SUMMARY
Alternative splicing (AS) plays a critical role in cell fate transitions, development, and disease. Recent studies have shown that AS also influences pluripotency and somatic cell reprogramming. We profiled transcriptome-wide AS changes that occur during reprogramming of fibroblasts to pluripotency. This analysis revealed distinct phases of AS, including a splicing program that is unique to transgene-independent induced pluripotent stem cells (iPSCs). Changes in the expression of AS factors Zcchc24, Esrp1, Mbnl1/2, and Rbm47 were demonstrated to contribute to phase-specific AS. RNA-binding motif enrichment analysis near alternatively spliced exons provided further insight into the combinatorial regulation of AS during reprogramming by different RNA-binding proteins. Ectopic expression of Esrp1 enhanced reprogramming, in part by modulating the AS of the epithelial specific transcription factor Grhl1. These data represent a comprehensive temporal analysis of the dynamic regulation of AS during the acquisition of pluripotency.
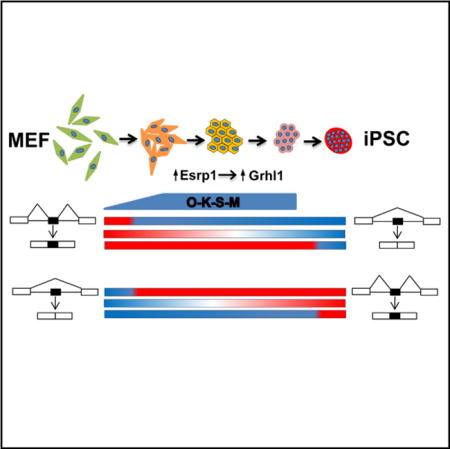
Graphical abstract
INTRODUCTION
Alternative splicing (AS) is a versatile post-transcriptional mechanism that expands protein diversity required for numerous cell fate transitions during development (Kalsotra and Cooper, 2011). In addition, alterations in AS patterns can lead to disease, in many cases through dysregulation of key splicing regulators (Cieply and Carstens, 2015). The regulation of AS involves the assembly of both ubiquitous and cell-type-specific splicing factors on RNA cis elements within or flanking regulated exons that can influence splicing positively or negatively (Chen and Manley, 2009). Discerning how different splicing factors collaborate to regulate key programs of AS remains a challenge but one that can now be addressed using new genomic technologies.
The discovery that exogenous factors can reprogram somatic cells into induced pluripotent stem cells (iPSCs) has opened up new fields of investigation that hold great promise for new therapeutic applications (Lengner, 2010). Only recently have roles for AS in pluripotency and reprogramming begun to emerge, including the identification of roles for several splicing regulators such as muscleblind-like splicing factors Mbnl1/2, RBFOX2, Srsf3, and U2af1 (Gabut et al., 2011; Han et al., 2013; Ohta et al., 2013; Salomonis et al., 2010; Venables et al., 2013; Wu et al., 2010; Yeo et al., 2009). However, it is likely that additional splicing regulators play a role during the acquisition of pluripotency. A temporal analysis of transcriptional dynamics during reprogramming of mouse embryonic fibroblasts (MEFs) to iPSCs revealed that it is a multistep process with distinct temporal phases, but AS was not assessed (Samavarchi-Tehrani et al., 2010). It is therefore essential to define the AS programs that accompany these transitional phases and to identify splicing factors driving these dynamic changes. Interestingly, an early phase of reprogramming from MEFs to iPSCs is characterized by a mesenchymal-to-epithelial transition (MET), which includes activation of the epithelial splicing regulatory proteins 1 and 2 (Esrp1 and Esrp2), suggesting that they may promote splicing changes at this critical phase of reprogramming (Li et al., 2010; Samavarchi-Tehrani et al., 2010).
We conducted a comprehensive temporal analysis of AS during reprogramming using RNA sequencing (RNA-seq). This revealed temporally dynamic and complex patterns of AS. By correlating AS patterns with differential expression of a broad panel of RNA-binding proteins (RBPs) and known splicing factors, we were able to identify several examples of splicing regulators that influence AS during reprogramming. Ectopic expression of Esrp1 substantially enhanced reprogramming efficiency, in part through a splicing switch in the epithelial-specific transcription factor Grhl1. These data highlight the importance of regulated AS during cell fate transitions.
