Abstract
Autophagy mediates resistance to various anticancer agents. In melanoma, resistance to targeted therapy has been linked to expression of Wnt5A, an intrinsic inhibitor of β-catenin, which also promotes invasion. In this study, we assessed the interplay between Wnt5A and autophagy by combining expression studies in human clinical biopsies with functional analyses in cell lines and mouse models. Melanoma cells with high Wnt5A and low β-catenin displayed increased basal autophagy. Genetic blockade of autophagy revealed an unexpected feedback loop whereby knocking down the autophagy factor ATG5 in Wnt5Ahigh cells decreased Wnt5A and increased β-catenin. To define the physiological relevance of this loop, melanoma cells with different Wnt status were treated in vitro and in vivo with the potent lysosomotropic compound Lys05. Wnt5Ahigh cells were less sensitive to Lys05 and could be reverted by inducing β-catenin activity. Our results suggest the efficacy of autophagy inhibitors might be improved by taking the Wnt signature of melanoma cells into account.
Keywords: Wnt5A, autophagy, melanoma, lysosome, cellular homeostasis
Introduction
Autophagy is a catabolic process that maintains cellular homeostasis through the degradation of cellular constituents and the generation of basic building blocks for the synthesis of new macromolecules (1). Initially described as a key survival feature for cancer cells, autophagy has raised great attention for its potential ability to promote cell death (2). The molecular basis that define these pro- vs. anti-tumorigenic roles of autophagy are not well defined (3,4). A better knowledge of genetic and pharmacological modulators of autophagy would have important basic and clinical implications for cutaneous melanoma. Histological studies revealed a great intra and inter-tumoral variability in multiple autophagy genes (5). One of the most intriguing changes in expression relates to the autophagy factor, autophagy related gene 5 (ATG5), a key player in autophagosome formation. Studies in large cohorts of melanoma patients indicate that ATG5 is downregulated in the transition from benign nevi to primary melanomas (6). Loss of ATG5 at this early stage of melanoma progression was required for bypassing senescence associated with oncogene activation in primary melanocytes. Further analyses in large datasets of human melanomas demonstrated that this downregulation of ATG5 was a result of a partial heterozygous loss (7). Moreover, newly-developed animal models revealed that this heterozygosity was physiologically relevant in the context of melanoma progression. Knockdown of ATG5 decreased melanoma cell survival in these mouse models (7), and in other settings related to cellular or environmental stress (8). The relevance of dissecting autophagy in the context of lysosomal-associated functions is further emphasized by the discovery of lineage markers that selectively deregulate lysosomal pathways in melanoma cells (9–11), as well as by pharmacological responses in the context of lysosomotropic agents (12–14). Yet, the extent to which oncogenic cascades impinge on the regulation and execution of autophagy in melanoma remain unclear.
We and others have previously demonstrated key roles of the Wnt pathway in melanoma progression as reviewed in (15–19). The canonical Wnt pathway, driven by β-catenin, is critical for the bypassing of melanocyte senescence and melanoma growth. The non-canonical Wnt ligand, Wnt5A, drives a highly metastatic phenotype (20) via the activation of PKC/Ca2+ signaling cascades that result in the reorganization of actin and filamin in the cytoskeleton (21). Interestingly, in conditions of stress such as irradiation or therapy, Wnt5A also drives a senescent-like phenotype, where cells go into an arrest that bears the hallmarks of senescence, yet remain highly invasive in vitro and in vivo (22). In melanoma, Wnt5A has been shown to inhibit β-catenin, via a GSK3β independent, SIAH2 mediated degradation (23). Of interest, autophagy-deficient breast epithelial cells display reduced levels of several invasive markers, among them, Wnt5A (24). Additionally, β-catenin is a negative regulator of basal and induced autophagy both in vivo and in vitro in colon cancer models (25). Together these studies suggested a feedback loop between Wnt signaling and autophagy, but whether this is the case in melanoma is unknown. Therefore, if Wnt5A suppresses β-catenin, we hypothesized that activated Wnt5A signaling would lead to increased autophagy levels in melanoma, and in this study we test this hypothesis.
In addition to affecting melanoma progression, loss of heterozygosity of ATG5 was found to affect response to the inhibition of the v-raf murine sarcoma viral oncogene homolog B (BRAF). The BRAF gene is mutated in over 50% of human melanomas, and this mutation is considered a driver mutation for melanoma. BRAF activates signaling within the mitogen activated protein kinase (MAPK) pathway, driving proliferation of melanoma cells. Given the prevalence of this mutation, a large effort was placed into developing inhibitors to the mutant BRAF protein, and the downstream pathway. These inhibitors have large effects in the clinic; however the effects are only temporary, making the development of other drugs necessary. Given that ATG5 loss can affect response to BRAF inhibitors, there is interest in examining the combination of autophagy inhibition with BRAF inhibition. Recently, dimeric chloroquines such as Lys05 have been developed that target the lysosome more effectively than the parent compound hydroxychloroquine, (HCQ) (13). Lys05 augments the efficacy of BRAF inhibition in a BRAF inhibitor resistant melanoma model (14). Lys05 has also shown activity in models of ovarian cancer (26) and hepatocellular carcinoma (27). We asked whether the canonical vs. non-canonical Wnt status of melanoma cells could dictate their level of response to autophagy inhibition. Given the importance of the Wnt signaling pathway in melanoma progression and therapy resistance, understanding the effects of Wnt signaling on autophagy will be critical for the successful translation of these studies.
Materials and Methods
Cell Culture
Human melanoma cells (FS5, FS4, FS13, FS14) obtained from Franklin Square Hospital were maintained in RPMI (Invitrogen) supplemented with 10% FBS, 4mM L-glutamine and 100 units/ml penicillin and streptomycin. Human melanoma cells (WM793, WM35, WM164, and 1205Lu) part of the Wistar Institute Collection (https://wistar.org/lab/meenhard-herlyn-dvm-dsc/page/melanoma-cell-lines-0) were maintained in MCDB153 (Sigma)/ Liebovitz L-15 (Cellgro) (4:1 ratio) supplemented with 2% FBS and 1.6mM CaCl2. Murine melanoma cells (Yumm 1.7, Yumm 1.7 over-expressing WNT5A, and Yumm 2.1 CTNNB1) were maintained in DMEM (Invitrogen), supplemented with 10% FBS, 4 mM L-glutamine and 100 units/ml penicillin and streptomycin. All cells were maintained at 37°C in 5% CO2 in a tissue culture incubator. These cells were used in experiments upto five to ten passages from thawing (between 2014 and 2017). Cell stocks were fingerprinted using AmpFLSTR® Identifiler® PCR Amplification Kit from Life Technologies TM at The Wistar Institute Genomics Facility. Although it is desirable to compare the profile to the tissue or patient of origin, our cell lines were established over the course of 40 years, long before acquisition of normal control DNA was routinely performed. However, each STR profile is compared to our internal database of over 200 melanoma cell lines, as well as control lines, such as HeLa and 293T. STR profiles are available upon request. The supernatants of cells are routinely collected and tested for mycoplasma (monthly) using a Lonza MycoAlert assay at the University of Pennsylvania Cell Center Services.
Treatments
Cells were treated with 100ng/mL or 200ng/mL of recombinant Wnt5A (rWnt5A, R&D Systems, cat. no. 645WN010CF) for 16 hours. Treatment with bafilomycin A1 (Sigma, cat. no. B1793) was performed at a final concentration of 100nM for 3–4 hours. The autophagy inhibitor Lys05 (obtained from the laboratory of Dr. Ravi Amaravadi) was used at various concentrations in vitro (1, 3, 5, and 10μM) for 16–24 hours and in vivo in mice at 20mg/kg daily for 14 days total. Cells were treated with the GSK3 inhibitor, lithium chloride (Sigma, cat. no. 203637), at various concentrations (2 mM, 5 mM and 10 mM) for 8 hours. Treatment with the GSK3 inhibitor, LY2090314 (Selleckchem cat. no. S7063) was performed at various concentrations (2, 5, and 10 nM) for 8 hours. For combination treatments, cells were first treated with rWnt5A for 16 hours, then bafilomycin A1 was added for 3 hours or Lys05 for 16 hours. For treatments combining GSK3 inhibitors with Lys05, cells were first treated with lithium chloride or LY2090314 for 8 hours followed by Lys05 treatment for 16 hours.
Cell Viability Assays
Following drug treatment, cell viability was assessed using trypan blue exclusion assay or using a MTS cell proliferation assay kit. Using the trypan blue exclusion assay method, cells were counted in the presence of trypan blue and percent cell viability was determined and normalized to control. For the MTS assay, following drug treatment, MTS reagent (Cell titer Aqueous One Solution Reagent, Promega) was added to the cells. The cells were then incubated at 37°C in 5% CO2 for 1–4 hours; absorbance was recorded every hour at 490 nm. Finally, percent cell proliferation was determined and normalized to control.
