Abstract
The lung is an architecturally complex organ comprised of a heterogeneous mixture of various epithelial and mesenchymal lineages. We have used single-cell RNA-sequencing and signaling lineage reporters to generate a spatial and transcriptional map of the lung mesenchyme. We find that each mesenchymal lineage has a distinct spatial address and transcriptional profile leading to unique niche regulatory functions. The Mesenchymal Alveolar Niche Cell is Wnt responsive, expresses Pdgfrα, and is critical for alveolar epithelial cell growth and self-renewal. In contrast, the Axin2+ Myofibrogenic Progenitor cell preferentially generates pathologically deleterious myofibroblasts after injury. Analysis of the secretome and receptome of the alveolar niche reveals functional pathways that mediate growth and self-renewal of alveolar type 2 progenitor cells including IL-6/Stat3, Bmp, and Fgf signaling. These studies define the cellular and molecular framework of lung mesenchymal niches and reveal the functional importance of developmental pathways promoting self-renewal versus pathological response to tissue injury.
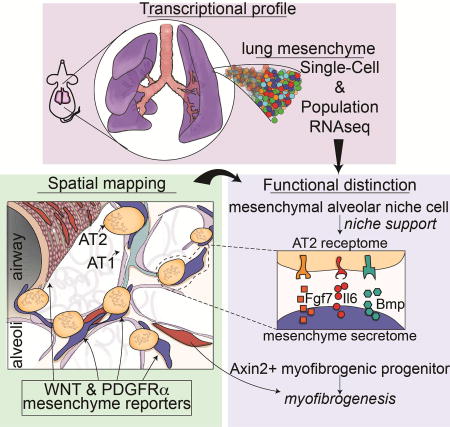
Graphical abstract
INTRODUCTION
In adult tissues, epithelial progenitors receive paracrine signals from the surrounding mesenchymal niche, which can modulate their ability to proliferate and differentiate. The mammalian lung is comprised of a myriad of specialized epithelial cells surrounded by a poorly defined heterogeneous mesenchyme. The lung mesenchymal compartment includes airway smooth muscle (ASM), vascular smooth muscle (VSM), endothelium, and poorly defined interstitial mesenchymal cells. In the lung alveolus, the alveolar type 2 (AT2) cell population, or subpopulations within it, is thought to be the predominant epithelial progenitor cell, capable of self-renewal and generating the alveolar type 1 (AT1) lineage after injury (Barkauskas et al., 2013; Rock et al., 2011). AT2 and AT1 cells are thought to receive signals from the adjoining lung mesenchyme, which are important for both homeostasis and regeneration after injury. Different epithelial-mesenchymal interactions are likely to occur in the conducting airways, including those between basal stem cells, secretory cells and the adjoining mesenchyme. However, as in most tissues, information on the identity, spatial distribution, and behavior of these mesenchymal lineages within the various progenitor cell niches in the lung is limited.
The lung alveolar compartment is the primary site of gas exchange, the most fundamental function of the respiratory system. The lung alveolus forms a niche of interacting and communicating cell types including the AT1 and AT2 epithelial lineages, vascular endothelial cells, and poorly understood mesenchymal lineages. Previous work has suggested that AT2 cells are located near Pdgfrα+ mesenchymal cells, which promote AT2 self-renewal and differentiation into AT1 cells in ex vivo three-dimensional organoid cultures (Barkauskas et al., 2013). These observations suggest that spatially restricted alveolar niche signaling derived from the mesenchyme may influence AT2 cell behavior.
Defining the identity and cell behavior of lung mesenchymal lineages is complicated by the architectural complexity of the respiratory system. Moreover, there are few cell specific marker genes that demarcate different mesenchymal lineages within the lung or most other adult tissues. Recent evidence has revealed a critical role for an epithelial derived hedgehog signal in regulation of airway-adjacent mesenchyme homeostasis and repair in the lung (Peng et al., 2015). However, there is little understanding or characterization of the different mesenchymal lineages and their respective abilities to promote homeostasis and regeneration in the lung alveolus or in most other tissue specific niches.
To identify and characterize the mesenchymal lineages that participate in and provide preferential signals to promote alveolar self-renewal and regeneration versus linages and signals involved in an ineffective injury response such as myofibroblast expansion, we have isolated and sorted the entirety of lung mesenchyme into five different cell lineages based on signal transduction pathway readouts. The use of signaling reporters allows for the characterization of cell lineages based on a functional readout and combined with an unbiased single-cell RNA sequencing approach, we have begun to reveal mechanistic insights into the identified lineages. Each of these lineages has a specific spatial address and distinct transcriptional profile. From these studies, we identify a Mesenchymal Alveolar Niche Cell (MANC) that has unique capabilities in supporting alveolar growth and regeneration. We also identify a distinct and separate lineage called the Axin2+ Myofibrogenic Progenitor (AMP) cell, which is a major contributor to pathologically deleterious myofibroblasts, as well as airway smooth muscle cells after injury. Using high-resolution transcriptome data, we identify key signal transduction pathways that regulate lung alveolar epithelial self-renewal and differentiation including IL6-Stat3, Bmp, and Fgf signaling. These studies help to define the cellular, molecular, and spatial complexity and heterogeneity of two lung mesenchymal niches and identify paracrine pathways critical for promoting lung alveolar self-renewal versus deleterious tissue remodeling after injury, thus providing a roadmap for understanding the epithelial-mesenchymal interactions critical for adult tissue homeostasis and regeneration.
RESULTS
Signaling pathway reporter systems can identify and characterize distinct adult lung mesenchymal lineages
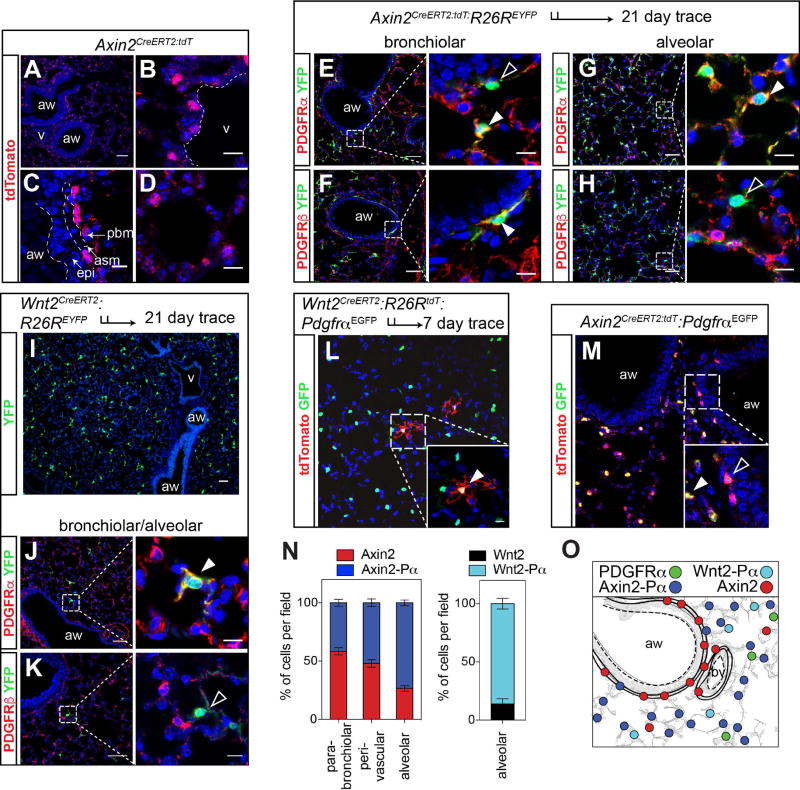
We used a recently described Axin2CreERT2:tdT reporter line generated in our laboratory (Frank et al., 2016), along with a Wnt2CreERT2 line (Peng et al., 2013), and a PdgfrαEGFP reporter line (Hamilton et al., 2003), to de-convolute the complexity of the mesenchymal compartment in the adult lung by identifying the relative location and distribution of mesenchymal cells displaying Wnt or Pdgfr signaling activity and expression. Wnt-responsive, Axin2+ cells can be found throughout the adult lung mesenchyme, in the alveolar regions as well as surrounding the conducting airways and blood vessels (Fig. 1A–D and Supplemental Fig. 1A–D). Many of the Axin2-derived cells in the alveolar niche expressed Pdgfrα whereas Axin2+ lineage-traced cells surrounding the airways primarily expressed Pdgfrβ (Fig. 1E–H). Using the Wnt2CreERT2 line crossed to a R26REYFP reporter (Wnt2CreERT2:R26REYFP), we demonstrate that Wnt2+ cells are located exclusively in the alveolar region of the lung (Fig. 1I). Moreover, using immunostaining and Wnt2creERT2:R26RtdT:PdgfraEGFP mice, we show that approximately 90% of Wnt2 expressing cells express Pdgfrα but not Pdgfrβ (Fig. 1J, K, L, N). Finally, using the PdgfrαEGFP reporter line, we show that the majority of Pdgfrα+ cells are located in the alveolar region (Fig. 1L, M and N). Approximately 74% of the Axin2 cells located in the alveolar region express Pdgfrα, while a majority of the Axin2 cells surrounding the airways do not (Fig. 1M and N). These data suggest that alveolar mesenchymal cells are regionally distributed and can be divided into several distinct types based on their expression of Axin2, Wnt2, and Pdgfrα. Overall, the Axin2+/Pdgfrα+ (Axin2-Pα) cells and Wnt2+/Pdgfrα+ (Wnt2-Pα) cells are found in the alveolar region whereas the Axin2+/Pdgfrα− (Axin2+) cells are found primarily surrounding airways or blood vessels, giving each of these different lineages a unique spatial address in the lung (Fig. 1O).
Figure 1. Signaling pathway reporter activity identified distinct mesenchymal cells in the adult mouse lung.
(A–D) Axin2CreERT2:tdT reporter mice express tdTomato throughout the lung mesenchyme. (E–H) Axin2-lineage traced cells shows that Axin2+ positive cells surrounding the airways rarely express Pdgfrα (E, black arrowhead) whereas they do express Pdgfrβ+ (F, white arrowhead). In contrast, Axin2+ cells in the alveolar region are predominantly Pdgfrα positive (E and G, white arrowheads) and generally do not express Pdgfrβ (H, black arrowhead). (I) Wnt2-expressing mesenchymal cells are only present in the alveolar compartment. (J and K) Immunostaining shows that Wnt2-derived cells are Pdgfrα+ (white arrowhead) and not Pdgfrβ+ (black arrowhead). (L) Wnt2creERT2:R26RtdTomato:PdgfrαEGFP mice reveal that the majority of Wnt2+ cells are PdgfrαEGFP positive. (M) Axin2CreERT2:tdT:PdgfrαEGFP mice reveal Axin2-Pα double positive and Axin2-tdT single positive cells, with the Axin2-Pα cells found primarily in the alveolar region (inset). (N) Quantification of the spatial distribution of Axin2 and PdgfrαEGFP positive cells (left graph) and quantification of the alveolar distribution of Wnt2 mesenchymal cells (right graph). (O) Schematic showing the mesenchymal populations and their general spatial distributions. Data in N are means ± SEM. Scale bars A–K low magnification are 50 µm, high magnification are 10 µm. aw=airway, v=blood vessel, asm= airway smooth muscle, pbm= peri-bronchial mesenchyme and epi=epithelium.
