Abstract
Somatic mosaicism refers to the fact that cells within an organism have different genomes. It is now clear that somatic mosaicism occurs in all brains and that somatic mutations in a subset of cells can cause various rare neurodevelopmental disorders. However, for most individuals, the extent and consequences of somatic mosaicism are largely unknown. The complexity and unique features of the brain suggest that somatic mosaicism can play an important role in behavior and cognition. Here we review recent manuscripts showing instances of somatic mosaicism in the brain and estimating its extent and possible biological consequences. The consequences of somatic mosaicism span vast dimensions -from a single-locus variant, to genes and gene networks, to cells, to the interactions of the mosaic cells via neural networks affecting behavior and cognition. We highlight how systems biology approaches are particularly well suited for the complex emerging field of brain somatic mosaicism.
Keywords: Somatic mosaicism, Brain development, Mutation, Retrotransposition
Introduction
Somatic mosaicism results from de novo DNA changes within cells of a body. Each individual cell within an organism has a history of growth, cell division, differentiation, exposure to chemical insults/metabolic stresses, DNA damage and repair that leads to the accumulation of mutations in its DNA. It is inevitable that genomic changes will accumulate within somatic cells during the life of an organism, but somatic mosaicism is of particular interest in the brain because of some of the brain’s unique features. For the most part, once the mammalian brain is developed, the neuronal population is not replenished, with the exception of two regions harboring adult neurogenesis (the dentate gyrus of the hippocampus and the subventricular zone). Thus, individual somatic mutations persist throughout the lifetime of the neuron, which can coincide with the lifetime of the organism. Mosaic DNA mutations can potentially alter the physiological properties of each neuron, contributing to overall brain function. The importance of different circuits to the immediate behavior of the organism is modulated over time as the state of the brain changes. Thus, small groups of neurons or even single neurons can influence the behavior of the organism.
The multi-dimensional nature of somatic mosaicism and brain function presents multiple challenges and opportunities for understanding their relationship. In this article, we review the studies, techniques and data that underlie our current understanding of somatic mosaicism. We then explore multiple dimensions of somatic mosaicism data and discuss perspectives on holistic and data integrative approaches that shed light on the role of somatic mosaicism in the brain.
Variant types and mechanisms generating mosaicism
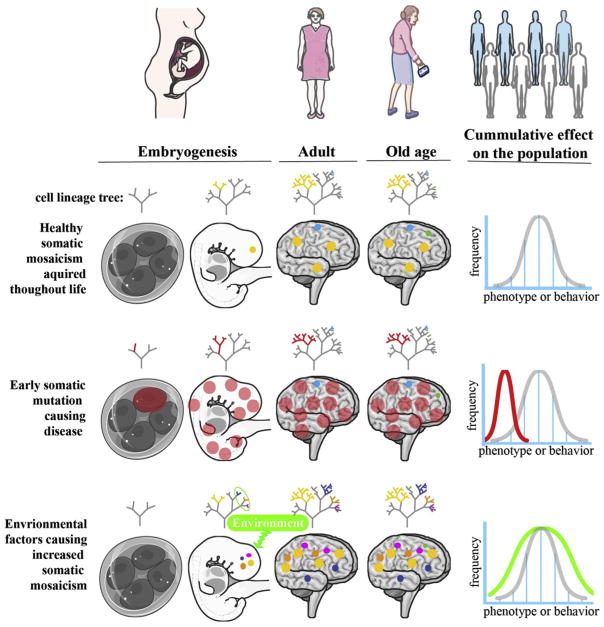
Surprising levels of genomic variation occur within the brain. Structural variations (SV), including copy number variations (CNV), LINE-1 retrotransposon insertions, deletions associated with LINE-1, along with single nucleotide variants (SNVs), create somatic genomic variation within neurons of the human brain. The spectrum of somatic variants in the brain is the aggregation of the different types of variants that occur over the life of all cells within the brain (Figure 1). By definition, somatic variants are restricted to a subset of cells within the body and have been traditionally difficult to identify in an unbiased manner. High-throughput sequencing, and especially single cell DNA sequencing, has enabled the identification of different types of somatic variations in the healthy human brain. These studies have revealed that every cell in the brain has somatic mutations and that, compared to inherited variants, somatic variants often cause a more drastic change.
Figure 1.
Somatic variants being acquired during the lifetime of individuals. Trees represent cell lineage and colors represent somatic variants. For healthy individuals (top panel), a moderate level of somatic variants occur during development and also throughout the life of the individual. On a population wide level, this moderate somatic mosaicism leads to a moderate level of variation of phenotypes or behavior, creating intangible variance [15]. For the early somatic mutation causing a disease (middle panel), a somatic variant occurs early in development, resulting in a large proportion of cells carrying a detrimental variant. The bottom panel represents the hypothesis that environmental factors influence the rates of somatic mutations throughout the life of an individual. On an organismal level, we speculate that this level of mosaicism may result in a wider variance of phenotypes or behaviors (green line).