Bliss synergy assay
We seeded 2000 cells per well in 100μl in 96 well format. Cells were allowed to adhere overnight, then incubated for 7 hours with LY2090314 or LiCl before the addition of Lys05. LY2090314 or LiCl compounds were added in a 1:2 dilution series running vertically with 20 nM and 5 mM respectively as the highest concentration. Lys05 compound was added in 1:1.5 dilution series starting at 10 μM in horizontal orientation. Plates were incubated for an additional 72 hours and cell growth was assessed using alamar blue assay. Each plate was normalized to media and doxorubicin controls, single compound concentration curves were calculated and used for Bliss synergy calculations. Samples were subsequently analyzed with an EPICS XL (Beckman-Coulter, Inc., Brea, CA) apparatus. We used the effect-based Bliss Independence Model since the compounds act independently and are mutually non-exclusive (28–30). Normalized data was converted to fraction affected values (F). We then calculated the predicted inhibition values (P) if the two compounds exhibit additive effects based on fraction affected observed for each single compound at the corresponding concentrations being compared. [P=Fa + Fb Fab 0<P>1]. Predicted F equals the fraction affected by compound “a” (Fa) at concentration x plus the fraction affected by compound “b” (Fb) at concentration y minus the product of the two (Fab). The difference between the predicted additive fraction affected and the experimentally observed fraction affected is the Bliss number where a positive value indicates synergy, a negative value indicates antagonism and an overlap of predicted and observed combination effects gives a Bliss number of zero and indicates additivity. Due to the nature of the assay, we detected a background noise level of +/− 10%. To decrease the possibility of pursuing false positives, we only considered combinations whose Bliss value was greater than x the background, i.e., a value greater than 30.
Western Blot
Briefly 20–30 μg of protein lysate was run on 4–12% NuPAGE Bis Tris gel (Invitrogen cat. no. NW04122BOX) or 14% Tris glycine Novex WedgeWell (Invitrogen cat. no. XP00145BOX) gel and transferred onto a PVDF membrane using an iBlot2 transfer machine manufactured by Life technologies. The membranes were then blocked with 5% TBST/milk or 5% TBST/BSA (bovine serum albumin) for 1 hour at room temperature and incubated with the appropriate primary antibodies at 4°C overnight. The following primary antibodies and concentrations were used: ATG5 (1:1000, Cell Signaling cat. no. 2630), LC3B (1:1000, Novus Biologicals, cat. no. NB100-2220), HSP90 (1:4000, Cell Signaling, cat. no. 4877S), non-phospho active CTNNB1 (β-catenin) (1:1000, Cell Signaling cat. no. 8814S), Histone H3 (1:1000, Cell Signaling, cat. no. 9715S), Phospho-GSK-3α/β (1:1000, Cell Signaling, cat. no. 9331S), GAPDH (1:1000, Cell Signaling, 2118S), SQSTM1 (D3) p62 (1:500, Santa Cruz Biotech, cat. no. sc28359), SQSTM1 (1:1000, Cell Signaling cat. no. 5114S), biotinylated Wnt5A (500 ng/ml, R&D Systems, cat. no. BAF645), β tubulin (1:1000, Cell Signaling, 2146S), PKCα (1:1000 Cell Signaling, cat. no. 2056S), phospho-PKCα/β (1:1000, Cell Signaling, cat. no. 9375S), CaMKII pan (1:1000, Cell Signaling, cat. no. 3362S), and p-CaMKII (1:1000, Cell Signaling, cat. no. 3356S). Following overnight incubation with the primary antibodies, the membranes were washed in TBST and incubated with the appropriate secondary antibodies (anti-mouse HRP, anti-rabbit HRP or streptavidin at 0.2–0.02ug/ml). Luminata Crescendo (Millipore, MA) was used to visualize the proteins on an ImageQuant 4000 scanner.
SiRNA assays
Negative control (Qiagen, cat. no. 1027281) and siRNA against human CTNNB1 (Qiagen, cat. no. SI04379662) or siRNA against human WNT5A (Qiagen, cat. no. s103025596) were transfected in melanoma cells at the concentration of 20 nM using Lipofectamine 2000 (Thermo Fisher Scientific), as per the manufacturer’s protocol.
Generation of shATG5 clones
shRNA constructs against ATG5 and a non-target control shRNA were obtained from the laboratory of Dr. Maureen Murphy. The clones used are Control (non-target): Shc002 lot 11151021; ATG5 (construct #1): NM004849-1-40s1c1; ATG5 (construct #2): NM004849- 1170s1c1. Briefly 293T cells were transfected with ATG5 shRNA vector and lentiviral packaging plasmids (PVSVG, PLP2, and PLP1). The supernatant containing virus was collected 48 hours and 72 hours post-transfection. The supernatants were then combined, filtered using 0.45 μm filters, and stored at −80C until needed. Melanoma cells were transduced with the virus in the presence of polybrene at the final concentration of 5 μg/ml; 24 hours post-transduction, the melanoma cells were fed with regular media and selected with puromycin at a final concentration of 0.5 μg/ml.
GFP-LC3 Transient Transfection
Melanoma cells were transfected with 0.3 μg of GFP-LC3 (2 μg/ml) using Opti-MEM and Fugene; 24 hours post-transfection, the cells were visualized using a Leica TCS SP5 II scanning laser confocal microscope.
TOPFlash Assay
Cells were assayed according to the manufacturer’s protocol. TOPFlash vectors were obtained from Addgene (M51 Super 8x FOPFlash/TOPFlash mutant, #12457; M50 Super 8x TOPFlash, 12456). Melanoma cells were plated to achieve 70% confluency in 6-well plates. Cells were co-transfected with pTK-RLuc (Green Renilla Luciferase) along with either Topflash or Fopflash vectors. After 48 hours, cells were harvested and luciferase activity was measured using the Dual-Luciferase® Reporter (DLR ) Assay System (Promega, E1910), where firefly luciferase signal was normalized to its corresponding Renilla Luciferase signal. TOPflash/FOPflash signal was determined from each treatment and graphed using GraphPad Prism.
Nuclear and cytoplasmic extraction
Cells were assayed according to the manufacturer’s protocol. Nuclear and cytoplasmic extraction reagents were obtained from Thermoscientific (NE-PER, Product # 78833). Nuclear and cytoplasmic lysates were stored at −80°C.
Organotypic 3D skin reconstructs
3D skin reconstructs were made as previously described (31) . Briefly, fibroblasts (6.4 x 104) were plated on top of the acellular layer of each insert (BD cat.no. 355467 and Falcon cat. no. 353092) and incubated for 45 minutes at 37°C in 5% CO2 in a tissue culture incubator. DMEM containing 10% FBS was added to the tissue culture trays and incubated for 4 days. The reconstructs were then incubated in HBSS containing 1% dialized FBS (wash media) for 1 hour. After removal of the wash media, reconstruct media was added. Melanoma cells (8.3 x 104) and keratinocytes (4.17 x 105) were added inside each insert. Until day 18, the media was changed every other day. Reconstructs where then fixed in 10% formalin, paraffin embedded, and stained. Melanoma cells at different states of invasion were used: RGP (radial growth phase) VGP (vertical growth phase) and metastatic.
2D Boyden Chamber assay
Melanoma cells grown in 10% RPMI media were plated in cell permeable inserts (Corning) nested in multi-well plates containing 20% RPMI. The cell invasion chambers were then placed in a 37°C in 5% CO2 tissue culture incubator overnight to allow cell migration through the membrane of the inserts. Finally, the cells at the bottom of the invasion chambers were fixed with methanol, stained with crystal violet, and counted.
3D Spheroids assay
Melanoma spheroids were generated as previously described (32). Briefly, melanoma cells were grown on agar for 3–5 days to allow the formation of spheroids. The spheroids were then embedded in a collagen plug and allowed to invade.
Electron Microscopy
Electron microscopy analysis was performed using FS13 cells stably transfected with vector alone or CMV-driven HA-tagged WNT5A. Average area of AV (autophagy vesicle) per section was calculated using ImageJ software.
Immunofluorescence (IF)
IF was performed as previously described (32). For skin reconstructs, paraffin embedded sections were first deparaffinized using multiple xylene washes and then re-hydrated using multiple alcohol washes and a water rinse. Following antigen retrieval (20 minutes steaming using antigen retrieval buffer from Vector Labs), the sections were incubated in hydrogen peroxide, washed in PBS and then blocked using IF blocking buffer. SQSTM1 (D-3) (1:100, Santa Cruz biotech, cat. no. sc28359) antibody was diluted in IF blocking buffer and added to the slides. Samples were then incubated overnight at 4°C in a humidified chamber. The next day, the slides were washed in PBS and probed with the appropriate secondary antibody (1:2000, Alexa fluor 568 goat anti-mouse IgG H+L, Life technologies, cat. no. A1103) for 1 hour at room temperature. Slides were then washed with PBS and mounted in Prolong Gold anti-fade reagent containing DAPI (Invitrogen). A Leica TCS SP5 II scanning laser confocal microscope was used to capture the images. For cell monolayers, 2–4×104 cells were seeded onto glass cover slips and placed in a 37°C in 5% CO2 tissue culture incubator overnight. Cells were then fixed using 95% methanol. Primary antibodies: LC3B (1:1000, Novus Biologicals, cat. no. NB100-2220) and β-Catenin (L54E2) (Cell Signaling, cat. no. 2677S) were diluted in blocking buffer and added to the cells overnight at 4°C. Cells were then washed with PBS and incubated with the appropriate secondary antibodies (1:2000 Alexa fluor 647 goat anti-rabbit IgG, Invitrogen, cat. no. A21244 and Alexa fluor 555 goat anti-mouse, Invitrogen, cat. no. A21422) for 1 hour at room temperature. Cells were then washed in PBS and mounted with Prolong Gold containing DAPI (Invitrogen). A Leica TCS SP5 II scanning laser confocal microscope was used to capture the images.
