SlORRM4 is necessary for timely fruit ripening, mitochondrial health, and RNA editing in tomato.
Abstract
RNA editing plays a key posttranscriptional role in gene expression. Existing studies on cytidine-to-uridine RNA editing in plants have focused on maize (Zea mays), rice (Oryza sativa), and Arabidopsis (Arabidopsis thaliana). However, the importance and regulation of RNA editing in several critical agronomic processes are not well understood, a notable example of which is fruit ripening. Here, we analyzed the expression profile of 33 RNA editing factors and identified 11 putative tomato (Solanum lycopersicum) fruit ripening-related factors. A rapid virus-induced gene silencing assay indicated that the organelle RNA recognition motif-containing protein SlORRM4 affected tomato fruit ripening. Knocking out SlORRM4 expression using a clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 genome editing strategy delayed tomato fruit ripening by lowering respiratory rate and ethylene production. Additionally, the expression of numerous genes associated with fruit ripening and mitochondrial functions changed significantly when SlORRM4 was knocked out. Moreover, the loss of SlORRM4 function significantly reduced RNA editing of many mitochondrial transcripts, leading to low-level expression of some core subunits that are critical for mitochondrial complex assembly (i.e. Nad3, Cytc1, and COX II). Taken together, these results indicate that SlORRM4 is involved in RNA editing of transcripts in ripening fruit that influence mitochondrial function and key aspects of fruit ripening.
In flowering plants, cytidine-to-uridine (C-to-U) RNA editing is a posttranscriptional modification that occurs only in plastids and mitochondrial transcripts. The number of RNA editing sites is not conserved among species, varying from over 3,000 editing sites in Selaginella uncinata plastids to only 11 editing sites in Physcomitrella patens mitochondria (Rüdinger et al., 2009; Oldenkott et al., 2014). RNA editing generally converts a set of amino acids to conserved residues that are important for the proper function of organelle-specific proteins (Liu et al., 2013). In addition, RNA editing can affect essential gene translation processes, generating new translation initiation or termination codons and removing stop codons, leading to larger functional proteins (Kadowaki et al., 1995; Grewe et al., 2009). As cellular powerhouses that convert chemical energy into ATP, mitochondria require appropriate DNA maintenance and expression to generate proteins necessary for normal functions, such as respiratory chain complexes, cytochrome c maturation complexes, and translation apparatuses. RNA editing factors are required for mitochondrial RNA editing, and their loss of function impairs mitochondrial functions, causing phenotype defects in plants. These include slower growth, delayed flowering time, and hampered ethylene sensitivity in Arabidopsis (Arabidopsis thaliana) orrm4 and slo2 mutants (Shi et al., 2016), kernels with empty pericarps in maize (Zea mays) dek35, dek36, dek10, emp9, emp7, and emp5 mutants (Liu et al., 2013; Sun et al., 2015; Chen et al., 2017; Qi et al., 2017; Wang et al., 2017; Yang et al., 2017), as well as white-striped leaves/panicles in rice (Oryza sativa) wsp1 mutants (Zhang et al., 2017).
Currently, four families of RNA editing factors are known to be involved in C-to-U editing: pentatricopeptide repeat (PPR) proteins, multiple organellar RNA editing factor (MORF)/RNA editing factor interacting proteins (RIPs), organelle zinc finger1 (OZ1), and organelle RNA recognition motif-containing proteins (ORRMs; Sun et al., 2016). Typically, PPR proteins function as site-specific factors in RNA editing, and over 400 members are present in flowering plants (Small and Peeters, 2000). Numerous PPR genes have been characterized during the past decade, revealing their molecular functions in plant growth and development (Barkan and Small, 2014). Similarly, MORF/RIP members, such as RIP1 protein, are required for efficient editing at most mitochondrial sites, and Arabidopsis rip1 mutants exhibit a dwarf phenotype (Bentolila et al., 2012). Recent data have shown that ORRM proteins influence plant growth and flowering times. One notable member in Arabidopsis, ORRM4, is a major mitochondrial editing factor, with the Atorrm4 mutant exhibiting slower growth and delayed flowering time (Shi et al., 2016). Additionally, ORRM5 participates in both C-to-U editing and RNA splicing (Shi et al., 2017). Previously, it was shown that PSII activity, green pigmentation, and plant/leaf size were decreased by mutation of the AtORRM6 gene, which is required for photosynthetic psbF-C77 site editing. (Hackett et al., 2017). The combined actions of these RNA editing factors exert profound effects on plant organelle biogenesis and, consequently, on photosynthesis or respiration, two processes that play essential roles in plant development and environmental responses (Takenaka et al., 2013).
RNA editing factors have been demonstrated to influence maize seed development, which provides nearly 70% of the food intake for humankind and raw materials for industry (Wang et al., 2012; Liu et al., 2013). Because seeds are highly active in metabolic terms, an impaired energy supply to seeds often results in embryo and endosperm arrest. Indeed, mutants of some RNA editing factors, such as emp7 and dek36, showed defective kernels with empty pericarps in maize (Barkan and Small, 2014; Sun et al., 2015). Fruit ripening is a very complicated process of organ transformation from the unripe stage to the ripe stage, which requires substantial energy to yield an attractive edible fruit (Brady, 1987). However, very few studies have characterized the role of RNA editing factors in fresh fruit ripening. Tomato (Solanum lycopersicum) is the fourth most economically important crop in the world and a classic model for studying fleshy fruit ripening (Lin et al., 2014). Therefore, to further improve our understanding of the various roles that RNA editing factors play in plants, it is necessary to identify and characterize their functions in tomato fruit ripening.
In this study, we identified 11 putative ripening-related RNA editing factors. Transient silencing of these RNA editing factors in tomato fruits suggested that SlORRM4 (of the ORRM family) was involved in fruit ripening and was localized to the mitochondria. Compared with wild-type tomato, clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-mediated SlORRM4 knockout mutants showed dramatic delays in fruit ripening, with significantly decreased respiratory rates and ethylene production. Moreover, loss of SlORRM4 function disrupted RNA editing for several key mitochondrial genes and affected the accumulation of core subunits in the respiratory chain complex. These results shed light on the poorly understood and complicated relationship between RNA editing factors and fruit ripening.
RESULTS
Silencing of the Ripening-Related Protein SlORRM4 Affects Tomato Fruit Ripening, and It Encodes a Mitochondria-Localized RNA Editing Factor
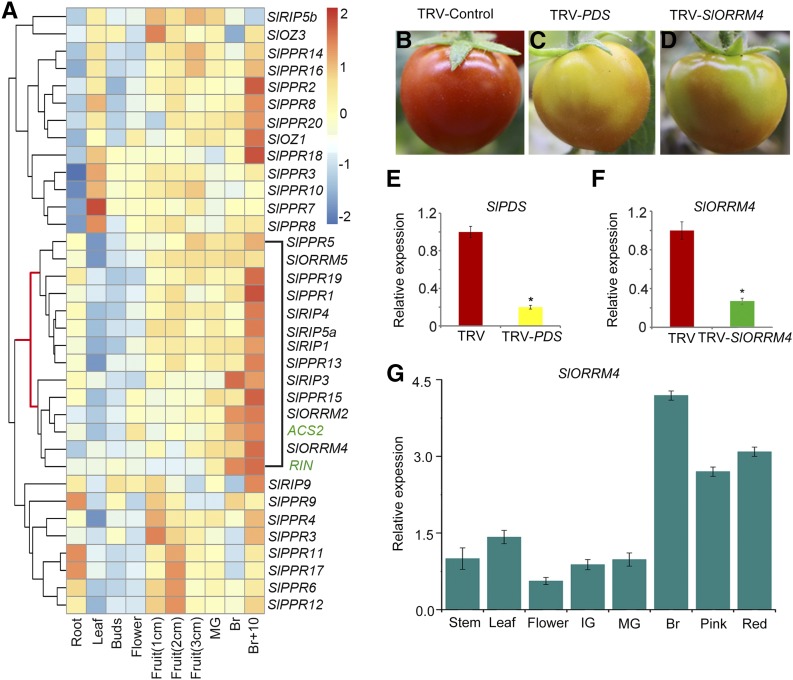
To characterize putative RNA editing factors in tomato, we used the sequences of known Arabidopsis editosome proteins to search the tomato protein database in the SOL Genomics Network (http://solgenomics.net/). We identified 392 putative RNA editing factors in tomato (Supplemental Data Set S1; Supplemental Fig. S2A). Through expression analysis and hierarchical clustering analysis of the putative RNA editing factors, we successfully identified 11 genes as putative ripening-related RNA editing factors in tomato whose transcript levels were associated with fruit ripening (Supplemental Fig. S1), including SlPPR1, SlPPR13, SlPPR15, SlPPR19, SlRIP1, SlRIP3, SlRIP4, SlRIP5a, SlRIP9, SlORRM2, and SlORRM4 (Fig. 1A). Quantitative real-time (qRT)-PCR of the 11 genes (along with ACS2) at three stages of fruit ripening (i.e. the mature green [MG], breaker [Br], and 10 d after breaker [Br+10] stages) further revealed that the transcript levels of the 11 putative RNA editing factors were ripening related (Supplemental Fig. S2B). To further verify the role of the RNA editing factors in fruit ripening, we performed a virus-induced gene silencing (VIGS) assay for these 11 putative ripening-related RNA editing factors (data not shown). VIGS of the phytoene desaturase gene (PDS) was used as a positive control. Interestingly, compared with Tobacco rattle virus (TRV)-infected control fruits, fruits infiltrated with TRV-PDS and TRV-SlORRM4 exhibited patchy color with different shades of yellow and orange (Fig. 1, B–D). The PDS and SlORRM4 mRNA levels in the yellow areas of the TRV-PDS and TRV-SlORRM4 fruits decreased by approximately 80% and 70%, respectively, compared with the red areas in the TRV control fruits (Fig. 1, E and F). These results suggested that transient silencing of SlORRM4 affected fruit ripening.