RESULTS
Comprehensive Temporal Analysis of Dynamic Changes in Splicing during Somatic Cell Reprogramming
In order to profile AS during induction of pluripotency (using the Yamanaka factors Oct4, Klf4, Sox2, and cMyc or OKSM), we used MEFs from mice harboring a doxycycline (Dox)-inducible polycistronic OKSM cassette (Stadtfeld et al., 2010). MEFs isolated from these mice can be efficiently reprogrammed with Dox, and using this induced pluripotency model, we conducted a time course analysis of AS during reprogramming including the isolation of three stable transgene-independent iPSC clones. The iPSC clones expressed Nanog at levels similar to embryonic stem cells (ESCs) and displayed an ESC-like morphology (Figures S1A and S1B). At each time point following Dox induction, RNA was isolated from Ssea1-positive cells using magnetic activated cell sorting (MACS) in order to enrich for cells at an intermediate stage of reprogramming and exclude cells that will never enter the pluripotent state (Brambrink et al., 2008). RNA from triplicate samples was subjected to 101 base pair paired-end sequencing. AS analysis was conducted using replicate multivariate analysis of transcript splicing (rMATS) (Shen et al., 2014). Whereas this analysis identified changes in cassette exons (also referred to as skipped exons [SEs]), alternative 3′ and 5′ splice sites, retained introns, as well as mutually exclusive exons, we subsequently focused primarily on cassette exons. Our analysis revealed substantial changes in AS at each time point following OKSM induction, revealing distinct temporal patterns of AS during reprogramming (Figure 1A; Table S1). To further delineate these patterns, we conducted an unbiased temporal cluster analysis of all differentially spliced exons. Briefly, we performed a co-splicing network analysis of differentially regulated alternative exons during the time course, used a permutation-based procedure to assess the significance of correlation between pairs of exons, and then partitioned the network with appropriate correlation threshold to identify clusters of exons corresponding to distinct temporal splicing patterns. This defined 18 clusters corresponding to distinct patterns of AS changes across the time course (Figures 1A and 1B; Table S2). These clusters included, for example, groups of exons with increased inclusion or skipping in a graded manner (clusters 1 and 2), those that change very early (clusters 3 and 8), and those with the most-pronounced changes in splicing from day 20 to transgene-independent iPSC clones (clusters 4 and 5). For several of these clusters, we used RT-PCR to validate these distinct patterns of splicing changes and determine quantitatively the values for exon inclusion, or percent spliced in (PSI) at each time point (Figure 1C).
Figure 1. Genome-wide Analysis of Alternative Splicing during Induced Pluripotency Reveals Distinct Phases of Regulation.
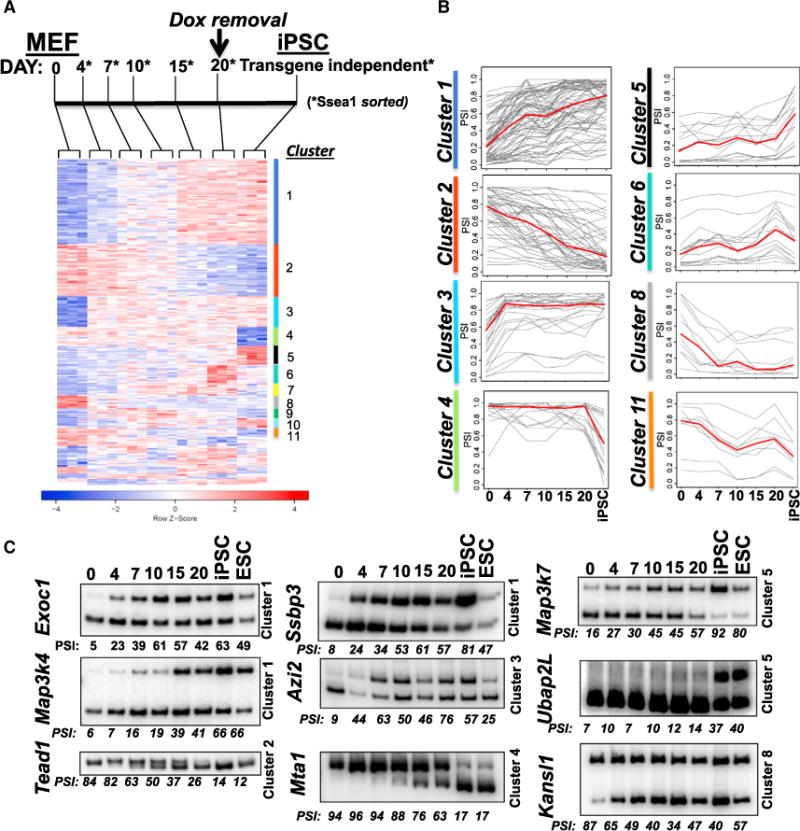
(A) Experimental design and heatmap of genome-wide alternative splicing changes; blue and red represent decreased and increased PSI, respectively, relative to the mean of each transcript across the time course.
(B) Examples of clusters with the median PSI of three replicates at each time point of each exon within selected clusters are graphed in gray and the median PSI for the cluster in red.
(C) Radiolabeled RT-PCR validations of phase-specific AS during reprogramming as well as in ESCs (last lane). For supplemental data, see also Figure S1.
The Regulation of AS during Reprogramming Involves Complex and Dynamic Combinatorial Contributions by Numerous Splicing Factors
The temporal patterns of splicing changes suggested coordinated regulation by different splicing factors at defined stages of the process. We used the RNA-seq data to evaluate total gene expression changes of all mouse genes at all stages across the time course (Table S3). Then, to identify candidate regulators of splicing during reprogramming, we conducted a temporal cluster analysis using total gene expression levels for a list of 226 genes encoding RBPs with known or inferred functions in splicing regulation. From this list, we identified 95 RBPs as candidate regulators of AS during reprogramming using three criteria: (1) expressed in at least one of the seven time points (average fragments per kilobase of transcript per million mapped reads [FPKM] > 5.0); (2) significant change in FPKM values across the seven time points (ANOVA p < 0.01); and (3) at least 2-fold change in FPKM values between the two time points with the highest and lowest average expression levels. These analyses separated the 95 RBPs into nine distinct clusters (Figure 2A; Table S2). In addition to defining clusters of RBPs with similar temporal patterns of expression, we also conducted a Jackknife version Pearson correlation coefficient between the expression values of the 95 RBPs and the PSI values for all regulated exons that changed across the time course (Table S2).
