Abstract
Purpose of review
Only recently has it become clear that epithelial ovarian cancer (EOC) is comprised of such distinct histotypes--with different cells of origin, morphology, molecular features, epidemiologic factors, clinical features, and survival patterns—that they can be thought of as different diseases sharing an anatomical location. Herein, we review opportunities and challenges in studying EOC heterogeneity,
Recent findings
The 2014 World Health Organization diagnostic guidelines incorporate accumulated evidence that high- and low-grade serous tumors have different underlying pathogenesis, and that, on the basis of shared molecular features, most high grade tumors, including some previously classified as endometrioid, are now considered to be high-grade serous. At the same time, several studies have reported that high-grade serous EOC, which is the most common histotype, is itself made up of reproducible subtypes discernable by gene expression patterns.
Summary
These major advances in understanding set the stage for a new era of research on EOC risk and clinical outcomes with the potential to reduce morbidity and mortality. We highlight the need for multidisciplinary studies with pathology review using the current guidelines, further molecular characterization of the histotypes and subtypes, inclusion of women of diverse racial/ethnic and socioeconomic backgrounds, and updated epidemiologic and clinical data relevant to current generations of women at risk of EOC.
Keywords: Epithelial ovarian cancer, histotype, gene expression subtype, survival, pathology
Introduction
Ovarian cancer is the most deadly gynecologic cancer, with only 46% of women surviving five years after diagnosis [1]. Over the past decade it has become clear that invasive epithelial ovarian cancer (EOC) represents a group of tumor types that, despite arising in a similar anatomical location, have different cells of origin, morphology, molecular features, epidemiologic factors, clinical characteristics and survival [2–5]. Furthermore, reproducible gene expression-based molecular subtypes of the most common histotype of EOC, high grade serous cancers (HGSC), have been observed in multiple studies [6–8]. Understanding similarities and differences in epidemiologic, pathological, molecular, and clinical features by biologically relevant subtypes is essential in order to reduce morbidity and mortality from EOC, as has been achieved with breast cancer [9,10]. For breast cancer, subtype-specific risk factors have been described [11–14], targeted treatments are effective in improving clinical outcomes [15,16], and differences in the incidence and mortality of subtypes [17,18] has set the stage to identify causes of racial, ethnic and socioeconomic disparities [19]. Similar research in EOC is in its infancy partially because it is a rare cancer (incidence 11.9 per 100,000) [20] but also because the recent major paradigm shifts in the understanding of EOC point to the need to approach EOC research in new ways.
Major Paradigm Shifts in EOC Histotype Classification
Invasive EOC has traditionally been separated according to histologic appearance, including serous, endometrioid (EC), clear cell (CCC), and mucinous (MC) histotypes. Immunohistochemical (IHC) markers have recently enabled the refinement of traditional histologic categorization schemes into more homogeneous “molecular” subgroups. As well, IHC markers have served to highlight differences between low- and high-grade tumors within each of the serous and EC groups [21,22] such that 1) HGSC and low grade serous (LGSC) are understood to develop along different pathways and are not part of a continuum of disease as was previously believed, and 2) many high-grade EC tumors have close similarities to HGSC. These distinctions have been included in the new 2014 World Health Organization (WHO) diagnostic classification guidelines [23]. Also, converging lines of research support that most HGSC arise from the fallopian tubal fimbriae [24,25], while EC and CCC likely arise from endometriotic lesions [26–29], and a high proportion of advanced stage MC are now considered to be metastases from other primary tumor sites [5,22]. The new diagnostic guidelines are considerably more reproducible across pathologists and may more accurately reflect biological differences since there are clearer survival differences between histotypes. This is well-illustrated in the study by Kommoss et al. [30] where an expert pathologist used the 2014 WHO guidelines to re-review diagnostic slides from a 2002 clinical trial for which he had originally classified the tumors, with only 54% concordance between his two reviews. Another pathologist independently reviewed the same slides using the 2014 WHO guidelines, and concordance between the two pathologists’ reviews was 98%. While there was originally no survival difference between histotypes, the MC and CCC histotypes classified in the later review had markedly worse survival [30]. Conceivably, epidemiologic research that defined histotypes using prior guidelines may have failed to identify histotype-specific etiologic differences that may yet exist when more accurate histotypes are defined by the 2014 WHO guidelines. Since evidence was accumulating over time to support the new classification for many years before it was published (e.g., reviewed in [5]), and as early as 2010 was demonstrated to be highly reproducible across trained pathologists [31], the extent of misclassification in existing EOC studies likely depends on the years of diagnoses, and whether cases were reviewed by expert gynecologic pathologists.
Characteristics of EOC Histotypes
Molecular Features
Nearly all HGSC have TP53 mutations [32] and frequent homologous recombination deficiency, largely explained by somatic/germline BRCA1/2 alterations (33% in The Cancer Genome Atlas (TCGA)[7]). Thus, HGSC is genetically highly unstable as reflected by widespread copy number alterations (CNA) [7] with considerable tumor heterogeneity [33,34]. In contrast LGSC has intact TP53 function and very few CNA; LGSC also has much higher frequencies of KRAS and BRAF mutations than HGSC [35], but mutation rates may differ by stage [36]. The other histotypes rarely harbor TP53 mutations, and instead have mutations in KRAS (MC), CTNNB1 (EC), PTEN (EC) and PIK3CA (CCC) [5,22,37].
Pathology review of cell type is now highly reproducible and is considered the “gold standard” for histotype classification. In some cases, IHC markers can help with classification. Extending this work, an algorithm using eight IHC markers (WT1, TP53 (p53), CDKN2A (p16), HNF1B, PGR (PR), TFF3, ARID1A, and VIM (Vimentin)) assessed using tumor microarrays (TMA) has been developed to predict the five major histotypes [21,38]. The authors report that it correctly classified tumors 93% of the time based on expert pathology review. The algorithm designated most high-grade carcinomas (including many high-grade EC) as HGSC, reflecting the tumor’s immunophenotype as well as underlying molecular abnormalities. As well, in that study population reduced marker sets, which may be easier to implement clinically, were reported to have reasonable prediction accuracy; a four marker panel including WT1, TP53, NAPSA, and PGR had 87% accuracy, and a six marker panel additionally including CDKN2A and TFF3 had 91% accuracy [21].
While there is no question that there are well-defined molecular differences between EOC histotypes, it is also becoming clear that there are similarities across cancers arising in different organ sites. Molecular similarities between serous fallopian tube and ovarian cancers, and to a lesser extent serous primary peritoneal carcinoma have been noted [39] (While serous fallopian tube and ovarian cancer have comparable epidemiologic factor profiles, primary peritoneal cases tend to be older, more obese, and have higher parity [39–41]). Also, HGSC is similar to basal-like breast cancer and a serous-like subtype of endometrial cancer in terms of somatic CNA, TP53 mutations, BRCA mutations and epigenetic silencing, and CCNE1 and MYC amplification [42]. Understanding the molecular similarities between these cancer types may provide additional opportunities to decipher causal associations for modifiable factors and possible treatment options.
