Abstract
Deep sequencing has been revolutionizing biology and medicine in recent years, providing single base-level precision for our understanding of nucleic acid sequences in high throughput fashion. Sequencing of RNA, or RNA-Seq, is now a common method to analyze gene expression and to uncover novel RNA species. Aspects of RNA biogenesis and metabolism can be interrogated with specialized methods for cDNA library preparation. In this study, we review current RNA-Seq methods for general analysis of gene expression and several specific applications, including isoform and gene fusion detection, digital gene expression profiling, targeted sequencing and single-cell analysis. In addition, we discuss approaches to examine aspects of RNA in the cell, technical challenges of existing RNA-Seq methods, and future directions.
INTRODUCTION
RNA molecules are essential components of all living cells. Understanding the identity and abundance of each RNA molecule in a given cell under a specific condition is the ultimate goal of RNA research. Much of what we know about RNA comes from studies using biochemical methods, where a small number of specific molecules are analyzed. High-throughput approaches that enable interrogation of RNA sequences on a large scale emerged in the early 1990s. The expressed sequence tag (EST) method developed by Adams et al. examines gene expression by partially sequencing complementary DNA (cDNA) clones, revealing both the sequence and the abundance of corresponding RNAs.1 EST data played a pivotal role in identification of new genes in genomes in the 1990s. However, the high sequencing cost of the method limited its use in expression analysis, and the data is largely believed to be semi-quantitative. The Serial Analysis of Gene Expression (SAGE) method developed by Velculescu et al. significantly cut down the cost of expression analysis on a per gene basis,2 thanks to sequencing only a short tag region per cDNA (15 bp for the short SAGE method and 21 bp for the long SAGE method). However, the emergence of DNA microarray technology in the mid-1990s superseded EST and SAGE methods for gene expression analysis, largely due to its much better affordability for large scale studies.3,4 DNA microarray analysis of gene expression is based on hybridization of fluorescently labeled targets that are derived from transcripts to probes that are attached to a solid surface through printing or in situ synthesis. However, while the method enables interrogation of transcripts genome-wide, the requirement that a priori sequence information or reference genomes/transcriptomes be available for designing the microarray probes limited the development and application of this technology in discovery applications. In addition, cross-hybridization and background signals often lead to low specificity or low sensitivity for some genes.
The first decade of this millennium witnessed the advent of massive parallel sequencing, also known as deep sequencing or Next Generation Sequencing (NGS). Lauded as revolutionary in biology and medicine for its ability to acquire an unprecedented amount of data in a short time, deep sequencing quickly transformed RNA research. RNA-Seq is now the method of choice to study gene expression and identify novel RNA species. Compared to DNA microarray-based methods, RNA-Seq offers less background noise and a greater dynamic range for detection. Most importantly, RNA-Seq directly reveals sequence identity, crucial for analysis of unknown genes and novel transcript isoforms. Several different technologies have been developed for RNA-Seq.5–9 Here we review general aspects of RNA-Seq and applications of RNA-Seq to study specific problems. We discuss issues and remedies related to bias and sensitivity in these methods, two paramount concerns in an RNA-Seq experiment. We also discuss specialized methods that investigate aspects of RNA biogenesis and metabolism.
GENERAL ASPECTS OF RNA-Seq
While direct sequencing of RNA molecules is possible,10 most RNA-Seq experiments are carried out on instruments that sequence DNA molecules due to the technical maturity of commercial instruments designed for DNA-based sequencing. Therefore, cDNA library preparation from RNA is a required step for RNA-Seq. Each cDNA in an RNA-Seq library is composed of a cDNA insert of certain size flanked by adapter sequences, as required for amplification and sequencing on a specific platform. The cDNA library preparation method varies depending on the RNA species under investigation, which can differ in size, sequence, structural features and abundance. Major considerations include (1) how to capture RNA molecules of interest; (2) how to convert RNA to double-stranded cDNAs with defined size ranges; and (3) how to place adapter sequences on the cDNA ends for amplification and sequencing. These are discussed in the following sections.
Selection of Poly(A) + Transcripts
Sequencing of polyadenylated RNA is perhaps the most common application of RNA-Seq. In eukaryotic organisms, most protein-coding RNAs (mRNAs) and many long noncoding RNAs (lncRNAs) (>200 nt) contain a poly(A) tail. The poly(A) tail provides technical convenience for enrichment of poly(A) + RNAs from total cellular RNA, in which they account for approximately 1–5% of the pool. Poly(A) + RNA selection can be carried out with magnetic or cellulose beads coated with oligo-dT molecules. Alternatively, polyadenylated RNAs can be selected using oligo-dT priming for reverse transcription (RT). While efficiently incorporating both poly(A) selection and RT in one step, oligo-dT priming-based methods can exhibit 3′ bias, resulting in sequencing reads enriched for the 3′ portion of the transcript. In addition, oligo-dT can frequently prime at internal A-rich sequences of transcripts, a phenomenon called internal poly(A) priming,7,11 leading to biased RT. Therefore, poly(A) purification is a preferred method to select poly(A) + RNA unless a very low amount of RNA is available (see next).
rRNA Depletion
Non-polyadenylated RNAs, such as prokaryotic mRNAs, fragmented mRNAs from formalin-fixed, paraffin-embedded (FFPE) samples, and poly(A)-transcripts in eukaryotic cells are often the subject of investigation. A major issue in sequencing these RNAs is how to eliminate ribosomal RNAs (rRNAs), which are the most abundant RNA species in the cell but of little interest in most studies. Several approaches have been developed to deplete them from the RNA pool.
One approach to eliminate rRNAs is based on sequence-specific probes that can hybridize to rRNAs. Unwanted rRNAs or their cDNAs are hybridized with biotinylated DNA or locked nucleic acid (LNA) probes, followed by depletion with streptavidin beads. Alternatively, rRNAs are targeted by anti-sense DNA oligos and digested by RNase H, a method also known as probe-directed degradation (PDD). While this approach is less laborious than hybridization, it requires continuous coverage of rRNAs and unique probe sets designed for different species. A noncontinuous sequence-based method was recently developed which has addressed some of these issues.12,13 In this method, all cDNAs, including those of rRNAs and other RNAs, are circularized, and are hybridized to rRNA probes. The hybridized sequences are then digested by duplex-specific nuclease (DSN), making them unusable for amplification. However, this approach requires high input amounts of total RNA, which can be challenging when dealing with clinical samples.
Another approach for rRNA reduction uses specific, not-so-random (NSR) primers which bind to the RNA molecules of interest during RT, thus avoiding rRNAs. This method, commercialized by NuGEN under the name Ovation RNA-Seq, uses hexamer or heptamer primers whose sequences are absent from rRNAs.14,15 Similar to this approach, one study used 44 heptamers to avoid both rRNAs and highly-expressed transcripts.16 A recent report used only 40 primers for RT instead of 700 NSR primers commonly used in other studies.17 A key advantage of this approach is that NSR primers work well with partially degraded RNA and low-input samples. However, like other sequence targeting methods, this approach suffers from off-target priming, and is species dependent. Nevertheless, NSR primers are frequently used in prokaryotic species, for which poly(A) purification is not an option.
