Abstract
Acetaminophen toxicity is a leading cause of acute liver failure (ALF). We found that miRNA-122 (miR-122) is down-regulated in liver biopsy specimens of patients with ALF and in acetaminophen-treated mice. A marked decrease in the primary miR-122 expression occurs in mice on acetaminophen overdose because of suppression of its key transactivators, hepatocyte nuclear factor (HNF)-4α and HNF6. More importantly, the mortality rates of male and female liver-specific miR-122 knockout (LKO) mice were significantly higher than control mice when injected i.p. with an acetaminophen dose not lethal to the control. LKO livers exhibited higher basal expression of cytochrome P450 family 2 subfamily E member 1 (CYP2E1) and cytochrome P450 family 1 subfamily A member 2 (CYP1A2) that convert acetaminophen to highly reactive N-acetyl-p-benzoquinone imine. Upregulation of Cyp1a2 primary transcript and mRNA in LKO mice correlated with the elevation of aryl hydrocarbon receptor (AHR) and mediator 1 (MED1), two transactivators of Cyp1a2. Analysis of ChIP-seq data in the ENCODE (Encyclopedia of DNA Element) database identified association of CCCTC-binding factor (CTCF) with Ahr promoter in mouse livers. Both MED1 and CTCF are validated conserved miR-122 targets. Furthermore, depletion of Ahr, Med1, or Ctcf in Mir122−/− hepatocytes reduced Cyp1a2 expression. Pulse-chase studies found that CYP2E1 protein level is upregulated in LKO hepatocytes. Notably, miR-122 depletion sensitized differentiated human HepaRG cells to acetaminophen toxicity that correlated with upregulation of AHR, MED1, and CYP1A2 expression. Collectively, these results reveal a critical role of miR-122 in acetaminophen detoxification and implicate its therapeutic potential in patients with ALF.
Although acetaminophen is a widely used antipyretic and analgesic and is generally safe, it is a dose-related toxin in all mammals and causes approximately 1000 cases of acute liver failure (ALF) annually in the United States.1, 2 N-acetylcysteine (NAC) is a useful antidote for acetaminophen toxicity, but it is effective only if given within first 24 hours of acetaminophen ingestion.1, 3
Conjugation as a glucuronide or sulfate group is the main acetaminophen clearance pathway; a small fraction is metabolized by cytochrome P450 family 2 subfamily E member 1 (CYP2E1) and cytochrome P450 family 1 subfamily A member 2 (CYP1A2) to the highly reactive metabolite N-acetyl-p-benzoquinone imine (NAPQI).4 CYP2E1 is considered the key enzyme involved in acetaminophen bioactivation. However, metabolomics studies in the wild-type and Cyp2e1−/− mice surprisingly revealed that the contribution of CYP2E1 in acetaminophen metabolism decreases with increasing acetaminophen doses.5 Most of the NAPQI is detoxified by conjugation with glutathione (GSH); however, with GSH depletion, NAPQI covalently binds to protein thiols; these (acetaminophen-cysteine) adducts result in activation of Bax, which translocates to the mitochondrial outer membrane, where it forms pores with the help of other proapoptotic Bcl-2 family members, such as Bad, truncated Bid, and Bak.6 Intermembrane proteins, such as cytochrome c, Smac/DIABLO, apoptosis-inducing factor, and endonuclease G, are released through these pores.7 Formation of NAPQI adducts also causes oxidative and nitrative stress, which results in mitochondrial membrane permeability transition.2, 4 Mitochondrial membrane permeability transition causes uncoupling of oxidative phosphorylation, which amplifies oxidative and nitrative stress and results in depletion of ATP reserves. Endonuclease G cleaves cellular DNA; however, because of low ATP reserve, it initiates necrosis instead of apoptosis.4 Reactive oxidative and nitrative stress, as well as damage associated molecular patterns released from necrotic hepatocytes, can contribute to the induction of proinflammatory cytokines, which attract neutrophils to aggravate liver damage.4, 8, 9
miRNA-122 (miR-122) is a liver-specific conserved miRNA that regulates metabolic homeostasis and hepatitis B and C virus replication.10, 11 However, miR-122 loss of function predisposes mice to develop spontaneous steatohepatitis and hepatocellular cancer,12, 13 implicating its essential role in maintaining liver homeostasis. Being abundantly expressed in adult mammals and regulating the expression levels of many targets in hepatic cells,12, 13, 14 it is tempting to speculate that miR-122 also modulates drug toxicity, a major function of the liver.
Circulating miR-122 has been well established as an early and sensitive biomarker for liver injury caused by acetaminophen overdose.15, 16 In fact, serum miR-122, high-mobility group box protein 1 (HMGB1), and cytokeratin-18 are specific biomarkers for acetaminophen toxicity in ALF.15 However, the role of miR-122 in acetaminophen toxicity has not been studied in vivo. In this study, we investigated the role of miR-122 in acetaminophen-related liver toxicity using an experimental model of liver-specific miR-122 knockout (LKO) mice.
Materials and Methods
Mouse Strains, Animal Husbandry, and Treatment with Acetaminophen
The following inbred strains in the C57BL6J background were used: Mir122 LKO (Mir122loxP/loxP albumincyclization recombination+), Mir122 germline knockout (KO) (Mir122−/−), and Mir122loxP/loxP (control).12 All animals were housed in a temperature-controlled room under a 12 hours light/12 hours dark cycle and under helicobacter-free conditions and fed normal chow diet. All animal studies were reviewed and approved by the Ohio State University Institutional Laboratory Animal Care and Use Committee. Both male and female mice were used for experiments. For acetaminophen treatment, 10- to 12-week-old mice were fasted overnight followed by i.p. injection of acetaminophen at indicated dose dissolved in phosphate-buffered saline (PBS) at 37°C or PBS alone. Mice were allowed to have food and euthanized at indicated time points by CO2 asphyxiation. Blood was collected by cardiac puncture, and livers were isolated for histologic and molecular analysis. Unless mentioned otherwise, three to five male mice of each genotype were used for each treatment and time point.
Serologic Analysis, Histologic Analysis, and Scoring Necrosis
Alanine aminotransferase (ALT) level was measured in serum and primary hepatocyte culture supernatant using an ALT kit (catalog number A524-150; Teco Diagnostics, Anaheim, CA) following the manufacturer's protocol.
For histologic analysis, tissues were fixed in 4% paraformaldehyde and embedded in paraffin. Hematoxylin and eosin (H&E) staining of liver sections was performed as described,12 and H&E-stained slides were scored for necrosis in using the following scoring system: 0, no necrosis; 1, mild necrosis; 2, moderate necrosis; 3, severe necrosis; 4, 70% to 80% liver is necrotic; and 5, whole liver is necrotic. A pathologist expert (W.M.L.) in drug-induced liver injury blinded to the experimental design performed necrosis scoring. Fisher exact tests were used to verify whether acetaminophen treatment caused significantly higher necrosis scores in LKO mice than in control mice at different time points. Holm's procedure was used for the adjustment of multiple comparisons.
NAPQI Quantification
NAPQI-cysteine adducts in livers and sera of control and LKO mice were measured using the high-performance liquid chromatography–electrochemical detection method.17
RNA Isolation and Northern Blot Analysis
The whole liver and serum RNAs (for Northern blot analysis) were isolated using TRIzol (catalog number 15596026; Thermo Fisher Scientific, Pittsburgh, PA) following the manufacturer's protocol and subjected to Northern blot analysis using 32P-labeled antisense miR-122 or an antisense 5S-rRNA oligo as described before.18
Real-Time RT-PCR
TaqMan miRNA Assay (Applied Biosystems, Foster City, CA) was used to quantify mature (catalog number 002245) and primary miR-122 expression (catalog number Mm03306556_pri) in DNase 1–treated liver, hepatocyte, or cellular RNA according to the manufacturer's instructions. Normalization was performed with Rnu6B (for miR-122) and 18S rRNA (for pri–miR-122). For mRNA expression analysis, DNase 1–treated total RNA was reverse transcribed into cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems), and real-time PCR was performed using the SYBR Green assay method. The expression was normalized to that of Gapdh, α-actin, or 18S rRNA. All real-time reactions, including controls without cDNA, were run in triplicate in a thermocycler (Bio-Rad, Hercules, CA). Relative expression was calculated using the comparative threshold cycle method. Primer sequences are provided in Table 1. Mir-122loxP/loxP and cyclization recombination genotyping primers and PCR conditions are described earlier.12
Table 1.
RT-PCR Primer Used in This Study
| Gene symbol | Primer sequence |
|---|---|
| Mouse | |
| Ctcf | Forward: 5′-GCCCTCTCTCTTGATCGTAAAC-3′ |
| Reverse: 5′-GTCCTTCCCAAGTCTGCATTTA-3′ | |
| Cyp2e1 | Forward: 5′-ATAGAAGTTGGAACCTGCCCCC-3′ |
| Reverse: 5′-AGCACCTCCTTGACAGCCTTGTAG-3′ | |
| Cyp1a1 | Forward: 5′-TGAGGCAGGGAGATGACTTCAAG-3′ |
| Reverse: 5′-TGACATTGGCTACTGACACGACC-3′ | |
| hnCyp1a2 | Forward: 5′-GATAAACGGAAGGCTCAGAATCC-3′ |
| Reverse: 5′-CCAGAACACTAAACAGAAGATGGCAG-3′ | |
| Cyp1a2 | Forward: 5′-CCGCTCGAGGCAGGGAGATGACTTCAAGGG-3′ |
| Reverse: 5′-AACTATGTTTAAACTGATGGCTGTGGTGACTGTGTC-3′ | |
| hnAhr | Forward: 5′-GGCCAGGACCAGTGTAGAGC-3′ |
| Reverse: 5′-CTTCTAATCCCATCTGCCAGG-3′ | |
| Ahr | Forward: 5′-GAATGGCTTTGTGCTGGTTGTC-3′ |
| Reverse: 5′-TCCGTCCTTCCCTTTCTTGTTC-3′ | |
| Med1 | Forward: 5′-TATGCCTCTCCATCTGACCTGCTG-3′ |
| Reverse: 5′-CAAACAACGGGATTCCTGTGC-3′ | |
| Actin | Forward: 5′-ACAACGGCTCCGGCATGT-3′ |
| Reverse: 5′-TCTTGCTCTGGGCCTCGTCAC-3′ | |
| Gapdh | Forward: 5′-TCCTGCACCACCAACTGCTTAG-3′ |
| Reverse: 5′-TGCTTCACCACCTTCTTGATGTC-3′ | |
| 18S | Forward: 5′-TGACGGAAGGGCACCACCAG-3′ |
| Reverse: 5′-TCGCTCCACCAACTAAGAACGGC-3′ | |
| Human | |
| CTCF | Forward: 5′-TACAAACACACCCACGAGAAG-3′ |
| Reverse: 5′-CTCCAGTATGAGAGCGAATGTG-3′ | |
| AHR | Forward: 5′-TCAAGTCAAATCCTTCCAAGCG-3′ |
| Reverse: 5′-CCGTTTCTTTCAGTAGGGGAGG-3′ | |
| MED1 | Forward: 5′-GGAAACCGAGGAGTCAGAAAAGC-3′ |
| Reverse: 5′-TGAGATGAGAGCCCAGTCCATTC-3′ | |
| CYP1A2 | Forward: 5′-TTCATCCTGGAGACCTTCCGAC-3′ |
| Reverse: 5′-TTGACCTGCCACTGGTTTACG-3′ | |
| CYP2E1 | Forward: 5′-GAACTTCCACCTACTCAGCACTCC-3′ |
| Reverse: 5′-TTGGGGTCCAGAGATTGATGG-3′ | |
miRNA Mimics and Inhibitors
miRNA mimics and inhibitors used in this study are as follows: miRIDIAN mimic negative control (NC) (catalog number CN-001000-01), miRIDIAN mimic mmu–miR-122 mimic (catalog number C-310464-07), miRIDIAN hairpin inhibitor NC (catalog number IN-001005-01), and miRIDIAN hairpin inhibitor mmu–miR-122 antisense (catalog number IH-310464-08; Dharmacon, Lafayette, CO).
siRNAs
siRNAs used in this study are as follows: si-Ahr (catalog number M-044066-01) and si-Med1 and si-NC (catalog number D-001206-13; Dharmacon).
