Abstract
Multiple myeloma (MM) is a frequently incurable hematological cancer in which overactivity of MYC plays a central role, notably through up-regulation of ribosome biogenesis and translation. To better understand the oncogenic program driven by MYC and investigate its potential as a therapeutic target, we screened a chemically diverse small-molecule library for anti-MM activity. The most potent hits identified were rocaglate scaffold inhibitors of translation initiation. Expression profiling of MM cells revealed reversion of the oncogenic MYC-driven transcriptional program by CMLD010509, the most promising rocaglate. Proteome-wide reversion correlated with selective depletion of short-lived proteins that are key to MM growth and survival, most notably MYC, MDM2, CCND1, MAF, and MCL-1. The efficacy of CMLD010509 in mouse models of MM confirmed the therapeutic relevance of these findings in vivo and supports the feasibility of targeting the oncogenic MYC-driven translation program in MM with rocaglates.
INTRODUCTION
Multiple myeloma (MM) is a hematological malignancy characterized by a clonal proliferation of plasma cells in the bone marrow micro-environment (1). According to the National Cancer Institute (NCI), the prevalence of MM was estimated at 89,650 people in the United States in 2012, with an annual incidence of 6.3 new cases per 100,000 individuals and a 5-year overall survival (OS) of 46.6% (2). Despite notable therapeutic advances, MM remains a frequently incurable hematological malignancy. Therefore, there is an urgent unmet need for the development of therapeutic options with new mechanisms of action in MM.
The transcription factor MYC plays a central role in the progression of the disease. About two-thirds of patients who are newly diagnosed with MM harbor MYC activation, which correlates with adverse clinical outcome (3). In about 20 to 40% of the patients, this is explained by translocations that involve the MYC locus (4, 5). Another 10% of patients harbor a gain of 8q24 comprising MYC (6). MYC is centrally positioned in cell growth and cancer regulatory networks. MYC heterodimerizes with MAX, to bind the E-box element CACGTG. Functional categories of MYC-induced genes include cell growth (ribosome biogenesis and protein synthesis), cell cycle control, energy production (glycolysis, glutaminolysis, and mitochondrial biogenesis), anabolic metabolism (synthesis of amino acids, nucleotides, and lipids), and DNA replication (7).
Alterations in ribosome synthesis and translation activity are a defining feature of most cancer cells, satisfying the increased anabolic demands associated with malignant transformation and tumor growth. MYC directly enhances transcription genes encoding ribosomal proteins, ribosomal RNA, and proteins of the translation initiation complex. The ribosome and translation initiation complexes have historically been perceived as passive complexes, supporting protein synthesis without having an active role in directing functional expression of the genome. However, recent studies have revealed that ribosome biogenesis and translation activity are highly regulated to drive specific translation programs, independent of the genetic makeup of the cancer (8–10), and represent a crucial node for hyperactivation of oncogenic downstream signaling in cancer cells (11). Accordingly, ribosome biogenesis and translation initiation potentially provide very attractive targets for cancer therapy.
Here, we sought to identify chemical compounds that disrupt ribosome biogenesis and translation activation in MM. We also describe analogs of the rocaglate natural product class as very potent compounds, which inhibit the oncogenic translation program supporting MM.
RESULTS
MYC and translation activation in MM
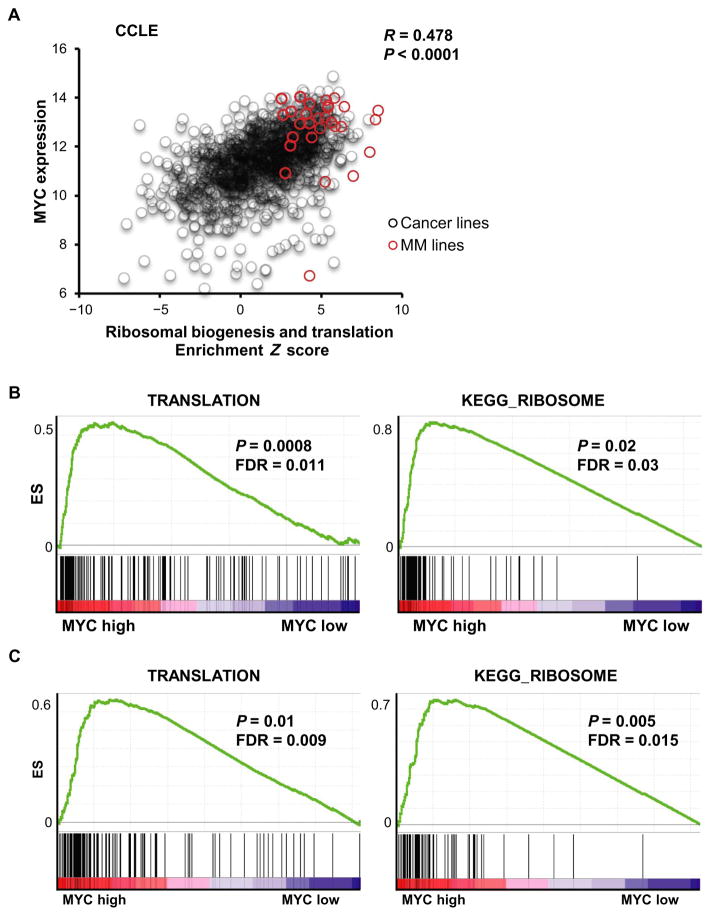
To examine potential correlation between MYC expression and translation activation in MM, we analyzed the Cancer Cell Line Encyclopedia (CCLE), a large database of gene expression profiling for more than 1000 human cancer cell lines. We first generated a Z score for each cell line by combining two KEGG (Kyoto Encyclopedia of Genes and Genomes) canonical pathway gene sets: ribosomal biogenesis and translation. We found a significant correlation between MYC expression and translation activation across the ~1000 CCLE cell lines (R = 0.478, P < 0.001) (Fig. 1A). In particular, many MM cell lines had increased MYC expression and an enrichment of ribosomal biogenesis and translation (high Z score), which was also observed in other hematological malignancies (fig. S1). We used MM patient tumor cell–derived gene expression profiling (GSE6477 and GSE16558) to further confirm the enrichment of translation (Fig. 1B) and ribosomal biogenesis (Fig. 1C) in the context of high MYC expression. These results establish a strong correlation between MYC expression and translation activity in MM.
Fig. 1. Correlation of MYC expression with ribosomal biogenesis and translation activity.
(A) Correlation analysis of MYC expression on the y axis and the Z-score enrichment across over 1000 cell lines from the CCLE database. A Z score was generated for each cell line by combining two KEGG canonical pathway gene sets: ribosomal biogenesis and translation. Red dots indicate MM cell lines, and black circles indicate all CCLE cell lines except MM. A significant correlation between MYC expression and the translation activation was observed; R = 0.478, P < 0.0001. Gene Set Enrichment Analysis (GSEA) of MM patient tumor cell-derived gene expression profiling (GSE6477 and GSE16558) shown in (B) and (C), confirming enrichment of translation and ribosomal biogenesis in context of high expression of MYC. ES, enrichment score; FDR, false discovery rate.