In addition to the sequence-based approaches mentioned above, some methods take advantage of certain features of rRNAs for their elimination. The C0T-hybridization method is based on heat denaturation, re-annealing and selective degradation by DSN. Double-stranded cDNAs originated from abundant sequences are preferentially degraded because of their more rapid annealing kinetics compared to less abundant ones.18 Selective degradation has also been achieved by using the enzyme terminator 5′-phosphate-dependent exonuclease (TEX), which recognizes RNA molecules with 5′-monophosphate, as with rRNAs and tRNAs.19
In summary, the selection of an approach for enriching RNA transcripts of interest for sequencing depends on the goal of the experiment and many technical factors. Several studies have compared protocols for removal of rRNA by depletion- and priming-based methods.7,20–23 In eukaryotic cells, oligo-dT bead-based purification of poly(A) + RNA is the method of choice for most applications, because of its ease of use and relatively low cost. For low-input samples, however, oligo-dT priming generally offers better results. Both poly(A) selection methods can effectively address intron sequence contamination. If an RNA sample is partially degraded or the user is interested in noncoding RNAs, depletion of rRNAs by PDD or NSR-priming is typically necessary. PDD works better for high-input samples whereas NSR-priming is primarily used for lower inputs of RNA.
Fragmentation
After poly(A) + selection or rRNA depletion, RNA samples are typically subject to RNA fragmentation to a certain size range before RT. This is necessary because of the size limitation of most current sequencing platforms, e.g., <600 bp on Illumina sequencers. RNAs can be fragmented with alkaline solutions, solutions with divalent cations, such Mg++, Zn++, or enzymes, such RNase III. Fragmentation with alkaline solutions or divalent cations is typically carried out at an elevated temperature, such as 70°C, to mitigate the effect of RNA structure on fragmentation. Nevertheless, RNA fragmentation by chemical fragmentation is not completely random. Similarly, the RNase III-based method can also introduce bias because of the enzyme’s preference for double-stranded RNA sequences.24 Thus, uneven fragmentation of RNA can be a source of bias, leading to differential representation of specific regions of RNA.
Alternatively, intact RNAs can be reverse transcribed, and full-length cDNA can be fragmented. A traditional method to fragment cDNA requires the use of acoustic shearing, which is less amenable to automation than RNA fragmentation. Alternatively, full-length double-stranded cDNAs can be fragmented by DNases. Recent development in using transposon-based, so-called tagmentation method has made it simple to fragment cDNA and add adapter sequences at the same time.25 In this method, an active variant of the Tn5 transposase mediates the fragmentation of double-stranded DNA and ligates adapter oligonucleotides at both ends in a quick reaction (~5 min). However, it is notable that Tn5 and other enzyme-based cDNA fragmentation methods require a precise enzyme:DNA ratio, making method optimization less straightforward than RNA fragmentation. Consequently, fragmenting RNA is currently still the most frequently used approach in RNA-Seq library preparation.
Adapters and Directionality
In a standard RNA-Seq library protocol, cDNAs of a desired size generated from RT of fragmented RNAs with random hexamer primers or from fragmented full-length cDNAs are ligated to DNA adapters before amplification and sequencing. While simple, this approach loses the information about which DNA strand corresponds to the sense strand of RNA. Lack of strand specificity would make it difficult to identify antisense and novel RNA species and cause inaccurate measurement of sense RNA expression. Several methods have been developed to capture the directionality of RNA in cDNA libraries.26
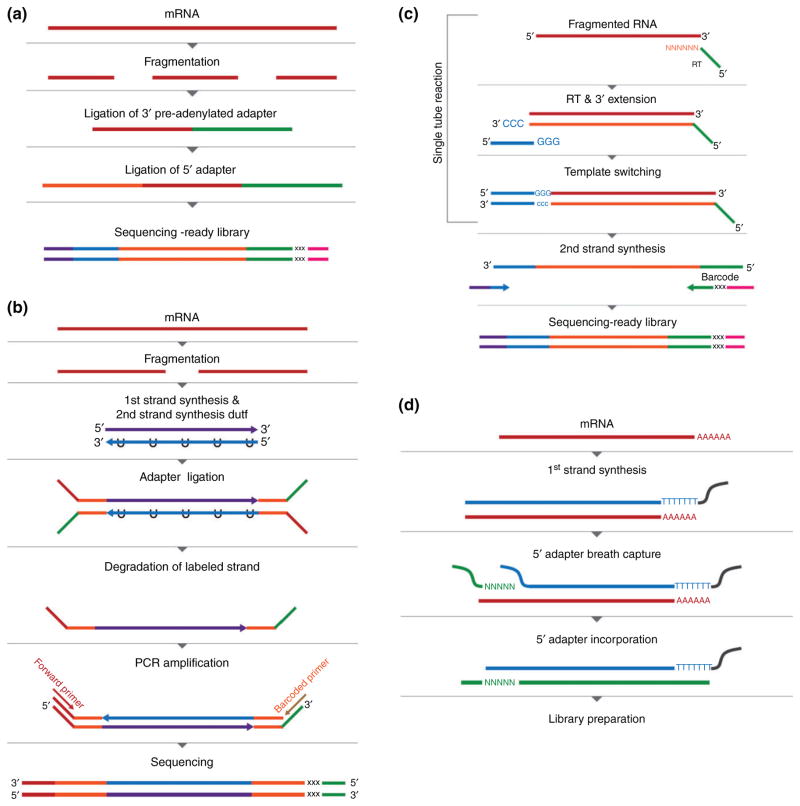
The first approach involves attaching different adapters directly to the 5′ and 3′ ends of the RNA molecule (Figure 1(a)). Originally designed for small RNA-Seq,27 this method begins with removal of 3′ phosphate group from fragmented RNA and addition of a 5′ phosphate group. This is followed by sequential ligations of a 5′ adenylated 3′ adapter using a truncated RNA ligase II and a 5′ adapter ligation using RNA ligase I. The sequence difference between 5′ and 3′ adapters preserves RNA strandedness. While simple to implement, this approach suffers from substantial biases due to the influence of both 5′ and 3′ end sequences on the ligation steps. This issue, however, has recently been mitigated significantly by using random nucleotides at the ligation end of each adapter.28,29
FIGURE 1.
Methods for strand-specific RNA-Seq. (a) Ligation of the 3′ preadenylated and 5′ adapters. “xxx” indicates barcode. (b) Labeling of the second strand with dUTP, followed by enzymatic degradation. (c) The Peregrine method involves template-switch attachment of the 3′ adapter. (d) BrAD-Seq captures the 3′ adapter by taking advantage of terminal breathing of double-stranded DNA.
A second approach incorporates dUTP into the second strand of cDNA (Figure 1(b)). The labeled strand can be degraded before PCR amplification with uracil DNA glycosylase (UDG), an enzyme that cleaves the uracil base in dUTP-containing DNA. In addition, the U-containing strand is a very poor template for thermostable polymerases, such as the Phusion polymerase, making it essentially not amplifiable. As such, only the first strand cDNA with defined adapter sequences is amplified, conferring directional information to the sequencing reads. A systematic comparison of different protocols for strand-specific RNA-Seq indicated that the dUTP-based method was the most effective in terms of evenness of coverage.30 Indeed, this method is currently the most frequently used in commercial directional RNA-Seq library preparation protocols. However, since it requires extra enzymatic and purification steps that are laborious and can cause material loss, it is generally not suitable for low-input samples.
There have been several attempts to develop new methods based on adding adapters to fragmented RNA. One of these methods, Pedegrine, uses template-switching after priming with a random-hexamer that contains a small tag (Figure 1(c)).31 Another method, BrAD-Seq, takes advantages of temporary strand separation in double-stranded DNA, called ‘breathing,’ to introduce a tag sequence (Figure 1(d)).32 A third example is sequential ligation of tags to RNA that has been fragmented to 200 nt, preserving even small RNAs.33,34 A DSN-mediated normalization step is used to remove rRNA products. A similar protocol for stranded RNA-Seq libraries from all RNA species has also been reported,35 where an oligo containing a tag sequence for RT is ligated to the 3′ end of RNA. A second tag is introduced by the RT primer. First strand synthesis is then followed by circularization of cDNA by DNA ligase and direct amplification of the library using the two tags.