Epidemiologic Factors
Misclassification of histotypes and differences between studies in their categorization may interfere with the ability of studies to identify true differences in risk factors across histotypes; still, histotype-specific risk factor associations have been uncovered even using prior classification guidelines in large pooled studies. Table 1 provides a summary of results from the Ovarian Cancer Association Consortium (OCAC) [43–46], the Ovarian Cancer Cohort Consortium (OC3) [47], the Collaborative Group on Epidemiological Studies of Ovarian Cancer [48–52], and the Million Women’s Study [53]. Because these are generally based on earlier histotype definitions, many of the studies were not able to present results separately by LGSC and HGSC. For some factors, there is an association with EOC overall which is stronger among specific histotypes (e.g., for parity and tubal ligation, inverse associations are more pronounced for EC and CCC). For other factors, associations are only present for specific histotypes (e.g., cigarette smoking and increased risk of MC, and suggestive decreased risk of CCC and EC; and estrogen-only hormone therapy and increased risk of serous and EC). Differences in the incidence of histotypes have been observed by race, with a higher incidence of CCC in Asian women [54]. Genetic risk varies by histotype as well, though genome wide association studies of the rarer histotypes are generally underpowered [55]. The HNF1B locus represents an intriguing example of genetic heterogeneity; different SNPs in this gene are associated with serous versus CCC tumors [56].
Table 1.
Selected epidemiologic factors and EOC risk by histotype in three ovarian cancer consortia: Ovarian Cancer Cohort Consortium (OC3), Ovarian Cancer Association Consortium (OCAC) and the Collaborative Group on Epidemiological Studies of Ovarian Cancer
| Risk Factors | Serous | Endometrioid | Clear Cell | Mucinous | ||||
|---|---|---|---|---|---|---|---|---|
| Direction of Associatio n |
RR or OR (95% CI)a |
Direction of Associatio n |
RR or OR (95% CI)a |
Direction of Associatio n |
RR or OR (95% CI)a |
Direction of Associatio n |
RR or OR (95% CI)a |
|
| Cigarette smoking (Current vs. never smokers) [44,47,48] | Null | 0.99 (0.94–1.05) | Modest Inverse | 0.93 (0.79–1.09) | Modest Inverse | 0.95 (0.74–1.21)b | Positive | 1.27 (1.01–1.59) |
| 0.89 (0.76–1.04) | 0.84 (0.69–1.02) | 0.74 (0.56–0.98) | 1.31 (1.03–1.65) | |||||
| 0.99 (0.91–1.08) | 0.81 (0.70–0.94) | 0.80 (0.63–1.01) | 1.79 (1.47–2.17) | |||||
| Tubal ligation [43,47,53] | Weak Inverse | 0.91 (0.79–1.06) | Strong Inverse | 0.60 (0.41–0.88) | Strong Inverse | 0.35 (0.18–0.69) | Inconsistent | 1.01 (0.60–1.71) |
| 0.81 (0.74–0.89) | 0.48 (0.40–0.59) | 0.52 (0.40–0.67) | 0.68 (0.52–0.89) | |||||
| 0.84 (0.77–0.92) | 0.54 (0.43–0.69) | 0.55 (0.39–0.77) | 0.99 (0.84–1.18) | |||||
| BMI (Per 5 kg/m2 increase) [45,47,50] | Null | 0.97 (0.93–1.01) | Positive | 1.07 (0.99–1.16) | Null | 1.04 (0.92–1.17) | Positive | 1.08 (0.96–1.20) |
| 0.98 (0.94–1.02) | 1.17 (1.11–1.23) | 1.06 (0.96–1.17) | 1.19 (1.06–1.32) | |||||
| 1.00 (0.96–1.04) | 1.07 (1.01–1.13) | 1.05 (0.95–1.15) | 1.15 (1.07–1.23) | |||||
| Endometriosis [26,47] | Null (High- grade)c | 1.11 (0.70–1.74) | Positive | 2.32 (1.36–3.95) | Positive | 2.87 (1.53–5.39) | Null | 1.62 (0.58–4.51) |
| 1.13 (0.97–1.32) | 2.04 (1.67–2.48) | 3.05 (2.43–3.84) | 1.02 (0.69–1.50) | |||||
| N/A | N/A | N/A | N/A | |||||
| Estrogen-only hormone therapy (Ever vs. never use) [46,49] | Positive | N/Ad | Positive | N/Ad | Null | N/Ad | Null | N/Ad |
| 1.57 (1.23–2.00) | 1.82 (1.10–3.03) | 0.80 (0.38–1.68) | 0.80 (0.38–1.69) | |||||
| 1.59 (1.45–1.75) | 1.42 (1.19–1.69) | 0.88 (0.71–1.10) | 0.91 (0.66–1.24) | |||||
| Oral contraceptive use (Duration per 5-year increase of use) [47,51] | Inverse | 0.85 (0.81–0.89) | Inverse | 0.86 (0.77–0.95) | Inverse | 0.86 (0.74–1.00) | Null | 1.02 (0.80–1.31) |
| N/A | N/A | N/A | N/A | |||||
| 22.1% (SE 2.9) reduction in risk | 27.1% (SE 4.8) reduction in risk | 21.3% (SE 7.3) reduction in risk | 6.7% (SE 5.8) reduction in risk | |||||
| Parity (Per term pregnancy) [47,52] | Weak Inverse | 0.93 (0.92–0.95) | Inverse | 0.78 (0.74–0.83) | Strong Inverse | 0.68 (0.61–0.76) | Weak Inverse | 0.91 (0.84–0.99) |
| N/A | N/A | N/A | N/A | |||||
| 0.87 (0.83–0.91) | 0.72 (0.66–0.77) | 0.56 (0.49–0.65) | 0.85 (0.77–0.93) | |||||
| Breastfeeding Duration (Per year for OC3 and per month for the Collaborative Group) [47,52] | Inverse | 0.94 (0.86–1.03) | Inverse | 0.85 (0.69–1.05) | Null | 1.03 (0.81–1.33) | Inverse | 0.88 (0.63–1.23) |
| N/A | N/A | N/A | N/A | |||||
| 0.98 (0.97–0.99) | 0.98 (0.98–1.00) | 1.00 (0.97–1.03) | 0.97 (0.95–1.00) | |||||
RR: relative risk, OR: odds ratio, CI: confidence interval, SE: standard error, N/A: not applicable
The order of the RRs from top to bottom is OC3, OCAC, and the Collaborative Group on Epidemiological Studies of Ovarian Cancer. For tubal ligation, the Million Women Study was included instead of the Collaborative Group. For parity and breastfeeding, a pooled analysis of nine studies in the Collaborative Group was used instead of the full set of studies in the Collaborative Group on Epidemiological Studies of Ovarian Cancer.
Although cigarette smoking overall was not associated with risk of clear cell ovarian cancer in OC3, an inverse association was observed for pack-years (Per 20 pack-years: RR=0.68, 95% CI=0.53–0.89).
Among LGSC, a strong positive association was observed for women with a history of endometriosis (OC3: RR=3.77, 95% CI=1.24–11.48; OCAC: OR=2.11, 95% CI=1.39–3.20).
