ABSTRACT
Development of vaccines that are both safe and effective remains a costly and time-consuming challenge. To accelerate the pace of development and improve the efficacy and safety of candidate vaccines for both existing and emerging infectious agents, we have used a distributed development approach. This features the managed integration of individual expert groups having the requisite vaccine platforms, pre-clinical models, assays, skills and knowledge pertinent to a specific pathogen into a single, end-to-end development team capable of producing a new vaccine tailored to that particular agent. Distributed development focuses on integrating existing effort across multiple institutions rather than developing new capabilities or consolidating resources within an individual organization. Previously we have used the distributed development strategy to generate vaccine candidates for emerging viral diseases. Coxiella burnetii is a highly infectious and resilient bacterium and the causative agent of Q fever. Treatment for Q fever can require months of antibiotics. The current vaccine for Q-fever is only approved in Australia and requires prescreening due to the potential for severe reactogenicity in previously exposed individuals. Here we discuss Q-VaxCelerate, a distributed development consortium for the development of a new vaccine to prevent Q fever.
KEYWORDS: vaccine, Coxiella burnetii, Qfever, subunit, peptide, distribute development
Background
The complexity of vaccine development, which requires diverse expertise, infrastructure, and access to key reagents and technologies, presents scientific and logistical challenges to investigators involved in this process. One strategy for addressing the multiple requirements for a successful vaccine program is the use of a distributed development model, where academic and industry groups partner to integrate relevant domain expertise and technological assets to complete a vaccine development program. Previously we described how a distributed development model was used to meet the unique needs of rapid response vaccines for emerging infectious diseases (EID).1 Here we apply our previous experience to address the need for a less reactogenic and efficacious vaccine for Coxiella burnetii.
C. burnetii is a gram negative obligate intracellular bacterium and the causative agent of Q fever in humans. C. burnetii can form a spore-like small cell variant that exhibits remarkable resistance to heat, pressure, and ultraviolet inactivation. Infection with C. burnetii can result from as few as 1–10 bacteria, often by inhalation of fomites from infected animals, although ticks may also act as transmission vectors. Within 9–40 d post exposure, approximately half of infected individuals exhibit Q fever symptoms that include profound headache, chest and joint pain, fever, and fatigue, along with respiratory and gastrointestinal symptoms.2,3 Treatment for Q fever requires weeks to months of doxycycline or selected fluoroquinolone antibiotics.4 Patients who are pregnant or are infected with strains resistant to first line antibiotic therapies require more carefully managed care. A subset of patients develop persistent Q fever, typified by endocarditis or other localized infection, fatigue or malaise, and requiring antibiotic treatment that may range from 1.5 y to lifelong.5,6 Consequently, the preferred means of infection control for C. burnetii is vaccination of at risk populations. The current vaccine, Q-VAX® (Commonwealth Serum Laboratories Ltd.), is approved for human use only in Australia and requires pre-screening as vaccination can elicit severe reactogenic responses in individuals previously exposed to the bacterium.7-9 Acellular vaccine formulations generated via chloroform-methanol or trichloroacetic acid extraction can also elicit adverse reactions, though more recent formulations exhibit improved safety profiles in pre-clinical testing.8 An outbreak of Q fever centered in the Noord Brabant province of the Netherlands underscored the challenges associated with Q fever treatment and vaccination. As many as half of infected individuals may experience mild symptoms and not seek medical attention, thereby precluding a diagnosis of Q fever.10 As a result the application of a broad vaccination program with Q-VAX® to curb the outbreak faced logistical hurdles due to the need to pre-screen all prospective vaccinees. In addition, compliance with long term antibiotic treatment can be challenging to monitor. Thus, there is a need for a new Q fever vaccine with an improved safety profile that elicits a robust and protective immune response.7,8,11
The United States Defense Threat Reduction Agency identified the need for a new Q fever vaccine since service personnel deployed to endemic regions are at particular risk. To address this need, we undertook an approach informed by our prior distributed development efforts with a rapid response platform targeting influenza and Lassa fever.1 Q-VAX® is an inactivated whole cell preparation, components of which, likely contribute to the reactogenicity observed in some patients.8 In lieu of an attenuated or inactivated vaccine, we opted for a platform to deliver epitopes that we predicted would generate C. burnetii specific T-cells and potentially protect against Q fever. In addition, the epitopes presented can also be selected to minimize cross or self reactive epitopes and thereby limit one class of potential adverse reactions to vaccination. The design and assessment of candidate vaccines also anticipates factors important to regulatory approval, including data management, the use of 2 animal model test systems and vaccine platform selection.
