Abstract
Gram-positive bacteria use sortase cysteine transpeptidase enzymes to covalently attach proteins to their cell wall and to assemble pili. In pathogenic bacteria sortases are potential drug targets, as many of the proteins that they display on the microbial surface play key roles in the infection process. Moreover, the Staphylococcus aureus Sortase A (SaSrtA) enzyme has been developed into a valuable biochemical reagent because of its ability to ligate biomolecules together in vitro via a covalent peptide bond. Here we review what is known about the structures and catalytic mechanism of sortase enzymes. Based on their primary sequences, most sortase homologs can be classified into six distinct subfamilies, called class A–F enzymes. Atomic structures reveal unique, class-specific variations that support alternate substrate specificities, while structures of sortase enzymes bound to sorting signal mimics shed light onto the molecular basis of substrate recognition. The results of computational studies are reviewed that provide insight into how key reaction intermediates are stabilized during catalysis, as well as the mechanism and dynamics of substrate recognition. Lastly, the reported in vitro activities of sortases are compared, revealing that the transpeptidation activity of SaSrtA is at least 20-fold faster than other sortases that have thus far been characterized. Together, the results of the structural, computational, and biochemical studies discussed in this review begin to reveal how sortases decorate the microbial surface with proteins and pili, and may facilitate ongoing efforts to discover therapeutically useful small molecule inhibitors.
1. INTRODUCTION
Bacteria display a variety of proteins on their surface that to enable them to effectively interact with their environment. Gram-positive bacteria use sortase cysteine transpeptidase enzymes to covalently attach proteins to their cell wall, and to assemble pili. Sortases in pathogenic bacteria are frequently important virulence factors, as many of the proteins that they display have key roles in the infection process, such as promoting bacterial adhesion, nutrient acquisition, and the evasion and suppression of the immune response (Cascioferro, Totsika, & Schillaci, 2014; Schneewind & Missiakas, 2012, 2014; Siegel, Liu, & Ton-That, 2016; Spirig, Weiner, & Clubb, 2011). As a result, a significant amount of effort has been put forth to elucidate the mechanism of sortase-mediated catalysis and to discover small-molecule sortase inhibitors that could function as potent antiinfective agents (Bradshaw et al., 2015; Cascioferro et al., 2014; Clancy, Melvin, & McCafferty, 2010; Maresso & Schneewind, 2008; Suree, Jung, & Clubb, 2007). Moreover, sortases have been developed into valuable biochemical reagents to ligate distinct biomolecules together via a covalent peptide bond. This in vitro transpeptidation activity has been harnessed for a variety of useful applications, including among others, covalently attaching proteins to cells, attaching fluorophores or drugs to antibodies, cyclizing proteins, and immobilizing peptides on solid surfaces (Antos, Truttmann, & Ploegh, 2016; Popp & Ploegh, 2011; Ritzefeld, 2014; Schmidt, Toplak, Quaedflieg, & Nuijens, 2017; Schmohl & Schwarzer, 2014; Voloshchuk, Liang, & Liang, 2016).
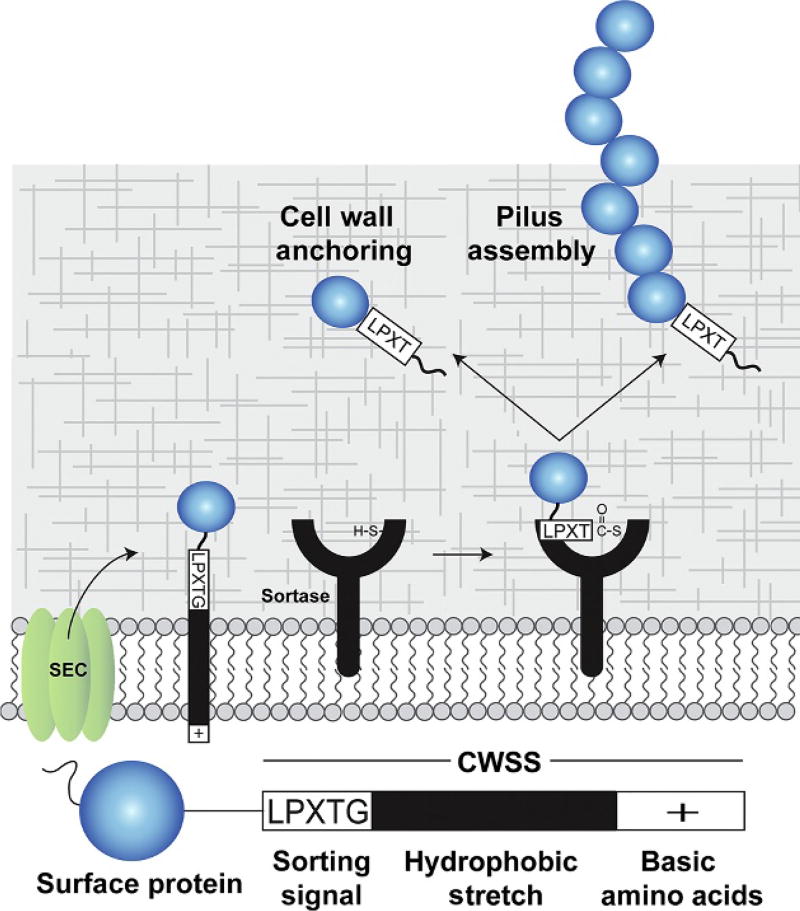
Sortases perform two distinct functions in bacteria: (1) attach proteins directly to the cell wall or (2) assemble pili, long proteinaceous fibers that project from the microbial surface (Fig. 1). Both reactions are mechanistically related and operate on secreted proteins that contain a C-terminal cell wall sorting signal (CWSS). The CWSS contains a five-residue sortase recognition motif, frequently LPXTG, that is followed by a hydrophobic domain and a positively charged cytoplasmic anchor that retains the protein substrate in the membrane (Schneewind, Model, & Fischetti, 1992). The Sortase A enzyme from Staphylococcus aureus (SaSrtA) has been studied in detail and is archetypal (Mazmanian, Liu, Ton-That, & Schneewind, 1999; Ton-That, Liu, Mazmanian, Faull, & Schneewind, 1999). SaSrtA attaches surface proteins to the cell wall by recognizing an LPXTG motif within the CWSS of its protein substrate (Fig. 2A). Catalysis begins when SaSrtA’s active site cysteine residue nucleophilically attacks the carbonyl carbon in the peptide bond between the Thr and Gly residues in the sorting signal (Fig. 2A, step 1) (Clancy et al., 2010; Connolly & Clubb, 2005). This generates a tetrahedral intermediate that quickly collapses to form a semi-stable thioacyl intermediate in which sortase is covalently linked via its cysteine residue to its protein substrate (Fig. 2A, step 2). SaSrtA then recognizes a second substrate, the cell wall precursor, lipid II (Fig. 2A, step 3) (Perry, Ton-That, Mazmanian, & Schneewind, 2002; Ruzin et al., 2002) and catalyzes a reaction in which the N-terminal primary amine group within the cross-bridge peptide nucleophilically attacks the carbonyl carbon atom within the thioacyl bond. This second transient tetrahedral intermediate resolves into the protein–lipid II product in which the components are joined via a peptide bond (Fig. 2A, step 4) (Perry et al., 2002; Schneewind, Fowler, & Faull, 1995; Ton-That, Faull, & Schneewind, 1997). The lipid II-linked protein is then incorporated into the peptidoglycan via the transpeptidation and glycosylation reactions that synthesize the cell wall. In contrast to attaching proteins to the cell wall, a second type of sortase, frequently called “pilin polymerases,” construct bacterial pili by polymerizing pilin protein subunits (Fig. 2B) (Hendrickx, Budzik, Oh, & Schneewind, 2011; Kline, Dodson, Caparon, & Hultgren, 2010; Mandlik, Swierczynski, Das, & Ton-That, 2008; Siegel et al., 2016; Spirig et al., 2011; Ton-That & Schneewind, 2003). These pilin-assembling enzymes employ a similar transpeptidation reaction as SaSrtA, but instead of using lipid II as a nucleophile to attach proteins to the cell wall, a lysine amino group located within a protein pilin subunit is used as a secondary substrate to attack the sortase–protein thioacyl intermediate (Fig. 2B, steps 3 and 4). This reaction constructs pili by covalently linking protein subunits together via lysine–isopeptide bonds. Both types of sortase-catalyzed processes occur on the extracellular membrane, where the enzyme and its substrate are membrane associated (Cozzi et al., 2011; Spirig et al., 2011; Wu et al., 2012).
Fig. 1.
Sortase enzymes attach proteins to the cell wall and assemble pili. (A) Overview of anchoring and pilus assembly reactions. A protein that is to be displayed (blue) contains an N-terminal secretion signal and a C-terminal cell wall sorting signal (CWSS). The CWSS contains an LPXTG-like sorting signal sequence that is processed by the sortase, a nonpolar polypeptide segment (black), and a C-terminal segment of positively charged residues (+). After secretion through the Sec translocon, the protein remains embedded in the lipid bilayer via the nonpolar segment within the CWSS. The sortase enzyme then cleaves between the threonine and glycine residues to form a sortase–protein thioacyl intermediate in which the active site cysteine is covalently linked to the carbonyl carbon atom of the threonine. There are two basic types of sortases: (1) cell wall anchoring sortases that attach protein to the crossbridge peptide of the cell wall and (2) pilin polymerase sortases that covalently link pilin subunits together via lysine–isopeptide bonds. In both cases, the enzymes function as transpeptidases. Some sortases are capable of performing both functions, attaching proteins to the cell wall and polymerizing pili.
Fig. 2.
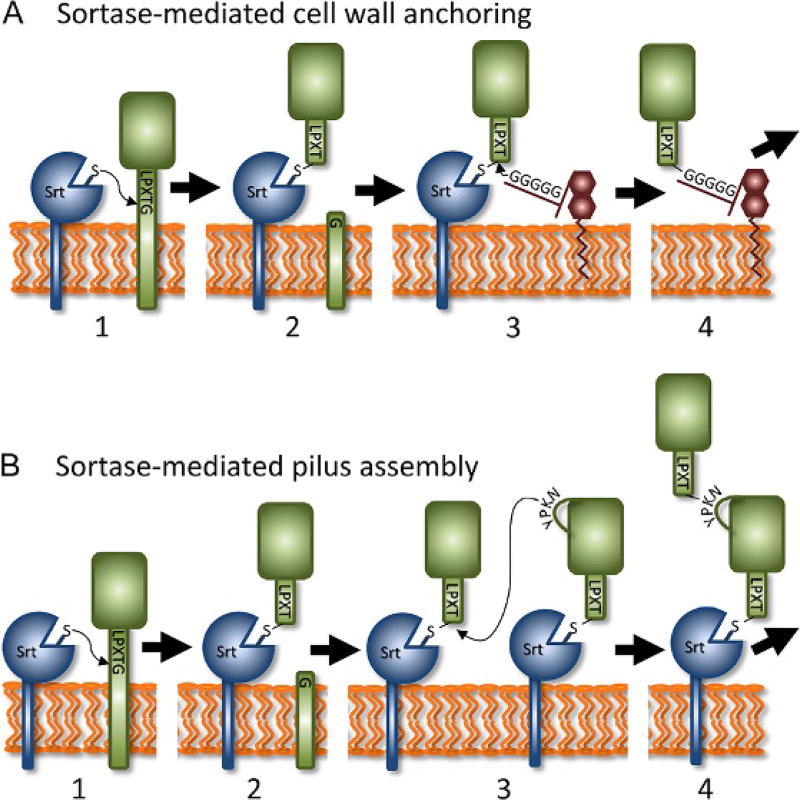
Mechanism of cell wall protein anchoring and pilus assembly. Sortases perform two basic functions in bacteria: (1) attach proteins to the cell wall and (2) join proteins together to construct pili. (A) In the cell wall anchoring reaction, the sortase and substrate are both membrane bound. The reaction occurs via four distinct steps. Sortase first recognizes a sorting signal motif within the CWSS and nucleophilically attacks the threonine’s carbonyl carbon atom via its active site cysteine residue (for demonstration purposes the LPXTG sorting signal recognized by class-A type enzymes is shown, step 1). The LPXTG sorting signal is then cleaved to produce a sortase–substrate thioacyl intermediate (step 2). Next, the crossbridge peptide from a lipid II molecule nucleophilically attacks the thioacyl intermediate (step 3). Lastly, a new peptide bond is formed between the lipid II molecule and surface protein to produce a protein–lipid II intermediate that is incorporated into cell wall by the transglycosylation and transpeptidation reactions that synthesize the peptidoglycan (step 4). (B) In the pilus assembly reaction, steps 1–2 produce a sortase–substrate thioacyl intermediate, similar to the cell wall anchoring reaction. In this reaction, the sortase recognizes a pilin protein that contains a CWSS. However, a lysine residue within the pilin motif from an adjacent pilin protein performs the nucleophilic attack on the thioacyl intermediate (step 3). A new protein–protein isopeptide bond is formed that covalently links the pilin proteins (step 4). This assembly process is repeated to build an isopeptide-linked pilus shaft that contains multiple pilin proteins. Depending on the type of pilus, distinct tip and base pilin proteins can be located at the ends of the pilus shaft, which are incorporated through a similar mechanism and involve covalent linkages via lysine-derived isopeptide bonds. Finally, the intact pilus is attached to the cell wall via sortase-catalyzed attachment of the pilus to lipid II, similar to cell wall protein display. Some sortases are capable of performing both functions, attaching proteins to the cell wall and functioning as pilin polymerases.
At present, 3330 gene sequences encoding sortase enzymes have been identified within 1098 species of bacteria (Finn et al., 2016). Sortases are primarily found in Gram-positive bacteria, but are also present to a lesser extent in some species of Gram-negative and archaebacteria. Based on their primary sequences, most sortase homologs can be classified into six distinct subfamilies, called class A–F enzymes (Dramsi, Trieu-Cuot, & Bierne, 2005; Spirig et al., 2011). Class A, B, and D enzymes are prevalent in bacteria within the Firmicutes phylum, while class E and F enzymes predominate in Actinobacteria. Class C enzymes are found in both Firmicutes and Actinobacteria. Similar to SaSrtA (a class A enzyme), all sortases contain a His–Cys–Arg catalytic triad (Ilangovan, Ton-That, Iwahara, Schneewind,& Clubb, 2001; Marraffini, Ton-That, Zong, Narayana, & Schneewind, 2004; Ton-That, Mazmanian, Alksne, & Schneewind, 2002), and their primary sequences harbor a highly conserved TLXTC motif that contains the catalytically essential cysteine residue (Clancy et al., 2010). All sortases characterized to date catalyze a transpeptidation reaction that joins an LPXTG-like sorting signal within the CWSS of their protein substrate to an amino nucleophile. However, their sorting signal and nucleophile substrate specificities can vary substantially. These distinct specificities enable microbes to utilize more than one type of sortase to elaborate their surface, with each sortase operating nonredundantly to display or assemble distinct proteins on the cell surface (Comfort & Clubb, 2004; Pallen, Lam, Antonio, & Dunbar, 2001).