Identification of rocaglate derivatives
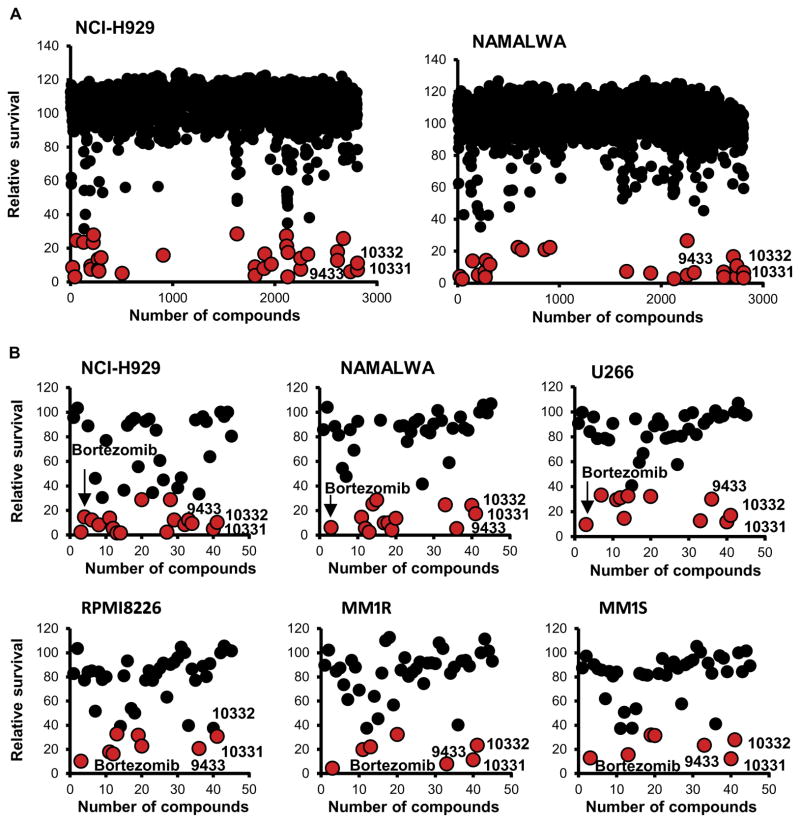
To identify a chemical biological probe with which to perturb lymphoid malignancies in the context of MYC overexpression, we screened a diverse small-molecule library (2812 compounds) provided by the Boston University Center for Molecular Discovery (BU-CMD). The BU-CMD screening collection is composed largely of compounds synthesized using diversity-oriented synthesis techniques, as well as synthetic analogs of bioactive natural product scaffolds, assembled with the goal of accessing greater structural complexity and a broader coverage of chemical space than is possible with conventional combinatorial chemistry–based libraries (12). Two lymphoid cell lines with high expression of MYC were used: NCI-H929 (MM) and NAMALWA (Burkitt lymphoma). We identified 45 compounds, which potently inhibited proliferation of at least one of the cell lines (Fig. 2A and fig. S2). We validated these 45 hits in five MM cell lines and one Burkitt lymphoma cell line (Fig. 2B). On the basis of their structure-activity relationships (SARs), this secondary validation highlighted three compounds (CMLD010331, CMLD010332, and CMLD009433), all totally synthetic analogs of the rocaglate natural product class (fig. S2B). These three compounds were the most active inhibiting proliferation of MM cells to a level similar to that of bortezomib (Fig. 2B).
Fig. 2. Chemical library screen.
(A) Small-molecule library generated by the Boston University Center (~3000 synthetic compounds) against NCI-H929 (MM) and NAMALWA (Burkitt lymphoma). We identified 45 compounds with a potent inhibition of proliferation for at least one of the cell lines. (B) Validation of 45 hits in six cell lines harboring diverse MM driver genomic features of MM, showing that the most effective compounds to inhibit proliferation were in the rocaglate class, namely, CMLD010331, CMLD010332, and CMLD09433 (red dots). These compounds had similar potency to bortezomib.
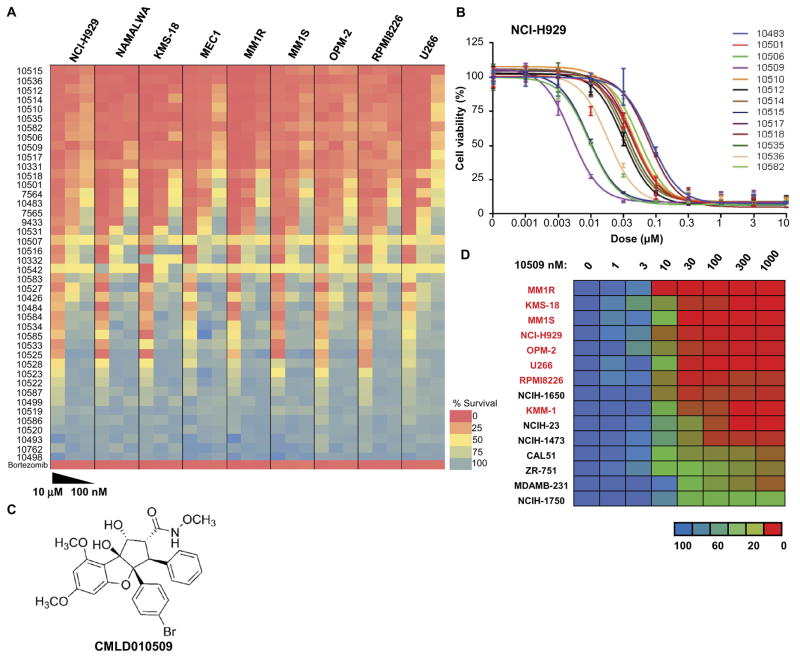
Rocaglate (flavagline) natural products (and synthetic analogs) are recognized to inhibit protein synthesis (13, 14). To better define the activity of rocaglate derivatives against MM cells, we performed a follow-up SAR study involving 40 additional structurally related compounds (Fig. 3A and fig. S2C). These included synthetic samples of both natural and nonnatural rocaglates. Of the set, the compound CMLD010509 [SDS-1-021 (13, 15)] was identified as the most potent compound, with a median inhibitory concentration (IC50) below 10 nM for most MM cell lines tested (Fig. 3, B and C). The compound was less effective against lung and breast cancer cell lines, with an IC50 of ~30 nM (Fig. 3D).
Fig. 3. High potency of rocaglate inhibitors in MM.
(A) Heat map of the effects of 40 rocaglate derivatives on relative survival of MM cells (NCI-H929, KMS-18, MM1R, MM1S, OPM-2, RPMI8226, and U266) along with lymphoma cell lines (NAMALWA and MEC1) shows strong activity with low doses of the rocaglate compounds in these cell lines. (B) IC50 of these 40 compounds in NCI-H929 showing that CMLD010509 was the most potent compound, with an IC50 below 10 nM. (C) CMLD010509 is a synthetic rocaglate derivative containing the cyclopenta[b]tetrahydrobenzofuran core structure. (D) IC50 of CMLD010509 showing an IC50 below 10 nM for most MM cell lines tested (indicated in red), whereas it was relatively resistant in lung and breast cancer cell lines (in black) with an IC50 of ~30 nM.
Gene expression altered by CMLD010509 inhibiting translation in MM
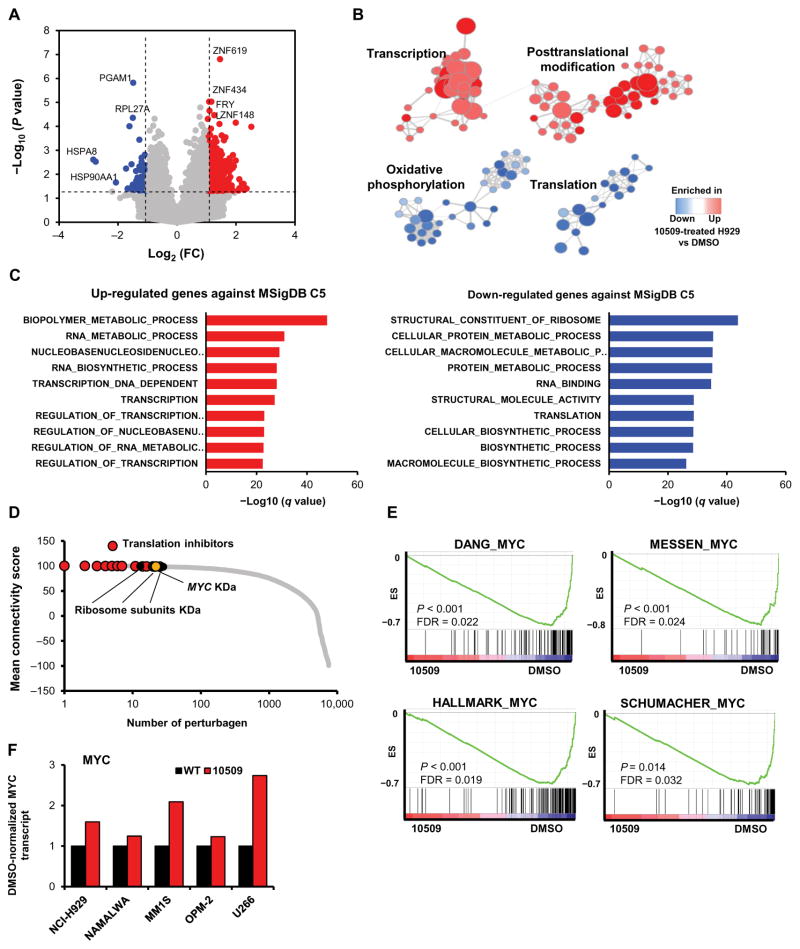
To further define the mode of action for CMLD010509 against MM, we performed RNA sequencing (RNA-seq) on drug-versus dimethyl sulfoxide (DMSO)–treated cells in five different MM cell lines. We identified 845 and 475 genes that were significantly up-regulated (table S1A) and down-regulated (table S1B), respectively, with a fold change higher than 2 and a P value of <0.05 (Fig. 4A). Several zinc finger transcription factors were among the top up-regulated genes, whereas ribosomal proteins and heat shock proteins were among the most significantly down-regulated (Fig. 4A). Furthermore, we defined four clusters by arranging the significantly altered gene sets in a network enrichment map (16). The transcription and posttranslational clusters were enriched in CMLD010509-treated cells, whereas oxidative phosphorylation and translation clusters were enriched in control cells, indicating down-regulation in CMLD010509-treated cells (Fig. 4B and table S1C). The most significantly up-regulated genes were highly enriched for transcriptional activation, and the most significantly down-regulated genes were highly enriched for ribosome components and translation (Fig. 4C and table S1, D and E). This transcriptome profile has been associated with low translational flux and consequent inactivation of heat shock factor 1 (HSF1) (17). Here, inhibition of translation reverses constitutive HSF1 activation in cancers, thereby leading to extensive remodeling of the transcriptome. We confirmed that CMLD010509 inhibits the heat shock response in MM cells, as indicated by decreased HSP70 and HSP90 mRNA and down-regulation of the HSF1 program in CMLD010509-treated cells by pathway analysis (fig. S3A). To assess whether HSF1 is constitutively activated in MM, we used a previously published HSF1 activation signature (18) to interrogate publicly available data sets. We observed that this HSF1 activation signature was increased in MM cells compared to normal plasma cells (19, 20) and correlated with poor clinical outcome in MM (fig. S3, B and C) (20, 21).