TaqMan Probes for Real-Time RT-PCR
TaqMan probes for real-time RT-PCR used in this study are as follows: mmu–miR-122 (catalog number 002245) and RNU6b (catalog number 001093; Life Technologies, Carlsbad, CA).
Luciferase Assay
Amplification and cloning of 3′-untranslated regions (3′-UTRs) to psiCHECK2 (Promega, Madison, WI) were performed as previously described.1 CTCF and MED1 3′-UTRs harboring miR-122–binding sites were cloned into psiCHECK-2 (catalog number C8021; Promega) at the 3′-UTR region of Renilla luciferase at XhoI and NotI sites. psiCHECK-2 constructs were co-transfected with either control scramble RNA or miR-122 mimic RNA (25 nmol/L) by Lipofectamine 3000 in Hepa1-6 or HEK293T cells. Luciferase activities were measured by the Dual-Luciferase Reporter Assay System (catalog number E1980; Promega) 48 hours after transfection. Primer sequences used for luciferase assay are provided in Table 2.
Table 2.
Primers Used for Luciferase Assay
| Gene symbol | Primer sequence |
|---|---|
| Ctcf-luc | Forward: 5′-CCGCTCGAGTTGCTCGGCACCAGGACTATTG-3′ |
| Reverse: 5′-AAGAATGCGGCCGCAAGGTTCAAACGCTGGCTCC-3′ | |
| Ctcf-mut-luc | Forward: 5′-CTCTCTCTTGATCGTAAACCTGAGGTGAACGGCCACGGGTTTCCC-3′ |
| Reverse: 5′-GGGAAACCCGTGGCCGTTCACCTCAGGTTTACGATCAAGAGAGAG-3′ | |
| Med1-luc | Forward: 5′-CCGCTCGAGAAAGTCTACATCCCAGCATT-3′ |
| Reverse: 5′-ATAAGAATGCGGCCGCTTTTAAGTGAACTATGGCTG-3′ | |
| Med1-mut-luc | Forward: 5′-TTCAGACTTAAATGTGAGGTCTGTATGCTC-3′ |
| Reverse: 5′-GGGGAGCATACAGACCTCACATTTAAGTCT-3′ |
RNA Isolation and Northern Blot Analysis
The whole liver and serum RNAs (for Northern blot analysis) were isolated using TRIzol (catalog number 15596026; Thermo Fisher Scientific, Pittsburgh, PA) following the manufacturer's protocol and subjected to Northern blot analysis using 32P-labeled antisense miR-122 or an antisense 5S-rRNA oligo as previously described.18
Western Blot Analysis
Whole cell or tissue extracts were prepared in the lysis buffer [50 mmol/L Tris HCl (pH 8.1), 10 mmol/L EDTA (pH 8.0), and 1% SDS] containing protease inhibitor cocktails (Sigma, St. Louis, MO) followed by sonication to shear DNA. Liver nuclei were isolated using sucrose density gradient centrifugation as previously described,19 and nuclear proteins were extracted in SDS lysis buffer. These extracts (20 to 50 μg of protein) were subjected to immunoblotting with specific antibodies, and the signal was developed with ECL reagent (Thermo Fisher Scientific) on X-ray film or the Odyssey CLx Imaging System (LI-COR, Lincoln, NE) after incubation with appropriate secondary antibodies. Western blot signals were quantified by ImageJ software version 1.48 (NIH, Bethesda, MD; http://imagej.nih.gov/ij) following the online manual for X-ray film or Image Studio Software version 5.2 for the Odyssey CLx Imaging System (LI-COR). The catalog number and source of antibodies used are provided in Table 3. Protein concentrations were measured using Protein Assay Dye Reagent (catalog number 5000006; Bio-Rad) with bovine serum albumin as the standard.
Table 3.
Antibodies Used in This Study
| Antibody | Vendor | Catalog no. |
|---|---|---|
| CYP2E1 | Abcam, Cambridge, MA | ab28146 |
| GAPDH | EMD Millipore, Billerica, MA | MAB374 |
| CYP1A2 | Santa Cruz Biotechnology, Dallas, TX | sc-53241 |
| AHR | Santa Cruz Biotechnology, Dallas, TX | sc-5579 |
| MED1 | Santa Cruz Biotechnology, Dallas, TX | sc-8998 |
| Actin | Santa Cruz Biotechnology, Dallas, TX | sc-8432 |
| HNF4α | Santa Cruz Biotechnology, Dallas, TX | sc-8987 |
| HNF6 | Santa Cruz Biotechnology, Dallas, TX | sc-13050 |
| Ku-86 | Santa Cruz Biotechnology, Dallas, TX | sc-1484 |
| Ki-67 | Cell Signaling Technology, Danvers, MA | 12202 |
| Anti-mouse IgG, HRP linked | GE Life Sciences, Pittsburgh, PA | NA931V |
| Anti-rabbit IgG, HRP linked | GE Life Sciences, Pittsburgh, PA | NA934V |
AHR, aryl hydrocarbon receptor; CYP1A2, cytochrome P450 family 1 subfamily A member 2; CYP2E1, cytochrome P450 family 2 subfamily E member 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HNF, hepatocyte nuclear factor; HRP, horseradish peroxidase; MED1, mediator 1.
Cell Lines and Transfection
HepG2, Hep3B, and H293T cell lines were obtained from ATCC (Manassas, VA) and cultured as recommended by the supplier. Drs. Gretchen Darlington and James Taylor provided Hepa1-6 and Huh7 cells, respectively, and Dr. Bo Yu provided MHCCLM3 and MHCC97H cells. Cells culture were transfected with 50 nmol/L miRIDIAN mimic mmu–miR-122 (catalog number C-310464-07) or NC RNA (catalog number CN-001000-01) using Lipofectamine 3000 (catalog number L3000015; Thermo Fisher Scientific). Transfections were performed as described for siRNA. Hepa1-6 cells were transfected with 50 nmol/L miRIDIAN hairpin inhibitor specific for mmu–miR-122 (catalog number IH-310464-08) and control inhibitor (catalog IN-001005-01). Cells were harvested 48 to 72 hours after transfection for luciferase assay or RNA and protein isolation.
Immunohistochemical Analysis
For immunohistochemical analysis, slides were dewaxed and exposed to 95°C for 30 minutes for antigen retrieval. After incubation of liver sections with antibodies, the color was developed by the diaminobenzidine method and counterstained with hematoxylin.
Tissues from Patients with ALF
Frozen liver tissues from patients with ALF, all with nonacetaminophen causes (8 unknown, 1 mushroom), were obtained from the Department of Gastroenterology, Hepatology, and Endocrinology, Hannover Medical School, Hannover, Germany. Human specimens (blood and tissues) were collected after informed consent from patients with ALF. The study was performed as per the guidelines of and was approved by the ethics committee of Hannover Medical School. Four normal human liver biopsy specimens served as controls. The serum ALT, aspartate transaminase, and international normalized ratio values of these patients are provided in Supplemental Table S1.
Hepatocyte Isolation, in Vitro Culture, Transfection, and Treatment with Acetaminophen
Hepatocytes were isolated as described previously.20 Briefly, miR-122 KO or wild-type mice (25 to 30 g) were anesthetized with ketamine and xylazine was injected i.p. Livers were perfused with 30 mL of perfusion buffer (catalog number 17701038; Thermo Fisher Scientific) (5 mL/minute) and then with 50 mL of warm (37°C) liver digestion buffer (catalog number 17703034; Thermo Fisher Scientific) via the portal vein. The livers were aseptically removed to a sterile Petri dish that contained Dulbecco's modified Eagle's medium at 4°C to stop digestion. The hepatocytes were released by peeling off the hepatic capsule and dispersed by shaking the digested liver in Dulbecco's modified Eagle's medium at 4°C, followed by passing through a 70-μm strainer and collected by centrifugation at 50 × g for 3 minutes at 4°C. Hepatocytes were suspended in hepatocyte wash media (catalog number 17704024; Thermo Fisher Scientific) and subjected to purification on 90% Percoll density gradient at 200 × g for 10 minutes. Purified hepatocytes were washed twice in hepatocyte wash media before suspending in Williams' E medium supplemented with 5% serum and 1 mmol/L glutamine. The hepatocytes were counted, and viability was determined by trypan blue dye exclusion. The cells were plated on 6-well plate coated with rat tail type I collagen (BD Bioscience, San Jose, CA) at a density of 1 × 106 cells per well. For gene transfection, hepatocytes were washed the next day with PBS and incubated in HepatoZYME-SFM media (catalog number 17705021; Thermo Fisher Scientific). Lipofectamine 3000 (catalog number L3000015; Thermo Fisher Scientific) was used to transfect primary hepatocytes with scrambled siRNA (catalog number D-001206-13-05; Thermo Fisher Scientific) or gene-specific siRNAs. After 6 to 8 hours, media was replaced with HepatoZYME-SFM supplemented with 1.25 μg/cm2 of collagen I to provide a sandwich matrix. For acetaminophen treatment, hepatocytes were washed with PBS after 4 hours of plating and incubated with Williams' E media. Primary hepatocytes were treated with acetaminophen or PBS for different time points.
Estimation of Concentrations of GSH and Oxidized GSH Metabolites in the Liver by Nuclear Magnetic Resonance
Each frozen liver tissue with exact weight (within the 150- to 200-mg range) was recorded for nuclear magnetic resonance (NMR) data normalization. Frozen liver tissues were thawed on ice in 300 μL of cold acetonitrile and 300 μL of cold milliQ water for 15 minutes and homogenized at 4°C in Bullet Blender (Next Advance Model Gold BB24-AU), with 300 μL of 0.5-mm zirconium oxide beads at level 8 for 3 minutes and repeated once. Next, 200 μL of cold water was added to each sample and subjected to centrifugation at 13,000 × g for 15 minutes at 4°C to remove tissue debris and beads. The supernatant was then transferred to a 15-mL conical tube, and 700 μL of cold acetonitrile was added to each tube. Residual tissue debris was pelleted by centrifugation at 5000 × g for 20 minutes at 4°C in a swinging rotor. The supernatant was then transferred to a fresh 15-mL tube, and an additional 1 mL of cold acetonitrile and 0.55 mL of cold water were added. A total of 2 mL of cold chloroform was then added to each tube followed by vigorous vortexing and centrifugation at 5000 × g for 20 minutes at 4°C for phase separation. The aqueous phase was then transferred to a fresh tube and temporarily stored at −20°C. A total of 1.1 mL of cold water was added to the organic phase (to ensure the complete recovery of polar metabolites), followed by vortexing and centrifugation with the same parameters used for phase separation. The aqueous phase was then transferred and combined with the corresponding previous aqueous phase, which was then lyophilized and resuspended in 600 μL of 50 mmol/L sodium phosphate buffer (pH 7.4) in D2O and transferred to a 5-mm NMR tube.
Two-dimensional 13C-1H HSQC spectra were collected at 298 K on a Bruker 850 MHz AVANCE III spectrometer equipped with a cryogenically cooled TCI probe. A total of 512 × 2048 (N1 × N2) complex points were collected with 16 scans per increment. All NMR data were zero-filled, Fourier transformed, and phase-corrected using NMRPipe. Identification of metabolites from the two-dimensional NMR spectra was performed using the online COLMAR database query (http://spin.ccic.ohio-state.edu/index.php/colmar, last accessed June 28, 2017). The concentrations of GSH and oxidized GSH (GSSG) in the NMR tube (total volume of 600 μL) were calculated from peak intensities by comparison with standard spectra of 1 mmol/L GSH and 1 mmol/L GSSG, respectively. The absolute amounts of GSH and GSSG were then calculated and divided by the weight of each tissue sample as reported.
Treatment of Hepatocytes with Cycloheximide, Puromycin, and MG-132
Primary hepatocytes were isolated from WT and LKO mice and plated in collagen-coated 12-well plates at 80% confluence in Williams' E media supplemented with 5% serum and 1 mmol/L glutamine. The next day cells were washed with PBS and incubated in HepatoZYME-SFM media. Cells were treated with PBS, cycloheximide (20 μg/mL) or puromycin (40 μg/mL), and dimethyl sulfoxide or MG-132 (10 μg/mL) and harvested at different time points for protein extraction and immunoblotting.