A number of excellent reviews have been written that describe the overall function of sortases in bacteria, their development as biochemical reagents, and efforts to discover therapeutically useful sortase inhibitors (Antos et al., 2016; Bradshaw et al., 2015; Cascioferro et al., 2014; Clancy et al., 2010; Maresso & Schneewind, 2008; Popp & Ploegh, 2011; Ritzefeld, 2014; Schmidt et al., 2017; Schmohl & Schwarzer, 2014; Schneewind & Missiakas, 2012, 2014; Siegel et al., 2016; Spirig et al., 2011; Suree et al., 2007; Voloshchuk et al., 2016). In this chapter, we review what is known about their atomic structures and the molecular basis of substrate recognition and catalysis.
2. STRUCTURAL BIOLOGY: ENZYME STRUCTURE AND CLASS-SPECIFIC VARIATIONS
2.1 The Archetypal SaSrtA Enzyme
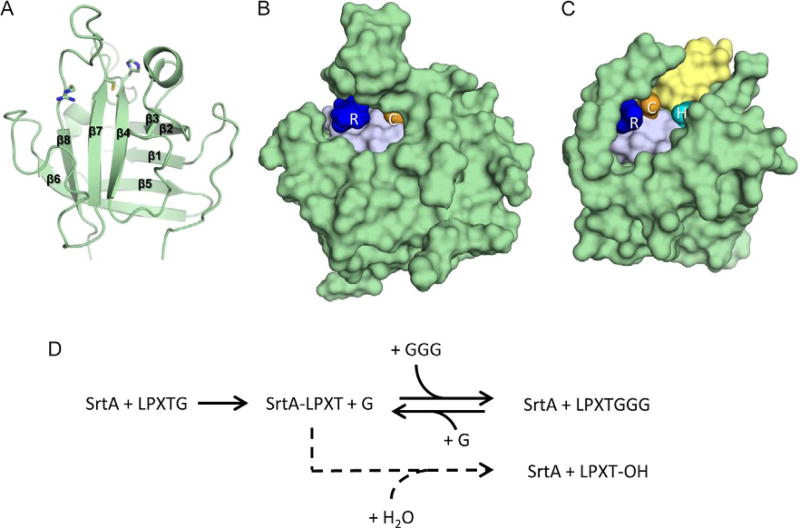
The NMR structure of SaSrtA determined by the Clubb and Schneewind groups was the first reported structure of a sortase enzyme (Ilangovan et al., 2001) (Fig. 3A). The primary sequence of this class A enzyme exhibits features that are generally conserved in other sortases. It has three components: (i) an N-terminal signal sequence that enables it to be transported across the membrane through the Sec translocon, (ii) a nonpolar segment of amino acids that embed the enzyme in the bilayer, (iii) and a conserved, water-soluble C-terminal catalytic domain that contains the His–Cys–Arg triad (Marraffini et al., 2004; Ton-That et al., 2002). The structure of the catalytic domain was determined, residues 60–206 of SaSrtA (SaSrtAΔ59). This structure revealed the now canonical “sortase fold” that contains a closed eight-stranded β-barrel architecture (Fig. 3A). The atomic coordinates of SaSrtAΔ59 were precisely defined, with the exception of a 19 amino acid flexible loop that connects strands β6 to β7 (the β6/β7 loop). Subsequently, a crystal structure of SaSrtAΔ59 was determined at 2.0 Å resolution that is very similar to the solution structure; the Cα coordinates in structurally ordered parts of the crystal and NMR structures have an RMSD of 1.97 Å (Zong, Bice, Ton-That, Schneewind, & Narayana, 2004). In both structures, residues in the His–Cys–Arg triad are positioned adjacent to one another within the active site (His120, Cys184, and Arg197 in SaSrtA). As described in detail later, subsequent studies revealed that the LPXTG sorting signal substrate of SaSrtA binds to a pocket that is positioned adjacent to the active site cysteine (Fig. 3B) (Suree et al., 2009). The base of the pocket in SaSrtA is formed by residues in strands β4 and β7, while the walls are formed by surface loops that connect strand β6 to β7 (β6/β7 loop), strand β3 to β4 (β3/β4 loop), and strand β2 to helix H2 (β2/H2 loop). All sortases are thought to utilize similarly positioned sorting signal binding grooves. Interestingly, SaSrtA requires Ca2+ for efficient catalysis, as the removal of this ion reduces activity fivefold (Ilangovan et al., 2001; Naik et al., 2006); other divalent cations also affected enzyme activity, but to a lesser extent. NMR chemical shift mapping and biochemical studies revealed that Ca2+ binds to a pocket located between the β3/β4 and β6/β7 loops, where it is likely coordinated by the side chains of Glu105, Glu108, Asp112, Glu171, and a backbone carbonyl from Asn114 (Ilangovan et al., 2001; Naik et al., 2006). NMR dynamics experiments indicate that Ca2+ binding alters the mobility and structure of the β6/β7 active site loop, thereby allosterically regulating the enzyme’s affinity for the sorting signal. Interestingly, this mechanism of metal-dependent regulation appears to be unique to SaSrtA, as no other structurally characterized member of the sortase-superfamily contains a structurally similar Ca2+ binding pocket.
Fig. 3.
Structure and transpeptidation reaction of representative class A sortases. (A) S. aureus SrtA (SaSrtA) NMR structure (cartoon), showcasing an eight-stranded β-barrel with active site His120, Cys184, and Arg197 residues (sticks). (B) SaSrtA NMR structure (green surface) with active residues Arg (blue) and Cys (orange). The active site His is occluded by a closed β7/β8 loop, and there is no obvious groove for a full-length peptide to exit the active site. (C) S. pyogenes SrtA (SpySrtA) structure (green surface) with active site Arg (blue), Cys (orange), and His (cyan) residues. An open β7/β8 loop creates a clear channel that can be seen running between active Cys and His residues, indicating the potential exit channel (yellow) for the full-length peptide substrate. (D) In vitro, SaSrtA catalyzes a reversible transpeptidation reaction (top, indicated by solid arrows) in which it joins LPXTG and (Gly)3 peptides. In the absence of glycine oligopeptide, SaSrtA acts a protease and cleaves the LPXTG peptide between its threonine and glycine residues (bottom, indicated by a dashed arrow). In this spurious pathway, a water molecule, instead of lipid II, performs the second nucleophilic attack to cleave the thioacyl bond between sortase and substrate, thereby hydrolyzing the peptide. On the cell surface, hydrolysis is presumably undesirable, as proteolysis separates the protein from its membrane anchor, releasing it from the microbe. Transpeptidation occurs faster than the rate of proteolysis in vitro, making SaSrtA a valuable bioconjugation reagent (kcat = 0.28 ± 0.02 and 0.086 ± 0.015 s−1, respectively). Although all sortases are thought to catalyze transpeptidation reactions on the cell surface, this activity has only been reconstituted in vitro for a few sortases in addition to SaSrtA (listed in Table 2).
2.2 Class-Specific Variations
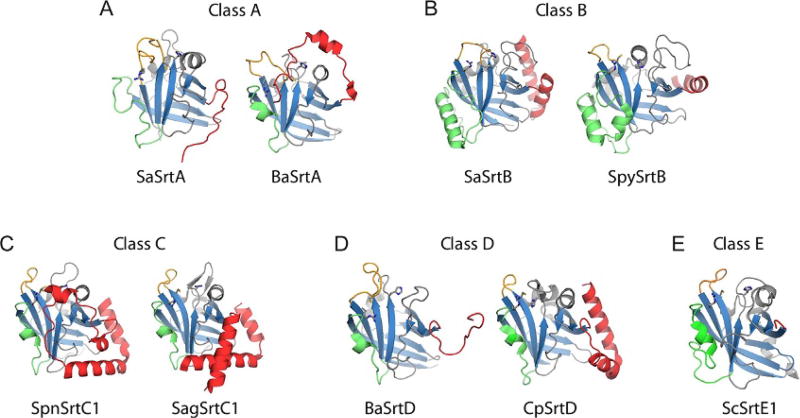
Structures of class A, B, C, D, and E sortases have been reported, revealing unique class-specific variations that likely impact function and modulate substrate specificity. Table 1 lists the structures of sortases that have thus far been determined, and Fig. 4 displays representative class A–E structures for comparison. As expected, all types of sortases contain a conserved catalytic domain that adopts a sortase fold (Fig. 4, blue). However, there are significant class-specific variations that are localized to four distinct structural foci: (i) the N-terminal segment that precedes the catalytic domain (red), (ii) the loop between strands β6 and β7 (the β6/β7 loop) (green), (iii) the loop between strands β7 and β8 (the β7/β8 loop) (yellow), and (iv) the C-terminal polypeptide segment that follows the catalytic domain (Fig. 4). Below, we discuss these differences and highlight their functional implications if they are known.
Table 1.
Structurally Characterized Sortase Enzymes
| Organism and Sortase | PDB | Bound Ligands or Substrates | Method |
|---|---|---|---|
| Class A | |||
| S. aureus SrtA (SaSrtA) | 1IJA | N/A | NMR |
| S. aureus SrtA-C184A (SaSrtA) | 1T2O | N/A | X-ray |
| S. aureus SrtA (SaSrtA) | 1T2P | N/A | X-ray |
| S. aureus SrtA (SaSrtA) | 1T2W | LPETG | X-ray |
| S. aureus SrtA (SaSrtA) | 2KID | Cbz–LPAT*, Ca2+ | NMR |
| S. aureus SrtA (SaSrtA) | 2MLM | Benzo[d]isothiazol-3-one based inhibitor | NMR |
| B. anthracis SrtA (BaSrtA) | 2KW8 | N/A | NMR |
| B. anthracis SrtA (BaSrtA) | 2RUI | Boc–LPAT* | NMR |
| S. pyogenes SrtA | 3FN5 | N/A | X-ray |
| S. pyogenes SrtA | 3FN6 | Cys in suflphenic acid form | X-ray |
| S. pyogenes SrtA | 3FN7 | N/A | X-ray |
| S. agalactiae SrtA | 3RCC | Zn2+ | X-ray |
| S. pneumoniae SrtA | 4O8L | N/A | X-ray |
| S. pneumoniae SrtA-C207A | 4O8T | N/A | X-ray |
| S. mutans SrtA | 4TQX | Chalcone | X-ray |
| Class B | |||
| S. aureus SrtB (SaSrtB) | 1NG5 | N/A | X-ray |
| S. aureus SrtB (SaSrtB) | 1QWZ | MTSET | X-ray |
| S. aureus SrtB (SaSrtB) | 1QX6 | E-64 | X-ray |
| S. aureus SrtB (SaSrtB) | 1QXA | Gly3 | X-ray |
| S. aureus SrtB (SaSrtB) | 4FLD | Cbz–NPQT* | X-ray |
| B. anthracis SrtB | 1RZ2 | N/A | X-ray |
| B. anthracis SrtB | 2OQW | AAEK1 | X-ray |
| B. anthracis SrtB | 2OQZ | AAEK2 | X-ray |
| S. pyogenes SrtB | 3PSQ | Zn2+, Cl− | X-ray |
| C. difficile SrtB | 4UX7 | N/A | X-ray |
| Class C | |||
| A. oris SrtC-1 | 2XWG | Ca2+ | X-ray |
| S. pneumoniae SrtC-1 | 2W1J | Glycerol | X-ray |
| S. pneumoniae SrtC-3 | 2W1K | N/A | X-ray |
| S. pneumoniae SrtC-1-H131D | 2WTS | Alanine | X-ray |
| S. pneumoniae SrtC-2 | 3G66 | N/A | X-ray |
| S. pneumoniae SrtC-2 | 3G69 | SO42− | X-ray |
| S. agalactiae SrtC-1Pilus Island-2a | 3O0P | N/A | X-ray |
| S. agalactiae SrtC-1 Pilus Island-1 “Type III” | 3RBI | N/A | X-ray |
| S. agalactiae SrtC1- Pilus Island-1 C184A; KDPYS to IPNTG | 3RBJ | N/A | X-ray |
| S. agalactiae SrtC-1 Pilus Island-1 “Type II” | 3RBK | N/A | X-ray |
| S. agalactiae SrtC1 Pilus Island-1 “Type I”—open lid | 3TB7 | N/A | X-ray |
| S. agalactiae SrtC-1 Pilus Island-1 | 3TBE | MTSET | X-ray |
| S. agalactiae SrtC-2 Pilus Island-1 | 4G1H | Ca2+ | X-ray |
| S. agalactiae SrtC-1 Pilus Island-1 | 4G1J | N/A | X-ray |
| S. suis SrtC-1 | 3RE9 | N/A | X-ray |
| Class D | |||
| B. anthracis SrtD | 2LN7 | N/A | NMR |
| C. perfringens SrtD | 4D70 | None | X-ray |
| Class E | |||
| S. coelicolor SrtE-1 | 5CUW | N/A | X-ray |
Fig. 4.
Structural variation by class of sortase. Sortases representative of the major themes seen for each class are displayed (cartoon) with active site residues (sticks). The hallmark sortase β-barrel (blue) and major sources of structural variability are highlighted, including N-terminus (red), β6/β7 loop (green), and β7/β8 loop (orange). Panels A–E show representative class A–E enzymes, respectively.
2.2.1 Class A Housekeeping Enzymes: Variable Active Site Loops and N-Termini May Modulate Substrate Recognition
These enzymes are typified by the aforementioned SaSrtA sortase (Fig. 4A). They have been proposed to perform a housekeeping role in the cell by anchoring a large number of functionally distinct proteins to the cell wall (Comfort & Clubb, 2004). Bioinformatics and biochemical analyses indicate that they recognize sorting signals that contain an LPXTG consensus, where X is any amino acid. In addition to SaSrtA, class A enzyme structures have been reported for Streptococcus pyogenes (Race et al., 2009), Bacillus anthracis (Weiner et al., 2010), Streptococcus agalactiae (Khare, Krishnan, et al., 2011), and Streptococcus mutans (Wallock-Richards et al., 2015). A common feature is the presence of a short helix within the β6/β7 loop, which has been shown to contact the sorting signal upon substrate binding (Chan et al., 2015; Suree et al., 2009). However, the dynamics of the loop can vary substantially. In the apo-form of SaSrtA, the loop is structurally disordered and mobile, therefore, lacking the β6/β7 helix (Ilangovan et al., 2001; Naik et al., 2006). However, sorting signal binding triggers a disordered-to-ordered transition, resulting in helix formation and helix–substrate interactions in the holo-enzyme (described below) (Suree et al., 2009). In contrast, all other class A enzyme structures contain a preformed binding pocket for the sorting signal in which the β6/β7 loop adopts an ordered state that contains the short helix.
