Abstract
The gut microbiota have both direct and indirect effects on drug and xenobiotic metabolism and this can have consequences for both efficacy and toxicity. Indeed microbiome-driven drug metabolism is essential for the activation of certain prodrugs such as e.g., azo drugs such as prontosil and neoprontosil resulting in the release of sulphanilamide. In addition to providing a major source of reductive metabolizing capability the gut microbiota provide a suite of additional reactions including acetylation/deacylation decarboxylation, dehydroxylation, demethylation, dehalogenation and importantly, in the context of certain types of drug-related toxicity, conjugate hydrolysis reactions. In addition to direct effects the gut microbiota can affect drug metabolism and toxicity indirectly via e.g., the modulation of host drug metabolism and disposition and competition of bacterial-derived metabolites for xenobiotic metabolism pathways. And, of course, the therapeutic drugs themselves can have effects, both intended and unwanted, which can impact on the health and composition of the gut microbiota with unforeseen consequences.
Keywords: Microbiome, Deconjugation, drug metabolism, prodrug activation, toxicity, efficacy
Introduction
To state the obvious, the aim of very many studies in drug metabolism and toxicity is ultimately to understand the factors that cause compounds to be ineffective therapeutically or cause toxicity in patients and, by using this knowledge to design better compounds, provide safe and effective treatments to patients. Whilst the presence of the gut microbiota has been acknowledged for many years it was nevertheless generally ignored by those working in drug metabolism and toxicology as being largely an irrelevance (albeit an interesting one). However, there has been a revolution in our understanding of the complexity, and system-wide, effects of this forgotten organ, brought about in no small measure by advances in molecular biology. This has revealed the diversity of the gut ecosystem, leading to a major re-evaluation of role of the gut microbiota in human health and disease. Thus, in adult humans the gut microbiota comprises up to ca. 1 Kg of bacteria, the majority of which are obligate anaerobes from the genera Bacteriodes, Clostridium, Lactobacillus, Escherichia and Bifidobacteria together with an assortment of yeasts and other microorganisms, to say nothing of the many viruses. The result is a complex and dynamic ecology comprising of at least 2000 species, with the composition varying depending upon the region of the gut examined. These microbes then provide benefits to the host via enhanced energy recovery from undigested food, defence against pathogens and interactions with both immune and nervous systems. These insights have led to a reaffirmation of the view that these microorganisms are not mere passengers but crew, providing multiple benefits for the host and, as a by-product of their symbiotic relationship with the host, directly and indirectly affecting the pharmacological/toxicological effects of numerous drugs. The rediscovery of the impact that the microbes that go to form this important “external” organ can have has led to a reawakened interest in their study. Further, there is now an increasing appreciation that the microbiome represents a “drugable target” as there is clear potential for altering the composition, and therefore metabolic capability, of the microbiome using a range of approaches, including pharmaceuticals. Such manipulation might be intentional, aimed at beneficially modifying the activities of the gut microbiota to improve the health and wellbeing of the host such as those claimed for pre- and probiotic interventions etc. Alternatively, changes wrought to the microbiome might also cause unintentional “collateral damage” resulting from e.g., exposure to antibiotics, and these modifications may bring with them adverse consequences. As such changes can be long lasting, the effect of alterations in the composition and functionality of the gut microbiota, given its symbiotic role, should now perhaps be more actively considered as part of the risk assessment process for new drugs. That said, it has been clear for a long time that the sheer complexity of the host-gut microbiome interaction means that modelling the various interactions between host and gut microbiota in such a way as to adequately predict the outcome of an intervention will require both novel approaches and the generation of much new knowledge1–3.
However, for the drug metabolism and toxicology communities, despite many early studies showing its importance in some instances of xenobiotic biotransformation (e.g., see refs4,5), the gut microbiota have not been a focus. Nevertheless, increased awareness is important not only because the microbiota perform a range of important metabolic reactions but because the gut microbiome also represents a source of physiological variability between both individuals and populations. Such variability can affect the disposition and toxicity of drugs and their metabolites. These effects can either be direct or through secondary interactions mediated through e.g., the metabolic exchange and the co-metabolism and processing of many diverse endogenous and dietary substrates6. These “metabolome–metabolome” interactions7 are still poorly understood, but it is clear that some bacterially-derived metabolites have the potentially to modulate the hosts’ drug metabolising systems as discussed below4. There is however, reason to believe, from the increasing number of research papers and reviews8–16 on the topic, that the gut microbiota are moving out of the shadows and are moving towards centre stage in drug safety studies and personalized health care.
Direct Drug Metabolism by the Gut Microbiota
The gut microbiota have the capability of preforming a wide range of metabolic reactions on drugs, drug metabolites and other xenobiotics. As summarized below, by far the most important biotransformations involve reductive metabolism and hydrolytic reactions (particularly on conjugates). In addition decarboxylations, dehydroxylations dealkylations, dehalogenations and deaminations have also been described.
Reductive Metabolism
The “classic” examples of gut microbial metabolism of therapeutic drugs are to be found in the reduction of the azo-antibacterial pro-drugs based on sulphanilamide such e.g., prontosil17,18 and neoprontosil17. Reductive metabolism of these, and a range of 5-aminosalicylic acid pro-drugs used in the treatment of ulcerative colitis and inflammatory bowel conditions, is mediated largely by the gut microbiota. So, the therapeutic activity of compounds such as sulfasalazine19,20, olsalazine21, ipsalazide and balsalazide22 depends upon the release of aminosalicylic acid to treat the inflammation. This ability to perform reductive metabolism on azo dyes and nitropolycyclic aromatic hydrocarbons was shown for bacteria of the genera Clostridia and Eubacteria by Rafii and Cerniglia23. Their investigation on the azo- and nitroreductases found in three strains of Clostridia and one Eubacterium and who concluded that, whilst the enzymes from different bacteria had different electrophoretic mobilities, both azo- and nitroreductase activities resided in the same enzyme.
Gut microbial nitroreductase activity, where nitro-groups are reduced to amines, has been shown to be important for benzodiazepines such as nitrazepam, clonazepam and bromezepam which contain this functional group. Thus nitroreduction as a result of the gut microflora has been demonstrated both in vitro and in vivo for the production of 7-aminonitrazepam, 7-aminoclonazepam and 2-(2-amino-5-bromobenzoyl) pyridine respectively from their respective parent drugs24–26. In orally dosed pregnant rats ca. 30% of the excreted nitrazepam metabolites were in the form of the nitro-reduced metabolites 7-aminonitrazepam and 7-acetylaminonitrazepam. However, the production of reduced metabolites fell to 2% if the animals were pre-treated with antibiotics, which also led to the almost complete abolition of nitroreductase activity. Reductive metabolism of nitro groups can have unwanted toxicological consequences such as nitrazepam-related teratogenicity24. Thus, in the rat, antibiotic treatment led to both a reduction in this microbiota-driven metabolism and a concomitant reduction in nitrazepam-induced teratogenicity. When [14C]-clonazepam was dosed orally to germ-free rats the reduced metabolites of the drug accounted for only 15% of the urinary excreted radioactivity but colonisation with an intestinal flora caused a large increase in nitroreduction to 77%25with the major metabolite identified as 7-acetamido-clonazepam. Gut microbiome-derived nitroreduction has also been shown for metronidazole in vitro and in vivo in the rat27–29 producing the amino metabolite 1-(2-aminoimidazol-1-yl)-3 methoxypropanol-2-ol and acetamide (a known rat carcinogen). As well as the toxicity associated with the reduction of nitrazepam described above, the reductive metabolism of the nitro-containing antibiotic chloramphenicol may be the cause of drug-induced bone marrow injury30. The metabolite responsible has been suggested as being p-aminophenyl-2-amino-1,3-propanediol31, generated in a small percentage of patients following oral administration. These p-aminophenyl-2-amino-1,3-propanediol-producing patients were reported to have a high percentage of coliform bacteria with the ability to metabolize the drug to this metabolite. However, other metabolites30–32 including p-nitrophenyl-2-dicloroacetamido-1,3-propanediol and 2-dichloroacetamid-3-hydroxypropio-p-nitrophenone have also been proposed as being responsible for this toxicity. Another interesting example of nitroreduction is provided by studies on the effects of the gut microbiota on the radiation sensitizer misonidazole33. Misonidazole was shown to be converted to its amino derivative [l-(2-aminoimidazol-l-yl)-3-methoxypropan-2-ol in both pure and mixed cultures of intestinal microbiota and, crucially, was seen in the excreta of normal but not germfree rats. In another study it was seen that mice treated with penicillin for a week before misonidazole administration showed both increased drug exposure and decreased neurotoxicity as well as a range of other effects34.
Hydrazone linkages in drugs are also susceptible to reductive cleavage by the gut microbiota as evidenced by studies in dogs35 and humans36 on the drug levosimendan, and in humans in the case of eltrombopag37. Reductive metabolism also occurs for sulphur-containing compounds as seen in the microbiota-driven reduction of the sulphoxide-containing drugs sulphinpyrazone and sulindac38 via the formation of sulphides on incubation with faeces (human or rabbit). Omeprazole, has also been shown in vitro to be reduced to its sulphide metabolite by the intestinal microbiota39. The reductive metabolism of the benzisoxazole ring of the anticonvulsant Zonisamide40,41 by several strains of gut bacteria has been demonstrated with Clostridium sporogenes showing the highest activity40. The reductive metabolism benzisoxazole ring of the antipsychotic drug risperidone by the gut microflora has also been shown for both rat and dog. In the case of rat cleavage of the ring of both the unchanged drug and some of its hydroxy-metabolites occurred in vivo, and in the presence of cecal contents42. Similar biotransformations were seen in vivo for dogs42. The in vitro reductions of the N-oxide prodrug loperamideoxide to loperamide43 and the H2 receptor antagonists ranitidine44 and nitazidine45 (but not cimetidine or famotidine45) have also been demonstrated. Studies on an antitumor combination therapy containing potassium 1,2,3,4-tetrahydro-2,4-dioxo-1,3,5-triazine-6-carboxylate (potassium oxonate, which acts on orotate phosphoribosyl-transferase to inhibit the conversion of 5-fluorouracil to its active form and so reduces gut toxicity) showed that the compound was metabolized to cyanuric acid. This biotransformation occurred in the GI tract and was shown, at least in part, to be due to the action of the gut microbiota in the cecum46.
Perhaps the most interesting example of the complex way in which microbiota-driven reductive drug metabolism can affect the fate of drugs remains the reduction of the drug digoxin at the hands of the gut microbiota47–53. This biotransformation first came to light in studies by Lindenbaum and co-workers47 who found that the production of reduced metabolites, such as dihydrodigoxin etc., was subject dependent. This was revealed in studies of the drugs bioavailability when analysis of the urinary excretion of these relatively inactive reduced metabolites by 131 normal subjects was performed. In the case of ca. one-third of these subjects the reduced metabolites formed over 5% of the excreted drug-related material and the amount produced varied inversely with bioavailability. This phenomenon, seen after either single or multiple doses, appeared to be stable over time but when some subjects were administered erythromycin the excretion of metabolites such as dihydrodigoxin was no longer seen. Another clue to microbial involvement was seen with lower urinary excretion of the reduced metabolite following intravenous dosing leading to the conclusion that the observed reductive metabolism was due to “the activity of a variable component of the intestinal flora”. Later48,49 the organism responsible for the reductive metabolism of digoxin was identified as E. lentum, although it was noted that its presence did not always correlate with the production of the reduced metabolites. Additionally, an inverse relationship between the concentration of arginine in the growth medium and formation of the reduced metabolites was observed. Interestingly in babies of less than 8 months in age reduced digoxin metabolites were not found50, despite the presence of E. lentum, but after 16 months a third of the children studied did produce reduced metabolites of the drug (although the amounts seen in the adult population (ca. 10%) were not found in patients less than 9 years old). Another investigation demonstrated an inverse relationship between the presence of C. difficile and the digoxin-reducing E. lentum with the latter less prevalent in the fecal samples of 77 nursing home residents infected with C. difficile who had previously undergone enteral feeding or received antibiotics treatment51. As well as effects of age these authors also observed that the extent of digoxin reduction varied both within and between populations showing e.g., that within an Indian population significant differences existed between rural villagers, who produced ca. 5% of the reduced metabolites compared to 23% formed by urban dwellers. Between population effects were seen on comparing a group of North American subjects who formed some 36% of the reduced metabolites with a South Indian population who only produced 13.7%. Such differences remained even after Indian subjects emigrated to the US52.