Amplification and Molecular Labels
Due to the detection limit of most sequencers, cDNA libraries need to be amplified by PCR before sequencing. While only a small number of amplification cycles (8–12) are used during PCR, variations in cDNA size and composition can result in uneven amplification. Amplification of some cDNAs plateau while others continue to amplify exponentially. To correct for PCR amplification bias, methods that eliminate PCR duplicates from sequencing results have been introduced. In one method, under the assumption of random RNA fragmentation, final sequencing reads having the same start and stop coordinates are considered as PCR duplicates and are merged.36,37 However, a drawback of this approach is elimination of reads derived from frequently fragmented regions due to not-so-stochastic fragmentation.38 Another method is to use molecular labels, also known as unique molecular identifiers (UMIs), to distinguish PCR products.39–41 Molecular labels are typically introduced within the adapter sequence, prior to PCR amplification. In a modified protocol for making cDNAs from single cells, molecular labels were introduced by the Tn5 transposase during fragmentation of double-stranded, amplified cDNA42. However, in some applications, such as digital counting of targeted RNAs, molecular labels are added during RT.40,43,44 Molecular labels differ in size (number of bases) and complexity. In principle, they comprise either defined sequences or random nucleotides. Defined sequences, chosen for their even distribution in final libraries, are more technically challenging to make because of sequence selection and manufacturing complexity. By contrast, random sequences, while easy to implement, give high variability among molecular labels. Molecular labeling is particularly valuable in situations where input RNA is scarce and a large number of PCR cycles is required for sequencing, such as single-cell RNA-Seq (see next).45
RNA-Seq METHODS FOR SPECIFIC GOALS
Tag-Based Methods for Gene Expression Profiling
DGE-Seq
Digital gene expression (DGE)-Seq, or Tag-Seq, is a deep sequencing method derived from SAGE.46,47 As in SAGE, the method involves attachment of mRNA to beads via the poly(A) tail, first and second strand cDNA syntheses on the beads, and digestion of double-stranded cDNA with a frequent cutting restriction enzyme. The remaining 3′ fragment attached to the beads is then ligated to its 5′ end adapter with a recognition site for another restriction enzyme, called tagging enzyme. The tagging enzyme cleaves the cDNA and generates a short 21 bp tag, which is then ligated to a second adapter at its 3′ end. The cDNA is then amplified by PCR, followed by sequencing. Because only a short tag is sequenced from the whole transcript, DGE-Seq is more economical than traditional RNA-Seq for a given depth of sequencing and can provide a higher dynamic range of detection when the same number of reads is generated. By design, DGE-Seq preserves RNA strandedness. This method has been commercialized by several companies and is useful especially when simple gene expression profiling is the goal.48 It is also the method of choice when the complete genome or transcriptome is not available for full alignment of RNA-Seq reads.
3′ End Sequencing
A number of methods specifically sequence the 3′ end region of transcripts. Most of these methods were first developed to interrogate alterative cleavage and polyadenylation sites, a widespread phenomenon in all eukaryotes.49 As in DGE-Seq, the data from these methods can also be used to study gene expression. Some of these methods use oligo(dT) to prime RT or sequencing, such as PAS-Seq,50 polyA-Seq,51 3′T-fill,52 and so on. One concern with this approach is that internal poly(A) priming can generate a high frequency of truncated cDNA.11 Other methods, such as 3P-Seq53 and 3′READS,54 use RNA-based ligation to capture the 3′ end fragments. While these methods successfully address the internal priming issue, sequence preference of RNA ligases can introduce bias. A more detailed review of these methods can be found in Ref 55. Also notable are methods that can examine both the 3′ end region and the poly(A) tail length at the same time. TAIL-Seq adds an adapter to the 3′ end of the poly(A) tail and carries out sequencing from both ends of the insert (paired-end sequencing) to reveal both the length of poly(A) tail and the sequence near the poly(A) site.56 A related method, PAT-Seq, uses Klenow polymerase to add an adapter sequence to the 3′ end of the poly(A) tail, and uses single-read sequencing to obtain the sequence near the poly(A) site, as well as part of or the whole poly(A) tail.57
In essence, DGE-Seq and 3′ end sequencing are tag-based approaches that use one fragment to represent a transcript. While efficient for gene expression analysis, they can have higher variability than ‘shotgun’ style RNA-Seq, where one transcript is represented by multiple fragments. Biases from fragmentation, adapter ligation and PCR can make tag-based data more prone to batch effects.
Sequencing to Reveal Alternative Splicing and Gene Fusion
Almost all multi-exon genes display alternative splicing (AS).58 AS plays an important role in regulation of cellular processes, and aberrations of the process are associated with many human diseases.59 Some RNA-Seq reads cover exon-exon junctions, providing direct evidence of AS. In addition, reads mapped to internal exonic regions can be used to predict the AS pattern using statistical inference methods.60 A more direct approach to examine AS is to sequence the exon-exon junction region directly. Using oligo pairs targeted to specific exon-exon junction sequences, the Fu lab developed RASL-Seq,61 which provides analysis of specific splice junction regions. However, prior knowledge of the exon–exon junction sequences is required for the oligo design.
Similar to splicing, gene fusion events can place two noncontinuous genomic regions together in a single transcript. Created by chromosomal rearrangements, gene fusions are present in approximately 20% of cancer.62 Fusion events can be detected using RNA-Seq data along with specific bioinformatic methods.63–65 Detection of a fusion event is typically revealed by reads containing fusion junctions or by differences in expression between the 5′ and 3′ ends of genes that are fused. Regular RNA-Seq methods are typically not sufficiently sensitive to detect fusion junctions. Several methods have been developed, including (1) enrichment of RNA-Seq reads for genes of interest, (2) exon capture, and (3) amplicon sequencing. In a recent study, exon capture of 467 cancer-related genes was successfully employed for the detection of their fusion events.66 Amplicon targeting requires primer design at the 5′ and 3′ ends of a transcript, and allows quantitative analysis of fusion events. This strategy was recently employed for the detection of ALK fusion events in lung cancer samples.67 Two similar amplicon-based methods have been developed and commercialized by NuGene and ArcherDX,68,69 where expected fusion events were examined by two sequence-specific primers together with a common primer that targets adapter sequence.
The ultimate solution to unravel the complexity of alternatively splicing and gene fusion isoforms is to sequence each transcript from the beginning to the end. Two strategies have been established to this end. Single-molecule real-time sequencing (SMRT) on the PacBio sequencing platform offers long reads up to 5 kb.70 However, this method is costly, and has a high error-rate and low multiplexing capacity. Hybrid sequencing methods have been introduced which combine SMRT data with short, standard RNA-Seq reads.71–73 A second approach, named synthetic long-read-RNA sequencing (SLR-RNA-Seq), is based on the Illumina MOLECULO system.74 With SLR RNA-Seq, an RNA substrate is diluted to no more than 1000 transcripts per well so that the probability that two transcripts from the same gene are in the same well is very low. As such, reads for a gene from each well can be assembled to cover the entire length of a single transcript of the gene. After fragmentation, barcoding, library preparation, and sequencing on the Illumina MOLECULO platform, the structures of the original transcripts in the pools can be delineated. Compared to PacBio sequencing, SLR-RNA-Seq was found to deliver longer transcripts and a greater number of detected isoforms.74 These methods are particularly valuable for analysis of AS and aberrant fusion isoforms.