Interestingly, class A enzymes exhibit structural variations near their catalytic histidine residue, causing some structures to contain a second groove that leads into the active site. This structural variation was first highlighted in the crystal structure of S. pyogenes SrtA (SpySrtA) (Race et al., 2009). Its catalytic domain adopts the same canonical eight-stranded β-barrel sortase fold typified by SaSrtA, but the positioning of its Cys-S sulfhydryl group differs. In SpySrtA, this sulfhydryl group points toward the active site His-δN and is separated by a distance of 5.4 Å, whereas in SaSrtA it points away from His-δN such that they are separated by 6.5 Å. These differences are illustrated by Fig. 3B and C, which show the structures of SaSrtA and SpySrtA, respectively. The side chain positioning in the SpySrtA structure results in the formation of a unique groove that leads into the active site; the walls are defined by residues in helix H1 and the β7/β8 loop, and the base is defined by residues in the β4/β5 loop (Fig. 3C, yellow). This groove is positioned adjacent to the active site, opposite the sorting signal binding groove, providing a potential binding site for amino acids located C-terminal to the LPXTG motif in the protein substrate, or for the lipid II substrate. In contrast, this groove is partially masked in the structure of apo-SaSrtA because of interactions between residues in the β7/β8 loop and helix H1. The structure of the S. agalactiae SrtA (SagSrtA) has also been determined, and like SpySrtA, contains the same secondary groove adjacent to the active site. The SagSrtA structure is unique for the conformation of its β6/β7 loop; however, it seems likely that this difference may be a byproduct of the buffer used to crystallize the protein, as residues in the loop coordinate a Zn2+ ion in conjunction with an adjacent protein in the asymmetric unit (Khare, Krishnan, et al., 2011).
Some class A enzymes contain a flexible, N-terminal appendage that may modulate substrate binding. In the NMR structure of the B. anthracis SrtA (BaSrtA) sortase, the appendage, formed by residues Asp57 to Val79, wraps around the body of the protein to contact the enzyme’s active site (Fig. 4A, colored red) (Weiner et al., 2010). The first eight residues, Asp57 to Pro64, adopt an extended conformation and partially shield the active site His126 residue from the solvent, while the remainder of the appendage contains a short alpha helix and wraps around the surface of the catalytic domain to contact helix H2 and the β2/H2 loop. Recent studies suggest that the N-terminal appendage modulates substrate access to the enzyme, possibly increasing the efficiency of protein display by reducing unproductive hydrolytic cleavage of enzyme–protein covalent intermediates that form during the cell wall-anchoring reaction (Chan et al., 2015). Conformational plasticity in a related N-terminal segment has also recently been observed in the structure of the class A sortase from S. mutans SrtA (SmSrtA) (Wallock-Richards et al., 2015). The SmSrtA crystal structure reveals a dimer in which an extended N-terminal helix preceding the catalytic domain interacts with residues in the active site of a symmetry-related molecule. Although these intermolecular interactions may be an artifact of crystallization, they highlight the proclivity of amino acids preceding the catalytic domain to interact with the enzyme’s active site, which has now been observed in BaSrtA and several class C enzymes (described below). NMR studies also revealed that the SaSrtA and BaSrtA enzymes exhibit distinct active site conformational dynamics even though they recognize sorting signals that contain an LPXTG motif (Ilangovan et al., 2001; Naik et al., 2006; Weiner et al., 2010). In BaSrtA, the β6/β7 and β7/β8 loops adopt rigid and mobile states prior to engaging the sorting signal, respectively; however, these loop dynamics are reversed in SaSrtA (the β6/β7 and β7/β8 loops are mobile and structured, respectively). The coordinates for the class A enzyme from Streptococcus pneumoniae (SpnSrtA) have also been deposited in the PDB, but its biological significance is unclear as the protein adopts an unusual β-strand swapped dimer and a paper describing this structure has not been published.
2.2.2 Mixed Function Class B Enzymes: An Extended β6/β7 Loop Is Used to Recognize Noncanonical Sorting Signals
Class B enzymes have diverse functions, with members of this subfamily either anchoring proteins to the cell wall or acting as pilin polymerases that assemble pili (Kang, Coulibaly, Proft, & Baker, 2011; Lazzarin et al., 2015; Mazmanian, Ton-That, Su, & Schneewind, 2002). The sorting signals recognized by class B enzymes vary, but are predicted to have a NPX[T/S] [N/G/S] consensus instead of LPXTG (Comfort & Clubb, 2004). Five crystal structures of class B sortases have been reported. When compared to the canonical SaSrtA structure, two major differences are apparent as class B enzymes contain: (i) additional helices located N-terminal to the catalytic domain (Fig. 4B, colored red) and (ii) a much longer β6/β7 loop that contains an additional α-helix (Fig. 4B, colored green). Structures of class B enzymes that attach proteins to the cell wall were determined first. In 2004, structures of the S. aureus SrtB (SaSrtB) enzyme bound to several nonspecific sulfhydryl modifiers were elucidated (Zong, Mazmanian, Schneewind, & Narayana, 2004). As described below, subsequent studies of SaSrtB bound to its signal peptide revealed that the extended β6/β7 loop is involved in recognizing its distinct NPQTN sorting signal substrate (Jacobitz et al., 2014). The role of the additional N-terminal helices remains unknown, but they may be important for dictating this class B enzyme’s preference for anchoring substrates to buried, uncrosslinked portions of the cell wall (Marraffini & Schneewind, 2005; Mazmanian et al., 2003). The apo-structures of SaSrtB and B. anthracis SrtB (BaSrtB) were also reported in 2004 and are structurally similar (Cα coordinate RMSDs 3.2 Å) (Zhang et al., 2004). These structures differ in that a portion of the β7/β8 loop in BaSrtB appears to be dynamic, as electron density for this region is missing. Minor conformational variations also occur in the β6/β7 loop and the short loop that connects helices H1 and H2 in these enzymes. A structure of BaSrtB bound to a aryl (beta-amino) ethyl ketone inhibitor has also been determined, revealing only small structural differences with the apo-form of the enzyme (Maresso et al., 2007). Very recently, the atomic structure of the SrtB enzyme from Clostridium difficile (CdSrtB) was determined by X-ray crystallography (Chambers, Roberts, Shone, & Acharya, 2015) and is nearly identical in structure to SaSrtB (RMSD = 1.93 Å for all Cα atoms). Interestingly, unlike the SaSrtB and BaSrtB proteins that attach heme-binding proteins to the cell wall, CdSrtB appears to play a more generalized function, as a genetic analysis has predicted that it attaches seven proteins to the cell wall, none of which are thought to be involved in iron acquisition (Chambers et al., 2015).
Some members of the class B subfamily function as pilin polymerases, instead of attaching proteins to the cell wall. Structures of two polymerizing class B sortases have been determined, S. pyogenes (SpySrtB) and S. pneumoniae (SpnSrtB) (also referred to as SrtG-1) (Kang et al., 2011; Shaik et al., 2015). This work revealed that class B enzymes that attach proteins to the cell wall or assemble pili adopt generally similar tertiary structures. However, the polymerizing enzymes are unique because they contain an additional short helix and β-strand within the extended β6/β7 loop (Fig. 4B, SpySrtB). The latter alteration adds an uncommon ninth β-strand to the protein that is not inserted into the conserved β-barrel core, but instead paired with a portion of strand β6 on the protein’s surface. Understanding how class B sortases can have similar structures yet distinct functions is a major unresolved question.
2.2.3 Class C Pilin Polymerases: An N-Terminal “Lid” Regulates Sorting Signal Substrate Access
Class C sortases can function as polymerases that link pilin proteins together via lysine–isopeptide bonds to construct pili (Fig. 2) (Ton-That & Schneewind, 2003). In some instances, a single class C enzyme can also perform double duty, acting as both a pilin polymerase and a cell wall anchoring sortase that attaches proteins to the peptidoglycan. Members of this class recognize proteins with sorting signals that contain the consensus [L/I]PXTG (Comfort & Clubb, 2004). Because of their unique polymerizing function, class C enzymes have been actively studied with a total of 15 structures being reported to date. Interestingly, nearly all of these enzymes contain a “lid” structure, an elongated N-terminal region that occludes the active site (Fig. 4C, red) (Mandlik et al., 2008; Manzano et al., 2008; Manzano, Izoré, Job, Di Guilmi, & Dessen, 2009; Spirig et al., 2011). Members of this group also harbor a unique C-terminal nonpolar helix that is important for function and likely embedded in the membrane (Cozzi et al., 2011; Wu et al., 2012).
The first class C enzymes to be structurally characterized were SrtC-1 and SrtC-3 from S. pneumoniae (SpnSrtC1 and SpnSrtC3) by Manzano et al. (2008). This work revealed the presence of an N-terminal lid that contains a conserved DP(F/W/Y) motif. The aspartic acid residue in the lid motif favorably contacts the conserved active site arginine residue in the His–Cys–Arg triad. The lid is also bound to the active site via sulfur–aromatic interactions between the active site cysteine and aromatic residues in the lid (Phe, Trp, or Tyr depending upon the enzyme). Owing to these key interactions, the Asp and aromatic residues in the lid motif are referred to as “anchors” (Manzano et al., 2008, 2009). Interestingly, the B-factors for residues comprising the lid are elevated, suggesting that they are mobile in solution and transiently detach from the enzyme’s active site. Subsequent structures of class C sortases from S. agalactiae (Cozzi et al., 2011, 2012; Khare, Fu, Huang, Ton-That, & Narayana, 2011), A. oris (Persson, 2011), and S. pneumoniae (Neiers et al., 2009) supported this idea, revealing similar patterns of elevated B factors or stretches of missing electron density in regions flanking the lid anchor residues. Several other class C structures have been determined, which reveal only small shifts in lid positioning or subtle differences in the amount of electron density that define their lids.
It has been proposed that the lid regulates enzyme activity (Manzano et al., 2008, 2009) (add references Mazmanian et al., 2002; Zong, Mazmanian, et al., 2004). When it adopts the closed state observed in nearly all crystal structures, the lid occludes the binding site for the sorting signal and holds the enzyme in an inactive state. The enzyme can then become activated by partial dislodgement of the lid, enabling binding of the sorting signal, and formation of the enzyme–substrate thioacyl intermediate. This notion is compatible with modeling studies of the sortase–signal complex, as well as in vitro data that have demonstrated that mutants harboring alterations in the lid exhibit increased rates of sorting signal hydrolysis (Cozzi et al., 2011) and, in some instances, reduced stability (Manzano et al., 2009). However, the role of the lid in catalysis is not fully understood, since cellular studies of pilin polymerases containing mutations that should presumably dislodge the lid exhibit wild-type transpeptidation activity in vivo (Cozzi et al., 2011; Wu et al., 2012). Moreover, structural data have shown that the lid does not completely block access to the enzyme active site, since the crystal structure of S. agalactiae SrtC-1 (SagSrtC1) shows the enzyme bound to the nonspecific sulfhydryl modifier (MTSET) despite adopting a closed-lid state (Khare, Fu, et al., 2011).
Recent NMR and computational studies of SpnSrtC1 suggest that the lid in class C enzymes prefers to adopt a closed and rigid state (Jacobitz et al., 2016). This work revealed that the lid in SpnSrtC1 adopts a rigid conformation in solution that is devoid of large magnitude conformational excursions that occur on mechanistically relevant timescales. Additionally, point mutations in the lid were shown to induce dynamic behavior that correlates with increased hydrolytic activity and sorting signal substrate access to the active site cysteine. These results support the notion that the lid in this class C enzyme has a negative regulatory function, and imply that a significant energetic barrier must be surmounted to dislodge it from the active site and initiate pilus biogenesis. Presumably, an as of yet unidentified factor(s) must pry the lid open to hold the enzyme in a catalytically active state that can assemble pili.
One structure of the SagSrtC1 enzyme appears to have captured the lid in an “open” conformation, providing insight into the mechanism of lid-opening that is expected to precede binding of the sorting signal (Fig. 4C, compare left to right) (Khare, Fu, et al., 2011). The structure of SagSrtC1 was determined from multiple crystalline forms, and one of these structures, solved in space group C2, showcases the lid in an “open” conformation. The enzyme maintains the typical sortase fold and, excluding the N-terminal extension preceding the β-barrel core, is extremely similar to SagSrtC1 structures previously solved in space groups P212121 and P312 with an average backbone RMSD of 0.72 Å (Khare, Fu, et al., 2011). However, in space group C2, residues A38–E71, which typically form the lid structure, instead form an extended helical structure with the aromatic lid anchor residue of the conserved DP(F/W/Y) motif (Y51) displaced from the active site by over 30 Å to a position where it stacks against the backbone of helix H2. An additional crystal structure of Streptococcus suis SrtC-1 (SsSrtC1) also maintains a similar “open” conformation of the lid, with the same extended helix replacing what was expected to be a closed lid (Lu et al., 2011). As nearly all structures of class C enzymes possess a closed lid, which NMR and computational studies suggest is immobile, it is tempting to speculate that the unique open structure observed for SagSrtC1 and SsSrtC1 arose from the solvent conditions used to obtain this crystal form of the protein. Finally, when regions outside of the lid are compared in other class C structures, perhaps the most significant deviation from the norm is the addition of a short C-terminal α-helix opposite the active site in the structure of SpnSrtC3; however, its functional importance has not been determined (Manzano et al., 2008).