Lineage-specific and Single-Cell transcriptome analysis reveals distinct mesenchymal lineages within the adult lung
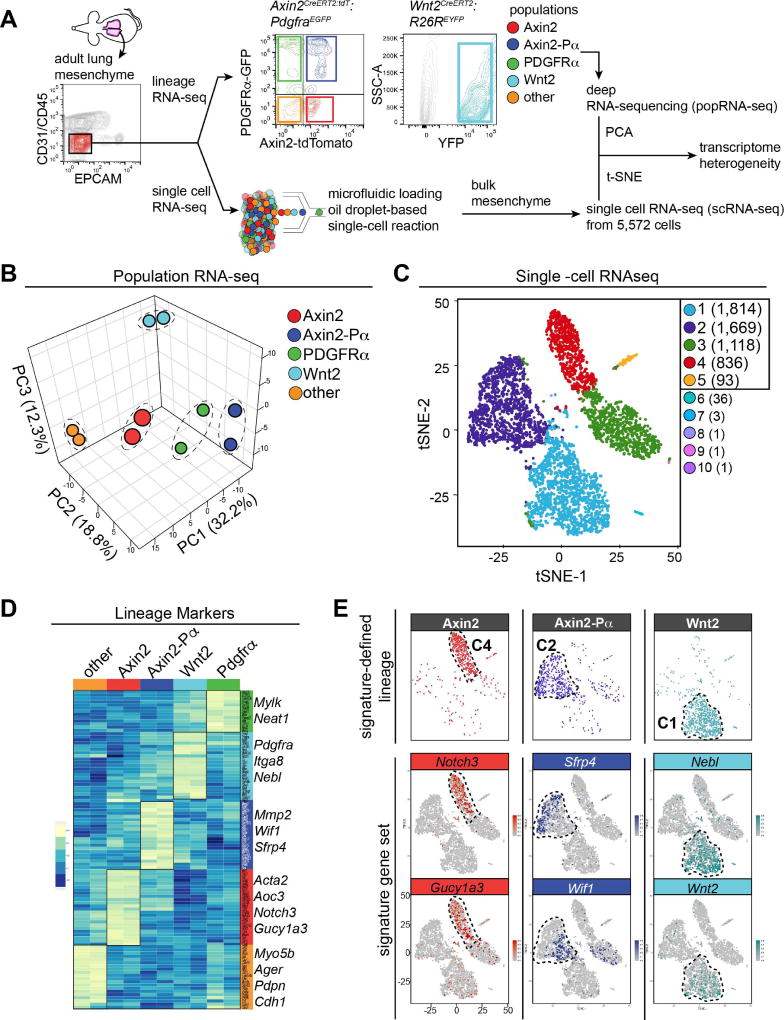
To further assess the various mesenchymal cell types defined above, we performed population RNA-seq (popRNA-seq) analysis and single-cell RNA sequencing (scRNA-seq) analysis. In order to obtain deep sequencing of the five different cell populations identified above, we used fluorescence activated cell sorting (FACS) to isolate these cells based on presence or absence of Axin2, Pdgfrα, and Wnt2 expression (Fig. 2A and Supplemental Fig. 1E–G). Of note, using these methods we were able to isolate greater than 90% of the live Epcam-/CD31-/CD45- (lineage-negative) cell population in the lung, which comprises the vast majority of the lung mesenchymal compartment (Supplemental Figure 1H and I). Principal Component Analysis (PCA) of the popRNA-seq data revealed that all five populations exhibited significantly different transcriptomes (Fig. 2B).
Figure 2. Lung mesenchymal cells have distinct transcriptomes.
(A) Schematic of strategy to construct and compare transcriptomic data from lineage reporter popRNA-seq versus scRNA-seq. The sorting strategy for mesenchyme from Axin2CreERT2:tdT:PdgfraEGFP and Wnt2creERT2:R26REYFP traced cells is shown. Representative flow-cytometry plots are shown. (B) Three-dimensional principal component analysis (PCA) from popRNA-seq of the 5 mesenchymal populations, n=2 mice. (C) tSNE plot derived from scRNA-seq of 5,572 cells, colored by K means of 10 clustering. Clusters 1–5 were used for analysis as cluster 6–10 contained only a few cells each. (D) Heatmap of selected lineage enriched genes. An abbreviated list of genes specific for each of the five different cell populations are shown to the right of the heatmap. (E) Overlay plots of gene signatures derived from (D) projected onto the scRNA-seq reveal distinct patterns correlating with the identified lineage reporters.
An analysis of known mesenchymal markers including signaling and structural genes in the popRNA-seq data reveals that the “other” and Axin2+ cells exhibit characteristics of a smooth muscle, myofibroblast or pericyte-like cell with enriched expression of genes such as Acta2, Tagln and Des (Fig. 2D and Supplemental Fig. 2A). Since less than 10% of the Axin2+ traced cells surrounding the airways or blood vessels express the smooth muscle marker SM22α or αSMA by IHC, these data suggest that the Axin2+ cells may be primed for smooth muscle or myofibroblast gene expression but is not yet committed to these differentiated lineages (Supplemental Figure 1A–C). In contrast, the Pdgfrα+, Axin2-Pα+, and Wnt2+ cells exhibited enriched expression of genes associated with matrix-producing fibroblasts found in the alveoli such as Elastin (Eln), Pdgfrα (Pdgfra) and Collagen (Col1a1) (Fig. 2D and Supplemental Fig. 2A). Of note, cells in the ‘other’ population express epithelial markers such as Ager, Pdpn and Cdh1 suggesting that this population may include cells that were not removed during the Epcam/CD31/CD45 negative sort to purify mesenchymal cells (Fig. 2D).
We also subjected the entire lung mesenchymal population to In-Drop scRNA-seq analysis (Fig. 2A). Assessment of the scRNA-seq data from 5,572 lung mesenchymal cells reveals a tSNE based clustering into five distinct groups, each consisting of more than 90 cells (Fig. 2C and Supplemental Fig. 2B). Expression of some candidate mesenchymal markers showed that clusters 4 and 5 contained the majority of Pdgfrα+ cells while clusters 2 and 3 contained the majority of Acta2, Tagln, and Des expressing cells (Supplemental Fig. 2B and C). Pdgfrβ was expressed in both clusters 2, 3, and 4 and Col1a1 was broadly expressed in all clusters (Supplemental Fig. 2C). Analysis of the most highly expressed genes in each cluster shows that cluster 5 represents epithelial cells that were not removed using FACS isolation (Supplemental Fig. 2D).
To test whether these five groups from the scRNA-seq were analogous to those found in our popRNA-seq analysis, we identified hallmark genes from the popRNA-seq dataset that clearly demarcated the five different groups of cells (Fig. 2D). The Axin2-Pα+ cells express multiple inhibitors of Wnt signaling including Wif1 and Sfrp4, indicating that Wnt signaling is finely regulated in these cells and their neighboring cells. In contrast, Axin2+ cells were enriched for genes indicative of a myofibroblast lineage including Acta2 and Aoc3, a myofibroblast marker expressed in dermal mesenchyme (Hsia et al., 2016). Aoc3 expression is confined to the mesenchyme surrounding the parabronchial airways (Supplemental Figure 2E). Expression of Notch3 and Gucy1a3, a receptor and Notch target gene highly expressed in pericytes and vascular smooth muscle (Fig. 2D) (Chang et al., 2011; Wang et al., 2014), are also enriched in Axin2+ cells. The Wnt2+ cells displayed unique expression of Nebl, a cardiac Z-disk protein that has also been shown to be significantly down-regulated in human IPF lung tissue, and may represent vestigial remnants of the primordial cardio-pulmonary progenitor ontogeny of the Wnt2 expressing lineage (Bauer et al., 2015; Moncman and Wang, 1995; Peng et al., 2013). We overlaid these signature genes onto the scRNA-seq analysis using the 10× Genomics software. This analysis revealed that the distinct mesenchymal cell types identified by functional reporter readouts represent distinct clusters derived from the unbiased parallel scRNA-seq approach with Axin2+ cells represented by cluster 4 (C4), Axin2-Pα+ cells represented by cluster 2 (C2), and Wnt2+ cells represented by cluster 1 (C1) (Fig. 2E). Thus, the Axin2-Pα+, Axin2+, and Wnt2+ cells exhibit unique transcriptomes identifiable through the use of both scRNA-seq and popRNA-seq analysis, which suggest that these cells represent distinct lineages with potentially unique cell behaviors in the lung.
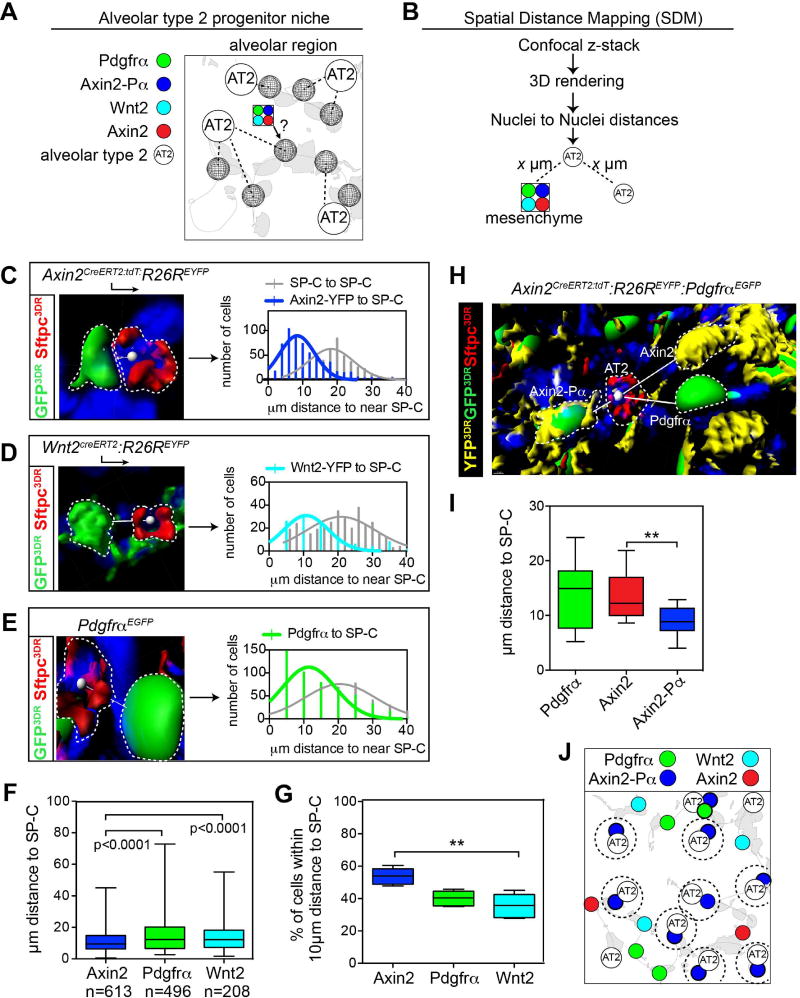
Development of a Spatial Distance Mapping (SDM) algorithm to map the epithelial-mesenchymal interactions in the alveolar niche
The AT2 lineage is thought to harbor a resident epithelial progenitor of the lung alveolus due to its capability to self-renew and differentiate into AT1 cells after injury (Barkauskas et al., 2013; Desai et al., 2014; Rock et al., 2011). To determine which of the mesenchymal lineages defined above helps to form the alveolar niche with the AT2 cell population, we developed a spatial distance mapping (SDM) algorithm using the AT2 cell as an anchor point (Fig. 3A). This allowed us to define a distinct spatial address for each of the mesenchymal lineages we have identified. SDM is based on identification of the relative location of lineage reported cell types to Sftpc-expressing AT2 cells using confocal microscopy–based three-dimensional rendering to measure nuclei-nuclei distance using DAPI staining (Fig. 3B). Using SDM, the Axin2+ lineage was found to be preferentially located closest to AT2 cells versus either the Pdgfrα+ or Wnt2+ lineages (Fig. 3C–G). We next tested whether Axin2-Pα+ lineage, which has a distinct transcriptome, were found in a unique spatial arrangement in relation to AT2 cells. These analyses revealed that the Axin2-Pα+ lineage is located closer to AT2 cells, compared to the Axin2+/Pα− lineage (Fig. 3H and I). Given their preferential close proximity to AT2 cells, these data suggest that the Axin2-Pα+ mesenchymal lineage is a critical component of the alveolar niche that communicates preferentially to AT2 cells (Fig. 3J).