Measuring the number and types of somatic mutations within healthy and diseased contexts is essential to understand the role of somatic mosaicism in health and disease. Studies over the past 5 years have provided the first estimates of the rate of different mutations per cell. Depending on the study and detection method, there are differences in rate estimates, but the current estimates provide essential upper and lower bounds to understand the somatic mutational landscape of an average single neuron. Within the healthy human brain, a single neuron is estimated to contain on average ~800–2000 SNVs, with 80% of the SNVs being C>T transitions enriched in actively transcribed genes when analyzed by whole genome sequencing of single cells amplification in vitro [11]. A study of single mouse neurons using nuclear transfer for whole genome amplification estimates ~100 SNVs per mouse neuron with ~40% of the SNVs being C>T transitions [8].
Single cell sequencing studies identified structural genomic variants in human neurons. Variations of DNA larger than 1 kb in size are classified as structural variants, including copy number gains and losses of sequence. Hinting that somatic structural variants could have significant phenotypic impacts, germline structural variants are important contributors to neurological and neuropsychiatric disease and often disrupt multigene regions. [13] reported that 13–41% of frontal cortex neurons have at least one megabase-size de novo CNV. Also using single cell sequencing, another study identified megabase-size CNVs in non-diseased human brain [2]. A recent study sequencing single cloned mouse neurons also identified complex chromothripsis events. Interestingly, these megabase-size de novo CNVs are much larger than germline CNVs found in healthy humans, suggesting that large CNVs, while tolerated within the brain, would probably be lethal if present in every cell of the body.
A study published by [14] showed that a the mobile DNA element LINE-1 (L1) reporter can actively create neuronal mosaicism by somatic retrotransposition, a process whereby the L1 sequence inserts into a new location by a copy and paste mechanism. This finding led to the hypothesis that somatic mosaicism could play a role in generating neuronal diversity and potentially expanding the range of behavior of the individual [15]. However, it was not clear how representative of endogenous retrotransposition the reporter system was, and several studies shifted the focus to endogenous L1. Since these initial reports, single cell sequencing approaches identified the genomic location of individual insertions within brain cells and confirmed that L1 retrotransposition occurred in neurons. But the frequency of these events remains somewhat controversial, with estimates ranging from ~1 event in every five cells to ~13 events per cell [1,6,17]. A recent study, using targeted L1 sequencing on single cells, identified and validated somatic L1 insertions in neurons and glia of non-diseased individuals. It also identified somatic deletions associated with the L1 sequence and estimated a rate of 0.58–1 L1-associated somatic variants per cell [5].
Proper neural development requires active DNA repair. Maintaining genome integrity is essential for all cells to suppress cancer and to faithfully propagate genetic information, but the brain is particularly vulnerable to defective DNA repair. Defects in DNA repair components such as the non-homologous end-joining (NHEJ) pathway, LIG4 and XRCC4, result in neuro-developmental defects and microcephaly. Mutations in DNA damage response genes ataxia telangiectasia mutated (ATM), ataxia telangiectasia related (ATR), and ATR-interacting protein (ATRIP) cause neuronal degeneration [3,9,12]. Similarly, other DNA repair pathways, such as transcription coupled repair, homologous recombination, and nucleotide excision repair, are required for proper neural development [10,16]. In neural progenitors, a small group of long neuronal genes are prone to developing double-stranded DNA breaks, which are related to the transcription of these genes that are often mutated in cancers or neuropsychiatric disorders [19]. Neural progenitors undergo a period of rapid expansion correlated with a short cell cycle and gamma H2AX staining, a hallmark of double stranded DNA damage. At the end of this progenitor expansion, approximately 50% of cells undergo apoptosis. In sum, these studies have proven that somatic variations occur in healthy brain cells and highlight the potential importance of somatic mosaicism to brain function. But it is largely unknown if different cell types within the healthy or diseased brain harbor different levels or specific somatic mutations.
Dimensions of somatic mosaicism
Collecting data
At present, the amount of data on somatic variants in the brain is relatively scarce, and several research groups are developing and improving laboratory and analytical techniques to accurately detect somatic variants. The NIMH-funded Brain Somatic Mosaicism Network (BSMN) aims to collect a large volume of genomic data on individuals affected by neuropsychiatric diseases and controls. These datasets will be a shared community resource and will include whole genome sequencing (WGS) at different depths, whole exome sequencing (WES) and targeted sequencing for mobile element insertion (MEI) profiling. Sequencing will be performed on single cells, pools of cells and bulk tissue samples. Single cell WGS provides the most comprehensive assessment of somatic mutations, but at a high cost per cell. Targeted sequencing enables sampling many cells for the same cost, restricting sequencing to a small subset of the genome. Bulk tissue sequencing captures data for many cells all at once and is effective in detecting mutations present in a large fraction of the cells but lacks power to confidently detect mutations present in one or a few cells. Single cell sequencing overcomes this problem but can also introduce sequence artifacts due to genome amplification, which pose an extra challenge to analysis. What mix of sequencing strategies would yield the maximum amount of information is not yet known. As data from many different sequencing strategies are being generated by the BSMN and other somatic mosaicism studies, this picture will become clearer.