2.2.4 Class D Enzymes: Specialized Sortases That Attach Cell Wall Proteins That Contain an LPXTA Sorting Signal
Class D sortases predominate in Bacilli species and recognize an unusual LPXTA motif consensus, in which an alanine (underlined) replaces the canonical glycine residue (Comfort & Clubb, 2004). Currently, only two structures of class D enzymes have been reported. In 2012, the NMR structure of the B. anthracis SrtD enzyme was published (BaSrtD, and also previously referred to as SrtC) (Robson et al., 2012). BaSrtD anchors proteins required for efficient sporulation to the surface of the cell wall. The catalytic domain of BaSrtD adopts the conserved eight-stranded β-barrel sortase fold (Fig. 4D). Structurally, it is most similar to members of the class A subfamily as it contains an ordered 310 helix within the β6/β7 loop and lacks the elongated β6/β7 loop and lid that are found in class B and C enzymes, respectively (Fig. 4). Interestingly, ultracentrifugation studies indicate that isolated BaSrtD forms a dimeric structure with a KD of 89 µM (Robson et al., 2012). Based on resonance line broadening effects observed in its NMR spectrum, BaSrtD dimerization is potentially mediated by residues in the structurally disordered β2/β3 and β4/H1 surface loops that are positioned adjacent to the active site histidine residue. It has been suggested that this disordered surface may mediate interactions with lipid II or other factors on the cell surface, but this has not been demonstrated experimentally. Recently, the structure of the Clostridium perfringens SrtD (CpSrtD) sortase was determined at 1.99 Å (Suryadinata et al., 2015). Similar to BaSrtD, CpSrtD adopts a β-barrel sortase fold that contains a short helix within the β6/β7 loop. However, CpSrtD also contains two alpha helices at its N-terminus, and unlike BaSrtA, its β2/β3 and β4/H1 loops are structurally ordered (the β2/β3 loop also contains a two-turn alpha helix). Furthermore, CpSrtD exists as a monomer according to crystallographic and dynamic light scattering studies. Interestingly, CpSrtD exhibits catalytic activity in vitro that is enhanced in a magnesium-dependent manner, making it one of only two known sortases (the other being SaSrtA) whose activity is modulated by metal ions. The origin of this stimulatory effect is not known in CpSrtD, but in SaSrtA, metal binding increases enzyme activity by modulating the structure and dynamics of the β6/β7 loop (described below).
2.2.5 Class E Enzymes: A Novel LAXTG Sorting Signal for Anchoring of Surface Proteins in Actinobacteria
Class E enzymes predominate in soil and freshwater-dwelling Actinobacteria and have not been studied extensively. Bioinformatic predictions suggest that members of this group recognize an unusual LAXTG sorting signal motif in which the highly conserved proline residue is replaced with alanine (underlined) (Comfort & Clubb, 2004). Two class E sortases from Streptomyces coelicolor, ScSrtE1 and ScSrtE2, have been shown to display chaplin proteins to promote aerial hyphae development (Duong et al., 2012). In vitro studies indicate that ScSrtE1 and ScSrtE2 can hydrolyze LAETG- and LAHTG-containing peptides, cleaving the peptide bond after the threonine residue. The enzymes exhibit promiscuous activity, as they also cleave an LAETG peptide at a secondary site following the alanine (Duong et al., 2012). Very recently, we reported the first crystal structure of a class E sortase, the 1.93 Å resolution structure of ScSrtE1 (Fig. 4E) (Kattke et al., 2016). The structure is similar to class A enzymes, as its β6/β7 loop contains a single 310 helix. However, variations in the conformation of its β3/β4 and β6/β7 loops are evident. ScSrtE1 contains a 21 amino acid insertion immediately following the 310 helix in the β6/β7 loop. This long insertion is similar in length to that observed in class B sortases, but is distinctly devoid of secondary structure, whereas class B sortases contain an additional alpha helix. In the ScSrtE1 structure, the active site is bound to a tripeptide that is presumably a proteolytic protein fragment that was present in the crystallization buffer. The coordinates of the tripeptide and computational modeling with substrate mimics suggest that ScSrtE1 and other members of this group may use a class E-specific tyrosine residue present within their β3/β4 loops to recognize the alanine backbone within the LAXTG substrate. It is possible that the tyrosine participates in a hydrogen bond with the amide nitrogen of the alanine residue within the signal, an interaction that is not possible in substrates containing a proline at this position. However, the role of the conserved tyrosine in dictating substrate specificity was not experimentally determined because single amino acid mutants of ScSrtE1 that altered this position in the protein were unstable. Class F enzymes are also prevalent in Actinobacteria, but they have not been structurally or biochemically characterized.
3. STRUCTURAL BIOLOGY: MOLECULAR BASIS OF SUBSTRATE RECOGNITION
All sortases characterized to date catalyze a transpeptidation reaction that joins an LPXTG-like sorting signal within the CWSS of their protein substrate to an amino nucleophile (Fig. 2). However, depending upon the type of sortase, the chemical structure of these substrates can vary substantially. For example, biochemical and bioinformatics analyses suggest that class A, B, C, D, and E enzymes have evolved specificities for distinct LPXTG, NPX[T/S][N/G/S], [L/I]PXTG, LPXTA, and LAXTG sorting signals, respectively (unambiguous differences from LPXTG are underlined) (Comfort & Clubb, 2004). In addition, they can either recognize nucleophiles that originate from lipid II (sortases that anchor proteins to the cell wall), or a lysine residue located within another protein (sortases that function as pilin polymerases) (Fig. 2). Typically, microbes encode genes for more than one sortase enzyme (Pallen et al., 2001). Their distinct substrate specificities enable multiple sortases to nonredundantly operate, with different types of sortases “sorting” distinct proteins to the cell surface or assembling pili. Below, we summarize what is currently known about the molecular basis of substrate recognition.
3.1 Sorting Signal Recognition
The sorting signal recognized by sortases is located within the CWSS of the protein substrate, which in turn is embedded in the bilayer via its hydrophobic domain (Fig. 1) (Schneewind et al., 1992). Our structural studies have shed light onto how class A and B enzymes recognize LPATG and NPQTN sorting signals, respectively (Chan et al., 2015; Jacobitz et al., 2014; Suree et al., 2009). Since sortases can hydrolyze their cognate sorting signals and bind to them weakly in vitro, these structural studies made use of a substrate analog developed by the Jung group in which the threonine moiety contains a sulfhydryl group in place of its carbonyl atom (Jung et al., 2005). The analog (hereafter called T*), mimics the threonine residue in the native sorting signal substrate, but forms a disulfide bond with the active site cysteine residue. This leads to the production of a stable enzyme–substrate complex that is suitable for structural studies.
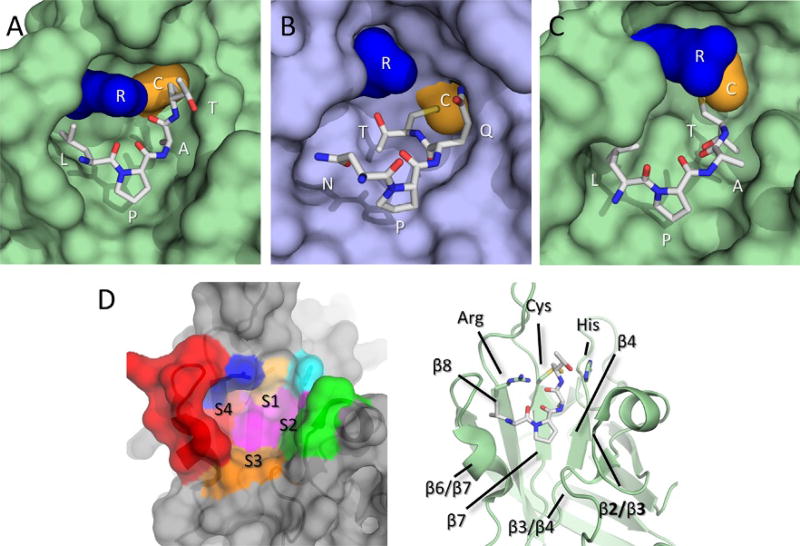
Using T*-containing sorting signal analogs the atomic structures of three sortase–substrate complexes have been determined by the Clubb group: the class A SaSrtA–LPXT* (Fig. 5A), class B SaSrtB–NPQT* (Fig. 5B), and class A BaSrtA–LPAT* (Fig. 5C) complexes (Chan et al., 2015; Jacobitz et al., 2014; Suree et al., 2009). To facilitate a discussion of the binding interactions that govern signal recognition, we henceforth refer to residues in each sorting signal in relation to their positioning relative to the scissile bond. Residues in an L–P–X–T–G sorting signal are referred to as P4–P3–P2–P1–P1′ and their corresponding binding sites on the enzyme as subsites S4–S3–S2–S1–S1′, respectively.
Fig. 5.
Sorting signal recognition. (A) The SaSrtA–LPAT* complex. (B) The SaSrtB–NPQT* complex. (C) The BaSrtA–LPAT* complex, shown with N-terminal appendage removed from view for clarity. Enzymes are shown as surface representations with SrtA types in light green and SaSrtB in light blue, substrate mimics are shown as gray sticks. Active site Cys and Arg residues are shown as gold and blue surfaces, respectively. (D) Conserved recognition sites for sortase enzymes. Left, SaSrtA shown as a transparent surface representation with recognition subsites determined from the combination of sortase structures color coded as follows: S4 is shown in red, S3 in orange, S2 in green, and S1 in magenta, and active site Arg in blue, Cys in gold, and His in cyan. Right, Cartoon diagram of SaSrtA with secondary structure elements that contribute to substrate binding labeled for clarity.
3.1.1 Sorting Signal Recognition by Class A Enzymes
The NMR structure of the SaSrtA–LPAT* complex provided the first insight into the molecular basis of sorting signal recognition (Suree et al., 2009) (Fig. 5A). In the structure of the complex, the peptide substrate binds to a pocket adjacent to the active site cysteine whose base is formed by residues located in strands β4 and β7, and whose walls are formed by surface loops that connect strand β6 to β7 (β6/β7 loop), strand β3 to β4 (β3/β4 loop), and strand β2 to helix H2 (β2/H2 loop). The location of this binding site is consistent with chemical shift mapping studies (Liew et al., 2004). Binding of the signal to SaSrtA causes a major reorganization of the active site, including a disordered-to-ordered transition of the β6/β7 loop to create a short 310 helix that contacts the signal, as well as displacement of the β7/β8 loop. The latter change may have a regulatory role, exposing the active site histidine to the solvent, and possibly facilitating the binding of the secondary lipid II substrate only after the sorting signal has first bound to the enzyme. Additional 15N relaxation analyses revealed that the β6/β7 loop, which is highly dynamic in the unbound state, rigidifies upon peptide binding (Naik et al., 2006; Suree et al., 2009). The peptide binding mode in the SaSrtA–LPAT* complex differs substantially from a previously reported crystal structure of SaSrtA noncovalently bound to a LPETG peptide (Zong, Bice, et al., 2004). In the crystal structure, the peptide is presumably nonspecifically bound, which is not surprising as the sorting signal substrate binds to SaSrtA with very weak affinity (Km = 7.33mM) (Frankel et al., 2005).
The structure of the SaSrtA–LPAT* complex reveals how class A enzymes recognize the P4 and P3 residues within the LPXTG sorting signal, Leu and Pro, respectively (Suree et al., 2009). The P4 leucine side chain is positioned within a large hydrophobic S4 pocket that is formed by residues V161, V166, V168, and L169 in the reordered β6/β7 loop, and I199 on strand β8. The P3 Pro residue plays a key architectural role, forming a kink in the sorting signal that enables it to adopt an “L-shaped” structure in which the C-terminal end is positioned toward the active site cysteine. Numerous hydrophobic contacts to the proline are formed by the S3 site. Surprisingly, in the structure of the SaSrtA–LPAT* complex, the positioning of the P1 and P2 side chains is incompatible with biochemical and bioinformatics data (Comfort & Clubb, 2004; Fischetti, Pancholi, & Schneewind, 1990; Kruger, Otvos, et al., 2004). Subsequent structural and computational studies revealed the origin of this discrepancy and are described in Section 3.1.3.
The recently determined structure of the class A BaSrtA–LPAT* complex revealed a generally similar binding mode for the LPXTG sorting signal, but also indicated fundamental differences in the conformational dynamics and structure of the active site (Chan et al., 2015). Similar to SaSrtA, the peptide adopts an L-shaped conformation in the BaSrtA–LPAT* complex by virtue of a kink at position P3, and the β6/β7 loop is instrumental in building the S4 site that dictates specificity. However, four significant differences in the binding mechanism are apparent. First, in contrast to SaSrtA, the β6/β7 loop in BaSrtA is structured in the absence of substrate (Weiner et al., 2010) and only experiences modest changes upon signal binding (the coordinates of the Cα backbone atoms in the loop in the apo- and signal-bound forms of BaSrtA have an RMSD of 0.76 Å). Thus, the β6/β7 loop forms a preformed binding pocket for the P4 residue in BaSrtA, whereas in SaSrtA, the loop is flexible prior to signal binding (Naik et al., 2006; Suree et al., 2009). Second, the β7/β8 loop, which was disordered in the unmodified BaSrtA structure, undergoes a disordered-to-ordered transition as a result of binding to the substrate. A comparison of the apo- and bound-forms of BaSrtA reveals that the active site cysteine residue is displaced ~7 Å upon peptide binding (Chan et al., 2015; Weiner et al., 2010). This movement allows the β7/β8 loop to form new intraprotein interactions with residues within the β4/β5 loop, which presumably stabilize the β7/β8 loop and cause it to become ordered. In contrast, the β7/β8 loop of SaSrtA is structurally ordered in the apo-state and is displaced when the signal binds. Third, unlike SaSrtA, the BaSrtA enzyme contains an N-terminal appendage that partially encapsulates the sorting signal (Chan et al., 2015; Suree et al., 2009). The N-terminal appendage precedes the catalytic domain in the primary sequence and wraps around the body of the protein to contact the active site. The side chain of Ile61 in the appendage forms extensive contacts with the active site histidine, while the hydroxyl group of Ser59 is positioned to form a hydrogen bond with the backbone carbonyl oxygen of the proline residue in the bound peptide. Interestingly, NMR and in vitro kinetics data suggest that the appendage transiently detaches from the isolated enzyme when the sorting signal is bound. A model of the thioacyl intermediate constructed from the coordinates of the complex suggests that the N-terminal appendage may obstruct nucleophile access to the active site, potentially increasing the efficiency of protein display by reducing the unproductive hydrolytic cleavage of enzyme–protein covalent intermediates. A fourth difference between the BaSrtA–LPAT* and SaSrtA–LPAT* complexes is the positioning of the P1 and P2 side chains, which are described in Section 3.1.3.
