidermal patterning through the WEREWOLF pathway in Arabidopsis thaliana. Plant Signal Behav 10: e1103407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labra M, Ghiani A, Citterio S, Sgorbati S, Sala F, Vannini C, Ruffini-Castiglione M, Bracale M (2002) Analysis of cytosine methylation pattern in response to water deficit in pea root tips. Plant Biol 4: 694–699 [Google Scholar]
- Lan P, Li W, Lin WD, Santi S, Schmidt W (2013) Mapping gene activity of Arabidopsis root hairs. Genome Biol 14: R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P, Li W, Schmidt W (2012) Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol Cell Proteomics 11: 1156–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2008) WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A (2000) Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science 289: 2350–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WY, Lee D, Chung WI, Kwon CS (2009) Arabidopsis ING and Alfin1-like protein families localize to the nucleus and bind to H3K4me3/2 via plant homeodomain fingers. Plant J 58: 511–524 [DOI] [PubMed] [Google Scholar]
- Li WF, Perry PJ, Prafulla NN, Schmidt W (2010) Ubiquitin-specific protease 14 (UBP14) is involved in root responses to phosphate deficiency in Arabidopsis. Mol Plant 3: 212–223 [DOI] [PubMed] [Google Scholar]
- Li M, Qin C, Welti R, Wang X (2006) Double knockouts of phospholipases Dzeta1 and Dzeta2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol 140: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394–408 [DOI] [PubMed] [Google Scholar]
- Meaney MJ, Szyf M (2005) Environmental programming of stress responses through DNA methylation: Life at the interface between a dynamic environment and a fixed genome. Dialogues Clin Neurosci 7: 103–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Sato A, Ohta M, Furukawa J (2011) Increased tolerance to salt stress in the phosphate-accumulating Arabidopsis mutants siz1 and pho2. Planta 234: 1191–1199 [DOI] [PubMed] [Google Scholar]
- Mora-Macías J, Ojeda-Rivera JO, Gutiérrez-Alanís D, Yong-Villalobos L, Oropeza-Aburto A, Raya-González J, Jiménez-Domínguez G, Chávez-Calvillo G, Rellán-Álvarez R, Herrera-Estrella L (2017) Malate-dependent Fe accumulation is a critical checkpoint in the root developmental response to low phosphate. Proc Natl Acad Sci USA 114: E3563–E3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Toev T, Heisters M, Teller J, Moore KL, Hause G, Dinesh DC, Bürstenbinder K, Abel S (2015) Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev Cell 33: 216–230 [DOI] [PubMed] [Google Scholar]
- Nakada S, Tai I, Panier S, Al-Hakim A, Iemura S, Juang YC, O’Donnell L, Kumakubo A, Munro M, Sicheri F, et al. (2010) Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466: 941–946 [DOI] [PubMed] [Google Scholar]
- Nakamura Y. (2013) Phosphate starvation and membrane lipid remodeling in seed plants. Prog Lipid Res 52: 43–50 [DOI] [PubMed] [Google Scholar]
- R Core Team (2015) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Radjacommare R, Usharani R, Kuo CH, Fu H (2014) Distinct phylogenetic relationships and biochemical properties of Arabidopsis ovarian tumor-related deubiquitinases support their functional differentiation. Front Plant Sci 5: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T (2014) deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42: W187–W191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem 78: 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Celma J, Pan IC, Li W, Lan P, Buckhout TJ, Schmidt W (2013) The transcriptional response of Arabidopsis leaves to Fe deficiency. Front Plant Sci 4: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Henao JE, Vélez-Bermúdez IC, Schmidt W (2016) The regulation and plasticity of root hair patterning and morphogenesis. Development 143: 1848–1858 [DOI] [PubMed] [Google Scholar]
- Secco D, Wang C, Shou H, Schultz MD, Chiarenza S, Nussaume L, Ecker JR, Whelan J, Lister R (2015) Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife 4: 10.7554/eLife.09343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AP, Jain A, Deal RB, Nagarajan VK, Poling MD, Raghothama KG, Meagher RB (2010) Histone H2A.Z regulates the expression of several classes of phosphate starvation response genes but not as a transcriptional activator. Plant Physiol 152: 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanisić V, Malovannaya A, Qin J, Lonard DM, O’Malley BW (2009) OTU domain-containing ubiquitin aldehyde-binding protein 1 (OTUB1) deubiquitinates estrogen receptor (ER) alpha and affects ERalpha transcriptional activity. J Biol Chem 284: 16135–16145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer WR, Walsh RC, Kalfayan LJ (1989) Sequence and structure of the Drosophila melanogaster ovarian tumor gene and generation of an antibody specific for the ovarian tumor protein. Mol Cell Biol 9: 5726–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel I, Ischebeck T, König S, Hołubowska A, Sporysz M, Hause B, Heilmann I (2008) The type B phosphatidylinositol-4-phosphate 5-kinase 3 is essential for root hair formation in Arabidopsis thaliana. Plant Cell 20: 124–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole JM, Vermeer JE, Zhang Y, Gadella TW Jr., Nielsen E (2008) Root hair defective4 encodes a phosphatidylinositol-4-phosphate phosphatase required for proper root hair development in Arabidopsis thaliana. Plant Cell 20: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticconi CA, Abel S (2004) Short on phosphate: Plant surveillance and countermeasures. Trends Plant Sci 9: 548–555 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez SM, Ricardi MM, Dorosz JG, Fernandez PV, Nadra AD, Pol-Fachin L, Egelund J, Gille S, Harholt J, Ciancia M, et al. (2011) O-glycosylated cell wall proteins are essential in root hair growth. Science 332: 1401–1403 [DOI] [PubMed] [Google Scholar]
- Wada Y, Kusano H, Tsuge T, Aoyama T (2015) Phosphatidylinositol phosphate 5-kinase genes respond to phosphate deficiency for root hair elongation in Arabidopsis thaliana. Plant J 81: 426–437 [DOI] [PubMed] [Google Scholar]
- Wang X, Yan X, Liao H (2010) Genetic improvement for phosphorus efficiency in soybean: a radical approach. Ann Bot 106: 215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong-Villalobos L, González-Morales SI, Wrobel K, Gutiérrez-Alanis D, Cervantes-Peréz SA, Hayano-Kanashiro C, Oropeza-Aburto A, Cruz-Ramírez A, Martínez O, Herrera-Estrella L (2015) Methylome analysis reveals an important role for epigenetic changes in the regulation of the Arabidopsis response to phosphate starvation. Proc Natl Acad Sci USA 112: E7293–E7302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, et al. (2013) Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell 152: 642–654 [DOI] [PMC free article] [PubMed] [Google Scholar]