Abstract
Background
Folate-dependent one-carbon metabolism provides one-carbon units for several biological processes. This pathway is highly compartmentalized in eukaryotes, with the mitochondrial pathway producing formate for use in cytoplasmic processes. The mitochondrial enzyme MTHFD2 has been reported to use NAD+ as a cofactor while the isozyme MTHFD2L utilizes NAD+ or NADP+ at physiologically relevant conditions. Because MTHFD2 is highly expressed in many cancer types, we sought to determine the cofactor preference of this enzyme.
Results
Kinetic analysis shows that purified human MTHFD2 exhibits dual redox cofactor specificity, utilizing either NADP+ or NAD+ with the more physiologically relevant pentaglutamate folate substrate.
Conclusion
These results show that the mitochondrial folate pathway isozymes MTHFD2 and MTHFD2L both exhibit dual redox cofactor specificity. Our kinetic analysis clearly supports a role for MTHFD2 in mitochondrial NADPH production, indicating that this enzyme is likely responsible for mitochondrial production of both NADH and NADPH in rapidly proliferating cells.
Keywords: MTHFD2, NADH, NADPH, One-carbon metabolism
Background
One-carbon (1C) metabolism is a universal folate-dependent pathway that generates 1C units for de novo purine and thymidylate synthesis, interconversion of several amino acids, production of universal methyl donors, and regeneration of redox cofactors. Because these metabolic processes play critical roles in cancer cells [1, 2], 1C metabolism has long been an important target for the development of chemotherapeutic drugs.
One-carbon metabolism is highly compartmentalized in eukaryotes [3], and mitochondria play a critical role in cellular 1C metabolism. Mitochondria import 1C donors such as serine and glycine and oxidize the 1C units to formate, which is exported to the cytoplasm as a 1C unit for use in purine and thymidylate synthesis and homocysteine remethylation (Fig. 1a) [4–8]. Interconversion of 1C units in mammalian mitochondria is catalyzed by three distinct members of the MTHFD (methylene-tetrahydrofolate dehydrogenase) family of enzymes: MTHFD2L, MTHFD2, and MTHFD1L. MTHFD1L is a monofunctional 10-CHO-THF synthetase [9]. It catalyzes the final step in the mitochondrial pathway to produce formate, thus controlling the flux of 1C units from mitochondria into cytoplasmic processes.
Fig. 1.
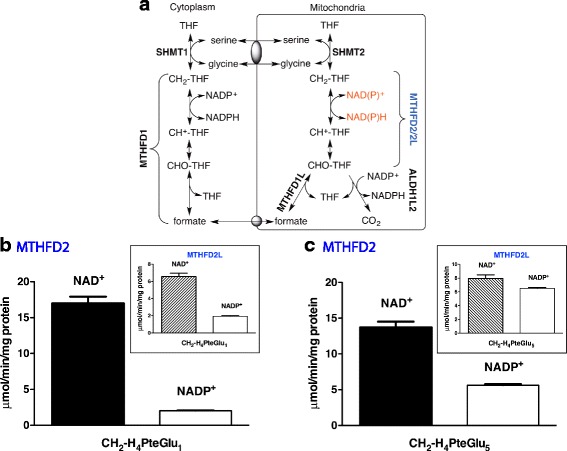
MTHFD2 exhibits NADP+-dependent dehydrogenase activity with 5,10-CH2-THF pentaglutamate. a Compartmentation of mammalian one-carbon metabolism. MTHFD1 is the cytoplasmic trifunctional C1-THF synthase that catalyzes 10-formyl-THF synthetase, 5,10-methenyl-THF cyclohydrolase, and 5,10-methylene-THF dehydrogenase activities. In mammalian mitochondria, bifunctional MTHFD2 or MTHFD2L enzymes catalyze 5,10-methenyl-THF cyclohydrolase and 5,10-methylene-THF dehydrogenase activities, and monofunctional MTHFD1L catalyzes the 10-formyl-THF synthetase reaction. SHMT1 and SHMT2 represent cytoplasmic and mitochondrial serine hydroxymethyltransferase isozymes, respectively. Gray ovals represent putative metabolite transporters. b, c Purified MTHFD2 was assayed for NAD+- and NADP+-dependent 5,10-CH2-THF dehydrogenase activity with saturating concentrations of CH2-H4PteGlu1 or CH2-H4PteGlu5 (insets, 5,10-CH2-THF dehydrogenase activity of MTHFD2L; data from ref. 11). CH2-H4PteGlu1 and CH2-H4PteGlu5 concentrations were 354 and 429 μM, respectively. NAD+ and NADP+ concentrations were 1.0 and 6.0 mM, respectively. NAD+-dependent reactions also included 5 mM MgCl2 and 25 mM Pi. NADP+-dependent reactions included only 5 mM MgCl2. Each column represents the mean ± S.E. of triplicate determinations