3.1.2 Sorting Signal Recognition by Class B Enzymes
The structure of the SaSrtB–NPQT* complex provided the first-ever insight into signal recognition by class B enzymes (Jacobitz et al., 2014), which binds an NPQTN sorting signal in the case of SaSrtB (Fig. 5B) (Mazmanian et al., 2002). SaSrtB uses a rigid pocket to bind to the “L”-shaped peptide. Specifically, the P4 asparagine residue is primarily recognized by contacts to the β6/β7 loop. However, as compared to class A enzymes, the P4 residue is more solvent exposed, concordant with the more hydrophilic nature of the asparagine side chain. The binding mode of the sorting signal in class A and B enzymes is compatible with biochemical data from the McCafferty group. This data demonstrated that replacement of the β6/β7 loop in SaSrtA with the corresponding loop from SaSrtB resulted in a chimeric protein that could recognize the SaSrtB-specific NPQTN sorting signal motif (Bentley et al., 2007). The P3 proline residue also appears to have the same function as in the BaSrtA–LPAT* and SaSrtA–LPAT* structures, altering the trajectory of the peptide so that its C-terminal end points toward the active site. As with BaSrtA, the β6/β7 loop in SaSrtB forms a rigid, preformed binding pocket for the sorting signal; the structures of BaSrtA in the free- and bound-states are nearly identical (the Cα coordinates have an RMSD = 0.44 Å). Moreover, within the BaSrtA–LPAT* and SaSrtB–NPQT* complexes, the P1 and P2 side chains in the bound substrates adopt similar positions relative to the enzyme active site that are distinct from their positioning observed in the structure of the SaSrtA–LPAT* complex (see Section 3.1.3).
3.1.3 Sorting Signal Conformational Heterogeneity: Thr-In vs Thr-Out
A comparison of the structures of the SaSrtA–LPAT* and SaSrtB–NPQT* complexes revealed that there were major differences in the conformation of the P1 and P2 residues of the bound sorting signals (Jacobitz et al., 2014). In particular, the P2 glutamine residue in the SaSrtB–NPQT* complex rests along the wall of the pocket and points out toward the solvent, while in the SaSrtA–LPAT* structure, the analogous P2 residue (alanine) points toward the base of the active site. The P1 threonine residues in these complexes also adopt fundamentally distinct conformations. In the SaSrtB–NPQT* structure, the threonine is buried in the active site (Thr-in position) where it forms two hydrogen bonds with the active site arginine residue, whereas in the SaSrtA–LPAT* structure, it is projected toward solvent and hydrogen bonds with the active site histidine residue (Thr-out position). Interestingly, an inspection of the recently determined structure of the BaSrtA–LPAT* complex reveals a P1 and P2 positioning that is similar to what is seen in the SaSrtB–NPQT* substrate complex; the P1 threonine side chain adopts a Thr-in conformation in which it is buried within the active site. Thus, the Thr-in conformation of the peptide can be adopted by signal peptides bound to both class A and B enzymes.
Several lines of evidence suggest that the Thr-in conformer observed in the structures of the BaSrtA–LPAT* and SaSrtB–NPQT* complexes represents the catalytically active form of the bound substrate. First, in these structures, the side chain of the P2 residue projects into the solvent and is not recognized. This makes sense as the P2 residue within the sorting signals, also called the “X” position, is not recognized by sortases according to bioinformatics (Comfort & Clubb, 2004) and biochemical data (Kruger, Otvos, et al., 2004). This is in contrast to the SaSrtA–LPAT* complex in which the peptide adopts the Thr-out conformer and the side chain at the P2 “X” position projects into the active site where it contacts A118 and I182. Second, the high degree of sequence conservation at site P1, which contains a threonine residue in ~95%of predicted sorting signals, suggests functional relevance of the Thr-in sorting signal conformer observed in the BaSrtA–LPAT* and SaSrtB–NPQT* complexes. This is in marked contrast to the Thr-out conformer observed in the SaSrtA–LPAT* complex that projects into the solvent and is not recognized by the enzyme. Finally, as described in Section 4, computational modeling of sortase reaction intermediates indicates that the Thr-in conformer likely facilitates the creation of an oxyanion hole for substrate stabilization during thioacyl intermediate formation.
3.2 Nucleophile Recognition
Unlike the mechanism of sorting signal recognition, sortase binding to their secondary substrate, the amino nucleophile to which the sorting signal is joined, remains poorly understood. Depending upon the type of sortase, cell wall-anchoring vs pilin polymerizing, the nucleophile can either originate from a lipid II molecule or a lysine residue within a pilin protein substrate, respectively. Thus far, only lipid II recognition by sortases has been explored, but the mechanism of binding still remains enigmatic. Experimental studies have investigated how the SaSrtA enzyme interacts with polyglycine peptides that mimic the pentaglycine crossbridge peptide moiety within the intact S. aureus lipid II molecule, as large quantities of the intact, water-soluble portion of lipid II are difficult to obtain. NMR chemical shift mapping of the SaSrtA–LPAT* complex using a triglycine peptide titrant revealed a low affinity, continuous interaction surface on the enzyme that contains portions of the β7/β8 loop, β4/H2 loop, and an N-terminal segment of helix H1 (Suree et al., 2009). The mapping data coarsely define the interaction surface, but do not provide specific details about triglycine–enzyme interactions. Interestingly, significant chemical shift changes are not observed when apo-SaSrtA is titrated with the triglycine peptide, but only when added to the SaSrtA–LPAT* complex. This observation is compatible with the proposed bi–bi mechanism of catalysis (Frankel et al., 2005; Huang et al., 2003) and suggests that LPXTG sorting signal binding may direct catalysis forward by causing alterations in the β7/β8 loop that unmask the binding surface for lipid II. Crystals of SaSrtB modified with a small molecule sulfhydryl modifier were soaked with a triglycine peptide, and the structure was determined by molecular replacement. In this complex, the N-terminal amine of the triglycine peptide is 6.4 Å from the active site histidine, which is occluded from solvent by a closed β7/β8 loop (Zong, Mazmanian, et al., 2004). While the binding mode of the triglycine peptide to SaSrtB is generally compatible with the NMR chemical shift data from the SaSrtA–LPAT* complex, it does not fit the accepted view of the mechanism that would have the incoming nucleophile deprotonated by the active site histidine residue.
Interestingly, other class A sortases appear to contain a preformed binding site for the crossbridge peptide in lipid II. In the crystal structures of the SpySrtA, SagSrtA, and SmSrtA sortases, their β7/β8 loops are displaced in the absence of the sorting signal, creating a surface-exposed groove adjacent to the active site histidine and cysteine residues (Fig. 3C, yellow) (Khare, Krishnan, et al., 2011; Race et al., 2009; Wallock-Richards et al., 2015). Recent docking studies of BaSrtA highlight a potential binding site for the amino component of lipid II in B. anthracis (diaminopimelic acid) (Chan et al., 2015). This site is positioned adjacent to the active site histidine, between the β7/β8 and β4/H3 loops, and would position the nucleophile near the electrophilic carbonyl carbon atom in the thioacyl bond. Interestingly, exposure of this site requires partial displacement of the N-terminal appendage, which has been postulated to mask the acyl-linked reaction intermediate from hydrolysis. Conclusive determination of the molecular basis of amino nucleophile selectivity will require structure determination of sortases bound to their secondary substrates. The mechanism of pilin protein substrate recognition by pilin polymerases also remains to be determined.
4. COMPUTATIONAL STUDIES
4.1 The Thr-In Conformation Enables Sortases to Employ a Substrate-Stabilized Active Site
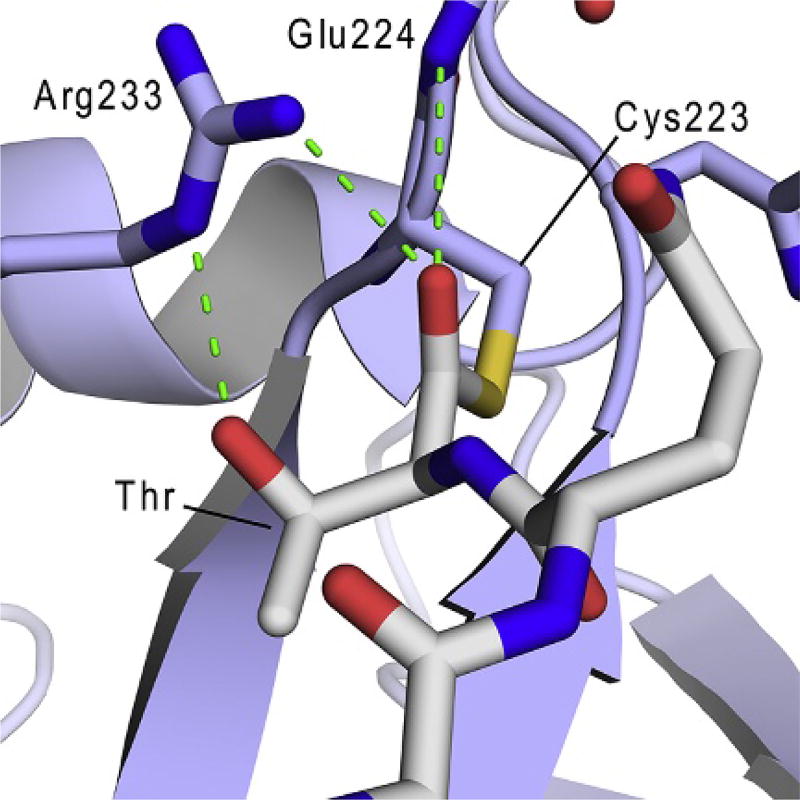
Computational studies have leveraged the experimentally determined structures of sortase–substrate complexes to gain insight into the mechanism of catalysis. Jacobitz et al. generated an energy minimized model of the thioacyl intermediate using the coordinates of the SaSrtB–NPQT* complex (Jacobitz et al., 2014). The model of the thioacyl intermediate is shown in Fig. 6 and reveals that the Thr-in conformation adopted by the highly conserved P1 threonine residue (see Section 3.1.3) enables it to hydrogen bond to the active site arginine residue. This interaction positions the active site arginine proximal to the thioacyl bond, where it presumably can stabilize the two high-energy tetrahedral oxyanion intermediates that form during catalysis (Jacobitz et al., 2014). In this capacity, threonine–arginine interactions facilitate the formation of an oxyanion hole in which the arginine neutralizes the negative charge of the oxyanion. Further stabilization of the oxyanion is achieved by interactions from the backbone amide of E224 that is located immediately C-terminal to the active site cysteine. This novel “substrate-stabilized oxyanion hole” presumably increases substrate specificity for a threonine residue at position P1 and is compatible with biochemical studies that have shown that the enzyme is unable to utilize sorting signals that contain conservative mutations at the P1 position (e.g., Ser or Val instead of Thr) (Jacobitz et al., 2014). A similar computational strategy was used to model the structure of the BaSrtA thioacyl intermediate using the experimentally determined coordinates of the BaSrtA–LPAT* complex (Chan et al., 2015). This model displayed an analogous hydrogen bonding interaction between the P1 threonine residue and the active site arginine. As in the SaSrtB model, the P1 threonine carbonyl atom in the thioacyl bond is in close proximity to the active site arginine guanidino group, suggesting that both enzymes employ-related substrate-stabilized oxyanion holes to facilitate catalysis (Chan et al., 2015). However, it should be noted that the active site conformations of the two models differ slightly, since in the model of the BaSrtA thioacyl intermediate, the active site arginine adopts a more extended structure that allows it to form an additional hydrogen bond to the P3 proline residue.
Fig. 6.
The substrate-stabilized oxyanion hole. The energy minimized model of the SaSrtB-NPQT thioacyl intermediate displayed with SaSrtB (light blue cartoon), residues in the active site and oxyanion hole (sticks), and NPQT substrate (gray sticks). The side chain hydroxyl of the substrate’s P1 Thr residue and backbone carbonyl participate in a hydrogen bonding network with the active site Arg, and the backbone amide of Glu224 that together build an oxyanion hole to stabilize the high energy tetrahedral reaction intermediates. Reproduced from Jacobitz, A. W., Wereszczynski, J., Yi, S. W., Amer, B. R., Huang, G. L., Nguyen, A. V., et al. (2014). Structural and computational studies of the Staphylococcus aureus Sortase B-substrate complex reveal a substrate-stabilized oxyanion hole, The Journal of Biological Chemistry, 289, 8891–8902, p. jbc.M113.509273.
4.2 Sorting Signals Bound to SaSrtA Can Interchange Between Thr-Out and Thr-In Conformers
As described in Section 3.1.3, a comparison of the experimentally determined structures of the SaSrtA–LPAT*, SaSrtB–NPQT*, and BaSrtA–LPAT* revealed fundamental differences in the positioning of the P1 and P2 residues in the bound sorting signal (Chan et al., 2015; Jacobitz et al., 2014; Suree et al., 2009).
To rectify the discrepancies between the SaSrtA–LPAT* Thr-out and SaSrtB–NPQT* Thr-in conformations, Jacobitz et al. performed molecular dynamics (MD) simulations for three thioacyl intermediate systems: SaSrtA–LPAT, SaSrtB–NPQT, and SaSrtB–NPAT (Jacobitz et al., 2014). Through the use of umbrella sampling calculations, the free energy landscape of transitions between the Thr-out and Thr-in states was mapped for each complex. These studies indicated that SaSrtA–LPAT could adopt both the Thr-in and Thr-out states with equal probability, while for both NPQT and NPAT substrates, SaSrtB could only sample the Thr-in state. Based on these results, it appears as if the Thr-in conformation is likely the more evolutionarily conserved state, and that the inherent flexibility of SaSrtA allows for the Thr-out conformation with an LPAT substrate that was captured by Suree et al. (2009).
4.3 Dynamic Sorting Signal Recognition by SaSrtA
MD studies of SaSrtA have provided additional insight into the sorting signal recognition process. By performing conventional and accelerated MD simulations of both the sorting signal free and bound states, Kappel et al. proposed that sorting signal binding is a mixture of conformational selection and induced fit mechanisms (Kappel, Wereszczynski, Clubb, & McCammon, 2012). For example, the β6/β7 loop appears to follow the conformational selection paradigm: it sampled a range of stable conformations in the apostate, some of which were relatively close to the bound configurations. In contrast, the β7/β8 “open” state from the NMR structure was only stable in the presence of a bound sorting signal, suggesting an induced fit mechanism. In addition, analysis of the sorting signal-bound conformations showed that an allosteric network runs throughout the protein, linking the calcium ion, sorting signal, and proposed lipid-II binding regions to one another. In a complementary study, Moritsugu et al. used the multiscale enhanced sampling method to probe the allosteric effects of the calcium ion and sorting signal (Moritsugu, Terada, & Kidera, 2012). Simulations of each combination of bound states showed that binding of both molecules is required to stabilize the β6/β7 and β7/β8 loops in conformations observed in the NMR–LPAT* structure. Overall, these simulations point toward a mechanism in which calcium, sorting signal, and potentially lipid II binding are modulated by a dynamic network that includes the β6/β7 loop region in SaSrtA.