The solution to the problem of why E lenta can be present but digoxin reduction did not take place and the relationship between this reduction, when it occurred, and arginine has recently been resolved by some elegant studies by Haiser et al53. This reinvestigation of the metabolism of digoxin by E lenta, combining transcript profiling, comparative genomics, and culture-based assays, revealed a cytochrome-encoding operon (a “cardiac glycoside reductase” (cgr)). This cgr operon, found in the type strain of E. lenta but not in nonreducing strains was up-regulated by the drug but inhibited by arginine. The authors showed that “the abundance of the cgr operon predicts digoxin reduction by the human gut microbiome”, and predicted its inactivation. Studies in gnotobiotic mice colonised with either non-reducing or digoxin-reducing E lenta showed that high dietary protein reduced the reductive metabolism of digoxin (with changes in serum pharmacokinetics and urinary excretion) but did not affect mice having the non-reducing strain. The authors suggested that this was “likely through inhibitory effects of elevated luminal arginine on cgr operon expression”54. These studies nicely illuminate a complex microbiome-drug interaction that requires colonisation of the host with an appropriate strain of the microorganism involved and having the potential for dietary modulation as well.
Demethylations, deaminations, dehydroxylations, deacylations and decarboxylations and oxidations
Important as reductive metabolism is the gut microbiota are also capable of a range of additional biotransformations including those involving Demethylation, deamination, dehydroxylation, deacylation, decarboxylation or oxidation. The microbial demethylation of drugs such as methamphetamine and 4’-hydroxy methamphetamine has been shown in vitro55. O- and N-demethylation of drugs incubated with rat-derived microbiota has been investigated with the observation that the gut microbial metabolism of model compounds and drugs56. N-dealklyation did not occur for any of the compounds studied and O-dealkylation was only seen only for relatively simple aromatic compounds56. Microbial O-dealkyation has also been shown to be part of the metabolism of the spleen tyrosine kinase inhibitor fostamatinib57 In this case a 3,5-benzene diol metabolite produced by the O-demethylation and dehydroxylation of one of the metabolites of the drug,“ R529”, was ascribed to the action of anaerobic gut bacteria. This biotransformation was supported by studies involving the incubation of R529 with human-derived faeces. Thus metabolism of the drug to the 3,5-benzene diol metabolite involved a series of host and bacterial reactions with hepatic cytochrome P450-mediated p-O-demethylation of the drug followed by further O-demethylations and dihydroxylation by gut bacteria. The removal of the acetyl moiety from N-acetylated drugs, such as phenacetin, bucetin, and acetaminophen (paracetamol), and related compounds such as acetanilide, showed that they were all subject to N-deacylation to reveal phenetidine, aniline and p-aminophenol56, all of which have potential for toxicity
The biotransformation of 5-fluorocytosine to 5-fluorouracil by deamination has also been ascribed to the action of the gut microbiota58. In in vitro studies, a semi-continuous culture system was shown to be capable of converting 5-fluorocytosine to 5-fluorouracil after both acute and chronic (2 week exposure) to the drug. A lag in production of up to 8h was noted when using acute, but not chronic, exposure of the system to 5-fluorocytosine implying that the enzyme/s needed to perform the deamination reaction required induction. More recent in vitro studies used both viable and nonviable E. coli as well as patient-derived fecal samples from neutropenic patients. In the case of the patients, samples were taken before beginning 5-fluorocytosine-based antimicrobial/antifungal prophylaxis and then after 1 week of treatment)59. On incubation with viable E. coli for 48 h the amount of the drug had fallen by an average of ca.70%, and this was accompanied by a corresponding increase 5-fluorouracil concentrations. In incubations conducted with nonviable E. coli a 44% decrease in 5-fluorocytosine concentrations was observed. Incubation of 5-fluorocytosine with human feces obtained prior to, but not after, antimicrobial/antifungal prophylaxis resulted in “significant” 5-fluorocytosine deamination59.
The dehydroxylation/decarboxylation reactions represent another of the biotransformation capabilities of the gut microbiota. A well-known example of such reactions involves the dehydroxylation/decarboxylation of L-dopa (levodopa, L-3,4-dihydroxyphenylalanine). That the gut microbiota might be involved in the metabolism of the drug was first suggested by studies by Sandler et al60,61 who noted that, on treating patients suffering from Parkinson’s disease with L-dopa, the urinary excretion of m-hydroxyphenylacetic acid was increased. In addition concentrations of m-hydroxyphenylacetic acid were significantly reduced in quantity after the administration of neomycin, suggesting that some microbial dehydroxylation of dopamine or L-dopa occurs. In rats dosed with either L-dopa or dopamine the metabolite m-hydroxyphenylacetic acid was present in the urine of control rats but absent from that of germ free animals62. Studies on the fate of [14C]-DL-dopa, and potential metabolites, incubated with rat cecal contents63 suggested that microbial metabolism was via 3,4-dihydroxyphenylacetic acid and decarboxylation or dehydroxylation to 4-methylcatechol or 3-hydroxyphenylacetic acid. Decarboxylation of 3-hydroxyphenylacetic acid was seen to give rise to m-cresol and 3-hydroxyphenylpropionic acid was also detected. The decarboxylation of Dopa by gut bacteria was suggested as a mechanism for reducing the exposure of the drug in the brain64. In the dog65 the bioavailability of L-dopa after either hepatoportal or IV dosing was similar but the AUC for L-dopa was reduced, and that of dopamine increased following duodenal administration. Antibiotic administration to suppress the gut microbiota abolished this effect in the treated animals. However, reductions in the bioavailability of L-dopa have also been ascribed to the presence of infection by H. pylori (e.g.66,67) with concomitant reductions in clinical effects of the drug on Parkinsons symptoms. Improved adsorption and pharmacokinetic profiles for the drug were seen upon elimination of the infection. A possible reason for this H. pylori-related reduced bioavailability was suggested based on the observation that solutions of L-dopa incubated with H. pylori showed a decrease in concentration over time67. Further, bacteria pre-incubated with L-dopa showed significantly reduced adhesion to gastric epithelial cells. The authors concluded that these results demonstrated a direct interaction of L-dopa with the adhesins (proteins present on the outer membranes of the bacteria) that enable H. pylori to bind to these cells.
An example of oxidative metabolism by the gut microbiota is provided by studies on the anaerobic incubation in vitro of the anthelmintic drug levamisole which resulted in several thiazole ring-opened metabolites69 including levametabol I, which may possess anti-tumour activity. The bacteria responsible for these biotransformations were mainly derived from the Bacteriodes and Clostridia. The authors noted that “the formation of the hydroxamic lactam functionality from levamisole must involve an oxidation step, despite the anaerobic conditions required for the bacterial activity”69. Whilst descriptions of gut microbiotal oxidation/dehydrogenation are rare the example of the biotransformation dietary carcinogen 2-amino-3,6-dihydro-3H-imidazo [4,5-f]quinolone(IQ) to its 7-hydroxy metabolite70 was highlighted as a further example in support of the hypothesis that the bacterial metabolism of levamisole was oxidative. A further example of a drug where oxidative metabolism via hydroxylation was observed can be found in a study on a potential gut microbiota-mediated drug-drug interaction between lovastatin and antibiotics in the rat71. On incubation of the drug in vitro with human and rat fecalase preparations four metabolites were produced. These comprised the demethylbutyryl metabolite (designated as M4), and 3 ring opened species, including the active hydroxyacid (M8). Two of the ring opened metabolites (M4 and M9) also appeared to have been hydroxylated. The authors noted that, following antibiotic treatment, the systemic exposure of the active hydroxyacid metabolite was significantly reduced, with the amounts present in feces also reduced by ca 60%. These result prompted the authors to suggest that, where patients taking lovastatin to control plasma cholesterol concentrations are placed on long term antibiotic treatment, the concomitant suppression of the gut microbiota “might lead to serious outcomes due to a failure to control serum cholesterol levels”71.
Gut Microbial Mediated Hydrolysis of Drugs, Prodrugs and Xenobiotic Conjugates
As well as reductions the gut microflora are adept at hydrolytic reactions which can occur on the drugs themselves, prodrugs or conjugated metabolites. An early example of drug biotransformation via hydrolysis was the observation that methotrexate was metabolized by the intestinal flora of normal mice72. Subsequent studies involving the incubation of radiolabelled [3H]-methotrexate with CDF1 mouse cecal contents73. At least three metabolites were formed, the principal one being identified by the authors as 4-amino-4-deoxy-N10-methylpteroic acid (APA). The metabolite was also found in urine and feces of mice administered the drug73.
The problems of trying to deliver peptidic drugs with respect to degradation by the gut flora are well recognized and e.g., when the metabolism of insulin and calcitonin by microorganisms was examined in rat cecal contents both were rapidly degraded, with the latter more prone to proteolysis74. Subsequent studies investigated the use of protease inhibitors as a means of improving the stability and bioavailability of these peptides75 and, if inhibitors such as camostat and aprotinin, were present in incubations of insulin and calcitonin with rat cecal contents degradation could indeed be inhibited. In the case of the metabolic fate of the peptidic drug azetirelin, a thyrotropin-releasing hormone analogue, it was found that plasma concentrations of the drug were maintained in rats following the administration of antibiotics76. Incubation of azetirelin with rat, dog and human fecal suspensions confirmed that the drug was indeed subject to metabolism by anaerobic bacteria, and that this was inhibited by antibiotics. Subsequently an enteric capsule was prepared where azetirelin was formulated with n-lauryl-beta-D-maltopyranoside as a formulation enhancer and citric acid as potential inhibitor of bacterial degradation77. When tested in fasted dogs over 40% bioavailability for the drug was achieved using the new formulation (compared with ca. 15% when not formulated in this way)
To improve of poor biopharmaceutical properties, particularly solubility, drugs can be administered as e.g., phosphate or sulfate ester prodrugs and these can be acted on by hydrolytic enzymes produced by the gut microbiota. Indeed in the case of the laxative sodium picosulfate, which is administered as a disulfate, efficacy depends on its conversion to the 4,4'-dihydroxydiphenyl-(2 pyridyl)-methane by gut bacteria78. The desulfation appeared to be catalysed by a novel sulfotransferase, rather than the action of a sulfatase, and required the presence phenolic compounds such as e.g., phenol, acetaminophen, tannic acid or flavonoids in the incubations.
For many drugs and their metabolites that are subject to conjugation to form sulfates, glucuronides or glycosides the bile provides a major route of excretion and, once these conjugates come into contact with the gut microbiota there is obvious potential for deconjugation to occur. Hydrolytic enzymes capable of deconjugating drug metabolites are widely distributed across a range of species (see., e.g. ref 79) and can also have effects on the bioavailability of many natural products present in food and health products as glucuronides/glucosides (e.g., the flavone glucuronide biacalin80 or the Soy isoflavones that give rise to phytoestrogens such as equol are well known81,82). A major effect of these enzymes on drugs and their metabolites is the hydrolysis of biliary excreted conjugated metabolites (e.g., glucosides, glucuronides and sulfates). The liberation of the aglycones by microbial enzymes enables their resorption (enterohepatic recycling) by the host and as such can increase the exposure of the organism to the drug itself or bioactive metabolites. However, hydrolysis of conjugates, particularly glucuronides, by bacterial glucuronidases also results in the direct exposure of the gut to the pharmacological effects of the drug/bioactive metabolites potentially resulting in toxicity. The benefits of preventing the deconjugation of glucuronides with respect to reducing the toxicity of the DNA topoisomerase I inhibitor irinotecan, have provided an excellent example of this approach84–88. Irinotecan, a camptothecin, derivative, is commonly used for treating colon cancer but the dose limiting side effect is the severe diarrhoea caused by exposure of the gut following the hydrolysis of an otherwise inactive glucuronide conjugate of a metabolite (“SN-38”) of the drug. An assessment of the damage to the GI tract in rats exposed to the drug was found to correlate with the β-glucuronidase activity present. Reduction of this glucuronidase activity by the use of antibiotics administered via the drinking water not only reduced the diarrhoea and cecal toxicity but also prevented the deconjugation of the glucuronide of 7-ethyl-10-hydroxycamptothecin. Subsequent studies84–86 showed that antibiotic treatment affected the distribution of the active metabolite, markedly reducing exposure of the large intestine tissue, without effect on the parent drug, and completely inhibiting the deconjugation of the 7-ethyl-10-hydroxy-camptothecin glucuronide in the luminal contents. In an exploration of this phenomenon the effects of various treatment regimens, including using a preparation (“TJ-14”) containing the β-glucuronidase inhibitor biacalin and antibiotic administration, were examined to minimize toxicity in the rat model86. This study found that dosing antibiotic mixtures composed of either streptomycin/penicillin or neomycin/bacitracin almost completely eliminated fecal β-glucuronidase activity, whilst TJ-14 administration also had similar effects in moderating weight loss and delaying the drug-associated diarrhoea. Dosing animals with activated charcoal had lesser, but still significant, effects on toxicity. In addition, experiments on tumour-bearing rats using TJ-14, neomycin/bacitracin, and charcoal reduced intestinal toxicity but did not reduce efficacy. In contrast the administration of either the P-glycoprotein and cMOAT/MRP2 inhibitor of cyclosporin A or the UDP-glucuronosyltranferase inhibitor valproic acid increased intestinal toxicity, though not at the expense of the drugs’ efficacy. These results clearly highlighted a number of various ways in which the intestinal toxicity of irinotecan might be modulated to the benefit of patients by reducing the exposure of the gut to its deconjugated and toxic metabolite. Another study examined the in vitro effects of antibiotics (levofloxacin, streptomycin, ampicillin and amoxicillin/clavulanate) on the deconjugation of the glucuronide of SN-38 by bacterial β-glucuronidase86. Ciprofloxacin, enoxacin, gatifloxacin, but not the other antibiotics, inhibited the conversion of the SN-38-G glucuronide to the aglycone. In the same study incubation with phenolphthalein-β-D-glucuronide, used as a typical β-glucuronidase substrate, also reduced the deconjugation of the SN-38-G glucuronide, and it was presumed that this was by competitive inhibition86.