OC3 evaluated hormone therapy use overall but not by type of therapy.
Clinical Potential
Patterns of histotype-specific incidence and survival differ considerably by stage. The majority of HGSC and LGSC are diagnosed at an advanced stage, whereas the majority of EC, CCC and MC are diagnosed at an early stage [5]. Sixty percent of women with EOC present with distant disease, with a median five year survival of only 29% [57]. For distant stage, survival for MC and CCC is dramatically low (particularly in the years directly after diagnosis), with better survival for EC and LGSC, than HGSC [58–60]. Five-year survival for localized/regional disease is considerably better at 82% [57], with the worst survival for HGSC followed by CCC and MC, and the best survival for EC and LGSC [5]. African American women experience worse survival than women of European ancestry, while women of Asian ancestry tend to have better survival [61]. This could in part be due to differences in distributions of histotypes across racial/ethnic groups but is also likely impacted by disparities in access to care and treatment [62].
At present, the standard of care for EOC is surgical debulking, or removal of the tumor burden from throughout the peritoneal cavity, and combination platinum/taxane-based chemotherapy [63]. MC and CCC appear to be less responsive to these regimens and CCC are also more likely to recur than the other histotypes given the same treatment [64–68]. Nevertheless, the clinical care model is still a “one size fits all” approach. A gene expression analysis across LGSC, EC, MC and CCC has also revealed two classes of tumors significantly correlated with progression free survival; the better outcome group had a higher proportion of low-grade, early-stage disease and of MC and a lower proportion of CCC [69]. It is imperative that histotype is taken into account in EOC clinical trials so that histotype-specific clinical care guidelines can be developed to improve treatment effectiveness. In breast cancer, the major molecular subtypes (luminal A, luminal B, triple negative/basal-like, normal-like and HER2 type) have different response patterns to available therapies [18,70,71], and targeted treatments have been developed (e.g., the anti-HER2 monoclonal antibody trastuzumab for the HER2 subtype and poly adenosine diphosphate-ribose polymerases (PARP) inhibitors for the basal-like subtype) [15,16]. While these treatments have garnered some success, their effectiveness is not widespread [16,72], indicating the need for additional research to identify novel subtype-specific molecular targets that can be used alone or in combination with current therapies to improve survival. The question of whether the molecular features of histotypes remain constant through treatment is important; remarkably, it is possible for lung adenocarcinomas to transform to small-cell lung cancers after developing resistance to EGFR inhibitors [73]. It is unknown whether such transformations occur in EOC histotypes.
Characteristics of HGSC Molecular Subtypes
Gene Expression Subtypes
Several studies have reported that HGSC tumors separate into four distinct molecular subtypes based on mRNA expression patterns [6–8]. In the only study to date that examined epidemiologic factors and risk of these subtypes, differences in associations were observed for age at diagnosis, race, breast-feeding, and first-degree family history of breast or ovarian cancers [74]. To cleanly separate tumors into subtypes, some researchers have removed samples that were difficult to cluster [6,7], so the reported subtypes may not capture the full complexity of the disease. The subtypes are commonly referred to as mesenchymal, immunoreactive, proliferative and differentiated [7]. Generally, the mesenchymal subtype has the worst survival, and the immunoreactive subtype has the most favorable survival. Hofree et al. defined genetic subtypes of HGSC by performing network-based clustering on germ-line and somatic variant data from TCGA, but these subtypes are not concordant with the gene expression subtypes [75]. A recent analysis of genotyping accuracy has raised questions about the quality of sequencing-based variant calls in TCGA’s HGSC samples [76] which may affect the findings in Hofree et al. [75]. Given the exploratory nature of molecular clustering and limitations of the approaches used, more research is needed about how many underlying molecular subtypes exist [77–79] and if they are consistent across populations [80].
Tumor Micro-environment
Patterns of gene expression are thought to arise because of transcriptional programs active in cancer cells as a consequence of the amplification of a gene or genes in a pathway. However, alternative explanations also exist. It is possible that subtypes are merely approximations of tumor subgroups sharing similar molecular mechanisms [81]. It is also possible that some tumors efficiently recruit tumor microenvironment substrate while others do not. In many cancer types, RNA expression profiling of tumor tissue is influenced by different stromal cell-types in the tumor micro-environment [82–84]. Still another possibility has little to do with the cancer itself: maybe cancer cells express the same markers but certain individuals have a variable response based on constitutive factors. This may have important implications for treatment. For example, a tumor of the mesenchymal subtype arising through epithelial-to-mesenchymal transition (EMT) might be treated by EMT inhibitors, while a tumor arising from an increased recruitment of stromal support cells could focus on disabling this recruitment.
In HGSC, mesenchymal and immunoreactive tumors have significantly lower tumor content [85]. This is consistent with a model where these signatures arise at least in part from stromal gene expression. Mesenchymal tumors are associated with a strong stromal reaction and increased desmoplasia while immunoreactive tumors have high levels of infiltrating T-cells [6]. Recently, a pathology-based scoring system recapitulated the TCGA-defined subtypes [86]. The authors propose a scoring scheme for proliferative as tumors with “proliferative and solid growth architecture,” and differentiated as tumors with “papillary growth and glandular architecture.” This classification scheme was highly reproducible, with a reported overall average consistency of 74% across scores from six pathologists.
HGSC subtypes should be considered at both the tissue and cellular level. It may be possible that a single tumor expresses the signatures of multiple subtypes across different cells, which is masked when observed in bulk. New experimental techniques provide the opportunity to interrogate such hypotheses directly. This phenomenon has been observed in 66 single cells in a HGSC tumor [87] and a subset of glioblastoma tumors by single cell profiling [88]. However, it has been recently shown that glioblastoma tumors are inherently heterogeneous by chance [89], and more single cell data is required to fully explore this phenomenon. Single cell RNAseq can reveal the types of cells in a complex mixture [90]. Measuring single cells sidesteps the need for deconvolution methods and overcomes many difficulties inherent to estimating differential cell type proportions in bulk tumor mixtures. Droplet-based methods can profile tens of thousands of single cells from a sample [91], but require fresh tissue and are costly [92]. For deep characterization of many samples, reductions in sequencing costs and analytical approaches that use single cell sequencing to inform deconvolution of bulk tumors will continue to improve the feasibility of these methods for large studies.
Clinical Potential
There is some evidence that response to treatment varies by HGSC gene expression signatures. A recent study reported that women with the proliferative and mesenchymal subtypes benefit from experimental treatment (Bevacizumab) over standard chemotherapy [93], which raises the possibility that similar subtype-specific variations in response to other actively developing therapeutics for EOC, such as PARP inhibition and immune therapies [94,95] might exist. For example, immunoreactive tumors may be good candidates for immunotherapy [79]; an open-label single-arm Phase II trial has been initiated to evaluate efficacy and safety of pembrolizumab anti-PD-1 monotherapy in these cases [96]. For mesenchymal tumors, TGF-beta inhibition may be considered since the TGF-beta pathway is significantly up-regulated compared to other subtypes, which aligns well with other supervised expression studies reporting worse clinical outcomes associated with this pathway [97–100].