Targeted RNA-Seq
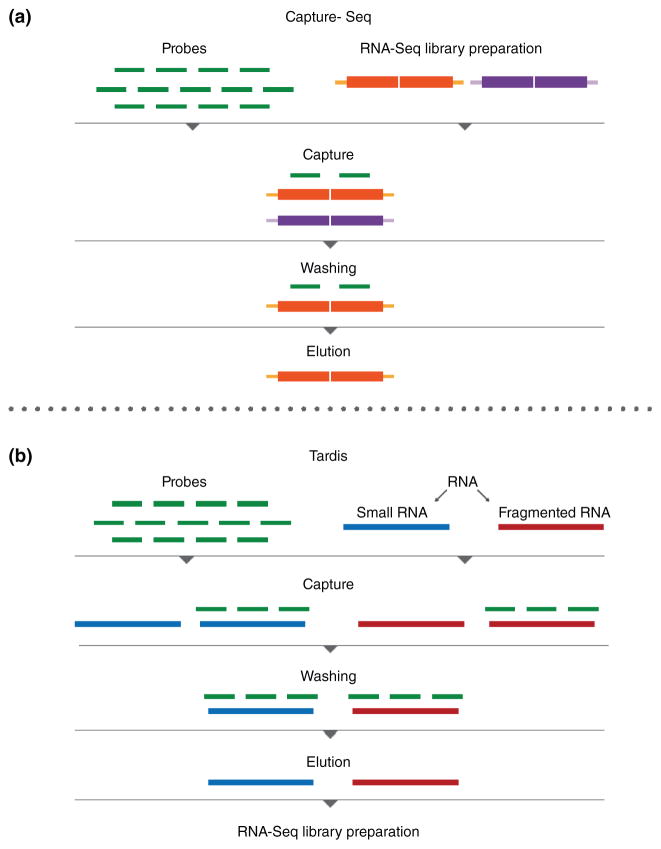
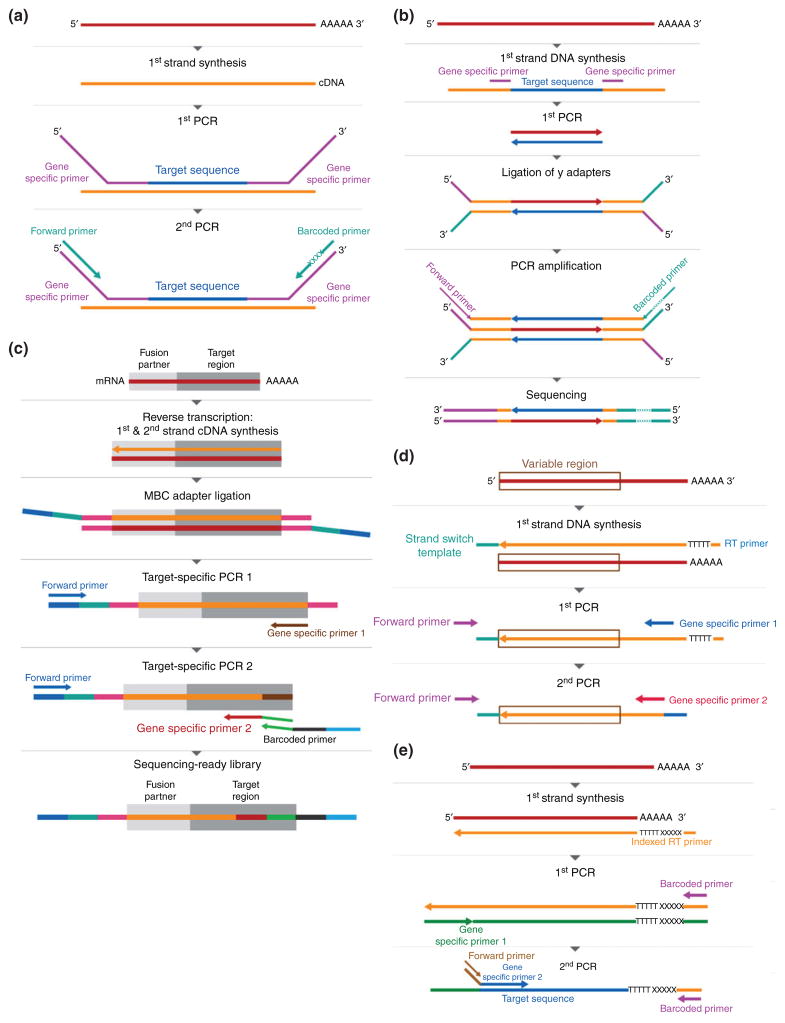
Selection a specific set of transcripts for sequencing is often desirable when a defined group of genes is of interest. Lowly expressed genes that cannot be readily analyzed using whole transcriptome sequencing can also be detected using targeted RNA-Seq methods. Two general approaches have been used, namely, target capture and amplicon sequencing. The target capture approach involves selection of specific genes using a set of biotinylated probes which bind cDNA,66,75,76 or RNA77 (Figure 2). By contrast, amplicon sequencing employs gene-specific primers for the amplification of cDNA targets (Figure 3). Approaches differ in the amplicon design, including two specific primers after cDNA synthesis by template switch,78 nested PCR with one specific primer and one common primer for adapter,69 and specific targeting primers in combination with a primer for poly(A) tail priming.45
FIGURE 2.
Targeted RNA-Seq by target capture. (a) The Capture-Seq method is based on capture of regions of interest by hybridization of RNA-Seq libraries to DNA oligonucleotide probes. (b) TARDIS is based on hybridization of input RNA to DNA oligonucleotide probes. The enrichment step is followed by the construction of a directional RNA-Seq library by ligation of 3′ and 5′ adapters.
FIGURE 3.
Targeted RNA-Seq by amplicon sequencing. (a) PCR method using gene specific primers with overhangs containing sequences for common primers. (b) PCR methods using a pair of gene-specific primers followed by ligation of adapters with sequences necessary for sequencing. (c) Archer methods for detection of gene fusion. Sequence for a common primer is introduced by adapter ligation and is followed by nested PCR with gene-specific primers. MBC, Molecular Barcoded. (d) Detection of the TCR variable region. The sequence of a common primer is introduced during reverse transcription, and RT is followed by nested PCR with gene-specific primers. (e) Digital encoding of targeted mRNAs. Reverse transcription will introduce molecular indexes and sequences for common primers. Nested PCR follows, where gene-specific primers capture targeted sequences.