Other aspects of the SaSrtA recognition and catalytic processes have also been explored by computational studies. Biswas et al. used a hybrid MD and biochemical experimental approach to probe the roles of the conserved sorting signal Leu and Pro residues in substrate binding (Biswas, Pawale, Choudhury, & Roy, 2014). Comparative simulations with LPAT, APAT, and LAAT substrates demonstrated that contacts between the leucine side chain and SaSrtA contribute to stabilize the β6/β7 loop, whereas the kink that is induced by the proline appears to be essential for recognition. In another study, Tian and Eriksson performed simulations in which His120 and Cys184 were in their zwitterionic and neutral forms (Tian & Eriksson, 2011). Their results showed that Arg197 adopts distinct conformations based upon the charged state of the protein, which helps to stabilize the catalytically active form. It should be noted that each of these studies was performed with the sorting signal in the Thr-out state. Although the global effects of the Thr-in and Thr-out states on the induced fit/conformational selection process, allosteric networks, and recognition processes are likely similar, subtle differences may exist that influence some of the fine details that resulted from these simulations.
5. CATALYTIC MECHANISM
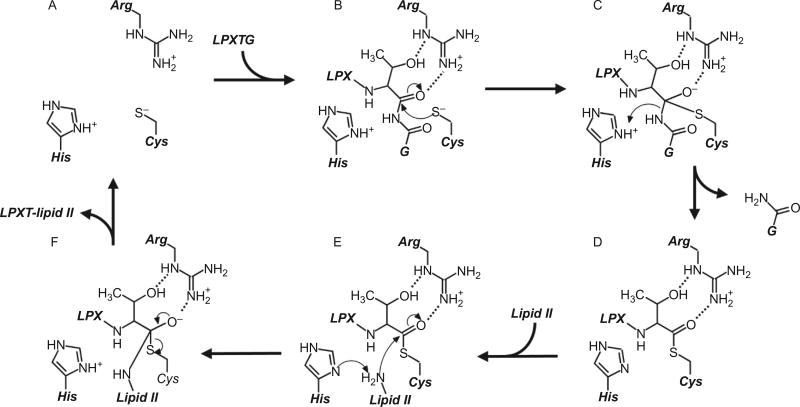
The current model of the molecular mechanism of the SaSrtA enzyme is presented in Fig. 7. Kinetic studies indicate that catalysis occurs through a ping-pong mechanism that begins when the sortase recognizes the CWSS of a membrane anchored protein (Frankel et al., 2005; Huang et al., 2003). The LPXTG-type sorting signal within the CWSS binds to a groove on the sortase whose base is formed by residues in the β6/β7 loop, strands β4, β7, β8, and whose walls are formed by residues in the β2/β3 and β3/β4 loops (Fig. 4D). Here the sorting signal’s L-shaped structure dictated by the highly conserved proline residue at P3 (>90% conserved) orients the residue P4 for recognition in subsite S4 on the β6/β7 loop, as the C-terminal end of the sorting signal is directed toward the active site (Suree et al., 2009). In order for catalysis to proceed, the enzyme must contain a properly charged active site in which the cysteine and histidine residues are in their thiolate and imidazolium forms, respectively. Based on pKa measurements of their active site residues (Connolly et al., 2003; Frankel et al., 2005; Weiner et al., 2010), when removed from the cell surface, in isolation less than 1% of the SaSrtA and BaSrtA sortases possess an appropriately charged active site that can perform catalysis. This explains why sortases in isolation catalyze reactions very slowly, as presumably only a small fraction of these enzymes are active (Section 6). The notion that the active site is predominantly dormant is substantiated by structural analyses of SaSrtA, which revealed that its active site cysteine and histidine residues are not close enough to form a thiolate-imidazolium ion-pair (the Cys-S-His-δN distance is 6.5 and 7.6 Å in the NMR and crystal structures, respectively) (Ilangovan et al., 2001; Zong, Bice, et al., 2004). In the rare instance that the cysteine and histidine residues are appropriately ionized, the cysteine thiolate nucleophilically attacks the carbonyl carbon on the P1 residue (Fig. 7B). This leads to the formation of the first tetrahedral intermediate, which is likely stabilized by an oxyanion hole that is formed by the active site arginine residue and a backbone amide in the β7/β8 loop (Jacobitz et al., 2014). In nearly all sorting signals a threonine occurs at the P1 position, which based on structural and MD simulations helps to stabilize the oxyanion hole by forming a hydrogen bond to the active site arginine (Jacobitz et al., 2014) (Fig. 7C). The first oxyanion is a transient intermediate, and quickly collapses to form a semi-stable thioacyl intermediate as the scissile peptide bond is broken. In the thioacyl intermediate, the substrate’s P1-Thr and the active site cysteine are joined via a thioacyl bond (Fig. 7D) (Aulabaugh et al., 2007). This process is presumably assisted by the conserved active site histidine residue, which may act as a general acid to protonate the amino leaving group. Beyond stabilizing the oxyanion intermediate, the side chain of the active site arginine residue may orient the substrate in the active site by forming a hydrogen bond to the backbone carbonyl atom in the P2 residue (Chan et al., 2015; Suree et al., 2009; Tian & Eriksson, 2011). In the next step of the reaction, a secondary substrate bearing an amine group enters the active site and is presumably deprotonated by the active site histidine residue to facilitate its nucleophilic attack on the thioacyl bond (Fig. 7E). A second tetrahedral intermediate then forms which may also be stabilized by an oxyanion hole that is constructed with the assistance of the sorting signal’s P1 residue (Fig. 7F). The second tetrahedral intermediate then quickly collapses to form the peptide bond linked product. Sortases that anchor proteins to the cell wall join the protein substrate to the cell wall precursor lipid II, whereas pilin polymerases join the protein to a lysine amine located within another pilin protein. These enzymes are believed to use a similar mechanism to catalyze transpeptidation.
Fig. 7.
Molecular mechanism of sortase enzymes. The active site of sortase consists of a His–Cys–Arg triad, and in its active form, the His and Cys residues form a thiolate–imidazolium ion-pair (A). The reaction begins with recognition of an appropriate sorting signal (here, the LPXTG sorting signal for SrtA types is shown), and the active site Cys residue performs nucleophilic attack on the carbonyl carbon at the substrate’s P1 position (B). An oxyanion tetrahedral intermediate is stabilized by the nearby Arg residue that is likely oriented by interactions with the side chain of the substrate’s P1 residue, which is a threonine in over 95% of all substrates (C). The active His residue concomitantly donates a proton to the leaving group, and the tetrahedral transition state then collapses to form a semistable, thioacyl intermediate between the substrate’s P1 residue and the active site Cys (D). Next, the secondary substrate (here shown as lipid II used by cell wall anchoring sortases) enters the active site, where its terminal amine is deprotonated by the active His residue before performing nucleophilic attack on the carbonyl carbon in the thioacyl bond (E); this second tetrahedral intermediate (F) collapses to form a peptide bond between the two substrates, and the product is finally released to leave the regenerated active site (A).
6. IN VITRO TRANSPEPTIDATION ACTIVITY
Many sortase enzymes exhibit in vitro transpeptidation and/or proteolytic activity. In vitro activity was first demonstrated for the prototypical SaSrtA enzyme by Ton-That et al. (1999). Their assay utilized FRET-based detection of activity using a reporter LPXTG sorting signal peptide that contained donor and quencher fluorophores at each end. Cleavage of this peptide by SaSrtA, liberates the donor from the quencher enabling enzyme activity to be detected as an increase in fluorescence. Initial studies demonstrated that SaSrtA catalyzes an in vitro transpeptidation that joins two peptides, one that contains an LPXTG sorting signal motif and a second peptide that contains N-terminal glycine residues (Huang et al., 2003; Ton-That, Mazmanian, Alksne, & Schneewind, 2000). Studies using this assay also demonstrated that the active site histidine, cysteine, and arginine residues are important for catalysis (Marraffini et al., 2004; Ton-That et al., 2002). Although the FRET-based assay is easy to employ, inner filter effects can occur at high substrate concentrations leading to inaccurate measurements of the enzyme’s kinetic parameters (Kruger, Dostal, et al., 2004). Subsequent development of a medium-throughput HPLC enzyme assay enabled more accurate measurement of the kinetic parameters and revealed that transpeptidation occurs with a kcat = 0.28 ± 0.02s−1, and Km values for its LPXTG and secondary Gly5 peptide substrates of 7.33 ± 1.01mM and 196 ± 64µM, respectively (Frankel et al., 2005). In the absence of the Gly5 peptide SaSrtA acts as a protease, hydrolytically cleaving the sorting signal between the threonine and glycine residues with a kcat = 0.086 ± 0.015s−1. As hydrolysis occurs much slower than transpeptidation, it can be largely avoided when the Gly5 peptide is in excess. As described in Section 5, when sortase is purified and removed from the cell surface only a small fraction (~0.06%) contains a properly ionized active site that can catalyze transpeptidation. This small subpopulation is much more enzymatically active with an estimated kcat/Km greater than 105 M−1 (Frankel et al., 2005). On the cell surface, the transpeptidation reaction catalyzed by membrane-associated SaSrtA may occur at a rate that is faster than the rate of the reconstituted in vitro transpeptidation reaction that employs short peptide substrates. This is because pulse-chase labeling experiments using intact cells indicate that the reaction is complete in <3 min, and it is likely that individual sortase enzymes attach several proteins to the cell wall during this time (Schneewind et al., 1992). The increased in vivo rate may result from the fact that the sortase and both of its substrates are embedded in the membrane. It is also possible that there exists yet to be identified factors on the cell surface that facilitate sortase association with its substrates and/or the conversion of its active site into the properly ionized state.
Since the original work on SaSrtA, the in vitro enzymatic activities of several other sortases have been characterized (summarized in Table 2). Interestingly, none of these enzymes are as active as SaSrtA and in many instances only their proteolytic activity has been demonstrated. Generally, their activities have not been rigorously characterized as only the amount of product generated by the sortase has been measured after a specific incubation time. At present, only the SaSrtA, SpySrtA, and SaSrtB sortases have been shown to catalyze a transpeptidation reaction that joins two peptides together via a peptide bond. The native SaSrtA enzyme catalyzes transpeptidation ~20–500-fold faster than the other enzymes making it a useful bioconjugation reagent. Moreover, the activity of SaSrtA has been improved using directed evolution approaches, resulting in tetramutant enzyme that is ~140-fold more active than the native SaSrtA (Chen et al., 2011). Additional rate enhancements have been achieved by altering the reaction conditions and by fusing the nucleophile substrate to SaSrtA (Amer, Macdonald, Jacobitz, Liauw, & Clubb, 2016). The reader is referred to a number of excellent reviews describing the use of sortase as a bioconjugation reagent (Proft, 2010; Tsukiji & Nagamune, 2009; Wu & Guo, 2012).
Table 2.
Activity of Sortase Enzymes In Vitro
| Sortase | Primary Substrate (Km mM) | Secondary Substrate (Km µM) |
Cleavage kcat (s−1) | Transpeptidation kcat (s−1) |
|---|---|---|---|---|
| Class A | ||||
| SaSrtAΔ24 (Frankel, Kruger, Robinson, Kelleher, & McCafferty, 2005) | Abz-LPETG-Dap(Dnp) (7.33 ± 1.01) | Gly5 (196 ± 64) | 0.086 ± 0.015 | 0.28 ± 0.02 |
| SaSrtAΔ24 (Kruger, Dostal, & McCafferty, 2004) | Abz-LPETG-Dap(Dnp) (5.5 mM) | Gly5 (140) | NR | 0.27 |
| SaSrtAΔ24 (Bentley, Lamb, & McCafferty, 2008) | Abz-LPETGG-Dap(Dnp) (8.76 ± 0.78)a | Gly5 (NR) | NR | 1.10 ± 0.06a |
| SaSrtAΔ59 (Chen, Dorr, & Liu, 2011) | Abz-LPETGK-(Dnp) (7.6 ± 0.5) | Gly3 (140 ± 30) | NR | 1.5 ± 0.2 |
| SaSrtAΔ59 Evolved tetramutant (Chen et al., 2011) | Abz-LPETGK-(Dnp) (0.17 ± 0.03) | Gly3 (4800 ± 700) | NR | 4.8 ± 0.8 |
| SpySrtAΔ81 (Race et al., 2009) | Abz-LPETGG-Dap(Dnp) (0.83 ± 0.11) | Ala2 (NR) | NR | 0.0136 ± 0.0011 |
| BaSrtAΔ56 (Weiner, Robson, Marohn, & Clubb, 2010) | Abz-LPETG-Dap(Dnp) (0.038 ± 4)b | m-DAPc | 0.0004 ± 0.0001b | c |
| BaSrtAΔ56 (Chan et al., 2015) | Abz-LPETG-Dap(Dnp) (0.306 ± 0.023) | m-DAPc | 3.6 ± 0.2 × 10−5 | c |
| BaSrtAΔ64 (Chan et al., 2015) | Abz-LPETG-Dap(Dnp) (0.173 ± 0.011) | m-DAPc | 5.7 ± 0.2 × 10−5 | c |
| SmutSrtAΔ40 (Wallock-Richards et al., 2015) | Dabcyl-QALPETGEE-Edans (0.0904 ± 0.0047)b | NR | Yes | NR |
| Class B | ||||
| SaSrtBΔ21 (Bentley, Gaweska, Kielec, & McCafferty, 2007) | Abz-KVENPQTNAGT-Dap(DNP) (7.8 ± 2) | Gly5 (NR) | NR | 5.4 ± 0.5 × 10−4 |
| SaSrtBΔ31 (Jacobitz et al., 2014) | SNKDKVENPQTNAGT (1.8) | Gly5 (NR) | NR | 1.010−4 |
| BaSrtBΔ37 (Maresso, Chapa, & Schneewind, 2006) | Abz -KTDNPKTGDEA-Dap(DNP) | NR | Yes | NR |
| CdSrtBΔ26 (van Leeuwen et al., 2014) | KIVKSPKTGDETQLMK KPPVPPKTGDSTTIGK | Gly4/5 or Ala-d-Glu-DAP | NR | NR |
| Class C | ||||
| SpnSrtC117–228 (Manzano et al., 2008) | IPQTG in RrgB30–633 | YPKN in RrgB30–633 | No | Yes |
| SagSrtC143–254 from PI-2a (Cozzi et al., 2011) | Dabcyl-KKVTIPQTGGIGT-Edans (0.0138) | NR | Yes | NR |
| SagSrtC142–305 from PI-1 (Cozzi et al., 2012) | Dabcyl-RPPGVFPKTGGIG-Edans (0.0 1358 ± 0.00063) | NR | 1.16 ± 0.044 × 10−3 | NR |
| Dabcyl-RPSIPNTGGIG-Edans (0.03100 ± 0.00462) | NR | 1.77 ± 0.101 × 10−3 | NR | |
| Dabcyl-RGGLIPKTGEQQ-Edans (0.01639 ± 0.00250) | NR | 0.77 ± 0.038 × 10−3 | NR | |
| SagSrtC242–283 from PI-1 (Cozzi et al., 2012) | Dabcyl-RPPGVFPKTGGIG-Edans (0.006385 ± 0.00142) | NR | 1.04 ± 0.058 × 10−3 | NR |
| Dabcyl-RGGLIPKTGEQQ-Edans (0.02733 ± 0.00435) | NR | 4.36 ± 0.256 × 10−4 | NR | |
| Dabcyl-RPSIPNTGGIG-Edans (0.05715 ± 0.00354) | NR | 5.56 ± 0.174 × 10−3 | NR | |
| Class D | ||||
| BaSrtDΔ55 (Robson, Jacobitz, Phillips, & Clubb, 2012) | VQGEKLPNTASNN | m-DAP (NR) | Yes | NR |
| BaSrtDΔ55 (Marraffini & Schneewind, 2006) | Abz-GEKLPNTASNN-Dnp | m-DAP (NR) | Yes | NR |
| CpSrtDΔ23–187 (Suryadinata, Seabrook, Adams, Nuttall, & Peat, 2015) | Aβ1–16-LPQTGS | NR | Yes | NR |
Values reported from fluorescence assay and subject to inner filter effect and are likely underestimates of true parameters.
