Abstract
Background
Bloodstream infection (BSI) in children causes significant morbidity and mortality. There are few studies describing the epidemiology of BSI in South African children.
Methods
A retrospective descriptive cohort study was conducted at a paediatric referral hospital in Cape Town, South Africa. The National Health Laboratory Service (NHLS) microbiology database was accessed to identify positive blood culture specimens during the period 2011–2012. Demographic and clinical details, antimicrobial management and patient outcome information were extracted from medical and laboratory records. Antibiotic susceptibility results of identified organisms were obtained from the NHLS database.
Results
Of the 693 unique bacterial and fungal BSI episodes identified during the study period, 248 (35.8%) were community-acquired (CA), 371 (53.5%) hospital-acquired (HA) and 74 (10.7%) healthcare-associated (HCA). The overall risk was 6.7 BSI episodes per 1000 admissions. Escherichia coli, Staphylococcus aureus and Streptococcus pneumoniae were the most frequent causes of CA-BSI and Klebsiella pneumoniae, Acinetobacter baumanii and S.aureus were most commonly isolated in HA-BSI. On multivariable analysis, severe underweight, severe anaemia at the time of BSI, admission in the ICU at the time of BSI, and requiring ICU admission after BSI was diagnosed were significantly associated with 14-day mortality.
Conclusion
This study adds to the limited literature describing BSI in children in Africa. Further studies are required to understand the impact that BSI has on the paediatric population in sub-Saharan Africa.
Keywords: Bloodstream infections, Children, Africa, Antimicrobial resistance
Background
Bloodstream infection (BSI) causes significant morbidity and mortality in children and increase healthcare expenditure [1, 2]. In Africa, community-acquired (CA)-BSI was identified in 5.8% of children presenting to hospital in Tanzania and up to 19.9% in rural Ghana [3, 4], while hospital-acquired (HA) BSI was less frequently described. In a recent study among children hospitalised in a Kenyan district hospital, the overall risk of acquiring a nosocomial BSI was 5.9 per 1000 admissions. [5]. South Africa has a paucity of literature describing BSI in children. In one study, the risk of CA-BSI and HA-BSI was 16 and 5 per 1000 admissions, respectively [6]. More recent studies have documented lower pathogen yields and high contamination rates in blood culture specimens. The spectrum of pathogens described was similar to elsewhere in Africa with the exception of lower rates of infection caused by non-typhoidal Salmonella species [7, 8].
Clinical manifestations of childhood BSI in sub-Saharan Africa (SSA) are also poorly documented. Fever may be the only manifestation of childhood invasive infection. In SSA with the added burden of malaria, HIV-infection, tuberculosis and malnutrition, the treatment of fever is often syndromic, targeting multiple possible aetiologies, without necessarily considering BSI as a potential cause [9]. Not all fever is due to malaria, as shown in a recent Tanzanian study where viruses and bacteria caused more than 90% of childhood fevers. In that study only 6.4% of fever episodes were caused by malaria [10]. In HIV-infected children treated with antiretroviral therapy (ART), the risk of BSI is highest in the first 3 months after ART initiation with a steady decline thereafter [11].
Antimicrobial resistance is an important global public health threat. A systematic review on antimicrobial resistance among gram-negative bacteria in developing countries including 15 studies from Africa showed that in children, 50% and 11% of Escherichia coli isolates were resistant to ampicillin and gentamicin, respectively and 30% of Klebsiella pneumoniae isolates were resistant to ceftriaxone [12]. These antimicrobials are used empirically to treat ill children with invasive infection. None of the studies in this review included children from South Africa but Dramowski and colleagues recently showed that more than 75% of K. pneumoniae isolates at their hospital in Cape Town were extended spectrum beta-lactamase (ESBL) producing organisms [7].
Despite the growing body of literature there remains many gaps in our knowledge about the clinical and laboratory aspects of BSI in SSA. To address some of these aspects of BSI in SSA we conducted an observational study aimed at describing the epidemiology, clinical manifestations and antimicrobial management of BSI at Red Cross War Memorial Children’s Hospital (RCWMCH).
Methods
Study design and setting
This retrospective descriptive cohort study was completed at RCWMCH on children with culture-confirmed BSI, diagnosed between 1 January 2011 and 31 December 2012. RCWMCH is a 273-bed tertiary level paediatric hospital in Cape Town, Western Cape province, South Africa. The hospital has emergency, general paediatric, specialised paediatric, surgical, burns, trauma and intensive care facilities serving as a referral centre for the Western Cape and surrounding provinces. Dedicated early neonatal care linked to obstetric services is provided by other hospitals in Cape Town. However, neonates requiring immediate surgical care, or medical care after the first 7 days of life are admitted to RCWMCH.
Study population
Most children hospitalised in RCWMCH originate from poor, peri-urban communities in the Western Cape. During the study period there were 43,663 admissions to the hospital and 16,951 blood culture specimens were processed from patients with suspected invasive infection, i.e. 1 blood culture specimen per 2.6 hospital admissions. The medical microbiology database of the National Health Laboratory Service (NHLS) was accessed, and 2084 positive blood culture results of children hospitalised at RCWMCH during the study period were identified. Organisms isolated from these blood cultures were stratified into pathogens and non-pathogens. Coagulase-negative staphylococci, Staphylococcus epidermidis, Bacillus spp., Micrococcus spp., viridans streptococci, and coryneform bacteria were regarded as contaminants and excluded from the analysis unless they were isolated in two or more independent blood culture specimens from the same patient within a 48-h period, in which case they were included as a pathogen [13]. Within this group of blood culture results, repeat blood cultures within 14 days of the initial blood culture and yielding an identical pathogen were regarded as part of the same potential BSI episode, and therefore excluded. A recurrent BSI episode was defined as the isolation of the same or different organism on blood culture more than 14 days after the initial/previous BSI episode. To verify each BSI episode (new or recurrent) identified during this laboratory review, the medical records of all patients linked to these events were reviewed. During this process 20 episodes were excluded as they had been misclassified as BSI episodes. Thus the study population comprised 548 children who experienced 693 unique bacterial or fungal BSI episodes (Fig. 1).