The target capture approach has been shown to provide greater complexity and uniformity than the amplicon-based approach in a recent whole exome study.79 However, target capture methods are more costly. Therefore, for studies that do not involve complex analysis, amplicon-based methods are preferred, as exemplified by analysis of T-cell and B-cell receptor repertoires.78,80,81
Single-Cell RNA-Seq
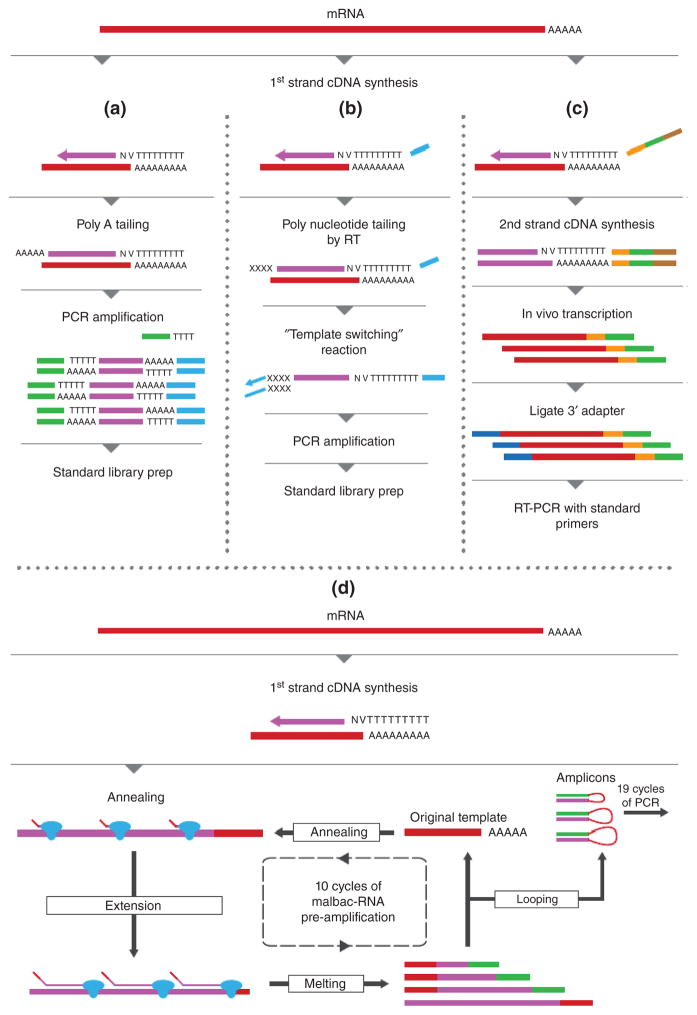
RNA-Seq of cell populations gives rise to expression profiles that are averaged across cells. However, a cell bulk often contains different types or subtypes which are impossible to dissect using population-based analysis. In addition, the co-expression patterns between genes in a cell are lost when aggregating cells. Thus, understanding gene expression at the single-cell level is important for obtaining the full picture of gene regulation in cells. Major challenges in single-cell analysis include isolation of single cells, sensitive methods to prepare cDNA libraries with very low inputs of RNA, and computational methods tailored for single-cell analysis. Several recent reviews have summarized these technologies and issues related to single-cell research.82–86 Here, we focus on the library preparation aspect for single-cell RNA-Seq (Figure 4).
FIGURE 4.
RNA-Seq of single cells. (a) Reverse transcription with oligo-dT primers and a universal primer sequence is followed by poly(A) tailing. After PCR amplifications, standard RNA-Seq libraries are prepared. (b) Reverse transcription incorporates a universal primer sequence. Template switching of reverse transcription is followed by annealing of the oligonucleotide with the sequence for a second PCR primer. (c) cDNA synthesis introduces the T7 promoter sequence at the 5′ end. After second strand cDNA synthesis, cRNA copies are generated by in vitro transcription. Finally, the second adapter is ligated to the 3′ end of the cRNA and libraries are constructed by PCR amplification. (d) Single-cell MALBAC RNA-Seq. Primers with seven random nucleotides at the 3′ end are annealed to cDNA and extended. Amplicons are looped to protect them from being further amplified. Ten cycles of quasilinear amplification are followed by exponential PCR.
A single mammalian cell contains approximately 5–15 pg of RNA. However, the RNA-Seq methods described above are generally not suitable for sequencing RNA with less than 1 ng. Thus special RNA/DNA amplification or enhanced efficiency for sample processing is needed for single-cell RNA-seq. Some methods, such as CEL-Seq and MARS-Seq, introduce a T7 promoter sequence with oligo(dT) during RT, which enables linear amplification of input RNA by in vitro transcription.87,88 Second strand cDNA synthesis has been enhanced by template-switching during RT (SMART-Seq)89 and poly(A) tailing of cDNA.90,91 These methods, however, do not provide strand-specific information, because the cDNA is subsequently fragmented and ligated to a second set of adapters. Recently, the multiple annealing and looping-based cycles (MALBAC) method, originally designed for single-cell genomic DNA amplification, was applied to RNA-Seq.92
Because of a greater extent of amplification, molecular labels are particularly important for single-cell RNA-Seq to detect over-amplified products. In addition, barcoding is typically used to label cells, enabling simultaneous preparation of libraries from multiple cells.40,42,93 On this note, recent high throughput single-cell methods have used oligonucleotide beads to deliver barcodes to cells and mRNAs at the same time (CytoSeq),45 or have separated cells with different barcodes in aqueous droplets (inDrop and Drop-seq).94,95
State-of-the-art technologies are in development to characterize transcriptomes inside the cells, providing spatial information of RNA expression.96 One method, TIVA, is based on introduction of biotinylated tags to cells in tissue, followed by targeted activation of these tags by a laser in selected cells.97 The laser activates poly(U) tracts on the biotinylated tags, enabling them to bind mRNAs in the targeted cell. The bound mRNAs are then selected by streptavidin, cloned, and sequenced. Another method, fluorescent in situ RNA sequencing (FISSEQ), combines in situ amplification with sequencing by oligonucleotide ligation.98
SEQUENCING OF OTHER RNA SPECIES
Small RNAs
Small noncoding RNAs below 30 nt, such as miRNAs, piRNAs, and endosiRNAs, are processed from primary transcripts. miRNAs can be efficiently captured by direct ligation with adapters (see above).27 Thanks to the 5′ phosphate group and 3′ OH group of miRNAs, no additional processing of RNA is necessary before the ligations. As discussed above, this method naturally preserves the strandedness of RNA but introduces substantial biases due to influence of sequence on ligation. Using degenerate random nucleotides at the ligation ends of adapters can effectively mitigate bias.28,29 Alternatively, the 5′ adapter ligation step, which appears to be more prone to bias than the 3′ adapter ligation (personal observation), can be eliminated if the single-stranded cDNA is circularized by DNA ligase and amplified by PCR.99
Because of the short insert size of the cDNA library for small RNAs, it is necessary to use a specific method to separate them from contaminant DNAs, such as PCR products without any inserts. These methods include electrophoresis separation or adding the RT primer to block the 3′ adapter from ligating with the 5′ adapter.
Circular RNA
One recent surprising finding has been the discovery of circular RNAs,100 which are generated by back-splicing.101 Circular RNAs can be sequenced by digesting away linear RNAs using exonuclease R, followed by regular RNA-Seq methods involving fragmentation, RT and PCR.
Specifically Generated RNA Fragments
A growing number of methods have been developed in the past few years to interrogate RNAs at different stages of biogenesis and metabolism or interactions with proteins or other RNAs, and RNA structural features. Some of these methods are summarized in Table 1. While these methods differ widely at the RNA-capturing step, ranging from RNase protection to immunoprecipitation to metabolic labeling, the cDNA library preparation methods are similar.
TABLE 1.
Methods to Interrogate Different Aspects of RNA Life Cycle
| Purpose | Methods | RNA Species Sequenced |
|---|---|---|
| Transcribing RNA polymerase | GRO-Seq102 | Nascent RNA transcribed in vitro |
| NET-Seq103–105 | Nascent RNA associated with RNA polymerase in vivo | |
| RNA synthesis, processing and degradation | 4sU-Seq106–108 | Newly synthesized RNA labeled with 4-thiouridine (4sU) in vivo |
| Translational status | Ribosome profiling (Riboseq)99 | Ribosome-bound mRNA fragments protected from RNase digestion |
| RNA–protein interactions | RIP-Seq109–111 | RNA in immunoprecipitated ribonucleoprotein complex |
| HIT-CLIP/CLIP-seq,112–114 iCLIP115 | RNA fragments crosslinked to interacting proteins by UV (254 nm) | |
| PAR-CLIP116 | RNA labeled with 4sU and crosslinked to interacting proteins by UV (365 nm) | |
| RNA structure probing | PARS,117 FragSeq118 | RNA fragments generated by RNases digesting double-stranded regions (V1) or single-stranded regions (S1 or P1) |
| DMS-Seq119; SHAPE-S120 icSHAPE121 | Unstructured RNA regions labeled with reactive chemicals | |
| RNA–RNA interactions | RAP-RNA122 | RNA isolated by antisense probe |
Most methods generate short RNA fragments that are often made into cDNAs using the small RNA sequencing approach (see above).