These values calculated assuming a hydrolytic shunt mechanism.
The enzyme reportedly does not perform this reaction in vitro.
Sorting signals for all substrates are highlighted in bold.
Errors are reported where published.
“Yes” indicates the reaction was performed in vitro but kinetics parameters were not reported.
NR, not reported.
m-DAP, mesodiaminopimelic acid.
7. CONCLUSIONS AND FUTURE DIRECTIONS
Sortase enzymes are ubiquitous in Gram-positive bacteria where they attach proteins to the cell wall and construct pili. Their important role in displaying virulence factors makes them promising drug targets (Bradshaw et al., 2015; Cascioferro et al., 2014; Clancy et al., 2010; Maresso & Schneewind, 2008; Suree et al., 2007), while their ability catalyze in vitro transpeptidation has made them useful bioconjugation reagents (Antos et al., 2016; Bentley et al., 2008; Maresso et al., 2006; Popp & Ploegh, 2011; Ritzefeld, 2014; Schmidt et al., 2017; Schmohl & Schwarzer, 2014; Tsukiji & Nagamune, 2009; Voloshchuk et al., 2016). Considerable effort has been put forth to elucidate the molecular mechanism of catalysis, class-specific structural features that dictate function, and the molecular basis of substrate recognition. Structural and computational studies of three sortase–peptide complexes have provided insight into the initial steps of catalysis—binding of the sorting signal to the active site and formation of the first tetrahedral and thioacyl intermediates (Chan et al., 2015; Jacobitz et al., 2014; Suree et al., 2009). This work has shown that class A and B sortases recognize their sorting signal substrates in a similar manner. The side chains of the P1, P3, and P4 residues are recognized. The bound signal adopts an L-shaped conformation as a result of kink introduced at the proline P3 residue, positioning the side chain of the P4 residue within a pocket formed by the β6/β7 loop, and directing the P1 and P2 residues toward the active site cysteine. The positioning of P1 and P2 exhibit conformational heterogeneity, with substrate assisted catalysis occurring when the threonine P1 side chain contacts the active arginine residue to stabilize the oxyanion hole (Jacobitz et al., 2014). Other classes of sortases may bind their sorting signals in a generally similar manner, with class Cpilin polymerases requiring unlatching of a lid structure to enable signal access (Manzano et al., 2008), and class E enzymes using a unique surface to recognize alanine at position P3 instead of proline (Kattke et al., 2016). The second-half of the transpeptidation reaction remains poorly understood—nucleophilic attack of the thioacyl enzyme–substrate intermediate by an amine group and peptide bond formation. No conclusive evidence has emerged to pinpoint the location of the secondary substrate binding site for lipid II or pilin proteins in cell wall anchoring or pilin assembling sortases, respectively. Deciphering how some sortases function as polymerases, while others attach proteins to the cell wall will require the development of robust biochemical assays to monitor pilus assembly and novel substrate analogs to visualize nucleophile recognition. Finally, although many small molecule sortase inhibitors have been identified, they have yet to be developed into a drug to treat bacterial infections. Given the rising prevalence of antibiotic-resistant bacteria, pressure to develop viable sortase inhibitors as therapeutics is growing and will undoubtedly lead to the discovery and characterization of more potent and specific compounds.
References
- Amer BR, Macdonald R, Jacobitz AW, Liauw B, Clubb RT. Rapid addition of unlabeled silent solubility tags to proteins using a new substrate-fused sortase reagent. Journal of Biomolecular NMR. 2016;64(3):197–205. doi: 10.1007/s10858-016-0019-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antos JM, Truttmann MC, Ploegh HL. Recent advances in sortase-catalyzed ligation methodology. Current Opinion in Structural Biology. 2016;38:111–118. doi: 10.1016/j.sbi.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulabaugh A, Ding W, Kapoor B, Tabei K, Alksne L, Dushin R, et al. Development of an HPLC assay for Staphylococcus aureus sortase: Evidence for the formation of the kinetically competent acyl enzyme intermediate. Analytical Biochemistry. 2007;360(1):14–22. doi: 10.1016/j.ab.2006.10.021. [DOI] [PubMed] [Google Scholar]
- Bentley ML, Gaweska H, Kielec JM, McCafferty DG. Engineering the substrate specificity of Staphylococcus aureus Sortase A. The beta6/beta7 loop from SrtB confers NPQTN recognition to SrtA. The Journal of Biological Chemistry. 2007;282(9):6571–6581. doi: 10.1074/jbc.M610519200. [DOI] [PubMed] [Google Scholar]
- Bentley ML, Lamb EC, McCafferty DG. Mutagenesis studies of substrate recognition and catalysis in the Sortase A transpeptidase from Staphylococcus aureus. The Journal of Biological Chemistry. 2008;283(21):14762–14771. doi: 10.1074/jbc.M800974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas T, Pawale VS, Choudhury D, Roy RP. Sorting of LPXTG peptides by archetypal Sortase A: Role of invariant substrate residues in modulating the enzyme dynamics and conformational signature of a productive substrate. Biochemistry. 2014;53(15):2515–2524. doi: 10.1021/bi4016023. [DOI] [PubMed] [Google Scholar]
- Bradshaw WJ, Davies AH, Chambers CJ, Roberts AK, Shone CC, Acharya KR. Molecular features of the sortase enzyme family. The FEBS Journal. 2015;282(11):2097–2114. doi: 10.1111/febs.13288. [DOI] [PubMed] [Google Scholar]
- Cascioferro S, Totsika M, Schillaci D. Sortase A: An ideal target for anti-virulence drug development. Microbial Pathogenesis. 2014;77:105–112. doi: 10.1016/j.micpath.2014.10.007. [DOI] [PubMed] [Google Scholar]
- Chambers CJ, Roberts AK, Shone CC, Acharya KR. Structure and function of a Clostridium difficile sortase enzyme. Scientific Reports. 2015;5:9449. doi: 10.1038/srep09449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AH, Yi SW, Terwilliger AL, Maresso AW, Jung ME, Clubb RT. Structure of the Bacillus anthracis Sortase A enzyme bound to its sorting signal: A flexible amino-terminal appendage modulates substrate access. The Journal of Biological Chemistry. 2015;290(42):25461–25474. doi: 10.1074/jbc.M115.670984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I, Dorr BM, Liu DR. A general strategy for the evolution of bond-forming enzymes using yeast display. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(28):11399–11404. doi: 10.1073/pnas.1101046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy KW, Melvin JA, McCafferty DG. Sortase transpeptidases: Insights into mechanism, substrate specificity, and inhibition. Biopolymers. 2010;94(4):385–396. doi: 10.1002/bip.21472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comfort D, Clubb RT. A comparative genome analysis identifies distinct sorting pathways in Gram-positive bacteria. Infection and Immunity. 2004;72(5):2710–2722. doi: 10.1128/IAI.72.5.2710-2722.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly K, Clubb R. No title. In: Waksman G, Caparon M, Hultgren C, editors. Structural biology of bacterial pathogenesis. Washington, DC: ASM Press; 2005. pp. 101–127. [Google Scholar]
- Connolly KM, Smith BT, Pilpa R, Ilangovan U, Jung ME, Clubb RT. Sortase from Staphylococcus aureus does not contain a thiolate-imidazolium ion pair in its active site. The Journal of Biological Chemistry. 2003;278(36):34061–34065. doi: 10.1074/jbc.M305245200. [DOI] [PubMed] [Google Scholar]
- Cozzi R, Malito E, Nuccitelli A, D’Onofrio M, Martinelli M, Ferlenghi I, et al. Structure analysis and site-directed mutagenesis of defined key residues and motives for pilus-related sortase C1 in group B Streptococcus. The FASEB Journal. 2011;25(6):1874–1886. doi: 10.1096/fj.10-174797. [DOI] [PubMed] [Google Scholar]
- Cozzi R, Prigozhin D, Rosini R, Abate F, Bottomley MJ, Grandi G, et al. Structural basis for group B Streptococcus pilus 1 Sortases C regulation and specificity. PLoS One. 2012;7(11):e49048. doi: 10.1371/journal.pone.0049048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dramsi S, Trieu-Cuot P, Bierne H. Sorting sortases: A nomenclature proposal for the various sortases of Gram-positive bacteria. Research in Microbiology. 2005;156(3):289–297. doi: 10.1016/j.resmic.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Duong A, Capstick DS, Di Berardo C, Findlay KC, Hesketh A, Hong H-J, et al. Aerial development in Streptomyces coelicolor requires sortase activity. Molecular Microbiology. 2012;83(5):992–1005. doi: 10.1111/j.1365-2958.2012.07983.x. [DOI] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Research. 2016;44(D1):D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti V, Pancholi V, Schneewind O. Conservation of a hexapeptide sequence in the anchor region of surface proteins from Gram-positive cocci. Molecular Microbiology. 1990;4(9):1603–1605. doi: 10.1111/j.1365-2958.1990.tb02072.x. [DOI] [PubMed] [Google Scholar]
- Frankel BA, Kruger RG, Robinson DE, Kelleher NL, McCafferty DG. Staphylococcus aureus sortase transpeptidase SrtA: Insight into the kinetic mechanism and evidence for a reverse protonation catalytic mechanism. Biochemistry. 2005;44(33):11188–11200. doi: 10.1021/bi050141j. [DOI] [PubMed] [Google Scholar]
- Hendrickx APA, Budzik JM, Oh S-Y, Schneewind O. Architects at the bacterial surface—Sortases and the assembly of pili with isopeptide bonds. Nature Reviews. Microbiology. 2011;9(3):166–176. doi: 10.1038/nrmicro2520. [DOI] [PubMed] [Google Scholar]
- Huang X, Aulabaugh A, Ding W, Kapoor B, Alksne L, Tabei K, et al. Kinetic mechanism of Staphylococcus aureus sortase SrtA. Biochemistry. 2003;42(38):11307–11315. doi: 10.1021/bi034391g. [DOI] [PubMed] [Google Scholar]
- Ilangovan U, Ton-That H, Iwahara J, Schneewind O, Clubb RT. Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proceedings of the National Academy of Sciences. 2001;98(11):6056–6061. doi: 10.1073/pnas.101064198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobitz AW, Wereszczynski J, Yi SW, Amer BR, Huang GL, Nguyen AV, et al. Structural and computational studies of the Staphylococcus aureus Sortase B-substrate complex reveal a substrate-stabilized oxyanion hole. The Journal of Biological Chemistry. 2014;289:8891–8902. doi: 10.1074/jbc.M113.509273. p. jbc.M113.509273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobitz AW, Naziga EB, Yi SW, McConnell SA, Peterson R, Jung ME, et al. The ‘lid’ in the Streptococcus pneumoniae SrtC1 sortase adopts a rigid structure that regulates substrate access to the active site. The Journal of Physical Chemistry B. 2016;120(33):8302–8312. doi: 10.1021/acs.jpcb.6b01930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung ME, Clemens JJ, Suree N, Liew CK, Pilpa R, Campbell DO, et al. Synthesis of (2R,3S) 3-amino-4-mercapto-2-butanol, a threonine analogue for covalent inhibition of sortases. Bioorganic & Medicinal Chemistry Letters. 2005;15(22):5076–5079. doi: 10.1016/j.bmcl.2005.07.073. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Coulibaly F, Proft T, Baker EN. Crystal structure of Spy0129, a Streptococcus pyogenes Class B Sortase involved in pilus assembly. PLoS One. 2011;6(1):e15969. doi: 10.1371/journal.pone.0015969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappel K, Wereszczynski J, Clubb RT, McCammon JA. The binding mechanism, multiple binding modes, and allosteric regulation of Staphylococcus aureus Sortase A probed by molecular dynamics simulations. Protein Science. 2012;21(12):1858–1871. doi: 10.1002/pro.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattke MD, Chan AH, Duong A, Sexton DL, Sawaya MR, Cascio D, et al. Crystal structure of the Streptomyces coelicolor Sortase E1 transpeptidase provides insight into the binding mode of the novel class E sorting signal. PLoS One. 2016;11(12):e0167763. doi: 10.1371/journal.pone.0167763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare B, Krishnan V, Rajashankar KR, I-Hsiu H, Xin M, Ton-That H, et al. Structural differences between the Streptococcus agalactiae housekeeping and pilus-specific sortases: SrtA and SrtC1. PloS One. 2011a;6(8):e22995. doi: 10.1371/journal.pone.0022995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare B, Fu Z-Q, Huang I-H, Ton-That H, Narayana SVL. The crystal structure analysis of group B Streptococcus sortase C1: A model for the ‘lid’ movement upon substrate binding. Journal of Molecular Biology. 2011b;414(4):563–577. doi: 10.1016/j.jmb.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline KA, Dodson KW, Caparon MG, Hultgren SJ. A tale of two pili: Assembly and function of pili in bacteria. Trends in Microbiology. 2010;18(5):224–232. doi: 10.1016/j.tim.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger RG, Otvos B, Frankel BA, Bentley M, Dostal P, McCafferty DG. Analysis of the substrate specificity of the Staphylococcus aureus Sortase transpeptidase SrtA†. Biochemistry. 2004a;43(6):1541–1551. doi: 10.1021/bi035920j. [DOI] [PubMed] [Google Scholar]
- Kruger RG, Dostal P, McCafferty DG. Development of a high-performance liquid chromatography assay and revision of kinetic parameters for the Staphylococcus aureus sortase transpeptidase SrtA. Analytical Biochemistry. 2004b;326(1):42–48. doi: 10.1016/j.ab.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Lazzarin M, Cozzi R, Malito E, Martinelli M, D’Onofrio M, Maione D, et al. Noncanonical sortase-mediated assembly of pilus type 2b in group B Streptococcus. The FASEB Journal. 2015;29(11):4629–4640. doi: 10.1096/fj.15-272500. [DOI] [PubMed] [Google Scholar]
