An unusual β-amylase from Arabidopsis (BAM2) is catalytically active in the presence of K+, exhibits sigmoidal kinetics, functions as a tetramer, and has a putative secondary carbohydrate-binding site.
Abstract
The Arabidopsis (Arabidopsis thaliana) genome contains nine β-amylase (BAM) genes, some of which play important roles in starch hydrolysis. However, little is known about BAM2, a plastid-localized enzyme reported to have extremely low catalytic activity. Using conservation of intron positions, we determined that the nine Arabidopsis BAM genes fall into two distinct subfamilies. A similar pattern was found in each major lineage of land plants, suggesting that these subfamilies diverged prior to the origin of land plants. Moreover, phylogenetic analysis indicated that BAM2 is the ancestral member of one of these subfamilies. This finding, along with the conservation of amino acids in the active site of BAM2, suggested that it might be catalytically active. We then identified KCl as necessary for BAM2 activity. Unlike BAM1, BAM3, and BAM5, three Arabidopsis BAMs that all exhibited hyperbolic kinetics, BAM2 exhibited sigmoidal kinetics with a Hill coefficient of over 3. Using multi-angle light scattering, we determined that BAM2 was a tetramer, whereas BAM5 was a monomer. Conserved residues from a diverse set of BAM2 orthologs were mapped onto a homology model of the protein, revealing a large, conserved surface away from the active site that we hypothesize is a secondary carbohydrate-binding site. Introduction of bulky methionine for glycine at two points on this surface reduced catalytic activity significantly without disrupting the tetrameric structure. Expression analysis indicated that BAM2 is more closely coexpressed with other starch degradation enzymes than any other BAM, suggesting that BAM2 may play an important role in starch degradation in plants.
β-Amylases (EC 3.2.1.2; CAZy Family 14) hydrolyze α-1,4-glycosidic bonds releasing maltose from the nonreducing ends of starch and are found in plants, green algae, and some bacteria. Most flowering plant genomes, including that of Arabidopsis (Arabidopsis thaliana), contain nine or more β-amylase (BAM) family members divided into four clades based on amino acid sequence alignments (Fulton et al., 2008). Some of these BAMs play important roles in hydrolyzing starch in various cell types, including BAM3 (referring to the Arabidopsis genes) in mesophyll cell chloroplasts mostly at night (Lao et al., 1999; Kaplan and Guy, 2005; Fulton et al., 2008; Monroe et al., 2014) and BAM1 in guard cell chloroplasts at the beginning of the day (Valerio et al., 2011; Horrer et al., 2016). Some BAMs are catalytically inactive pseudoenzymes, including BAM4 and BAM9, which are both plastidic and probably regulatory (Fulton et al., 2008; Li et al., 2009; J.D. Monroe, unpublished data), and BAM7 and BAM8, which contain N-terminal DNA-binding domains and are nuclear transcription factors (Reinhold et al., 2011; Soyk et al., 2014). Because of its relative abundance, BAM5 was the first Arabidopsis β-amylase purified and characterized (Monroe and Preiss, 1990). However, its cytosolic location in phloem tissue precludes the prediction of a logical function (Wang et al., 1995), and bam5 mutants lack an apparent phenotype (Laby et al., 2001). BAM6 has an N-terminal extension similar in length to those of plastid-targeted BAM proteins, is closely related to BAM5 (Fulton et al., 2008), and may be involved in starch metabolism in older plants (Monroe et al., 2014).
Little is known about BAM2 other than that it is plastid localized and that the purified enzyme was found to have extremely low catalytic activity (Fulton et al., 2008; Li et al., 2009). Observing no phenotype in leaves of 4-week-old bam2 plants, Fulton et al. (2008) suggested that BAM2 might lack a function and may have been recently derived from the BAM7 gene following the loss of its N-terminal DNA-binding domain. BAM2 was reported to have low starch-binding capacity, and it was speculated that this could be due to a unique four-amino acid insertion in a surface loop near the active site that is not present in the active BAMs (Li et al., 2009). In contrast to these previous findings, we observed a starch excess phenotype in 8-week-old bam2 leaves suggesting that BAM2 may play a role in older leaves (Monroe et al., 2014).
Detailed biochemical and structural analysis of a soybean (Glycine max) seed BAM revealed that it is a monomeric (β/α)8-fold protein with a deep catalytic cleft where two Glu residues act as a general acid and base during hydrolysis (Mikami et al., 1994). Thirteen additional residues in the active site of the soybean BAM form hydrogen bonds with starch (Laederach et al., 1999). Barley (Hordeum vulgare) and sweet potato (Ipomea batatas) BAMs were also crystallized; the barley enzyme is monomeric (Rejzek et al., 2011) but the sweet potato enzyme is a homotetramer, although the sweet potato monomer was catalytically active (Thoma et al., 1963; Cheong et al., 1995). Sequence alignments indicate that all of the crystallized plant BAMs are orthologs of Arabidopsis BAM5. Fulton et al. (2008) used these crystal structures to model the active sites of BAM3 and BAM4 and noticed that the catalytically active BAM3 shares all of the critical residues involved in starch binding and catalysis; however, there were numerous changes in BAM4 that might explain its catalytic inactivity.
Here, we describe a phylogenetic analysis of the BAM gene family, based on conserved intron positions and the occurrence of specific BAM orthologs in diverse land plant lineages, that sheds new light on the evolutionary history of BAM2. After finding that the activity of BAM2 requires KCl, we sought to characterize some of the properties of this highly unusual BAM protein. This analysis, coupled with coexpression data, suggests that BAM2 may play an important role in starch degradation.
RESULTS
Land Plant Genomes Contain Two BAM Subfamilies
Sequence alignments are typically used to reveal gene orthology among closely related species, but sequence changes over long spans of time or due to divergence of protein function can make it difficult to decipher the early history of gene families. Most eukaryotic genes contain introns, and the positions of introns within coding sequences can be conserved for long spans of time, making them excellent tools for understanding the evolution of gene families (Henricson et al., 2010).
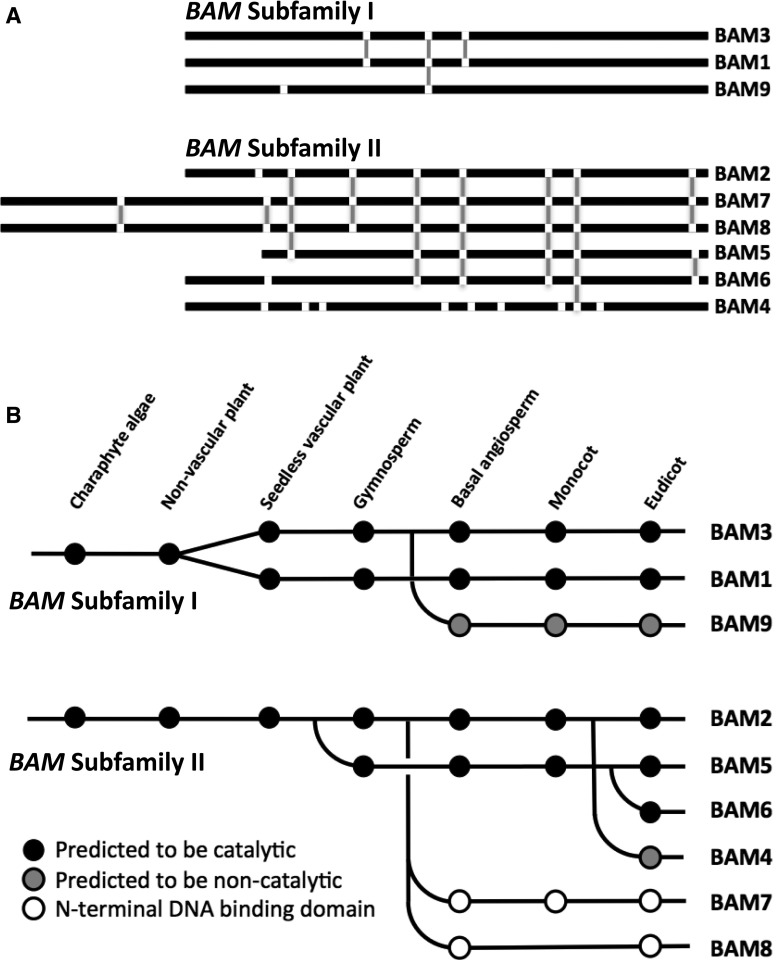
Intron positions within the nucleotide sequences of the nine Arabidopsis BAM genes were identified using the National Center for Biotechnology Information (NCBI) Gene pages (www.ncbi.nlm.nih.gov/gene/) and then mapped onto an amino acid sequence alignment generated using Clustal Omega (Sievers et al., 2011; Supplemental Fig. S1). Among the Arabidopsis BAM genes, two distinct patterns of conserved intron positions were apparent. BAM1, BAM3, and BAM9 each contain only two or three introns; BAM1 and BAM3 share all three of their intron positions, and one of these introns is shared by all three genes (Fig. 1A). The remaining six BAM genes contain six to nine introns, none of which are shared with any of the first set of BAM genes. However, five of these six BAM genes share five intron positions, three genes share six intron positions, and all six genes share one intron position (Fig. 1A). We refer to these clusters of genes as BAM subfamily I (containing BAM1) and BAM subfamily II (containing BAM2). Interestingly, the two plastid-localized pseudoenzymes, BAM4 and BAM9, are in different subfamilies. This is in contrast to their placement in the same clade based on multiple sequence alignments (Fulton et al., 2008). We believe that the intron position data are a more reliable indicator of ancestry/homology and that the placement of these BAMs in the same clade may have been a consequence of both of their sequences having diverged significantly from the catalytically active BAMs in both subfamilies.