CONCLUSION AND FUTURE DIRECTIONS
Since its inception about 8 years ago, RNA-Seq has become a widely used and indispensable approach for studying gene expression and interrogating aspects of RNA biogenesis and metabolism. As sequencing technologies continue to advance, many methods have been developed and new RNA-Seq methods are expected to emerge in the future. We believe several areas are particularly relevant to RNA analysis. First, current sequencing platforms have size limitations. Instruments that can handle long reads and offer high read output would be particularly beneficial for quantitative analysis of transcript isoforms, including those generated by alternative initiation, AS, and alternative polyadenylation, and gene fusions. Second, current sequencing chemistry cannot handle homopolymers well. This is particularly relevant for sequencing the poly(A) tail region, which plays important roles in transcript metabolism. Third, the sensitivity of sequencing needs to be further improved so that amplification can be reduced or eliminated. This will also be important for single-cell analysis, where currently only a small fraction of genes can be examined. Finally, interrogation of spatial information of RNA expression in the cell, showing much promise in recent studies, is a new frontier for RNA-Seq.
Acknowledgments
We would like to thank Jonathon Kirkbride for designing the illustrations in this manuscript and members of BT laboratory and Michael B. Mathews for helpful discussions. This work was partially funded by grants from the NIH (GM084089) to BT and (GM105178) to MT.
Footnotes
Conflict of interest: Masoud Toloue and Radmila Hrdlickova work at Bioo Scientific, a private corporation.
References
- 1.Adams MD, Dubnick M, Kerlavage AR, Moreno R, Kelley JM, Utterback TR, Nagle JW, Fields C, Venter JC. Sequence identification of 2,375 human brain genes. Nature. 1992;355:632–634. doi: 10.1038/355632a0. [DOI] [PubMed] [Google Scholar]
- 2.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 3.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 4.Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 5.Han Y, Gao S, Muegge K, Zhang W, Zhou B. Advanced applications of RNA sequencing and challenges. Bioinform Biol Insights. 2015;9:29–46. doi: 10.4137/BBI.S28991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nookaew I, Papini M, Pornputtapong N, Scalcinati G, Fagerberg L, Uhlen M, Nielsen J. A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40:10084–10097. doi: 10.1093/nar/gks804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adiconis X, Borges-Rivera D, Satija R, DeLuca DS, Busby MA, Berlin AM, Sivachenko A, Thompson DA, Wysoker A, Fennell T, et al. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat Methods. 2013;10:623–629. doi: 10.1038/nmeth.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S, Tighe SW, Nicolet CM, Grove D, Levy S, Farmerie W, Viale A, Wright C, Schweitzer PA, Gao Y, et al. Multi-platform assessment of transcriptome profiling using RNA-seq in the ABRF next-generation sequencing study. Nat Biotechnol. 2014;32:915–925. doi: 10.1038/nbt.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Dijk EL, Jaszczyszyn Y, Thermes C. Library preparation methods for next-generation sequencing: tone down the bias. Exp Cell Res. 2014;322:12–20. doi: 10.1016/j.yexcr.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, Thompson JF, Bowers J, Jarosz M, Milos PM. Direct RNA sequencing. Nature. 2009;461:814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 11.Nam DK, Lee S, Zhou G, Cao X, Wang C, Clark T, Chen J, Rowley JD, Wang SM. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc Natl Acad Sci USA. 2002;99:6152–6156. doi: 10.1073/pnas.092140899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Archer SK, Shirokikh NE, Preiss T. Probe-directed degradation (PDD) for flexible removal of unwanted cDNA sequences from RNA-Seq libraries. Curr Protoc Hum Genet. 2015;85:111511–111536. doi: 10.1002/0471142905.hg1115s85. [DOI] [PubMed] [Google Scholar]
- 13.Archer SK, Shirokikh NE, Preiss T. Selective and flexible depletion of problematic sequences from RNA-seq libraries at the cDNA stage. BMC Genomics. 2014;15:401. doi: 10.1186/1471-2164-15-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armour CD, Castle JC, Chen R, Babak T, Loerch P, Jackson S, Shah JK, Dey J, Rohl CA, Johnson JM, et al. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat Methods. 2009;6:647–649. doi: 10.1038/nmeth.1360. [DOI] [PubMed] [Google Scholar]
- 15.Adomas AB, Lopez-Giraldez F, Clark TA, Wang Z, Townsend JP. Multi-targeted priming for genomewide gene expression assays. BMC Genomics. 2010;11:477. doi: 10.1186/1471-2164-11-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhargava V, Ko P, Willems E, Mercola M, Subramaniam S. Quantitative transcriptomics using designed primer-based amplification. Sci Rep. 2013;3:1740. doi: 10.1038/srep01740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnaud O, Kato S, Poulain S, Plessy C. Targeted reduction of highly abundant transcripts with pseudo-random primers. bioRxiv. 2016;60:169–174. doi: 10.2144/000114400. [DOI] [PubMed] [Google Scholar]
- 18.Ko MS. An ‘equalized cDNA library’ by the reassociation of short double-stranded cDNAs. Nucleic Acids Res. 1990;18:5705–5711. doi: 10.1093/nar/18.19.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 20.Sultan M, Amstislavskiy V, Risch T, Schuette M, Dokel S, Ralser M, Balzereit D, Lehrach H, Yaspo ML. Influence of RNA extraction methods and library selection schemes on RNA-seq data. BMC Genomics. 2014;15:675. doi: 10.1186/1471-2164-15-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He S, Wurtzel O, Singh K, Froula JL, Yilmaz S, Tringe SG, Wang Z, Chen F, Lindquist EA, Sorek R, et al. Validation of two ribosomal RNA removal methods for microbial metatranscriptomics. Nat Methods. 2010;7:807–812. doi: 10.1038/nmeth.1507. [DOI] [PubMed] [Google Scholar]
- 22.Sun Z, Asmann YW, Nair A, Zhang Y, Wang L, Kalari KR, Bhagwate AV, Baker TR, Carr JM, Kocher JP, et al. Impact of library preparation on downstream analysis and interpretation of RNA-Seq data: comparison between Illumina PolyA and NuGEN Ovation protocol. PLoS One. 2013;8:e71745. doi: 10.1371/journal.pone.0071745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao W, He X, Hoadley KA, Parker JS, Hayes DN, Perou CM. Comparison of RNA-Seq by poly (A) capture, ribosomal RNA depletion, and DNA microarray for expression profiling. BMC Genomics. 2014;15:419. doi: 10.1186/1471-2164-15-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholson AW. Ribonuclease III mechanisms of double-stranded RNA cleavage. WIREs RNA. 2014;5:31–48. doi: 10.1002/wrna.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picelli S, Bjorklund AK, Reinius B, Sagasser S, Winberg G, Sandberg R. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 2014;24:2033–2040. doi: 10.1101/gr.177881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borodina T, Adjaye J, Sultan M. A strand-specific library preparation protocol for RNA sequencing. Methods Enzymol. 2011;500:79–98. doi: 10.1016/B978-0-12-385118-5.00005-0. [DOI] [PubMed] [Google Scholar]
