Abstract
RpfB is multidomain protein that is crucial for Mycobacterium tuberculosis resuscitation from dormancy. This protein cleaves cell-wall peptidoglycan, an essential bacterial cell wall polymer formed by glycan chains of β-(1-4)-linked-N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) cross-linked by short peptide stems. RpfB is structurally complex being composed of five distinct domains, namely a catalytic, a G5 and three DUF348 domains. Here, we have undertaken a combined experimental and computation structural investigations on the entire protein to gain insights into its structure-function relationships. CD spectroscopy and light scattering experiments have provided insights into the protein fold stability and into its oligomeric state. Using the available structure information, we modeled the entire protein structure, which includes the two DUF348 domains whose structure is experimentally unknown, and we analyzed the dynamic behavior of RpfB using molecular dynamics simulations. Present results highlight an intricate mutual influence of the dynamics of the different protein domains. These data provide interesting clues on the functional role of non-catalytic domains of RpfB and on the mechanism of peptidoglycan degradation necessary to resuscitation of M. tuberculosis.
Keywords: Tuberculosis, resuscitation, dormancy, Molecular Dynamics
Introduction
Proteins are frequently characterized by intricate molecular organizations as they may contain several distinct structural/functional domains. Interestingly, some modular domains are frequently shared by proteins endowed with different biochemical properties and/or involved in unrelated biological processes. Multi-domain proteins may be endowed with different levels of flexibility. Clearly, the overall rigidity/flexibility of these proteins depends on the strength of the mutual interactions that the different domains establish. Therefore, the definition of the structural and dynamic properties of multi-domain proteins represents an important step for a full understanding of the molecular mechanisms underlying their functions. One intriguing example of a functionally important multi-domain protein is represented by the Resuscitation-promoting factor B (RpfB) isolated from the pathogen Mycobacterium tuberculosis which is responsible of Tuberculosis (TB), one of the world’s deadliest transmissible diseases with about 1.5 million deaths per year. Several investigations have shown that RpfB, along with other four homologues (RpfA, RpfC, RpfD, and RpfE) encoded by the pathogen, are involved in M. tuberculosis resuscitation from the non-replicative dormant state.
Bacterial resuscitation from dormancy is a complex and multi-step phenomenon, which involves both cell wall hydrolases and kinases (Hett, Chao, & Rubin, 2010; Ruggiero, De Simone, Smaldone, Squeglia, & Berisio, 2012; Ruggiero et al., 2011; Shah, Laaberki, Popham, & Dworkin, 2008; Squeglia et al., 2011; Squeglia, Ruggiero, & Berisio, 2015). However, this process is mainly triggered by the modelling of the peptidoglycan (PGN), an essential cell wall envelope component present in most bacteria that confers the characteristic shape and size, protection against environmental stress and plays a key role in growth and division (Alvarez, Espaillat, Hermoso, de Pedro, & Cava, 2014; Meroueh et al., 2006; Squeglia et al., 2011; Squeglia et al., 2015; Vollmer, Blanot, & de Pedro, 2008). Therefore, enzymes involved in modelling of PGN are important targets for the development of antimicrobial agents (Correale, Ruggiero, Capparelli, Pedone, & Berisio, 2013; Ruggiero et al., 2012; Silva et al., 2016).
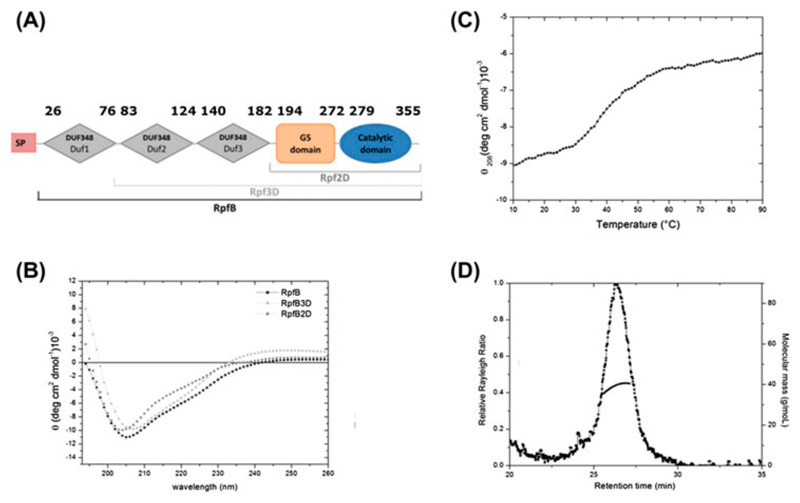
Although the five genes encoding for Rpfs are collectively dispensable for growth in vitro (Kana et al., 2008), only the deletion the specific gene encoding for RpfB is not dispensable for resuscitation in vivo (Tufariello et al., 2006). This makes RpfB is an attractive target for innovative therapeutical approaches (Hett, Chao, Deng, & Rubin, 2008; Hett et al., 2007; Mukamolova et al., 2006). Not surprisingly, RpfB shows among Rpfs the highest structural complexity, as it contains four extra domains in addition to the catalytic domain: the G5 cell-wall adhesive domain and three N-terminal repetitive domains of unknown function (DUF348) (Figure 1A). Previous structural studies have elucidated the structures of various portions of RpfB, including its catalytic domain (Cohen-Gonsaud et al., 2005; Ruggiero et al., 2013; Squeglia et al., 2013), the G5 and the first of the three DUF348 domains (Ruggiero et al., 2015; Ruggiero et al., 2009). The catalytic domain adopts the fold typical of c-type lysozyme and lytic transglycosylases, hydrolases known to cleave