Mitochondrial MTHFD2 and MTHFD2L are both bifunctional enzymes possessing 5,10-methenyl-THF (CH+-THF) cyclohydrolase and 5,10-methylene-THF (CH2-THF) dehydrogenase activities. MTHFD2 was initially identified as an NAD+-dependent 5,10-CH2-THF dehydrogenase [10]. The Mthfd2 gene is expressed only in transformed mammalian cells and embryonic or nondifferentiated adult tissues [10, 11]. MTHFD2L, identified in 2011, is homologous to MTHFD2, sharing 60–65% amino acid sequence identity among various mammals. The Mthfd2l gene is expressed in adult mammals (highest expression in the brain and lung) and also at all stages of embryogenesis [11, 12].
Mthfd2 has been identified in a screen of 19 cancer cell types as one of the 50 most commonly overexpressed genes [13]. Increased MTHFD2 expression is associated with acute myeloid leukemia, breast cancer, lung cancer, and liver cancer [14–20], and MTHFD2 is considered a novel target for anticancer therapy [21, 22]. A number of recent studies have shown that the mitochondrial 1C pathway is often reprogrammed in cancer cells and is especially critical for maintaining NADPH/NADP+ redox homeostasis [19, 23–27]. MTHFD2 is generally regarded as the enzyme responsible for this mitochondrial NADPH production, although ALDH1L2 has also been invoked [28]. Whereas ALDH1L2 clearly uses NADP+ [29], MTHFD2 has been considered an NAD+-dependent methylenetetrahydrofolate dehydrogenase since its discovery and early characterization [10, 30]. This raises the question whether MTHFD2 is in fact involved in maintaining mitochondrial NADPH/NADP+ redox homeostasis.
We previously showed that MTHFD2L can use either NAD+ or NADP+ at physiologically relevant substrate levels [11]. The use of NAD+ versus NADP+ will have a dramatic effect on the rate and direction of flux of 1C units in mitochondria, by affecting the equilibrium between 5,10-CH2-THF and 10-CHO-THF (and thus formate), depending on the relative levels of NAD+ and NADP+ in the mitochondrial matrix [11, 31]. Given the importance of MTHFD2 as a potential chemotherapeutic drug target, we have reinvestigated the redox cofactor specificity of the enzyme under more physiologically relevant conditions. We show here that MTHFD2, like MTHFD2L, possesses dual redox cofactor specificity for its CH2-THF dehydrogenase activity at physiologically relevant substrate levels.
Methods
Chemicals and reagents
NAD+ and NADP+ were purchased from US Biological (Swampscott, MA) and Sigma (St. Louis, MO), respectively. THF was prepared by the hydrogenation of folic acid (Sigma) using platinum oxide as a catalyst and purification of the THF product on a DEAE cellulose column (Sigma) [32]. CH2-THF was prepared nonenzymatically from THF and formaldehyde (Fisher, Waltham, MA) [33]. The yield of CH2-THF was determined by solving the equilibria of THF, formaldehyde, and β-mercaptoethanol [34]. Tetrahydropteroylpentaglutamate (H4PteGlu5) was prepared by a modified NaBH4 reduction from the corresponding pteroylpentaglutamate (PteGlu5) (Schircks Laboratories, Jona, Switzerland), as described previously [35]. Further preparation of 5,10-CH2-H4PteGlu5 was accomplished by incubation with formaldehyde as described previously [33].