Fig. 4. Transcription activation and translation inhibition induced by the rocaglate derivative CMLD010509.
(A) Volcano plot of RNA-seq of drug- versus DMSO-treated cells in five different MM cell lines showing 845 and 475 genes significantly up- and down-regulated, respectively, with a fold change (FC) higher than 2. (B) Network enrichment map identifying two clusters enriched in CMLD010509-treated cells (transcription and posttranslational modification clusters) and two clusters enriched in control cells (oxidative phosphorylation and translation clusters). (C) Bar graphs showing the most significantly up- and down-regulated genes in CMLD010509-treated cells determined by querying the Molecular Signatures Database (MSigDB) C5 gene sets. (D) Connectivity score using LINCS NIH program with the gene signature of CMLD010509 against 10,000 “pertubagen” signatures (corresponding to short hairpin RNA, open reading frame, and compounds). (E) GSEA analysis of CMLD010509 using RNA-seq data. Several MYC gene sets were among the most significantly enriched compared to DMSO control cells. (F) Bar graphs of MYC transcript expression in MM cell lines and NAMALWA in CMLD010509-treated cells, normalized to DMSO control. WT, wild type; KD, knockdown.
To further characterize the link between CMLD010509 and translation inhibition, we used our CMLD010509 expression signature to query the Library of Integrated Network–Based Cellular Signatures (LINCS) National Institutes of Health (NIH) program (www.lincscloud.org). The LINCS database is a large catalog of perturbational gene expression profiles across 77 cell lines. It includes over 20,000 chemical compounds and more than 22,000 genetic perturbations (knockdown and overexpression of genes). The most significant positive correlations with the CMLD010509 signature were represented by known translation inhibitors such as emetine, omacetaxine, and cephaeline, as well as knockdown of ribosome subunits and translation initiation factors (Fig. 4D and table S1F). These results are consistent with the potent activity of CMLD010509 as a translation inhibitor.
MYC knockdown was one of the perturbations with the most positive correlation with our compound signature. The GSEA of our compounds’ effects using the RNA-seq data revealed that several MYC gene sets were among the most significantly enriched compared to the DMSO control cells, indicating a down-regulation of the entire MYC pathway in CMLD010509-treated cells (Fig. 4E). Several MYC target genes were down-regulated by the drug; however, the amounts of MYC transcript were not decreased in CMLD010509-treated cells, indicating posttranscriptional regulation of MYC activity (Fig. 4F). Together, these results reinforce the role of MYC in translation activation in MM and the mechanistic action of CMLD010509 as a translation inhibitor in MM cells.
Disruption of the oncogenic translation program in MM by CMLD010509
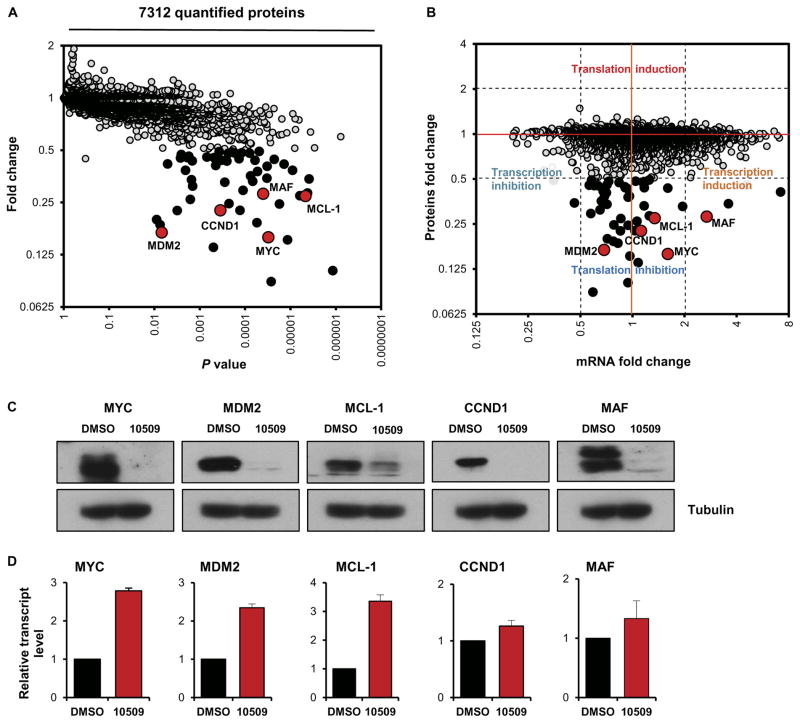
To better define the consequences of CMLD010509-induced translation inhibition, we performed an unbiased, proteome-wide experiment in a quantitative and highly parallel format (22). We compared the immediate impact of CMLD010509 treatment (100 nM) to that of vehicle control in NCI-H929 cells. Exposure for 2 hours was selected to identify primary, immediate consequences of compound action and to mitigate expected, confounding effects of suppressed CMLD010509 target proteins. Each treatment condition was prepared in three biological replicates and individually labeled using isobaric tagging. We identified 7312 proteins, of which 54 were significantly depleted by more than twofold after CMLD010509 exposure (P < 0.05; fold change, >2) (Fig. 5A and table S2A). Several key oncoproteins in MM were among the proteins most depleted by the compound, including MYC, MDM2, CCND1, MCL-1, and MAF. MYC is overexpressed in about 40% of MM by either translocations or gain of 8q24, among other potential epigenetic regulations (4, 5). CCND1 is overexpressed in about 20% of MM in the case of t(11;14) or focal gain in 11q13 (4, 23). MDM2 is an ubiquitin ligase (E3) that acts as a negative regulator of p53 (24). MCL-1 belongs to the Bcl-2 family and is one of the driver oncogenes in gain-of-1q cancers (25). MAF is frequently overexpressed in MM in relation to t(14;16) (26).
Fig. 5. Regulation of the oncogenic translation program of MM by CMLD010509.
(A) Expression proteomics showing the impact of CMLD010509 treatment (100 nM) relative to vehicle control in NCI-H929 cells after a 2-hour incubation (three biological replicates were used and individually tagged using isobaric tagging). We identified 7312 proteins, of which 54 were significantly depleted by more than twofold through CMLD010509 exposure (P < 0.05; fold change, >2). The most depleted proteins included MYC, MDM2, CCND1, MCL-1, and MAF (red dots). (B) Comparison of the protein fold changes to the transcript fold changes in CMLD010509- versus vehicle-treated cells. A large majority of depleted proteins had a transcript fold change lower than twofold, suggesting a specific translational mechanism of action for CMLD010509. (C) Immunoblots and (D) qRT-PCR analysis of MYC, MDM2, CCND1, MCL-1, and MAF in compound-treated (100 nM, 3 hours) or vehicle-treated NCI-H929. All five genes were depleted at the protein level, whereas their transcripts were unchanged or slightly overexpressed.