the sugar chains in the PGN polymer (Cohen-Gonsaud et al., 2005; Squeglia et al., 2013). Also, crystallographic and MD simulations on the complexes between RpfB and sugar ligands provided the first representation at atomic level of the key enzyme-carbohydrate interactions. A c-type lysozyme fold was also observed in RpfB homologues RpfE (Mavrici, Prigozhin, & Alber, 2014) and RpfC (Chauviac et al., 2014). X-ray studies have also evidenced a novel fold of the G5 domain, constituted by two β-sheets connected by a small triple helix motif, and denominated as β-TH-β (Ruggiero et al., 2009). The β-TH-β fold exhibits typical feature of adhesive proteins (Richardson & Richardson, 2002), a finding which has suggested that G5 domains may be important for PGN adhesion (Ruggiero et al., 2009). Further studies have shown that this protein acts on glycan chains of PGN using a transglycosylase catalytic mechanism (Nikitushkin et al., 2015). DUF348 domains are widely spread in proteins and normally occur as tandem repeats or, like in RpfB, in conjunction with G5 domains. Interestingly, DUF348 structure displays an unexpected ubiquitin-like fold (Ruggiero et al., 2015). Based on the structural similarity of the DUF348 domain to ubiquitin and frequent association to the G5 domain, we previously proposed to name this domain as G5-linked-Ubiquitin-like domain, UBLG5 (Ruggiero et al., 2015).
Figure 1.
(A) Domain organization of RpfB, based on the Pfam database(Finn et al., 2008). (B) CD spectra of RpfB (residues 30-362), RpfB2D (183-362) and RpfB3D (115-362) measured in 10 mM sodium phosphate. (C) Thermal unfolding curve of RpfB (0.2 mg mL-1) at 208 nm. (D) Analytical SEC-MALS; The black curve represents the Rayleigh ratio (left scale) against the elution time. Molecular masses (right scale), reported in grey, correspond to a monomeric state.
Despite the several partial structural information collected on isolated domains (Maione et al., 2015; Ruggiero et al., 2013; Ruggiero et al., 2015; Ruggiero et al., 2009; Squeglia et al., 2013), structural features of the entire multi-domain RpfB have never been elucidated. In this work, we recombinantly produced RpfB deprived of its signal peptide (residues 30-362) and carried out its structural characterization in solution, by CD spectroscopy and light scattering studies. In addition, using the available structural data, we modeled the structure of the entire RpfB and analyzed its dynamic behavior using molecular dynamics simulations. These data provided further insights into the function of the non-catalytic RpfB domains.
Materials and Methods
2.1. Cloning, expression and purification of RpfB
A full-length version of RpfB omitting its signal peptide sequence (residues 30-362) was readily expressed in E. coli as a soluble, His6-tagged cleavable protein at expression levels of 8 mg/L. After protein expression, purification was carried out using similar protocols previously adopted (Ruggiero et al., 2009). EDTA-free Protease Inhibitor Cocktail Tablets (Roche Diagnostics, Mannheim, Germany) were used during protein purification to prevent degradation of the protein. After removal of the cell debris by centrifugation (21,000 × g for 35 min), the supernatant was purified using an Äkta Explorer fast protein liquid chromatography system (GE Healthcare, UK) with a Ni-NTA affinity column (GeHealthCare) in a buffer containing 50 mM Tris-HCl, 300 mM NaCl, and 10% (v/v) glycerol (pH 8.0). A linear gradient of imidazole (10-500 mM) was applied to elute the protein. The His6-tag cleavage using recombinant TEV protease was undertaken under dialysis in 50 mM TrisHCl, 150 mM NaCl, 10% (v/v) glycerol, pH 8.0 at 4°C. After TEV removal, the cleaved protein was further purified on gel-filtration chromatography (Superdex 200 16/60) (50 mM TrisHCl, 150 mM NaCl, 10% (v/v) glycerol, pH 8.0). Protein separation and purity were determined with sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The molecular mass of the purified protein was checked by mass spectrometry (36 kDa).
2.2. Circular Dichroism
To analyze the conformational state of RpfB, far-UV circular dichroism (CD) spectra were registered at 10°C. All CD spectra were recorded with a Jasco J-715 spectropolarimeter equipped with a Peltier temperature control system (Model PTC-423-S). Molar ellipticity per mean residue, [θ] in deg cm2•dmol-1, was calculated from the equation: [θ] = [θ]obs•mrw•(10•l•C)-1, where [θ]obs is the ellipticity measured in degrees, mrw is the mean residue molecular mass (109,8 Da), C is the protein concentration in g•L-1 and l is the optical path length of the cell in cm. Far-UV measurements (195-250 nm) were carried out at 20 °C using a 0.1 cm optical path length cell and a protein concentration of 0.2 mg mL-1.