Preparation of MTHFD2 and MTHFD2L
Purified human MTHFD2 was a gift from Dr. Vipin Suri (Raze Therapeutics). Briefly, 6× histidine-tagged human MTHFD2 was expressed in Escherichia coli and purified using size exclusion chromatography. The resulting protein corresponded to the molecular weight of 36.7 kDa with the tag. Cloning, expression, and purification of rat MTHFD2L were conducted as described previously [11].
5,10-Methylene-THF dehydrogenase assay
A microplate assay was used for determination of kinetic parameters as described previously [36]. CH2-THF dehydrogenase activity was determined by an end-point assay. The reaction buffer consisted of 50 mM HEPES (pH 8.0), 100 mM KCl, 5 mM MgCl2, 0.4 mM CH2-THF, 40 mM β-mercaptoethanol, and either NAD+ (1 mM) or NADP+ (6 mM). Potassium phosphate (25 mM) was also included for the NAD+-dependent activity. Sixty microliters of reaction mixture without CH2-THF and 20 μl of purified MTHFD2 or MTHFD2L were mixed, and the enzyme reaction was initiated by the addition of 20 μl of CH2-THF followed by incubation at 30 °C for 5 min. The reaction was quenched with 200 μl of 3% perchloric acid, and the plate was read at 350 nm on FlexStation 3 (Molecular Devices, Sunyvale, CA). The path length was corrected using near-infrared measurements [37]. For the determination of kinetic parameters, initial rate data was fitted to the Michaelis-Menten equation by non-linear regression using Prism (GraphPad, La Jolla, CA).
Results and discussion
Although the bifunctional MTHFD2 is widely known as a NAD+-dependent 5,10-CH2-THF dehydrogenase, it has been reported to use NADP+ at low efficiency [31]. However, that study did not use physiologically relevant substrate concentrations. To verify redox cofactor specificity of MTHFD2, we first compared 5,10-CH2-THF dehydrogenase activities of the enzyme with NAD+ and NADP+ under standard saturating substrate conditions. In order to allow direct comparisons between MTHFD2 and MTHFD2L, MTHFD2L activity assays were conducted in parallel. The MTHFD2L data were virtually identical to our previously published results [11]; data from [11] are included in the figures for comparison. With 5,10-CH2-H4PteGlu1 substrate, NAD+-dependent dehydrogenase activity of MTHFD2 was 8.5-fold higher than its NADP+-dependent activity (Fig. 1b). NAD+-dependent dehydrogenase activity of MTHFD2L was 3.4-fold higher than its NADP+-dependent activity under saturating substrate conditions (Fig. 1b; inset).
To explore redox cofactor specificity in MTHFD2 under more physiologically relevant substrate conditions, we repeated the assay with CH2-H4PteGlu5. Folylpolyglutamate specificity is one of the characteristic features of enzymes in the one-carbon metabolism [38], and the folate coenzymes typically found in mammalian mitochondria contain chain lengths of 6–9 glutamates [39]. While the NAD+-dependent activity of MTHFD2 slightly decreased, its maximal NADP+-dependent activity considerably increased with CH2-H4PteGlu5 compared to CH2-H4PteGlu1 (Fig. 1c). By comparison, MTHFD2L exhibits an even more dramatic increase in NADP+-dependent activity with the pentaglutamate coenzyme (Fig. 1c; inset).
To further investigate redox cofactor specificity of MTHFD2, steady-state kinetic parameters for CH2-THF dehydrogenase activity were determined using CH2-H4PteGlu1. MTHFD2 showed higher specific activity than MTHFD2L with both NAD+ and NADP+ (Fig. 2a, b). With the monoglutamate folate substrate, MTHFD2 exhibited a k cat/KM ratio eightfold higher for NAD+ than for NADP+, indicating a strong preference for NAD+ at saturating substrate concentrations (Table 1). In comparison, MTHFD2L has only a twofold higher k cat/KM for NAD+ versus NADP+ [11].
Fig. 2.