Figure 3. Spatial distance mapping to define the alveolar niche in the lung.
(A) Schematic showing the alveolar niche, consisting of AT2 cells near mesenchymal cells. (B) Flow-chart for spatial distance mapping. (C–E) Representative images showing mesenchymal cell nuclei to nearest AT2 nuclei vectors from the indicated mice. Distance vectors were plotted in frequency distribution plots that show the frequency of distance relationships within 2µm bin sizes along the x-axis. (F) Average distance between each mesenchymal lineage and an AT2 cell. (G) Percent of each mesenchymal lineage population found within 10µm of an AT2 cell. (H) Reconstructed z-stack from Axin2CreERT2:tdT:R26REYFP:PdgfrαEGFP lungs indicating distances between AT2 cells (Sftpc+) and Axin2, Axin2-Pα and Pdgfrα cell lineages. (I) Average distance between each mesenchymal lineage and an AT2 cell indicates that the Axin2-Pα cell resides closest in proximity to AT2 cells. (J) Schematic showing Axin2-Pα cells are spatially distributed near Sftpc+ AT2 cells. Box and whisker plots, bars are min and max values. Asterisks * or ** indicates p<0.05, 0.01 respectively by one-way ANOVA.
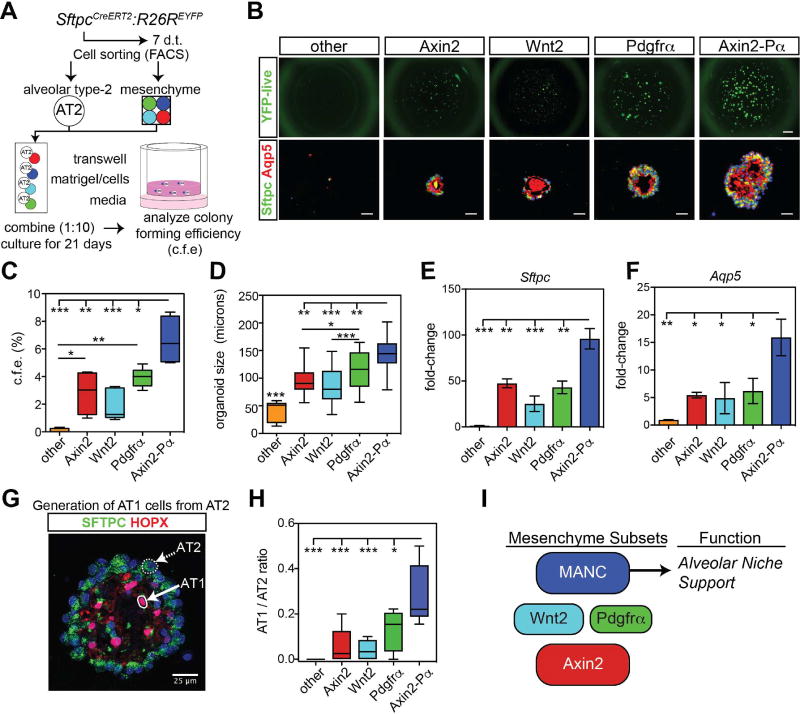
Axin2-Pα cells define a mesenchymal alveolar niche cell (MANC) that promotes AT2 cell self-renewal
Previous studies have shown that mesenchymal cells are necessary for alveolar organoid development in vitro, indicating the importance of mesenchymal-epithelial signaling in alveolar growth (Barkauskas et al., 2013). To determine which of the mesenchymal lineages identified preferentially promote alveolar cell growth and self-renewal, alveolar organoid assays were performed using mature AT2 cells isolated from SftpcCreERT2:R26REYFP adult mice (Barkauskas et al., 2013; Chapman et al., 2011). The five different mesenchymal lineages or populations were combined with AT2 cells to form organoids and were subsequently counted, sectioned for IHC, and isolated for gene expression analysis (Fig. 4A). While organoids visibly formed in the presence of all the different mesenchymal cell types except for the lineage-negative “other” cell population, the Axin2-Pα lineage generated a significantly greater number of organoids than any other lineage (Fig. 4B and C). The organoids generated using the Axin2-Pα lineage were also larger in size than those generated by the other mesenchymal cell types (Fig. 4D). Consistent with the increased colony formation, the expression of Sftpc and Aqp5 was also significantly increased in the Axin2-Pα co-culture condition (Fig. 4E and F). In order to evaluate AT2 to AT1 differentiation, we analyzed the ratio of AT1 (Hopx+ nuclei) to AT2 (Sftpc+) cells within the mature organoids. These data reveal a greater AT1/AT2 ratio in the presence of the Axin2-Pα+ lineage versus the other lineages, suggesting that this lineage not only promotes better growth but also differentiation into AT1 cells (Fig. 4G and H). Together, these data identify the Axin2-Pα+ lineage as a Mesenchymal Alveolar Niche Cell or MANC, which is preferentially capable of promoting AT2 cell self-renewal and differentiation in AT1 cells (Fig. 4I).
Figure 4. The mesenchymal alveolar niche cell promotes AT2 cell self-renewal and differentiation.
(A) Experimental outline to test the ability of mesenchymal lineages to promote alveolar organoid growth. (B) Live SftpcCreERT2:R26REYFP labeled cells showing EYFP native fluorescence (top row) or immunostaining for the indicated markers from fixed organoids (bottom row). Scale bars= 1mm (top), 25 µm (bottom). (C) Colony forming efficiency of alveolar organoids with the different mesenchymal lineages. (D) Size of alveolar organoids. (E and F) Q-PCR data from organoid cultures shows an increased expression of the AT2 genes Sftpc and the AT1 gene Aqp5 using the Axin2-Pα MANCs. (G) Example of AT2 and AT1 cell generation in an alveolar organoid stained with the AT1 cell marker Hopx and the AT2 cell marker Sftpc. The image shown is a max projection. (H) Quantification data of the number of AT1/AT2 cells generated in each co-culture condition showing a higher ratio in the Axin2-Pα co-cultured assays. (I) Diagram modeling mesenchymal lineages and how the MANC lineage supports the alveolar niche. Representative data are shown from n=4–6 mice, box and whiskers indicate min and max, error bars are means ± SEM. Asterisks, *, ** and *** indicate p<0.05, 0.01 and 0.001 respectively by one-way ANOVA.
Axin2+ cells define an Axin2 myofibrogenic progenitor (AMP) that contributes to the deleterious fibrotic response in the lung after injury
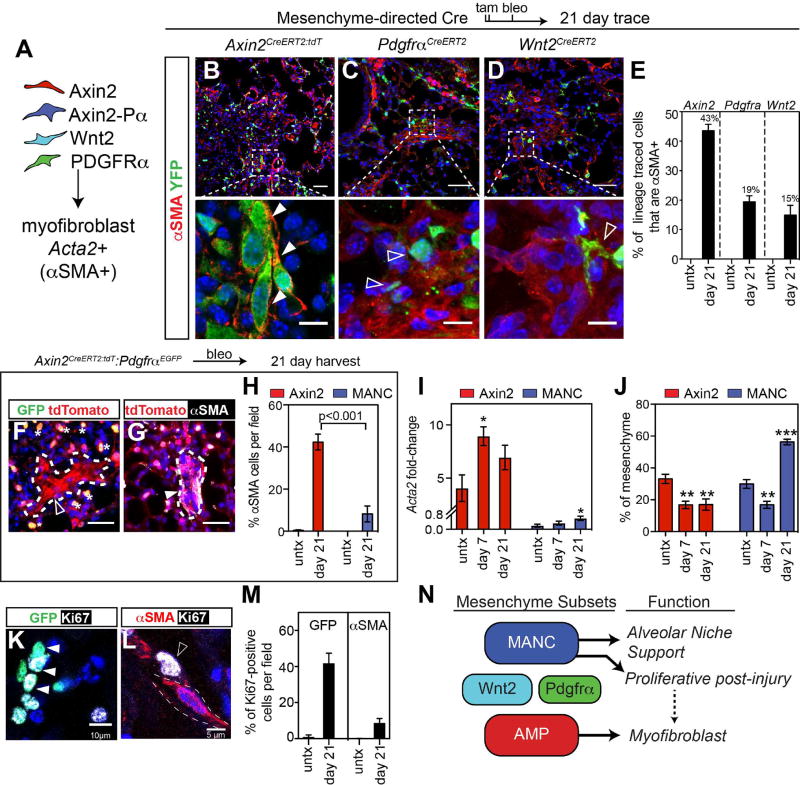
Lung fibrosis is a chronic and progressive disease that ultimately leads to respiratory failure and death in the absence of organ transplant. While our data show that the MANC lineage is a critical component of the alveolar niche that preferentially promotes AT2 cell growth, we wanted to identify which of the identified lineages preferentially contributed towards the deleterious fibrotic response to injury. While there are no mouse models of lung fibrosis that recapitulate all of the hallmarks of the human disease, bleomycin injury does lead to a transient fibrotic response consisting of increased numbers of αSMA+ myofibroblasts in the alveolar region, similar to some of the pathology observed in human lung fibrosis (Adamson and Bowden, 1974; Degryse and Lawson, 2011; Kapanci et al., 1992; Rock et al., 2011). To determine which of the lineages we identified preferentially contribute towards the αSMA+ myofibroblast response after bleomycin injury, we performed lineage tracing analysis using the Axin2CreERT2:tdT, PdgfraCreERT2 and Wnt2CreERT2 mice and the R26REYFP reporter. Recombination was induced in all three lineages and intra-tracheal instillation of bleomycin was performed to injure the lungs and induce fibrotic remodeling. After 21 days, αSMA+ myofibroblasts were quantified from each of these lineage-tracing experiments (Fig. 5A). While all three lineages generated αSMA+ cells, the Axin2 lineage contributed to nearly half of the myofibroblasts whereas less than 20% were derived from either the Wnt2 or Pdgfrα lineages (Fig. 5B–E). Of note, there were no detectable αSMA positive lineage-traced myofibroblasts in the alveolar region of control animals using any of these reporters (Fig. 5E). Importantly, while many myofibroblasts maintain expression of Axin2, they express little or no Pdgfrα (Fig. 5F–H).