Fig. 1.
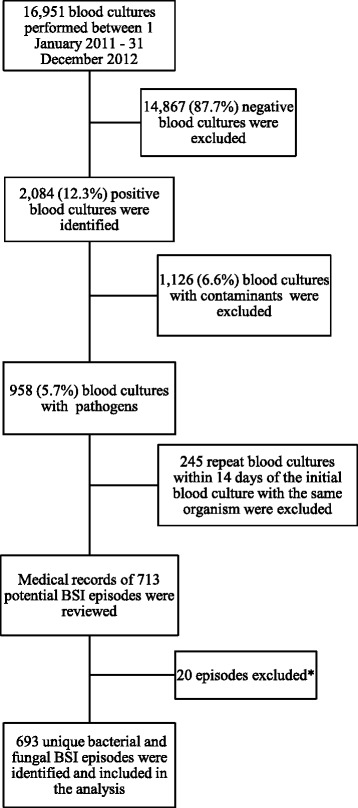
Selection of BSI episodes for data analysis at Red Cross War Memorial Children’s Hospital (RCWMCH). *14 post-mortem specimens and 6 non-blood specimens (peritoneal fluid [3], eye fluid aspirate [1], pus [1] and lymph node aspirate [1]) inoculated in blood culture bottles
Data collection
Paper-based medical records of patients with BSI episodes were reviewed and relevant data was extracted and manually transferred onto standardised data collection forms. HIV status was determined using HIV test results obtained from patient medical and laboratory records. Microbiological results obtained from the NHLS database were also entered on the data collection forms.
Microbiological procedures
All microbiology testing was conducted at the Groote Schuur National Health Laboratory Service (NHLS) microbiology laboratory. The laboratory uses the BACTEC™ 9240 automated blood culture system (Becton Dickinson, Sparks, Maryland). For blood culture bottles flagging positive with mono-morphic Gram-negative bacilli seen on Gram stain, a locally validated method of direct inoculation into the automated Vitek®2 system (bioMérieux, Inc., France) was employed, using Vitek®2 ID-GNB and AST-N133 cards for identification and susceptibility testing respectively. This direct method was supplemented, where necessary with repeat testing from bacterial colonies subcultured onto agar plates, using either Vitek®2, disk diffusion or gradient diffusion E-test (bioMérieux, Marcy l’Etoile, France) methods. Standard biochemical methods and disk diffusion and gradient diffusion antibiotic susceptibility tests were used to evaluate Gram-positive organisms, while yeasts were tested with the Vitek®2’s YST identification and AST-YS07 cards.
Susceptibility results were interpreted according to the Clinical Laboratory Standards Institute (CLSI) criteria for 2011 and 2012 [14, 15]. Ertapenem minimum inhibitory concentration (MIC) breakpoints for Gram-negative organisms were revised in 2012, as were the ciprofloxacin breakpoints for Salmonella species. For Pseudomonas species, the breakpoints for meropenem, imipenem and piperacillin-tazobactam were revised in 2012.
ESBLs were detected by the Vitek®2 Advanced Expert system or by the double disk synergy method if using disk diffusion testing. Despite the changes to reporting of cephalosporin susceptibility in ESBL-producing organisms introduced by CLSI in 2010, the laboratory, in line with the contemporary national practice, continued to report ESBL- producing Enterobacteriaceae as resistant to all cephalosporins.
Study definitions
BSI was classified as (1) community-acquired (CA) if the pathogen was isolated within the first 48 h after admission and did not meet the definition of healthcare-associated infection, (2) hospital-acquired (HA) if the pathogen was isolated from a blood culture obtained more than 48 h after hospitalisation without the patient having been a resident of a long-term care facility or hospitalised during the preceding 28 days, or (3) healthcare-associated (HCA) if the pathogen was isolated in a patient within the first 48 h of being hospitalised from a long-term care facility, or readmitted within 28 days of discharge from a hospital or birth facility [5, 13]. BSI with a clinical focus refers to a laboratory confirmed BSI event accompanied by one of more clinical sites of infection such as pneumonia, meningitis, soft tissue infection or line infection as extracted from the patient’s medical records.
The HIV status was defined as follows: (1) HIV-infected: a child < 18 months old with a positive HIV DNA PCR result confirmed by either a quantitative HIV RNA PCR or repeat HIV DNA PCR test, or a child ≥ 18 months old with 2 positive serological test results (HIV ELISA or HIV Rapid test) or a positive HIV DNA PCR result confirmed by either a quantitative HIV RNA PCR or repeat HIV DNA PCR test, (2) HIV-uninfected: a child with a negative HIV serological or HIV DNA PCR result and (3) Unknown HIV status: a child with unknown maternal HIV status and who was not tested for HIV infection.
Moderate and severe underweight for age (UWFA) were defined as weight-for-age z score (WAZ) between −2 and −3 standard deviations (SD) below the median World Health Organisation (WHO) growth reference standards, and a WAZ < −3 SD respectively [16].
Antimicrobial management at the time of the BSI was considered to be effective if the organism isolated was susceptible to the prescribed antimicrobial agent.
Statistical analysis
All data was entered anonymously into an Excel spreadsheet and analysed using Stata release 12.0 statistical software package (STATACorp, College Station, Texas). Continuous variables were expressed as medians (interquartile range, IQR). Frequencies and proportions were used to describe categorical variables. Chi-squared and Kruskal-Wallis tests of association were used to assess associations between categorical and numerical variables across the type of BSI for the baseline patient characteristics and BSI with or without a clinical focus for blood marker variables. Cox proportional hazards regression was used to assess factors associated with 14-day mortality, adjusting for the baseline demographic, clinical and blood marker variables. P-values < 0.05 were considered significant.