The benefits of reducing irinotecan-induced toxicity by preventing glucuronide hydrolysis have been elegantly demonstrated by the synthesis of a specific inhibitor of bacterial glucuronidase. One of these, (1-((6,8-dimethyl-2-oxo-1,2-dihydroquinolin-3-yl)-3-(4-ethoxyphenyl)-1-(2-hydroxyethyl)thiourea)87.also termed “Inhibitor 1” was shown to be highly effective in abolishing irinotecan-induced toxicity in mice. A subsequent study88 showed that the use of the inhibitor in mice had no effect on the PK of either irinotecan or its metabolites. This investigation also obtained the crystal structures of the β-glucuronidase’s found in the bacteria S. agalactiae, C. perfringens, E. coli and characterized the B. fragilis enzyme. The study demonstrated that whilst these β-glucuronidase’s were structurally similar there were significant differences in their catalytic properties and susceptibility to inhibition. Following this ground breaking innovation an alternative approach that looked at the properties of a library of existing, US FDA-approved, drugs determined that five therapeutic compounds were also inhibitors of purified bacterial β-glucuronidase89. These included the monoamine oxidase inhibitors nialamide, isocarboxazid, and phenelzine the tricyclic antidepressant amoxapine and the antimalarial drug mefloquine. Of these the drugs nialamide, isocarboxazide, and amoxapine were seen to have no significant activity against purified mammalian β-glucuronidase but were active in an assay that employed E. coli. The authors suggested that these three drugs could be “repurposed” to reduce irinotecan toxicity. In a follow on study90 the interaction of amoxapine and its metabolites 7-hydroxyamoxapine and 8-hydroxyamoxapine, together with a control drug loxapine, with bacterial β-glucuronidase were modelled using computational methods (docking and molecular dynamics simulation). This work indicated that both amoxapine and its metabolites could bind to the active site of the bacterial glucuronidase and this was also demonstrated by enzyme and cell based assays against E. coli β-glucuronidase and live E. coli cell-based assay. Further, the administration of amoxapine to tumor-bearing mice treated with irinotecan also resulted in reduced toxicity90.
Irinotecan is not the only therapeutic agent to produce inactive glucuronides that, on hydrolysis, release aglycones capable of inducing gut toxicity. The approach of β-glucuronidase inhibition therefore has more general applications than controlling irinotecan toxicity, and was shown to be very effective in eliminating the small intestinal injury caused by non-steroidal anti-inflammatory drugs (NSAIDs)91,92. The structures of many NSAIDs includes a carboxylic acid moiety and this is frequently the site for metabolism via the formation of acyl (ester) glucuronides which, once formed, are often excreted via the bile. Once in contact with the gut-microbiota these ester glucuronides are rapidly hydrolysed to release the aglycone and it has been demonstrated in animals that the toxic action of the liberated NSAID is responsible for damage to the intestinal mucosa. As with irinotecan-induced toxicity these adverse side effects are no longer seen following the administration of the bacterial glucuronidase inhibitor as shown in studies with diclofenac91, indomethacin and ketoprofen92 in mice. These effects on reduced gut toxicity could still be seen if the inhibitor was dosed sometime after diclofenac itself had been dosed91. Such data clearly point to the bacterial β-glucuronidase-mediated cleavage of glucuronides to liberate the NSAID (and/or bioactive metabolites) as cause of the observed enteropathy.
The potential benefits for this type of approach to reducing drug-related toxicity are clear and discovery of specific inhibitors of bacterial β-glucuronidase remains an active area of research with further recent reports of the use of virtual screening to identify specific inhibitors of this enzyme93.
An interesting microbiome-driven drug-drug interaction, with serious consequences for the patient, has been highlighted in studies on sorivudine an antiviral, used to treat infections of varicella-zoster virus and herpes simplex virus type 194. The drug (1-β-D-arabinofuranosyl-5-(E)-(2-bromovinyl) uracil) is metabolised by gut bacterial phosphorolytic enzymes to (E)-5-(2-bromovinyl) uracil (BVU), with high hydrolytic activity seen in the contents of the large intestine and caecum of the rat95. High activity for the conversion of the drug to BVU was found for the Bacteroides species B. vulgatus, B. thetaiotaomicron, B. fragilis, B. uniformis and B. eggerthii. Treating rats with the antibiotics ampicillin, metronidazole or a cocktail of bacitracin, neomycin and streptomycin) resulted in low concentrations of BVU in the circulation whereas they were elevated on administration of kanamycin (which is selective for aerobes over anaerobes). Based on these data it appeared that BVU was the result of hydrolysis by anaerobic bacteria, particularly species of Bacteroides. Where the production of BVU becomes problematic is if the drug is co-administered with the anticancer drug 5-fluorouracil (5-FU), or prodrugs of such as tegafur. In such circumstances 5-FU is seen to accumulate in the systemic circulation with increased toxicity, including death, as a consequence. The enhanced exposure of patients to 5-FU appears to be the result of the inactivation by BVU of the hepatic enzyme dihydropyrimidine dehydrogenase (DPD) which would otherwise inactivate 5-FU.
Microbial Processing of Xenobiotic Glutathione Conjugates
Many xenobiotics, drugs, agrochemicals, natural products and industrial chemicals are subject to metabolism, generally through P450-related biotransformations, resulting in the formation of reactive, and potentially toxic, metabolites to varying degrees. The glutathione conjugates formed in during the detoxication process in the liver are subsequently excreted in the bile where the gut microflora acts on them through bacterial C-S-lyases. Studies (on agrochemicals)96 have demonstrated the formation of large numbers of metabolites resulting from metabolism of the glutathione moiety with, in cases, the glutathione conjugate reduced to a free thiol group on the drug. The formation of such a downstream thiol metabolites of acetaminophen in this way was then followed by methylation by the host to give the methylthio adduct of the drug97. Indeed, even the regeneration of the parent compound itself from its glutathione conjugate has been described 96.
Bacterial acetylation
Reports of conjugation reactions performed by the gut microbiota are comparatively rare but not completely unknown. Both N- and O-acetylation by bacterial N-acetyl transferases (NATs) have been shown, with the former highlighted as potentially important for the bioactivation of genotoxic aromatic amines98,99. In addition the conversion of 5-aminosalyclic acid to N-acetyl-5-aminosalicylic acid by bacterial N-acetylation activity was demonstrated for a number of species100,101. 4-Aminosalicylic acid was less efficiently acetylated and p-aminobenzoic acid was a poor substrate. With respect to both substrate spectrum and catalytic efficiency Pseudomonas aeruginosa was seen to be the most efficient at performing this reaction of the 11 species investigated102. This N-acetylation may be important given the suggestion that the pancreatitis sometimes observed in children following treatment with olsalazine or sulfasalazine, both 5-aminosalyclic acid-producing drugs, may result from the toxicity due to N-acetyl-5-aminosalycylic acid102. In addition to the production of 5-aminosalyclic acid the metabolism of sulfasalazine produces both 5-aminosalyclic acid and sulfapyridine. The latter is also a substrate for bacterial N-acetylation, resulting in the formation of N-acetylsulfapyridine (together with the aforementioned N-acetyl-5-aminosalycylic acid), via the gut microbiota of species such as rat, guinea pig, dog and humans101.
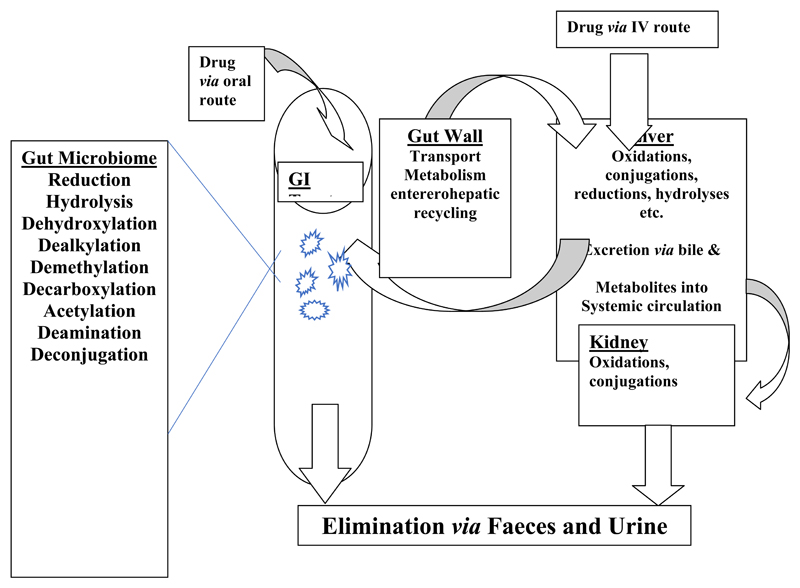
It will be clearly from the above that the gut microbiota are capable of making a wide range of biotransformations to synthetic drugs (see Figure 1 for a schematic representation) and those described here are summarised in Table 1. It is however, very likely that the extent of gut microbial metabolism is underestimated as this aspect of drug biotransformation is not routinely investigated.
Figure 1.
Sites and types of metabolism for drugs following oral or intravenous administration.
Table 1.