Figure 5. Myofibroblasts are derived from an Axin2+ myogenic progenitor cell.
(A) Diagram showing the experimental design to determine the lineage of origin for myofibroblasts generated after bleomycin lung injury. (B–D) Axin2CreERT2:tdT lineage traced mice show robust differentiation into αSMA positive myofibroblasts, while PdgfraCreERT2 and Wnt2CreERT2 lineage traced cells often form near αSMA+ cells but are rarely αSMA+, scale bars low and high magnification are 50µm and 10µm respectively (E) Quantification of the lineage tracing from B–D. (F–G) Reduced PdgfrαEGFP expression is observed in myofibroblasts, however these cells are positive for Axin2-tdTomato. Asterisks indicate Axin2-Pα cells. Scale bar=25µm (H) Quantification of the αSMA staining in Axin2-tdTomato single positive and MANC (Axin2-Pα) cells. (I) Axin2+ and MANC cells were sorted from treated mice and Q-PCR was performed. (J) Quantification from flow cytometry data on live cells gated on mesenchyme for the AMP and MANC cells from treated Axin2CreERT2:tdT:PdgfraEGFP mice, n=3–5 mice per time point for I and J. (K and L) PdgfrαEGFP (MANC) cells co-express the proliferation marker Ki67 at day 21 post bleomycin injury. (M) Quantification of proliferation data from K–L. (N) Model diagram showing that the AMP lineage is responsive to injury by up-regulating a myofibroblast gene expression pattern. Error bars are means ± SEM, ***, ** and * indicates p<0.001, 0.01 by one-way ANOVA.
To determine whether bleomycin induced lung injury would cause trans-differentiation of the lung mesenchymal lineages we have identified, we assessed Acta2 expression and changes in population numbers during early myofibrogenic (day 7 post bleomycin) and later resolution time points (day 21). At day 7 post-bleomycin, the FACS purified Axin2+ cells significantly up-regulated the expression of Acta2 while the MANCs did not (Fig. 5I). Consistent with the significant tissue damage, the overall numbers of Axin2+ cells and MANCs were reduced at day 7 (Fig. 5J). However, by day 21, the MANCs but not the Axin2+ cells rebounded in cell number (Fig. 5J). Consistent with this we observed increased proliferation in Pdgfrα expressing cells relative to the αSMA+ cells (Fig. 5K–M). To further address cell plasticity amongst the identified lineages, we tested whether bleomycin injury caused Wnt2+ cells to acquire increased Axin2 expression. Prior to injury approximately 30% of Wnt2+ cells express Axin2 using the Axin2lacZ reporter (Supplemental Fig. 3A–C). After bleomycin injury, this percentage was not significantly altered (Supplemental Fig. 3C). These data suggest that Wnt2+ cells do not significantly alter Axin2 expression after acute lung injury. The specific ability of Axin2+ cells to up-regulate Acta2 and differentiate into myofibroblasts after bleomycin injury and their relative inability to support AT2 cell growth in organoids suggests that they are an Axin2+ myofibrogenic progenitor (AMP) cell. Together, these data demonstrate the existence of at least two distinct mesenchyme lineages within the alveolar region; the MANC which is a critical component of the alveolar niche that promotes AT2 cell self-renewal and proliferates after acute lung injury and the AMP that contribute towards the deleterious myofibroblast trans-differentiation after injury (Fig. 5N).
The AMP lineage includes an airway smooth muscle cell progenitor that is activated after injury
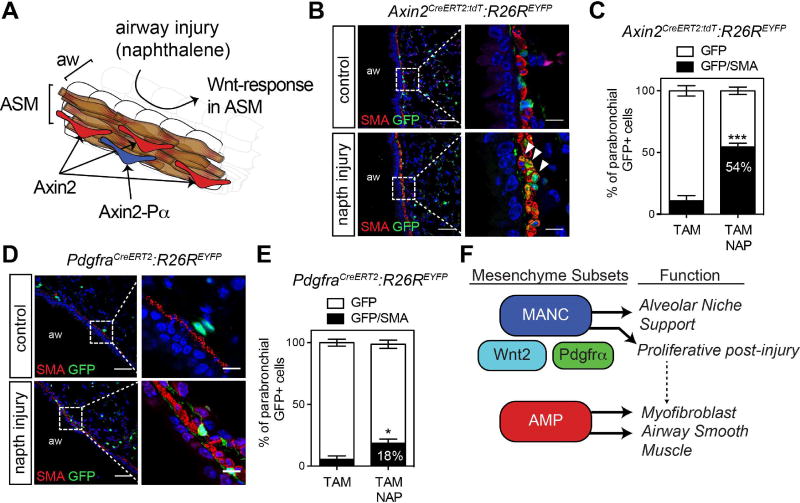
The Axin2+ cells surrounding the airways are interdigitated within the parabronchial smooth muscle layer, yet most do not express αSMA (Fig. 6A and B and Supplemental Fig. 1A). Despite their lack of αSMA immunoreactivity, transcriptome analysis of the AMP lineage indicates their preferential expression of myogenic genes, suggesting that they may act as progenitors of smooth muscle. To determine whether the AMPs that are located underlying the bronchiolar airways could generate parabronchial smooth muscle after airway injury, we performed lineage tracing using the Axin2CreERT2:tdT and PdgfrαCreERT2 mouse lines in the context of naphthalene based airway epithelial injury. Naphthalene treatment induces significant airway secretory cell depletion, causing an epithelial regenerative response within three weeks after injury with a concomitant and progressive Wnt-mediated response in the underlying proliferative parabronchial smooth muscle (Volckaert et al., 2013; Volckaert et al., 2011) (Fig. 6A, Supplemental Fig. 4). Three weeks after naphthalene injury, greater than 50% of parabronchial Axin2+ cells acquired αSMA expression, a dramatic increase over uninjured controls (Fig. 6B and C). In contrast, Pdgfrα+ cells responded less robustly, with only 18% of lineage labeled cells generating αSMA+ parabronchial smooth muscle cells (Fig. 6D and E). These data suggest that the AMP cell lineage contains a parabronchial smooth muscle progenitor responsive to airway injury (Fig. 6F).
Figure 6. AMP cells generate airway smooth muscle following airway epithelial injury by naphthalene.
(A) Axin2+ cells (red) are the predominant Axin2 lineage residing near the airway smooth muscle (ASM-brown). Axin2+/Pdgfrα+ cells (blue) comprise a small (42%) of the overall Axin2+ cells surrounding the airways. In the naphthalene injury model airway epithelial cells are selectively ablated and a Wnt-dependent ASM proliferative response occurs. (B and C) Lineage traced Axin2+ cells, EYFP-positive and αSMA was analyzed by immunostaining and quantified. (D and E) Lineage traced Pdgfrα+ cells, EYFP+ and Acta2 was analyzed by immunostaining and quantified. (F) Model diagram indicating that the Axin2+ AMP lineage generates both parabronchial smooth muscle and myofibroblasts after injury. Representative images are shown from n=3 mice per treatment group. Error bars are means ± SEM. *=p<0.05 and ***=p<0.001 by U-test. Aw=airway. Scale bars, low magnification =50µm, high magnification =10 µM.
Dose dependent response to Wnt signaling delineates the MANC and AMP lineages
FACS analysis shows that there are two distinct populations of Axin2+ mesenchymal cells in the lung, one with very high levels of Axin2 expression (Axin2bright) and one with lower levels (Axin2dim) (Supplemental Fig. 4A). Characterization of these two populations shows that the Axin2bright population preferentially expresses Pdgfrα, a distinguishing gene of the MANC lineage, whereas the Axin2dim population preferentially expresses Pdgfrβ and Acta2, which are hallmark genes of the AMP lineage (Supplemental Fig. 4B and C). Since Axin2 expression quantitatively reports Wnt responsiveness in cells (Al Alam et al., 2011; Frank et al., 2016), we examined whether genetic activation of Wnt signaling could convert the AMP lineage into the MANC lineage which would have beneficial effects on promoting alveolar regeneration over fibrotic remodeling after injury. Treatment of AMPs and MANCs in culture with a Gsk3β inhibitor (CHIR) to activate Wnt signaling resulted in an increase in Acta2 and Notch3 expression specifically in the AMP population (Supplemental Fig. 4D). Next, we used Axin2-Cre-directed activation of a β-catenin gain of function allele (Ctnnb1Flox(Ex3)/wt) referred to as Ctnnb1GOF in vivo to assess the response to increased Wnt/β-catenin signaling. At 21 days post tamoxifen induction, the lungs were harvested and MANC and AMP cells were isolated using FACS. The AMP population exhibited up-regulation of Acta2, Pdgfrb and Notch3 whereas the MANCs did not (Supplemental Fig. 4E). These data indicate that while MANC cells do not alter their AMP gene signature, the AMP cells do. Furthermore, activation of Wnt/β-catenin activity in vivo led to increased numbers of lineage-traced αSMA expressing myofibroblasts but did not affect the percentage of Axin2-lineage traced Pdgfrα+ MANCs in the lung (Supplemental Fig. 4F and G). These data suggest that AMP cells are uniquely sensitive to the genetic activation of β-catenin with only little changes indicating MANC to AMP plasticity in this model.