Figure 1.
Conserved intron positions and evolutionary history of Arabidopsis BAM genes. A, Black lines represent aligned amino acid sequences, and white breaks indicate intron positions. Vertical gray lines represent conserved intron positions within multiple genes. B, Dots represent the presence of BAM genes within the genomes of species listed in the text. BAM genes in a single row were judged to be orthologous based on sequence alignments and domain structure. Predictions of catalytic activity or inactivity were based on the conservation of active site residues.
We then attempted to determine when the two subfamilies became separated, by mapping intron positions onto an alignment of each of the BAM proteins from a diverse set of land plant genomes, including those from four angiosperms: the basal species (Amborella trichopoda), a monocot (Sorghum bicolor), and two eudicots (Arabidopsis and Vitis vinifera; Supplemental Fig. S2), a seedless vascular plant (Selaginella moellendorffii) and a gymnosperm (Picea abies; Supplemental Fig. S3), two nonvascular plants (Physcomitrella patens and Marchantia polymorpha), and a charaphyte alga (Klebsormidium flaccidum; Supplemental Fig. S4), all obtained from public databases. K. flaccidum is considered to be closely related to the algal ancestor of all land plants (Leliaert et al., 2012). Reference sequences were used when possible. BAM sequences from Pinus taeda were included in the alignment shown in Supplemental Figure S3, but intron positions were not available. As in the Arabidopsis BAM genes, a similar pattern of two distinct subfamilies of BAM genes distinguishable by shared intron positions was found in each of the land plant genomes, suggesting that the two subfamilies were separated prior to the origin of land plants around 450 million years ago.
In order to hypothesize the origins of each of the Arabidopsis BAM genes, we used a combination of approaches, including the intron positions, amino acid sequence alignments (Supplemental Figs. S2–S4), phylogenetic trees (Supplemental Figs. S5–S7), and close examination of domains and sequence motifs that define a given clade. Most of the eudicot genomes examined contain at least one each of the BAM sequences found in Arabidopsis (Fig. 1B). Monocot genomes appear to lack BAM4, BAM6, and BAM8 but possess all of the remaining BAMs. The basal angiosperm, A. trichopoda, contains a BAM8 sequence but lacks BAM4 and BAM6. Two gymnosperm genomes were analyzed: P. taeda and Picea alba, each of which contains two clusters of three or four BAMs that are closely related to BAM1 and BAM3. The pine genome also contains one additional BAM gene that aligns most closely with BAM5, while the spruce genome contains one additional BAM gene that aligns most closely with BAM2 (Supplemental Fig. S3).
Alignments of BAM sequences from the seedless vascular plant S. moellendorffii revealed seven BAMs, two pairs in subfamily I that aligned with BAM1 and BAM3 and three BAMs in subfamily II that aligned most closely with BAM2. Two nonvascular plant genomes from the moss P. patens and the liverwort M. polymorpha also were examined. P. patens contains seven BAMs, four of which are in subfamily I and align most closely with the BAM1/BAM3 clade and three that are most closely related to BAM2. M. polymorpha has five BAMs, but only three of these appear to be similar to any of the Arabidopsis clades. Four of the M. polymorpha genes are in subfamily I, and two of these resemble the BAM1/3 clade. The fifth M. polymorpha BAM is in subfamily II, and it resembles BAM2 in terms of intron positions and sequence conservation. K. flaccidum contains five BAM sequences, two of which resemble BAM1/3 in sequence and intron position and two of which resemble BAM2.
These findings were used to trace the origin of each Arabidopsis BAM gene back to the early land plant ancestors. Starting with subfamily I, proteins similar to BAM1 or BAM3 were found in each genome examined, indicating that the BAM1/BAM3 clade is ancient (Fig. 1B). The split of this clade into two distinct forms was observed in the seedless vascular plant but not in either of the nonvascular plant genomes, suggesting that the split of BAM1 and BAM3 occurred prior to the origin of vascular plants. BAM9-like proteins were found in each of the angiosperms but not in any other lineage of land plants, indicating that BAM9 evolved from the BAM1/3 clade prior to the origin of angiosperms. We could not determine whether BAM9 originated from BAM1 or BAM3.
Subfamily II contains the remaining six Arabidopsis BAMs, and they fall into three distinct clades based on sequence alignment (Fulton et al., 2008). The clade containing BAM2 also includes the nuclear transcription factors BAM7 and BAM8. Each of the genomes examined contains at least one member of this clade, but proteins like BAM7 and BAM8 that contain the N-terminal DNA-binding domain are found only in angiosperms and probably evolved relatively recently (Fig. 1B). Proteins resembling BAM5 are found only in one of the gymnosperms examined and in angiosperms. Proteins resembling BAM6 are difficult to distinguish from BAM5 and might be found only in the Brassicaceae. Taken together, this analysis reveals that, prior to the origin of seed plants, the BAM proteins from subfamily II are structurally similar to BAM2, indicating that BAM2 is probably the founding member of subfamily II in land plants.
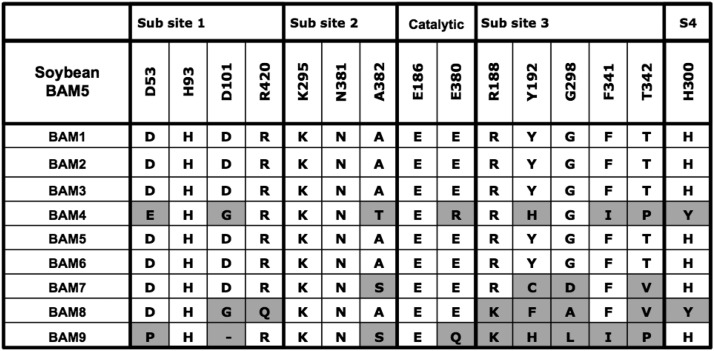
Identification of the active site residues that bind starch was then used to predict whether Arabidopsis BAM2 was likely to be catalytically active or not. Each of the residues identified as binding starch in the soybean BAM active site (Laederach et al., 1999) was identified in an alignment of the Arabidopsis BAMs (Fig. 2). As was determined previously, BAM4 and BAM9 do not share all of these residues, and BAM4 is known to be catalytically inactive (Fulton et al., 2008; Li et al., 2009). BAM7 and BAM8 have low or no catalytic activity (Reinhold et al., 2011), and some of the active site residues that bind starch are not conserved in these proteins. In the BAMs that are known to be catalytically active, BAM1, BAM3, and BAM5 contain all of the conserved active site residues. Those residues also are perfectly conserved in BAM2. Because BAM2 appears to have given rise to the catalytically active BAM5 (Fig. 1B), and because it shares all of the active site residues that are known to bind starch in the catalytically active forms (Fig. 2), we suspected that BAM2 was catalytically active under certain conditions.
Figure 2.
Conservation in the nine Arabidopsis BAMs of 15 starch-binding active site residues identified in soybean BAM5 (BMY1) by Laederach et al. (1999). Subsites 1 to 4 refer to the four Glc residues at the nonreducing end of the substrate. Residues in each Arabidopsis BAM that differ from the corresponding residues in the soybean BAM are shaded gray.