Q-VaxCelerate consortium
Q-VaxCelerate Consortium partners each address specific project requirements and objectives, and include 3 academic groups and 2 biomedical industry groups (Fig. 1). Epivax is an immunoinformatics company with demonstrated expertise in the identification, selection, and evaluation of target epitopes. InnatOss, a biotechnology company based in Oss, Netherlands, near the geographic epicenter of the 2007–2011 Q fever outbreak, has developed a diagnostic test for cell mediated immunity against C. burnetii and provides access to human donors previously exposed to C. burnetii. 21st Century Biochemicals provides high quality peptide synthesis for immunoassays and provides technical expertise regarding the design and validation of peptide concatemers. The Vaccine and Immunotherapy Center (VIC) at Massachusetts General Hospital incorporates the use of cytometry by time of flight mass spectroscopy (CyTOF) to conduct high dimensional analysis of both human and murine mononuclear cell samples. Dr. Richard Bucala's laboratory from Yale provides significant experience with RNA replicon vaccine platforms that can be tailored for specific antigens. Dr. Richard Bowen's laboratory from Colorado State University (CSU) is proficient in the development of animal models to study the pathology, vaccination, and treatment of infectious agents under biosafety containment. VIC also provides an experienced project management team to coordinate scientific and administrative tasks.
Figure 1.
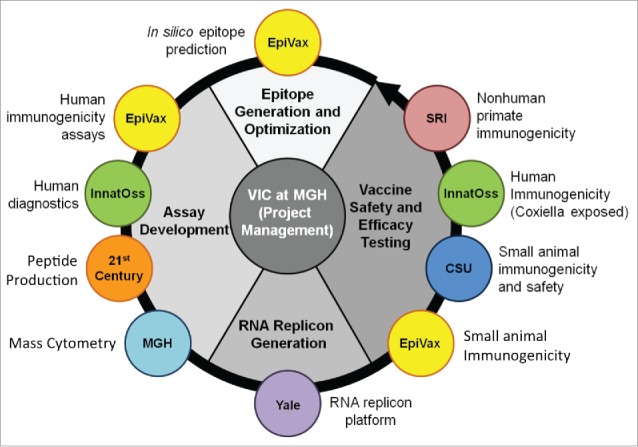
Organization of the Q-VaxCelerate Program.
Vaccine development approach
The mechanism of the immune response to C. burnetii remains an active area of investigation, with a consensus view that T-cell mediated immunity is essential to controlling the pathogen and that B-cell responses can augment T cell responses to expedite control of infection.12 To develop a novel C. burnetii vaccine with an improved safety profile, the Q-VaxCelerate consortium is pursuing a peptide based vaccine using parallel evaluation of candidates in both human donor samples and a mouse model of C. burnetii infection (Fig. 2). An epitope targeted T-cell vaccine approach was selected to eliminate the presence of intact lipopolysaccharide proteins that are thought to contribute to whole cell C. burnetii vaccine reactogenicity and, through downselection of homologous epitopes, to minimize cross reactivity with host and microbiome proteins.13,14
Figure 2.
Schematic representation of the Q-VaxCelerate Program workflow.
Immunoinformatic prediction of candidate epitopes
To identify candidate T-cell epitopes Epivax first conducted in silico analysis of available C. burnetii genome sequences and identified peptides predicted to bind either human MHC I or MHC II. Predicted epitopes were curated to eliminate peptides with significant homology to human or commensal bacteria sequences. The resulting selected set of epitopes were then synthesized and screened for in vitro MHC binding to further refine the candidate pool for advanced testing in biological systems.
