Abstract
Single molecule sequencing has recently been used to obtain full-length cDNA sequences that improve genome annotation and reveal RNA isoforms. Here, we used one such method called isoform sequencing (Iso-Seq) from Pacific Biosciences (PacBio) to sequence a cDNA library from the Asian malaria mosquito Anopheles stephensi. More than 600,000 full length cDNAs, referred to as reads of insert, were identified. Due to the inherently high error-rate of PacBio sequencing, we tested different approaches for error-correction. We found that error-correction using Illumina RNA-seq generated more data than using the default SMRT pipeline. The full-length error-corrected PacBio reads greatly improved the gene annotation of Anopheles stephensi: 4867 gene models were updated and 1785 alternatively-spliced isoforms were added to the annotation. In addition, six trans-splicing events, where exons from different primary transcripts were joined together, were identified in An. stephensi. All six trans-splicing events appear to be conserved in Culicidae, as they are also found in An. gambiae and Aedes aegypti. The proteins encoded by trans-splicing events are also highly conserved and the orthologs of these proteins are cis-spliced in outgroup species, indicating that trans-splicing may arise as a mechanism to rescue genes that broke up during evolution.
Introduction
Single molecule real-time (SMRT) sequencing developed by Pacific Biosciences (PacBio) is a third-generation sequencing method that provides long reads that go well beyond kilobases. It has been used to facilitate assemblies and analyses of genomes of various species including a few insects (Jiang et al. 2014; Berlin et al. 2015; Hall et al. 2016). The long reads offered by SMRT sequencing has also been used, in the form of Iso-Seq, to obtain full-length cDNA sequences that could improve genome annotation and reveal RNA isoforms (Sharon et al. 2013; Abdel-Ghany et al. 2016). It is for this purpose that we used Iso-Seq to sequence a cDNA library from the adult males of the Asian malaria mosquito Anopheles stephensi.
In addition to significantly improving the annotation of the recently published An. stephensi genome (Jiang et al., 2014), full-length or long-read cDNA sequencing revealed a very interesting evolutionary phenomena, namely trans-splicing. RNA splicing is the process that forms a mature messenger RNA (mRNA) by joining exons and removing introns from the precursor mRNA (pre-mRNA). For majority of eukaryotic genes, splicing is mediated in cis by the spliceosome. The spliceosome brings the exons on both sides of an intron into close proximity and then cleaves the 5’ splice site and ligates the 5’ splice site to the branch point in the intron. This produces a lariat structured RNA. The spliceosome then cleaves the 3’ splice site, ligates exons and releases the lariat. Intriguingly, splicing can also occur in trans, where exons from more than one separate pre-mRNAs are joined. Trans-splicing is well studied in trypanosomes and nematodes, where a spliced leader RNA is spliced to the 5’ ends of the first exon on many pre-mRNAs (Douris, Telford, and Averof 2010).
In higher eukaryotes, trans-splicing does not involve spliced leaders. Trans-splicing has been observed in fruit flies, rodents, humans and many other organisms (Shao et al. 2012; Lasda and Blumenthal 2011; Horiuchi, Giniger, and Aigaki 2003; Caudevilla et al. 1998; Dorn, Reuter, and Loewendorf 2001; Herai and Yamagishi 2010). Based on the relationship of the two pre-mRNAs joined in trans-splicing, trans-splicing can be grouped into three categories: inter-allelic, intragenic and intergenic. A well-known example of inter-allelic trans-splicing is the lola gene, which is essential for the nervous system development in Drosophila (Horiuchi, Giniger, and Aigaki 2003). Trans-splicing of lola was inferred from interallelic complementation tests on lethal mutations in lola exons and verified by allelic SNP makers in Drosophila hybrids. Later, a study utilizing RNA-seq data from Drosophila hybrids identified more trans-splicing between homologous alleles, suggesting inter-allelic trans-splicing occurs commonly. Intragenic trans-splicing is the scenario where splicing occurs between two pre-mRNAs from the same genetic loci. The two pre-mRNAs can come from the same strand, and an example is the Carnitine O-octanoyltransferase gene in the rat liver where the exons are duplicated in the mRNA (Caudevilla et al. 1998). They can also come from opposite strand, like the fruit fly mod (modifier of mdg4) genes (Dorn, Reuter, and Loewendorf 2001). Intergenic trans-splicing occurs when the pre-mRNAs come from different genes. These genes can be located at distant genomic loci on different chromosomes. For example, the bursicon gene in Anopheles gambiae is trans-spliced from three exons on chromosome arm 2L and one exon on chromosome arm 2R (Robertson et al. 2006).
The understanding of trans-splicing has been significantly improved by the advent of next generation sequencing technology. Trans-splicing events are generally identified by finding non-co-linear transcripts, which are RNA-seq reads that fail to align to the corresponding DNA sequences in the reference genome in a linear pattern. Although this approach cannot detect inter-allelic and other trans-splicing that generate co-linear transcripts, a significant number of trans-splicing have been detected (Davidson, Majewski, and Oshlack 2015; Liu et al. 2015). For example, a recent study of eight insect species across five orders detected 1,627 trans-splicing events (Kong et al. 2015). Some of the trans-splicing events are conserved across species, indicating that trans-splicing is not transcriptional noise and is likely to be functionally important (Kong et al. 2015). Moreover, the previous notion that fusion transcripts are the markers of tumor cells has been called into question, as several studies and the Encyclopedia of DNA Elements (ENCODE) project demonstrated that chimeric RNAs are common in normal tissues and cell lines (Gingeras 2009).
The SMRT isoform sequencing (Iso-Seq) technology from Pacific Biosciences has been applied to discover trans-splicing or fusion genes (Weirather et al. 2015). Iso-Seq can generate full-length transcript sequences from the polyA-tail to the 5′ end, providing isoform-level resolution of transcriptome data. Full-length or long read cDNA sequences by Iso-Seq provide significant advantage in the identification and characterization of trans-splicing events compared with short RNA-seq data by Illumina, which can only provide information on the small segment around the trans-spliced site. Furthermore, the structure of full trans-spliced mRNA is hard to infer from short RNA-seq data, due to the fact that majority of the reads generated from the trans-spliced mRNAs cannot be differentiated from the ones from cis-spliced mRNA. This will not be an issue for Iso-seq data, which provide reads representing full-length transcripts.