BAM2 Activity Requires High Ionic Strength
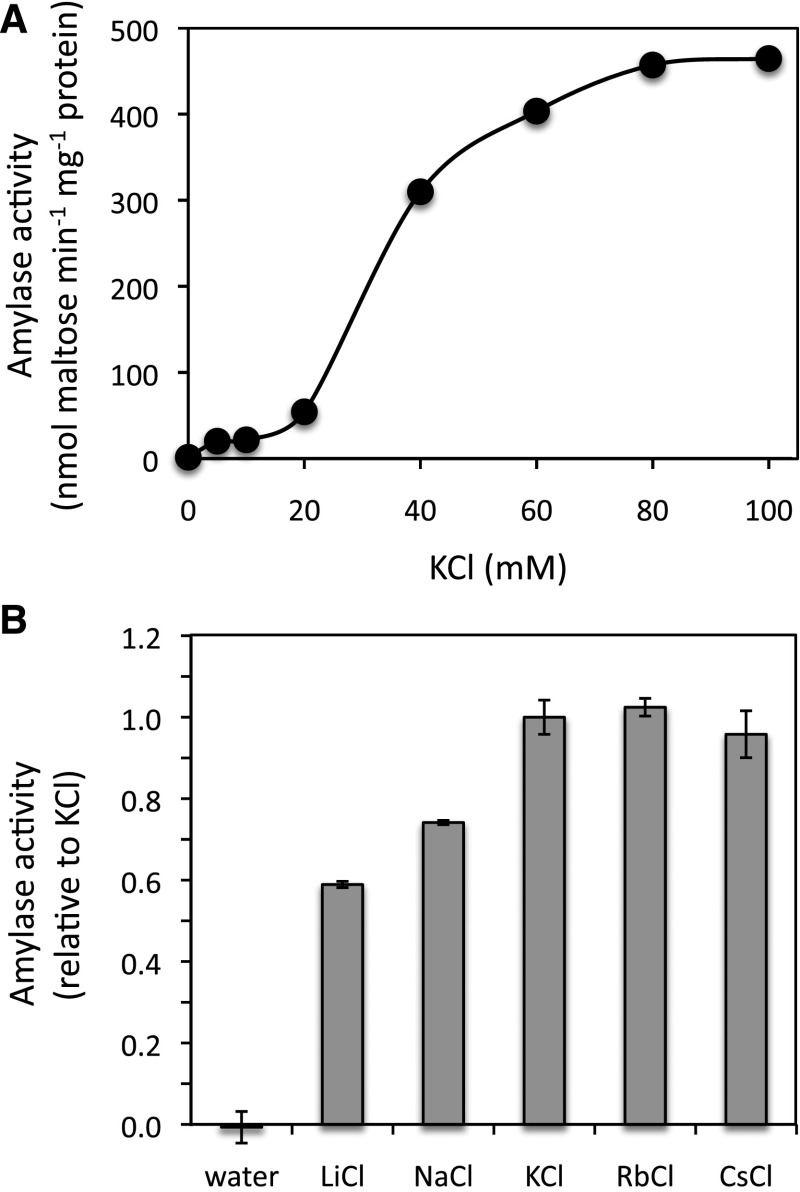
In order to characterize Arabidopsis BAM2, a cDNA clone was initially amplified using PCR from cDNA synthesized from mRNA that we isolated from Arabidopsis plants (Columbia ecotype). The BAM2 cDNA was inserted into pEZT-NL for the purpose of protein localization. We then amplified the coding sequence from that plasmid, such that the predicted 55-amino acid N-terminal chloroplast transit peptide (Fulton et al., 2008) would be omitted and inserted the coding sequence for the mature protein into pETDuet-2 for expression with an N-terminal 6-His tag. The clone was then sequenced, and the sequence encoding the mature protein was found to be identical to NM_116273.5. The expression of BAM1 and BAM3 from pET29a with C-terminal 6-His tags was described previously (Monroe et al., 2014). A full-length BAM5 cDNA was synthesized de novo based on the reference sequence NM_117609 and expressed in pET29a with a C-terminal 6-His tag. After purification to near homogeneity (Supplemental Fig. S8), the enzyme activities were measured using soluble starch (Acros Organics; no. 42449) as the substrate in 50 mm MES buffer, pH 6, with no additional salts. The maltose produced was quantified using the Somogyi-Nelson method (Nelson, 1944). We used this substrate because the low solubility of both amylopectin and amylose precluded the measurement of kinetic parameters (see below). Compared with active BAMs, the activity of BAM2 was negligible, as was observed by others (Fulton et al., 2008; Li et al., 2009). After testing a variety of additions to the assay mixture, including reduced and oxidized DTT and various salts, we discovered that the enzyme was active in the presence of KCl, reaching maximum activity with about 80 mm KCl when the enzyme was assayed with 100 mg mL−1 soluble starch (Fig. 3A). Activity without KCl was negligible. The products of the reactions of BAM2 and BAM5 with soluble starch were both confirmed to be maltose as determined by thin-layer chromatography (TLC; Supplemental Fig. S9).
Figure 3.
Effect of salts on the activity of purified BAM2 using 100 mg mL−1 soluble starch as the substrate. A, Effect of KCl on the specific activity of BAM2. B, Effect of 100 mm salts on the activity of BAM2 using 100 mg mL−1 soluble starch. Activity is relative to that with KCl, and columns represent means ± sd (n = 3).
Many enzymes are reported to require or to have elevated activity in the presence of monovalent ions, and these effects can be either specific, in which the ion(s) form part of the enzyme’s structure, or general, in which the enzyme simply requires an elevated ionic strength for activity (Gohara and Di Cera, 2016). None of the crystallized plant BAMs contained any ions, and the active site residues of BAM2 are well conserved, so it seemed likely that the ion effect on BAM2 was general in nature. If this were the case, then other ions should produce similar effects. We tested a variety of cations at a concentration of 100 mm, including the chloride salts of Li+, Na+, K+, Rb+, and Cs+, and all activated BAM2 to varying degrees, suggesting that the effect was not specific (Fig. 3B). Similar concentrations of CaCl2 and MgCl2 were much less effective at activating BAM2 than KCl, but KSO4 and K+ acetate were as effective as KCl, suggesting that the important ion was a cation (Supplemental Fig. S10). In contrast, the activity of BAM5 was unaffected by any of the salts tested (Supplemental Fig. S11). Potassium is the probable physiological activator due to its higher concentration (120–200 mm) in the chloroplast stroma (Finazzi et al., 2015). With the discovery of BAM2’s catalytic activity, we next wanted to compare the kinetic properties of BAM2 with those of the three other active BAM enzymes.