Immunohistochemistry (IHC)
IHC was performed on paraffin embedded slides as previously described (32) . Paraffin-embedded sections were deparaffinized and rehydrated using the same process described in the IF section. For nuclear proteins: following antigen retrieval and blocking in IF blocking buffer, the appropriate primary antibody Sox10 (1:100, Santa Cruz, cat. no. sc17342), or β-Catenin (L54E2) (1:200, Cell Signaling, cat. no. 2677S was diluted in blocking buffer and added to slides for overnight incubation at 4°C in a humidified chamber. On day 2, the slides were washed with PBS and incubated for 1 hour at room temperature with the appropriate secondary antibody (donkey peroxidase anti-goat, 1:100, Jackson ImmunoResearch or goat anti-polyvalent, ThermoScientific, cat. no. TP-060-BN). All slides were then washed with PBS and incubated in AEC (3-Amino-9-Ethyl-l-Carboazole, Thermoscientific, TA-060-SA) chromogen. Finally, the slides were rinsed briefly in H2O, incubated in Meyer’s hematoxylin for 60 seconds and rinsed with H2O. The slides were there mounted using Aquamount. For cytoplasmic proteins: following antigen retrieval, slides were first blocked with hydrogren peroxidase block (Thermoscientific, TA-060_H202Q), washed and blocked once more with Ultra V block (Thermoscientific, TA-060-PBQ) to reduce non-specific background. Slides were then incubated overnight with the appropriate primary antibody (mCherry 1:100, Novus biologicals, cat. no. NBP2-25157, LC3B 1:1000, Novus Biologicals, cat. no. NB100-2220, or biotinylated Wnt5A 500 ng/ml, R&D Systems, cat. no. BAF645) at 4°C in a humidified chamber. On Day 2, slides were washed and incubated in the appropriate secondary antibody (anti-rabbit biotinylated, TR-060-BN, or goat anti-polyvalent, ThermoScientific, cat. no. TP-060-BN) for 1 hour at room temperature. Next, slides were washed and incubated in streptavidin solution (Thermoscientific cat. no. TS-060-HR). This was followed by incubation in AEC and a rinse in H2O. Finally, slides were incubated in Meyer’s hematoxylin for 60 seconds, rinsed with H2O, and mounted with Aquamount.
TCGA Analysis
Clinical data relating to melanoma tumor stage and relative mRNA levels of Wn5A, ATG5 and SQSTM1 were collected from the TCGA (The Cancer Genome Atlas) database (https://cancergenome.nih.gov/). The analysis was conducted using data from a total of 472 patients (103 patients with primary melanoma and 369 patients with metastatic melanoma).
Reverse Phase Protein Array (RPPA) analysis
Whole cell protein lysates were extracted and RPPA analysis was performed at the MDACC Functional Proteomics Core Facility. Data was processed as described previously (33,34). Normalized and median centered data of significantly different (P<0.05) proteins between control and shATG5 treatments was subjected to unsupervised clustering, and organized into a heatmap. Antibodies used for RPPA are listed at the core facility’s website (http://www.mdanderson.org/education-and-research/resources-for-professionals/scientific-resources/core-facilities-and-services/functional-proteomics-rppa-core/index.html).
Tissue microarrays (TMA)
TMA containing 0.9-mm cores of 22 nevi, 30 invasive, and 130 metastatic malignant melanomas were constructed at the Fox Chase Cancer Center (Philadelphia, PA); full details about these samples are previously described (35). In addition 14 samples of other tumor types were also included. These tissue arrays were provided by Dr. George Xu of the University of Pennsyvania, and stained at Wistar under the IRB Exemption # EX21205258-2. The median of each biomarker’s measured values was used as a cutoff to separate its values to high and low. For Wnt5A to LC3 comparisons, all melanoma samples were used. Next, in samples (melanoma and non-melanoma, but all tumor) with both high Wnt5a (>100) and high LC3 (>80), Spearman’s correlation analyses were performed to test the correlation between Wnt5a and LC3, and between LC3 and β-catenin.
Real Time PCR
As previously described RNA was extracted using Trizol (Invitrogen) and the RNeasy Mini kit (Qiagen) (36). cDNA was then prepared using the iscript DNA synthesis kit (Bio-Rad, cat no. 1708891). SYBR Green dye-based PCR amplification was used to measure gene expression; qPCR was performed using the ABI StepOnePlus apparatus. The mRNA levels of samples were normalized with the mRNA levels of 18S using Universal 18S primers (Invitrogen, cat. No. AM1718). mRNA expression was determined using the standard curve method recommended by the manufacturer’s protocol (Perkin Elmer, Waltham, MA). Primers used are: ATG5 Forward, 5’ GGC CAT CAA TCG GAA ACT CAT 3’; ATG5 Reverse, 5’ AGC CAC AGG ACG AAA CAG CTT 3’; ATG12 Forward, 5’ TAG AGC GAA CAC GAA CCA TCC 3’; ATG12 Reverse, 5’ CAC TGC CAA AAC ACT CAT AGA GA 3’; WNT5A Forward, 5’ ATT CTT GGT GGT CGC TAG GTA 3’; WNT5A Reverse, 5’ CGC CTT CTC CGA TGT ACT GC 3’.
In vivo assay
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) and conducted at the Wistar Institute, an AAALAC (Association for the Assessment and Accreditation of Laboratory Animal Care) accredited facility. Yumm 1.7 parental, Yumm 1.7 overexpressing WNT5A or Yumm 2.1 CTNNB1 in which β catenin is stabilized were injected subcutaneously onto 6–8 week old C57/BL6 mice (Charles River). Tumor sizes were measured every 2 days using digital calipers. Tumor volumes were calculated using the formula 0.5 x (length x width)2. Once tumors reached 150 mm3 in volume, mice were treated with Lys05 at 20mg/kg daily for a total of 14 days. At the end of the experiment, mice were euthanized and tumors and lungs were harvested for further analysis.
Statistical Analysis
Student t-test was used for in vitro assays involving the comparison of two groups; analysis of variance was performed using the Welch’s correction test. For multiple comparisons, ANOVA statistical analysis was used. All graphs and statistical analysis were made using GraphPad Prism 6. Statistical significance was annotated as follows: ns, p>0.05; *, p≤0.05; **, p≤0.01; ***, p≤0.001; and ****, p≤0.0001
Results
Wnt5A correlates with autophagy levels in melanoma
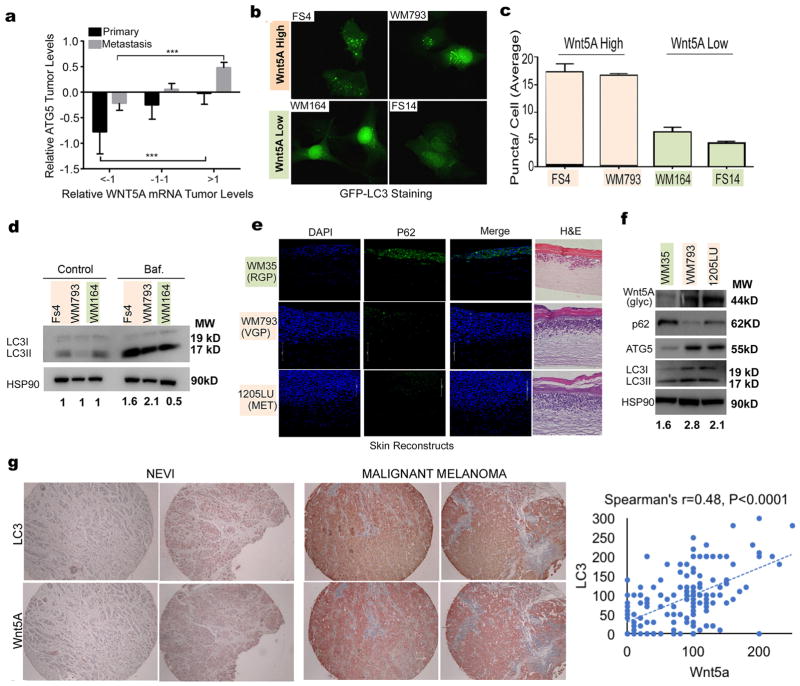
We analyzed the correlation between WNT5A and ATG5 expression as measured in primary and metastatic tumors from 472 melanoma patients (103 patients with primary melanoma and 369 patients with metastatic melanoma) using the cancer genome atlas (TCGA). Our analysis confirmed previous observations that ATG5 is high in metastatic melanoma and low in primary melanoma (Figure 1A, Supplementary Figure 1A). ATG5 expression significantly correlated with WNT5A expression. To further investigate these findings, we first measured autophagy in a panel of Wnt5Ahigh and Wnt5Alow melanoma cells. Microtubule-associated protein 1A/1B-light chain 3 (LC3) is a marker of autophagy, where the cytosolic form of the protein (LC3I) becomes conjugated to phosphatidylethanolamine to form LC3II, which associates with the outer and inner autophagosomal membranes. During the process of autophagy, ATG4 cleaves off LC3 on the outer membrane and lysosomal enzymes degrade LC3 in the autolysosome resulting in low LC3 in the autolysosome (37). Thus LC3 is considered a marker of autophagy. Melanoma cells were transfected with the GFP-LC3B construct which produces a diffuse fluorescence in the absence of autophagy and a punctate fluorescence that represents autophagosomes (37). We observed a diffuse fluorescence in Wnt5Alow melanoma cells (WM164, FS14) indicative of low autophagy activity as compared to a punctate fluorescence in FS4, and WM793 cells, which have higher Wnt5A (Figure 1B, quantitated in 1C). We confirmed this observation by measuring autophagic flux in FS4, WM793 and WM164 cells using a bafilomycin clamp which blocks the fusion of the autophagosome with the lysosome, allowing for the measurement of accumulated LC3II which is a marker of autophagosome synthesis (38). We found that cells with higher levels of Wnt5A as shown in supplemental Figure 1B had higher LC3II levels compared to Wnt5Alow cells (Figure 1D). Additional cell lines are shown in Supplementary Figure 1C. Densitometry for all Western blots in this manuscript is included in Supplementary Table 1.