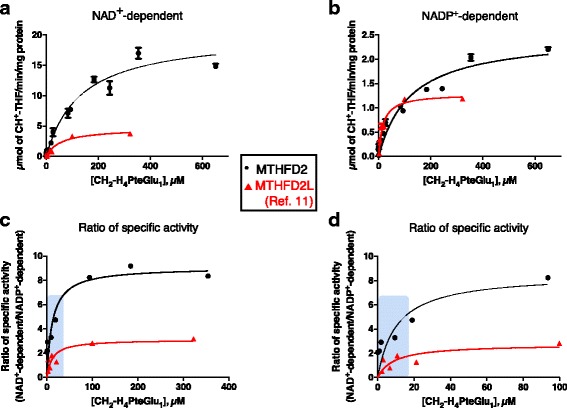
Redox cofactor specificity of MTHFD2 with CH2-H4PteGlu1. CH2-THF dehydrogenase activity of purified MTHFD2 was assayed with respect to CH2-H4PteGlu1 concentration using NAD+ (1.0 mM) (panel a) or NADP+ (6.0 mM) (panel b). NAD+-dependent reactions also included 25 mM Pi. The data were fit to the Michaelis-Menten equation. c The ratio of NAD+- to NADP+-dependent activity plotted as a function of CH2-H4PteGlu1 concentration. The 0–100 μM CH2-H4PteGlu1 range is magnified in panel (d). Data for MTHFD2L from ref. [11]. The shaded boxes in c and d indicate the reported mitochondrial matrix concentration ranges for 5,10-CH2-THF (2.5–25 μM) [41–43]
Table 1.
Kinetic parameters for MTHFD2 5,10-CH2-THF dehydrogenase activity. 5,10-CH2-THF kinetic parameters were determined using saturating concentrations of NAD+ (1.0 mM) or NADP+ (6.0 mM). When NAD+ was used, potassium phosphate (25 mM) and MgCl2 (5 mM) were also included
| CH2-H4PteGlu1 | CH2-H4PteGlu5 | |||||
|---|---|---|---|---|---|---|
| KM (μM) | kcat (s−1) | kcat/KM (s−1 μM−1) | KM (μM) | kcat (s−1) | kcat/KM (s−1 μM−1) | |
| NAD+-dependent | 133 ± 20 | 12.4 ± 0.71 | 0.093 | 359 ± 32 | 15.4 ± 0.55 | 0.043 |
| NADP+-dependent | 123 ± 24 | 1.5 ± 0.11 | 0.012 | 302 ± 35 | 6.4 ± 0.29 | 0.021 |
To better understand the cofactor preference of the MTHFD2 dehydrogenase activity, the ratio of NAD+-dependent specific activity versus NADP+-dependent specific activity was calculated at each CH2-H4PteGlu1 concentration (Fig. 2c). At high CH2-H4PteGlu1 concentrations, both MTHFD2 and MTHFD2L clearly preferred NAD+. However, as the folate substrate concentration was lowered into the physiological range (2.5–25 μM reported mitochondrial matrix CH2-THF concentration range), the ratio of NAD+- to NADP+-dependent activity for both enzymes decreased. MTHFD2L approached a ratio of 1, whereas the ratio for MTHFD2 dropped from 8 to 2–4 in the physiological folate range (Fig. 2d).
With the more physiologically relevant pentaglutamate substrate (CH2-H4PteGlu5), MTHFD2’s preference for NAD+ is dramatically decreased (Fig. 3a, b). MTHFD2 exhibited a k cat/KM ratio only twofold higher for NAD+ than for NADP+ (Table 1). Moreover, importantly, as the pentaglutamate substrate concentration was lowered into the physiological range, the ratio of NAD+- to NADP+-dependent activity for MTHFD2 approached 1 (Fig. 3c, d), similar to MTHFD2L.
Fig. 3.