Results
Classification and risk of bloodstream infection
Overall, 248 (35.8%) CA-, 371 (53.5%) HA- and 74 (10.7%) HCA-BSI episodes were identified. Fewer blood cultures were processed and fewer BSI episodes occurred in 2012 compared to 2011. The overall risk of CA-BSI, HA-BSI and HCA-BSI was 5.7, 8.5 and 1.7 episodes per 1000 admissions, respectively (Table 1).
Table 1.
Classification and risk of bloodstream infection (BSI), 2011–2012
| 2011 | 2012 | 2011–2012 | |
|---|---|---|---|
| Hospital admissions | 22,685 | 20,978 | 43,663 |
| Blood cultures processed | 9108 | 7843 | 16,951 |
| Classification of bloodstream infection (n/N(%)) | |||
| Community-acquired | 151/413 (36.6) | 97/280 (34.6) | 248/693 (35.8) |
| Hospital-acquired | 214/413 (51.8) | 157/280 (56.1) | 371/693 (53.5) |
| Healthcare-associated | 48/413 (11.6) | 26/280 (9.3) | 74/693 (10.7) |
| Risk of BSI per 1000 hospital admissions | |||
| Community-acquired | 6.7 | 4.6 | 5.7 |
| Hospital-acquired | 9.4 | 7.5 | 8.5 |
| Healthcare-associated | 2.1 | 1.2 | 1.7 |
Baseline characteristics of study participants
Table 2 describes the baseline characteristics of the study participants at the time of each BSI episode and disaggregates the information according to the type of BSI. The median age at the time of BSI was 11.5 months (IQR 3.6–50). Furthermore, 40.7% (273/670) of episodes occurred in children who were moderately or severely underweight. Of the 524 children (75.6%) whose HIV status was known at the BSI event, 13.4% (70/524) were HIV-infected.
Table 2.
Baseline patient characteristics
| CA-BSI (N = 248a) |
HA-BSI (N = 371a) |
HCA-BSI (N = 74) |
Total BSIs (N = 693a) |
|
|---|---|---|---|---|
| Age median (IQR) | 9.3 (3.1–39.3) | 13.0 (4.0–59.7) | 7.1 (1.4–67.0) | 11.5 (3.6–50.0) |
| Age categories, n/N(%) | ||||
| 0–28 days | 31 (12.5) | 28 (7.5) | 16 (21.6) | 75 (10.8) |
| 1 - < 2 months | 17 (6.9) | 17 (4.6) | 6 (8.1) | 40 (5.8) |
| 2 - < 12 months | 88 (35.5) | 132 (35.6) | 22 (29.7) | 242 (34.9) |
| 12 months – < 5 years | 67 (27) | 106 (28.6) | 11 (14.9) | 184 (26.5) |
| > 5 years | 45 (18.1) | 88 (23.7) | 19 (25.7) | 152 (21.9) |
| WAZ median (IQR) | −1.2 (−2.5–0.1) | −1.8 (−3.4–0.6) | −1.9 (−3.3–1.0) | −1.6 (−3–0.3) |
| WAZ categories, n/N (%) | ||||
| Severe underweight | 44/240 (18.3) | 103/361 (28.5) | 22/69 (31.9) | 169/670 (25.2) |
| Moderate underweight | 33/240 (13.8) | 60/361 (16.6) | 11/69 (15.9) | 104/670 (15.5) |
| Mild-normal | 163,240 (67.9) | 198/361 (54.8) | 36/69 (52.2) | 397/670 (59.3) |
| HIV-infected, n/N (%) | 33 (13.3) | 33 (8.9) | 4 (5.4) | 70 (10.1) |
| HIV-uninfected, n/N (%) | 172 (69.3) | 232 (62.5) | 49 (66.2) | 454 (65.5) |
| HIV not tested, n/N (%) | 43 (6.2) | 106 (28.6) | 21 (28.4) | 169 (24.4) |
| Receiving ART at the time of the BSI in HIV infected child, n/N(%) | 11/33 (33.3) | 29/33 (87.9) | 3/4 (75) | 43/70 (61.4) |
a N denominator used unless otherwise stated, IQR interquartertile range, BSI bloodstream infection, CA-BSI community-acquired bloodstream infection, HA-BSI hospital-acquired bloodstream infection, HCA-BSI healthcare-associated bloodstream infection, WAZ weight-for-age z-score, HIV-infected human immunodeficiency virus infected, ART antiretroviral therapy
Clinical spectrum of bloodstream infections
A clinical site of infection in addition to the bloodstream was identified in 481/663 (72.5%) of the BSI episodes, of which 327/481(68.0%) had a single identifiable site of infection. Two or more clinical sites of infection were identified in the remaining 154 BSI episodes (Table 3). Pneumonia and gastroenteritis were commonly diagnosed at the time of the BSI episode. There was no significant difference in the proportions of BSI episodes with and without a clinical focus associated with a temperature ≥ 38° C; 60% (270/450) vs 62.8% (108/172), RR = 0.99 (95% CI: 0.86–1.14).
Table 3.