- 27.Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008;44:3–12. doi: 10.1016/j.ymeth.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jayaprakash AD, Jabado O, Brown BD, Sachidanandam R. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 2011;39:e141. doi: 10.1093/nar/gkr693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun G, Wu X, Wang J, Li H, Li X, Gao H, Rossi J, Yen Y. A bias-reducing strategy in profiling small RNAs using Solexa. RNA. 2011;17:2256–2262. doi: 10.1261/rna.028621.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, Gnirke A, Regev A. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langevin SA, Bent ZW, Solberg OD, Curtis DJ, Lane PD, Williams KP, Schoeniger JS, Sinha A, Lane TW, Branda SS. Peregrine: a rapid and unbiased method to produce strand-specific RNA-Seq libraries from small quantities of starting material. RNA Biol. 2013;10:502–515. doi: 10.4161/rna.24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Townsley BT, Covington MF, Ichihashi Y, Zumstein K, Sinha NR. BrAD-seq: breath adapter directional sequencing: a streamlined, ultra-simple and fast library preparation protocol for strand specific mRNA library construction. Front Plant Sci. 2015;6:366. doi: 10.3389/fpls.2015.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller DF, Yan PS, Buechlein A, Rodriguez BA, Yilmaz AS, Goel S, Lin H, Collins-Burow B, Rhodes LV, Braun C, et al. A new method for stranded whole transcriptome RNA-seq. Methods. 2013;63:126–134. doi: 10.1016/j.ymeth.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller DF, Yan PX, Fang F, Buechlein A, Ford JB, Tang H, Huang TH, Burow ME, Liu Y, Rusch DB, et al. Stranded Whole Transcriptome RNA-Seq for All RNA Types. Curr Protoc Hum Genet. 2015;84:111411–111423. doi: 10.1002/0471142905.hg1114s84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heyer EE, Ozadam H, Ricci EP, Cenik C, Moore MJ. An optimized kit-free method for making strand-specific deep sequencing libraries from RNA fragments. Nucleic Acids Res. 2015;43:e2. doi: 10.1093/nar/gku1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R Genome Project Data Processing S. The sequence alignment/map format and SAM tools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu GK, Xu W, Wilhelmy J, Mindrinos MN, Davis RW, Xiao W, Fodor SP. Molecular indexing enables quantitative targeted RNA sequencing and reveals poor efficiencies in standard library preparations. Proc Natl Acad Sci USA. 2014;111:1891–1896. doi: 10.1073/pnas.1323732111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casbon JA, Osborne RJ, Brenner S, Lichtenstein CP. A method for counting PCR template molecules with application to next-generation sequencing. Nucleic Acids Res. 2011;39:e81. doi: 10.1093/nar/gkr217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kivioja T, Vaharautio A, Karlsson K, Bonke M, Enge M, Linnarsson S, Taipale J. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods. 2012;9:72–74. doi: 10.1038/nmeth.1778. [DOI] [PubMed] [Google Scholar]
- 41.Shiroguchi K, Jia TZ, Sims PA, Xie XS. Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proc Natl Acad Sci USA. 2012;109:1347–1352. doi: 10.1073/pnas.1118018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lonnerberg P, Linnarsson S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 43.Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc Natl Acad Sci USA. 2011;108:20166–20171. doi: 10.1073/pnas.1110064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu GK, Wilhelmy J, Stern D, Fan HC, Fodor SP. Digital encoding of cellular mRNAs enabling precise and absolute gene expression measurement by single-molecule counting. Anal Chem. 2014;86:2867–2870. doi: 10.1021/ac500459p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan HC, Fu GK, Fodor SP. Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015;347:1258367. doi: 10.1126/science.1258367. [DOI] [PubMed] [Google Scholar]
- 46.Asmann YW, Klee EW, Thompson EA, Perez EA, Middha S, Oberg AL, Therneau TM, Smith DI, Poland GA, Wieben ED, et al. 3′ tag digital gene expression profiling of human brain and universal reference RNA using Illumina Genome Analyzer. BMC Genomics. 2009;10:531. doi: 10.1186/1471-2164-10-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morrissy AS, Morin RD, Delaney A, Zeng T, McDonald H, Jones S, Zhao Y, Hirst M, Marra MA. Next-generation tag sequencing for cancer gene expression profiling. Genome Res. 2009;19:1825–1835. doi: 10.1101/gr.094482.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen EQ, Bai L, Gong DY, Tang H. Employment of digital gene expression profiling to identify potential pathogenic and therapeutic targets of fulminant hepatic failure. J Transl Med. 2015;13:22. doi: 10.1186/s12967-015-0380-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian B, Manley JL. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci. 2013;38:312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilkening S, Pelechano V, Jarvelin AI, Tekkedil MM, Anders S, Benes V, Steinmetz LM. An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res. 2013;41:e65. doi: 10.1093/nar/gks1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods. 2013;10:133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng D, Tian B. RNA-binding proteins in regulation of alternative cleavage and polyadenylation. Adv Exp Med Biol. 2014;825:97–127. doi: 10.1007/978-1-4939-1221-6_3. [DOI] [PubMed] [Google Scholar]
- 56.Chang H, Lim J, Ha M, Kim VN. TAIL-seq: genomewide determination of poly(A) tail length and 3′ end modifications. Mol Cell. 2014;53:1044–1052. doi: 10.1016/j.molcel.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 57.Harrison PF, Powell DR, Clancy JL, Preiss T, Boag PR, Traven A, Seemann T, Beilharz TH. PAT-seq: a method to study the integration of 3′-UTR dynamics with gene expression in the eukaryotic transcriptome. RNA. 2015;21:1502–1510. doi: 10.1261/rna.048355.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 60.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, Qiu J, Fu XD. RASL-seq for massively parallel and quantitative analysis of gene expression. Curr Protoc Mol Biol. 2012;Chapter 4(Unit 4.13):11–19. doi: 10.1002/0471142727.mb0413s98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mertens F, Antonescu CR, Mitelman F. Gene fusions in soft tissue tumors: Recurrent and overlapping pathogenetic themes. Genes Chromosomes Cancer. 2015;55:291–310. doi: 10.1002/gcc.22335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carrara M, Beccuti M, Lazzarato F, Cavallo F, Cordero F, Donatelli S, Calogero RA. State-of-the-art fusion-finder algorithms sensitivity and specificity. Biomed Res Int. 2013;2013:340620. doi: 10.1155/2013/340620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu J, Zhang W, Huang S, He Z, Cheng Y, Wang J, Lam TW, Peng Z, Yiu SM. SOAPfusion: a robust and effective computational fusion discovery tool for RNA-seq reads. Bioinformatics. 2013;29:2971–2978. doi: 10.1093/bioinformatics/btt522. [DOI] [PubMed] [Google Scholar]
- 65.Liu C, Ma J, Chang CJ, Zhou X. FusionQ: a novel approach for gene fusion detection and quantification from paired-end RNA-Seq. BMC Bioinformatics. 2013;14:193. doi: 10.1186/1471-2105-14-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Levin JZ, Berger MF, Adiconis X, Rogov P, Melnikov A, Fennell T, Nusbaum C, Garraway LA, Gnirke A. Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol. 2009;10:R115. doi: 10.1186/gb-2009-10-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moskalev EA, Frohnauer J, Merkelbach-Bruse S, Schildhaus HU, Dimmler A, Schubert T, Boltze C, Konig H, Fuchs F, Sirbu H, et al. Sensitive and specific detection of EML4-ALK rearrangements in non-small cell lung cancer (NSCLC) specimens by multiplex amplicon RNA massive parallel sequencing. Lung Cancer. 2014;84:215–221. doi: 10.1016/j.lungcan.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 68.Scolnick JA, Dimon M, Wang IC, Huelga SC, Amorese DA. An efficient method for identifying gene fusions by targeted RNA sequencing from fresh frozen and FFPE samples. PLoS One. 2015;10:e0128916. doi: 10.1371/journal.pone.0128916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Otazu IB, Zalcberg I, Tabak DG, Dobbin J, Seuanez HN. Detection of BCR-ABL transcripts by multiplex and nested PCR in different haematological disorders. Leuk Lymphoma. 2000;37:205–211. doi: 10.3109/10428190009057647. [DOI] [PubMed] [Google Scholar]