Pulse-Chase Studies in Hepatocytes in Culture to Measure the Half-Life of CYP2E1
For measuring the half-life of Cyp2e1, primary hepatocytes were isolated from WT and LKO mice. Cells were plated in collagen-coated 6-well plate at 80% confluency in Williams' E media supplemented with 5% serum. The next day cells were washed with PBS three times and incubated for 2 hours in methionine- and cysteine-free DMEM that contained 10% dialyzed fetal bovine serum. A total of 100 μCi of 35S-methionine was added to each well, and after 1 hour cells were washed with PBS and harvested (0 hours) or incubated with unlabeled media and harvested at different time points. Whole cell lysates in radioimmunoprecipitation assay buffer were subjected to immunoprecipitation with a Cyp2e1 antibody. Immunoprecipitated proteins were resolved by SDS-PAGE, transferred to nitrocellulose membrane, and subjected to autoradiography and immunoblotting.
TUNEL Assay
Paraffin embedded liver sections were deparaffinized and rehydrated for antigen retrieval. Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay was performed following manufacturer's protocol (catalog number 0-17-141; EMD Millipore, Billerica, MA).
JNK Activity Assay
c-Jun–N-terminal protein kinase (JNK) activity was measured in the liver extracts using GST-c-JUN peptide and Ɣ32P-ATP as substrates as previously described.21
Statistical Analysis
Real-Time RT-PCR and transfection assays were performed in triplicate. The data are presented as means ± SD. Most of the experiments were performed at least three times with multiple replicates. Two-sample t-tests were used for the other two group comparisons. Statistical analysis of mouse survival was performed using the log-rank test after taking into consideration multiple comparisons with Holm's procedure. P < 0.05 is considered significant.
Results
Acetaminophen Overdose Suppresses mir-122 Expression
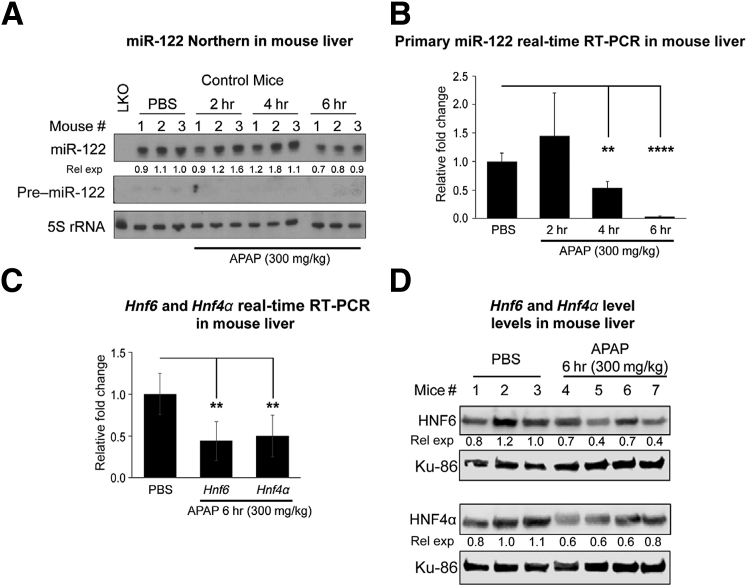
To determine whether acute liver injury modulates the miR-122 expression, we first measured its level in the livers of patients with ALF and controls. Significant down-regulation of miR-122 levels was observed in ALF biopsy specimens compared with non-ALF controls (Supplemental Figure S1A), suggesting that acute liver injury suppresses miR-122 expression. To determine whether drug toxicity modulates liver miR-122 expression in mice, we measured hepatic miR-122 levels in miR-122loxP/loxP (control) mice treated with acetaminophen (300 mg/kg). Six hours after administration of acetaminophen, there was an approximately 40% reduction of miR-122 levels (Figure 1A and Supplemental Figure S1B), whereas highly unstable precursor miR-122 was detectable only in PBS-injected mice. Acetaminophen toxicity also down-regulated primary miR-122 levels as early as 4 hours after injection (Figure 1B), which correlated with a decrease in expression of hepatocyte nuclear factor (HNF)-4α and HNF6, two key transcriptional activators of the Mir122 gene at the RNA (Figure 1C) and protein levels (Figure 1D). The decrease in their protein levels, especially of HNF6, was evident as early as 2 hours after acetaminophen overdose (Supplemental Figure S1C). These results suggest that acetaminophen overdose inhibits miR-122 expression by suppressing these two key transcription factors.
Figure 1.
miRNA-122 (mir-122) expression is down-regulated in mouse livers on acetaminophen (APAP) overdose. A: Northern blot analysis of precursor and mature miR-122 in livers of control (Mir122loxP/loxP) mice injected with acetaminophen or phosphate-buffered saline (PBS). B: Normalized primary miR-122 expression in livers of PBS- or acetaminophen-injected animals for indicated hours. C and D: Relative expression (rel exp) of Hnf6 and Hnfα RNA normalized to Gapdh (C) and proteins normalized to Ku-86 (D) in mice treated with acetaminophen for 6 hours. Data are expressed as means ± SEM. n = 3 to 5 mice of each genotype in each treatment group. ∗∗P < 0.01, ∗∗∗∗P < 0.0001 (two-tailed t-test). HNF, hepatocyte nuclear factor; LKO, liver specific miR-122 knockout.
As reported earlier,22 we also noted that the increase in serum miR-122 occurred as early as 2 hours of acetaminophen toxicity and was more pronounced than the elevation in serum ALT levels (Supplemental Figure S1D). To determine whether all three variants of miR-122 (21 to 23 nucleotides) are released into circulation after acetaminophen overdose, we performed Northern blotting that revealed the presence of all three forms, among which the 23 nucleotides variant was most abundant in the serum of acetaminophen-injected control mice (Supplemental Figure S1E).
Loss of miR-122 Sensitizes Mice to Acetaminophen Overdose
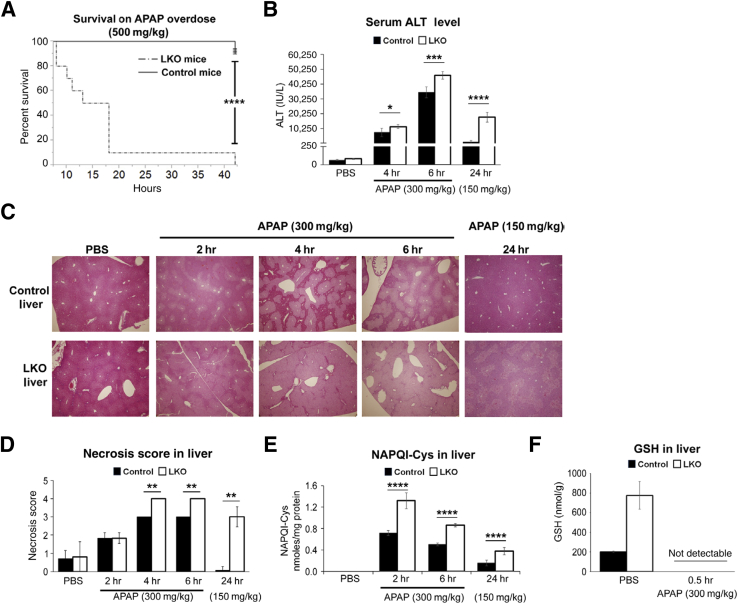
Because the liver is the major organ involved in drug metabolism and acetaminophen overdose suppresses miR-122 expression, we were curious to know whether the loss of miR-122 would modulate sensitivity to acetaminophen overdose. To this end, we treated ten 12-week-old male mice with acetaminophen (500 mg/kg), which is lethal only to LKO mice, and monitored their survival. Nine LKO mice died within 19 hours of acetaminophen injection, and the remaining one died within 43 hours, whereas only 1 of 10 control mice died within this period (Figure 2A). We also observed higher mortality of male LKO mice at lower dose of acetaminophen (300 mg/kg) compared with control male mice given the same dose (Supplemental Figure S2A). Higher mortality rate (P < 0.0001) of LKO mice correlated with increased liver damage, as demonstrated by elevated serum ALT levels in LKO mice after 4 hours (P = 0.04) and 6 hours (P = 0.002) of acetaminophen (300 mg/kg) exposure, which was more remarkable (12.7-fold more than the control mice) (P = 1.84 × 10–6) after 24 hours of drug (150 mg/kg) treatment (Figure 2B). A reduced dose of acetaminophen was used for 24 hours of treatment because LKO mice exhibited mortality within 24 hours at 300 mg/kg if acetaminophen (data not shown). Higher serum ALT levels in acetaminophen-treated LKO mice correlated with increased centrilobular necrosis compared with control mice (representative H&E-stained liver sections shown in Figure 2C). Scoring necrotic areas in liver sections of mice treated with acetaminophen (300 mg/kg) showed excessive hepatocyte degeneration in LKO mice compared with the controls at 4 and 6 hours (Figure 2D). More pronounced necrosis occurred in LKO mice compared with control mice treated with acetaminophen (150 mg/kg) for 24 hours correlating with increased serum ALT levels (Figure 2B). Furthermore, significant elevation of NAPQI-cysteinyl adducts with cellular proteins both in the liver (Figure 2E) and serum of the mutant mice (Supplemental Figure S2B) correlated with increased susceptibility of the mutant mice to acetaminophen overdose.
Figure 2.
Liver-specific miRNA-122 (miR-122) knockout (LKO) mice are more susceptible to acetaminophen (APAP)-induced mortality and hepatotoxicity. A: Kaplan-Meier survival curve of control and LKO mice injected with acetaminophen. The mortality rate of overnight fasted mice injected with acetaminophen was monitored. B: Serum alanine aminotransferase (ALT) levels in acetaminophen overdosed animals. C: Representative images of hematoxylin and eosin (H&E)–stained liver sections of mice treated with acetaminophen for different hours. D: Necrosis scores were evaluated from H&E-stained liver sections. E and F: The levels of the hepatic N-acetyl-p-benzoquinone imine (NAPQI)–cysteine (Cys) adduct (E) and glutathione (GSH) (F) in acetaminophen overdosed mice. Data are expressed as means ± SEM. n = 10 of each genotype (A); n = 4 to 5 of each genotype (B and D–F). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 (two-tailed t-test). Original magnification, ×40 (C). PBS, phosphate-buffered saline.
We also quantified hepatic GSH levels by NMR in LKO and control mice treated with acetaminophen (Supplemental Figure S2C). NAPQI is detoxified primarily by reacting with GSH, thereby depleting cellular GSH. To our surprise, we found that the basal GSH level is more than threefold higher in the PBS-injected LKO mice than in wild-type mice. However, it gets depleted in both genotypes within 30 minutes of acetaminophen treatment (Figure 2F and Supplemental Table S2), suggesting rapid depletion of GSH in the LKO livers, which correlates with higher NAPQI levels in these mice (Figure 2E). These results indicate that although LKO livers have substantially higher GSH levels, GSH gets rapidly depleted because of increased NAPQI formation.
The increase in the phosphorylation of JNK, a marker of cellular stress response,23 was also observed in LKO livers in acetaminophen overdosed mice (Supplemental Figure S3A). Notably, JNK activity remained higher in LKO mice even after 24 hours of acetaminophen treatment (Supplemental Figure S3B), indicating hepatocyte injury and death. TUNEL assay also revealed a greater number of hepatocytes with DNA strand breaks in LKO mice treated with acetaminophen for 24 hours (Supplemental Figure S4A). Increased hepatocyte death also resulted in more facilitated proliferation in LKO mice, as demonstrated by the higher population of Ki-67–positive hepatocytes in acetaminophen-treated LKO mice (Supplemental Figure S4B). We also observed higher mortality of female LKO mice (P < 0.0016) compared with control female mice subjected to acetaminophen toxicity (Supplemental Figure S5A). Lethal acetaminophen dose (750 mg/kg) in female LKO mice was higher than in males (500 mg/kg) because female mice are relatively resistant to the drug.24 Taken together, these results suggest a protective role of miR-122 against acute acetaminophen hepatotoxicity.