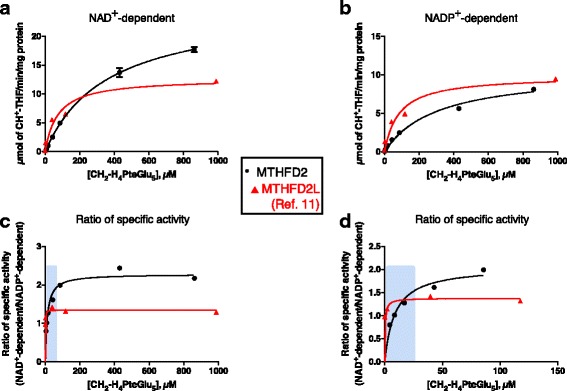
Redox cofactor specificity of MTHFD2 with CH2-H4PteGlu5. CH2-THF dehydrogenase activity of purified MTHFD2 was assayed with respect to CH2-H4PteGlu5 concentration using NAD+ (1.0 mM) (panel a) or NADP+ (6.0 mM) (panel b). NAD+-dependent reactions also included 25 mM Pi. The data were fit to the Michaelis-Menten equation. c The ratio of NAD+- to NADP+-dependent activity plotted as a function of CH2-H4PteGlu5 2concentration. The 0–150 μM CH2-H4PteGlu5 range is magnified in panel (d). Data for MTHFD2L from ref. [11]. The shaded boxes in c and d indicate the reported mitochondrial matrix concentration ranges for 5,10-CH2-THF as described in Fig. 2
Conclusion
The role of MTHFD2 in 1C metabolism and generation of redox coenzymes places this enzyme in two pathways of central metabolic importance. Understanding how MTHFD2 is involved in both of these biological processes is of critical importance to effectively develop therapeutics targeting this enzyme for cancer treatment. As previously reported [31, 40], the methylenetetrahydrofolate dehydrogenase activity of MTHFD2 exhibits a higher preference for NAD+ than for NADP+ with monoglutamylated THF (Figs. 1b and 2). However, using pentaglutamylated THF, a physiologically relevant substrate that the enzyme would encounter in mitochondria, MTHFD2 shows increased NADP+-dependent activity (Figs. 1b and 3). Indeed, at the lowest CH2-H4PteGlu5 concentrations, MTHFD2 is more active with NADP+ than with NAD+ (Fig. 3d). These data reveal that MTHFD2, like MTHFD2L, is a dual redox cofactor-specific methylenetetrahydrofolate dehydrogenase, active with both NAD+ and NADP+ under physiological conditions.
The mitochondrial 1C pathway is now understood to be especially critical for maintaining NADPH/NADP+ redox homeostasis [19, 23–27]. Despite the fact that MTHFD2 has been consistently described as an NAD+-dependent 5,10-CH2-THF dehydrogenase since its initial description in 1960 [30], several of these studies invoked MTHFD2 as the source of mitochondrial NADPH production. The kinetic analyses reported here clearly reveal the ability of MTHFD2 to use NADP+ in vitro and provide a mechanistic basis for these flux analyses that implicate MTHFD2 in mitochondrial NADPH production [24, 26].
Both Mthfd2 and Mthfd2l are expressed during embryogenesis but differ in timing of expression. Mthfd2l expression is low in early developmental stages but begins to increase at embryonic day 10.5 and remains elevated through birth while Mthfd2 is expressed more abundantly during early developmental stages and begins to taper off, with little or no expression observed in most adult tissues [4, 11]. Due to the similarity of the enzymatic activities of MTHFD2 and MTHFD2L, we propose that MTHFD2 may be expressed to boost flux through the mitochondrial folate pathway during early periods of embryogenesis when MTHFD2L alone is not sufficient to support high rates of cell proliferation. Likewise, enhanced expression of MTHFD2 in cancer cells is predicted to enable increased flux through the mitochondrial 1C metabolic pathway, enabling unregulated proliferation.
Acknowledgements
We thank Dr. Vipin Suri (Raze Therapeutics) for supplying purified human MTHFD2 protein.
Funding
This work was supported in part by NIH F32HD074428 to JM and GM086856 and HD083809 to DRA.
Availability of data and materials
The datasets used and/or analyzed in this study are available from the corresponding author on a reasonable request.
Abbreviations
- 10-CHO-THF
10-Formyl-tetrahydrofolate
- 1C
One-carbon
- CH+-THF
5,10-Methenyl-tetrahydrofolate
- CH2-H4PteGlu1
5,10-Methylene-tetrahydropteroyl monoglutamate
- CH2-H4PteGlu5
5,10-Methylene-tetrahydropteroyl pentaglutamate
- CH2-THF
5,10-Methylene-tetrahydrofolate
- HEPES
4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid
- THF
Tetrahydrofolate
Authors’ contributions
MS, JM, and DRA designed the research. MS performed the research. MS, JM, and DRA analyzed the data. MS, JM, and DRA wrote the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Minhye Shin, Email: alsgp01@gmail.com.