Biotransformation of Drugs/Drug Metabolites Performed by the Gut Microbiota*
| Biotransformation | Drug/Metabolite | Comments | Ref |
|---|---|---|---|
| Reduction | |||
| Balsalazide | Azo bond reduction | 22 | |
| Bromezepam | Nitro-reduction | 26 | |
| Clonazepam | Nitro-reduction | 25 | |
| Chloramphenicol | Nitro-reduction | 30 | |
| Digoxin | Double bond reduction | 47–54 | |
| Eltrombopag | Hydrazone cleavage | 37 | |
| Ipsalazide | Azo bond reduction | 22 | |
| levosimendan | Hydrazone cleavage | 35–36 | |
| Loperamideoxide | N-oxide reduction | 43 | |
| Metronidazole | Nitro reduction | 27–29 | |
| Misonidazole | Nitro-reduction | 33 | |
| Neoprontosil | Azo bond reduction | 18 | |
| Nitrazepam | Nitro-reduction | 24 | |
| Nizatidine | N-oxide reduction | 45 | |
| Olsalazine | Azo bond reduction | 22 | |
| Omeprazole | Sulphoxide reduction | 39 | |
| Potassium 1,2,3,4-tetrahydro-2,4-dioxo-1,3,5-triazine-6-carboxylate (potassium oxonate) | 46 | ||
| Prontosil | Azo bond reduction | 17,18 | |
| Ranitidine | N-oxide reduction | 44 | |
| Risperidone | benzisoxazole ring reduction | 42 | |
| Sulfasalazine | Azo bond reduction | 19,20 | |
| Sulfinpyrazone | Sulphoxide reduction | 38 | |
| Sulindac | Sulphoxide reduction | 38 | |
| Zonisamide | Benzisoxazole ring reduction | 40,41 | |
| Hydrolysis | |||
| azetirelin | Proteolysis | 76,77 | |
| calcitonin | Proteolysis | 74 | |
| Diclofenac glucuronide | Hydrolysis to diclofenac | 91,92 | |
| indomethacin glucuronide | Hydrolysis to indomethacin | 92 | |
| insulin | Proteolysis | 74–75 | |
| Irinotecan metabolite SN-38 glucuronide | Glucuronide hydrolysis | 83–90 | |
| Ketoprofen glucuronide | Hydrolysis to ketoprofen | 92 | |
| methotrexate | Production of 4-amino-4-deoxy-N10 -methylpteroic acid | 72–73 | |
| sodium picosyulphate, | Desulfation to 4,4'-dihydroxy -diphenyl-(2 pyridyl)-methane | 78 | |
| Sorivudine (1-beta-D-arabinofuranosyl-5-(E)-(2-bromovinyl)uracil) | Hydrolysis to (E)-5-(2-bromo -vinyl)uracil | 94 | |
| Deacylation | |||
| bucetin | Formation of phenitidine | 56 | |
| Phenacetin, | Formation of phenitidine | 56 | |
| Acetaminophen (paracetamol) | Formation of p-aminophenol | 56 | |
| Demethylation | |||
| methamphetamine | N-Demethylation | 55 | |
| 4’-hydroxy methamphetamine | N-Demethylation | 55 | |
| O-Dealkylation | |||
| Fostamatinib | O-Demethylation of the metabolite R529 | 57 | |
| Dehydroxylation | |||
| Fostamatinib | Dehydroxylation of the metabolite R529 | 57 | |
| L-Dopa (levodopa, L-3,4-dihydroxy-phenylalanine). | Dehydroxylation | 60,61 | |
| Decarboxylation | |||
| L-Dopa (levodopa, L-3,4-dihydroxy-phenylalanine) | 64,65 | ||
| Deamination | |||
| 5-Fluorocytosine | Deamination to 5-fluorouracil | 58,59 | |
| Oxidation | |||
| Levamisole | Thiazole ring-opening | 69 | |
| Lovastatin | Hydroxylated metabolites | 71 | |
| Acetylation | |||
| 5-Aminosalicylic acid | Production of N-acetyl-5-amino salicylic acid | 100–103 | |
| Sulfapyridine | Production of N-acetyl -sulfapyridine | 101 |
This table represents a summary of the examples discussed in this review and is not designed to be, nor is it, comprehensive as e.g., any drug (or its metabolites) secreted into the bile as a sulfate or glucuronide will be liable to deconjugation by the gut microbiota.
Microbiome-conditional Effects and Consequences.
The direct effects that the gut microbiota can exert on the metabolism and toxicity of drugs, their metabolites and related xenobiotics summarised above in all likelihood represents only the tip of the iceberg as the contribution made by this forgotten organ is not routinely assessed. However, the influence of the gut microbiota extends beyond these direct effects and a number of indirect mechanisms, whereby the microbiome affect the metabolism, disposition and (potentially) the toxicity of xenobiotics, have been identified. And, whilst our knowledge of these is even more limited it is clear from the indirect effects that have been described so far in the literature, that the effects of the gut microbiota may include the modulation of host metabolic enzymes/ transporters, direct competition for metabolism via particular host metabolic routes/enzymes and enhancement of toxicity as a result of other effects on host biochemistry.
Competition
The host organism has to pay a price for the benefits accruing from having an active and healthy gut microbiota and one of these is the need to detoxify and dispose of myriad microbial waste products. Indeed it is arguable that one (of many) factors resulting in the development of the range of host xenobiotic metabolising systems that we see today was the need to eliminate unwanted microbiota-derived metabolites such as e.g. ethanol, benzoic acid and p-cresol etc. Indeed the detoxication and removal of p-cresol has recently been shown to have direct consequences for the metabolic fate of acetaminophen (paracetamol)104. The production of p-cresol by Clostridia 105 during the metabolism of tyrosine and phenylalanine is potentially damaging to the host in two ways. Firstly, as an aromatic phenol, the preferred means for the metabolism of p-cresol is via sulfation. However, when large amounts of phenolic compounds are present the limited capacity of sulfation results in their glucuronidation and, when this is no longer effective then may lead to oxidative metabolism via CYP450s. The oxidative metabolism of any p-cresol that evades conjugation results in the formation of highly reactive metabolites (RMs) that are subject to detoxication via conjugation/reaction with glutathione. The RMs formed by this the P450-mediated metabolism consist of both a quinone methide (CYP2D6, 2C19, 1A2, 1A1, and 2E1) and, in a recently described alternative route of bioactivation, aromatic oxidation (mainly via CYP2E1 but with a contribution from P450s such as CYP1A1, 1A2, and 2D6) to a 4-methyl-O-hydroquinone that is further oxidized to 4-methyl-[1,2]benzoquinone106. And, although this will likely place some stress on the glutathione system, under normal circumstances the formation of these RMs should not represent a problem for the host. However, in cases where the host is subject to a high baseline load of p-cresol, there will be direct competition between it and other phenols for both sulfation and glutathione conjugation. Unfortunately, as with sulfation, the capacity of glutathione conjugation is limited and its depletion by p-cresol will potentially reduce the ability of the host to detoxify other phenols, including drugs such as acetaminophen, with potentially adverse consequences. It is therefore to be expected that exposure to both by p-cresol and RM-forming phenolic drugs, such as acetaminophen, will result in enhanced toxicity due to competition for glutathione-dependent detoxication. As indicated above, some evidence in support of this hypothesis is provided via studies on acetaminophen104 in humans where, after consumption of 1 g of the drug, it was seen that the glucuronide to sulfate conjugate ratio of the drug present in urine was clearly affected by competition for sulfation by p-cresol.
Modulation
Whilst there is no evidence that acetaminophen itself is metabolised to any great extent by the gut microbiota (although N-acetylated drugs such as phenacetin and acetaminophen etc., have been shown to be deacetylated to some extent in vitro56) effects on the pharmacokinetics of the drug in rats dosed orally with the drug have been reported following antibiotic treatment. Rats, administered bacitracin, streptomycin and neomycin to eliminate the gut microbiota107, together with control animals, were dosed with acetaminophen and concentrations of drug and six metabolites in the plasma determined via LC-MS/MS. In the antibiotic treated animals the AUCs of the drug and its glutathione conjugate were higher than those of the controls whilst, in contrast, the ratio of the AUC of the sulfate conjugate to acetaminophen was lower. Such effects may have resulted from a range of factors including the modulation of the of the xenobiotic metabolizing enzyme systems of the host. Indeed changes in the drug metabolizing capabilities of gut and liver have been reported with effects on the expression of e.g., CYPs and conjugating enzyme systems. For example, microbiome-driven effects obtained by comparing hepatic preparations from germ free and microbiota-containing rats revealed differences in the expression of P450s capable of bioactivating of mutagenic heterocyclic aromatic amines108. Effects on acetaminophen toxicity have also been noted as a result of host exposure to gut microbial metabolites derived from dietary components. Recently one such metabolite, 3,4-dihydroxyphenylacetic acid (a metabolite of e.g., quercetin109) was shown to have protective effects on acetaminophen-induced liver in the mouse following intragastric administration110. The mechanism for this 3,4-dihydroxyphenylacetic acid-related hepatoprotective effect was suggested as being related to increased nuclear factor erythroid 2-related factor 2 (Nrf-2) translocation to the nucleus and expression of enzymes responsible for glucuronidation, sulfation and glutathione synthesis/ metabolism). Modulation of drug metabolizing enzymes following the colonisation of germ free mice, using either specific strains of bacteria or microbiota from conventionally raised mice, has been shown via DNA microarray analysis. This study highlighted a range of responses for xenobiotic metabolizing systems in the intestine. In the case of animals colonized with B. thetaiotaomicron111 decreases in the xenobiotic metabolizing enzymes glutathione S-transferase (GST) and CYP2D2 were noted, together with the transporter protein Mdr 1a. Conversely, the use of E. coli and B.infantis were associated with increased expression whilst a conventional gut microbiota produced no change. Further studies have shown that the presence or absence of the gut microbiota influences liver gene expression112. A comparison of germfree and control mice detected some 112 genes, many of which were related to xenobiotic metabolism, that were differentially expressed between them. Administration of pentobarbital to mice of both types showed that its metabolism, as measured by length of anaesthesia, was significantly more efficient in the germ free animals. Toda T, et al113 studied the effects of the intestinal microbiota on hepatic CYP P450 and CYP mRNA expression for control and germ-free (GF) mice finding that the major CYP isozymes were lower in the livers of the former. This higher CYP expression in the control animals was correlated with higher expression of nuclear factors such as the pregnane-receptor (PXR) and constitutive androstane receptor (CAR), transporters and conjugation enzymes involved in the detoxication of the bile acid lithocholic acid. The authors postulated that these differences came about because, in mice with gut microbiota, exposure to lithocholic acid caused the activation of PXR and CAR thereby increasing CYP expression. The gut microbial metabolism of tryptophan and indole has been highlighted as providing ligands for the aryl hydrocarbon receptor AHR114–116 and the metabolite indole 3- propionic acid has also been demonstrated to act as a ligand for PXR115.
The effects of the presence or absence of the gut microflora on the metabolism of steroids has been known for many years117 and indeed studies on liver microsomes have shown that the hydroxylation of a range of sterols was up to twice as efficient in germ-free animals compared to conventional rats. This was associated with greater amounts of cytochrome P-450 present in germ-free animals (2.53 + 0.45 nmol/ mg of protein) compared to conventional animals (1.72 + 0.04 nmol/ mg of protein). However, reduced activity compared to conventional animals was also seen in germ free rats which the authors considered to be “in accordance with the slower cholesterol and bile acid turnover in germ-free compared to conventional rats”. Gut microbial-conditional effects resulting from the biotransformation of Soy to produce endocrinologically active phytoestrogens have been demonstrated to affect host endogenous steroid118 metabolism. These effects were suggested as perhaps being the result of changes in expression of the estrogen-hydroxylating CYPs, leading to changes in the amounts of 4-hydroxyestrogen (reduced) and 2-hydroxyestrogen (increased) excreted by post-menopausal women118. Given the widespread effects of hormones on the regulation of metabolizing enzymes wider effects on drug metabolism would also be anticipated.
Effects on hepatic Cyp8b1 expression and the subsequent alteration of bile acid profiles, including effects on taurocholate and tauromuricholate, were detected using a metabonomic approach during the time course of the colonization of axenic mice119. In addition the expression and activity of both Cyp3a11 and Cyp2c29 were also increased. When these data were subjected to statistical modelling of hepatic metabolite profiles and microbial composition (based on 16S RNA gene pyrosequencing) strong associations for the Coriobacteriaceae family with hepatic triglyceride, glucose, and glycogen levels and the metabolism of xenobiotics were observed119.
In a study in male and female germ free and rats recolonised with the microbiota of normal animals (or humans) the impact of the presence or absence gut microbiota on intestinal and hepatic xenobiotic conjugating enzymes in the rat was investigated 120. Effects were noted on the glutathione transferases (GSTs), glutathione peroxidase (GPX2), epoxide hydrolases (EPHXs), N-acetyltransferases (NATs) and sulphotransferases (SULTs), with the sex of the animals also representing a variable in some cases. Thus hepatic SULT1A1, SULT1C1, and SULT1C2 were seen to be elevated in germ-free animals in both male and females (1.5- to 2.6-fold) whilst for SULT1B1 and SULT1C2 the increases were 0.4/0.6 and 1.3/1.6-fold respectively. NAT2 was 1.4/1.5-fold higher for male and female germ-free rats. Similarly GSTA1/2 were elevated 4.0/5.0-fold, GSTA4 between 1.5/1.9-fold and GSTM1 1.1/1.5-fold in male and female germ-free animals respectively compared to controls. The epoxide hydrolases, EPHX1 and EPHX2 were 3.5/2.4 and 1.4/2.1-fold higher in male and female germ free rats respectively. Some enzymes showed organ–specific, or regional, expression with e.g., NAT2 only detected in the large bowel and the SULTs expressed in liver and large intestine but absent from the large intestine. Recolonization with human gut microbiota resulted in smaller effects on the expression of these enzymes in the colon compared to the use of the gut microbiota of rat. Effects on glucuronidating (UGT) and GST enzymes were seen in germ-free and human gut microbiota colonised rats dosed with (+)-catechin or (-)-epicatechin (with humanized rats showing reduced CYP2C11 expression compared to germ-free animals,)121.