Candidate epitope evaluation
The in silico analysis of C. burnetii genomic sequence identified putative MHC I and MHC II epitopes. Synthesis of peptides containing the candidate epitopes allows for evaluation in biological systems to identify those peptides capable of binding cognate receptors and eliciting C. burnetii-specific responses. Three specific assessments are performed. Peptides are screened for antigenic ex vivo recall responses in lymphocytes from donors with known prior exposure to C. burnetii. Donors from a well described cohort from the village of Herpen, where the first cases of the 2007–2011 Q fever outbreak were reported, provide blood samples from both C. burnetii exposure positive and negative donors as determined by antibody and IFNγ production.4,11,15-17 Concurrently, candidate peptides are evaluated in a guinea pig model of reactogenicity to minimize the probability of adverse reaction to the selected peptide epitopes.18 Candidate peptides are then selected for immunogenicity testing in mice on the basis of donor peptide stimulation efficacy and reactogenicity profile in guinea pigs. These selected peptides are then evaluated for immunogenicity and protection using a humanized murine model of C. burnetii.
Assessment of murine models of C. burnetii vaccination and infection
To further explore the immune response to C. burnetii infection and vaccination, CSU and VIC work together to investigate and compare 2 murine model systems. The infection and vaccination studies are conducted with C57Bl/6 mice, both wild-type and a transgenic strain in which the MHC II has been replaced by human HLA-DR3. As the epitope predictions are on the basis of human MHC binding, pre-clinical testing in a transgenic humanized MHC murine module enables peptide evaluation to validate immunogenicity and guide downstream selection. Though C57Bl/6 mice are less sensitive to C. burnetii infection than Balb/c mice, both immune responses and pathological measures can be quantified.19 In addition both whole-cell inactivated and sub-unit vaccines elicit protection from C. burnetii infection in C57Bl/6 mice.8,20,21
Serial blood sample collection through the course of vaccination and infection enables CyTOF profiling of the immune response in mice. A particular advantage to this technology is the capacity to assay >37 parameters simultaneously, which allows for the profiling of numerous immune cell types across a range of cytokine and activation markers from <200 microliters of blood. The comparison of the wild-type and HLA-DR3 mice provides important validation of the model systems and highlights any differences resulting from the transgenic expression of the human MHC II allele that may influence outcomes of subsequent assessments. In addition, the capacity to conduct serial sampling and broad immunoprofiling from study animals may provide important insights into key timepoints and markers associated with successful vaccination and enhanced control of infection.
Assessment of human donor immune responses to C. burnetii exposure
The immunostimulative capacity of candidate peptides can be assessed by incubation of donor blood with peptide. Samples from C. burnetii exposure positive donors and naïve donors are incubated with candidate peptides and responses measured in an interferon-γ ELIspot assay to evaluate epitope antigenicity and possible correlations to patient outcome. Importantly, the donor pool is well defined, with characterization through measurement of C. burnetii antibodies and ex vivo interferon-γ production in response to stimulation with killed whole C. burnetii. Donors can then be selected to best represent the diversity of HLA genotypes. To investigate the heterogeneity of human immune responses, the ex vivo immune responses of donors with diverse clinical outcomes can be interrogated using traditional flow cytometry and CyTOF, and the resting and challenge response of donors can be compared.
The data from peptide stimulation and inactivated C. burnetii stimulation of donor blood provide insight into the immunogenicity of candidate peptides and the diversity of immune responses from donors with a history of sub-clinical and clinical infections or no infection. The results of these studies may identify immune hallmarks unique to individuals who exhibited robust control of C. burnetii infection as compared to patients who required clinical intervention. Identification of peptide antigens or cell responses that correlate to particular clinical outcomes can inform the design of a vaccine candidate to promote an immune response that correlates with superior responses to C. burnetii infection. Investigation of these donors may also reveal allele specific responses that correspond to a more or less favorable protective immune profile.