Clinical classification of bloodstream infection (BSI)
| CA-BSI (N = 242a) |
HA-BSI (N = 353a) |
HCA-BSI (N = 68a) |
Total BSIs (N = 663a) |
|
|---|---|---|---|---|
| n/N (%)) | n/N (%) | n/N (%) | n/N (%) | |
| BSI with no clinical focus | 28 (11.6) | 131(37.1) | 23 (33.8) | 182 (27.5) |
| Hypothermiab | 5/28 (17.9) | 2/131 (1.5) | 0/23 (0) | 7/182 (3.8) |
| Feverc | 14/28 (50) | 77/131 (58.8) | 17/23 (74) | 108/182 (59.3) |
| BSI with a clinical focus | 214(88.4) | 222(62.9) | 45(66.2) | 481 (72.5) |
| Hypothermiab | 11/214 (5.1) | 4/222 (1.8) | 5/45 (11.1) | 20/481 (4.2) |
| Feverc | 112/214 (52.3) | 136/222 (61.3) | 22/45 (48.9) | 270/481 (56.1) |
| Pneumonia | 65/214 (30.4) | 91/222 (41) | 16/45 (35.6) | 172/481 (35.8) |
| Gastroenteritis | 67/214 (31.3) | 28/222 (12.6) | 10/45 (22.2) | 105/481 (21.8) |
| Soft tissue infectiond | 18/214 (8.4) | 50/222 (22.5) | 4/45 (8.9) | 72/481 (15) |
| Meningitis | 39/214 (18.2) | 1/222 (0.5) | 13/45 (28.9) | 53/481 (11) |
| UTI | 29/214 (13.6) | 20/222 (9) | 4/45 (8.9) | 53/481 (11) |
| Line sepsis | 4/214 (1.9) | 29/222 (13.1) | 3/45 (6.7) | 36/481 (7.4) |
| URTI | 20/214 (9.3) | 8/222 (3.6) | 3/45 (6.7) | 31/481 (6.4) |
| Empyema | 4/214 (1.9) | 5/222 (2.3) | – | 9/481 (1.9) |
| Septic arthritis | 6/214 (2.8) | 3/222 (1.4) | – | 9/481 (1.9) |
| Bronchiolitis | 4/214 (1.9) | – | 1/45 (2.2) | 5/481 (1) |
| Pleural effusion | – | 3/222 (1.4) | – | 3/481 (0.6) |
BSI bloodstream infection, CA-BSI community-acquired bloodstream infection, HA-BSI hospital-acquired bloodstream infection, HCA-BSI healthcare-associated bloodstream infection, aN = denominator used unless otherwise stated; bBody temperature ≤ 35.5 ° C around the time of BSI; cBody temperature ≥ 38 °C around the time of BSI; dSoft tissue infection includes skin abscesses, burn wound infections, wound sepsis and infected eczema or scabies, UTI urinary tract infection, URTI upper respiratory tract infection
Spectrum of organisms
Fifty two different organisms were isolated from the 693 BSI episodes including 11 fungi. Fifty eight percent of the BSI episodes were caused by gram-negative bacteria, 36% by gram-positive bacteria and 6% by fungi. Candida albicans (19/42) was the dominant fungal isolate. Cryptococcus neoformans was isolated in one HIV-infected child with severe immunodeficiency i.e. an absolute CD4 count of < 20 × 109/L and a CD4 percentage of < 1% (Table 4).
Table 4.
Spectrum of organisms identified during the study period 2011–2012
| CA-BSI n/N (%) |
HA-BSI n/N (%) |
HCA-BSI n/N (%) |
Total BSIs n/N (%) |
|
|---|---|---|---|---|
| (N = 248) | (N = 371) | (N = 74) | (N = 693) | |
| Gram-negative bacteria | 117 (47.2) | 240 (64.7) | 43 (58.1) | 400 (57.7) |
| Escherichia coli | 49 (19.8) | 33 (8.9) | 10 (13.5) | 92 (13.3) |
| Klebsiella pneumoniae | 5 (2) | 78 (21) | 9 (12.2) | 92 (13.3) |
| Acinetobacter baumanii | 4 (1.6) | 54 (14.6) | 1 (1.4) | 59 (8.5) |
| Pseudomonas aeruginosa | 3 (1.2) | 19 (5.1) | 4 (5.4) | 26 (3.8) |
| Non-typhoidal Salmonella | 11 (4.4) | 6 (1.6) | 3 (4.1) | 20 (2.9) |
| Enterobacter cloacae | 3 (1.2) | 12 (3.2) | 3 (4.1) | 18 (2.6) |
| Serratia marcescens | 0 (0) | 17 (4.6) | 1 (1.4) | 18 (2.6) |
| Neisseria meningitidis | 12 (4.8) | 0 (0) | 1 (1.4) | 13 (1.9) |
| Haemophilus influenza B | 6 (2.4) | 1 (0.3) | 2 (2.7) | 9 (1.3) |
| Salmonella typhi | 4 (1.6) | 0 (0) | 0 (0) | 4 (0.6) |
| Other | 20 (8.1) | 20 (5.4) | 9 (12.2) | 49 (7.1) |
| Gram-positive bacteria | 124 (50) | 97 (26.1) | 28 (37.8) | 249 (35.9) |
| Staphylococcus aureus | 55 (22.2) | 37 (10) | 11 (14.9) | 103 (14.9) |
| Streptococcus pneumoniae | 45 (18.1) | 6 (1.6) | 6 (8.1) | 57 (8.2) |
| Enterococcus faecalis | 4 (1.6) | 23 (6.2) | 5 (6.8) | 32 (4.6) |
| Streptococcus group B | 13 (5.2) | 0 (0) | 4 (5.4) | 17 (2.5) |
| Enterococcus faecium | 2 (0.8) | 14 (3.8) | 0 (0) | 16 (2.3) |
| Other | 5 (2) | 17 (4.6) | 2 (2.7) | 24 (3.5) |
| Mycobacterium tuberculosis | 1 (0.4) | 1 (0.3) | 0 (0) | 2 (0.3) |
| Fungi | 6 (2.4) | 33 (8.9) | 3 (4.1) | 42 (6.1) |
| Candida albicans | 1 (0.4) | 16 (4.3) | 2 (2.7) | 19 (2.7) |
| Candida parapsilosis | 1 (0.4) | 9 (2.4) | 1 (1.4) | 11 (1.6) |
| Other Candida species | 4 (1.6) | 8 (2.2) | 0 (0) | 12 (1.7) |
N = denominator used unless otherwise stated
Susceptibility to antimicrobial agents
The majority of Staphylococcus aureus isolates 81/103 (78.6%) were susceptible to cloxacillin. The remaining 22 isolates were methicillin-resistant of which 17 were HA- BSI, accounting for 17/371 (5%) of all HA-BSI. Of the 57 S. pneumoniae isolates, 26.3% (15/57) were resistant to penicillin according to meningitis criteria with MIC > 0.06 μg/ml. Susceptibility testing for ceftriaxone and vancomycin was conducted in 52 and 21of the S. pneumoniae isolates, respectively with100% susceptibility for both antimicrobials. Of the 48 Enterococcus isolates, 54.2% (26/48) were susceptible to ampicillin and all were susceptible to vancomycin.