Disease, and the presence or absence of a gut microflora, has been shown to modulate the distribution of alpha, mu, and pi class glutathione GSTs in the colons of conventional and germ-free (GF) mice with induced experimental colitis122.
Most recently a study examined the effects on host hepatic drug metabolizing enzymes of colonisation of germ free mice and normal mice with probiotics and exogenous bacteria123. Five groups of mice were studied including conventional mice, germ free mice, germ free mice exposed to colonization by environmental exposure for 2 months and two groups composed of conventional and germ free mice administered a probiotic containing 8 strains of bacteria. In the case of germ free animals the Cyp3a genes were down regulated and the Cyp4a cluster was upregulated. Changes in the Cyp3a expression correlated with alterations in PXR expression, whilst peroxisome proliferator-activated receptor α-DNA binding correlated with that of Cyp4a gene expression. Conventional mice administered the probiotic responded with an increase in the amounts of the mRNAs for Cyp4v3, alcohol dehydrogenase 1, and carboxyesterase 2a, combined with a decrease in the mRNAs for a number of glutathione-S-transferases whereas the response of germ-free animals was a decrease in the mRNAs of UDP-glucuronosyltransferases 1a9 and 2a3.
The modulation of the activity of drug metabolizing enzymes/transporters, via induction, inhibition or competition for individual drug metabolizing pathways clearly has the potential to result in drug-microbiome interactions (DMI’s) which may be either beneficial or injurious to the host. Such factors may eventually be found to be significant variables in patient outcomes with consequences for the practice of personalized medicine and the minimization of adverse drug reactions, particularly “idiosyncratic” drug toxicity.
Drug effects on the Gut Microbiota
Given the interplay between host and gut microbiota permanent changes in the composition of the latter resulting from drug treatment may have important long term consequences for the host. So, in considering the effects of the microbiota on drug metabolism there is also the need to consider the potential for the administration of drugs to radically alter its composition either directly on the microorganisms themselves, or as a result of toxic or pharmacological effects on the gut. Clearly the most obvious category of drugs to impinge on the microbiota are antibiotics and, whilst it is not possible here to fully review the topic, many studies have shown antibiotic administration to have both short and long term effects on its composition in animals and humans. In particular, incomplete recovery of the microflora in response to repeated exposure to ciprofloxacin has been shown for the distal gut microbiota of humans124. Whilst these observations were based on a relatively small sample (3 volunteers) the effect of antibiotic administration on the gut microbiota was “profound” with a rapid decrease in diversity and changes in community composition taking place within a few days of beginning administration. Although, following cessation of dosing, the microbiota recovered somewhat, this recovery was often incomplete. And, whilst the changes observed in bacterial communities in response to ciprofloxacin was noted as being broadly similar it differed between both subjects and between the two courses of antibiotic treatment and, at the end of the experiment the composition of the gut microbiota was different from what it had been at the start. As the authors noted, “Antibiotic perturbation may cause a shift to an alternative stable state, the full consequences of which remain unknown124.” One obvious consequence is of course the selection and persistence of antibiotic resistance in the gut and studies have revealed both ecological disturbances in the human gut microbiota after antibiotic administration and the long-term persistence of antibiotic resistance genes125.
As discussed above the presence or absence of the gut microbiota appears to have effects on CYP expression related to the lithocholic acid exposure to the host113. In a study on the effects of ciprofloxacin126 Cyp3a expression was suppressed in mouse liver by reducing lithocholic acid-producing intestinal flora. The authors noted that hepatic Cyp3a11 expression and triazolam metabolism were significantly reduced by treating SPF mice with the antibiotic, but that such changes were not seen when germ-free mice were dosed. In addition there was a reduction in both lithocholic acid-producing bacteria in the feces and the amount of its taurine conjugate in the livers of the SPF mice administered ciprofloxacin. Further support for the hypothesis that these effects were driven by the production of lithocholic acid, which is known to activate both the farnesoid X and PXR receptors, was provided by the response of germ free mice that, when treated with this bile acid, showed increased expression of Cyp3a11.
Whilst the effects of antibiotics on the gut microbiota, if unwanted, are hardly unexpected the increasing evidence that the very widely used proton pump inhibitors (PPI) cause changes in the microbiota (apart from those that can be anticipated for H. pylori), including reducing diversity, perhaps represents a less obvious consequence of therapy. However, numerous studies (of which a selection is given here) have associated PPI use and C. difficile incidence as well as changes in the ecology of the gut microbiota128–131. In a small scale study128 the use of these drugs resulted in decreases to observed operational taxonomic unit (OTU) counts after both 1 week and 1 month of dosing. These effects were partly reversible after a 1 month recovery period, supporting the hypothesis that PPIs disrupt the healthy human gut microbiome, and were suggested as a potential explanation for the association between prolonged PPI usage and the incidence of C. difficile. A much larger study, that examined fecal samples obtained from 1827 healthy twins, also revealed effects of PPIs on the gut microbiota129 showing significantly lower abundance and microbial diversity in those treated with such drugs. Concomitantly, there was a significant increase in the abundance of oral and upper GI tract species in fecal samples. These observations were confirmed by an independent interventional study and a paired analysis between 70 monozygotic twin pairs discordant for PPI use. These findings indicated a significant impact of PPIs on the gut microbiome and led the authors to caution against their over-use129.
In a further large scale in humans the effect of PPI use on the gut microbiota was undertaken on some 1815 individuals, in three cohorts, with 211 of the subjects using PPIs at the time of stool sampling130. PPI use was found to be associated with a significant decline in diversity, with changes in 20% of the bacterial taxa. As with the other large scale study described above129 species of oral bacteria were seen to be over-represented in the faecal microbiome of PPI-users. The authors suggested that the differences resulting from PPI use were “consistently associated with changes towards a less healthy gut microbiome” and in line with changes predisposing users to infection with C. difficile. On a population level, the effects of PPI were considered to be more prominent than the effects of antibiotics or other commonly used drugs.
A comparison of the faecal microbiomes of 32 of subjects with ≥5 years of continuous PPI use, compared with 29 non-users, found that changes in bacterial populations had occurred at the both species and phylum level with, in the case of the latter, decreased Bacteroidetes and increased Firmicutes131. The authors suggested that this alteration in the Firmicutes: Bacteroidetes ratio might pre-dispose PPI-treated subjects to C. difficile infection.
Another class of widely used compounds with a clear ability to affect gut physiology via toxicity, as described above, is composed of the NSAIDS. Various effects of exposure to NSAIDs on the composition of the gut microbiota have been described some of which are considered below132–134. An examination of the effects of age and administration of NSAIDs on the intestinal microbiota in a group of subjects aged between 70 and 85 years compared to that of much younger individuals (mean age 28yr) found “remarkable changes” 132 in composition. In terms of age-related differences it was found that the overall number of microbes was reduced in elderly compared to younger subjects but, interestingly, was higher in the elderly NSAID users compared to non-users of the same age group. Whilst many changes seemed to be associated with age the authors noted that the Actinobacteria group showed a reduction in Collinsella spp. in elderly subjects using NSAIDs in comparison to both the non-users and young adults. Similarly, the numbers of Lactobacilli seen the elderly NSAIDs users was reduced compared to non-users, leading the authors to suggest “that the use of NSAID along with age may also influence the composition of intestinal microbiota”. In a separate study133 the effect on the gut microbiota of exposure to NSAIDs was examined in a group of over 150 subjects. It was noted that the type of NSAID being used by these individuals had a significant influence on the composition of the gut microbiota with individual NSAIDs associated with distinct microbial populations. Thus the investigators found that aspirin users could be discriminated from those taking no medication via four OTUs namely Prevotella sp., Bacteroides sp., family Ruminococcaceae, and Barnesiella sp.), whilst the bacterial profiles seen for celecoxib and ibuprofen users both showed enrichment in the Acidaminococcaceae and Enterobacteriaceae. In the case of ibuprofen users the families Propionibacteriaceae, Pseudomonadaceae, Puniceicoccaceae and Rikenellaceae also showed greater abundance compared to either non-users or those taking naproxen. Individuals taking a combination of NSAIDs and proton-pump inhibitors differed from those taking only NSAIDs in species of Bacteroides and Erysipelotrichaceae. Further, Bacteroides species and a bacterium of family Ruminococcaceae differed between those only taking NSAIDs and those combining them with antidepressants and laxatives from those using NSAIDs alone. The authors concluded from this investigation, not unreasonably, that “bacteria in the gastrointestinal tract reflect the combinations of medications that people ingest”132.
An investigation of the interactions between the microbiota and the NSAID indomethacin at “clinically relevant doses” in mice, using both acute and chronic exposures, resulted in damage to the intestine described as “reminiscent of the upper and lower GI complications induced by NSAIDs in humans”134. Dosing with indomethacin was also associated with alterations in the intestinal microbiota in these mice, particularly expansion of pro-inflammatory bacteria. When treated with antibiotics changes, in both the pharmacokinetics and pharmacodynamics of the drug, were noted that were ascribed to the prevention of glucuronide hydrolysis by bacterial β-glucuronidases (as would be anticipated based on the results of the inhibition of this enzyme described earlier91,92). Given that both PPI’s and NSAID’s have been shown to alter the composition of the gut microbiota it is hardly surprising that the use of these drugs on combination has been the subject of increasing interest.
A recent review135 on the topic of combined PPI and NSAID use concluded that, whilst PPIs were effective as a means of reducing damage to the stomach resulting from NSAID use they were “without proven benefit in preventing NSAID-related damage in the rest of the GI tract” and that the “frequent use of PPIs can exacerbate NSAID-induced small intestinal injury by altering intestinal microbiota”. Positive benefit has been seen from the use of probiotics in the prevention of NSAID-induced damage in patients receiving PPI and NSAIDs136.
Clearly, the use of therapeutic drugs that are designed to directly act on bacteria such as antibiotics, or those that the affect gut physiology via intended pharmacology, e.g., PPI’s, or accidentally through unintended toxicity, thereby causing intestinal damage, including the NSAID’s, have an obvious potential to result in changes to the environment that lead to compositional changes in the gut microbiota. It is however, less clear what the effects of other drugs might be on the biochemistry of the gut microbiota. Recent studies in mice137 have shown significant changes to the physiology, structure, and gene expression of the active gut microbiome following short-term exposure to a panel of xenobiotics (which included antibiotics). A range of bacterial genes were found to respond to drug exposure across, with changes seen in e.g., those responsible for antibiotic resistance, drug metabolism and response to stress. These effects were seen across a range of phyla. The authors suggested that the “results demonstrate the power of moving beyond surveys of microbial diversity to better understand metabolic activity, highlight the unintended consequences of xenobiotics, and suggest that attempts at personalized medicine should consider interindividual variations in the active human gut microbiome”137. Certainly, in e.g., the light of the differential responses of the microbiome seen for the various NSAIDs described above132, it would be of great interest to see this type of study expanded to cover a larger number of compounds and therapeutic classes.
Summary
The range of effects that the gut microbiota can have on drugs, and vice versa, have obvious implications for drug toxicity testing, where differences in outcome may reflect not only strain of animal but microbiome-specific effects. Clearly, when moving from animals to patients such effects also have the potential to produce unexpected, and potentially unwelcome, variability in response to the administration of drugs to both individual patients and populations. The resurgence in interest in this, no longer, “forgotten organ” of metabolism is however, promising and may lead to a reversal of the current situation where little real consideration is given to the gut microbiota and its effects on drug absorption, disposition, metabolism, pharmacology or toxicity in either the drug discovery or drug development phases of research programs. Currently there is little evidence that regulatory bodies are aware of the potential importance of the gut microbiome and this may, potentially, represent something of an oversight. However, this situation may change as we obtain a better understanding of these complex interactions which, in our view, could provide novel insights for drug discovery and development and significant benefits for personalized medicine. The microbiome undoubtedly represents a “drugable target”, and there is no doubt that it is possible to modulate both its composition and metabolic activity. What is less clear is what represents an appropriate “drugable” target, and what the effects of drugging the microbiome might have on the overall composition the gut microbiota and the downstream consequences for both it and the host. Irrespective of this, with respect to drug efficacy and toxicity, the potential for these microorganisms to affect ADMET clearly deserves increased awareness and attention from the drug metabolism community.