Delivery of peptide vaccine
Several considerations influence the selection of a delivery method for peptide vaccines including requirement for adjuvant, capacity to deliver multiple peptides, risk of vector integration, and durability of peptide presentation. The prototypical peptide vaccine delivery in model systems utilizes some version of a prime/boost vaccination protocol with an adjuvant such as complete or incomplete Freund's adjuvant or alum. However, this approach is not amenable to regulatory review. An alternative to the delivery of synthesized peptides is the use of vector platforms that incorporate nucleic acid sequence to deliver target peptide(s) and may be comprised of purified DNA or RNA, or a modified viral genome.22 RNA vectors are less stable DNA platforms though they eliminate concerns of DNA integration into the host genome. RNA replicon systems based on RNA virus genomes are modified to remove the pathogenic potential while retaining replicative capacity to extend the length of peptide presentation. A challenge in the use of nucleic acid vaccines seen in vaccines for both microbes and cancer arises from the need to deliver multiple epitopes to elicit effective responses.20,22-25 The identification and selection of peptides that are both effectively processed and presented (antigenic) and elicit protective immune responses (immunogenic) often relies on bioinformatic approaches that are continuously refined.26-28 In addtion, the delivery platform must effectively produce peptides that can then be presented. EpiVax has developed a robust set of tools to address epitope prediction and selection.13,14,29 Dr. Bucala's laboratory at Yale has made extensive use of RNA replicons to develop candidate malaria vaccines. In this effort, a concatemer of selected C. burnetii peptides are integrated into the RNA replicon, which can be delivered both in vitro and in vivo to evaluate peptide production, toxicity, stability and immunogenicity.
Conclusion/next steps
To date the Q-VaxCelerate consortium has completed the in silico selection and HLA binding assessments of candidate peptides. Concurrently human donors were characterized for the presence of anti-C. burnetii antibodies and cytokine production in response to ex vivo stimulation. Ongoing work is now evaluating the response of donor samples to stimulation with candidate peptides. Simultaneously, we have determined the infectious dose of C. burnetii for C57Bl/6 and HLA-DR3 mice and conducted killed whole bacteria vaccination and challenge studies to confirm protection and establish baseline responses. Data from ongoing analysis and future experiments will be used to assess the immunogenicity and protection conferred by candidate peptides in the mouse model and to further investigate responses from human donors that represent the cross-section of clinical groups associated with C. burnetii infection.
From the work completed, we have come to appreciate that a distributed development consortium research requires frequent, clear, critical and open communication between partners, particularly with respect to studies dependent on coordinated execution at 2 or more sites. In addition there are risks posed by application of innovative technologies such as CyTOF or novel applications of existing technologies such as in silico epitope prediction and replicon vaccine platforms previously used for expression of protein rather than peptide concatemers. These challenges must be discussed openly and continuously, and carefully managed with clear inclusion/exclusion metrics and alternative approaches in place. Otherwise, the ability to deliver within the constraints of budget and time can be compromised.
The Q-VaxCelerate consortium comprises more than 20 individuals from more than 6 academic and industrial organizations with a range of expertise and experiences that would be difficult to recapitulate in a single laboratory. The inclusion of various groups and role of individual researchers is primarily on the basis of their expertise. However, a clear benefit is also derived from areas of shared experience: immunoassay design, animal models of infectious disease, or constraints of working in biocontainment. Shared areas of experience can prevent silo-ing, reduce project risk, In particular, we have found that a multi-center multi-disciplinary team improves the real-time troubleshooting, facilitates critical assessment and