Table 5 describes antimicrobial susceptibility of the most commonly isolated Gram-negative organisms. Only 9.9% (9/91) of E.coli isolates were susceptible to ampicillin while 70.3% (64/91) were susceptible to gentamicin. Thirty four percent (31/92) of all E.coli isolates were extended-spectrum beta lactamase (ESBL)-producing organisms of which 10 and 16 were community-acquired and hospital-acquired respectively, accounting for 4% (10/248) and 4.3% (16/371) of CA- and HA-BSI, respectively. By contrast, 77.3% (68/88) of K. pneumoniae isolates were ESBL producers which accounted for 16.7% (62/371) of HA-BSI. No carbapenem-resistant Enterobacteriacae (CRE) isolates were identified on blood culture during the study period. Resistance to both aminoglycosides and carbapenems was present in 22% (13/59) of A. baumanii isolates. All A. baumanii isolates were susceptible to colistin. All non-typhoidal Salmonella ssp. isolates were susceptible to ceftriaxone, with only a single isolate not susceptible to ampicillin and ciprofloxacin. Candida albicans and parapsilosis isolates showed a 100% susceptibility to fluconazole.
Table 5.
Rates of antimicrobial susceptibility for the common gram-negative organisms identified
| Acinetobacter baumanii (N = 59) | Escherichia coli (N = 92) | Klebsiella pneumoniae (N = 92) | Pseudomonas aeruginosa (N = 26) | Non-typhoidal Salmonella (N = 20) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Antimicrobial | No isolates tested | No (%) Susceptible | No isolates tested | No (%) Susceptible | No isolates tested | N (%) Susceptible | No isolates tested | N (%) Susceptible | No isolates tested | N (%) Susceptible |
| Ampicillin | – | – | 91 | 9 (9.9) | 92 | – | – | – | 20 | 19 (95) |
| Co-amoxyclava | – | – | 89 | 43 (48.3) | 91 | 27 (29.7) | – | – | 1 | 1 (100) |
| Ceftriaxone | – | – | 90 | 59 (65.6) | 88 | 20 (22.7) | – | – | 20 | 20 (100) |
| Ceftazidime | 59 | 24 (40.7) | 91 | 60 (65.9) | 91 | 20 (22) | 26 | 23 (88.5) | 1 | 1 (100) |
| Ciprofloxacin | 59 | 45 (76.3) | 87 | 70 (80.5) | 85 | 52 (61.2) | 26 | 15 (57.7) | 19 | 18 (94.7) |
| Piperacillin-tazobactam | 58 | 10 (17.2) | 6$ | 5 (83.3) | 49$ | 34 (69.4) | 7$ | 1 (14.3) | 1$ | 1 (100) |
| Gentamicin | 59 | 27 (45.8) | 91 | 64 (70.3) | 89 | 25 (28.1) | 25 | 15 (60) | – | – |
| Amikacin | 58 | 26 (44.8) | 92 | 77 (83.7) | 91 | 70 (76.9) | 26 | 16 (61.5) | – | – |
| Meropenem | 56 | 26 (46.4) | 92 | 92 (100) | 92 | 92 (100) | 26 | 22 (84.6) | 20 | 20 (100) |
| Imipenem | 49 | 27 (55.1) | 90 | 90 (100) | 90 | 90 (100) | 26 | 21 (80.8) | 20 | 20 (100) |
| Ertapenem | 92 | 92 (100) | 91 | 91 (100) | – | – | 20 | 20 (100) | ||
| Colistin | 59 | 59 (100) | 90 | 90 (100) | 89 | 89 (100) | 26 | 23 (88.5) | – | – |
aCo-amoxyclav = Amoxycillin-clavulanic acid co-formulation $ due to technical limitations with the Vitek®2 AST card piperacillin-tazobactam was infrequently reported in 2011–2012; No number
Antimicrobial therapy
Information on antimicrobial therapy was available for 92% (638/693) of BSI episodes. The median (IQR) time to effective antibiotic therapy for bacterial isolates of CA-, HA- and HCA- BSI was 0 (0–1) day, 1 (0–2) day and 1 (0–3) day, respectively. The median (IQR) time to effective antifungal therapy for fungal isolates was 4.5 (4–5) days, 3 (0.5–4) days and 2.5 (2–3) days for CA-, HA- and HCA-BSI, respectively. Of the 638 episodes, 592 (92.8%) `received effective antimicrobial therapy based on the susceptibility profile of the isolate. In the remaining 46 episodes, 42 were treated with an ineffective antimicrobial, and 4 fungal BSI episodes did not receive antifungal therapy. Of the 42 bacterial episodes not treated with an effective antimicrobial, 10 (23.8%) resulted in a fatal outcome with the median (IQR) time to death of 4 (2–9) days, and the remaining 32 (76.2%) episodes resolved on ineffective antibiotic therapy. In 2 of the 4 fungal episodes that were not treated with antifungal therapy the patients died before the culture results were known. The remaining 2 children recovered uneventfully.