Figure 1.
Silencing of the ripening-related protein SlORRM4 affects tomato fruit ripening. A, Expression analysis of 33 RNA editing factors across different tomato organs and fruit ripening stages. Internal control genes for fruit ripening are ACS2 and RIN. Each column represents a tomato organ. Three-week sand-grown seedlings were harvested for roots and leaves. Plants were harvested for unopened flower buds (Buds) and fully open flowers (Flowers). Additional flowers were allowed to self-pollinate. The bar on the right represents normalized expression data from high to low (blue to red). Species names are abbreviated as follows: At, Arabidopsis; Sl, tomato. B to D, Tomato infiltrated with vectors TRV-control, TRV-PDS, and TRV-SlORRM4, respectively. E, qRT-PCR analysis of PDS transcripts in TRV-control (red sections) and TRV-PDS (yellow sections) tomato fruits. The internal reference was Actin. Values are means ± sd of three independent replicates. The asterisk indicates P < 0.05 (Student’s t test). F, qRT-PCR analysis of SlORRM4 transcripts in TRV-control (red sections) and TRV-SlORRM4 (yellow sections) tomato fruits. The internal reference was Actin. Values are means ± sd of three independent replicates. The asterisk indicates P < 0.05 (Student’s t test). G, SlORRM4 expression in vegetative and reproductive tomato organs, analyzed with qRT-PCR. Actin was the internal control. Values are means ± sd of three independent replicates. IG, Immature green.
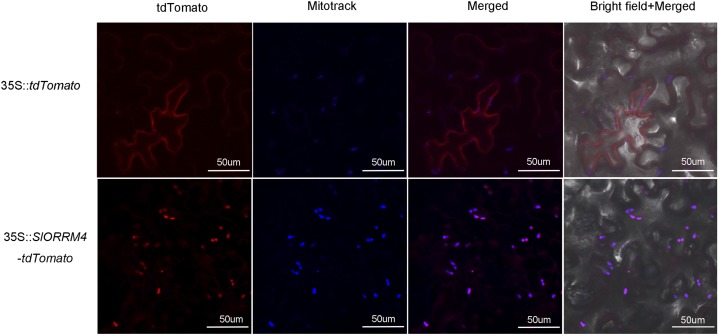
SlORRM4 was predicted to encode an ∼27-kD (252-amino acid) ORRM protein, with a conserved RNA recognition motif (RRM) at the N terminus and a putative Gly-rich (GR) domain at the C terminus (Supplemental Fig. S3). SlORRM4 was expressed in all major tissues during tomato growth and development (Fig. 1G), although the abundance of SlORRM4 mRNA was relatively higher in ripening fruits than in roots, stems, leaves, and young fruits. In particular, the mRNA levels of SlORRM4 were highest at the Br stage of ripening (Fig. 1G). To investigate the subcellular localization of the SlORRM4 protein, the full-length SlORRM4 protein was epitope tagged with the fluorescent tdTomato protein at the C-terminal end and expressed transiently in tobacco leaves. The respective red and green fluorescent signals of SlORRM4:tdTomato and MitoTracker colocalized to the mitochondria (Fig. 2). Thus, SlORRM4 was targeted to the mitochondria.
Figure 2.
Subcellular localization of SlORRM4 in mitochondria. Tobacco leaf epidermal cells were used for subcellular localization. 35::SlORRM4-tdTomato represents the tdTomato fusion protein at the C terminus of the full-length SlORRM4 protein. 35::tdTomato is the control. Mitochondrial signals are marked with MitoTracker Green. Bars = 50 µm.
Generation of Stable Loss-of-Function SlORRM4 Mutants Using the CRISPR/Cas9 Gene Editing System
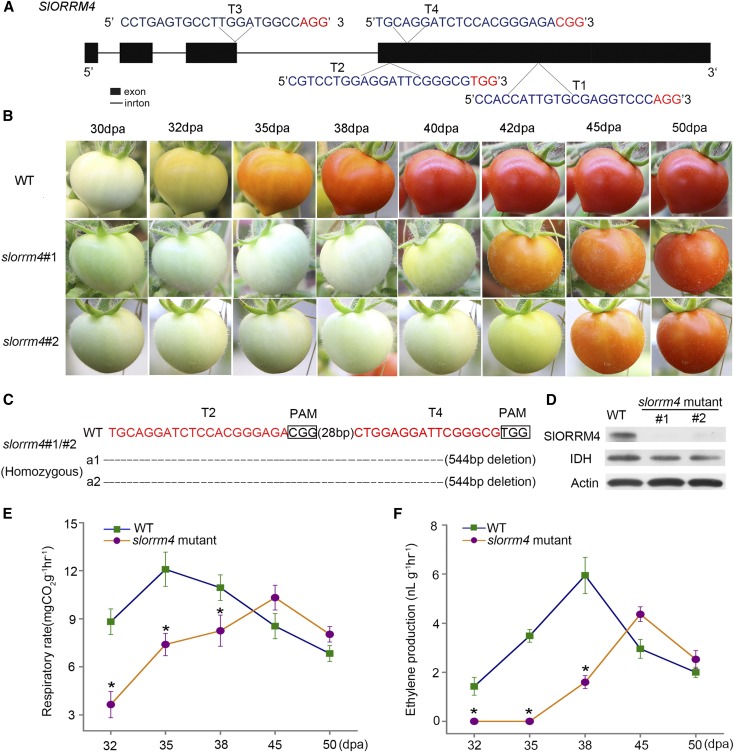
To better define the function of SlORRM4 in tomato fruit ripening, we generated slorrm4 mutants using CRISPR/Cas9 gene editing technology. Four target sites were designed for SlORRM4 (Fig. 3A), and 11 T0-independent transgenic lines were obtained through Agrobacterium tumefaciens-mediated transformation (Supplemental Fig. S4A). Four lines were genome edited, including heterozygous, biallelic, and chimeric mutants (Supplemental Fig. S4, B and C). We found a large 544-bp deletion on one allele at the SlORRM4 locus in the CR-SlORRM4#4 line (Supplemental Fig. S4A). In addition, off-target analysis suggested that no off-target activity occurred in the CR-SlORRM4 T0 lines. For each target, two of the most likely off-target sites were tested (Supplemental Table S3).
Figure 3.
Loss of SlORRM4 function caused great delay of tomato fruit ripening. A, Schematic illustration of four targets in the SlORRM4 genomic sequence. B, slorrm4#1 and slorrm4#2 displayed delayed fruit ripening compared with the wild type (WT). C, Editing analysis of T2 and T4 sites of slorrm4#1 and slorrm4#2 mutants. Red letters indicate the target site, dashes represent deletions, and small rectangular frames indicate the protospacer adjacent motif (PAM). D, SlORRM4 protein expression was inhibited in the slorrm4 mutant. Isocitrate dehydrogenase (IDH) and Actin were the internal controls for mitochondrial protein and nucleocytoplasmic protein, respectively. E, Respiratory rate was reduced and delayed in slorrm4 mutants. Asterisks indicate P < 0.05 (Student’s t test). F, Ethylene production was reduced and delayed in slorrm4 mutants. Asterisks indicate P < 0.05 (Student’s t test).