Challenges and Limitations in Studying Epidemiology of Cancer Subtypes
Potential Biases in Tissue-based Studies
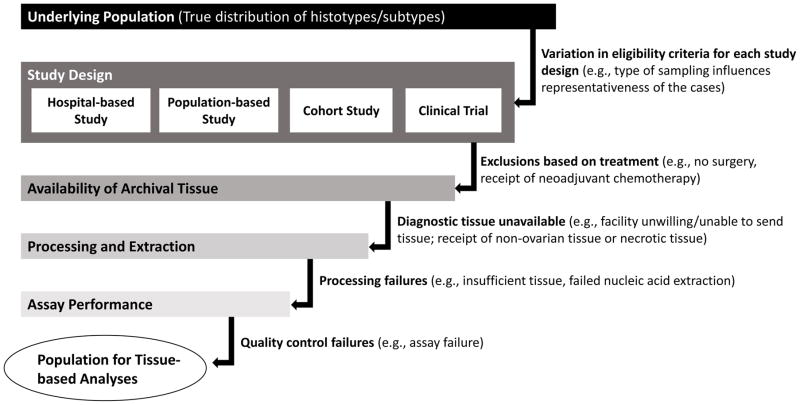
Given the demonstrated heterogeneity of EOC, it is critical to perform studies that have accurate histotyping and to acquire tissue for HGSC subtyping and other molecular studies. It is logistically difficult to obtain fresh frozen tissue for large studies, but advances in molecular assays that can be performed using archival formalin-fixed paraffin-embedded tumors provide the opportunity to obtain a more representative case group. Still, there are several potential sources of bias that are important to recognize, and if possible, quantify (Figure 1). As many as 18% of women with EOC do not receive surgery [101] (although biopsy tissue or ascitic fluid are sometimes available). Use of neoadjuvant therapy prior to surgery, which is likely to influence the populations of tumor cells that remain for surgical removal, has been increasing from approximately 9% in 2004 to 23% in 2013 [102]. Archival specimens typically must be requested from a large number of hospitals, and some proportion of the specimens will not be available for various reasons (e.g., the hospital does not retain the blocks, or does not have the staffing necessary to identify, pull and send the blocks to researchers). Even if blocks are available, the slide/block that was used for the original diagnosis may not be available or the hospital may not be willing to release that block for research purposes. In fact, sometimes only blocks from other sites, most commonly the omentum, are available, and the degree to which the tumor is similar when it is collected from metastatic sites is not well-characterized. As with most cancer types, tumor heterogeneity comes into play but since EOC tumors are typically very large, appropriate sampling of the tumor is an important consideration, particularly with respect to necrosis and cellularity of the sample. For TMAs, it is important to take cores from different parts of the tumor and have multiple replicates to try to assess heterogeneity. Additional attrition occurs through the processing and extraction processes, and failure of quality control measures specific to the assay of interest. The extent to which the cases lost at each of these steps differ from the cases that are included introduces bias.
Figure 1.
Sources of bias in tissue-based studies of ovarian cancer
Study Design Considerations
Beyond collection of the tumor samples themselves, demographics, lifestyle behaviors, and clinical factors are important for multivariate analyses of risk, progression and survival. Population-based case-control studies are essential for studying EOC risk because it is a rare disease. Since they rely on population–based cancer registries, which identify essentially all cases in a well-defined geographic region, they have the potential to produce a representative distribution of histotypes in the population of interest. Also, accurate response proportions for EOC cases can be calculated and differences between responders and non-responders can be evaluated using data from cancer registries (e.g., age, histotype, geographic region, receipt of surgery and chemotherapy, residual disease, time between diagnosis and death). Older and more advanced EOC patients are most difficult to enroll and tend to be underrepresented in case-control studies. Rapid case ascertainment allows for the identification of eligible cases as early as one to two months after diagnosis, though representation of rapidly fatal cancers is still problematic. In the North Carolina Ovarian Cancer Study [103], even with rapid case ascertainment, approximately 4% of eligible cases were deceased at ascertainment suggesting that the most aggressive cases were not enrolled. Further examination showed that ~15% of African American cases were deceased at ascertainment, highlighting the need to understand the underlying basis of this disparity [104]. While selection bias is a concern for case-control studies, for tissue-based studies, to the extent that it is approved by Institutional Review Boards, tissue can be requested even for non-responders.
Most existing EOC population-based case-control studies in the U.S. have focused on disease risk and many do not have clinical and outcome data. Also, nearly all were completed before 2010 [105], with epidemiologic data less representative of current exposures that may affect risk and survival (e.g., differences in oral contraceptive formulations, changes in the prevalence of obesity). Because these studies are often conducted in a large geographic region with many hospitals where diagnoses have occurred, requesting medical records and abstracting them for clinical and prognostic variables is time-consuming and costly, and often there is an unavoidably large proportion of missing data.
Hospital-based studies have the potential to increase inclusion of aggressive EOC cases and provide improved access to medical records and clinical data, and collection of serial samples pre- and post-treatment. Women can be approached for enrollment when they are being evaluated for suspected EOC. However, it is difficult to accrue large numbers of cases from a single institution so a multi-institutional effort with related complexities is required. Prospective cohort studies, to the extent that they have complete follow-up and identification of EOC cases, are able to collect data on the most aggressive cases prior to diagnosis. However, it is very difficult to obtain a sizable number of cases of EOC. Since cohort studies are not typically disease-focused, important risk factor data are often incomplete, and critical pathologic data, clinical and prognostic factors, as well as tumor tissue are often not available. Although clinical trials have detailed pathologic, prognostic and outcome data, their eligibility criteria typically result in a highly selected patient population which is unlikely to be representative of the spectrum of histotypes. Racial/ethnic minorities also tend to be underrepresented in clinical trials. For any of these study designs, careful consideration about the degree of accuracy of the existing histotypes is important to determine whether re-classification or re-review is needed. Adding tumor marker assays could help with updating tumor classification but likely results in a loss of sample size as described above.
Importance of Consortia and Interdisciplinary Approaches
Because of the low incidence of EOC and the need to study risk and survival separately by histotype and molecular subtype, efforts to pool existing data from many studies are essential. In addition to the previously noted consortia, OCAC and OC3, the newly formed Ovarian Cancer in Women of African Ancestry (OCWAA) consortium addresses disparities in risk and survival in African American women by pooling data from existing case-control and cohort studies. Work emerging from this group is likely to provide new insights into risk, prognosis and treatment of EOC because of differences in genetic background and exposures to epidemiologic factors. Formation of these consortia do not remedy possible misclassification from earlier histotype assignments and the lack of available clinical and prognostic variables as well as tumor tissue. The Ovarian Tumor Tissue Consortium (OTTA) includes tissue-based studies and focuses on tumor biomarkers and survival [106,107], with several large efforts underway. Compilations of systematically harmonized public gene expression datasets such as curatedOvarianData [108] are complementary resources. Although these consortia and compendia represent large numbers of studies, analyses of the rarer histotypes, MC, LGSC, and CCC, remain underpowered. Bringing together multidisciplinary teams including pathologists, epidemiologists, bioinformaticians, biostatisticians, genome biologists, and clinicians to leverage existing data and conceptualize novel and more powerful approaches will accelerate advances.