To further study the mechanism of action of CMLD010509, we compared the protein fold changes to the transcript fold changes in CMLD010509- versus vehicle-treated cells (Fig. 5B). The large majority of depleted proteins had a change in transcript amount of less than twofold, indicative of a selective translation inhibitory mode of action for CMLD010509. To validate these findings, we measured MYC, MDM2, CCND1, MCL-1, and MAF by immunoblotting and quantitative real-time polymerase chain reaction (qRT-PCR) after compound treatment as above (100 nM, 2 hours). All five genes were depleted at the protein level, whereas the amounts of each transcript were unchanged or slightly increased (Fig. 5, C and D), consistent with post-transcriptional effects of CMLD010509. Similar results were observed in six additional MM cell lines (fig. S4, A to G). To assess whether the targeted proteins were directly depleted by inhibiting their synthesis, we pulse-labeled cells with 35S-cysteine/methionine and immunoprecipitated the proteins of interest. In the presence of CMLD010509, we observed selective inhibition of new translation for MYC, CCND1, and MCL-1 as opposed to housekeeping proteins such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (fig. S4H).
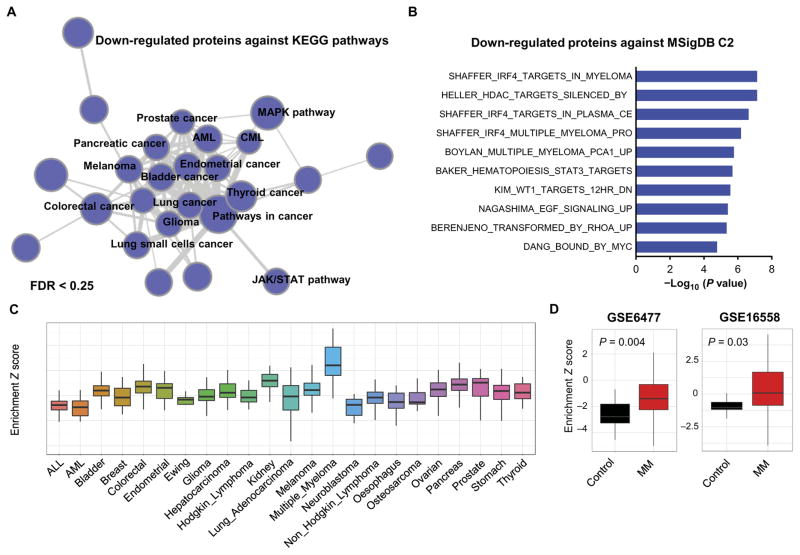
To further establish pathways affected by CMLD010509, we queried the 54 proteins with more than twofold depletion against the KEGG database. Cancer pathways were highly enriched among these depleted proteins, representing a distinct cluster by network enrichment analysis (Fig. 6A and table S2B). Moreover, when interrogating the whole MSigDB C2 canonical pathway database (representing 1330 gene sets), we found that the most highly enriched pathways consisted of MM pathways: IRF4 and MYC targets. This result indicates that a significant number of CMLD010509-depleted proteins are present in the IRF4 and MYC pathways (P < 0.001), as defined by the GSEA transcriptional pathways. IRF4 is a key transcription factor essential for plasma cell differentiation, upon which MM cells are dependent on (Fig. 6B and table S2C) (27), suggesting that CMLD010509 inhibits translation of key oncoproteins in MM. To determine whether this translational program is specific for MM, we used the signature of 54 proteins to calculate Z scores for over 1000 cell lines across the CCLE database. The CMLD010509 signature was enriched in MM cell lines at the transcriptional level compared to other cancer cell lines (Fig. 6C). Furthermore, the signature was also significantly enriched in plasma cells of patients with MM compared to that of healthy donors (P < 0.05), in two independent gene expression profiling experiments including patients at diagnosis (GSE16558) and at relapse (GSE6477) (Fig. 6D). Together, these results demonstrate that CMLD010509 inhibits the translation of key oncoproteins, supporting the proliferation and survival of MM.
Fig. 6. Pathway analysis of CMLD010509.
(A) Network enrichment analysis of 54 depleted proteins with more than twofold change analyzed in the KEGG database, showing that cancer pathways were highly enriched among these depleted proteins. AML, acute myeloid leukemia; CML, chronic myeloid leukemia; MAPK, mitogen-activated protein kinase; JAK/STAT, Janus kinase/signal transducer and activator of transcription. (B) The 54 depleted proteins were queried against MSigDB C2 canonical pathway database, showing an enrichment of MM specific oncoproteins including IRF4 pathway. (C) Z scores of the 54 proteins evaluated on 1000 cell lines of the CCLE database. (D) Z scores of the 54 proteins evaluated on two independent gene expression profiles including patients at diagnosis (GSE16558) and at relapse (GSE6477).
Induction of apoptosis in MM cells by CMLD010509
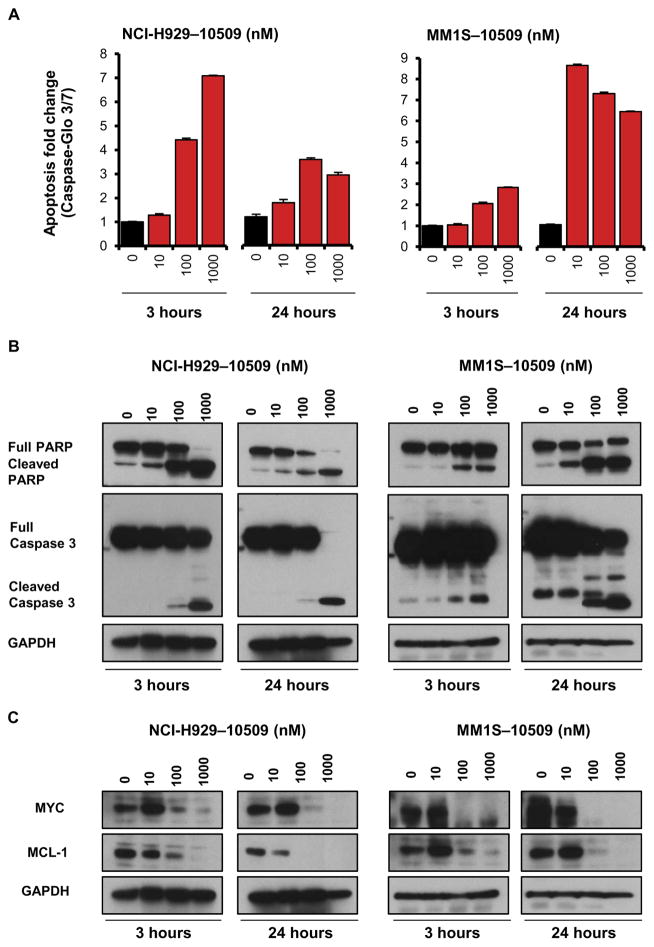
We next explored the antiproliferative consequences of inhibiting the oncogenic translation program in MM cells with CMLD010509. The drug was associated with a strong induction of an apoptotic response in NCI-H929 and MM1S cells, as measured by caspase-3 and caspase-7 activation (Fig. 7A) and cleavage of both poly[adenosine 5′-diphosphate–ribose] polymerase (PARP) and caspase-3 (Fig. 7B). Kinetic studies revealed a rapid apoptotic response to CMLD010509 at 3 hours, associated with depletion of MYC and MCL-1 even at low concentrations (10 nM) (Fig. 7C). We next assessed the in vitro activity of CMLD010509 against human peripheral blood mono-nuclear cells (PBMCs) and found no cytotoxicity, suggesting the potential for a useful therapeutic window for this agent against MM (fig. S5).
Fig. 7. Induction of apoptosis by CMLD010509.
(A) Apoptosis assay measured by caspase-3 and caspase-7 activation (Caspase-Glo 3/7) at 3 and 24 hours in NCI-H929 and MM1S cells treated with three concentrations of CMLD010509 versus vehicle control. (B) Immunoblots for cleavage of both PARP and caspase-3 in response to three concentrations of CMLD010509 in NCI-H929 and MM1S cells at 3 and 24 hours. (C) Immunoblots for MYC and MCL-1 in response to three concentrations of CMLD010509 in NCI-H929 and MM1S cells at 3 and 24 hours.