Jessica Momb, Email: jessicamomb@gmail.com.
Dean R. Appling, Phone: 512-471-5842, Email: dappling@austin.utexas.edu
References
- 1.Mattaini KR, Sullivan MR, Vander Heiden MG. The importance of serine metabolism in cancer. J Cell Biol. 2016;214:249–257. doi: 10.1083/jcb.201604085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ducker GS, Rabinowitz JD. One-carbon metabolism in health and disease. Cell Metab. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tibbetts AS, Appling DR. Compartmentalization of mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr. 2010;30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- 4.Pike ST, Rajendra R, Artzt K, Appling DR. Mitochondrial C1-THF synthase (MTHFD1L) supports flow of mitochondrial one-carbon units into the methyl cycle in embryos. J Biol Chem. 2010;285:4612–4620. doi: 10.1074/jbc.M109.079855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Momb J, Lewandowski JP, Bryant JD, Fitch R, Surman DR, Vokes SA, Appling DR. Deletion of Mthfd1l causes embryonic lethality and neural tube and craniofacial defects in mice. Proc Natl Acad Sci U S A. 2013;110:549–554. doi: 10.1073/pnas.1211199110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nikkanen J, Forsström S, Euro L, Paetau I, Kohnz RA, Wang L, Chilov D, Viinamäki J, Roivainen A, Marjamäki P, et al. Mitochondrial DNA replication defects disturb cellular dntp pools and remodel one-carbon metabolism. Cell Metab. 2016;23:635–648. doi: 10.1016/j.cmet.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 7.Meiser J, Tumanov S, Maddocks O, Labuschagne CF, Athineos D, Van Den Broek N, Mackay GM, Gottlieb E, Blyth K, Vousden K, et al. Serine one-carbon catabolism with formate overflow. Sci Adv. 2016;2:e1601273. doi: 10.1126/sciadv.1601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao XR, Ong S-E, Goldberger O, Peng J, Sharma R, Thompson DA, Vafai SB, Cox AG, Marutani E, Ichinose F, et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. elife. 2016;5:e10575. doi: 10.7554/eLife.10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walkup AS, Appling DR. Enzymatic characterization of human mitochondrial C1-tetrahydrofolate synthase. Arch Biochem Biophys. 2005;442:196–205. doi: 10.1016/j.abb.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Mejia NR, MacKenzie RE. NAD-dependent methylenetetrahydrofolate dehydrogenase is expressed by immortal cells. J Biol Chem. 1985;260:14616–14620. [PubMed] [Google Scholar]
- 11.Shin M, Bryant JD, Momb J, Appling DR. Mitochondrial MTHFD2L is a dual redox cofactor-specific methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase expressed in both adult and embryonic tissues. J Biol Chem. 2014;289:15507–15517. doi: 10.1074/jbc.M114.555573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolusani S, Young BA, Cole NA, Tibbetts AS, Momb J, Bryant JD, Solmonson A, Appling DR. Mammalian Mthfd2l encodes a mitochondrial methylenetetrahydrofolate dehydrogenase isozyme expressed in adult tissues. J Biol Chem. 2011;286:5166–5174. doi: 10.1074/jbc.M110.196840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, Huang J, Asplund A, Mootha VK. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pikman Y, Puissant A, Alexe G, Furman A, Chen LM, Frumm SM, Ross L, Fenouille N, Bassil CF, Lewis CA, et al. Targeting MTHFD2 in acute myeloid leukemia. J Exp Med. 2016;213:1285–1306. doi: 10.1084/jem.20151574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Huang Y, Jiang C, Ou H, Guo B, Liao H, Li X, Yang D. Methylenetetrahydrofolate dehydrogenase 2 overexpression is associated with tumor aggressiveness and poor prognosis in hepatocellular carcinoma. Dig Liver Dis. 2016;48:953–960. doi: 10.1016/j.dld.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 16.Moran DM, Trusk PB, Pry K, Paz K, Sidransky D, Bacus SS. Kras mutation status is associated with enhanced dependency on folate metabolism pathways in non-small cell lung cancer cells. Mol Cancer Ther. 2014;13:1611–1624. doi: 10.1158/1535-7163.MCT-13-0649. [DOI] [PubMed] [Google Scholar]