Identification of the mesenchymal niche derived paracrine signals that promote growth and regeneration in the lung alveolus
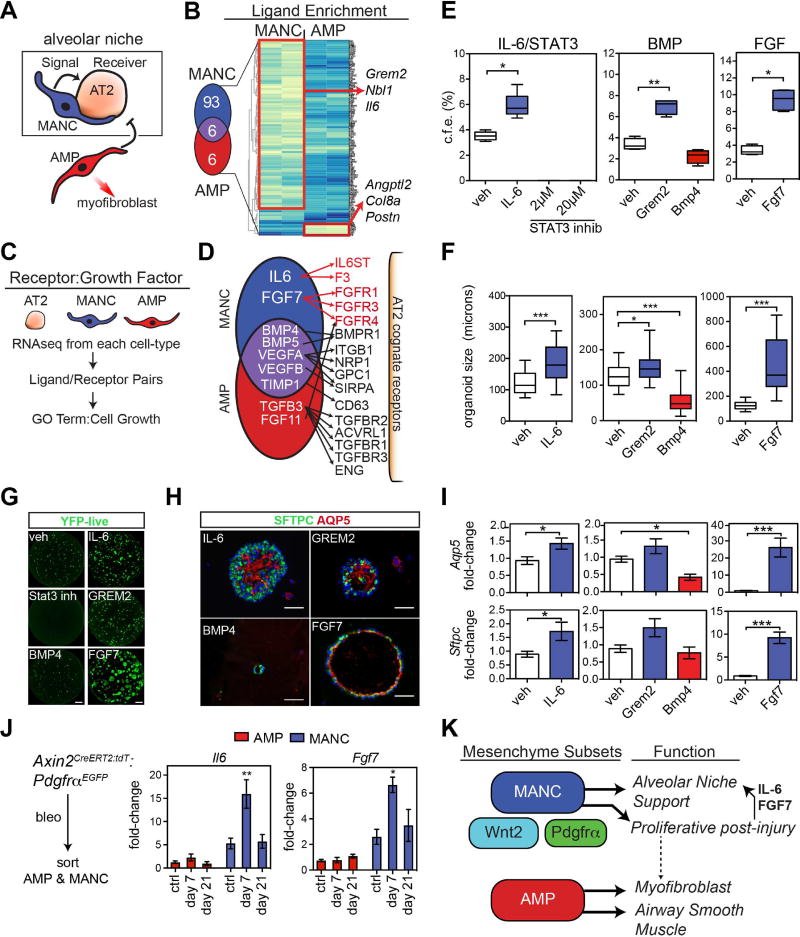
To define the paracrine signaling pathways that induce alveolar growth through activation of AT2 cells, we utilized two separate methods. First, we identified signaling ligands preferentially expressed in MANCs versus AMPs (Fig. 7A and B). Second, we utilized a method for identifying ligand and cognate receptor expression in various cell lineages using our RNA-seq transcriptome data and annotations derived from the FANTOM5 database (Ramilowski et al., 2015) (Fig. 7C and 7D). Using these two methods, we developed a secretome for MANCs and AMPs and a receptome for AT2 cells to assess mesenchymal-epithelial crosstalk in the lung alveolus. Since the MANC lineage preferentially promotes the self-renewal of AT2 cells, we chose to focus on the pathways identified in the secretome that were specific to the MANC lineage. Examination of ligand enrichment revealed preferential expression of IL6 and Grem2, a Bmp signaling inhibitor (Fig. 7B). Analysis of the secretome-receptome pairing from the FANTOM5 database shows that in addition to Il6 and Grem2, Fgf7 was also preferentially expressed in the MANC lineage (Fig. 7D). Several ligands were expressed at approximately equal levels in the MANC and AMP lineage including Bmp4 and Bmp5. However, only the MANC lineage expressed the Bmp inhibitor Grem2, suggesting important modulation of Bmp signaling in the alveolar niche by the MANC.
Figure 7. Defining the lung alveolar niche through characterization of the MANC–specific AT2 receptome.
(A) MANC cells are intimately associated with AT2 cells produce signals that likely direct AT2 growth or quiescence. (B) MANC and AMP enriched genes for secreted proteins were identified from the popRNA-seq data. The Venn diagram shows the number of unique genes and the genes listed to the right are unique to either the MANC or AMP lineage as indicated. (C) Schematic for cell type ligand/receptome analysis (D) Summary of acquired data as outlined in C, showing IL6:IL-6ST and Fgf7:Fgfr identified as unique ligand:receptor pairs in the MANC:AT2 interactions. (E) SftpcCreERT2:R26REYFP cells were combined with MANC as outlined in Figure 4 and colony forming efficiency was calculated after 21 days in culture. (F) In the same experiments, organoid size was calculated, error bars are min and max values. (G) Native EYFP fluorescence of representative wells from the control (veh) and treated AT2 cells cultured with MANCs, scale bar is 1 mm (H) Immunostaining of the organoids from the indicted treatment groups for expression of Sftpc and Aqp5, scale bar is 50µm (I) Q-PCR from the indicated conditions showing differential expression of AT1 differentiation marker Aqp5 (top row) and the AT2 cell marker Sftpc (bottom row). (J) Assessment of the in vivo relevance of MANC produced growth factors Il6 and Fgf7, showing changes due to bleomycin injury. (K) Overview model showing that regionally distributed mesenchymal lineages are distinct with MANCs spatially located next to AT2 cells to promote AT2 cell growth and differentiation while a distinct AMP lineage is poised to generate myofibroblasts following tissue damage. N=4–5 mice per condition, the same veh conditions were used for Bmp and Fgf experiments and are re-plotted for each pathway graph to aid visualization. Error bars are means ± SEM. *, **, ** are p<0.05, 0.01 and 0.001 respectively by U-test or one-way ANOVA. Scale bars=25µm.
To determine the functional relevance of these pathways in AT2 self-renewal, alveolar growth, and AT1 cell differentiation, we utilized the alveolar organoid assay to test the effects of their activation or inhibition. Inhibition of Stat3 results in a profound loss of organoid formation while activation using recombinant IL6 increased organoid formation and size (Fig. 7E–H). Addition of recombinant Grem2 caused an increase in organoid formation and size, while addition of recombinant Bmp4 lead to a reduction organoid formation and significantly decreased their size (Fig. 7E–H). Addition of recombinant Fgf7 lead to a dramatic increase in organoid formation and a robust increase in the size of the organoids (Fig. 7E–H). In addition to increasing the number and size of alveolar organoids, IL-6, Grem2 and Fgf7 enhanced differentiation as noted by increased Aqp5 and Sftpc expression (Fig. 7I). In contrast, Bmp4 treatment decreased alveolar epithelial differentiation as noted by decreased Aqp5 expression (Fig. 7I). Taken together, these data point to a careful balance of IL6/Stat3, Bmp, and Fgf signaling in alveolar growth with IL6 and Fgf7 promoting AT2 cell self-renewal while Bmp signaling inhibiting AT2 growth and AT1 differentiation. To further define the relevance of the MANC-produced AT2 growth factors in vivo, we measured expression of Il6 and Fgf7 in MANCs versus AMP cells after bleomycin treatment. Shortly after bleomycin instillation, at day 7, MANCs specifically up-regulated the expression of these growth factors, implicating the importance of MANC cells in promoting the alveolar regenerative response (Fig. 7J). These data define an alveolar niche with diverse mesenchymal lineages including MANC and AMP cells, each with a distinct role in promoting alveolar growth and regeneration versus the deleterious fibrotic remodeling response after injury (Fig. 7K).
DISCUSSION
Despite the growing appreciation of the importance of epithelial-mesenchymal signaling in adult tissue homeostasis and regeneration, the identity of tissue specific mesenchymal lineages and the molecular pathways that regulate their niche promoting behavior in comparison to deleterious disease remodeling processes remains unclear. Our data provide a comprehensive de-convolution of the lung mesenchyme, which allowed us to identify a critical alveolar niche supporting cell lineage called the MANC, and a lineage called the AMP that is a major contributor towards the myofibrotic remodeling process after acute lung injury. Using a combined secretome-receptome analysis, we show that key paracrine pathways originating in the MANC play a critical role in promoting alveolar growth and regeneration versus the fibrotic response after injury. Together, this study provides a comprehensive examination of a tissue specific niche through the analysis of the various mesenchymal lineages and their relative contribution towards a pro-regenerative process versus a deleterious disease remodeling process.
Recent studies have begun to reveal that epithelial-mesenchymal signaling plays a critical role in tissue homeostasis and cancer development. In pancreatic cancer, sonic hedgehog has been shown to promote mesenchymal growth (Rhim et al., 2014). Loss of Shh leads to increased aggressiveness of pancreatic tumors suggesting that during cancer development, mesenchyme can restrain tumor growth. In contrast, Shh signaling inhibits mesenchymal growth and proliferation in the adult lung during normal homeostasis (Peng et al., 2015). Loss of Shh or hedgehog signaling in general leads to increased proliferation in both hedgehog receptive mesenchyme as noted by Gli1 expression as well as in the adjoining airway epithelium. While such studies provided important information regarding epithelial-mesenchymal cross-talk in cancer as well as normal homeostasis, there remains a dearth of information regarding the relative contribution of mesenchymal lineage heterogeneity in either a regenerative process or a deleterious disease remodeling process. Important limitations in previous studies have been the lack of markers for different mesenchymal lineages within a given tissue, a definition of mesenchymal heterogeneity at the single cell level, and comparison of different mesenchymal lineages within the same tissue. Our studies were specifically designed to overcome these limitations by utilizing multiple mesenchymal cell lineage reporters focused on important known paracrine pathways active in the developing and adult lung combined with a comprehensive single cell transcriptome analysis to dissect the functional heterogeneity within the lung mesenchyme. Using spatial distance mapping or SDM, our data has defined at high resolution the placement of specific mesenchymal lineages throughout the entire lung. This analysis has revealed the existence of multiple distinct lineages marked by combinations of Axin2, Pdgfrα, and Wnt2 expression, with each of these occupying a unique spatial address, suggesting differing functions in supporting either the alveolar or airway niches in the lung. Importantly, the single-cell RNA transcriptome analysis revealed a previously underappreciated heterogeneity in the lung mesenchyme. While scRNA-seq analysis does not provide the depth or sensitivity to detect all expressed genes in a given cell, combined with our popRNA-seq approach using the lineage specific reporters, we were able to resolve these differing cell types with specific gene expression signatures, confirming their unique lineage demarcation. Complimentary findings reported by Lee et. al., 2017, reveal that anatomically discreet mesenchymal cells, are transcriptionally programmed with specialized niche-support capabilities, depending on their spatial identity, for airway or alveolar epithelial self-renewal.
Several important considerations are revealed by our study. While the adult lung is a normally quiescent organ with little cellular turnover at homeostasis, there is a high level of active Wnt signaling as noted by the finding that more than two-thirds of the adult lung mesenchyme exhibits Wnt-signaling pathway responsiveness as reported by the Axin2CreERT2:tdT mouse line. This line faithfully reports Axin2 gene expression and thus Wnt signaling responsiveness in a pattern identical to the Axin2lacZ line (Choi et al., 2013; Frank et al., 2016). The broad extent of active Wnt signaling during homeostasis in the mesenchyme of a quiescent organ such as the lung suggests that this pathway is playing a role in maintaining quiescence or priming the tissue for an injury response. In the lung, there are two populations of Axin2 expressing mesenchymal cells which have differing characteristics. The Axin2bright cells encompass the MANC lineage, whereas the Axin2dim cells encompass the AMP cell lineage, indicating that these two lineages can be isolated based on their relative levels of Wnt responsiveness. Genetic activation of β-catenin in these two lineages leads to aberrant expression of mesenchymal markers and precocious alveolar myofibroblast differentiation in the absence of injury. These data suggest that increased Wnt signaling can lead to a deleterious response in the lung mesenchyme, possibly promoting a fibrotic phenotype. This is supported by findings that Wnt signaling is activated in idiopathic pulmonary fibrosis lesions in humans and in the murine bleomycin model (Cao et al., 2016; Chilosi et al., 2003; Konigshoff et al., 2008). However, other data also suggests that Wnt signaling may be important for AT2 cell self-renewal in vitro (Flozak et al., 2010; Liu et al., 2015), indicating a fine tune balance of Wnt signaling activity for homeostasis and regeneration in the lung. This is consistent with data showing that MANCs express several Wnt signaling inhibitors in addition to exhibiting high levels of Wnt responsiveness.