Figure 2. Complex Regulation of AS during Reprogramming Involving Multiple Splicing Factors.
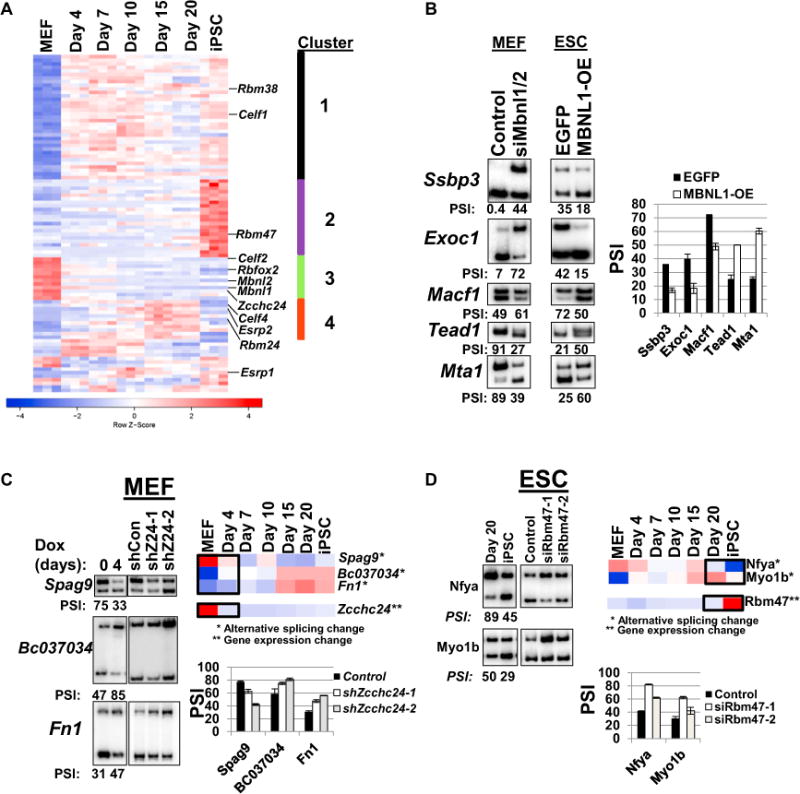
(A) Heatmap of gene expression changes of 95 RBPs selected based on the criteria described in the text. Examples of RBPs addressed in subsequent figures are listed to the right.
(B) RT-PCR analysis of AS events that occur during reprogramming and are induced by Mbnl1/2 knockdown in MEFs (left column) or are reverted by ectopic MBNL1 in V6.5 ESCs (right column and graph, which is the average PSI of biological duplicate with error bars representing SD from the mean [SDM]).
(C) RT-PCR analysis of AS events that occur by day 4 and are induced by two independent Zcchc24 small hairpin RNAs (shRNAs) in MEFs.
(D) RT-PCR analysis of AS events that change between day 20 and iPSC and are reverted by two independent siRNAs for Rbm47 in ESCs (for C and D, heatmaps of RNA-seq data are shown [upper right] and RT-PCR [left] with quantitation of triplicate RT-PCRs in the graph and error bars representing SDM). For supplemental data, see also Figures S2 and S3.
The genome-wide analysis of AS and RBP expression changes during the reprogramming time course suggested that the unique temporal patters of splicing are coordinated by multiple splicing factors, and we aimed to experimentally identify examples of these. In the case of muscleblind-like 1 and 2, we noted progressive downregulation of Mbnl1 and Mbnl2 at each of the phases of reprogramming (Figure S2A). Mbnl1 and Mbnl2 were previously shown to regulate AS programs that differ between differentiated cells and pluripotent stem cells (Han et al., 2013; Venables et al., 2013). In agreement with this, we found that either Mbnl1/2 knockdown in MEFs or forced expression in ESCs induced reciprocal splicing changes in the Ssbp3, Exoc1, Macf1, Tead1, and Mta1 transcripts consistent with the changes observed during reprogramming (Figures 2B and S2A). Interestingly, these Mbnl-regulated AS events represent multiple temporal clusters (1, 2, and 4), which exhibit contrasting temporal patterns of change (Figures 1B and 1C). Taken together, our findings support previous studies indicating an important role for Mbnl1/2 in regulation of splicing during reprogramming and also suggest that the progressive downregulation of these factors contributes to the changes in AS that occur across the time course.