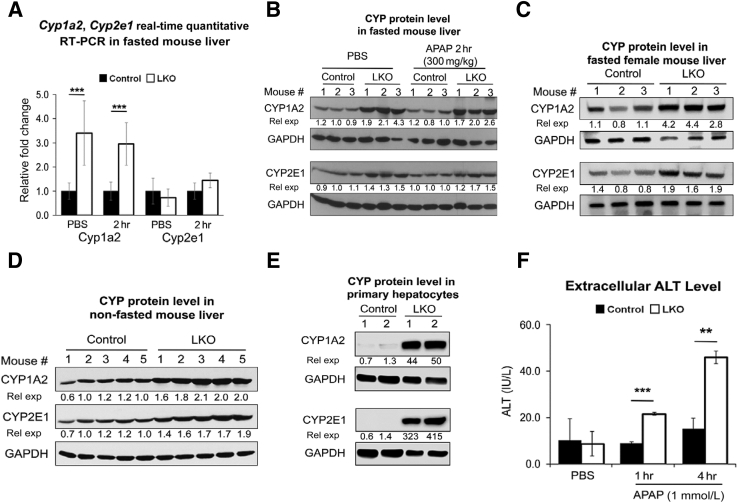
Hepatic Cyp1a2 and Cyp2e1 Levels Are Increased in miR-122 LKO Mice
Next, we sought to elucidate the mechanism of increased susceptibility of LKO mice to acetaminophen overdose initially focusing on CYP2E1 and CYP1A2, two key enzymes that oxidize acetaminophen to toxic NAPQI. To this end, we first looked at the microarray data of 8- to 10-week-old mouse livers (https://www.ncbi.nlm.nih.gov/geo; accession number GSE20610), which revealed higher basal Cyp1a2 mRNA level in LKO mice compared with controls (1.685 fold, P = 3.075 × 10−7) (GSE20610) without significant changes in Cyp2e1 (1.071-fold, P = 0.13). Indeed, Cyp1a2 but not Cyp2e1 RNA level was higher in LKO mice compared with control mice injected with PBS or acetaminophen (300 mg/kg) for 2 hours (Figure 3A). Surprisingly, both CYP1A2 and CYP2E1 proteins were elevated in LKO livers relative to controls injected with PBS and remained high even after acetaminophen treatment (Figure 3B and Supplemental Figure S5B), suggesting posttranscriptional regulation of CYP2E1 in the mutant livers. Basal levels of these two Cyp450s were also higher in the livers of nonfasted LKO male mice and in fasted female LKO mice compared with respective controls, suggesting that miR-122 regulates their expression in both sexes irrespective of the food intake (Figure 3, C and D, and Supplemental Figure S5, C and D). Furthermore, LKO hepatocytes in culture also showed higher CYP2E1 and CYP1A2 levels compared with the controls (Figure 3E), and were more susceptible to acetaminophen toxicity, as demonstrated by elevated ALT levels in culture supernatants (Figure 3F) and increased hepatocyte death (Supplemental Figure S5E). These results suggest that upregulation of these two Cyp450s that increases NAPQI level (Figure 2E) is involved in the higher susceptibility of LKO mice to acetaminophen overdose.
Figure 3.
Basal cytochrome P450 family 2 subfamily E member 1 (CYP2E1) and cytochrome P450 family 1 subfamily A member 2 (CYP1A2) levels are higher in liver-specific miRNA-122 (miR-122) knockout (LKO) livers compared with the controls liver. A and B: Hepatic Cyp1a2 and Cyp2e1 mRNA (A) and protein (B) levels in fasted male mice. C and D: Hepatic CYP protein levels in fasted female (C) and nonfasted male (D) mice. E: CYP expressions in hepatocytes in culture for 12 hours. F: Alanine aminotransferase (ALT) levels in culture supernatants of hepatocytes treated with phosphate-buffered saline (PBS) or acetaminophen for 6 hours. Hepatocytes from 2 mice of each genotype (age and sex-matched) were used. Data are expressed as means ± SEM. n = 3 to 5 of each genotype (A). ∗∗P < 0.01, ∗∗∗P < 0.001 (two-tailed t-test). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; rel exp, relative expression.
Derepression of Aryl Hydrocarbon Receptor and Mediator 1 Contributes to the Upregulation of Hepatic Cyp1a2 in miR-122 LKO Mice
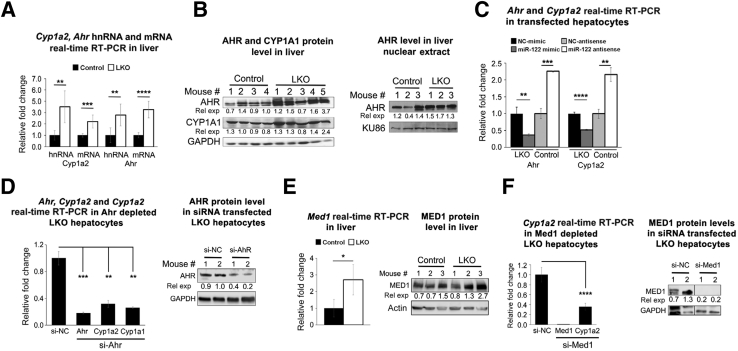
To gain insight into the underlying mechanism of higher basal CYP1A2 and CYP2E1 levels in miR-122 depleted liver, we initially searched different miRNA target prediction databases. However, none of those predicted these two Cyp450s as miR-122 targets, suggesting their indirect regulation by miR-122. Hence, we hypothesized that the loss of miR-122 activates the Cyp1a2 gene by modulating expression or activity of a key transcription factor(s) and up-regulates the CYP2E1 protein by enhancing its translation or stability. To test this notion, we initially measured primary transcript or heteronuclear RNA (hnRNA), which showed upregulation of Cyp1a2 hnRNA in LKO livers relative to the controls (Figure 4A), suggesting transcriptional activation of Cyp1a2 and/or stabilization of the primary transcript.
Figure 4.
Expressions of aryl hydrocarbon receptor (AHR), a key transactivator of cytochrome P450 family 1 subfamily A member 2 (CYP1A2), and mediator 1 (MED1), a transcriptional co-activator are upregulated in liver-specific miRNA-122 (miR-122) knockout (LKO) livers. A: Expression of primary transcript (hnRNA) and mRNA of Cyp1a2 and Ahr in livers. B: AHR and cytochrome P450 family 1 subfamily A member 1 (CYP1A1) protein levels in liver lysates (left panel) and AHR and Ku-86 (normalizer) levels in nuclear extracts (right panel). C: Relative expression (rel exp) of Ahr and Cyp1a2 in Mir122−/− hepatocytes transfected with miR-122 mimic (122-S) or scrambled (NC-S) RNA and control hepatocytes transfected with miR-122 inhibitor or NC inhibitor 48 hours after transfection. D:Ahr and Cyp1a2 expression (left panel) and AHR protein level (right panel) in Mir122−/− hepatocytes transfected with Ahr siRNA (50 nmol/L). Expression of Ahr hnRNA and Cyp1a1 mRNA (normalized to Gapdh) and miR (normalized to Rnu6b). E:Med1 mRNA expression (left panel) and protein level (right panel) in livers. F:Cyp1a2 expression (left panel) and MED1 protein level (right panel) in miR-122−/− hepatocytes transfected with Med1 siRNA. Hepatocytes from 2 mice of each genotype (age and sex matched) were used for Western and real-time RT-PCR(C, D, and F). Data are expressed as means ± SEM. n = 4 to 5 of each genotype (A and E). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 (two-tailed t-test).
To elucidate the underlying mechanism of up-regulation of Cyp1a2 in LKO livers, we revisited our microarray data, which revealed a small but significant increase in RNA levels of aryl hydrocarbon receptor (AHR), a widely studied transactivator of Cyp1a2 and Cyp1a1,25 in LKO livers than in control livers (1.3-fold, P = 0.0055). On binding to its ligands (endogenous or xenobiotics), AHR gets translocated to the nucleus and dimerizes with the co-activator, AHR nuclear translocator, to occupy Ahr response elements on target gene promoters to induce their expression. AHR activates both Cyp1a1 and Cyp1a2 genes on binding to several Ahr response elements located between these two divergent genes. We hypothesized that upregulation of Ahr in miR-122 depleted livers might play a causal role in Cyp1a2 activation. Indeed, both RNA (3.3-fold) and protein (1.74-fold) levels of AHR (Figure 4, A and B), as well as CYP1A1 (1.4-fold) (Figure 4B), were elevated in LKO livers compared with controls. As expected, AHR levels in liver nuclear extracts were also elevated in LKO mice (Figure 4B). To find out whether miR-122 can modulate Ahr and Cyp1a2 expression, we transfected Mir-122−/− hepatocytes in culture with miR-122 mimic or scrambled mimic and control hepatocytes with anti–miR-122 or anti-NC (NC-AS) 6 hours after plating and measured their expression after 48 hours. We observed that ectopic miR-122 suppressed Ahr and Cyp1a2 RNA level in Mir-122−/− hepatocytes, whereas both were up-regulated in the control hepatocytes depleted of miR-122 (Figure 4C). To evaluate AHR's role in Cyp1a2 regulation, we knocked down its expression in Mir-122−/− hepatocytes by transfecting siRNA, which resulted in 82% and 68% decreased Ahr and Cyp1a2 RNA levels after 48 hours (Figure 4D). As expected, Cyp1a1 expression was also reduced by 74% in Ahr depleted hepatocytes. Although the AHR protein level was reduced by approximately 45% in siRNA transfected cells (Figure 4D), Cyp450 proteins could not be detected consistently in hepatocytes in culture for >48 hours.
Because it has been reported that mediator 1 (MED1), a transcriptional co-activator, upregulates Cyp1a2 expression by interacting with constitutive androstane receptor, a nuclear hormone receptor,26 we entertained the possibility that MED1 could play a role in Cyp1a2 induction in LKO mice. Indeed, microarray data showed upregulation of Med1 (1.3-fold, P = 0.0007) in miR-122 depleted livers, which was confirmed both at the RNA and protein levels (Figure 4E). Moreover, knock down of Med1 reduced Cyp1a2 expression in LKO hepatocytes (Figure 4F). Collectively, these results demonstrate that upregulation of Ahr and Med1 plays a key role in Cyp1a2 induction in LKO livers.
miR-122 Regulates Expression of Ahr and Med1 by Distinct Mechanisms
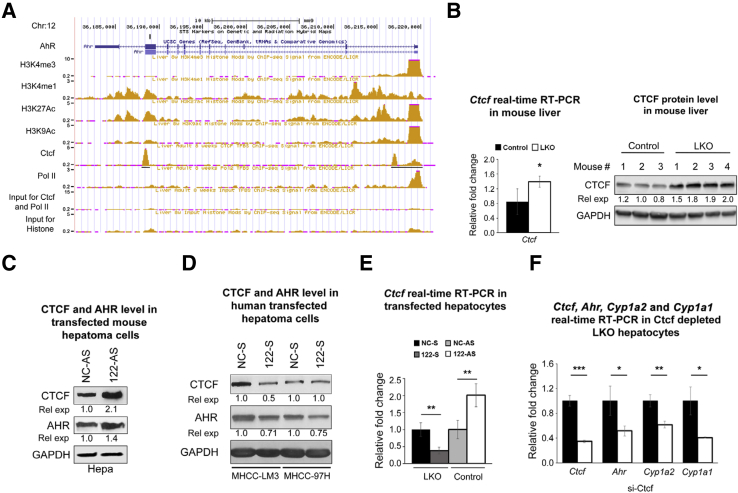
Next, the obvious question was how miR-122 regulates Ahr and Med1 expression. Lack of miR-122–binding sites in mouse Ahr 3′-UTR or coding exons indicated that it is not directly regulated by miR-122. Upregulation of both Ahr hnRNA and mRNA levels suggested that Ahr, like Cyp1a2, is also transcriptionally activated in miR-122 depleted livers (Figure 4A). To identify a potential transactivator(s) of Ahr, we searched the ENCODE (Encyclopedia of DNA Element) database in the University of California, San Francisco genome browser.27 Indeed, ChIP-seq data28 identified the strong association of Ctcf (CCCTC-binding factor) with the promoter, immediate downstream region of the transcription start site and within intron 8 of Ahr gene in 8-week-old mouse livers (Figure 5A). Strong occupancy of RNA polymerase II and histone H3K4Me1/Me3 and H3K9Ac/K27Ac confirmed that Ahr is transcriptionally active in mouse livers (Figure 5A). CTCF, a validated miR-122 target in human,29 is a zinc finger protein that functions as a transcriptional activator, repressor, or insulator, depending on the chromosomal location of its cognate sites.30 Indeed, CTCF protein and RNA levels are elevated in LKO livers (Figure 5B). Moreover, depletion of miR-122 in mouse Hepa1-6 cells by transfecting anti–miR-122 oligo up-regulated CTCF and AHR expression (Figure 5C and Supplemental Figure S6A), whereas miR-122 mimic transfection suppressed their expression in human hepatoma (MHCCLM3 and MHCC-97H) cells (Figure 5D and Supplemental Figure S6, B–D). Similar results were observed in control and LKO hepatocytes transfected with anti–miR-122 and miR-122 mimic oligos, respectively (Figure 5E). To determine whether CTCF has any role in Ahr expression, we depleted it in LKO hepatocytes by transfecting siRNA, which resulted in the inhibition of not only Ahr but also Cyp1a2 expression (Figure 5F).