Acknowledgments
The authors confirm that after reading the journal's policy on disclosure of potential conflicts of interest they have none to declare and in addition they confirm that they have read the journal's authorship agreement and that they have reviewed and approved the manuscript.
References
- 1.Nicholson JK, Holmes E, Lindon JC, Wilson ID. The challenges of modelling mammalian biocomplexity. Nature Biotechnology. 2004;10:1268–74. doi: 10.1038/nbt1015. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–7. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 3.Holmes E, Kinross J, Gibson GR, Burcelin R, Jia W, Pettersson S, et al. Therapeutic Modulation of Microbiota-Host Metabolic Interactions. Science Translational Medicine. 2012;4:137. doi: 10.1126/scitranslmed.3004244. [DOI] [PubMed] [Google Scholar]
- 4.Illing HPA. Techniques for microfloral and associated metabolic studies in relation to the absorption and enterohepatic circulation of drugs. Xenobiotica. 1981;11:815–30. doi: 10.3109/00498258109045319. [DOI] [PubMed] [Google Scholar]
- 5.Boxenbaum HG, Bekersky I, Jack MJ, Kaplan SA. Influence of gut microflora on bioavailability. Drug Met Rev. 1979;9:259–79. doi: 10.3109/03602537908993894. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson JK, Wilson ID. Understanding 'global' systems biology: metabonomics and the continuum of metabolism. Nat Rev Drug Discov. 2003;2:668–76. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- 7.Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nature Reviews Microbiology. 2005;3:431–8. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 8.Haiser HJ, Turnbaugh PJ. Is It Time for a Metagenomic Basis of Therapeutics? Science. 2012;336:1253–5. doi: 10.1126/science.1224396. [DOI] [PubMed] [Google Scholar]
- 9.Saad R, Rizkallah MR, Aziz RK. Gut Pharmacomicrobiomics: the tip of an iceberg of complex interactions between drugs and gut-associated microbes. Gut Pathol. 2012;4 doi: 10.1186/1757-4749-4-16. Published online 2012 Nov 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang MJ, Kim HG, Kim JS, et al. The effect of gut microbiota on drug metabolism. Expert Opinion Drug Met Tox. 2013;9:1295–308. doi: 10.1517/17425255.2013.807798. [DOI] [PubMed] [Google Scholar]
- 11.Ursell LK, Knight R. Xenobiotics and the Human Gut Microbiome: Metatranscriptomics Reveal the active players. Cell metabolism. 2013;17:317–8. doi: 10.1016/j.cmet.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Jia W. Cometabolism of microbes and host: Implications for drug metabolism and drug-induced toxicity. Clin Pharmacol Therapeutics. 2013;94:574–8. doi: 10.1038/clpt.2013.157. [DOI] [PubMed] [Google Scholar]
- 13.Jeong HG, Kang MJ, Kim HG. Role of intestinal microflora in xenobiotic-induced toxicity. Molecular Nutrition Food Res. 2013;57:84–99. doi: 10.1002/mnfr.201200461. [DOI] [PubMed] [Google Scholar]
- 10.Rachel N Carmody, Turnbaugh Peter J. Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. Clin Invest. 2014;124:4173–81. doi: 10.1172/JCI72335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klaassen CD, Cui JY. Mechanisms of How the Intestinal Microbiota Alters the Effects of Drugs and Bile Acids. Drug Met Disp. 2015;43:1505–21. doi: 10.1124/dmd.115.065698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swanson HI. Drug Metabolism by the Host and Gut Microbiota: A Partnership or Rivalry? Drug Metab Disp. 2015;43:1499–504. doi: 10.1124/dmd.115.065714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gingel R, Bridges JW, Williams RT. The role of the gut flora in the metabolism of prontosil and neoprontasil in the rat. Xenobiotica. 1971;1:143–56. doi: 10.3109/00498257109044386. [DOI] [PubMed] [Google Scholar]
- 18.Gingel R, Bridges JW. Intestinal azo-reduction and glucuronide conjugation of prontosil. Xenobiotica. 1973;9:599–604. doi: 10.3109/00498257309151548. [DOI] [PubMed] [Google Scholar]
- 19.Peppercorn MA, Goldman P. The role of intestinal bacteria in the metabolism of salicylazosulfapyridine. J Pharmacol Exp Ther. 1972;181:151–62. [PubMed] [Google Scholar]
- 20.Schroder H, Gustafsson BE. Azo reduction of salicyl-azosulphapyridine in germ free and conventional rats. Xenobiotica. 1973;3:225–31. doi: 10.3109/00498257309151518. [DOI] [PubMed] [Google Scholar]
- 21.Truelove SC. Evolution of olsalazine. Scandinavian J Gastroenterology. 1988:3–6. doi: 10.3109/00365528809101538. 23s148. [DOI] [PubMed] [Google Scholar]
- 22.Chan RP, Pope DJ, Gilbert AP, Sacra PJ, Baron JH, Lennard-Jones JE. Studies of two novel sulfasalazine analogs, ispsalazide and balsalazide. Digestive Disease Science. 1983;28:609–15. doi: 10.1007/BF01299921. [DOI] [PubMed] [Google Scholar]
- 23.Rafii F, Cerniglia CE. Reduction of azo dyes and nitroaromatic compounds by bacterial enzymes from the human intestinal tract. Environ Health Perspect. 1995;103(Suppl 5):17–9. doi: 10.1289/ehp.95103s417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeno S, Sakai T. Involvement of the intestinal microflora in nitrazepam-induced teratogenicity in rats and its relationship to nitroreduction. Teratology. 1991;44:209–14. doi: 10.1002/tera.1420440209. [DOI] [PubMed] [Google Scholar]
- 25.Elmer GW, Remmel RP. Role of the intestinal microflora in clonazepam metabolism in the rat. Xenobiotica. 1984;14:829–40. doi: 10.3109/00498258409151481. [DOI] [PubMed] [Google Scholar]
- 26.Fujii J, Inotsume N, Nakanoi M. Degradation of Bromazepam by the Intestinal Microflora. Chem Pharm Bull. 1987;35:4338–41. doi: 10.1248/cpb.35.4338. [DOI] [PubMed] [Google Scholar]
- 27.Koch RL, Chrystal EJT, Beaulieu BB, Jr, Goldman P. Acetamide a metabolite of metronidazole formed by the intestinal flora. Biochem Pharmacol. 1979;28:3611–5. doi: 10.1016/0006-2952(79)90407-6. [DOI] [PubMed] [Google Scholar]
- 28.Koch RL, Beaulieu BB, Goldman P. Role of the intestinal flora in the metabolism of misonidazole. Biochem Pharmacol. 1980;29:3281–4. doi: 10.1016/0006-2952(80)90304-4. [DOI] [PubMed] [Google Scholar]
- 29.Koch RL, Goldman P. The anaerobic metabolism of metronidazole forms n-(2-hydroxyethyl)-oxamic acid. J Pharmacol Exp Ther. 1979;208:406–410. [PubMed] [Google Scholar]
- 30.Yunis AA. Chloramphenicol toxicity: 25 years of research. Amer J Med. 1989;87(3N):44N–48N. [PubMed] [Google Scholar]
- 31.Holt R. The bacterial degradation of chloramphenicol. Lancet. 1967:11259–60. doi: 10.1016/s0140-6736(67)92720-1. [DOI] [PubMed] [Google Scholar]
- 32.Jimenez JJ, Arimura GK, Abou-Khalil WH, lsildar M, Yunis AA. Chloramphenicol-induced bone marrow injury: potential role of bacterial metabolites of chloramphenicol. Blood. 1987;70:1180–5. [PubMed] [Google Scholar]
- 33.Koch R, Bealieu BB, Jr, Goldmans P. Role of the intestinal flora in the metabolism of misonidazole. Biochem Pharmacol. 1980;29:3281–4. doi: 10.1016/0006-2952(80)90304-4. [DOI] [PubMed] [Google Scholar]
- 34.Sheldon PW, Clarke C, Dawson KB, Simpson W, Simmons DJC. Intestinal microflora as potential modifiers of sensitizer activity in vivo. International Journal of Radiation Oncology*Biology*Physics. 1984;10:1371–5. doi: 10.1016/0360-3016(84)90351-1. [DOI] [PubMed] [Google Scholar]
- 35.Antila S, Huuskonen H, Nevalainen T, Kanerva H, Vanninen P, Lehtonen L. Site dependent bioavailability and metabolism of levosimendan in dogs. Eur J Pharm Sci. 1999;9:85–91. doi: 10.1016/s0928-0987(99)00048-2. [DOI] [PubMed] [Google Scholar]
- 36.Antila S, Pesonen U, Lehtonen L, et al. Pharmacokinetics of levosimendan and its active metabolite OR-1896 in rapid and slow acetylators. Eur J Pharm Sci. 2004;23:213–22. doi: 10.1016/j.ejps.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 37.Deng Y, Rogers M, Sychterz C, et al. Investigations of Hydrazine Cleavage of Eltrombopag in Humans, Investigations of hydrazine cleavage of eltrombopag in humans. Drug Metabolism and Disposition. 2011;39:1747–54. doi: 10.1124/dmd.111.040188. [DOI] [PubMed] [Google Scholar]
- 38.Strong HA, Renwick AG, George CF, Liub YF, Hill MJ. The reduction of sulphinpyrazone and sulindac by intestinal bacteria. Xenobiotica. 1987;17:685–96. doi: 10.3109/00498258709043976. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe K, Yamashita S, Furruno H, Kawasaki H, Gomita Y. Metabolism of omemprazole by gut flora in rats. J Pharm Sci. 1995;84:516–7. doi: 10.1002/jps.2600840425. [DOI] [PubMed] [Google Scholar]
- 40.Kitamura S, Sugihara K, Kuwasako M, Tatsumi K. The role of mammalian intestinal bacteria in the reductive metabolism of zonisamide. J Pharm Pharmacol. 1997;49:253–6. doi: 10.1111/j.2042-7158.1997.tb06790.x. [DOI] [PubMed] [Google Scholar]
- 41.Stiff DD, Robicheau JT. Reductive metabolism of the anticonvulsant agent zonisamide, a 1,2-benzisoxazole derivative. Xenobiotica. 1992;22:1–11. doi: 10.3109/00498259209053097. [DOI] [PubMed] [Google Scholar]
- 42.Meuldermans W, Hendrickx J, Mannens G, et al. The metabolism and excretion of risperidone after oral administration to rats and dogs. Drug Met Dispos. 1994;22:129–38. [PubMed] [Google Scholar]
- 43.Lavrijsen K, Van Dyck D, Van Houdt J, et al. Reduction of the prodrug loperamide oxide to its active drug loperamide in the gut of rats, dogs, and humans. Drug Metab Dispos. 1995;23:354–62. [PubMed] [Google Scholar]
- 44.Basit AW, Lacey LF. Colonic metabolism of ranitidine; implications for its delivery and absorption. Int J Pharmacy. 2001;227:157–65. doi: 10.1016/s0378-5173(01)00794-3. [DOI] [PubMed] [Google Scholar]
- 45.Basit AW, Newton JM, Lacey LF. Susceptibility of the H2-receptor antagonists cimetidine, famotidine and nizatidine, to metabolism by the gastrointestinal microflora. Int J Pharmacy. 2002;237:23–33. doi: 10.1016/s0378-5173(02)00018-2. [DOI] [PubMed] [Google Scholar]
- 46.Yoshisue K, Masuda H, Matsushima E, Ikeda K, Nagayama S, Kawaguchi Y. Tissue distribution and biotransformation of potassium oxonate after oral administration of a novel antitumor agent (drug combination of tegafur, 5-chloro-2,4-dihydroxypyridine, and potassium oxonate) to rats. Drug Metab Dispos. 2000;28:1162–7. [PubMed] [Google Scholar]
- 47.Lindenbaum J, Tse-Eng D, Butler VP, Jr, Rund DG. Urinary excretion of reduced metabolites of digoxin. Am J Med. 1981;71:67–74. doi: 10.1016/0002-9343(81)90260-6. [DOI] [PubMed] [Google Scholar]
- 48.Dobkin JF, Saha JR, Butler VP, Jr, Neu HC, Lindenbaum J. Inactivation of digoxin by Eubacterium lentum, an anaerobe of the human gut flora. Trans Assoc Am Physicians. 1982;95:22–9. [PubMed] [Google Scholar]
- 49.Saha JR, Butler VP, Jr, Neu HC, Lindenbaum J. Digoxin-inactivating bacteria: identification in human gut flora. Science. 1983;220:325–7. doi: 10.1126/science.6836275. [DOI] [PubMed] [Google Scholar]