Conclusions and Future Directions
Despite considerable effort, progress in identifying modifiable factors to prevent EOC and reduce mortality from this disease has been elusive. Approaching research with the understanding that EOC comprises biologically-relevant histotypes with distinct cells of origin provides an opportunity to more accurately identify etiologic risk factors, prognostic relationships and appropriate treatment strategies. The rarity of the disease and the degree of heterogeneity requires large sample sizes and multidisciplinary initiatives. While the molecular features of HGSC have been studied more than those of the other histotypes, in-depth characterization of all of the histotypes, and more precise characterization of HGSC subtypes, is needed to further reduce misclassification and increase power to detect subtype-specific associations.
Acknowledgments
We thank Dr. Mary Anne Rossing for her thoughtful insights on this manuscript.
Funding: The research reported in this publication was supported by Huntsman Cancer Foundation and the National Cancer Institute of the National Institutes of Health under Award Number P30 CA042014, as well as R01 CA168758 (to JAD and MAR) and R01 CA142081 (to JMS). It was also supported in part by a career enhancement award from the Mayo Clinic Ovarian Cancer SPORE to CW (P50 CA136393), and by a grant from the Gordon and Betty Moore Foundation (GBMF4552) to CSG. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Conflict of Interest
Jennifer Anne Doherty, Lauren Cole Peres, Chen Wang, and Gregory P. Way each declare no potential conflicts of interest.
Casey S. Greene reports grants from Gordon and Betty Moore Foundation during the conduct of the study and personal fees from SomaLogic outside the submitted work.
Joellen M. Schildkraut reports grants from National Cancer Institute during the conduct of the study.
Human and Animal Rights and Informed Consent
This article contains no studies with human or animal subjects performed by any of the authors.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Kobel M, Kalloger SE, Boyd N, et al. Ovarian carcinoma subtypes are different diseases: Implications for biomarker studies. PLoS Med. 2008;5:1749–60. doi: 10.1371/journal.pmed.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilks CB, Ionescu DN, Kalloger SE, et al. Tumor cell type can be reproducibly diagnosed and is of independent prognostic significance in patients with maximally debulked ovarian carcinoma. Hum Pathol. 2008;39:1239–51. doi: 10.1016/j.humpath.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Vaughan S, Coward JI, Bast RC, Jr, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer Nature Publishing Group. 2011;11:719–25. doi: 10.1038/nrc3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCluggage WG. Morphological subtypes of ovarian carcinoma: a review with emphasis on new developments and pathogenesis. Pathology. 2011;43:420–32. doi: 10.1097/PAT.0b013e328348a6e7. [DOI] [PubMed] [Google Scholar]
- 6.Tothill RW, Tinker AV, George J, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 7.The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konecny GE, Wang C, Hamidi H, et al. Prognostic and therapeutic relevance of molecular subtypes in high-grade serous ovarian cancer. J Natl Cancer Inst. 2014;106:dju249. doi: 10.1093/jnci/dju249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reis-Filho JS, Pusztai L. Gene expression profiling in breast cancer: Classification, prognostication, and prediction. Lancet. 2011;378:1812–23. doi: 10.1016/S0140-6736(11)61539-0. [DOI] [PubMed] [Google Scholar]
- 10.Prat A, Pineda E, Adamo B, et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast. 2015;24:S26–35. doi: 10.1016/j.breast.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 11.Sisti JS, Collins LC, Beck AH, Tamimi RM, Rosner BA, Eliassen AH. Reproductive risk factors in relation to molecular subtypes of breast cancer: Results from the nurses’ health studies. Int J Cancer. 2016;138:2346–56. doi: 10.1002/ijc.29968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamimi RM, Colditz GA, Hazra A, et al. Traditional breast cancer risk factors in relation to molecular subtypes of breast cancer. Breast Cancer Res Treat. 2012;131:159–67. doi: 10.1007/s10549-011-1702-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang XR, Sherman ME, Rimm DL, et al. Differences in risk factors for breast cancer molecular subtypes in a population-based study. Cancer Epidemiol Biomarkers Prev. 2007;16:439–43. doi: 10.1158/1055-9965.EPI-06-0806. [DOI] [PubMed] [Google Scholar]
- 14.Yang XR, Chang-Claude J, Goode EL, et al. Associations of breast cancer risk factors with tumor subtypes: A pooled analysis from the breast cancer association consortium studies. J Natl Cancer Inst. 2011;103:250–63. doi: 10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rios J, Puhalla S. PARP inhibitors in breast cancer: BRCA and beyond. Oncology. 2011;25:1014–25. [PubMed] [Google Scholar]
- 16.Brenton JD, Carey LA, Ahmed A, Caldas C. Molecular classification and molecular forecasting of breast cancer: Ready for clinical application? J Clin Oncol. 2005;23:7350–60. doi: 10.1200/JCO.2005.03.3845. [DOI] [PubMed] [Google Scholar]
- 17.Kohler BA, Sherman RL, Howlader N, et al. Annual Report to the Nation on the Status of Cancer, 1975–2011, Featuring Incidence of Breast Cancer Subtypes by Race/Ethnicity, Poverty, and State. J Natl Cancer Inst. 2015;107:djv048. doi: 10.1093/jnci/djv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desmedt C, Haibe-Kains B, Wirapati P, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14:5158–65. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- 19.Palmer JR, Ambrosone CB, Olshan AF. A collaborative study of the etiology of breast cancer subtypes in African American women: The AMBER consortium. Cancer Causes Control. 2014;25:309–19. doi: 10.1007/s10552-013-0332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howlader N, Noone A, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2013. National Cancer Institute; Bethesda, MD: based on November 2015 SEER data submission, posted to the SEER web site, April 2016. [Internet]. [cited 2016 Nov 2]. Available from: http://seer.cancer.gov/csr/1975_2013/ [Google Scholar]
- 21.Köbel M, Rahimi K, Rambau PF, et al. An Immunohistochemical Algorithm for Ovarian Carcinoma Typing. Int J Gynecol Pathol. 2016;35:430–41. doi: 10.1097/PGP.0000000000000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soslow RA. Histologic Subtypes of Ovarian Carcinoma. Int J Gynecol Pathol. 2008;27:161–74. doi: 10.1097/PGP.0b013e31815ea812. [DOI] [PubMed] [Google Scholar]
- 23.Kurman RJ, Carcangiu ML, Herrington CS, Young RH. WHO Classification of Tumours of Female Reproductive Organs. 4. Lyon: IARC; 2014. The new diagnostic guidelines for epithelial ovarian cancer encompass a large body of evidence that results in more reproducible diagnosis across pathologists, and potentially more biologically-relevant histotype classifications. [Google Scholar]