We sought to identify additional examples of select RBPs, whose expression changes were observed in a phase-specific manner, that may contribute to the AS patterns observed during reprogramming. Two such examples are Zcchc24, which displays splicing activity in a reporter assay (R.P.C. and B.C., unpublished data) and is transcriptionally inactivated by day 4, and Rbm47, a recently identified splicing factor that we noted was upregulated specifically between day 20 and iPSCs (Vanharanta et al., 2014; Figures 2C and 2D). We used RNAi-mediated depletion in MEFs or ESC/iPSCs, respectively, and screened a panel of early or late AS events by RT-PCR. This uncovered three early events regulated by Zcchc24 and two late events regulated by Rbm47 (Figures 2C, 2D, and S2B–S2D), suggesting that these factors contribute to phase-specific patterns of AS during reprogramming. Whereas these examples demonstrate that this resource can be leveraged to identify novel regulators of AS during reprogramming, future efforts will be necessary to more comprehensively characterize the multifactorial regulation of AS during induced pluripotency.
RBPs/splicing factors influence AS patterns by binding to pre-mRNA in the introns flanking alternative exons or in the exons themselves. Identifying enriched RBP-binding motifs near alternative exons is a bioinformatic method that can implicate specific factors in the regulation of splicing. We therefore carried out motif enrichment analysis near alternative exons to provide further insights into other splicing factors potentially involved in AS regulation during reprogramming. Examples of known RBP-binding motifs enriched in the introns relative to alternatively spliced exons were identified (Figure S3A). As expected, Esrp1 (addressed in Figure 3)-binding sites were enriched upstream and Mbnl1-binding sites were enriched downstream of exons that undergo skipping during reprogramming, respectively, consistent with the position-dependent RNA maps for these splicing factors and their reciprocal expression patterns during reprogramming (Dittmar et al., 2012; Han et al., 2013). Interestingly, we identified two AS events that were antagonistically regulated by depletion of Mbnl1/2 or Esrp1 in MEFs or ESCs, respectively (Figures S3B and S3C; Dittmar et al., 2012; Han et al., 2013). Rbfox2 is involved in the regulation of AS during ESC differentiation into mesoderm, and it is downregulated as MEFs acquire pluripotency, with binding motifs enriched downstream of iPSC-silenced exons (Figure S3A; Table S3). Also, this analysis identified enrichment for other splicing-factor-binding sites such as Ptbp1, Rbm24, and Rbm38. We also mapped all enriched 6mers, which is likely to include the binding sites of AS regulators for which the cognate-binding sites have yet to be defined (Figure S3D). These motif enrichment data can inform future studies aimed at further characterizing the multifactorial regulation of AS in reprogramming.
Figure 3. Esrp1 Regulates MET Phase AS and Enhances Reprogramming.
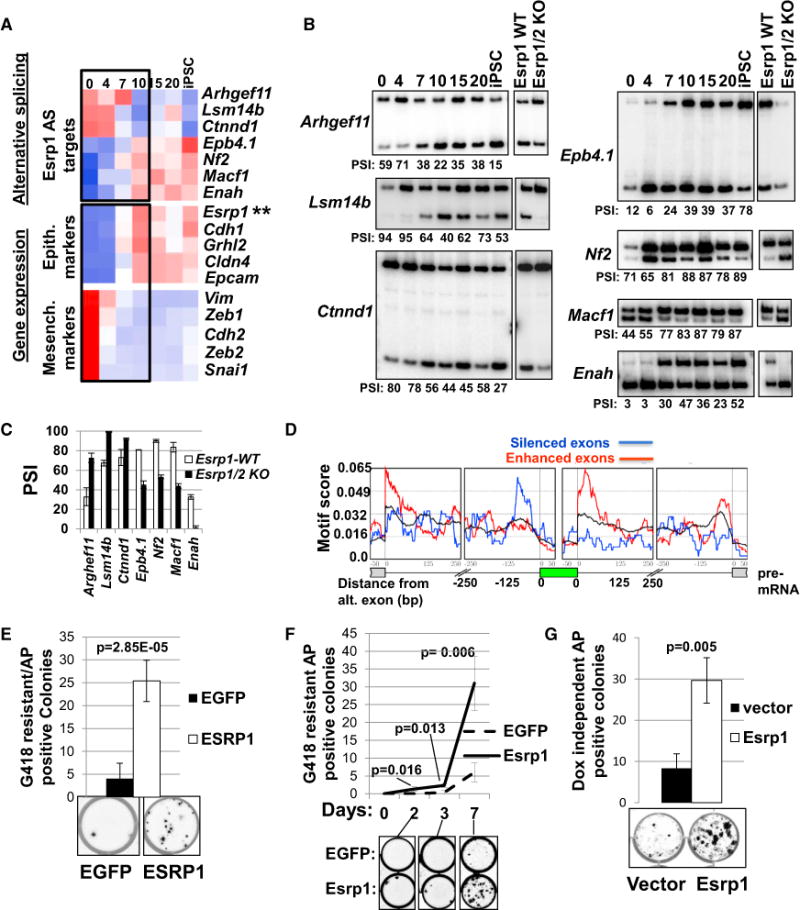
(A) Heatmap of AS events that coincide with the expression changes in epithelial and mesenchymal marker genes during reprogramming.
(B) RT-PCR validation of MET-phase AS events that are reverted by Esrp1/2 knockout in ESCs.