BAM2 Exhibits Cooperative Kinetics and Is Tetrameric
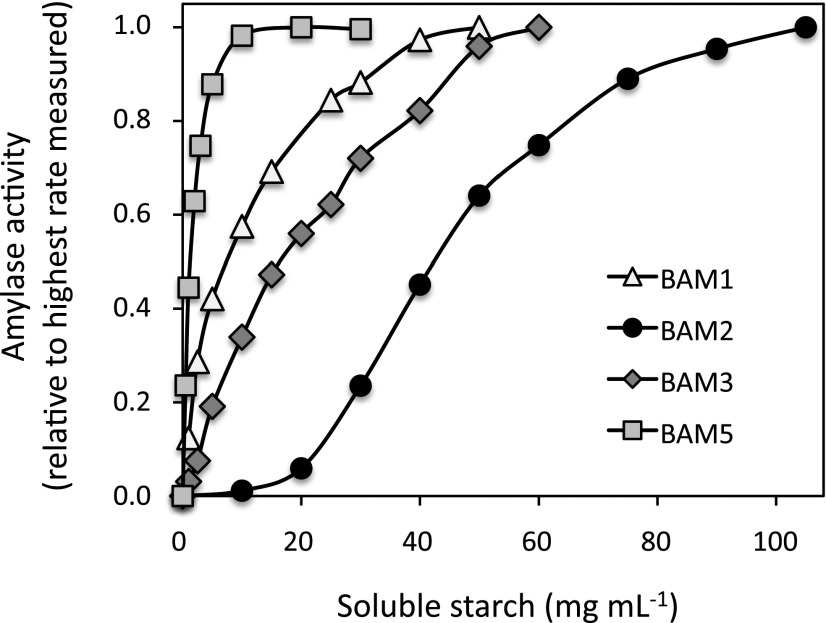
We compared the catalytic activities of His-tagged forms of BAM1, BAM2, BAM3, and BAM5 using soluble starch as the substrate (Fig. 4), measuring their Vmax, Km, and Hill coefficients as determined by fitting the data to the Michaelis-Menten equation using Excel Solver (Gadagkar and Call, 2015; Table I). Of the four enzymes, BAM1 and BAM3 had Vmax values that were about 3-fold higher than those of BAM2 and BAM5. The Km of BAM2 was the highest, at about 46 mg mL−1 soluble starch, whereas BAM5’s Km was the lowest, at 2.3 mg mL−1 soluble starch. Comparing BAM1 and BAM3, BAM1 had a higher affinity for the substrate, with a Km of about 12 mg mL−1 soluble starch, whereas BAM3 had a Km of about 26 mg mL−1 soluble starch. When the activities of these two enzymes were measured at lower substrate concentrations, BAM1 had higher specific activity than BAM3 despite its lower Vmax, as was observed earlier using amylopectin as the substrate (Li et al., 2009). BAM1, BAM3, and BAM5 all had Hill coefficients of about 1.3 or less, indicating a lack of cooperativity (Table I). Surprisingly, BAM2 displayed very different kinetics than that of the other BAMs and had a Hill coefficient of over 3 (Fig. 4; Table I). BAM2 was essentially inactive at substrate levels below 10 mg mL−1 and only became saturated at over 100 mg mL−1 soluble starch. The viscosity of the substrate precluded the use of higher concentrations of soluble starch.
Figure 4.
Effect of soluble starch concentration on the activity of purified Arabidopsis BAM1, BAM2, BAM3, and BAM5. All points are relative to the highest rate measured for each enzyme and represent duplicate assays.
Table I. Kinetic parameters (Vmax, Km, and Hill coefficient) of purified BAM1, BAM2, BAM3, and BAM5 from Arabidopsis.
The substrate was soluble starch. Values are means ± sd from three replicate data sets. Within each parameter, means that are not significantly different (P > 0.05) share the same letter (Student’s t test).
| Enzyme | Vmax | Km | Hill Coefficient |
|---|---|---|---|
| nmol min−1 mg−1 protein | mg mL−1 soluble starch | ||
| BAM1 | 892 ± 59 a | 11.9 ± 2.9 a | 0.91 ± 0.09 a |
| BAM2 | 393 ± 52 b | 45.8 ± 2.8 b | 3.11 ± 0.21 c |
| BAM3 | 1,119 ± 91 c | 26.3 ± 8.8 c | 1.13 ± 0.12 b |
| BAM5 | 207 ± 23 d | 2.3 ± 0.1 d | 1.32 ± 0.07 a,b |
The observation that BAM2 exhibits cooperativity suggested that it either functions as a multimeric protein in which the subunits interact, analogous to phosphofructokinase (Berg et al., 2002), or that it possesses a secondary binding site (SBS) away from the active site and that a ligand binding to the SBS influences the active site, similar to the monomeric Bacillus circulans xylanase (Ludwiczek et al., 2007). It is also possible that both predictions are true. The BAM5 ortholog from sweet potato was crystallized as a homotetramer, but that enzyme did not exhibit cooperative kinetics and the protein was active as a monomer (Cheong et al., 1995). SBSs are found on some polysaccharide-active enzymes (Møller and Svensson, 2016), where they can serve a variety of functions, including localizing the enzyme near its substrate, guiding the substrate into the active site, enhancing processivity, or causing allosteric regulation (Cuyvers et al., 2012). A crystallized β-amylase from Bacillus cereus var mycoides was shown to contain a C-terminal carbohydrate-binding module (CBM) and several SBSs. Mutational analysis showed that the CBM and one of the SBSs facilitated binding to raw starch and enhanced catalytic activity (Ye et al., 2004). However, to date, no CBM or SBS has been documented in any plant β-amylase, and no β-amylase has been observed to display cooperative kinetics.
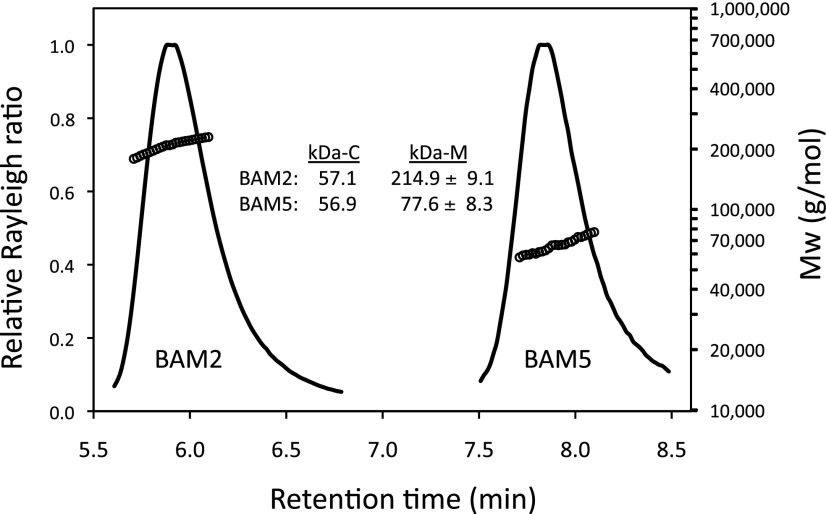
In order to determine the multimeric state of BAM2, we subjected the purified protein to size-exclusion chromatography-multi-angle light scattering (SEC-MALS). The calculated molecular mass of the recombinant BAM2 monomer is 57.1 kD. However, the average molecular mass from the SEC-MALS analysis of purified BAM2 in a solution of 10 mm MOPS, pH 7, and 250 mm KCl was 214.9 ± 9.1 kD (n = 6), so, under conditions in which BAM2 was active, the ratio of the apparent mass to the predicted mass was 3.8:1, suggesting that it was a tetramer (Fig. 5). In contrast, purified BAM5, having a calculated molecular mass of 56.9 kD, was monomeric in solution, with an average measured molecular mass of 77.6 ± 8.3 kD (n = 4).
Figure 5.
SEC-MALS elution profiles (representative) and masses of BAM2 and BAM5 from Arabidopsis. Purified proteins were in 10 mm MOPS, pH 7, and 250 mm KCl. kDa-C is the calculated mass from the mature amino acid sequence of the expressed proteins, and kDa-M is the average mass of the active protein measured using MALS ± sd. For BAM2, n = 6; for BAM5, n = 4.