Our popRNA-seq transcriptome analysis of the various mesenchymal lineages in the adult lung allowed us to identify critical paracrine signals generated by the MANC lineage that promote alveolar growth and regeneration. Some of these pathways are known to play an important role in lung development, however, their role in adult lung homeostasis or regeneration has remained unclear. In particular, the IL6-Stat3 pathway appears to be essential to AT2 cell self-renewal as a recent study identified that IL-6 can promote AT2 cell growth (Liang et al., 2016). We also show that Fgf7 signaling plays an important role in AT2 cell self-renewal and alveolar growth. Fgf7, previously known as keratinocyte growth factor or Kgf, was known to promote AT2 cell proliferation in in vitro cell culture (Cardoso et al., 1997; Panos et al., 1993; Yano et al., 1996). However, the origin of Fgf7 expression in the lung and how it relates to AT2 cell function in vivo has remained unclear. Our data show that the specialized MANC lineage preferentially expresses Fgf7 to promote AT2 cell proliferation as well as differentiation into AT1 cells. Given the pro-growth signals provided by IL6-Stat3 and Fgf7 arising in MANCs, these pathways are clearly critical for regeneration of AT2 cells after lung injury. While IL6 and Fgf7 play important roles through their activation of Stat3 and Fgf receptor signaling, there is likely redundancy in the inputs for these two pathways that remains to be uncovered.
In contrast to IL6 and Fgf7, Bmp signaling appears to inhibit AT2 differentiation into AT1, a mechanism that involves expression of secreted Bmp inhibitors such as Grem2 in the MANC lineage. What role Bmp signaling plays in the adult lung is unclear. In the developing lung, loss of Bmp signaling leads to decreased Sox9+ distal endoderm progenitor development whereas increased Bmp4 expression causes a loss of Sox2+ lung endoderm progenitor development (Bellusci et al., 1996; Eblaghie et al., 2006; Wang et al., 2013; Weaver et al., 2000; Weaver et al., 1999). The finding that Bmp signaling inhibits AT2 cell expansion and differentiation into AT1 cells suggests that the role for Bmp signaling in the adult alveolus may be different than during lung development. A similar finding was shown for hedgehog signaling where in the developing lung it promotes growth and differentiation of early lung endoderm progenitors while in the adult lung, hedgehog maintains quiescence in both the endoderm and mesenchyme (Peng et al., 2015). It is important to note that our SDM measurements indicate a significant difference in the proximity of the AMP and MANC to the AT2 cell. The relevance of the proximity of niche cells may have implications for bioavailability of ligands, the mechanisms of which are important lines for future investigation. These findings suggest that the roles for paracrine signaling pathways in adult tissue maintenance and regeneration may not consistently be extrapolated from their roles in organ development.
The present study reveals important mesenchymal lineage heterogeneity in the adult lung and that the adult lung mesenchyme is composed of spatially, transcriptionally, and functionally discreet lineages that each play an important role in the response to injury. These data also show that the important contribution of the lung mesenchyme towards regeneration of the alveolus can be separated from the lineage that promotes myofibrotic remodeling after injury. These findings will be important when compared to niche specific mesenchymal lineages in other adult tissues as they are identified and may provide a new paradigm on how developmental signaling pathways are co-opted in different ways to promote or inhibit tissue regeneration.
STAR METHODS
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
The generation of and genotyping information regarding the R26REYFP, R26RtdTomato, PdgfraEGFP, PdgfraCreERT2 and Axin2LacZ mouse lines have been previously described and were all purchased from Jackson Laboratories. The Axin2CreERT2-tdT mouse line was generated as described (Frank et al., 2016). The Wnt2CreERT2 mouse was described previously (Peng et al., 2013). The SftpcCreERT2 mouse line was generously provided by Dr. Hal Chapman (Chapman et al., 2011). The Ctnnb1lox(ex3) allele was described previously (Harada et al., 1999). Unless otherwise indicated all mice were maintained on a mixed background and were 6–8 weeks of age for all experiments described in this study. All animal procedures were performed under the guidance of the University of Pennsylvania Institutional Animal Care and Use Committee.
Tamoxifen induction of cell-lineage tracing
Tamoxifen (Sigma-Aldrich) was dissolved in corn oil at a concentration of 20 mg/ml. Mice were administered tamoxifen (200 mg/kg) as indicated for each experiment by oral gavage.
Bleomycin and Naphthalene Lung injury
One week post tamoxifen induction, pharmaceutical grade Bleomycin (Hospira) was administered to anesthetized mice via non-surgical intratracheal route at a concentration of 2U/kg while control animals were instilled with PBS. Naphthalene was prepared at a concentration of 20 mg/ml dissolved in corn-oil, mice received a single intraperitoneal injection of 200 mg/kg, control animals received corn-oil only via IP injection. Quantification in naphthalene model, was performed by counting the number of YFP/αSMA co-stained cells within 50µm of the airway epithelium.
METHOD DETAILS
Histology
At the time of tissue harvest mice were euthanized by CO2 inhalation. Chest cavity was exposed and lungs cleared of blood by perfusion with cold PBS via the right ventricle. Lungs were inflated with 2% paraformaldehyde under constant pressure of 30 cm water and allowed to fix overnight. Tissue was dehydrated through a series of ethanol washes after which they were embedded in paraffin and sectioned. Immunohistochemistry was performed following heat antigen retrieval methods and stained with the following antibodies. GFP (chicken, Aves, 1:500), GFP (goat, Abcam, 1:100), RFP (rabbit, Rockland, 1:250), Pdfgrα (rabbit, Cell Signaling, 1:50), Pdgfrα (goat, R&D Systems, 1:50), Pdgfrβ (rabbit, Cell Signaling, 1:100), Pdgfrβ (R&D Systems, 1:400), Sftpc (rabbit, Millipore, 1:250), Sftpc (goat, Santa Cruz, 1:50), Aqp5 (rabbit, Abcam, 1:100), Ki67 (rabbit, Abcam, 1:50) and PO4-Stat3 (y727) (rabbit, Cell Signaling, 1:50). Whole-mount staining was used for data shown in Figure 2 for Axin2CreERT2:PdgfrαEGFP:R26REYFP lineage traced for 7 days. Tissue was fixed as outlined above however the following day the lung tissue was washed several times in PBS then it was embedded in 4% low melt agarose. Afterwards lung tissue was sectioned by vibratome into 150µm thick slices. These were further incubated with antibody to SP-C (goat, Santa Cruz, 1:50) and secondary antibody in PBS with 0.5% Triton X-100 for two days. The stained slices were cleared with Scale A2 and Scale B4 for one week, then mounted on coverslips and imaged by confocal microscopy. RNAScope (Advanced Cell Diagnostics) was performed on 4% PFA fixed wild-type adult lung tissue according to the manufacture’s protocol using a probe against mm-Aoc3.
Single Cell Suspension and Staining
Single cell suspensions were generated as follows. First the lung was removed and minced with a razor blade. Minced lung was placed in a digestion solution containing 480 U/ml Collagenase Type I (Life Technologies) 50 U/ml Dispase (Collaborative Biosciences) and 0.33 U/ml DNase (Roche), this was allowed to incubate in a 37° water bath with frequent agitation for 45 minutes. Cell solution was filtered through 100µm and 40µm cell strainer (BD Falcon). ACK lysis was used to remove blood cells. Cell pellets were resuspended in FACS buffer, containing sterile PBS with 1% FBS and 1mM EDTA. Antibodies used for flow cytometry are as follows, Epcam-APC, Epcam-FITC, CD31-PeCy7, CD45-PeCy7 and PDGFRα-APC all from eBiosciences. Cells were stained for approximately 30 minutes in the dark, after which they were washed once with FACS Buffer followed by addition of a viability dye, DAPI to label dead cells.
Flow Cytometry and sorting for RNA isolation
Single cell suspensions were gated on viability or Live cells by Dapi exclusion, then gated on singlets. The cell analyzer used was an LSR Fortessa (BD Biosciences). Cell sorting was performed on a MoFlo Astrios Eq (Beckman Coulter). For cell culture experiments, cells were sorted in FACS buffer containing 10% FBS. For RNA, cells of interest (150k per tube) were collected in Trizol-LS reagent (ThermoFisher). The RNA was isolated according to the manufacture’s protocol. Following RNA purification, cDNA was generated using the SuperScript IV First-Strand Synthesis System (ThermoFisher). Quantitative Real-Time PCR was then performed on a QuantStudio 7 Flex using SYBR green reagents (ThermoFisher).
Population RNA Sequencing and Analysis
RNA was extracted using Trizol-LS (Life Technologies) and MinElute RNA Cleanup Kit (Qiagen). Library prep was conducted using Illumina truSeq stranded mRNA kit and Clontech SMARTer RNA-seq amplification kit. Fastq files were assessed for quality control using the FastQC program. Fastq files were aligned against the mouse reference genome (mm9) using the STAR aligner (Dobin et al., 2013). Duplicate reads were flag using the MarkDuplicates program from Picard tools. Per gene read counts for Ensembl (v67) gene annotations were computed using the R package with duplicate reads removed. Gene counts represented as counts per million (CPM) were first nominalized using TMM method in the edgeR R package and genes with 25% of samples with a CPM < 1 were removed and deemed low expressed. The data was transformed using the VOOM function from the limma R package (Law et al., 2014). Differential gene expression was performed using a linear model with the limma package. Heatmaps and PCA plots were generated in R. Gene Ontology enrichment analysis was performed using the ToppGene Suite (http://toppgene.cchmc.org/) (Chen et al., 2009). The GEO accession number for the popRNA-seq data is GSE92699. Cell type ligand/receptome data was derived from AT2 cell popRNA-seq and MANC or AMP popRNA-seq data, genes with expression of FPKM=1 or greater were selected followed by analysis using ligand/receptor annotations derived from the FANTOM5 project. Gene ontology was performed on ligand:receptor pairs and Cell Growth category was interrogated for potential candidates.
Single-Cell RNA-sequencing using In-Drop and the GemCode platform
Axin2CreERT2:tdT:PdgfraEGFP mouse lung was dissected and single-cell preparation was sorted as described above. Cells were negatively sorted for Live, EPCAM-, CD31-, CD45-cells, herein referred to as bulk mesenchyme. The sorted cells were loaded onto a GemCode instrument (10× Genomics, Pleasanton, CA, USA) to generate single-cell barcoded droplets (GEMs) according to the manufacture’s protocol using the 10× Single Cell 3’ v1 chemistry. The resulting libraries were sequenced across two lanes on an Illumina HiSeq2500 instrument in High-output mode. Reads were aligned and subsequent analyses performed using the Cell Ranger (Pipeline). We obtained 68k reads per cell with a median genes per cell of 1,017 and median UMI count per cell of 2,040. The GEO accession number for the scRNA-seq data is GSE99714.