interpretation of data, and improves experimental design and optimization, particularly where portions are completed by more than one research site. Our prior experience with consortium approaches is consistent with Q-VaxCelerate, where integration of multiple experimental systems and research groups with established expertise provides efficiencies of time and cost that would not readily be attainable in a single center. The primary goal of the consortium is to generate, under a constrained timeline, a vaccine candidate that exhibits a favorable response profile with respect to both immunogenicity and safety in pre-clinical models. In parallel, we seek to generate novel insights into the immune response to C. burnetii vaccination and infection in both humans and small-animal model systems. Finally, the implementation of a distributed consortium approach may serve as a template for projects involving other multi-center multi-disciplinary teams.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Leblanc P, Moise L, Luza C, Chantaralawan K, Lezeau L, Yuan J, Field M, Richer D, Boyle C, Martin WD, et al.. VaxCelerate II: Rapid development of a self- assembling vaccine for Lassa fever VaxCelerate II: Rapid development of a self-assembling vaccine for Lassa fever. 2014;5515(October 2016):1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Derrick EH. “Q” fever, a new fever entity: clinical features, diagnosis and laboratory investigation. Rev Infect Dis. 1983;5(4):790-800. doi: 10.1093/clinids/5.4.790. PMID:6622891 [DOI] [PubMed] [Google Scholar]
- [3].Maurin M, Raoult D. Q fever. Clin Microbiol Rev. 1999;12(4):518-553. PMID:10515901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Dijkstra F, Riphagen-Dalhuisen J, Wijers N, Hak E, Van der Sande MAB, Morroy G, Schneeberger PM, Schimmer B, Notermans DW, Van der Hoek W. Antibiotic therapy for acute Q fever in The Netherlands in 2007 and 2008 and its relation to hospitalization. Epidemiol Infect. 2011;139(9):1332-1341. doi: 10.1017/S0950268810002621. PMID:21087542 [DOI] [PubMed] [Google Scholar]
- [5].Parker NR, Barralet JH, Bell AM. Seminar Q fever. Lancet. 2006;367:679-688. doi: 10.1016/S0140-6736(06)68266-4. PMID:16503466 [DOI] [PubMed] [Google Scholar]
- [6].Million M, Raoult D. No Such Thing as Chronic Q Fever. Emerg Infect Dis. 2017;23(5):856-857. doi: 10.3201/eid2305.151159. PMID:28418317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kazar J, Brezina R, Palanova A, Tvrdá B, Schramek S. Immunogenicity and reactogenicity of a Q fever chemovaccine in persons professionally exposed to Q fever in Czechoslovakia. Bull World Health Organ. 1982;60(3):389-394. PMID:6982774 [PMC free article] [PubMed] [Google Scholar]
- [8].Waag DM, England MJ, Bolt CR, Williams JC. Low-dose priming before vaccination with the phase I chloroform-methanol residue vaccine against Q fever enhances humoral and cellular immune responses to Coxiella burnetii. Clin Vaccine Immunol. 2008;15(10):1505-1512. doi: 10.1128/CVI.00119-08. PMID:18701647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Biotherapies CSL, Road P. A guide to Q fever and Q fever vaccination. Marmion B, Spelman D, eds. Med J Aust. 2009. Parkville, ABN 66120398067. [Google Scholar]
- [10].Wielders CCH, Wuister AMH, De Visser VL, De Jager-Leclercq MG, Groot CAR, Dijkstra F, et al. Characteristics of hospitalized acute Q fever patients during a large epidemic, the Netherlands. PLoS One. 2014;9(3). PMID:24614585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Madariaga MG, Rezai K, Trenholme GM, Weinstein RA. Q fever: A biological weapon in your backyard. Lancet Infect Dis. 2003;3(11):709-721. doi: 10.1016/S1473-3099(03)00804-1. PMID:14592601 [DOI] [PubMed] [Google Scholar]
- [12].Andoh M, Zhang G, Russell-Lodrigue KE, Shive HR, Weeks BR, Samuel JE. T cells are essential for bacterial clearance, and gamma interferon, tumor necrosis factor alpha, and B cells are crucial for disease development in Coxiella burnetii infection in mice. Infect Immun. 2007;75(7):3245-3255. PMID:17438029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Moise L, Gutierrez AH, Bailey-Kellogg C, Terry F, Leng Q, Abdel Hady KM, VerBerkmoes NC, Sztein MB, Losikoff PT, Martin WD, et al.. The two-faced T cell epitope: examining the host-microbe interface with JanusMatrix. Hum Vaccin Immunother. 