- 17.Liu F, Liu Y, He C, Tao L, He X, Song H, Zhang G. Increased MTHFD2 expression is associated with poor prognosis in breast cancer. Tumour Biol. 2014;35:8685–8690. doi: 10.1007/s13277-014-2111-x. [DOI] [PubMed] [Google Scholar]
- 18.Lehtinen L, Ketola K, Makela R, Mpindi JP, Viitala M, Kallioniemi O, Iljin K. High-throughput RNAi screening for novel modulators of vimentin expression identifies MTHFD2 as a regulator of breast cancer cell migration and invasion. Oncotarget. 2013;4:48–63. doi: 10.18632/oncotarget.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koufaris C, Gallage S, Yang T, Lau CH, Valbuena GN, Keun HC. Suppression of MTHFD2 in MCF-7 breast cancer cells increases glycolysis, dependency on exogenous glycine, and sensitivity to folate depletion. J Proteome Res. 2016;15:2618–2625. doi: 10.1021/acs.jproteome.6b00188. [DOI] [PubMed] [Google Scholar]
- 20.Samanta D, Park Y, Andrabi SA, Shelton LM, Gilkes DM, Semenza GL. PHGDH expression is required for mitochondrial redox homeostasis, breast cancer stem cell maintenance, and lung metastasis. Cancer Res. 2016;76:4430–4442. doi: 10.1158/0008-5472.CAN-16-0530. [DOI] [PubMed] [Google Scholar]
- 21.Tedeschi PM, Vazquez A, Kerrigan JE, Bertino JR. Mitochondrial methylene tetrahydrofolate dehydrogenase (MTHFD2) overexpression is associated with tumor cell proliferation and is a novel target for drug development. Mol Cancer Res. 2015;13:1361–1366. doi: 10.1158/1541-7786.MCR-15-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gustafsson R, Jemth A-S, Gustafsson NMS, Färnegårdh K, Loseva O, Wiita E, Bonagas N, Dahllund L, Llona-Minguez S, Häggblad M, et al. Crystal structure of the emerging cancer target MTHFD2 in complex with a substrate-based inhibitor. Cancer Res. 2017;77:937–948. doi: 10.1158/0008-5472.CAN-16-1476. [DOI] [PubMed] [Google Scholar]
- 23.Ye J, Fan J, Venneti S, Wan Y-W, Pawel BR, Zhang J, Finley LWS, Lu C, Lindsten T, Cross JR, et al. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discovery. 2014;4:1406–1417. doi: 10.1158/2159-8290.CD-14-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X, Kang Y, Rabinowitz JD. Reversal of cytosolic one-carbon flux compensates for loss of the mitochondrial folate pathway. Cell Metab. 2016;23:1140–1153. doi: 10.1016/j.cmet.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Baba T, Miyabayashi K, Sato T, Shima Y, Ichinose T, Miura D, Ohkawa Y, Suyama M, Morohashi KI. Role of Ad4-binding protein/steroidogenic factor 1 in regulating NADPH production in adrenocortical Y-1 cells. Endocr J. 2017;64:315–324. doi: 10.1507/endocrj.EJ16-0467. [DOI] [PubMed] [Google Scholar]
- 27.Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, Leitch AM, Johnson TM, DeBerardinis RJ, Morrison SJ. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527:186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krupenko NI, Dubard ME, Strickland KC, Moxley KM, Oleinik NV, Krupenko SA. ALDH1L2 is the mitochondrial homolog of 10-formyltetrahydrofolate dehydrogenase. J Biol Chem. 2010;285:23056–23063. doi: 10.1074/jbc.M110.128843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scrimgeour KG, Huennekens FM. Occurrence of a DPN-linked N5,N10-methylene tetrahydrofolic dehydrogenase in ehrlich ascites tumor cells. Biochem Biophys Res Comm. 1960;2:230–233. doi: 10.1016/0006-291X(60)90019-X. [DOI] [Google Scholar]