Alveolar Organoid Assay
Alveolar organoid assays were performed as described previously (Frank et al., 2016; Peng et al., 2015) with some modifications from the original protocol (Barkauskas et al., 2013). Briefly, SftpcCreERT2:R26REYFP mice were induced with tamoxifen, one week later mice were euthanized and single cell suspensions were made from the lung tissue as described above. At the same time the indicated mouse strains reporting lung mesenchyme were also harvested and single cell suspensions were made from lungs. Following red blood cell lysis with ACK buffer the cell suspensions were filtered through 40µm cell strainers (BD Falcon) and stained with antibodies for EPCAM-APC, CD31-PeCy7 and CD45-PeCy7. After staining, cells were resuspended in FACS buffer containing DAPI as a viability stain. Sorted cells were collected in DMEM+10%FBS, after which cells were centrifuged and counted by trypan exclusion. Approximately 5 × 103 SP-C cells were mixed with 5 × 104 mesenchymal cells in 50% Matrigel (growth factor reduced, phenol-red free) (Corning) in cell media, small airway growth media (SABM, Lonza) with the following additives; 1× insulin/transferrin, 0.1 µg/ml Cholera Toxin (Sigma), 25 ng/ml EGF (Peprotech), 30 µg/ml bovine pituitary extract (Lonza), 0.01µM Retinoic acid (Sigma & Lonza) and 5% FBS (Denville). Rock inhibitor, Y27632 (Sigma) was included in the media for the first two days and fresh media was added every two days. In conditions using Wnt2CreERT2:R26REYFP mesenchyme, the imaging using fluorescence stereomicroscopy reveals primarily Sftpc+ EYFP+ cells as they are the major cellular constituent in the organoids and thus comprising the vast majority of the visible fluorescence. In these conditions, the majority of Wnt2CreERT2:R26REYFP mesenchyme attaches to the bottom of the culture well. RNA was isolated on the final day of the experiment by Trizol (Life Technologies) isolation followed by cDNA synthesis using the Superscript IV RT kit (Life Technologies).
Ligand treatments of organoids were performed using the following reagents at the indicated concentrations, mIL-6 50ng/ml (R&D Systems), STATTIC 2 and 20 µM (Selleck BioChemicals), mGREM2 25 ng/ml (R&D Systems), mBMP4 50 ng/ml (R&D Systems), FGF7 25 ng/ml (R&D Systems). Ligands were added at the time of the first media change (after removal of Rock inhibitor), new ligands were added upon each media change or every other day.
QUANTIFICATION AND STATISTICAL ANALYSIS
Spatial Distance Mapping
For each mouse line indicated confocal z-stacks were acquired using Leica TCS SP8 or Leica STED 3× Super-resolution scopes, from at least five different sections from three individual mice per genotype tested. Data were analyzed using Imaris software (Bitplane) following 3D-rendering. Distance vectors, calculated in Imaris software, were drawn from the cell of interest to the nearest Sftpc cell using the measurements tool setting the DAPI channel (for nuclei) as the centroid for each paired point. Distances were mapped across each section by a blinded technician.
Statistical Analysis
Statistical analysis was performed in Prism 7 for Mac and R. A two-tailed non-parametric Mann-Whitney U-test was used for the comparison between two experimental groups and a one-way ANOVA was used for multiple comparisons. Data were considered significant if p < 0.05.
Supplementary Material
(A) Immunostaining shows that Axin2+ cells residing near airways do not express the smooth muscle marker SM22α. (B) Immunostaining shows that a small number of Axin2+ cells residing near blood vessels do express the smooth muscle marker αSMA. (C) Quantitation of the percent of Axin2+ cells surrounding airways or blood vessels that are αSMA+ or SM22+. (D) Efficiency of lineage tracing marks appoximately 44% of the Axin2+ cells (n=3 mice). (E-G) For FACS isolation of lung mesenchyme, single cell suspensions were prepared for each lung. The suspensions were gated on live cells based on exclusion of DAPI stain and then singlets were selected (E) to obtain single cells, followed by FSC-A and SSC-A gating as shown in F. (G) Cells that are negative for endothelial/hematopoietic (CD31-/CD45-) and epithelial (Epcam-) are considered mesenchymal cell types. Shown are representative flow plots derived from a single mouse. (H-I) Axin2-tdTomato labels approximately 36.6% of all cells with the adult murine lung,. Flow cytometry percentages from Axin2CreERT2-tdT:PdgfraEGFP mice, n=4 mice. The sum of all the four categories is ~90%. ASM=airway smooth muscle, VSM=blood vessel smooth muscle, aw=airway, bv=blood vessel.
(A) Relative expression of the indicated genes derived from the popRNA-seq shows that each isolated cell population expresses different levels of mesenchymal and myofibroblast related genes. X-axis to the left of the Eln data is in FPKM and is the same for all of the data, error bars are mean ± SEM. (B) The K clustering method showing the top 5 clusters obtained from the In-drop scRNA-seq. (C) Expression distribution of some of the same marker genes show in A, projected onto the scRNA-seq data. (D) Heatmap generated from the top genes in the clusters 1-5 outlined to the right, log2-fold-change greater than 2 and p-value less than 0.05. (E) In Situ staining for an AMP-lineage gene, Aoc3, shows spatially distinct localization in the peri-bronchiolar mesenchyme (pbm-speckled). Red blood cells (rbc), airway (aw), blood vessel (bv), alveolar region (alv).
(A) Wnt2CreERT2:R26REYFP:Axin2LacZ lineage traced were induced to assess Wnt2 and Axin2 lineages in parallel in control and bleomycin injured lungs. YFP lineage traced fluorescence images were merged with bright phase image and pseudo-colored red to facilitate visualization. (B) Quantification data from histology for LacZ and YFP costaining, n=3 mice. Error bars represent means ± SEM.
(A and B) Flow cytometry analysis from Axin2CreERT2:tdT gated on live CD31/CD45/Epcam-negative cells showing the Axin2-bright and dim cell populations and that the Axin2-bright cells are the Pdgfrα-positive MANC population whereas the tdTomato single positive encompass the Pdgfrα- AMP lineage. (C) RNA was generated from sorted MANC and AMP cells and Q-PCR was used to assess expression of the indicated genes associated with MANCs or AMPs. (D) AMP and MANC cells were cultured in matrigel and treated with the GSK3β inhibitor CHIR (CHIR99021) to activate β-catenin. Q-PCR was performed 48 hours later for Axin2, Acta2 and Notch3. (E) Activation of Wnt/β-catenin activity in Axin2+ in vivo, reveals increased expression of myofibroblast markers, in AMPs but little change in MANCs, n=3-4 mice per group. (E) Representative image from an induced β-catenin mutant mouse showing the presence of alveolar interstitial myofibroblasts that is not observed in wild-type controls. (F) Quantitation of the change in percentage of aSMA+ myofibroblasts after activation of Wnt/β-catenin activity in Axin2+ cells. Error bars are means ± SEM, * or ** indicates a p-value < 0.05 and 0.01, respectively by U-test.
(A) Axin2CreERT2:tdT:R26REYFP mice were induced with tamoxifen and one week later administered naphthalene by intraperitoneal injection. (B-F) Time course analysis showing the appearance of αSMApositive AMP cells following naphthalene injury. Arrows indicate αSMA-positive smooth muscle bearing the AMP/EYFP lineage. (B and C) Large airways similar to those shown in Figure 6. (D-F) small airways show a similar smooth muscle cell response during the 21 days post-injury time course. Representative images are shown from n=3 mice per time-point and treatment group. Aw=airway.
Highlights.
Single Cell and Population-based RNA sequencing stratifies lung mesenchymal subsets
Identification of functionally distinct myofibrogenic and alveolar niche cell types
Enriched Fgf/Il6/Bmp signaling defines the alveolar niche secretome:receptome
A single cell approach reveals the functional pathways that define the cellular and molecular framework of lung mesenchymal niches and reveal the functional importance of developmental pathways in promoting self-renewal versus pathological response to tissue injury.
Acknowledgments
These studies were supported by funding form the NIH to E.E.M. (R01-HL132999, R01-HL132349, R01-HL087825, U01-HL134745, U01-HL110942) and J.A.Z. (T32-HL007843). The authors would like acknowledge Florin Tuluc and his staff at the Flow Cytometry Core Laboratory of Children’s Hospital of Philadelphia for their assistance with FACS, and Collin Stabler and Renata Pellegrino and her staff at the Center for Applied Genomics at Children’s Hospital of Philadelphia for their assistance with single-cell RNA sequencing. The authors are also grateful for the technical assistance provided by the Penn Cardiovascular Histology Core for their histological services.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.A.Z and E.E.M designed experiments and wrote the manuscript with input from co-authors. J.A.Z, W.J.Z., D.B.F., C.A.C., S.Z., M.P.M. performed experiments and data analysis. M.P.M. performed bioinformatics analysis.