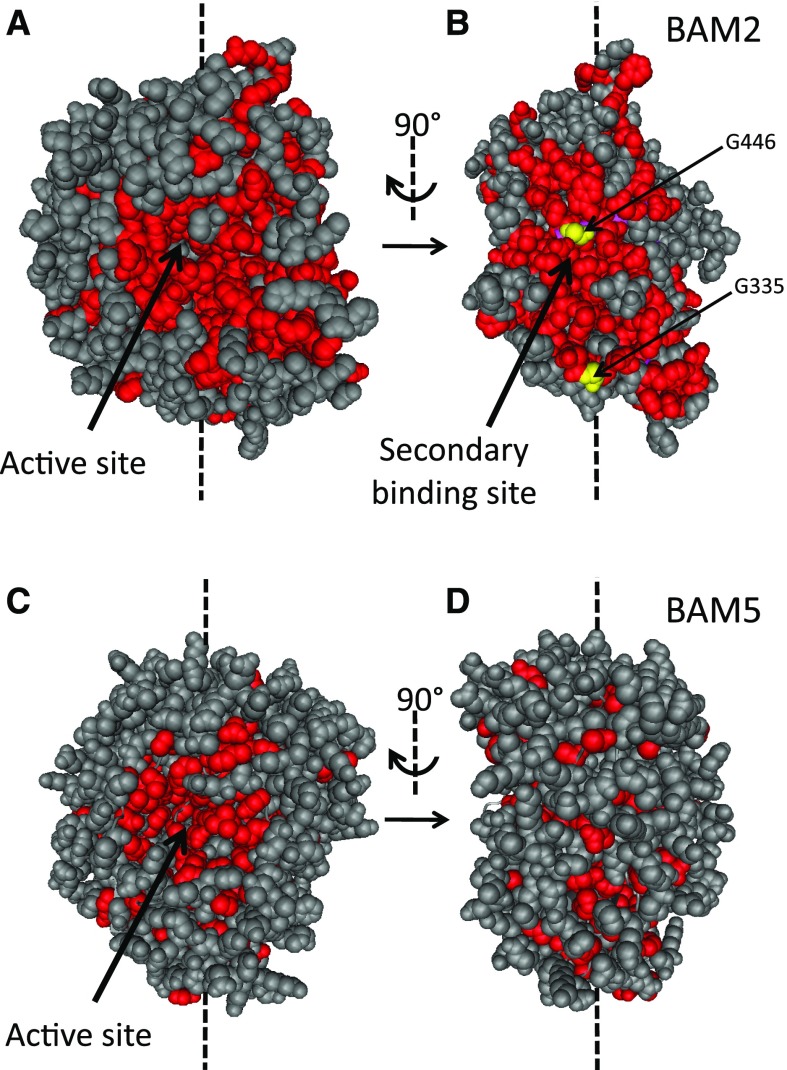
SBSs are usually discovered by observing ligands bound to crystallized proteins (Møller and Svensson, 2016). Once a bound ligand is observed, a predicted SBS can then be confirmed using mutagenesis to disrupt ligand binding followed by activity or substrate-binding assays. Lacking a crystal structure for BAM2, we used an alternative method, based on the prediction that the SBS for binding a polysaccharide would be a relatively large surface that is conserved among BAM2 orthologs. Smaller surfaces involved in subunit contact sites are typically not highly conserved (Caffrey et al., 2004). We aligned the amino acid sequences of BAM2 orthologs from Arabidopsis, five other flowering plants, P. patens, and S. moellendorffii and then identified all of the perfectly conserved residues. These residues were highlighted in a homology model of Arabidopsis BAM2 generated using I-TASSER (Yang et al., 2015) and viewed using Cn3D (version 4.3; Wang et al., 2000; Fig. 6, A and B). The conserved residues (colored red) with significant solvent exposure were visualized using space-fill mode. Conserved amino acids in the active site region of BAM2 were readily apparent (Fig. 6A). After rotating the model 90°, another large conserved surface was visible on the side of the protein (Fig. 6B). A similar analysis of BAM5, which is the closest catalytically active Arabidopsis paralog of BAM2 (Fig. 1B), lacked conservation in the same region (Fig. 6D), although the active site was conserved (Fig. 6C). We hypothesize that this secondary conserved surface in BAM2 is an SBS and that it binds starch or a product of starch degradation. Efforts to detect starch binding to BAM2 using starch-containing PAGE or pull-down assays using insoluble starch were inconclusive, perhaps because the affinity of the putative SBS of BAM2 for starch was too low or because the native ligand is soluble. Therefore, we used mutagenesis to test for the presence of a functionally important SBS in BAM2.
Figure 6.
Homology structures of Arabidopsis BAM2 and BAM5 illustrating conserved, surface-exposed residues. A, Structure of BAM2, viewing the active site in which nonconserved residues are colored gray and conserved residues are colored red. B, Same structure as in A but rotated 90°. Two conserved Gly residues that were mutated to Met are colored yellow. C, Structure of BAM5, viewing the active site colored as in A. D, Same structure as in C but rotated 90°.
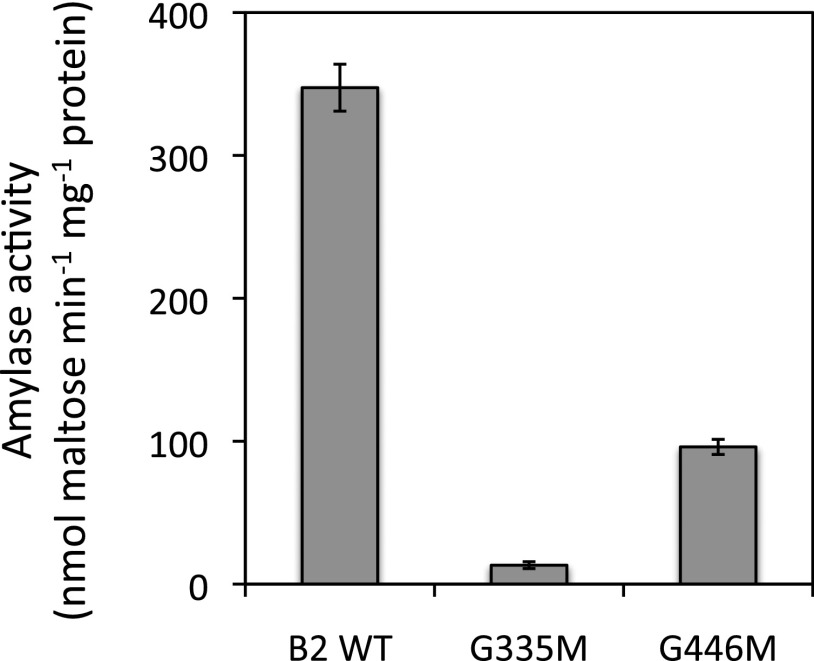
If the conserved surface of BAM2 is an SBS, we predicted that mutations introducing bulky side chains into the SBS would disrupt starch binding and prevent activation of the enzyme. Two conserved Gly residues, one in the middle of the putative SBS (Gly-446) and one at its edge (Gly-335; highlighted yellow in Fig. 6B), were replaced with Met residues, thus avoiding the insertion of charged or hydrophobic residues that might have secondary effects. The mutant proteins were purified and assayed using 100 mg mL−1 soluble starch hydrolysis in the presence of 250 mm KCl. When assayed with nearly saturating levels of soluble starch, the activity of both mutants was lower than the wild-type activity: G335M by over 95% and G446M by about 70% (Fig. 7). SEC-MALS analysis revealed that both mutant proteins were tetrameric (Supplemental Fig. S12), indicating that the mutations did not interfere with the protein’s quaternary structure. This was expected based on the homology model of BAM2, in which both Gly side chains appear to be exposed to the solvent.
Figure 7.
Catalytic activity of wild-type (WT) BAM2 and two mutants of BAM2 generated by site-directed mutagenesis in which Gly residues at positions 335 and 446 were changed to Met. Assays were conducted with 100 mg mL−1 soluble starch and 250 mm KCl. Bars represent activity ± sd (n = 3).
BAM2 Is Coexpressed with Most of the Genes Involved in Starch Degradation
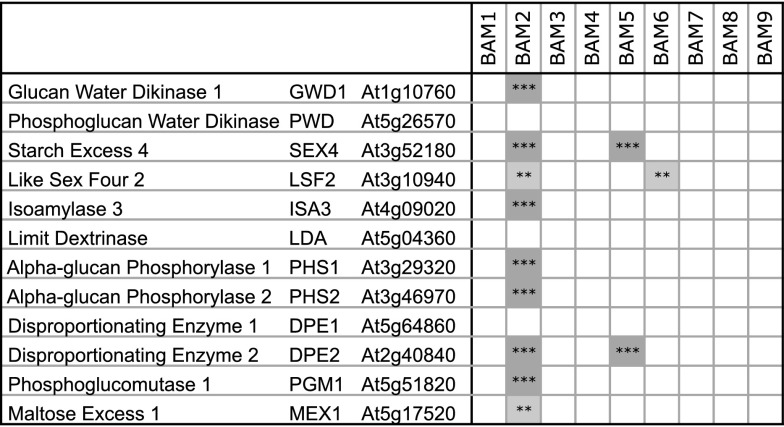
If BAM2 plays a role in starch degradation, we reasoned that its expression might be coordinated with that of other enzymes involved in starch degradation. Twelve Arabidopsis genes have been mentioned in several recent reviews (Zeeman et al., 2010; Smirnova et al., 2015) as being involved directly in starch degradation (Fig. 8). Using the ATTED-II database (http://atted.jp/; Aoki et al., 2016), we queried the lists of genes coexpressed with each of the Arabidopsis BAMs across plant development, looking for each of the 12 starch degradation-associated genes, and the results were striking. Neither BAM1 nor BAM3, the only two BAMs for which there is strong phenotypic evidence for roles in starch degradation, is significantly coexpressed with any of the 12 starch degradation genes, indicating that their roles in starch degradation may be specific to only certain cell types. BAM5 is coexpressed with two of the 12 genes with high significance (P < 0.001, as defined by ATTED-II), despite it being cytosolic and without a known role in plastidic starch degradation. BAM6 is coexpressed with one of the 12 genes at a lower significance level (P < 0.01). In contrast, BAM2 is coexpressed with nine of the 12 genes, seven at the highest significance level (Fig. 8). While protein levels do not always correlate with transcript levels (Skeffington et al., 2014), the fact that BAM2 is broadly coexpressed with most genes involved in starch metabolism does suggest a functional role for BAM2.