- 24.Crum CP, Drapkin R, Miron A, et al. The distal fallopian tube: a new model for pelvic serous carcinogenesis. Curr Opin Obstet Gynecol. 2007;19:3–9. doi: 10.1097/GCO.0b013e328011a21f. [DOI] [PubMed] [Google Scholar]
- 25.Piek JMJ, Van Diest PJ, Zweemer RP, et al. Dysplastic changes in prophylactically removed Fallopian tubes of women predisposed to developing ovarian cancer. J Pathol. 2001;195:451–6. doi: 10.1002/path.1000. [DOI] [PubMed] [Google Scholar]
- 26.Pearce CL, Templeman C, Rossing MA, et al. Association between endometriosis and risk of histological subtypes of ovarian cancer: A pooled analysis of case-control studies. Lancet Oncol Elsevier Ltd. 2012;13:385–94. doi: 10.1016/S1470-2045(11)70404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A Mutations in Endometriosis- Associated Ovarian Carcinomas. N Engl J Med. 2010;363:1532–43. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones S, Wang T-L, Shih I-M, et al. Frequent Mutations of Chromatin Remodeling Gene ARID1A in Ovarian Clear Cell Carcinoma. Science (80-) 2010;330:228–31. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurman RJ, McConnell TG. Characterization and comparison of precursors of ovarian and endometrial carcinoma: parts I and II. Int J Surg Pathol. 2010;18:181S–189S. doi: 10.1177/1066896910370881. [DOI] [PubMed] [Google Scholar]
- 30.Kommoss S, Gilks CB, du Bois A, Kommoss F. Ovarian carcinoma diagnosis: the clinical impact of 15 years of change. Br J Cancer Nature Publishing Group. 2016;115:1–7. doi: 10.1038/bjc.2016.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobel M, Kalloger S, Baker P, et al. Diagnosis of ovarian carcinoma cell type is highly reproducible: a transcanadian study. Am J Surg Pathol. 2010;34:984–93. doi: 10.1097/PAS.0b013e3181e1a3bb. [DOI] [PubMed] [Google Scholar]
- 32.Ahmed AA, Etemadmoghadam D, Temple J, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 2010;221:49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bashashati A, Ha G, Tone A, et al. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J Pathol. 2013;231:21–34. doi: 10.1002/path.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paracchini L, Mannarino L, Craparotta I, et al. Regional and temporal heterogeneity of epithelial ovarian cancer tumor biopsies: implications for therapeutic strategies. Oncotarget. 2016:5. doi: 10.18632/oncotarget.10505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vang R, Shih I-M, Kurman RJ. Ovarian Low-grade and High-grade Serous Carcinoma: Pathogenesis, Clinicopathologic and Molecular Biologic Features, and Diagnostic Problems. Adv Anat Pathol. 2009;16:267–82. doi: 10.1097/PAP.0b013e3181b4fffa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong K-K, Tsang YTM, Deavers MT, et al. BRAF Mutation Is Rare in Advanced-Stage Low-Grade Ovarian Serous Carcinomas. Am J Pathol American Society for Investigative Pathology. 2010;177:1611–7. doi: 10.2353/ajpath.2010.100212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuo K-T, Mao T-L, Jones S, et al. Frequent Activating Mutations of PIK3CA in Ovarian Clear Cell Carcinoma. Am J Pathol. 2009;174:1597–601. doi: 10.2353/ajpath.2009.081000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalloger SE, Köbel M, Leung S, et al. Calculator for ovarian carcinoma subtype prediction. Mod Pathol. 2011;24:512–21. doi: 10.1038/modpathol.2010.215. [DOI] [PubMed] [Google Scholar]
- 39.Sørensen RD, Schnack TH, Karlsen MA, Høgdall CK. Serous ovarian, fallopian tube and primary peritoneal cancers: A common disease or separate entities - A systematic review. Gynecol Oncol Elsevier Inc. 2015;136:571–81. doi: 10.1016/j.ygyno.2015.01.534. [DOI] [PubMed] [Google Scholar]
- 40.Jordan SJ, Green AC, Whiteman DC, et al. Serous ovarian, fallopian tube and primary peritoneal cancers: a comparative epidemiological analysis. Int J cancer. 2008;122:1598–603. doi: 10.1002/ijc.23287. [DOI] [PubMed] [Google Scholar]
- 41.Grant DJ, Moorman PG, Akushevich L, Palmieri RT, Bentley RC, Schildkraut JM. Primary peritoneal and ovarian cancers: An epidemiological comparative analysis. Cancer Causes Control. 2010;21:991–8. doi: 10.1007/s10552-010-9525-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Getz G, Gabriel SB, Cibulskis K, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sieh W, Salvador S, McGuire V, et al. Tubal ligation and risk of ovarian cancer subtypes: A pooled analysis of case-control studies. Int J Epidemiol. 2013;42:579–89. doi: 10.1093/ije/dyt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faber MT, Kjær SK, Dehlendorff C, et al. Cigarette smoking and risk of ovarian cancer: a pooled analysis of 21 case-control studies. Cancer Causes Control. 2013;24:989–1004. doi: 10.1007/s10552-013-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olsen CM, Nagle CM, Whiteman DC, et al. Obesity and risk of ovarian cancer subtypes: Evidence from the Ovarian Cancer Association Consortium. Endocr Relat Cancer. 2013;20:251–62. doi: 10.1530/ERC-12-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee AW, Ness RB, Roman LD, et al. Association Between Menopausal Estrogen-Only Therapy and Ovarian Carcinoma Risk. Obstet Gynecol. 2016;127:828–36. doi: 10.1097/AOG.0000000000001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wentzensen N, Poole EM, Trabert B, et al. Ovarian cancer risk factors by histologic subtype: An analysis from the Ovarian Cancer Cohort Consortium. J Clin Oncol. 2016;34:2888–98. doi: 10.1200/JCO.2016.66.8178. This large pooled analysis of cohort studies of invasive epithelial ovarian cancer reports differences in associations with several well-characterized epidemiologic factors by histotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calle EE, Gapstur SM, Patel AV, et al. Ovarian cancer and smoking: Individual participant meta-analysis including 28–114 women with ovarian cancer from 51 epidemiological studies. Lancet Oncol Elsevier Ltd. 2012;13:946–56. doi: 10.1016/S1470-2045(12)70322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collaborative Group on Epidemiological Studies of Ovarian Cancer. Menopausal hormone use and ovarian cancer risk: Individual participant meta-analysis of 52 epidemiological studies. Lancet. 2015;385:1835–42. doi: 10.1016/S0140-6736(14)61687-1. Collaborative Group on Epidemiological Studies of Ovarian Cancer Open Access article distributed under the terms of CC BY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collaborative Group on Epidemiological Studies of Ovarian Cancer. Ovarian cancer and body size: individual participant meta-analysis including 25,157 women with ovarian cancer from 47 epidemiological studies. PLoS Med. 2012;9:e1001200. doi: 10.1371/journal.pmed.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beral V, Doll R, Hermon C, Peto R, Reeves G. Ovarian cancer and oral contraceptives: collaborative reanalysis of data from 45 epidemiological studies including 23,257 women with ovarian cancer and 87,303 controls. Lancet. 2008;371:303–14. doi: 10.1016/S0140-6736(08)60167-1. [DOI] [PubMed] [Google Scholar]