(C) Quantitation of the PSI changes induced by Esrp1/2 KO in ESCs in biological duplicate; error bars are SDM.
(D) Enrichment for Esrp-binding motifs near exons regulated at the MET phase (day 7). Esrp-binding motif enrichment is seen upstream of day 7 alternative exons that decrease in PSI (blue) and downstream of those that increase in PSI (red); the pre-mRNA is shown below with the alternative exon represented in green and coordinates in base pairs (bps).
(E) Doxycycline-reprogrammable MEFs with an Oct4-NEO-Resistance allele were transduced with EGFP or Esrp1-emerald retroviral vectors and treated with Dox (2 μg/ml) for 7 days followed by G418 selection for 1 week and then AP stain. Average colonies per well of five biological replicates with error bars representing SDM (above) and representative images of AP-positive colonies (below) are shown.
(F) Time course of Dox treatment prior to G418 selection for 1 week using the same MEF line as (E). Error bars are SDM of biological triplicate at each time point.
(G) AP-positive colonies that persisted after 10 days of Dox treatment followed by its removal and 10 days of Dox-free culture, conducted in biological triplicate.
For supplemental data, see Figures S3 and S4.
Previous studies identified induction of epithelial cell markers, including Esrp1 and Esrp2, early during reprogramming consistent with a MET phase (Li et al., 2010; Samavarchi-Tehrani et al., 2010). Although our analysis for SE exon changes did not reveal an obvious cluster that corresponded to this pattern, we further examined the role of the Esrps in regulating splicing primarily at the time period corresponding to MET based on epithelial and mesenchymal marker expression (Figure 3A). We validated seven splicing switches for a panel of alternative exons that included previously defined Esrp-regulated events as well as several that were identified using RNA-seq analysis of Esrp1/Esrp2 double knockout ESCs (Figures 3B, 3C, and S3C; Table S4; Dittmar et al., 2012; Warzecha et al., 2010). Binding motif analysis near exons that change in splicing at day 7, where Esrp1 expression is first observed, revealed enrichment for Esrp-binding sites upstream of SEs and downstream of included exons, consistent with our previously identified RNA map for Esrp (Figure 3D). These data indicate that the upregulation of Esrp1 and Esrp2 promotes AS changes during the MET phase of reprogramming.
Enhancement of Reprogramming by Ectopic Esrp1
Inducing changes in AS through alterations in splicing factor expression can impact reprogramming as shown in studies where depletion of Mbnl1/2 enhanced reprogramming efficiency (Han et al., 2013). Because the induction of MET was previously shown to enhance reprogramming efficiency and Esrp1 is highly upregulated during this phase, we hypothesized that ectopic expression of Esrp1 in MEFs might similarly enhance reprogramming (Samavarchi-Tehrani et al., 2010; Li et al., 2010; Figure 3A). To test this, we used MEFs from a transgenic mouse that is heterozygous for the Tet-OP-OKSM transgene that also harbors a targeted Oct4-Neomycin-resistance (Oct4-NeoR) knockin allele that allowed us to use G418 selection to score reprogramming efficiency based on activation of the endogenous Oct4 locus (Wernig et al., 2007). Ectopic Esrp1 expression substantially enhanced reprogramming based upon the number of alkaline-phosphatase-positive colonies at day 10 of Dox induction (Figures S4A and S4B) as well as the number of G418 resistant/alkaline phosphatase (AP)-positive colonies and earlier acquisition of G418 resistance (Figures 3E and 3F). The number of Dox-independent, AP-positive colonies and Nanog-positive colonies was also enhanced by Esrp1 (Figures 3G, S4C, and S4D). This effect was not simply due to increased proliferation because ectopic Esrp1 induced a modest but significant decrease in the overall cell proliferation rate (Figure S4E).
One of the most-robust switches in AS from day 4 to 10 of reprogramming (corresponding to an MET phase) is the activation of Esrp1-dependent exon 5 in the transcription factor Grainyhead-like 1 (Grhl1) transcript. This exon was previously identified as an Esrp-regulated event, and it showed skipping after Esrp1 ablation in ESCs (Figure 4A; Table S4; Bebee et al., 2015). This Esrp-regulated AS event is critical for the expression of Grhl1 because skipping of this exon induces a frameshift that results in a premature termination codon (PTC) upstream of the CP2 DNA-binding domain, thereby producing a truncated protein predicted to be functionally impaired as a transcription factor (Figure 4B). Furthermore, the PTC would generate a predicted target for nonsense-mediated mRNA decay (NMD). We noted that ectopic Esrp1 induced robust inclusion of Grhl1 exon 5 at 3 and 5 days after induction of reprogramming (Figure 4C). We therefore tested whether Grhl1 was a functionally relevant Esrp1 target in the context of reprogramming. Indeed, ectopic expression of the full-length isoform of Grhl1 enhanced reprogramming whereas the shorter isoform did not (Figures 4D, S4F, and S4G). Esrp-regulated splicing of Grhl1 thus provides an example of a functional link between AS and enhanced reprogramming efficiency. Grhl1 is also activated at the level of transcription during reprogramming (Table S3) and hence represents an interesting example where transcriptional activation of a gene needs to be coupled with AS in order to produce a functional protein. However, there are almost surely other Esrp-regulated splicing events that also functionally contribute to the ability of Esrp1 to promote reprogramming. We also suspect that other splicing factors whose expression is regulated during reprogramming play key roles in the process, and the comprehensive analysis of AS provided will serve to inform future investigations to modulate splicing during reprogramming and cell differentiation. A model for the complex regulation of AS in reprogramming that is supported by our data is summarized in Figure 4E. However, we note that this model is incomplete and the roles of additional splicing factors and key regulated AS events require further investigation.