Outcomes
85 % (590/693) of BSI episodes were successfully treated and the children were discharged from hospital after these episodes. During the study period, 103 (14.9%) children died during or after a BSI episode prior to hospital discharge. Seventy of these deaths occurred within 14 days as a direct result of the BSI and were included in the survival analysis as there was a high chance of correctly identifying relevant risk factors in these children. The median time (IQR) to death of these 70 children was 3 (1–8) days. Of the 33 deaths that occurred after 14 days, the median time (IQR) to death was 46 (23–67) days.
Table 6 describes risk factors associated with 14-day inpatient mortality in children with BSI. On multivariable analysis, severe UWFA, severe anaemia at the time of BSI, admission in the ICU at the time of BSI, and requiring ICU admission after BSI was diagnosed were significantly associated with 14-day mortality. HIV-infection was not associated with increased mortality risk.
Table 6.
Risk factors associated with 14-day inpatient mortality in children with BSI
| Factor | Univariate HR (95% CI) | P value | Adjusted HR (95%CI) | P value |
|---|---|---|---|---|
| Age | N = 693 | |||
| 0–28 days | 0.9 (0.4–2) | 0.834 | 1 | |
| 1 - < 2 months | 2.2 (1.1–4.6) | 0.035 | 1 | |
| 2 - < 12 months | 1.2 (0.8–2) | 0.376 | 1 | |
| 12 months – 5 years | 0.7 (0.4–1.2) | 0.219 | 1 | |
| > 5 years | 1 | 1 | ||
| Nutritional status | N = 693 | |||
| Severe underweight for age | 2.4 (1.5–3.9) | 0.0001 | 2.0 (1.2–3.3) | 0.007 |
| Moderate underweight for age | 1.1 (0.6–2) | 0.851 | 1 | |
| Mild-normal weight for age | 1 | 1 | ||
| Weight not recorded | 2.4 (1–5.9) | 0.060 | 4.9 (1.2–20.8) | 0.029 |
| Haemoglobin | N = 693 | |||
| Severe anaemia (< 7 g/dL) | 3.3 (1.7–6.4) | 0.001 | 2.7 (1.3–5.6) | 0.006 |
| Mild anaemia (7–11 g/dL) | 1 (0.6–1.7) | 0.909 | 1 | |
| No anaemia (≥ 11 g/dL) | 1 | 1 | ||
| Result not available | 3.7 (1.2–11.9) | 0.025 | 4.2 (1.3–13.7) | 0.018 |
| ICU management | N = 693 | |||
| ICU resident at time of BSI | 1.5 (0.89–2.7) | 0.124 | 6.5 (3.7–11.6) | 0.0001 |
| BSI requiring ICU management | 5.2 (3.3–8.4) | 0.0001 | 3.5 (1.8–6.7) | 0.0001 |
| BSI not requiring ICU management | 1 | 1 | ||
| Type of BSI | N = 693 | |||
| CA | 1 | 1 | ||
| HA | 1.2 (0.8–2) | 0.377 | 1 | |
| HCA | 0.9 (0.4–1.9) | 0.806 | 1 | |
| HIV infection | N = 693 | |||
| Yes | 1.5 (0.8–2.9) | 0.221 | 1 | |
| No | 1 | 1 | ||
| Unknown status | 0.8 (0.5–1.5) | 0.523 | 1 |
Discussion
The current study describes the aetiology and quantifies the risk of BSI at our hospital. Gram-negative organisms were the predominant cause of BSI. The risk was lower for CA-BSI but higher for HA-BSI than rates obtained in a previous South African study conducted 20 years previously [6]. The isolation of 5.7% pathogens from blood culture specimens in the present study is similar to that from another referral hospital in Cape Town [7]. Possible reasons for the low pathogen yield in our setting may include inadequate volumes of blood inoculated into blood culture specimen bottles, lack of clear clinical indications for performing blood cultures potentially inflating the number of negative cultures and the prior use of antimicrobials, particularly ceftriaxone, as part of the syndromic case management of sick children at primary health care facilities [5, 9, 17]. The contaminant prevalence of 6.6% from 16,951 blood cultures is more than double the internationally acceptable contamination rate of 2–3%, suggesting that urgent attention is needed to improve aseptic measures when obtaining specimens for blood culture [18, 19]. A previous analysis of blood culture results over a 5-year period (2008–2012) from the same institution highlighted problems of high contamination rates particularly in the less than one-year age group and low pathogen yield, and also documented the predominance of gram-negative pathogens. [8].
S. aureus, E.coli, K. pneumoniae and A. baumanii predominated in our study. This spectrum of organisms is similar to that documented in other African studies [5–7]. Non-typhoidal Salmonella is a much more frequent cause of BSI in African countries to the north of South Africa, especially those in malaria-endemic areas, but has been reported as an important cause of BSI in an early South African study [20].
At the time of BSI, 27.5% of the episodes were not associated with a clinical focus of infection and 65.1% had fever (≥ 38 °C) or hypothermia (< 35.5 °C). A study done in Guinea-Bissau showed that the sensitivity of fever for detecting BSI was low at 54% with a positive predictive value (PPV) of 12% [21]. However, a Tanzanian study showed that temperature ≥ 38.5 °C was a strong predictor of BSI (odds ratio (OR) 7, 95% CI: 2.2–14.8, P = 0.0001) although the number of patients enrolled in that study was small [22]. Furthermore, a Malawian study of 225 hospitalised children found that the presence of at least one of five clinical features (oral thrush, malnutrition, chronic cough, lethargy on history and lethargy on examination) predicted BSI with a sensitivity of 69% [23].