Figure 5.
miRNA-122 (miR-122) targets both mouse and human CCCTC-binding factor (CTCF) by binding at different sites. A: ChIP-seq data of polymerase II (Pol II), modified H3-histones associated with active promoters, and Ctcf on aryl hydrocarbon receptor (Ahr) promoter in 8-week-old mice livers downloaded from the ENCODE (Encyclopedia of DNA Element) database. B: Ctcf RNA and protein levels in mouse livers. C: CTCF and AHR levels in Hepa cells transfected with miR-122 inhibitor (122-AS) or scrambled inhibitor (NC-AS) for 48 hours (lysate pooled from 3 wells). D: CTCF and AHR levels in hepatocellular carcinoma cells transfected with miR-122 or NC mimic (lysate pooled from 3 wells). E: Relative expression (rel exp) of Ctcf and in Mir122−/− hepatocytes transfected with miR-122 mimic (122-S) or scrambled (NC-S) RNA and control hepatocytes transfected with 122-AS or NC-AS 48 hours after transfection. F: Ctcf, Ahr, and cytochrome P450 family 1 subfamily A member 2 (Cyp1a2) expression in liver-specific miR-122 knockout (LKO) hepatocytes transfected with Ctcf siRNA. E and F: Hepatocytes from 2 mice of each genotype (age and sex matched) were used. Data are expressed as means ± SEM. n = 3 controls (B); n = 4 LKO mice (B); n = 3 to 5 mice of each genotype (F). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus control (two-tailed t-test). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To find out whether miR-122 targets mouse CTCF, we searched different miRNA target prediction databases; TargetScan31 predicted a conserved miR-122–binding site in 3′-UTR of Ctcf. To confirm this, we performed luciferase reporter assay. For this purpose, we co-transfected the psi-CHECK2 vector harboring mouse Ctcf-3′-UTR region, including the miR-122–binding site (Supplemental Figure S6E) downstream of Renilla luciferase and miR-122 mimic or scrambled (NC RNA) in Hepa cells. Assay of Renilla luciferase activity after 48 hours showed its 24% reduction in cells that expressed ectopic miR-122 compared with the control, which was abrogated if the miR-122–binding site was mutated (Supplemental Figure S6F).
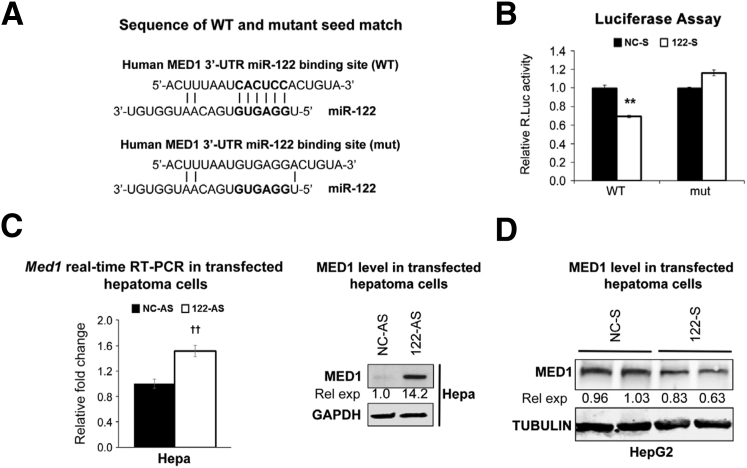
Because MED1 is also predicted as conserved in the miR-122 target (Figure 6A), we next sought to determine whether it is a true target. Indeed, co-transfection experiments showed that miR-122 mimic inhibited Renilla luciferase activity by 31% in cells expressing wild-type MED1 3′-UTR luciferase, which was reversed by transfecting psi-CHECK2 mutant MED1 3′-UTR (Figure 6B). Med1 RNA and protein expression were also up-regulated on transfecting anti–miR-122 in Hepa cells (Figure 6C). In contrast, ectopic miR-122 expression in HepG2 cells reduced the MED1 level (Figure 6D). However, we could not measure Cyp1a2/1a1 expression in these cells because endogenous Cyp1a2/1a1 genes are silent in these cells because they could not be induced even after 2,3,7,8-tetrachlorodibenzo-p-dioxin treatment (data not shown), a potent activator of Ahr.32 Taken together, these results demonstrate CTCF and MED1 are miR-122 direct targets involved in Cyp1a2 expression in LKO livers.
Figure 6.
miRNA-122 (miR-122) suppresses mediator 1 (MED1) by interacting with a conserved site in its 3′-untranslated region (3′-UTR). A: Wild-type (WT) and miR-122 cognate site mutant (mut) in Med1 3′-UTR. B: Luciferase activity in cells co-transfected with psi-CHECK2 MED1 3′-UTR WT or psi-CHECK2 MED1 3′-UTR mut with miR-122/NC mimic. C and D: MED1 level [normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH)] in hepatocellular carcinoma cells transfected with miR-122/NC inhibitor (D) and in HepG2 cells transfected with miR-122/NC mimic. Data are expressed as means ± SEM. n = 3. ∗∗P < 0.01 versus NC-S (two-tailed t-test); ††P < 0.01 versus NC-AS (two-tailed t-test). rel exp, relative expression; R.Luc, Renilla luciferase.
Cyp2e1 Protein Level Is Increased in miR-122 LKO Livers because of Enhanced Translation
Because CYP2E1 is upregulated in LKO livers only at the protein level (Figure 3B), we next attempted to address whether this is because of its increased translation and/or protein stability. Pulse-chase studies found increased 35S-methionine incorporation in Mir-122−/− depleted hepatocytes compared with controls (Supplemental Figure S7A); however, the stability of the de novo synthesized protein was not reduced during a chase with cold methionine for several hours, suggesting its long half-life. Furthermore, no significant changes in CYP2E1 stability were noted in hepatocytes of either genotype on treatment with cycloheximide, a protein biosynthesis inhibitor or MG-132, a proteasome inhibitor (Supplemental Figure S7B), supporting the notion that the increase in CYP2E1 level in LKO livers is not attributable to its differential turnover or stability. Collectively, these results suggest that CYP2E1 translation is enhanced in miR-122 LKO mice.
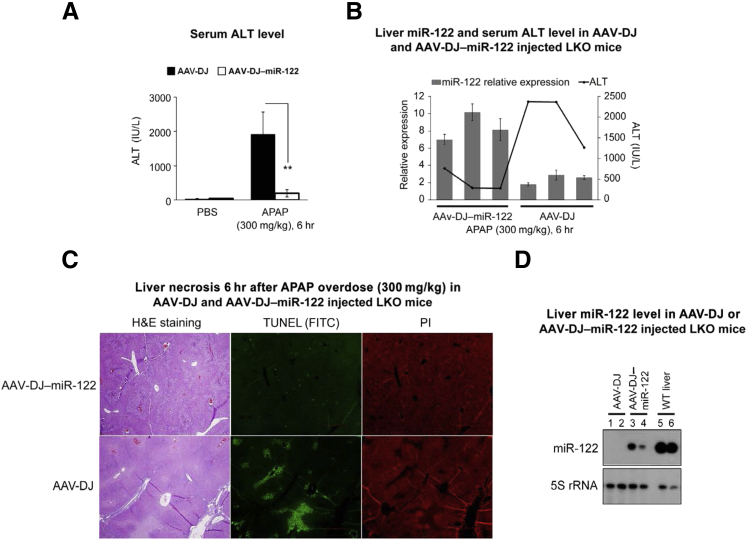
Delivery of miR-122 Suppresses Acetaminophen-Induced Hepatotoxicity in miR-122 LKO Mice
Next, we addressed whether miR-122 replenishment could rescue acetaminophen-induced liver toxicity in LKO livers. To test this, we delivered adenoassociated virus (AAV)–DJ virus harboring miR-122 or AAV-DJ12 in the neonatal LKO mice as previously described (Supplemental Figure S8).33 As expected, the acetaminophen-induced serum ALT level was significantly reduced in mice delivered miR-122 (Figure 7A). In addition, the ALT level inversely correlated with miR-122 expression; the miR-122 level was elevated by 3.4-fold (P = 0.0035) in mice delivered miR-122 compared with the controls (AAV-DJ) (Figure 7B). Of note, AAV-DJ delivery in LKO mice did not cause liver damage. Liver histologic analysis revealed that both groups developed liver necrosis on acetaminophen overdose; however, it was less pronounced in mice injected with AAV-DJ–miR-122 (Figure 7C). A lesser number of TUNEL-positive hepatocytes in AAV-DJ–miR-122 injected mice compared with those that received AAV-DJ indicates reduced DNA strand breaks on acetaminophen overdose (Figure 7C). The inability of exogenous miR-122 to completely block acetaminophen liver toxicity is probably because its expression was only 20% to 30% of the wild-type livers (Figure 7D). The results of this proof-of-concept experiment suggest that acetaminophen hepatotoxicity in LKO mice could be partially rescued by miR-122 delivery.
Figure 7.
Adenoassociated virus (AAV)–DJ virus–mediated miRNA-122 (miR-122) delivery reduces liver damage in acetaminophen overdosed liver-specific miR-122 knockout (LKO) mice. A: Mean serum alanine aminotransferase (ALT) level in mice injected with AAV-DJ or AAV-DJ–miR-122 and treated with phosphate-buffered saline (PBS) or acetaminophen. B: Representative relative expression of miR-122 in the liver (bar diagram) and serum ALT level (line diagram) in mice injected with AAV-DJ or AAV-DJ–miR-122. C: Representative hematoxylin and eosin (H&E) and terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) (green fluorescence), staining of liver sections of mice. Liver nuclei were stained with propidium iodide (PI) (red). D: Liver miR-122 level in LKO livers delivered miR-122 compared with control [wild-type (WT)] liver. Data are expressed as means ± SEM. n = 3 to 5 mice are used per genotype per treatment. ∗∗P < 0.01 (two-tailed t-test). Original magnification: ×40 (C). FITC, fluorescein isothiocyanate.
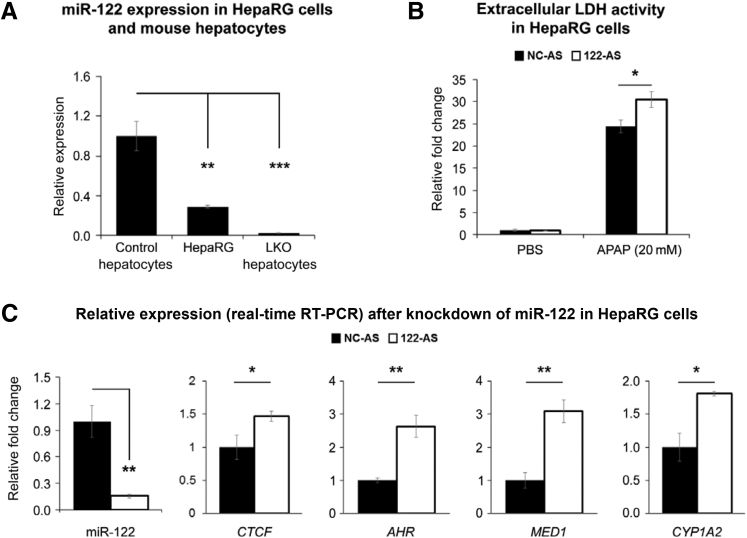
miR-122 Depletion Sensitizes Human HepaRG Cells to Acetaminophen Toxicity
Next, we extended this study to differentiated human HepaRG cells that are reported to exhibit acetaminophen toxicity.34 These cells express miR-122, albeit at a lower level than mouse hepatocytes (Figure 8A). To find out the role of miR-122, if any, in acetaminophen detoxification in these cells, we transfected them with anti–miR-122 followed by acetaminophen (20 mmol/L) treatment 48 hours after transfection and assayed lactate dehydrogenase (LDH) activity in culture supernatants 24 hours later. As reported earlier,34 we observed a significant increase in the extracellular LDH activity on acetaminophen exposure compared with PBS treatment (Figure 8B). Notably, extracellular LDH activity in anti–miR-122 transfected cells was approximately fivefold higher than those transfected with NC (NC-AS) RNA. Moreover, depletion of miR-122 in HepaRG cells resulted in upregulation of CTCF, AHR, MED1, and CYP1A2 expression (Figure 8C). In miR-122 mimic transfected cells, there was a trend in a decrease in acetaminophen-induced LDH release (Supplemental Figure S9A); however, it was not significant, probably because of the only approximately fourfold increase in the ectopic miR-122 level (Supplemental Figure S9B). Taken together, these results suggest that the sensitivity of HepaRG cells to acetaminophen could be modulated by miR-122. Of note, we did not find detectable LDH release from undifferentiated HepaRG cells after acetaminophen exposure probably because CYP1A2/2E1s were not detectable in these cells (not shown). Collectively, these results suggest that endogenous miR-122 protects HepaRG cells from acetaminophen toxicity.