- 70.Rhoads A, Au KF. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics. 2015;13:278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Au KF, Sebastiano V, Afshar PT, Durruthy JD, Lee L, Williams BA, van Bakel H, Schadt EE, Reijo-Pera RA, Underwood JG, et al. Characterization of the human ESC transcriptome by hybrid sequencing. Proc Natl Acad Sci USA. 2013;110:E4821–E4830. doi: 10.1073/pnas.1320101110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Au KF, Underwood JG, Lee L, Wong WH. Improving PacBio long read accuracy by short read alignment. PLoS One. 2012;7:e46679. doi: 10.1371/journal.pone.0046679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu Z, Peters RJ, Weirather J, Luo H, Liao B, Zhang X, Zhu Y, Ji A, Zhang B, Hu S, et al. Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J. 2015;82:951–961. doi: 10.1111/tpj.12865. [DOI] [PubMed] [Google Scholar]
- 74.Tilgner H, Jahanbani F, Blauwkamp T, Moshrefi A, Jaeger E, Chen F, Harel I, Bustamante CD, Rasmussen M, Snyder MP. Comprehensive transcriptome analysis using synthetic long-read sequencing reveals molecular co-association of distant splicing events. Nat Biotechnol. 2015;33:736–742. doi: 10.1038/nbt.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mercer TR, Clark MB, Crawford J, Brunck ME, Gerhardt DJ, Taft RJ, Nielsen LK, Dinger ME, Mattick JS. Targeted sequencing for gene discovery and quantification using RNA CaptureSeq. Nat Protoc. 2014;9:989–1009. doi: 10.1038/nprot.2014.058. [DOI] [PubMed] [Google Scholar]
- 76.Clark MB, Mercer TR, Bussotti G, Leonardi T, Haynes KR, Crawford J, Brunck ME, Cao KA, Thomas GP, Chen WY, et al. Quantitative gene profiling of long noncoding RNAs with targeted RNA sequencing. Nat Methods. 2015;12:339–342. doi: 10.1038/nmeth.3321. [DOI] [PubMed] [Google Scholar]
- 77.Portal MM, Pavet V, Erb C, Gronemeyer H. TARDIS, a targeted RNA directional sequencing method for rare RNA discovery. Nat Protoc. 2015;10:1915–1938. doi: 10.1038/nprot.2015.120. [DOI] [PubMed] [Google Scholar]
- 78.Mamedov IZ, Britanova OV, Zvyagin IV, Turchaninova MA, Bolotin DA, Putintseva EV, Lebedev YB, Chudakov DM. Preparing unbiased T-cell receptor and antibody cDNA libraries for the deep next generation sequencing profiling. Front Immunol. 2013;4:456. doi: 10.3389/fimmu.2013.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Samorodnitsky E, Jewell BM, Hagopian R, Miya J, Wing MR, Lyon E, Damodaran S, Bhatt D, Reeser JW, Datta J, et al. Evaluation of hybridization capture versus amplicon-based methods for whole-exome sequencing. Hum Mutat. 2015;36:903–914. doi: 10.1002/humu.22825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O’Connell AE, Volpi S, Dobbs K, Fiorini C, Tsitsikov E, de Boer H, Barlan IB, Despotovic JM, Espinosa-Rosales FJ, Hanson IC, et al. Next generation sequencing reveals skewing of the T and B cell receptor repertoires in patients with wiskott-Aldrich syndrome. Front Immunol. 2014;5:340. doi: 10.3389/fimmu.2014.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fang H, Yamaguchi R, Liu X, Daigo Y, Yew PY, Tanikawa C, Matsuda K, Imoto S, Miyano S, Nakamura Y. Quantitative T cell repertoire analysis by deep cDNA sequencing of T cell receptor alpha and beta chains using next-generation sequencing (NGS) Oncoimmunology. 2014;3:e968467. doi: 10.4161/21624011.2014.968467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- 83.Saliba AE, Westermann AJ, Gorski SA, Vogel J. Single-cell RNA-seq: advances and future challenges. Nucleic Acids Res. 2014;42:8845–8860. doi: 10.1093/nar/gku555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 85.Saadatpour A, Lai S, Guo G, Yuan GC. Single-cell analysis in cancer genomics. Trends Genet. 2015;31:576–586. doi: 10.1016/j.tig.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58:610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 87.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 88.Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 91.Sasagawa Y, Nikaido I, Hayashi T, Danno H, Uno KD, Imai T, Ueda HR. Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol. 2013;14:R31. doi: 10.1186/gb-2013-14-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chapman AR, He Z, Lu S, Yong J, Tan L, Tang F, Xie XS. Single cell transcriptome amplification with MALBAC. PLoS One. 2015;10:e0120889. doi: 10.1371/journal.pone.0120889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grun D, Kester L, van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat Methods. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- 94.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Avital G, Hashimshony T, Yanai I. Seeing is believing: new methods for in situ single-cell transcriptomics. Genome Biol. 2014;15:110. doi: 10.1186/gb4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lovatt D, Ruble BK, Lee J, Dueck H, Kim TK, Fisher S, Francis C, Spaethling JM, Wolf JA, Grady MS, et al. Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nat Methods. 2014;11:190–196. doi: 10.1038/nmeth.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SS, Li C, Amamoto R, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang L. Splicing noncoding RNAs from the inside out. WIREs RNA. 2015;6:651–660. doi: 10.1002/wrna.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell. 2015;161:541–554. doi: 10.1016/j.cell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nojima T, Gomes T, Grosso AR, Kimura H, Dye MJ, Dhir S, Carmo-Fonseca M, Proudfoot NJ. Mammalian NET-Seq reveals genome-wide nascent transcription coupled to RNA processing. Cell. 2015;161:526–540. doi: 10.1016/j.cell.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol. 2011;29:436–442. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Miller C, Schwalb B, Maier K, Schulz D, Dumcke S, Zacher B, Mayer A, Sydow J, Marcinowski L, Dolken L, et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol. 2011;7:458. doi: 10.1038/msb.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zeisel A, Kostler WJ, Molotski N, Tsai JM, Krauthgamer R, Jacob-Hirsch J, Rechavi G, Soen Y, Jung S, Yarden Y, et al. Coupled pre-mRNA and mRNA dynamics unveil operational strategies underlying transcriptional responses to stimuli. Mol Syst Biol. 2011;7:529. doi: 10.1038/msb.2011.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jayaseelan S, Doyle F, Tenenbaum SA. Profiling post-transcriptionally networked mRNA subsets using RIP-Chip and RIP-Seq. Methods. 2014;67:13–19. doi: 10.1016/j.ymeth.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zambelli F, Pavesi G. RIP-Seq data analysis to determine RNA-protein associations. Methods Mol Biol. 2015;1269:293–303. doi: 10.1007/978-1-4939-2291-8_18. [DOI] [PubMed] [Google Scholar]
- 111.Wessels HH, Hirsekorn A, Ohler U, Mukherjee N. Identifying RBP targets with RIP-seq. Methods Mol Biol. 2016;1358:141–152. doi: 10.1007/978-1-4939-3067-8_9. [DOI] [PubMed] [Google Scholar]
- 112.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. PAR-CliP—a method to identify transcriptome-wide the binding sites of RNA binding proteins. J Vis Exp. 2010;141:129–141. doi: 10.3791/2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, Schroth GP, Pachter L, Doudna JA, Arkin AP. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc Natl Acad Sci USA. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. RNA–RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]