- 31.Yang X-M, MacKenzie RE. NAD-dependent methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase is the mammalian homolog of the mitochondrial enzyme encoded by the yeast mis1 gene. Biochemistry. 1993;32:11118–11123. doi: 10.1021/bi00092a022. [DOI] [PubMed] [Google Scholar]
- 32.Curthoys NP, Rabinowitz JC. Formyltetrahydrofolate synthetase. Binding of adenosine triphosphate and related ligands determined by partition equilibrium. J Biol Chem. 1971;246:6942–6952. [PubMed] [Google Scholar]
- 33.Appling DR, West MG. Monofunctional NAD-dependent, 5,10-methylenetetrahydrofolate dehydrogenase from saccharomyces cerevisiae. Methods Enzymol. 1997;281:178–188. doi: 10.1016/S0076-6879(97)81024-5. [DOI] [PubMed] [Google Scholar]
- 34.Kallen RG, Jencks WP. The mechanism of the condensation of formaldehyde with tetrahydrofolic acid. J Biol Chem. 1966;241:5851–5863. [PubMed] [Google Scholar]
- 35.Suliman HS, Sawyer GM, Appling DR, Robertus JD. Purification and properties of cobalamin-independent methionine synthase from Candida albicans and Saccharomyces cerevisiae. Arch Biochem Biophys. 2005;441:56–63. doi: 10.1016/j.abb.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 36.Wagner W, Breksa AP, III, Monzingo AF, Appling DR, Robertus JD. Kinetic and structural analysis of active site mutants of monofunctional NAD-dependent 5,10-methylenetetrahydrofolate dehydrogenase from Saccharomyces cerevisiae. Biochemistry. 2005;44:13163–13171. doi: 10.1021/bi051038x. [DOI] [PubMed] [Google Scholar]
- 37.Palmer KF, Williams D. Optical-properties of water in near-infrared. J Opt Soc Am. 1974;64:1107–1110. doi: 10.1364/JOSA.64.001107. [DOI] [Google Scholar]
- 38.Schirch V, Strong WB. Interaction of folylpolyglutamates with enzymes in one-carbon metabolism. Arch Biochem Biophys. 1989;269:371–380. doi: 10.1016/0003-9861(89)90120-3. [DOI] [PubMed] [Google Scholar]
- 39.Lin B-F, Huang R-FS, Shane B. Regulation of folate and one-carbon metabolism in mammalian cells. III. Role of mitochondrial folylpoly-γ-glutamate synthetase. J Biol Chem. 1993;268:21674–21679. [PubMed] [Google Scholar]
- 40.Christensen KE, Mirza IA, Berghuis AM, Mackenzie RE. Magnesium and phosphate ions enable NAD binding to methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase. J Biol Chem. 2005;280:34316–34323. doi: 10.1074/jbc.M505210200. [DOI] [PubMed] [Google Scholar]
- 41.Horne DW, Patterson D, Cook R. Effect of nitrous oxide inactivation of vitamin B12-dependent methionine synthetase on the subcellular distribution of folate coenzymes in rat liver. Arch Biochem Biophys. 1989;270:729–733. doi: 10.1016/0003-9861(89)90556-0. [DOI] [PubMed] [Google Scholar]
- 42.Seither RL, Trent DF, Mikulecky DC, Rape TJ, Goldman ID. Folate-pool interconversions and inhibition of biosynthetic processes after exposure of L1210 leukemia cells to antifolates. Experimental and network thermodynamic analyses of the role of dihydrofolate polyglutamates in antifolate action in cells. J Biol Chem. 1989;264:17016–17023. [PubMed] [Google Scholar]
- 43.Nijhout HF, Reed MC, Lam SL, Shane B, Gregory JF, 3rd, Ulrich CM. In silico experimentation with a model of hepatic mitochondrial folate metabolism. Theor Biol Med Model. 2006;3:40. doi: 10.1186/1742-4682-3-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed in this study are available from the corresponding author on a reasonable request.