In vivo suppression of MM by CMLD010509
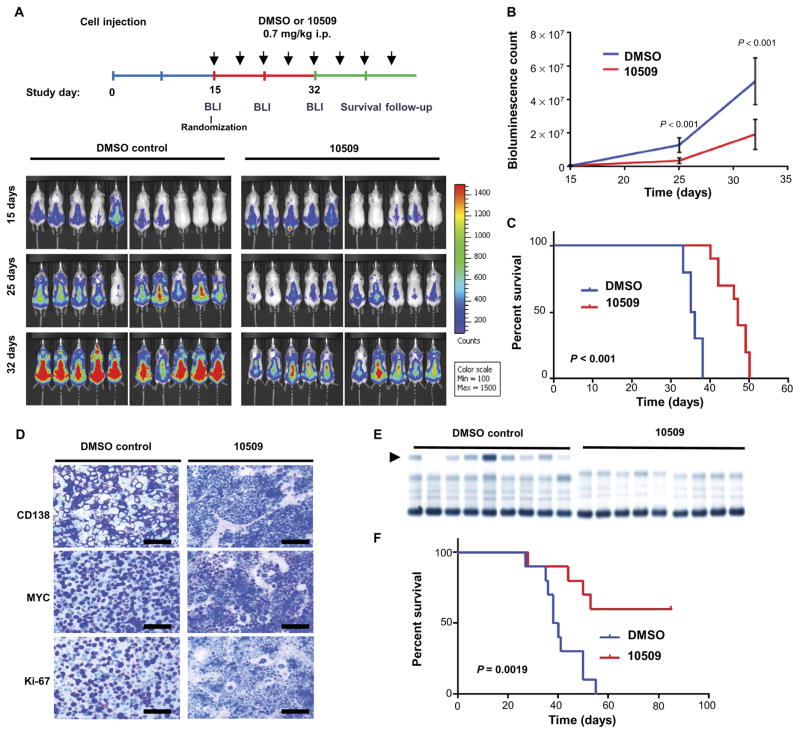
On the basis of the mechanism of action of CMLD010509 in MM, we assessed whether the drug would be effective and tolerable in animal models. We first established a xenograft model by intravenous injection of MM1S cells expressing luciferase (luc) into severe combined immunodeficient (SCID) mice. After engraftment, the mice were randomized to vehicle and compound treatment groups, with equal amounts of bioluminescence imaging (BLI) intensity in each group. CMLD010509 or vehicle control was administered via intraperitoneal injection at 0.7 mg/kg twice a week (Fig. 8A). CMLD010509 caused a marked reduction in tumor burden despite the very rapid growth of this aggressive MM cell line, as monitored by BLI and prolonged survival (median OS, 35 versus 47 days; P < 0.001) (Fig. 8, B and C). Notably, 4 weeks of CMLD010509 treatment were well tolerated with preservation of body weight and normal complete blood counts (fig. S6, A to C). To confirm the in vivo efficacy of CMDL010509, we used a second xenograft model consisting of SCID mice inoculated intravenously with KMS-11 cells. The mice were treated with intraperitoneal injections of either CMLD010509 at 0.7 mg/kg or vehicle control twice a week, starting at week 4, and were sacrificed at week 8. Immunohistochemistry studies were conducted on bone marrow samples to assess the presence of CD138+ tumor cells as well as amounts of MYC and Ki-67 protein. A decrease in CD138+ plasma cells, depletion of MYC, and inhibition of proliferation (Ki-67 staining) were observed in mice treated with CMLD010509 compared to those treated with vehicle control (Fig. 8D), further validating the activity of CMLD010509 on MM cell lines in vivo.
Fig. 8. In vivo suppression of MM by CMLD010509.
(A) Diagram of in vivo studies performed. SCID mice were injected with MM1S GFP (green fluorescent protein)/ Luc+. After engraftment, the mice were randomized to two groups based on BLI, and CMLD010509 or vehicle control was administered by intraperitoneal (i.p.) injections at 0.7 mg/kg two times a week. Tumor growth was assessed by weekly bioluminescence (BLI) counts, (B) showing a significant difference in tumor growth between the DMSO-treated group and the CMLD010509 group; P < 0.001. (C) Kaplan-Meier curve showing a significant survival difference between vehicle- and drug-treated mice, with a median OS of 35 versus 47 days, P < 0.001. (D) Immunohistochemistry for CD138, Ki-67, and MYC in bone marrow samples from the KMS-11–injected mice. Scale bars, 50 μm. (E) Electrophoresis of serum proteins from transplantable Vk*Myc mice indicating the presence of a γ-globulin (M-spike) in the DMSO control group (arrow) at 4 weeks. C57BL/6 syngeneic mice were intravenously injected with 3.5 × 106 Vk*Myc cells. At day 7 after tumor cells injection, mice were randomized into two groups and received DMSO (n = 10) or CMLD010509 drug (n = 10, 0.7 mg/kg per mouse) through intraperitoneal injections twice a week. (F) Kaplan-Meier curve showing a significant survival difference between vehicle- and drug-treated mice, with a median OS of 39 days versus not reached, P = 0.0019. Statistical analyses were performed using log-rank test.
Finally, to complete the assessment of CMLD010509’s effects on MYC-driven MM cells in vivo, we used an immunocompetent mouse model, in which transplantable mouse MM cells were injected into C57BL/6 mice. The tumor cells are driven by plasma cell–specific MYC overexpression (Vk*Myc cells) (28). Mice were treated with either CMLD010509 at 0.7 mg/kg or vehicle control intraperitoneally twice a week, beginning 1 week after cell tumor inoculation. Treatment was well tolerated, as indicated by normal transthyretin and albumin serum concentrations at 6 weeks (fig. S6D). Serum electrophoresis proteins (SPEPs) revealed a robust decrease of M-spike in CMLD010509-treated mice as shown at week 4 (Fig. 8E), which was associated with a significantly prolonged survival—median survival of 39 days in the control group versus not reached in the CMLD010509-treated group at this end of the experiment (90 days) (P = 0.0019) (Fig. 8F). Together, these data indicate that CMLD010509 is highly active and well tolerated in vivo in several mouse models of MM.
DISCUSSION
The oncogenic translation program of tumors is supported by aberrant activation of ribosome biogenesis and dysregulation of the translation initiation machinery (8–10). Here, we report the correlation between MYC activation and the translation of an oncogenic program in MM, which can be inhibited by the rocaglate derivative CMLD010509. We identified the rocaglate derivative CMLD010509 (SDS-1-021) as a highly specific inhibitor of the oncogenic translation program supporting MM—including key oncoproteins such as MYC, MDM2, CCND1, MAF, and MCL-1.
Although ribosome biogenesis and translation initiation complex function have historically been perceived as passive processes, recent studies have revealed that they are actively regulated to support specific translation programs (8, 9). Here, using a proteome-wide approach, we report that targeting translation initiation in MM selectively depletes multiple oncoproteins while leaving other housekeeping proteins largely unperturbed. The selective inhibition of translation initiation for oncogenic mRNA in the context of cancer can be explained by several mechanisms. First, some messengers are translated in a cap-dependent manner, whereas others are translated through the internal ribosome entry site process. Cap-dependent translation relies on the ability of the eukaryotic translation initiation factor 4F (eIF4F) complex to bind to the 5′ 7-methylguanosine cap present on mature mRNAs (29). Moreover, the presence of complex secondary structures within the 5′ untranslated region (5′UTR) influences mRNA translation. Transcripts with complex 5′UTR secondary structures are subject to greater dependence on the initiation machinery and therefore might be more sensitive to its inhibition. Subsequent studies have shown that the limiting factor is often eIF4E, mainly through its ability to recruit the eIF4A helicase (30–33). Complex 5′UTR structures have been identified in several prooncogenic mRNAs such as those encoding MYC, MDM2, and cyclins (10).
Rocaglate natural products, including silvestrol and rocaglamide A, comprise a class of protein synthesis inhibitors (14, 17) that interact with the eIF4A—a protein subunit of the translation initiation complex (14, 34). These compounds increase the affinity between eIF4A and mRNA and specifically clamp eIF4A onto polypurine sequences in mRNA 5′UTR, thus blocking ribosome subunit scanning and reducing protein expression from transcripts bearing the rocaglamide A–eIF4A target sequence (35). The compound CMLD010509 is a potent and selective translation inhibitor through an eIF4E phosphorylation–independent mechanism (15). Here, we demonstrate that CMLD010509 inhibits translation of a key suite of oncoproteins in MM that constitute an oncogenic translation program supporting MM oncogenesis. Moreover, this compound is highly effective in vivo in several mouse models, suggesting an effective therapeutic approach toward targeting MYC in MM.
The present work was aimed to assess the translational potential of rocaglates in MM. We did not focus on the specific relationship between MYC and translation activation in MM. Moreover, the specific effect of CMLD010509 on the translational oncogenic program in MM could be further explored to fully determine its mechanism of action.