Figure 8.
Differentiated HepaRG cells depleted of miRNA-122 (miR-122) are sensitive to acetaminophen toxicity. A: miR-122 expression in HepaRG cells and mouse hepatocytes. B: Lactate dehydrogenase (LDH) activity in culture supernatants of cells transfected with miR-122 (122-AS) or scrambled inhibitor (NC-AS) for 48 hours followed by acetaminophen treatment for 24 hours. C: Expressions of miR-122, CTCF, AHR, MED1, and CYP1A2 in cells transfected with 122-AS or NC-AS. Data are expressed as means ± SEM. n = 3. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 (two-tailed t-test). LKO, liver-specific miR-122 knockout; PBS, phosphate-buffered saline.
Discussion
In the present study, we found that miR-122 expression is significantly reduced in human ALF livers and in mouse livers after acetaminophen overdose. Notably, the Mir-122 gene is transcriptionally suppressed in mouse livers early in acetaminophen toxicity probably because of a decrease in two key transcription factors, HNF4α and HNF6. In addition, miR-122 regulates acetaminophen metabolism at least in part by controlling the expression of CYP1A2 and CYP2E1, two key enzymes involved in xenobiotic metabolism. Because of the low level of miR-122, derepressed CYP1A2 and CYP2E1 rapidly oxidize acetaminophen to NAPQI, deplete GSH, and increase NAPQI-cysteinyl protein adduct formation, causing oxidative stress that culminates in hepatocyte death and liver toxicity. miR-122 may also regulate acetaminophen detoxification via glucuronidation and sulfation and conjugation of NAPQI with GSH. Quantification of acetaminophen metabolites by NMR revealed high acetaminophen-glucuronide and acetaminophen-GSH levels in LKO livers compared with control livers (V.C. and B.Z., unpublished data), suggesting that miR-122 regulates its metabolism with multiple mechanisms. Microarray data revealed deregulation of expression of several Ugts (UDP-glucuronyltransferases), Sults (sulfotransferases), and Gstp2 (GSH-S-transferase p2) in miR-122 depleted livers, which may contribute to the altered acetaminophen metabolism and increased mortality rate of LKO mice to the drug overdose.
Our results unraveled a novel but complicated mechanism by which miR-122 regulates CYP1A2 expression through CTCF, AHR, and MED1. A key finding is that miR-122 regulates expression of AHR, a transcriptional activator, and MED1, a co-activator, in mouse and human hepatic cells. On the basis of our findings, we propose a model that depicts regulation of acetaminophen metabolism by miR-122 (Supplemental Figure S10).
Our study also found that miR-122 regulates CYP2E1 protein synthesis without affecting its mRNA level or protein stability. It has been reported that the regulation of CYP2E1 expression occurs at the translational and posttranslational levels.35, 36 However, the precise mechanism of its regulation by miR-122 is yet to be understood. It is likely that a key factor(s) that promotes translation of Cyp2e1 mRNA may be upregulated in the miR-122 depleted liver. Alternatively, Cyp2e1 mRNA could undergo some posttranscriptional RNA modifications, such as methylation/demethylation, enhancing its translation in LKO livers.
After the discovery of NAC as a therapy for drug-induced liver injury in 1977, several agents, such as metformin,37 inhibitors of JNK,38 and S-nitrosoglutathione reductase,39 were found to have a protective role in experimental models; however, none were found to be more effective than NAC. Unfortunately, NAC therapy is effective only if administered within first few hours of the onset of liver toxicity. Thus, drugs that could be effective at later hours of toxicity are urgently needed. Recently, by screening a miRNA mimic library, Yang et al40 identified seven miRNAs, including miR-194, miR-125b, and miR-122, that protected mouse hepatocytes from acetaminophen-induced cell death. Moreover, miR-125b mimic delivery 1 hour after acetaminophen overdose increased survival of mice. We believe that certain miR-122 mimic alone or in combination with NAC could be therapeutically effective in restoring metabolic homeostasis that will inhibit necrosis and promote liver regeneration. Most of these new agents are tested in rodent models. It will be more informative to test these chemical inhibitors along miRNA-based therapeutics in the newly developed models, such as pig41 and monkey,42 and ex vivo human liver model43 that mimic acetaminophen toxicity in humans.
Several mitogenic signaling pathways, such as IL-6/STAT-3 and EGFR/c-Met/MAPK, Wnt/β-catenin, NF-κB, and ILK, are activated to promote liver regeneration after acetaminophen injury in mice.44 Significant increase in Ki-67–positive hepatocytes in LKO mice after acetaminophen overdose (Supplemental Figure S4B) could be a compensatory mechanism to overcome excessive necrosis or because of inherent ability of Mir122−/− hepatocytes to proliferate because of induction of growth-promoting factors (eg, β-catenin, CcnD1, CcnG1, c-Myc) that are up-regulated in miR-122 depleted livers.9, 45 Toxicogenomic studies have found that c-MYC and AP-1 are induced by acetaminophen and that c-Myc/AP-1–centered interactomes are the most significant networks of proteins associated with liver injury in humans.46 Increased basal c-MYC and E2F1 expression in LKO livers45 might also promote hepatocyte proliferation after acetaminophen overdose–induced hepatocyte death in these mice.
In summary, we found miR-122 as a novel regulator of acetaminophen-induced ALF. Our study established that miR-122 modulates drug metabolism at least in part by regulating CTCF, AHR, MED1, CYP1A2, and CYP2E1. Our in vitro and in vivo studies revealed the promising therapeutic potential of miR-122 against acetaminophen toxicity; hence, it can be explored as an antidote against drug-induced ALF. Because several anticancer drugs exhibit hepatotoxicity, it likely that this abundant miRNA in the liver might be involved in their metabolism by regulating many drug metabolizing enzymes. Moreover, regulation of CYP1A2 and CYP2E1 by miR-122 can be exploited in designing drugs for patients with hepatocellular carcinoma.
Acknowledgments
We thank Dr. Huban Kutay for technical assistance; Dr. Bo Wang and Juan Barajas for generating psi-CHECK2 CTCF 3′-UTR vector and psi-CHECK2 MED1 3′-UTR vector, respectively; Dr. Hemant K. Bid for Ki-67 immunohistochemistry data; and Dr. Joseph Luna for critically reading the manuscript.
V.C. designed, performed, and analyzed all experiments unless stated otherwise, organized the figures, and was involved in writing; K.-Y.T. designed, performed, and analyzed the experiments shown in Figure 6 and Supplemental Figure S5; C.-H.L. performed and analyzed Figures 1D, 5C, and 6D; B.Z., L.B.-L. and R.B. helped in generating NMR data for liver metabolites; N.W. injected AAV-DJ and AAV-DJ–miR-122; X.Z. performed statistical analysis of the data; D.Y. and N.J. generated data in patients with ALF; L.J. designed, performed, and analyzed Figure 2E and Supplemental Figure S2A; W.M.L. scored H&E slides for necrosis and was involved in writing the manuscript; K.G. conceived, coordinated the study, and wrote the manuscript; S.T. performed initial studies on the viability of mice on acetaminophen treatment and critically read the manuscript.
Footnotes
Supported in part by NIH grants DK088076 (K.G.) and R01A193244 (K.G.).
Dislcosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2017.08.026.
Contributor Information
William M. Lee, Email: william.lee@utsouthwestern.edu.
Kalpana Ghoshal, Email: kalpana.ghoshal@osumc.edu.
Supplemental Data
A: miRNA-122 (miR-122) relative expression (Rel. Exp.) (normalized to U6 snRNA) in nine patients with acute liver failure (ALF) and four normal human livers. B: Quantification of miR-122 Northern blot data in livers of phosphate-buffered saline (PBS)– or acetaminophen (APAP)–treated mice. C: Western blot analysis of hepatocyte nuclear factor (HNF)-4α, HNF6, and Ku-86 in liver nuclear extracts (50 mg of protein) from control mice treated with APAP (300 mg/kg) for 2 hours. D: Comparison of serum alanine aminotransferase (ALT) and miR-122 [ real-time RT-PCR in control mice treated with PBS or APAP (300 mg/kg) for 2 hours. E: Northern blot analysis of total serum RNA from control (C1 to C4) and liver-specific miR-122 knockout (LKO) mice treated with APAP (150 mg/kg) for 24 hours. RNAs from 3 mice were pooled for the assay. Data are expressed as means ± SEM. ∗P < 0.05 (two-tailed t-test).
A: Kaplan-Meier survival curve of control and liver-specific miRNA-122 (miR-122) knockout (LKO) mice injected with acetaminophen (APAP) (300 mg/kg). The mortality rate of overnight fasted mice injected with acetaminophen was monitored. B:N-acetyl-p-benzoquinone imine (NAPQI)–cysteine adducts in the serum of control and LKO mice treated with phosphate-buffered saline (PBS) or APAP. C: Two-dimensional 13C-1H HSQC nuclear magnetic resonance (NMR) method applied to liver tissue to identify and quantify metabolites. The depicted spectrum was collected from wild-type mice 0.5 hours after PBS treatment (control). Each spectral feature (cross-peak) corresponds to a C-H pair in a metabolite, and the peak amplitude is proportional to its concentration. Circles indicate the cross-peak that belong to glutathione (GSH) and oxidized glutathione (GSSG). The amplitudes from unique peaks were used for the determination of the normalized concentration index of Figure 2F. Data are expressed as means ± SEM. n = 5 of each genotype (A). ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 versus control (two-tailed t-test).
A: Western blot analysis of phospho–c-Jun–N-terminal protein kinase (p-JNK) in the control and liver-specific miRNA-122 (miR-122) knockout (LKO) liver lysates. B: JNK activity in the control and LKO liver lysates from 6 mice of each genotype treated with acetaminophen (APAP) (150 mg/kg) for 24 hours. JNK activity was measured by the incorporation of the 32P group from γ32P-ATP to c-Jun N-terminal peptide conjugated to glutathione S-transferase (32P-c-Jun-GST), pulled down with glutathione-sepharose beads, separated in SDS-PAGE, and subjected to autoradiography. Of note, JNK activity was not detected in phosphate-buffered saline (PBS)–injected liver lysates of control and LKO mice (data not shown). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; t-JNK, total c-Jun N-terminal Kinase; WT, wild type.
A: Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay in the control and liver-specific miRNA-122 (miR-122) knockout (LKO) mice treated with acetaminophen (APAP) demonstrated increased nuclear DNA fragmentation in LKO livers (green fluorescence). Hepatocyte nuclei were visualized with propidium iodide (red fluorescence). B: Immunohistochemical (IHC) analysis of liver sections with Ki-67 antibody revealed an increased number of proliferating hepatocytes in LKO livers treated with APAP. Data are expressed as means ± SEM. n = 6. ∗∗P < 0.01 versus control (two-tailed t-test). Original magnification: ×100 (A and B, main images); ×200 (B, insets).