We then chose two representative T0 lines (CR-SlORRM4#4 and CR-SlORRM4#11) for further analysis. CR-SlORRM4#4 was a chimeric mutant with a 544-bp deletion on one allele, and the CR-SlORRM4#11 mutant had a 1-bp insertion and a 3-bp deletion in both alleles at the T1 site (Supplemental Fig. S4E). Compared with fruit ripening in wild-type plants, fruit ripening in the CR-SlORRM4#4 mutant was delayed dramatically by approximately 8 d. Typically, wild-type fruit changes color around 32 DPA. However, the fruit of the CR-SlORRM4#4 line remained green during that time, and a similar effect was observed in the CR-SlORRM4#11 line (Supplemental Fig. S4D). To further confirm that SlORRM4 promotes tomato fruit ripening, we next identified homogenous mutants with a 544-bp deletion at both alleles of the SlORRM4 locus in the T1 generation of CR-SlORRM4#4 plants (Supplemental Fig. S5A); those mutants were termed slorrm4 mutants (Fig. 3C). Moreover, western-blot data indicated that SlORRM4 was knocked out in the slorrm4 mutants (Fig. 3D).
Loss of SlORRM4 Function Extensively Delayed Tomato Fruit Ripening
The slorrm4 mutants showed comparable plant height, leaf size, flower number, and fruit weight to those of wild-type plants (Supplemental Fig. S5C). However, the slorrm4 mutants showed a more severe flower abscission and a lower fruit-setting ratio compared with wild-type plants, which might have resulted from the abnormal development of floral organs (Supplemental Fig. S5B). Fruit ripening time was much slower in the slorrm4 mutants than in the wild type (Fig. 3B). A comparison of wild-type and slorrm4 mutant fruits at 30, 32, 35, 40, 42, 45, and 50 DPA revealed that fruit color change in the slorrm4 mutant was postponed for approximately 10 d compared with the wild-type fruits (Fig. 3B). Even at 60 DPA, the slorrm4#1 and slorrm4#2 fruits remained orange, while the wild-type fruits were fully red (Supplemental Fig. S6).
We next tested whether fruit ripening delays in slorrm4 mutants correlated with delays in elevated respiratory rates and ethylene production. Notably, fruits from slorrm4 mutants showed significantly decreased respiratory rates compared with wild-type levels at 32, 35, and 38 DPA (Fig. 3E). Similarly, the respiratory peak in slorrm4 mutants was much lower and slower than that in wild-type fruits, suggesting that SlORRM4 knockout inhibited the respiration in ripening tomato fruits (Fig. 3E). Compared with wild-type fruits, ethylene production of slorrm4 fruits also dropped considerably during ripening, and the ethylene peak was significantly lower and occurred later (Fig. 3F). Taken together, these results suggest that the loss of SlORRM4 function delayed fruit ripening in tomato by inhibiting respiration and ethylene production.
SlORRM4 Modulates Key Genes Involved in Fruit Ripening and Mitochondrial Functions
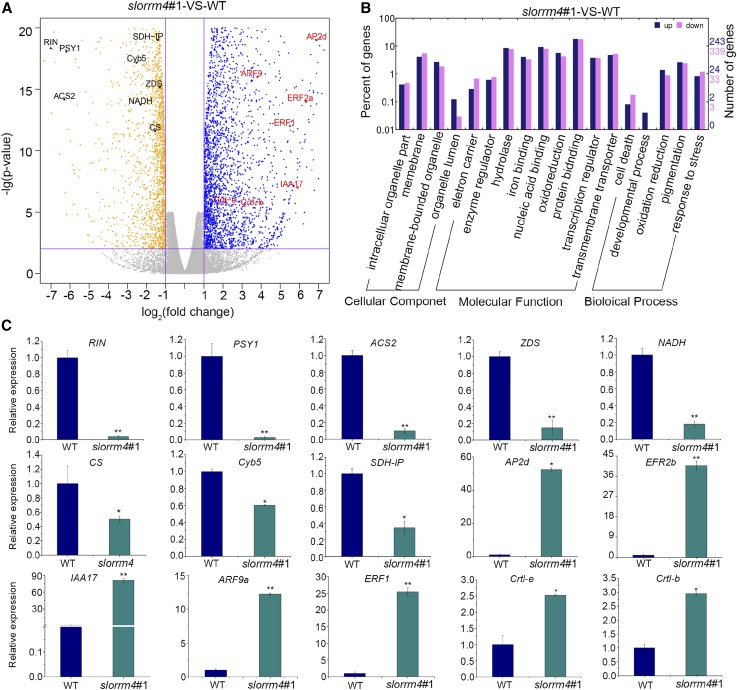
To elucidate how SlORRM4 affects fruit ripening, we performed RNA sequencing (RNA-seq) with total RNA from wild-type and slorrm4#1 fruits at 32 DPA (three biological replicates for each sample). Approximately 14 million clean reads were obtained, which were all well aligned to the tomato genome (Tomato Genome Consortium, 2012; Supplemental Fig. S7A). For each sample, the reads from three biological replicates were highly correlated (based on Pearson’s correlation coefficient), indicating a strong reproducibility and reliability of the RNA-seq data (Supplemental Fig. S7B). Bioinformatic analysis revealed 2,435 differentially expressed genes (DEGs). Among them, 1,138 (46.74%) genes were up-regulated and 1,297 (53.29%) genes were down-regulated compared with those genes in wild-type fruits (Supplemental Fig. S7C; Supplemental Data Set S2). Down-regulated genes include key genes associated with the Krebs cycle and mitochondrial function (citrate synthase [CS], succinate dehydrogenase iron-sulfur protein [Sdh-IP], niacinamide dehydrogenase [NADH], and cytochrome 450 [Cyb5]) and ethylene and lycopene biosynthesis (ACS2, phytoene synthase [PSY1], ζ-carotene desaturase [ZDS], and the transcription factor RIN; Fig. 4A). In contrast, core genes associated with lycopene decomposition and other genes related to fruit ripening were up-regulated, including lycopene β-cyclase (Crtl-b), lycopene ε-cyclase (Crtl-e), indoleacetic acid17 (IAA17), ethylene response factor 1 (ERF1), ethylene response factor 2a (ERF2a), auxin response factor 9 (ARF9), and APETALA2d (AP2d; Fig. 4A).
Figure 4.
SlORRM4 regulates the expression of fruit ripening and mitochondria function-related genes. A, Volcano plots were used to visualize RNA-seq data. Each point corresponds to a DEG. Blue plots represent up-regulated genes, and orange plots represent down-regulated genes, in slorrm4#1 compared with the wild type. Eight representatively up-regulated genes are marked in black text, and seven significantly down-regulated genes are labeled in red text. Fold change > 2 and P < 0.01 (indicating DEGs) are marked with purple lines. B, Functional categories of genes that differed in abundance between slorrm4#1 and the wild type (WT; both at 32 DPA). Dark blue and purple represent up- and down-regulated genes, respectively. C, qRT-PCR validation of RNA-seq results showing the relative expression of 15 significant DEGs. Values are means ± sd of three independent experiments in three biological repeats. Asterisks indicate P < 0.05 (*) and P < 0.01 (**; Student’s t test).
Gene Ontology (GO) analysis revealed significant enrichment of terms associated with mitochondria function, including electron carrier, iron binding, cell death, and oxidation reduction under molecular function (Fig. 4B). In addition, DEGs also were enriched significantly for terms related to fruit ripening, such as pigmentation in biological process and transcription regulator in molecular function (Fig. 4B). qRT-PCR validation of 15 genes (eight up-regulated and seven down-regulated) was highly correlated with the RNA-seq data (Fig. 4C; Supplemental Fig. S8, A and B). In summary, slorrm4 mutation delayed fruit ripening through direct or indirect regulation of key genes related to mitochondrial function and fruit ripening.