Figure 8.
Coexpression analysis from the ATTED-II database (http://atted.jp/; Aoki et al., 2016) for each Arabidopsis BAM gene with 12 genes involved in starch degradation. Significance levels as provided by the ATTED-II database are P < 0.001 (***) and P < 0.01 (**).
DISCUSSION
The BAM gene family in Arabidopsis contains nine members: five are catalytically active, and four appear to be pseudoenzymes that were likely derived from catalytically active ancestors. After gene duplication, pseudoenzymes adopt new noncatalytic functions, and residues that were once essential for catalytic activity often change by mutation (Jackson et al., 2015). BAM2 was proposed to have been derived relatively recently from BAM7 (Fulton et al., 2008), one of the catalytically inactive BAMs in which four of the 15 starch-binding active site residues are different from those in the active BAMs (Fig. 2). However, BAM2 has all of the conserved active site residues found in active BAMs, and it is unlikely that all residues lacking in BAM7 would have simultaneously mutated back to those necessary for catalytic activity. Analysis of intron positions (Fig. 1A) and the occurrence of orthologs in diverse land plant genomes (Fig. 1B) revealed that BAM2 originated much earlier than BAM7 and is probably the founding member of one of two subfamilies of BAM genes that became separated prior to the origin of land plants. Together, these observations led us to suspect that BAM2 is probably catalytically active.
After testing a variety of common laboratory reagents, we discovered that increasing the concentration of KCl stimulated BAM2 activity (Fig. 3A). Assays using other monovalent and divalent ions revealed that the effect was strongest with KCl, RbCl, and CsCl, but other salts also were effective (Fig. 3B), suggesting that the effect probably does not involve a specific ion-binding site but is associated with the ionic strength of the assay solution in general. Replacing K+ with cations other than Rb+ and Cs+ resulted in lower activity (Fig. 3B), but replacing Cl− with other anions had no effect (Supplemental Fig. S10), so the activating ion is likely to be a cation. Many other enzymes have been shown to behave similarly (for review, see Gohara and Di Cera, 2016; Okur et al., 2017). In chloroplasts, the cation composition is dominated by K+, which may be as high as 200 mm (Finazzi et al., 2015), so K+ is likely to be the in vivo activator of BAM2. KCl levels in chloroplasts fluctuate under stress conditions (Schröppel-Meier and Kaiser, 1988), so it is possible that BAM2 activity is regulated in vivo by ionic strength. Alternatively, a certain ionic strength may be needed simply to stabilize the active conformation of BAM2.
Compared with BAM5, which has a low Km for soluble starch, the affinity of BAM2 for starch is about 20 times lower (Table I). However, being located in the phloem cytosol and with an unknown function, the natural substrate for BAM5 may actually be a small glucan (Wang et al., 1995); thus, it is understandable that BAM2 differs significantly from BAM5. The affinities of BAM1 and BAM3 for soluble starch are only 4- and 2-fold higher than that of BAM2, respectively (Table I). The fact that starch is insoluble, with a high density of nonreducing ends on its surface, makes it difficult to interpret the significance of Km values for these hydrolases, but the plastidic, catalytically active BAMs do not seem to be fundamentally different in their affinity for starch. However, BAM2 is unique among characterized BAMs in exhibiting cooperative kinetics with a Hill coefficient of over 3 (Fig. 4; Table I). Cooperativity in enzyme activity can result from interactions between subunits (Berg et al., 2002) or from the binding of a substrate to an SBS (Ludwiczek et al., 2007). Our results indicate that BAM2 is a tetramer (Fig. 5), and indirect evidence suggests that it may have an SBS (Figs. 6 and 7). The size of the conserved surface and the large distance between Gly-335 and Gly-446 (21.8 Å) is consistent with the idea that this SBS may bind starch and not a small molecule, but confirmation will require more detailed structural analysis.
Another enzyme involved in leaf starch metabolism was recently reported to exhibit cooperativity. The kinetics of Arabidopsis starch branching enzyme2 (BE2.2), which introduces α-1,6 linkages in the formation of amylopectin, was sigmoidal, with a Hill coefficient of 3.4 (Wychowski et al., 2017). Unlike BAM2, which was tetrameric in the absence of its substrate (Fig. 5), BE2.2 was monomeric in the absence of amylopectin but is clearly altered by, and may become oligomeric by, binding to its substrate.
Unlike the well-known cooperative binding of allosteric effectors to phosphofructokinase tetramers that regulate glycolysis under varying cellular energy demands (Berg et al., 2002), the physiological benefit of cooperativity in BAM2 is less clear. In contrast, there are many described benefits of an SBS in enzymes that act on polysaccharides. Binding of an enzyme’s substrate to the SBS can enhance catalytic activity by a variety of mechanisms, including keeping the enzyme in the vicinity of its insoluble substrate or by guiding the substrate to the active site (Cuyvers et al., 2012). Notably, β-agarase acts on double-helical agarose chains in which a large SBS on the side of the enzyme was proposed to facilitate the unwinding of the substrate (Allouch et al., 2004). Amylopectin also contains double helical chains of about 12 to 15 Glc units at the surface of starch granules (Zeeman et al., 2010) and, in that state, they are recalcitrant to hydrolysis. The currently accepted model to explain how hydrolases gain access to amylopectin chains is that two kinases, glucan water dikinase (GWD) and phosphoglucan water dikinase, phosphorylate amylopectin at C6- and C3-hydroxyls. The resulting ionic repulsion separates the strands, allowing hydration and access by hydrolases (Hejazi et al., 2008). Before the glucan chains can be completely hydrolyzed, however, two phosphatases, Starch Excess4 (SEX4) and Like Sex Four2, are required to remove the phosphates (Wilkens et al., 2016). It is possible that the SBS on BAM2 allows it to act alone to unwind and hydrolyze pairs of amylopectin chains. However, this seems unlikely, given that BAM2 is highly coexpressed with GWD1 and SEX4 (Fig. 8). Alternatively, as BAMs cannot hydrolyze starch processively without releasing the substrate to allow phosphatases to remove the phosphates, having an SBS may prevent BAM2 from diffusing away from the starch granule, effectively increasing its catalytic rate.
Despite the progress made here in characterizing the catalytic activity of BAM2, its physiological role remains a significant question. BAM2-deficient mutants accumulate starch in older leaves (Monroe et al., 2014), suggesting that BAM2 either has a minor role throughout leaf development that only becomes apparent in older leaves due to a slow, continual starch buildup or that it plays an important role only in older leaves. Microarray data from leaves harvested throughout development indicate that BAM2 expression is highest in younger leaves and then declines over time (Breeze et al., 2011), supporting the first hypothesis. Other microarray experiments suggest that BAM2 is expressed throughout the plant but especially in leaves at the onset of the night, in guard cells, in the central region of the root cap, and in seed endosperm (Winter et al., 2007), tissues that are known to accumulate starch. Unlike BAM1 and BAM3 (Monroe et al., 2014), publicly available microarray data suggest that BAM2 expression is not strongly affected by abiotic stress. Additionally, analysis of coexpression data revealed that BAM2 is highly coexpressed with most of the genes currently associated with starch degradation across plant development and tissues (Fig. 8). These expression patterns and the conservation of BAM2 across land plants indicate that BAM2 probably plays an important role in starch degradation, but that role is unknown at present.
Although the role of BAM2 in Arabidopsis is undetermined, what is clear is that BAM2 should not be dismissed as an essentially inactive enzyme resulting from a recent gene duplication. Intron conservation and sequence alignments indicate that BAM2 is an ancient enzyme found in all land plants, and we found that Arabidopsis BAM2 has significant catalytic activity when given certain physiologically compatible conditions. The presence of BAM2 in early land plants that contained fewer BAMs indicates that it probably played an essential role in starch degradation. The many unique features of Arabidopsis BAM2 activity and structure reported here point to a novel mechanism of action that deserves further study.