2013;9(7):1577-1586. doi: 10.4161/hv.24615. PMID:23584251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Moise L, Gutierrez A, Kibria F, Martin R, Tassone R, Liu R, Terry F, Martin B, De Groot AS. iVAX: An integrated toolkit for the selection and optimization of antigens and the design of epitope-driven vaccines. Hum Vaccin Immunother. 2015;11(9):2312-2321. doi: 10.1080/21645515.2015.1061159. PMID:26155959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schoffelen T, Limonard GJ, Bleeker-Rovers CP, Bouwman JJ, van der Meer JW, Nabuurs-Franssen M, et al.. Diagnosis of Coxiella burnetii infection: comparison of a whole blood interferon-gamma production assay and a Coxiella ELISPOT. PLoS One. 2014;9(8):e103749. doi: 10.1371/journal.pone.0103749. PMID:25084353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Limonard GJM, Peters JB, Besselink R, Groot CAR, Dekhuijzen PNR, Vercoulen JH, Nabuurs-Franssen MH. Persistence of impaired health status of Q fever patients 4 years after the first Dutch outbreak. Epidemiol Infect. 2016;144(6):1142-1147. doi: 10.1017/S0950268815002216. PMID:26508155 [DOI] [PubMed] [Google Scholar]
- [17].Morroy G, Van Der Hoek W,Nanver ZD, Schneeberger PM, Bleeker-Rovers CP, Van Der Velden J, Coutinho RA. The health status of a village population, 7 years after a major Q fever outbreak. Epidemiol Infect. 2016;144(6):1153-1162. doi: 10.1017/S0950268815002472. PMID:26560803 [DOI] [PubMed] [Google Scholar]
- [18].Wilhelmsen CL, Waag DM. Guinea pig abscess/hypersensitivity model for study of adverse vaccination reactions induced by use of Q fever vaccines. Comp Med. 2000;50(4):374-378. PMID:11020154 [PubMed] [Google Scholar]
- [19].Scott GH, Williams JC, Stephenson EH. Animal models in Q fever: pathological responses of inbred mice to phase I Coxiella burnetii. J Gen Microbiol. 1987;133(3):691-700. PMID:3655728 [DOI] [PubMed] [Google Scholar]
- [20].Chen C, Dow C, Wang P, Sidney J, Read A, Harmsen A, Samuel JE, Peters B. Identification of CD4+ T cell epitopes in C. burnetii antigens targeted by antibody responses. PLoS One. 2011;6(3):e17712. doi: 10.1371/journal.pone.0017712. PMID:21423609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhang G, Peng Y, Schoenlaub L, Elliott A, Mitchell W, Zhang Y. Formalin-inactivated coxiella burnetii phase i vaccine-induced protection depends on b cells to produce protective igm and igg. Infect Immun. 2013;81(6):2112-2122. PMID:23545296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Li W, Joshi M, Singhania S, Ramsey K, Murthy A. Peptide vaccine: progress and challenges. Vaccines. 2014;2(3):515-536. doi: 10.3390/vaccines2030515. PMID:26344743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Martin SD, Brown SD, Wick DA, Nielsen JS, Kroeger DR, Twumasi-Boateng K, Holt RA, Nelson BH. Low mutation burden in ovarian cancer may limit the utility of neoantigen-targeted vaccines. PLoS One. 2016;11(5):1-22. doi: 10.1371/journal.pone.0155189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Castle JC, Kreiter S, Diekmann J, Löwer M, van de Roemer N, de Graaf J, Selmi A, Diken M, Boegel S, Paret C, et al.. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72(5):1081-1091. doi: 10.1158/0008-5472.CAN-11-3722. PMID:22237626 [DOI] [PubMed] [Google Scholar]
- [25].Lin S, Cheng C-W, Su E. Prediction of B-cell epitopes using evolutionary information and propensity scales. BMC Bioinformatics. 2013;14(Suppl 2):S10. doi: 10.1186/1471-2105-14-S2-S10. PMID:23484214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Soria-Guerra RE, Nieto-Gomez R, Govea-Alonso DO, Rosales-Mendoza S. An overview of bioinformatics tools for epitope prediction: Implications on vaccine development. J Biomed Inform. 2015;53:405-414. doi: 10.1016/j.jbi.2014.11.003. PMID:25464113 [DOI] [PubMed] [Google Scholar]
- [27].Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: Application to the MHC class i system. Bioinformatics. 2015;32(4):511-517. doi: 10.1093/bioinformatics/btv639. PMID:26515819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nielsen M, Lundegaard C, Worning P, Lauemøller SL, Lamberth K, Buus S, Brunak S, Lund O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12(5):1007-1017. doi: 10.1110/ps.0239403. PMID:12717023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Messitt TJ, Terry F, Moise L, Martin W, De Groot AS. A comparison of two methods for T cell epitope mapping: “cell free” in vitro versus immunoinformatics. Immunome Res. 2011;7(2):1577-1586. doi: 10.4172/1745-7580.1000045 [DOI] [PMC free article] [PubMed] [Google Scholar]