Figure 1. Wnt5A expression correlates with high autophagy in melanoma.
A. Analysis of the relative mRNA levels of ATG5 and WNT5A in patients with primary (N=103) and metastatic (N=369) melanoma (N=369) using the TCGA data base (one-way ANOVA, ns= p>0.05, **= p ≤0.01, ***= p ≤ 0.001, ****= p<0.0001). B. Wnt5Ahigh (FS4, WM793) and Wnt5Alow (WM164, FS14) melanoma cell lines transfected with GFP-LC3 producing a punctate fluorescence as autophagy vesicles accumulate. C. Quantitation of average puncta per cell in B. D. Western blot of LC3I/II levels in melanoma cells using a bafilomycin (Baf.) clamp in Wnt5Ahigh (FS4, WM793) and Wnt5Alow (WM164) melanoma cells; loading control: HSP90. Autophagy flux quantification: ratio of Baf. (LC3II/HSP90) to control (LC3II/HSP90). E. Immunofluorescence analysis of p62 (green) in organotypic 3D reconstruct models of melanoma cells in RGP (RGP-WM35), VGP (VGP-WM793), and metastatic (MET-1205Lu) stages. RGP, radial growth phase; VGP, vertical growth phase; DAPI, 4,6-diamidino-2-phenylindole (blue). F. Western blot of Wnt5A, p62, ATG5, and LC3II/I levels in WM35, WM793, and 1205Lu melanoma cells; loading control: HSP90. Numbers represent ratio of LC3II to loading control. G. LC3 and Wnt5A IHC staining of tissue microarrays; Spearman’s analysis shows the correlation between LC3 and Wnt5A (p<0.0001).
We further investigated the correlation between autophagy and Wnt5A expression by staining 3D skin reconstructs with p62. The autophagy cargo adaptor p62 (also known as sequestosome1- SQSTM1) is degraded when autophagy is induced (39). The 3D skin reconstructs shown represent radial growth phase (early-stage) melanoma (WM35), vertical growth phase (intermediate) stage melanoma (WM793) and metastatic melanoma (1205Lu). Our results show that reconstructs made with 1205Lu and WM793 have low p62 levels compared to reconstructs made with early-stage WM35 cells (Figure 1E). However, data from several groups also implicate p62 in autophagy-independent signaling as well (40,41), and on its own, p62 is not considered a robust marker of autophagy. Therefore, to further confirm the implication of increased autophagy in our skin reconstructs, we performed Western blot analysis of the protein levels of p62, ATG5, LC3I/II and Wnt5A in the cells used to make the reconstructs. Wnt5A is increased in VGP (WM793) and metastatic (1205LU) melanoma (Figure 1F); we observe decreased p62, increased ATG5 and the accumulation of LC3II in the melanoma cells with high Wnt5A. Additional melanoma cell lines are shown in Supplementary Figure 1C. To confirm this in melanoma patient samples, we stained a melanoma tissue microarray (n=196 samples) with Wnt5A and LC3, and then compared the relationship between LC3 and Wnt5A in these samples. Wnt5A staining correlated in intensity to that of LC3, to a significance of p<0.0001. Examples of staining in corresponding tumor samples, and the Spearman’s analysis is shown in Figure 1G, full data is provided in Supplementary Table 2. Overall, we found that high Wnt5A expression correlates with high autophagy in melanoma cells, suggesting that Wnt5A signaling may either contribute to, or be affected by, autophagy.
Autophagy is regulated by and regulates Wnt signaling
To determine whether Wnt5A increases the autophagy levels of melanoma cells, we genetically manipulated WNT5A in melanoma cells and measured autophagy activity in isogenic clones. ATG5 is a major autophagy gene required for autophagosome synthesis (1). To determine if WNT5A could affect levels of ATG5, we tested WNT5A-overexpressing isogenic clones of FS13 melanoma cells and found that WNT5A overexpression led to a significant increase in ATG5 mRNA levels, as well as of ATG12, in concordance with increased Wnt5A (Figure 2A). ATG12 is conjugated to ATG5; this conjugation is essential for autophagosome synthesis (42). Overexpression of Wnt5A in Wnt5Alow melanoma cells (FS13) led to a significant increase in autophagy as measured by GFP-rich puncta (Figure 2B, arrows) and electron microscopy of autophagic vacuoles (Figure 2C). Treatment of Wnt5A low melanoma cells (WM164) with rWnt5A increased Wnt5A and led to a decrease in p62 (Figure 2D). Autophagy flux can be measured by LC3II turnover in the presence and absence of bafilomycin A1 (BAF) a lysosomal degradation inhibitor, known as a BAF clamp assay (38), which allows for the detection of accumulated LC3II when autophagy is high. rWnt5A treatment also increases accumulated LC3II showing that Wnt5A increases autophagy in melanoma cells (Figure 2E). To determine whether WNT5A knockdown would decrease autophagy, we knocked down WNT5A in FS4 Wnt5Ahigh cells using shRNA (Figure 2F). WNT5A knockdown results in a decrease in LC3II accumulation (Figure 2G). To support these data, we also used siRNA in the same cell line, and observed the same result (Supplementary Figure 2A,B). Finally we performed shRNA knockdown in an additional cell line (1205LU) and confirmed the observation that WNT5A knockdown decreases the accumulation of LC3II (Supplementary Figure 2C,D). Overall, our results show that increasing WNT5A increases autophagy, and that knockdown of WNT5A can decrease autophagy in melanoma cells.
Figure 2. Wnt5A increases autophagy in melanoma.
A. qRT-PCR analysis of WNT5A, ATG5, and ATG12 mRNA levels in FS13 control cells (EV) and FS13 overexpressing WNT5A (WNT5A), (left to right); (two-tailed unpaired t-test, **= p ≤0.01, ***= p ≤ 0.001). B. FS13 control (EV) and FS13 overexpressing WNT5A (WNT5A) transfected with GFP-LC3 produce a punctate fluorescence as autophagy vesicles accumulate; graph represents average of puncta per cell; (two-tailed unpaired t-test, ****=p<0.0001). C. Electron microscopy of FS13 control (EV) and FS13 overexpressing WNT5A (WNT5A); arrowheads point to autophagy vesicles that appear following Wnt5A overexpression. Dark granules represent melanin; graph represents average autophagy vesicles per cell (two-tailed unpaired t-test, **= p< 0.01). D. Western blot of p62 and Wnt5A in WM164 melanoma cells upon treatment with rWnt5A at the indicated doses for 16 hours; loading control: HSP90. E. Western blot of LC3I/II in Wm164 melanoma cells upon treatment with rWnt5A at the indicated doses for 16 hours using a bafilomycin clamp; loading control: HSP90. Autophagy flux quantification: ratio of Baf. (LC3II/HSP90) to control (LC3II/HSP90). F. Western blot of Wnt5A levels in FS4 melanoma cells upon shRNA-mediated knockdown of Wnt5A; loading control: HSP90. G. Western blot of LC3II/I levels in FS4 control and two different FS4 sh_Wnt5A (sh1 and sh2) clones upon bafilomycin treatment; loading control: HSP90. Autophagy flux quantification: ratio of Baf. (LC3II/HSP90) to control (LC3II/HSP90).
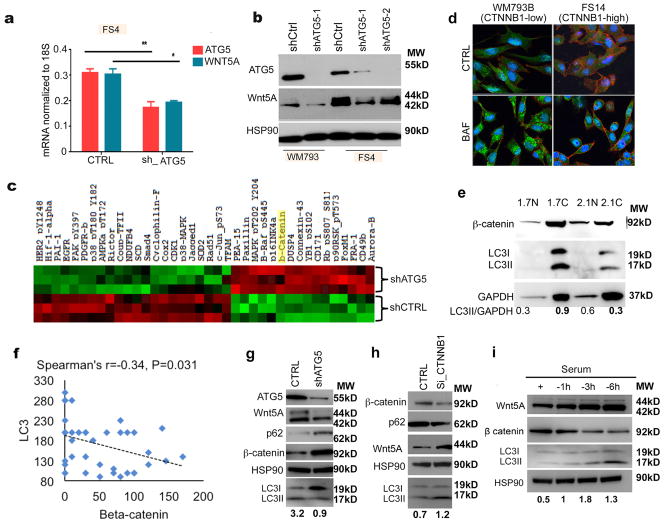
To determine whether autophagy can in turn affect Wnt signaling in melanoma cells, we knocked down ATG5 in Wnt5A high melanoma cells. We found that knocking down ATG5 results in a significant decrease in Wnt5A at the mRNA and protein levels (Figures 3A,B). To determine what other effects ATG5 knockdown might have on melanoma cells, we performed an RPPA analysis on melanoma cells with ATG5 intact, or knocked down; the Wnt5A target β–catenin was significantly upregulated (Figure 3C). We first confirmed the inverse relationship between β– catenin function, and autophagy by performing immunofluorescent co-staining of β– catenin, and LC3 in WM793 and FS14 cells. We found that when autophagy was high, β-catenin was relegated to the cell membrane, indicative of non-transcriptional activity, but when autophagy was low, β-catenin was expressed all over the cell (Figure 3D); additional melanoma cell lines are shown in supplemental Figure 2E. Next, we used nuclear and cytoplasmic fractions to correlate the localization of β-catenin to autophagy. Yumm 2.1 melanoma cells have stabilized β-catenin as compared to Yumm 1.7 cells that are derived from the same background, but do not have stabilized β-catenin. Nuclear and cytoplasmic westerns showed that high nuclear β-catenin correlated to low autophagy as measured by LC3I/II ratios (Figure 3E). These data suggested that the transcriptional activity of β-catenin may be critical for its roles in autophagy. Next, to confirm the observation that LC3 and Wnt5A were inversely correlated to β-catenin, we analyzed the expression of β-catenin in patient samples with high LC3 and Wnt5A (n=80). We found that there was a negative correlation between β-catenin and Wnt5A and LC3, to a statistically significant level (p=0.031). This included all melanoma samples, but we also included other tumors present on the array, that stained positive for both LC3 and Wnt5A in order to achieve the numbers necessary for statistical significance (Figure 3F, Supplementary Table 2). In melanoma alone, when trying to compare all three proteins, we approached but did not quite reach statistical significance (p=0.07).