2.3. Light scattering
Purified protein was analyzed by size-exclusion chromatography connected to a triple-angle light scattering detector equipped with a QELS module (quasi-elastic light scattering). Protein samples of 500 µg were loaded on a S200 10/30 column, equilibrated in 50 mM Tris-HCl (pH 8.0) and 200 mM NaCl, and 5% (v/v) glycerol. A constant flow rate of 0.5 mL/min was applied. Elution profiles were detected by a Shodex interferometric refractometer and analyzed using a miniDawn TREOS light scattering system (Wyatt Instrument Technology Corp.). Data were processed using the Astra 5.3.4.14 software package.
2.4. Notations and molecular modeling
RpfB domains have been defined throughout the text as follows: DUF1 (residues 36 to 81), DUF2 (residues 82 to 138), DUF3 (residues 139 to 193), G5 (residues 194 to 271), and CAT (residues 272 to 355). Consequently, hinge regions connecting the various domains are: DUF1-DUF2 (residues 78 to 85), DUF2-DUF3 (residues 135 to 142), DUF3-G5 (residues 190 to 197), and G5-CAT (residues 268 to 275). Structural models for DUF1 and DUF2, that have never been experimentally characterized, were generated by MODELLER 9V9 (Eswar, Eramian, Webb, Shen, & Sali, 2008) using the crystal structure of the DUF348 domain, DUF3, as a template (Figure 1A). The stereo-chemical quality of these models was improved by energy minimization using the GROMACS package (Van Der Spoel et al., 2005).
2.5. Molecular dynamics
Molecular dynamics (MD) simulations were performed using the region 30-362 as a starting model, with the GROMACS package, with the all-atom AMBER99sb ILDN force field (Lindorff-Larsen et al., 2010) in combination with the tip3p explicit water model(Horn et al., 2004). A rectangular box was used to accommodate the structural model, which included 163795 protein atoms and 52947 water molecules. After energy-minimization, the solvent was equilibrated for 100 ps during which the protein atoms were restrained to the energy-minimized Cartesian coordinates by a 1000 kJ·mol-1·nm-2 spring constant. Subsequently, unrestrained MD simulations were carried out for 250 ns using periodic boundary conditions, an integration time step of 2.0 fs and by adopting LINCS as a constraint algorithm (Hess, Bekker, Berendsen, & Fraaije, 1998). Simulations were performed in the NPT ensemble by coupling the system to weak external pressure and temperature baths (1 atm and 300 K), with coupling constants of 0.1 ps and 1.0 ps, respectively. V-rescale algorithm was applied for the temperature coupling and Berendsen for pressure coupling (Berendsen, Postma, van Gunsteren, DiNola, & Haak, 1984; Bussi, Donadio, & Parrinello, 2007). The particle-mesh Ewald (PME) method was used to account for the electrostatic contribution to non-bonded interactions (grid spacing of 0.12 nm). The simulations were analyzed using in-house programs and analysis tools from the GROMACS package (Van Der Spoel et al., 2005). RMSF were calculated on the equilibrated part of the trajectory in each individual domain. To this end we disclosed the initial 50ns of the simulation in the RMSF analysis. Protein bending angles were defined across center of masses of individual domains.
3. Results and discussion
3.1. Solution studies
Solution structural features of RpfB, which was deprived only of the signal peptide were checked using circular dichroism (CD) and light scattering studies. The CD spectrum resembles those calculated for antiparallel β-sheets(Micsonai et al., 2015), with a deep minimum at 208 nm and shoulder at 222 nm (Figure 1B). This spectrum is similar to those recorded for the two truncated forms of the protein RpfB2D and RpfB3D (Figure 1B). To investigate the heat-induced changes in the protein secondary structure, thermal unfolding curves were achieved by following the CD signal at 208 nm as a function of temperature. The thermal unfolding takes place gradually without a steep transition (with a midpoint temperature close to 40°C). This non-cooperative thermal-induced unfolding indicates that the different protein domains possess different thermal stabilities (Figure 1C). Analytical size-exclusion chromatography (SEC), coupled with multi-angle light scattering (MALS) was carried out to investigate the oligomerization states of RpfB variants in solution. The on-line measurement of the intensity of the Rayleigh scattering as a function of the angle as well as the differential refractive index of the eluting peak in SEC was used to determine the molecular weight (MW). This analysis produced MW values which correspond to a monomeric species (Figure 1D). This result differs from those recently observed for RpfB homologue in Streptomyces coelicolor (sequence identity 34.8%), in which the full length enzyme is reported to form dimers whereas the enzyme deprived of DUF348 domains does not (Sexton et al., 2015). A possible dimerization in S. coelicolor indicates a different regulation mechanism in this bacterium, as compared to M. tuberculosis. Indeed, RpfB from M. tuberculosis was shown to synergize with the RipA endopeptidase (Hett et al., 2008; Ruggiero et al., 2010; Squeglia, Ruggiero, Romano, Vitagliano, & Berisio, 2014) whereas there is no RipA homologue encoded by the streptomycetes (Sexton et al., 2015). However, it should be kept in mind that estimates of molecular weights by filtration analyses may be risky for elongated proteins as those here described (Ruggiero et al., 2009). Therefore, the possibility that of RpfB isolated from M. tuberculosis and S. coelicolor adopt different oligomeric states needs further assessments.