In this research, we use both SMRT Iso-Seq data and Illumina RNA-seq data to detect trans-splicing events in Asian malaria mosquito Anopheles stephensi. To eliminate false positive discoveries due to PCR chimeras and transcriptional noise, only trans-splicing events supported by both datasets are used. In total, we identified six trans-splicing events in Anopheles stephensi, all of which are also found and conserved in Aedes aegypti. The proteins encoded by the trans-spliced mRNAs are also highly conserved and their orthologs are co-linearly transcribed in Culicidae out-groups. This finding indicates the need to preserve the mRNA completeness and protein function of genes broken-up during the course of evolution may be the driving force behind trans-splicing. As indicated earlier, we have also used the Iso-seq data to improve the An. stephensi genome annotation.
Results
Error correction with Illumina RNA-seq outperforms SMRT pipeline in both data accuracy and data quantity
Three Iso-Seq libraries with insert sizes of 1–2kb, 2–3kb, and 3–6kb were sequenced with four PacBio SMRT cells each (SRA accession: SRP081051). Each SMRT cell produced around 50,000 to 60,000 reads of insert. Full-length transcripts were defined by the presence of 5' primer, 3' primer, and the polyA tail in the reads of insert. Approximately 38%, 31%, and 9% of the reads of insert were identified as full length, non-chimeric reads for the 1–2kb, 2–3kb, and 3–6kb insert size libraries, respectively (Table 1). During the clustering process of the SMRT pipeline, on average 2 to 3 full length, non-chimeric reads could be clustered as one consensus isoform for the 1–2kb and 2–3kb libraries. For the 3–6kb size library, most consensus isoforms were only represented by a single full length, non-chimeric read. Therefore, in order to obtain sufficient number of long reads for analysis, larger insert sizes require deeper sequencing depth. Less than 17% of total consensus isoforms were polished as high-quality isoforms by Quiver. This indicates that in order to obtain enough high accuracy data through the SMRT pipeline alone, the number of cells sequenced for each library should be higher to provide enough coverage, particularly for long-insert libraries. All polished isoforms were used for the further analysis to avoid discarding useful information.
Table 1.
PacBio SMRT pipeline outputs metrics
| Data type | 1–2kb | 2–3kb | 3–6kb |
|---|---|---|---|
| Number of reads of insert | 248903 | 210594 | 202440 |
| Number of five prime reads | 136440 | 100147 | 54463 |
| Number of three prime reads | 146668 | 109776 | 65571 |
| Number of poly-A reads | 142375 | 106746 | 61281 |
| Number of filtered short reads | 13209 | 8876 | 7165 |
| Number of non-full-length reads | 138909 | 135425 | 175629 |
| Number of full-length reads | 96785 | 66293 | 19646 |
| Number of full-length non-chimeric reads | 96170 | 65955 | 19094 |
| Average full-length non-chimeric read length | 1388 | 1948 | 4357 |
| Number of consensus isoforms | 35248 | 30793 | 17405 |
| Average consensus isoforms read length | 1465 | 2044 | 4376 |
| Number of polished high-quality isoforms | 7414 | 6075 | 636 |
| Number of polished low-quality isoforms | 27834 | 24718 | 16769 |
As an alternative, we used RNA-seq data to error-correct the Iso-Seq data. Of 1,321 million base pairs of reads of insert, 668 million base pairs were corrected by Proovread with a high accuracy (Table 2). The mean of the average quality scores of each read of insert improved from 14.73 to 36. 4 after correction, indicating a significant improvement of accuracy. Compared with polished high-quality isoforms from the SMRT pipeline, 30 times more base pairs were corrected, although the mean value of the median quality scores of high accuracy corrected reads was slightly lower. In addition, the mean value of median quality scores of high accuracy corrected reads was 37.85, equivalent to an accuracy above 99.98%. This result demonstrates that it is favorable to use high-quality short reads for correction of Iso-Seq reads. This is also more economical as the RNA-seq data needed for the analysis is significantly cheaper than sequencing additional Iso-Seq SMRT Cells.
Table 2.
Comparisons of processed Iso-Seq data
| polished isoforms |
polished high-quality isoforms |
polished low-quality isoforms |
high accuracy corrected reads |
complete corrected reads |
reads of insert |
||
|---|---|---|---|---|---|---|---|
| Mean Quality Score |
min | 0 | 0.01 | 0 | 16.01 | 0.11 | 0.98 |
| max | 17.36 | 40 | 12.74 | 39.81 | 37.36 | 27.31 | |
| average | 12.7 | 37.15 | 7.72 | 36.4 | 28.25 | 14.73 | |
| median | 13.67 | 39.84 | 8.34 | 37.85 | 30.87 | 15.45 | |
| Length (base pairs) |
Total (million) |
190.6 | 23.8 | 166.8 | 668.6 | 1254.9 | 1321.4 |
| min | 330 | 567 | 330 | 70 | 249 | 11 | |
| max | 25502 | 5673 | 25502 | 8098 | 31415 | 31532 | |
| average | 2284.59 | 1686.06 | 2406.55 | 1284.73 | 2005.91 | 1996.32 | |
| median | 1829 | 1630 | 1891 | 1145 | 1636 | 1641 | |
| N25 | 4325 | 2005 | 4404 | 1939 | 4074 | 4214 | |
| N50 | 2244 | 1742 | 2471 | 1444 | 2234 | 2342 | |
| N75 | 1720 | 1416 | 1780 | 1046 | 1565 | 1624 | |
| N90 | 1358 | 1193 | 1407 | 727 | 1181 | 1206 | |
| N95 | 1213 | 1113 | 1244 | 613 | 919 | 928 |
Proteins encoded by trans-splicing are conserved
490 trans-splicing events were detected based on RNA-seq processed by MapSplice. 3,359 trans-splicing events were found by the PacBio pbtranscript-tofu package. In both RNA-seq and Iso-Seq technology, PCR chimeras could cause a large number of false positive results. Therefore, we set a criterion that splice junctions must be supported by both datasets to be considered as valid. In the end, six pairs of splice junctions were identified. All these six trans-splicing events are inter-chromosomal (Table 3).
Table 3.