Dramatic Defects of Mitochondrial Morphology and RNA Editing in Fruits of slorrm4 Mutants
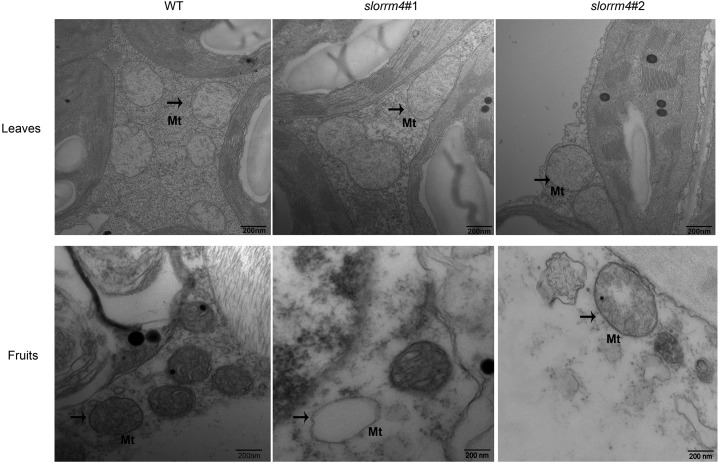
Because SlORRM4 localized to the mitochondria and because the expression levels of many genes related to mitochondrial function were significantly altered in slorrm4 mutants, we performed transmission electron microscopy (TEM) to study changes in mitochondrial structure between wild-type and slorrm4 mutant leaves and fruits. We found that the mitochondria in the leaves of the wild type and slorrm4 mutants were comparable and appeared healthy (Fig. 5). However, in contrast to the wild-type mitochondrial structure in tomato fruits, the mitochondria of slorrm4 mutant fruits lacked cristae and had empty internal structures (Fig. 5), indicating at least partial dysfunction of mitochondria in slorrm4 fruits.
Figure 5.
Ultrastructure of mitochondria in leaves and fruits of the wild type (WT) and the slorrm4 mutant. Black arrows point to the morphology of the enlarged mitochondria (Mt). Bar = 200 nm.
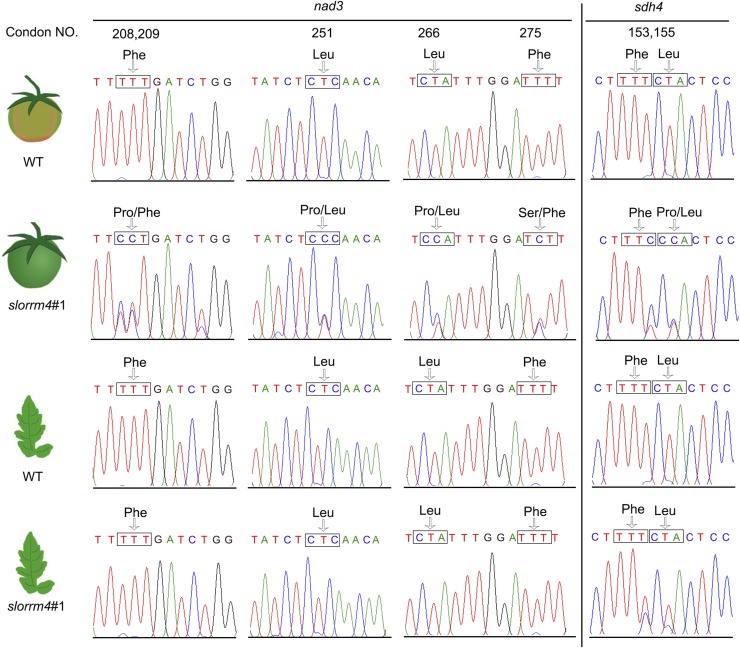
AtORRM4 was reported to be involved in mitochondrial RNA editing (Shi et al., 2016). To better understand the mechanism whereby SlORRM4 influences mitochondria functions and fruit ripening, we annotated tomato mitochondria genes (Supplemental Data Set S3), obtaining 37 predicted mitochondrial transcripts for sequencing (Supplemental Fig. S9A). By comparison with the RNA editing in wild-type ripening fruits, the slorrm4#1 mutant fruits showed significant deficiencies in C-to-U editing at five positions on nad3 (a mitochondrial gene encoding subunit 3 of NADH dehydrogenase): nad3-208, -209, -251, -266, and -275; these deficiencies caused changes in Nad3 residues from Phe to Pro, two Leu to Pro, and Phe to Ser (Fig. 6). Next, we analyzed the amino acid sequences flanking these five consecutive editing sites of Nad3 among several plant and animal species (Supplemental Fig. S10A). We found that the Phe (nad3-208 and -209) residue was conserved (Supplemental Fig. S10B). These results suggested that the RNA editing of nad3 residues 208 and 209 might be more pivotal for Nad3 stability and activity. Moreover, a dramatically lowered editing efficiency at the sdh4-153 and sdh4-155 loci was found in slorrm4#1 fruits (Fig. 6), but it resulted in only one amino acid change (Leu to Pro) for SDH4 (a mitochondrial gene encoding subunit 4 of SDH dehydrogenase; Fig. 6). In addition, RNA editing in slorrm4#1 mutant fruits also was affected at the nad4-953, attB-181, and rps4-289 sites (Supplemental Fig. S9B). Considering the different transcript abundances of SlORRM4 in leaves and ripening fruits, we next examined the RNA editing efficiency in nad3 and sdh4 transcripts from wild-type and slorrm4#1 leaves and found that it was not significantly affected in mutant leaves (Fig. 6). Overall, our data indicated that SlORRM4 strongly contributed to the editing of these mitochondrial transcript sites in tomato fruits.
Figure 6.
RNA editing of nad3 and sdh4 transcripts in fruits and leaves of the wild type (WT) and the slorrm4 mutant. Small rectangular frames indicate the codons of the amino acids.
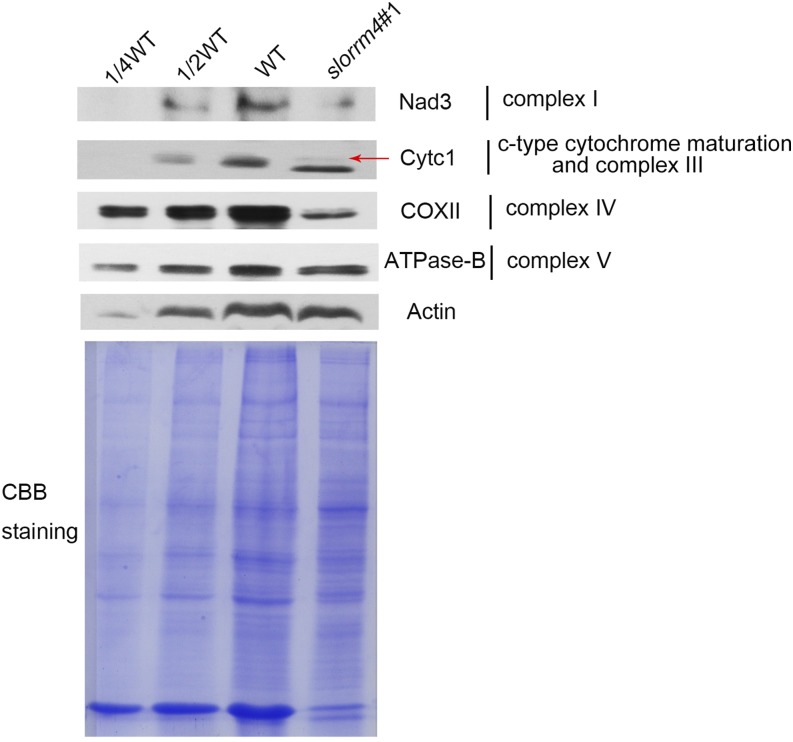
To verify whether defects in RNA editing in slorrm4#1 mutant fruits can affect the mitochondrial respiratory chain, we performed western blotting to detect the levels of Nad3 in complex I, cytochrome c1 (Cytc1) in complex III, cytochrome c oxidase subunit 2 (COX II) in complex IV, and B subunit of ATPase (ATPase-B) in complex V. Due to the lack of appropriate antibody, we could not examine the level of SDH4 in complex II. The accumulation of the above proteins was essential for the assembly of corresponding complexes, which function in the mitochondrial respiratory chain. In slorrm4 mutant fruits, the accumulation of Nad3, Cytc1, and COX II proteins, but not ATPase-B protein, decreased sharply compared with that observed in wild-type fruits (Fig. 7). These results suggested that loss of SlORRM4 function might impair the mitochondrial respiratory chain.
Figure 7.
Levels of core subunits of the mitochondrial complex were reduced in the slorrm4#1 mutant. Nad3, Cytc1, COXII, and ATPase-B are core subunits of complexes I, III, IV, and V. Actin was used as a loading control. Total proteins were extracted from fruits at 32 DPA and stained with Coomassie Brilliant Blue (CBB staining). WT, Wild type.
DISCUSSION
SlORRM4 Is Involved in Tomato Fruit Ripening
In addition to ethylene (Yang and Hoffman, 1984), transcription factors (Giovannoni, 2007), and DNA demethylases (Lang et al., 2017), recent evidence has emerged showing that noncoding RNAs, such as microRNAs and long noncoding RNAs, are additional key regulators of fruit ripening (Moxon et al., 2008; Zhu et al., 2015; Wang et al., 2016). Given the increasing number of fruit ripening regulators that are being identified, this study was focused on the function of RNA editing factors in fleshy fruit ripening. Our data clearly indicated a role for SlORRM4 in promoting tomato fruit ripening. SlORRM4 expression occurred at very low levels in stems, leaves, and young developing fruits but increased dramatically in ripening fruits (Fig. 1G). Indeed, compared with the time of the color change in wild-type fruits, a slorrm4 mutant with a 544-bp deletion exhibited great delay of fruit ripening about 10 d later (Fig. 3B). Moreover, even at 60 DPA, fruits of the slorrm4 mutants remained orange (Supplemental Fig. S6).