Figure 4. Esrp1 Promotes Expression of a Grhl1 Isoform that Enhances Reprogramming.
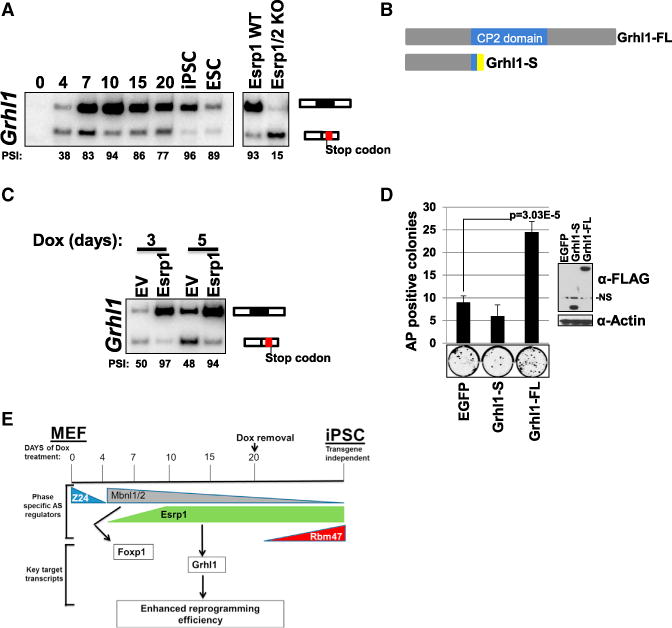
(A) RT-PCR analysis of Grhl1 exon 5 splicing during reprogramming and effect of Esrp1 KO in ESCs.
(B) Diagram of the full-length isoform of Grhl1 (Grhl1-FL) including the CP2 DNA-binding domain in blue and the truncated/short isoform (Grhl1-S) with exon 5 skipped and three out-of-frame amino acids from exon 6 that precede the PTC depicted in yellow.
(C) Dox-reprogrammable MEFs were transduced with empty vector or Esrp1-FLAG and treated with Dox for the indicated time followed by RNA isolation and RT-PCR to assess Grhl1 exon 5 inclusion.
(D) Dox-reprogrammable MEFs were transduced with EGFP, Grhl1-S, or Grhl1-FL and assayed for AP-positive colonies after 10 days of Dox treatment in biological quadruplicate; representative AP-positive colonies shown below; error bars are SDM.
(E) Schematic of regulators of alternative splicing during induced pluripotency.
For supplemental data, see Figure S4.
DISCUSSION
Although previous studies have revealed regulated AS in pluripotent stem cells and in reprogramming, the work presented here constitutes a detailed temporal analysis of AS during the acquisition of pluripotency and implicates complex and phasic regulation by different splicing factors. Many of these dynamic changes in AS and splicing factor expression that we identified using our network level analysis of temporal changes in splicing would elude detection through analysis limited to differences between somatic cells (including MEFs) and pluripotent stem cells. In addition to the splicing factors characterized here, our data provide a tool that can be further leveraged to characterize other splicing factors that are important for induced pluripotency as well as differentiation into distinct cell lineages.
We determined that Esrp1 dramatically enhances and accelerates cellular reprogramming, an effect that can be attributed in part to the AS regulation of Grhl1. However, the Esrps are not required for maintenance of pluripotency, as we could maintain Esrp1/Esrp2 double KO ESCs indefinitely without a loss of pluripotent cell markers. This observation is similar to that for Klf4, which promotes reprogramming yet is not required for pluripotency based on analysis of the Klf4 KO phenotype (Segre et al., 1999). Esrp1 is an example of a splicing factor that is activated during reprogramming and can enhance efficiency, an effect that is in contrast to that of the Mbnl1/2 splicing factors, which inhibit reprogramming. Our studies suggest that a splicing switch in the transcription factor Grhl1 transcript is one Esrp-regulated AS event during reprogramming that supports the acquisition of pluripotency. Few examples exist of alternatively spliced genes that affect reprogramming in an isoform-specific manner as we have shown for Grhl1. An additional example is Foxp1, where an ESC-specific isoform was shown to alter DNA binding specificity toward pluripotency-associated genes (Gabut et al., 2011). It will thus be of interest to further investigate the transcriptional program influenced by Grhl1 at the MET phase of reprogramming and in pluripotent stem cells.