- 52.Kurian AW, Balise RR, McGuire V, Whittemore AS. Histologic types of epithelial ovarian cancer: Have they different risk factors? Gynecol Oncol. 2005;96:520–30. doi: 10.1016/j.ygyno.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 53.Gaitskell K, Green J, Pirie K, Reeves G. Tubal ligation and ovarian cancer risk in a large cohort: Substantial variation by histological type. Int J Cancer. 2016;138:1076–84. doi: 10.1002/ijc.29856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fuh KC, Shin JY, Kapp DS, et al. Gynecol Oncol. Vol. 136. Elsevier B.V; 2015. Survival differences of Asian and Caucasian epithelial ovarian cancer patients in the United States; pp. 491–7. [DOI] [PubMed] [Google Scholar]
- 55.Phelan CM, Kuchenbaecker KB, Tyrer JP, et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat Genet. 2017 doi: 10.1038/ng.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen H, Fridley BL, Song H, et al. Epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer. Nat Commun. 2013;4:1628. doi: 10.1038/ncomms2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.National Cancer Institute. Cancer Stat Facts: Ovarian Cancer. Bethesda, MD: [Google Scholar]
- 58.Winter WE, Maxwell GL, Tian C, et al. Prognostic factors for stage III epithelial ovarian cancer: A Gynecologic Oncology Group study. J Clin Oncol. 2007;25:3621–7. doi: 10.1200/JCO.2006.10.2517. [DOI] [PubMed] [Google Scholar]
- 59.Seidman JD, Yemelyanova A, Cosin JA, Smith A, Kurman RJ. Survival Rates for International Federation of Gynecology and Obstetrics Stage III Ovarian Carcinoma by Cell Type. Int J Gynecol Cancer. 2012;22:367–71. doi: 10.1097/IGC.0b013e31823c6f80. [DOI] [PubMed] [Google Scholar]
- 60.MacKay HJ, Brady MF, Oza AM, et al. Prognostic relevance of uncommon ovarian histology in women with stage III/IV epithelial ovarian cancer. Int J Gynecol Cancer. 2010;20:945–52. doi: 10.1111/IGC.0b013e3181dd0110. [DOI] [PubMed] [Google Scholar]
- 61.Jemal A, Ward EM, Johnson CJ, et al. Annual Report to the Nation on the Status of Cancer, 1975–2014, Featuring Survival. J Natl Cancer Inst. 2017;109:1–22. doi: 10.1093/jnci/djx030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bandera EV, Lee VS, Rodriguez-Rodriguez L, Powell CB, Kushi LH. Racial/Ethnic Disparities in Ovarian Cancer Treatment and Survival. Clin Cancer Res [Internet] 2016;22:5909–14. doi: 10.1158/1078-0432.CCR-16-1119. Available from: http://clincancerres.aacrjournals.org/cgi/doi/10.1158/1078-0432.CCR-16-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: Ovarian Cancer Including Fallopian Tube Cancer and Primary Peritoneal Cancer [Internet] 2016 Available from: https://www.nccn.org/professionals/physician_gls/pdf/ovarian.pdf.
- 64.Brown J, Frumovitz M. Mucinous tumors of the ovary: Current thoughts on diagnosis and management. Curr Oncol Rep. 2014:16. doi: 10.1007/s11912-014-0389-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hess V, A’Hern R, Nasiri N, et al. Mucinous epithelial ovarian cancer: A separate entity requiring specific treatment. J Clin Oncol. 2004;22:1040–4. doi: 10.1200/JCO.2004.08.078. [DOI] [PubMed] [Google Scholar]
- 66.Pectasides D, Pectasides E, Psyrri A, Economopoulos T. Treatment issues in clear cell carcinoma of the ovary: a different entity? Oncologist. 2006;11:1089–94. doi: 10.1634/theoncologist.11-10-1089. [DOI] [PubMed] [Google Scholar]
- 67.Takano M, Tsuda H, Sugiyama T. Clear cell carcinoma of the ovary: Is there a role of histology-specific treatment? J Exp Clin Cancer Res. 2012;31:53. doi: 10.1186/1756-9966-31-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jelovac D, Armstrong DK. Recent progress in the diagnosis and treatment of ovarian cancer. CA Cancer J Clin. 2011;61:183–203. doi: 10.3322/caac.20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang C, Winterhoff BJ, Kalli KR, et al. Expression signature distinguishing two tumour transcriptome classes associated with progression-free survival among rare histological types of epithelial ovarian cancer. Br J Cancer Nature Publishing Group. 2016;114:1412–20. doi: 10.1038/bjc.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11:5678–85. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 71.Carey LA, Dees EC, Sawyer L, et al. The triple negative paradox: Primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–34. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 72.Guha M. PARP inhibitors stumble in breast cancer. Nat Biotechnol. 2011;29:373–4. doi: 10.1038/nbt0511-373. [DOI] [PubMed] [Google Scholar]
- 73.Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation from non-small-cell lung cancer to small-cell lung cancer: Molecular drivers and cells of origin. Lancet Oncol. 2015;16:e165–72. doi: 10.1016/S1470-2045(14)71180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schildkraut J, Iversen E, Akushevich L, et al. Molecular Signatures of Epithelial Ovarian Cancer: Analysis of Associations with Tumor Characteristics and Epidemiologic Risk Factors. Cancer Epidemiol Biomarkers Prev. 2013;22:1709–21. doi: 10.1158/1055-9965.EPI-13-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hofree M, Shen JP, Carter H, Gross A, Ideker T. Network-based stratification of tumor mutations. Nat Methods. 2013;10:1108–15. doi: 10.1038/nmeth.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Buckley AR, Standish KA, Bhutani K, et al. Pan-cancer analysis reveals technical artifacts in TCGA germline variant cells. bioRxiv. 2016:91263. doi: 10.1186/s12864-017-3770-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tan TZ, Miow QH, Huang RY-J, et al. Functional genomics identifies five distinct molecular subtypes with clinical relevance and pathways for growth control in epithelial ovarian cancer. EMBO Mol Med. 2013;5:1051–66. doi: 10.1002/emmm.201201823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Planey CR, Gevaert O. CoINcIDE: A framework for discovery of patient subtypes across multiple datasets. Genome Med. 2016;8:27. doi: 10.1186/s13073-016-0281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang C, Armasu SM, Kalli KR, et al. Pooled clustering of high-grade serous ovarian cancer gene expression leads to novel consensus subtypes associated with survival and surgical outcomes. Clin Cancer Res. 2017 doi: 10.1158/1078-0432.CCR-17-0246. This paper reports that the mesenchymal high grade serous gene expression subtype that is associated with poor survival is also associated with a higher rate of residual disease after surgical debulking compared to the other subtypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Way GP, Rudd J, Wang C, et al. Comprehensive Cross-Population Analysis of High-Grade Serous Ovarian Cancer Supports No More Than Three Subtypes. G3 Genes, Genomes, Genet. 2016 doi: 10.1534/g3.116.033514. g3.116.033514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ogino S, Fuchs CS, Giovannucci E. How many molecular subtypes? Implications of the unique tumor principle in personalized medicine. Expert Rev Mol Diagn. 