3.2. Modeling of RpfB and MD simulations
Molecular modelling
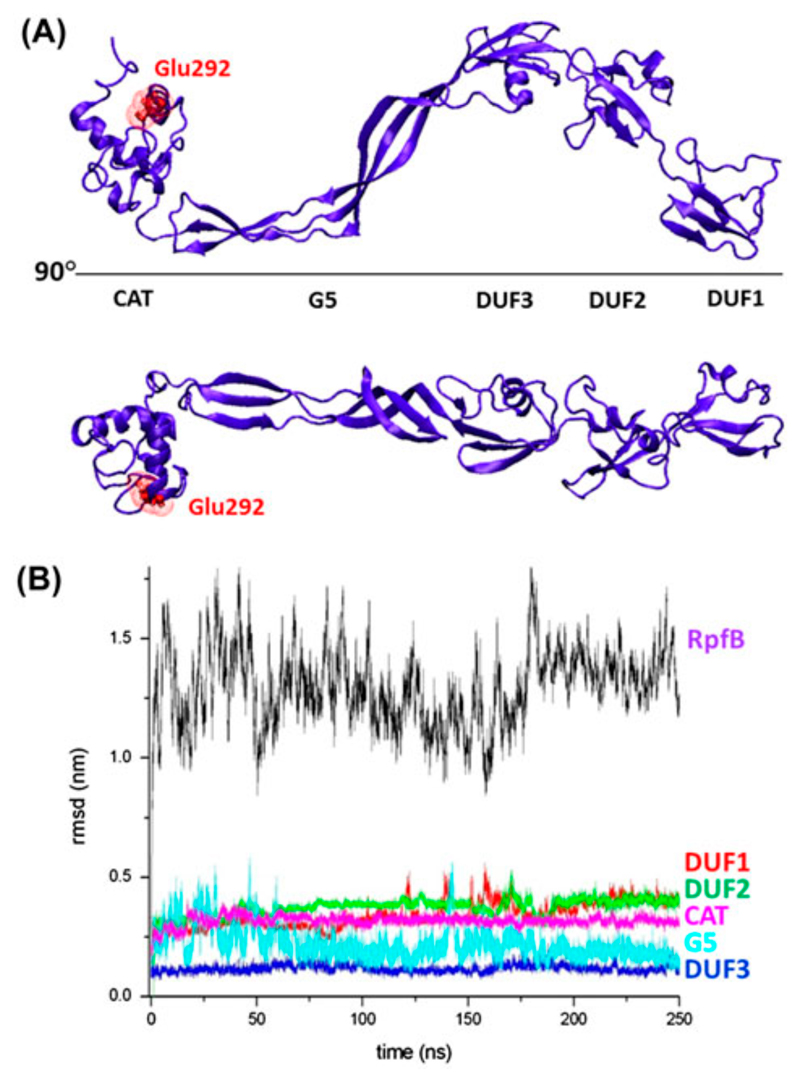
Taking into account (a) the monomeric state of RpfB highlighted by the solution studies reported above and (b) the recently solved crystal structure of RpfB3D, which contains all three types of the domains present in RpfB, a complete three-dimensional model of the protein, deprived only of its signal peptide was built. Specifically, we preliminarily generated homology models of the two missing N-terminal DUF348 domains of RpfB (DUF1 and DUF2, Figure 1) using the structure of DUF3, embedded in RpfB3D crystal structure, as a template. The reliability of this approach is assured by the sequence identity of DUF3 with DUF1 (26.8%) and DUF2 (36.2%), by the conservation of key residues forming hydrophobic and electrostatic interactions (Figure 2), and by the analysis of the stereo-chemical parameters of the generated models. These homology models were then linked to the N-terminal end of RpfB3D to generate RpfB. The minimized model is an S-shaped molecule, whose five domains occupy the same plane (Figure 3A).
Figure 2.
Sequence alignment between DUF3, DUF2 and DUF1 domains. Conserved glycines and aspartic residues are highlighted in yellow and red, respectively. Conserved hydrophobic residues are highlighted in green.
Figure 3.
(A) Two views of the minimised model of RpfB; the catalytic Glu292 is reported in red stick representation; (B) RMSD values along the trajectory computed for the entire molecule and for isolated domains.
Molecular dynamics – structure stability
To further validate this model and to gain insights into the local and global RpfB flexibility we performed Molecular Dynamics calculations. To assess the evolution of the structure in the simulation timescale (250 ns), a number of stereo-chemical parameters (gyration radius, secondary structure and deviations from the starting model) were monitored along the trajectories. As expected on the basis of the sequence similarities, the analysis of secondary structure evolution throughout the trajectory shows a rather conserved secondary structure composition in all five domains of the simulated model (Figure S1). The evaluation of root-mean-square deviations (RMSD) (calculated on the Cα atoms) between the starting model and the trajectory structures, which present an average RMSD value of 1.3 nm, suggests that significant rearrangements of the protein structure occur (Figure 3B). The RMSD values are more than 4-fold smaller when they are separately computed for the five individual domains. This indicates that main component of this rearrangement is represented by variations of the relative orientation of the RpfB domains (see also below).