Trans-splicing sites in three Culicidae
| An. stephensi | ||||||||
| donor | acceptor | |||||||
| contig | position | strand | sequence around splice site* | Contig | position | strand | sequence around splice site* | |
| Tm1.1 | stI-e1 | 8284604 | − | ATCAAGAAGGATAATCATAACTGCA|GT | stI-e2 | 29650741 | − | AC|TGTTGGTTGGGAGGGTGAACAACGG |
| Tm1.2 | stI-e3 | 29650494 | − | CGCAATGCTGCTGAAGCGTGTTGCG|GT | stI-e2 | 49700929 | + | AG|GAGTTGCAAAATCAGGTTGATTTTC |
| Tm2 | stI-e1 | 10533884 | − | ATCTGTTCGATGATGATCGAAAGTT|GT | stI-e2 | 13176261 | + | AG|TATCGCACACGGTACAAGATTTGGT |
| Tm3 | stI-e4 | 31171572 | + | CATCACTACTCCTGCCATCTGTGTC|GT | stI-e1 | 4936658 | − | AG|GGCGTATTTATCTTCAACATCGTGC |
| Tm4 | stI-e4 | 31171572 | + | CATCACTACTCCTGCCATCTGTGTC|GT | stI-e1 | 4948239 | + | AG|GGCGTATTTATCTTCAACATCGTGC |
| Tm5 | stI-e1 | 14042259 | + | AAAGCTGAAAGATGTCGTTGATCAG|GT | stI-e2 | 31153557 | − | AG|CAAAGTTCCTTCGCCTGCTGGCTAC |
| An. gambiae | ||||||||
| donor | acceptor | |||||||
| contig | position | strand | sequence around splice site* | Contig | position | strand | sequence around splice site* | |
| Tm1.1 | X | 14803808 | − | ATCAAGAAGGACAATCACAACTGCA|GT | 2L | 40170764 | + | AG|TTGAGCGGCGTCGTCGATTCAACAT |
| Tm1.2 | 2L | 40171011 | + | CGCAATGCTGCAGAAGCGTGTCGCG|GT | 2R | 56727316 | + | AG|GAGTTGCAAAACCAAGTTGACTTTC |
| Tm2 | X | 4334405 | − | ATCTGCTCGATGATGATCGAAAGTT|GT | 2R | 13216641 | − | AG|TATCGCACACGGTACAGGATTTGGT |
| Tm3 | 3R | 42647718 | + | CATCACCACTCCTGCCATCTGTGTC|GT | X | 8704053 | − | AG|GGCGTATTCATCTTCAACATCGTGC |
| Tm4 | 3R | 42647718 | + | CATCACCACTCCTGCCATCTGTGTC|GT | X | 8717291 | + | AG|GGCGTATTCATCTTCAACATCGTGC |
| Tm5 | X | 1025131 | + | AAAGCTGAAAGATGTCGTTGATCAG|GT | 2R | 27239887 | + | AG|CAAAGTTCCTTCGCCTGCTGGCTAC |
| An. gambiae | ||||||||
| donor | acceptor | |||||||
| contig | position | strand | sequence around splice site* | Contig | position | strand | sequence around splice site* | |
| Tm1.1 | supercont1.30 | 2525719 | + | ATCAAGAAGGATAACCATAACTGCA|GT | supercont1.497 | 69990 | − | AG|TCGAGAGGCGTCGTCGTTTCAATAT |
| supercont1.322 | 325659 | − | ATCAAGAAGGATAACCATAACTGCA|GT | supercont1.497 | 69990 | − | AG|TCGAGAGGCGTCGTCGTTTCAATAT | |
| Tm1.2 | supercont1.497 | 69743 | − | CATCATTCTGCAGCACAAACTGGCG|GT | supercont1.541 | 339301 | − | AG|GAAATGCAAAAGCAGCTCGACTATT |
| Tm2 | supercont1.75 | 2116294 | − | GTTTGCTCGATGATGATCGAAAGTT|GT | supercont1.187 | 287309 | − | AG|TATCCCACACCGTTCAGGATTTGGT |
| Tm3/Tm4 | supercont1.496 | 104246 | − | TATCACGACTCCGACGATCTGTGTT|GT | supercont1.715 | 64386 | + | AG|GGCGTATTCATCTTCAACCTGGTGC |
| Tm5 | supercont1.54 | 128027 | − | GAAGCTGAAGGACGTAGTCGATCAG|GT | supercont1.179 | 490627 | − | AG|CAAAGTTCCTTTGCCTGCTGGTTAC |
splice junctions are indicated as “|”. 25 bp upstream of donor site and 25 bp downstream of acceptor site are shown.
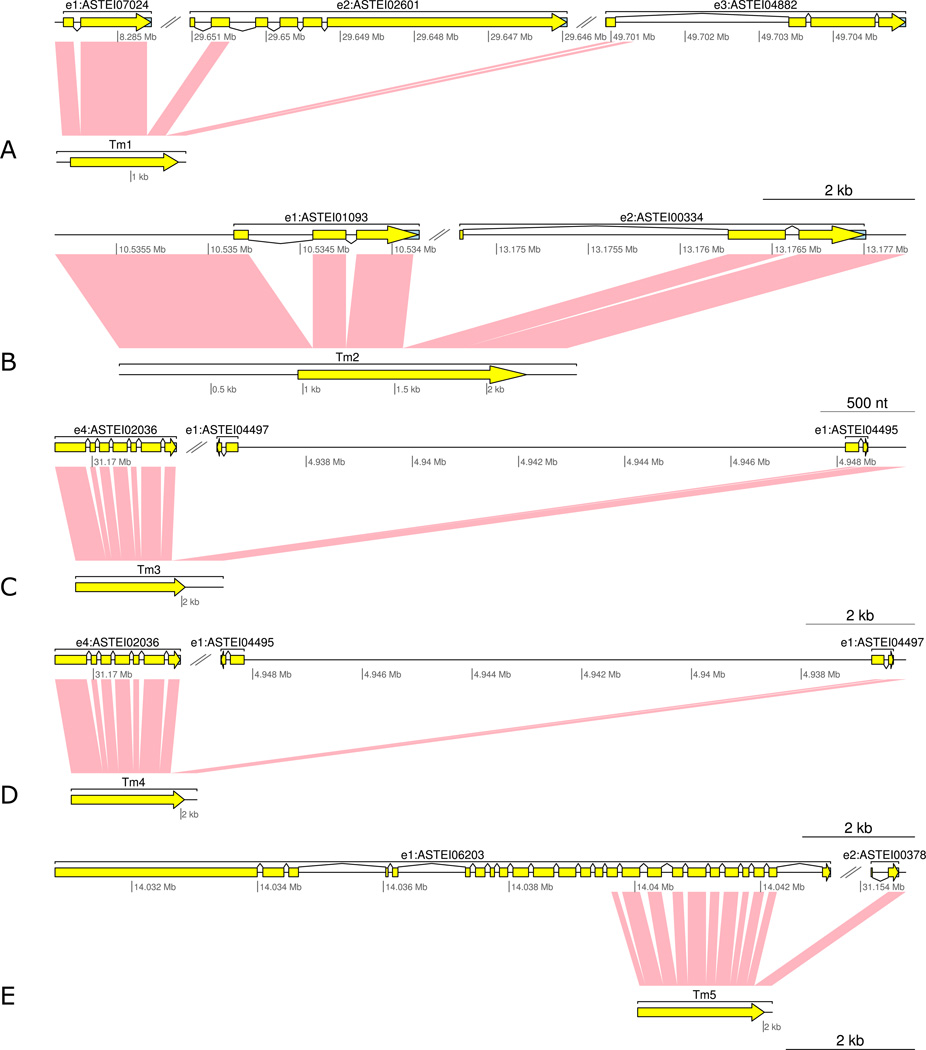
Trans-spliced mRNA 1 (Tm1) is one mRNA created from two trans-splicing events (Figure 2 A). The Pre-mRNAs of Tm1 are located in chromosome elements 1, 2 and 3. This mRNA has five exons: two shared with gene ASTEI07024, one shared with gene ASTEI02601, and one shared with the intron of gene ASTEI04882. The mRNA encoded a 475 aa peptide with two domains. The first domain encodes MiT/TFE transcription factors, N-terminal (IPR031867), which is shared with gene ASTEI07024. The second domain is Myc-type, basic helix-loop-helix domain (IPR011598), shared with gene ASTEI02601. ASTEI07024 is a mosquito specific gene. Alignment of peptide sequences of ASTEI02601 to its Drosophila ortholog FBgn0041164 revealed that the exon utilized by the Tm1 trans-splicing event contributes to amino acid sequences that do not exist in their Drosophila orthologs. This is also the case for the exon shared between Tm1 and ASTEI04882 