Severe underweight was present in 25.2% of children in our study and was a significant predictor of 14-day mortality. These findings are in agreement with previous African studies showing that severe malnutrition significantly increases the risk of BSI, and that nutritional status is a significant determinant of survival in children with BSI [5, 24]. Although HIV infection was known to be present in 13.4% of the study cohort it did not influence 14-day mortality. Approximately 60% of the HIV-infected group was on ART at the time of BSI. This may partly explain why HIV infection did not influence mortality.
Our results show that ESBL-producing K. pneumoniae and E. coli have become important causes of CA- and HA-BSI. Of particular concern was that 30% of E.coli isolates in the CA-BSI group were ESBL-producing organisms. A recent study from another Cape Town hospital documented lower prevalence of CA-BSI caused by ESBL-producing E.coli (11.2%) and HA-BSI caused by ESBL-producing K.pneumoniae (78.3%) [7]. Although carbapenem-resistant Enterobacteriaceae infection was not observed during the study period, carbapenem over-utilisation as observed in the present study (data not shown) may with time lead to the emergence of carbapenem resistant BSI isolates.
Study strengths and limitations
The study describes the spectrum of organisms causing BSI in children. The results obtained highlight the growing concern of antibiotic resistance developing particularly in CA–BSI. One limitation was the lack of susceptibility data for piperacillin-tazobactam due to technical limitations at the time of the study. With on-going surveillance of organisms causing BSI and their susceptibility patterns, empiric antimicrobial choices may need to be altered in the future. Subsequent to the study period, updated definitions of reporting laboratory confirmed BSI were release by the CDC [25]. These definitions were not utilised in the current study and as a result, few of the HA-BSI episodes may have been misclassified. Organisms implicated in mucosal barrier injury BSI were also not stratified.
Due to the retrospective study design, there are limitations in the completeness and availability of clinical and laboratory data. The true burden of community-acquired BSI may have been underestimated. Before sick children are referred from primary and secondary health facilities to our hospital, they are often administered broad-spectrum antibiotics. This practice is consistent with the World Health Organization Integrated Management of Childhood Illness guidelines that have been implemented in South Africa, and frequently results in no growth on blood culture [9]. Even though the study cohort included neonates, the results obtained would not be an accurate reflection of the burden of BSI in this special group of infants. We were only able to explore a limited number of potential risk factors of BSI mortality. Further studies with larger sample sizes are required to provide a more in depth understanding of this aspect of BSI. Furthermore, it was not possible to explore risk factors associated with the acquisition of a BSI due to the lack of a suitable control group of children without BSI.
Conclusion
Our study provides insights about the risk, aetiology, clinical manifestations, antimicrobial therapy and outcomes of BSI in a sub-Saharan African setting characterized by high HIV prevalence. Further studies are required to gain a comprehensive understanding of the impact of BSI on the childhood population in SSA.
Acknowledgements
Spasina King is acknowledged for assisting with data collection from the medical records.
Funding
No funding was required for the completion of this study.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Abbreviations
- ART
Antiretroviral therapy
- BSI
Bloodstream infection
- CA
Community-acquired
- CLSI
Clinical Laboratory standards institute
- CRE
Carbapenem-resistant Enterobacteriacae
- CRP
C-reactive protein
- ESBL
Extended-spectrum beta lactamase
- FBC
Full blood count
- HA
Hospital-acquired
- HCA
Healthcare-associated
- HIV infection
Human immunodeficiency virus infection
- IQR
Interquartile range
- NHLS
National health laboratory service
- PCT
Procalcitonin
- SSA
Sub-Saharan Africa
- UWFA
Underweight for age
- WAZ
Weight for age z-score
- WCC
White cell count
Authors’ contributions
HL, BE and JN conceived and designed the study. CB was responsible for the overseeing the microbiological analysis at the NHLS and interpreting susceptibility results. VP contributed to the statistical analysis of the results. HL wrote the paper with contributions from BE, CB and JN. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Ethics approval was obtained from the Human Research Ethics Committee, Faculty of Health Sciences, University of Cape Town; reference number: HREC 619/2012. Informed consent was not obtained from individual patients as the data was collected and collated retrospectively, and patient details were anonymised before data analysis.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Harsha Lochan, Email: harshalochan@gmail.com.
Vashini Pillay, Email: Vash.pillay@uct.ac.za.
Colleen Bamford, Email: Colleen.bamford@uct.ac.za.
James Nuttall, Email: James.nuttall@uct.ac.za.
Brian Eley, Email: Brian.eley@uct.ac.za.