Whereas we identified large-scale changes in splicing, further studies are needed to characterize the functional impact of these isoform switches at the protein level. Most AS events remain poorly characterized in terms of their impact on protein function and often require detailed case-by-case analysis. Nonetheless, among the events with substantial changes in splicing during reprogramming, we identified several cases where isoform-specific differences may be functionally related to cellular reprogramming and pluripotency. The switch-like AS of transcriptional co-regulators metastasis associated 1 (Mta1), TEA domain family member 1 (Tead1), and nuclear transcription factor-Y alpha (Nfya) during reprogramming involves exons with previously characterized roles in modulating the protein functions by encoding a nuclear localization sequence (NLS), altering DNA binding specificity and directing protein-protein interactions with other transcription factors, respectively (Dolfini et al., 2012; Jiang et al., 2000; Liang et al., 2008; Roder et al., 1999; Yaguchi et al., 2005). However, there are surely changes in AS during reprogramming and cell differentiation that generate different protein isoforms that are functionally related to cell fate decisions.
Our results highlight the complex and dynamic role that AS plays in reprogramming and pluripotency and the need to consider the combinatorial functions of numerous splicing factors in directing these cellular transitions. In addition to defining temporal patterns of AS associated with acquisition of pluripotency, these studies also provide a rationale and draft blueprint for harnessing splicing factors to direct pluripotent cells down defined lineages into distinct differentiated cell types useful for regenerative medicine and disease modeling.
EXPERIMENTAL PROCEDURES
Mice and Primary Cells
Dox-inducible MEFs were isolated from embryos harboring a Col1a1-tetO-OKSM and Rosa-26- rtTA (Stadtfeld et al., 2010). ESCs were from either Esrp1flox/flox; Esrp2−/− or Esrp1 wild-type; Esrp2−/− mice or V6.5. All mouse use was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Pennsylvania.
Induction of Pluripotency
MEFs that were homozygous for the Col1a1-OKSM and Rosa26-rtTA alleles were seeded onto gelatin-coated plates in ESC media and 2 μg/ml Dox for the indicated time points. The cultures were passaged as needed and in the presence of irradiated feeder MEFs from day 10 through iPSC. At each time point, cultures were purified via Ssea1-MACS according to the manufacturer’s protocol (Miltenyi Biotec), and the Ssea1-positive cells were then lysed in Trizol reagent (Invitrogen) and stored at −80°C. At day 20, Dox was removed and clones that maintained an ESC-like morphology independent of transgene expression for at least 2 weeks were mechanically isolated and expanded.
Retro and Lentiviral Transduction
For ectopic expression experiments, cDNAs were introduced into MEFs using retroviral transduction as described previously (Warzecha et al., 2009). shRNAs targeting Zcchc24 were introduced using the lentiviral vector pLKO.1; packaging was conducted in 293T cells by transfecting 1 μg pLKO-shRNA; 0.7 μg pSPAX2, and 0.3 μg cytomegalovirus (CMV)-vesicular stomatitis virus G protein (VSV-G) per well of a 6-well plate. MEFs were infected overnight with a 1:5 ratio of viral supernatant to MEF media. AP staining was carried out according to the manufacturer’s protocol (Vector Labs). Colonies were imaged using the Typhoon fluorescence imager and quantified using Image Quant colony counter tool.
siRNA Transfection
MEFs were transfected with control, Mbnl1, and Mbnl2 siRNAs (Dharmacon Smartpools; Han et al., 2013) using siRNA-Max transfection reagent (Life Technologies) according to the manufacturer’s protocol. ESCs and iPSCs were transfected with control and Rbm47 siRNAs (QIAGEN) also with siRNA-Max after pre-plating feeder-depleted single-cell suspensions onto gelatin-coated plates. Feeder MEFs were then added after 5 hr.
Computational Analysis of RNA-Seq Data
Statistical Methods
For RNA-seq analysis, we used Cuffdiff (v2.2.0) to calculate RNA-seq-based gene expression levels using the FPKM metric and identified differential gene expression between the two time points at FDR < 5% for a >2-fold difference in gene expression. To identify differential AS events between day 0 and other time points, we used the statistical parameters of rMATS v3.0.8.
For the temporal cluster analysis of time course iPSC RNA-seq data, we analyzed changes in RBP gene expression for significant changes in FPKM across the seven time points (ANOVA p < 0.01). A similar approach was used for AS changes at a Jackknife correlation coefficient threshold of 0.5 and a minimum of five exons per cluster (FDR = 0.02 based on permutation test).
For motif enrichment analysis, we scanned for motif occurrences separately in exons or their 250 base pairs upstream or downstream introns. For each motif, after we counted the number of occurrences in the differentially spliced exons and the control exons, we calculated the p value for enrichment via the Fisher’s exact test (right-sided) and used Benjamini-Hochberg FDR correction to adjust for multiple testing and identified enriched motifs at FDR < 5% and p value < 0.01.
For additional details on statistical methods, see Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Distinct phases of alternative splicing occur during induced pluripotency
Phases of alternative splicing are regulated by multiple RNA-binding proteins
The splicing factor Esrp1 enhances reprogramming efficiency through Grhl1
In Brief.