References
- Adamson IY, Bowden DH. The pathogenesis of bleomycin-induced pulmonary fibrosis in mice. The American journal of pathology. 1974;77:185–197. [PMC free article] [PubMed] [Google Scholar]
- Al Alam D, Green M, Tabatabai Irani R, Parsa S, Danopoulos S, Sala FG, Branch J, El Agha E, Tiozzo C, Voswinckel R, et al. Contrasting expression of canonical Wnt signaling reporters TOPGAL, BATGAL and Axin2(LacZ) during murine lung development and repair. PloS one. 2011;6:e23139. doi: 10.1371/journal.pone.0023139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, Hogan BL. Type 2 alveolar cells are stem cells in adult lung. The Journal of clinical investigation. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer Y, Tedrow J, de Bernard S, Birker-Robaczewska M, Gibson KF, Guardela BJ, Hess P, Klenk A, Lindell KO, Poirey S, et al. A novel genomic signature with translational significance for human idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol. 2015;52:217–231. doi: 10.1165/rcmb.2013-0310OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellusci S, Henderson R, Winnier G, Oikawa T, Hogan BL. Evidence from normal expression and targeted misexpression that bone morphogenetic protein (Bmp-4) plays a role in mouse embryonic lung morphogenesis. Development. 1996;122:1693–1702. doi: 10.1242/dev.122.6.1693. [DOI] [PubMed] [Google Scholar]
- Cao Z, Lis R, Ginsberg M, Chavez D, Shido K, Rabbany SY, Fong GH, Sakmar TP, Rafii S, Ding BS. Targeting of the pulmonary capillary vascular niche promotes lung alveolar repair and ameliorates fibrosis. Nature medicine. 2016;22:154–162. doi: 10.1038/nm.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso WV, Itoh A, Nogawa H, Mason I, Brody JS. FGF-1 and FGF-7 induce distinct patterns of growth and differentiation in embryonic lung epithelium. Dev Dyn. 1997;208:398–405. doi: 10.1002/(SICI)1097-0177(199703)208:3<398::AID-AJA10>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Chang AC, Fu Y, Garside VC, Niessen K, Chang L, Fuller M, Setiadi A, Smrz J, Kyle A, Minchinton A, et al. Notch initiates the endothelial-to-mesenchymal transition in the atrioventricular canal through autocrine activation of soluble guanylyl cyclase. Dev Cell. 2011;21:288–300. doi: 10.1016/j.devcel.2011.06.022. [DOI] [PubMed] [Google Scholar]
- Chapman HA, Li X, Alexander JP, Brumwell A, Lorizio W, Tan K, Sonnenberg A, Wei Y, Vu TH. Integrin alpha6beta4 identifies an adult distal lung epithelial population with regenerative potential in mice. The Journal of clinical investigation. 2011;121:2855–2862. doi: 10.1172/JCI57673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilosi M, Poletti V, Zamo A, Lestani M, Montagna L, Piccoli P, Pedron S, Bertaso M, Scarpa A, Murer B, et al. Aberrant Wnt/beta-catenin pathway activation in idiopathic pulmonary fibrosis. The American journal of pathology. 2003;162:1495–1502. doi: 10.1016/s0002-9440(10)64282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Zhang Y, Xu M, Yang Y, Ito M, Peng T, Cui Z, Nagy A, Hadjantonakis AK, Lang RA, et al. Distinct functions for Wnt/beta-catenin in hair follicle stem cell proliferation and survival and interfollicular epidermal homeostasis. Cell stem cell. 2013;13:720–733. doi: 10.1016/j.stem.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degryse AL, Lawson WE. Progress toward improving animal models for idiopathic pulmonary fibrosis. The American journal of the medical sciences. 2011;341:444–449. doi: 10.1097/MAJ.0b013e31821aa000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 2014;507:190–194. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eblaghie MC, Reedy M, Oliver T, Mishina Y, Hogan BL. Evidence that autocrine signaling through Bmpr1a regulates the proliferation, survival and morphogenetic behavior of distal lung epithelial cells. Dev Biol. 2006;291:67–82. doi: 10.1016/j.ydbio.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Flozak AS, Lam AP, Russell S, Jain M, Peled ON, Sheppard KA, Beri R, Mutlu GM, Budinger GR, Gottardi CJ. Beta-catenin/T-cell factor signaling is activated during lung injury and promotes the survival and migration of alveolar epithelial cells. J Biol Chem. 2010;285:3157–3167. doi: 10.1074/jbc.M109.070326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DB, Peng T, Zepp JA, Snitow M, Vincent TL, Penkala IJ, Cui Z, Herriges MJ, Morley MP, Zhou S, et al. Emergence of a Wave of Wnt Signaling that Regulates Lung Alveologenesis by Controlling Epithelial Self-Renewal and Differentiation. Cell Rep. 2016;17:2312–2325. doi: 10.1016/j.celrep.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Molecular and cellular biology. 2003;23:4013–4025. doi: 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia LT, Ashley N, Ouaret D, Wang LM, Wilding J, Bodmer WF. Myofibroblasts are distinguished from activated skin fibroblasts by the expression of AOC3 and other associated markers. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E2162–2171. doi: 10.1073/pnas.1603534113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapanci Y, Ribaux C, Chaponnier C, Gabbiani G. Cytoskeletal features of alveolar myofibroblasts and pericytes in normal human and rat lung. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1992;40:1955–1963. doi: 10.1177/40.12.1333502. [DOI] [PubMed] [Google Scholar]
- Konigshoff M, Balsara N, Pfaff EM, Kramer M, Chrobak I, Seeger W, Eickelberg O. Functional Wnt signaling is increased in idiopathic pulmonary fibrosis. PloS one. 2008;3:e2142. doi: 10.1371/journal.pone.0002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Tammela T, Hofree M, Choi J, Marjanovic N, Han S, Canner D, Paschini M, Bhang DH, Jacks T, Regev, Kim CF. Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell. 2017 doi: 10.1016/j.cell.2017.07.028. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Zhang Y, Xie T, Liu N, Chen H, Geng Y, Kurkciyan A, Mena JM, Stripp BR, Jiang D, et al. Hyaluronan and TLR4 promote surfactant-protein-C-positive alveolar progenitor cell renewal and prevent severe pulmonary fibrosis in mice. Nature medicine. 2016;22:1285–1293. doi: 10.1038/nm.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kumar VS, Zhang W, Rehman J, Malik AB. Activation of type II cells into regenerative stem cell antigen-1(+) cells during alveolar repair. Am J Respir Cell Mol Biol. 2015;53:113–124. doi: 10.1165/rcmb.2013-0497OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncman CL, Wang K. Nebulette: a 107 kD nebulin-like protein in cardiac muscle. Cell Motil Cytoskeleton. 1995;32:205–225. doi: 10.1002/cm.970320305. [DOI] [PubMed] [Google Scholar]
- Panos RJ, Rubin JS, Csaky KG, Aaronson SA, Mason RJ. Keratinocyte growth factor and hepatocyte growth factor/scatter factor are heparin-binding growth factors for alveolar type II cells in fibroblast-conditioned medium. The Journal of clinical investigation. 1993;92:969–977. doi: 10.1172/JCI116673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T, Frank DB, Kadzik RS, Morley MP, Rathi KS, Wang T, Zhou S, Cheng L, Lu MM, Morrisey EE. Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature. 2015;526:578–582. doi: 10.1038/nature14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T, Tian Y, Boogerd CJ, Lu MM, Kadzik RS, Stewart KM, Evans SM, Morrisey EE. Coordination of heart and lung co-development by a multipotent cardiopulmonary progenitor. Nature. 2013;500:589–592. doi: 10.1038/nature12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski JA, Goldberg T, Harshbarger J, Kloppmann E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, Rost B, et al. A draft network of ligand-receptor-mediated multicellular signalling in human. Nature communications. 2015;6:7866. doi: 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, Dekleva EN, Saunders T, Becerra CP, Tattersall IW, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer cell. 2014;25:735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Barkauskas CE, Cronce MJ, Xue Y, Harris JR, Liang J, Noble PW, Hogan BL. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E1475–1483. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert T, Campbell A, De Langhe S. c-Myc regulates proliferation and Fgf10 expression in airway smooth muscle after airway epithelial injury in mouse. PloS one. 2013;8:e71426. doi: 10.1371/journal.pone.0071426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert T, Dill E, Campbell A, Tiozzo C, Majka S, Bellusci S, De Langhe SP. Parabronchial smooth muscle constitutes an airway epithelial stem cell niche in the mouse lung after injury. The Journal of clinical investigation. 2011;121:4409–4419. doi: 10.1172/JCI58097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Pan L, Moens CB, Appel B. Notch3 establishes brain vascular integrity by regulating pericyte number. Development. 2014;141:307–317. doi: 10.1242/dev.096107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tian Y, Morley MP, Lu MM, Demayo FJ, Olson EN, Morrisey EE. Development and regeneration of Sox2+ endoderm progenitors are regulated by a Hdac1/2-Bmp4/Rb1 regulatory pathway. Dev Cell. 2013;24:345–358. doi: 10.1016/j.devcel.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver M, Dunn NR, Hogan BL. Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development. 2000;127:2695–2704. doi: 10.1242/dev.127.12.2695. [DOI] [PubMed] [Google Scholar]
- Weaver M, Yingling JM, Dunn NR, Bellusci S, Hogan BL. Bmp signaling regulates proximal-distal differentiation of endoderm in mouse lung development. Development. 1999;126:4005–4015. doi: 10.1242/dev.126.18.4005. [DOI] [PubMed] [Google Scholar]
- Yano T, Deterding RR, Simonet WS, Shannon JM, Mason RJ. Keratinocyte growth factor reduces lung damage due to acid instillation in rats. Am J Respir Cell Mol Biol. 1996;15:433–442. doi: 10.1165/ajrcmb.15.4.8879176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Immunostaining shows that Axin2+ cells residing near airways do not express the smooth muscle marker SM22α. (B) Immunostaining shows that a small number of Axin2+ cells residing near blood vessels do express the smooth muscle marker αSMA. (C) Quantitation of the percent of Axin2+ cells surrounding airways or blood vessels that are αSMA+ or SM22+. (D) Efficiency of lineage tracing marks appoximately 44% of the Axin2+ cells (n=3 mice). (E-G) For FACS isolation of lung mesenchyme, single cell suspensions were prepared for each lung. The suspensions were gated on live cells based on exclusion of DAPI stain and then singlets were selected (E) to obtain single cells, followed by FSC-A and SSC-A gating as shown in F. (G) Cells that are negative for endothelial/hematopoietic (CD31-/CD45-) and epithelial (Epcam-) are considered mesenchymal cell types. Shown are representative flow plots derived from a single mouse. (H-I) Axin2-tdTomato labels approximately 36.6% of all cells with the adult murine lung,. Flow cytometry percentages from Axin2CreERT2-tdT:PdgfraEGFP mice, n=4 mice. The sum of all the four categories is ~90%. ASM=airway smooth muscle, VSM=blood vessel smooth muscle, aw=airway, bv=blood vessel.
(A) Relative expression of the indicated genes derived from the popRNA-seq shows that each isolated cell population expresses different levels of mesenchymal and myofibroblast related genes. X-axis to the left of the Eln data is in FPKM and is the same for all of the data, error bars are mean ± SEM. (B) The K clustering method showing the top 5 clusters obtained from the In-drop scRNA-seq. (C) Expression distribution of some of the same marker genes show in A, projected onto the scRNA-seq data. (D) Heatmap generated from the top genes in the clusters 1-5 outlined to the right, log2-fold-change greater than 2 and p-value less than 0.05. (E) In Situ staining for an AMP-lineage gene, Aoc3, shows spatially distinct localization in the peri-bronchiolar mesenchyme (pbm-speckled). Red blood cells (rbc), airway (aw), blood vessel (bv), alveolar region (alv).
(A) Wnt2CreERT2:R26REYFP:Axin2LacZ lineage traced were induced to assess Wnt2 and Axin2 lineages in parallel in control and bleomycin injured lungs. YFP lineage traced fluorescence images were merged with bright phase image and pseudo-colored red to facilitate visualization. (B) Quantification data from histology for LacZ and YFP costaining, n=3 mice. Error bars represent means ± SEM.
(A and B) Flow cytometry analysis from Axin2CreERT2:tdT gated on live CD31/CD45/Epcam-negative cells showing the Axin2-bright and dim cell populations and that the Axin2-bright cells are the Pdgfrα-positive MANC population whereas the tdTomato single positive encompass the Pdgfrα- AMP lineage. (C) RNA was generated from sorted MANC and AMP cells and Q-PCR was used to assess expression of the indicated genes associated with MANCs or AMPs. (D) AMP and MANC cells were cultured in matrigel and treated with the GSK3β inhibitor CHIR (CHIR99021) to activate β-catenin. Q-PCR was performed 48 hours later for Axin2, Acta2 and Notch3. (E) Activation of Wnt/β-catenin activity in Axin2+ in vivo, reveals increased expression of myofibroblast markers, in AMPs but little change in MANCs, n=3-4 mice per group. (E) Representative image from an induced β-catenin mutant mouse showing the presence of alveolar interstitial myofibroblasts that is not observed in wild-type controls. (F) Quantitation of the change in percentage of aSMA+ myofibroblasts after activation of Wnt/β-catenin activity in Axin2+ cells. Error bars are means ± SEM, * or ** indicates a p-value < 0.05 and 0.01, respectively by U-test.
(A) Axin2CreERT2:tdT:R26REYFP mice were induced with tamoxifen and one week later administered naphthalene by intraperitoneal injection. (B-F) Time course analysis showing the appearance of αSMApositive AMP cells following naphthalene injury. Arrows indicate αSMA-positive smooth muscle bearing the AMP/EYFP lineage. (B and C) Large airways similar to those shown in Figure 6. (D-F) small airways show a similar smooth muscle cell response during the 21 days post-injury time course. Representative images are shown from n=3 mice per time-point and treatment group. Aw=airway.