MATERIALS AND METHODS
Production and Purification of Recombinant Proteins
For the expression of BAM2, a cDNA clone was initially isolated from cDNA that was synthesized from mRNA isolated from Arabidopsis (Arabidopsis thaliana) ecotype Columbia leaves for the purpose of localizing the protein. Total RNA was transcribed into cDNA with the iScript cDNA synthesis system (Bio-Rad). From this template, a full-length BAM2 clone was isolated using the primers 5′-GGGAATTCATGGCGATTAGGTTGAATCATAGTG-3′ and 5′-ATGGATCCATCTCGGGGTTGGTCTCTTGTGT-3′ and ligated into pEZT-NL. We then used this plasmid as the template for a PCR to generate a clone lacking the predicted 55-amino acid N-terminal chloroplast transit peptide (Fulton et al., 2008). These primers were 5′-AAGGATCCAGCAGAGAGTACTGAGGAAGATCGAG-3′ and 5′-TTGTCGACTCACTCGGGGTTGGTCTCTTG-3′. After cloning the PCR product into pMOSBlue (GE Life Sciences), the resulting plasmid was digested with BamHI and SalI and ligated into pETDuet-1 for protein expression with an N-terminal 6-His tag. The resulting plasmid was sequenced to verify that the insert was identical to NM_116273.5. Plasmids for expressing BAM1 and BAM3 in Escherichia coli were a gift from H. Reinhold and were described by Monroe et al. (2014). A full-length BAM5 cDNA was synthesized de novo based on the reference sequence NM_117609 (GeneScript) in pET29a, so that the expressed protein had a C-terminal 6-His tag. Each plasmid was then transformed into BL21+ Escherichia coli cells for protein expression. Protein expression and purification were as described by Monroe et al. (2014). The concentration of the purified proteins was determined using the Bio-Rad Protein Assay Kit with BSA as the standard.
Amino acid substitutions were generated using the QuikChange mutagenesis strategy (Agilent Technologies). Mutagenesis primers contained the desired mutation flanked on either side by ∼20 nucleotides complementary to the template sequence. Primer sequences were as follows: B2_G335M, 5′-GGACTGGATTTTTTCGCGATATGGGCGATTATGACAGCTACTATG-3′, and B2_G446M, 5′-AGGCTCTGGCAGATCCAGAAATGCTAGTTTGGCAGGTGCTGAATG-3′, along with their complementary sequences. The pETDuet-1 plasmid containing wild-type BAM2 was used as the template. The PCR protocol, used to amplify the entire plasmid, was as follows: 95°C for 5 min; cycle of 95°C for 30 s, 60°C for 30 s, and 72°C for 4 min repeated 20 times; and final extension at 72°C for 10 min. PCR products were digested with DpnI for 1.5 h at 37°C and then transformed into DH5α E. coli competent cells. Amino acid substitutions were confirmed by sequencing of the plasmid insert at Eurofins Genomics.
Enzyme Activity Assays
Purified BAM enzymes were diluted with 50 mm MOPS, pH 7, containing 1 mg mL−1 BSA or porcine gelatin. Amylase assays were conducted in 0.5 mL containing 50 mm MES (pH 6) and various amounts of soluble starch (Acros Organics). Unless noted, BAM2 assays also included 250 mm KCl. Reactions were stopped after 20 min by immersion in a boiling water bath for 3 min; then, reducing sugars were measured by the Somogyi-Nelson assay (Nelson, 1944) with maltose as the standard. Vmax, Km, and Hill coefficients were determined by fitting the data to the Hill equation using Excel Solver (Gadagkar and Call, 2015). Products of the reactions of BAM2 and BAM5 with soluble starch were analyzed by TLC according to the method of Seibold et al. (2006).
MALS
Proteins at a concentration of 1 to 2 mg mL−1 were injected into a 4.6- × 300-mm size-exclusion column with a particle size of 5 µm and a pore size of 300 Å at a flow rate of 0.5 mL min−1 in filter-sterilized (0.2 µm) 10 mm MOPS, pH 7, with or without 250 mm KCl. Absorbance data were collected on an Agilent G1315B Diode Array detector at 280 and 212 nm, and light scattering was measured on a miniDAWN-TREOS (Wyatt Technologies). Data were analyzed using ASTRA (version 6.1.5.22). The predicted molecular mass and extinction coefficient of each monomer were determined using the Protein Identification and Analysis Tools on the ExPASy Server (Gasteiger et al., 2005).
Phylogenetic Analysis
BAM reference sequences were obtained from the NCBI database. Picea abies BAM sequences were obtained from the Spruce Genome Project (http://congenie.org/). Intron positions within the coding sequences were identified using NCBI Gene pages and located in amino acid sequences that were aligned using Clustal Omega (Sievers et al., 2011) and visualized using BOXSHADE version 3.21. Phylogenetic trees were drawn using FigTree version 1.4.0.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment and intron positions within the nine Arabidopsis BAM proteins.
Supplemental Figure S2. Alignment and intron positions of the nine Arabidopsis BAM proteins with BAM proteins from A. trichopoda, V. vinifera, and S. bicolor.
Supplemental Figure S3. Alignment and intron positions of the nine Arabidopsis BAM proteins with BAM proteins from P. abies, P. taeda, and S. moellendorffii.
Supplemental Figure S4. Alignment and intron positions of the nine Arabidopsis BAM proteins with BAM proteins from P. patens, M. polymorpha, and K. flaccidum.
Supplemental Figure S5. Unrooted phylogenetic tree of aligned BAM proteins from Arabidopsis, A. trichopoda, V. vinifera, and S. bicolor.
Supplemental Figure S6. Unrooted phylogenetic tree of aligned BAM proteins from Arabidopsis, P. abies, P. taeda, and S. moellendorffii.
Supplemental Figure S7. Unrooted phylogenetic tree of aligned BAM proteins from Arabidopsis, P. patens, M. polymorpha, and K. flaccidum.
Supplemental Figure S8. SDS-PAGE of purified BAM1, BAM2, BAM3, and BAM5.
Supplemental Figure S9. TLC of the products of BAM2 and BAM5 after acting on soluble starch.
Supplemental Figure S10. Effect of 100 mm of various salts on the activity of BAM2.
Supplemental Figure S11. Effect of 100 mm salts on the activity of purified BAM5.
Supplemental Figure S12. SEC-MALS elution profiles of BAM2 wild type, G335M, and G446M.
Acknowledgments
We thank Heike Reinhold for the BAM1 and BAM3 plasmids and Nathan Wright for helpful discussions.
Footnotes
This work was supported by a National Science Foundation Research at Undergraduate Institutions grant to J.D.M.
Articles can be viewed without a subscription.