Figure 3. ATG5 affects Wnt signaling in melanoma.
A. qRT-PCR analysis of ATG5 and WNT5A levels in FS4 control cells (CTRL) and FS4 with stable knockdown of ATG5 (sh_ATG5), (two-tailed unpaired t-test, *=p ≤ 0.05, **= p ≤ 0.01). B. Western blot of ATG5 and Wnt5A in FS4 and WM793 control (shCTRL) and FS4 and WM793 cells with stable knockdown of ATG5 (shATG5-1 and shATG5-2); loading control: HSP90. C. RPPA (reverse phase protein microarray) analysis of FS4 control cells (shCTRL) and FS4 with stable knockdown of ATG5 (sh-ATG5). D. Immunofluorescence analysis of β-catenin (CTNNB1) (red) and LC3 (green) in WM793 (CTNNB1low) and FS14 (CTNNB1high) upon treatment with bafilomycin ; DAPI, 4,6-diamidino-2-phenylindole (blue). E. Western blot of β-catenin, LC3I/II, and GAPDH in Yumm 1.7 and Yumm 2.1 nuclear (N) and cytoplasmic (C) lysates; loading control: GAPDH. F. Spearman’s analysis of LC3/Wnt5A high samples and β-catenin IHC staining in tissue microarrays showing the correlation between LC3 and β–catenin (p =0.031). G. Western blot of ATG5, Wnt5A, p62, β-catenin and LC3I/II in FS4 shCTRL and FS4 shATG5; loading control: HSP90. Numbers represent ratio of LC3II to loading control. H. Western blot of β-catenin, p62, Wnt5A and LC3I//II in WM164 control (CTRL) and in WM164 with siRNA-mediated knockdown of β-catenin (Si-CTNNB1); loading control: HSP90. Numbers represent ratio of LC3II to loading control. I. Western blot of Wnt5A, β-catenin, and LC3II/I in WM164 showing changes in Wnt signaling upon autophagy induction by serum starvation for the indicated time points; loading control: HSP90. Numbers represent ratio of LC3II to loading control.
These data were consistent with the observation that Wnt5A is decreased upon ATG5 knockdown, since Wnt5A is a negative regulator of β–catenin (23). We confirm this by showing that knocking down Wnt5A in melanoma cells restores β–catenin levels (Supplementary Figure 3A). To confirm changes in β–catenin after ATG5 knockdown, we performed Western analysis to corroborate the RPPA data. By Western blot, ATG5-knockdown cells also had increased β–catenin, confirming β–catenin is upregulated upon autophagy inhibition (Figure 3G, additional cell line, Supplementary Figure 3B). To determine whether β–catenin was involved in the regulation of autophagy in melanoma cells; we knocked down β–catenin by siRNA and found that decreasing β–catenin in melanoma cells leads to an increase in autophagy and an increase in Wnt5A (Figure 3H). This is concomitant with an increase in the accumulation of LC3II, using a BAF clamp assay (Supplementary Figure 3C). This is consistent with previous observations that β–catenin can inhibit autophagy, and our observation that decreasing ATG5 can decrease Wnt5A. These data prompted us to further investigate whether there is a feedback loop between Wnt signaling and autophagy in melanoma. To test this, we induced autophagy in WM164 melanoma cells, which have low Wnt5A and high β–catenin levels by serum starvation. We found that activating autophagy in these cells leads to a switch from canonical to non-canonical Wnt signaling as indicated by the increase in Wnt5A and the decrease in β–catenin (Figure 3I). We confirmed these data in an additional β-catenin low line (Supplementary Figure 3D). These data suggest that autophagy signaling may involve antagonism between the canonical (β–catenin-driven) and non-canonical (Wnt5A-driven) Wnt signaling pathways.
Inhibition of Autophagy in Wnt5Ahigh cells requires higher doses of Lys05
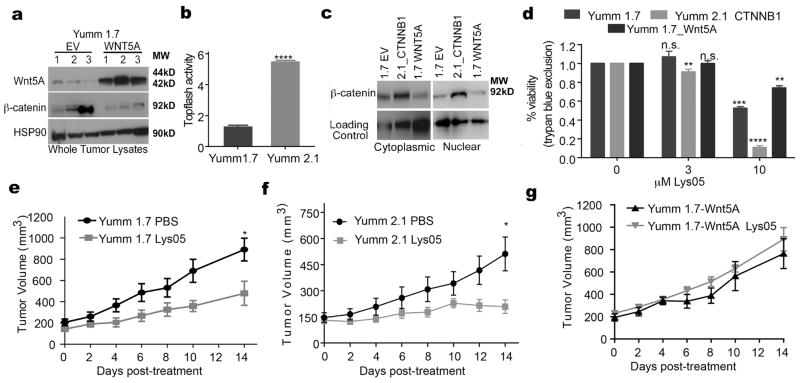
The rationale for autophagy inhibition has previously been based on the notion that autophagyhigh cells will respond better to autophagy inhibitors. However, we were curious as to whether a threshold effect could occur, such that cells with very high levels of autophagy would require higher doses of autophagy inhibitors. We used the new autophagy inhibitor Lys05, which has been shown to effectively target autophagy in melanoma cells (13). To perform these experiments in vivo, we used an allograft model of melanoma. Yumm1.7 cells are derived from the BRAFV600EPTEN−/−/CDKN2A−/− mouse model of melanoma, and can be allogeneically injected into C57BL/6 mice. Yumm 1.7 cells have low Wnt5A, and transfecting these cells with Wnt5A decreases their levels of β–catenin as measured in in vivo tumors (Figure 4A). In addition, we used Yumm 2.1_CTNNB1 in which β-catenin has been stabilized. β–catenin levels were measured by TopFlash® activity and Western analysis of nuclear and cytoplasmic levels of β–catenin, respectively (Figure 4B,C). We treated the Yumm 1.7, Yumm 2.1 CTNNB1 and Yumm 1.7-Wnt5A cells with Lys05 and measured cell viability by trypan blue exclusion assay. Melanoma cells in which β–catenin is stabilized (Yumm 2.1) are more sensitive to Lys05 than melanoma cells with lower β–catenin expression (Yumm 1.7). Wnt5A high cells (Yumm 1.7 Wnt5A) showed the lowest sensitivity of all (Figure 4D), but ultimately respond at higher doses.
Figure 4. The Wnt status of melanoma cells affects their response to autophagy inhibition in vitro and in vivo.
A. Western blot of Wnt5A and β-catenin using lysates from Yumm 1.7 tumors (N=3) and Yumm 1.7_WNT5A (Yumm 1.7 overexpressing WNT5A) tumors (N=3); loading control: HSP90. B. Topflash luciferase assay for the measurement of active β-catenin in Yumm 1.7 cells and Yumm 2.1 in which β-catenin is stabilized (two-tailed unpaired t-test, ****=p<0.0001). C. Western blot of β-catenin using cytoplasmic and nuclear lysates from Yumm 1.7 control cells (1.7 EV), Yumm 2.1 in which β-catenin is stabilized (2.1_CTNNB1) and Yumm 1.7 overexpressing WNT5A (1.7 WNT5A); loading control: β-tubulin for cytoplasmic extracts and histone H3 for the nuclear extracts. D. Measurement of cell viability by trypan blue assay in Yumm 1.7, Yumm 2.1 CTNNB1, and Yumm 1.7_WNT5A treated with Lys05 at the indicated doses for 24 hours, (two-tailed unpaired t-test, ns= p>0.05, **= p ≤ 0.01, ***= p ≤ 0.001, ****=p<0.0001). E–G Tumor volume in mice bearing Yumm 1.7 tumors, Yumm 2.1 CTNNB1 tumors or Yumm 1.7-WNT5A tumors, mice were treated with 20mg/kg Lys05 by I.P. or with equimolar amount of PBS for 14 days (two-tailed unpaired t-test at day 14 of treatment, ns= p>0.05, *= p ≤ 0.05).