Tomato fruits are climacteric and show a sudden increase in respiration at the onset of ripening, usually in concert with increased production of the gaseous hormone ethylene (Rhodes, 1980; Yang and Hoffman, 1984). The respiration rate and ethylene production were significantly reduced in many transcription factor mutants during fruit ripening (Vrebalov et al., 2002; Manning et al., 2006). In addition, the decreased expression of some other fruit ripening-related factors by RNA interference in tomato, such as vacuolar invertase inhibitor (SlVIF) or zinc finger transcription factor 2 (SlZPF2), delays the rise in the respiratory rate and ethylene production during fruit ripening (Weng et al., 2015; Qin et al., 2016). In our study, the increased respiratory rates and ethylene production were similarly greatly delayed during fruit ripening in slorrm4 mutants (Fig. 3, E and F). Thus, the observed ripening delays in slorrm4 mutants resulted from lower respiration rates and ethylene production relative to wild-type fruits at the same stage of fruit ripening. Moreover, the peak of the respiratory rate and ethylene production of slorrm4 mutants was lower than that in wild-type fruits (Fig. 3, E and F), which was likely responsible for the orange fruit at 60 DPA in slorrm4 mutants. In addition, the mRNA abundance of some key fruit ripening-related genes was significantly up-regulated or down-regulated in slorrm4-mutant fruits in comparison with the wild type, implying that SlORRM4 might influence fruit ripening by regulating those ripening-related genes indirectly (Fig. 4A). Furthermore, the expression of many core mitochondrial enzyme genes, such as NADH, CS, and SDH-IP, was decreased greatly in the slorrm4 mutants, which underscored the crucial role of mitochondria in regulating fruit ripening (Fig. 4A). However, it is likely that the altered expression of genes related to mitochondrial function in the RNA-seq experiment resulted from the abnormal mitochondria in slorrm4 mutants. Therefore, the specific functional mechanism whereby SlORRM4 promotes fruit ripening was not clearly determined in this study, and further investigation is required to elucidate a potential direct link between mitochondrial function and tomato fruit ripening.
SlORRM4 Is Required for RNA Editing and Mitochondrial Function
The ORRM family of proteins in Arabidopsis is involved in the editing of many mitochondrial transcript sites (Shi et al., 2015). Results from a previous study on ORRM4 in Arabidopsis proved that the RRM domain provides the editing activity to AtORRM4, whereas the GR domain has a minor effect on editing and is required for its interaction with ORRM3 and with itself, suggesting that the RRM and GR domains have different functions (Shi et al., 2016). Currently, a complementation assay has been developed to investigate RNA editing versus tomato fruit ripening. In our study, knockout of SlORRM4 significantly reduced RNA editing of several mitochondrial genes. RNA editing of nad3 led to several amino acid changes, notably to Pro (Fig. 6). Because Pro can disrupt the secondary structure of proteins, its presence probably directly affected the Nad3 conformation and impaired its protein expression level in mitochondria (Fig. 7). Our findings corroborated recent studies on RNA editing factors in two maize mutants, in which it was reported that Nad3 protein expression decreases significantly by altering the editing and that the lowered NADH dehydrogenase activity negatively affects mitochondrial function (Yuan and Liu, 2012; Qi et al., 2017). Together, these results provide convincing evidence that RNA editing of nad3 is critical for mitochondrial function and biogenesis.
The slorrm4 mutants also exhibited considerably decreased sdh4 transcript editing, a fairly novel finding, as few studies have focused on sdh4 transcript editing. Plant mitochondrial sdh4 has a limited number of editing sites. Only seven editing sites have been found in tomato, two of which were silent (Adams et al., 2001). Here, we demonstrated that RNA editing of sdh4-153 and sdh4-155 was reduced dramatically in slorrm4 fruits, although we were unable to examine SDH4 accumulation, as appropriate antibodies were unavailable. Thus, more research is necessary to demonstrate the effect of the sdh4 transcript on mitochondrial function.
Our slorrm4 mutant fruits exhibited abnormal mitochondrial morphologies that could be linked to the reduction of complex subunits in the mitochondrial respiratory chain (Fig. 7). The mitochondria respiratory chain is composed of five major complexes (complex I–complex V), which are critical for proper cristae morphology (Logan, 2006). Thus, the reduced complex I maize mutants of PPR proteins impaired the mitochondrial ultrastructure (Keren et al., 2012; Chen et al., 2017; Qi et al., 2017; Wang et al., 2017). In addition, the lack of complex III in maize ppr mutants strongly decreased or completely abolished cristae formation (Sosso et al., 2012). To confirm that the mitochondrial respiratory chain was less functional in the slorrm4#1 mutant, we examined the protein levels of the core subunits of complex III, complex IV, and complex V (namely Cytc1, COX II, and ATPase-B). Cytc1 and COX II expression were decreased significantly, indicating that the assembly of complex III and complex IV might be disrupted (Fig. 7). Previously, the loss of cytochrome c expression in Arabidopsis cytc1/cytc2 double mutants abolished complex IV activity and arrested mitochondria function (Welchen et al., 2012). In addition, a much darker band appeared just below the Cytc1 band in the sloorm4 mutant (Fig. 7), which we hypothesized to belong to another unknown factor that was activated with decreasing levels of Cytc1 in the slorrm4 mutant. Previous data have demonstrated that, with dysfunction of the cytochrome c pathway, other alternative and compensatory respiration pathways can be activated (Kühn et al., 2015). Further confirmation of the unknown band warrants further study. In contrast, the ATPase-B level did not change in slorrm4#1 mutant fruits (Fig. 7), suggesting that continued synthesis occurred in mitochondria, but perhaps with reduced capability. Taken together, these results indicated that SlORRM4 was critical for RNA editing and mitochondrial function in tomato fruits.
The ORRM4 Protein Displayed Species- and Tissue-Specific Functions in Tomato and Arabidopsis
Despite being homologs, RNA editing factors from different plant species may have distinct functions. Here, we compared RNA editing defects in tomato and Arabidopsis orrm4 mutants. As expected, ORRM4 function in regulating RNA editing differed between tomato and Arabidopsis (Shi et al., 2016). Notably, C-to-U editing of the five nad3 loci decreased significantly in slorrm4 mutant fruits but only slightly in Arabidopsis (Fig. 6). In contrast, an Arabidopsis orrm4 mutant exhibited dramatically decreased RNA editing of rpl5, whereas we observed nearly identical RNA editing of rpl5 between the slorrm4 mutant and wild-type fruit (data not shown). Several previous reports also have described interspecific variations. For example, a comparative study on maize versus Arabidopsis revealed interspecies differences in editing of the atp4 transcript. In maize ppr mutants, atp4-59 editing was completely abolished, whereas in Arabidopsis ppr mutants, atp4-89 and atp4-250 editing was only slightly impaired and atp4-59 was unaffected (Wang et al., 2017). Data from another study demonstrated that ccmB-43 editing of a ppr mutant in maize and Arabidopsis was divergent (Yang et al., 2017). Together, these results revealed that SlORRM4 showed species-specific function in plants.
In the maize ppr mutant, RNA editing did not differ across kernels and seedlings (Wang et al., 2017). Here, however, the RNA editing of two mitochondrial transcripts differed between the leaves and fruits of the slorrm4 mutant. Specifically, slorrm4 mutant fruits were significantly deficient in C-to-U editing of nad3, but leaves were not (Fig. 6). The editing efficiency at the sdh4-153 and sdh4-155 loci also was decreased dramatically in mutant fruits, but less so in leaves (Fig. 6). These differences probably resulted from tissue variations in SlORRM4 gene expression patterns: SlORRM4 had low expression in leaves and high expression in ripening fruits (Fig. 1G). Consistent with this differential SlORRM4 expression and tissue-specific RNA editing between tomato leaves and fruits, the mitochondrial morphology of slorrm4 mutant leaves was that of a healthy organelle, but the mitochondria of slorrm4 mutant fruits had empty internal structures (Fig. 5), indicating that the SlORRM4-regulated mitochondria health was strongly correlated with fruit ripening. Another study found that RNA editing in three transcripts (ndhB, ndhF, and ndhD) was altered in tomato fruits compared with leaves (Kahlau and Bock, 2008). Moreover, tomato fruits in five different ripening stages also exhibited fluctuating editing efficiency at specific sites within these three different mRNAs for subunits of plastid NAD(P)H dehydrogenase (Kahlau and Bock, 2008). These results indicated that tomato fruit ripening was closely associated with the fluctuating editing efficiency at specific sites within ndhB, ndhF, and ndhD mRNAs (Kahlau and Bock, 2008). These results suggested that SlORRM4 showed tissue-specific function in tomato leaves and fruits.