- van Leeuwen HC, Klychnikov OI, Menks MAC, Kuijper EJ, Drijfhout JW, Hensbergen PJ. Clostridium difficile sortase recognizes a (S/P)PXTG sequence motif and can accommodate diaminopimelic acid as a substrate for transpeptidation. FEBS Letters. 2014;588(23):4325–4333. doi: 10.1016/j.febslet.2014.09.041. [DOI] [PubMed] [Google Scholar]
- Liew CK, Smith BT, Pilpa R, Suree N, Ilangovan U, Connolly KM, et al. Localization and mutagenesis of the sorting signal binding site on Sortase A from Staphylococcus aureus. FEBS Letters. 2004;571(1–3):221–226. doi: 10.1016/j.febslet.2004.06.070. [DOI] [PubMed] [Google Scholar]
- Lu G, Qi J, Gao F, Yan J, Tang J, Gao GF. A novel ‘open-form’ structure of sortase C from Streptococcus suis. Proteins. 2011;79(9):2764–2769. doi: 10.1002/prot.23093. [DOI] [PubMed] [Google Scholar]
- Mandlik A, Swierczynski A, Das A, Ton-That H. Pili in Gram-positive bacteria: Assembly, involvement in colonization and biofilm development. Trends in Microbiology. 2008;16(1):33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano C, Contreras-Martel C, El Mortaji L, Izoré T, Fenel D, Vernet T, et al. Sortase-mediated pilus fiber biogenesis in Streptococcus pneumoniae. Structure (London, England: 1993) 2008;16(12):1838–1848. doi: 10.1016/j.str.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Manzano C, Izoré T, Job V, Di Guilmi AM, Dessen A. Sortase activity is controlled by a flexible lid in the pilus biogenesis mechanism of Gram-positive pathogens. Biochemistry. 2009;48(44):10549–10557. doi: 10.1021/bi901261y. [DOI] [PubMed] [Google Scholar]
- Maresso AW, Schneewind O. Sortase as a target of anti-infective therapy. Pharmacological Reviews. 2008;60(1):128–141. doi: 10.1124/pr.107.07110. [DOI] [PubMed] [Google Scholar]
- Maresso AW, Chapa TJ, Schneewind O. Surface protein IsdC and sortase B are required for heme-iron scavenging of Bacillus anthracis. Journal of Bacteriology. 2006;188(23):8145–8152. doi: 10.1128/JB.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresso AW, Wu R, Kern JW, Zhang R, Janik D, Missiakas DM, et al. Activation of inhibitors by sortase triggers irreversible modification of the active site. The Journal of Biological Chemistry. 2007;282(32):23129–23139. doi: 10.1074/jbc.M701857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Schneewind O. Anchor structure of staphylococcal surface proteins V. Anchor structure of the sortase B substrate IsdC. The Journal of Biological Chemistry. 2005;280(16):16263–16271. doi: 10.1074/jbc.M500071200. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Schneewind O. Targeting proteins to the cell wall of sporulating Bacillus anthracis. Molecular Microbiology. 2006;62(5):1402–1417. doi: 10.1111/j.1365-2958.2006.05469.x. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Ton-That H, Zong Y, Narayana SVL, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus A conserved arginine residue is required for efficient catalysis of Sortase A. The Journal of Biological Chemistry. 2004;279(36):37763–37770. doi: 10.1074/jbc.M405282200. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285(5428):760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Ton-That H, Su K, Schneewind O. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(4):2293–2298. doi: 10.1073/pnas.032523999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, Jelenska J, et al. Passage of heme-iron across the envelope of Staphylococcus aureus. Science (New York, NY) 2003;299(5608):906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- Moritsugu K, Terada T, Kidera A. Disorder-to-order transition of an intrinsically disordered region of sortase revealed by multiscale enhanced sampling. Journal of the American Chemical Society. 2012;134(16):7094–7101. doi: 10.1021/ja3008402. [DOI] [PubMed] [Google Scholar]
- Naik MT, Suree N, Ilangovan U, Liew CK, Thieu W, Campbell DO, et al. Staphylococcus aureus Sortase A transpeptidase. Calcium promotes sorting signal binding by altering the mobility and structure of an active site loop. The Journal of Biological Chemistry. 2006;281(3):1817–1826. doi: 10.1074/jbc.M506123200. [DOI] [PubMed] [Google Scholar]
- Neiers F, Madhurantakam C, Fälker S, Manzano C, Dessen A, Normark S, et al. Two crystal structures of pneumococcal pilus sortase C provide novel insights into catalysis and substrate specificity. Journal of Molecular Biology. 2009;393(3):704–716. doi: 10.1016/j.jmb.2009.08.058. [DOI] [PubMed] [Google Scholar]
- Pallen MJ, Lam AC, Antonio M, Dunbar K. An embarrassment of sortases—A richness of substrates? Trends in Microbiology. 2001;9(3):97–101. doi: 10.1016/s0966-842x(01)01956-4. [DOI] [PubMed] [Google Scholar]
- Perry AM, Ton-That H, Mazmanian SK, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. The Journal of Biological Chemistry. 2002;277(18):16241–16248. doi: 10.1074/jbc.M109194200. [DOI] [PubMed] [Google Scholar]
- Persson K. Structure of the Sortase AcSrtC-1 from Actinomyces oris. Acta Crystallographica. Section D, Biological Crystallography. 2011;67(Pt. 3):212–217. doi: 10.1107/S0907444911004215. [DOI] [PubMed] [Google Scholar]
- Popp MW-L, Ploegh HL. Making and breaking peptide bonds: Protein engineering using Sortase. Angewandte Chemie International Edition. 2011;50(22):5024–5032. doi: 10.1002/anie.201008267. [DOI] [PubMed] [Google Scholar]
- Proft T. Sortase-mediated protein ligation: An emerging biotechnology tool for protein modification and immobilisation. Biotechnology Letters. 2010;32(1):1–10. doi: 10.1007/s10529-009-0116-0. [DOI] [PubMed] [Google Scholar]
- Race PR, Bentley ML, Melvin JA, Crow A, Hughes RK, Smith WD, et al. Crystal structure of Streptococcus pyogenes Sortase A. The Journal of Biological Chemistry. 2009;284(11):6924–6933. doi: 10.1074/jbc.M805406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzefeld M. Sortagging: A robust and efficient chemoenzymatic ligation strategy. Chemistry. 2014;20(28):8516–8529. doi: 10.1002/chem.201402072. [DOI] [PubMed] [Google Scholar]
- Robson SA, Jacobitz AW, Phillips ML, Clubb RT. Solution structure of the Sortase required for efficient production of infectious Bacillus anthracis spores. Biochemistry. 2012;51(40):7953–7963. doi: 10.1021/bi300867t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin A, Severin A, Ritacco F, Tabei K, Singh G, Bradford PA, et al. Further evidence that a cell wall precursor [C(55)-MurNAc-(peptide)-GlcNAc] serves as an acceptor in a sorting reaction. Journal of Bacteriology. 2002;184(8):2141–2147. doi: 10.1128/JB.184.8.2141-2147.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Toplak A, Quaedflieg PJ, Nuijens T. Enzyme-mediated ligation technologies for peptides and proteins. Current Opinion in Chemical Biology. 2017;38:1–7. doi: 10.1016/j.cbpa.2017.01.017. [DOI] [PubMed] [Google Scholar]
- Schmohl L, Schwarzer D. Sortase-mediated ligations for the site-specific modification of proteins. Current Opinion in Chemical Biology. 2014;22:122–128. doi: 10.1016/j.cbpa.2014.09.020. [DOI] [PubMed] [Google Scholar]
- Schneewind O, Missiakas DM. Protein secretion and surface display in Grampositive bacteria. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 2012;367(1592) doi: 10.1098/rstb.2011.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneewind O, Missiakas D. Sec-secretion and sortase-mediated anchoring of proteins in Gram-positive bacteria. Biochimica et Biophysica Acta. 2014;1843(8):1687–1697. doi: 10.1016/j.bbamcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70(2):267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- Schneewind O, Fowler A, Faull KF. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268(5207):103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- Shaik MM, Lombardi C, Maragno Trindade D, Fenel D, Schoehn G, Di Guilmi AM, et al. A structural snapshot of type II pilus formation in Streptococcus pneumoniae. Journal of Biological Chemistry. 2015;290:22581–22592. doi: 10.1074/jbc.M115.647834. p. M115.647834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel SD, Liu J, Ton-That H. Biogenesis of the Gram-positive bacterial cell envelope. Current Opinion in Microbiology. 2016;34:31–37. doi: 10.1016/j.mib.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirig T, Weiner EM, Clubb RT. Sortase enzymes in Gram-positive bacteria. Molecular Microbiology. 2011;82(5):1044–1059. doi: 10.1111/j.1365-2958.2011.07887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suree N, Jung ME, Clubb RT. Recent advances towards new anti-infective agents that inhibit cell surface protein anchoring in Staphylococcus aureus and other Grampositive pathogens. Mini Reviews in Medicinal Chemistry. 2007;7(10):991–1000. doi: 10.2174/138955707782110097. [DOI] [PubMed] [Google Scholar]
- Suree N, Liew CK, Villareal VA, Thieu W, Fadeev EA, Clemens JJ, et al. The structure of the Staphylococcus aureus Sortase-substrate complex reveals how the universally conserved LPXTG sorting signal is recognized. The Journal of Biological Chemistry. 2009;284(36):24465–24477. doi: 10.1074/jbc.M109.022624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadinata R, Seabrook SA, Adams TE, Nuttall SD, Peat TS. Structural and biochemical analyses of a Clostridium perfringens sortase D transpeptidase. Acta Crystallographica. Section D, Biological Crystallography. 2015;71(7):1505–1513. doi: 10.1107/S1399004715009219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B-X, Eriksson LA. Catalytic mechanism and roles of Arg197 and Thr183 in the Staphylococcus aureus Sortase A enzyme. The Journal of Physical Chemistry. B. 2011;115(44):13003–13011. doi: 10.1021/jp2058113. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Molecular Microbiology. 2003;50(4):1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Faull KF, Schneewind O. Anchor structure of staphylococcal surface proteins. A branched peptide that links the carboxyl terminus of proteins to the cell wall. The Journal of Biological Chemistry. 1997;272(35):22285–22292. doi: 10.1074/jbc.272.35.22285. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(22):12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-That H, Mazmanian SK, Alksne L, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH2-GLY3 substrates. The Journal of Biological Chemistry. 2000;275(13):9876–9881. doi: 10.1074/jbc.275.13.9876. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Mazmanian SK, Alksne L, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus Cysteine 184 and histidine 120 of sortase form a thiolate-imidazolium ion pair for catalysis. The Journal of Biological Chemistry. 2002;277(9):7447–7452. doi: 10.1074/jbc.M109945200. [DOI] [PubMed] [Google Scholar]
- Tsukiji S, Nagamune T. Sortase-mediated ligation: A gift from Gram-positive bacteria to protein engineering. Chembiochem. 2009;10(5):787–798. doi: 10.1002/cbic.200800724. [DOI] [PubMed] [Google Scholar]
- Voloshchuk N, Liang D, Liang J. Sortase A mediated protein modifications and peptide conjugations. Current Drug Discovery Technologies. 2016;12(4):205–213. doi: 10.2174/1570163812666150903115601. [DOI] [PubMed] [Google Scholar]
- Wallock-Richards DJ, Marles-Wright J, Clarke D, Maitra A, Dodds M, Hanley B, et al. Molecular basis of Streptococcus mutans Sortase A inhibition by the flavonoid natural product trans-chalcone. Chemical Communications. 2015;51:10483–10485. doi: 10.1039/c5cc01816a. [DOI] [PubMed] [Google Scholar]
- Weiner EM, Robson S, Marohn M, Clubb RT. The Sortase A enzyme that attaches proteins to the cell wall of Bacillus anthracis contains an unusual active site architecture. The Journal of Biological Chemistry. 2010;285(30):23433–23443. doi: 10.1074/jbc.M110.135434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Guo Z. Sortase-mediated transpeptidation for site-specific modification of peptides, glycopeptides, and proteins. Journal of Carbohydrate Chemistry. 2012;31(1):48–66. doi: 10.1080/07328303.2011.635251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Mishra A, Reardon ME, Huang I-H, Counts SC, Das A, et al. Structural determinants of Actinomyces sortase SrtC2 required for membrane localization and assembly of type 2 fimbriae for interbacterial coaggregation and oral biofilm formation. Journal of Bacteriology. 2012;194(10):2531–2539. doi: 10.1128/JB.00093-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Wu R, Joachimiak G, Mazmanian SK, Missiakas DM, Gornicki P, et al. Structures of sortase B from Staphylococcus aureus and Bacillus anthracis reveal catalytic amino acid triad in the active site. Structure. 2004;12(7):1147–1156. doi: 10.1016/j.str.2004.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y, Bice TW, Ton-That H, Schneewind O, Narayana SVL. Crystal structures of Staphylococcus aureus Sortase A and its substrate complex. The Journal of Biological Chemistry. 2004a;279(30):31383–31389. doi: 10.1074/jbc.M401374200. [DOI] [PubMed] [Google Scholar]
- Zong Y, Mazmanian SK, Schneewind O, Narayana SV. The structure of Sortase B, a cysteine transpeptidase that tethers surface protein to the Staphylococcus aureus cell wall. Structure. 2004b;12(1):105–112. doi: 10.1016/j.str.2003.11.021. [DOI] [PubMed] [Google Scholar]