when we aligned the peptide sequence of ASTEI04882 to that of the ortholog FBgn0034176. No obvious Drosophila ortholog for the complete Tm1 protein has been observed. There is some similarity between the protein of FBgn0263112 to the peptide sequences coded by the first three exons, particularly the third exon of Tm1 (28.87% identify). Interestingly, the complete 474 aa peptide sequence coded by Tm1 is highly homologous to some dipteran out-groups. The examples include gene XP_011304746 in Fopius arisanus (33.17% identity) and XP_012252483 in Athalia rosae (31.53% identity). The mRNAs of these genes in the outgroup species are co-linear to the genome, and thus likely to be cis-spliced.
Figure 2.
Trans-splicing events in Anopheles stephensi. In each panel, the top section stands for the genomic regions to which the trans-spliced mRNA aligns. Related gene annotation is also provided. The bottom section stands for the full-length mRNA sequence. The pink blocks in the middle represent matches between genomic sequence and mRNA. Yellow bar represents coding region.
The exons of Tm2 come from two cis-spliced genes ASTEI01093 and ASTEI00334 (Figure 2 B). Both genes don't have orthologs outside of mosquitos. The protein encoded by the trans-spliced Tm2 consists of 515 amino acids, which belongs to the neurotransmitter-gated ion-channel (IPR006201) family. This Tm2 protein is orthologous to the Drosophila gene FBgn0037950 with high similarity (83.57% identity). This trans-spliced protein is conserved in Insecta. All of its non-mosquito orthologs appear to be cis-spliced.
The donor sites of the trans-splicing event of Tm3 and Tm4 are identical (Figure 2 C and D). The genes on the acceptor site of these two events are paralogous to each other. The paralogs are in close proximity but of different orientation, probably due to a tandem duplication. The coding sequences of the two paralogs were identical, and consequently Tm3 and Tm4 encode identical protein. The 3’UTR is different in Tm3 and Tm4. Trans-splicing exists in both ASTEI004497 and ASTEI004495, as supported by full length transcripts covering the 3’ UTR in both genes. The encoded protein is a neurotransmitter symporter (IPR000175). The Drosophila gene FBgn0181657 is annotated as an ortholog to ASTEI02036 but in fact, sequence alignment showed that this is only a partial match, and FBgn0181657 is more similar and aligns over its full length to the fusion protein of ASTEI02036 and ASTEI004497/ASTEI004495. This fusion protein is highly conserved across Insecta. Like Tm2, Tm3 and Tm4 orthologs outside mosquitoes are cis-spliced.
The longest read we obtained from Tm5 is 2,142 bp (Figure 2 E). This read likely represents an mRNA with an incomplete 5’ end, because the start codon is missing. Nevertheless, this read covers the trans-splicing site between chromosome elements 1 and 2. Tm5 joins exons from ASTEI06203 and ASTEI00378. The protein Tm5 encodes is uncoordinated protein 13 (IPR027080). It is conserved across Insecta. FBpp0300963 is annotated as an ortholog to ASTEI06203. Interestingly, the last 53 amino acids encoded by FBpp0300963 is 76% identical to the 54 amino acids encoded by the last exon of ASTEI00378, while it is only 7% identical to amino acid encoded by the last exon of ASTEI06203. This implies that the fusion protein is ancestral and trans-splicing between exons of ASTEI006203 and ASTEI00378 is a way to keep the protein intact.
Trans-splicing is highly conserved in Culicidae
To investigate whether the above trans-splicing events are An. stephensi specific or are conserved, we checked the transcriptome data of Anopheles gambiae and Aedes aegypti. We predicted trans-splicing sites using MapSplice (Wang et al. 2010) with RNA-seq data as described in the methods.