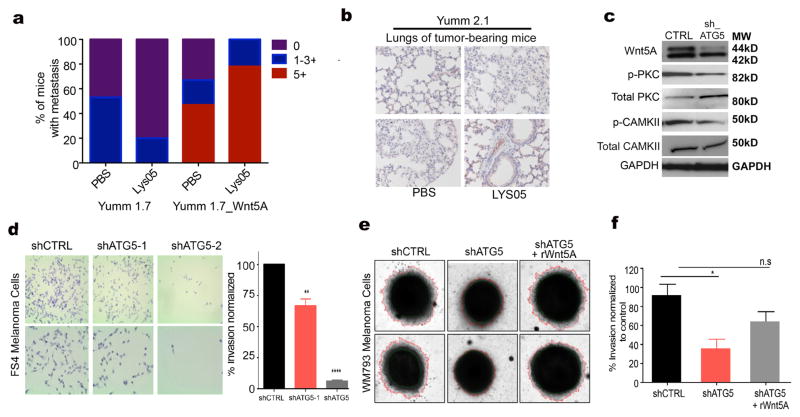
Next, we tested in vivo whether Wnt5A and β–catenin modulation would affect the ability of Lys05 to inhibit melanoma tumor growth. We subcutaneously injected 6–8 week old C57BL/6 mice with 2.0x105 of Yumm 1.7, Yumm 1.7-Wnt5A or Yumm 2.1 CTNNB1. Once the tumors reached 150mm3 in size, mice were treated with Lys05, administered by intraperitoneal route (I.P.) at 20mg/kg daily for 14 days. Control mice for each cell type were injected with PBS for 14 days. We found that Lys05 significantly decreased tumor growth in the mice bearing Yumm 1.7 (Figure 4E) and Yumm 2.1 β–catenin high tumors (Figure 4F), while Yumm 1.7-Wnt5A tumor-bearing mice did not respond to Lys05 treatment at the given dose (Figure 4G). When the lungs of the mice were examined for metastasis, Yumm1.7 tumor-bearing mice showed a reduction in metastasis after Lys05 treatment, where Yumm 1.7_Wnt5A tumor-bearing mice did not (Figure 5A). Yumm 2.1 CTNNB1 tumor-bearing mice showed no evidence of metastases at all, in any condition (Figure 5B), confirming previous data from our lab and others that increased β–catenin inhibits melanoma metastasis (43,44). We further corroborated these studies in vitro. Since Wnt5A promotes melanoma cell invasion, and data in figure 3 show that ATG5 knockdown decreases Wnt5A, we tested whether inhibiting autophagy by ATG5 knockdown would affect Wnt5A-mediated invasion. Our results show that ATG5 knockdown leads to a significant decrease in Wnt5A, and its downstream signaling (Figure 5C), and in invasion in FS4 melanoma cells in a 2D Boyden chamber invasion model (Figure 5D). Using a 3D spheroid model, we found that knocking down ATG5 in WM793 melanoma cells also results in a significant decrease in invasion. Treating ATG5-knockdown cells with rWnt5A rescues the invasion of melanoma cells (Figure 5E,F). These data indicate that disrupting autophagy not only inhibits tumor growth, it also inhibits metastatic spread.
Figure 5. Autophagy inhibition affects Wnt5A-mediated melanoma invasion.
A. Graph of metastatic colonies in mice bearing Yumm 1.7 and Yumm 1.7_WNT5A melanoma tumors; metastases were identified using sox10 or mcherry antibodies by immunohistochemistry (IHC); mice were treated with PBS or the autophagy inhibitor Lys05. Graph represents percent of mice with metastasis. B. Immunohistochemical analysis of lung metastasis using sox10 antibody in mice bearing Yumm 2.1 tumors; mice were treated with PBS or Lys05. C. Western blot of Wnt5A, phospho-PKC, total PKC, phospho-CAMKII, and total CAMKII in FS4 cells upon shRNA-mediated knockdown of ATG5; loading control: GAPDH. D. Boyden chamber assay for the measurement of invasion in FS4 control (shCTRL) and FS4 cells with stable knockdown of ATG5 (shATG5-1 and shATG5-2); Graph represents invasion as a percent of control in FS4 control (shCTRL) and FS4 cells with stable knockdown of ATG5 (shATG5-1 and shATG5-2), (two-tailed unpaired t-test, **= p ≤ 0.01, ****= p<0.0001). E. 3D spheroid invasion assay for the measurement of invasion in WM793 control (shCTRL) and WM793 cells with stable knockdown of ATG5 (shATG5, and shATG5 cells treated with rWnt5A). F. Graph represents invasion as a percent of control in WM793 control (shCTRL) and WM793 cells with stable knockdown of ATG5 (shATG5) and shATG5 cells treated with rWnt5A (one way ANOVA ns= p>0.05, *= p ≤ 0.05).
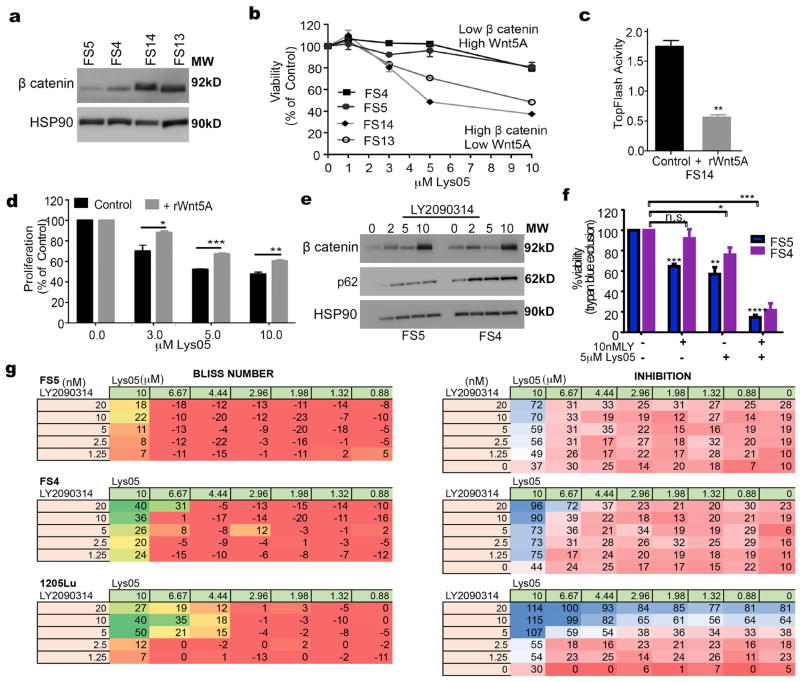
Overall, our results suggested that melanoma cells with low Wnt5A and high β–catenin have increased sensitivity to Lys05-mediated autophagy inhibition. To confirm this we treated Wnt5Alow, β–catenin high melanoma cells (FS13 and FS14) and melanoma cells with high Wnt5A and low β–catenin (FS4, FS5) with Lys05 (Figure 6A,B). Indeed, Wnt5Ahigh cells are less sensitive to Lys05 both in terms of cell viability (Figure 6B, Supplementary Figure 4A) and in terms of apoptosis as measured by Annexin V staining (Supplementary Figure 4B). Next, we treated melanoma cells that are high in β– catenin (FS14) with rWnt5A to induce Wnt5A-mediated β–catenin degradation. β–catenin activity was significantly decreased by Wnt5A as measured by Topflash® assay (figure 6C). We found that treating FS14 cells with rWnt5A makes the cells less sensitive to Lys05-mediated autophagy inhibition, suggesting that Wnt5A-mediated decreases in β–catenin may decrease sensitivity to Lys05 (figure 6D). These data suggest that, as with many other signaling pathways, continued activation of the pathway, in this case by a switch from canonical to non-canonical Wnt signaling can render cells less sensitive to drugs targeted to inhibit this pathway. Given these results, therefore, we next tested whether pharmacological modulation of β– catenin expression could sensitize melanoma cells that are high in Wnt5A and low in β–catenin to Lys05 mediated autophagy inhibition. We used two different commercially available β–catenin activators (LY2090314 and lithium chloride) to test whether combining β–catenin activation with Lys05 would induce more cell death than using Lys05 alone. Both LY2090314 and lithium chloride activate β–catenin through the inhibition of GSK3β, a negative regulator of β–catenin. Our results show that treating melanoma cells that are high in Wnt5A and low in β–catenin with LY2090314 increases β–catenin levels (Figure 6E). The observed increase in β-catenin was also induced by LiCl (Supplementary Figure 4C) and autophagy is decreased (Supplementary Figure 4D). Treating Wnt5A high cells with LY2090314 significantly increases their sensitivity to Lys05 mediated autophagy inhibition (Figure 6F). To test if LY2090314 has synergy with Lys05, we performed a series of inhibition assays. We used the effect-based Bliss Independence Model since the compounds act independently and are mutually non-exclusive (28–30) .The results are displayed as a grid-like design of constant ratio combinations, which resulted in 35 data points for each combination. A total of 5 melanoma cell lines were assessed to identify synergistic potential of the β–catenin activator LY2090314 in combination with autophagy inhibition. Of these, the lines FS4, FS5 and 1205Lu, which have high Wnt5A, showed synergy with the combination of LY2090314 and Lys05 (Figure 6G). The two Wnt5A low lines (WM35 and WM164) demonstrated no synergy, potentially because they already have high β–catenin (Supplementary Figure 4E). Overall, our results show that increasing β–catenin by genetic stabilization of β–catenin or pharmacological inhibition of GSK3 significantly improves the response of aggressive melanoma cells (low β–catenin and high Wnt5A) to autophagy inhibition, lowering the dose of drug required to eradicate these otherwise resistant cells.
Figure 6. β-catenin increases the sensitivity of melanoma cells to autophagy inhibition.