In summary, we present an indirect but highly effective mechanism for targeting an oncogenic translation program frequently driven by MYC in MM. Even with the most recent advances in the management of plasma cell dyscrasias, MM remains a frequently incurable disease. Therefore, there is an urgent need for additional treatment options with new modes of activity. As shown here, rocaglates inhibit a specific translation oncogenic program related to high expression of MYC, with a very potent activity in vitro and in vivo. Thus, targeting dysregulated translation initiation with rocaglates rather than targeting the elongation machinery with other translation inhibitors might be less toxic to normal tissues. Several characteristics of CMLD010509 support its further preclinical development. First, the compound can be made by total synthesis from simple starting materials, which avoids reliance on limited bioresources (36). Second, its mode of action suggests a simple strategy for therapeutic drug monitoring to minimize toxicity in patients based on routinely available measurements of short-lived serum proteins such as prealbumin (transthyretin) or factor VII. Finally, successful clinical development might be improved by use of the oncogenic signature we have proposed as a potential companion diagnostic to identify patients most likely to benefit from rocaglate treatment. The targeting of protein degradation in MM with proteasome inhibitors has become a mainstay in the management of this disease. On the basis of the preclinical data presented here, we suggest that targeting protein homeostasis in the opposite way, through selective inhibition of protein synthesis, is worthy of further development and may provide an equally effective approach in the management of MM and possibly other hematological malignancies.
MATERIALS AND METHODS
Study design
The objectives of this study were as follows: (i) to identify compounds able to interfere with MYC activation in MM, (ii) to assess the mechanistic role of rocaglate compounds in MM, and (iii) to determine the preclinical efficacy of CMLD010509 as an effective treatment against MM. CellTiter-Glo was used to evaluate the efficacy of a small-molecule screen against two cell lines with high expression of MYC, to validate the best hits against six cell lines, and to generate IC50 values of rocaglate compounds. Functional evaluation of CMLD010509 was performed by RNA-seq of five MM cell lines and tandem mass tag (TMT) mass spectrometry using NCI-H929 exposed to CMLD010509 or DMSO in both cases. Further, qRT-PCR and Western blotting were performed to validate the data. We used GSEA and the LINCS cloud databases for pathway analyses. The effect of CMLD010509 in vitro was assessed on MM cell lines by Caspase-Glo 3/7 and Western blotting for caspase-3 and PARP. To determine the preclinical efficacy of CMLD010509, we used three different mouse models. The first one was a xenograft model where MM1S luc/GFP cells were injected intravenously into SCID mice. After 2 weeks of engraftment, tumor formation was monitored by BLI. To allocate animals to experimental groups, we measured BLI using Living Image 4.0 software and randomized mice to obtain equal amounts of BLI intensity in each group. Mice were treated with either CMLD010509 or DMSO and monitored weekly for BLI, blood count, and body weight and for survival. The second xenograft model consisted of injecting KMS-11 cells into SCID mice and treating them with either CMLD010509 or DMSO. Mice were all sacrificed at week 6 and studied for immunohistochemistry of the bone marrow with CD138, MYC, and Ki-67 antibodies. Finally, we developed an immunocompetent mouse model by injecting Vk*Myc cells into C57BL/6 mice. After 1 week of engraftment, mice were randomized into two groups and treated with either CMLD010509 or DMSO. Mice were monitored for SPEP and survival. The experiment was terminated after 3 months of follow-up.
Cell lines and primary cells
NCI-H929, NAMALWA, MM1S, MM1R, U266, and RPMI8226 were purchased from American Type Culture Collection, OPM-2 was from DSMZ, and KMS-11 and KMM-1 were from Japanese Collection of Research Bioresources Cell Bank. MM1S luc/GFP cells were gifts from A. Kung (Dana-Farber Cancer Institute), and CAL51, NCI-1650, NCI-1750, NCI-23, DU4475, and ZR-751 cells were gifts from the Jänne Lab (Dana-Farber Cancer Institute). Vk*Myc cells used in this study were isolated from late-stage Vk*Myc mice (gift from L. Bergsagel and M. Chesi) and expanded in vivo. All cells were maintained under 5% CO2 in a medium according to their specifications. PMBCs were obtained from the peripheral blood of healthy volunteers by Ficoll gradient centrifugation.
Small-molecule screen
MM cell lines were treated with 2812 compounds provided by the BU-CMD (www.bu.edu/cmd). A microplate dispenser, the Matrix WellMate (Thermo Fisher Scientific), was used to dispense 2000 adherent cells or 5000 suspension cells per well into 384-well microplates. Compounds were added using Biomek FX pin tool (Beckman Coulter). After treatment, cytotoxicity was measured by CellTiter-Glo (Promega) according to the manufacturer’s protocol, and luminescence signals were read using EnSpire (PerkinElmer), a plate reader.
Cell viability assay
Relative cell growth and survival were measured in a 96-well microplate format by using the luminescent detection of CellTiter-Glo or Caspase-Glo as the end point. For IC50, 5000 adherent cells and 30,000 suspension cells were plated and incubated with compounds for 72 hours. Each analysis was performed three times.
Protein and RNA isolation
Harvested cells were lysed using lysis buffer (Cell Signaling) supplemented with 5 mM NaF and 1 mM phenylmethylsulfonyl fluoride for 30 min on ice, and lysates were centrifuged at 14,000g for 30 min. Protein concentration was measured using Bio-Rad Protein Assay kit. Total RNA was isolated from cells using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol and evaluated for quantity and quality by NanoDrop spectrophotometer.
Quantitative real-time PCR
For qRT-PCR, 100 ng of total RNA per sample was used for reverse transcription using the SuperScript III First-Strand Synthesis Kit (Invitrogen). qRT-PCR was done on a StepOnePlus Real-Time PCR System (Applied Biosystems) using SYBR Green I Master Mix. Gene expression was normalized to 18S. The primer sequences are as follows: human MYC, TCCGTCCTCGGATTCTCTGCTCT (forward) and GCCTCCAGCAGAAGGTGATCCA (reverse); human MDM2, CCCAAGACAAAGAAGAGAGTGTGG (forward) and CTGGGCAGGGCTTATTCCTTTTCT (reverse); human CCND1, GCTGCGAAGTGGAAACCATC (forward) and CCTCCTTCTGCACACATTTGAA (reverse); human MAF, CTGGCAATGAGCAACTCCGA (forward) and AGCCGGTCATCCAGTAGAGT (reverse); human MCL-1, TGCTTCGGAAACTGGACATCA (forward) and TAGCCACAAAGGCACCAAAAG (reverse); and human 18S, TCAACTTTCGATGGTAGTCGCCGT (forward) and TCCTTGGATGTGGTAGCCGTTTCT (reverse).
RNA sequencing
NCI-H929, NAMALWA, U266, MM1S, and OPM-2 cells were treated with 50 nM CMLD010509 or vehicle control for 6 hours. RNA was prepared as mentioned earlier. A starting amount of 500 ng of RNA was used to prepare polyadenylate-enriched, single bar–coded libraries using the NEBNext Kit. Quality control of the libraries was evaluated by Bioanalyzer analysis with High Sensitivity chips (Agilent Technologies). Sequencing was performed on a HiSeq 2500 (Illumina) by 2× 50–base pair paired-end reads at the Biopolymers Facility of Harvard Medical School. We used Bcbio_nextgen (https://github.com/chapmanb/bcbio-nextgen/) to process the RNA-seq data. Briefly, cutadapt (https://github.com/marcelm/cutadapt/) was used to trim adapters; trimmed reads were aligned to human reference genome (GRCh37) with to-phat2; read count for each gene was calculated by HTSeq. Genes with low expression (fragments per kilobase million of <1 across all samples) were filtered out. GSEA was used to identify significantly enriched pathways (16), with an FDR of <0.25 and a P value of <0.05. Gene sets were downloaded from the Broad Institute’s MSigDB (www.broadinstitute.org/gsea/index.jsp).
Immunoblotting
Cell lysates (50 μg) were subjected to SDS–polyacrylamide gel electrophoresis (PAGE), transferred to a polyvinylidene fluoride membrane, blocked with 5% nonfat dry milk, and incubated with primary antibodies overnight at 4°C. Primary antibodies used included anti-MYC, anti–MCL-1, anti-CCDN1 (all Cell Signaling), anti-MDM2, anti-MAF (all Santa Cruz Biotechnology), and anti-tubulin (Sigma-Aldrich).
TMT spectrometry
NCI-H929 cells were treated in triplicate with CMLD010509 (100 nM) or vehicle control for 2 hours. Proteins were isolated and further processed for quantitative proteomic analysis by TMT (Thermo Fisher Scientific) as per the manufacturer’s protocol, with mass spectrometry.