All the trans-splicing events in An. stephensi also exist in An. gambiae (Table 3). In addition, the chromosomal assignment of the orthologs involved in trans-splicing are the same and the sequences around the splice sites are highly similar between these two species. In Ae. aegypti, the supercontigs are not assigned to chromosomes and thus chromosomal position cannot be inferred. Based on supercontigs, the orthologs of the trans-splicing An. stephensi events are observed with a few differences. First, the trans-spliced gene may share the exons with a different cis-spliced gene. For example, the ASTEI02601 orthologs AAEL010693 and AAEL010696 do not share exons with Tm1 in Aedes aegypti. Instead, the shared exon is in their neighboring gene AAEL010700. Second, duplication events are different between Anophelinae and Culicinae: the ASTEI07024 orthologs were duplicated and located on different supercontigs in Ae. aegypti, while the ortholog of gene ASTEI004495/ASTEI004497 is a single gene AAEL012596 in Ae. aegypti. Interestingly, trans-splicing was kept during duplications of these genes. In 17 of the 18 trans-splicing events in all three species, the 5’ and 3’ termini of the introns follow the GU-AG rule. The only exception is the first trans-splicing site of Tm1 in An. stephensi, where the acceptor site is AC instead of AG.
Improvement of genome annotation
Comparisons of the existing gene annotation of An. stephensi and the updated annotation by PASA with error-corrected high-accuracy Iso-Seq data are provided in the link http://tu07.fralin.vt.edu/cgi-bin/PASA_r20140417/cgi-bin/status_report.cgi?db=ECRItr. The previous gene annotation is based on gene annotation software Maker (Holt and Yandell 2011), where protein homology based and ab inito prediction were applied. Transcriptomes were not used by this Maker annotation. In the previous annotation, 11,789 protein coding genes were annotated. Each gene has only one isoform, which indicates that alternative splicing is largely ignored. In addition, genes were mostly annotated with UTRs missing. The updated annotation enhanced the existing one by adding UTRs, identifying alternative spliced isoforms, and adjusting exon boundaries. In total, 3,323 genes were updated with addition of UTRs, 1,785 genes were updated with alternatively spliced isoforms and 1,923 genes were updated with exons adjusted or gene merging. These structural changes of genes altered 1,878 protein sequences (Table 4).
Table 4.
Annotation improvement in Anopheles stephensi using PASA
| Num Gene Model Updates |
Num Alt Splice isoforms to Add |
|
|---|---|---|
| EST assembly extends UTRs. | 3323 | 0 |
| EST assembly alters protein sequence, passes validation. | 697 | 0 |
| EST assembly properly stitched into gene structure. | 1065 | 0 |
| EST assembly stitched into Gene model requires alternative splicing isoform. |
0 | 1785 |
| EST-assembly found capable of merging multiple genes. | 161 | 0 |
| Totals (some models in multiple classes) | 4867 | 1785 |
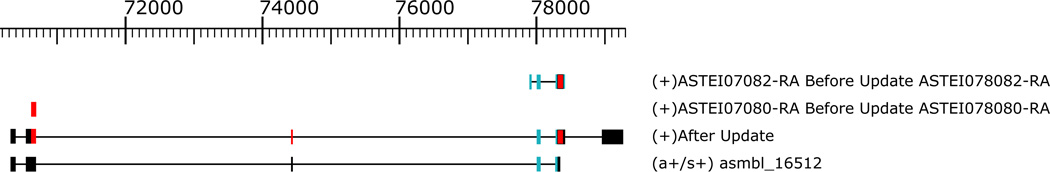
One example demonstrating the improvement of gene annotation can be reflected in the annotation of the gene doublesex (Suzuki et al. 2001). doublesex is a gene essential for sexual dimorphism and it contains male-specific and female-specific isoforms. In our analysis, the Iso-Seq data was obtained from males only, so we would expect to observe only the male isoform. The gene doublesex in mosquitos spans a region of 90,000 base pairs with another gene inserted in one of its introns. As a result, in the majority of Anophelinae, this gene is mis-annotated as two genes. After the annotation updating by PASA (Figure 3), the two parts of doublesex ASTEI07080 and ASTEI07082 were merged into one complete model. This model is the complete male isoform of doublesex as expected.
Figure 3.
Updated annotation for the doublesex gene in Anopheles stephensi. The first row and second row below the genomic size ruler represent gene ASTEI07082 and ASTI07080. The third row is an updated annotation from PASA which merges the two genes. The forth row is the evidence from Iso-Seq transcripts that supports the updated annotation.
Discussion
Two separate gene breakup events are necessary to form the trans-splicing of gene of Tm1. The first one which separated the third and fourth exons of Tm1 happened before the formation of Diptera. In Culicidae, trans-splicing was used to join these two separated exons, while this trans-splicing either did not happen or was later lost in Drosophila. The second gene breakup which separated exon2 and exon3 happened only in Culicidae. In Aedes, the region transcribing the first pre-mRNA of Tm1 was duplicated and both copies maintained their ability to be trans-spliced. The breakup of the ancestor genes of the other trans-spliced mRNAs described in this paper happened after the formation of Diptera but before Culicidae. All their Drosophila orthologs remained as canonical genes that utilize cis-splicing, while the formation of the complete mRNAs in Anophelinae and Aedes relies on trans-splicing. The high conservation of the trans-splicing sites across three divergent mosquito species indicates single origin for each trans-splicing event.
Although trans-splicing has been observed in many higher eukaryotes, its mechanism remains largely unclear. One well known model is that trans-splicing happens through mutually complementary intron sequences. The introns of two separate pre-mRNAs will pair bringing the two molecules together promoting trans-splicing (Wally, Murauer, and Bauer 2012). However, a recent study in Drosophila showed that two intronic RNA sequence motifs are critical and perhaps sufficient to initiate trans-splicing in the mod gene (Gao et al. 2015). In both models, the nucleotide sequences of pre-mRNAs effect the conformation of the RNA-spliceosome complex and then influence splicing, which is essentially the same as cis-splicing. It is reasonable to assume that the splicing machinery is no different for trans-splicing. Trans-splicing is observed across a wide range of eukaryotes and likely exists in all eukaryotes, just like cis-splicing (Douris, Telford, and Averof 2010). In addition, the only factor differentiating cis-splicing and trans-splicing is whether there are more than one pre-mRNAs. As a process in three-dimension, splicing requires spatial proximity of splice sites (Hiller et al. 2007; Warf and Berglund 2010). It may not matter whether the two separate pre-mRNAs directly interact with each other through base pairing or through binding to the spliceosome using specific sequence motifs. As long as the splice sites are spatially close and accessible, trans-splicing reactions may occur just as cis-splicing.