Integrating data
For the expanding somatic mosaicism datasets, an overarching goal will be to identify biological consequences from somatic mosaicism and its potential role in neuropsychiatric disorders. Achieving this goal is particularly challenging due to the multidimensional nature of somatic mosaicism. As illustrated in Figure 1, with individuals acquiring somatic variants during their lives, some of these dimensions are related to cells (cell identity, cell lineage, cell type, brain region it belongs to), individuals (genetic background, medical history, behavior), mosaic variants (type of variant, genomic regions affected) and developmental time. The impact of somatic mosaicism on cognition and behavior depends not only on these dimensions but also on interactions between cells carrying different genomic variants. Much insights can be gained by integrating mosaicism data with brain connectivity studies and meta-analytic databases on connectivity. While such integration studies are challenging, they can spark the development of a new set of analytical tools delving into the complexity of the genome and structural and functional brain connectivity.
Some special cases of somatic mosaicism are particularly interesting as they can provide a more direct link between mosaic genotype and phenotype and can serve as a basis to calibrate analytical strategies. One such case is focal cortical dysplasia, an epilepsy-causing malformation of some cortical regions caused by somatic mutations in genes from the mTOR pathway. Figure 1 middle panel, is an example in which a relatively large proportion of cells carry the phenotype-causing variant. Regions containing the variant are easily identifiable through altered morphology in the cortex, and sequencing of bulk tissue samples from these regions is likely to detect the variant. Cancer constitutes another such instance, in which accumulated mutations confer on cells the ability to escape control mechanisms and undergo rapid proliferation. Overgrowth makes cancer cells easily identifiable and their clonal nature allows cancer-related mutations to be discovered with bulk tissue sequencing. In these cases, a clear morphological phenotype and the presence of the same somatic variants in a large cell population make these cases more tractable experimentally and analytically.
A more general case of somatic mosaicism as illustrated by Figure 1, top and bottom panels. In these hypothetical examples, each different somatic variant is present in a small number of cells and can have a subtle phenotype on each cell. Moreover, the phenotype may be different from cell to cell depending on the variant, the cell type and its current state. The overall levels of mutations may be affected by environmental factors. This case is clearly much more challenging to study from both the experimental and analytical standpoints. Due to low allele frequency, these somatic variants are unlikely to be confidently detected in bulk tissue sequencing, requiring single-cell sequencing approaches. The presence of multiple variants, each affecting each cell in a different way, makes analysis more challenging. Yet the collective contribution of all the variants to the behavior of a cell population may be significant and observable.
Given the complexity of the brain and the genome, a systems biology approach is well suited to provide insight into the relationships between somatic mosaicism and brain connectivity and function.
Studies based on magnetic resonance imaging (MRI) have been successful in delineating brain areas related to cognitive function and infer connectivity between them. Using multi-modal MRI data from the Human Connectome Project (HCP), a recent study generated such a parcellation of the human cerebral cortex and developed a machine-learning classifier to locate each of these cortical areas in new subjects, even if they have atypical parcellation [7].
Functional connectivity between brain areas has been inferred through temporal correlation analysis of their activity patterns. Co-activation meta-analysis or meta-connectomics combines activity data from primary studies in the published literature to infer connectivity and answer questions that were not posed originally in the primary studies [4]. These connectivity studies identified a “rich club” of highly connected brain areas that are involved in many cognitive functions. Deficiency in rich-club connectivity has been associated with schizophrenia [18], suggesting a lower level of brain communication capacity may have a key role in this disorder.
Somatic variants can directly influence properties of each cell but can also be consequences of altered physiology leading to DNA mutation. The genomics literature has documented a vast set of properties or genomic regions that include sequence content, chromatin state, physical proximity, co-expression, gene function and relative replication time, among many others. We envision that a graph-theoretical representation of the genome, linking different genomic regions to each other based on their properties, will be instrumental in identifying meaningful information in somatic variant data, by connecting each variant with the properties of its genomic context. Rich metadata on the biological samples and clinical information on the individuals will provide links to brain regions across studies and related patient groups. Such a graph-theoretical representation is amenable to data-driven discovery of holistic properties of brain function and somatic mosaicism. Predictive models based on machine-learning or statistical techniques are key to test the robustness of these findings, which then will translate into new hypotheses and directions for research.