In conclusion, our study highlights an important role for an RNA editing factor, SlORRM4, in tomato fruit ripening and demonstrates that SlORRM4 plays a key role in mitochondrial function and RNA editing in tomato fruit. However, further studies on the mechanism of how SlORRM4 influences fruit ripening and if there exists a link between fruit ripening and RNA editing still need to be done in the future.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Tomato plants (Solanum lycopersicum ‘Micro-Tom’ [MT]) were grown in a greenhouse under standard conditions (16 h of light at 26°C followed by 8 h of darkness at 20°C). Leaf samples were obtained 12 weeks after transplantation. Flowers were tagged at anthesis to follow fruit ages accurately through development. Transgenic and wild-type lines were collected at set ripening stages (MG, Br, and Br+10) based on DPA. Samples were immediately frozen in liquid nitrogen after collection and stored at −80°C.
Phylogenetic Analysis
Protein sequences from the RIP, ORRM, and OZ1 families in Arabidopsis (Arabidopsis thaliana) and tomato were aligned. The alignment was performed using ClustalX 2.1 with default parameters (Larkin et al., 2007) and then imported into MEGA 5.2 to construct the phylogenetic tree. The neighbor-joining method with 1,000 bootstrap replicates was used (Tamura et al., 2011).
Heat Map Analysis
To visualize the expression patterns of the RNA editing factors, which is based on RNA-seq data of tomato (Tomato Genome Consortium, 2012) in different tomato, excluding the RPKM below 15 in the Br+10 stage, a heat map was created using R (http://www.r-project.org/). First, the RPKM of tomato RNA editing factors and fruit ripening marker genes were transformed to log2 (value) and normalized to reveal the differences in expression levels among tomato organs and during fruit ripening. RNA editing factors with a similar expression pattern to fruit ripening maker genes were selected for the following experiments.
RNA Isolation and qRT-PCR Analysis
Total RNA was isolated from fruit samples using TRIzol reagent prepared in our laboratory (Zhu et al., 2015); RNA concentration was measured using a NAS-99 spectrophotometer (ATCGene). After DNase treatment to remove genomic DNA, RNA integrity was assessed with agarose gel electrophoresis. A TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix kit (Trans) was used to synthesize cDNA from a 2-μg aliquot of total RNA. Next, qRT-PCR was performed using SYBR Green PCR Master Mix with a real-time PCR system (CFX96; Bio-Rad). The reaction volume was 20 μL, and the thermocycling program was as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. Actin was selected as the reference gene for normalization. Every experiment included three biological repeats, each with three technical replicates. Oligonucleotide primers for this analysis are listed in Supplemental Table S1.
VIGS
TRV was used to perform VIGS on MT fruits following the protocol from a previous study (Yan et al., 2012). About 300- to 500-bp fragments of 11 RNA editing factors and PDS were designed using the VIGS tool (http://solgenomics.net/tools/vigs) to avoid off-target silencing. Specific cDNA fragments corresponding to the RNA editing factors were individually amplified and inserted into pTRV2 vectors that were transferred to Agrobacterium tumefaciens strain GV3101, now containing pTRV1, pTRV2, and pTRV2 fragments (PDS and RNA editing factor cDNA). Bacteria were cultured at 28°C in Luria-Bertani medium (pH 5.6) containing 10 mm MES and 20 μm acetosyringone with three antibiotics (kanamycin, gentamycin, and rifampicin). After shaking for 12 h, cultures were harvested and resuspended in infiltration buffer (10 mm MgCl2 and 200 μm acetosyringone) to a final OD600 of 1. Vectors pTRV1 and pTRV2 (empty or with insert) were mixed at a 1:1 ratio and incubated at room temperature for 3 h. Using a vacuum pump, A. tumefaciens buffer containing pTRV2 (empty or with insert) was infiltrated into tomato seedlings. After incubation for 48 h at 28°C, these seedlings were sown. Tomato seedlings infiltrated with empty pTRV2 and pTRV2-PDS were negative and positive controls. Fruit ripening progress was observed daily. Oligonucleotide primers for this analysis are listed in Supplemental Table S2.
Subcellular Localization of SlORRM4
The open reading frame of SlORRM4 without stop codon was cloned into the tdTomato expression vector using a one-step cloning kit (Vazyme). tdTomato was a red fluorescent protein (Graewe et al., 2009). The control tdTomato vector and fusion SlORRM4-tdTomato vector harboring the constitutively expressed cauliflower mosaic virus 35S promoter were transferred to A. tumefaciens strain GV3101 and introduced into tobacco (Nicotiana tabacum) leaves for 48 h. MitoTracker Green FM (Thermo Fisher Scientific) was used as a mitochondria-selective fluorescent label. After 48 h of infiltration, the colocalization of tdTomato-MitoTracker signals was inspected with an Olympus Nikon A1RMPSi confocal microscope with excitation maximum at 554 nm.
Selection of Single Guide RNA Target Sequences and pYLCRISPR/Cas9Pubi-H-SlORRM4 Vector Construction
CRISPR-P (http://cbi.hzau.edu.cn/crispr/) was used to select four specific single guide RNAs (sgRNAs) for targeting SlORRM4 (Supplemental Table S4). Selection criteria aimed to maximize editing efficiency: GC content in target sites was more than 40%, and target sequences avoided four or more consecutive T nucleotides, as RNA polymerase III would recognize such regions as a termination signal. Because sgRNA secondary structure greatly influences editing efficiency, the RNA Folding program (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form2.3) was used to ensure that no more than 5 bp was between the target and sgRNA sequences. Upon selection, targeting sgRNAs were cloned using one round of Golden Gate ligation into the pYLCRISPR/Cas9Pubi-H binary plasmid (Ma et al., 2015). Oligonucleotide primers for this analysis are listed in Supplemental Table S5.
Plant Transformation
The final pYLCRISPR/Cas9Pubi-H-SlORRM4 binary vector was transformed into MT using A. tumefaciens (Eck et al., 2006). Transgenic tomato lines were selected through their hygromycin resistance. After in vitro regeneration, plants were transplanted into soil.
DNA Extraction and Mutation Analysis
DNA was extracted from fresh frozen leaves (1–5 mg) using a hi-DNA secure plant kit (Tiangen). Total DNAs from T0 and T1 transgenic plants were used as templates for PCR amplification with primers flanking the sgRNA target sites. The following standard PCR program was used: 94°C for 3 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and 72°C for 7 min. PCR products were sequenced directly or cloned into the pEasy-T1 (TransGen, Biotech) vector and then Sanger sequenced to identify mutations. Superimposed sequence chromatograms produced with biallelic and heterozygous mutations were deciphered using DSDecode (http://dsdecode.scgene.com/) and manual analyses. Oligonucleotide primers for this analysis are listed in Supplemental Tables S6 and S7.
Measurements of Respiratory Rate and Ethylene Production
Fruit from slorrm4 mutants and the wild type were harvested at 32, 35, 38, 45, and 50 DPA, then placed in an open box for 3 h to avoid measuring respiratory wounds and wound ethylene, both transiently generated as a consequence of harvesting. For respiratory rate measurements, the box was sealed and incubated at room temperature for 1 h. Subsequently, 1-mL gas samples were added to a respiration meter (CA-10; Sable System). Respiratory rates were calculated via comparing peak areas of the gas samples with known CO2 standards and normalizing for fruit weight.
For ethylene measurements, the box containing tomatoes was incubated at room temperature for 2 h before 1-mL gas samples were analyzed in a gas chromatograph (GC-14; Shimadzu) equipped with an activated alumina column and a flame ionization detector. Ethylene concentrations were calculated via comparing sample peak areas with known ethylene standards and normalizing for fruit weight. Samples for both measurements each contained three replicates with four fruits per replicate.