Splicing greatly diversifies the proteome by promoting the formation of new genes through alternative splicing. Allelic trans-splicing creates new combinations of alleles in mRNA (Horiuchi, Giniger, and Aigaki 2003). Intragenic trans-splicing can generate new transcripts by exon reuse (Caudevilla et al. 1998). Although intergenic trans-splicing in theory can produce new genes by joining exons from different genes, such a mechanism for novel gene formation does not appear to be favored, as the intergenic trans-splicing events observed are largely involved in ancient gene rescue rather than new gene creation. Thus, intergenic trans-splicing events likely evolved from ancestral cis-splicing or allelic trans-splicing; the scenario where two random unrelated distant segments of the genome acquire the ability to be trans-spliced together should be rare if any. In addition, all the proteins encoded by our trans-spliced events are highly conserved. It appears that strong purifying selection acts to maintain these trans-splicing events. If the protein is not essential, or other alternative strategies are adopted as in the case of Tm1 in Drosophila, intergenic trans-splicing may not occur or may be lost later during evolution. We can only speculate the possible reasons why intergenic trans-splicing appears to exist mainly as a gene rescue mechanism not a common facility to generate transcriptome/proteome diversity (Kong et al. 2015). First, unlike cis-splicing or the other two types of trans-splicing, it is difficult to ensure that two pre-mRNAs made from distant genomic loci share the same or overlapping temporal and spatial transcription (Gingeras 2009). Second, upon transcription the physical distance or locations of the pre-mRNAs could hinder trans-splicing. Finally, it is hard for the two pre-mRNAs to coevolve when their DNA templates are shaped by potentially different evolutionary forces.
Experimental Procedures
Library preparation and RNA sequencing
Fifteen 1–3 day old Anopheles stephensi (Indian Type Stain) adult male mosquitoes were homogenized in 900 ul of RNA lysis buffer and total RNA was isolated using the Quick-RNA MiniPrep kit (Zymo Research, Irvine, CA) according to the manufacturer’s protocol. Three hundred ul of the homogenate was used for total RNA isolation and total RNA was eluted with 30 ul H2O. Two samples of 3.2 ul each (300 ng/ul) total RNA was submitted to for PacBio sequencing at the Interdisciplinary Center for Biotechnology Research of University of Florida (Gainesville, FL). The RNA was reverse transcribed using the SMRTer PCR cDNA synthesis kit and amplified. Three sequencing libraries (1–2kb, 2–3kb, 3–6kb) were prepared according to the PacBio Iso-Seq protocol. The sequencing was performed on the PacBio RS II using P4-C2 chemistry. Four SMRT cells were run from each of the three libraries. Illumina RNA-seq data used in this study are from (Jiang et al. 2015).
SMRT pipeline analysis for Iso-Seq data
Analysis was performed using the PacBio SMRT-Analysis package v2.3 (http://www.pacb.com/devnet/). Analysis was run on the three libraries separately. The Iso-Seq bioinformatics pipeline consists of two major modules: classify and cluster. Reads of insert were obtained by identifying the adapter separator and then merging subreads into consensus sequence reads. Reads of insert were then classified into full-length, non-artificial-concatemer reads and non-full-length reads. Full-length, non-artificial-concatemer reads from the same isoform were clustered using the ICE algorithm and consensus isoforms were predicted. The consensus isoforms were then polished by Quiver utilizing the non-full-length reads. Default parameters were used when running Quiver, which means only consensus with more than 99% accuracy were binned into high-quality isoforms by Quiver. The low-quality isoforms binned by Quiver are generally the ones with low transcription level or low sequencing depth. Although less accurate, the low-quality isoforms also contain useful information and are used together with high-quality isoforms for our analysis.
Error correction of Iso-Seq data
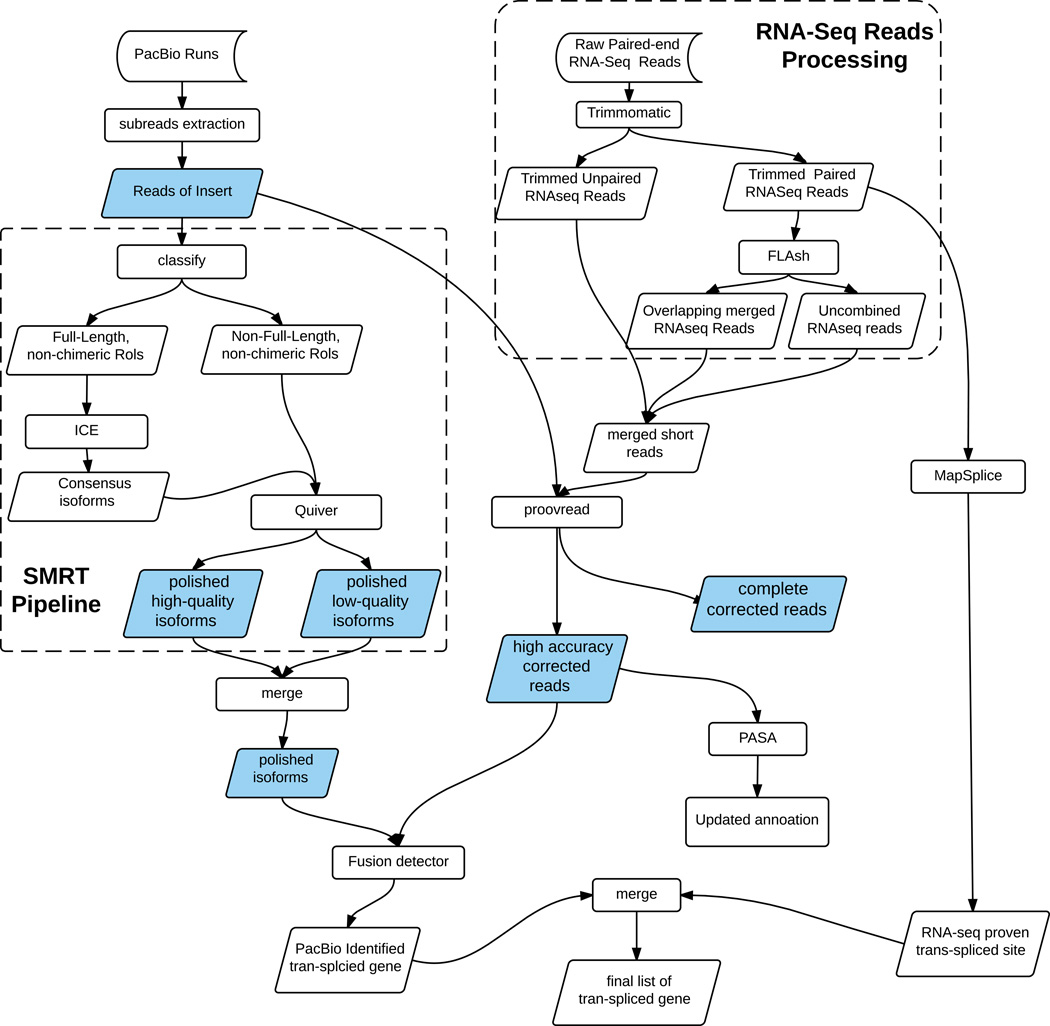
The reads of insert of the three libraries were combined and subjected to error correction. RNA-seq data was processed as shown in Figure 1 to achieve best performance for error correction. First, raw RNA-seq were trimmed with Trimmomatic with parameter “2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36” (Bolger, Lohse, and Usadel 2014). The resulting trimmed paired reads were merged with FLASH with default parameters (Magoč and Salzberg 2011). The merged reads along with reads that failed to merge and unpaired reads from Trimmomatic were combined into one fastq file. This fastq file was of 26 Gb in size and used as short reads to correct the reads of insert with proovread (Hackl et al. 2014). Proovread is a high accuracy PacBio correction tool, which works via iterative alignment of short reads to produce consensus sequences. Proovread outputted high accuracy PacBio reads with low quality regions trimmed as well as complete corrected PacBio reads including poorly corrected regions. Only the high accuracy trimmed Iso-Seq reads were used for our further analysis.