- 50.Butler VP, Jr, Saha JR, Lindenbaum J. Digoxin inactivation by the gut flora in infancy and childhood. Pediatrics. 1987;79:544–8. [PubMed] [Google Scholar]
- 51.Bennet RG, Beamer BA, Greenough WB, Lindenbaum J, Bartlett JG. Colonisation with digoxin reducing strains of Eubacterium lentum and Clostridium difficile infection in nursing home patients. J Diarrheol Dis Res. 1992;10:87–92. [PubMed] [Google Scholar]
- 52.Mathan VI, Wiederman J, Dobkin JF, Lindenbaum J. Geographic differences in digoxin inactivation, a metabolic activity of the human anaerobic gut flora. Gut. 1989;30:971–7. doi: 10.1136/gut.30.7.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–8. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haiser HJ, Seim KL, Balskus EP, Turnbaugh PJ. Mechanistic insight into digoxin inactivation by Eggerthella lenta augments our understanding of its pharmacokinetics. Gut Microbes. 2014;5:233–8. doi: 10.4161/gmic.27915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caldwell J, Hawksworth GM. The demethylation of methamphetamine by intestinal microflora. J Pharm Pharmacol. 1973;25:422–4. doi: 10.1111/j.2042-7158.1973.tb10043.x. [DOI] [PubMed] [Google Scholar]
- 56.Smith GE, Griffiths LA. Metabolism of N-acylated and O-alkylated drugs by the intestinal microflora during anaerobic incubation in vitro. Xenobiotica. 1974;4:477–87. doi: 10.3109/00498257409052100. [DOI] [PubMed] [Google Scholar]
- 57.Sweeny DJ, Li W, Clough J, et al. Metabolism of Fostamatinib, the Oral Methylene Phosphate Prodrug of the Spleen Tyrosine Kinase Inhibitor R406 in Humans: Contribution of Hepatic and Gut Bacterial Processes to the Overall Biotransformation. Drug Metab Dispos. 2010;38:1166–76. doi: 10.1124/dmd.110.032151. [DOI] [PubMed] [Google Scholar]
- 58.Harris E, Manning BW, Federle TW, Diasio RB. Conversion of 5-fluorocytosine to 5-fluorouracil by human intestinal microflora. Antimicrob Agents Chemother. 1986;29:44–8. doi: 10.1128/aac.29.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vermes A, Kuijper EJ, Guchelaar HJ, Dankert J. An in vitro study on the active conversion of flucytosine to fluorouracil by microorganisms in the human intestinal microflora. Chemotherapy. 2003;49:17–23. doi: 10.1159/000069784. [DOI] [PubMed] [Google Scholar]
- 60.Calne DB, Karoum F, Ruthven CRJ, Sandler M. The metabolism of orally administered l-DOPA in Parkinsonism. Br J Pharmacol. 1969;37:57–68. doi: 10.1111/j.1476-5381.1969.tb09522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sandler M, Goodwin BL, Ruthven CRJ, Calne DB. Therapeutic Implications in Parkinsonism of m-Tyramine Formation from L-Dopa in Man. Nature. 1971;229:414–6. doi: 10.1038/229414a0. [DOI] [PubMed] [Google Scholar]
- 62.Bakke OM. Degradation of DOPA by Intestinal Microorganisms in vitro. Acta Pharmacologica et Toxicologica. 1971;30:115–121. doi: 10.1111/j.1600-0773.1971.tb00640.x. [DOI] [PubMed] [Google Scholar]
- 63.Goldin BR, Peppercorn MA, Goldman P. Contribution of host and intestinal microflora in the metabolism of L-Dopa by the rat. J Pharmacol Exp Ther. 1973;186:160–6. [PubMed] [Google Scholar]
- 64.Peppercorn MA, Goldman P. Drug-bacteria interaction. Reviews in Drug Metabolism and Drug Interactions. 1976;11:75–88. [Google Scholar]
- 65.Sasahara K, Nitanai T, Habara T, et al. Dosage form design for mprovement of bioavailability of levodopa IV: Possible causes of low bioavailability of oral levodopa in dogs. J Pharm Sci. 1981;70:730–3. doi: 10.1002/jps.2600700705. [DOI] [PubMed] [Google Scholar]
- 66.Pierantozzi M, Pietroiusti A, Brusa L, et al. Helicobacter pylori eradication and l-dopa absorption in patients with PD and motor fluctuations. Neurology. 2006;66:1824–9. doi: 10.1212/01.wnl.0000221672.01272.ba. [DOI] [PubMed] [Google Scholar]
- 67.Hasriza Hashim, Azmin S, Razlan H, Yahya NW, et al. Eradication of Helicobacter pylori Infection Improves Levodopa Action, Clinical Symptoms and Quality of Life in Patients with Parkinson’s Disease. PLOS One. 2014;9:e112330. doi: 10.1371/journal.pone.0112330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niehues M, Hense A. In-vitro interaction of l-dopa with bacterial adhesins of Helicobacter pylori: an explanation for clinicial differences in bioavailability? J Pharm Pharmacol. 2009;61:1303–7. doi: 10.1211/jpp/61.10.0005. [DOI] [PubMed] [Google Scholar]
- 69.Shu YZ, Kingston DGI, Van Tassall RL, Wilkins TD. Metabolism of levamisole, an anti-colon cancer drug by human intestinal bacteria. Xenobiotica. 1992;21:737–50. doi: 10.3109/00498259109039513. [DOI] [PubMed] [Google Scholar]
- 70.Bashir M, Kingston DGI, Carman RJ, van Tassell RL, Wilkins TD. Anaerobic metabolism of 2-amino-3-methyl-3H-imidazo[4,5-f]quinoline (IQ) by human fecal flora. Mutation Res Lett. 1987;190:187–90. doi: 10.1016/0165-7992(87)90027-3. [DOI] [PubMed] [Google Scholar]
- 71.Yoo DH, Kim IS, Van Le TK, Jung I-H, Yoo HH, Kim D-H. Gut microbiota-mediated drug interactions between lovastatin and antibiotics. Drug Metab Dispos. 2014;42:1508–13. doi: 10.1124/dmd.114.058354. [DOI] [PubMed] [Google Scholar]
- 72.Zaharko DS, Bruckner H, Oliverio VT. Antibiotics alter methotrexate metabolism and excretion. Science. 1969;166:887–8. doi: 10.1126/science.166.3907.887. [DOI] [PubMed] [Google Scholar]
- 73.Valerino DM, Johns DG, Zaharko DS, et al. Studies of the metabolism of methotrexate by intestinal flora-I: Identification and study of biological properties of the metabolite 4-amino-4-deoxy-N10-methylpteroic acid. Biochem Pharmacol. 1972;21:821–31. doi: 10.1016/0006-2952(72)90125-6. [DOI] [PubMed] [Google Scholar]
- 74.Tozaki H, Emi Y, Horisaka E, et al. Metabolism of peptide drugs by the microorganisms in rat cecal contents. Biol Pharm Bull. 1995;18:929–31. doi: 10.1248/bpb.18.929. [DOI] [PubMed] [Google Scholar]
- 75.Tozaki H, Emi Y, Horisaka E, et al. Degradation of Insulin and Calcitonin and Their Protection by Various Protease Inhibitors in Rat Caecal Contents: Implications in Peptide Delivery to the Colon. J Pharm Pharmacol. 1997;49:164–68. doi: 10.1111/j.2042-7158.1997.tb06773.x. [DOI] [PubMed] [Google Scholar]
- 76.Sasaki I, Tamura T, Shibakawa T, et al. Metabolism of azetirelin, a new thyrotropin-releasing hormone (TRH) analogue, by intestinal microorganisms. Pharm Res. 1997;14:1004–7. doi: 10.1023/a:1012141025938. [DOI] [PubMed] [Google Scholar]
- 77.Sasaki I, Tozaki H, Matsumoto K, et al. Development of an oral formulation of azetirelin, using n-lauryl-beta-D-maltopyranoside as an absorption enhancer. Biol Pharm Bull. 1999;22:611–5. doi: 10.1248/bpb.22.611. [DOI] [PubMed] [Google Scholar]
- 78.Kim DH, Hyun SH, Shim SB, et al. The role of intestinal bacteria in the transformation of sodium picosulfate. Jpn J Pharmacol. 1992;59:1–5. doi: 10.1254/jjp.59.1. [DOI] [PubMed] [Google Scholar]
- 79.Cole CB, Fuller R, Mallet AK, et al. The influence of the host on expression of intestinal microbial enzyme activities involved in metabolism of foreign compounds. J Appl Bacteriology. 1985;59:549–553. doi: 10.1111/j.1365-2672.1985.tb03359.x. [DOI] [PubMed] [Google Scholar]
- 80.Akao T, Kawabata K, Yanagisawa E, et al. Baicalin, the predominant flavone glucuronide of scutellariae radix, is absorbed from the rat gastrointestinal tract as the aglycone and restored to its original form. J Pharm Pharmacol. 2000;52:1563–8. doi: 10.1211/0022357001777621. [DOI] [PubMed] [Google Scholar]
- 81.Bowey E, Adlercreutz H, Rowland I. Metabolism of isoflavones and lignans by the gut microflora: a study in germ-free and human flora associated rats. Food Chem Toxicol. 2003;41:631–6. doi: 10.1016/s0278-6915(02)00324-1. [DOI] [PubMed] [Google Scholar]
- 82.Turner NJ, Thomson BM, Shaw IC. Bioactive isoflavones in functional foods: the importance of gut microflora and bioavailability. Nutr Rev. 2003;6:204–13. doi: 10.1301/nr.2003.jun.204-213. [DOI] [PubMed] [Google Scholar]
- 83.Takasuna K, Hagiwara T, Hirohashi M, et al. Involvement of beta-glucuronidase in intestinal microflora in the intestinal toxicity of the antitumor camptothecin derivative irinotecan hydrochloride (CPT-11) in rats. Cancer Res. 1996;56:3752–7. [PubMed] [Google Scholar]
- 84.Takasuna K, Hagiwara T, Hirohashi M, et al. Inhibition of intestinal microflora beta-glucuronidase modifies the distribution of the active metabolite of the antitumor agent, irinotecan hydrochloride (CPT-11) in rats. Cancer Chemother Pharmacol. 1998;42:280–6. doi: 10.1007/s002800050818. [DOI] [PubMed] [Google Scholar]
- 85.Takasuna K, Hagiwara T, Watanabe K, et al. Optimal antidiarrhea treatment for antitumor agent irinotecan hydrochloride (CPT-11)-induced delayed diarrhea. Cancer Chemotherapy Pharmacol. 2006;58:494–503. doi: 10.1007/s00280-006-0187-8. [DOI] [PubMed] [Google Scholar]
- 86.Kodawara T, Higashi T, Negoro Y, et al. The Inhibitory Effect of Ciprofloxacin on the β-Glucuronidase-mediated Deconjugation of the Irinotecan Metabolite SN-38-G. Basic Clin Pharmacol Toxicol. 2016;118:333–7. doi: 10.1111/bcpt.12511. [DOI] [PubMed] [Google Scholar]
- 87.Wallace BD, Wang H, Lane KT, et al. Alleviating Cancer Drug Toxicity by Inhibiting a Bacterial Enzyme. Science. 2010;330:831–35. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wallace BD, Roberts AB, Pollet RM, et al. Cell, Structure and Inhibition of Microbiome β-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity. Cell. 2015;22:1238–49. doi: 10.1016/j.chembiol.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ahmad S, Hughes MA, Yeh LA, et al. Potential repurposing of known drugs as potent bacterial β-glucuronidase inhibitors. J Biomol Screen. 2012;17:957–65. doi: 10.1177/1087057112444927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ahmad S, Williams AL, Phan AT, et al. Old drug new use--amoxapine and its metabolites as potent bacterial β-glucuronidase inhibitors for alleviating cancer drug toxicity. Clin Cancer Res. 2014;20:3521–30. doi: 10.1158/1078-0432.CCR-14-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.LoGuidice A, Wallace BD, Bendel L, et al. Pharmacologic Targeting of Bacteria-Glucuronidase Alleviates Nonsteroidal Anti-Inflammatory Drug-Induced Enteropathy in Mice. JPET. 2012;341:447–54. doi: 10.1124/jpet.111.191122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saitta KS, Carmen Z, Lee KK, et al. Bacterial β-glucuronidase inhibition protects mice against enteropathy induced by indomethacin, ketoprofen or diclofenac: mode of action and pharmacokinetics. Xenobiotica. 2014;44:28–35. doi: 10.3109/00498254.2013.811314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheng TC, Chuang KH, Roffler SR, et al. Discovery of Specific Inhibitors for Intestinal E. coli β-Glucuronidase through In Silico Virtual Screening. The Scientific World Journal. 2015 doi: 10.1155/2015/740815. Article ID 740815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nakayama H, Kinouchi T, Kataoka K, et al. Intestinal anaerobic bacteria hydrolyse sorivudine, producing the high blood concentration of 5-(E)-(2-bromovinyl)uracil that increases the level and toxicity of 5-fluorouracil. Pharmacogenetics. 1997;7:35–43. doi: 10.1097/00008571-199702000-00005. [DOI] [PubMed] [Google Scholar]