References
- 1.Berkley JA, Lowe BS, Mwangi I, Williams T, Bauni E, Mwarumba S, et al. Bacteremia among children admitted to a rural hospital in Kenya. N Engl J Med. 2005;352:39–47. doi: 10.1056/NEJMoa040275. [DOI] [PubMed] [Google Scholar]
- 2.Reddy EA, Shaw AV, Crump JA. Community-acquired bloodstream infections in Africa. Lancet Infect Dis. 2010;10:417–432. doi: 10.1016/S1473-3099(10)70072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crump JA, Ramadhani HO, Morrissey AB, Msuya LJ, Yang L-Y, Chow S-C, et al. Invasive bacterial and fungal infections among hospitalised HIV-infected and HIV-uninfected children and infants in northern Tanzania. Tropical Med and International health. 2011;16(7):830–837. doi: 10.1111/j.1365-3156.2011.02774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen MV, Sarpong N, Krumkamp R, Dekker D, Loag W, Amemasor S, et al. Incidence and characteristics of bacteremia among children in rural Ghana. PLoSONE. 2012;7(9):e44063. doi: 10.1371/journal.pone.0044063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aiken AM, Mturi N, Njuguna P, Mohammed S, Berkley JA, Mwangi I, et al. Risk and causes of paediatric hospital-acquired bacteraemia in Kilifi District Hopsital, Kenya: a prospective cohort study. Lancet. 2011;378:2021–2027. doi: 10.1016/S0140-6736(11)61622-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotton MF, Burger PJ, Bodenstein WJM. Bacteraemia in children in the south-western cape. A hospital-based survey. S Afr MJ. 1992;81:87–90. [PubMed] [Google Scholar]
- 7.Dramowski A, Cotton MF, Rabie H, Whitelaw A. Trends in paediatric bloodstream infections at a south African referral hospital. BMC Paediatrics. 2015;15:33–44. doi: 10.1186/s12887-015-0354-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lochan H, Bamford C, Eley B. Blood cultures in sick children. S Afr Med J. 2013;103(12):918–920. doi: 10.7196/SAMJ.6979. [DOI] [PubMed] [Google Scholar]
- 9.WHO/UNICEF . Integrated Management of Childhood Illness Chart Booklet. Geneva: WHO/UNICEF; 2008. [Google Scholar]
- 10.D’Acremont V, Kilowoko M, Kyungu E, Philipina S, Sangu W, Kahama-Moro J, et al. Beyond malaria – causes of fever in outpatient Tanzanian children. N Engl J Med. 2014;370:809–814. doi: 10.1056/NEJMoa1214482. [DOI] [PubMed] [Google Scholar]
- 11.Musiime V, Cook A, Bakeera-Kitaka S, Vhembo T, Lutakome J, Keishanyu R, et al. Bacteremia, causative agents and antimicrobial susceptibility among HIV-1–infected children on antiretroviral therapy in Uganda and Zimbabwe. Pediatr Infect Dis J. 2013;32:856–862. doi: 10.1097/INF.0b013e31828c3991. [DOI] [PubMed] [Google Scholar]
- 12.Le Doare K, Bielicki J, Heath PT, Sharland M. Systematic review of antibiotic resistance rates among gram-negative bacteria in children with sepsis in resource-limited countries. J Pediatric Infect Dis Society. 2015;4(1):11–20. doi: 10.3233/JPI-2009-0146. [DOI] [PubMed] [Google Scholar]
- 13.Centres for Disease Control and Prevention and the National Healthcare Safety Network. CDC/NHSN Surveillance Definitions for Specific Types of Infections. Atlanta: CDC, 2014. http://www.socinorte.com/wpcontent/uploads/2014/06/17pscNosInfDef_current.pdf. Accessed 1 Dec 2017.
- 14.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-First Infromational Supplement. CLSI document M100-S21. Wayne, PA. Clinical and Laboratory Standards Institute; 2011
- 15.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Approved Standard - Eleventh Edition. CLSI document M02-A11. Wayne, PA. Clinical and Laboratory Standards Institute; 2012
- 16.World Health Organization. The WHO child growth standards. Geneva: World Health Organization 2006. http://apps.who.int/gho/data/imr.jsp?id=72/RefSource> Accessed 7 July 2014.
- 17.Connell TG, Rele M, Cowley D, Buttery JP, Curtis N. How reliable is a negative blood culture result? Volume of blood submitted for culture in routine practice in a children’s hospital. Pediatrics. 2007;119(5):891–896. doi: 10.1542/peds.2006-0440. [DOI] [PubMed] [Google Scholar]
- 18.Hall KK, Lyman JA. Updated review of blood culture contamination. Clin Microbiol Rev. 2006;19(4):788–802. doi: 10.1128/CMR.00062-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavlovsky M, Press J, Peled N, Yagupsky P. Blood culture contamination in pediatric patients: young children and young doctors. Pediatr Infect Dis. 2006;25(7):611–614. doi: 10.1097/01.inf.0000220228.01382.88. [DOI] [PubMed] [Google Scholar]
- 20.Berkowitz FE. Bacteraemia in hospitalised black south African children. Am J Dis Child. 1984;138:551–556. doi: 10.1001/archpedi.1984.02140440035008. [DOI] [PubMed] [Google Scholar]
- 21.Isendahl J, Manjuba C, Rodrigues A, Xu W, Henriques-Normack B, Giske CG, et al. Prevalence of community-acquired bacteraemia in Guinea-Bissau: an observational study. BMC Infect Dis. 2014;14:3859. doi: 10.1186/s12879-014-0715-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christopher A, Mshana SE, Kidenya BR, Hokororo A, Morona D. Bacteremia and resistant gram-negative pathogens among under-fives in Tanzania. Ital J Pediatr. 2013;39:27–36. doi: 10.1186/1824-7288-39-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norton EB, Archibald LK, Nwanyanwu OC, Kazembe PN, Dobbie H, Reller B, et al. Clinical predictors of bloodstream infections and mortality in hospitalized Malawian children. Paediatr Infect Dis J 2004;23:145-151.10.1542/peds.2006-0440 Accessed 23 November 2016. [DOI] [PubMed]
- 24.Sigaúque B, Roca A, Mandomando I, Morais L, Quintó L, Sacarlal J, et al. Community-acquired bacteraemia among children admitted to a rural hospital in Mozambique. Pediatr Infect Dis J. 2009;28:108–113. doi: 10.1097/INF.0b013e318187a87d. [DOI] [PubMed] [Google Scholar]
- 25.Centers for Disease Control and prevention and the National Healthcare safety Network. CDC/NHSH Surveillance. Bloodstream Infection Event (Central Line-Associated Bloodstream Infection and non-central line-associated Bloodstream Infection. CDC January 2017. https://www.cdc.gov/nhsn/pdfs/pscmanual/4psc_clabscurrent.pdf. Accessed 18 June 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.