Figure 1.
Data processing and analysis pipelines for both RNA-Seq data and Iso-Seq data. Processed Iso-Seq data that are highlighted in blue are compared in (Table 2).
Fusion transcripts detection with both Iso-Seq data and RNA-seq data
The Anopheles stephensi Indian strain genome version2 was downloaded from Vectorbase (Giraldo-Calderon et al. 2015). Based on our previous research (data not shown), we are able to include majority of Anopheles stephensi Indian genome in five fasta sequences, with each of the sequences representing one chromosomal arm. The five fasta sequences were used as the genome sequence in the fusion transcripts detection analysis. The high accuracy trimmed Iso-Seq reads were aligned to the genome and then processed by the fusion_finder.py script of PacBio pbtranscript-tofu package (https://github.com/PacificBiosciences/cDNA_primer.git). Proovread could potentially remove trans-spliced transcripts when it removed PCR chimeric reads during correcting. Therefore, the unpolished consensus isoforms were added to the above analysis to supplement more reads. MapSplice, a splice junction discovery software was used to predict fusion genes in the RNA-seq data (Wang et al. 2010). With the “—fusion” option, MapSplice performed canonical and semi-canonical fusion junction detection after the RNA-seq reads were aligned to the genome. Only fusion junctions supported by both Iso-Seq and RNA-seq data were used. In total, six splice junctions were identified.
Genome annotation updates
Genome version2 and annotations version2.2 of the Anopheles stephensi Indian strain were downloaded from Vectorbase (Giraldo-Calderon et al. 2015). PASA (Program to Assemble Spliced Alignments) Release r20140417 (http://pasapipeline.github.io/) was used to update the existing annotations using evidence generated from the high accuracy trimmed Iso-Seq reads, and then compared the updated annotation to existing gene structure annotations (Haas et al. 2003). As the high accuracy trimmed Iso-Seq reads kept the transcribed orientation, option “--transcribed_is_aligned_orient” were added when the PASA pipeline were launched.
Acknowledgments
This work was supported by NIH grants AI121284, AI077680, and AI105575, and by the Virginia Agricultural Experimental Station.
References
- Abdel-Ghany Salah E, Hamilton Michael, Jacobi Jennifer L, Ngam Peter, Devitt Nicholas, Schilkey Faye, Ben-Hur Asa, Reddy Anireddy SN. A Survey of the Sorghum Transcriptome Using Single-Molecule Long Reads. Nature Communications. 2016;7 doi: 10.1038/ncomms11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin Konstantin, Koren Sergey, Chin Chen-Shan, Drake James P, Landolin Jane M, Phillippy Adam M. Assembling Large Genomes with Single-Molecule Sequencing and Locality-Sensitive Hashing. Nat Biotech. 2015;33:623–630. doi: 10.1038/nbt.3238. [DOI] [PubMed] [Google Scholar]
- Bolger Anthony M, Lohse Marc, Usadel Bjoern. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudevilla Concha, Serra Dolors, Miliar Angel, Codony Carles, Asins Guillermina, Bach Montserrat, Hegardt Fausto G. Natural Trans-Splicing in Carnitine Octanoyltransferase Pre-mRNAs in Rat Liver. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(21):12185–12190. doi: 10.1073/pnas.95.21.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson Nadia M, Majewski Ian J, Oshlack Alicia. JAFFA: High Sensitivity Transcriptome-Focused Fusion Gene Detection. Genome Medicine. 2015;7(1):43. doi: 10.1186/s13073-015-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn Rainer, Reuter Gunter, Loewendorf Andrea. Transgene Analysis Proves mRNA Trans-Splicing at the Complex mod(mdg4) Locus in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(17):9724–9729. doi: 10.1073/pnas.151268698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douris Vassilis, Telford Maximilian J, Averof Michalis. Evidence for Multiple Independent Origins of Trans-Splicing in Metazoa. Molecular Biology and Evolution. 2010;27(3):684–693. doi: 10.1093/molbev/msp286. [DOI] [PubMed] [Google Scholar]
- Gao Jun Li, Fan Yu Jie, Wang Xiu Ye, Zhang Yu, Pu Jia, Li Liang, Shao Wei, Zhan Shuai, Hao Jianjiang, Xu Yong Zhen. A Conserved Intronic U1 snRNP-Binding Sequence Promotes Trans-Splicing in Drosophila. Genes and Development. 2015;29(7):760–771. doi: 10.1101/gad.258863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras Thomas R. Implications of Chimaeric Non-Co-Linear Transcripts. Nature. 2009;461(7261):206–211. doi: 10.1038/nature08452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo-Calderón Gloria I, Emrich Scott J, MacCallum Robert M, Maslen Gareth, Emrich Scott, Collins Frank, Dialynas Emmanuel, et al. VectorBase: An Updated Bioinformatics Resource for Invertebrate Vectors and Other Organisms Related with Human Diseases. Nucleic Acids Research. 2015;43(D1):D707–D713. doi: 10.1093/nar/gku1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas Brian J, Delcher Arthur L, Mount SM Stephen M, Wortman Jennifer R, Smith Roger K, Hannick Linda I, Maiti Rama, et al. Improving the Arabidopsis Genome Annotation Using Maximal Transcript Alignment Assemblies. Nucleic Acids Research. 