Cieply et al. show that posttranscriptional gene regulation during induced pluripotency of mouse embryo fibroblasts involves temporally coordinated changes in alternative splicing that is mediated by multiple splicing factors. One such factor, Esrp1, which is activated at the critical MET phase, and its target gene, Grhl1, enhance reprogramming efficiency.
Acknowledgments
We thank the Penn and UCLA Sequencing cores for RNA-seq. Funding: F32GM109630 (to B.C.); P20GM103436 (to J.W.P.); RO1GM08809 (to R.P.C.); R01AR066741 (to R.P.C.), The Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA Award (to Y.X.); GM088342 (to Y.X.); GM105431 (to Y.X.); U01HG007912 (to Y.X.); Chinese National Natural Science Foundation grants 61332014 (to X.S.); 61272121 (to X.S.); China Scholarship Council fellowship (to Y.G.); and Alfred P. Sloan Foundation fellowship (to Y.X.).
Footnotes
ACCESSION NUMBERS
The accession number for the RNA-seq data reported in this paper is GEO: GSE76233.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, four figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.03.025.
AUTHOR CONTRIBUTIONS
Experimental Design, B.C., C.J.L., R.P.C., J.W.P, Y.G., and Y.X.; Acquisition of Data, B.C., J.W.P., C.J.L., A.N.-D., and T.W.B.; Data Analysis, B.C., J.W.P., Y.G., X.S., Y.X., and R.P.C.; Preparation of Figures, B.C., J.W.P., Y.G., Y.X., and R.P.C.; Writing and/or Editing of Manuscript, B.C., J.W.P., T.W.B., C.J.L., Y.G., Y.X., and R.P.C.
References
- Bebee TW, Park JW, Sheridan KI, Warzecha CC, Cieply BW, Rohacek AM, Xing Y, Carstens RP. The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development. eLife. 2015;4:e08954. doi: 10.7554/eLife.08954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieply B, Carstens RP. Functional roles of alternative splicing factors in human disease. Wiley Interdiscip Rev RNA. 2015;6:311–326. doi: 10.1002/wrna.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Jiang P, Park JW, Amirikian K, Wan J, Shen S, Xing Y, Carstens RP. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol Cell Biol. 2012;32:1468–1482. doi: 10.1128/MCB.06536-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfini D, Minuzzo M, Pavesi G, Mantovani R. The short isoform of NF-YA belongs to the embryonic stem cell transcription factor circuitry. Stem Cells. 2012;30:2450–2459. doi: 10.1002/stem.1232. [DOI] [PubMed] [Google Scholar]
- Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O’Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO, et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN, et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498:241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SW, Trujillo MA, Sakagashira M, Wilke RA, Eberhardt NL. Novel human TEF-1 isoforms exhibit altered DNA binding and functional properties. Biochemistry. 2000;39:3505–3513. doi: 10.1021/bi991048w. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ. iPS cell technology in regenerative medicine. Ann N Y Acad Sci. 2010;1192:38–44. doi: 10.1111/j.1749-6632.2009.05213.x. [DOI] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Z. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- Ohta S, Nishida E, Yamanaka S, Yamamoto T. Global splicing pattern reversion during somatic cell reprogramming. Cell Rep. 2013;5:357–366. doi: 10.1016/j.celrep.2013.09.016. [DOI] [PubMed] [Google Scholar]
- Roder K, Wolf SS, Larkin KJ, Schweizer M. Interaction between the two ubiquitously expressed transcription factors NF-Y and Sp1. Gene. 1999;234:61–69. doi: 10.1016/s0378-1119(99)00180-8. [DOI] [PubMed] [Google Scholar]
- Salomonis N, Schlieve CR, Pereira L, Wahlquist C, Colas A, Zambon AC, Vranizan K, Spindler MJ, Pico AR, Cline MS, et al. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci USA. 2010;107:10514–10519. doi: 10.1073/pnas.0912260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat Methods. 2010;7:53–55. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanharanta S, Marney CB, Shu W, Valiente M, Zou Y, Mele A, Darnell RB, Massagué J. Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer. eLife. 2014;3:e02734. doi: 10.7554/eLife.02734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Lapasset L, Gadea G, Fort P, Klinck R, Irimia M, Vignal E, Thibault P, Prinos P, Chabot B, et al. MBNL1 and RBFOX2 cooperate to establish a splicing programme involved in pluripotent stem cell differentiation. Nat Commun. 2013;4:2480. doi: 10.1038/ncomms3480. [DOI] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S, Guo W, Xing Y, Carstens RP. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 2010;29:3286–3300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Habegger L, Noisa P, Szekely A, Qiu C, Hutchison S, Raha D, Egholm M, Lin H, Weissman S, et al. Dynamic transcriptomes during neural differentiation of human embryonic stem cells revealed by short, long, and paired-end sequencing. Proc Natl Acad Sci USA. 2010;107:5254–5259. doi: 10.1073/pnas.0914114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi M, Wada Y, Toh Y, Iguchi H, Kono A, Matsusue K, Takiguchi S. Identification and characterization of the variants of metastasis-associated protein 1 generated following alternative splicing. Biochim Biophys Acta. 2005;1732:8–14. doi: 10.1016/j.bbaexp.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