RNA Deep Sequencing Data Analysis
Total RNA was isolated from wild-type and slorrm4#1 fruits at 32 DPA using TRIzol reagent. mRNA was then enriched using oligo(dTs) coupled with magnetic beads before being cut into 300-bp fragments (Novogene). Next, RNA-seq libraries were sequenced on HiSeq PE150 (Illumina) with approximately 14 million 150-bp paired-end reads. All raw reads were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/) under accession number SRP104094. Clean reads were quality checked in FastQC (version 0.11.3). Reads were aligned to the tomato reference genome (version SL2.50) using Tophat (version 1.4.6), and fragments were assigned to genes using Feature Counts (version 2.0.14; Trapnell et al., 2012; Liao et al., 2014). Differential gene expression between the wild type and the slorrm4 mutant was identified by the deseq2 package (Anders, 2010) using the criteria of fold change > 2 and Q value < 0.01 (Supplemental Data Set S2). Volcano plots were used to visualize the DEGs above (Li, 2012).
GO Enrichment Analysis
GO-TermFinder (version 0.86) was used to analyze DEG enrichment of GO terms. Next, the generated GO annotations were plotted in WEGO (http://wego.genomics.org.cn/). Proteins were filtered based on their grouping to cellular components, molecular functions, and biological functions.
TEM Analysis
For TEM analysis, the young leaves and pericarps of wild-type and slorrm4#1 50-DPA fruits were fixed with 2.5% glutaraldehyde for over 2 h and washed three times with 0.1 mL of phosphate buffer. Then, the sample was fixed with 1% osmic acid for 2 h and washed three times with 0.1 mL of phosphate buffer. Dehydration of the sample was reached with different concentrations of acetone (30%, 50%, 70%, 90%, and 100%). Next, the sample was embedded and aggregated with ethoxyline resin and sliced up with a Leica UC6 ultramicrotome. After dying with both uranyl acetate and lead citrate, the prepared sample was observed with a JEM-1230 transmission electron microscope.
Mitochondria Gene Annotation in Tomato
Tomato mitochondria genome sequences (GCA_000325825.1) were BLAST aligned (version 2.4.0 + x64-linux) with Swiss-Prot (version 20150715) and the Mitochondria Protein Database (version 20151027, Map). The E-value was 1e-7, and the query gencode was 10. Alignment results were output in GenBank format and processed in Maptools (version 1.0). Annotated transcripts are shown in Supplemental Data Set S3. The final 30 valid tomato mitochondria annotated sequences were deposited in NCBI under GenBank accession number MF766469 using Sequin (version 9.20) software.
Mitochondrial RNA Editing Analysis
For the analysis, RNA was isolated from 32-DPA wild-type and slorrm4#1 leaves and fruits. Next, it was reverse transcribed into cDNA using a TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix kit (Trans) with random primers. The mitochondrial genes were amplified using KOD PLUS DNA polymerase (Toyobo) and sequenced. Mutant and wild-type sequences were compared for C-to-T differences resulting from RNA editing. Oligonucleotide primers used for the analysis are listed in Supplemental Table S8.
Protein Extraction and Western Blot
Protein extraction was performed according to Li et al. (2017) with slight modifications. For tomato leaves and fruits, protein extractions were performed in an extraction buffer containing 100 mm Tris-HCl (pH 7.5), 1 mm EDTA, 10% (v/v) glycerol, and a 1% (v/v) protease inhibitor cocktail. Four volumes of extraction buffer (v/w) was added to the fruit sample before homogenization. Two volumes of extraction buffer (v/w) was added to the leaf sample before homogenization. The supernatant was used for western blot after centrifugation at 20,000g for 20 min at 4°C. Protein concentration was determined using the Bradford kit (Solarbio), and 40 μg of proteins was subjected to SDS-PAGE. Proteins were then transferred to a polyvinylidene difluoride membrane and incubated overnight at 4°C with various primary antibodies. The monoclonal antibody against the SlORRM4 protein was prepared by screening an antibody library of peptide antigen, and the effective peptide antibody sequence is GGEEQLSADQGTES (Abmart). The peptide antibody against the SlORRM4 protein was at a concentration of 1:500. The antibody against Nad3 and COX II was at a concentration of 1:1,000 (Agrisera), while the antibody against Cytc1, ATPase-B, and α-IDH was diluted to 1:5,000 (Agrisera). The concentration of the polyclonal antibody against actin was 1:5,000 (EASYBIO). An enhanced chemiluminescence kit (Absin) was used for detection after incubation with the horseradish peroxidase-conjugated secondary antibody (B&M).
Statistical Analysis
Significance analysis of the data in this study was conducted using SPSS (version 20.0) software. For two data sets, statistical significance between the two data sets was computed using Student’s t test. It is displayed with one asterisk when the difference is significant (P < 0.05), and if the difference is highly significant, it is displayed with two asterisks (P < 0.01).
Accession Numbers
Raw sequencing data from the RNA-seq experiment are stored at the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/sra/) under accession number SRP104094. The tomato mitochondria annotated sequences were deposited to NCBI GenBank under accession number MF766469.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Detailed diagram of the pipeline for the identification of ripening-related RNA editing factor in tomato.
Supplemental Figure S2. Identification of fruit ripening-related RNA editing factors using expression pattern analysis.
Supplemental Figure S3. Alignment of ORRM protein sequences in Arabidopsis and tomato.
Supplemental Figure S4. CR-SlORRM4 mutants were generated using the CRISPR/Cas9 system in T1 transgenic plants.
Supplemental Figure S5. Agricultural traits of slorrm4 mutants.
Supplemental Figure S6. Phenotypes of wild-type, slorrm4#1, and slorrm4#2 fruits at 60 DPA.
Supplemental Figure S7. RNA-seq data were high quality, and multiple DEGs were found.
Supplemental Figure S8. Correlation coefficient between RNA-seq and qRT-PCR.
Supplemental Figure S9. SlORRM is required for mitochondria RNA editing in tomato fruit.
Supplemental Figure S10. SlORRM4 plays a key role in RNA editing of the nad3 transcript.
Supplemental Table S1. Primers used for qRT-PCR.
Supplemental Table S2. Primers used for VIGS.
Supplemental Table S3. Detection of mutations on putative off-target sites.
Supplemental Table S4. Sequences of target sites.
Supplemental Table S5. Primers used for recombinant pYLCRISPR/Cas9Pubi-H-SlORRM4 vector construction.
Supplemental Table S6. Primers used for target site mutation analysis.
Supplemental Table S7. Primers used for off-target site mutation analysis.
Supplemental Table S8. Primers used for cloning and sequencing of mitochondrial transcripts.
Supplemental Data Set S1. Expression profiles of tomato PPR, MORF/RIP, OZ, and ORRM gene families.
Supplemental Data Set S2. DEGs between wild-type fruits and slorrm4 mutant fruits.
Supplemental Data Set S3. Annotations of the tomato mitochondrial genome.
Acknowledgments
We thank Yaoguang Liu (South China Agriculture University) for providing the binary vector pYLCRISPR/Cas9 system and Tian Wang for technical assistance. We also thank Baocai Tan (Shandong University, Jinan, China) and Rentao Song (China Agricultural University, Beijing, China) for technical guidance.
Footnotes
This work was supported by the National Key R&D Program of China (2016YFD0400901) and the National Natural Science Foundation of China (31672208, 31622050, and 91540118) to H.Z.