Acknowledgments
We thank M.L. Gage and the BSMN network for critical reading and discussions related to the manuscript. The Gage Laboratory was partially funded by NIH TR01 MH095741, NIH U01 MH106882, The G. Harold & Leila Y. Mathers Foundation (Grant #2012-PG-00), The Leona M. and Harry B. Helmsley Charitable Trust, JPB Foundation, Annette C. Merle-Smith, and Bob and Mary Jane Engman. Some figures use images from the Servier Medical Art PowerPoint Image Bank.
References
- 1.Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai X, Evrony GD, Lehmann HS, Elhosary PC, Mehta BK, Poduri A, Walsh CA. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell Rep. 2014;8:1280–1289. doi: 10.1016/j.celrep.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coufal NG, Garcia-Perez JL, Peng GE, Marchetto MC, Muotri AR, Mu Y, Carson CT, Macia A, Moran JV, Gage FH. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc Natl Acad Sci U S A. 2011;108:20382–20387. doi: 10.1073/pnas.1100273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crossley NA, Fox PT, Bullmore ET. Meta-connectomics: human brain network and connectivity meta-analyses. Psychol Med. 2016;46:897–907. doi: 10.1017/S0033291715002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erwin JA, Paquola AC, Singer T, Gallina I, Novotny M, Quayle C, Bedrosian TA, Alves FI, Butcher CR, Herdy JR, et al. L1-associated genomic regions are deleted in somatic cells of the healthy human brain. Nature Neuroscience. 2016;19:1583–1591. doi: 10.1038/nn.4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, et al. A multi-modal parcellation of human cerebral cortex. Nature. 2016;536:171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazen JL, Faust GG, Rodriguez AR, Ferguson WC, Shumilina S, Clark RA, Boland MJ, Martin G, Chubukov P, Tsunemoto RK, et al. The complete genome sequences, unique mutational spectra, and developmental potency of adult neurons revealed by cloning. Neuron. 2016;89:1223–1236. doi: 10.1016/j.neuron.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iourov IY, Vorsanova SG, Liehr T, Kolotii AD, Yurov YB. Increased chromosome instability dramatically disrupts neural genome integrity and mediates cerebellar degeneration in the ataxia-telangiectasia brain. Hum Mol Genet. 2009;18:2656–2669. doi: 10.1093/hmg/ddp207. [DOI] [PubMed] [Google Scholar]
- 10.Laugel V. Cockayne syndrome: the expanding clinical and mutational spectrum. Mech Ageing Dev. 2013;134:161–170. doi: 10.1016/j.mad.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Lodato MA, Woodworth MB, Lee S, Evrony GD, Mehta BK, Karger A, Lee S, Chittenden TW, D’Gama AM, Cai X, et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015;350:94–98. doi: 10.1126/science.aab1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McConnell MJ, Kaushal D, Yang AH, Kingsbury MA, Rehen SK, Treuner K, Helton R, Annas EG, Chun J, Barlow C. Failed clearance of aneuploid embryonic neural progenitor cells leads to excess aneuploidy in the Atm-deficient but not the Trp53-deficient adult cerebral cortex. J Neurosci. 2004;24:8090–8096. doi: 10.1523/JNEUROSCI.2263-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, Shumilina S, Lasken RS, Vermeesch JR, Hall IM, et al. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 15.Muotri AR, Gage FH. Generation of neuronal variability and complexity. Nature. 2006;441:1087–1093. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- 16.Shen J, Gilmore EC, Marshall CA, Haddadin M, Reynolds JJ, Eyaid W, Bodell A, Barry B, Gleason D, Allen K, et al. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat Genet. 2010;42:245–249. doi: 10.1038/ng.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Upton KR, Gerhardt DJ, Jesuadian JS, Richardson SR, Sanchez-Luque FJ, Bodea GO, Ewing AD, Salvador-Palomeque C, van der Knaap MS, Brennan PM, et al. Ubiquitous L1 mosaicism in hippocampal neurons. Cell. 2015;161:228–239. doi: 10.1016/j.cell.2015.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Heuvel MP, Sporns O, Collin G, Scheewe T, Mandl RC, Cahn W, Goni J, Hulshoff Pol HE, Kahn RS. Abnormal rich club organization and functional brain dynamics in schizophrenia. JAMA Psychiatry. 2013;70:783–792. doi: 10.1001/jamapsychiatry.2013.1328. [DOI] [PubMed] [Google Scholar]
- 19.Wei PC, Chang AN, Kao J, Du Z, Meyers RM, Alt FW, Schwer B. Long neural genes harbor recurrent DNA break clusters in neural stem/progenitor cells. Cell. 2016;164:644–655. doi: 10.1016/j.cell.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]