References
- Allouch J, Helbert W, Henrissat B, Czjzek M (2004) Parallel substrate binding sites in a β-agarase suggest a novel mode of action on double-helical agarose. Structure 12: 623–632 [DOI] [PubMed] [Google Scholar]
- Aoki Y, Okamura Y, Tadaka S, Kinoshita K, Obayashi T (2016) ATTED-II in 2016: a plant coexpression database towards lineage-specific coexpression. Plant Cell Physiol 57: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JM, Tymoczko JL, Stryer L (2002) Biochemistry, Ed 5 WH Freeman, New York [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey DR, Somaroo S, Hughes JD, Mintseris J, Huang ES (2004) Are protein-protein interfaces more conserved in sequence than the rest of the protein surface? Protein Sci 13: 190–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong CG, Eom SH, Chang C, Shin DH, Song HK, Min K, Moon JH, Kim KK, Hwang KY, Suh SW (1995) Crystallization, molecular replacement solution, and refinement of tetrameric β-amylase from sweet potato. Proteins 21: 105–117 [DOI] [PubMed] [Google Scholar]
- Cuyvers S, Dornez E, Delcour JA, Courtin CM (2012) Occurrence and functional significance of secondary carbohydrate binding sites in glycoside hydrolases. Crit Rev Biotechnol 32: 93–107 [DOI] [PubMed] [Google Scholar]
- Finazzi G, Petroutsos D, Tomizioli M, Flori S, Sautron E, Villanova V, Rolland N, Seigneurin-Berny D (2015) Ions channels/transporters and chloroplast regulation. Cell Calcium 58: 86–97 [DOI] [PubMed] [Google Scholar]
- Fulton DC, Stettler M, Mettler T, Vaughan CK, Li J, Francisco P, Gil M, Reinhold H, Eicke S, Messerli G, et al. (2008) β-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active β-amylases in Arabidopsis chloroplasts. Plant Cell 20: 1040–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadagkar SR, Call GB (2015) Computational tools for fitting the Hill equation to dose-response curves. J Pharmacol Toxicol Methods 71: 68–76 [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In Walker JM, ed, The Proteomics Protocols Handbook. Humana Press, Totowa, NJ, pp 571–607 [Google Scholar]
- Gohara DW, Di Cera E (2016) Molecular mechanisms of enzyme activation by monovalent cations. J Biol Chem 291: 20840–20848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejazi M, Fettke J, Haebel S, Edner C, Paris O, Frohberg C, Steup M, Ritte G (2008) Glucan, water dikinase phosphorylates crystalline maltodextrins and thereby initiates solubilization. Plant J 55: 323–334 [DOI] [PubMed] [Google Scholar]
- Henricson A, Forslund K, Sonnhammer ELL (2010) Orthology confers intron position conservation. BMC Genomics 11: 412–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrer D, Flütsch S, Pazmino D, Matthews JS, Thalmann M, Nigro A, Leonhardt N, Lawson T, Santelia D (2016) Blue light induces a distinct starch degradation pathway in guard cells for stomatal opening. Curr Biol 26: 362–370 [DOI] [PubMed] [Google Scholar]
- Jackson BC, Thompson DC, Charkoftaki G, Vasiliou V (2015) Dead enzymes in the aldehyde dehydrogenase gene family: role in drug metabolism and toxicology. Expert Opin Drug Metab Toxicol 11: 1839–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Guy CL (2005) RNA interference of Arabidopsis β-amylase8 prevents maltose accumulation upon cold shock and increases sensitivity of PSII photochemical efficiency to freezing stress. Plant J 44: 730–743 [DOI] [PubMed] [Google Scholar]
- Laby RJ, Kim D, Gibson SI (2001) The ram1 mutant of Arabidopsis exhibits severely decreased β-amylase activity. Plant Physiol 127: 1798–1807 [PMC free article] [PubMed] [Google Scholar]
- Laederach A, Dowd MK, Coutinho PM, Reilly PJ (1999) Automated docking of maltose, 2-deoxymaltose, and maltotetraose into the soybean beta-amylase active site. Proteins 37: 166–175 [DOI] [PubMed] [Google Scholar]
- Lao NT, Schoneveld O, Mould RM, Hibberd JM, Gray JC, Kavanagh TA (1999) An Arabidopsis gene encoding a chloroplast-targeted β-amylase. Plant J 20: 519–527 [DOI] [PubMed] [Google Scholar]
- Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, De Clerck O (2012) Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci 31: 1–46 [Google Scholar]
- Li J, Francisco P, Zhou W, Edner C, Steup M, Ritte G, Bond CS, Smith SM (2009) Catalytically-inactive β-amylase BAM4 required for starch breakdown in Arabidopsis leaves is a starch-binding-protein. Arch Biochem Biophys 489: 92–98 [DOI] [PubMed] [Google Scholar]
- Ludwiczek ML, Heller M, Kantner T, McIntosh LP (2007) A secondary xylan-binding site enhances the catalytic activity of a single-domain family 11 glycoside hydrolase. J Mol Biol 373: 337–354 [DOI] [PubMed] [Google Scholar]
- Mikami B, Degano M, Hehre EJ, Sacchettini JC (1994) Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis. Biochemistry 33: 7779–7787 [PubMed] [Google Scholar]
- Møller MS, Svensson B (2016) Structural biology of starch-degrading enzymes and their regulation. Curr Opin Struct Biol 40: 33–42 [DOI] [PubMed] [Google Scholar]
- Monroe JD, Preiss J (1990) Purification of a β-amylase that accumulates in Arabidopsis thaliana mutants defective in starch metabolism. Plant Physiol 94: 1033–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe JD, Storm AR, Badley EM, Lehman MD, Platt SM, Saunders LK, Schmitz JM, Torres CE (2014) β-Amylase1 and β-amylase3 are plastidic starch hydrolases in Arabidopsis that seem to be adapted for different thermal, pH, and stress conditions. Plant Physiol 166: 1748–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N. (1944) A photometric adaptation of the Somogyi method for the determination of glucose. J Biol Chem 153: 375–380 [Google Scholar]
- Okur HI, Hladílková J, Rembert KB, Cho Y, Heyda J, Dzubiella J, Cremer PS, Jungwirth P (2017) Beyond the Hofmeister series: ion-specific effects on proteins and their biological functions. J Phys Chem B 121: 1997–2014 [DOI] [PubMed] [Google Scholar]
- Reinhold H, Soyk S, Simková K, Hostettler C, Marafino J, Mainiero S, Vaughan CK, Monroe JD, Zeeman SC (2011) β-Amylase-like proteins function as transcription factors in Arabidopsis, controlling shoot growth and development. Plant Cell 23: 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejzek M, Stevenson CE, Southard AM, Stanley D, Denyer K, Smith AM, Naldrett MJ, Lawson DM, Field RA (2011) Chemical genetics and cereal starch metabolism: structural basis of the non-covalent and covalent inhibition of barley β-amylase. Mol Biosyst 7: 718–730 [DOI] [PubMed] [Google Scholar]
- Schröppel-Meier G, Kaiser WM (1988) Ion homeostasis in chloroplasts under salinity and mineral deficiency. I. Solute concentrations in leaves and chloroplasts from spinach plants under NaCl or NaNO3 salinity. Plant Physiol 87: 822–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibold G, Auchter M, Berens S, Kalinowski J, Eikmanns BJ (2006) Utilization of soluble starch by a recombinant Corynebacterium glutamicum strain: growth and lysine production. J Biotechnol 124: 381–391 [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeffington AW, Graf A, Duxbury Z, Gruissem W, Smith AM (2014) Glucan, water dikinase exerts little control over starch degradation in Arabidopsis leaves at night. Plant Physiol 165: 866–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova J, Fernie AR, Steup M (2015) Starch degradation. In Nakamura Y, ed, Starch: Metabolism and Structure. Springer Japan, pp 239–290 [Google Scholar]
- Soyk S, Simková K, Zürcher E, Luginbühl L, Brand LH, Vaughan CK, Wanke D, Zeeman SC (2014) The enzyme-like domain of Arabidopsis nuclear β-amylases is critical for DNA sequence recognition and transcriptional activation. Plant Cell 26: 1746–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma JA, Koshland DE Jr, Ruscica J, Baldwin R (1963) The number of binding sites of sweet potato beta amylase. Biochem Biophys Res Commun 12: 184–188 [DOI] [PubMed] [Google Scholar]
- Valerio C, Costa A, Marri L, Issakidis-Bourguet E, Pupillo P, Trost P, Sparla F (2011) Thioredoxin-regulated β-amylase (BAM1) triggers diurnal starch degradation in guard cells, and in mesophyll cells under osmotic stress. J Exp Bot 62: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Monroe J, Sjölund RD (1995) Identification and characterization of a phloem-specific β-amylase. Plant Physiol 109: 743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH (2000) Cn3D: sequence and structure views for Entrez. Trends Biochem Sci 25: 300–302 [DOI] [PubMed] [Google Scholar]
- Wilkens C, Auger KD, Anderson NT, Meekins DA, Raththagala M, Abou Hachem M, Payne CM, Gentry MS, Svensson B (2016) Plant α-glucan phosphatases SEX4 and LSF2 display different affinity for amylopectin and amylose. FEBS Lett 590: 118–128 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wychowski A, Bompard C, Grimaud F, Potocki-Véronèse G, D’Hulst C, Wattebled F, Roussel X (2017) Biochemical characterization of Arabidopsis thaliana starch branching enzyme 2.2 reveals an enzymatic positive cooperativity. Biochimie 140: 146–158 [DOI] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER suite: protein structure and function prediction. Nat Methods 12: 7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Miyake H, Tatsumi M, Nishimura S, Nitta Y (2004) Two additional carbohydrate-binding sites of beta-amylase from Bacillus cereus var. mycoides are involved in hydrolysis and raw starch-binding. J Biochem 135: 355–363 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Kossmann J, Smith AM (2010) Starch: its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol 61: 209–234 [DOI] [PubMed] [Google Scholar]