35S labeling
NCI-H929 and MM1S cells were preincubated in methionine-free RPMI medium supplemented with dialyzed fetal bovine serum (FBS) and penicillin-streptomycin (Thermo Fisher Scientific) for 10 min. 35S-methionine was added to the medium at a final concentration of 100 μCi/ml (~9 μM methionine), and the cells were labeled for 20 min at 37°C. An equal volume of complete RPMI medium (containing an excess of unlabeled methionine; ~50 μM final) was added, and the cells were incubated for an additional 10 min at 37°C. The cells were then collected by centrifugation and washed with cold 1× phosphate-buffered saline. Whole-cell extracts were prepared by resuspending the cells in 1× Cell Lysis Buffer (Cell Signaling) supplemented with Halt protease and phosphatase inhibitors (Thermo Fisher Scientific) followed by a 30-min incubation on ice. The extracts were centrifuged for 15 min at 18,000g to remove the insoluble material. For the immunoprecipitation (IP) experiments, the whole-cell extracts were incubated with primary antibodies [anti-MYC (Abcam #ab32), anti-cyclin D1 (Abcam #ab134175), anti–MCL-1 (Cell Signaling #94296), anti-GAPDH (Abcam #ab181602), or anti–control immunoglobulin G (Cell Signaling #3900)] overnight at 4°C with rotation. The following morning, 40 μl of DynaBeads Protein G (Invitrogen) was added, and the samples were rotated for an additional hour. The beads were separated using a magnetic rack and washed three times with Cell Lysis Buffer. The immunoprecipitated material was eluted in 25 μl of 1× SDS sample buffer. The whole-cell extracts and the IP samples were resolved by SDS-PAGE and transferred to a nitrocellulose membrane (GE Healthcare). The membrane was stained with Ponceau S to visualize total proteins in samples. The membrane was then air-dried and exposed to a phosphorimager screen to visualize the 35S-labeled proteins.
In vivo studies
SCID mice (n = 10 per group) used for xenograft experiments were injected intravenously with 5 × 106 MM1S GFP-Luc+ and monitored by blood cell count, body weight, BLI, and survival. After engraftment, the mice were randomized into two groups on the basis of BLI, and CMLD010509 or vehicle control was administered by intraperitoneal injection at 0.7 mg/kg twice a week. Similar studies were performed in SCID mice (n = 10 per group) by injecting 7 × 106 KMS-11, but because these cells are not luc+, the mice were sacrificed at week 6, and the bone marrow of femurs was subjected to immunohistochemistry as described below. For the immunocompetent mouse model, C57BL/6 mice were purchased from the Jackson Laboratory. The transplantable Vk*MYC cell line, which was a gift from M. Chesi and P. L. Bergsagel (Mayo Clinic, Scottsdale, AZ), was maintained and expanded in vitro using RPMI 1640 containing 10% FBS (Sigma-Aldrich), 2 mM L-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml) (GIBCO). Vk*Myc cells (3.5 × 106) were injected intravenously into C57BL/6 mice, and the peripheral blood from the mice was collected for SPEP measurement at serial time points by submandibular bleeding. SPEP was performed using the QuickGel Protein Kit (Helena Laboratories) according to the manufacturer’s protocol. After 1 week of Vk*Myc cell injection, the mice were randomized to therapy or vehicle control (intraperitoneal injection at 0.7 mg/kg twice a week, n = 10 mice per group).
Study approval
All mice were treated, monitored, and sacrificed in accordance with an approved protocol of the Dana-Farber Cancer Institute Animal Care and Use Committee.
Immunohistochemistry
Murine tissues were fixed overnight in 10% buffered formalin and stored in 70% ethanol before proceeding to paraffin-embedding, sectioning, and staining with hematoxylin and eosin. Immunohistochemistry was performed using anti-CD138, anti–Ki-67, and anti–c-Myc, as previously reported (37).
Statistical analysis
Unpaired Student’s t test was used to compare two independent groups. One-way analysis of variance (ANOVA) was used when three or more independent groups were compared. For survival data, Kaplan-Meier curves were plotted and compared using a log-rank test. All tests were two-sided. A P value of <0.05 was considered statistically significant. Pearson coefficient was used to assess the correlation between two groups. Analysis was performed with GraphPad Prism Software (GraphPad Software Inc.). Mann-Whitney U test was used to quantify the enrichment of a given signature in a ranked list of genes, which was represented as a Z score. GSEA was performed as described previously using GSEA v2.0.10 (www.broadinstitute.org/gsea/) (16). MSigDB collections (http://software.broadinstitute.org/gsea/msigdb/index.jsp) and specific gene sets (KEGG Translation and Ribosome and several MYC gene sets) were tested for their enrichment in data sets—MM patients with high expression of MYC versus low expression or MM cell lines treated with CMLD010509 versus DMSO. Investigate_GeneSets (http://software.broadinstitute.org/gsea/msigdb/annotate.jsp) was used to investigate pathways enriched in a list of genes against MSigDB collections. The analysis was performed by applying two-tailed Fisher’s test as implemented in the Investigate_GeneSets module at MSigDB. For network enrichment mapping, gene sets with significant enrichment in CMLD010509- or DMSO-treated cells by GSEA were selected on the basis of P < 0.05 and FDR < 0.25 and visualized with Cytoscape v3.3.0 (www.cytoscape.org/cy3.html). This software organizes the significant gene sets into a network, where nodes correspond to gene sets, and the edges reflect significant overlap between the nodes according to Fisher’s test. The size of the nodes is proportional to the number of genes in the gene set, as previously described (38).
Supplementary Material
Fig. S1. Correlation between MYC expression and translation activation in different hematologic malignancies.
Fig. S2. Three potent rocaglate derivatives identified by an initial drug screen of a small-molecule compound library.
Fig. S3. Association of HSF1 activation with poor outcomes in MM.
Fig. S4. RNA-seq and TMT proteomic data validation in several MM cell lines.
Fig. S5. Assessment of CMLD010509 toxicity on PBMCs.
Fig. S6. Evaluation of CMLD010509 toxicity in vivo.
Table S1. RNA-seq of MM cells treated with CMLD010509 (provided as an Excel file).
Table S2. TMT proteomic analysis of NCI-H929 treated with CMLD010509 (provided as an Excel file).
Acknowledgments
Acknowledgments: We are thankful to M. Mendillo (Northeastern University, Boston, MA) for analysis of the HSF1 activation signature in MM data sets, to A. Kung (Dana-Farber Cancer Institute, Boston, MA) for providing the MM1S luc/GFP cells, to P. Jänne (Dana-Farber Cancer Institute, Boston, MA) for providing lung and breast cell lines, and to L. Bergsagel and M. Chesi (Mayo Clinic, Scottsdale, AZ) for providing Vk*Myc cells.
Funding: This work was supported, in part, by NCI R01 CA181683-01A1. Work at Boston University is supported by R35GM118173 and R24GM111625. We thank S. Stone for the initial synthesis of CMLD010509. R.Y.E. was supported by the National Institute of General Medical Sciences T32GM007753.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: RNA-seq data have been deposited to the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession no. GSE94827. All compounds tested are from J.A.P. under a material transfer agreement with Boston University.
Author contributions: S.M., N.G., J.B., L.W., and I.M.G. designed the study. S.M., D.H., Y.J.S., K.Z.S., R.Y.E., S.V.G., T.Y., and J.Z. performed the experiments. S.M., D.H., J.P., J.S., L.W., and I.M.G. analyzed the data. W.G.D., L.E.B., J.K.S., and J.A.P. generated reagents. I.M.G. provided funding. All authors wrote, reviewed, and edited the manuscript.