A: Survival of 12-week-old control and liver-specific miRNA-122 (miR-122) knockout (LKO) female mice (10 per genotype) treated with acetaminophen (APAP) after overnight fasting. B–D: Quantification of cytochrome P450 family 1 subfamily A member 2 (CYP1A2) and cytochrome P450 family 2 subfamily E member 1 (CYP2E1) protein levels measured by immunoblotting after 2 hours of APAP treatment in fasted male mice, fasted female mice, and nonfasted male mice, respectively. E: Phase contract microscopic image of control and Mir122−/− hepatocytes treated with APAP in culture for 6 hours. Data are expressed as means ± SEM. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 (two-tailed t-test). PBS, phosphate-buffered saline.
A: miRNA-122 (miR-122), CCCTC-binding factor (Ctcf), and aryl hydrocarbon receptor (Ahr) expression in mouse Hepa cells 48 hours after transfection with 50 nmol/L miR-122 inhibitor (122-AS) or scrambled inhibitor (NC-AS). Each transfection assay was performed in triplicate. B–D: miR-122, Ctcf, and Ahr expression in human hepatocellular carcinoma (MHCC-LM3 and Huh7) cells, respectively, after 48 hours of transfection with 50 nmol/L miR-122 mimic (122-S) or scrambled mimic (NC-S). E: miR-122 binding site in the 3′-untranslated region (3′-UTR) of Ctcf conserved across mouse and humans. F: Luciferase activity in cells co-transfected with psi-CHECK2 Ctcf 3′-UTR wild type (WT) or psi-CHECK2 Ctcf 3′-UTR mutant (mut) with miR-122/NC mimic. Data are expressed as means ± SEM. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus control (two-tailed t-test).
A: Pulse-chase studies found that de novo synthesis of cytochrome P450 family 2 subfamily E member 1 (CYP2E1) protein is higher in liver-specific miRNA-122 (miR-122) knockout (LKO) hepatocytes. Hepatocytes from the control and LKO mice after overnight culture were incubated at 37°C in 5% CO2 incubator with 35S-methionine in methionine-free medium that contained dialyzed fetal bovine serum for 1 hour followed by incubation in the regular media for different time periods. Cellular lysates in radioimmunoprecipitation assay buffer were immunoprecipitated with CYP2E1 antibody and pulled-down proteins were separated by SDS-PAGE, transferred to nitrocellulose membrane, and exposed to film for 48 hours at −80°C to detect 35S-methionine (35S-met)–labeled polypeptides. The membrane was then subjected to CYP2E1 immunoblotting. B: Control and LKO hepatocytes after overnight culture were treated with cycloheximide (CHX) (20 μg/mL), puromycin (40 mg/mL), or MG-132 (10 μg/mL) for indicated periods, and cell extracts in were subjected to Western blot analysis with indicated antibodies. CYP2E1 levels were normalized to those of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). DMSO, dimethyl sulfoxide; PBS, phosphate-buffered saline.
Schematic representation of adenoassociated virus (AAV)-8–mediated miRNA-122 (miR-122) delivery in liver-specific miRNA-122 knockout (LKO) mice to determine whether exogenous miR-122 can rescue acetaminophen (APAP)–induced liver injury in LKO mice. PBS, phosphate-buffered saline.
A: Lactate dehydrogenase (LDH) activity in the culture supernatant of HepaRG cells transfected with miRNA-122 (miR-122) mimic (122-S) or negative control mimic (50 nmol/L) (NC-S) for 48 hours followed by treatment with acetaminophen (APAP) (20 mmol/L) for 24 hours. B: miR-122 expression. PBS, phosphate-buffered saline.
A model depicting regulation of acetaminophen (APAP) metabolism by miRNA-122 (miR-122) based on our results. miR-122 may also regulate expression of Ugts, Sults, and Gstps, which are involved in APAP detoxification and renal excretion. Ahr, aryl hydrocarbon receptor; Ctcf, CCCTC-binding factor; Cyp1A2, cytochrome P450 family 1 subfamily A member 2; Cyp2E1, cytochrome P450 family 2 subfamily E member 1; GSH, glutathione; Med1, mediator 1; NAC, N-acetylcysteine; NAPQI, N-acetyl-p-benzoquinone imine.
References
- 1.Lee W.M. Drug-induced acute liver failure. Clin Liver Dis. 2013;17:575–586. doi: 10.1016/j.cld.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du K., Ramachandran A., Jaeschke H. Oxidative stress during acetaminophen hepatotoxicity: sources, pathophysiological role and therapeutic potential. Redox Biol. 2016;10:148–156. doi: 10.1016/j.redox.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dear J.W., Antoine D.J., Park B.K. Where are we now with paracetamol? BMJ. 2015;351 doi: 10.1136/bmj.h3705. h3705. [DOI] [PubMed] [Google Scholar]
- 4.Hinson J.A., Roberts D.W., James L.P. Mechanisms of acetaminophen-induced liver necrosis. Handb Exp Pharmacol. 2010:369–405. doi: 10.1007/978-3-642-00663-0_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C., Krausz K.W., Idle J.R., Gonzalez F.J. Identification of novel toxicity-associated metabolites by metabolomics and mass isotopomer analysis of acetaminophen metabolism in wild-type and Cyp2e1-null mice. J Biol Chem. 2008;283:4543–4559. doi: 10.1074/jbc.M706299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bajt M.L., Farhood A., Lemasters J.J., Jaeschke H. Mitochondrial bax translocation accelerates DNA fragmentation and cell necrosis in a murine model of acetaminophen hepatotoxicity. J Pharmacol Exp Ther. 2008;324:8–14. doi: 10.1124/jpet.107.129445. [DOI] [PubMed] [Google Scholar]
- 7.Bajt M.L., Cover C., Lemasters J.J., Jaeschke H. Nuclear translocation of endonuclease G and apoptosis-inducing factor during acetaminophen-induced liver cell injury. Toxicol Sci. 2006;94:217–225. doi: 10.1093/toxsci/kfl077. [DOI] [PubMed] [Google Scholar]
- 8.Woolbright B.L., Jaeschke H. Role of the inflammasome in acetaminophen-induced liver injury and acute liver failure. J Hepatol. 2017;66:836–848. doi: 10.1016/j.jhep.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGill M.R., Jaeschke H. Mechanistic biomarkers in acetaminophen-induced hepatotoxicity and acute liver failure: from preclinical models to patients. Expert Opin Drug Metab Toxicol. 2014;10:1005–1017. doi: 10.1517/17425255.2014.920823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thakral S., Ghoshal K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Curr Gene Ther. 2015;15:142–150. doi: 10.2174/1566523214666141224095610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szabo G., Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. 2013;10:542–552. doi: 10.1038/nrgastro.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu S.H., Wang B., Kota J., Yu J., Costinean S., Kutay H., Yu L., Bai S., La Perle K., Chivukula R.R., Mao H., Wei M., Clark K.R., Mendell J.R., Caligiuri M.A., Jacob S.T., Mendell J.T., Ghoshal K. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai W.C., Hsu S.D., Hsu C.S., Lai T.C., Chen S.J., Shen R., Huang Y., Chen H.C., Lee C.H., Tsai T.F., Hsu M.T., Wu J.C., Huang H.D., Shiao M.S., Hsiao M., Tsou A.P. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luna J.M., Scheel T.K., Danino T., Shaw K.S., Mele A., Fak J.J., Nishiuchi E., Takacs C.N., Catanese M.T., de Jong Y.P., Jacobson I.M., Rice C.M., Darnell R.B. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 2015;160:1099–1110. doi: 10.1016/j.cell.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antoine D.J., Dear J.W., Lewis P.S., Platt V., Coyle J., Masson M., Thanacoody R.H., Gray A.J., Webb D.J., Moggs J.G., Bateman D.N., Goldring C.E., Park B.K. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology. 2013;58:777–787. doi: 10.1002/hep.26294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubin P.H., Yuan H., Devine R.K., Hynan L.S., Jain M.K., Lee W.M. Micro-RNA-122 levels in acute liver failure and chronic hepatitis C. J Med Virol. 2014;86:1507–1514. doi: 10.1002/jmv.23987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muldrew K.L., James L.P., Coop L., McCullough S.S., Hendrickson H.P., Hinson J.A., Mayeux P.R. Determination of acetaminophen-protein adducts in mouse liver and serum and human serum after hepatotoxic doses of acetaminophen using high-performance liquid chromatography with electrochemical detection. Drug Metab Dispos. 2002;30:446–451. doi: 10.1124/dmd.30.4.446. [DOI] [PubMed] [Google Scholar]
- 18.Kutay H., Bai S., Datta J., Motiwala T., Pogribny I., Frankel W., Jacob S.T., Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Wilkie G.S., Schirmer E.C. Purification of nuclei and preparation of nuclear envelopes from skeletal muscle. Methods Mol Biol. 2008;463:23–41. doi: 10.1007/978-1-59745-406-3_2. [DOI] [PubMed] [Google Scholar]
- 20.Wang A.Y., Lian L.H., Jiang Y.Z., Wu Y.L., Nan J.X. Gentiana manshurica Kitagawa prevents acetaminophen-induced acute hepatic injury in mice via inhibiting JNK/ERK MAPK pathway. World J Gastroenterol. 2010;16:384–391. doi: 10.3748/wjg.v16.i3.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakurai T., Maeda S., Chang L., Karin M. Loss of hepatic NF-κB activity enhances chemical hepatocarcinogenesis through sustained c-Jun N-terminal kinase 1 activation. Proc Natl Acad Sci U S A. 2006;103:10544–10551. doi: 10.1073/pnas.0603499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K., Zhang S., Marzolf B., Troisch P., Brightman A., Hu Z., Hood L.E., Galas D.J. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanawa N., Shinohara M., Saberi B., Gaarde W.A., Han D., Kaplowitz N. Role of JNK translocation to mitochondria leading to inhibition of mitochondria bioenergetics in acetaminophen-induced liver injury. J Biol Chem. 2008;283:13565–13577. doi: 10.1074/jbc.M708916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McConnachie L.A., Mohar I., Hudson F.N., Ware C.B., Ladiges W.C., Fernandez C., Chatterton-Kirchmeier S., White C.C., Pierce R.H., Kavanagh T.J. Glutamate cysteine ligase modifier subunit deficiency and gender as determinants of acetaminophen-induced hepatotoxicity in mice. Toxicol Sci. 2007;99:628–636. doi: 10.1093/toxsci/kfm165. [DOI] [PubMed] [Google Scholar]
- 25.Zhou S.F., Wang B., Yang L.P., Liu J.P. Structure, function, regulation and polymorphism and the clinical significance of human cytochrome P450 1A2. Drug Metab Rev. 2010;42:268–354. doi: 10.3109/03602530903286476. [DOI] [PubMed] [Google Scholar]
- 26.Jia Y., Guo G.L., Surapureddi S., Sarkar J., Qi C., Guo D., Xia J., Kashireddi P., Yu S., Cho Y.W., Rao M.S., Kemper B., Ge K., Gonzalez F.J., Reddy J.K. Transcription coactivator peroxisome proliferator-activated receptor-binding protein/mediator 1 deficiency abrogates acetaminophen hepatotoxicity. Proc Natl Acad Sci U S A. 2005;102:12531–12536. doi: 10.1073/pnas.0506000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenbloom K.R., Sloan C.A., Malladi V.S., Dreszer T.R., Learned K., Kirkup V.M., Wong M.C., Maddren M., Fang R., Heitner S.G., Lee B.T., Barber G.P., Harte R.A., Diekhans M., Long J.C., Wilder S.P., Zweig A.S., Karolchik D., Kuhn R.M., Haussler D., Kent W.J. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raney B.J., Dreszer T.R., Barber G.P., Clawson H., Fujita P.A., Wang T., Nguyen N., Paten B., Zweig A.S., Karolchik D., Kent W.J. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 2014;30:1003–1005. doi: 10.1093/bioinformatics/btt637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu H., He J.H., Xiao Z.D., Zhang Q.Q., Chen Y.Q., Zhou H., Qu L.H. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology. 2010;52:1431–1442. doi: 10.1002/hep.23818. [DOI] [PubMed] [Google Scholar]