- 95.Okuda H, Ogura K, Kato A, Takubo H, Watabe T. A possible mechanism of eighteen patient deaths caused by interactions of sorivudine, a new antiviral drug, with oral 5-fluorouracil prodrugs. J Pharmacol Exp Ther. 1998;287:791–9. [PubMed] [Google Scholar]
- 96.Bakke JE, Gustafsson JA. Role of intestinal flora in metabolism of agrochemicals conjugated with glutathione. Xenobiotica. 1986;16:1047–56. doi: 10.3109/00498258609038982. [DOI] [PubMed] [Google Scholar]
- 97.Mikov M, Caldwell J, Dolphin CT, Smit RL. The role of intestinal microflora in the formation of the methylthio adduct metabolites of paracetamol. Studies in neomycin-pretreated and germ-free mice. Biochem Pharmacol. 1988;37:1445–9. doi: 10.1016/0006-2952(88)90005-6. [DOI] [PubMed] [Google Scholar]
- 98.Grant DM, Josephy PD, Lord HL, Morrison LD. Salmonella typhimurium strains expressing human arylamine N-acetyltransferases: metabolism and mutagenic activation of aromatic amines. Cancer Res. 1992;52:3961–4. [PubMed] [Google Scholar]
- 99.Hein DW, Doll MA, Rustan TD, Gray K, Feng Y, Ferguson RJ. Metabolic activation and deactivation of arylamine carcinogens by recombinant human NAT1 and NAT2 acetyltransferases. Carcinogenesis (Land.) 1993;14:1633–8. doi: 10.1093/carcin/14.8.1633. [DOI] [PubMed] [Google Scholar]
- 100.Dull BJ, Salata K, Goldman P. Role of the intestinal flora in the acetylation of sulfasalazine metabolites. Biochem Pharmacol. 1987;36:3772–4. doi: 10.1016/0006-2952(87)90034-7. [DOI] [PubMed] [Google Scholar]
- 101.van Hogezand RA, Kennis HM, van Schaik A, et al. Bacterial acetylation of 5-aminosalicylic acid in faecal suspensions cultured under aerobic and anaerobic conditions. Eur J Clin Pharmacol. 1992;43:189–2. doi: 10.1007/BF01740669. [DOI] [PubMed] [Google Scholar]
- 102.Delome’nie C, Fouix S, Longuemaux S, et al. Identification and Functional Characterization of Arylamine N-Acetyltransferases in Eubacteria: Evidence for Highly Selective Acetylation of 5-Aminosalicylic Acid. J Bacteriology. 2001;183:3417–27. doi: 10.1128/JB.183.11.3417-3427.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Garau P, Orenstein SR, Neigut DA, et al. Pancreatitis associated with olsalazine and sulfasalazine in children with ulcerative colitis. J Pediatric Gastroenterology and Nutrition. 1994;18:481–5. doi: 10.1097/00005176-199405000-00015. [DOI] [PubMed] [Google Scholar]
- 104.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmaco-metabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Nat Acad Sci US. 2009;106:14728–33. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dawson LF, Donahue EH, Cartman ST, et al. The analysis of para-cresol production and tolerance in Clostridium difficile 027 and 012 strains. BMC Microbiology. 2011;11:86. doi: 10.1186/1471-2180-11-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yan Z, Zhong HM, Maher N, et al. Bioactivation of 4-methylphenol (p-cresol) via cytochrome P450-mediated aromatic oxidation in human liver microsomes. Drug Met Disp. 2005;33:1867–6. doi: 10.1124/dmd.105.006387. [DOI] [PubMed] [Google Scholar]
- 107.Lee SH, An JH, Lee HJ, Jung BH. Evaluation of pharmacokinetic differences of acetaminophen in pseudo germ-free rats. Biopharmaceutics Drug Disp. 2012;33:292–3. doi: 10.1002/bdd.1799. [DOI] [PubMed] [Google Scholar]
- 108.Rowland IR, Mallet AK, Cole CB, Fuller R. Mutagen activation by hepatic fractions from conventional, germ free and monoassociated rats. Arch Toxicol. 1987;11(Suppl):261–3. doi: 10.1007/978-3-642-72558-6_47. [DOI] [PubMed] [Google Scholar]
- 109.Aura AM, O’Leary KA, Williamson G, Ojala M, Bailey M, Puupponen-Pimiä R. Quercetin derivatives are deconjugated and converted to hydroxyphenylacetic acids but not methylated by human fecal flora in vitro. J Agric Food Chem. 2002;50:1725–30. doi: 10.1021/jf0108056. [DOI] [PubMed] [Google Scholar]
- 110.Xue H, Xie W, Jiang Z. 3,4-Dihydroxy- phenylacetic acid, a microbiota-derived metabolite of quercetin, attenuates acetaminophen (APAP)-induced liver injury through activation of Nrf-2. Xenobiotica. 2016 doi: 10.3109/00498254.2016.1140847. [DOI] [PubMed] [Google Scholar]
- 111.Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–3. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 112.Bjorkholm B, Bok CM, Lundin A, Rafter J, Hibberd ML, Pettersson S. Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One. 2009;4:e6958. doi: 10.1371/journal.pone.0006958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Toda T, Saito N, Ikarashi N, Ito K, Yamamoto M, Ishige A, et al. Intestinal flora induces the expression of Cyp3a in the mouse liver. Xenobiotica. 2009;39:323–34. doi: 10.1080/00498250802651984. [DOI] [PubMed] [Google Scholar]
- 114.Hubbard TD, Murray IA, Bisson WH, et al. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Scientific Reports. 2015;5:12689. doi: 10.1038/srep12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hubbard TD, Murray IA, Perdew GH. Indole and Tryptophan Metabolism: Endogenous and Dietary Routes to Ah Receptor Activation. Drug Metab Dispos. 2015;43:1522–35. doi: 10.1124/dmd.115.064246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP. Symbiotic Bacterial Metabolites Regulate Gastrointestinal Barrier Function via the Xenobiotic Sensor PXR and Toll-like Receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Einarsson K, Gustafsson J-E, Gustafsson BE. Differences between Germ-free and Conventional Rats in Liver Microsomal Metabolism of Steroids. J Biol Chem. 1973;248:3623–30. [PubMed] [Google Scholar]
- 118.Duncan AM, Wangen KE, Kurzer MS. Soy consumption alters endogenous estrogen metabolism in postmenopausal women. Cancer Epidemiol Biomarkers Prevent. 2000;9:781–6. [PubMed] [Google Scholar]
- 119.Claus SP, Ellero SL, Berger B, et al. Colonization-Induced Host-Gut Microbial Metabolic Interaction. MBio. 2011;2:e00271–10. doi: 10.1128/mBio.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Meinl W, Sczesny S, Brigelius-Flohé R, Blaut M, Glatt H. Impact of gut microbiota on intestinal and hepatic levels of phase 2 xenobiotic-metabolizing enzymes in the rat. Drug Met Disp. 2009;37:1179–86. doi: 10.1124/dmd.108.025916. [DOI] [PubMed] [Google Scholar]
- 121.Lhoste EF, Ouriet V, Bruel S, et al. The human colonic microbiota influences the alterations of xenobiotic-metabolizing enzymes by catechins in male F344 rats. Food Chem Toxicol. 2003;41:695–703. doi: 10.1016/s0278-6915(03)00010-3. [DOI] [PubMed] [Google Scholar]
- 122.Edalat M, Mannervik B, Axelsson LG. Selective expression of detoxifying glutathione transferases in mouse colon: effect of experimental colitis and the presence of bacteria. Histochem Cell Biol. 2004;122:151–9. doi: 10.1007/s00418-004-0688-7. [DOI] [PubMed] [Google Scholar]
- 123.Selwyn FP, Cheng SL, Klaassen CD, Cui JY. Regulation of Hepatic Drug-Metabolizing Enzymes in Germ-Free Mice by Conventionalization and Probiotics. Drug Metab Dispos. 2016;44:262–74. doi: 10.1124/dmd.115.067504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Nat Acad Sci US. 2011;108(suppl. 1):4554–61. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156:3216–23. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- 126.Toda T, Ohi K, Kudo T, Ikarashi N, Ito K, Sugiyama K. Ciprofloxacin suppresses Cyp3a in mouse liver by reducing lithocholic acid-producing intestinal flora. Drug Metab Pharmacokinet. 2009;24:201–8. doi: 10.2133/dmpk.24.201. [DOI] [PubMed] [Google Scholar]
- 127.Vesper BJ, Jawdi A, Altman KW, Haines GK, 3rd, Tao L, Radosevich JA. The effect of proton pump inhibitors on the human microbiota. Curr Drug Metab. 2009;10:84–9. doi: 10.2174/138920009787048392. [DOI] [PubMed] [Google Scholar]
- 128.Seto CT, Jeraldo P, Orenstein R, Chia N, DiBaise JK. Prolonged use of a proton pump inhibitor reduces microbial diversity: implications for Clostridium difficile susceptibility. Microbiome. 2014;2:42. doi: 10.1186/2049-2618-2-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jackson MA, Goodrich JK, Maxan M-E, et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut. 2016;65:749–56. doi: 10.1136/gutjnl-2015-310861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Imhann F, Bonder MJan, Vila AV, et al. Proton pump inhibitors affect the gut microbiome. Gut. 2016;65:740–8. doi: 10.1136/gutjnl-2015-310376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Clooney AG, Bernstein CN, Leslie WD, et al. A comparison of the gut microbiome between long-term users and non-users of proton pump inhibitors. Aliment Pharmacol Ther. 2016;43:974–84. doi: 10.1111/apt.13568. [DOI] [PubMed] [Google Scholar]
- 132.Makivuokko H, Tiihonen K, Tynkkynen S, Rautonen N. The effect of age and non-steroidal anti-inflammatory drugs on human intestinal microbiota composition. Brit J Nutrition. 2010;103:227–34. doi: 10.1017/S0007114509991553. [DOI] [PubMed] [Google Scholar]
- 133.Rogers MA, Aronoff DM. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin Microbiol Infect. 2016;22:178.e1–9. doi: 10.1016/j.cmi.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liang X, Bittinger K, Li X, Abernethy DR, Bushman FD, FitzGeral GA. Bidirectional interactions between indomethacin and the murine intestinal microbiota. eLife. 2015;4:e08973. doi: 10.7554/eLife.08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Marlicz W, Loniewski I, Grimes DS, Quigley EM. Nonsteroidal anti-inflammatory drugs, proton pump inhibitors, and gastrointestinal injury: Contrasting interactions in the stomach and small intestine. Mayo Clin Proc. 2014;89:1699–709. doi: 10.1016/j.mayocp.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 136.Endo H, Higurashi T, Hosono K, et al. Efficacy of Lactobacillus casei treatment on small bowel injury in chronic low-dose aspirin users: a pilot randomized controlled study. J Gastroenterol. 2011;46:894–5. doi: 10.1007/s00535-011-0410-1. [DOI] [PubMed] [Google Scholar]
- 137.Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]