A. Western blot of β catenin in FS5, FS4, FS14 and FS13 melanoma cells; loading control: HSP90. B. Measurement of cell viability by MTS assay in low β catenin/high Wnt5A melanoma cells (FS5, FS4) and high β-catenin/low Wnt5A melanoma cells (FS14, FS13) upon treatment with Lys05 at the indicated doses; graph represents viability as a percent of control. C. Topflash luciferase assay for the measurement of active β-catenin in FS14 control cells and FS14 treated with rWnt5A at 200ng/mL for 16 hours (two-tailed unpaired t-test, **= p ≤ 0.01). D. Measurement of cells viability by MTS assay in melanoma cells treated with Lys05; FS14 control and FS14 cells were pre-treated with 200ng/ml rWnt5A at the indicated doses (two-tailed unpaired t-test *= p ≤ 0.05, **= p ≤ 0.01, ***= p ≤ 0.001). E. Western blot of β-catenin, and p62 in FS5 and FS4 melanoma cells upon treatment with the GSK3 inhibitor (LY2090314)) for the indicated doses for 8 hours; loading control: HSP90. F. Measurement of cell viability by trypan blue exclusion assay in FS5 and FS4 melanoma cells treated with the GSK3 inhibitor, LY2090314 (10nM), Lys05 (5μM), and the combination treatment (Lys05 and LY2090314) at the indicated doses (one-way ANOVA, ns= p>0.05, **= p ≤ 0.01, ***= p ≤ 0.001, ****= p<0.0001) . G. Melanoma cells (FS5, FS4, and 1205Lu) were treated with the indicated doses of LY2090314 (in nM) for 7 hours followed by the addition of Lys05 (in μM) for a total 72 hours incubation; cell growth was assessed by Alamar Blue staining. Synergy was calculated using the Bliss formula; the bliss numbers for each combination are color coded from red to green. Red represents no synergy (additive) effects of the two compounds. Green values are combinations with synergy greater than 20. The inhibition seen with each drug combination is color coded as a gradient from red (0%) through white (50%) to blue (100%) inhibition.
Discussion
Autophagy is a process that can be initiated by multiple stresses in the tumor microenvironment, including hypoxia, nutrient deprivation, and therapeutic stress. Autophagy may play multiple roles in tumor progression, from therapy resistance to metastastic progression. Moreover, autophagy may also play a critical role in tumor dormancy and conversely, modulate programs of premature senescence in response to aberrant oncogenic signaling (45). Perhaps due to observations such as this, and the need to suppress autophagy to bypass senescence, autophagy was initially thought to be an anti- tumorigenic process. However, recent data suggest that autophagy may also be pro-tumorigenic, specifically in terms of allowing tumor cells to resist therapy (46). Consequently, in recent years many pre-clinical studies have been undertaken to explore the potential of autophagy as a target for melanoma therapy. Tumor specific deletion of Atg7 in a genetically engineered mouse model with BRAFv600E and Pten-deficient melanomas inhibited tumor growth and improved animal survival; moreover, the deletion of Atg7 combined with dabrafenib treatment significantly decreased tumor growth (47). Complete deletion of Atg5 in a similar mouse model compromised melanoma metastasis and enhanced the response to dabrafenib (7). The effects of Atg5, however, were dose dependent, as heterozygous losses of this gene (recapitulating a finding in human melanoma lesions), worsened tumor progression and drug response (7). Independent analyses of autophagy in 3D culture models and in tumor biopsies obtained from metastatic patients enrolled in phase II trial of sorafenib and temozolomide revealed a direct correlation between tumor autophagy activity and poor clinical response (48). The aforementioned preclinical findings and the observation of melanomas having a particular wiring of lysosomal-associated pathways (9) support the use of lysosomotropic agents as alternative treatments to improve chemo-sensitivity. These findings have prompted clinical trials involving pharmacological autophagy inhibition with the goal of improving chemo-sensitivity (46). Here we report that sensitivity to autophagy inhibitors is highly dependent on the status of the Wnt pathway: thus shifting non-canonical to canonical Wnt signaling may further sensitize cells to autophagy inhibition.
The Wnt signaling pathways play critical roles in melanoma development and progression. While the canonical Wnt signaling pathway, specifically β–catenin, is responsible for the bypassing of melanocyte senescence, transformation and proliferation, the non-canonical Wnt signaling pathway, characterized by Wnt5A, drives metastatic progression (49). In this study we show that both pathways feedback to control autophagy, where β–catenin signals to decrease autophagy and Wnt5A signals to increase it, confirming separate studies in different cancers from other groups. In melanoma, our studies indicate that the interplay of the canonical and non-canonical Wnt pathways result in a dichotomous control of autophagy. Increasing autophagy by increasing Wnt5A ultimately results in a decreased sensitivity (but not complete resistance) to autophagy inhibition. These results open the exciting possibility of Wnt5A acting in drug sensitivity beyond previously described functions in the control of efflux pumps such as the ATP-binding cassette sub-family B member 1 (ABCB1) (50). This leads to an increase in drug resistance in general due to increased efflux of drug from the cell, which is a mechanism we cannot exclude here. Together these data suggest that to disable autophagy in Wnt5Ahigh cells, higher doses of inhibitors should be used in patients whose melanoma tumors have high Wnt5A. Alternatively, strategies to increase β–catenin, prior to treatment with autophagy inhibitors may also prove useful. However, the caveat with this approach is the fact that β–catenin increases melanocyte transformation and growth, so those studies would have to be very carefully considered.
Perhaps one of the most unexpected results of this study is the finding of the impact of the Wnt5A-autophagy loop in invasion and metastasis, a link not previously made. The particular efficacy of the Lys05 compound inhibiting the invasion of Wnt5Alow cells, suggest that lysosome blockers may be of great use not just in rendering cells more sensitive to BRAF inhibitors, but also of use in the treatment of patients who do not bear the BRAF mutation. The BRAF mutation is increasingly rarer with increasing age of diagnosis, such that at ages 60 and above, less than a third of melanoma patients bear the BRAF mutation, so other therapeutic options are needed (51). Moreover, even those aged patients expressing BRAFV600E have an enhanced resistance to targeted therapy (36). Therefore, autophagy inhibition may be particularly useful in this group of patients, and this is a topic of ongoing investigation in our laboratory. Overall, our studies reveal novel roles for autophagy inhibitors in the inhibition of metastasis, and reveal new mechanisms by which melanoma cells may resist autophagy inhibitors. Further research is required to fully harness the potential of these very promising drugs.
Supplementary Material
Acknowledgments
FINANCIAL SUPPORT: A.T. Weeraratna, A. Kaur, and R. Behera are supported by R01CA174746. A. Ndoye, A. Budina-Kolomets, V.W. Rebecca, M. Murphy, R. Amaravadi and A.T. Weeraratna. are supported by P01 CA114046 and A Ndoye, A.T. Weeraratna and R Amaravadi are also supported by P50CA174523. CH Kugel III is supported by T32CA009171. M.R. Webster is supported by K99 CA208012-01. R. Amaravadi is supported by R01CA169134M. Soengas and AT Weeraratna are also supported by a Melanoma Research Alliance/ L’Oréal Paris-USA Women in Science Team Science Award. A. Budina-Kolomets and M. Murphy are supported by R01 CA139319. M. Herlyn and G.B. Mills are supported by a gift from the Adelson Medical Research Foundation. Core facilities used in this grant are supported by P30CA010815 and NCI #CA16672.