- 30.Kim S., Yu N.-K., Kaang B.-K. CTCF as a multifunctional protein in genome regulation and gene expression. Exp Mol Med. 2015;47:e166. doi: 10.1038/emm.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mimura J., Fujii-Kuriyama Y. Functional role of AhR in the expression of toxic effects by TCDD. Biochim Biophys Acta. 2003;1619:263–268. doi: 10.1016/s0304-4165(02)00485-3. [DOI] [PubMed] [Google Scholar]
- 33.Hsu S.H., Delgado E.R., Otero P.A., Teng K.Y., Kutay H., Meehan K.M., Moroney J.B., Monga J.K., Hand N.J., Friedman J.R., Ghoshal K., Duncan A.W. MicroRNA-122 regulates polyploidization in the murine liver. Hepatology. 2016;64:599–615. doi: 10.1002/hep.28573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGill M.R., Yan H.M., Ramachandran A., Murray G.J., Rollins D.E., Jaeschke H. HepaRG cells: a human model to study mechanisms of acetaminophen hepatotoxicity. Hepatology. 2011;53:974–982. doi: 10.1002/hep.24132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S.M., Wang Y., Nabavi N., Liu Y., Correia M.A. Hepatic cytochromes P450: structural degrons and barcodes, posttranslational modifications and cellular adapters in the ERAD-endgame. Drug Metab Rev. 2016;48:405–433. doi: 10.1080/03602532.2016.1195403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez F.J. The 2006 Bernard B. Brodie Award Lecture: Cyp2e1. Drug Metab Dispos. 2007;35:1–8. doi: 10.1124/dmd.106.012492. [DOI] [PubMed] [Google Scholar]
- 37.Du K., Farhood A., Jaeschke H. Mitochondria-targeted antioxidant Mito-Tempo protects against acetaminophen hepatotoxicity. Arch Toxicol. 2017;91:761–773. doi: 10.1007/s00204-016-1692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gunawan B.K., Liu Z.X., Han D., Hanawa N., Gaarde W.A., Kaplowitz N. c-Jun N-terminal kinase plays a major role in murine acetaminophen hepatotoxicity. Gastroenterology. 2006;131:165–178. doi: 10.1053/j.gastro.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 39.Cox A.G., Saunders D.C., Kelsey P.B., Jr., Conway A.A., Tesmenitsky Y., Marchini J.F., Brown K.K., Stamler J.S., Colagiovanni D.B., Rosenthal G.J., Croce K.J., North T.E., Goessling W. S-nitrosothiol signaling regulates liver development and improves outcome following toxic liver injury. Cell Rep. 2014;6:56–69. doi: 10.1016/j.celrep.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang D., Yuan Q., Balakrishnan A., Bantel H., Klusmann J.H., Manns M.P., Ott M., Cantz T., Sharma A.D. MicroRNA-125b-5p mimic inhibits acute liver failure. Nat Commun. 2016;7:11916. doi: 10.1038/ncomms11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee K.C., Palacios Jimenez C., Alibhai H., Chang Y.M., Leckie P.J., Baker L.A., Stanzani G., L Priestnall S., Mookerjee R.P., Jalan R., Davies N.A. A reproducible, clinically relevant, intensively managed, pig model of acute liver failure for testing of therapies aimed to prolong survival. Liver Int. 2013;33:544–551. doi: 10.1111/liv.12042. [DOI] [PubMed] [Google Scholar]
- 42.Tamai S., Iguchi T., Niino N., Mikamoto K., Sakurai K., Sayama A., Shimoda H., Takasaki W., Mori K. A monkey model of acetaminophen-induced hepatotoxicity; phenotypic similarity to human. J Toxicol Sci. 2017;42:73–84. doi: 10.2131/jts.42.73. [DOI] [PubMed] [Google Scholar]
- 43.Schreiter T., Sowa J.P., Schlattjan M., Treckmann J., Paul A., Strucksberg K.H., Baba H.A., Odenthal M., Gieseler R.K., Gerken G., Arteel G.E., Canbay A. Human ex-vivo liver model for acetaminophen-induced liver damage. Sci Rep. 2016;6:31916. doi: 10.1038/srep31916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Apte U., Singh S., Zeng G., Cieply B., Virji M.A., Wu T., Monga S.P. Beta-catenin activation promotes liver regeneration after acetaminophen-induced injury. Am J Pathol. 2009;175:1056–1065. doi: 10.2353/ajpath.2009.080976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang B., Hsu S.H., Wang X., Kutay H., Bid H.K., Yu J., Ganju R.K., Jacob S.T., Yuneva M., Ghoshal K. Reciprocal regulation of microRNA-122 and c-Myc in hepatocellular cancer: role of E2F1 and transcription factor dimerization partner 2. Hepatology. 2014;59:555–566. doi: 10.1002/hep.26712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gunawan B.K., Kaplowitz N. Mechanisms of drug-induced liver disease. Clin Liver Dis. 2007;11:459–475. doi: 10.1016/j.cld.2007.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: miRNA-122 (miR-122) relative expression (Rel. Exp.) (normalized to U6 snRNA) in nine patients with acute liver failure (ALF) and four normal human livers. B: Quantification of miR-122 Northern blot data in livers of phosphate-buffered saline (PBS)– or acetaminophen (APAP)–treated mice. C: Western blot analysis of hepatocyte nuclear factor (HNF)-4α, HNF6, and Ku-86 in liver nuclear extracts (50 mg of protein) from control mice treated with APAP (300 mg/kg) for 2 hours. D: Comparison of serum alanine aminotransferase (ALT) and miR-122 [ real-time RT-PCR in control mice treated with PBS or APAP (300 mg/kg) for 2 hours. E: Northern blot analysis of total serum RNA from control (C1 to C4) and liver-specific miR-122 knockout (LKO) mice treated with APAP (150 mg/kg) for 24 hours. RNAs from 3 mice were pooled for the assay. Data are expressed as means ± SEM. ∗P < 0.05 (two-tailed t-test).
A: Kaplan-Meier survival curve of control and liver-specific miRNA-122 (miR-122) knockout (LKO) mice injected with acetaminophen (APAP) (300 mg/kg). The mortality rate of overnight fasted mice injected with acetaminophen was monitored. B:N-acetyl-p-benzoquinone imine (NAPQI)–cysteine adducts in the serum of control and LKO mice treated with phosphate-buffered saline (PBS) or APAP. C: Two-dimensional 13C-1H HSQC nuclear magnetic resonance (NMR) method applied to liver tissue to identify and quantify metabolites. The depicted spectrum was collected from wild-type mice 0.5 hours after PBS treatment (control). Each spectral feature (cross-peak) corresponds to a C-H pair in a metabolite, and the peak amplitude is proportional to its concentration. Circles indicate the cross-peak that belong to glutathione (GSH) and oxidized glutathione (GSSG). The amplitudes from unique peaks were used for the determination of the normalized concentration index of Figure 2F. Data are expressed as means ± SEM. n = 5 of each genotype (A). ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 versus control (two-tailed t-test).
A: Western blot analysis of phospho–c-Jun–N-terminal protein kinase (p-JNK) in the control and liver-specific miRNA-122 (miR-122) knockout (LKO) liver lysates. B: JNK activity in the control and LKO liver lysates from 6 mice of each genotype treated with acetaminophen (APAP) (150 mg/kg) for 24 hours. JNK activity was measured by the incorporation of the 32P group from γ32P-ATP to c-Jun N-terminal peptide conjugated to glutathione S-transferase (32P-c-Jun-GST), pulled down with glutathione-sepharose beads, separated in SDS-PAGE, and subjected to autoradiography. Of note, JNK activity was not detected in phosphate-buffered saline (PBS)–injected liver lysates of control and LKO mice (data not shown). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; t-JNK, total c-Jun N-terminal Kinase; WT, wild type.
A: Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay in the control and liver-specific miRNA-122 (miR-122) knockout (LKO) mice treated with acetaminophen (APAP) demonstrated increased nuclear DNA fragmentation in LKO livers (green fluorescence). Hepatocyte nuclei were visualized with propidium iodide (red fluorescence). B: Immunohistochemical (IHC) analysis of liver sections with Ki-67 antibody revealed an increased number of proliferating hepatocytes in LKO livers treated with APAP. Data are expressed as means ± SEM. n = 6. ∗∗P < 0.01 versus control (two-tailed t-test). Original magnification: ×100 (A and B, main images); ×200 (B, insets).
A: Survival of 12-week-old control and liver-specific miRNA-122 (miR-122) knockout (LKO) female mice (10 per genotype) treated with acetaminophen (APAP) after overnight fasting. B–D: Quantification of cytochrome P450 family 1 subfamily A member 2 (CYP1A2) and cytochrome P450 family 2 subfamily E member 1 (CYP2E1) protein levels measured by immunoblotting after 2 hours of APAP treatment in fasted male mice, fasted female mice, and nonfasted male mice, respectively. E: Phase contract microscopic image of control and Mir122−/− hepatocytes treated with APAP in culture for 6 hours. Data are expressed as means ± SEM. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 (two-tailed t-test). PBS, phosphate-buffered saline.
A: miRNA-122 (miR-122), CCCTC-binding factor (Ctcf), and aryl hydrocarbon receptor (Ahr) expression in mouse Hepa cells 48 hours after transfection with 50 nmol/L miR-122 inhibitor (122-AS) or scrambled inhibitor (NC-AS). Each transfection assay was performed in triplicate. B–D: miR-122, Ctcf, and Ahr expression in human hepatocellular carcinoma (MHCC-LM3 and Huh7) cells, respectively, after 48 hours of transfection with 50 nmol/L miR-122 mimic (122-S) or scrambled mimic (NC-S). E: miR-122 binding site in the 3′-untranslated region (3′-UTR) of Ctcf conserved across mouse and humans. F: Luciferase activity in cells co-transfected with psi-CHECK2 Ctcf 3′-UTR wild type (WT) or psi-CHECK2 Ctcf 3′-UTR mutant (mut) with miR-122/NC mimic. Data are expressed as means ± SEM. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus control (two-tailed t-test).
A: Pulse-chase studies found that de novo synthesis of cytochrome P450 family 2 subfamily E member 1 (CYP2E1) protein is higher in liver-specific miRNA-122 (miR-122) knockout (LKO) hepatocytes. Hepatocytes from the control and LKO mice after overnight culture were incubated at 37°C in 5% CO2 incubator with 35S-methionine in methionine-free medium that contained dialyzed fetal bovine serum for 1 hour followed by incubation in the regular media for different time periods. Cellular lysates in radioimmunoprecipitation assay buffer were immunoprecipitated with CYP2E1 antibody and pulled-down proteins were separated by SDS-PAGE, transferred to nitrocellulose membrane, and exposed to film for 48 hours at −80°C to detect 35S-methionine (35S-met)–labeled polypeptides. The membrane was then subjected to CYP2E1 immunoblotting. B: Control and LKO hepatocytes after overnight culture were treated with cycloheximide (CHX) (20 μg/mL), puromycin (40 mg/mL), or MG-132 (10 μg/mL) for indicated periods, and cell extracts in were subjected to Western blot analysis with indicated antibodies. CYP2E1 levels were normalized to those of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). DMSO, dimethyl sulfoxide; PBS, phosphate-buffered saline.
Schematic representation of adenoassociated virus (AAV)-8–mediated miRNA-122 (miR-122) delivery in liver-specific miRNA-122 knockout (LKO) mice to determine whether exogenous miR-122 can rescue acetaminophen (APAP)–induced liver injury in LKO mice. PBS, phosphate-buffered saline.
A: Lactate dehydrogenase (LDH) activity in the culture supernatant of HepaRG cells transfected with miRNA-122 (miR-122) mimic (122-S) or negative control mimic (50 nmol/L) (NC-S) for 48 hours followed by treatment with acetaminophen (APAP) (20 mmol/L) for 24 hours. B: miR-122 expression. PBS, phosphate-buffered saline.
A model depicting regulation of acetaminophen (APAP) metabolism by miRNA-122 (miR-122) based on our results. miR-122 may also regulate expression of Ugts, Sults, and Gstps, which are involved in APAP detoxification and renal excretion. Ahr, aryl hydrocarbon receptor; Ctcf, CCCTC-binding factor; Cyp1A2, cytochrome P450 family 1 subfamily A member 2; Cyp2E1, cytochrome P450 family 2 subfamily E member 1; GSH, glutathione; Med1, mediator 1; NAC, N-acetylcysteine; NAPQI, N-acetyl-p-benzoquinone imine.