Structure and dynamics of the individual domains
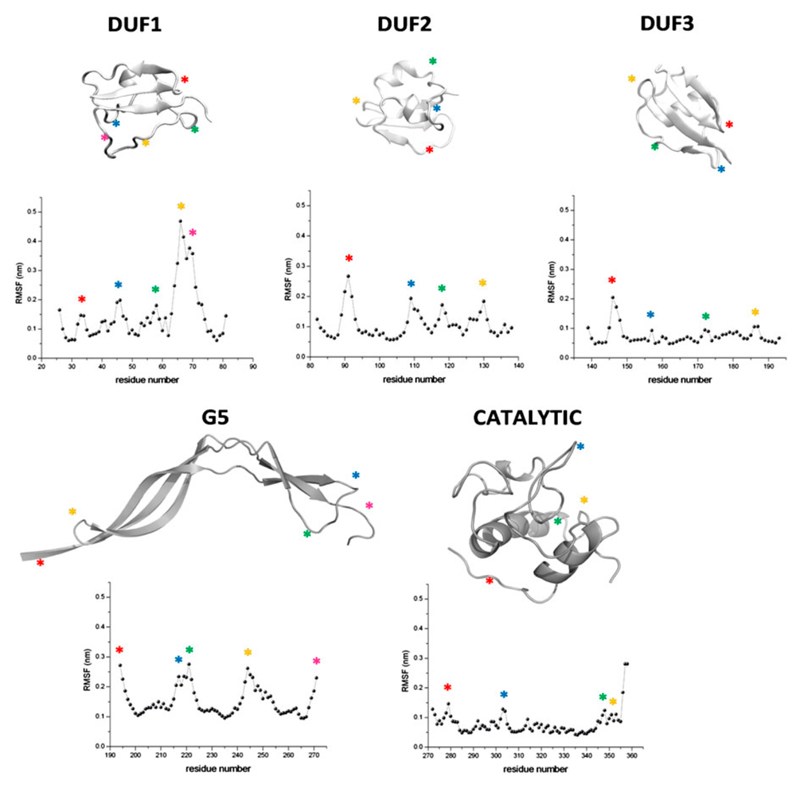
The stability and the dynamics of the individual domains along the trajectory was evaluated by considering RMSD values, secondary structure and root mean square fluctuation (RMSF) values. As anticipated above, individual domain display rather low RMSD values. This observation, along with the substantial conservation of the secondary structure elements, indicates that all five domains of the protein present folds that are well preserved in the simulation. The analysis RMSF values for each individual domain, computed in the equilibrated region of trajectory (50 to 250 ns), shows a low conformational mobility for the catalytic domain, whereas the G5 and DUF348 domains are highly flexible, with spikes corresponding to loops and low RMSF values for regions embedded in secondary structure elements (Figure 4). Interestingly, the analysis of the conformational space sampled by protein residues clearly indicates that the three strictly conserved Gly residues (Gly 33, 90, and 146, Figure 2) are endowed with a distinctive behavior. Indeed, all of them show a strong tendency to populate (phi,psi) regions that are unusual or even forbidden for other residues. In particular, left-handed helix conformation is observed in 100% of the number of conformers of in the cases of Gly33 and Gly146 and 38.5% for Gly90. Therefore, the peculiar conformation of this residue is important for preserving the structural integrity of the ubiquitin-like fold. In this scenario, the conservation of a Gly residue in DUF348 sequences is likely related to this structural requirement.
Figure 4.
Evolution of RSMF values along the trajectory computed for the five isolated domains. Maxima in the plots and their correspondences in the cartoon sketches are indicated with asterisks.
Inter-domain motions
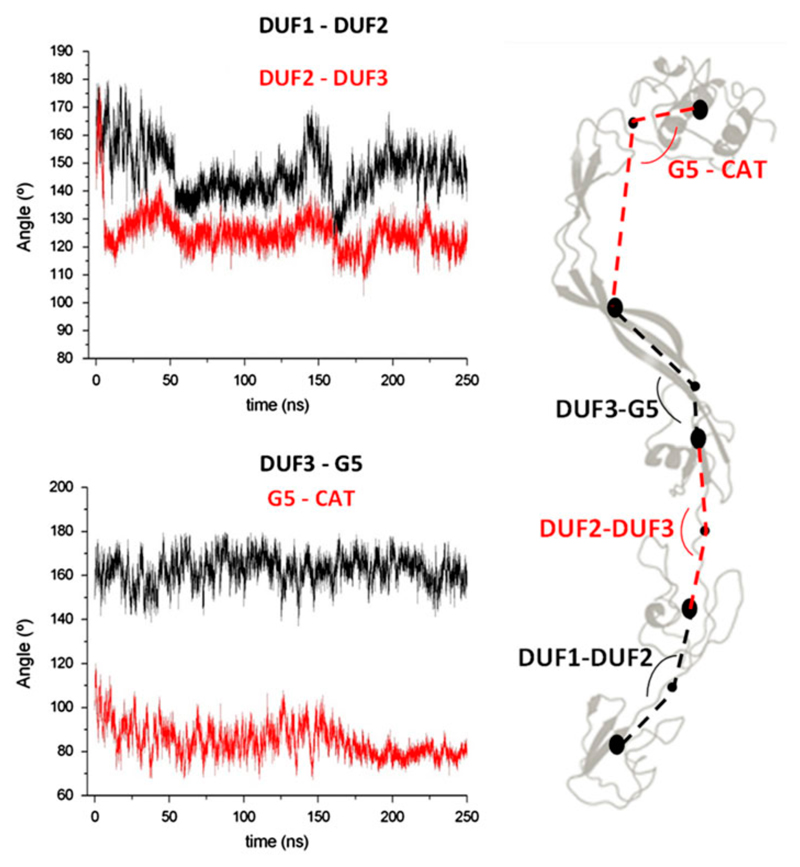
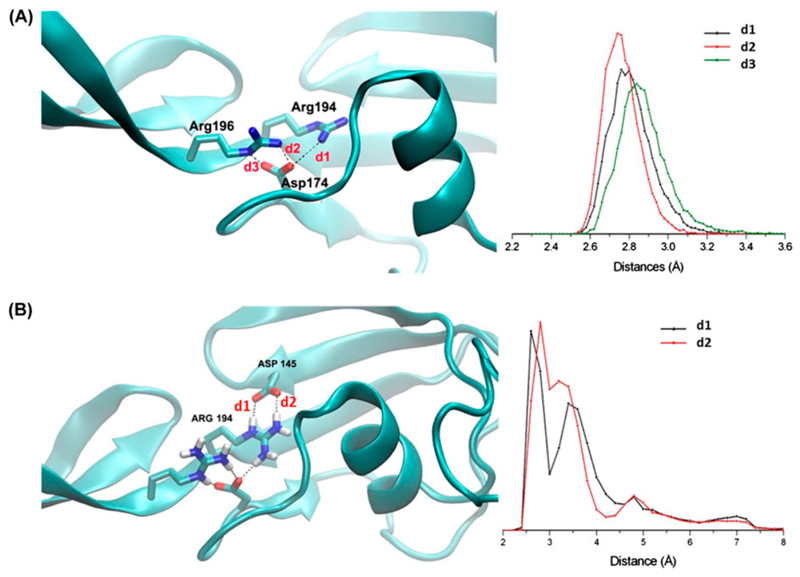
Bending angles between domains were computed along the trajectory (See Methods for hinge definition). This analysis shows that lowest motions are observed in the relative orientation between the third DUF348 and the G5 domain (DUF3-G5), followed by those between the catalytic domain and the G5 domain (G5-CAT, Figure 5). Differently, large inter-domain motions are