References
- 1.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–73. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 2.Tsujimoto Y, Shimizu S. Another way to die: autophagic programmed cell death. Cell Death Differ. 2005;12(Suppl 2):1528–34. doi: 10.1038/sj.cdd.4401777. [DOI] [PubMed] [Google Scholar]
- 3.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–34. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 4.Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10:1533–41. doi: 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Checinska A, Soengas MS. The gluttonous side of malignant melanoma: basic and clinical implications of macroautophagy. Pigment Cell Melanoma Res. 2011;24:1116–32. doi: 10.1111/j.1755-148X.2011.00927.x. [DOI] [PubMed] [Google Scholar]
- 6.Liu H, He Z, von Rütte T, Yousefi S, Hunger RE, Simon H-U. Down-regulation of autophagy-related protein 5 (ATG5) contributes to the pathogenesis of early-stage cutaneous melanoma. Sci Transl Med. 2013;5:202ra123. doi: 10.1126/scitranslmed.3005864. [DOI] [PubMed] [Google Scholar]
- 7.García-Fernández M, Karras P, Checinska A, Cañón E, Calvo GT, Gómez-López G, et al. Metastatic risk and resistance to BRAF inhibitors in melanoma defined by selective allelic loss of ATG5. Autophagy. 2016;12:1776–90. doi: 10.1080/15548627.2016.1199301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marino ML, Pellegrini P, Di Lernia G, Djavaheri-Mergny M, Brnjic S, Zhang X, et al. Autophagy is a protective mechanism for human melanoma cells under acidic stress. J Biol Chem. 2012;287:30664–76. doi: 10.1074/jbc.M112.339127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alonso-Curbelo D, Riveiro-Falkenbach E, Pérez-Guijarro E, Cifdaloz M, Karras P, Osterloh L, et al. RAB7 controls melanoma progression by exploiting a lineage-specific wiring of the endolysosomal pathway. Cancer Cell. 2014;26:61–76. doi: 10.1016/j.ccr.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 10.Ploper D, De Robertis EM. The MITF family of transcription factors: Role in endolysosomal biogenesis, Wnt signaling, and oncogenesis. Pharmacol Res. 2015;99:36–43. doi: 10.1016/j.phrs.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Rambow F, Job B, Petit V, Gesbert F, Delmas V, Seberg H, et al. New Functional Signatures for Understanding Melanoma Biology from Tumor Cell Lineage-Specific Analysis. Cell Rep. 2015;13:840–53. doi: 10.1016/j.celrep.2015.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma X-H, Piao S, Wang D, McAfee QW, Nathanson KL, Lum JJ, et al. Measurements of tumor cell autophagy predict invasiveness, resistance to chemotherapy, and survival in melanoma. Clin Cancer Res. 2011;17:3478–89. doi: 10.1158/1078-0432.CCR-10-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McAfee Q, Zhang Z, Samanta A, Levi SM, Ma X, Piao S, et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci U S A. 2012;109:8253–8. doi: 10.1073/pnas.1118193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma X-H, Piao S-F, Dey S, McAfee Q, Karakousis G, Villanueva J, et al. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest. 2014;124:1406–17. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paluncic J, Kovacevic Z, Jansson PJ, Kalinowski D, Merlot AM, Huang MLH, et al. Roads to melanoma: Key pathways and emerging players in melanoma progression and oncogenic signaling. Biochim Biophys Acta - Mol Cell Res. 2016:770–84. doi: 10.1016/j.bbamcr.2016.01.025. [DOI] [PubMed] [Google Scholar]
- 16.Larue L, Delmas V. The WNT/Beta-catenin pathway in melanoma. Front Biosci. 2006;11:733–42. doi: 10.2741/1831. [DOI] [PubMed] [Google Scholar]
- 17.O’Connell MP, Weeraratna AT. Hear the Wnt Ror: how melanoma cells adjust to changes in Wnt. Pigment Cell Melanoma Res. 2009;22:724–39. doi: 10.1111/j.1755-148X.2009.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Webster MR, Kugel CH, Weeraratna AT. The Wnts of change: How Wnts regulate phenotype switching in melanoma. Biochim Biophys Acta. 2015;1856:244–51. doi: 10.1016/j.bbcan.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webster MR, Weeraratna AT. A Wnt-er Migration: The Confusing Role of - Catenin in Melanoma Metastasis. Sci Signal. 2013;6:pe11–pe11. doi: 10.1126/scisignal.2004114. [DOI] [PubMed] [Google Scholar]
- 20.Weeraratna AT, Jiang Y, Hostetter G, Rosenblatt K, Duray P, Bittner M, et al. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1:279–88. doi: 10.1016/s1535-6108(02)00045-4. [DOI] [PubMed] [Google Scholar]
- 21.O’Connell MP, Fiori JL, Baugher KM, Indig FE, French AD, Camilli TC, et al. Wnt5A activates the calpain-mediated cleavage of filamin A. J Invest Dermatol. 2009;129:1782–9. doi: 10.1038/jid.2008.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webster MR, Xu M, Kinzler KA, Kaur A, Appleton J, O’Connell MP, et al. Wnt5A promotes an adaptive, senescent-like stress response, while continuing to drive invasion in melanoma cells. Pigment Cell Melanoma Res. 2015;28:184–95. doi: 10.1111/pcmr.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Topol L, Jiang X, Choi H, Garrett-Beal L, Carolan PJ, Yang Y. Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent beta-catenin degradation. J Cell Biol. 2003;162:899–908. doi: 10.1083/jcb.200303158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lock R, Kenific CM, Leidal AM, Salas E, Debnath J. Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov. 2014;4:466–79. doi: 10.1158/2159-8290.CD-13-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petherick KJ, Williams AC, Lane JD, Ordóñez-Morán P, Huelsken J, Collard TJ, et al. Autolysosomal β-catenin degradation regulates Wnt-autophagy-p62 crosstalk. EMBO J. 2013;32:1903–16. doi: 10.1038/emboj.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeVorkin L, Hattersley M, Kim P, Ries J, Spowart J, Anglesio MS, et al. Autophagy Inhibition Enhances Sunitinib Efficacy in Clear Cell Ovarian Carcinoma. Mol Cancer Res. 2017;15:250–8. doi: 10.1158/1541-7786.MCR-16-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gade TPF, Tucker E, Nakazawa MS, Hunt SJ, Wong W, Krock B, et al. Ischemia Induces Quiescence and Autophagy Dependence in Hepatocellular Carcinoma. Radiology. 2017:160728. doi: 10.1148/radiol.2017160728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greco W, Unkelbach H-D, Pöch G, Sühnel J, Kundi M, WB Consensus on concepts and terminology for combined-action assessment: The Saariselkä Agreement. Arch Complex Environ Stud. 1992;4:65–9. [Google Scholar]
- 29.Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol. 2006;2:458–66. doi: 10.1038/nchembio817. [DOI] [PubMed] [Google Scholar]
- 30.Goldoni M, Johansson C. A mathematical approach to study combined effects of toxicants in vitro: evaluation of the Bliss independence criterion and the Loewe additivity model. Toxicol In Vitro. 2007;21:759–69. doi: 10.1016/j.tiv.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Li L, Fukunaga-Kalabis M, Herlyn M. The three-dimensional human skin reconstruct model: a tool to study normal skin and melanoma progression. J Vis Exp. 2011 doi: 10.3791/2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Connell MP, Marchbank K, Webster MR, Valiga AA, Kaur A, Vultur A, et al. Hypoxia induces phenotypic plasticity and therapy resistance in melanoma via the tyrosine kinase receptors ROR1 and ROR2. Cancer Discov. 2013;3:1378–93. doi: 10.1158/2159-8290.CD-13-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies MA, Stemke-Hale K, Lin E, Tellez C, Deng W, Gopal YN, et al. Integrated Molecular and Clinical Analysis of AKT Activation in Metastatic Melanoma. Clin Cancer Res. 2009;15:7538–46. doi: 10.1158/1078-0432.CCR-09-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gopal YNV, Deng W, Woodman SE, Komurov K, Ram P, Smith PD, et al. Basal and treatment-induced activation of AKT mediates resistance to cell death by AZD6244 (ARRY-142886) in Braf-mutant human cutaneous melanoma cells. Cancer Res. 2010;70:8736–47. doi: 10.1158/0008-5472.CAN-10-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu X-Z, Garcia MV, Li T, Khor L-Y, Gajapathy RS, Spittle C, et al. Cytoskeleton alterations in melanoma: aberrant expression of cortactin, an actin-binding adapter protein, correlates with melanocytic tumor progression. Mod Pathol. 2010;23:187–96. doi: 10.1038/modpathol.2009.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaur A, Webster MR, Marchbank K, Behera R, Ndoye A, Kugel CH, et al. sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature. 2016;532:250–4. doi: 10.1038/nature17392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klionsky D, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun J-A, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 40.Moscat J, Karin M, Diaz-Meco MT. p62 in Cancer: Signaling Adaptor Beyond Autophagy. Cell. 2016;167:606–9. doi: 10.1016/j.cell.2016.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, et al. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13:343–54. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Otomo C, Metlagel Z, Takaesu G, Otomo T. Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy. Nat Struct Mol Biol. 2013;20:59–66. doi: 10.1038/nsmb.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arozarena I, Bischof H, Gilby D, Belloni B, Dummer R, Wellbrock C. In melanoma, beta-catenin is a suppressor of invasion. Oncogene. 2011;30:4531–43. doi: 10.1038/onc.2011.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chien AJ, Moore EC, Lonsdorf AS, Kulikauskas RM, Rothberg BG, Berger AJ, et al. Activated Wnt/beta-catenin signaling in melanoma is associated with decreased proliferation in patient tumors and a murine melanoma model. Proc Natl Acad Sci U S A. 2009;106:1193–8. doi: 10.1073/pnas.0811902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gewirtz DA. Autophagy, senescence and tumor dormancy in cancer therapy. Autophagy. 2009;5:1232–4. doi: 10.4161/auto.5.8.9896. [DOI] [PubMed] [Google Scholar]
- 46.Ndoye A, Weeraratna AT. Autophagy- An emerging target for melanoma therapy. F1000Research. 2016;5:1–9. doi: 10.12688/f1000research.8347.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie X, Koh JY, Price S, White E, Mehnert JM. Atg7 Overcomes Senescence and Promotes Growth of BrafV600E-Driven Melanoma. Cancer Discov. 2015;5:410–23. doi: 10.1158/2159-8290.CD-14-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma X-H, Piao S, Wang D, McAfee QW, Nathanson KL, Lum JJ, et al. Measurements of tumor cell autophagy predict invasiveness, resistance to chemotherapy, and survival in melanoma. Clin Cancer Res. 2011;17:3478–89. doi: 10.1158/1078-0432.CCR-10-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Connell MP, Weeraratna AT. Hear the Wnt Ror: how melanoma cells adjust to changes in Wnt. Pigment Cell Melanoma Res. 2009;22:724–39. doi: 10.1111/j.1755-148X.2009.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hung T-H, Hsu S-C, Cheng C-Y, Choo K-B, Tseng C-P, Chen T-C, et al. Wnt5A regulates ABCB1 expression in multidrug-resistant cancer cells through activation of the non-canonical PKA/β-catenin pathway. Oncotarget. 2014;5:12273–90. doi: 10.18632/oncotarget.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Libra M, Malaponte G, Navolanic PM, Gangemi P, Bevelacqua V, Proietti L, et al. Analysis of BRAF mutation in primary and metastatic melanoma. Cell Cycle. 2005;4:1382–4. doi: 10.4161/cc.4.10.2026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.