References
- Adams KL, Rosenblueth M, Qiu YL, Palmer JD (2001) Multiple losses and transfers to the nucleus of two mitochondrial succinate dehydrogenase genes during angiosperm evolution. Genetics 158: 1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S. (2010) Analysing RNA-Seq data with the DESeq package. Mol Biol 43: 17 [Google Scholar]
- Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415–442 [DOI] [PubMed] [Google Scholar]
- Bentolila S, Heller WP, Sun T, Babina AM, Friso G, van Wijk KJ, Hanson MR (2012) RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc Natl Acad Sci USA 109: E1453–E1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady C. (1987) Fruit ripening. Annu Rev Plant Physiol 38: 155–178 [Google Scholar]
- Chen X, Feng F, Qi W, Xu L, Yao D, Wang Q, Song R (2017) Dek35 encodes a PPR protein that affects cis-splicing of mitochondrial nad4 intron 1 and seed development in maize. Mol Plant 10: 427–441 [DOI] [PubMed] [Google Scholar]
- Eck JV, Kirk DD, Walmsley AM (2006) Tomato (Lycopersicum esculentum). Methods Mol Biol 343: 459–474 [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ. (2007) Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol 10: 283–289 [DOI] [PubMed] [Google Scholar]
- Graewe S, Retzlaff S, Struck N, Janse CJ, Heussler VT (2009) Going live: a comparative analysis of the suitability of the RFP derivatives RedStar, mCherry and tdTomato for intravital and in vitro live imaging of Plasmodium parasites. Biotechnol J 4: 895–902 [DOI] [PubMed] [Google Scholar]
- Grewe F, Viehoever P, Weisshaar B, Knoop V (2009) A trans-splicing group I intron and tRNA-hyperediting in the mitochondrial genome of the lycophyte Isoetes engelmannii. Nucleic Acids Res 37: 5093–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JB, Shi X, Kobylarz AT, Lucas MK, Wessendorf RL, Hines KM, Bentolila S, Hanson MR, Lu Y (2017) An organelle RNA recognition motif protein is required for photosystem II subunit psbF transcript editing. Plant Physiol 173: 2278–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlau S, Bock R (2008) Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell 20: 856–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Tal L, des Francs-Small CC, Araújo WL, Shevtsov S, Shaya F, Fernie AR, Small I, Ostersetzer-Biran O (2012) nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. Plant J 71: 413–426 [DOI] [PubMed] [Google Scholar]
- Kadowaki K, Ozawa K, Kazama S, Kubo N, Akihama T (1995) Creation of an initiation codon by RNA editing in the coxl transcript. Curr Genet 28: 422. [DOI] [PubMed] [Google Scholar]
- Kühn K, Yin G, Duncan O, Law SR, Kubiszewski-Jakubiak S, Kaur P, Meyer E, Wang Y, Small CC, Giraud E, et al. (2015) Decreasing electron flux through the cytochrome and/or alternative respiratory pathways triggers common and distinct cellular responses dependent on growth conditions. Plant Physiol 167: 228–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang Z, Wang Y, Tang K, Tang D, Datsenka T, Cheng J, Zhang Y, Handa AK, Zhu JK (2017) Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc Natl Acad Sci USA 114: 4511–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li R, Li R, Li X, Fu D, Zhu B, Tian H, Luo Y, Zhu H (2017) Multiplexed CRISPR/Cas9-mediated metabolic engineering of gamma-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol J (in press) [DOI] [PMC free article] [PubMed]
- Li W. (2012) Volcano plots in analyzing differential expressions with mRNA microarrays. J Bioinform Comput Biol 10: 1231003. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Lin T, Zhu G, Zhang J, Xu X, Yu Q, Zheng Z, Zhang Z, Lun Y, Li S, Wang X, et al. (2014) Genomic analyses provide insights into the history of tomato breeding. Nat Genet 46: 1220–1226 [DOI] [PubMed] [Google Scholar]
- Liu YJ, Xiu ZH, Meeley R, Tan BC (2013) Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell 25: 868–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan DC. (2006) The mitochondrial compartment. J Exp Bot 57: 1225–1243 [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T (2008) Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res 18: 1602–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenkott B, Yamaguchi K, Tsuji-Tsukinoki S, Knie N, Knoop V (2014) Chloroplast RNA editing going extreme: more than 3400 events of C-to-U editing in the chloroplast transcriptome of the lycophyte Selaginella uncinata. RNA 20: 1499–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Tian Z, Lu L, Chen X, Chen X, Zhang W, Song R (2017) Editing of mitochondrial transcripts nad3 and cox2 by Dek10 is essential for mitochondrial function and maize plant development. Genetics 205: 1489–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G, Zhu Z, Wang W, Cai J, Chen Y, Li L, Tian S (2016) A tomato vacuolar invertase inhibitor mediates sucrose metabolism and influences fruit ripening. Plant Physiol 172: 1596–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes M., JC (1980) Respiration and senescence of plant organs. In DD Davies, ed, The Biochemistry of Plants, Vol 2. Academic Press, New York, pp 419–462 [Google Scholar]
- Rüdinger M, Funk HT, Rensing SA, Maier UG, Knoop V (2009) RNA editing: only eleven sites are present in the Physcomitrella patens mitochondrial transcriptome and a universal nomenclature proposal. Mol Genet Genomics 281: 473–481 [DOI] [PubMed] [Google Scholar]
- Shi X, Castandet B, Germain A, Hanson MR, Bentolila S (2017) ORRM5, an RNA recognition motif-containing protein, has a unique effect on mitochondrial RNA editing. J Exp Bot 68: 2833–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Germain A, Hanson MR, Bentolila S (2016) RNA recognition motif-containing protein ORRM4 broadly affects mitochondrial RNA editing and impacts plant development and flowering. Plant Physiol 170: 294–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Hanson MR, Bentolila S (2015) Two RNA recognition motif-containing proteins are plant mitochondrial editing factors. Nucleic Acids Res 43: 3814–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small ID, Peeters N (2000) The PPR motif: a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25: 46–47 [DOI] [PubMed] [Google Scholar]
- Sosso D, Mbelo S, Vernoud V, Gendrot G, Dedieu A, Chambrier P, Dauzat M, Heurtevin L, Guyon V, Takenaka M, et al. (2012) PPR2263, a DYW-subgroup pentatricopeptide repeat protein, is required for mitochondrial nad5 and cob transcript editing, mitochondrion biogenesis, and maize growth. Plant Cell 24: 676–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Wang X, Bonnard G, Shen Y, Xiu Z, Li X, Gao D, Zhang Z, Tan BC (2015) Empty pericarp7 encodes a mitochondrial E-subgroup pentatricopeptide repeat protein that is required for ccmFN editing, mitochondrial function and seed development in maize. Plant J 84: 283–295 [DOI] [PubMed] [Google Scholar]
- Sun T, Bentolila S, Hanson MR (2016) The unexpected diversity of plant organelle RNA editosomes. Trends Plant Sci 21: 962–973 [DOI] [PubMed] [Google Scholar]
- Takenaka M, Zehrmann A, Verbitskiy D, Härtel B, Brennicke A (2013) RNA editing in plants and its evolution. Annu Rev Genet 47: 335–352 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wang G, Wang G, Wang F, Song R (2012) A transcriptional roadmap for seed development in maize. In Seed Development: Omics Technologies toward Improvement of Seed Quality and Crop Yield. Springer, Dordrecht, The Netherlands, pp 81–97 [Google Scholar]
- Wang G, Zhong M, Shuai B, Song J, Zhang J, Han L, Ling H, Tang Y, Wang G, Song R (2017) E+ subgroup PPR protein defective kernel 36 is required for multiple mitochondrial transcripts editing and seed development in maize and Arabidopsis. New Phytol 214: 1563–1578 [DOI] [PubMed] [Google Scholar]
- Wang X, Ai G, Zhang C, Cui L, Wang J, Li H, Zhang J, Ye Z (2016) Expression and diversification analysis reveals transposable elements play important roles in the origin of Lycopersicon-specific lncRNAs in tomato. New Phytol 209: 1442–1455 [DOI] [PubMed] [Google Scholar]
- Welchen E, Hildebrandt TM, Lewejohann D, Gonzalez DH, Braun H-P (2012) Lack of cytochrome c in Arabidopsis decreases stability of complex IV and modifies redox metabolism without affecting complexes I and III. Biochim Biophys Acta 1817: 990–1001 [DOI] [PubMed] [Google Scholar]
- Weng L, Zhao F, Li R, Xu C, Chen K, Xiao H (2015) The zinc finger transcription factor SlZFP2 negatively regulates abscisic acid biosynthesis and fruit ripening in tomato. Plant Physiol 167: 931–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan HX, Fu DQ, Zhu BZ, Liu HP, Shen XY, Luo YB (2012) Sprout vacuum-infiltration: a simple and efficient agroinoculation method for virus-induced gene silencing in diverse solanaceous species. Plant Cell Rep 31: 1713–1722 [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35: 155–189 [Google Scholar]
- Yang YZ, Ding S, Wang HC, Sun F, Huang WL, Song S, Xu C, Tan BC (2017) The pentatricopeptide repeat protein EMP9 is required for mitochondrial ccmB and rps4 transcript editing, mitochondrial complex biogenesis and seed development in maize. New Phytol 214: 782–795 [DOI] [PubMed] [Google Scholar]
- Yuan H, Liu D (2012) Functional disruption of the pentatricopeptide protein SLG1 affects mitochondrial RNA editing, plant development, and responses to abiotic stresses in Arabidopsis. Plant J 70: 432–444 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Cui X, Wang Y, Wu J, Gu X, Lu T (2017) The RNA editing factor WSP1 is essential for chloroplast development in rice. Mol Plant 10: 86–98 [DOI] [PubMed] [Google Scholar]
- Zhu B, Yang Y, Li R, Fu D, Wen L, Luo Y, Zhu H (2015) RNA sequencing and functional analysis implicate the regulatory role of long non-coding RNAs in tomato fruit ripening. J Exp Bot 66: 4483–4495 [DOI] [PMC free article] [PubMed] [Google Scholar]