observed between DUF1-DUF2 and DUF2-DUF3 (Figure 5). This finding well agrees with the observation that the G5 and DUF3 domains are strictly connected by a long β-strand (Figure 3). A deeper analysis of interactions between G5 and DUF domains evidence that three H-bonds exist between Asp174 (belonging to the DUF3 domain) and Arg196 of the G5 domain. These H-bonding interactions remain stable throughout the simulation, as highlighted by the H-bonding distance plot in Figure 6A. Similarly stable are two H-bonds between Arg194 and Asp145 of the DUF3 domain (Figure 6B). Therefore, Asp174 plays a key role in domain-domain interactions between G5 and DUF3. Consistently, sequence alignment among the three DUF domains (Figure 2) shows that the aspartic residue at position 145 is also conserved in the other two DUF348 domains. Analysis of trajectories shows that these aspartic residues are also involved in salt bridges in the regions connecting DUF domains, namely between Asp60 and Arg80 (DUF1-DUF2) and between Asp114 and Lys139 (DUF2-DUF3). Both interactions are conserved in the trajectories (Figure S2).
Figure 5.
Evolution of angles computed between (A) domain DUF1-DUF2 and DUF2-DUF3 and (B) domains DUF3-G5 and G5-CAT. Definition of domains is schematized in panel C and reported more precisely in the Methods section.
Figure 6.
Inter-domain salt bridge interactions involving (A) Asp174 and Arg194/Arg196 and (B) Asp145 and Arg194. Left panels report cartoon and ball-and-stick representations of the protein whereas right panels report plots of H-bonding distances throughout the MD simulation.
This fine balance between rigidity and plasticity, e.g. the ability to expose flexible surfaces centered around rigid hotspots, was recorded as a possible reason for the molecular recognition capacities of ubiquitin like folds (Lange et al., 2008). In this scenario, it is likely that, similar to ubiquitin, DUF348 domains may be instrumental to RpfB for the molecular recognition of its target, PGN (Figure S3). The model of the entire RpfB obtained in this study goes along this line, as its planarity well agrees with the role of all non-catalytic domains to help proper positioning of the catalytic site of RpfB on the glycosidic part of PGN, so to improve efficiency of cell wall hydrolysis during resuscitation.