2012;12:621–8. doi: 10.1586/erm.12.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Finak G, Bertos N, Pepin F, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–27. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 83.Conti J, Thomas G. The Role of Tumour Stroma in Colorectal Cancer Invasion and Metastasis. Cancers (Basel) 2011;3:2160–8. doi: 10.3390/cancers3022160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gentles AJ, Bratman SV, Lee LJ, et al. Integrating Tumor and Stromal Gene Expression Signatures With Clinical Indices for Survival Stratification of Early-Stage Non–Small Cell Lung Cancer. JNCI J Natl Cancer Inst. 2015:107. doi: 10.1093/jnci/djv211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang S, Jing Y, Zhang M, et al. Stroma-associated master regulators of molecular subtypes predict patient prognosis in ovarian cancer. Sci Rep Nature Publishing Group. 2015;5:16066. doi: 10.1038/srep16066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Murakami R, Matsumura N, Mandai M, et al. Establishment of a Novel Histopathological Classification of High-Grade Serous Ovarian Carcinoma Correlated with Prognostically Distinct Gene Expression Subtypes. Am J Pathol American Society for Investigative Pathology. 2016;186:1103–13. doi: 10.1016/j.ajpath.2015.12.029. [DOI] [PubMed] [Google Scholar]
- 87.Winterhoff BJ, Maile M, Mitra AK, et al. Single cell sequencing reveals heterogeneity within ovarian cancer epithelium and cancer associated stromal cells. Gynecol Oncol. 2017;144:598–606. doi: 10.1016/j.ygyno.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science (80-) 2014;344:1396–401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang Q, Hu X, Hu B, et al. Tumor evolution of glioma intrinsic gene expression subtype associates with immunological changes in the microenvironment. bioRxiv. 2016:52076. doi: 10.1016/j.ccell.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jaitin DA, Kenigsberg E, Keren-Shaul H, et al. Massively Parallel Single-Cell RNA-Seq for Marker-Free Decomposition of Tissues into Cell Types. Science (80-) 2014;343:776–9. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Macosko EZ, Basu A, Satija R, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–14. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Y, Navin NE. Advances and Applications of Single-Cell Sequencing Technologies. Mol Cell. 2015;58:598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kommoss S, Winterhoff B, Oberg AL, et al. Bevacizumab may differentially improve ovarian cancer outcome in patients with proliferative and mesenchymal molecular subtypes. Clin Cancer Res. 2017 doi: 10.1158/1078-0432.CCR-16-2196. This paper provides supportive evidence for differences in response to treatment by high grade serous gene expression subtypes, which is evidence against a “one size fits all” approach to treatment of ovarian cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fong PC, Yap TA, Boss DS, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol Off J Am Soc Clin Oncol. 2010;28:2512–9. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 95.Hamanishi J, Mandai M, Ikeda T, et al. Safety and Antitumor Activity of Anti-PD-1 Antibody, Nivolumab, in Patients With Platinum-Resistant Ovarian Cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2015;33:4015–22. doi: 10.1200/JCO.2015.62.3397. [DOI] [PubMed] [Google Scholar]
- 96.Merck Sharp & Dohme Corp. Efficacy and Safety Study of Pembrolizumab (MK-3475) in Women with Advanced Recurrent Ovarian Cancer (MK-3475/KEYNOTE-100) [Internet] ClinicalTrials.gov. [cited 2017 Jun 14]. Available from: https://clinicaltrials.gov/ct2/show/NCT02674061.
- 97.Yeung T-L, Leung CS, Wong K-K, et al. TGF-β modulates ovarian cancer invasion by upregulating CAF-derived versican in the tumor microenvironment. Cancer Res. 2013;73:5016–28. doi: 10.1158/0008-5472.CAN-13-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cheon D-J, Tong Y, Sim M-S, et al. A collagen-remodeling gene signature regulated by TGF-β signaling is associated with metastasis and poor survival in serous ovarian cancer. Clin Cancer Res An Off J Am Assoc Cancer Res. 2014;20:711–23. doi: 10.1158/1078-0432.CCR-13-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Karlan BY, Dering J, Walsh C, et al. POSTN/TGFBI-associated stromal signature predicts poor prognosis in serous epithelial ovarian cancer. Gynecol Oncol. 2014;132:334–42. doi: 10.1016/j.ygyno.2013.12.021. [DOI] [PubMed] [Google Scholar]
- 100.Riester M, Wei W, Waldron L, et al. Risk prediction for late-stage ovarian cancer by meta-analysis of 1525 patient samples. J Natl Cancer Inst. 2014:106. doi: 10.1093/jnci/dju048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shalowitz DI, Epstein AJ, Ko EM, Giuntoli RL. Non-surgical management of ovarian cancer: Prevalence and implications. Gynecol Oncol Elsevier Inc. 2016;142:30–7. doi: 10.1016/j.ygyno.2016.04.026. [DOI] [PubMed] [Google Scholar]
- 102.Melamed A, Hinchcliff EM, Clemmer JT, et al. Gynecol Oncol [Internet] Vol. 143. Elsevier Inc; 2016. Trends in the use of neoadjuvant chemotherapy for advanced ovarian cancer in the United States; pp. 236–40. Available from: http://dx.doi.org/10.1016/j.ygyno.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 103.Moorman PG, Calingaert B, Palmieri RT, et al. Hormonal risk factors for ovarian cancer in premenopausal and postmenopausal women. Am J Epidemiol. 2008;167:1059–69. doi: 10.1093/aje/kwn006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schildkraut JM, Abbott SE, Alberg AJ, et al. Association between Body Powder Use and Ovarian Cancer: the African American Cancer Epidemiology Study (AACES) Cancer Epidemiol Biomarkers Prev [Internet] 2016;25:1411–8. doi: 10.1158/1055-9965.EPI-15-1281. Available from: http://cebp.aacrjournals.org/content/early/2016/05/12/1055-9965.EPI-15-1281.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cannioto RA, Trabert B, Poole EM, Schildkraut JM. Ovarian cancer epidemiology in the era of collaborative team science. Cancer Causes Control Springer International Publishing. 2017;0:0. doi: 10.1007/s10552-017-0862-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sieh W, Kobel M, Longacre TA, et al. Hormone-receptor expression and ovarian cancer survival: An Ovarian Tumor Tissue Analysis consortium study. Lancet Oncol. 2013;14:853–62. doi: 10.1016/S1470-2045(13)70253-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kobel M, Kalloger SE, Lee S, et al. Biomarker-based ovarian carcinoma typing: A histologic investigation in the ovarian tumor tissue analysis consortium. Cancer Epidemiol Biomarkers Prev. 2013;22:1677–86. doi: 10.1158/1055-9965.EPI-13-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ganzfried BF, Riester M, Haibe-Kains B, et al. curatedOvarianData: clinically annotated data for the ovarian cancer transcriptome. Database. 2013;2013:bat013–bat013. doi: 10.1093/database/bat013. [DOI] [PMC free article] [PubMed] [Google Scholar]