REFERENCES AND NOTES
- 1.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Noone AM, Yu M, Cronin KA. Use of imputed population-based cancer registry data as a method of accounting for missing information: Application to estrogen receptor status for breast cancer. Am J Epidemiol. 2012;176:347–356. doi: 10.1093/aje/kwr512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chng WJ, Huang GF, Chung TH, Ng SB, Gonzalez-Paz N, Troska-Price T, Mulligan G, Chesi M, Bergsagel PL, Fonseca R. Clinical and biological implications of MYC activation: A common difference between MGUS and newly diagnosed multiple myeloma. Leukemia. 2011;25:1026–1035. doi: 10.1038/leu.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker BA, Wardell CP, Murison A, Boyle EM, Begum DB, Dahir NM, Proszek PZ, Melchor L, Pawlyn C, Kaiser MF, Johnson DC, Qiang YW, Jones JR, Cairns DA, Gregory WM, Owen RG, Cook G, Drayson MT, Jackson GH, Davies FE, Morgan GJ. APOBEC family mutational signatures are associated with poor prognosis translocations in multiple myeloma. Nat Commun. 2015;6:6997. doi: 10.1038/ncomms7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Affer M, Chesi M, Chen WD, Keats JJ, Demchenko YN, Tamizhmani K, Garbitt VM, Riggs DL, Brents LA, Roschke AV, Van Wier S, Fonseca R, Bergsagel PL, Kuehl WM. Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia. 2014;28:1725–1735. doi: 10.1038/leu.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B, Stewart JP, Zhan F, Khatry D, Protopopova M, Protopopov A, Sukhdeo K, Hanamura I, Stephens O, Barlogie B, Anderson KC, Chin L, Shaughnessy JD, Jr, Brennan C, Depinho RA. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006;9:313–325. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 7.Kress TR, Sabò A, Amati B. MYC: Connecting selective transcriptional control to global RNA production. Nat Rev Cancer. 2015;15:593–607. doi: 10.1038/nrc3984. [DOI] [PubMed] [Google Scholar]
- 8.Truitt ML, Ruggero D. New frontiers in translational control of the cancer genome. Nat Rev Cancer. 2016;16:288–304. doi: 10.1038/nrc.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Truitt ML, Conn CS, Shi Z, Pang X, Tokuyasu T, Coady AM, Seo Y, Barna M, Ruggero D. Differential requirements for eIF4E dose in normal development and cancer. Cell. 2015;162:59–71. doi: 10.1016/j.cell.2015.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nat Rev Drug Discov. 2015;14:261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 11.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown LE, Chih-Chien Cheng K, Wei W-G, Yuan P, Dai P, Trilles R, Ni F, Yuan J, MacArthur R, Guha R, Johnson RL, Su X-z, Dominguez MM, Snyder JK, Beeler AB, Schaus SE, Inglese J, Porco JA., Jr Discovery of new antimalarial chemotypes through chemical methodology and library development. Proc Natl Acad Sci USA. 2011;108:6775–6780. doi: 10.1073/pnas.1017666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodrigo CM, Cencic R, Roche SP, Pelletier J, Porco JA., Jr Synthesis of rocaglamide hydroxamates and related compounds as eukaryotic translation inhibitors: Synthetic and biological studies. J Med Chem. 2012;55:558–562. doi: 10.1021/jm201263k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bordeleau M-E, Robert F, Gerard B, Lindqvist L, Chen SMH, Wendel H-G, Brem B, Greger H, Lowe SW, Porco JA, Jr, Pelletier J. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest. 2008;118:2651–2660. doi: 10.1172/JCI34753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu J, Cencic R, Wang W, Porco JA, Jr, Pelletier J. Translation inhibition by rocaglates is independent of eIF4E phosphorylation status. Mol Cancer Ther. 2016;15:136–141. doi: 10.1158/1535-7163.MCT-15-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santagata S, Mendillo ML, Tang Y-c, Subramanian A, Perley CC, Roche SP, Wong B, Narayan R, Kwon H, Koeva M, Amon A, Golub TR, Porco JA, Jr, Whitesell L, Lindquist S. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science. 2013;341:1238303. doi: 10.1126/science.1238303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, Fraenkel E, Ince TA, Whitesell L, Lindquist S. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150:549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agnelli L, Bicciato S, Mattioli M, Fabris S, Intini D, Verdelli D, Baldini L, Morabito F, Callea V, Lombardi L, Neri A. Molecular classification of multiple myeloma: A distinct transcriptional profile characterizes patients expressing CCND1 and negative for 14q32 translocations. J Clin Oncol. 2005;23:7296–7306. doi: 10.1200/JCO.2005.01.3870. [DOI] [PubMed] [Google Scholar]
- 20.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, Epstein J, Yaccoby S, Sawyer J, Burington B, Anaissie E, Hollmig K, Pineda-Roman M, Tricot G, van Rhee F, Walker R, Zangari M, Crowley J, Barlogie B, Shaughnessy JD., Jr The molecular classification of multiple myeloma. Blood. 2006;108:2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulligan G, Mitsiades C, Bryant B, Zhan F, Chng WJ, Roels S, Koenig E, Fergus A, Huang Y, Richardson P, Trepicchio WL, Broyl A, Sonneveld P, Shaughnessy JD, Jr, Bergsagel PL, Schenkein D, Esseltine D-L, Boral A, Anderson KC. Gene expression profiling and correlation with outcome in clinical trials of the proteasome inhibitor bortezomib. Blood. 2007;109:3177–3188. doi: 10.1182/blood-2006-09-044974. [DOI] [PubMed] [Google Scholar]
- 22.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, Sougnez C, Knoechel B, Gould J, Saksena G, Cibulskis K, McKenna A, Chapman MA, Straussman R, Levy J, Perkins LM, Keats JJ, Schumacher SE, Rosenberg M, Getz G, Golub TR Multiple Myeloma Research Consortium. Widespread genetic heterogeneity in multiple myeloma: Implications for targeted therapy. Cancer Cell. 2014;25:91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho Y-J, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao M-S, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hurt EM, Wiestner A, Rosenwald A, Shaffer AL, Campo E, Grogan T, Bergsagel PL, Kuehl WM, Staudt LM. Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell. 2004;5:191–199. doi: 10.1016/s1535-6108(04)00019-4. [DOI] [PubMed] [Google Scholar]
- 27.Shaffer AL, Emre NCT, Lamy L, Ngo VN, Wright G, Xiao W, Powell J, Dave S, Yu X, Zhao H, Zeng Y, Chen B, Epstein J, Staudt LM. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chesi M, Robbiani DF, Sebag M, Chng WJ, Affer M, Tiedemann R, Valdez R, Palmer SE, Haas SS, Stewart AK, Fonseca R, Kremer R, Cattoretti G, Bergsagel PL. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell. 2008;13:167–180. doi: 10.1016/j.ccr.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5' noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40:515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- 32.Koromilas AE, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5' non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 1992;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feoktistova K, Tuvshintogs E, Do A, Fraser CS. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc Natl Acad Sci USA. 2013;110:13339–13344. doi: 10.1073/pnas.1303781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, Mavrakis KJ, Jiang M, Roderick JE, Van der Meulen J, Schatz JH, Rodrigo CM, Zhao C, Rondou P, de Stanchina E, Teruya-Feldstein J, Kelliher MA, Speleman F, Porco JA, Jr, Pelletier J, Ratsch G, Wendel H-G. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwasaki S, Floor SN, Ingolia NT. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature. 2016;534:558–561. doi: 10.1038/nature17978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lajkiewicz NJ, Roche SP, Gerard B, Porco JA., Jr Enantioselective photocycloaddition of 3-hydroxyflavones: Total syntheses and absolute configuration assignments of (+)-ponapensin and (+)-elliptifoline. J Am Chem Soc. 2012;134:13108–13113. doi: 10.1021/ja305342f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akbay EA, Moslehi J, Christensen CL, Saha S, Tchaicha JH, Ramkissoon SH, Stewart KM, Carretero J, Kikuchi E, Zhang H, Cohoon TJ, Murray S, Liu W, Uno K, Fisch S, Jones K, Gurumurthy S, Gliser C, Choe S, Keenan M, Son J, Stanley I, Losman JA, Padera R, Bronson RT, Asara JM, Abdel-Wahab O, Amrein PC, Fathi AT, Danial NN, Kimmelman AC, Kung AL, Ligon KL, Yen KE, Kaelin WG, Jr, Bardeesy N, Wong K-K. D-2-hydroxyglutarate produced by mutant IDH2 causes cardiomyopathy and neurodegeneration in mice. Genes Dev. 2014;28:479–490. doi: 10.1101/gad.231233.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lane AA, Chapuy B, Lin CY, Tivey T, Li H, Townsend EC, van Bodegom D, Day TA, Wu S-C, Liu H, Yoda A, Alexe G, Schinzel AC, Sullivan TJ, Malinge S, Taylor JE, Stegmaier K, Jaffe JD, Bustin M, te Kronnie G, Izraeli S, Harris MH, Stevenson KE, Neuberg D, Silverman LB, Sallan SE, Bradner JE, Hahn WC, Crispino JD, Pellman D, Weinstock DM. Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation. Nat Genet. 2014;46:618–623. doi: 10.1038/ng.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Correlation between MYC expression and translation activation in different hematologic malignancies.
Fig. S2. Three potent rocaglate derivatives identified by an initial drug screen of a small-molecule compound library.
Fig. S3. Association of HSF1 activation with poor outcomes in MM.
Fig. S4. RNA-seq and TMT proteomic data validation in several MM cell lines.
Fig. S5. Assessment of CMLD010509 toxicity on PBMCs.
Fig. S6. Evaluation of CMLD010509 toxicity in vivo.
Table S1. RNA-seq of MM cells treated with CMLD010509 (provided as an Excel file).
Table S2. TMT proteomic analysis of NCI-H929 treated with CMLD010509 (provided as an Excel file).