Concluding Remarks
In the present investigation, we have unveiled the structural and dynamic features of the multi-domain resuscitation promoting factor RpfB. We generated the structure of the entire enzyme starting from the available structural data and modelling the missing regions. MD simulations of this model evidenced interesting information, which suggest functional properties of non-catalytic domains. Although simulations evidence large inter-domain motions, the five domains of RpfB mostly occupy the same plane. According to the currently accepted structural model of PGN, glycan chains are arranged perpendicular to the cell wall plane and every strand is connected to others by peptide crosslinks placed within the cell wall plane (Meroueh et al., 2006). We previously suggested that the presence in the G5 domain of solvent exposed β-sheets is important for PGN adhesion (Ruggiero et al., 2009). Present data indicate that UBLG5 domains play a similar role in the adhesion of the enzyme to polymeric peptidoglycan. Consistently, these domains share the dynamical properties of ubiquitin which have been considered important for its adaptability during protein targeting (Lange et al., 2008).
In this framework, non-catalytic domains of RpfB may concur to cell wall association of the protein by combining different structural mechanisms. In particular, the adhesive properties of natural beta-sheet domains like G5 and molecular adaptability of UBLG5 domains may confer to RpfB the ability of both efficient binding and cell wall adaptation.
Supplementary Material
Acknowledgements
Authors acknowledge the EU Indigo Linking Programme Italy-UK-India (GA 609535).
References
- Alvarez L, Espaillat A, Hermoso JA, de Pedro MA, Cava F. Peptidoglycan remodeling by the coordinated action of multispecific enzymes. Microbial drug resistance. 2014;20(3):190–198. doi: 10.1089/mdr.2014.0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. The Journal of chemical physics. 2007;126(1) doi: 10.1063/1.2408420. 014101. [DOI] [PubMed] [Google Scholar]
- Chauviac FX, Robertson G, Quay DH, Bagneris C, Dumas C, Henderson B, et al. Cohen-Gonsaud M. The RpfC (Rv1884) atomic structure shows high structural conservation within the resuscitation-promoting factor catalytic domain. Acta crystallographica. Section F, Structural biology communications. 2014;70(Pt 8):1022–1026. doi: 10.1107/S2053230X1401317X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Gonsaud M, Barthe P, Bagneris C, Henderson B, Ward J, Roumestand C, Keep NH. The structure of a resuscitation-promoting factor domain from Mycobacterium tuberculosis shows homology to lysozymes. Nat Struct Mol Biol. 2005;12(3):270–273. doi: 10.1038/nsmb905. [DOI] [PubMed] [Google Scholar]
- Correale S, Ruggiero A, Capparelli R, Pedone E, Berisio R. Structures of free and inhibited forms of the L,D-transpeptidase LdtMt1 from Mycobacterium tuberculosis. Acta crystallographica. Section D, Biological crystallography. 2013;69(Pt 9):1697–1706. doi: 10.1107/S0907444913013085. [DOI] [PubMed] [Google Scholar]
- Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods in molecular biology. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, et al. Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry. 1998;18(12):1463–1472. [Google Scholar]
- Hett EC, Chao MC, Deng LL, Rubin EJ. A mycobacterial enzyme essential for cell division synergizes with resuscitation-promoting factor. PLOS Pathogens. 2008;4(2):e1000001. doi: 10.1371/journal.ppat.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hett EC, Chao MC, Rubin EJ. Interaction and modulation of two antagonistic cell wall enzymes of mycobacteria. PLOS Pathogens. 2010;6(7):e1001020. doi: 10.1371/journal.ppat.1001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hett EC, Chao MC, Steyn AJ, Fortune SM, Deng LL, Rubin EJ. A partner for the resuscitation-promoting factors of Mycobacterium tuberculosis. Molecular microbiology. 2007;66(3):658–668. doi: 10.1111/j.1365-2958.2007.05945.x. [DOI] [PubMed] [Google Scholar]
- Horn HW, Swope WC, Pitera JW, Madura JD, Dick TJ, Hura GL, Head-Gordon T. Development of an improved four-site water model for biomolecular simulations: TIP4P-Ew. The Journal of chemical physics. 2004;120(20):9665–9678. doi: 10.1063/1.1683075. [DOI] [PubMed] [Google Scholar]
- Kana BD, Gordhan BG, Downing KJ, Sung N, Vostroktunova G, Machowski EE, et al. Mizrahi V. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Molecular microbiology. 2008;67(3):672–684. doi: 10.1111/j.1365-2958.2007.06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KF, Becker S, et al. de Groot BL. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320(5882):1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78(8):1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maione V, Ruggiero A, Russo L, De Simone A, Pedone PV, Malgieri G, et al. Isernia C. NMR Structure and Dynamics of the Resuscitation Promoting Factor RpfC Catalytic Domain. PloS one. 2015;10(11):e0142807. doi: 10.1371/journal.pone.0142807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrici D, Prigozhin DM, Alber T. Mycobacterium tuberculosis RpfE crystal structure reveals a positively charged catalytic cleft. Protein science : a publication of the Protein Society. 2014;23(4):481–487. doi: 10.1002/pro.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroueh SO, Bencze KZ, Hesek D, Lee M, Fisher JF, Stemmler TL, Mobashery S. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc Natl Acad Sci U S A. 2006;103(12):4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micsonai A, Wien F, Kernya L, Lee YH, Goto Y, Refregiers M, Kardos J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(24):E3095–3103. doi: 10.1073/pnas.1500851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamolova GV, Murzin AG, Salina EG, Demina GR, Kell DB, Kaprelyants AS, Young M. Muralytic activity of Micrococcus luteus Rpf and its relationship to physiological activity in promoting bacterial growth and resuscitation. Mol Microbiol. 2006;59(1):84–98. doi: 10.1111/j.1365-2958.2005.04930.x. [DOI] [PubMed] [Google Scholar]