2003;31(19):5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackl Thomas, Hedrich Rainer, Schultz Jörg, Förster Frank. Proovread: Large-Scale High-Accuracy PacBio Correction through Iterative Short Read Consensus. Bioinformatics (Oxford, England) 2014;30(21):1–8. doi: 10.1093/bioinformatics/btu392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall Andrew Brantley, Papathanos Philippos-Aris, Sharma Atashi, Cheng Changde, Akbari Omar S, Assour Lauren, Bergman Nicholas H, et al. Radical Remodeling of the Y Chromosome in a Recent Radiation of Malaria Mosquitoes. Proceedings of the National Academy of Sciences. 2016;113(15):201525164. doi: 10.1073/pnas.1525164113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herai Roberto Hirochi, Beleza Yamagishi Michel E. Detection of Human Interchromosomal Trans-Splicing in Sequence Databanks. Briefings in Bioinformatics. 2010;11(2):198–209. doi: 10.1093/bib/bbp041. [DOI] [PubMed] [Google Scholar]
- Hiller Michael, Zhang Zhaiyi, Backofen Rolf, Stamm Stefan. Pre-mRNA Secondary Structures Influence Exon Recognition. PLoS Genetics. 2007;3(11):2147–2155. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt Carson, Yandell Mark. MAKER2: An Annotation Pipeline and Genome-Database Management Tool for Second-Generation Genome Projects. BMC Bioinformatics. 2011 doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi Takayuki, Giniger Edward, Aigaki Toshiro. Alternative Trans-Splicing of Constant and Variable Exons of a Drosophila Axon Guidance Gene, Lola. Genes and Development. 2003;17(20):2496–2501. doi: 10.1101/gad.1137303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Xiaofang, Biedler James K, Qi Yumin, Hall Andrew Brantley, Tu Zhijian. Complete Dosage Compensation in Anopheles Stephensi and the Evolution of Sex-Biased Genes in Mosquitoes. Genome Biology and Evolution. 2015;7(7):1914–1924. doi: 10.1093/gbe/evv115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Xiaofang, Peery Ashley, Hall A Brantley, Sharma Atashi, Chen Xiao-Guang, Waterhouse Robert M, Komissarov Aleksey, et al. Genome Analysis of a Major Urban Malaria Vector Mosquito, Anopheles Stephensi. Genome Biology. 2014;15(9):459. doi: 10.1186/s13059-014-0459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Yimeng, Zhou Hongxia, Yu Yao, Chen Longxian, Hao Pei, Li Xuan. The Evolutionary Landscape of Intergenic Trans-Splicing Events in Insects. Nature Communications. 2015;6 doi: 10.1038/ncomms9734. Nature Publishing Group: 8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasda Erika L, Blumenthal Thomas. Trans-Splicing. Wiley Interdisciplinary Reviews: RNA. 2011;2(3):417–434. doi: 10.1002/wrna.71. [DOI] [PubMed] [Google Scholar]
- Liu Silvia, Tsai Wei-Hsiang, Ding Ying, Chen Rui, Fang Zhou, Huo Zhiguang, Kim SungHwan, et al. Comprehensive Evaluation of Fusion Transcript Detection Algorithms and a Meta-Caller to Combine Top Performing Methods in Paired-End RNA-Seq Data. Nucleic Acids Research. 2015 doi: 10.1093/nar/gkv1234. gkv1234 –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč Tanja, Salzberg Steven L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson Hugh M, Navik Julia A, Walden KKO, Willi Honegger Hans. The Bursicon Gene in Mosquitoes: An Unusual Example of mRNA Trans-Splicing. Genetics. 2007;176(2):1351–1353. doi: 10.1534/genetics.107.070938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Wei, Zhao Qiong-yi, Wang Xiu-ye, Xu Xin-yan, Tang Qing, Li Muwang, Li Xuan, Xu Yong-zhen. Alternative Splicing and Trans -Splicing Events Revealed by Analysis of the Bombyx Mori Transcriptome. 2012:1395–1407. doi: 10.1261/rna.029751.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon Donald, Tilgner Hagen, Grubert Fabian, Snyder Michael. A Single-Molecule Long-Read Survey of the Human Transcriptome. Nature Biotechnology. 2013;31(October):1009–1014. doi: 10.1038/nbt.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Masataka G, Ohbayashi Fumi, Mita Kazuei, Shimada Toru. The Mechanism of Sex-Specific Splicing at the Doublesex Gene Is Different between Drosophila Melanogaster and Bombyx Mori. Insect Biochemistry and Molecular Biology. 2001;31(12):1201–1211. doi: 10.1016/s0965-1748(01)00067-4. [DOI] [PubMed] [Google Scholar]
- Wally Verena, Murauer Eva M, Bauer Johann W. Spliceosome-Mediated Trans-Splicing: The Therapeutic Cut and Paste. Journal of Investigative Dermatology. 2012;132(8) doi: 10.1038/jid.2012.101. Nature Publishing Group: 1959–66. [DOI] [PubMed] [Google Scholar]
- Wang Kai, Singh Darshan, Zeng Zheng, Coleman Stephen J, Huang Yan, Savich Gleb L, He Xiaping, et al. MapSplice: Accurate Mapping of RNA-Seq Reads for Splice Junction Discovery. Nucleic Acids Research. 2010;38(18):e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf M Bryan, Berglund J Andrew. Role of RNA Structure in Regulating Pre-mRNA Splicing. Trends in Biochemical Sciences. 2010;35(3):169–178. doi: 10.1016/j.tibs.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirather Jason L, Afshar Pegah Tootoonchi, Clark Tyson A, Tseng Elizabeth, Powers Linda S, Underwood Jason G, Zabner Joseph, Korlach Jonas, Wong Wing Hung, Au Kin Fai. Characterization of Fusion Genes and the Significantly Expressed Fusion Isoforms in Breast Cancer by Hybrid Sequencing. Nucleic Acids Research. 2015;43(18) doi: 10.1093/nar/gkv562. gkv562 –. [DOI] [PMC free article] [PubMed] [Google Scholar]