- Nikitushkin VD, Demina GR, Shleeva MO, Guryanova SV, Ruggiero A, Berisio R, Kaprelyants AS. A product of RpfB and RipA joint enzymatic action promotes the resuscitation of dormant mycobacteria. The FEBS journal. 2015 doi: 10.1111/febs.13292. [DOI] [PubMed] [Google Scholar]
- Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci U S A. 2002;99(5):2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero A, De Simone P, Smaldone G, Squeglia F, Berisio R. Bacterial cell division regulation by Ser/Thr kinases: a structural perspective. Current protein & peptide science. 2012;13(8):756–766. doi: 10.2174/138920312804871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero A, Marasco D, Squeglia F, Soldini S, Pedone E, Pedone C, Berisio R. Structure and functional regulation of RipA, a mycobacterial enzyme essential for daughter cell separation. Structure. 2010;18(9):1184–1190. doi: 10.1016/j.str.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Ruggiero A, Marchant J, Squeglia F, Makarov V, De Simone A, Berisio R. Molecular determinants of inactivation of the resuscitation promoting factor B from Mycobacterium tuberculosis. Journal of biomolecular structure & dynamics. 2013;31(2):195–205. doi: 10.1080/07391102.2012.698243. [DOI] [PubMed] [Google Scholar]
- Ruggiero A, Squeglia F, Marasco D, Marchetti R, Molinaro A, Berisio R. X-ray structural studies of the entire extracellular region of the serine/threonine kinase PrkC from Staphylococcus aureus. The Biochemical journal. 2011;435(1):33–41. doi: 10.1042/BJ20101643. [DOI] [PubMed] [Google Scholar]
- Ruggiero A, Squeglia F, Romano M, Vitagliano L, De Simone A, Berisio R. The structure of Resuscitation promoting factor B from M. tuberculosis reveals unexpected ubiquitin-like domains. Biochimica et biophysica acta. 2015 doi: 10.1016/j.bbagen.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Ruggiero A, Tizzano B, Pedone E, Pedone C, Wilmanns M, Berisio R. Crystal Structure of the Resuscitation-Promoting Factor (Delta DUF)RpfB from M. tuberculosis. Journal of Molecular Biology. 2009;385(1):153–162. doi: 10.1016/j.jmb.2008.10.042. [DOI] [PubMed] [Google Scholar]
- Sexton DL, St-Onge RJ, Haiser HJ, Yousef MR, Brady L, Gao C, et al. Elliot MA. Resuscitation-promoting factors are cell wall-lytic enzymes with important roles in the germination and growth of Streptomyces coelicolor. Journal of bacteriology. 2015;197(5):848–860. doi: 10.1128/JB.02464-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135(3):486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JR, Bishai WR, Govender T, Lamichhane G, Maguire GE, Kruger HG, et al. Alves CN. Targeting the cell wall of Mycobacterium tuberculosis: a molecular modeling investigation of the interaction of imipenem and meropenem with L,D-transpeptidase 2. Journal of biomolecular structure & dynamics. 2016;34(2):304–317. doi: 10.1080/07391102.2015.1029000. [DOI] [PubMed] [Google Scholar]
- Squeglia F, Marchetti R, Ruggiero A, Lanzetta R, Marasco D, Dworkin J, et al. Silipo A. Chemical basis of peptidoglycan discrimination by PrkC, a key kinase involved in bacterial resuscitation from dormancy. Journal of the American Chemical Society. 2011;133(51):20676–20679. doi: 10.1021/ja208080r. [DOI] [PubMed] [Google Scholar]
- Squeglia F, Romano M, Ruggiero A, Vitagliano L, De Simone A, Berisio R. Carbohydrate recognition by RpfB from Mycobacterium tuberculosis unveiled by crystallographic and molecular dynamics analyses. Biophysical journal. 2013;104(11):2530–2539. doi: 10.1016/j.bpj.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squeglia F, Ruggiero A, Berisio R. Exit from mycobacterial dormancy: a structural perspective. Current medicinal chemistry. 2015;22(14):1698–1709. doi: 10.2174/0929867322666150209153027. [DOI] [PubMed] [Google Scholar]
- Squeglia F, Ruggiero A, Romano M, Vitagliano L, Berisio R. Mutational and structural study of RipA, a key enzyme in Mycobacterium tuberculosis cell division: evidence for the L- to-D inversion of configuration of the catalytic cysteine. Acta crystallographica. Section D, Biological crystallography. 2014;70(Pt 9):2295–2300. doi: 10.1107/S1399004714013674. [DOI] [PubMed] [Google Scholar]
- Tufariello JM, Mi K, Xu J, Manabe YC, Kesavan AK, Drumm J, et al. Chan J. Deletion of the Mycobacterium tuberculosis resuscitation-promoting factor Rv1009 gene results in delayed reactivation from chronic tuberculosis. Infect Immun. 2006;74(5):2985–2995. doi: 10.1128/IAI.74.5.2985-2995.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS microbiology reviews. 2008;32(2):149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.